Abstract
One of the earliest steps in pre-mRNA recognition involves binding of the splicing factor U2 snRNP auxiliary factor (U2AF or MUD2 in Saccharomyces cerevisiae) to the 3′ splice site region. U2AF interacts with a number of other proteins, including members of the serine/arginine (SR) family of splicing factors as well as splicing factor 1 (SF1 or branch point bridging protein in S. cerevisiae), thereby participating in bridging either exons or introns. In vertebrates, the binding site for U2AF is the pyrimidine tract located between the branch point and 3′ splice site. Many small introns, especially those in nonvertebrates, lack a classical 3′ pyrimidine tract. Here we show that a 59-nucleotide Drosophila melanogaster intron contains C-rich pyrimidine tracts between the 5′ splice site and branch point that are needed for maximal binding of both U1 snRNPs and U2 snRNPs to the 5′ and 3′ splice site, respectively, suggesting that the tracts are the binding site for an intron bridging factor. The tracts are shown to bind both U2AF and the SR protein SRp54 but not SF1. Addition of a strong 3′ pyrimidine tract downstream of the branch point increases binding of SF1, but in this context, the upstream pyrimidine tracts are inhibitory. We suggest that U2AF- and/or SRp54-mediated intron bridging may be an alternative early recognition mode to SF1-directed bridging for small introns, suggesting gene-specific early spliceosome assembly.
Pre-mRNA splicing is a conserved process occurring in a wide variety of eucaryotes with differing exon/intron architectures (reviewed in references 4, 6, 9, 15, 20, and 26). Vertebrates typically have small exons and large introns. Nonmetazoans frequently have the opposite genetic organization, with introns smaller than the minimum permissible for splicing of a vertebrate intron. Drosophila melanogaster possesses a mixture of these two classes of intron sizes (16, 23). In addition, more than half of the small introns in Drosophila are missing a prominent vertebrate splicing signal, the 3′ polypyrimidine tract (23). For these reasons, Drosophila provides a model system in which to study potential mechanistic variations operating during recognition of splicing signals.
In the general model of early vertebrate spliceosome complex assembly, U1 snRNP binds to the 5′ splice site and U2 snRNP auxiliary factor (U2AF) binds to the 3′ polypyrimidine tract, thereby facilitating U2 snRNP interaction with the branch point. Various members of the serine/arginine (SR) family of proteins may participate by promoting or stabilizing these interactions (reviewed in references 13, 22, and 31). This family of proteins may also act as exon or intron bridging factors via their SR-mediated interaction with SR domains on the small subunit of U2AF (U2AF35) and the U1 70K protein (32, 33, 38). SF1, originally discovered as an essential splicing factor in reconstitution assays (19), has also been observed to bind to the branch point (7, 8). In yeast, BBP (branch point bridging protein), the ortholog to SF1, functions as an intron bridging factor via interactions with U1 snRNP-associated proteins and the large subunit of U2AF (U2AF65) (1, 2). It is assumed that vertebrate SF1 can play a similar role, although the mammalian equivalents to the yeast U1 snRNP proteins that interact with BBP have not yet been identified. Furthermore, the relationship between bridging by SR proteins and that afforded by SF1 is unclear.
We have previously examined the cis-acting sequences required for efficient splicing of a constitutively spliced small (59-nucleotide [nt]) intron from the D. melanogaster mle gene that lacks a well-defined pyrimidine tract between the branch point and 3′ splice site (18, 29). Assembly of initial ATP-dependent spliceosomes (complex A) on the mle intron requires both the 5′ and 3′ splice sites, suggesting concerted recognition of the entire intron (29). Instead of a classic pyrimidine tract, the mle intron contains two C-rich tracts located between the 5′ splice site and branch point that are necessary for efficient splicing of this intron (18). In addition to a requirement for maximal splicing efficiency, the pyrimidine stretches are also necessary for binding of U2AF, interaction of factors with the 5′ splice site, and proper assembly of the active spliceosome, suggesting that these sequences affect early assembly events at both ends of this small intron. Interestingly, the upstream C-rich tracts are inhibitory if a classical 3′ pyrimidine tract is introduced between the branch point and 3′ splice site (18). This observation suggests competing pathways of factor binding to this substrate and also raises the possibility of alternative gene-specific modes of association of constitutive factors with introns.
Here we demonstrate that both U2AF and an SR protein, SRp54, interact with the C-rich tracts in the mle intron. The central location of the pyrimidine tracts, their importance for maximal splicing, and the ability of human SRp54 to interact with U2AF65 instead of U2AF35 (37) suggested that the binding of SRp54 to the tracts could replace SF1 in bridging this intron. Immunoprecipitation studies using an antibody specific for SF1 indicated that SF1 did not contact mle precursor RNA unless a pyrimidine tract was introduced downstream of the branch point. Furthermore, antibodies against either SRp54 or U2AF immunoprecipitated both halves of a precleaved mle splicing substrate, suggesting that these factors either directly or indirectly interact with both the 5′ and 3′ splice sites. We suggest that SRp54 participates in bridging the small mle intron via its ability to bind both the C-rich tracts and the large subunit of U2AF.
MATERIALS AND METHODS
Plasmids and oligonucleotides.
The following minigenes contained all or part of the first intron from the D. melanogaster mle gene, along with 56 nt of the first exon and 96 nt of the second exon. The wild-type mle substrate contained the entire 59-nt intron. Mle-py1,2, mle+py, and mle-py1,2+py were identical to the wild-type pre-mRNA except for the introduced mutations indicated in Fig. 1, which have been described previously (18). Mutants were constructed by mutagenic PCR. The deletion mutants 5′ss+py and 5′ss-py contained 56 nt of exon 1 plus the first 38 nts of the intron, with wild-type and mutant C-rich tracts, respectively. MleΔ5′ lacked nt +1 to +8 of the intron in the context of the wild-type mle substrate. Mle5′mt contained the 5′ splice site nucleotides GGTA (−1 to +3) mutated to TTCG. The series of mle intron expansion substrates was constructed by the addition of a NcoI site at either +39 (BP65) or +10 (5′65) of the wild-type intron via PCR. Insertion of 15 or 30 nt of Drosophila troponin T intron sequences (between the TaqI and NlaIII sites in intron 7) into the NcoI site of BP65 or 5′65, respectively, made BP80 and BP95 or 5′80 and 5′95. Readdition of C-rich tracts to the expansion constructs was accomplished by insertion of a fragment from the mle intron (nt +1 to +38) containing both tracts and the natural sequence between them into the NcoI site. Both wild-type (ResWT) and mutant (ResMT) versions of the intron were inserted. The sequences of all plasmids were verified by using a Thermo Sequenase Radiolabeled Terminator Cycle Sequencing kit (Amersham Life Science, Inc.). The A62 substrate was created by deleting +8 to +65 of the MINX splicing substrate (27) to shorten the 120-nt intron to 62 nt.
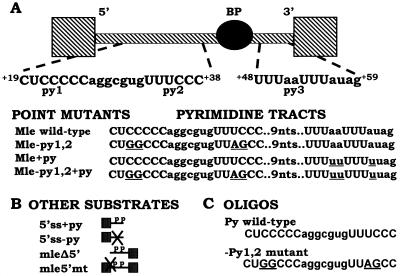
FIG. 1.
Sequence of the mle intron. (A) Top, diagram of the mle intron depicting the locations of various splicing signals. The two upstream C-rich pyrimidine tracts located between the 5′ splice site and branch point are designated py1 and py2. The poor pyrimidine tract located downstream of the branch point is labeled py3. Bottom, sequences of the wild-type and mutant pyrimidine tracts. The introduced point mutations are underlined. Mutation of the upstream tracts is denoted −py1,2; improvement of the downstream pyrimidine tract is denoted +py. (B) Other substrates used in immunoprecipitation and/or UV cross-linking experiments. The C-rich tracts are represented by the letter p, while mutations are indicated by an X. (C) Sequences of both the wild-type and mutant RNA oligonucleotides used in competition and immunoprecipitation experiments.
The following DNA or RNA oligonucleotides containing sequences from the mle intron were obtained from Gibco BRL: a DNA oligonucleotide complementary to +7 to +17 of the mle intron, and RNA oligonucleotides corresponding to the wild-type (CUCCCCCaggcgugUUUCCC) or mutant (CUGGCCCaggcgugUUAGCC) C-rich tracts. Reverse transcriptase (RT)-mediated PCR (RT-PCR) oligonucleotides included 5′ primers complementary to nt 131 to 148 or 183 to 200 of the Drosophila or human SRp54 N-terminal RNA recognition motif (RRM), respectively. Drosophila 3′ primers were complementary to nt 683 to 700 (spacer region) or 860 to 877 (a portion of the antigenic peptide). The human 3′ primer corresponded to nt 708 to 728, which also encode a portion of the peptide to which the anti-SRp54 antibody was raised.
In vitro spliceosome assembly.
Splicing reaction mixtures consisting of 6.5 fmol of radiolabeled substrate, 50% D. melanogaster Schneider 2 (S2) nuclear extract, 1.6 mM MgCl2, 20 mM phospho-l-arginine, 1.2 mM dithiothreitol, 1.2% polyethylene glycol, and 2 mM ATP were incubated at 22°C as described previously (18). Aliquots were taken at the indicated time points. Spliceosome complexes were analyzed by the addition of heparin to a final concentration of 0.2 mg/ml and electrophoresis on native RNP gels (29).
UV cross-linking and competition.
In vitro assembly reaction mixtures were incubated at 22°C for 7 min before the addition of heparin to a final concentration of 2 mg/ml. Mixtures were transferred to ice and UV irradiated immediately for 10 min. Reactions were subsequently digested with RNase A at 37°C for 30 min. Labeled proteins were displayed on sodium dodecyl sulfate–10% polyacrylamide gels and visualized by autoradiography. Competition of cross-linking used unlabeled competitor RNA oligonucleotides, corresponding to either wild-type or mutant C-rich tracts, described above, added at time zero. The amount of competitor added was 0, 50, 100, or 200 pmol.
Immunoprecipitations and immunoblotting.
Immunoprecipitations of cross-linked proteins, assembled spliceosomes, and RNA oligonucleotides were performed with antibodies against the 50-kDa subunit of Drosophila U2AF, provided by D. Rio (17), antipeptide antibodies A and B against the human SRp54, provided by N. Chaudhary (12), or anti-SF1 antibodies. The SF1 antibodies were two polyclonal rabbit sera raised against a His-tagged recombinant fragment of human SF1 that includes amino acids 2 to 320, a region of SF1 in which the human and Drosophila proteins are highly homologous (22a). Immunoprecipitation of precleaved radiolabeled pre-mRNA substrates was conducted by incubating the assembly reactions at 22°C for 7 min before the addition of RNase H and a DNA oligonucleotide complementary to nt +7 to +17 of the mle intron. This mixture was then incubated at 30°C for 3 min before the addition of anti-SRp54 or anti-U2AF antibody. All immunoprecipitations were conducted by using protein A-Sepharose (Pharmacia) in a buffer consisting of 20 mM NaCl, 50 mM Tris-HCl (pH 7.5), and 0.05% Nonidet P-40. Immunoprecipitates were washed in this same buffer before elution in protein or RNA sample buffer. For Western blotting, transfer and detection procedures were performed as described in the PolyScreen instruction manual (Dupont).
5′ splice site protection.
To detect protein interactions at the mle 5′ splice site, an oligonucleotide complementary to this region was added in excess along with RNase H after splicing complex assembly had been allowed to proceed for the indicated amount of time. After incubation at 30°C for 20 min to direct cleavage of the substrate, RNA was prepared for electrophoresis on a 5% denaturing polyacrylamide gel.
Cloning of Drosophila SRp54.
Drosophila SRp54 cDNA was cloned by obtaining an EST (epitope sequence tag) plasmid (LD 13325) from the Berkley Drosophila Genome Project/HHMI EST Project. This clone was then fully sequenced by using a Thermo Sequenase Radiolabeled Terminator Cycle Sequencing kit (Amersham Life Science), and its amino acid sequence was directly compared to both the human SRp54 amino acid sequence and the entire database, using the Blast+Beauty, Align, or ClustalW computer program.
RT-PCR.
RT-PCRs were performed on total RNA isolated from either S2 cells, Drosophila third-instar larvae (male and female), or HeLa cells. The 5′ primer used in the reactions containing S2 or larval RNA corresponded to nt 131 to 148 of the Drosophila cDNA (dSRp54 RRM), and the 3′ primer was complementary to either nt 683 to 700 (dSRp54 spacer region) or 860 to 877 (dSRp54 antigenic peptide) of the cDNA. Primers used to amplify the HeLa RNA were complementary to nucleotides 183 to 200 (hSRp54 RRM) and nucleotides 708 to 728 (antigenic peptide) of the human SRp54 cDNA. Reverse transcription was performed with 1 μg of both the appropriate RNA and 3′ primer, 10 nmol of deoxynucleoside triphosphates and avian myeloblastosis virus RT (Promega) at 50°C for 30 min under standard buffer conditions. To begin the PCR, 1 μg of the 5′ primer as well as recombinant Taq polymerase (Perkin-Elmer) was added. The reaction cycled 30 times between 94°C for 40 s, 55°C for 40 s, and 72°C for 1 min, and the reaction product was then subjected to electrophoresis on an ethidium bromide-stained 1% agarose gel.
Nucleotide sequence accession number.
The Drosophila SRp54 sequence has been assigned GenBank accession no. AF055719.
RESULTS
The SR protein SRp54 cross-links to the mle intron, and its binding is dependent on wild-type C-rich tracts located upstream of the branch point.
We have previously observed that two short pyrimidine tracts located upstream of the branch point within the small mle intron are necessary for maximal spliceosome assembly and association of U2AF (18). The sequences of these tracts (designated py1 and py2) and available mutations (−py1,2, +py, and −py1,2+py) are indicated in Fig. 1. Despite the requirement of the tracts for maximal U2AF binding, the tract sequences (CUCCCCC and UUUCCC) do not resemble optimal binding sites for U2AF (10, 28), suggesting the participation of another factor during early pre-mRNA recognition.
We used UV cross-linking to identify additional proteins binding to the upstream pyrimidine tracts. UV cross-linking revealed a protein of approximately 75 kDa (p75) whose cross-linking tracked with the presence of the upstream C-rich tracts. This protein cross-linked to the wild-type mle pre-mRNA (Fig. 2A, lane 1). Mutation of the upstream pyrimidine tracts depressed cross-linking of p75 in the context of either the full-length intron or a partial intron containing only the 5′ splice site and upstream C-rich tracts (Fig. 2A, lanes 6 and 3, respectively). Interestingly, interaction of p75 with the substrate was not dependent on the presence of either a 5′ splice site (Fig. 2A, lanes 4 and 5) or a 3′ splice site (Fig. 2A, lane 2). Instead, observation of cross-linking required only the presence of the wild-type C-rich pyrimidine tracts (Fig. 2A; compare lanes 2 and 3). Improving the 3′ splice site to provide a more classical pyrimidine tract downstream of the branch point did not eliminate association of p75 (Fig. 2A, lane 7), although it did cause the appearance of another interacting protein, p70 (see below). Cross-linking of p75 to the mle substrate occurred early in the reaction prior to assembly of complex A and did not require ATP (data not shown). Thus, it appears that there is a factor, p75, interacting with the C-rich pyrimidine tracts during the earliest steps of spliceosome assembly.
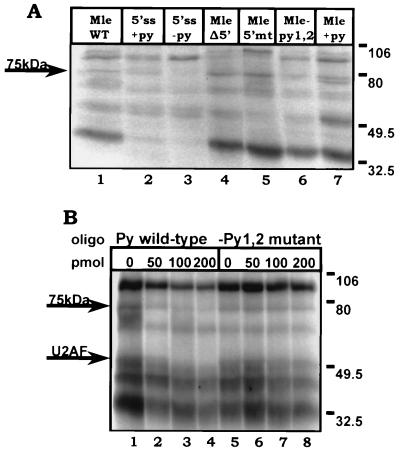
FIG. 2.
A 75-kDa protein interacts with the upstream C-rich tracts. (A) UV cross-linking of proteins to the wild-type (WT) and mutant mle introns. The position of a 75-kDa protein (p75) whose cross-linking is dependent on the presence of wild-type upstream pyrimidine tracts is marked by an arrow. The precursor RNAs used are diagrammed in Fig. 1. A second band appearing below p75 in lane 7 and designated p70 in the text is discussed with reference to Fig. 5. Positions of markers here and in all other relevant figures are indicated in kilodaltons. (B) Competition of cross-linking of p75 by an oligonucleotide containing wild-type but not mutant C-rich pyrimidine tracts. Cross-linking of p75 to mle wild-type precursor RNA was monitored in the presence of increasing amounts of RNA oligonucleotides corresponding to either wild-type or mutant C-rich tracts. The amount of competitor used is indicated above the corresponding lane, and the competitor sequences are shown in Fig. 1C. The bands corresponding to p75 and U2AF are indicated, and positions of markers are shown on the right.
The necessity of the pyrimidine tracts for p75 binding was tested in competition experiments in which UV cross-linking of p75 to radiolabeled wild-type mle precursor RNA was assessed in the presence of increasing amounts of unlabeled competitor RNA oligonucleotides corresponding to either wild-type or mutant C-rich tracts (Fig. 2B). UV cross-linking of p75 to the mle substrate was competed by a wild-type RNA oligonucleotide but not by equivalent concentrations of a competitor oligonucleotide containing the py1,2 mutations (Fig. 2B; compare lanes 1 to 4 with 5 to 8). Therefore, the loss of p75 cross-linking to the mle pre-mRNA containing mutated pyrimidine tracts reflected a loss of binding and not an inability to detect association due to the absence of nucleotides necessary for establishing the chemical cross-link. It should be noted that the wild-type RNA oligonucleotide, but not the mutant oligonucleotide, also competed cross-linking of U2AF. This observation agrees with previous published results demonstrating that the elements influence association of U2AF with the mle intron (18). Therefore, the upstream pyrimidine tracts appear to interact with two trans-acting factors, U2AF and a protein of approximately 75 kDa.
Several proteins, including SF1 (2), SRp54 (37), and other members of the SR family of proteins, are known to interact with U2AF. The identity of p75 was thus investigated by conducting immunoprecipitation experiments using antibodies specific for proteins known to interact with U2AF. One such antibody was an antipeptide antibody raised against human SRp54 and specific for a region of the protein between the RRM and SR domains (12). This antibody recognized two bands of 70 to 75 kDa on Western blots of Drosophila S2 or HeLa extract (Fig. 3B, lane 1 or 4), indicating the presence of SRp54 in Drosophila. The multiple bands presumably reflect differential levels of phosphorylation of SRp54 because phosphatase treatment or purification of extract proteins resulted in increased protein mobility (data not shown and Fig. 3B, lanes 2 and 3).
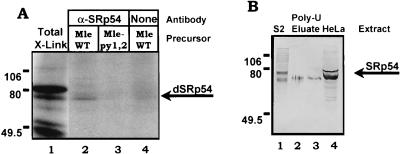
FIG. 3.
p75 is Drosophila SRp54. (A) Immunoprecipitation of cross-linked proteins with an antipeptide antibody specific for SRp54. Products of UV cross-linking reactions using wild-type (WT) or mutant mle precursor RNAs were immunoprecipitated from S2 nuclear extract with anti-SRp54 antibodies specific for amino acids 209 to 225 of the human protein (antibody A [12]). The positions of Drosophila SRp54 and of the markers are indicated. Here and in all other relevant figures, total X-link (lane 1) indicates all of the proteins that UV cross-link to mle wild-type precursor mRNA. (B) Western blot of Drosophila and HeLa nuclear extracts using the antipeptide anti-SRp54 antibody. Lane 1, Drosophila S2 nuclear extract proteins; lanes 2 and 3, HeLa nuclear extract proteins partially purified on poly(U)-Sepharose; lane 4, HeLa nuclear proteins. The multiple bands presumably reflect differential phosphorylation of the SR domain. Similar results for both blotting and immunoprecipitation were observed with a different anti-SRp54 antipeptide antibody (antibody B [12]). Markers are indicated.
Use of the antibody to immunoprecipitate UV cross-linked proteins binding to the mle substrate indicated that SRp54 binds to the mle intron. The anti-SRp54 antibody recognized a UV cross-linked protein of 75 kDa when wild-type mle was used as the splicing substrate (Fig. 3A). p75 comigrated with the band identified in Fig. 2A, suggesting that p75 is SRp54. No band was evident when mle-py1,2 was used as the substrate, indicating that cross-linking of Drosophila SRp54 required the upstream pyrimidine tracts and also further confirmed the identity of the protein being monitored in our mutation experiments in Fig. 2A as SRp54. These results support the idea that the C-rich pyrimidine tracts bind SRp54.
Antibodies against SRp54 and U2AF immunoprecipitate RNA oligonucleotides corresponding to the C-rich tracts.
To more fully determine which factors interact with the upstream pyrimidine stretches, further immunoprecipitation experiments were carried out. In these studies, RNA oligonucleotides containing mle intron sequence from nt +19 to +38, either wild type or mutated for the upstream pyrimidine tracts (py1,2 mutant), were incubated in S2 extract and anti-SRp54 or anti-U2AF antibodies were used to immunoprecipitate the radiolabeled oligonucleotides. As can be seen in Fig. 4, the wild-type RNA oligonucleotide was immunoprecipitated with either antibody (lanes 3 and 5). This ability was lost when the C-rich tracts were mutated (lanes 4 and 6). The results from this experiment support the hypothesis that both SRp54 and U2AF interact with the C-rich tracts located between the 5′ splice site and branch point in the mle intron.
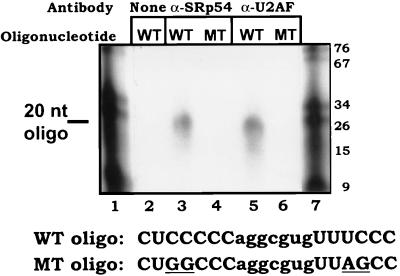
FIG. 4.
Both SRp54 and U2AF interact with the upstream C-rich tracts. RNA oligonucleotides corresponding to either a wild-type (WT; lanes 2, 3, and 5) or mutant (MT; lanes 4 and 6) C-rich tract (diagrammed at the bottom) were incubated in S2 extract for 7 min and then immunoprecipitated with either anti-SRp54 (lanes 3 and 4) or anti-U2AF (lanes 5 and 6) antibody. Immunoprecipitated RNA was displayed on a 10% denaturing polyacrylamide gel. The position of the immunoprecipitated oligonucleotide is indicated. Markers (nucleotide lengths indicated) are in lanes 1 and 7.
The Drosophila homolog of SRp54 is similar to human SRp54 within the RNA binding domain.
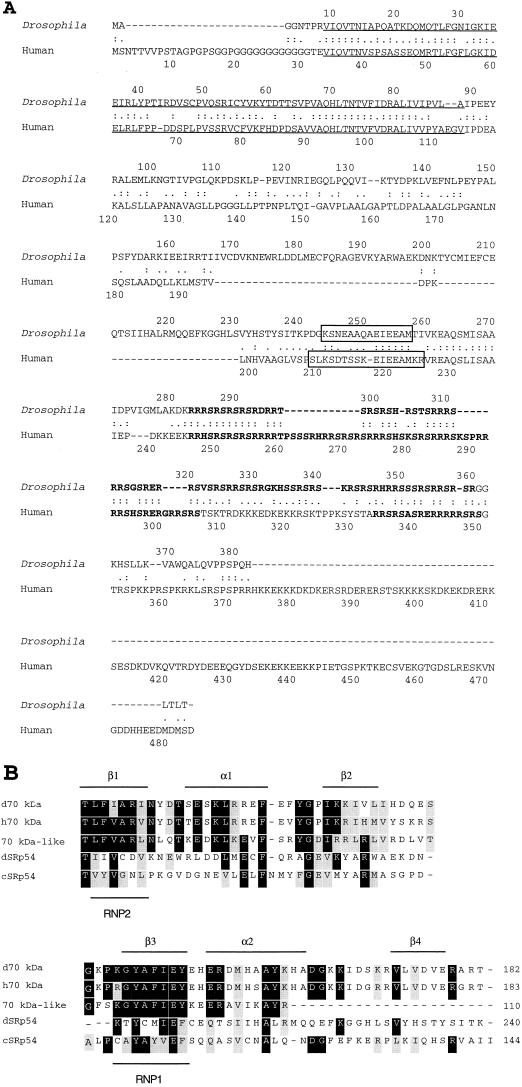
Only the human version of SRp54 has been reported to date (12). A computer search of the Berkley Drosophila Genome Project revealed an EST clone that showed homology with the human SRp54 RRM region. The clone (LD 13325) was provided, and sequencing revealed that it contained a likely cDNA for Drosophila SRp54. Figure 5A compares the sequences of the human and Drosophila proteins. The RRMs located at the N termini of the two proteins exhibited 60% identity and 83.8% similarity, indicating that the Drosophila and human proteins could exhibit very similar RNA binding specificity. Such strong conservation of the RRM between the human and Drosophila proteins is striking, given the observation that the RRM of human SRp54 shares only 17 to 21% identity with other human SR proteins (37). The SR regions of the two proteins were also similar.
FIG. 5.
Drosophila SRp54. (A) The sequence of Drosophila SRp54 compared with the sequence of human SRp54. The N-terminal RRM is underlined; the SR region is in bold, and the peptide epitope for the anti-SRp54 antibody used in this study is boxed in both proteins. (B) Sequence comparison of the central putative RRM in Drosophila SRp54 (dSRp54) compared to a central putative RRM in a candidate SRp54 gene from C. elegans (cSRp54; GenBank accession no. 1813908) and the N-terminal RRMs of the closest RNA binding domains in the sequence database from a U1 snRNP 70K-like protein (3), Drosophila U1 snRNP 70K (d70 kDa), and human U1 snRNP 70K (h70 kDa). Black shading indicates amino acid identity; gray shading depicts conservation.
There were two differences between the proteins. The human SRp54 contains 106 amino acids downstream of the SR domain not present in the Drosophila protein. The function of this domain is unknown. In addition, the two proteins differed by the sequences within the space separating the RRM and the SR domain. Within this region of the Drosophila protein, however, was a sequence with strong similarity to the domain in the human protein that was the peptide epitope for the anti-SRp54 antibody used in the immunoprecipitation experiments in Fig. 3 and 4 (peptide sequence is boxed in Fig. 5A). Thus, the isolated cDNA is Drosophila SRp54, and the protein being identified by immunoprecipitation using the peptide antibody is likely to be that coded for by the identified cDNA.
Interestingly, a computer search also revealed a putative Caenorhabditis elegans SRp54 (GenBank accession no. 1813946 and 1813908). Sequence analysis showed that its N-terminal RRM was 34.5% identical (70.2% similar) to the Drosophila RRM and 36.6% identical (73.2% similar) to the human SRp54 RRM. This proposed homolog also contained a substantial SR domain. The potential presence of SRp54 in divergent organisms implies an important role in RNA processing.
When the spacer regions of the two proteins were aligned, it became apparent that the Drosophila protein contains an additional 60 amino acids in the region between the RRM and the peptide epitope compared to the human protein. Comparison of this sequence to the database indicated that this region could be considered a second RRM. Sequence comparison indicated that the closest related RRM sequences were those found in human and Drosophila U1 snRNP 70K proteins. The Drosophila SRp54 sequence is 15.0 or 17.5% identical and 43.8 or 48.8% similar to Drosophila or human U1 70K protein, respectively. It also shows 19.4% identity and 39.5% similarity to a human U1 70K-like protein (3) within this region. The proposed SRp54 from C. elegans also contains a second putative RRM with 31.3% identity and 57.5% similarity to the Drosophila second RRM domain (Fig. 5B).
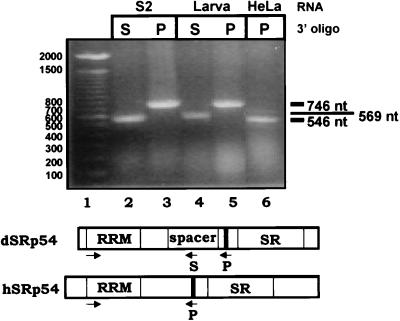
It is unusual to observe such a substantial sequence difference between the human and Drosophila versions of an SR protein. Such a difference raised the possibility of multiple forms of either the human or Drosophila SRp54 protein, with or without the second RRM. To address this question, we performed RT-PCR analysis of mRNA from Drosophila S2 cells, Drosophila third-instar larvae (male and female), and HeLa cells. We used a 5′ primer specific for the N-terminal RRM of each protein and a 3′ primer specific for the common antigenic peptide. We also performed a second amplification of the Drosophila RNAs, using a downstream primer within the second RRM. All of the reactions yielded a single strong amplification product (Fig. 6). Sequencing of the RT-PCR products confirmed their identities. The Drosophila RNA was larger than the human RNA, using the peptide-specific primers reflecting the extra internal domain in the Drosophila protein. While the RT-PCRs all yielded a single product, the possibility remains that other forms of SRp54 exist, possibly in tissues not tested here. We deduce from this experiment that there is a single SRp54 mRNA in the tested RNA populations and that the Drosophila SRp54 mRNA has an additional RRM compared to the human protein.
FIG. 6.
RT-PCR analysis of SRp54 RNA in D. melanogaster and human cells reveals only a single SRp54 mRNA species. Whole-cell RNA from S2 cells, Drosophila third-instar larvae (male and female), or HeLa cells was subjected to RT-PCR amplification using primers that flank the region between the N-terminal RRMs and the epitopes of both proteins to detect potential variants within the spacer region. The 5′ primers were complementary to a region within the N-terminal RRM domain of each gene. Two sets of 3′ primers were used. The first was complementary to nucleotides which encode the peptide against which the anti-SRp54 antibody was raised (labeled P). This region is present in both Drosophila and human SRp54 (dSRp54 and hSRp54) and was used to amplify RNAs from all sources. The expected products are 746 nt (using Drosophila RNA) or 546 nt (using HeLa RNA). The second 3′ primer was specific for the Drosophila RNA and was complementary to the central putative RRM domain (marked S). This amplification product is 569 nt. Marker sizes are given in nucleotides, positions of the primers are diagrammed below, and the sizes of the RT-PCR products are indicated at the right.
SF1 does not bind the wild-type mle intron.
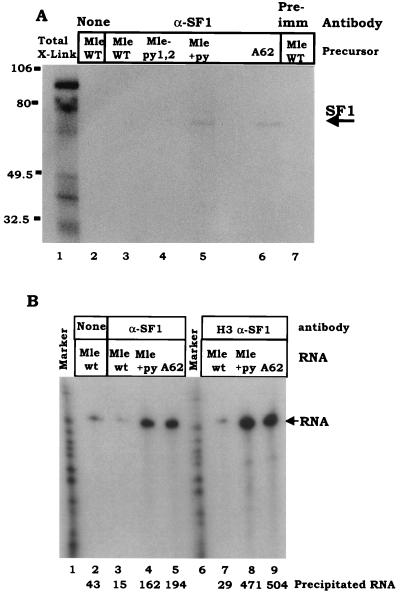
SF1 has been postulated to be a branch point binding protein that bridges introns via binding to U2AF and U1 snRNP-associated proteins (2, 7, 8). Observation of SRp54 binding to the centrally located C-rich pyrimidine tracts in the mle intron raised the question of whether SRp54 obviated the need for participation of SF1 as a splice site bridging factor for this small intron. To investigate whether SF1 is used during mle splicing, we immunoprecipitated UV cross-linked proteins and in vitro-assembled spliceosomes, using antibodies specific for SF1 that were raised against an N-terminal fragment (amino acids 2 to 320) of human SF1 (5) that is highly conserved between humans and Drosophila (22a). As a positive control, an adenovirus-based substrate known to require SF1 (A62) was used. SF1 did interact with the A62 substrate (Fig. 7A, lane 6), as revealed by an immunoprecipitated, cross-linked band of 70 kDa. In contrast, when mle wild-type pre-mRNA was used in the reaction, no detectable cross-linked and immunoprecipitated band was observed (Fig. 7A, lane 3). Therefore, SF1 does not appear to UV cross-link to the mle intron. When the 3′ splice site of the mle intron was strengthened by the addition of a more classical 3′ pyrimidine tract downstream of the branch point, SF1 cross-linking was detected (Fig. 7A, lane 5).
FIG. 7.
SF1 does not UV bind to the wild-type (WT) mle splicing substrate. (A) UV cross-linking of SF1. The radiolabeled precursor RNAs indicated above the gel were subjected to UV cross-linking as described for Fig. 2. mle precursor RNAs are diagrammed in Fig. 1; the A62 construct is an adenovirus-based substrate that has had its single intron internally deleted to 62 nt (see Materials and Methods). Polyclonal antibodies against human SF1 protein were then added to immunoprecipitate cross-linked proteins. Immunoprecipitated proteins were displayed on a sodium dodecyl sulfate–10% polyacrylamide gel. The band corresponding to SF1 is indicated. Markers are as shown. Pre-imm, preimmune serum. (B) Immunoprecipitation of spliceosomes with anti-SF1 antibodies. The indicated precursor RNAs were incubated under standard splicing conditions in S2 extract for 7 min to permit assembly of complex A. Reaction mixtures were immunoprecipitated with the two anti-SF1 peptide antibodies indicated. Radiolabeled RNAs within the immunoprecipitates were displayed on denaturing urea gels. The amount of immunoprecipitated RNA was quantified in the PhosphorImager; relative density units are indicated below the lanes.
This experiment also addresses the appearance of a second band in the UV cross-linking studies represented in Fig. 2A. When mle+py was used as the cross-linking substrate, a 70-kDa protein was detected as interacting with the pre-mRNA that was not present in assays using wild-type RNA (Fig. 2A, lane 7; Fig. 7A, lane 5). The experiment in Fig. 7A suggests that this protein was SF1. Thus, these experiments suggest that SF1 does not cross-link to the wild-type first intron from the mle gene unless a downstream pyrimidine tract has been introduced. Addition of a downstream pyrimidine tract does not affect mle splicing. In this context, however, the upstream pyrimidine tracts are deleterious; i.e., the intron assembles and splices better with a downstream pyrimidine tract if the upstream tracts are mutated (18). Combined with the cross-linking results, this observation suggests that simultaneous association of both SRp54 and SF1 with the mle intron is not optimal.
The absence of UV cross-linking of SF1 did not rule out the presence of SF1 within the mle spliceosome. To better assess this possibility, complexes assembled on the wild-type and mutant mle precursor RNAs were directly immunoprecipitated with two SF1 antipeptide antibodies (Fig. 7B). Antibodies against SF1 were able to immunoprecipitate considerable complex assembled on either the adenovirus-based small intron (A62) or the mle intron to which a strong pyrimidine tract downstream of the branch point had been added. In contrast, little to no complex was immunoprecipitated with the complexes assembled on the wild-type mle intron. Quantification of the amount of radiolabeled RNA precipitated indicated a 10- to 15-fold difference between the ability of the antibody to immunoprecipitate wild-type complexes versus complexes formed on a mutant precursor with a strong downstream pyrimidine tract. This observation suggests that SF1 is not a component of the mle spliceosome.
SRp54 and U2AF contact both the 5′ and 3′ ends of the mle intron.
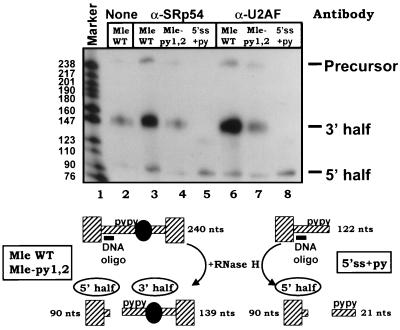
The association of SRp54, but not SF1, with the mle pre-mRNA raised the possibility that either SRp54 or U2AF65 bound to the centrally located C-rich elements functions as a bridging factor(s) for this intron. To address this question, immunoprecipitation of precleaved substrates was used. In these experiments, radiolabeled substrates were incubated in S2 extract for a short period of time sufficient for the assembly of complex A but not subsequent assemblies and subjected to RNase H-mediated cleavage using a DNA oligonucleotide complementary to a region between the 5′ splice site and C-rich pyrimidine tracts. Following cleavage, antibodies against SRp54 or U2AF were used to immunoprecipitate RNA associated with the factors. If the protein that the antibody recognizes interacts with both sides of the intron, directly or indirectly, then both halves of the cleaved substrate should be immunoprecipitated. If, on the other hand, the factor makes contact with one of the ends, then only that part of the substrate should be present in the immunoprecipitate. Figure 8 (lanes 3 and 6) shows that anti-SRp54 and anti-U2AF antibodies immunoprecipitated both the 3′ and the 5′ halves of the mle wild-type intron. This ability was lost and the signal dropped to background levels when the upstream elements were mutated (Fig. 8, lanes 4 and 7). These results support the idea of SRp54 and/or U2AF performing a bridging function in the mle intron, such that one or both proteins contact sequences and/or factors bound to the 5′ and 3′ splice sites. When a substrate that contained only intron sequence from nt +1 to +38 (down to and including the C-rich tracts) was used, both anti-SRp54 and anti-U2AF antibodies were still able to immunoprecipitate the 5′ half of the construct. This observation indicates that the association of SRp54 and U2AF with the 5′ end of the mle intron is independent of any contribution from splicing signals present in the 3′ half of the intron (branch point, 3′ pyrimidine tract, 3′ splice site, or others). As neither a deletion of the 5′ splice site nor a mutation of this signal affected SRp54 cross-linking (Fig. 2, lanes 4 and 5), it is possible that SRp54 and U2AF interact with the upstream C-rich tracts first and then make contact with other factors binding to the 5′ splice site.
FIG. 8.
SRp54 and U2AF contact both ends of the mle intron. Radiolabeled RNA substrates were incubated in S2 extract for 7 min. Following the addition of RNase H and a DNA oligonucleotide (GCAACATAACC) complementary to the region between the 5′ splice site and upstream pyrimidine stretches (nt +7 to +17 of the intron; diagrammed at the bottom), the reaction mixtures were incubated at 30°C for 3 min. They were next subjected to immunoprecipitation using anti-SRp54 or anti-U2AF antibody. Immunoprecipitated RNAs were prepared for electrophoresis on a 10% denaturing polyacrylamide gel. The bands corresponding to precursor RNA or RNAs matching the 5′ or 3′ halves of the substrate following RNase H-mediated cleavage are indicated. Predicted products from each precursor used are depicted below the gel. Marker sizes are given in nucleotides. WT, wild type.
The C-rich tracts need to be located near both the branch point and the 5′ splice site in order for spliceosome formation to proceed.
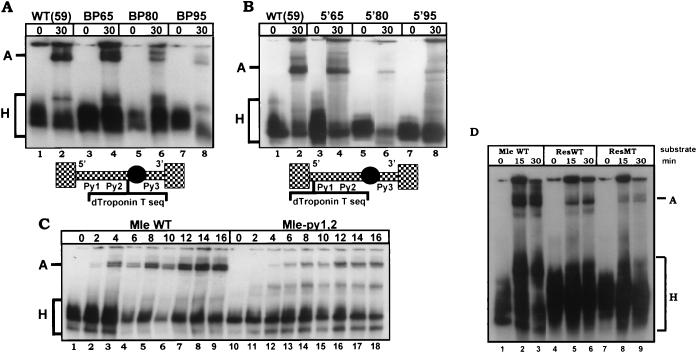
Another possible outcome of a bridging model using centrally located intron signals is that the distance between the interacting cis elements might be critical. To test this possibility, internal expansions were made in the 59-nt mle intron. An NcoI site was introduced either between the C-rich tracts and the branch point (BP65) or between the tracts and the 5′ splice site (5′65). The Drosophila troponin T intron sequence was inserted at this site in BP65 or 5′65 to create BP80 and BP95 or 5′80 and 5′95, respectively. These introns have total lengths of 80 or 95 nt, compared to the 59 nt of the wild-type mle intron. The ability of these various splicing substrates to be recognized was then tested in spliceosome assembly assays. Figure 9A depicts the results obtained with the substrates containing an expansion between the upstream pyrimidine-rich elements and the branch point in an assembly assay. Even a moderate expansion of 6 nt began to inhibit complex A formation, while larger insertions of 21 or 36 nt completely abolished all assembly. Figure 9B displays the phenotypes of the 5′ splice site-pyrimidine tract expansions. These substrates also lose the ability to form complex A when expanded. Thus, there is a requirement for the C-rich tracts to be in close proximity to both the 5′ splice site and branch point for maximal assembly of complex A.
FIG. 9.
Expanding the distance between the C-rich sequences and either the 5′ splice site or branch point inhibits spliceosome complex assembly. (A and B) Spliceosome assembly of expanded mle introns. The 59-nt wild-type mle intron [WT (59)] was expanded to a total length of 65, 80, or 95 nt by additions between the upstream pyrimidine tracts and the branch point (A, the BP series) or between the 5′ splice site and the pyrimidine tracts (B, the 5′ series). All constructs were incubated in S2 extract, aliquots were taken at the indicated time points, and heparin was added to a final concentration of 0.2 mg/ml. The reactions were then displayed on native polyacrylamide gels in order to visualize spliceosome complex assembly. Complexes A and H are indicated. dTroponin T seq, Drosophila troponin T sequence. (C) Spliceosome assembly of the mle intron mutated for the upstream pyrimidine tracts. Assembly of the mle-py1,2 mutant (right) was compared to that of the wild type (left) in a native assembly assay under the conditions described for panel A. (D) Spliceosome assembly of expanded mle introns in which two copies of the wild-type (ResWT) or mutant (ResMT) C-rich tracts were inserted into the expansion cassette. This construct effectively created an expanded intron containing four copies of the C-rich tracts between the 5′ splice site and branch point by duplicating the region containing the C-rich tracts.
The assay used in Fig. 9 indicates a phenotype for the expansion mutants during the assembly of complex A. We previously reported assembly phenotypes for the upstream pyrimidine tract mutations on assembly of complex B, not complex A (18). Our previous experiments used a high concentration of heparin (2.0 mg/ml) to prevent nonspecific associations prior to native gel electrophoresis, whereas the present experiments used a lower concentration (0.2 mg/ml). Reanalysis of the assembly phenotype of upstream pyrimidine tract mutations on spliceosome assembly with the lower heparin concentration revealed that the major effect of the mutations was on complex A, not complex B, formation (Fig. 9C). The lower-heparin conditions also caused the disappearance of a wild-type complex running just above complex H that is dependent on the upstream C-rich tracts, ATP, and U1 and U2 snRNPs for formation (data not shown), and which presumably reflects a form of complex A that has lost U2 snRNPs. This observation suggests that complex A formed on the mle intron partially disassociates in high heparin, perhaps reflecting the small size of the intron and the restrictions such size imposes on stable complex assembly. Furthermore, it indicates that the upstream pyrimidine tracts are required for complex A formation and the introduction of U2 snRNPs into the spliceosome.
To see if the intron expansions could be rescued by the introduction of additional C-rich elements, we created a set of constructs in which two copies of the repeat, both wild type and mutant, were introduced into one of the mle intron expansions. This addition created a minimally expanded intron with four copies of the element between the 5′ splice site and branch point. Analysis of spliceosome assembly of the two constructs indicated that the additional repeats provided better assembly than a mutant construct but did not restore assembly to the wild-type level (Fig. 9D). This result is what would be expected if factor(s) bound to the C-rich tracts were required to simultaneously contact unique factors bound on either side. This result also supports earlier reported observations that assembly of complex A on the mle intron requires both a 5′ and 3′ splice site (29) and suggested that the intron is recognized as a unit of splice sites and internal C-rich pyrimidine tracts.
Expanding the distance between the upstream pyrimidine tracts and either the branch point or 5′ splice site depresses factor association with the 5′ splice site.
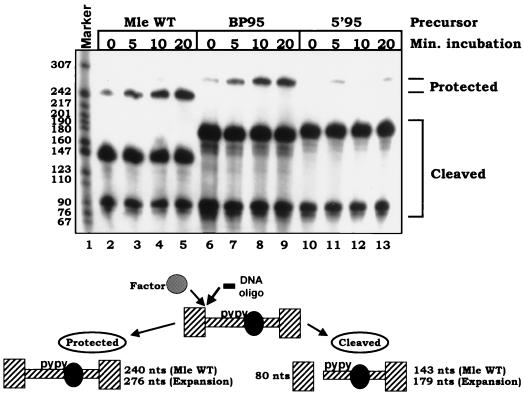
The assembly results discussed above indicate that the upstream pyrimidine tracts are necessary for the association of U2 snRNPs with the spliceosome so as to form complex A. To assess the requirement for the upstream tracts on events at the 5′ splice site, we analyzed the ability of splicing factors (e.g., U1 snRNP) to bind to and protect the 5′ splice site. Expansion of the mle intron on either side of the upstream pyrimidine tracts lowered the ability of factors to protect the 5′ splice site against digestion with RNase H and a complementary oligonucleotide (Fig. 10). Expansion upstream of the C-rich tracts severely inhibited binding of factors to the 5′ splice site (22% protection compared to the wild type), whereas expansion downstream of the tracts afforded 50% of the protection given by the wild-type substrate. We have previously reported that mutation of the upstream tracts severely inhibited protection of the 5′ splice site and that the protection was U1 dependent (18). Therefore, the upstream tracts are necessary for the binding of both U2 and U1 snRNPs to the ends of the intron.
FIG. 10.
Association of factors with the 5′ splice site is depressed by expanding the distance between the upstream pyrimidine tracts and either the 5′ splice site or branch point. To monitor 5′ splice site protection, RNase H along with an oligonucleotide complementary to the 5′ splice site (nt −4 to +13) was added during an assembly reaction using the depicted expansion constructs. Time points indicate time (minutes) of addition of the oligonucleotide. RNA products were displayed on a 5% denaturing polyacrylamide gel. Protection is indicated by the appearance of a band of the length of precursor RNA and a reduction in the amount of cleavage product. Precursor RNAs are as in Fig. 7. Anticipated cleavage and protection products are diagrammed below the gel. Marker sizes are given in nucleotides. WT, wild type.
DISCUSSION
Examination of the factors involved in pre-mRNA splicing has revealed an amazing conservation from yeasts to humans. Both snRNPs and other factors are conserved, suggesting that constitutive recognition of splicing signals may be very similar for all introns. U2AF is one such conserved factor participating in one of the earliest steps of recognition by binding to sequences at the 3′ end of the intron and facilitating binding of U2 snRNPs to the branch point (17, 30, 35, 36). Another conserved protein is SF1, which binds to the branch point region (5, 7). U2AF and SF1 cooperatively interact to recognize the branch point and pyrimidine tract (8). In the yeast Saccharomyces cerevisiae, genetic interactions have established that SF1 acts to bridge the intron by making contacts with both U2AF and proteins bound to U1 snRNPs (2). Because the U1 snRNP proteins implicated in SF1-mediated bridging have not yet been observed in vertebrates, it is unclear if SF1 plays this role in assembly of the mammalian spliceosome. The conservation between the yeast and mammalian proteins, however, argues that SF1-mediated bridging should be universal.
Intron size and sequence diverges greatly across the eucaryotic kingdom. Typical vertebrate introns are large and contain noticeable pyrimidine tracts in the region between the branch point and 3′ splice site (16). These tracts are the binding site for U2AF (35). In contrast, many nonvertebrates have small introns, many of which are so small that they cannot function in a vertebrate system (14, 29). In addition, they frequently lack recognizable pyrimidine tracts downstream of the branch point (23). Here we suggest that one of these introns from the Drosophila mle gene may utilize a mode of early assembly different from that defined for other introns, in either yeasts or vertebrates.
The mle intron has unusual C-rich pyrimidine tracts located in the short interval between the 5′ splice site and branch point that are required for maximal spliceosome assembly and the binding of U2AF and the SR protein SRp54. Unlike most SR proteins, SRp54 contacts U2AF via its large subunit (U2AF65) rather than its small subunit (U2AF35) (37). Our data suggest that this mode of interaction could potentially serve to create a bridge for small introns, obviating the need for the involvement of SF1. Bridging was revealed by the requirement for the sequence and location of the C-rich tracts for the association of U1 and U2 snRNPs, as well as the ability of SRp54 and U2AF to contact both the 5′ and 3′ splice sites, either directly or indirectly. This bridging appeared to operate in the absence of SF1 because, in contrast to spliceosomes formed with precursor RNAs with normal 3′ pyrimidine tracts, mle spliceosomes could not be immunoprecipitated with anti-SF1 antibodies.
The mle intron lacks a classical pyrimidine tract between the branch point and 3′ splice site. When such a site was added, the intron functioned; but in this context, the upstream C-rich tracts were inhibitory for assembly and activity (18), suggesting alternative competitive modes of early recognition. Addition of the downstream tracts was accompanied by the ability to detect 10- to 15-fold higher levels of immunoprecipitation of the mle spliceosome with antibodies specific for SF1, suggesting that SF1 contacts the mle intron only in the presence of downstream tracts. Furthermore, these results suggest that simultaneous association of SRp54 and SF1 with the intron is not optimal for assembly or activity.
Taken together, these observations suggest that when introns are very small, U2AF and SRp54 may function to provide early recognition and intron bridging. SR proteins have been observed to provide both cross-exon and cross-intron interactions in a number of vertebrate pre-mRNAs (reviewed in references 9, 13, 22, and 25). Models for these interactions invoke associations with U2AF that involve the SR domain of the small subunit of U2AF. In contrast, SF1 bridging models invoke an interactions with the large subunit of U2AF (2). The relationship between the two modes of bridging has been unclear.
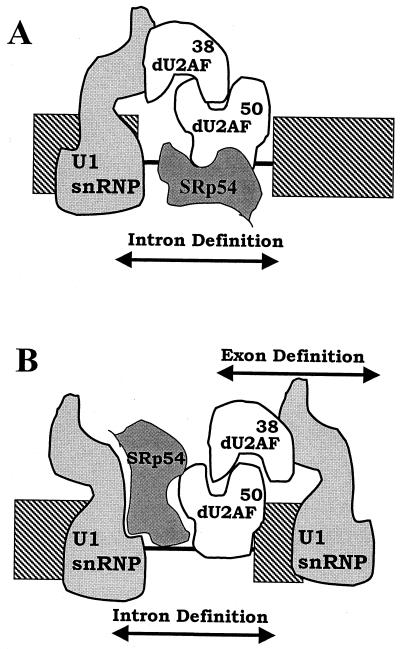
Our results suggest that individual introns may differ in the mechanism of bridging employed and raise the possibility of gene-specific early assembly. In S. cerevisiae, branch points are well conserved but pyrimidine tracts are relatively rare and play only a weak role in intron recognition. Accordingly, MUD2 (the gene encoding the U2AF65 homolog) is inessential for viability; instead SF1 (BBP) plays a pivotal role (2). In contrast, in vertebrates, U2AF is required and binds to a prominent splicing signal, the 3′ polypyrimidine tract (36) and also contacts the branch site (30). Bridging is afforded by SR proteins and presumably by SF1. Our data suggest a third scenario operating in small introns such as those in Drosophila or C. elegans. In these cases, U2AF and SRp54 function to bind to pyrimidine tracts in the vicinity of the branch point, thereby providing intron bridging. One scenario for this bridging is that SRp54, via its ability to bind the upstream pyrimidine tracts in the mle intron, facilitates U2AF binding upstream of the branch point and positions the small subunit of U2AF to make optimal interactions with the U1 snRNP 70K protein (Fig. 11A). This model presents the mle intron as a recognition unit equivalent to the SR-mediated recognition units postulated in exon definition in vertebrates.
FIG. 11.
Suggested models for participation of U2AF and SRp54 in splice site bridging. In model A, the SR-rich small subunit of Drosophila U2AF (dU2AF) contacts the U1 snRNP 70K protein to affect interactions across a small intron. Use of pyrimidine tracts upstream of the branch point is postulated to position U2AF such that participation of SF1 or other normal bridging proteins is unnecessary. In model B, the SR protein SRp54 is used to make contacts with factors bound to the 5′ splice site, whereas the small subunit of U2AF is used to bridge the downstream exon. This model suggests how both intron and exon definition could operate in adjacent areas of a single pre-mRNA.
Also possible, however, is an alternative mechanism whereby U2AF participates in simultaneous interactions across a neighboring exon and intron. In recent years, both intron and exon bridging models for recognition of splice sites in multiexon RNAs have been proposed (1, 2, 6, 9, 25, 27, 38). In general, organisms with small introns are thought to operate via intron bridging, whereas in organisms with large introns, exon bridging is proposed. D. melanogaster presents an example of an organism with a split exon/intron organization such that small and large introns are mixed within individual transcription units. The mle gene is one such split unit. The intron studied in this report is quite small; the adjacent intron is considerable larger. Such mixed architecture presents problems for models of splice site pairing that use either consistent exon or intron bridging. The results in this study suggest a mechanism whereby both could occur simultaneously (Fig. 11B). As this model depicts, a small upstream intron using a bridging factor such as SRp54 or SF1 that contacts the large subunit of U2AF would permit simultaneous interaction of the small subunit of U2AF with factors binding the downstream exon.
As part of this study, we sequenced the Drosophila homolog of SRp54. While considerable conservation was found within both the RRM and SR domains, the Drosophila protein contained extra sequence within the spacer region. Analysis of this sequence revealed that it resembled an RRM. In fact, computer searches attempting to find motifs similar to this sequence yielded the RRM regions of both the human and Drosophila U1 snRNP 70-kDa proteins. The proposed C. elegans SRp54 also contains this second RRM domain, suggesting that in these nonvertebrates, the protein may possess some unique functions. The exact role of this second RRM is unknown. It could affect the affinity and specificity of the first RRM, it could function in protein-protein interactions, or it could perform a completely unique activity related to small-intron recognition (11, 21, 24, 34). Experiments comparing and contrasting the Drosophila and human proteins should lend insight into the function of this protein.
ACKNOWLEDGMENTS
We thank N. Chaudhary for generous gifts of SRp54-specific antibodies and discussions about the protein, and we thank R. Sierra for excellent technical experience and for sequencing Drosophila SRp54.
C.K. and S.M.B. were supported by a fellowship from the Robert A. Welch Foundation and NIH RO1 GM38526.
REFERENCES
- 1.Abovich N, Liao X C, Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- 2.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 3.Adams D S, Li Q, Szabo T, Tan X, Pero S, Czop J K. Cloning and characterization of a family of cDNAs from human histiocyte macrophage cells encoding a glycine/arginine-rich basic protein related to the highly conserved 70 kD U1-snRNP protein. 1996. GenBank accession no. 1174217 annotation. [DOI] [PubMed] [Google Scholar]
- 4.Adams M D, Rudner D Z, Rio D C. Biochemistry and regulation of pre-mRNA splicing. Curr Opin Cell Biol. 1996;8:331–339. doi: 10.1016/s0955-0674(96)80006-8. [DOI] [PubMed] [Google Scholar]
- 5.Arning S, Gruter P, Bilbe G, Krämer A. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA. 1996;2:794–810. [PMC free article] [PubMed] [Google Scholar]
- 6.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;2:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 7.Berglund J A, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 8.Berglund J A, Abovich N, Rosbash M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black D L. Finding splice sites in a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 10.Champion-Arnaud P, Gozani O, Palandijian L, Reed R. Accumulation of a novel spliceosomeal complex on pre-mRNAs containing branch site mutations. Mol Cell Biol. 1995;15:5750–5756. doi: 10.1128/mcb.15.10.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandler S D, Mayeda A, Yeakley J M, Krainer A R, Fu X D. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc Natl Acad Sci USA. 1997;94:3596–3601. doi: 10.1073/pnas.94.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhary N, McMahon C, Blobel G. Primary structure of a human arginine-rich nuclear protein that colocalizes with spliceosome components. Proc Natl Acad Sci USA. 1991;88:8189–8193. doi: 10.1073/pnas.88.18.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu X-D. The superfamily of arginine/serine rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 14.Guo M, Lo P C H, Mount S M. Species-specific signals for the splicing of a short Drosophila intron in vitro. Mol Cell Biol. 1993;13:1104–1118. doi: 10.1128/mcb.13.2.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991;253:157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- 16.Hawkins J D. A survey on intron and exon lengths. Nucleic Acids Res. 1988;16:9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanaar R, Roche S E, Beall E L, Green M R, Rio D C. The conserved pre-mRNA splicing factor U2AF from Drosophila: requirement for viability. Science. 1993;262:569–572. doi: 10.1126/science.7692602. [DOI] [PubMed] [Google Scholar]
- 18.Kennedy C F, Berget S M. Pyrimidine tracts between the 5′ splice site and branch point facilitate splicing and recognition of a small Drosophila intron. Mol Cell Biol. 1997;17:2774–2780. doi: 10.1128/mcb.17.5.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krämer A. Purification of splicing factor SF1, a heat-stable protein that functions in the assembly of a presplicing complex. Mol Cell Biol. 1992;12:4545–4552. doi: 10.1128/mcb.12.10.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 21.Lu J, Hall K B. An RBD that does not bind RNA: NMR secondary structure determination and biochemical properties of the C-terminal RNA binding domain from the human U1A protein. J Mol Biol. 1995;247:739–752. doi: 10.1006/jmbi.1995.0177. [DOI] [PubMed] [Google Scholar]
- 22.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:16569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 22a.Mazroui, R., and A. Krämer. Unpublished data.
- 23.Mount S M, Burks C, Hertz G, Stormo G D, White O, Fields C. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 1992;20:4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez I, McAfee J G, Patton J G. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry. 1997;36:11881–11890. doi: 10.1021/bi9711745. [DOI] [PubMed] [Google Scholar]
- 25.Reed R. Initial splice site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- 26.Rio D C. Splicing of pre-mRNA: mechanism, regulation and role in development. Curr Opin Genet Dev. 1993;3:574–584. doi: 10.1016/0959-437x(93)90093-5. [DOI] [PubMed] [Google Scholar]
- 27.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh R, Valcárcel J, Green M R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine-tract binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 29.Talerico M, Berget S M. Intron definition in splicing of small Drosophila introns. Mol Cell Biol. 1994;14:3434–3445. doi: 10.1128/mcb.14.5.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valcárcel J, Gaur R K, Singh R, Green M R. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 31.Valcárcel J, Green M R. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- 32.Wang Z, Hoffman H M, Grabowski P J. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 34.Xu R M, Jokhan I, Cheng X, Mayeda A, Krainer A R. Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA recognition motifs. Structure. 1997;5:559–570. doi: 10.1016/s0969-2126(97)00211-6. [DOI] [PubMed] [Google Scholar]
- 35.Zamore P D, Green M R. Identification, purification and characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci USA. 1989;86:9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zamore P D, Green M R. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intracellular distribution. EMBO J. 1991;10:207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W-J, Wu J Y. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol Cell Biol. 1996;16:5400–5408. doi: 10.1128/mcb.16.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]