Abstract
The IκBα protein is able both to inhibit nuclear import of Rel/NF-κB proteins and to mediate the export of Rel/NF-κB proteins from the nucleus. We now demonstrate that the c-Rel–IκBα complex is stably retained in the cytoplasm in the presence of leptomycin B, a specific inhibitor of Crm1-mediated nuclear export. In contrast, leptomycin B treatment results in the rapid and complete relocalization of the v-Rel–IκBα complex from the cytoplasm to the nucleus. IκBα also mediates the rapid nuclear shuttling of v-Rel in an interspecies heterokaryon assay. Thus, continuous nuclear export is required for cytoplasmic retention of the v-Rel–IκBα complex. Furthermore, although IκBα is able to mask the c-Rel-derived nuclear localization sequence (NLS), IκBα is unable to mask the v-Rel-derived NLS in the context of the v-Rel–IκBα complex. Taken together, our results demonstrate that IκBα is unable to inhibit nuclear import of v-Rel. We have identified two amino acid differences between c-Rel and v-Rel (Y286S and L302P) which link the failure of IκBα to inhibit nuclear import and DNA binding of a mutant c-Rel protein to oncogenesis. Our results support a model in which loss of IκBα-mediated control over c-Rel leads to oncogenic activation of c-Rel.
The Rel/nuclear factor κB (NF-κB) family of eukaryotic transcription factors regulates the expression of genes involved in immune and inflammatory responses (for reviews, see references 4 and 26). Rel family members are characterized by the presence of a 300-amino-acid domain termed the Rel homology domain, which encompasses the sequences required for DNA binding, dimerization, and nuclear translocation. The Rel family of proteins includes NF-κB1 (p50/p105), NF-κB2 (p52/p100), p65 (RelA), RelB, and c-Rel. The activity of Rel proteins is modulated in large part through association with one or more members of the inhibitor-of-κB (IκB) family of proteins (for reviews, see references 4 and 26). The IκB family of proteins is characterized by the presence of multiple copies of ankyrin repeats and includes IκBα, IκBβ, IκBγ, IκBɛ, IκBR, Bcl-3, NF-κB1 (p105), and NF-κB2 (p100).
The IκBα protein efficiently controls the nuclear-cytoplasmic distribution of dimeric Rel complexes that contain either c-Rel or p65 (RelA). In unstimulated cells, IκBα sequesters the dimeric Rel complex in the cytoplasm, presumably through masking of the nuclear localization sequence (NLS) within Rel proteins (5, 23, 44, 60). Upon exposure of cells to a variety of extracellular stimuli, IκBα becomes phosphorylated at two amino-terminal serine residues by the IκB kinase complex (15, 34, 41, 58, 61). Signal-induced phosphorylation of IκBα targets IκBα for ubiquitin-dependent degradation by the 26S proteasome (1, 11, 17, 49, 54, 59). Degradation of IκBα enables the free Rel dimer to translocate to the nucleus and activate κB-dependent gene expression. One of the target genes of Rel proteins is the IκBα gene itself, resulting in the rapid induction of newly synthesized IκBα protein (31, 32, 48, 52). Newly synthesized IκBα is able to enter the nucleus, bind to Rel proteins, and direct the nuclear export of the Rel-IκBα complex (2, 3, 46). The ability of IκBα to both inhibit nuclear import of Rel proteins and export Rel proteins from the nucleus provides an effective mechanism for ensuring that transcriptional activation of gene expression by Rel proteins occurs in a regulated and transient manner.
The importance of tight regulation of the transcriptional activation property of Rel proteins is highlighted by the involvement of Rel proteins in tumorigenesis. For example, Ras-induced activation of NF-κB is required to suppress apoptosis and thereby facilitate Ras-mediated oncogenic transformation (19, 33). C-terminal rearrangements of the NF-κB2 gene, resulting in mutant p100-related proteins that display increased nuclear localization and transcriptional activation properties, have been implicated in human lymphomas (10). Taken together, these results suggest that constitutive nuclear activation of Rel proteins contributes to oncogenic processes.
The v-Rel oncoprotein has been a prototype for understanding how Rel family members are able to mediate oncogenic transformation (for a review, see reference 26). We have previously shown that v-Rel-mediated oncogenic transformation requires a threshold level of nuclear v-Rel (45). However, the mechanism by which this nuclear threshold level of v-Rel is established or maintained is not known. The v-Rel protein is predominantly cytoplasmic in v-Rel-transformed avian lymphocytes, presumably due to its association with avian IκB proteins, including IκBα, NF-κB1 (p105), and NF-κB2 (p100) (13, 25, 30, 50). Ectopic expression of IκBα is unable to inhibit v-Rel-mediated transformation of avian lymphoid cells or to prevent tumor formation in transgenic mice expressing v-Rel (8, 45). These results present the hypothesis that the threshold level of nuclear v-Rel is established by the inability of IκB proteins to efficiently regulate v-Rel. Consistent with this notion, IκBα is unable to efficiently inhibit the DNA-binding property of v-Rel (8, 16). In this report, we demonstrate that IκBα is also unable to inhibit nuclear import of v-Rel. Rather, IκBα-mediated partitioning of v-Rel between the cytoplasm and the nucleus is accomplished by an export-dependent process.
The v-Rel oncoprotein, a mutant form of c-Rel transduced by the avian retrovirus Rev-T, contains a number of differences from c-Rel (57). In particular, v-Rel lacks the C-terminal 118 amino acids, which removes a potent transcriptional activation domain (42). Furthermore, v-Rel contains 13 single-amino-acid changes and three in-frame deletions relative to c-Rel (57). We have identified two amino acid differences between c-Rel and v-Rel that are responsible for the failure of IκBα to inhibit nuclear import of v-Rel. The same two amino acid differences are also responsible for the inability of IκBα to displace v-Rel from the v-Rel–DNA complex. Our results establish a clear link between failure of IκBα to regulate nuclear import and DNA binding of c-Rel and oncogenic activation of c-Rel.
MATERIALS AND METHODS
Construction of recombinant DNA molecules.
Many of the recombinant DNA molecules used in this study have been described previously (16, 44). The construction of additional recombinant DNA molecules was performed by standard techniques (47). An EcoRI fragment containing the avian IκBα cDNA was used as the progenitor for all of the mutant IκBα genes utilized in this study (14). All point mutations were constructed from phagemid single-stranded DNA. The presence of each mutation within the respective cDNAs was confirmed by nucleotide sequence analysis. The epitope-tagged IκBα protein (IκBα-LBD) contains an 18-amino-acid peptide derived from the ligand binding domain (LBD) of platelet-derived growth factor. The LBD epitope tag consists of the sequence EVIVVPHSLPFML. A plasmid containing a segment of DNA encoding the LBD epitope tag and antipeptide serum against the LBD epitope tag were provided by Dan Donoghue (University of California). The IκBα genes were expressed in chicken embryo fibroblasts (CEF) by using a retroviral vector derived from pJD214 (18) and in monkey COS-1 cells by using either a spleen necrosis virus (SNV)-derived or a cytomegalovirus (CMV)-derived vector (6). The chicken c-Rel and turkey v-Rel cDNAs have been described previously (16).
Dual-expression vectors containing an internal ribosome entry site (IRES) were constructed from a Rev-T-derived plasmid, pVV, and an IRES-containing plasmid (24, 53). The c-Rel-540 or v-Rel genes were inserted in the appropriate orientation 5′ of the IRES as XbaI fragments. The wild-type or mutant IκBα genes were inserted in the appropriate orientation 3′ of the IRES as NcoI fragments. Details of all plasmid constructions and primer sequences are available upon request.
Cell culture and transfection.
CEF were obtained from Spafas and grown in M199 containing 10% tryptose phosphate and 10% fetal calf serum (FCS). DNA transfections into CEF were typically performed with a total of 10 μg of the retroviral expression plasmids (cotransfections were performed with 5 μg of each plasmid) and 0.3 μg of a replication-competent DNA clone of SNV, pSW253 (18). Transfections were performed with calcium phosphate coprecipitates onto 2 × 105 to 2.5 × 105 CEF per 60-mm-diameter dish. The biochemical properties and cellular localization of the Rel or IκBα proteins were typically analyzed 4 to 5 days after transfection of CEF with the appropriate plasmids.
COS-1 cells were grown in Dulbecco’s modified Eagle’s medium containing 10% FCS. Transfections were performed with 2 μg of the retroviral plasmids or with 2 μg of pCMV4-based expression plasmids (cotransfections were performed with 1 μg of each plasmid) and 15 μl of Lipofectamine (GIBCO BRL) per sample onto 2.5 × 105 COS-1 cells per 35-mm-diameter dish. The biochemical experiments and the cellular localization of the Rel proteins were typically analyzed 36 to 48 h after transfection of COS-1 cells with the appropriate plasmids.
Rel-transformed cells were grown in RPMI 1640 containing 15% FCS.
Indirect immunofluorescence.
Indirect immunofluorescence from CEF, COS-1 cells, or interspecies heterokaryons was conducted on coverslips with the appropriate antisera as previously described (25). Polyclonal rabbit antiserum directed against v-Rel (R2) or monoclonal mouse ascitic fluid directed against c-Rel (3C1; provided by Henry R. Bose, Jr., University of Texas) was used to detect the ectopically expressed Rel proteins. Polyclonal rabbit antiserum directed against IκBα (R1807) was used to detect the ectopically expressed IκBα proteins. The appropriate anti-rabbit or anti-mouse fluorescein isothiocyanate-conjugated secondary antibody (Jackson Laboratories) or anti-rabbit Cy5-conjugated secondary antibody (Jackson Laboratories) was used for detection of the ectopically expressed proteins. Indirect immunofluorescence from Rel-transformed cells was conducted with the R2 antiserum as previously described (25). In some experiments, leptomycin B (provided by Minoru Yoshida, University of Tokyo) was added to the culture medium at the indicated time at a concentration of 5 nM prior to fixation of the cells for indirect immunofluorescence. The coverslips were mounted on glass slides by using Mowiol containing 2.5% DABCO (Sigma).
Immunoprecipitation analyses of Rel and IκBα proteins.
Cell lysates for the immunoprecipitation experiments were prepared in ELB (50 mM Tris-HCl [pH 7.9], 250 mM NaCl, 0.1% Triton X-100, 5 mM EDTA, and 1 mM dithiothreitol). The protease inhibitors used were 1 mM phenylmethylsulfonyl fluoride; antipain, aprotinin, leupeptin, and soybean trypsin-chymotrypsin inhibitor (5 μg/ml each); and pepstatin (0.5 μg/ml). The phosphatase inhibitors used were 0.4 mM sodium orthovanadate and 1 mM sodium fluoride. The expression of the Rel and IκBα proteins within the ELB cell lysates was confirmed by immunoblot analysis with the enhanced chemiluminescence (ECL) system (Amersham) with polyclonal rabbit antiserum directed against v-Rel (R3) or against IκBα (R1807). Equivalent amounts of protein were used for the immunoprecipitation analyses. Immunoprecipitation of LBD-tagged IκBα proteins was performed with 3 μl of affinity-purified anti-LBD serum per sample. The presence of c-Rel or v-Rel in anti-LBD immunoprecipitates was determined by immunoblot analysis. A hybridoma supernatant directed against c-Rel (monoclonal antibody HY87; provided by Henry R. Bose, Jr.) was used at a dilution of 1:75 as the primary antiserum. The secondary antiserum was anti-mouse immunoglobulin G conjugated to horseradish peroxidase (New England Biolabs) used at a dilution of 1:4,000. In some experiments, cell lysates were incubated either with 3 μl of the mouse anti-NLS serum or with 3 μl of the mouse anti-NLS serum that had been preincubated with 30 μg of the NLS peptide prior to addition to cell lysates.
Immunoprecipitation of Rel proteins was performed with 3 μl of mouse anti-NLS peptide serum per sample. A peptide encompassing the Rel-derived NLS (GNKAKRQRSTLAWQKC) was coupled to keyhole limpet hemocyanin via the C-terminal cysteine residue and was injected into mice for production of the anti-NLS serum. The presence of the Rel proteins in anti-NLS immunoprecipitates was determined by immunoblot analysis with the R3 antiserum used at a dilution of 1:4,000. The secondary antiserum was anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (Amersham) used at a dilution of 1:4,000. Preliminary experiments were conducted to confirm that all immunoprecipitations were performed with an antibody excess to ensure quantitative immunoprecipitation of the respective proteins.
DNA binding by Rel proteins.
DNA binding by Rel proteins was assayed by solution UV cross-linking (16). Typically, 3 μl of ELB cell lysate from COS-1 cells singly transfected with the appropriate CMV-derived expression plasmid was equilibrated in binding buffer (20 mM HEPES [pH 7.9], 50 mM KCl, 1.0 mM EDTA, 5% glycerol) for 10 min in the presence of 2 μg of poly(dI/dC) prior to the addition of a double-stranded oligonucleotide containing a palindromic κB site. Primer extension was used to incorporate bromodeoxyuridine (BrdU) and 32P-labeled deoxynucleotides (dGTP and dCTP) into the double-stranded oligonucleotide. The cell lysates were incubated with the BrdU- and 32P-labeled κB oligonucleotide for 10 min prior to exposure to UV radiation for 5 min. The samples were boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer for 3 min prior to SDS-PAGE. Hexahistidine-tagged IκBα was purified by nickel-chelate chromatography from insect cells (Trichoplusia sp.) infected with a baculovirus expression vector encoding the IκBα protein. To measure the ability of IκBα to displace the Rel proteins from DNA, increasing amounts of the purified IκBα protein were added to the DNA-binding reaction mixtures 10 min after the addition of the BrdU- and 32P-labeled κB oligonucleotide. The amount of Rel proteins that remained bound to DNA after a 20-min incubation with exogenously added IκBα was determined by solution UV cross-linking and subsequent SDS-PAGE.
Transformation of avian lymphoid cells.
Soft-agar transformation assays were conducted as previously described (45). Equivalent expression of Rel proteins in the transfected CEF was confirmed by immunoblot analysis of CEF lysates.
Transient-transfection interspecies heterokaryon assay.
Interspecies heterokaryon assays were performed essentially as described previously (35). NIH 3T3 mouse fibroblasts were cultured in Dulbecco’s modified Eagle’s medium containing 10% FCS. In brief, CEF grown in 60-mm-diameter dishes were transfected with the appropriate dual vector expression plasmid encoding both Rel and either wild-type or mutant IκBα proteins. At 96 h posttransfection, 2.5 × 105 to 3 × 105 transfected CEF were seeded onto glass coverslips in 30-mm-diameter dishes. After overnight incubation, these cultures were seeded with 2.5 × 105 to 3 × 105 NIH 3T3 cells in M199 containing 10% tryptose phosphate and 10% FCS and were incubated for an additional 3 h in a 37°C CO2 incubator. At 30 min prior to fusion, 100 μg of cycloheximide (CHX) per ml was added to each of the cocultures. For fusion, the coverslips were placed cell side down onto prewarmed polyethylene glycol 8000 (Sigma) (50% [wt/vol] in Hanks’ balanced salt solution lacking calcium and magnesium). After 2 min, the coverslips were removed and washed extensively with Hanks’ balanced salt solution lacking calcium and magnesium. The coverslips were then transferred to prewarmed M199 containing 10% tryptose phosphate, 10% FCS, 100 μg of CHX per ml, and 10 μM cytosinarabinoside and were incubated for 30 min in a 37°C CO2 incubator. In some experiments, leptomycin B (provided by Minoru Yoshida) was included during the 30-min postfusion period at a concentration of 5 nM. The cells were fixed and stained for the localization of Rel proteins by indirect immunofluorescence with the R2 antiserum. Hoechst 33258 (Sigma) was included at a concentration of 5 μg/ml during the secondary antibody incubations to preferentially stain the mouse nuclei. A heterokaryon was scored as positive for nuclear shuttling if approximately equivalent staining of the Rel protein was detected in both nuclei of the heterokaryon.
RESULTS
Cytoplasmic localization of the v-Rel–IκBα complex is sensitive to leptomycin B and requires NES-like motifs in IκBα.
We have previously demonstrated that the avian c-Rel and v-Rel proteins differ markedly in their in vivo interactions with the avian IκBα protein (43). In particular, we identified mutant IκBα proteins that were able to associate with and retain the c-Rel protein in the cytoplasm but were completely deficient for association with and cytoplasmic retention of the v-Rel oncoprotein (43). Differential regulation of c-Rel and v-Rel by mutant IκBα proteins led us to put forth the hypothesis that the wild-type IκBα protein might retain c-Rel and v-Rel in the cytoplasm by distinct mechanisms. Specifically, we proposed that the cytoplasmic localization of the c-Rel–IκBα complex is derived from the ability of IκBα to inhibit nuclear import of c-Rel, while the cytoplasmic localization of the v-Rel–IκBα complex is derived from continuous IκBα-mediated nuclear export of the v-Rel–IκBα complex.
To further investigate the mechanism(s) responsible for cytoplasmic retention of Rel proteins by IκBα, we examined the cellular distributions of the individual proteins and of the respective Rel-IκBα complexes in transfected CEF. As the full-length c-Rel protein is predominantly cytoplasmic in CEF (7), we utilized a C-terminally truncated c-Rel protein (c-Rel-546 [Fig. 1]). The c-Rel-546 protein, like v-Rel, is predominantly nuclear when ectopically expressed in CEF (44) (Table 1). Likewise, the IκBα protein is predominantly nuclear when ectopically expressed in CEF (12, 36, 46). The use of c-Rel-546 and v-Rel in our experiments permitted us to examine functional differences in IκBα-mediated control over the nuclear localization of c-Rel and v-Rel following brief treatment with leptomycin B. Leptomycin B is a specific inhibitor of Crm1, a recently identified protein which mediates the nuclear export of leucine-rich nuclear export sequences (NESs) (21, 22, 39, 40, 51). Leptomycin B treatment did not alter the predominantly nuclear localization of the individual Rel and IκBα proteins (data not shown).
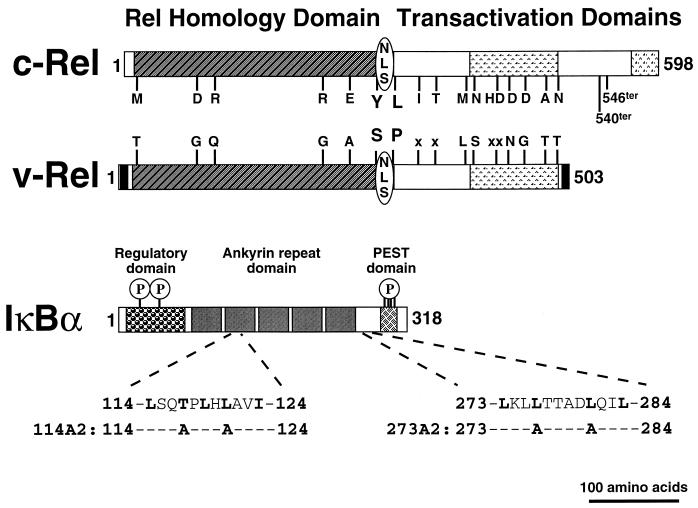
FIG. 1.
Domain organization of Rel and IκBα proteins. The c-Rel (top), v-Rel (middle), and IκBα (bottom) proteins are represented by rectangular boxes. The numbers to the left of each box indicate the first amino acid of each protein, and the numbers to the right of each box indicate the total number of amino acids in the respective proteins. The Rel homology domain is located at the N terminus of c-Rel and v-Rel. Both c-Rel and v-Rel contain an NLS located at the C terminus of the Rel homology domain. The c-Rel protein contains at least one additional transactivation domain located at its C terminus that is not present in v-Rel. The env-derived sequences uniquely present in v-Rel are indicated by the black boxes at the N and C termini of v-Rel. The v-Rel protein is identical in amino acid sequence throughout the entire Rel open reading frame except for the indicated amino acid substitutions, denoted in the single-letter code, and several small deletions, denoted by x to indicate the absence in v-Rel of the corresponding amino acid that is present in c-Rel. Termination codons (ter) were introduced into the full-length c-Rel protein following amino acid residue 540 or 546 to construct the c-Rel-540 or c-Rel-546 protein. The IκBα protein contains an N-terminal regulatory domain, a central domain containing multiple ankyrin-related repeats, and an acidic serine-rich (PEST) domain near the C terminus. The two sites of N-terminal cytokine-inducible serine phosphorylation and the four sites of constitutive serine phosphorylation within the C-terminal PEST domain of IκBα are indicated by the circled P’s. The amino acid sequences of two clusters of hydrophobic residues within the avian IκBα protein are indicated in the single-letter code below the rectangle representing IκBα. The critical residues relevant to the work in this study are in boldface, and the mutations introduced into IκBα are indicated.
TABLE 1.
Localization of Rel proteins in CEFa
| Rel proteinb | IκBα protein | LMB treatment | % of cellsc
|
||
|---|---|---|---|---|---|
| N | N-C | C | |||
| c-Rel-546 | None | None | 60 | 21 | 19 |
| WT IκBα | None | 1 | 9 | 90 | |
| WT IκBα | 30 min | 2 | 85 | 13 | |
| WT IκBα | 4 h | 8 | 90 | 2 | |
| A2/A2 | None | 3 | 9 | 88 | |
| c-Rel-546-SP | None | None | 68 | 29 | 3 |
| WT IκBα | None | 8 | 11 | 81 | |
| WT IκBα | 30 min | 58 | 42 | <1 | |
| WT IκBα | 4 h | 95 | 4 | 1 | |
| A2/A2 | None | 40 | 56 | 4 | |
| v-Rel-YL | None | None | 64 | 22 | 14 |
| WT IκBα | None | 3 | 9 | 88 | |
| WT IκBα | 30 min | 23 | 76 | 1 | |
| WT IκBα | 4 h | 23 | 77 | <1 | |
| A2/A2 | None | 5 | 16 | 79 | |
| v-Rel | None | None | 92 | 8 | <1 |
| WT IκBα | None | 15 | 14 | 71 | |
| WT IκBα | 30 min | 92 | 8 | <1 | |
| WT IκBα | 4 h | 90 | 9 | 1 | |
| 114A2 | None | 52 | 10 | 38 | |
| 273A2 | None | 39 | 15 | 46 | |
| A2/A2 | None | 90 | 10 | <1 | |
CEF were cotransfected with retroviral expression vectors encoding the indicated Rel protein and either wild-type (WT) or mutant IκBα proteins (Fig. 1). In some experiments, leptomycin B (LMB) was added to the culture medium at a concentration of 5 nM either 30 min or 4 h prior to fixation of the CEF for indirect immunofluorescence. The cellular localization of the indicated Rel protein was determined by indirect immunofluorescence. For each treatment, a total of 200 cells that were positive for expression of the respective Rel proteins in the absence or in the presence of the indicated IκBα protein were examined.
SNV-derived expression vectors encoding the indicated Rel proteins were constructed (Fig. 1).
Percentages of cells that displayed predominantly nuclear (N), whole-cell (N-C), and cytoplasmic (C) staining relative to the total number of cells that displayed staining for the respective Rel proteins.
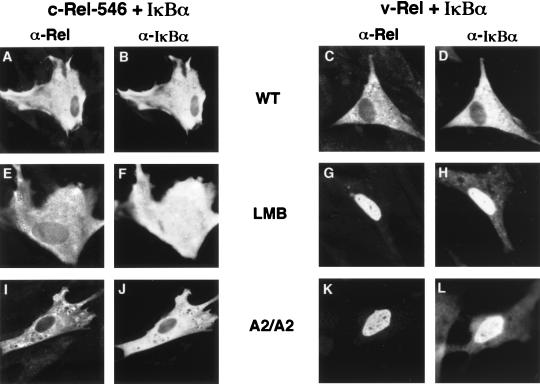
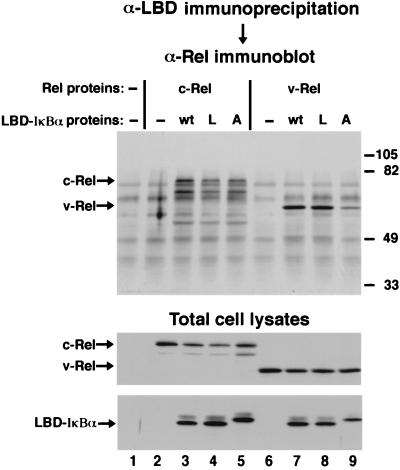
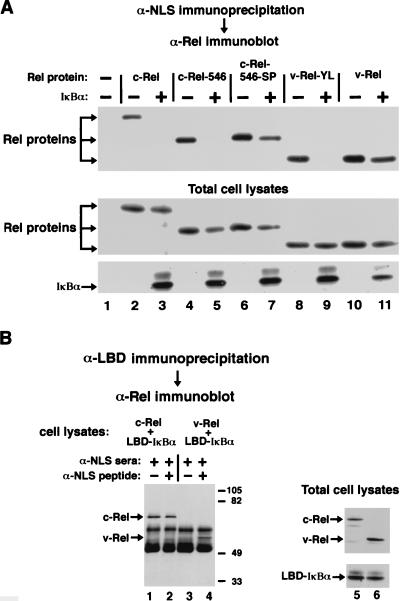
In contrast to the predominantly nuclear localization of the Rel and IκBα proteins when singly expressed in CEF, both the c-Rel-546–IκBα and v-Rel–IκBα complexes were predominantly cytoplasmic when coexpressed in CEF by using separate but otherwise equivalent retroviral expression vectors encoding the respective proteins (Fig. 2A to D; Table 1). Exposure of cotransfected CEF to leptomycin B for 30 min resulted in a slight shift in the distribution of the c-Rel-546–IκBα complex from predominantly cytoplasmic to whole cell (Fig. 2E and F; Table 1). A 4-h treatment with leptomycin B did not markedly increase the nuclear localization of the c-Rel-546–IκBα complex in CEF (Table 1). In contrast, significant nuclear localization of v-Rel was detected within 5 min of leptomycin B treatment (data not shown), while a 30-min treatment with leptomycin B resulted in the complete redistribution of the v-Rel–IκBα complex from the cytoplasm to the nucleus in both CEF (Fig. 2G and H; Table 1) and COS-1 cells (data not shown). Leptomycin B treatment did not significantly alter the stability of the respective Rel-IκBα complexes in either CEF or COS-1 cells (data not shown). Furthermore, leptomycin B treatment did not disrupt the ability of IκBα to associate with either c-Rel (Fig. 3, upper panel, compare lanes 3 and 4) or v-Rel (Fig. 3, upper panel, compare lanes 7 and 8). The fact that cytoplasmic retention of the v-Rel–IκBα complex, but not the c-Rel–IκBα complex, is disrupted by leptomycin B suggests that cytoplasmic retention of v-Rel by IκBα requires continuous nuclear export mediated by Crm1.
FIG. 2.
Cytoplasmic localization of the v-Rel–IκBα complex is sensitive to leptomycin B and requires the integrity of two NES-like motifs in IκBα. CEF were cotransfected with retroviral vectors encoding (i) both c-Rel-546 and either wild-type (WT) or mutant IκBα proteins or (ii) both v-Rel and either wild-type or mutant IκBα proteins, as indicated. The cellular localization of c-Rel-546 (A, E, and I) and cotransfected IκBα protein (B, F, and J) or of v-Rel (C, G, and K) and cotransfected IκBα protein (D, H, and L) within the same cell was determined by double-label indirect immunofluorescence with anti-Rel and anti-IκBα sera. In some experiments, leptomycin B (LMB) was added to the culture medium at a concentration of 5 nM 30 min prior to fixation of the CEF for indirect immunofluorescence. The cells shown are representative of more than 50 cells that were positive for the expression of both of the indicated Rel and IκBα proteins.
FIG. 3.
Coimmunoprecipitation analysis. COS-1 cells were either singly transfected with CMV-derived expression vectors encoding c-Rel (lane 2) or v-Rel (lane 6) or were cotransfected with CMV-derived expression vectors encoding either c-Rel or v-Rel and either wild-type (wt) LBD-tagged IκBα (lanes 3, 4, 7, and 8) or the mutant (A) LBD-tagged A2/A2 IκBα protein (lanes 5 and 9). In some experiments, leptomycin B (L) was added to the culture medium at a concentration of 5 nM 30 min prior to preparation of the cell lysates (lanes 4 and 8). (Upper panel) Equivalent aliquots of cell lysates were subjected to immunoprecipitation with anti-LBD serum. The immunoprecipitated proteins were electrophoresed through an SDS–8% polyacrylamide gel, and the proteins were transferred to nitrocellulose. The relative amounts of c-Rel or v-Rel that coimmunoprecipitated with either the wild-type or mutant A2/A2 IκBα proteins were determined by ECL immunoblot analysis with a monoclonal anti-Rel hybridoma supernatant. The arrows on the left indicate the positions of coimmunoprecipitated c-Rel or v-Rel proteins. The positions of molecular size markers (in thousands) are indicated on the right. (Middle and lower panels) The expression of c-Rel, v-Rel, and LBD-tagged IκBα proteins in the total cell lysates was confirmed by ECL immunoblot analysis with anti-Rel serum (middle panel) and anti-IκBα serum (lower panel). The arrows on the left of the middle panel indicate the positions of the transfected c-Rel and v-Rel proteins. The arrow on the left of the lower panel indicates the position of the transfected LBD-tagged IκBα proteins.
IκBα contains two clusters of hydrophobic residues that resemble previously described NESs (20, 56), an N-terminal cluster located within the second ankyrin repeat (amino acids 114 to 124 in avian IκBα [Fig. 1]) and a C-terminal cluster located between the ankyrin repeat domain and the PEST domain (amino acids 273 to 284 in avian IκBα [Fig. 1]). To determine if these NES-like motifs contribute to the ability of IκBα to retain either c-Rel or v-Rel in the cytoplasm, we constructed mutant IκBα proteins containing two alanine substitutions within either the 114–124 region (114A2 [Fig. 1]), the 273–284 region (273A2 [Fig. 1]), or both of these regions (A2/A2). The cellular distributions and stabilities of these mutant IκBα proteins when singly expressed in CEF were not significantly different from those of the wild-type IκBα protein (data not shown). The ability of the mutant IκBα proteins to retain c-Rel-546 and v-Rel in the cytoplasm following coexpression in CEF was characterized by indirect immunofluorescence. The mutant 114A2 and 273A2 proteins were still able to efficiently retain c-Rel-546 in the cytoplasm but had reduced ability to retain v-Rel in the cytoplasm (Table 1). The double mutant IκBα protein (A2/A2) was also able to efficiently retain c-Rel-546 in the cytoplasm (Fig. 2I and J; Table 1) but was completely defective for cytoplasmic retention of v-Rel (Fig. 2K and L; Table 1). The inability of the mutant A2/A2 protein to relocalize v-Rel to the cytoplasm was not a function of reduced stability of the A2/A2 protein, as mutations within both leucine-rich clusters in IκBα did not significantly alter the stability of IκBα when coexpressed with either c-Rel or v-Rel (data not shown). Furthermore, alanine substitutions within the leucine-rich clusters in IκBα did not disrupt the ability of IκBα to associate with either c-Rel or v-Rel, either in COS-1 cells (Fig. 3, upper panel, lanes 3 and 6) or in the Saccharomyces cerevisiae two-hybrid system (data not shown). These results indicate that the integrity of both the 114–124 and the 273–284 leucine-rich NES-like clusters in IκBα is required for cytoplasmic localization of the v-Rel–IκBα complex.
IκBα inhibits nuclear import of c-Rel but mediates nuclear shuttling of v-Rel in the interspecies heterokaryon assay.
To directly examine nuclear export of v-Rel by the wild-type IκBα protein, we utilized an interspecies heterokaryon assay. The interspecies heterokaryon assay requires that a protein be exported from one nucleus and imported into the other nucleus (35). Thus, the interspecies heterokaryon assay provides a sensitive in vivo assay for nuclear export and nuclear import processes.
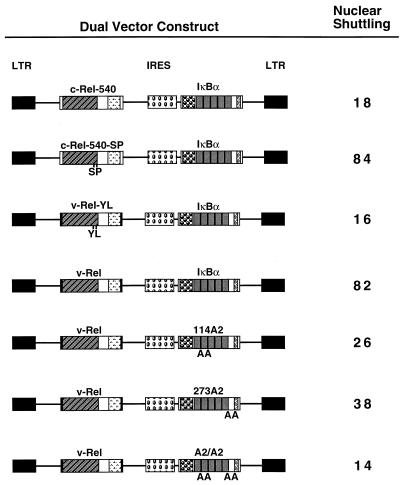
To ensure that Rel and IκBα proteins were coexpressed in a single CEF, a series of dual-expression vectors containing an IRES were constructed that allowed for the expression of either c-Rel or v-Rel and wild-type or mutant IκBα proteins from a single mRNA transcript (Fig. 4, upper panel). Coexpression of a truncated c-Rel protein (c-Rel-540) with the wild-type IκBα protein in this dual-expression vector resulted in a significant, though not complete, relocalization of the c-Rel-540 protein from the nucleus to the cytoplasm (compare Fig. 4A and D). The inability of IκBα to completely relocalize the c-Rel-540 protein from the nucleus to the cytoplasm is due to reduced levels of IκBα expression when the IκBα open reading frame is placed 3′ of the IRES (data not shown). In contrast to the partial relocalization obtained by coexpression of c-Rel and IκBα in this dual-expression vector, v-Rel remained predominantly nuclear when coexpressed with IκBα (compare Fig. 4G and J). A quantitative measure of the localization of the Rel proteins when coexpressed with IκBα was obtained by confocal laser scanning microscopy. This quantitation revealed that approximately 10% of the c-Rel-540 protein was present in the nuclei of CEF when coexpressed with IκBα, while greater than 50% of the v-Rel protein was present in the nuclei of CEF when coexpressed with IκBα (data not shown). The stability of IκBα in the presence of c-Rel-540 was essentially equivalent to the stability of IκBα in the presence of v-Rel, as determined by pulse-chase analysis of transfected CEF (data not shown).
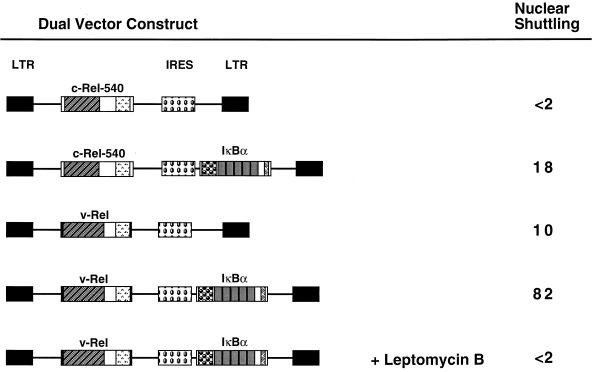
FIG. 4.
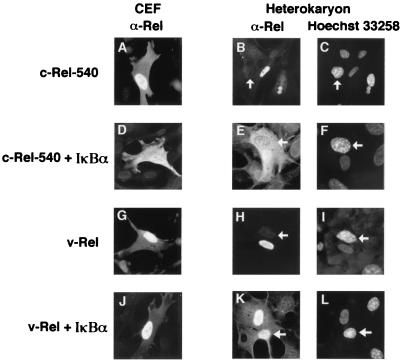
IκBα-mediated nuclear shuttling of v-Rel. (Upper panel) Dual-expression vectors that enable coexpression of Rel and IκBα proteins from a single mRNA transcript. The rectangles representing the SNV-derived long terminal repeat (LTR), the Rel and IκBα open reading frames, and the IRES are indicated by the different fill patterns. Nuclear shuttling was measured by the ability of the Rel protein encoded by each vector to accumulate in the mouse nucleus of the chicken-mouse heterokaryon and is indicated on the right. The numbers represent the percentages of heterokaryons that were positive for Rel staining in both the chicken nucleus and the mouse nucleus relative to the total number of heterokaryons examined that were positive for Rel expression in the chicken nucleus. A total of at least 50 heterokaryons from a minimum of three independent experiments were scored for each construct. In some experiments, leptomycin B was included during the 30-min postfusion period at a concentration of 5 nM. (Lower panels) CEF were transfected with retroviral expression vectors that encoded either c-Rel-540 (A to C) or v-Rel (G to I). CEF were also transfected with retroviral expression vectors that encoded either both c-Rel-540 and IκBα (D to F) or both v-Rel and IκBα (J to L). The cellular localization of the Rel proteins in the transfected CEF was determined by indirect immunofluorescence with anti-Rel serum (A, D, G, and J). The cells shown are representative of more than 200 cells that were positive for the expression of the indicated Rel protein. The localization of the Rel proteins was also determined following fusion of the transfected CEF with mouse NIH 3T3 fibroblasts (B, E, H, and K). Hoechst 33258 was used to identify the mouse nucleus of the heterokaryon (C, F, I, and L). The mouse nucleus of each heterokaryon is indicated by a white arrow. The heterokaryons shown are representative of more than 50 heterokaryons that were positive for expression of the indicated Rel protein.
CEF transfected with these dual vectors were fused with mouse NIH 3T3 cells to form heterokaryons. CHX was included in the culture medium for 30 min prior to fusion and throughout a 30-min postfusion incubation to inhibit protein synthesis. The stability of IκBα was not altered by CHX treatment (data not shown). The mouse and chicken nuclei were distinguished by differential staining with Hoechst 33258 dye, while the presence of Rel proteins in the heterokaryon nuclei was detected by indirect immunofluorescence with anti-Rel serum. Expression of either c-Rel-540 or v-Rel proteins alone did not result in significant accumulation of the Rel proteins in the mouse nucleus (Fig. 4B and H). Likewise, coexpression of IκBα with the c-Rel-540 protein did not result in significant accumulation of the c-Rel-540 protein in the mouse nucleus. Rather, the c-Rel-540 protein remained in the cytoplasm of the heterokaryon (Fig. 4E), consistent with the hypothesis that IκBα is able to efficiently inhibit nuclear import of c-Rel.
In contrast, coexpression of IκBα with v-Rel resulted in significant accumulation of v-Rel in the mouse nucleus (Fig. 4K). Importantly, the inclusion of leptomycin B in the culture medium during the 30-min postfusion period completely abolished IκBα-mediated accumulation of v-Rel in the mouse nucleus of the heterokaryon (Fig. 4, upper panel). Coexpression of v-Rel with either the 114A2 or 273A2 protein reduced nuclear shuttling of v-Rel, and nuclear shuttling of v-Rel was not observed in the presence of the A2/A2 protein (Fig. 5). No significant differences in the stabilities of the wild-type or mutant IκBα proteins were observed when they were coexpressed with v-Rel (data not shown). The ability of v-Rel to accumulate in the mouse nucleus of the heterokaryon in an IκBα-dependent manner provides direct evidence that IκBα is able to mediate the nuclear export of v-Rel. Furthermore, these results suggest that IκBα does not inhibit nuclear import of v-Rel.
FIG. 5.
Nuclear shuttling of Rel proteins. Dual-expression vectors that enable coexpression of Rel and IκBα proteins from a single mRNA transcript are diagramed. The rectangles representing the SNV-derived long terminal repeat (LTR), the Rel and IκBα open reading frames, and the IRES are indicated by the different fill patterns. The mutant Rel and IκBα genes are indicated by name. The amino acid substitutions within the mutant Rel or IκBα genes are denoted in the single-letter code below the rectangles representing the respective Rel or IκBα genes. Nuclear shuttling was measured by the ability of the Rel protein encoded by each vector to accumulate in the mouse nucleus of the chicken-mouse heterokaryon and is indicated on the right. The numbers represent the percentages of heterokaryons that were positive for Rel staining in both the chicken nucleus and the mouse nucleus relative to the total number of heterokaryons examined that were positive for Rel expression in the chicken nucleus. A total of at least 50 heterokaryons from a minimum of three independent experiments were scored for each dual-expression vector construct.
The Rel-derived NLS is not masked within the v-Rel–IκBα complex.
As the ability of IκB proteins to inhibit nuclear import of their cognate Rel proteins is thought to derive from masking of the Rel-derived NLS within the Rel-IκB complex (5, 23, 60), we asked whether IκBα is able to mask the c-Rel-derived or v-Rel-derived NLS within the respective Rel-IκBα complexes. The ability of antipeptide serum specific for the Rel-derived NLS to immunoprecipitate c-Rel or v-Rel was determined. The ectopically expressed c-Rel, c-Rel-546, and v-Rel proteins were immunoprecipitated by anti-NLS serum from COS-1 cell lysates (Fig. 6A, upper panel, lanes 2, 4, and 10). However, coexpression of IκBα with either c-Rel or c-Rel-546 abolished the ability of the anti-NLS serum to immunoprecipitate either c-Rel or c-Rel-546 (Fig. 6A, upper panel, lanes 3 and 5). Leptomycin B treatment of cotransfected cells or alanine substitutions within the two NES-like motifs of IκBα (A2/A2) did not increase the ability of the anti-NLS serum to immunoprecipitate the c-Rel protein when coexpressed with IκBα (data not shown), consistent with the inability of these experimental conditions to markedly alter cytoplasmic retention of the c-Rel–IκBα complex in either CEF (Fig. 2) or COS-1 cells (data not shown). In contrast to the case for c-Rel, the anti-NLS serum was able to efficiently immunoprecipitate the v-Rel protein from COS-1 cells cotransfected with v-Rel and IκBα (Fig. 6A, upper panel, lane 11).
FIG. 6.
Exposure of the Rel-derived NLS. (A) COS-1 cells were either mock transfected (lane 1), singly transfected with CMV-derived expression vectors encoding the indicated Rel proteins (lanes 2, 4, 6, 8, and 10), or cotransfected with CMV-derived expression vectors encoding the indicated Rel proteins and wild-type IκBα (lanes 3, 5, 7, 9, and 11). In the upper panel, equivalent aliquots of cell lysates were subjected to immunoprecipitation with mouse anti-NLS (α-NLS) serum. The immunoprecipitated proteins were electrophoresed through an SDS–8% polyacrylamide gel, and the proteins were transferred to nitrocellulose. The relative amounts of the indicated Rel proteins that immunoprecipitated with the anti-NLS serum were determined by ECL immunoblot analysis with a polyclonal rabbit anti-Rel (α-Rel) serum. The arrows on the left indicate the positions of the immunoprecipitated Rel proteins. In the middle and lower panels, the expression of the indicated Rel and wild-type IκBα proteins in the total cell lysates was confirmed by ECL immunoblot analysis with anti-Rel serum (middle panel) and anti-IκBα serum (lower panel). The arrows on the left of the middle panel indicate the positions of the transfected Rel proteins. The arrow on the left of the lower panel indicates the position of the transfected LBD-tagged IκBα protein. (B) COS-1 cells were either cotransfected with CMV-derived expression vectors encoding both c-Rel and wild-type LBD-tagged IκBα (lanes 1 and 2) or cotransfected with CMV-derived expression vectors encoding both v-Rel and wild-type LBD-tagged IκBα (lanes 3 and 4). In the left panel, equivalent aliquots of cell lysates were incubated with anti-NLS (α-NLS) serum for 30 min prior to immunoprecipitation with anti-LBD serum (lanes 1 and 3). In some samples, the anti-NLS serum was preincubated with 10 μg of the NLS peptide per μl of anti-NLS serum for 30 min prior to addition of the anti-NLS serum to the cell lysates. The immunoprecipitated proteins were electrophoresed through an SDS–8% polyacrylamide gel, and the proteins were transferred to nitrocellulose. The relative amounts of c-Rel or v-Rel that coimmunoprecipitated with the wild-type IκBα proteins were determined by ECL immunoblot analysis with a monoclonal anti-Rel hybridoma supernatant. The arrows on the left indicate the positions of coimmunoprecipitated c-Rel or v-Rel proteins. The positions of molecular size markers (in thousands) are indicated on the right. In the right panel, the expression of c-Rel (lane 5), v-Rel (lane 6), and LBD-tagged IκBα proteins (lanes 5 and 6) in the total cell lysates was confirmed by ECL immunoblot analysis with anti-Rel serum (upper panel) and anti-IκBα serum (lower panel). The arrows on the left of the upper panel indicate the positions of the transfected c-Rel and v-Rel proteins. The arrow on the left of the lower panel indicates the position of the transfected wild-type LBD-tagged IκBα proteins.
Although the anti-NLS serum was able to immunoprecipitate v-Rel from cells cotransfected with v-Rel and IκBα, IκBα was not detected in the anti-NLS immunoprecipitates (data not shown). However, v-Rel was efficiently coimmunoprecipitated with IκBα by using antiserum directed against an epitope tag placed on the C terminus of IκBα (Fig. 3). We therefore examined the ability of the respective Rel proteins to coimmunoprecipitate with epitope-tagged IκBα proteins in the presence of the anti-NLS serum. Incubation of cell lysates containing the c-Rel–IκBα complex with anti-NLS serum did not alter the ability of c-Rel to coimmunoprecipitate with the epitope-tagged IκBα protein (Fig. 6B, lane 1). In contrast, incubation of cell lysates containing the v-Rel–IκBα complex with anti-NLS serum markedly reduced the ability of v-Rel to coimmunoprecipitate with the epitope-tagged IκBα protein (Fig. 6B, lane 3). Preincubation of the anti-NLS serum with an excess of NLS peptide restored the ability of v-Rel to coimmunoprecipitate with the epitope-tagged IκBα protein (Fig. 6B, lane 4).
Taken together, these results indicate that the v-Rel-derived NLS is accessible within the v-Rel–IκBα complex. Furthermore, these results are consistent with a model in which binding of an immunoglobulin molecule to the v-Rel-derived NLS disrupts the v-Rel–IκBα complex.
IκBα is unable to efficiently displace v-Rel from DNA.
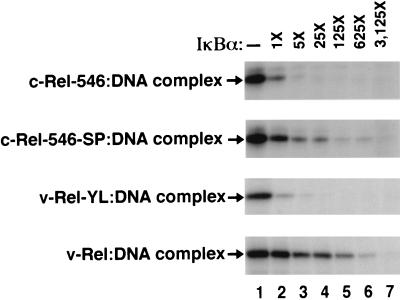
Previous reports have indicated that IκBα does not efficiently inhibit DNA binding by v-Rel (8, 16). However, as the ability of IκBα to control Rel-dependent transcription in vivo likely derives from its ability to displace Rel proteins from DNA (55), we examined the ability of IκBα to displace either c-Rel or v-Rel from preformed Rel-DNA complexes in vitro. The amount of either c-Rel-546 or v-Rel that remained bound to a palindromic κB binding site following addition of increasing amounts of IκBα was determined by solution UV cross-linking. Equivalent amounts of the c-Rel-546 and v-Rel proteins were bound to DNA in the absence of exogenous IκBα (Fig. 7, lane 1). Exogenously added IκBα markedly reduced the amount of the c-Rel-546 protein that was bound to DNA (Fig. 7, compare lanes 1 and 2). A 125-fold excess of IκBα was required to achieve an equivalent reduction in the amount of v-Rel that was bound to DNA (Fig. 7, compare lanes 1 and 6). Thus, IκBα is unable to efficiently displace v-Rel from a preformed v-Rel–DNA complex.
FIG. 7.
IκBα-mediated displacement of Rel proteins from Rel-DNA complexes. COS-1 cells were singly transfected with CMV-derived expression vectors encoding the indicated Rel proteins. Equivalent amounts of either c-Rel-546-, c-Rel-546-SP-, v-Rel-YL-, or v-Rel-containing lysates were incubated with a 32P-labeled oligonucleotide containing a palindromic κB site for 10 min at room temperature. Once the Rel-DNA complexes had formed, the cell lysates were incubated in either the absence (lane 1) or presence (lanes 2 to 7) of increasing amounts of purified baculovirus-expressed IκBα for 20 min at room temperature. The 1× amount of IκBα corresponds to 1.6 ng of purified baculovirus-expressed IκBα. The relative amounts of each of the Rel proteins that remained bound to the 32P-labeled oligonucleotide following incubation with the purified IκBα were determined by solution UV cross-linking. The protein-DNA adducts were electrophoresed through an SDS–8% polyacrylamide gel and visualized by autoradiography. The arrows on the left indicate the positions of the respective Rel-DNA adducts.
Two amino acid differences between c-Rel and v-Rel account for the differential ability of IκBα to control the nuclear functions of c-Rel and v-Rel.
The results described above identify critical differences in IκBα-mediated control over nuclear localization and DNA binding by c-Rel and v-Rel. As the failure of IκBα to control nuclear localization of v-Rel is likely due to the inability of IκBα to mask the v-Rel-derived NLS, we focused our attention on two amino acid differences between c-Rel and v-Rel that flank the Rel-derived NLS (amino acids 288 to 294 [Fig. 1]). The c-Rel protein contains a tyrosine residue at position 286 and a leucine residue at position 302, while the v-Rel protein contains a serine residue in place of Y286 and a proline residue in place of L302 (Fig. 1). We therefore constructed two recombinant proteins in which these two amino acids were interchanged between c-Rel and v-Rel (c-Rel-546-SP and v-Rel-YL). The c-Rel-546-SP and v-Rel-YL proteins were expressed in CEF and in COS-1 cells, and their functional and physical interactions with IκBα were characterized.
Cytoplasmic localization of the c-Rel-546-SP–IκBα complex was significantly more sensitive to leptomycin B treatment than that of the wild-type c-Rel-546–IκBα complex, while cytoplasmic localization of the v-Rel-YL–IκBα complex was less sensitive to leptomycin B treatment than that of the wild-type v-Rel–IκBα complex (Table 1). Although the leptomycin B sensitivities of the c-Rel-546-SP–IκBα and v-Rel-YL–IκBα complexes were intermediate relative to those of the c-Rel-546–IκBα and v-Rel–IκBα complexes, complete restoration of the phenotypic differences between c-Rel-546 and v-Rel was observed in the interspecies heterokaryon assay (Fig. 5) and in the anti-NLS immunoprecipitation experiment (Fig. 6A). Thus, two amino acid differences between c-Rel and v-Rel, which flank the Rel-derived NLS, are primarily responsible for the failure of IκBα to inhibit nuclear import of v-Rel.
The role of these two amino acid differences in differential regulation of DNA binding was also examined. A 25-fold excess of IκBα was required to achieve an equivalent reduction in the amount of the c-Rel-546-SP protein that was bound to DNA, relative to the amount of IκBα required to displace c-Rel-546 from DNA (Fig. 7). Conversely, 125-fold less IκBα was required to achieve an equivalent reduction in the amount of v-Rel-YL protein that was bound to DNA, relative to v-Rel (Fig. 7). Taken together, these results demonstrate that these two amino acid differences between c-Rel and v-Rel are primarily responsible for the functional and physical differences in the responsiveness of c-Rel and v-Rel to the inhibitory functions of IκBα.
Reduced control by IκBα over the nuclear functions of c-Rel correlates with oncogenic activation.
To determine if failure of IκBα to inhibit nuclear import and DNA binding of Rel proteins correlates with increased oncogenic activation of Rel proteins, the ability of the wild-type and mutant Rel proteins to transform primary avian lymphocytes was determined. The c-Rel-546-SP protein was able to transform avian lymphoid cells with markedly greater efficiency than the c-Rel-546 protein (Table 2). Furthermore, cell lines could readily be established from avian lymphocytes infected with the c-Rel-546-SP virus, whereas avian lymphocytes infected with the c-Rel-546 virus grew very poorly in liquid culture (Table 2). Consistent with our previous results (42), the ability of the v-Rel-YL protein to transform avian lymphoid cells was reduced relative to that of v-Rel (Table 2). Therefore, our results demonstrate that Rel-derived amino acids that are primarily responsible for reduced control by IκBα over nuclear import and DNA binding by Rel proteins correlate with differences in the oncogenic properties of Rel proteins.
TABLE 2.
Loss of IκBα-mediated inhibition of nuclear import and DNA binding correlates with increased transformation
| Rel proteina | Leptomycin B sensitivityb | IκBα-mediated displacementc | Transformationd (%) | Establishment of cell linese (%) |
|---|---|---|---|---|
| c-Rel-546 | 2 | 1 | 6 | <1 |
| c-Rel-546-SP | 58 | 25 | 23 | 65 |
| v-Rel-YL | 23 | 1 | 29 | 65 |
| v-Rel | 92 | 125 | 100 | 90 |
SNV-derived expression vectors encoding the indicated Rel proteins were constructed (Fig. 1).
CEF were transfected with retroviral expression vectors that encoded the indicated Rel protein and wild-type IκBα. Leptomycin B was added to the culture medium at a concentration of 5 nM 30 min prior to fixation of the CEF for indirect immunofluorescence. Values are percentages of cells that displayed predominantly nuclear staining relative to the total number of cells that displayed staining for the transfected Rel proteins. For each treatment, a total of 200 cells that were positive for expression of the respective Rel proteins were examined.
COS-1 cells were singly transfected with CMV-derived expression vectors encoding the indicated Rel proteins. Equivalent amounts of cell lysates were incubated with a 32P-labeled oligonucleotide containing a palindromic κB site for 10 min at room temperature. The relative amounts of each of the Rel proteins that remained bound to the 32P-labeled oligonucleotide following incubation with increasing amounts of baculovirus-purified IκBα were determined by solution UV cross-linking. Values are fold excesses of IκBα required to displace the indicated Rel proteins from DNA, relative to the amount of IκBα required to displace the c-Rel-546 protein from the c-Rel-546–DNA complex.
Avian lymphoid cells were infected with viruses encoding the indicated Rel proteins. The ability of avian lymphoid cells infected with viruses encoding the indicated Rel proteins to form colonies in soft agar was determined. The relative efficiency of transformation by the indicated Rel proteins is derived from the mean of six independent experiments, in which the total number of colonies obtained with each virus within a single experiment was normalized to the total number of colonies obtained with virus encoding the wild-type v-Rel protein (set at 100%). The actual numbers of colonies that formed in soft agar for the six independent experiments were as follows: for c-Rel-546, 1, 2, 13, 7, 9, and 0; for c-Rel-546-SP, 3, 5, 47, 8, 67, and 0; for v-Rel-YL, 11, 26, 17, 45, 14, and 3; for v-Rel, 59, 45, 130, 99, 85, and 15.
Transformed cell lines could not be established following infection of avian lymphoid cells with virus encoding the c-Rel-546 protein. In contrast, cell lines could be established from more than 65% of colonies obtained following infection with virus encoding c-Rel-546-SP or v-Rel-YL, and from more than 90% of colonies obtained following infection with virus encoding the wild-type v-Rel protein.
A threshold level of nuclear v-Rel is required for v-Rel-mediated transformation of avian lymphocytes (45). However, the v-Rel protein is predominantly cytoplasmic in v-Rel-transformed avian lymphocytes (13, 25, 30, 50). To determine the role of Crm1-mediated nuclear export in cellular partitioning of v-Rel in v-Rel-transformed avian lymphocytes, we examined the localization of v-Rel in avian lymphocytes following leptomycin B treatment. While v-Rel was distributed throughout both the cytoplasm and the nucleus in the absence of leptomycin B (Fig. 8A), brief leptomycin B treatment resulted in a significant nuclear accumulation of v-Rel in avian lymphocytes (Fig. 8B). Furthermore, consistent with our observations for cotransfected CEF (Table 1), cytoplasmic localization of the c-Rel-546-SP protein was also sensitive to leptomycin B treatment (data not shown), while cytoplasmic localization of the v-Rel-YL protein was not affected by leptomycin B treatment of avian lymphocytes transformed by the respective Rel proteins (data not shown). These results are consistent with the notion that loss of IκBα-mediated control contributes to oncogenic transformation of avian lymphoid cells.
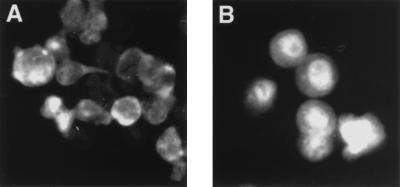
FIG. 8.
Cytoplasmic localization of v-Rel in v-Rel-transformed avian lymphocytes is sensitive to leptomycin B. v-Rel-transformed cell lines were established following infection of avian lymphocytes with virus encoding v-Rel. The cellular localization of v-Rel was determined by indirect immunofluorescence with anti-Rel serum. Leptomycin B was either not added (A) or added to the culture medium at a concentration of 5 nM 60 min prior to fixation of the Rel-transformed cells for indirect immunofluorescence (B). The cells shown are representative of more than 200 cells that were positive for the expression of v-Rel.
DISCUSSION
Cytoplasmic sequestration of the v-Rel–IκBα complex requires continuous IκBα-mediated nuclear export of v-Rel.
The ability of IκBα to inhibit nuclear import of dimeric Rel complexes provides an effective mechanism for cytoplasmic sequestration of Rel proteins (5, 23, 60). Consistent with this notion, our results indicate that cytoplasmic retention of the c-Rel–IκBα complex is not disrupted by leptomycin B treatment. In marked contrast, our results demonstrate that cytoplasmic localization of the v-Rel–IκBα complex is disrupted by brief leptomycin B treatment. Thus, cytoplasmic sequestration of v-Rel by IκBα requires continuous nuclear export of the v-Rel–IκBα complex.
We have utilized nuclear shuttling of v-Rel in the interspecies heterokaryon assay as an in vivo assay for nuclear export. Since nuclear shuttling requires that a protein be exported from one nucleus and imported into the heterologous nucleus, nuclear shuttling in the interspecies heterokaryon assay is a function of both nuclear export and subsequent nuclear import (35). Our observation that nuclear shuttling of v-Rel is markedly increased by coexpression of IκBα is consistent with the previous observation that IκBα can mediate the nuclear export of NF-κB following coinjection of IκBα and NF-κB into Xenopus oocyte nuclei (3). In addition, nuclear shuttling of v-Rel is abolished by leptomycin B treatment, demonstrating the involvement of Crm1 in IκBα-mediated nuclear export of Rel proteins.
The C-terminal NES of IκBα has previously been implicated in both IκBα-mediated nuclear export and protein-protein interactions with Crm1 (3, 40). We find that alanine substitutions in the C-terminal NES of IκBα reduce, but do not eliminate, nuclear shuttling of v-Rel. It has previously been demonstrated that alanine substitutions within the C-terminal NES of IκBα reduce, but do not completely abolish, IκBα-mediated nuclear export of the p65 subunit of NF-κB (3). We find that mutations within both the C-terminal NES and a hydrophobic cluster of residues in the second ankyrin repeat are necessary to completely abolish IκBα-mediated nuclear shuttling of v-Rel. Our results suggest that hydrophobic amino acids within both the C-terminal NES and the second ankyrin repeat of IκBα are required for protein-protein interactions between Crm1 and IκBα.
Failure of IκBα to mask the v-Rel-derived NLS enables v-Rel to evade inhibition of nuclear import by IκBα.
The ability of leptomycin B to disrupt cytoplasmic retention of the v-Rel–IκBα complex is consistent with a transient nuclear localization of the v-Rel–IκBα complex. Furthermore, although IκBα is required for nuclear export of v-Rel in the interspecies heterokaryon assay, IκBα is unable to prevent subsequent nuclear import of v-Rel into the heterologous nucleus of the heterokaryon. Our results demonstrate that the v-Rel-derived NLS, but not the c-Rel-derived NLS, is accessible to the anti-NLS peptide serum within the context of the respective Rel-IκBα complexes. Taken together, these results are consistent with the hypothesis that failure of IκBα to mask the v-Rel-derived NLS is responsible for the inability of IκBα to inhibit nuclear import of v-Rel.
Two models can be proposed to understand how nuclear import of v-Rel is accomplished despite association with IκBα. The fact that the v-Rel-derived NLS is accessible to anti-NLS serum within the context of the v-Rel–IκBα complex is consistent with a model in which the v-Rel-derived NLS is accessible to the importin-α/β receptor complex (for reviews, see references 27 and 38). In this model, binding of the importin-α/β complex to the v-Rel-derived NLS in the context of the v-Rel–IκBα complex would enable nuclear import of the v-Rel–IκBα complex.
An alternate model is that the v-Rel-derived NLS is not exposed within the context of the v-Rel–IκBα complex but that the v-Rel-derived NLS is transiently exposed due to rapid association and dissociation of the v-Rel–IκBα complex. The free v-Rel protein would then be transported to the nucleus by the importin-α/β receptor complex, while the free IκBα protein would then be transported to the nucleus by virtue of its own nuclear import sequence (46) or would be degraded. The v-Rel–IκBα complex can be readily immunoprecipitated from cells, and our previous analysis of Rel-IκBα interactions in the S. cerevisiae two-hybrid system suggests that the affinity of the v-Rel–IκBα complex does not significantly differ from the affinity of the c-Rel–IκBα complex (43). However, our current results, which indicate that the anti-NLS serum is able to disrupt the v-Rel–IκBα complex, are consistent with a model in which the v-Rel–IκBα complex undergoes rapid association and dissociation in vitro. If the v-Rel–IκBα complex undergoes similar rapid association and dissociation reactions in vivo, the resultant transient exposure of the v-Rel-derived NLS might enable its recognition by the importin-α/β receptor complex.
Relationship between the inhibitory properties of IκBα and Rel-mediated oncogenesis.
Expression of v-Rel as the oncoprotein of the avian retrovirus Rev-T leads to oncogenic transformation of avian lymphocytes (for a review, see reference 26). Likewise, targeted expression of v-Rel in murine thymocytes leads to the development of aggressive T-cell lymphoma or leukemias in transgenic mice (9). In marked contrast, retrovirus-mediated expression of c-Rel in avian lymphocytes does not lead to oncogenic transformation, and transgenic expression of c-Rel is not tumorigenic in mice (9, 26). Clearly, amino acid differences between c-Rel and v-Rel are responsible for the marked differences in their biological properties. However, the identification of critical biochemical differences between c-Rel and v-Rel that can be linked to specific amino acid differences between c-Rel and v-Rel and that are responsible for the potent oncogenic properties of v-Rel has been a difficult task.
Previous studies have demonstrated marked differences between c-Rel and v-Rel in terms of how IκBα controls their distribution between the nucleus and the cytoplasm (30, 43). For example, following infection of avian lymphocytes with a retroviral vector encoding c-Rel, the c-Rel protein is efficiently relocalized from the nucleus to the cytoplasm, presumably by the action of endogenous IκB proteins (30). In contrast, significantly larger amounts of v-Rel remain in the nucleus following retrovirus-mediated expression of v-Rel in avian lymphocytes (30). Since a threshold nuclear level of v-Rel is required for oncogenic transformation of avian lymphocytes (45), our current results are consistent with a model in which the failure of IκBα to inhibit nuclear import of v-Rel enables this critical nuclear level of v-Rel to be established.
One experimental concern regarding the inability of IκBα to inhibit nuclear import of v-Rel is that the behavior of proteins ectopically expressed in CEF or COS-1 cells may not accurately reflect the behavior of endogenous proteins. To address this concern, we examined the cellular distribution of v-Rel in v-Rel-transformed avian lymphocytes in the absence and presence of leptomycin B. Brief leptomycin B treatment of v-Rel-transformed avian lymphocytes resulted in a significant nuclear accumulation of v-Rel. These results demonstrate that continuous nuclear export is required for cytoplasmic retention of v-Rel, presumably by the action of endogenous IκBα (30).
Previous studies have demonstrated marked differences between c-Rel and v-Rel in terms of how IκBα controls their ability to bind DNA (8, 16). We now demonstrate that a 125-fold excess of IκBα must be added to the v-Rel–DNA complex compared to the c-Rel–DNA complex in order to displace equivalent amounts of c-Rel and v-Rel from their respective DNA-bound complexes. Thus, IκBα is deficient in control over both nuclear localization of and DNA binding by v-Rel. Consequently, IκBα is unable to fully repress v-Rel-dependent transcriptional activation of a κB-dependent reporter gene (46a). It is clear that transcriptional activation of target genes by v-Rel is required for v-Rel-mediated oncogenesis, as forced cytoplasmic localization of v-Rel by fusion of a cis-acting NES to v-Rel is sufficient to prevent v-Rel-mediated transformation of avian lymphocytes (45). However, ectopic expression of IκBα is not sufficient to abolish v-Rel-mediated transformation of avian or murine lymphocytes, consistent with the hypothesis that v-Rel is resistant to the inhibitory properties of IκBα (8, 45). The resistance of v-Rel to the inhibitory properties of IκBα will certainly contribute to the ability of v-Rel to activate transcription of specific target genes in v-Rel-transformed cells.
We have identified two amino acid differences between c-Rel and v-Rel, a serine substitution for tyrosine 286 in c-Rel (Y286S) and a proline substitution for leucine 302 in c-Rel (L302P), which are primarily responsible for the differential control over nuclear localization and DNA binding by IκBα. Importantly, the introduction of these two v-Rel-derived amino acids into an otherwise nontransforming c-Rel protein markedly increased the ability of the c-Rel protein to transform avian lymphocytes. Our results are consistent with the hypothesis that the failure of IκBα to control the nuclear functions of c-Rel enables manifestation of the oncogenic potential of c-Rel.
However, the introduction of these two c-Rel-derived amino acids into v-Rel (v-Rel-YL) reduced, but did not abolish, the ability of the v-Rel-YL protein to transform avian lymphocytes. It is likely that the introduction of these two c-Rel-derived amino acids into v-Rel did not fully restore responsiveness of the v-Rel-YL protein to inhibition by IκBα. Consistent with this notion, cytoplasmic retention of the v-Rel-YL–IκBα complex remained partially sensitive to leptomycin B treatment, and nuclear v-Rel-YL protein could readily be detected from v-Rel-YL-transformed avian lymphocytes (46a). Furthermore, although equivalent amounts of IκBα were required to displace the v-Rel-YL and c-Rel-546 proteins from DNA when these proteins were derived from COS-1 cell lysates, IκBα was unable to efficiently displace the v-Rel-YL protein from nuclear extracts derived from v-Rel-YL-transformed avian lymphocytes (46a). Failure of IκBα to fully regulate the v-Rel-YL protein may account for the ability of the v-Rel-YL protein to transform avian lymphoid cells.
An alternative though not mutually exclusive hypothesis is that the ability of the v-Rel-YL protein to transform avian lymphocytes reflects additional biochemical differences between c-Rel and v-Rel. In particular, the v-Rel-YL protein still contains multiple amino acid substitutions relative to c-Rel, including amino acid differences within the amino terminus of the Rel homology domain. These amino-terminal differences are responsible for differences in the sequence-specific DNA-binding properties of c-Rel and v-Rel and have been shown to contribute to the oncogenic properties of v-Rel (28, 29, 37). Taken together, the available experimental data are most consistent with a model in which differences in the oncogenic properties of c-Rel and v-Rel reflect both the ability of v-Rel to evade the inhibitory properties of IκBα and the ability of v-Rel to regulate a distinct set of target genes.
ACKNOWLEDGMENTS
We thank Andrew Chappell and Michelle Wald for technical assistance and David J. Pintel and Peter Wilden for critical review of the manuscript. We thank Henry R. Bose, Jr., for his gift of the HY87 and 3C1 anti-Rel monoclonal antibodies, Dan Donoghue for his gift of the anti-LBD serum, and Minoru Yoshida for his generous gift of leptomycin B.
This work was supported by American Cancer Society grant RPG-98-097-01-MGO, by Public Health Service grant CA-55027 from the National Cancer Institute, by USDA NRICGP award 95-04073, by University of Missouri Research Board grant RB97-175, and by the University of Missouri Molecular Biology Program.
REFERENCES
- 1.Alkalay I, Yaron A, Hatzubai A, Orian A, Ciechanover A, Ben-Neriah Y. Stimulation-dependent IκBα phosphorylation marks the NF-κB inhibitor for degradation via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1995;92:10599–10603. doi: 10.1073/pnas.92.23.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arenzana-Seisdedos F, Thompson J, Rodriguez M S, Bachelerie F, Thomas D, Hay R T. Inducible nuclear expression of newly synthesized IκBα negatively regulates DNA-binding and transcriptional activities of NF-κB. Mol Cell Biol. 1995;15:2689–2696. doi: 10.1128/mcb.15.5.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arenzana-Seisdedos F, Turpin P, Rodriguez M, Thomas D, Hay R T, Virelizier J L, Dargemont C. Nuclear localization of IκBα promotes active transport of NF-κB from the nucleus to the cytoplasm. J Cell Sci. 1997;110:369–378. doi: 10.1242/jcs.110.3.369. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin A S., Jr The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 5.Beg A A, Ruben S M, Scheinman R I, Haskill S, Rosen C A, Baldwin A S., Jr IκB interacts with the nuclear localization sequences of the subunits of NF-κB: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 6.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Coupling of a signal response domain in IκBα to multiple pathways for NF-κB activation. Mol Cell Biol. 1995;15:2809–2816. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capobianco A J, Simmons D L, Gilmore T D. Cloning and characterization of a chicken c-Rel cDNA: unlike p59v-rel, p68c-rel is a cytoplasmic protein in chicken embryo fibroblasts. Oncogene. 1990;5:257–266. [PubMed] [Google Scholar]
- 8.Carrasco D, Perez P, Lewin A, Bravo R. IκBα overexpression delays tumor formation in v-rel transgenic mice. J Exp Med. 1997;186:279–288. doi: 10.1084/jem.186.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrasco D, Rizzo C A, Dorfman K, Bravo R. The v-rel oncogene promotes malignant T-cell leukemia/lymphoma in transgenic mice. EMBO J. 1996;15:3640–3650. [PMC free article] [PubMed] [Google Scholar]
- 10.Chang C C, Zhang J, Lombardi L, Neri A, Dalla-Favera R. Rearranged NFKB-2 genes in lymphoid neoplasms code for constitutively active nuclear transactivators. Mol Cell Biol. 1995;15:5180–5187. doi: 10.1128/mcb.15.9.5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 12.Cressman D E, Taub R. IκBα can localize in the nucleus but shows no direct transactivation potential. Oncogene. 1993;8:2567–2573. [PubMed] [Google Scholar]
- 13.Davis N, Bargmann W, Lim M Y, Bose H., Jr Avian reticuloendotheliosis virus-transformed lymphoid cells contain multiple pp59v-rel complexes. J Virol. 1990;64:584–591. doi: 10.1128/jvi.64.2.584-591.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis N, Ghosh S, Simmons D L, Tempst P, Liou H C, Baltimore D, Bose H R., Jr rel-associated pp40: an inhibitor of the rel family of transcription factors. Science. 1991;253:1268–1271. doi: 10.1126/science.1891714. [DOI] [PubMed] [Google Scholar]
- 15.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature (London) 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 16.Diehl J A, McKinsey T A, Hannink M. Differential pp40IκB-β inhibition of DNA binding by rel proteins. Mol Cell Biol. 1993;13:1769–1778. doi: 10.1128/mcb.13.3.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diehl J A, Tong W, Sun G, Hannink M. TNF-α-dependent activation of a RelA homodimer in astrocytes: increased phosphorylation of RelA and MAD-3 precede activation of RelA. J Biol Chem. 1995;270:2703–2707. doi: 10.1074/jbc.270.6.2703. [DOI] [PubMed] [Google Scholar]
- 18.Dougherty J P, Temin H M. High mutation rate of a spleen necrosis virus-based retrovirus vector. Mol Cell Biol. 1986;6:4387–4395. doi: 10.1128/mcb.6.12.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finco T S, Westwick J K, Norris J L, Beg A A, Der C J, Baldwin A S., Jr Oncogenic Ha-Ras-induced signaling activates NF-κB transcriptional activity, which is required for cellular transformation. J Biol Chem. 1997;272:24113–24116. doi: 10.1074/jbc.272.39.24113. [DOI] [PubMed] [Google Scholar]
- 20.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 21.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 22.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 23.Ganchi P A, Sun S C, Greene W C, Ballard D W. IκB/MAD-3 masks the nuclear localization signal of NF-κB p65 and requires the transactivation domain to inhibit NF-κB p65 DNA binding. Mol Biol Cell. 1992;3:1339–1352. doi: 10.1091/mbc.3.12.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghattas I R, Sanes J R, Majors J E. The encephalomyocarditis virus internal ribosome entry site allows efficient coexpression of two genes from a recombinant provirus in cultured cells and in embryos. Mol Cell Biol. 1991;11:5848–5859. doi: 10.1128/mcb.11.12.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilmore T D, Temin H M. Different localization of the product of the v-rel oncogene in chicken fibroblasts and spleen cells correlates with transformation by REV-T. Cell. 1986;44:791–800. doi: 10.1016/0092-8674(86)90845-7. [DOI] [PubMed] [Google Scholar]
- 26.Gilmore T D, Koedood M, Piffat K A, White D W. Rel/NF-κB/IκB proteins and cancer. Oncogene. 1996;13:1367–1378. [PubMed] [Google Scholar]
- 27.Görlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 28.Hrdlickova R, Nehyba J, Humphries E H. In vivo evolution of c-rel oncogenic potential. J Virol. 1994;68:2371–2382. doi: 10.1128/jvi.68.4.2371-2382.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hrdlickova R, Nehyba J, Bose H R., Jr Mutations in the DNA-binding and dimerization domains of v-Rel are responsible for altered κB DNA-binding complexes in transformed cells. J Virol. 1995;69:3369–3380. doi: 10.1128/jvi.69.6.3369-3380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hrdlickova R, Nehyba J, Roy A, Humphries E H, Bose H R., Jr The relocalization of v-Rel from the nucleus to the cytoplasm coincides with induction of expression of Ikba and nfkb1 and stabilization of IκB-α. J Virol. 1995;69:403–413. doi: 10.1128/jvi.69.1.403-413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito C Y, Kazantsev A G, Baldwin A S., Jr Three NF-κB sites in the IκB-α promoter are required for induction of gene expression by TNF-α. Nucleic Acids Res. 1994;22:3787–3792. doi: 10.1093/nar/22.18.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Bail O, Schmidt-Ullrich R, Israel A. Promoter analysis of the gene encoding the I κBα/MAD-3 inhibitor of NF-κB: positive regulation by members of the Rel/NF-κB family. EMBO J. 1993;12:5043–5049. doi: 10.1002/j.1460-2075.1993.tb06197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayo M W, Wang C-Y, Cogswell P C, Rogers-Graham K S, Lowe S W, Der C J, Baldwin A S., Jr Requirement of NF-κB activation to suppress p53-independent apoptosis induced by oncogenic Ras. Science. 1998;278:1812–1815. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 34.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J, Young D B, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 35.Michael W M, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 36.Morin P J, Gilmore T D. The C-terminus of the NF-κB p50 precursor and an IκB isoform contain transcription activation domains. Nucleic Acids Res. 1992;20:2453–2458. doi: 10.1093/nar/20.10.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nehyba J, Hrdlickova R, Bose H R., Jr Differences in κB DNA-binding properties of v-Rel and c-Rel are the result of oncogenic mutations in three distinct functional regions of the Rel protein. Oncogene. 1997;14:2881–2897. doi: 10.1038/sj.onc.1201150. [DOI] [PubMed] [Google Scholar]
- 38.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 39.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 40.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 41.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Identification and characterization of an IκB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 42.Richardson P M, Gilmore T D. v-Rel is an inactive member of the Rel family of transcriptional activating proteins. J Virol. 1991;65:3122–3130. doi: 10.1128/jvi.65.6.3122-3130.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rottjakob E M, Sachdev S, Leanna C A, McKinsey T A, Hannink M. PEST-dependent cytoplasmic retention of v-Rel by IκB-α: evidence that IκB-α regulates cellular localization of c-Rel and v-Rel by distinct mechanisms. J Virol. 1996;70:3176–3188. doi: 10.1128/jvi.70.5.3176-3188.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sachdev S, Rottjakob E M, Diehl J A, Hannink M. IκB-α-mediated inhibition of nuclear transport and DNA-binding by Rel proteins are separable functions: phosphorylation of C-terminal serine residues of IκB-α is specifically required for inhibition of DNA-binding. Oncogene. 1995;11:811–823. [PubMed] [Google Scholar]
- 45.Sachdev S, Diehl J A, McKinsey T A, Hans A, Hannink M. A threshold nuclear level of the v-Rel oncoprotein is required for transformation of avian lymphocytes. Oncogene. 1997;14:2585–2594. doi: 10.1038/sj.onc.1201108. [DOI] [PubMed] [Google Scholar]
- 46.Sachdev S, Hoffmann A, Hannink M. Nuclear localization of IκBα is mediated by the second ankyrin repeat: the IκBα ankyrin repeats define a novel class of cis-acting nuclear import sequences. Mol Cell Biol. 1998;18:2524–2534. doi: 10.1128/mcb.18.5.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.Sachdev, S., and M. Hannink. Unpublished data.
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 48.Schatzle J D, Kralova J, Bose H R., Jr Avian IκBα is transcriptionally induced by c-Rel and v-Rel with different kinetics. J Virol. 1995;69:5383–5390. doi: 10.1128/jvi.69.9.5383-5390.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scherer D C, Brockman J A, Chen Z, Maniatis T, Ballard D W. Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simek S L, Stephens R M, Rice N R. Localization of the v-rel protein in reticuloendotheliosis virus strain T-transformed lymphoid cells. J Virol. 1986;59:120–126. doi: 10.1128/jvi.59.1.120-126.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 52.Sun S C, Ganchi P A, Ballard D W, Greene W C. NF-κB controls expression of inhibitor κBα: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 53.Sylla B S, Temin H M. Activation of oncogenicity of the c-rel proto-oncogene. Mol Cell Biol. 1986;6:4709–4716. doi: 10.1128/mcb.6.12.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Traenckner E B, Pahl H L, Henkel T, Schmidt K N, Wilk S, Baeuerle P A. Phosphorylation of human IκB-α on serines 32 and 36 controls IκB-α proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tran K, Merika M, Thanos D. Distinct functional properties of IκBα and IκBβ. Mol Cell Biol. 1997;17:5386–5399. doi: 10.1128/mcb.17.9.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 57.Wilhelmsen K C, Eggleton K, Temin H M. Nucleic acid sequences of the oncogene v-rel in reticuloendotheliosis virus strains T and its cellular homolog, the proto-oncogene c-rel. J Virol. 1984;52:172–182. doi: 10.1128/jvi.52.1.172-182.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 59.Yaron A, Gonen H, Alkalay I, Hatzubai A, Jung S, Beyth S, Mercurio F, Manning A M, Ciechanover A, Ben-Neriah Y. Inhibition of NF-κB cellular function via specific targeting of the IκB-ubiquitin ligase. EMBO J. 1997;16:6486–6494. doi: 10.1093/emboj/16.21.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zabel U, Henkel T, Silva M S, Baeuerle P A. Nuclear uptake control of NF-κB by MAD-3, an IκB protein present in the nucleus. EMBO J. 1993;12:201–211. doi: 10.1002/j.1460-2075.1993.tb05646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]