Abstract
Peroxisome proliferator-activated receptor gamma (PPARγ) is a master regulator of adipogenesis. Our previous study revealed that chicken PPARγ has 3 alternative promoters named as P1, P2, and P3, and the DNA methylation of promoter P3 was negatively associated with PPARγ mRNA expression in abdominal adipose tissue (AAT). However, the methylation status of promoters P1 and P2 is unclear. Here we assessed promoter P1 methylation status in AAT of Northeast Agricultural University broiler lines divergently selected for abdominal fat content (NEAUHLF). The results showed that promoter P1 methylation differed in AAT between the lean and fat lines of NEAUHLF at 7 wk of age (p < 0.05), and AAT expression of PPARγ transcript 1 (PPARγ1), which was derived from the promoter P1, was greatly higher in fat line than in lean line at 2 and 7 wk of age. The results of the correlation analysis showed that P1 methylation was positively correlated with PPARγ1 expression at 7 wk of age (Pearson's r = 0.356, p = 0.0242), suggesting P1 methylation promotes PPARγ1 expression. To explore the underlying molecular mechanism of P1 methylation on PPARγ1 expression, bioinformatics analysis, dual-luciferase reporter assay, pyrosequencing, and electrophoresis mobility shift assay (EMSA) were performed. The results showed that transcription factor NRF1 repressed the promoter activity of the unmethylated P1, but not the methylated P1. Of all the 4 CpGs (CpG48, CpG49, CpG50, and CpG51), which reside within or nearby the NRF1 binding sites of the P1, only CpG49 methylation in AAT was remarkably higher in the fat line than in lean line at 7 wk of age (3.18 to 0.57, p < 0.05), and CpG49 methylation was positively correlated with PPARγ1 expression (Pearson's r = 0.3716, p = 0.0432). Furthermore, EMSA showed that CpG49 methylation reduced the binding of NRF1 to the P1. Taken together, our findings illustrate that P1 methylation promotes PPARγ1 expression at least in part by preventing NRF1 from binding to the promoter P1.
Key words: chicken, PPARγ, promoter methylation, abdominal adipose tissue, NRF1
INTRODUCTION
The chicken is an economically important domestic avian species, and after pork, is the second most consumed meat in China (Aslam et al., 2020; Lee et al., 2020). Due to the extensive genetic selection, the meat yield and growth rate have been tremendously increased in broilers. However, these gains are accompanied by excessive fat deposition (Abdalla et al., 2018). The excessive fat deposition has been a critical concern for broiler industry because it reduces feed efficiency, meat quality and yield (Abdalla et al., 2018; Ma et al., 2020).
Adipogenesis is a very complicated process in which preadipocytes differentiate into adipocytes. Adipogenesis is controlled by many factors such as transcription factors, coregulators, and epigenetic regulators (Lee et al., 2019; Kuri-Harcuch et al., 2019; Squillaro et al., 2020). Peroxisome proliferator-activated receptor gamma (PPARγ) is an indispensable factor for adipogenesis in mammals and birds (Park et al., 2017; Lee et al., 2019). PPARγ controls the expression of hundreds of genes during adipogenesis, resulting in terminal differentiation of preadipocytes to mature adipocytes (Cristancho and Lazar, 2011). PPARγ gene expression is regulated by alternative promoters in mammals. For example, mouse and human PPARγ genes are regulated by 2 and 4 alternative promoters, respectively (Zhu et al., 1995; Al-Shali et al., 2004). DNA methylation was considered as an epigenetic mark that remarkably regulates gene expression (Kim and Costello, 2017). Previous studies in mammals have shown that DNA methylation regulates PPARγ expression during adipogenesis (Fujiki et al., 2009; Huang et al., 2018). In mouse 3T3-L1 preadipocytes, the alternative promoter PPARγ2 was hypermethylated, however, during 3T3-L1 preadipocyte differentiation, it was gradually demethylated, accompanied with the increase of PPARγ2 expression (Fujiki et al., 2009). Reporter gene assay showed that DNA methylation inhibited PPARγ2 promoter activity (Fujiki et al., 2009). An in vivo study revealed that DNA methylation of PPARγ2 promoter in visceral adipose tissues (VAT) was higher in obese diabetic mice than in wild-type controls, whereas PPARγ2 mRNA expression in VAT was lower in obese diabetic mice than in wild-type controls (Fujiki et al., 2009). All these results indicated that PPARγ2 promoter methylation inhibits PPARγ2 mRNA expression (Fujiki et al., 2009).
Our previous study demonstrated chicken PPARγ gene is transcriptionally regulated by 3 alternative promoters designed as P1, P2, and P3, and produces 5 transcript isoforms (PPARγs1-5), due to alternative promoter usage and alternative splicing (Duan et al., 2015). Among the 3 alternative promoters (P1, P2, and P3), P1 had strongest promoter activity and the P1-derived transcript isoform PPARγ1 was remarkably expressed in AAT (Cui et al., 2018). We had demonstrated that the promoter P3 was differentially methylated in AAT of Northeast Agricultural University broiler lines divergently selected for abdominal fat content (NEAUHLF) (Sun et al., 2014). However, the promoter P1 methylation status is unclear.
In this study, we found that the promoter P1 methylation in AAT was higher in the fat line than in lean line of NEAUHLF, and that the P1 methylation was correlated positively with PPARγ1 expression at 7 wk of age. Further study demonstrated that DNA methylation promoted PPARγ1 expression at least in part via reducing the binding of transcription factor NRF1 to the promoter P1. Our findings provide insight into how DNA methylation controls PPARγ expression in chicken adipose tissue development and adipogenesis.
MATERIAL AND METHODS
Animal and Tissues
The AAT were collected from 60 male birds (5 birds per line at 2 and 3 wk of age and twenty birds per line at 7 wk of age) derived from 19th generation of NEAUHLF, which have been divergently selected for abdominal fat content in our laboratory since 1996. The lean and fat broiler lines have similar body weight, but have striking phenotypic differences in abdominal fat weight (AFW), the abdominal fat percentage (AFP), feed conversion ratio (FCR), and the residual feed intake (RFI), as well as adopocyte size and number (Guo et al., 2011; Zhang et al., 2020; Chen et al., 2021).
Cell Culture
DF1 cells and an immortalized chicken preadipocyte cell line (ICP1) (Wang et al., 2017), were cultured in DMEM/F12 (Gibco, Waltham, MA) and DMEM (Gibco, Waltham, MA) with 10% FBS in a humidified atmosphere at 37°C and 5% CO2, respectively.
Bioinformatics Analysis
The promoter P1 sequence, which covers the first 1,891 bp upstream and the first 108 bp downstream of the transcription start site of chicken PPARγ1 was obtained from UCSC (https://genome.ucsc.edu). The CpG island analysis was performed by using EMBOSS online software (http://www.ebi.ac.uk/Tools/seqstats/emboss_cpg plot/) and the transcription factor binding sites (TFBS) within the P1 was predicted by using JASPAR (http://jaspar.genereg.net).
Sequenom MassARRAY Methylation Assays
The quantitative methylation analysis was conducted by Oebiotech (Shanghai, China). Genomic DNA was obtained from the AATs of NEAUHLF as previously described (Lam et al., 2018). The concentration and quality of DNA samples were determined by NanoDrop spectrophotometer (Uppsala, Sweden), and DNA bisulfite conversion was obtained by EpiTect Bisulfite Kit (Qiagen, Germany). Two pairs of primers were determined by EpiDesigner (Supplementary Table S1) for detection of DNA methylation of each CpG unit containing either individual or combinations of CpG sites. The bisulfite converted DNA was expanded by PCR using the 2 pairs of primers designed above and the PCR products were cleaned with SAP enzyme using the MassCLEAVE kit (Sequenom, San Diego, CA). Then, the products were subjected to T cleavage, and spotted onto a 384-well SpectroCHIP bioarray (Sequenom, San Diego, CA) using MassARRAY Nanodispenser 1000 (Sequenom, San Diego, CA) and analyzed by the MassARRAY Analyzer 4.0 (Sequenom, San Diego, CA). DNA methylation levels were calculated using MassARRAY EpiTYPER software (Sequenom, San Diego, CA) (Xu et al., 2021).
Pyrosequencing Methylation Assays
Pyrosequencing was used for quantitative methylation of individual CpG sites in the promoter P1. Genomic DNA isolation and DNA bisulfite conversion were performed using the same procedure as above. DNA was amplified by PCR using PyroMark PCR kit. Pyrosequencing reactions were subsequently determined by the PyroMarkGold Q96 ID. The percentage of DNA methylation of individual CpG48, CpG49, CpG50, and CpG51 site within the promoter P1 was quantified using PyroMark CpG software, respectively.
qRT-PCR Aassays
Total RNAs were isolated from the AAT using TRIzol (Invitrogen, Carlsbad, CA). These cDNA was acquired by the HiScript Reverse Transcriptase (Vazyme, China). The qRT-PCR was decided by SYBR Green PCR ReadyMix (Roche, WGC, UK). The NONO was applied as an internal control and the 2−ΔΔCT approach was adopted to compute relative expression. These primers used for qRT-PCR are represented in supplementary Table S1.
Plasmid Construction
The pCMV-HA-NRF1 and pGL3P1-327/+108 vectors were constructed previously by our laboratory (Cui et al., 2018). Both unmethylated and methylated pGL3P1-327/+108 vectors were prepared as described previously (Cui et al., 2021).
Promoter Luciferase-Reporter Assays
DF1 and ICP1 cells were cultured at 80% confluence in 24-well plates and co-transfected with the designated reporter vectors with either pCMV-HA or pCMV-HA-NRF1 added pRL-TK with Lipofectamine 8,000 (Beyotime, China). The activities were decided by the Dual Luciferase reporter-system (Promega, Madison, WI) at 48 h after co-transfection. The promoter activity was expressed as the ratio of Firefly to Renilla luciferase activity (FLU/RLU).
Electrophoretic Mobility Shift Assay
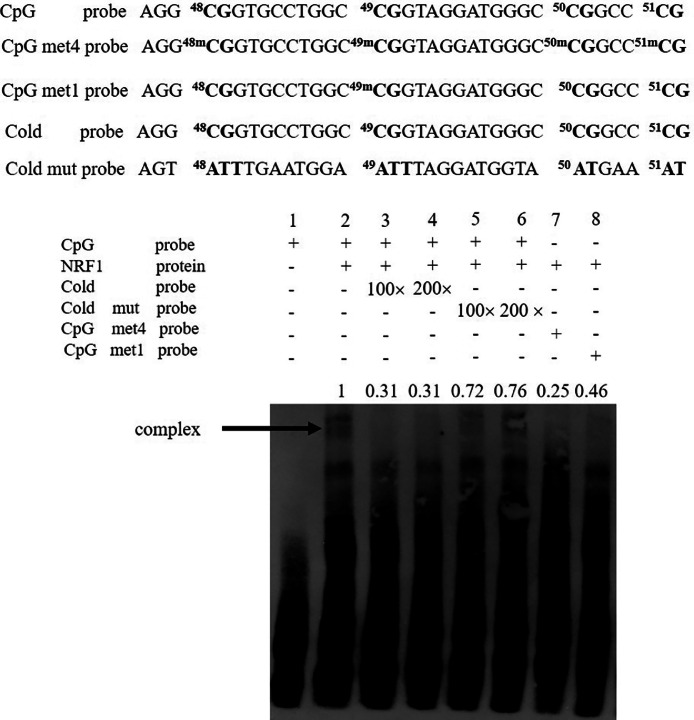
The nuclear extraction kit (Thermo Fisher, Carlsbad, CA) was employed to obtain the nuclear proteins from the DF1 cells transfected with pCMV-HA-NRF1. The NRF1 binding to the P1 promoter was decided by the EMSA Kit (Thermo Fisher, Carlsbad, CA). All the CpG sites (CpG48, CpG49, CpG50 and CpG51) were unmethylated in the unmethylated biotin-labeled P1 probe (CpG probe). All the 4 CpG sites were methylated in the methylated biotin-labeled P1 probe (CpG met4 probe). The partially methylated biotin-labeled P1 probe was named the CpG met1 probe, in which only CpG49 was methylated. The double-stranded P1 probes were cultured with the nuclear extract proteins for 20 min. A 100- or 200-fold cold probes or cold mutated probes were added for competition assays. All these probes were composed by Genewiz (Beijing, China). The detailed sequences of P1 probes were represented in supplementary Table S1.
Statistical Analysis
Data are showed as the mean ± SEM. The unpaired 2-tailed Student's t-tests were adopted to determined comparisons between group with Graph Pad Prism 7. The correlation between the promoter P1 methylation and PPARγ1 expression was analyzed using Pearson's r. The p values < 0.05 were regarded as a significant difference (*p < 0.05), and p values < 0.01 were considered as a tendency towards difference (**p < 0.01).
RESULTS
The P1 Promoter Methylation of Chicken PPARγ gene in AAT
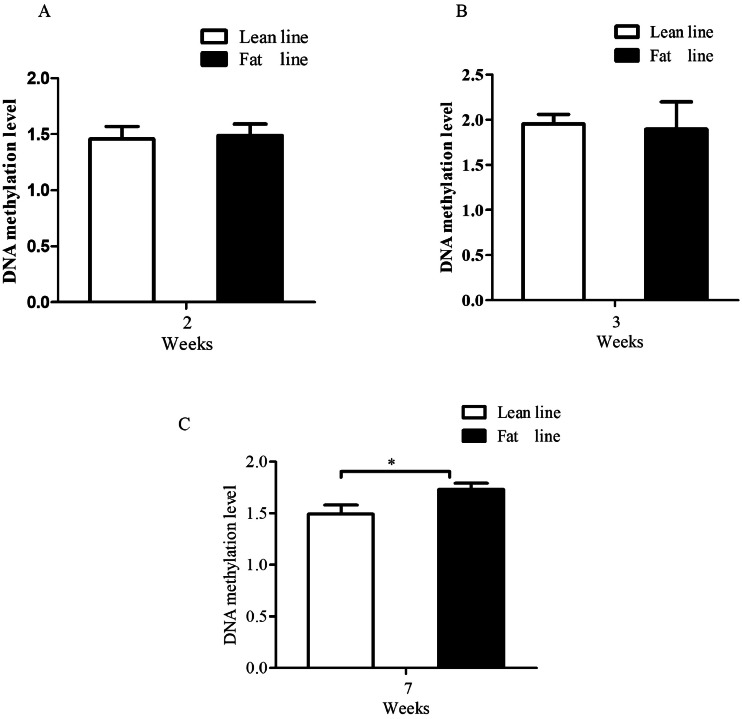
To investigate the promoter P1 methylation status, we first performed CpG island prediction in the promoter P1 sequence using EMBOSS. The results showed that there was a typical CpG island (385 bp, −333 to +52 bp, TSS = +1), containing 51 CpG sites (CpGs 1–51) within the promoter P1. Then, we detected DNA methylation of the predicted CpG island of the promoter P1 in AAT of NEAUHLF by using Sequenom MassARRAY platform. For technical reasons, individual or combinations of CpG sites was presented as one CpG unit in this assay (Wu et al., 2019), and all the 51 CpG sites were divided into 22 CpG units. Of these 22 CpG units, the DNA methylation of 20 CpG units was successfully determined, while the methylation of the other 2 CpG units (CpG10 and CpG20.21.22.23.24.25.26.27) was not detected due to the limitation of the Sequenom MassARRAY. The Sequenom MassARRAY analysis revealed that the P1 promoter was methylated in chicken AAT, and its methylation was 16.08% higher in fat line than in lean line at 7 wk of age (1.733 ± 0.06 vs.1.493 ± 0.09, p < 0.05, Figure 1C), but no difference in the P1 promoter methylation was observed between the lean and the fat lines at both 2 wk of age (1.458 ± 0.1107 vs. 1.488 ± 0.1028, p > 0.05) and 3 wk of age (1.952 ± 0.1067 vs. 1.894 ± 0.3035, p > 0.05) (Figures 1A and 1B).
Figure 1.
Comparison of the CpG island methylation of PPARγ gene promoter P1 in AAT of NEAUHLF. The CpG island methylation of PPARγ gene promoter P1 in AAT of NEAUHLFF at 2 (A), 3 (B), and 7 (C) wk of age was assessed using Sequenom Mass ARRAY platform.
Correlation Between the P1 Promoter Methylation and PPARγ1 Expression in Chicken AAT
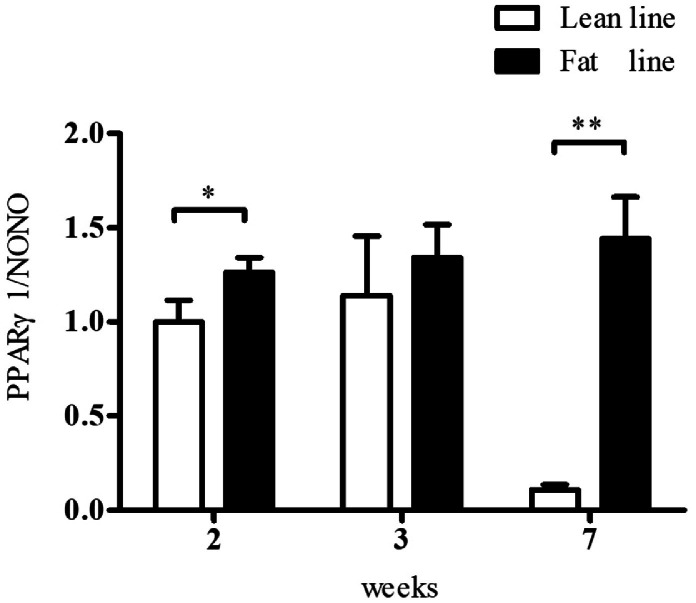
To determine whether DNA methylation controls the promoter P1 activity, we investigated PPARγ1 expression in the AAT of NEAUHLF using real-time RT-PCR and evaluated the correlation between the P1 promoter methylation and PPARγ1 expression. As shown in Figure 2, PPARγ1 expression displayed a trend towards high expression in fat line relative to the lean line at all 3 ages tested. In particular, PPARγ1 expression was higher in fat line than in lean line at 2 and 7 wk of age (p < 0.05). The results of the correlation analysis showed that there was no significant correlation between the P1 methylation and PPARγ1 expression in AAT of the lean and fat lines at 2 and 3 wk of age (Pearson's r = −0.16, p = 0.6588 and Pearson's r = 0.4344, p = 0.2097, respectively), but there was a remarkably positive correlation at 7 wk of age (Pearson's r = 0.356, p = 0.0242), suggesting that the P1 promoter methylation promotes PPARγ1 expression in AAT at 7 wk of age.
Figure 2.
Comparison of the PPARγ1 expression in AAT between the lean and fat lines of NEAUHLF at 2, 3, and 7 wk of age. PPARγ1 expression was assessed using qRT-PCR and normalized to NONO mRNA. All data are represented as the mean ± SEM. Student's t-test was used to compare the means between 2 groups. *p < 0.05, **p < 0.01.
DNA Methylation Abrogates NRF1-Mediated Inhibition of PPARγ1 Expression
To investigate the molecular mechanism underlying the promoting effect of the promoter P1 methylation on PPARγ1 expression, the JASPAR database was used to predict transcription factor binding sites (TFBSs) within the predicted CpG island of the promoter P1. The JASPAR database analysis showed that a total of 8 different TFBSs (E2F1, ZF5, HIC1, NRF1, MYB, AP2, USF, and EGR) overlapped with the CpG sites within the predicted P1 CpG island. Among these 8 TFBSs, NRF1 caught our attention. We previously demonstrated showed that NRF1 inhibited the P1 promoter activity and PPARγ1 expression (Cui et al., 2021). In addition, it has been shown that NRF1 is a methyl-sensitive transcription factor and DNA methylation inhibits its DNA binding and transactivation activity (Wang et al., 2017). These findings prompted us to hypothesize that the P1 methylation may promote PPARγ1 expression by interfering with negative regulation of the promoter P1 by NRF1.
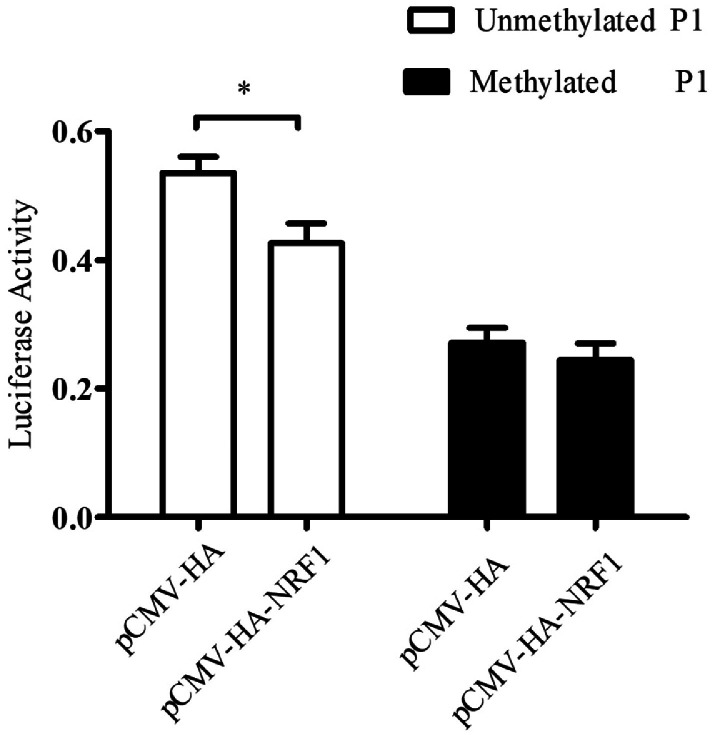
To test this hypothesis, we first performed a reporter assay to decide whether DNA methylation interferes with the NRF1-mediated inhibition of P1 activity. The reporter gene assay showed that NRF1 significantly repressed the activity of the unmethylated promoter P1 (pGL3P1-327/+108), causing a 20.45% decrease in the unmethylated P1 activity compared with the negative control (pCMV-HA) (p < 0.05, Figure 3), while NRF1 had no significant influence on the activity of the methylated P1 promoter (pGL3P1-327/+108) (p > 0.05, Figure 3). These results indicated that P1 methylation abrogated the NRF1-mediated P1 activity inhibition.
Figure 3.
DNA methylation abrogates the NRF1-mediated suppression of the promoter P1 activity. DF1 cells were co-transfected with either pCMV-HA-NRF1 or pCMV-HA, along with unmethylated or methylated P1 reporter as well as pRL-TK as an internal control. All data are presented as the mean ± SEM, *p < 0.05.
CpG49 Methylation Positively Correlates With the PPARγ1 Expression
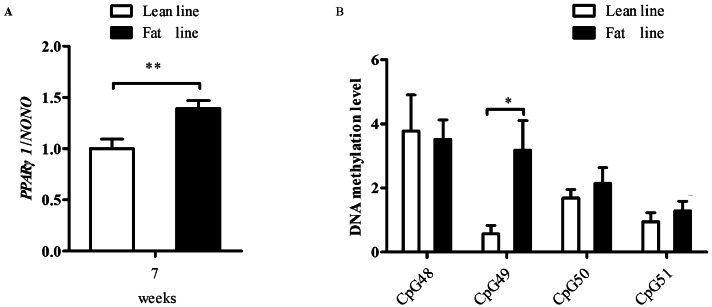
Three CpGs (CpG48 at +22, CpG50 at +45, and CpG51 at +51) resided within the NRF1 binding sites, and one CpG49 (at +32) was located in the vicinity of the 2 NRF1 binding sites. To further confirm that DNA methylation abrogates the NRF1-mediated suppression of the P1 activity, Pyrosequencing was used to quantify methylation levels of the 4 CpGs (CpG48, CpG49, CpG50, and CpG51) in 30 AAT samples of lean and fat lines of NEAUHLF (15 samples per line) at 7 wk of age. The results revealed that, of these 4 CpGs, only CpG49 (DNA methylation level was 0.57 in lean line, and 3.18 in fat line) was significantly differentially methylated in the AAT between the 2 lines of NEAUHLF (p < 0.05, Figure 4A). In parallel, PPARγ1 expression was assayed on the 30 abdominal adipose tissue samples using qRT-PCR. The gene expression results showed that PPARγ1 was differentially expressed in the AAT between the lean and fat lines (p < 0.01, Figure 4B). The correlation analysis showed that of these 4 CpGs, only CpG49 methylation was positively correlated with the PPARγ1 expression in AAT of NEAUHLF at 7 wk of age (Pearson's r = 0.3716, p = 0.0432), suggesting that CpG49 methylation abrogated the NRF1-mediated suppression of P1 promoter activity in vivo.
Figure 4.
The 4 individual CpG methylation and PPARγ1 expression in the AAT of lean and fat lines of NEAUHLF at 7 wk of age. (A) The DNA methylation analysis of the 4 individual CpG sites (CpG48, CpG49, CpG50 and CpG51) of P1 in AAT of the 2 broiler lines at 7 wk of age. (B) PPARγ1 expression in AAT of lean and fat lines of NEAUHLF at 7 wk of age. Student's t-test was used to compare the means between 2 groups. *p < 0.05, **p < 0.01.
CpG49 Methylation Prevent NRF1 From Binding to the Promoter P1
Last, to further verify that CpG49 methylation abrogates the NRF1-mediated suppression of the P1 promoter, we performed EMSA to determine the influence of CpG49 methylation on the binding of NRF1 to the P1 promoter. We synthesized 3 biotin-labeled probes (CpG, CpG met4, and CpG met1), a cold probe (unlabelled CpG probe), and a cold mutated probe (unlabelled mutant CpG probe). These 3 biotin-labeled probes shared the identical sequences, the CpG probe was unmethylated, CpG met4 probe contained four methylated CpGs (CpG48, CpG49, CpG50, and CpG51), and CpG met1 probe contained only 1 methylated CpG (CpG49). The EMSA results showed that as expected, NRF1 bound to CpG probe (Figure 5, lane 2) and its binding could be competitively inhibited by the addition of a 100- or 200-fold molar excess of the cold probe but not the cold mut probe (Figure 5, lanes 3, 4, 5, and 6). The binding affinity of NRF1 to both CpG met4 and CpG met1 probes was significantly lower than to the CpG probe (0.25 vs. 1 and 0.46 vs. 1, Figure 5, lanes 7 and 8). Comparatively, NRF1 had lower binding affinity for CpG met4 probe than for CpG met1 probe (0.25 vs. 0.46, Figure 5, lanes 7 and 8). Taken together, these results suggest that DNA methylation at CpG49 alone or in combination with other CpGs attenuates the binding of NRF1 to the P1 promoter.
Figure 5.
EMSA of the effects of the 4 individual CpG methylation on NRF1 binding to the promoter P1. The nuclear extracts were from the DF1 cells transfected with either control vector pCMV-HA or NRF1 expression vector pCMV-HA-NRF1, and EMSA was performed with the designated probes.
DISCUSSION
PPARγ is essential for adipogenesis and adipose tissue development. To date, no single factor has been demonstrated to drive adipogenesis in the absence of PPARγ (Lee and Ge, 2014). A number of studies, including our previous work, have shown that PPARγ expression is regulated by DNA methylation and that promoter methylation significantly decreases PPARγ expression. In the present study, we demonstrated that the methylation of the alternative promoter 1 promotes PPARγ1 expression, at least in part by preventing NRF1 binding to the promoter P1 of PPARγ gene in chicken adipose development.
DNA methylation is a critical epigenetic modification and plays crucial roles in a number of biological and pathological processes including gene expression, cell proliferation, differentiation, development, and disease (Cedar and Bergman, 2009; Wrzodek et al., 2012; Manzo et al., 2017; Cui et al., 2018). Generally, DNA methylation inhibits gene expression (Zhang et al., 2018; Jiang et al., 2020), but occasionally, it promotes gene expression (Spruijt et al., 2013; Halpern et al., 2014) . To date, there are 3 different mechanisms by which DNA methylation promotes gene expression. First, the binding of either chromatin protein or TFs to the methylated DNA results in chromatin remodeling from a highly condensed state to an open state, thus leading to increased gene expression (Zhu et al., 2016). For example, KLF4 binds to the methylated promoter of RHOC gene to remodel the chromatin of the RHOC promoter to an open state, thus leading to increased RHOC gene expression (Wan et al., 2017). Second, DNA methylation favors pre-mRNA processing such as RNA alternative splicing, and promotes gene expression (Wan et al., 2017). For example, hypermethylation of gene body promotes binding of MeCP2 and RNA alternative splicing, thus resulting in increased gene expression (Wan et al., 2017).
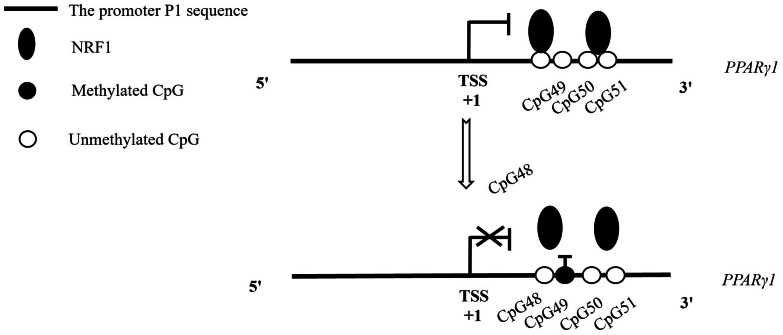
The third mechanism is that DNA methylation precludes the binding of negative regulatory transcription factors to target promoters, thereby leading to increased target gene expression (Wan et al., 2017). For example, Polycomb group protein can bind to FoxA2 promoter and inhibits FoxA2 expression, but promoter methylation prevents the Polycomb group protein from binding to FoxA2 promoter, leading to the increased expression of FoxA2 gene (Halpern et al., 2014). Our previous study showed that NRF1 repressed chicken PPARγ1 expression and the P1 promoter activity, and that the alternative promoter P1 harbored 6 NRF1 binding sites, and of these NRF1 binding sites, 2 NRF1 binding site mutations (N5 and N6) both almost completely abrogated the suppressive effect of NRF1 on the P1 promoter activity (Cui et al., 2018). In the present study, we demonstrated that the promoter P1 methylation abrogated NRF1-mediated inhibition of PPARγ1 expression, and DNA methylation at CpG49 alone (in the vicinity of N5 and N6) or in combination with other CpGs (CpG48, CpG50, CpG51 in N5 and N6) inhibited the binding of NRF1 to the P1 promoter. Based on previous and present findings, we propose a regulatory model depicted in Figure 6. NRF1 binds to the promoter P1 and inhibits PPARγ1 expression, but when DNA methylation occurs at CpG49 alone or in combination with other CpGs, it prevents NRF1 from binding to the promoter P1, thus leading to increased PPARγ1 expression.
Figure 6.
The proposed model for regulation of PPARγ1 expression by DNA methylation in chicken. NRF1 directly binds to the unmethylated P1 and inhibits PPARγ1 expression (upper panel). When CpG49 alone or in combination with the other CpGs is methylated, NRF1 cannot bind to the methylated promoter P1 and thus leading to derepressed expression of chicken PPARγ1 (lower panel). Open and filled circles represent the unmethylated and methylated CpGs, respectively.
In the present study, PPARγ1 mRNA expression was higher in fat line than in lean line at 7 wk of age, consistent with our previous study that PPARγ1 mRNA and PPARγ protein expression in AAT was higher in fat line than in lean line at 7 wk of age (Wang et al., 2012; Duan et al., 2015; Cui et al., 2018). Our previous study demonstrated that the promoter P3 methylation in AAT were higher in lean line than in fat line and that the P3 methylation and PPARγ mRNA level were negatively correlated (Sun et al., 2014), indicating that DNA methylation inhibits PPARγ expression. However, in this study, we found the promoter P1 methylation promoted PPARγ1 expression (Figures 4A and 4B). This difference indicates that these 2 alternative promoters of PPARγ gene are differentially epigenetically regulated in chicken adipogenesis and adipose development.
Besides DNA methylation, PPARγ expression is also regulated by histone modification, N6-methyladenosine (m6A), and post-translational modifications (PTMs) (Lee et al., 2019). Histone modifications can enhance or inhibit PPARγ transcription, for example, H4K20me1 modification in PPARγ gene body enhanced PPARγ transcription while H3K9me2 in PPARγ promoter inhibited PPARγ transcription (Okamura et al., 2010; Suzuki et al., 2023). The m6 A modification is the most abundant form of posttranscriptional RNA modification in eukaryotes. It has been shown that m6 A modification of PPARγ mRNA improved PPARγ mRNA stability and PPARγ protein expression (Guo et al., 2022). PPARγ is also regulated by a variety of post-translational modifications including phosphorylation, acetylation, and ubiquitination, for example, phosphorylation decreased PPARγ1 transcriptional activity (Pang et al., 2014), and ubiquitination inhibited PPARγ protein expression (Lee et al., 2018). Future study is needed to explore the roles and regulatory mechanisms mediated by epigenetic and post-translational modifications of PPARγ in chicken adipogenesis and adipose development.
CONCLUSIONS
We demonstrated that the promoter P1 of PPARγ is regulated by DNA methylation in chicken AAT, and its methylation promotes PPARγ1 expression, at least in part, through precluding the binding of NRF1 to the promoter P1.
ACKNOWLEDGMENTS
Support for this research was supported by the National Key Research and Development Program of China (2022YFF1000201), the National Natural Science Foundation of China (grant no.31572392), the Heilongjiang Province Education Department Fundamental Scientific Research Funds (grant no. 135509316) and Natural Scientific Foundation of Heilongjiang Province (LH2023C115).
Ethics Approval: This study was approved by the Northeast Agricultural University's Laboratory Animal Management Committee and was carried out in accordance with the guidelines on the care and use of experimental animals established by the Ministry of Science and Technology of the People's Republic of China (approval number 2006–398).
DISCLOSURES
None of the authors has any conflicts of interest to declare.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2024.103559.
Appendix. Supplementary materials
REFERENCES
- Aslam H.B., Alarcon P., Yaqub T., Iqba l.M., Häsler B. A value chain approach to characterize the chicken sub-sector in Pakistan. Front. Vet. Sci. 2020;7:361. doi: 10.3389/fvets.2020.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdalla B.A., Jie C., Nie Q., Zhang X. Genomic insights into the multiple factors controlling abdominal fat deposition in a chicken model. Front Genetics. 2018;9:262. doi: 10.3389/fgene.2018.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shali K., Cao H., Knoers N., Hermus A.R., Tack C.J., Hegele R.A. A single-base mutation in the peroxisome proliferator-activated receptor gamma4 promoter associated with altered in vitro expression and partial lipodystrophy. J. Clin. Endocrinol. Metab. 2004;89:5655–5660. doi: 10.1210/jc.2004-0280. [DOI] [PubMed] [Google Scholar]
- Cristancho A.G., Lazar M.A. Forming functional fat: a growing understanding of adipocyte differentiation. Nature Rev Mol Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Su Z., Li Y., Luan P., Wang S., Zhang H., Xiao F., Guo H., Cao Z., Li H., Leng L. Estimation of the genetic parameters of traits relevant to feed efficiency: result from broiler lines divergent for high or low abdominal fat content. Poult. Sci. 2021;100:461–466. doi: 10.1016/j.psj.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui T.T., Huang J.X., Sun Y., Ning B.L., Mu F., You X., Guo Y.Q., Li H., Wang N. KLF2 inhibits chicken preadipocyte differentiation at Least in part viadirectly repressing PPARγ transcript Variant 1 expression. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.627102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui T.T., Xing T., Huang J., Mu F., Jin Y.F., You X., Chu Y.K., Li H., Wang N. Nuclear respiratory factor 1 negatively regulates the P1 promoter of the peroxisome proliferator-activated receptor-γ gene and inhibits chicken adipogenesis. Front. Physiol. 2018;9:1823. doi: 10.3389/fphys.2018.01823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature Rev Genetics. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Duan K., Sun Y.N., Zhang X.F., Zhang T.M., Zhang W.J., Zhang J.Y., Wang G.H., Wang S.Z., Leng L., Li H., Wang N. Identification and characterization of transcript variants of chicken peroxisome proliferator-activated receptor gamma. Poult. Sci. 2015;94:2516–2527. doi: 10.3382/ps/pev229. [DOI] [PubMed] [Google Scholar]
- Fujiki K., Kano F., Shiota K., Murata M. Expression of the peroxisome proliferator activated receptor γ gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC. Biol. 2009;7:38. doi: 10.1186/1741-7007-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Sun B., Shang Z., Leng L., Wang Y., Wang N., Li H. Comparison of adipose tissue cellularity in chicken lines divergently selected for fatness. Poult. Sci. 2011;90:2024–2034. doi: 10.3382/ps.2010-00863. [DOI] [PubMed] [Google Scholar]
- Guo Y., Song W., Yang Y. Inhibition of ALKBH5-mediated m6 A modification of PPARG mRNA alleviates H/R-induced oxidative stress and apoptosis in placenta trophoblast. Environ. Toxicol. 2022;37:910–924. doi: 10.1002/tox.23454. [DOI] [PubMed] [Google Scholar]
- Huang Q., Ma C.Y., Chen L., Luo D., Chen R., Liang F.X. Mechanistic insights into the interaction between transcription factors and epigenetic modifications and the contribution to the development of obesity. Front Endocrinol. 2018;9:370. doi: 10.3389/fendo.2018.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern K.B., Vana T., Walker M. Paradoxical role of DNA methylation in activation of FoxA2 gene expression during endoderm development. J. Biol. Chem. 2014;289:23882–23892. doi: 10.1074/jbc.M114.573469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q.L., Lin D.D., Huang H.Y., Wang G.Z., Ye H.H. DNA methylation inhibits the expression of CFSH in mud crab. Front. Endocrinol. (Lausanne) 2020;11:163. doi: 10.3389/fendo.2020.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri-Harcuch W., Velez-delValle C., Vazquez-Sandoval A., Hernández-Mosqueira C., Fernandez-Sanchez V. A cellular perspective of adipogenesis transcriptional regulation. J. Cell Physiol. 2019;234:1111–1129. doi: 10.1002/jcp.27060. [DOI] [PubMed] [Google Scholar]
- Kim M., Costello J. DNA methylation: an epigenetic mark of cellular memory. Exp. Mol. Med. 2017;49:e322. doi: 10.1038/emm.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.Y., Song A.A., Loh T.C., Abdul R. Effects of lysine and methionine in a low crude protein diet on the growth performance and gene expression of immunity genes in broilers. Poult. Sci. 2020;9:2916–2925. doi: 10.1016/j.psj.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Schmidt H., Lai B., Ge K. Transcriptional and epigenomic regulation of adipogenesis. Mol. Cell Biol. 2019;39:e00601–e00618. doi: 10.1128/MCB.00601-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Ge K. Transcriptional and epigenetic regulation of PPARγ expression during adipogenesis. Cell Biosci. 2014;4:29. doi: 10.1186/2045-3701-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam D., Ancelin M.L., Ritchie K., Saffery R., Ryan J. DNA methylation and genetic variation of the angiotensin converting enzyme (ACE) in depression. Psychoneuroendocrinology. 2018;88:1–8. doi: 10.1016/j.psyneuen.2017.11.003. [DOI] [PubMed] [Google Scholar]
- Lee J.M., Choi S.S., Lee Y.H., Khim K.W., Yoon S., Kim B.G., Nam D., Suh P.G., Myung K., Choi J.H. The E3 ubiquitin ligase TRIM25 regulates adipocyte differentiation via proteasome-mediated degradation of PPARγ. Exp. Mol. Med. 2018;50:1–11. doi: 10.1038/s12276-018-0162-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Sun J., Zhu S., Du Z., Li D., Li W., Li Z., Tian Y., Kang X., Sun G. MiRNA and mRNAs analysis during abdominal preadipocyte differentiation in chickens. Animals. (Basel) 2020;10:468. doi: 10.3390/ani10030468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzo M.J., Wirz C., Ambrosi, Villaseor R., Baubec T. Isoform๕pecific localization of DNMT3A regulates DNA methylation fidelity at bivalent CpG islands. The EMBO Journal. 2017;36 doi: 10.15252/embj.201797038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura M., Inagaki T., Tanaka T., Sakai J. Role of histone methylation and demethylation in adipogenesis and obesity. Organogenesis. 2010;6:24–32. doi: 10.4161/org.6.1.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.K., Wang L., Giampietro A., Lai B.B., Lee J.E., Ge K. Distinct roles of transcription factors KLF4, Krox20, and peroxisome proliferator-activated receptor γ in adipogenesis. Mol. Cell Biol. 2017;37 doi: 10.1128/MCB.00554-16. e00554-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang X., Shu Y., Niu Z., Zheng W., Wu H., Lu Y., Shen P. PPARγ1 phosphorylation enhances proliferation and drug resistance in human fibrosarcoma cells. Exp. Cell Res. 2014;322:30–38. doi: 10.1016/j.yexcr.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Squillaro T., Peluso G., Galderisi U., Di Bernardo G. Long non-coding RNAs in regulation of adipogenesis and adipose tissue function. Elife. 2020;9:e59053. doi: 10.7554/eLife.59053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.N., Gao Y., Qiao S.P., Wang S.Z., Duan K., Wang Y.X., Li H., Wang N. Epigenetic DNA methylation in the promoters of Peroxisome Proliferator-Activated Receptor in chicken lines divergently selected for fatness. J. Anim. Sci. 2014;92:48–53. doi: 10.2527/jas.2013-6962. [DOI] [PubMed] [Google Scholar]
- Spruijt G.C., Gnerlich F., Smits A.H., Pfaffeneder T., Jansen P.W.T.C., Bauer C., Münzel M., Wagner M., Müller M., Khan F. Dynamic readers for 5-(Hydroxy) methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Komatsu T., Shibata H., Tanioka A., Vargas D., Kawabata-Iwakawa R., Miura F., Masuda S., Hayashi M., Tanimura-Inagaki K., et al. Crucial role of iron in epigenetic rewriting during adipocyte differentiation mediated by JMJD1A and TET2 activity. Nucleic. Acids. Res. 2023;51:6120–6142. doi: 10.1093/nar/gkad342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.Y., Wang Y.X., Gong P.F., Wang L.J., Liu C., Chen C., Jiang X.Y., Cheng B.H., Li H. Promoter methylation regulates ApoA-I gene transcription in chicken abdominal adipose tissue. J. Agric. Food Chem. 2019;67:4535–4544. doi: 10.1021/acs.jafc.9b00007. [DOI] [PubMed] [Google Scholar]
- Wang J., Chao T., Qian W. NRF1 coordinates with DNA methylation to regulate spermatogenesis. Faseb J Off Public Feder Am Soc Exp Biol. 2017;31:4959. doi: 10.1096/fj.201700093R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzodek C., Büchel F., Hinselmann G., Eichner J., Mittag F., Zell A. Linking the epigenome to the genome: correlation of different features to DNA methylation of CpG islands. PLoS. One. 2012;7:e35327. doi: 10.1371/journal.pone.0035327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhang T., Wu C., Wang S., Wang Y., Li H., Wang N. Immortalization of chicken preadipocytes by retroviral transduction of chicken TERT and TR. PLoS. One. 2017;12 doi: 10.1371/journal.pone.0177348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Su Y., Song Q., Tung B., Oyinlade O., Liu S., Ying M.Y., Ming G.L., Song H.J., Qian J. Methylated cis-regulatory elements mediate KLF4-dependent gene transactivation and cell migration. Elife. 2017;6:e20068. doi: 10.7554/eLife.20068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Na W., Wang Y.X., Wang Y.B., Wang N., Wang Q.G., Li Y.M., Li H. Characterization of chicken PPARγ expression and its impact on adipocyte proliferation and differentiation. Yi. Chuan. 2012;34:454–464. doi: 10.3724/sp.j.1005.2012.00454. [DOI] [PubMed] [Google Scholar]
- Xu S., Gao X., Ma Y., Deng J., Pan F. Association of methylation level and transcript level in TRAF5 gene with ankylosing spondylitis: a case-control study. Genes Immunity. 2021;22:101–107. doi: 10.1038/s41435-021-00135-7. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Qi C., Korenberg J.R., Chen X.N., Noya D., Rao M.S., Reddy J.K. Structural organization of mouse peroxisome proliferator-activated receptor gamma (mPPAR gamma) gene: alternative promoter use and different splicing yield two mPPAR gamma isoforms. Proc. Natl. Acad. Sci. u S. a. 1995;92:7921–7925. doi: 10.1073/pnas.92.17.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Liang Q., Wang N., Wang Q., Leng L., Mao J., Wang Y., Wang S., Zhang J. Microevolutionary dynamics of chicken genomes under divergent selection for adiposity. iScience. 2020;23 doi: 10.1016/j.isci.2020.101193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Guan Q., Chen G., Qian F., Liang J. DNA methylation of the CDC2L1 gene promoter region decreases the expression of the CDK11p58 protein and reduces apoptosis in keloid fibroblasts. Arch. Dermatol. Res. 2018;310:107–115. doi: 10.1007/s00403-017-1801-9. [DOI] [PubMed] [Google Scholar]
- Zhu H., Wang G., Jiang Q. Transcription factors as readers and effectors of DNA methylation. Nature Rev Genetics. 2016;17:551–565. doi: 10.1038/nrg.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.