Abstract
The MyoD family of basic helix-loop-helix (bHLH) proteins is required for myogenic determination and differentiation. The basic region carries the myogenic code and DNA binding specificity, while the N terminus contains a potent transcriptional activation domain. Myogenic activation is abolished when the basic region, bound to a myogenic E box, carries a mutation of Ala-114. It has been proposed that DNA binding of the MyoD basic region leads to recruitment of a recognition factor that unmasks the activation domain. Here we demonstrate that an A114N mutant exhibits an altered conformation in the basic region and that this local conformational difference can lead to a more global change affecting the conformation of the activation domain. This suggests that the deleterious effects of this class of mutations may result directly from defective conformation. Thus, the activation domain is unmasked only upon DNA binding by the correct basic region. Such a coupled conformational relationship may have evolved to restrict myogenic specificity to a small number of bHLH proteins among many with diverse functions yet with DNA binding specificities known to be similar.
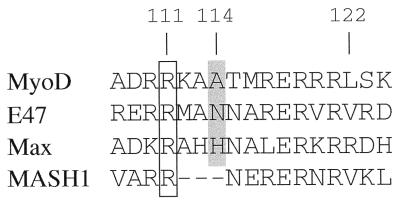
The skeletal muscle-specific basic helix-loop-helix (bHLH) transcription factors (including MyoD, Myf-5, myogenin, and MRF-4) are required for myogenic determination and differentiation (34). Previous work has identified Ala-114 and Thr-115 of MyoD as comprising a myogenic code—crucial basic-region residues of MyoD required for myogenic conversion and activation of muscle-specific genes (6, 8, 35). Introducing just these two residues from MyoD into corresponding positions in the ubiquitous bHLH protein E12 allows the altered E12 to function as a myogenic activator (9, 35) (see Table 1). Cooperative DNA binding was shown to be affected in some basic-region mutants (3), but lowered affinities or altered specificities of DNA binding by such mutants complicated the interpretation. Thus, the molecular mechanisms involved in deciphering the myogenic code remain unclear. Arg-111 of MyoD is highly conserved throughout the bHLH and bHLH/leucine zipper families (see Fig. 1). However, in protein-DNA cocrystals (12, 13, 21), it adopts a unique conformation due to the small size of Ala-114, which is specific to the myogenic bHLH proteins: Arg-111 is buried in the major groove and contacts the N-7 of the final G of CAGCTG. In other bHLH proteins, this arginine residue lies outside of the major groove and position 114 equivalents are occupied by bulkier amino acids, which themselves contact the G of CANNTG. These observations raise the possibility that the exposure of Arg-111 prevents myogenic function. This paper demonstrates the long-sought and fundamental mechanisms by which this important class of myogenic regulatory proteins activates transcription.
TABLE 1.
Myogenic activation in cells transfected with various plasmidsa
| Plasmid | % MHC+ cellsb | Relative activity ofc:
|
||
|---|---|---|---|---|
| −177HCAβ-Gal | 115MCKβ-Gal | 4RTKβ-Gal | ||
| Scribe | 0 | 1.1 | 2 | 1 |
| MyoD | 30 | 100 | 100 | 100 |
| A114N | 1.5 | 5 | 3 | 1.4 |
| A114H | 0 | 2.6 | NDe | 1.2 |
| A114G | 0 | 0.9 | 1 | 1 |
| T115N | 15 | 67 | ND | ND |
| E12 | 0 | 0.2 | 0.3 | 0.9 |
| E12-ATKd | 20 | 2.5 | 2.5 | 3.3 |
Two micrograms of reporter plasmids and 2 μg of various MyoD expression vectors were cotransfected into NIH 3T3 cells. The values are the averages of two independent experiments.
MHC+ cells were identified by MF20 staining, and their number was scored as a percentage of the total number of cells on the dish.
The β-Gal activity was measured and expressed relative to that for wild-type MyoD. β-Gal reporter genes were driven by the HCA promoter region (−177 to +68) in −177HCAβ-Gal, by the MCK enhancer region (−1207 to −1093) and promoter region (−82 to +7) in 115MCKβ-Gal, and by four copies of the MCK R-site and thymidine kinase gene minimal promoter in 4RTKβ-Gal (gift of S. Tapscott).
E12-ATK encodes E12 but with replacements at positions 114, 115, and 124 by the corresponding amino acids (alanine, threonine, and lysine) from MyoD.
ND, not determined.
FIG. 1.
Basic-region sequence alignment of the bHLH MyoD (mouse) and E47 (human) proteins, MASH1 (mouse), and the bHLH-LZ Max (mouse) protein. Amino acids are given in one-letter code; the numbering is according to full-length mouse MyoD. The conserved yet conformationally nonuniform arginines at the 111 positions are boxed; the residues at position 114, which likely determine the conformation of Arg-111, are shaded.
MATERIALS AND METHODS
Plasmid construction.
Amino acids are numbered according to full-length mouse MyoD throughout the text. PCR-based mutagenesis (15) was used to generate the mutations in which alanine at position 114 was replaced by asparagine (A114N), histidine (A114H), and glycine (A114G) in the backbone of pEMSV-MyoD (10), a eukaryotic expression vector for MyoD. Orientation was based on the sense strand of MyoD. ΔN-A114N was generated by replacing the ScaI-PmlI fragment of pA114N with the ScaI-AluI fragment from pEMSV-MyoD, which encodes two amino acids (aa) of MyoD. ΔN-A114N–VP16 and A114N–VP16 were generated by replacing the ScaI-StuI fragment of ΔN-MyoD–VP16 with the ScaI-StuI fragments from ΔN-A114N and A114N, respectively. To create bacterial expression vectors, an NdeI site encompassing the start codon for MyoD was introduced by PCR mutagenesis. The NdeI-HindIII fragment containing the coding region for MyoD was ligated into the backbone of pGM484 (gift from A. Klug, Medical Research Council) to generate pT7-MyoD. T7 vectors of MyoD basic-region mutations were constructed by replacing the PmlI-MluI fragment of MyoD in pT7-MyoD with those from the corresponding mutants in the eukaryotic expression vectors. Gal-MyoD and Gal-A114N fusion plasmids were generated by fusing the NdeI-HindIII fragments from T7 vectors in frame at the region encoding the carboxyl terminus of Gal4 DNA binding domain (aa 1 to 147) in the vector pGal0 (27). A PCR fragment encoding MyoD residues 1 to 75 was inserted between the NdeI and XbaI sites of pGal0 to generate Gal–MyoD(1–75). pG5-LacZ and pG5-Luc were created by inserting a PCR fragment, encoding five Gal4 DNA binding sites and the E1b TATA box from pG5CAT (Clontech), into pNNN–β-Gal (16) devoid of its promoter and pGL2-Basic (Promega), respectively.
Production of wild-type and mutant MyoD proteins.
Wild-type and mutant MyoD proteins were expressed in Escherichia coli BL21(DE3)/pLysS and purified to >90% homogeneity by standard protocols (29), and E47 protein was purified as described previously (30).
DNA binding assays (gel mobility shift assays).
Oligodeoxyribonucleotides were prepared on an automated DNA synthesizer (Applied Biosystems) using phosphoramidite chemistry. 1R contains the right (R) site E box of the muscle creatine kinase (MCK) enhancer, and 2R contains two copies of the R site in tandem repeats. One strand of the probe was end labeled with [γ-32P]ATP by using T4 polynucleotide kinase annealed to a 10-fold molar excess of the unlabeled complementary strand, and purified with a G-25 spin column (Boehringer Mannheim).
For standard gel mobility shift assays, 5 × 104 cpm of end-labeled DNA fragment (about 0.5 ng, or 10 to 35 fmol for a 20- to 30-bp probe) was incubated with 5 to 200 ng (as specified for separate experiments in the figures) of purified MyoD and/or E47 proteins for 15 min at room temperature in a solution containing 20 mM HEPES (pH 7.6), 50 mM KCl, 3 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 8% glycerol, and 1 μg of poly(dI-dC) in a final volume of 20 μl. The reaction mixtures were resolved on nondenaturing 4% polyacrylamide gel with 1× Tris-borate-EDTA at 10 V/cm and room temperature and were visualized by autoradiography.
Protease sensitivity assay.
Subtilisin (EC 3.4.21.14; Boehringer Mannheim) was added to proteins both in the presence and in the absence of specific DNA to final concentrations of 50 ng/μl and incubated at 37°C for different lengths of time. The proteolysis reactions were terminated by addition of 0.5 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma).
Cell culture and transfection assays.
Cell culture and transfection procedures were performed as described previously (16) except that transfections were done on 60-mm-diameter plates with 4 μg of total plasmid DNA unless otherwise specified. β-Galactosidase (β-Gal) and luciferase activities from cell lysates were assayed with chemiluminescent substrates from Tropix.
Immunofluorescent staining of cells.
Cells were washed, fixed in 4% paraformaldehyde in Tris-saline for 5 min, washed, permeabilized with 0.1% Triton X-100 in Tris-saline for 5 min, and washed again. Next, cells were stained with MF20 (mouse monoclonal antibody to myosin heavy chain [MHC] at a 1:10 dilution) for 1 h. After being washed, cells were stained with goat anti-mouse immunoglobulin G conjugated to fluorescein (1:500 dilution) for 1 h, washed, and examined by fluorescence microscopy. All washes were done two or three times in Tris-saline.
In vitro protein association assay.
Purified MyoD and A114N proteins were assayed for binding to glutathione S-transferase–p300 beads as previously described (28). Binding to six-histidine-tagged dTAFII40 was carried out with Ni-nitrilotriacetic acid resin (Qiagen) according to the manufacturer’s protocol.
RESULTS AND DISCUSSION
The MyoD A114N mutant fails to activate myogenic transcription upon binding to DNA.
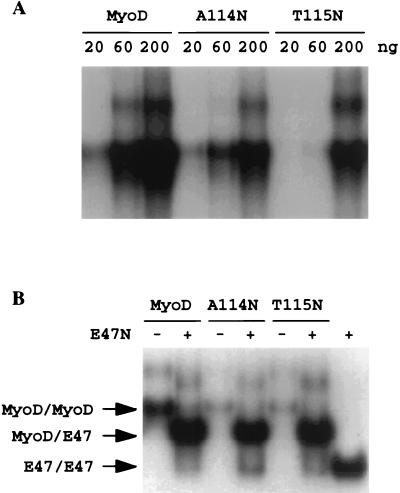
We sought to test the hypothesis that the exposure of Arg-111 prevents myogenic function by decreasing the ability of Arg-111 to easily fit into the major groove. We replaced Ala-114 in MyoD with bulky amino acids, asparagine and histidine, that occur naturally at the corresponding E-box-binding, basic-region positions of E47 and Max, respectively (Fig. 1). At most a threefold decrease in DNA binding was detected for the resulting A114N mutant homodimers (Fig. 2A). However, no differences in DNA binding were detected when the mutant proteins (heterodimerized with E47) were compared to the wild-type MyoD/E47 heterodimers for their affinities for specific DNA sequences (Fig. 2B). In contrast, when cotransfected with β-Gal reporter plasmids driven by muscle-specific promoters for either the MCK or human cardiac α-actin (HCA) genes, the A114N mutant activated the reporter gene to no more than 5% of the level achieved by wild-type MyoD (Table 1). The A114H mutant showed a defect similar to that of the A114N mutant. Another mutant, with Thr-115 of MyoD replaced by Asn (present at the corresponding positions in both E47 and Max [Fig. 1]) (T115N), exhibited a DNA-binding activity similar to that of the A114N mutant (Fig. 2) but retained at least 60% of the transcriptional activity of the wild-type protein (Table 1). Therefore, the detrimental effect of the A114N mutation on transcriptional activation is highly specific and is not likely engendered by subtle differences in DNA binding affinity but, rather, must result from the effects of the bulky amino acid substitutions at position 114. These mutant proteins were expressed in transfected cells at the same levels as the wild-type MyoD protein as determined by Western blot analysis (data not shown). Their nuclear localization was not affected as determined by immunohistochemistry with an antibody (Santa Cruz) specific to the carboxyl terminus of MyoD (data not shown). This result is in accord with previous findings that MyoD contains one nuclear localization signal in each of the basic region and the H1 region and that disruption of both is required to abolish nuclear localization (32).
FIG. 2.
DNA binding by wild-type and mutant MyoD proteins. (A) Increasing amounts of MyoD proteins were incubated with labeled 1R probe (0.5 ng) for 15 min at room temperature and analyzed by nondenaturing 4% polyacrylamide gel electrophoresis. (B) Three hundred nanograms each of MyoD protein and 20 ng of E47N (a truncated form of E47 with a deletion in the N-terminal region) protein, together or separately, were incubated at 37°C for 15 min prior to DNA binding reactions carried out at room temperature for 15 min.
With assays for myogenic activity, we found that the A114N mutant dramatically loses the ability to promote myogenesis in transiently transfected NIH 3T3 cells: it produced 1.5% conversion, in contrast to 30% conversion by MyoD. That the A114N protein has retained any residual myogenic activity may result from the heterogeneous conformation in this molecule (see below).
A114N mutation alters the conformation of the basic region.
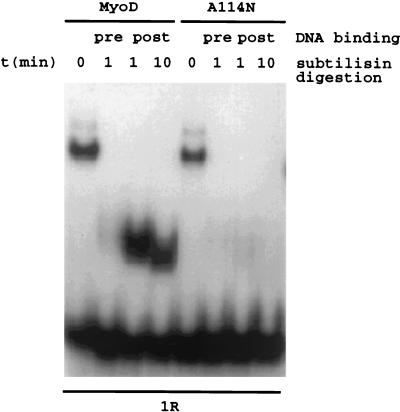
To examine the consequence of the A114N mutation for the conformation of Arg-111, the critical residue, we made use of the fact that the basic region of MyoD (which includes Arg-110 and Arg-111) happens to be a processing-site motif for subtilisin (2). We tested the subtilisin sensitivities of wild-type and mutant MyoD proteins (Fig. 3). Wild-type MyoD contains a core domain (consisting of the bHLH domain [unpublished result]) that is resistant to protease digestion in the presence of DNA, but in the A114N mutant molecule the core domain is much more sensitive to protease, even in the presence of DNA. These observations suggest that the basic domains of the mutant and wild-type proteins have different conformations. Although other possibilities remain, the simplest explanation is that in the mutant, unlike in the wild-type MyoD, revealed in the crystal structure (21), Arg-111 is no longer protected by DNA and therefore may not have direct contact with DNA. Accordingly, it is possible that in the A114N mutant Arg-111 is outside the major groove most or all of the time, due to the bulkiness of Asn at the 114 position, and thus becomes accessible for protease digestion.
FIG. 3.
Gel retardation assay. Six hundred forty nanograms of bacterially purified MyoD and A114N proteins were treated with subtilisin (50 ng/ml) both in the presence and in the absence of 0.5 ng of 1R DNA probe. pre, proteins were treated with subtilisin at 37°C for 1 min, the reaction was stopped by addition of PMSF, and then the proteins were incubated with 1R probe at 30°C for 30 min; post, proteins were incubated with 1R probe at 30°C for 30 min and then treated with subtilisin at 37°C for 1 or 10 min, and the reaction was stopped by addition of PMSF. Samples were analyzed by nondenaturing 4% polyacrylamide gel electrophoresis.
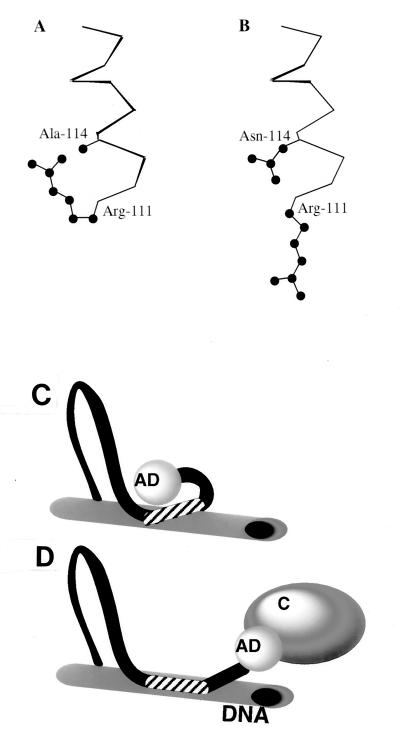
This model is consistent with the results of molecular modeling analysis with the determined structural coordinates of the MyoD-DNA cocrystal (21), which reveals that an Asn placed in the 114 position would impinge on Arg-111 (see Fig. 5, upper panel), and suggests that Asn at 114 might in reality force Arg-111 to remain out of the major groove, similar to the situation in the E47-DNA cocrystal (12, 21). In contrast, molecular modeling with the nondeleterious T115N mutant did not suggest such friction within the crystal structure. The final proof of the validity of this model must await X-ray crystallographic data for the mutant(s).
FIG. 5.
(A and B) Molecular modeling reveals different basic-domain conformations in wild-type MyoD (A) and the A114N mutant (B). α-Carbon traces of residues 111 and 114 are presented. Note the drastically different conformations of Arg-111. (C and D) Schematic representation of changes in MyoD structure following binding to DNA. Binding of MyoD basic domain (striped area) to a target DNA site (C) is associated with a distinctive change of conformational structure that is propagated to the activation domain (AD), allowing interaction with transcriptional coactivators (labeled as “C” in panel D). In panel C, the AD is sketched as interacting with the basic domain, an inference drawn from the inhibitory effects of basic domain elements shown in Table 2. DNA binding by the A114N mutant also keeps the AD in the conformation shown in panel C.
One prediction of the above-described model is that smaller amino acids at position 114 would accommodate Arg-111 in the major groove and allow for myogenic activity. Thus, we generated the A114G mutant. As is shown in Table 1, it failed to induce myogenic conversion, nor could it activate HCA, MCK, or 4R reporter constructs. Since glycine is known to be a frequent “α-helix breaker,” we speculate that it probably prevents the basic region from assuming the α-helical structure induced by cognate DNA sequences as occurs normally. Interestingly, when we compared the residues flanking Gly114 to the α-helix termination motifs deduced from protein folding studies (1), we found that the MyoD sequences perfectly matched the Schellman motif—with Met-116 at the C" position of the motif (requiring an apolar residue), Arg-110, Arg-111, and Lys-112 at C-4, C-3, and C-2 (requiring apolar Lys or Arg), and Ala-113 at C-1 (requiring a polar amino acid or Ala).
As a direct test of this model we attempted to obtain second-site mutations at position 111 that could suppress the myogenic defect in the A114N mutant. To ensure the presence of all 20 possible amino acid substitutions and to increase the throughput of the screening, we used PCR to make four libraries of site-directed random mutations at Arg-111 by using GNN, ANN, TNN, and CNN in place of the Arg codon. Each library has a limited complexity of 16, encoding five to seven different amino acids, and is represented by more than 200 independent transformants. Thus, the confidence of each different codon being present in the library is 99.999% (7). We collected transformants from each of the four libraries as a pool, transfected each pool of plasmid extracts into NIH 3T3 cells along with the HCA–β-Gal reporter plasmid, and assayed for transcriptional activation of the HCA promoter and myogenic conversion. If a library contained a mutation that can restore myogenic activity to A114N, it should have been easily detectable in this assay, because NIH 3T3 cells transfected by A114N plasmid spiked with 1% wild-type MyoD plasmid showed a level of activation of the HCA–β-Gal reporter significantly above the background level produced by the A114N plasmid alone, and muscle-like morphological changes of the transfected cells were visible. However, none of the four libraries showed elevated transcriptional activity or myogenic conversion.
Mammalian achaete-scute homologous protein 1 (MASH1), a neurogenic bHLH protein that is incapable of inducing myogenic conversion but can partially activate transcription from a single muscle promoter, the MCK E box (17), contains a bulky asparagine in its basic domain immediately adjacent to the Arg-111 site. However, steric considerations preclude a direct comparison between the basic-region peptides of MASH1 and MyoD. MASH1 has three fewer residues in its basic region, as it is missing amino acids corresponding to those at 112 to 114 of MyoD, which aligns the asparagine with T-115 of MyoD. We show in this paper that the MyoD T115N mutation not only transactivates but also allows myogenesis. It is possible that the activation of the MCK E box by MASH1 is fortuitous, and since activation of only one or a few muscle genes is not sufficient to cause myogenesis, such compromised specificity might not have been selected against in evolution. In fact, that MASH1 has no myogenic activity in vivo is most revealing.
These results suggest that the mechanism of myogenic activation at work in MyoD involves the ability of Arg-111 to lie within the major groove and make direct contact with the guanine of the E box. Since in all the revertants we created with various amino acids at position 111 there is still a bulky asparagine at position 114, it is unlikely that any amino acids at position 111 can make such a contact. Thus, it is not surprising that these mutants remain transcriptionally inactive. Furthermore, since Arg-111 is invariant throughout all known bHLH proteins, mutating it likely abolishes basic-region structure and/or DNA binding. In fact, some mutations of MyoD (8) and E47 (33) affecting this Arg have been shown to abolish DNA binding.
In the A114N mutant the altered basic-region conformation is propagated to the whole molecule to affect transcriptional activation.
In the basic region of the A114N mutant, an altered conformation which does not alter its DNA-binding specificities might lead to a transactivation defect in at least two ways. First, the altered basic-region conformation might lead to a profound change in the conformation of the whole molecule that affects the function of the activation domain(s). Second, the altered basic-region conformation might disrupt binding to the basic region of a recognition factor, such as myocyte enhancer factor 2C (MEF2C) (23). These two mechanisms do not need to be mutually exclusive.
To investigate whether the basic-region mutation affects the conformation of the whole molecule, we attempted to determine whether the transcription activation domain in the A114N mutant behaves differently than it does in MyoD by using two different assays. First, we fused full-length copies of either MyoD or the A114N mutant to the DNA-binding domain of Gal4 to generate Gal-MyoD and Gal-A114N and cotransfected each with a LacZ reporter gene driven by five Gal4 DNA binding sites into NIH 3T3 cells. Interestingly, Gal-A114N activated transcription 10 times better than Gal-MyoD (Table 2), even though the fusion proteins were expressed and appeared to bind to Gal4 DNA sites equally well (data not shown), indicating that the different conformation in the basic region of the A114N mutant affects the conformation and/or the accessibility of the activation domain(s). Secondly, we tested the ability of the activation domain in the A114N mutant to interact with molecules known to coactivate the MyoD activation domain. It has been shown previously that the MyoD coactivator p300 can target both the N-terminal activation domain (28) and the bHLH domain (11) (our unpublished data) of MyoD. There was no detectable difference between wild-type MyoD and the A114N mutant in the ability to bind to glutathione S-transferase–p300 (data not shown). In addition, dTAFII40 can also serve as a coactivator for MyoD, and it targets the amino-terminal activation domain of MyoD (unpublished data). The binding to 6His-TAFII40 of MyoD was comparable to that of A114N protein (data not shown). We found that while both p300 and dTAFII40 can potentiate the activation of myogenic promoters by MyoD in vivo, they do not potentiate the residual activation by the A114N mutant (Fig. 4B). Thus, the altered conformation of the basic domain of the mutant MyoD protein appears to be propagated to the activation domain at the N terminus, rendering the molecule incapable of responding to the coactivators. These effects are independent of any role of cofactors such as MEF2C (see below).
TABLE 2.
Different conformations in MyoD versus A114N mutant proteins inferred from Gal fusion experimentsa
| Gal4 fusion constructb | G5-LacZ activityc |
|---|---|
| Control | 1 |
| (1–318)MyoD | 20 |
| (1–318)A114N | 200 |
| (1–75)MyoD | 2,000 |
| (1–101)MyoD | 600 |
| (1–127)MyoD | 1 |
| (1–127)A114N | 1 |
| (1–161)MyoD | 0.3 |
| (1–161)A114N | 0.3 |
NIH 3T3 cells were transfected with 1.5 μg of G5-E1b-LacZ reporter plasmid and 1.5 μg of Gal4 fusion constructs on 60-mm-diameter duplicate plates.
Control, activity of the reporter plus empty fusion vector; (1–75)MyoD is a Gal4 fusion construct carrying the 75 amino-terminal residues of MyoD including the previously defined N-terminal activation domain (aa 1 to 53); (1–101), (1–127), and (1–161) indicate proteins that are truncated at the end of the Cys- and His-rich domain, the junction of the basic region and helix 1, and the end of helix 2, respectively; (1–318) indicates full-length proteins.
The results were derived from 5 to 10 independent experiments and are expressed as the ratio to the activity of the control.
FIG. 4.
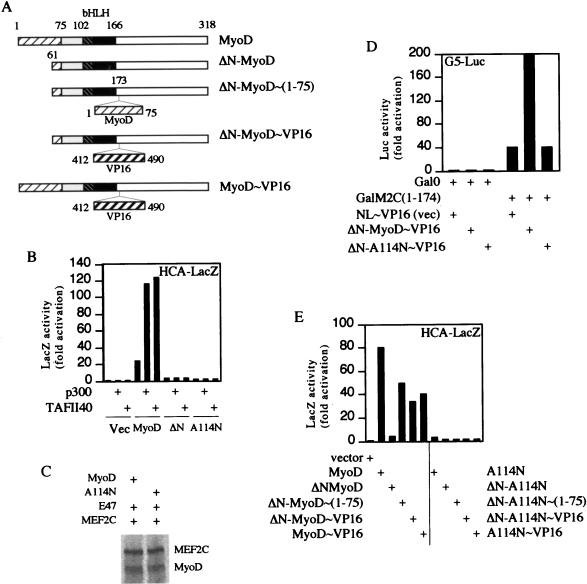
The A114N mutant fails to respond to coactivators of MyoD. (A) Schematic maps of the MyoD wild-type and mutated peptides used to acquire the data shown in panels B, D, and E. ▨, amino-terminal activation domain; ▧, DNA binding domain; ■, HLH dimerization domain. The grey box and white box do not symbolize any specific functional domains. (B) Residual transactivation activity of the A114N peptide cannot be potentiated by p300 or TAFII40; 1.5 μg of each plasmid was transfected. Vec, vector; ΔN, ΔN-MyoD∼(1-75). (C) The A114N mutant and MyoD interact with MEF2C equally well. Proteins synthesized by in vitro transcription-translation were mixed and immunoprecipitated with an antibody against the carboxyl terminus of MyoD. (D) Interaction of A114N with MEF2C [GalM2C(1–174)] is not reconstituted in a mammalian two-hybrid assay; 1 μg of each plasmid was used. (E) The A114N mutant fails to transactivate even when supplemented with VP16 activation domain. All transfections were done in NIH 3T3 cells and repeated at least three times; 2 μg of each plasmid was used. ΔN-MyoD and ΔN-A114N, MyoD and A114N proteins each lacking N-terminal aa 3 to 56; ΔN-MyoD∼VP16, ΔN∼MyoD with VP16 activation domain (aa 412 to 490) inserted between MyoD residues 173 and 174; ΔN-MyoD∼(1–75) and ΔN-A114N∼(1–75); ΔN-MyoD and ΔN-A114N with MyoD activation domain (aa 1 to 75) inserted between MyoD residues 173 and 174.
MEF2 proteins are known to be cofactors for myogenic bHLH proteins (18, 23). To test whether the basic-region conformation of the A114N mutant affects binding of MEF2C, we first examined whether MEF2C can be coimmunoprecipitated with the A114N protein. Either wild-type or A114N mutant MyoD was mixed with E47 protein and MEF2C protein and immunoprecipitated with an antibody against the carboxyl terminus of MyoD (Santa Cruz). All of the proteins were synthesized with an in vitro transcription-translation reagent (Promega). The wild-type protein and A114N mutant interact with MEF2C equally well in this assay (Fig. 4C). In a mammalian two-hybrid assay, MEF2C coding sequences were fused to the Gal4 DNA binding domain, while regions of MyoD or the A114N mutant were fused to the VP16 activation domain; these constructs were cotransfected with a luciferase reporter gene driven by five Gal4 DNA binding sites. We found that MyoD reconstituted the two-hybrid activation, presumably as a heterodimer with endogenous E proteins, as it was reported that E12 was required to support MyoD-MEF2C interaction (23), whereas the A114N mutant failed to do so (Fig. 4D). We obtained similar results with E12 overexpression (data not shown). Together, these data indicate that although MEF2C and the A114N mutant interact, the mutant locks up the VP16 activation domain (see below). Since MEF2C does not differentiate between wild-type MyoD and the A114N mutant, it is unlikely that MEF2C would be the postulated recognition factor (if there is any) that interprets the myogenic code in the basic region of MyoD. This is consistent with the fact that the MyoD-E12basic mutant can also interact with MEF2C in the presence of E12 (4).
Furthermore, the addition of the VP16 activation domain to the A114N mutant cannot rescue its diminished transactivation of muscle specific promoters (Fig. 4C), suggesting that, in the presence of the A114N mutation, neither the wild-type activation domain nor a substitution VP16 activation domain is functional. Note that this result is not to be confused either with the original finding that addition of the VP16 activation domain to the A114I mutant and the MyoD-E12basic protein restored some of their activities on an artificial, multimerized single E-box promoter, 4R-TK, (9, 34) or with recent results that confirmed such events on the artificial multimerized E-box promoter (reference 4 and our unpublished data). The 4R-TK promoter can be activated equally well by several nonmyogenic bHLH proteins, including E12, and, as such, does not appear to represent an appropriate reporter for assaying conformational requirements of myogenic activation. In fact, although addition of the VP16 activation domain rescues the basic-region mutants on a 4R-TK transcription reporter, its addition to the same mutant MyoD molecules fails to rescue them when myogenesis is used as an assay. In these instances it is possible that the A114I and MyoD-E12 basic proteins exhibit lowered affinity or cooperativity in binding to the 4R-TK promoter. However, even such low-level DNA interactions might well be compensated by the addition of the VP16 activation domain, whereas changes of the conformation of the protein in the A114N mutant are not restorable by addition of the VP16 activation domain. Thus, the activation domain cannot function properly unless there is an intact myogenic code in the basic region. Upon binding to the proper E-box DNA sequences the myogenic code determines the proper conformation of at least two separated domains of the molecule: the centrally located basic domain and the amino-terminal activation domain (Fig. 5C and D). In this light, it is important to note that when it is fused to Gal4, the basic domain is an extremely potent inhibitor of the N-terminal activation domain and that with the basic domain deleted, the activity of the activation domain fused to Gal4 increases remarkably (Table 2). This mechanism ensures that the activation domain is unmasked only upon DNA binding by the correct basic region. bHLH proteins exhibit specificity, but specificity based upon primary sequences is clearly not enough (5, 16, 24). Thus, cells likely use this clever unmasking strategy to control both the activity and the specificity of transcription factors. Such a mechanism may have evolved to restrict myogenic capability and specificity to a small number of bHLH proteins among many with diverse functions yet with DNA binding specificities known to be similar.
Intramolecular conformational regulation—a general theme?
Cellular factors in trans have previously been reported to negatively target the activation domains of transcriptional activators to affect cell physiology, e.g., Gal80 masking of the Gal4 activation domain when yeast cells grow in glucose medium (20) and MDM2 oncoprotein masking of the activation domain of the tumor suppressor protein p53 in certain tumors (22, 25). In cis, the unliganded steroid binding domain of glucocorticoid receptor (GR) was reported to inhibit both DNA binding and transcriptional activation by GR (14, 26). However, masking in cis of an activation domain by the DNA binding domain has not been documented, although it has been conjectured from previous work done with GR (19) and MyoD (35). Here we report an example of such intramolecular masking, mediated by the DNA binding domain of the activator, as a means of regulating the activation domain function and executing tissue specificity. This is also the first demonstration of a conformational difference in the DNA binding domain of a transcriptional regulator directly affecting the conformation and activity of the transcriptional activation domain. It is reminiscent of the properties of the yeast pheromone-receptor transcription factor, which changes conformation upon binding to a-specific but not to α-specific genes (31). It will be interesting to find out whether this strategy is also employed by other transcription factors that play crucial roles in controlling cell proliferation and differentiation.
ACKNOWLEDGMENTS
We thank D. Livingston, R. Tjian, L. Comai, G. Gill, and E. Olson for providing reagents, L. Nekludova and C. Pabo for a theoretical analysis of A114N mutant structure, and M. Stallcup, K. Yamamoto, and S. Tan for advice and discussion.
This work was supported in part by grants (to L.K.) from the National Institutes of Health.
Footnotes
J.H. dedicates this paper to the memory of Man-Hua Wu.
REFERENCES
- 1.Aurora R, Srinivasan R, Rose G D. Rules for alpha-helix termination by glycine. Science. 1994;264:1126–1130. doi: 10.1126/science.8178170. [DOI] [PubMed] [Google Scholar]
- 2.Barr P J. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell. 1991;66:1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- 3.Bengal E, Flores O, Rangarajan P N, Chen A, Weintraub H, Verma I M. Positive control mutations in the MyoD basic region fail to show cooperative DNA binding and transcriptional activation in vitro. Proc Natl Acad Sci USA. 1994;91:6221–6225. doi: 10.1073/pnas.91.13.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black B L, Molkentin J D, Olson E N. Multiple roles for the MyoD basic region in transmission of transcriptional activation signals and interaction with MEF2. Mol Cell Biol. 1998;18:69–77. doi: 10.1128/mcb.18.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackwell T K, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 6.Brennan T J, Chakraborty T, Olson E N. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc Natl Acad Sci USA. 1991;88:5675–5679. doi: 10.1073/pnas.88.13.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke L, Carbon J. A colony bank containing synthetic ColE1 hybrid plasmids representative of the entire E. coli genome. Cell. 1976;9:91. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- 8.Davis R L, Cheng P F, Lassar A B, Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- 9.Davis R L, Weintraub H. Acquisition of myogenic specificity by replacement of three amino acid residues from MyoD into E12. Science. 1992;256:1027–1030. doi: 10.1126/science.1317057. [DOI] [PubMed] [Google Scholar]
- 10.Davis R L, Weintraub H, Lassar A B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 11.Eckner R, Yao T-P, Oldread E, Livingston D M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 12.Ellenberger T, Fass D, Arnaud M, Harrison S C. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- 13.Ferre-D’Amare A R, Prendergast G C, Ziff E B, Burley S K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 14.Godowski P J, Picard D, Yamamoto K R. Signal transduction and transcriptional regulation by glucocorticoid receptor-LexA fusion proteins. Science. 1988;241:812–816. doi: 10.1126/science.3043662. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi R. Using PCR to engineer DNA. In: Erlich H A, editor. PCR technology. New York, N.Y: Stockton Press; 1989. pp. 61–70. [Google Scholar]
- 16.Huang J, Blackwell T K, Kedes L, Weintraub H. Differences between MyoD DNA binding and activation site requirements revealed by functional random sequence selection. Mol Cell Biol. 1996;16:3893–3900. doi: 10.1128/mcb.16.7.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson J E, Birren S J, Saito T, Anderson D J. DNA binding and transcriptional regulatory activity of mammalian achaete-scute homologous (MASH) proteins revealed by interaction with a muscle-specific enhancer. Proc Natl Acad Sci USA. 1992;89:3596–3600. doi: 10.1073/pnas.89.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaushal S, Schneider J W, Nadal Ginard B, Mahdavi V. Activation of the myogenic lineage by MEF2A, a factor that induces and cooperates with MyoD. Science. 1994;266:1236–1240. doi: 10.1126/science.7973707. [DOI] [PubMed] [Google Scholar]
- 19.Lefstin J A, Thomas J R, Yamamoto K R. Influence of a steroid receptor DNA-binding domain on transcriptional regulatory functions. Genes Dev. 1994;8:2842–2856. doi: 10.1101/gad.8.23.2842. [DOI] [PubMed] [Google Scholar]
- 20.Ma J, Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987;50:137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- 21.Ma P C, Rould M A, Weintraub H, Pabo C O. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 22.Mermod N, O’Neill E A, Kelly T J, Tjian R. The proline-rich transcriptional activator of CTF/NF-1 is distinct from the replication and DNA binding domain. Cell. 1989;58:741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- 23.Molkentin J D, Black B L, Martin J F, Olson E N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 24.Murre C, McCaw P S, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 25.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 26.Picard D, Salser S J, Yamamoto K R. A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell. 1988;54:1073–1080. doi: 10.1016/0092-8674(88)90122-5. [DOI] [PubMed] [Google Scholar]
- 27.Sadowski I, Ma J, Triezenberg S, Ptashne M. Gal4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 28.Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 30.Sun X H, Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991;64:459–470. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- 31.Tan S, Richmond T J. DNA binding-induced conformational change of the yeast transcriptional activator PRTF. Cell. 1990;62:367–377. doi: 10.1016/0092-8674(90)90373-m. [DOI] [PubMed] [Google Scholar]
- 32.Vandromme M, Cavadore J C, Bonnien A, Froeschle A, Lamb N, Fernandez A. Two nuclear localization signals present in the basic-helix 1 domains of MyoD promote its active nuclear translocation and can function independently. Proc Natl Acad Sci USA. 1995;92:4646–4650. doi: 10.1073/pnas.92.10.4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voronova A, Baltimore D. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc Natl Acad Sci USA. 1990;87:4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell T K, Turner D, Rupp R, Hollenberg S, Zhuang Y. The MyoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 35.Weintraub H, Dwarki V J, Verma I, Davis R, Hollenberg S, Snider L, Lassar A, Tapscott S J. Muscle-specific transcriptional activation by MyoD. Genes Dev. 1991;5:1377–1386. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]