ABSTRACT
Hypervirulent Klebsiella pneumoniae (hvKp) can cause infections in clinically healthy people, such as young and immunocompetent patients. Genes involved in the capsule synthesis or those encoding the siderophores have been adopted as predictors of hvKp. Certain sequence types, such as ST23 and ST86, have been associated with hvKp strains, too. The aim of this study was to investigate the presence of hvKp among 354 K. pneumoniae strains isolated from clinical samples of patients admitted to an Italian 900-bed hospital between 21 May 2021 and April 2022. All the isolates were screened by PCR for the amplification of virulence loci. Whole genome sequencing was performed in strains tested positive for at least one target gene. Thirteen out of 354 (3.7%) were hvKp. Five were wild type and belonged to the hypervirulent clones ST23, ST86, ST5, and ST375 and to the new clone ST6310. Six strains carried the blaKPC gene: three belonged to ST101, two to ST512, and one to ST395. Two isolates were ST147 and carried the blaNDM gene. Although hvKp isolation is not frequent, their presence should be systematically investigated to avoid the spreading of both virulent strains and strains with combined increase in virulence and resistance to antibiotics. PCR-based protocols are essential for surveillance of these strains, which do not always show a recognizable phenotype. Moreover, hvKp strains were isolated also from patients without history of recent foreign travels, indicating an increased spreading of these strains as well as an underestimated of their circulation so far.
IMPORTANCE
Klebsiella pneumoniae is a healthcare-associated pathogen frequently resistant to antibiotics. Hypervirulent strains of pneumoniae (hvKp) can spread from the primary site of infection to multiple sites causing life-threatening infections also in young otherwise healthy individuals. This study described the isolation of 13 isolates of K. pneumoniae with increased virulence in a large tertiary hospital over a 1-year period. Among them, eight strains were multidrug resistant and hypervirulent. Although these hypervirulent strains are still rare in Italy, their presence is particularly concerning since they can cause difficult-to-treat life-threatening infections. Moreover, not all the hypervirulent isolates were positive by the string test, so hvKp isolates were not always phenotypically detectable. Molecular biology techniques such as PCR amplification and next generation sequencing are therefore necessary for the detection of hvKp isolates, and surveillance programs exploiting molecular techniques are highly desirable.
KEYWORDS: hypervirulent Klebsiella pneumoniae, NDM, multidrug resistance, KPC, ST23, ST86, ST147, ST6310
INTRODUCTION
Klebsiella pneumoniae has been recognized by the World Health Organization as a critical priority healthcare-associated pathogen due to antibiotic resistance. Strains of K. pneumoniae with increased virulence have spread from Asia, where the first strain with this characteristic was first isolated in the 1980s (1) to Europe and the United States (2–5).
Hypervirulent strains of K. pneumoniae (hvKp) can cause infections in young otherwise healthy individuals, differently from nosocomial multidrug resistant (MDR) K. pneumoniae. hvKp can disseminate from the primary site of infection to multiple sites causing life-threatening infections, such as hepatic abscesses, pneumonia, necrotizing fasciitis, endophthalmitis, meningitis, and sepsis (4).
Currently, the diagnosis of hvKp infections is based on clinical criteria, which includes unusual metastatic spread of K. pneumoniae infections, clusters of K. pneumoniae infections with increased severity and mortality. On the other hand, no single microbiological characteristic can define hvKp (6). Hypermucoviscosity is one of the main characteristics of hvKp strains, as most of these strains overexpress genes for capsular polysaccharides such as rmpA and magA, resulting in hypermucoid phenotype and higher resistance to phagocytosis and intercellular killing by neutrophils. Hypermucoviscosity is detected by string test, in which the colony is stretched with a loop. The formation of a viscous string longer than 5 mm is considered a positive string test (7). However, recent studies (8, 9) have shown that hypermucosity is not an exclusive feature of hvKp, while conversely, not all hvKp isolates exhibit a hypermucoviscous phenotype. Most of hvKp isolates belong to sequence types ST23, ST25, ST65, ST86, and ST375 (6).
The presence of iron acquisition systems (e.g., aerobactin, salmochelin, yersiniabactin) involved in bacterial growth and survival is indicative of hvKp strains (4, 6). Detection of genes such as iutA and iroN, which regulate iron acquisition systems, by PCR has been used in several studies (5, 10).
Recently, the peg-344 gene has been included as a reliable, sensitive, and specific marker for the detection of hvKp such as aerobactin, salmochelin, rmpA, and rmpA2 (8, 11). Indeed, PEG-344, whose function is still unclear, was proved to be required for maximal virulence in a pneumonia model (12).
Most hvKp strains remain susceptible to several commonly used antibiotics. However, carbapenem-resistant K. pneumoniae strains belonging to ST11 became hypervirulent by acquiring a pLVPK-like virulence plasmid encoding aerobactin, salmochelin, and RmpA (13). On the other hand, some hypervirulent strains of ST25 and ST65 exhibit carbapenem resistance (14, 15).
The presence of both increased virulence and resistance in carbapenem-resistant hvKp limits the number of effective antimicrobial agents and therefore poses a serious challenge to treatment, infection control, and public health.
The aim of this study is to investigate the presence of kvKp isolates among K. pneumoniae strains isolated over a period of 1 year from patients at Fondazione IRCCS Policlinico San Matteo in Pavia (Italy).
MATERIALS AND METHODS
Samples
Fondazione IRCCS Policlinico San Matteo is a 900-bed hospital, located in Lombardy region of Italy, where Klebsiella pneumoniae carbapenemase (KPC)- producing K. pneumoniae has been endemic since 2013 (16). In the study period, 1,822 K. pneumoniae isolates were collected during the routines of the bacteriology laboratory from a total of 627 patients. In this study, we searched for hvKp strains in 354 non-repetitive clinical K. pneumoniae isolates (one per patient) collected from patients at Fondazione IRCCS Policlinico San Matteo between May 2021 and April 2022. The selected 354 strains included strains with characteristics associated with virulence and strains from invasive infections such as bloodstream infections. In addition, we wanted to evaluate the presence of strains with both virulence and resistance characteristics that had never been reported in our hospital. Therefore, extended spectrum beta-lactamase (ESBL)-producing and carbapenem-resistant strains, hereafter referred to as MDR, were also considered. In detail, the 354 strains included:
21 K. pneumoniae strains positive by string test and isolated from any clinical specimen;
225 K. pneumoniae MDR strains negative by string test and isolated from any clinical specimen except for blood cultures; and
108 K. pneumoniae wild-type isolates obtained from blood cultures.
The type of specimens and the type of patient from which the 354 K. pneumoniae isolates have been isolated, as well as the resistance profile of the isolates, are shown in Table 1.
TABLE 1.
Type of specimens and the type of patient from which the 354 K. pneumoniae isolates have been isolateda
| Criterion of selection | Number of samples for the criterion | Inpatients | Outpatients | Type of specimen | Number of samples | Wild type | ESBL | KPC | NDM | VIM | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|
| String test positive | 21 | 13 | 8 | Urine | 13 | 7 | 5 | 1 | NAb | NA | NA |
| Blood culture | 3 | 3 | NA | NA | NA | NA | NA | ||||
| Respiratory samples | 2 | 2 | NA | NA | NA | NA | NA | ||||
| Liver biopsy | 1 | 1 | NA | NA | NA | NA | NA | ||||
| Rectal swab | 1 | NA | NA | 1 | NA | NA | NA | ||||
| Urethral swab | 1 | NA | NA | 1 | NA | NA | NA | ||||
| String test negative and MDR | 225 | 221 | 4 | Rectal swab | 160 | NA | 8 | 144 | 4 | 1 | 3 (2 KPC + VIM; 1 KPC + NDM) |
| Respiratory samples | 30 | NA | 6 | 24 | NA | NA | NA | ||||
| Urine | 25 | NA | 2 | 23 | NA | NA | NA | ||||
| Wound/biopsy/dreinage samples | 6 | NA | NA | 5 | 1 | NA | NA | ||||
| Other | 4 | NA | NA | 3 | 1 | NA | NA | ||||
| Blood cultures | 108 | 80 | 28 | Blood culture | 108 | 57 | 19 | 30 | 1 | NA | 1 KPC + VIM |
The resistance profile of the isolates is also shown.
NA was used to indicate that zero strains have that characteristic.
MALDI-TOF (Bruker Daltonics GmbH, Bremen, Germany) equipped with Bruker Biotyper 3.1 database was used for species identification. The antibiotic resistance profile was determined by the BD-Phoenix instrument (Becton Dickinson, USA), using the NMIC-505 panel.
PCR amplification, sequencing, and genome analysis
Genomic DNA was extracted with a Blood and Tissue kit (QIAGEN, Düsseldorf, Germany) following the manufacturer’s instructions. PCR reactions were performed to investigate the presence of iutA, iroN loci (10), rmpA (17), and peg-344 (18).
The genomes of the isolates positive for at least one virulence gene were sequenced. Short reads were obtained by Illumina MiSeq with a 2 × 150 paired-end run, after Nextera XT library preparation (Illumina Inc., San Diego, USA). Genomes were assembled using Shovill 1.1. Kleborate (19) was used to assess the sequence type and to search for virulence and resistance genes. The presence of plasmids was evaluated with PlasmidFinder (20) and manually curated.
A coreSNP-based phylogeny was performed for isolates belonging to ST147, since an outbreak caused by K. pneumoniae ST147 started in Tuscany region of Italy in 2018 and continued until 2021 (21). The data set consisted of 181 strains of K. pneumoniae ST147 from the outbreak (21, 22) and the two isolates from this study. A genomic background was constructed by retrieving the 20 most similar high-quality genomes (N = 94) to those in the data set from the BV-BRC database (https://www.bv-brc.org/) according to k-mer content similarity (MASH). The P-DOR pipeline (https://github.com/SteMIDIfactory/P-DOR) was used to align all genomes in the data set to a reference (573.4026 in BV-BRC) and extract coreSNPs. Maximum likelihood phylogeny was inferred with RAxML (using 100 bootstrap resamples) on the resulting coreSNP alignment using the general time reversible model, as suggested by ModelTest-NG, with ascertainment bias correction (23).
RESULTS
K. pneumoniae was isolated from 627 patients during the study period. A total of 354 K. pneumoniae isolates were considered for this study, including 21 isolates with positive string test, 225 MDR isolates with negative string test, and 108 wild-type isolates from blood cultures with negative string test.
Twenty out of 354 isolates (5.6%) had at least one target gene of rmpA, iutA, iroN, and peg-344 gene amplified, and six isolates had all four genes amplified. The characteristics of the 20 patients from whom the 20 isolates were obtained are shown in Table 2. The median age of the 20 patients was 64.35 years (range: 37–81), and 11 were female.
TABLE 2.
Characteristics of the 20 patients from whom Klebsiella pneumoniae with increased virulence was isolateda
| Strain number | Isolation date | Sample | Sex | Age | Hospital ward | Outcome | String test | Resistance mechanisms | PCR rmpA | PCR iutA | PCR iroN | PCR peg-344 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5980 | 1May 2021 | Rectal swab | F | 74 | General medicine | NA | − | KPC | − | + | − | + |
| 5985 | 1 May 2021 | Bronchial aspirate | F | 63 | Intensive care unit | Recovered | − | KPC | − | + | − | + |
| 5999 | 6 May 2021 | Bronchoalveolar lavage | M | 74 | Intensive care unit | Deceased | − | KPC | − | + | − | + |
| 6002 | 8 May 2021 | Rectal swab | M | 52 | Intensive care unit | NA | − | KPC | − | + | − | + |
| 6007 | 12 May 2021 | Rectal swab | M | 56 | Hematology | NA | − | KPC | − | + | + | − |
| 6008 | 13 May 2021 | Rectal swab | M | 59 | General medicine | NA | − | KPC | − | + | − | − |
| 6082 | 8 June 2021 | Liver biopsy | F | 65 | Intensive care unit | Deceased | + | Wild type | + | + | + | + |
| 6114 | 22 June 2021 | Urine | M | 81 | Infectious diseases (outpatients) | NA | + | KPC | − | + | − | − |
| hvKp_2 | 3 Augurst 2021 | Bronchoalveolar lavage | M | 44 | Intensive care unit | Recovered | + | Wild type | + | + | + | + |
| hvKp_4 | 29 August 2021 | Blood culture | F | 71 | Obstetrics and gynecology | Recovered | + | Wild type | + | + | + | + |
| 6311 | 25 September 2021 | Rectal swab | M | 77 | Intensive care unit | NA | − | KPC | − | + | + | − |
| 6337 | 27 September 2021 | Dreinage | F | 49 | Obstetrics and gynecology | Recovered | − | NDM | − | + | + | + |
| 6452 | 21 October 2021 | Blood culture | M | 53 | Intensive care unit | Recovered | + | Wild type | + | + | + | + |
| 6399 | 27 October 2021 | Rectal swab | F | 75 | Pneumology | NA | + | KPC | − | + | + | − |
| 6403 | 27 October 2021 | Rectal swab | F | 76 | Emergency room | NA | − | KPC | − | + | + | − |
| 6445 | 11 November 2021 | Rectal swab | M | 37 | Otolaryngology | NA | − | NDM | + | + | + | + |
| 6465 | 12 November 2021 | Urethral swab | F | 76 | Pneumology | Recovered | + | KPC | − | + | − | − |
| 30715 | 24 November 2021 | Blood culture | M | 71 | Intensive care unit | Deceased | + | Wild type | − | + | − | + |
| hvKp_19 | 24 November 2021 | Urine | F | 80 | Outpatient sample collection center | NA | + | Wild type | + | + | + | + |
| 6843 | 14 April 2022 | Rectal swab | F | 54 | Pneumology | NA | − | KPC | − | + | + | − |
Outcome for the patients colonized by K. pneumoniae with increased virulence is indicated with NA (Not Applicable).
Nine strains (2.54%) were isolated from rectal swabs and were resistant to carbapenems, with eight strains bearing blaKPC and one bearing blaNDM encoding for the New Delhi metallo-beta-lactamase (NDM). Only one isolate from rectal swabs was string test positive. The remaining 11 strains were isolated mainly from blood cultures (n = 3; 0.8%), bronchoalveolar lavage (BAL) (n = 2; 0.56%), and urine (n = 2; 0.56%). Four of 11 strains had KPC carbapenemase and one strain had NDM. The characteristics of all the 20 genomes are shown in Table 3.
TABLE 3.
Characteristics of the genomes’ virulence score are attributed by Kleborate as follows: 0 = no yersinabactin, colibactin, or aerobactin; 1 = yersiniabactin only; 2 = yersiniabactin and colibactin (or colibactin only); 3 = aerobactin without yersiniabactin or colibactin; 4 = aerobactin with yersiniabactin (no colibactin); 5 = yersiniabactin, colibactin, and aerobactina
| Strain number | Sample | contig_count | N50 | largest_contig | total_size | ST | virulence_score | resistance_score |
|---|---|---|---|---|---|---|---|---|
| 5980 | Rectal swab | 163 | 203938 | 449385 | 5704928 | ST512 | 0 | 2 |
| 5985 | Bronchial aspirate | 161 | 161807 | 544125 | 5712389 | ST512 | 0 | 2 |
| 5999 | Bronchoalveolar lavage | 148 | 203931 | 449816 | 5708277 | ST512 | 0 | 2 |
| 6002 | Rectal swab | 184 | 109995 | 334526 | 5847712 | ST395 | 3 | 2 |
| 6007 | Rectal swab | 275 | 149647 | 331468 | 5702544 | ST512 | 0 | 2 |
| 6008 | Rectal swab | 118 | 157687 | 405925 | 5560881 | ST512 | 1 | 3 |
| 6082 | Liver biopsy | 100 | 195392 | 498479 | 5445960 | ST375 | 3 | 0 |
| 6114 | Urine | 140 | 158510 | 449810 | 5601209 | ST512 | 0 | 2 |
| hvKp_2 | Bronchoalveolar lavage | 115 | 116031 | 303700 | 5590050 | ST6310 | 3 | 0 |
| hvKp_4 | Blood culture | 85 | 316230 | 436906 | 5473727 | ST86 | 4 | 0 |
| 6311 | Rectal swab | 131 | 202114 | 426621 | 5757831 | ST101 | 4 | 3 |
| 6337 | Dreinage | 179 | 219779 | 823629 | 5748271 | ST147 | 3 | 2 |
| 6452 | Blood culture | 116 | 246519 | 788521 | 5655516 | ST23 | 5 | 0 |
| 6399 | Rectal swab | 202 | 136210 | 489606 | 5939332 | ST512 | 3 | 2 |
| 6403 | Rectal swab | 164 | 205081 | 380663 | 5818972 | ST101 | 4 | 3 |
| 6445 | Rectal swab | 228 | 173045 | 386555 | 5930012 | ST147 | 4 | 2 |
| 6465 | Urethral swab | 155 | 201170 | 523991 | 5941613 | ST512 | 3 | 2 |
| 30715 | Blood culture | 109 | 244625 | 449964 | 5425355 | ST727 | 1 | 0 |
| hvKp_19 | Urine | 61 | 323151 | 680484 | 5437774 | ST5 | 4 | 0 |
| 6843 | Rectal swab | 172 | 205082 | 520390 | 5820568 | ST101 | 4 | 3 |
Resistance score is attributed by Kleborate as follows: 1 = ESBL; 2 = carbapenemase; 3 = carbapenemase plus colistin resistance; 0 no resistance mechanism to beta-lactams.
Kleborate analyses showed that 13 (3.64%) strains had virulence score above 3 and seven of them were string test positive (Table 4). One strain isolated from a blood culture belonged to ST23 and had the highest virulence score. This strain is the only one in this study displaying aerobactin, salmochelin, yersiniabactin, and colibactin.
TABLE 4.
Characteristics of the 20 strains that were sequenceda
| Strain | String test | Resistance mechanisms | ST | Virulence score | Yersiniabactin | Colibactin | Aerobactin | Salmochelin | RmpADC | rmpA2 |
|---|---|---|---|---|---|---|---|---|---|---|
| 5980 | − | KPC | ST512 | 0 | − | − | − | − | − | − |
| 5985 | − | KPC | ST512 | 0 | − | − | − | − | − | − |
| 5999 | − | KPC | ST512 | 0 | − | − | − | − | − | − |
| 6002 | − | KPC | ST395 | 3 | − | − | iuc 1 | − | − | − |
| 6007 | − | KPC | ST512 | 0 | − | − | − | − | − | − |
| 6008 | − | KPC | ST512 | 1 | ybt 9; ICEKp3 | − | − | − | − | − |
| 6082 | + | Wild Type | ST375 | 3 | − | − | iuc 1 | iro 1 | rmp 1; KpVP-1 | rmpA2_3-47% |
| 6114 | + | KPC | ST512 | 0 | − | − | − | − | − | − |
| hvKp_2 | + | Wild Type | ST6310 | 3 | − | − | iuc 1 | iro 1 | rmp 1; KpVP-1 (truncated) | − |
| hvKp_4 | + | Wild Type | ST86 | 4 | ybt 9; ICEKp3 | − | iuc 1 | iro 1 | rmp 1; KpVP-1 | rmpA2_9 |
| 6311 | − | KPC | ST101 | 4 | ybt 9; ICEKp3 | − | iuc 1 | − | − | − |
| 6337 | − | NDM | ST147 | 3 | − | − | iuc 1 | − | rmp 1; KpVP-1 | rmpA2_6*−47% |
| 6452 | + | Wild Type | ST23 | 5 | ybt 1; ICEKp10 | clb 2 | iuc 1 | iro 1 | rmp 1; KpVP-1 | rmpA2_5-54% |
| 6399 | + | KPC | ST512 | 3 | − | − | iuc 1 | − | − | − |
| 6403 | − | KPC | ST101 | 4 | ybt 9; ICEKp3 | − | iuc 1 | − | − | − |
| 6445 | − | NDM | ST147 | 4 | ybt 9; ICEKp3 | − | iuc 1 | − | rmp 1; KpVP-1 | rmpA2_6*−47% |
| 6465 | + | KPC | ST512 | 3 | − | − | iuc 1 | − | − | − |
| 30715 | + | Wild Type | ST727 | 1 | ybt 4; plasmid (incomplete) | − | − | iro 1 | rmp unknown | − |
| hvKp_19 | + | Wild Type | ST5 | 4 | ybt 2; ICEKp1 | − | iuc 3 (truncated) | iro 3 (truncated) | rmp 3; ICEKp1 (truncated) | − |
| 6843 | − | KPC | ST101 | 4 | ybt 9; ICEKp3 | − | iuc 1 | − | − | − |
Resistance mechanism, sequence type, and the presence of virulence loci are displayed. A negative string test is indicated with “−”, while a positive one is indicated with “+”.
Eight strains meet the definition of convergent antimicrobial resistant and virulent strains as proposed by Lam et al. (19), having virulence score ≥3 and resistance score ≥1 (Table 3). Three strains belonging to ST101 isolated from rectal swabs and one strain ST147 isolated from a rectal swab had a virulence score of 4 and had aerobactin, salmochelin, and yersiniabactin. The three strains ST101 were KPC producing and the strain ST147 was NDM producing. The other four convergent antimicrobial resistant and virulent strains had a virulence score of 3 and belonged to ST147, ST395, and ST512.
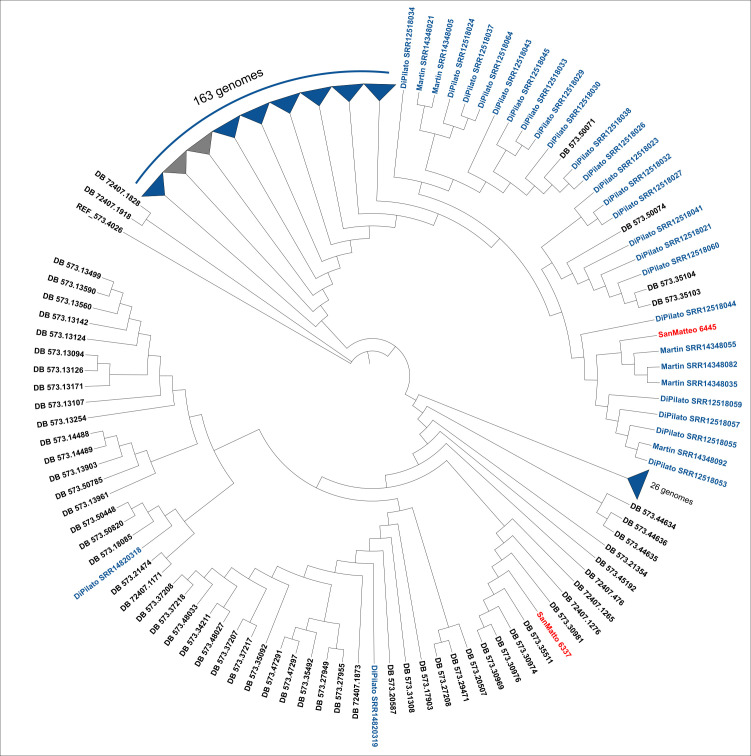
A phylogeny that included the two strains ST147 isolated in this study and the 181 ST147 strains from the outbreak that occurred in Tuscany (21, 22) is shown in Fig. 1. One of the two strains of this study (SanMatteo_6445) clustered in a monophyletic clade with three other strains from the study by Martin et al. (21) and the common branch is highly supported (100/100 bootstraps). Evaluation of clonal relatedness by core-genome SNPs reveals that clones were overall highly related to the cluster (SNP range: 19–20). As for other strains in the same cluster, the FIB(pQil)-type plasmid (pQil-NDM-147Tu; 56,064 bp; GenBank accession number CP071030) was identified. This plasmid carried the blaNDM-1 gene, along with other resistant determinants such as aacA4-cr, blaOXA-1, and blaCTX-M-15. In addition, the HIB-FIB(Mar) plasmid (pVIR-147Tu; 341,914 pb; GenBank accession number CP071028) was found. This one carried several resistant determinants such as armA, sul1 and sul2, blaCTX-M-15, and virulence ones such as rmpADC, iucABCD-iutA, and peg-344. On the contrary, the other strain (SanMatteo_6337) clustered with high branch support (94/100 bootstraps) with four strains from the BV-BRC DB isolated in Russia between 2018 and 2019 (SNP range: 41–104). The SanMatteo_6337 strain did not carry the FIB(pQil)-type plasmid and carried the HIB-FIB(Mar)-type plasmid, instead.
Fig 1.
Maximum likelihood phylogeny of Klebsiella pneumoniae ST147 based on SNP cores obtained using the P-DOR pipeline (https://github.com/SteMIDIfactory/P-DOR). The two strains of this study are indicated in red, while 181 genomes ST147 from the outbreak that occurred in Tuscany reported by Martin et al. (21) and Di Pilato et al. (22) are in blue.
DISCUSSION
This study reports the isolation of 13 isolates of hypervirulent K. pneumoniae, based on sequence type and the presence of virulence genes such as aerobactin and salmochelin. Six out of 13 strains were isolated from rectal swab (colonization), while seven strains caused infections.
The 13 strains had different characteristics. Two strains belonged to ST23 and hvKp ST86, respectively, and caused bloodstream infections in two patients who had not recently traveled abroad. hvKp ST23 was reported for the first time in Italy in 2014 (24), and in our hospital, it was isolated from urine and BAL samples between 2017 and 2018 (25). However, this sequence type is very uncommon in Italy and only five genomes of K. pneumoniae ST23 were found in the BV-BRC database as isolated in Italy, including the 6453 isolate of this study and the isolate from a BAL sample of the same patient (last accessed on 30 March 2023). ST23 strain isolated in this study was wild type but hvKp ST23 bearing carbapenemases were described in several European countries also from blood and respiratory samples (3, 24).
Equally rare is the ST86, which presence is documented in Italy so far only by the presence in BV-BRC databases of three genomes of K. pneumoniae ST86, including strain hvKp_4 of this study (reported in the BV_BRC database as 10028588). In Europe, ST86 hvKp strains have already been reported in Spain where three isolates out of 878 were found to be hypervirulent ST86 in a 7-year study (2) and in France where three out of 59 K. pneumoniae isolates were hypervirulent ST86 in a 5-year study (3). In France, carbapenem-resistant hvKp bearing OXA-48 have been isolated as well (26).
Other sequence types frequently associated with hypervirulence, such as ST5 and ST375, were found in this study. ST5 was previously isolated in Italy in a multicentric study (27). This strain, as well as hvKp_19 of this study, bears virulence factors such as ICEKp1, aerobactin, salmochelin, and rmpADC hypermucoidy locus. Only four genomes belonging to ST375 were found in the BV-BRC database as isolated in Europe, none in Italy. Hypervirulent ST375 was previously isolated in Japan (28) from liver abscesses of two patients. The ST375 isolate of this study was isolated from a liver biopsy, too, and as the isolates from Japan, it carried aerobactin and rmpA genes.
The new sequence type ST6310, a single locus variant of ST25 for mdh gene, was described for the first time in this study. Strain hvKp_2, which belonged to ST6310, had aerobactin, salmochelin, and rmpA loci and it was isolated from a patient who had lived in China.
hvKp isolates belonging to well-known MDR high-risk clones such as ST101, ST147, ST395, and ST512 were also found in this study. hvKp are usually susceptible to most antibiotics, but the ability of Klebsiella to acquire genetic elements leads to the emergence of MDR-hvKp clones (4, 13). Most MDR strains of this study did not have multiple virulence genes, except for three strains belonging to ST101 and one from ST147 that bore a wider range of virulence genes. The latter ST147 strain resulted closely related to the strains that caused a large hospital outbreak in the Tuscany region of Italy. Indeed, this isolate along with the strains described in the study of Di Pilato et al. are part of subclade of ST147, called ST147-vir, characterized by the presence of the ybt-encoding element ICEKpn3 and the pQil-NDM-147Tu-like and the pVIR-147Tu-like plasmids (22). The sequence type 147 is a high-risk clone associated with hospital-acquired infections worldwide and mediating the global spread of NDM-like metallo-beta-lactamase genes (29). Therefore, it is crucial to track the dissemination of these potential successful clones in hospital settings and understand their evolution to avoid the spread of resistant and virulent determinants, especially those determinants carried on plasmids, such as blaNDM-1.
In conclusion, this study described the isolation of 13 isolates of K. pneumoniae with increased virulence in a large tertiary hospital over a 1-year period. Although the isolation of these strains is not frequent, their presence is particularly concerning, as some of these isolates are not only hypervirulent but also MDR, effectively limiting the therapeutic options in case of infections.
Not all the hypervirulent isolates were positive by the string test, so hvKp are not always phenotypically detectable. Molecular biology techniques such as PCR amplification and next generation sequencing are therefore necessary for the detection of hvKp isolates. Surveillance programs exploiting PCR for the detection of hvKp isolates are desirable also because virulence genes are often carried on mobile genetic elements, which can be transferred within and between different bacterial species. In addition, the presence of hvKp should be systematically investigated in both wild-type and MDR strains, to avoid the spreading of strains with both increased virulence and resistance to antibiotics. Finally, hvKp strains were also isolated from patients with no history of recent travels abroad, suggesting an increased prevalence or an undetected circulation of hvKp isolates.
ACKNOWLEDGMENTS
Conceptualization: C.M. and M.C.; formal analysis: G.P. and S.G.; investigation: A.K., I.M., D.D.V., and M.A.; supervision: M.C. and P.C.; writing—original draft: C.M. and S.G; writing—review and editing: F.B. and P.C.
All authors have read and agreed to the published version of the manuscript.
AFTER EPUB
[This article was published on 30 January 2024 with inaccurate information in the legend of Fig. 1. The legend was updated in the current version, posted on 22 February 2024.]
Contributor Information
C. Merla, Email: c.merla@smatteo.pv.it.
Florence Claude Doucet-Populaire, University Paris-Saclay, AP-HP Hôpital Antoine Béclère, Service de Microbiologie, Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, France.
ETHICS APPROVAL
The study was designed and conducted in accordance with the Helsinki Declaration and approved by the Ethics Committee of Policlinico San Matteo in Pavia, Italy (Fasc. 2023-3.11/105).
DATA AVAILABILITY
Genome assembly data are available at NCBI under BioProject ID https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA814467PRJNA814467.
REFERENCES
- 1. Liu YC, Cheng DL, Lin CL. 1986. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med 146:1913–1916. doi: 10.1001/archinte.1986.00360220057011 [DOI] [PubMed] [Google Scholar]
- 2. Cubero M, Grau I, Tubau F, Pallarés R, Dominguez MA, Liñares J, Ardanuy C. 2016. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007-2013). Clin Microbiol Infect 22:154–160. doi: 10.1016/j.cmi.2015.09.025 [DOI] [PubMed] [Google Scholar]
- 3. Rafat C, Messika J, Barnaud G, Dufour N, Magdoud F, Billard-Pomarès T, Gaudry S, Dreyfuss D, Branger C, Decré D, et al. 2018. Hypervirulent Klebsiella pneumoniae, a 5-year study in a French ICU. J Med Microbiol 67:1083–1089. doi: 10.1099/jmm.0.000788 [DOI] [PubMed] [Google Scholar]
- 4. Russo TA, Marr CM. 2019. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 32:e00001-19. doi: 10.1128/CMR.00001-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parrott AM, Shi J, Aaron J, Green DA, Whittier S, Wu F. 2021. Detection of multiple hypervirulent Klebsiella pneumoniae strains in a New York city hospital through screening of virulence genes. Clin Microbiol Infect 27:583–589. doi: 10.1016/j.cmi.2020.05.012 [DOI] [PubMed] [Google Scholar]
- 6. Harada S, Doi Y. 2018. Hypervirulent Klebsiella pneumoniae: a call for consensus definition and international collaboration. J Clin Microbiol 56:e00959-18. doi: 10.1128/JCM.00959-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kumabe A, Kenzaka T. 2014. String test of hypervirulent Klebsiella pneumonia. QJM 107:1053. doi: 10.1093/qjmed/hcu124 [DOI] [PubMed] [Google Scholar]
- 8. Russo TA, Olson R, Fang C-T, Stoesser N, Miller M, MacDonald U, Hutson A, Barker JH, La Hoz RM, Johnson JR. 2018. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol 56:e00776-18. doi: 10.1128/JCM.00776-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Catalán-Nájera JC, Garza-Ramos U, Barrios-Camacho H. 2017. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes. Virulence 8:1111–1123. doi: 10.1080/21505594.2017.1317412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu F, Lv J, Niu S, Du H, Tang YW, Pitout JDD, Bonomo RA, Kreiswirth BN, Chen L. 2018. Multiplex PCR analysis for rapid detection of Klebsiella pneumoniae carbapenem-resistant (sequence type 258 [ST258] and ST11) and hypervirulent (ST23, ST65, ST86, and ST375) strains. J Clin Microbiol 56:e00731-18. doi: 10.1128/JCM.00731-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liao W, Long D, Huang Q, Wei D, Liu X, Wan L, Feng Y, Zhang W, Liu Y. 2020. Rapid detection to differentiate hypervirulent Klebsiella pneumoniae (hvKp) from classical K. pneumoniae by identifying peg-344 with loop-mediated isothermal amplication (LAMP). Front Microbiol 11:1189. doi: 10.3389/fmicb.2020.01189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bulger J, MacDonald U, Olson R, Beanan J, Russo TA. 2017. Metabolite transporter PEG344 is required for full virulence of hypervirulent Klebsiella pneumoniae strain hvKP1 after pulmonary but not subcutaneous challenge. Infect Immun 85:e00093-17. doi: 10.1128/IAI.00093-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW-C, Shu L, Yu J, Zhang R, et al. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/S1473-3099(17)30489-9 [DOI] [PubMed] [Google Scholar]
- 14. Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. 2015. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis 37:107–112. doi: 10.1016/j.ijid.2015.06.023 [DOI] [PubMed] [Google Scholar]
- 15. Zhang R, Lin D, Chan EW-C, Gu D, Chen G-X, Chen S. 2016. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother 60:709–711. doi: 10.1128/AAC.02173-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brolund A, Lagerqvist N, Byfors S, Struelens MJ, Monnet DL, Albiger B, Kohlenberg A, European Antimicrobial Resistance Genes Surveillance Network (EURGen-Net) capacity survey group . 2019. Worsening epidemiological situation of carbapenemase-producing Enterobacteriaceae in Europe, assessment by national experts from 37 countries, July 2018. Euro Surveill 24:1900123. doi: 10.2807/1560-7917.ES.2019.24.9.1900123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang HL, Chiang MK, Liou WJ, Chen YT, Peng HL, Chiou CS, Liu KS, Lu MC, Tung KC, Lai YC. 2010. Correlation between Klebsiella pneumoniae carrying pLVPK-derived loci and abscess formation. Eur J Clin Microbiol Infect Dis 29:689–698. doi: 10.1007/s10096-010-0915-1 [DOI] [PubMed] [Google Scholar]
- 18. Sanikhani R, Moeinirad M, Solgi H, Hadadi A, Shahcheraghi F, Badmasti F. 2021. The face of hypervirulent Klebsiella pneumoniae isolated from clinical samples of two Iranian teaching hospitals. Ann Clin Microbiol Antimicrob 20:58. doi: 10.1186/s12941-021-00467-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. 2021. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun 12:4188. doi: 10.1038/s41467-021-24448-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin MJ, Corey BW, Sannio F, Hall LR, MacDonald U, Jones BT, Mills EG, Harless C, Stam J, Maybank R, et al. 2021. Anatomy of an extensively drug-resistant Klebsiella pneumoniae outbreak in Tuscany, Italy. Proc Natl Acad Sci USA 118:e2110227118. doi: 10.1073/pnas.2110227118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Di Pilato V, Henrici De Angelis L, Aiezza N, Baccani I, Niccolai C, Parisio EM, Giordano C, Camarlinghi G, Barnini S, Forni S, et al. 2022. Resistome and virulome accretion in an NDM-1-producing ST147 sublineage of Klebsiella pneumoniae associated with an outbreak in Tuscany, Italy: a genotypic and phenotypic characterisation. Lancet Microbe 3:e224–e234. doi: 10.1016/S2666-5247(21)00268-8 [DOI] [PubMed] [Google Scholar]
- 23. Lewis PO. 2001. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst Biol 50:913–925. doi: 10.1080/106351501753462876 [DOI] [PubMed] [Google Scholar]
- 24. 2021. European centre for disease prevention and control. emergence of Hypervirulent Klebsiella pneumoniae St23 carrying Carbapenemase genes in EU/EEA countries. ECDC: Stockholm. [Google Scholar]
- 25. Thorpe HA, Booton R, Kallonen T, Gibbon MJ, Couto N, Passet V, López-Fernández S, Rodrigues C, Matthews L, Mitchell S, et al. 2022. A large-scale genomic snapshot of Klebsiella spp. isolates in Northern Italy reveals limited transmission between clinical and non-clinical settings. Nat Microbiol 7:2054–2067. doi: 10.1038/s41564-022-01263-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beyrouthy R, Dalmasso G, Birer A, Robin F, Bonnet R. 2020. Carbapenem resistance conferred by OXA-48 in K2-ST86 hypervirulent Klebsiella pneumoniae, France. Emerg Infect Dis 26:1529–1533. doi: 10.3201/eid2607.191490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arena F, Menchinelli G, Di Pilato V, Torelli R, Antonelli A, Henrici De Angelis L, Coppi M, Sanguinetti M, Rossolini GM. 2022. Resistance and virulence features of hypermucoviscous Klebsiella pneumoniae from bloodstream infections: results of a nationwide Italian surveillance study. Front Microbiol 13:983294. doi: 10.3389/fmicb.2022.983294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hoashi K, Harada S, Ishii Y, Aoki K, Ishikawa S, Oshiro Y, Shinzato T. 2019. Community-acquired liver abscess caused by capsular genotype K2-ST375 hypervirulent Klebsiella pneumoniae isolates. IDCases 17:e00577. doi: 10.1016/j.idcr.2019.e00577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lapp Z, Crawford R, Miles-Jay A, Pirani A, Trick WE, Weinstein RA, Hayden MK, Snitkin ES, Lin MY. 2021. Regional spread of blaNDM-1-containing Klebsiella pneumoniae ST147 in post-acute care facilities. Clin Infect Dis 73:1431–1439. doi: 10.1093/cid/ciab457 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genome assembly data are available at NCBI under BioProject ID https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA814467PRJNA814467.