Abstract
A variety of eukaryotic transcription factors, including the nuclear hormone receptors, Max-Mad, BCL-6, and PLZF, appear to mediate transcriptional repression through the ability to recruit a multiprotein corepressor complex to the target promoter. This corepressor complex includes the SMRT/N-CoR polypeptides, mSin3A or -B, and histone deacetylase 1 or 2. The presence of a histone-modifying activity in the corepressor complex has led to the suggestion that gene silencing is mediated by modification of the chromatin template, perhaps rendering it less accessible to the transcriptional machinery. We report here, however, that the corepressor complex actually appears to exhibit multiple mechanisms of transcriptional repression, only one of which corresponds with detectable recruitment of the histone deacetylase. We provide evidence instead of an alternative pathway of repression that may be mediated by direct physical interactions between components of the corepressor complex and the general transcription factor TFIIB.
Many transcription factors can exert both positive and negative effects on gene expression. The Ying-Yang-1 (YY-1) transcription factor, for example, can either repress or activate transcription at different promoters, whereas the Max transcription factor alternatively heterodimerizes with either a transcriptional activator (Myc) or a transcriptional repressor (Mad) to yield bimodal regulation of a given target gene (2, 16, 41, 49, 52, 57). Similarly, certain nuclear hormone receptors, such as thyroid hormone receptors (T3Rs), are capable of the alternative repression or activation of target gene expression, depending on hormone status, the constitution of the target promoter, and the cellular environment (4, 6–8, 12, 40). These dual regulatory properties arise, in part, from the ability of these transcription factors to physically recruit auxiliary polypeptides that help mediate the actual transcriptional response. T3Rs, for example, typically function as transcriptional silencers in the absence of hormone, a context in which these receptors bind to a class of corepressor proteins denoted SMRT/ N-CoR (SMRT and N-CoR, also known as TRAC and RIP13, are interrelated proteins produced by two distinct genetic loci) (10, 11, 15, 24, 26, 29, 31, 39, 42–44, 54–56, 58). Addition of hormone converts T3Rs into strong transcriptional activators, a process that is accompanied by the release of the SMRT/N-CoR corepressor polypeptides and the physical association of the receptors with a new set of proteins that function as transcriptional coactivators (10, 11, 24, 26, 29, 31, 39, 42–44, 54–56, 58; for a review, see reference 25).
Significant progress in understanding the role of SMRT/N-CoR proteins in transcriptional silencing has recently been made. The SMRT/N-CoR proteins can associate with other polypeptides to form a large corepressor complex containing mSin3A or -B, histone deacetylase-1 (HDAC-1) or HDAC-2, retinoblastoma protein-associated protein 46 (RbAp-46) or RbAp48, and at least two other polypeptides of unknown function (denoted SAP18 and -30, for silencer-associated proteins) (1, 3, 21, 22, 30, 31, 59; for reviews, see references 37 and 51). Although SMRT and N-CoR were initially identified as corepressors for the nuclear hormone receptors, the components of the SMRT/N-CoR–Sin3–HDAC complex can interact with, and appear to play a key role in transcriptional silencing by, a wide variety of nonreceptor transcription factors, including Mad-Max, YY-1, PLZF, BCL-6, and the retinoblastoma gene product (1, 3, 14, 21–23, 30, 31, 52, 59); orthologs of this complex have also been implicated in gene silencing in yeast (28, 34, 36, 45, 46, 48). Notably, different transcription factors can recruit the corepressor complex by interacting with different components of the complex: the nuclear hormone receptors appear to contact primarily the SMRT/N-CoR component, Mad-Max interacts principally with mSin3, and PLZF makes a combination of contacts with SMRT/N-CoR, mSin3, and HDAC (1, 10, 14, 19, 23, 24, 30, 32, 39, 42, 51a, 56).
Once tethered to a target promoter through interaction with a specific transcription factor, how does the SMRT corepressor complex actually silence transcription? The presence of HDAC-1 or -2 in the corepressor complex has led to the suggestion that transcription repression may reflect a deacetylation of the adjacent chromatin template (reviewed in references 37 and 51). This hypothesis provides a conceptual symmetry to the observation that many transcriptional coactivators possess reciprocal histone acetyltransferase activities and is consistent with the proposal that histone acetylation enhances, whereas deacetylation restricts, the accessibility of chromatin to the general transcriptional machinery (16, 49). However, chromatin remodeling is unlikely to be the sole mechanism of SMRT corepressor-mediated transcriptional silencing. For example, nuclear hormone receptors repress transcription in contexts (such as transient transfections and transcription assays in vitro) where the bulk of the DNA templates are not fully assembled into chromatin (see, e.g., references 5, 8, 12, 18, and 40). Furthermore, inhibition of HDAC activity in yeast, Drosophila, and vertebrate cells produces complex, mixed phenotypes involving both loss of repression and loss of activation (13, 27, 35, 38, 46, 47).
To better elucidate the mechanism(s) of repression, we sought to further dissect the nature and function of the SMRT-mSin3 corepressor complex. We report here that silencing by the corepressor complex appears to utilize multiple mechanisms of transcriptional repression, only one of which corresponds with recruitment of the HDAC. We also provide evidence of an alternative pathway of repression that may be mediated by direct physical interactions between components of the corepressor complex and the general transcription factor TFIIB.
MATERIALS AND METHODS
In vitro protein-protein binding assays.
The construction of the glutathione S-transferase (GST)–SMRT fusions was previously described (23, 39). Similar GST fusion constructs, representing various portions of the mSin3A-coding region, were constructed by cleavage of the pGEX-KG vector (20) and the target DNA at appropriate restriction sites and ligation by standard recombinant DNA methodology. The GST fusion proteins were expressed in Escherichia coli DH5α and were purified and immobilized to glutathione-agarose as previously described (20).
35S-radiolabeled mSin3A, TFIIB, or HDAC-1 was synthesized in vitro by use of the appropriate pSG5-, pT7β-, or pVZ-based plasmid in a coupled transcription-translation system (TnT kit; Promega). The 35S-labeled proteins were then incubated with a 50% slurry of the corresponding immobilized GST fusion protein in 200 to 300 μl of HEMG binding buffer (40 mM HEPES [pH 7.8], 50 mM KCl, 0.2 mM EDTA, 5 mM MgCl2, 0.1% Triton X-100, 10% glycerol, 1.5 mM dithiothreitol, 1× Complete Protease Inhibitor [Boehringer-Mannheim], and 0.5 mg of bovine serum albumin per ml) for 1 h at 4°C with gentle rocking. The agarose beads were then washed four times with 1 ml (each time) of HEMG buffer in the absence of protease inhibitor and bovine serum albumin. Bound proteins were eluted in 30 μl of 50 mM Tris-Cl (pH 6.8) containing 100 mM glutathione, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and visualized and quantified by PhosphorImager analysis (Molecular Dynamics Storm System) (23, 39).
Mammalian repression and two-hybrid assays.
The HindIII fragment of pACT2 (Clontech) was inserted between linker-modified EcoRI and BamHI sites of pSG5 (Stratagene) to create the pSG5-GAL4AD vector. The pSG5-GAL4DBD vector was created by transferring the HindIII-to BamHI portion of pGBT9 (Clontech) as a blunt-end fragment into the similarly treated EcoRI and BamHI sites of pSG5. The pSG5-GAL4DBD and pSG5-GAL4AD open reading frames in these pSG5 vectors were subsequently fused in frame to various subdomains of SMRT, mSin3A, or TFIIB by use of appropriate restriction sites and standard recombinant DNA subcloning techniques.
Transient transfections of CV-1 cells were performed by a calcium phosphate coprecipitation method (23). Each 60-mm-diameter plate, representing approximately 2.5 × 105 cells, was transfected with 500 ng of a pGAL-(17mer)-simian virus 40 (SV40) late promoter-luciferase reporter (23), 125 ng of the pSG5-GAL4DBD vector, 500 ng of the pSG5-GAL4AD vector, 500 ng of a pCH110 vector (Pharmacia) as an internal β-galactosidase control, and sufficient pUC19 to normalize the total DNA to 5 μg. Luciferase activity was determined after 46 h with a luciferase assay kit (Promega) and a TD 20/20 luminometer (Turner Design). The relative light units were normalized to the β-galactosidase activity.
RESULTS
The SMRT polypeptide possesses at least two distinct silencing domains that repress transcription when tethered to DNA, and one of these silencing domains strongly interacts with mSin3A in vivo and in vitro.
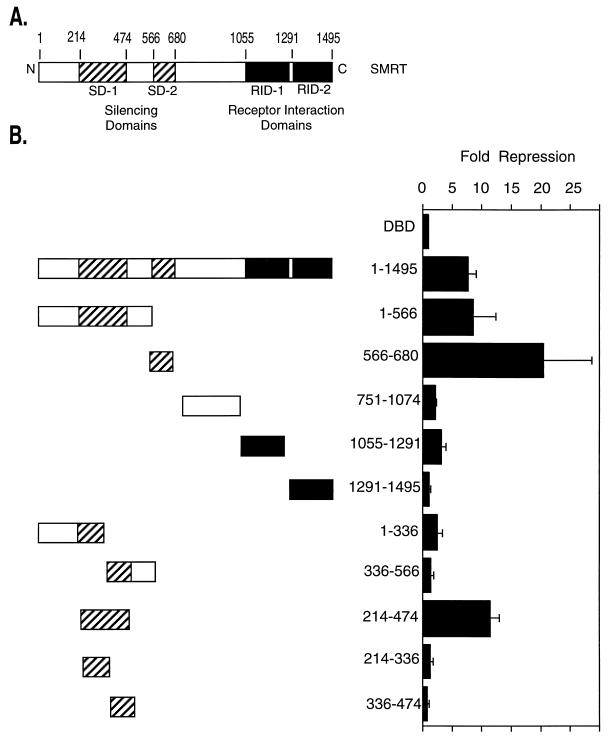
Prior work has indicated that the C-terminal half of SMRT serves to tether this protein to the nuclear hormone receptors, whereas the N-terminal half encompasses the domains that actually mediate transcriptional repression (see, e.g., references 10, 11, 24, 39, 42, and 54) (Fig. 1A). Our first goal was to more precisely determine the locations of these N-terminal silencing domains within SMRT. We therefore fused different segments of SMRT to an exogenous GAL4 DNA binding domain (GAL4-DBD) and then tested the ability of these fusion proteins to inhibit the expression of a promoter containing GAL4 binding sites linked to a suitable luciferase reporter gene. The constructs and reporter were introduced into CV-1 cells, and the luciferase activity relative to that of a β-galactosidase reporter introduced as an internal control was determined (Fig. 1B). Two separate and strong repression domains could be mapped in SMRT by this means: silencing domain 1 (SD-1), encompassing amino acids 214 to 474, and SD-2, encompassing amino acids 566 to 680 (Fig. 1B). These two domains represented the preponderance of the silencing activity of SMRT detectable in this fashion; constructs limited to the C-terminal half of SMRT exhibited little or no repression as determined by this form of assay (Fig. 1B) and in fact functioned as dominant negative inhibitors of native SMRT function.
FIG. 1.
Transcriptional silencing domains in SMRT. (A) Schematic representation of the SMRT corepressor. The regions involved in interactions with the nuclear hormone receptors are indicated (denoted receptor interaction domains, RID-1 and RID-2), as are the regions able to mediate transcriptional repression when expressed as GAL4-DBD fusions (SD-1 and SD-2). Numbers above the schematic refer to the relevant amino acid positions. (B) Domains of SMRT able to repress reporter gene expression in a transient-transfection assay. Different regions of SMRT, as depicted schematically on the left, were fused in frame to a GAL4-DBD-coding sequence and were expressed in CV-1 mammalian cells. The cells were simultaneously transfected with a reporter plasmid containing an SV40 late promoter bearing five binding sites (GAL4 17-mers) for the GAL4-DBD and driving the expression of a luciferase reporter gene. The cells were harvested 48 h later, and the luciferase activity was determined relative to that of a pCH110 β-galactosidase reporter, lacking GAL4 binding sites, employed as a negative control. Fold repression (right) was calculated as the reduction in luciferase expression mediated by a GAL4-DBD fusion construct relative to that mediated by an empty GAL4-DBD (DBD). The results presented represent the averages and standard deviations obtained from at least two duplicate experiments.
It has been suggested that SMRT-mediated repression requires the association of this protein with mSin3 (3, 22). We therefore sought to map the sites of interaction between SMRT and mSin3. We first tested the ability of radiolabeled mSin3A, synthesized by in vitro transcription-translation, to bind to a panel of different SMRT domains, expressed as GST fusions in E. coli and immobilized on a glutathione-agarose matrix (Fig. 2). Consistent with the hypothesis, the SD-1 of SMRT (amino acids 214 to 474) contains a strong binding site for mSin3 as determined by this in vitro assay, whereas mSin3A exhibited little or no binding to nonrecombinant GST or to sequences derived from the C-terminal half of SMRT (Fig. 2). A mammalian two-hybrid assay confirmed that these interactions also occurred in an in vivo context: GAL4 activation domain (AD) fusions containing the SMRT SD-1 displayed a strong functional interaction with an assortment of GAL4-DBD–mSin3A constructs when the two were coexpressed in CV-1 cells (Fig. 3C) (discussed in greater detail below). In contrast, regions of SMRT that lacked the SD-1 failed to detectably interact with mSin3A in the two-hybrid assay (Fig. 3C). Truncation of the SMRT SD-1 to amino acids 214 to 336 significantly impaired the ability to associate with mSin3 both in vitro and in vivo (Fig. 2 and 3C) and the ability of SD-1 to repress transcription (Fig. 1B), indicative of a correlation between these two functions. Intriguingly, however, we were unable to demonstrate an equivalent interaction between mSin3A and the SMRT SD-2 region (amino acids 566 to 680) (Fig. 2 and data not shown) despite the strong repression phenotype exhibited by the latter (Fig. 1B).
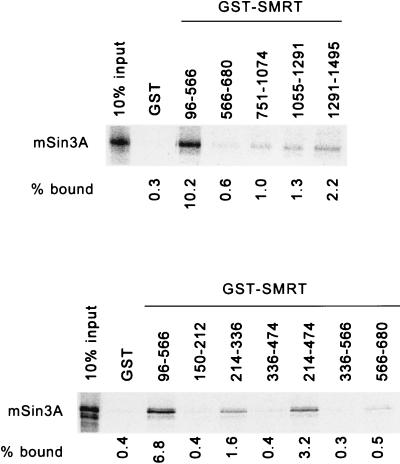
FIG. 2.
Localization of an interaction site for mSin3A within the SMRT polypeptide. Full-length, radiolabeled mSin3A was synthesized in vitro by a coupled transcription-translation protocol. The radiolabeled mSin3A protein was then incubated with various domains of SMRT, expressed in bacteria as GST fusion proteins, and immobilized on a glutathione-agarose column (the SMRT amino acids represented in each GST fusion are denoted above the panels; GST refers to a nonrecombinant GST construct employed as a negative control). The radiolabeled mSin3A protein bound by each GST-SMRT construct was eluted with soluble glutathione, resolved by SDS-PAGE, and visualized and quantified by PhosphorImager analysis. The percentage of the input mSin3A bound by each GST fusion is indicated below each lane.
FIG. 3.
Comparative mapping of repression and SMRT interaction domains located within mSin3A. (A) Schematic representation of mSin3A. The locations of four putative paired amphipathic helical domains, PAH1 through -4, are shown. Also depicted are the locations of two domains within mSin3A (SD-A and SD-B) that are able to repress transcription when expressed as GAL4-DBD fusions and the locations of two domains within mSin3A that are able to interact with SMRT. Numbers above the schematic refer to the relevant amino acid positions. (B) Domains of Sin3A able to repress transcription in a transient-transfection assay. Different domains of mSin3A, as depicted schematically on the left, were fused to a GAL4-DBD-coding frame and expressed in CV-1 mammalian cells. The cells were simultaneously transfected with a reporter plasmid containing an SV40 late promoter bearing five binding sites (GAL4 17-mers) for the GAL4-DBD and driving the expression of a luciferase reporter gene. The cells were harvested 48 h later, and the luciferase activity was determined relative to that of a pCH110 β-galactosidase reporter, lacking GAL4 binding sites, employed as an internal control. Fold repression (right) was calculated as the reduction in luciferase expression mediated by a GAL4-DBD fusion construct relative to that mediated by an empty GAL4-DBD (DBD). The results represent the averages and standard deviations from at least two duplicate experiments. (C) Domains of mSin3A able to interact with SMRT in a mammalian two-hybrid interaction. GAL4-DBD fusions representing different domains of mSin3A, as depicted schematically on the left of the panel, were introduced into CV-1 cells together with the GAL4 (17-mer) luciferase reporter and a series of GAL4-AD constructs. The GAL4-AD constructs included an empty GAL-AD construct, a GAL4-AD–SMRT (codons 96 to 566) construct, a GAL4-AD–SMRT (codons 751 to 1495) construct, a GAL4-AD–SMRT (codons 214 to 474) construct, and a GAL4-AD–SMRT (codons 214 to 336) construct. After 48 h, the cells were harvested and the luciferase activity was determined relative to that of pCH110 employed as an internal control (Relative Luc). The results represent the averages and standard deviations from at least two duplicate experiments.
The mSin3A polypeptide itself contains two autonomous transcriptional silencing domains, only one of which detectably interacts with HDAC-1.
The mSin3A molecule is comprised of four repetitive domains believed to represent paired amphipathic helices (PAH1 through -4), separated by regions of unique sequence (50) (Fig. 3A). We therefore employed the GAL4-DBD fusion technique to map the regions within this mSin3A structure capable of transcriptional repression. Two distinct mSin3A domains possessed strong silencing activity when assayed in this manner: mSin3A amino acids 57 to 215 (denoted SD-A and encompassing PAH-1 and a region of N-terminal flanking sequence) and amino acids 533 to 724 (denoted SD-B and located between PAH3 and PAH4) (Fig. 3B). In contrast, PAH2, PAH3, and PAH4 of mSin3A exhibited little or no silencing activity when tested in the same fashion (Fig. 3B).
We next mapped the regions of mSin3A responsible for its interaction with SMRT, using the mammalian two-hybrid assay (Fig. 3C). Two SMRT interaction domains were detected within mSin3A: the first coincided with PAH2 (mSin3 amino acids 272 to 404), and the second included PAH3 (mSin3 amino acids 404 to 545) (Fig. 3C). Significantly, the two SMRT association domains within mSin3A were fully separable from the two mSin3 silencing domains, strongly implying that mSin3A does not require a direct interaction with SMRT for transcription repression when tethered directly to a promoter as a GAL4-DBD fusion.
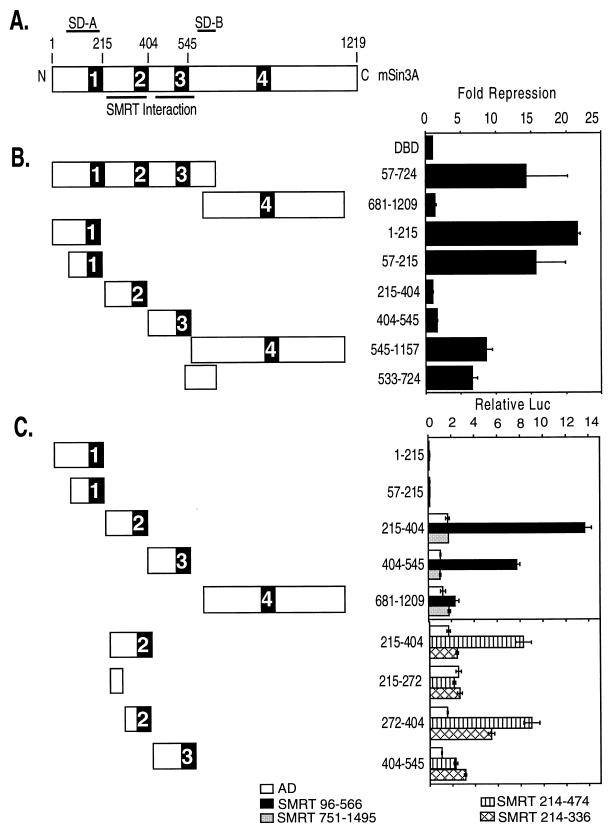
The ability of mSin3 to silence transcription has been proposed to be linked to its ability to recruit HDAC-1 and -2 and to thereby remodel chromatin into a transcriptionally nonpermissive structure (reviewed in references 37 and 51). Using an in vitro binding assay, we first confirmed and extended prior results that were obtained more indirectly (1, 3, 21, 22, 30, 59), namely, that HDAC-1 can bind to mSin3A (Fig. 4A). This HDAC-1 association site mapped to amino acids 545 to 1157 of mSin3A (Fig. 4B) and thus includes the SD-B of mSin3A, consistent with the concept that SD-B-mediated repression might operate through recruitment of the HDAC moiety. In contrast, however, the second, strong repression domain SD-A, located at the N terminus of mSin3A, exhibited no detectable interaction with HDAC-1 (compare Fig. 3B and 4A). These results suggest that both HDAC-1-dependent and -independent mechanisms of transcriptional repression may be operative in mSin3A. Consistent with the work of others, we did not observe any evidence of a direct interaction between HDAC-1 and any region of SMRT (data not shown).
FIG. 4.
Interaction of HDAC-1 with mSin3A and effects of trichostatin A on repression. (A) Binding of mSin3A to GST–HDAC-1. Full-length, radiolabeled mSin3A was synthesized in vitro by a coupled transcription-translation protocol. The radiolabeled mSin3A protein was then incubated with nonrecombinant GST (used as a negative control) or a GST fusion of HDAC-1, each expressed in bacteria and immobilized on a glutathione column. Radiolabeled mSin3A protein bound to the immobilized proteins was eluted with soluble glutathione, resolved by SDS-PAGE, and visualized and quantified by PhosphorImager analysis (left panel). The percentage of HDAC-1 bound to GST or to GST-HDAC-1 is depicted below the panel and was determined relative to the total radiolabeled mSin3A used in the assay (input). A parallel experiment using radiolabeled PLZF as a positive control for interaction with HDAC-1 is also presented (right panel). (B) Binding of HDAC-1 to GST-mSin3A. An experiment reciprocal to that in panel A was performed, using radiolabeled, full-length HDAC-1 and GST fusions representing various portions of mSin3A (as indicated above the lanes). Radiolabeled HDAC-1 protein bound to the different GST-mSin3A fusion constructs was eluted, resolved by SDS-PAGE, and visualized and quantified by PhosphorImager analysis, as described for panel A. (C) Lack of effect of trichostatin A on SMRT-mSin3A-mediated transcriptional repression. Various GAL4-DBD fusions representing the silencing domains of SMRT and mSin3A, as indicated below each panel, were transfected into CV-1 cells together with an SV40 late promoter (GAL4 17-mer) luciferase reporter in the presence or absence of 10 nM trichostatin A (TSA), an inhibitor of HDAC activity. The cells were harvested 48 h later, and the expression of the luciferase reporter, relative to that of the pCH100 internal control, was determined (“Relative Luc”). The results presented represent the averages and standard deviations obtained from at least two duplicate experiments.
As a different means of evaluating the role of histone modification in mSin3A-mediated gene silencing, we also tested the ability of a strong inhibitor of HDAC, trichostatin A, to counteract mSin3A-mediated repression (53). Levels of trichostatin A ranging up to near-toxic concentrations had no detectable effect on repression in our assays when tested with any of the silencing domains we elucided in our GAL4-DBD–mSin3A or GAL4-DBD–SMRT constructs (Fig. 4B). We conclude that HDAC activity is not a necessary prerequisite for mSin3A- or SMRT-mediated gene silencing in the transient-transfection assays described here.
General transcription factor TFIIB strongly interacts with components of the SMRT corepressor complex and may be a target for corepressor-mediated gene silencing.
Our studies described above indicated that gene silencing by corepressor includes mechanisms operating in addition to, or in conjunction with, the previously recognized ability of the SMRT-mSin3A complex to tether HDAC-1. Notably, many transcription factors, including the nuclear hormone receptors, interact directly with components of the general transcriptional machinery, and these interactions have been proposed to be an important mechanism of transcriptional regulation (see, e.g., reference 9 and references therein). Highly purified T3R, for example, is able to mediate transcriptional repression in vitro in the absence of detectable coregulators, apparently through direct inhibitory interactions between receptor and TFIIB and/or TFIID (5, 17, 18). We therefore investigated whether the SMRT corepressor might contribute to transcriptional silencing by also targeting components of the general transcriptional machinery, perhaps in synergy with the known inhibitory interactions between T3R and the preinitiation complex.
Indeed, we observed that SMRT strongly interacted with general transcription factor TFIIB in vitro (Fig. 5A). The interactions between SMRT and TFIIB were quite marked, being comparable to in strength or stronger than those observed between SMRT and T3R, and could also be observed in a reciprocal experiment using radiolabeled SMRT derivatives and a GST-TFIIB fusion polypeptide (data not shown). In contrast, little or no TFIIB bound to a nonrecombinant GST construct or to an assortment of other GST fusions employed as negative controls (Fig. 5A and data not shown).
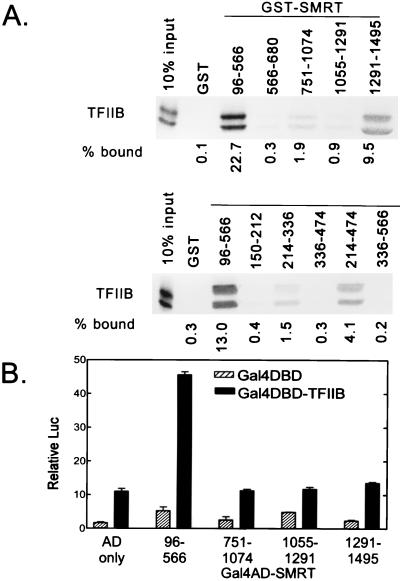
FIG. 5.
Interactions of TFIIB with SMRT. (A) Binding in vitro. Radiolabeled TFIIB was synthesized in vitro by a coupled transcription-translation protocol (input lane; the presence of a protein doublet is likely the result of translational initiations on both the correct codon and an internal AUG codon). The radiolabeled TFIIB protein was then incubated with various domains of SMRT (as denoted above the lanes), each expressed in bacteria as a GST fusion protein and immobilized on a glutathione-agarose column. Radiolabeled TFIIB bound to the different GST-SMRT agarose matrices was eluted with soluble glutathione, resolved by SDS-PAGE, and visualized and quantified by PhosphorImager analysis. The binding of TFIIB to nonrecombinant GST (GST) was also tested in parallel as a negative control. The percentage of the input TFIIB bound by each GST fusion is depicted below each lane. (B) Two-hybrid interaction between SMRT and TFIIB. Various subdomains of SMRT were fused to the GAL4-AD (as indicated below the panel) and were coexpressed in CV-1 cells together with either an empty GAL4-DBD or a GAL4-DBD–TFIIB fusion. The ability of the coexpressed constructs to stimulate expression of a luciferase reporter bearing GAL4-DBD binding sites, relative to that of a pCH110 reporter lacking GAL4-DBD binding sites (Relative Luc), was measured as for Fig. 3C.
Two distinct domains of SMRT bound to TFIIB in vitro. The strongest binding site mapped to amino acids 96 to 566 of SMRT (Fig. 5A) and was also be detected in vivo by the mammalian two-hybrid assay (Fig. 5B). Intriguingly, this strong TFIIB interaction domain encompassed the SMRT SD-1 region, and SMRT truncations that were severely impaired for the TFIIB interaction, such as the GST-SMRT(214-336) construct, were also severely impaired in transcriptional repression (compare Fig. 2A and 5A). It therefore appears that the SD-1 in SMRT strongly interacts with both mSin3A and TFIIB and that these interactions are both closely linked to the ability of this domain to repress transcription. A second, weaker TFIIB association domain was detected in vitro near the extreme C terminus of SMRT (amino acids 1291 to 1495) (Fig. 5A), but this second TFIIB interaction domain was apparently too weak for detection by two-hybrid analysis (Fig. 5B), and this domain of SMRT failed to display any notable silencing activity when expressed as a GAL4-DBD fusion (Fig. 3B).
Intriguingly, an additional strong interaction also occurred between TFIIB and the mSin3A component of the corepressor complex (Fig. 6). This interaction could be observed either with radiolabeled mSin3A and a GST-TFIIB construct (Fig. 6A) or, reciprocally, with radiolabeled TFIIB and a GST-mSin3A construct (Fig. 6B). The TFIIB interaction domain in mSin3A mapped to the same PAH3 region, mSin3A amino acids 404 to 545, as conferred interaction with SMRT (compare Fig. 6A and 3C). Thus, the potential for an extensive network of multiple interactions between TFIIB, mSin3A, SMRT, and the nuclear hormone receptors appears to exist. In contrast to the coincidence of the N-terminal silencing and TFIIB interaction domains on SMRT itself, however, the TFIIB interaction domain in mSin3A was neither necessary nor sufficient for transcriptional silencing by mSin3A GAL4-DBD fusion constructs (compare Fig. 6 and 3B).
FIG. 6.
TFIIB also interacts with mSin3A. (A) Interaction between GST-TFIIB and radiolabeled mSin3A. Full-length, radiolabeled mSin3A was synthesized in vitro by a coupled transcription-translation protocol. The radiolabeled mSin3A protein was then incubated either with immobilized nonrecombinant GST or with an immobilized GST-TFIIB fusion. Radiolabeled mSin3A protein bound to the GST or GST-TFIIB matrix was eluted with soluble glutathione, resolved by SDS-PAGE, and visualized and quantified by PhosphorImager analysis. The percentage of mSin3A bound is depicted numerically below the lanes and was determined relative to the total radiolabeled mSin3A used in the assay (input). (B) Localization of domains within mSin3A able to bind to TFIIB. Radiolabeled TFIIB was synthesized in vitro by a coupled transcription-translation protocol. The radiolabeled TFIIB protein was then incubated with various domains of mSin3A (as denoted above the lanes), each expressed in bacteria as a GST fusion protein and immobilized on a glutathione-agarose column. Radiolabeled TFIIB bound to the different GST-mSin3A agarose matrices was eluted with soluble glutathione, resolved by SDS-PAGE, and visualized and quantified by PhosphorImager analysis. The binding of TFIIB to nonrecombinant GST (GST) was also tested in parallel as a negative control. The percentage of the input TFIIB bound by each GST fusion is also depicted below each lane.
We wished to determine if the physical interaction between the SMRT SD-1 and TFIIB could be manifested as a functional interaction. We first examined whether TFIIB activity was altered by SMRT coexpression (Fig. 7A). TFIIB enhances transcription when tethered to a promoter through a GAL4 DBD; ectopic expression of SMRT resulted in a modest, but reproducible and dose-dependent, inhibition of this TFIIB-mediated activation, suggesting that SMRT can indeed counteract TFIIB function (Fig. 7A). Conversely, if SMRT mediates repression by interfering with TFIIB, one might predict that (i) TFIIB is normally limiting in target cells and (ii) overexpression of TFIIB might abrogate SMRT-mediated repression. Consistent with these concepts, elevated expression of TFIIB both enhanced basal promoter expression in CV-1 cells and reversed GAL4-DBD–SMRT-mediated repression in a dose-dependent manner (Fig. 7B). We suggest that TFIIB is potentially a target through which SMRT may mediate at least certain facets of transcriptional repression (see Discussion).
FIG. 7.
SMRT and TFIIB interact functionally. (A) SMRT expression inhibits transcriptional activation by TFIIB. A GAL4-DBD or GAL4-DBD–TFIIB construct (125 ng per plate) was introduced into CV-1 cells alone or with increasing amounts of a pSG5-SMRT derivative (1× = 125 ng/plate). The activity of the cointroduced GAL4 17-mer-luciferase (LUC) reporter was determined relative to that of the pCH110 lacZ internal control as for Fig. 2 and 3. (B) TFIIB expression counteracts SMRT-mediated repression. A GAL4-DBD or GAL4-DBD–SMRT fusion construct (125 ng per plate) was expressed in CV-1 cells alone or together with TFIIB (1× = 125 ng/plate). The activity of the cointroduced GAL4 (17-mer)-luciferase reporter was then determined relative to that of the pCH110 lacZ internal control as for Fig. 2 and 3.
N-CoR shares the ability of SMRT to interact with mSin3A and with TFIIB.
N-CoR is a second corepressor protein that is approximately 50% related to SMRT over regions of overlap but that also displays unique N-terminal sequences not present in SMRT. We therefore wished to extend our studies to an analysis of N-CoR. Although the silencing domains of N-CoR are divergent from those of SMRT, N-CoR shared the ability of SMRT to interact with mSin3A (Fig. 8A). N-CoR interacted primarily with the N terminus and PAH3 domains of mSin3A under our conditions and did not appear to interact detectably with the PAH2 domain (Fig. 8A). Notably, N-CoR also shared the ability of SMRT to bind to TFIIB (Fig. 8B), thus extending this observation to a second member of the corepressor family.
FIG. 8.
Interactions of N-CoR with mSin3A and TFIIB. (A) Interaction between N-CoR and mSin3A. Radiolabeled N-CoR, synthesized by in vitro transcription and translation, was tested for the ability to bind to GST fusions representing various domains of mSin3A or to nonrecombinant GST, as indicated above the lanes. Radiolabeled N-CoR bound to each GST construct was eluted with soluble glutathione, resolved by SDS-PAGE, and visualized and quantified by PhosphorImager analysis. The percentage of N-CoR bound in each assay is depicted below the lanes, and was determined relative to the total radiolabeled N-CoR used in the assay (input). (B) Interaction between N-CoR and TFIIB. The same protocol as in panel A was employed, but with a GST-TFIIB construct.
DISCUSSION
SMRT associates with an assortment of polypeptides that appear to participate in corepressor-mediated transcriptional silencing.
Efficient transcriptional silencing by nuclear hormone receptors in vivo appears to be predicated on the ability of these transcription factors to physically interact with the SMRT/N-CoR corepressor proteins (10, 11, 24, 26, 29, 31, 39, 42–44, 54–56, 58). The recognition that the SMRT/N-CoR proteins are part of a larger complex that includes mSin3A or -B, HDAC-1 or -2, RbAP46 or -48, and a panel of as-yet-uncharacterized other polypeptides establishes an important conceptual context with which to understand this mode of transcriptional silencing (1, 3, 21, 22, 30, 31, 59). Significantly, the SMRT–mSin3A–HDAC-1 complex also appears to be an important mediator of transcriptional silencing by a wide variety of nonreceptor transcription factors in vertebrates, and orthologs of Sin3 and HDAC have been implicated in transcriptional repression in yeast (13, 19, 28, 31–34, 36, 38, 45–48). Therefore, components of the SMRT-Sin3-HDAC corepressor complex appear to represent ancestral, and widely exploited, molecular effectors by which eukaryotes negatively regulate gene expression.
The presence of HDAC in the corepressor complex is particularly provocative and has led to the proposal that transcriptional repression may be, in part, mediated through the covalent modification of the chromatin template (see, e.g., references 37 and 51). We wished to better elucidate the manner in which the different components of the corepressor complex interact with one another and to establish which of these interactions are key to the transcriptional silencing phenotype. In the work presented here, we have confirmed that SMRT can interact with mSin3, which can interact, in turn, with HDAC-1. However, our detailed mapping of the determinants involved in these interactions indicates that transcriptional silencing is actually a multifaceted phenomenon, with both SMRT and mSin3 able to exhibit significant transcriptional repression in the absence of detectable association with each other and in the absence of detectable interaction with HDAC-1. Our work further suggests that one of these multiple modes of SMRT-mediated repression may operate through targeting of TFIIB. This observation appears to reconcile the actions of the corepressors with prior work implicating the transcriptional preinitiation complex itself as a target for nuclear hormone receptor-mediated gene regulation (5, 17, 18).
SMRT possess two transcriptional silencing domains, one of which confers interaction with mSin3A and the other of which mediates silencing by an as-yet-unelucidated mechanism.
Both silencing domains in SMRT are located within the N terminus and are physically and functionally distinct from the more C-terminal SMRT domains that confer interaction with the T3R, retinoic acid receptor, and retinoid X receptor (RXR) nuclear hormone receptors (summarized in Fig. 9). Although these silencing domains were mapped in our experiments by their ability, as GAL4-DBD fusions, to repress reporter gene transcription, the same SD-1 and SD-2 also appear to be critical for repression mediated by full-length SMRT. In fact, deletion of the SD-1 and SD-2 of SMRT converts the native protein from a corepressor into a dominant negative inhibitor of repression (10, 11, 39).
FIG. 9.
Summary of the interactions elucidated by the corepressor complex. Schematic representations of mSin3A (top) and SMRT (bottom) are shown. Arrows indicate the domains of each protein that mediate observable interactions with the domains of the other and/or with TFIIB. Also depicted are the locations of the domains of these proteins that can mediate transcriptional repression when expressed as GAL4-DBD fusions (SD-A and -B in mSin3A and SD-1 and -2 in SMRT). Numbers above or below each schematic refer to the relevant amino acid positions.
In our hands, the same SD-1 region of SMRT was also the site of interaction with mSin3A (summarized in Fig. 9); this SMRT-mSin3A interaction was detectable both by a protein-protein binding assay in vitro and by a mammalian two-hybrid characterization in vivo. Deletions that severely impaired the ability of SD-1 to bind to mSin3A also severely impaired the ability of SD-1 to repress, consistent with a correlation between mSin3A recruitment to SD-1 and transcriptional repression by this SMRT subdomain.
In contrast to this strong SD-1–mSin3A interaction, however, we were unable to observe a significant interaction between the SD-2 region of SMRT and mSin3A or, in fact, between SD-2 and any of the other corepressor proteins tested here. Some prior reports have suggested that both SD-1 and SD-2 can interact with mSin3 (24, 31). Notably, however, these reported interactions between mSin3 and SD-2 were much weaker than those observed with SD-1 and were detected only by two-hybrid analysis or by low-stringency immunoprecipitation of cell extracts; the interaction between SD-2 and mSin3 may, therefore, be an indirect one and appears to be disproportionally weak compared to the strong silencing phenotype of SD-2. Conversely, it might be argued that our own failure to observe an interaction between SD-2 and mSin3A by mammalian two-hybrid analysis may also reflect a technical limitation: strong silencing domains, such as SD-2, can be difficult to analyze by the two-hybrid approach, given that the repression domain can overwhelm the function of a tethered AD fusion. Significantly, however, we also observed no evidence for an SD-2–mSin3A interaction in vitro, using a protein-protein binding assay that is not subject to the limitations of the two-hybrid methodology. Our results are therefore most consistent with SMRT SD-2 mediating transcriptional silencing by a mechanism distinct from a strong, direct recruitment of mSin3A.
Transcriptional repression by mSin3A does not require a detectable interaction with either SMRT or HDAC-1.
At least two distinct domains of mSin3 are able to mediate transcriptional repression when expressed as GAL4-DBD fusion polypeptides: SD-A (located at the mSin3A N terminus and overlapping the PAH1 region) and SD-B (located between the PAH3 and PAH4 domains of mSin3A) (Fig. 9). Intriguingly, neither of these silencing domains in mSin3 coincides with the domains that interact with SMRT; the SMRT interaction domains map instead to regions of mSin3A encompassing the PAH2 and PAH3 domains (Fig. 9). Our results are consistent with prior reports that N-terminal truncations of mSin3 are impaired in repression (1, 22), but our detailed dissection suggests that it is the loss of the N-terminal SD-A region, rather than loss of the adjacent SMRT interaction domain, that is primarily responsible for this impairment. Notably, although unable to recruit SMRT, the N-terminal SD-A region of mSin3A is able to recruit a related corepressor protein, N-CoR (Fig. 8A) (1, 22). It is therefore possible that an interaction with N-CoR, and not SMRT, is necessary for mSin3A-mediated transcriptional repression. Alternatively, other, yet-to-be identified factors may interact with the mSin3A N terminus to mediate repression. Furthermore, SMRT itself appears likely to contribute to at least some aspects of gene silencing by the native corepressor complex through the actions of the SMRT SD-2 domain and/or through the ability of SMRT to interact with the general transcriptional machinery (see below).
In addition to interacting with SMRT and N-CoR, mSin3A was also able to bind to HDAC-1 in vitro; in agreement with prevailing models, the HDAC-1 interaction site included the SD-B region of mSin3A (1, 30). In contrast, HDAC-1 does not appear to interact directly with SMRT. The ability of mSin3A to recruit HDAC-1 through its SD-B may, as proposed elsewhere, be an important facet of mSin3A-mediated transcriptional silencing (37, 51). Our own results, however, indicate that HDAC-1 recruitment is not the only mechanism through which mSin3A mediates repression. Most provocative is the presence of the second strong silencing domain within mSin3A, SD-A, that fails to detectably recruit HDAC in vitro (our data) or in vivo (30) and thus appears to function in a HDAC-1 and -2-independent manner. Further suggestive is our observation that trichostatin A, a potent inhibitor of HDAC, has no detectable effect on transcriptional repression in any of our transient-transfection assays. Our results are consistent with previous reports demonstrating that inhibition of the ability of Sin3 to interact with HDAC-1 and -2, or inhibition of the activity of the deacetylase itself, often results in only a partial reversal of repression (3, 21, 22, 28, 30). In fact, different promoters appear to be subject to different mechanisms of repression; for example, the SV40 late promoter utilized in our own experiments has been reported to be refractory to the antirepression effects of trichostatin A (33). This work, taken as a whole, strongly indicates that corepressor-mediated transcriptional silencing is a multifaceted phenomenon and that different components of the corepressor complex contribute to different aspects of repression.
TFIIB represents a potential target for SMRT-mediated transcriptional repression.
What other mechanisms might participate in SMRT-mediated transcriptional repression? A substantial body of evidence indicates that nuclear hormone receptors can interfere with the formation of a transcriptional preinitiation complex in vitro, apparently through direct inhibitory contacts of receptor with components of the general transcriptional machinery (5, 17, 18). This form of autonomous repression in vitro, mediated by direct interactions between receptor and the preinitiation complex, has previously been difficult to reconcile with the apparent obligatory requirement for the corepressor complex for gene silencing in vivo. However, in this paper we report that the SMRT polypeptide is itself also capable of interaction with TFIIB; in fact, this TFIIB interaction is equal to or greater in magnitude than the extremely strong interaction observed between SMRT and the nuclear hormone receptors.
Significantly, the ability of SMRT to interact with TFIIB correlates closely with SMRT-mediated repression. For example, the determinants that define SD-1 within SMRT overlap with the determinants that confer the TFIIB interaction (Fig. 9). Similarly, overexpression of TFIIB can overcome at least some elements of SMRT-mediated repression. Intriguingly, TFIIB also demonstrates a specific and equally strong interaction with mSin3A; in this context, however, the TFIIB interaction maps to a separate site on mSin3A distinct from the mSin3A domains that confer repression. We propose that, in addition to possible modification of the chromatin template, transcriptional silencing by nuclear hormone receptors involves a network of concordant physical interactions between the nuclear hormone receptor, the corepressor complex, and the general transcriptional machinery that may interfere with formation of a functional preinitiation complex. Although our own data clearly implicate TFIIB as one plausible target through which repression may be mediated, it is important to note that neither the TFIIB interactions elucidated here nor interaction with HDAC-1 and -2 appears likely to account for all of the silencing properties of SMRT and mSin3A; presumably still more mechanisms remain to be elucidated.
When this paper was under review, we learned of work by others (G. E. O. Muscat, L. J. Burke, and M. Downes) reporting that, in common with SMRT, the N-CoR corepressor also interacts physically with TFIIB (35a), an observation that we have confirmed in our own work. Muscat et al. further report that N-CoR can inhibit the ability of TFIIB to recruit the TAFII-32 subunit of TFIID, an interaction important for transcriptional activation in certain promoter contexts. Thus, SMRT and N-CoR both have the potential to exert profound effects on the function of the general transcriptional machinery.
A recurring theme: multiple interactions through multiple, often overlapping, sites.
One outcome of these studies is the finding of an unanticipated multiplicity of interactions that can occur between different components of the transcriptional repression machinery (Fig. 9). A single domain within SMRT, for example, can interact both with mSin3A and with TFIIB. Conversely, one of the mSin3A domains that binds SMRT also binds TFIIB. This concept of multiple interactions extends to the contacts between the corepressor complex and the DNA-binding transcription factors that tether it. For example, T3Rs recruit corepressor by interacting with two different sites in the C terminus of SMRT (11, 39, 42, 54), whereas recruitment of corepressor by PLZF is mediated by multiple contacts between PLZF, the N and C termini of SMRT, and mSin3 (19, 23, 32, 51a). It is tempting to speculate that this multiplicity of interactions might generate both synergistic and antagonistic outcomes and thus might serve regulatory roles. For example, the interactions of TFIIB and mSin3A with the SD-1 of SMRT may be mutually exclusive and thus indicative of the existence of two alternative forms of SMRT complex, and two distinct modes of SMRT function, in the cell. Alternatively, certain of the interactions between different components of the corepressor machinery may occur simultaneously and be mutually enhancing or may occur sequentially in a temporally ordered fashion that leads to the step-by-step assembly of a functional corepressor complex on a target promoter. Future work will need to focus on these questions.
ACKNOWLEDGMENTS
We are indebted to D. Ayer, R. N. Eisenman, C. A. Hassig, M. A. Lazar, M. G. Rosenfeld, and S. L. Schreiber for generously providing molecular clones. We also thank Shelly Meeuson and Linfong Tzeng, who, as rotation students, assisted with some of the recombinant constructs, and Valentina Taryanik for technical help.
This work was supported by Public Health Service grant CA53394 from the National Cancer Institute.
REFERENCES
- 1.Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role of N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Ayer D E, Kretzner L, Eisenman R N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 3.Bagy L, Kao H-Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 4.Baniahmad A, Kohne A C, Renkawitz R. A transferable silencing domain is present in the thyroid hormone receptor, in the v-Erb A oncogene product, and in the retinoic acid receptor. EMBO J. 1992;11:1015–1023. doi: 10.1002/j.1460-2075.1992.tb05140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baniahmad A, Ha I, Reinberg D, Tsai S Y, Tsai M-J, O’Malley B W. Interaction of human thyroid hormone receptor beta with transcription factor TFIIB may mediate target gene derepression and activation by thyroid hormone. Proc Natl Acad Sci USA. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baniahmad A, Leng X, Burris T P, Tsai S Y, Tsai M J, O’Malley B W. The τ4 activation domain of the thyroid hormone receptor is required for release of a putative corepressor(s) necessary for transcriptional silencing. Mol Cell Biol. 1995;15:76–86. doi: 10.1128/mcb.15.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casanova J, Helmer E, Selmi-Ruby S, Qi J S, Au-Flieger M, Desai-Yajnik V, Koudinova N, Yarm F, Raaka B M, Samuels H H. Functional evidence for ligand-dependent dissociation of thyroid hormone and retinoid acid receptors from an inhibitory cellular factor. Mol Cell Biol. 1994;14:5756–5765. doi: 10.1128/mcb.14.9.5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H-W, Privalsky M L. The erbA oncogene represses the actions of both retinoid X and retinoid A receptors but does so by distinct mechanisms. Mol Cell Biol. 1993;13:5970–5980. doi: 10.1128/mcb.13.10.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H-W, Privalsky M L. Retinoid X and retinoic acid receptors interact with TFIIB by distinct mechanisms. Mol Cell Endocrinol. 1997;129:55–61. doi: 10.1016/s0303-7207(97)04040-9. [DOI] [PubMed] [Google Scholar]
- 10.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 11.Chen J D, Umesono K, Evans R M. SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proc Natl Acad Sci USA. 1996;93:7567–7571. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damm K, Thompson C C, Evans R M. Protein encoded by v-Erb A functions as a thyroid hormone receptor antagonist. Nature. 1989;339:593–597. doi: 10.1038/339593a0. [DOI] [PubMed] [Google Scholar]
- 13.DeRurbertis F, Kadosh D, Henchoz S, Pauli D, Reuter G, Struhl K, Spierer P. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature. 1991;384:589–591. doi: 10.1038/384589a0. [DOI] [PubMed] [Google Scholar]
- 14.Dhordain P, Albagli O, Lin R J, Ansieau S, Quief S, Leutz A, Kerckaert J P, Evans R M, Leprince D. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL-6 oncoprotein. Proc Natl Acad Sci USA. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Downes M, Burke L J, Bailey P J, Muscat G E. Two receptor interaction domains in the corepressor, N-CoR/RIP13, are required for an efficient interaction with Rev-erbA alpha and RVR: physical association is dependent on the E region of the orphan receptors. Nucleic Acids Res. 1996;2:4379–4386. doi: 10.1093/nar/24.22.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenfeld G. Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 17.Fondell J D, Roy A L, Roeder R G. Unliganded thyroid hormone receptor inhibits formation of a functional preinitiation complex: implications for active repression. Genes Dev. 1993;7:1400–1410. doi: 10.1101/gad.7.7b.1400. [DOI] [PubMed] [Google Scholar]
- 18.Fondell J D, Brunel F, Hisatake K, Roeder R G. Unliganded thyroid hormone receptor a can target TATA-binding protein for transcriptional repression. Mol Cell Biol. 1996;16:281–287. doi: 10.1128/mcb.16.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara F F, Zamir I, Seiser C, Grignani F, Lazar M A, Minucci S, Pelicci P G. Fusion proteins of the retinoic acid receptor-α recruit histone deacetylase in promyelocytic leukemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 20.Guan K L, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 21.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 22.Heinzel T, Lavinsky R M, Mullen T-M, Soderstrom M, Laherty C D, Torchia J, Yang W-M, Brard G, Ngo S G, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3, and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 23.Hong S-H, David G, Wong C W, Dejean A, Privalsky M L. SMRT corepressor interacts with PLZF, and with the PML-RARα and PLZF-RARα oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamel Y, Soderstrom M, Glass C K, Rosenfeld M G. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 25.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Nuclear hormone receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 26.Jackson T A, Richer J K, Bain D L, Takimoto G S, Tung L, Horwitz K B. The partial agonist activity of antagonist-occupied steroid receptors is controlled by a novel hinge domain-binding coactivator L7/SPA and the corepressors N-CoR or SMRT. Mol Endocrinol. 1997;11:693–705. doi: 10.1210/mend.11.6.0004. [DOI] [PubMed] [Google Scholar]
- 27.Johnston L A, Tapscott S J, Eisen H. Sodium butyrate inhibits myogenesis by interfering with transcriptional activation functions of MyoD and myogenin. Mol Cell Biol. 1992;12:5123–5130. doi: 10.1128/mcb.12.11.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 29.Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld M G, Glass C K. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 30.Laherty C D, Yang W-M, Sun J-M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3A corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Leo C, Schroen D J, Chen J D. Characterization of receptor interaction and transcriptional repression by the corepressor SMRT. Mol Endocrinol. 1997;11:2025–2037. doi: 10.1210/mend.11.13.0028. [DOI] [PubMed] [Google Scholar]
- 32.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Jr, Evans R M. Role of the histone deacetylase complex in acute promyelocytic leukemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 33.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 34.McKenzie E A, Kent N A, Dowell S J, Moreno F, Bird L E, Mellor J. The centromere and promoter factor 1, CPF1, of Saccharomyces cerevisiae modulates gene activity through a family of factors, including SPT21, RPD1(DIN3), RPD3, and CCR4. Mol Gen Genet. 1993;240:374–386. doi: 10.1007/BF00280389. [DOI] [PubMed] [Google Scholar]
- 35.McKnight G S, Hager L, Palmiter R D. Butyrate and related inhibitors of histone deacetylase block the induction of egg white genes by steroid hormones. Cell. 1980;22:469–477. doi: 10.1016/0092-8674(80)90357-8. [DOI] [PubMed] [Google Scholar]
- 35a.Muscat G E O, Burke L J, Downes M. The corepressor N-CoR and its variants RIP13a and RIPΔ1 directly interact with the basal transcription factors TFIIB, TAFII32, and TAFII70. Nucleic Acids Res. 1998;26:2899–2907. doi: 10.1093/nar/26.12.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nasmyth K, Stillman D, Kipling D. Both positive and negative regulators of HO transcription are required for mother-cell-specific mating type switching in yeast. Cell. 1987;48:579–589. doi: 10.1016/0092-8674(87)90236-4. [DOI] [PubMed] [Google Scholar]
- 37.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 38.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sande S, Privalsky M L. Identification of TRACs (T3 receptor-associating cofactors) a family of cofactors that associate with, and modulate the activity of, nuclear hormone receptors. Mol Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- 40.Sap J, Munoz A, Schmitt H, Stunnenberg H, Vennstrom B. Repression of transcription mediated by a thyroid hormone response element by the v-Erb A oncogene product. Nature. 1989;340:242–244. doi: 10.1038/340242a0. [DOI] [PubMed] [Google Scholar]
- 41.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultichi A I, DePinho R A. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 42.Seol W, Mahon M J, Lee Y K, Moore D D. Two receptor interacting domains in the nuclear hormone receptor corepressor RIP13/N-CoR. Mol Endocrinol. 1996;10:1646–1655. doi: 10.1210/mend.10.12.8961273. [DOI] [PubMed] [Google Scholar]
- 43.Shibata H, Nawaz Z, Tsai S Y, O’Malley B W. Gene silencing by COUP-TF is mediated by transcriptional corepressors, N-CoR and SMRT. Mol Endocrinol. 1997;11:714–724. doi: 10.1210/mend.11.6.0002. [DOI] [PubMed] [Google Scholar]
- 44.Smith C L, Nawaz Z, O’Malley B W. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogn, 4-hydroxytamoxifen. Mol Endocrinol. 1997;11:657–666. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- 45.Sternberg P W, Stern M J, Clark I, Herskowitz I. Activation of the yeast HO gene by release from multiple positive and negative controls. Cell. 1987;48:567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- 46.Stillman D J, Dorland S, Yu Y. Epistasis analysis of suppressor mutations that allow HO expression in the absence of the yeast SWI5 transcriptional activator. Genetics. 1994;136:781–788. doi: 10.1093/genetics/136.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sussel L, Vannier D, Shore D. Suppressors of defective silencing in yeast: effects on transcriptional repression at the HMR locus, cell growth, and telomere structure. Genetics. 1995;141:873–888. doi: 10.1093/genetics/141.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vidal M, Gaber R F. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wade P A, Pruss D, Wolffe A P. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 50.Wang H, Stillman D J. Transcriptional repression in Saccharomyces cerevisiae by a SIN3-LexA fusion protein. Mol Cell Biol. 1993;13:1805–1814. doi: 10.1128/mcb.13.3.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolffe A P. Sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 51a.Wong, C.-W., and M. L. Privalsky. Submitted for publication.
- 52.Yang W-M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulatory RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone deacetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 54.Zamir I, Harding H P, Atkins G B, Horlein A, Glass C K, Rosenfeld M, Lazar M A. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zamir I, Dawson J, Lavinsky R M, Glass C K, Rosenfeld M G, Lazar M A. Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Proc Natl Acad Sci USA. 1997;94:14400–14411. doi: 10.1073/pnas.94.26.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zamir I, Zhang J, Lazar M A. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev. 1997;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- 57.Zervos A S, Gyuris J, Brent R. Mxi 1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J, Zamir I, Lazar M A. Differential recognition of liganded and unliganded thyroid hormone receptor by retinoid X receptor-regulated transcriptional repression. Mol Cell Biol. 1997;17:6887–6897. doi: 10.1128/mcb.17.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18 a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]