Abstract
Objective
Cervical cancer (CC) is one of the most common gynecologic malignancies worldwide. Although rapid improvements have been made regarding its prevention and treatment, little is known about disease pathogenesis and the clinical relevance of reliable biomarkers. The present study evaluated the expression of cystatin B (CSTB) as a potential biomarker of CC.
Methods
Tissue microarray analysis and immunohistochemical staining were performed to detect CSTB expression, while CSTB mRNA and protein expression levels of freshly isolated CC tissue were measured by quantitative real-time PCR and western blot, respectively. Bioinformatics were used to analyze the CSTB co-expression network and functional enrichments.
Results
We observed high CSTB mRNA and protein expression levels in CC tissues, which was confirmed by tissue microarray in a comparison with paired adjacent non-cancerous cervical tissue samples. CSTB gene enrichments and associations with co-expressed genes were also observed. Further analysis showed that elevated CSTB expression was associated with pathological progress in CC.
Conclusion
Our data demonstrate that CSTB has the potential to be used as a tissue biomarker with clinical value in patients with CC, which may aid the development of intervention strategies.
Keywords: Cystatin B, lesion, pathological progress, tissue microarray, tumor, cervical cancer
Introduction
Cervical cancer (CC) is one of the most common gynecologic malignancies worldwide, with estimated new cases and deaths in 2020 of 604,000 (about 6.5% of all female cancer incidences) and 342,000 (about 7.7% of all female cancer deaths), respectively. 1 Typically, CC shows slow growth, and may not present with any signs or symptoms at an early stage. The main risk factor of CC is a sexually transmitted infection of human papillomavirus (HPV), and persistent infection with high-risk HPV can lead to cervical lesions and invasive cancer.2,3 However, HPV infection alone is not sufficient to cause precancerous lesions or CC. The human immune system usually clears more than 90% of infections within 2 years, and the time between HPV infection and the development of high-grade cervical lesions is usually 10 to 20 years.4,5 Other factors that may increase the risk of developing CC include smoking, multiple sexual partners, young age at first pregnancy, human immunodeficiency virus infection, and immunosuppression.6,7
During the past few decades, CC incidence and mortality rates have declined, largely because of the highly effective measures of primary (HPV vaccine) and secondary (screening by a regular Papanicolaou smear test, also termed Pap test) prevention.8,9 However, although HPV testing has a high level of sensitivity, it identifies many ‘transient’ infections, which can cause patients excessive anxiety and psychological stress and increase the economic burden of medical care. 10 A Pap test is an important tool in detecting CC, and early detection and intervention may reduce the morbidity and mortality of CC patients. 11 However, an abnormal Pap smear could be caused by mild inflammation or minor cell dysplasia, HPV or other infections, and cancer or precancerous lesions that include high-grade squamous intraepithelial lesion (HSIL) or low-grade SIL (LSIL). 12 More recently, the cutting-edge ThinPrep screening test has been used to detect CC cells. 13 Despite this progression in detection, the pathogenesis of CC remains unclear, so identifying a reliable biomarker for the early diagnosis of precancerous lesions and CC would be extremely valuable.
Cystatins are a cysteine proteinase inhibitor superfamily that can be classified into three types. 14 Cystatin B (CSTB) belongs to the type II family, and is a single-chain neutral protein encoded by a gene located at human chromosome 21q22.3.15,16 CSTB is an important player in the balance of protein synthesis and degradation involved in cell proliferation, differentiation, apoptosis, and immunomodulation.17,18 Moreover, previous studies have shown that aberrant expression of CSTB is involved in tumorigenesis.19,20 However, little is known about the association of CSTB expression with pathogenesis in CC.
The present study elucidated CSTB as a biomarker of CC and clarified its pathological value using extensive data analysis, with the aim of developing intervention strategies for CC.
Materials and methods
Human subjects and tissue sample preparation
Freshly isolated human tissues from three non-tumorous cervical tissues and nine cervical lesions (three LSIL, three HSIL, and three CC) were obtained from Jinshan Hospital (Fudan University, China) between January and December 2021, and were stored in liquid nitrogen before RNA and protein extraction. Informed consent was obtained from patients, and patient details were de-identified before public release. Ethical approval was obtained from the Ethics Committee of Jinshan Hospital. No patients underwent chemotherapy or radiotherapy before cytoreductive surgery. The pathological diagnosis of LSIL, HSIL, and CC was made by pathologists.
RNA extraction and quantitative real-time (qRT)-PCR
Total RNA from tissues was extracted using the RNA‐Quick Purification Kit (Yishan Biotechnology Co., Ltd., Shanghai, China). Reverse transcription (RT) of total mRNA was performed using the First Strand cDNA Synthesis Kit (Roche Diagnostics, Mannheim, Germany). cDNA was amplified and quantified using the 7300 Real‐Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with the FastStart Universal SYBR‐Green Master Kit (Roche Applied Science). Primer sequences were: 5′-CATTCAAGAGCCAGGTGGTC-3′ (Forward) and 5′-GGCTTTGTTGGTCTGGTAGTTAG-3′ (Reverse) for CSTB, and TCATCACCATTGGCAATGAG-3′ (Reverse) and 5′-CACTGTGTTGGCGTACAGGT-3′ (Reverse) for β-actin, and PCR conditions were initial denaturation at 95°C for 1 minute followed by 40 cycles of denaturation at 95°C for 10 s and annealing/extension at 60°C for 30 s. The 2−ΔΔCt method was used to measure relative levels of mRNA expression.
Protein extraction and western blot analysis
Tissues were lysed in SDS Lysis Buffer (Beyotime Biotechnology, Shanghai, China) supplemented with 1% phenylmethanesulfonyl fluoride (Beyotime Biotechnology) and 1% phosphatase inhibitor (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). Western blot analysis was performed as described previously 18 using a rabbit anti‐CSTB monoclonal antibody (1:10,000 dilution, Abcam, Cambridge, UK). Signals were detected using the BeyoECL Moon Kit (Beyotime Biotechnology) and normalized to β-actin.
Tissue microarray and immunohistochemical staining
A tissue microarray containing tumor tissues from 70 patients with CC was provided by Shanghai Outdo Biotech Co., Ltd. (Shanghai, China). Immunohistochemistry (IHC) was performed using the Mouse/Rabbit Specific HRP/DAB Detection IHC Kit (Abcam) as previously described. 21 De-paraffinized sections were reacted with rabbit polyclonal anti-CSTB antibodies (1:100 dilution, Abcam), using non-immune rabbit serum (1:200 dilution, Abcam) in phosphate-buffered saline instead of primary antibody as the negative control. Cells displaying positive staining were counted in at least 12 representative fields under the microscope and the mean percentage of positive cells was calculated. Immunostaining was assessed by two independent investigators blinded to clinical characteristics and outcomes. IHC scoring criteria were calculated as described previously. 18 First, the percentage scores of immuno‑positive cells were determined as 0 (no positive cells), 1 (≤25%), 2 (26%–50%), 3 (51%–75%), and 4 (>75%). Second, the staining intensity scores were determined as 0 (no coloration), 1 (pale brown), 2 (brown), and 3 (dark brown). Finally, IHC scores of CSTB expression were calculated by the sum point of the percentage scores and staining intensity scores.
The cervical cancer cohort from the cancer genome atlas (TCGA) and gene set enrichment analysis (GSEA)
Gene expression quantification data and clinical information of CC patients in the TCGA database were downloaded from the Genomic Data Commons Data Portal, and analyses were processed using TCGAbiolinks (https://portal.gdc.cancer.gov/projects/TCGA-CESC). The entire transcriptome expression profile was used for analysis and the number of permutations was set at 1000. Significant enrichments were determined at P < 0.05 and a false discovery rate <0.25. Hallmark gene sets with high and low expression of CSTB in CC were analyzed using GSEA (version 4.1.0; Broad Institute, Inc., Cambridge, MA, USA) from TCGA. Multiple GSEA plots were generated using “plyr” (https://cran.r-project.org/web/packages/plyr/), “ggplot2” (https://github.com/tidyverse/ggplot2), “grid”, and “gridExtra” (https://cran.r-project.org/web/packages/gridExtra/index.html) R packages (version 4.2.2, www.r-project.org). The University of Alabama Cancer Database (UALCAN) (http://ualcan.path.uab.edu) was used to assess the association of CSTB with patient age, tumor stage, and pathologic grade. 22
Analysis of gene–gene functional interaction networks, Kyoto encyclopedia of genes and genomes (KEGG) pathways, and gene ontology (GO) enrichment
Genes positively and negatively correlated with high and low expression of CSTB in CC from the TCGA database were predicted by LinkedOmics (https://www.linkedomics.org/login.php), a web-based platform for analyzing multiomics data and used for producing heatmaps. Gene–gene functional interactions of CSTB with the top 20 positively correlated genes were analyzed using an automatically selected weighting method with the GenMANIA prediction technique (http://genemania.org/search/homo-sapiens/CSTB/). 23 KEGG pathway and GO term enrichment analyses were performed to screen CSTB‑associated genes in CC samples from publicly available RNA-seq data (https://portal.gdc.cancer.gov/projects/TCGA-CESC).
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software Inc., La Jolla, CA, USA) and R software version 4.1.1 (www.r-project.org) ). For the analysis of quantitative data, the significance of normally distributed variables was analyzed using the Student’s t-test to compare two groups, and the significance of non-normally distributed variables was analyzed using the one-way analysis of variance followed by Tukey’s multiple comparisons test to compare ≥3 groups according to the type of experiment. Results are shown as the mean ± SD. A P-value <0.05 was considered significant.
Results
CSTB is highly expressed in cervical cancer
In UALCAN, the level of CSTB transcripts was found to be significantly higher in primary CC tissues than healthy cervical tissue (P < 0.0001; Figure 1a). qRT‐PCR also showed that CSTB mRNA expression tended to be higher in freshly isolated CC tissue samples compared with fresh healthy cervical tissue samples, although the difference was not statistically significant (Figure 1b). Western blot analysis revealed the levels of CSTB protein to be significantly higher in fresh CC tissues than healthy cervical and SIL tissues (n = 3) (P < 0.01; Figure 1c, d).
Figure 1.
Expression of CSTB in human cervical tissue. (a) Comparison of CSTB transcripts between healthy cervical tissues and primary tumor tissues of cervical squamous cell carcinoma from the cancer data analysis portal of the University of Alabama at Birmingham using TCGA samples. Analysis was performed using the Student’s t-test, and data are presented as the mean ± SD. ****, P < 0.0001. (b) CSTB mRNA detection by qRT-PCR. One-way ANOVA followed by Tukey’s multiple comparison test was used. Data are presented as the mean ± SD. (c) Representative image of western blot for the CSTB protein. (d) Semi-quantitative analysis of (c). One-way ANOVA followed by Tukey’s multiple comparisons test was used. Data are presented as the mean ± SD. CSTB, cystatin B; N, normal; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion; CC, cervical cancer; TCGA, the cancer genome atlas; qRT-PCR, quantitative real-time PCR; ANOVA, analysis of variance.
Using tissue microarray, we further confirmed the higher expression of CSTB protein in CC tissues compared with paired adjacent non-cancerous cervical tissue samples (Figure 2a). A comparison of IHC scores between paired healthy and CC tissues (n = 70) revealed that CSTB protein expression was higher in human CC tissues than in non-cancerous cervical tissues (Figure 2b). Moreover, IHC staining scores were significantly higher in CC tissues at all tumor stages analyzed compared with healthy cervical tissues (P < 0.0001; Figure 2c). These data suggest that CSTB is a potential tissue biomarker for CC. Analysis of the overall survival (OS) and time to progression (TTP) of patients with CC with tumors of different stages showed that tumors of a more advanced stage were associated with a worse OS (Figure 2d) and TTP (Figure 2e) than those of a lower stage.
Figure 2.
Detection of CSTB protein expression in tissue microarray. (a) Detection of CSTB protein expression in non-tumorous cervical tissues (Normal) and primary cervical cancer tissues (cancer) by immunohistochemistry staining. Representative images are shown. Original, magnification ×200; Amplified, magnification ×400. (b) Comparison of CSTB protein expression between 70 paired normal and cancer tissues after immunohistochemistry staining. The number of cases in each score group is indicated. (c) Analysis of the IHC score for different tumor stages. Data are presented as the mean ± SD. ****, P < 0.001 tumor (T) vs. normal (N); T1-4, tumor stage 1-4. (d) Analysis of the overall survival (OS) of patients with CC. (e) Analysis of the time to progression (TTP) of patients with CC. CSTB, cystatin B; IHC, immunohistochemistry; TNM, tumor, node and metastasis; I–IV, TNM stages I–IV; CC, cervical cancer.
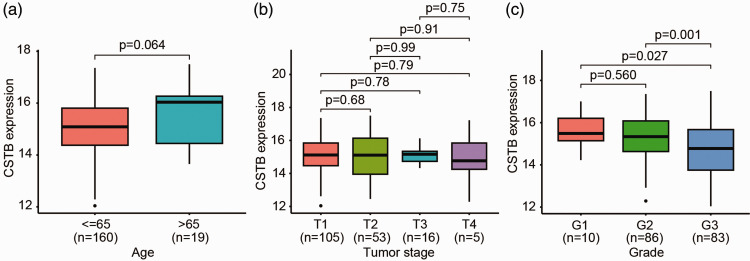
CSTB expression is not correlated with the tumor stage of cervical cancer
Subsequently, we examined the clinical relevance of CSTB in 179 patients with CC from TCGA. The expression of CSTB mRNA was not found to be correlated with patient age (comparing patients aged ≤65 years [n = 160] and >65 years [n = 19]) (Figure 3a) or tumor stage from T1 to T4 (Figure 3b), which is consistent with protein expression findings in tissue microarray analysis (Figure 2c). However, CSTB mRNA expression was significantly lower in tumors of pathological Grade 3 compared with those of Grades 1 and 2 (P < 0.05; Figure 3c).
Figure 3.
Association of CSTB mRNA with clinical features in patients with CC. (a) Comparison of CSTB mRNA expression between patients aged ≤65 and >65 years old using the Student’s t-test. (b) Comparison of CSTB mRNA expression between CC tumor stages from T1 to T4 using one-way ANOVA followed by Tukey’s multiple comparisons test. (c) Comparison of CSTB mRNA expression between the pathological grades of CC from G1 to G3 using one-way ANOVA followed by Tukey’s multiple comparisons test. Data are presented as the mean ± SD. CSTB, cystatin B; ANOVA, analysis of variance; CC, cervical cancer.
CSTB expression is correlated with that of other genes in cervical cancer co‐expression networks
Next, we examined whether CSTB is associated with co-expressed genes. Genes found to be positively and negatively correlated with high and low expression of CSTB are depicted on a heatmap (Figure 4a). GSEA analysis of CC cohorts from TCGA revealed that CSTB expression was enriched in arachidonic acid metabolism, focal adhesion, and neuroactive ligand–receptor interactions (Figure 4b). Gene–gene functional correlation analysis showed that CSTB expression had the strongest positive correlation with that of the following 20 genes: CTSH, CST3, DPP7, GSTT1, SARNP, CSTA, MAT2A, MYC, LRRK2, NUDC, FLNB, CST4, CST1, CST5, CST2, CST6, BAG3, KNG1, CTSL, and SPRY2 (Figure 4c). These networks are involved in physical interaction, genetic interaction, and co-expression.
Figure 4.
Association of CSTB expression with that of other genes. (a) Heatmap representing genes positively and negatively correlated with high and low expression of CSTB. (b) GSEA analysis showed functional enrichment of CSTB expression in CC cohorts from the TCGA database. (c) Gene–gene functional correlation analysis visualized the association of the top 20 positively related genes with CSTB expression. CSTB, cystatin B; GSEA, gene set enrichment analysis; TCGA, the cancer genome atlas; CC, cervical cancer.
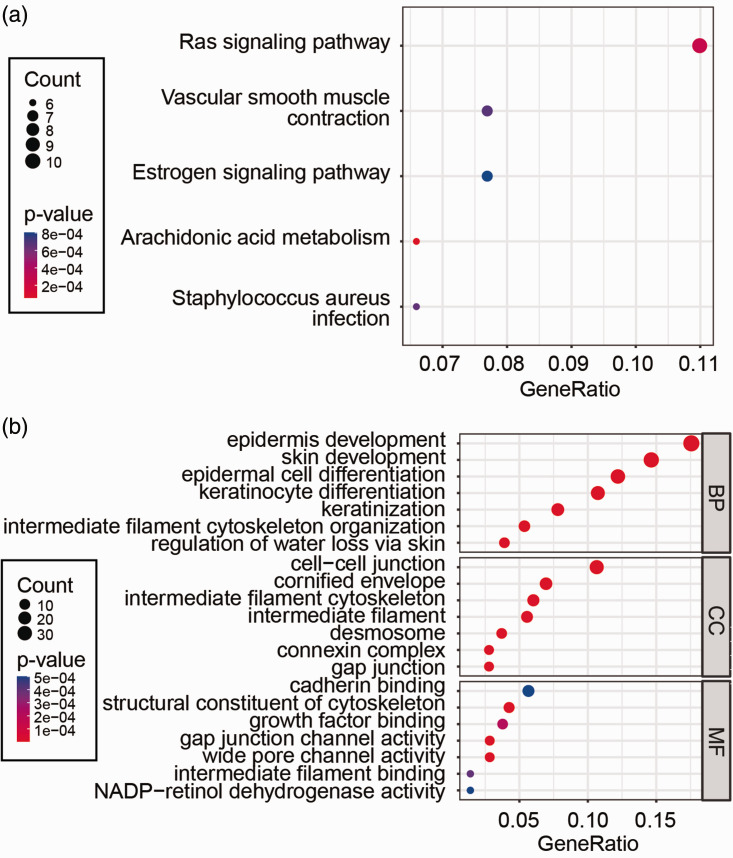
To elucidate the biological function of CSTB, KEGG pathway and GO term enrichment analyses were performed using human CC samples from publicly available RNA-seq data. KEGG pathway analysis showed that genes positively or negatively correlated with CSTB were mainly enriched in the Ras signaling pathway, vascular smooth muscle contraction, and the estrogen signaling pathway (Figure 5a). GO term annotation showed that CSTB co-expressed genes were primarily involved in epidermis development, cell–cell junctions, and cadherin binding (Figure 5b). These data indicate that the functions of CSTB in CC may be associated with those of correlated genes.
Figure 5.
Functional enrichment analyses of intersecting genes correlated with CSTB. (a) Five pathways identified from KEGG enrichment analysis. (b) GO term analysis. The bubble plot shows the top seven elements significantly enriched in GO categories BP, CC, and MF. GeneRatio refers to the ratio of the number of genes enriched in the term/pathway to the total number of genes in the terms/pathways.
CSTB, cystatin B; KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene ontology; BP, biological process; CC, cellular component; MF, molecular function.
Discussion
Among all female malignancies, the global incidence and death rate of CC is notably high, and it shows a trend of occurring in patients of an increasingly young age. Moreover, patients with advanced CC and those with metastatic disease have a particularly poor prognoses. Therefore, it is important to identify a biomarker correlated with tumor progression in human CC. The current study found that CSTB was expressed at significantly higher levels in CC tissues compared with healthy tissues and that it was associated with pathological processes in CC patients, suggesting that it has the potential to be a tissue biomarker for CC.
CSTB is a cysteine protease inhibitor that regulates degradation of the extracellular matrix and disruption of the basement membrane by modulating the activity of intracellular lysosomal proteases.24,25 Of the four types of proteases, aspartic proteases, matrix metalloproteases, serine proteases, and cysteine proteases, 26 the latter are the most abundant intracellular proteases in lysosomes, which are involved in the autophagic process and regulate metabolism under normal conditions. 27 CSTB functions in bone metabolism and protects cytoplasmic proteins from damage by cysteine proteases, and also regulates the physiological role of osteoclasts, thus inhibiting bone resorption. 28
There are two main pathways for protein degradation in eukaryotic cells: the lysosomal pathway and the ubiquitin–proteasome pathway.29,30 The most critical link of the lysosomal pathway is the fusion of autophagosomes and lysosomes, where multiple protein hydrolases (e.g., cysteine proteases) degrade proteins within the autophagosome. 31 Although cystatins lack signal sequences secreted outside the cell and are thought to act mainly intracellularly, cystatin protein expression changes have recently been detected in human ascites, serum, bronchial mucus, and saliva. 32,33 Variations in cysteine protease inhibitor activity have also been shown to affect tumor development. 33 Cysteine protease inhibitors such as CSTB can protect cells from endogenous or exogenous protease hydrolysis through the reversible competitive inhibition of tissue proteases, thus affecting various physiological and pathological processes.
Previous reports have identified CSTB as a serum biomarker of ovarian, bladder, and liver cancers.20,34,35 Indeed, the level of CSTB expression was found to be much higher in patients with hepatocellular carcinoma than in those with non-cancerous chronic liver disease, 36 while abnormal expression of CSTB is associated with metastasis and tumor progression.24,37 Moreover, CSTB expression is associated with prognosis in patients with squamous cell carcinoma of the head and neck, meningioma malignancy, and colorectal cancer.25,38,39 The current study observed a significantly higher level of CSTB expression in CC samples compared with healthy cervical tissue, and found that patients with advanced disease had a worse prognosis than those with tumors of a lower stage. These data indicate the clinical value of CSTB in patients with CC.
The dysregulation of CSTB expression has been shown to be mediated by various cytokines and growth factors, including tumor necrosis factor-α and transforming growth factor-β (TGF-β).40,41 We previously found that CSTB expression was regulated by the TGF-β signaling pathway in epithelial ovarian cancer. 18 CSTB was also shown to be downregulated by microRNA (miR)-143-3p, which was enhanced by TGF-β1 stimulation, 41 suggesting the existence of the TGF-β/miR-143-3p/CSTB axis in ovarian cancer cells. To further explore the biological function of CSTB in CC and its correlated genes, we performed KEGG and GO term enrichment analyses in the present study, which indicated that CSTB has a protumorigenic role, stimulates signaling pathways, and thus modulates cancer cell behavior. Analyses of public datasets revealed that CSTB co-expression with other positively related genes may function in the development of CC. However, validation in a larger cohort study is needed to confirm this.
In conclusion, CSTB is overexpressed in CC and could be used as a tissue biomarker for CC. The association of CSTB with pathological progress suggests that it may be valuable in developing clinically relevant intervention strategies for CC.
Acknowledgements
We thank the aforementioned databases in the text for data access.
Author contributions: Danjuan Ye: Conceptualization, data curation, formal analysis, validation, investigation, visualization, funding acquisition, and writing (original draft). Xiaoling Duan, Bin Guan, and Jia Yuan: Software, formal analysis, validation, investigation, and visualization. Yan Zhu, Jimin Shi, and Qi Lu: Validation, investigation, and methodology. Guoxiong Xu: Conceptualization, resources, data curation, supervision, funding acquisition, project administration, and writing (review and editing). All authors read and approved the final manuscript.
The authors declare that they have no competing interests.
Funding: This work was supported by grants from the Heath Commission of Shanghai Jinshan District (grant no. JSKJ-KTMS-2020-04) to D. Ye and the Natural Science Foundation of Shanghai (grant no. 23ZR1408900) to G. Xu.
Data availability statement
The names of the repositories and accession number(s) can be found in the article. The data used or analyzed during the study are available from the corresponding author upon reasonable request.
Informed consent statement
For qPCR and western blot experiments, written informed consent was obtained from patients. The need for informed consent for tissue microarray analysis was waived because the study was conducted on retrospective pathological tissue samples according to institutional requirements. The study was reviewed and approved by the Ethics Committee of Jinshan Hospital, Fudan University (approval no. JIEC-2021-S26).
ORCID iD
Guoxiong Xu https://orcid.org/0000-0002-9074-8754
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021; 71: 209–249. DOI: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev 2003; 16: 1–17. DOI: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hellsten C, Holmberg A, Astrom J, et al. Cervical cancer in Region Skane, Sweden 2017-2020 after the implementation of primary HPV screening: A quality assurance audit. Acta Obstet Gynecol Scand 2023; 103: 129–137. DOI: 10.1111/aogs.14691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saslow D, Solomon D, Lawson HW, et al. American Cancer Society, American Society for Colposcopy and Cervical Pathology, and American Society for Clinical Pathology screening guidelines for the prevention and early detection of cervical cancer. Am J Clin Pathol 2012; 137: 516–542. DOI: 10.1309/AJCPTGD94EVRSJCG. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez AC, Schiffman M, Herrero R, et al. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. J Natl Cancer Inst 2008; 100: 513–517. DOI: 10.1093/jnci/djn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowden SJ, Doulgeraki T, Bouras E, et al. Risk factors for human papillomavirus infection, cervical intraepithelial neoplasia and cervical cancer: an umbrella review and follow-up Mendelian randomisation studies. BMC Med 2023; 21: 274. DOI: 10.1186/s12916-023-02965-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma K, Machalek DA, Toh ZQ, et al. No woman left behind: achieving cervical cancer elimination among women living with HIV. Lancet HIV 2023; 10: e412–e420. DOI: 10.1016/S2352-3018(23)00082-6. [DOI] [PubMed] [Google Scholar]
- 8.Saslow D, Andrews KS, Manassaram‐Baptiste D, et al. Human papillomavirus vaccination 2020 guideline update: American Cancer Society guideline adaptation. CA Cancer J Clin 2020; 70: 274–280. DOI: 10.3322/caac.21616. [DOI] [PubMed] [Google Scholar]
- 9.Chor J, Davis AM, Rusiecki JM. Cervical cancer screening guideline for individuals at average risk. Jama 2021; 326: 2193–2194. DOI: 10.1001/jama.2021.13448. [DOI] [PubMed] [Google Scholar]
- 10.Wentzensen N, Schiffman M, Palmer T, et al. Triage of HPV positive women in cervical cancer screening. J Clin Virol 2016; 76: S49–S55. DOI: 10.1016/j.jcv.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padavu S, Aichpure P, Krishna Kumar B, et al. An insight into clinical and laboratory detections for screening and diagnosis of cervical cancer. Expert Rev Mol Diagn 2023; 23: 29–40. DOI: 10.1080/14737159.2023.2173580. [DOI] [PubMed] [Google Scholar]
- 12.Reid J. Women’s knowledge of Pap smears, risk factors for cervical cancer, and cervical cancer. J Obstet Gynecol Neonatal Nurs 2001; 30: 299–305. [PubMed] [Google Scholar]
- 13.Chen YC, Liang CN, Wang XF, et al. Follow-up study on ThinPrep cytology test-positive patients in tropical regions. World J Clin Cases 2022; 10: 12543–12550. DOI: 10.12998/wjcc.v10.i34.12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lalmanach G, Hoebeke J, Moreau T, et al. An immunochemical approach to investigating the mechanism of inhibition of cysteine proteinases by members of the cystatin superfamily. J Immunol Methods 1992; 149: 197–205. DOI: 10.1016/0022-1759(92)90251-n. [DOI] [PubMed] [Google Scholar]
- 15.Pennacchio LA, Lehesjoki AE, Stone NE, et al. Mutations in the gene encoding cystatin B in progressive myoclonus epilepsy (EPM1). Science 1996; 271: 1731–1734. [DOI] [PubMed] [Google Scholar]
- 16.Turk V, Stoka V, Turk D. Cystatins: biochemical and structural properties, and medical relevance. Front Biosci 2008; 13: 5406–5420. [DOI] [PubMed] [Google Scholar]
- 17.Keppler D. Towards novel anti-cancer strategies based on cystatin function. Cancer Lett 2006; 235: 159–176. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Gui L, Zhang Y, et al. Cystatin B is a progression marker of human epithelial ovarian tumors mediated by the TGF-beta signaling pathway. Int J Oncol 2014; 44: 1099–1106. DOI: 10.3892/ijo.2014.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leto G, Crescimanno M, Flandina C. On the role of cystatin C in cancer progression. Life Sci 2018; 202: 152–160. DOI: 10.1016/j.lfs.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 20.Feldman AS, Banyard J, Wu CL, et al. Cystatin B as a tissue and urinary biomarker of bladder cancer recurrence and disease progression. Clin Cancer Res 2009; 15: 1024–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren W, Zhang Y, Zhang L, et al. Overexpression of collagen type V alpha1 chain in human breast invasive ductal carcinoma is mediated by TGF-beta1. Int J Oncol 2018; 52: 1694–1704. DOI: 10.3892/ijo.2018.4317. [DOI] [PubMed] [Google Scholar]
- 22.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017; 19: 649–658. DOI: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010; 38: W214–W220. DOI: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian C, Ohlund D, Rickelt S, et al. Cancer cell-derived matrisome proteins promote metastasis in pancreatic ductal adenocarcinoma. Cancer Res 2020; 80: 1461–1474. DOI: 10.1158/0008-5472.CAN-19-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lah TT, Nanni I, Trinkaus M, et al. Toward understanding recurrent meningioma: the potential role of lysosomal cysteine proteases and their inhibitors. J Neurosurg 2010; 112: 940–950. DOI: 10.3171/2009.7.JNS081729. [DOI] [PubMed] [Google Scholar]
- 26.Verma S, Dixit R, Pandey KC. Cysteine Proteases: modes of activation and future prospects as pharmacological targets. Front Pharmacol 2016; 7: 107. DOI: 10.3389/fphar.2016.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaminskyy V, Zhivotovsky B. Proteases in autophagy. Biochim Biophys Acta 2012; 1824: 44–50. DOI: 10.1016/j.bbapap.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 28.Laitala-Leinonen T, Rinne R, Saukko P, et al. Cystatin B as an intracellular modulator of bone resorption. Matrix Biol 2006; 25: 149–157. DOI: 10.1016/j.matbio.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 29.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol 2005; 6: 79–87. DOI: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 30.Tai HC, Schuman EM. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat Rev Neurosci 2008; 9: 826–838. DOI: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- 31.Lorincz P, Juhasz G. Autophagosome-lysosome fusion. J Mol Biol 2020; 432: 2462–2482. DOI: 10.1016/j.jmb.2019.10.028. [DOI] [PubMed] [Google Scholar]
- 32.Ritonja A, Machleidt W, Barrett AJ. Amino acid sequence of the intracellular cysteine proteinase inhibitor cystatin B from human liver. Biochem Biophys Res Commun 1985; 131: 1187–1192. DOI: 10.1016/0006-291x(85)90216-5. [DOI] [PubMed] [Google Scholar]
- 33.Kos J, Mitrovic A, Mirkovic B. The current stage of cathepsin B inhibitors as potential anticancer agents. Future Med Chem 2014; 6: 1355–1371. DOI: 10.4155/fmc.14.73. [DOI] [PubMed] [Google Scholar]
- 34.Gashenko EA, Lebedeva VA, Brak IV, et al. Evaluation of serum procathepsin B, cystatin B and cystatin C as possible biomarkers of ovarian cancer. Int J Circumpolar Health 2013; 72. DOI: 10.3402/ijch.v72i0.21215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji NY, Kang YH, Park MY, et al. Development of a fluorescent microsphere immunoassay for cystatin B (CSTB) in serum of patients with hepatocellular carcinoma. Clin Chem Lab Med 2011; 49: 151–155. [DOI] [PubMed] [Google Scholar]
- 36.Lee MJ, Yu GR, Park SH, et al. Identification of cystatin B as a potential serum marker in hepatocellular carcinoma. Clin Cancer Res 2008; 14: 1080–1089. [DOI] [PubMed] [Google Scholar]
- 37.Butinar M, Prebanda MT, Rajkovic J, et al. Stefin B deficiency reduces tumor growth via sensitization of tumor cells to oxidative stress in a breast cancer model. Oncogene 2014; 33: 3392–3400. DOI: 10.1038/onc.2013.314. [DOI] [PubMed] [Google Scholar]
- 38.Anicin A, Gale N, Smid L, et al. Expression of stefin A is of prognostic significance in squamous cell carcinoma of the head and neck. Eur Arch Otorhinolaryngol 2013; 270: 3143–3151. DOI: 10.1007/s00405-013-2465-5. [DOI] [PubMed] [Google Scholar]
- 39.Kos J, Krasovec M, Cimerman N, et al. Cysteine proteinase inhibitors stefin A, stefin B, and cystatin C in sera from patients with colorectal cancer: relation to prognosis. Clin Cancer Res 2000; 6: 505–511. [PubMed] [Google Scholar]
- 40.Bidovec K, Bozic J, Dolenc I, et al. Tumor necrosis factor-alpha induced apoptosis in U937 cells promotes cathepsin D-independent stefin B degradation. J Cell Biochem 2017; 118: 4813–4820. DOI: 10.1002/jcb.26152. [DOI] [PubMed] [Google Scholar]
- 41.Guan W, Wang X, Lin Q, et al. Transforming growth factor‑beta/miR‑143‑3p/cystatin B axis is a therapeutic target in human ovarian cancer. Int J Oncol 2019; 55: 267–276. DOI: 10.3892/ijo.2019.4815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The names of the repositories and accession number(s) can be found in the article. The data used or analyzed during the study are available from the corresponding author upon reasonable request.