Abstract
Recent studies have indicated that the DNA replication machinery is coupled to silencing of mating-type loci in the budding yeast Saccharomyces cerevisiae, and a similar silencing mechanism may operate in the distantly related yeast Schizosaccharomyces pombe. Regarding gene regulation, an important function of DNA replication may be in coupling of faithful chromatin assembly to reestablishment of the parental states of gene expression in daughter cells. We have been interested in isolating mutants that are defective in this hypothesized coupling. An S. pombe mutant fortuitously isolated from a screen for temperature-sensitive growth and silencing phenotype exhibited a novel defect in silencing that was dependent on the switching competence of the mating-type loci, a property that differentiates this mutant from other silencing mutants of S. pombe as well as of S. cerevisiae. This unique mutant phenotype defined a locus which we named sng1 (for silencing not governed). Chromatin analysis revealed a switching-dependent unfolding of the donor loci mat2P and mat3M in the sng1− mutant, as indicated by increased accessibility to the in vivo-expressed Escherichia coli dam methylase. Unexpectedly, cloning and sequencing identified the gene as the previously isolated DNA repair gene rhp6. RAD6, an rhp6 homolog in S. cerevisiae, is required for postreplication DNA repair and ubiquitination of histones H2A and H2B. This study implicates the Rad6/rhp6 protein in gene regulation and, more importantly, suggests that a transient window of opportunity exists to ensure the remodeling of chromatin structure during chromosome replication and recombination. We propose that the effects of the sng1−/rhp6− mutation on silencing are indirect consequences of changes in chromatin structure.
During differentiation, switches in state of gene expression, i.e., from on to off and vice versa, are known to occur during cell division at discrete stages of development. These are exemplified by X-chromosome inactivation in mammals (39, 51), position effect variegation in Drosophila melanogaster (68), epigenetic imprinting of specific loci in mammals (35), and silencing at mating-type loci in yeasts (33, 48). Moreover, specific genes in cells of a particular differentiated state maintain the same state of expression and chromatin structure through numerous cell divisions. Such phenomena suggest the existence of molecular mechanisms that establish and maintain a particular state of gene expression during development and differentiation, through multiple rounds of DNA replication. Thus, DNA replication is obviously essential for cell proliferation as well as for altering and propagating particular states of gene expression during development and differentiation. Fission yeast is fast becoming an ideal model system for understanding the mechanisms underlying these processes. Earlier studies have shown that the establishment of silencing at the HMRa locus in Saccharomyces cerevisiae requires the passage of cells through S phase (41) and that a functional autonomous replication sequence (ARS) component of the cis-acting silencer element linked to the HMRa locus is essential (52). Moreover, mutations in the genes encoding the subunits of the origin recognition complex (ORC) at the ARS elements that flank the silent cassettes have been shown to be defective in mating-type silencing in S. cerevisiae (5, 20, 36, 40). These results suggest that the DNA replication machinery is somehow coupled to silencing of mating-type loci in S. cerevisiae. Furthermore, the finding of ORC1 and ORC2 homologs in fission yeast and Xenopus laevis (7, 24, 45) suggests that similar mechanisms may operate in Schizosaccharomyces pombe and higher eukaryotes. In addition, participation of chromatin structure in silencing is suggested by studies showing that mutations in the histone genes lead to loss of silencing (62) and the increased accessibility of the mating-type and telomeric loci in silencing-defective mutants to the endogenously expressed Escherichia coli dam methylase in budding yeast (23, 58).
In the mating-type switching system of S. pombe, the mat1 locus can switch between two alternately expressed alleles, while the closely linked donor loci (mat2 and mat3) contain the same genes present at mat1 but are silenced due to a repressive position effect control (4; see reference 33 for a review). Several mutations that affect mat2 and mat3 silencing, namely, clr1-4, swi6, and rik1, have recently been identified in S. pombe (12, 15, 63, 64) (Fig. 1). These mutations not only lead to derepression of the silent loci, mat2P and mat3M, but also allow the expression of a marker gene, such as ura4, when placed in vicinity of the donor loci (15, 63, 64). Moreover, these genes function to prohibit mitotic and meiotic recombination in the region between the mat2 and mat3 loci (12, 38, 63, 64) and therefore, appear to affect the mat2-mat3 region in a global fashion. In addition, these mutations affect the position effect control of expression of marker genes placed at the telomeric and centromeric regions (1). Interestingly, a 4.3-kb central segment of the 10.9-kb K region located between mat2 and mat3 has a strong homology with the cen2 and cen3 sequences (26). This region has been shown to play a role in establishing the cold spot of recombination and especially in epigenetic regulation of mating-type switching and directionality of switching (25, 26). Additionally, sequences flanking the mat2P locus also have ARS activity (46).
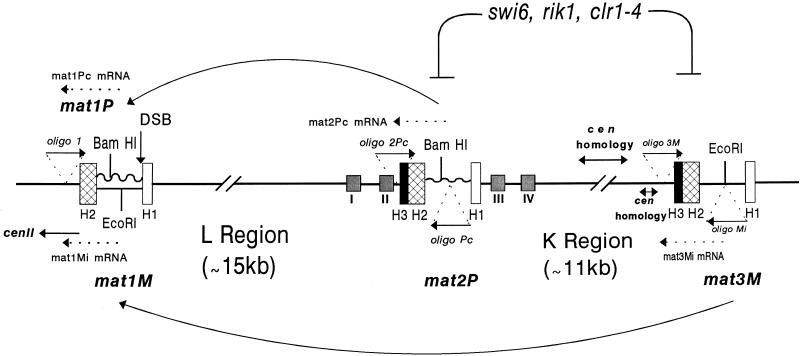
FIG. 1.
Cassette organization of the mating-type loci mat1, mat2P, and mat3M in S. pombe. All three cassettes contain the homology regions defined by H2 (135 bp) and H1 (59 bp), which are located at the two flanks of the Plus-specific (jagged line) and Minus-specific (straight line) regions. H3, the third region of homology, is present only at mat2 and mat3 loci. The arrows indicate the directional switching of Plus to Minus and Minus to Plus by transfer of information from mat2 and mat3 to mat1 by gene conversion-mediated transposition. The site of the in vivo DSB at the H1/allele junction in mat1 is indicated. The gene products of swi6, rik1, and clr1-4 are shown to be involved in silencing of the mat2 and mat3 loci. The locations of the four cis-acting silencer elements (I to IV) flanking the silent cassette mat2P are also indicated (14). The locations of oligonucleotide (oligo) 1, used for primer extension to detect the Pc and Mi transcripts from the mat1 locus (mat1Pc mRNA and mat1Mi RNA) for the experiment in Fig. 4, and those used to detect the RT-PCR products for mat2Pc (oligonucleotides 2Pc and Pc) and mat3Mi (oligonucleotides 3M and Mi) mRNAs for the experiment in Fig. 3 are indicated. The central 4.3-kb segment of the K region and some flanking regions which show strong homology to the centromeric sequences of cen2 and cen3 (26) are also indicated.
Therefore, working on the premise that there may be an underlying link between DNA replication, chromatin structure, and mating-type silencing, we screened for temperature-sensitive mutations in S. pombe that are also defective for silencing of the mating-type loci. A novel mutant which is temperature sensitive for growth and exhibits a defect in silencing of the donor cassettes only in efficiently switching strains was fortuitously isolated in such a screen (see below). In vivo analysis of the chromatin structure by using the E. coli dam methylase revealed a more accessible chromatin structure of the mating-type donor loci in this mutant; most interestingly, the increased accessibility of the donor loci is dependent on their switching competence. Because of this phenotype, the mutation is named sng1, for silencing not governed. The sng1− mutation is encoded by the gene rhp6, a RAD6 homolog in S. pombe, which is known to be involved in postreplication DNA repair, UV-induced mutagenesis, sporulation, and ubiquitination of histones (29, 49, 50). Thus, this study demonstrates a role of the RAD6/rhp6 gene in maintenance of silencing of mating-type loci and provides data suggesting that this function is mediated at the level of chromatin assembly.
MATERIALS AND METHODS
Plates and media.
To test for temperature sensitivity, strains (Table 1) were streaked for single-colony isolation on YEA plates, incubated for 4 days at 30 or 36°C, and then photographed. To monitor the haploid meiosis phenotype, cells were grown at 30°C for 4 days on sporulation medium and photographed with a phase-contrast microscope.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SPJ100 | h90 ura4−::dam/ura4 | This study |
| SPJ102 | h90 swi6− ura4−::dam/ura4 his2 ade6-216 | This study |
| SPJ187 | h90 leu1-32 ura4−::dam/ura4 rhp6−/sng1− ade6-216 | This study |
| SPJ189 | Msmto leu1-32 ura4−::dam/ura4 rhp6−/sng1− ade6-216 | This study |
| SPV12 | Msmto ura4−::dam/ura4 swi6− ade6-210 | This study |
| PG6 | h90 his2 leu1-32 ura4D18 ade6-210 swi6− | This study |
| SP837 | h90 leu1-32 ura4D18 ade6-216 | This study |
| SPJ110 | h90 leu1-32 ade6-216 swi7-1 rhp6−/sng1− ura4D18 | This study |
| SPJ113 | Msmto leu1-32 ura4D18 ade6-216 rhp6−/sng1− | This study |
| SPJ116 | PΔ17::leu2 ura4D18 ade6-216 rhp6−/sng1− | This study |
| SPJ117 | mat1M mat2,3Δ::leu2− leu1-32 rhp6−/sng1− ade6-216 | This study |
| SPJ123 | h90 ade6-216 ura4D18 rhp6−/sng1− swi5 | This study |
| SPJ119 | h90 leu1-32 lys1 ade6-210 swi2− rhp6−/sng1− | This study |
| PRZ119 | h+ leu1-32 ura4Δ18 lys1-131 ade6-210 rhp6Δ::ura4+ | L. Prakash |
RNA isolation, RT-PCR and Southern blotting.
RNA was prepared from cells grown at 30°C by the hot phenol method (57). Reverse transcription (RT)-PCRs were done in the logarithmic phase of amplification (8). First-strand cDNA synthesis was done with reverse transcriptase, using total RNA from cells grown in N+ or N− medium (31). The oligonucleotides used for cDNA synthesis were 2P (5′CAACGGATTACTAAAAACAGTTTAAATG3′), located outside the H3 box (Fig. 1) in the mat-2P locus, for the antisense Pc transcript; 3M (5′CCAATCAACTTAACATGAAGCAACTCCTGATAC3′), located outside the H3 box in the mat3M locus (Fig. 1), for the antisense Mi transcript; and polαas (5′CCATTGGTTGACTGTCAGACATTTTC3′) for the antisense polα transcript (59). The cDNA products were diluted 1:10 and 1 μl was used for PCR with the sense and antisense oligonucleotides. The sense-strand oligonucleotides used were Pc (5′GATTAAGAGCACCTATTTTCTTGCC3′) for Pc; Mi (5′CATACTAATAATGTCAGCAGAAGAC3′) for Mi, and polαs (5′GAAAGAAGATATATTCGACTTTAAAG3′) for polα. The cDNA products were resolved by agarose gel (1.2%) electrophoresis, transferred to nytran membranes, and hybridized with PCR products synthesized from the mat2P-, mat3M-, and polα-containing plasmids with the above sets of primers and labeled by the random primer method (18). The conditions for hybridization, washing, and autoradiography were as described elsewhere (55).
Gene cloning.
To clone the rhp6 gene, we transformed SPJ107, a sng1− mutant strain (h90 leu1− ura4D18 ade6-216 sng1-1), with the S. pombe partial HindIII genomic library cloned in the pWH5 vector (70). The vector contains the S. cerevisiae LEU2+ gene, which upon transformation can complement the leu1− defect of S. pombe. Two out of approximately 9,000 Leu+ transformants complemented the temperature-sensitive growth defect. The indicated fragments were subcloned into plasmid pWH5 or pIRT1 (54) for complementation assays.
Primer extension analysis.
For primer extension analysis, equivalent amounts of RNA (10 μg) isolated from different strains were used. The primer used was 5′GGGTAGGAAAGAGAGAGTAGTTGAAGG3′, which is located upstream of the H2 box of mat1 (oligonucleotide 1 [Fig. 1]) and allows analysis of the mat1-specific transcripts, Pc (529 nucleotides) from mat1P and Mi (315 nucleotides) from mat1M. Primer extension was carried out by the protocol of Triezenberg (65). The products were resolved on denaturing polyacrylamide gels and subjected to autoradiography.
Strain construction.
The S. pombe strain containing a chromosomally integrated E. coli dam methylase gene was constructed by homologous integration of plasmid pJS1, containing the dam and ura4 genes of S. pombe, into the ura4− locus. The dam gene was expressed from its own promoter. The general methods for DNA isolation, restriction digestion, Southern blotting, and hybridization have been described elsewhere (58). Genetic methods for constructing strains with required genotypes were as described by Moreno et al. (44).
RESULTS
Isolation of the sng1− mutant.
The sng1− mutant was fortuitously obtained during an attempt to generate temperature-sensitive mutants of the DNA polymerase (polα) gene by DNA-mediated transformation of yeast cells with the in vitro-mutagenized polα clone. However, we found that the sng1− mutation is genetically unlinked to the swi7/polα gene (data not shown) and presumably arose due to transformation-induced mutagenesis. We analyzed this mutant because it exhibited very interesting novel phenotypes. The sng1− mutant cells are temperature sensitive, as they fail to grow at 36°C (Fig. 2A). Microscopic analysis of the mutant showed elongated cells when grown at the restrictive temperature, and even at the semipermissive temperature (30°C) they appeared larger and more rounded than wild-type cells (data not shown). A more telling phenotype was that the h90 sng1− mutant cells, when placed in sporulation medium at the semipermissive temperature of 30°C, exhibited haploid meiosis, a phenotype suggestive of expression of P as well as M mating-type information in individual haploid cells that normally express one or the other type of information from mat1 (Fig. 2B). However, the donor-deleted (mat2,3Δ) sng1− strains did not exhibit the haploid meiosis phenotype (Table 2). More interestingly, the haploid meiosis defect was observed only in the efficiently switching background, h90, not in a variety of other genetic backgrounds in which switching is absent (Msmto and PΔ17; these strains carry a deletion of the cis-acting sequences near mat1 that are required for generation of double-strand breaks [DSB]) or drastically reduced (swi7−, swi2−, and swi5− [Table 2]). This property differentiates the sng1− mutant from the known silencing mutants of S. pombe (15, 38, 63, 64) and S. cerevisiae (28). In addition, the sng1− mutant did not show an elevation of expression of the normally silent mat2- or mat3-linked ura4 marker gene even in the switching (h90) strain (references 15, 38, 63, and 64 and data not shown). This genetic analysis showed that the haploid meiosis phenotype was dependent on both the presence of the donor loci and their utilization in switching. Thus, the phenotype must not be the result of a defect in the pat1 or mei2 gene, also known to confer this phenotype but without the requirement of mating-type information (10). We therefore inferred that switching allowed derepression of the donor loci that are actively engaged in switching in the sng1− mutant and that derepression did not extend up to the ura4 gene, which was placed distal to mat2 or mat3.
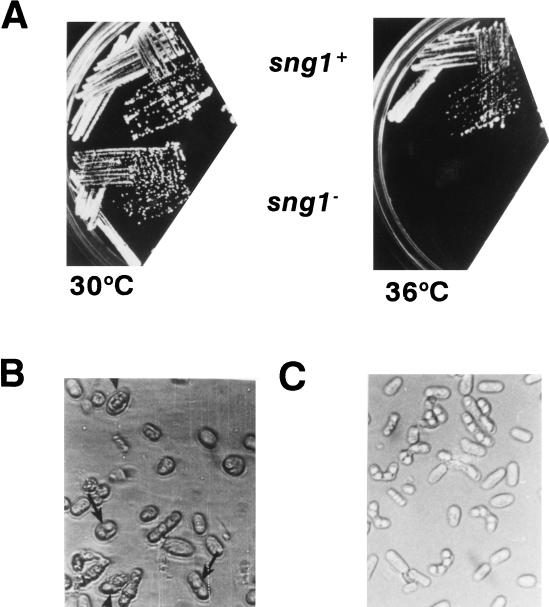
FIG. 2.
Different phenotypes of the sng1− mutant. (A) Temperature sensitivity. Strains used were wild-type strain SP837 (h90 leu1-32 ura4D18 ade6-M216) and mutant strain SPJ107 (h90 leu1-32 ura4D18 ade6-M216 sng1−). Temperatures at which strains were grown are indicated. h90 denotes homothallic strains that undergo sng1-1 efficient mating-type switching. (B) Haploid meiosis. The sng1− mutant cells exhibiting haploid meiosis are indicated by arrows. Wild-type cells produce asci containing four mature spores in zygotic diploid cells which result from switching and mating of cells of opposite mating type. In contrast, the sng1− mutant cells produce aberrant spores in a haploid cell.
TABLE 2.
The haploid meiosis phenotype of the sng1− mutant is dependent on both efficient switching and the presence of the donor loci
| Strain genotype | Switchability | Haploid meiosis phenotype |
|---|---|---|
| h90 | High | − |
| mat1M mat2,3Δ sng1− | Unswitchable | − |
| mat1P mat2,3Δ sng1− | Unswitchable | − |
| h90 sng1− | High | + |
| mat1Msmto sng1− | Unswitchable | − |
| mat1PΔ17 sng1− | Unswitchable | − |
| h90 swi7− sng1− | Reduced | − |
| h90 swi2− sng1− | Reduced | − |
| h90 swi5− sng1− | Reduced | − |
Derepression of the silent donor loci in the sng1− mutant and its dependence on their switching competence.
The above results suggested that in the sng1− mutant, the active process of switching may turn on the donor cassettes that are being copied in bringing about the switch. The sng1+ gene product may be required to establish the original state of silent gene expression existing before switching, and this restoration may not be achieved in the h90 sng1− mutant. To directly assess the inferred silencing defect in the sng1− mutant, we assayed for the synthesis of mat2- and mat3-specific transcripts in both the switching and nonswitching strains by the highly sensitive RT-PCR. PCR was carried out in the logarithmic phase of amplification (8), which allows quantitative comparison of the levels of transcripts. Supporting our interpretation above for the switching dependence of haploid meiosis phenotype of this mutant, high-level expression of silent loci transcripts was observed in a sng1− mutant in switching (h90 sng1−) background (mat2Pc transcript [Fig. 3A, lane 4]; mat3Mi transcript [Fig. 3B, lane 4]), but the expression level was very much reduced or absent in a nonswitching (Msmto sng1−) background (Fig. 3A and B, lane 8). Another nonswitching strain (PΔ17 sng1−) and a poorly switching strain (h90 swi5− sng1−) did not express the donor loci at a significant level and consequently did not exhibit the haploid meiosis phenotype (data not shown).
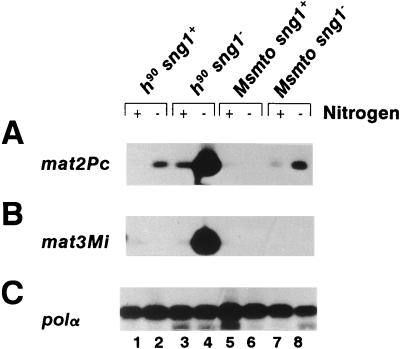
FIG. 3.
RT-PCR analysis of transcripts encoded by the donor loci. (A) mat2Pc; (B) mat3Mi; (C) polα. RNA was isolated from cells grown at the semipermissive temperature of 30°C in N+ (+; lanes 1, 3, 5, and 7) or N− (−; lanes 2, 4, 6, and 8) medium, a regimen that induces mat transcripts (31). RNA samples (10 μg) were treated with RNase-free DNase I and subjected to RT-PCR in the logarithmic phase of amplification (8) using oligonucleotides 2 Pc and Pc for monitoring mat2Pc transcription, oligonucleotides 3M and Mi (Fig. 1) for monitoring mat3Mi transcription, and oligonucleotides polαas and polαs for monitoring polα transcription. The products were resolved by agarose gel electrophoresis and subjected to Southern blotting and hybridization with homologous probes generated by PCR from the plasmid clones as described in Materials and Methods.
We also observed that a low level of leaky expression of the mat2Pc transcript was present in switching, sng1+ cells (Fig. 3A, lane 2) but not in a nonswitching strain (Fig. 3A, lane 6). One interpretation of this result is that the donor loci may have gained partial accessibility to the transcription machinery during recombination, thus yielding leaky expression of the donor cassette used during the switch (see Discussion).
This novel phenotype of expression of silent genes only in switching sng1− strains suggested the following explanation. During switching, a copy of the donor locus is transmitted to mat1 through recombination (33). We propose that during the replication required for recombination, the chromatin structure of donor loci is transiently perturbed. In wild-type cells, it must be reset to the same transcriptionally repressed structure as existed before DNA replication associated with switching. In contrast, we propose that in the sng1− mutant, the inactive chromatin structure is not reestablished, which leads to persistence of the perturbed open structure in switching cells and thus the partial expression of silent loci in the mutant. Next we tested whether this effect is limited to donor loci.
Regulation of the mat1-Mi transcript is altered in the sng1− mutant.
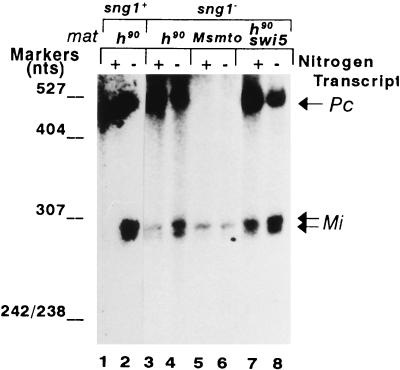
To check for any change in expression of the mat1-encoded transcripts, we carried out primer extension analysis. In wild-type cells, the mat1Mi transcript is completely repressed in N+ medium and nitrogen starvation leads to an induction of two transcripts differing at their 5′ ends by 20 nucleotides, as reported earlier (31) (Fig. 4, lanes 1 and 2). In the sng1− mutant, a low level of the two mat1Mi transcripts was detected even in N+ medium; while no further induction of these transcripts was observed upon nitrogen starvation in the nonswitching, Msmto background (lanes 5 and 6), in both h90 (lanes 3 and 4) and h90 swi5− backgrounds (lanes 7 and 8), the sng1− mutant exhibited further induction of nearly three- to fivefold. This result indicates that while the sng1− mutation causes the constitutive expression of the mat1Mi transcripts, it has no effect on pheromone inducibility of these transcripts, which is documented to occur in response to the Plus pheromone secreted by the cells of Plus (P) mating type in the homothallic, switching population (see below). (The level of mat1Pc transcript, which is known to be constitutively expressed [31], remained roughly constant in both N+ and N− media [lanes 1 to 4, 7, and 8] and served as a loading control.) Thus, the sng1− mutation alters the regulation of expression of all the three cassettes of the mating-type locus.
FIG. 4.
The sng1− mutation leads to constitutive expression of mat1Mi transcripts. Primer extension analysis for transcripts Pc (529 nucleotides [nts]) from mat1P and Mi (315 nucleotides) from mat1M, which specifically arise from the mat1 locus. Pc and Mi are indicated by arrows. RNA was isolated from cells of the indicated strains grown in N+ (lanes 1, 3, 5, and 7) and N− (lanes 2, 4, 6 and 8) media at 30°C and subjected to primer extension analysis as described previously (65), using oligonucleotide 1, which is able to reverse transcribe from both mat1Pc and mat1Mi RNAs, as these are known to extend beyond the H2 box (Fig. 1) (31).
The switching dependence of the sng1− phenotype requires chromosomal integrity and is independent of pheromone effect.
It is possible that in the nonswitching Msmto sng1− strain, the donor mating-type locus is not available for switching because of chromosomal folding and thus is also not available for derepression. To test this, we transformed an Msmto sng1− strain with the plasmid containing the mat2P locus and the S. pombe ars1 and ura4+ genes (plasmid pKE10 [14]) and tested whether the transformed cells would exhibit loss of silencing as indicated by haploid meiosis. However, we failed to notice any haploid meiosis in the transformed strain (data not shown). This finding rules out the possibility that the lack of silencing defect in the nonswitching sng1− mutant strain is due to nonavailability of the donor locus in the nonswitching background. On the other hand, the integrity of the mating-type region which brings about switching may be necessary for the derepression of the donor loci in the switching, sng1− mutant strain.
It is possible that the derepression of the silent loci in the sng1− mutation is due to the effect of pheromone elaborated by the cells of opposite mating type that are present in the h90 strains but not in the nonswitching (e.g., Msmto) strains. To test this, we mixed cells of the PΔ17 sng1− strain with either an Msmto strain or an Msmto sng1− strain on sporulation medium described earlier (13, 21). It has been shown (13, 21) that under such conditions, the cells of Plus (P) mating type respond to the pheromone produced by the cells of Minus (M) mating type by forming conjugation tubes leading to zygote formation, which leads subsequently to sporulation. We found that zygotes were indeed formed between PΔ17 sng1− strain and either the Msmto or Msmto sng1− strain. While mating of PΔ17 sng1− with Msmto sng1− cells produced zygotes which did not sporulate, mating of PΔ17 sng1− with the Msmto strain produced zygotic asci with four spores, though these were larger than normal wild-type asci. More important, however, no haploid meiosis was observed in either mating (data not shown). These results rule out the possibility that the derepression of the donor loci in the h90 sng1− strain was due to pheromone effect and also suggest that sng1− mutation does not affect pheromone responsiveness which is needed for zygote formation. It is also unlikely that sng1− mutation somehow triggers an alternate pheromone response pathway that, in turn, controls silencing.
We also tested whether the sng1− mutant is defective in healing the DSB at the mat1 locus (4), leading to persistence of the DSB for a longer time. However, this possibility was ruled out by Southern blot analysis indicating that the sng1− mutant exhibits quantitatively the same level of DSB at the mat1 locus as the wild-type cells (data not shown).
The sng1− mutant is encoded by rhp6, the RAD6 homolog in S. pombe.
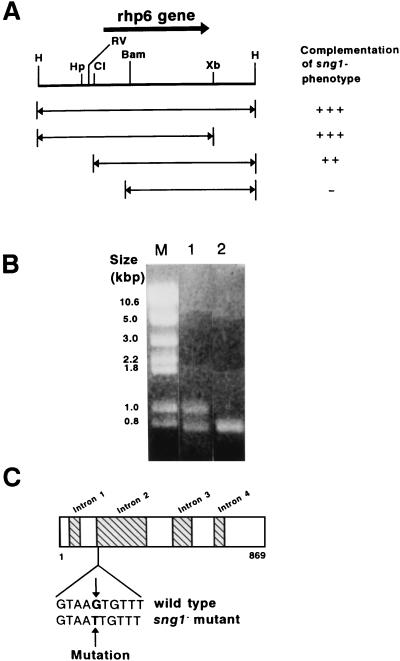
We isolated a single sng1+ clone from the partial HindIII genomic library of S. pombe by complementation of the temperature sensitivity and sporulation defect of the sng1− mutant (44, 70). The complementing plasmid containing a 3.2-kb HindIII insert, along with the S. cerevisiae LEU2 gene which complements the S. pombe leu1− defect (44), was integrated into the genome of the h90 leu1− sng1− strain by homologous recombination. The resulting strain showed complementation of both the haploid meiosis and temperature-sensitive growth defects of the sng1− mutation. When such an integrant was crossed with an h90 leu1− strain, each of the 14 asci analyzed gave a 2 Spo+ Leu+:2 Spo+ Leu− segregation pattern, with no segregant exhibiting the haploid meiosis or temperature-sensitive phenotypes. Thus, the plasmid was integrated at a site tightly linked to the sng1 locus, indicating that we had cloned the sng1+ locus. Subsequent subcloning showed that the region spanned by the EcoRV and BamHI sites was necessary and the region between the EcoRV and XbaI sites was sufficient for complementation (Fig. 5A). Sequencing of this region revealed that it encodes the gene rhp6, which is the S. pombe homolog of the RAD6 gene of S. cerevisiae (49). The rhp6 gene was previously isolated by Reynolds et al. (50).
FIG. 5.
The sng1− mutant is encoded by the rhp6+ gene. (A) Complementation of the sng1− genotype was tested by subcloning different fragments, as indicated, into pIRT1, transforming them into the h90 sng1− mutant strain, and checking for iodine staining and restoration of the temperature-sensitive phenotype. Restriction sites: H, HindIII; Hp, HpaI; RV, EcoRV; Cl, ClaI; Bam, BamHI; Xb, XbaI. (B) RT-PCR products of RNA for wild-type and mutant strains resolved by agarose gel electrophoresis. Lane M, molecular size markers; lane 1, rhp6−/sng1− mutant; lane 2, wild type. (C) Localization of the sng1-1 mutation in the second intron of the rhp6 gene. The gene map showing the intron and exon positions is based on data in reference 50.
To map the mutation in the rhp6−/sng1− mutant, we decided to amplify and sequence the rhp6 mRNA from the rhp6−/sng1− mutant by RT-PCR under conditions beyond the logarithmic phase of amplification. Gel analysis showed the expected band of 0.8 kb, which presumably originates from processed, intronless RNA in both the mutant and wild-type cells (Fig. 5B, lanes 1 and 2, respectively). Interestingly, however, the mutant also yielded an additional band of about 1.0 kb (lane 1). Since the rhp6 gene is known to contain four introns (50) (Fig. 5C), we thought that the larger 1.0-kb band may represent an incompletely spliced form of RNA. Sequencing of the two bands after cloning indicated that the 1.0-kb clone also contains the 200-bp second intron of rhp6 gene and revealed a transversion mutation (G to T) of the 5′ splice junction of the second intron at the fifth base position (GTAAG to GTAAT [Fig. 5C]). We believe that this mutation may affect the efficiency of splicing and thus reduce the cellular levels of rhp6 protein. The relative levels of the fully processed mRNA and the RNA still containing the second intron were quantitated by RT-PCR in the logarithmic phase of amplification (8). We find that the normal and unprocessed forms of rhp6 mRNA are present in the ratio of ∼1:10, indicating that the level of mature rhp6 mRNA is reduced to about 10% of the level found in wild-type cells (data not shown). It is surprising that a single-point mutation in the fifth base of the 5′ splice junction sequence has such a drastic effect on splicing. However, a similar inhibitory effect of a mutation in the fifth base of the 5′ splice site in the 12S RNA of the E1A gene on splicing has been shown earlier and found to be suppressed by a compensatory base change in the U1 snRNA (72).
Since disruption of the rhp6 gene is known to cause a defect in sporulation and UV-induced DNA repair (50), we checked for similar phenotypes in the rhp6−/sng1− mutant. We find that this mutant does indeed have a lower level of sporulation and greater UV sensitivity than those of wild-type cells, which are restored to normal levels upon homologous integration of the rhp6+ gene (data not shown). At different UV doses tested, the sng1−/rhp6− mutant exhibits a 10-fold-higher level of survival than rhp6Δ strain (data not shown). These data are understandable, given that the rhp6−/sng1− mutant exhibits a 10-fold reduction in the level of mature rhp6 mRNA.
The rhp6−/sng1− mutation alters the chromatin structure of the donor loci in a switching-dependent manner—a possible role of rhp6 in chromatin assembly.
Since the sng1 gene encodes the rhp6 protein, and the S. cerevisiae homolog RAD6 has been shown to be involved in conjugating ubiquitin to proteins including histones H2A and H2B (29), rhp6 may play a role in chromatin assembly of the donor mating-type loci. To check this possibility, the chromatin structure of the silent mating-type loci mat2P and mat3M was analyzed by using the in vivo-expressed E. coli dam methylase probe. We had shown earlier that the E. coli dam methylase activity, when expressed in S. cerevisiae (which also lacks DNA methylation), methylates the adenine residues in the Sau3AI sites (5′GATC3′) preferentially in the active genes compared to the inactive genes (58). Since S. pombe also lacks any DNA methylation (2), we presume that the E. coli dam methylase can serve as a probe for chromatin accessibility in this yeast as well. Therefore, we generated S. pombe strains in which the E. coli dam methylase is integrated at the ura4− locus (not shown). The resulting strains have normal growth rates and cellular phenotypes including sporulation, indicating that dam methylation has no deleterious effect on essential cellular functions. Moreover, a significant fraction of genomic DNA was found to be methylated in these strains (not shown).
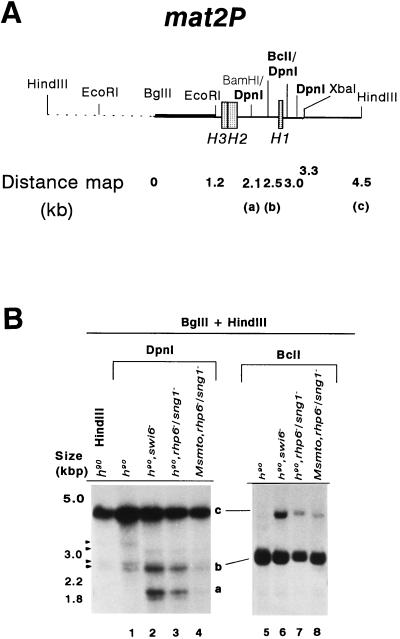
Methylation of mating-type loci was monitored in wild-type and mutant strains by using restriction enzymes that either are inhibited by methylation or are specific for methylated sequences followed by Southern blotting and indirect end labeling. Methylation of the adenine at the N-6 position of the GATC sequence renders it resistant to digestion with BclI and DpnII, while DpnI cleaves GATC only when dimethylated (where the adenine residues on both the DNA strands are methylated). The results show very little methylation of sites in wild-type cells (Fig. 6B, lane 1; Table 3), while in the swi6− mutant, a total of about 34% of the mat2P DNA is dimethylated at sites a and b, as indicated by cleavage with DpnI (Fig. 6B, lane 2; Table 3). The swi6− strain serves as a positive control for derepression of silent loci (14). The increased accessibility to dam methylase in the swi6− mutant provides the first direct evidence that the donor cassettes are silenced due to a repressive chromatin structure and that the swi6 protein is involved in establishing or stabilizing such a structure. Similarly, nearly 25% of the mat2P DNA is also dimethylated at the two sites in h90 rhp6−/sng1− strain, as indicated by increased DpnI digestion (Fig. 6B, lane 3; Table 3). Interestingly, in contrast to h90 rhp6−/sng1− strain, the nonswitching Msmto rhp6−/sng1− strain exhibits a much reduced level (3.1%) of total methylation of the sites (Fig. 6B, lane 4; Table 3). Quantitation of the data with DpnII yields a similar result (not shown). On the other hand, the BclI site (site b) is methylated to only a slightly higher level, as indicated by the level of the uncut band c, in the switching, h90 rhp6−/sng1− strain (13.5% [Fig. 6B, lane 7; Table 3]) compared to the nonswitching Msmto rhp6−/sng1− strain (8.3% [Fig. 6B, lane 8; Table 3]). This site was also significantly methylated in the swi6− strain (25.5% [Fig. 6B, lane 6; Table 3]). Thus, the difference of methylation between h90 rhp6−/sng1− and Msmto rhp6−/sng1− strains as indicated by BclI digestion is much less pronounced than the DpnI digestion. The basis of this difference may lie in the fact that DpnI measures only dimethylation, while BclI is sensitive to both hemi- and dimethylation; the component of hemimethylation is high but similar in both strains, and this may give an apparent reduction in difference in the level of methylation (hemi- and dimethylation) between expressed and repressed states, as monitored by BclI. Therefore, our data indicate that expression of the silent loci is better correlated with dimethylation and that the h90 rhp6−/sng1− mutant (where silent cassettes are expressed) shows an ∼8-fold-greater total dimethylation of sites than the Msmto rhp6−/sng1− mutant, where the silent cassettes are not expressed (Fig. 3A and B; Table 3). Likewise, it was shown earlier that in S. cerevisiae strains expressing the dam methylase, while the level of hemimethylation of sites within the promoter and coding region of the linked genes GAL1-10 was nearly the same in both the repressed and expressed states, the level of dimethylation of both the sites was approximately 10-fold greater in the expressed state than in the repressed state, indicating that the level of dimethylation was directly correlated with the state of expression of the gene (58).
FIG. 6.
Increased methylase accessibility of the mat2P locus in the rhp6−/sng1− mutant is dependent on its utilization for switching. (A) The restriction map of the 6.3-kb HindIII mat2 fragment shows the distance of various Sau3AI sites distal to the BglII site. (B) Southern blot analysis. Two DNA samples (1 to 2 μg) isolated from strains carrying the chromosomally integrated E. coli dam methylase gene were digested with BglII plus HindIII, followed by DpnI (lanes 1 to 4) or BclI (lanes 5 to 8). The first lane represents the DNA from the h90 strain digested with BglII plus HindIII alone. The samples were subjected to electrophoresis; after electrophoresis, the gel was blotted, and the samples were hybridized with the radiolabeled BglII-EcoRI fragment (thick bar in panel A) and subjected to autoradiography. Fragments labeled a, b, and c are defined in panel A. A low level of hybridization was observed consistently due to cross-hybridization elsewhere in the genome, possibly in the centromeric regions as indicated by the arrowheads. One of these cross-hybridizing bands also comigrates with the band at 2.5 kb. The level of methylation of band b was quantitated by subtracting the percent peak area corresponding to the band in the control lane from that determined for lanes 1 to 4 (Table 3). A faint band observed at roughly the position of band a in the BclI digests (lanes 5 to 8) may also arise due to cross-hybridization with centromeric regions, since there is no BclI site at the position of band a (26).
TABLE 3.
Increased methylation accessibility of the mat2P locus in the rhp6−/sng1− mutant depends on the switching competence of the strain
| Genotype | % Methylationa
|
|||
|---|---|---|---|---|
| Dimethylation (DpnI)
|
Hemi- + dimethylation (BclI), site b | |||
| Site a | Site bb | Sites a + b | ||
| h90 | 0.0 | 0.0 | 0.0 | 0.9 |
| h90 swi6− | 23.8 | 10.0 | 33.8 | 25.5 |
| Msmto swi6− | 31.2c | 8.9c | 40.1c | ND |
| h90 rhp6−/sng1− | 17.4 | 7.5 | 24.9 | 13.5 |
| Msmto rhp6−/sng1− | 1.5 | 1.6 | 3.1 | 8.3 |
Quantitation by densitometry of the autoradiograph shown in Fig. 6B.
The area of the band comigrating at the position of site b in the control lane in Fig. 6B was quantitated and subtracted from the areas determined for band b in lanes 1 to 4. ND, not determined.
Data from Fig. 8 were quantitated.
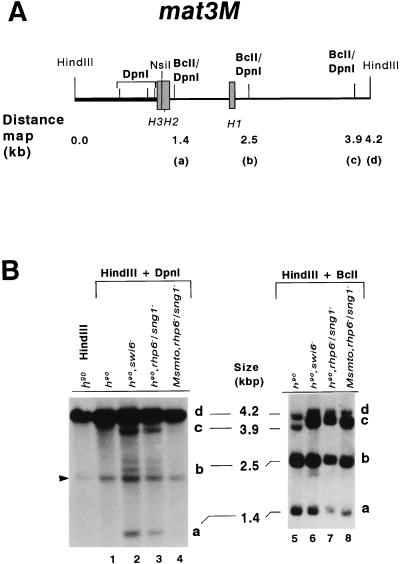
More dramatic results were obtained from the analysis of the mat3M locus. The two centromere-proximal sites (Fig. 7A) in the mat3M locus are unmethylated, as they were completely digested by the methylation-sensitive enzyme DpnII (data not shown) but not cleaved at all by DpnI in the mutant strains, as indicated by absence of any bands smaller than 1.2 kb (Fig. 7B, lanes 2 and 3). However, as indicated by the increased intensity of the bands, DpnI sites a and b, located within the mat3M allele-specific region, and centromere-distal site c are all significantly dimethylated in the h90 swi6− (27.9% [Fig. 7B, lane 2; Table 4]) and h90 rhp6−/sng1− (26.4% [Fig. 7B, lane 3; Table 4]) mutants, while no methylation is observed in the wild-type strain (Fig. 7B, lane 1; Table 4). More interestingly, like the results obtained for mat2P, these sites are significantly more dimethylated (nearly 26-fold) in the h90 rhp6−/sng1− strain (26.4% [Fig. 7B, lane 3; Table 4]) than in the Msmto rhp6−/sng1− strain, where no significant dimethylation is observed (∼1% [Fig. 7B, lane 4; Table 4]). Examination of the levels of hemi- and dimethylation for sites a plus b, calculated from the BclI digests, shows no difference of methylation between the h90 rhp6−/sng1− (51.8%) and Msmto rhp6−/sng1− (55.2%) strains (Fig. 7B, lanes 7 and 8, respectively; Table 4), though the overall level of methylation in these strains as well as the swi6− strain (50.2%) is higher than that in the wild-type strain (26.7% [Fig. 7B, lane 5; Table 4]). These data again indicate that derepression of the silent loci in h90 swi6− and h90 rhp6−/sng1− strains is directly correlated with dimethylation of the Sau3AI sites in the loci; these sites are not significantly dimethylated in the Msmto rhp6−/sng1− strain, which also does not exhibit a silencing defect.
FIG. 7.
Increased methylase accessibility of the mat3M locus in the rhp6−/sng1− mutant also depends on the switching competence. (A) The restriction map of the 4.2-kb HindIII mat3M DNA fragment shows the distances of different BclI and DpnI sites from the HindIII site. (B) Southern blot analysis. DNA samples (1 to 2 μg) isolated from strains carrying the chromosomally integrated E. coli dam methylase gene were digested sequentially with HindIII plus DpnI (lanes 1 to 4) and HindIII plus BclI (lanes 5 to 8). The first lane represents the DNA from the h90 strain digested with HindIII alone. The samples were subjected to electrophoresis; after electrophoresis, the gel was processed for blotting and hybridization with the NsiI HindIII fragment, indicated by the thick bar in panel A. The three bands corresponding to cleavage at the BclI sites are labeled a, b, and c; the uncut 4.2-kb HindIII band is labeled d. A common band (arrowhead in panel for DpnI digestions) arises from cross-hybridization with sequences elsewhere in the genome, possibly in the centromeric regions, and was not considered for quantitation. The arrow indicates the presence of an additional polymorphic Sau3AI site present in our strains but not found in published sequence. This band was also quantitated by densitometry and combined along with band b in Table 4. The centromere-proximal sites are also indicated on the left side of the map in panel A.
TABLE 4.
Increased methylation accessibility of the mat3M locus in the rhp6−/sng1− mutant is also switching dependent
| Genotype | % Methylationa
|
||||
|---|---|---|---|---|---|
| Hemi- + dimethylation (BclI lane), sites a + b + cb | Dimethylation (DpnI lane)
|
||||
| Site a | Site bb | Site c | Sites a + b + cb | ||
| h90 | 26.7 | 0.0 | 0.0 | 0.0 | 0.0 |
| h90 swi6− | 50.2 | 4.9 | 5.9 | 17.1 | 27.9 |
| Msmto swi6− | ND | 5.9c | 5.0c | 19.6c | 30.5c |
| h90 rhp6−/sng1− | 51.8 | 4.1 | 5.5 | 16.8 | 26.4 |
| Msmto rhp6−/sng1− | 55.2 | ∼0.2 | ∼0.4 | ∼0.4 | ∼1.0 |
Quantitation by densitometry of the autoradiograph shown in Fig. 8B.
For quantitation, the contribution of the slower-migrating band at 2.7 kb in Fig. 7B (lanes 2 and 3), indicated by the arrowhead, was also included. This site is not indicated in the map in Fig. 7 based on published sequence and may represent a polymorphic site. ND, not determined.
Data from Fig. 8 were quantitated.
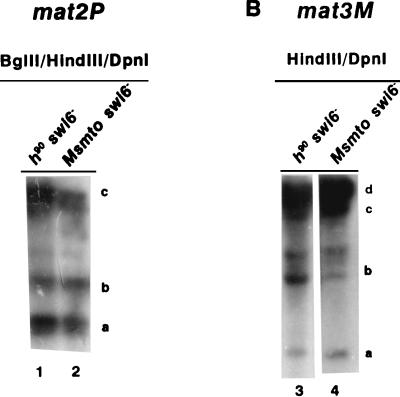
To check whether the methylation accessibility in the swi6− mutant was switching dependent, we carried out the same experiments within an Msmto swi6− strain. Results indicate that both for mat2P (Fig. 8A, lanes 1 and 2) and mat3M (Fig. 8B, lanes 3 and 4), the extents of DpnI digestion at different sites were roughly similar for h90 swi6− (Fig. 8, lanes 1 and 3) and Msmto swi6− (Fig. 8, lanes 2 and 4) strains (Tables 3 and 4). These results indicate that in contrast to the rhp6−/sng1− mutation, the swi6− mutation affects the chromatin structure at the donor loci independently of their switching status.
FIG. 8.
The methylation accessibility of donor loci in the swi6− mutant is independent of switching. DNA samples from h90 swi6− (lanes 1 and 3) and Msmto swi6− (lanes 2 and 4) mutants were digested with BglII, HindIII, and DpnI (A, lanes 1 and 2) or HindIII plus DpnI (B, lanes 3 and 4) and subjected to electrophoresis and Southern hybridization for mat2P (A) and mat3M (B) as described in the legends to Fig. 6 and 7, respectively.
The above results lead us to conclude that the rhp6−/sng1− mutant has an unfolded chromatin structure at the mat2P and the mat3M donor loci and, more importantly, that this unfolding is dependent on switching competence of these loci. We infer that rhp6 may affect chromatin assembly in the mat region subsequent to mating-type switching. In the absence of rhp6 protein, the initial inactive chromatin structure, which was presumably perturbed during switching, is not fully reestablished following switching, rendering it more accessible to dimethylation by the E. coli dam methylase.
DISCUSSION
A novel role of rhp6/RAD6 in gene regulation by modulating the chromatin structure of mating-type loci in fission yeast.
The present study makes several novel findings. First, we demonstrate for the first time that rhp6 regulates the expression of the silent mating-type loci in S. pombe. Second, this effect is brought about by alteration in the chromatin structure of the silent mating-type loci. Third and most important, we show that the silencing defect and chromatin unfolding in the rhp6−/sng1− mutant are switching dependent. Fourth, we show that swi6 regulates silencing by controlling the chromatin structure of donor loci. The above results suggest a role of rhp6 at the level of chromatin assembly. We offer the explanation that the chromatin structure of the donor loci is perturbed during switching; it must be fully reestablished in rhp6+ cells but not in the rhp6−/sng1− cells, thus leading to the derepression of the donor loci subsequent to their replication required for mat1 switching. Thus, rhp6 plays a role in propagating the chromatin structure and the expression status of mating-type loci from the mother to the daughter cells. The effect is unique to the mating-type loci since the expression of other genes such as hta1, htb1, nmt1, and pho1 was not affected in the mutant (data not shown). In addition, we show that dam methylase can also serve as an in vivo probe for chromatin structure in fission yeast (see below).
In recent years, several proteins involved in DNA repair have also been shown to regulate gene expression. For example, the human Rad3 and Rad25 proteins, which are involved in DNA excision repair, in fact either associate with (Rad25) or are the subunits (Rad3) of the general transcription factor TFIIH (17, 56, 67). Though rhp6 and its homologs in other species have been known for several years, its actual biochemical function has not been clear. Earlier reports have demonstrated elevation and randomization of Ty transposition in the vicinity of certain genes in rad6− mutants in S. cerevisiae (37, 47), suggesting that Rad6 may affect the chromatin structure of the genes so as to make them better targets for Ty transposition or recombination. Disruption of the HR6B homolog in mice has been shown to cause male infertility, presumably because of defective chromatin transition during spermatogenesis (53). Bryk et al. (6) have shown that deletion of RAD6 in S. cerevisiae leads to derepression of Ty1 RNA levels originating specifically from the copy of Ty1 integrated within the RDN1 locus. In an interesting study, a deubiquitinating enzyme was shown to interact with Sir4 protein, which regulates silencing of mating-type and telomeric loci in S. cerevisiae (42). Our recent results also show that at the nonpermissive temperature (36°C), the rhp6−/sng1− mutant as well as the rhp6Δ strain exhibit the cut (cells untimely torn) phenotype. The defect seems to be at the level of chromosome compaction and segregation, which is corroborated by fluorescence-activated cell sorting analysis (data not shown). However, experiments using hydroxyurea sensitivity indicate that the mutant is not defective in the replication check point control (data not shown). Thus, rhp6 may also be involved in global chromatin organization, integrity, and segregation during mitosis. Indeed, very recently it has been shown that rad6Δ causes a weak effect on the expression of hml::URA3 in heterothallic strain of S. cerevisiae, while a stronger effect was observed on expression of telomere-linked markers (27). It may be interesting to see if the rad6− mutant exhibits a stronger silencing defect at mating-type loci in switching strains of S. cerevisiae.
Biochemical functions of rhp6/Rad6.
The main biochemical function of the Rad6 protein is to conjugate ubiquitin to histones H2A and H2B (29). Ubiquitin is an evolutionarily conserved small polypeptide of 76 amino acids, and its conjugation to other proteins triggers their degradation by the N-end rule pathway (19, 43, 66). It was initially hypothesized that RAD6-mediated ubiquitination of chromosomal proteins, especially of histones H2A and H2B, is essential for chromatin remodeling and that this effect might explain the various phenotypes of rad6 mutants (29). However, subsequent work failed to establish the function of ubiquitination of H2A and H2B, since the defects of RAD6 deletion were not mimicked by the mutation of the conserved ubiquitination site in histones (61). It remains possible, however, that rad6 ubiquitinates some other nonhistone components of chromatin which participate in chromatin assembly. Because of the observed switching dependence for the derepression and chromatin unfolding of the silent donor loci in the rhp6−/sng1− mutant, we propose that certain proteins that participate in chromatin assembly may be transiently associated with chromatin during replication and assist in chromatin assembly. After chromatin assembly, these proteins may become ubiquitinated and channeled to the proteolytic degradation pathway, and possibly their removal leads to faithful reassembly of the repressed chromatin structure at the donor loci. Alternatively, ubiquitination may directly cause the dissociation of such a protein from the newly assembled chromatin structure. In rhp6−/sng1− mutants, because of reduction in the level of ubiquitination, the increased stability and persistently high levels of the protein may interfere with the maturation of proper chromatin structure. (The latter possibility is considered equally likely at this point since the UBR1 gene, which encodes the E3 enzyme involved in N-end rule degradation in S. cerevisiae, is not involved in silencing [27].) Such a hypothetical model is diagrammed in Fig. 9. This line of thinking obtains support from a recent report that UBP3, a deubiquitinating enzyme that interacts with one of the silencing proteins SIR4 in S. cerevisiae, is an inhibitor of silencing; UBP3 disruption causes a marked increase in silencing (42). All of these studies suggest that a fine balance of ubiquitination and deubiquitination of key regulatory proteins may constitute a key regulatory mechanism for control of chromatin structure of mating-type loci in S. cerevisiae and S. pombe.
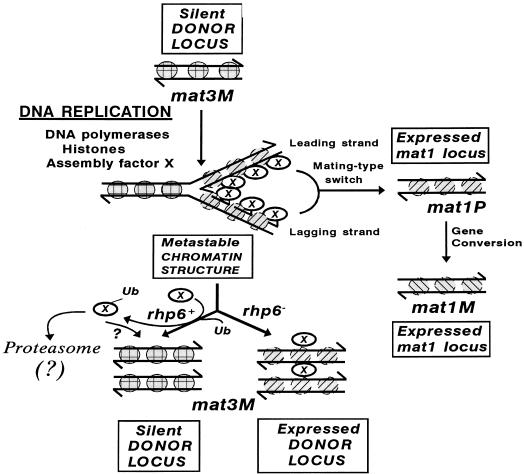
FIG. 9.
A hypothetical model for the role of rhp6 protein in chromatin assembly. The putative chromatin assembly protein (X), which is transiently expressed during cell cycle and associates with the newly assembled nucleosomes at the replication fork, may help in regulating the proper assembly of an inactive structure, for which its timely removal through ubiquitination by rhp6 and subsequent degradation is very critical. Alternatively, the ubiquitinated (Ub) protein X may remain associated with freshly assembling nucleosomes and play a structural role. In the absence of direct evidence for such a function in this system, ? indicates this possibility as an alternative to that of proteolysis by the proteasome. Nucleosomes of the freshly replicated daughter chromatids in the middle are shaded differently from the parental and the finally assembled inactive daughter chromatids and represent their perturbed state (metastable or potentially active). A close coordination of silencing with switching is indicated by depicting the transposition of the replicated mating-type donor locus to the mat1 locus.
The silencing defects of rhp6−/sng1− mutation are unique in comparison to other silencing mutants of S. pombe.
We find several aspects of the silencing phenotype of the rhp6−/sng1− mutant to be unique and intriguing. Unlike other silencing mutations, namely, swi6, clr1-clr4, and rik1 (14, 38, 63, 64), the rhp6−/sng1− mutation does not abolish the cold spot of recombination between the mat2 and mat3 loci and does not elevate the expression of the ura4 gene placed next to the mat2 and mat3 loci (data not shown). Even more interesting, the silencing defect in the rhp6−/sng1− mutant is dependent on the switching competence, whereas the defect in swi6, clr1-clr4, and rik1 mutants occurs in both switching and nonswitching strains (63, 64) or in strains where the expressed mat1 locus has been deleted (14), indicating their lack of dependence on switching. The structure of donor loci does become more accessible to dam methylase in both swi6− and rhp6−/sng1− mutants. However, there is one distinct difference: the unfolding and increased accessibility of chromatin of the mat2P and mat3M loci to dam methylation, especially dimethylation, in the rhp6−/sng1 mutant is greater in the switching background than in the nonswitching background, while in the swi6− mutant the increased chromatin accessibility is independent of switching. Therefore, the effects of swi6, clr1-clr4, and rik1 mutations may be exerted at the higher level of chromatin organization at the donor loci and the intervening K region, perhaps by a complex of these proteins, as suggested earlier (25, 63). It is pertinent to note here that the K region exhibits various degrees of homology to the centromeric sequences along its length: the central 4.3-kb region shows >90% homology, while the flanking regions show progressively decreasing levels of homology (26). It is a moot question whether this region plays a role analogous to that of centromere sequences in being organized into either a nonnucleosomal structure or a complex formed of products of swi6 and clr1-4 genes with nucleosomal histones (1, 26). However, rhp6 may play a different role: it may be required for coupling the process of switching, which involves DNA replication of the donor cassettes, with their chromatin assembly. A closer inspection of the methylation data reveals that GATC sequences at the two ends of the K region (sites marked 2.5 and 3.0 kb on the right side of mat2P locus in Fig. 6A and the three sites located on the left side of mat3M in Fig. 7A) are not dimethylated at all in the rhp6−/sng1− mutant. (These regions have been shown to have a weak homology to the centromeric sequences of cen2 and cen3 [26], which may account for the fainter bands in Fig. 6 and 7.) Thus, rhp6 does not affect the chromatin organization of the K region, which is consistent with lack of any effect on the recombinational and transcriptional cold spot in the rhp6−/sng1− mutant.
The apparent loss of repression of mat2Pc transcript in Msmto rhp6− strain but the lack of haploid meiosis phenotype in this strain may be explained as follows. The level of mat2Pc transcript expression in the Msmto rhp6−/sng1− mutant is similar to that in the wild-type h90 strain, which also lacks haploid meiosis (Fig. 3A, compare lanes 6 and 2). It is possible that the comparable but low levels of mat2Pc transcript in Msmto rhp6−/sng1− and h90 rhp6+ strains are not sufficient, as a certain threshold level of this transcript may be required for induction of meiosis. This level is elevated further >10-fold in the h90 rhp6−/sng1− strain (Fig. 3A, lane 4), which may exceed the threshold level needed to trigger meiosis. (Actually, it may not be mat1Pc as such that directly drives meiosis but rather mat2Pi, which probably is transcribed less but cannot be assayed in a mat2-specific manner.) Interestingly, the leaky expression of mat2Pc in the wild-type h90 strain is also switching dependent (Fig. 3A, lanes 2 and 8). It is worth mentioning here that this leaky expression of mat2Pc transcript has not been reported earlier because either heterothallic (64) or mat1Δ (14) strains had been used. A possible link of switching and silencing has recently been suggested by the finding of the gene DIS1, whose deletion drastically reduces the level of mating-type switching and whose overexpression interferes with silencing of mating-type loci in S. cerevisiae (71). Since rhp6−/sng1− mutation affects switching-dependent silencing, we tested whether it may also affect the directionality of switching. Our quantitative PCR data show that the relative levels of mat1P and mat1M sequences are similar in h90 and h90 rhp6−/sng1− strains, indicating that the rhp6−/sng1− mutation does not affect the directionality of switching (data not shown).
Development of dam methylation as an in vivo probe for monitoring chromatin structure in fission yeast.
We have shown earlier that the E. coli dam methylase, when expressed constitutively in strains of budding yeast (which lacks methylation of its own), preferentially dimethylates active rather than inactive genes. This approach has subsequently found widespread use for analyzing the structures of different chromatin regions in budding yeast (23, 32), since it offers the advantages of simpler experimental manipulation and lack of experimental artifacts associated with the in vitro approaches like micrococcal nuclease and DNase I analysis and, most important, it uniquely provides an in vivo assay for the accessibility of yeast genes. Our results showing increased dimethylation of sites in the mat2P and mat3M loci in the swi6− and rhp6−/sng1− mutants in comparison with the wild-type strains confirm that dam methylation can also serve as a probe for accessibility of expressed versus repressed regions of chromatin in fission yeast as well. Our successful application of this technique to two distantly related yeasts suggests that this approach may be applicable to other species as well.
A possible explanation for the roughly constant level of hemimethylation in both expressed and repressed states (this study and reference 58) may be the occurrence of a hypothetical narrow time window of S phase when freshly replicated hemimethylated DNA strands are generated from a dimethylated or unmethylated template. In repressed state, rapid reassembly of inaccessible structure prevents further methylation, but in the expressed state the more accessible structure allows methylation of the hemimethylated sites to yield the increased level of dimethylation.
Conclusions and significance.
Maintenance of the differentiated state of a eukaryotic cell requires that the chromatin structure and expression status of cell-type-specific genes be reestablished after DNA replication, when the nucleosome structure at the replication fork is known to be perturbed. Conceivably, the duplication of DNA must be coordinated in vivo by simultaneous assembly of nucleosomes at the replication fork (reviewed in reference 69). The transient perturbation of nucleosomes during replication is likely to alter the transcriptional activity of the gene. Thus, it is paramount that the original chromatin structure be reestablished in the differentiated cell following every replication cycle. The problem of reestablishment of the higher-order structure is particularly challenging in case of inactive genes that have a folded structure, which is inaccessible to transcription machinery and probes like DNase I and dam methylase (58). Therefore, it is likely that eukaryotic cells have a molecular mechanism that couples DNA replication to reestablishment of the chromatin structure. Notably, a recent study has shown a coupling of chromatin assembly at the templates undergoing DNA repair by the chromatin assembly factor CAF-I (22), which had previously been shown to play a role in assembly of nucleosomes on replicating DNA (60). More interestingly, an intimate role of the chromatin assembly genes CAC1 and CAC2 (30) and RLF2 (16) in telomere silencing has also been demonstrated in budding yeast.
Thus, our results provide support for the suggestion that a transient window of opportunity is available during chromosome replication to remodel chromatin structure, a general mechanism postulated for modulating gene expression in development (69). It is likely that rhp6 acts globally either directly or indirectly in reestablishment of chromatin structure of the mat1, mat2, and mat3 loci after DNA replication and switching. The rhp6-mediated ubiquitination of a key chromatin assembly protein may be a critical event in this process as proposed in our hypothetical model (see above and Fig. 9). Interesting work by Bailly et al. (3) has demonstrated that both RAD6 (of S. cerevisiae) and rhp6 (of S. pombe) interact physically with the UBR1 protein (required for N-end rule proteolysis [66]) and with RAD18 (which binds to single-stranded DNA). Such interactions may target RAD6 to proteolytic substrates to effect protein degradation during DNA replication, recombination, and repair, which may also affect chromatin assembly. Additionally, it is possible that other proteins involved in DNA repair, such as RAD18, also participate in switching-mediated chromatin alteration. Therefore, the observations in this paper point to an underlying link between mating-type switching by recombination and DNA replication and repair on the one hand and gene regulation by modulation of chromatin assembly through ubiquitination on the other. Further genetic and biochemical work is needed to identify the components that couple DNA replication to faithful chromatin assembly at the mating-type loci.
ACKNOWLEDGMENTS
This research was partly sponsored by the National Cancer Institute, DHHS, under contract with ABL. A major part of the work was carried out at the Institute of Microbial Technology, Chandigarh, India, with the support of the Department of Science and Technology and Council of Scientific and Industrial Research, New Delhi, India.
We thank Paul Young and David Beach for providing the S. pombe genomic library, Louise Prakash for the strains carrying the rhp6 disruption, Joey Hopkins and Shashi Batra for preparing the manuscript, Anne Arthur for editorial suggestions, K. Ekwall for plasmid pKE10, A. Naresh and K. Ganesan for critically reading the manuscript, R. N. Dubey for computer assistance, and K. Singh and Anil Theophilus for help with photography. The kind help of M. R. S. Rao, coordinator of the DBT National Sequencing Facility at the Indian Institute of Science, Bangalore, India, with sequencing is also gratefully acknowledged.
REFERENCES
- 1.Allshire R C, Nimmo E R, Ekwall K, Javerzat J P, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt centromeric segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 2.Anteguera F, Tamame P, Villanueva J, Santos T. DNA methylation in fungi. J Biol Chem. 1984;259:8033–8036. [PubMed] [Google Scholar]
- 3.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin conjugating activity to DNA damage sites. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 4.Beach D. Cell type switching by DNA transposition on fission yeast. Nature. 1983;305:682–688. [Google Scholar]
- 5.Bell S P, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcriptional silencing and DNA replication. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- 6.Bryk M, Banerjee M, Murphy M, Knudson K E, Garfinkel D J, Curcio M J. Transcriptional silencing of Ty1 element in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 7.Carpenter P B, Mueller P R, Dunphy W G. Role for a Xenopus ORC2-related protein in controlling DNA replication. Nature. 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- 8.Chelly J, Kahn A. RT-PCR and mRNA quantitation. In: Mullis K B, Ferre F, Gibbs R A, editors. The polymerase chain reaction. Boston, Mass: Birkhauser; 1994. pp. 97–109. [Google Scholar]
- 9.Egel R. Two tightly linked silent cassettes in the mating type region of Schizosaccharomyces pombe. Curr Genet. 1984;8:199–203. doi: 10.1007/BF00417816. [DOI] [PubMed] [Google Scholar]
- 10.Egel R. Mating-type genes, meiosis, and sporulation. In: Nasim A, Young P, Johnson B F, editors. Molecular biology of the fission yeast. New York, N.Y: Academic Press; 1989. pp. 31–73. [Google Scholar]
- 11.Egel R, Beach D, Klar A. Genes required for initiation and resolution steps of mating type switching in fission yeast. Proc Natl Acad Sci USA. 1994;81:3481–3485. doi: 10.1073/pnas.81.11.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egel R, Willer M, Nielsen O. Unblocking of meiotic crossing-over between the silent mating-type cassettes of fission yeast, conditioned by the recessive, pleiotropic mutant rik1. Curr Genet. 1989;15:407–410. [Google Scholar]
- 13.Egel R, Willer M, Kjaerulff S, Davey J, Nielsen O. Assessment of pheromone production and response in fission yeast by a halo test of induced sporulation. Yeast. 1994;10:1347–1354. doi: 10.1002/yea.320101012. [DOI] [PubMed] [Google Scholar]
- 14.Ekwall K, Nielsen O, Ruusala T. Repression of a mating type cassette in the fission yeast by four DNA elements. Yeast. 1991;7:745–755. doi: 10.1002/yea.320070709. [DOI] [PubMed] [Google Scholar]
- 15.Ekwall K, Ruusala T. Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics. 1994;136:53–64. doi: 10.1093/genetics/136.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enomoto S, McCune-Zierath P D, Gerami-Nejad M, Sanders M A, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 17.Feaver W J, Svesjstrup J Q, Bardwell L, Bardwell A J, Buratowsky S, Gulyas K D, Donahue T F, Friedberg E C, Kornberg R D. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell. 1993;75:1379–1387. doi: 10.1016/0092-8674(93)90624-y. [DOI] [PubMed] [Google Scholar]
- 18.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 19.Finley O, Varshavsky A. The ubiquitin system: functions and mechanisms. Trends Biochem Sci. 1985;10:343–347. [Google Scholar]
- 20.Foss M, McNally F J, Harwood J, Nasmyth K, Diffley J F X. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science. 1993;262:1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- 21.Fukui Y, Kaziro Y, Yamamoto M. Mating pheromone-like diffusible factor released by Schizosaccharomyces pombe. EMBO J. 1986;5:1991–1993. doi: 10.1002/j.1460-2075.1986.tb04454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaillard P-H L, Martini E M-D, Kaufman P D, Stillman B, Moustacchi C, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor-I. Cell. 1996;86:887–898. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 23.Gottschling D E. Telomere proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc Natl Acad Sci USA. 1992;89:4062–4065. doi: 10.1073/pnas.89.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grallert B, Nurse P. The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 1996;10:2644–2654. doi: 10.1101/gad.10.20.2644. [DOI] [PubMed] [Google Scholar]
- 25.Grewal S I S, Klar A J S. Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell. 1996;86:95–101. doi: 10.1016/s0092-8674(00)80080-x. [DOI] [PubMed] [Google Scholar]
- 26.Grewal S I S, Klar A J S. A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics. 1997;146:1221–1238. doi: 10.1093/genetics/146.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H, Kahana A, Gottschling D E, Prakash L, Liebman S. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6693–6699. doi: 10.1128/mcb.17.11.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivy J M, Klar A J S, Hicks J B. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jentsch S, McGrath J P, Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- 30.Kaufman P D, Kobayashi R, Cranston G. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 31.Kelly M, Burke J, Smith M, Klar A, Beach D. Four mating type genes control sexual differentiation in the fission yeast. EMBO J. 1988;7:1537–1544. doi: 10.1002/j.1460-2075.1988.tb02973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kladde M P, Simpson R T. Positioned nucleosomes inhibit dam methylation in vivo. Proc Natl Acad Sci USA. 1994;91:1361–1365. doi: 10.1073/pnas.91.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klar A J S. Molecular genetics of fission yeast cell type: mating type and mating-type interconversion. In: Jones E W, Pringle J R, Broach J, editors. The molecular biology of the yeast Saccharomyces: metabolism and gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 745–777. [Google Scholar]
- 34.Klar A J S, Bonaduce M J. swi6, a gene required for mating-type switching, prohibits meiotic recombination in the mat2-mat3 “cold spot.”. Genetics. 1991;129:1033–1042. doi: 10.1093/genetics/129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lalande M. Parental imprinting and human disease. Annu Rev Genet. 1997;30:173–195. doi: 10.1146/annurev.genet.30.1.173. [DOI] [PubMed] [Google Scholar]
- 36.Li J J, Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 37.Liebman S, Newnam G. A ubiquitin-conjugating enzyme, RAD6, affects the distribution of Ty1 retrotransposon integration positions. Genetics. 1993;133:499–508. doi: 10.1093/genetics/133.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorentz A L, Heim L, Schmidt H. The switching gene swi6 affects recombination and gene expression in the mating type region of Schizosaccharomyces pombe. Mol Gen Genet. 1992;233:436–442. doi: 10.1007/BF00265441. [DOI] [PubMed] [Google Scholar]
- 39.Lyon M F. Gene action in the X chromosome of the mouse (Mus musculus) Nature. 1961;48:39–46. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 40.Micklem G, Rowley A, Harwood J, Nasmyth K, Diffley J F X. Yeast origin recognition complex is involved in DNA replication and transcriptional silencing. Nature. 1993;366:87–89. doi: 10.1038/366087a0. [DOI] [PubMed] [Google Scholar]
- 41.Miller A M, Nasmyth K A. Role of DNA replication in the repression of silent mating type loci in yeast. Nature. 1984;312:247–251. doi: 10.1038/312247a0. [DOI] [PubMed] [Google Scholar]
- 42.Moazed D, Johnson A D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- 43.Monia B P, Ecker D J, Crooke S T. New perspectives on the structure and function of ubiquitin. Biotechnology. 1990;8:209–215. [Google Scholar]
- 44.Moreno S, Klar A, Nurse P. An introduction to molecular genetic analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1990;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 45.Muzi-Falconi M, Kelly T J. Orp1p, a member of cdc18/CDC6 family of S phase regulators, is homologous to a component of the origin recognition complex. Proc Natl Acad Sci USA. 1995;92:12475–12479. doi: 10.1073/pnas.92.26.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olsson T, Ekwall K, Ruusala T. The silent P mating type locus in fission yeast contains two autonomous replicating sequences. Nucleic Acids Res. 1993;21:855–861. doi: 10.1093/nar/21.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Picologlou S, Brown N, Liebman S W. Mutations in RAD6, a yeast gene encoding a ubiquitin-conjugating enzyme, stimulates retrotransposition. Mol Cell Biol. 1990;10:1017–1022. doi: 10.1128/mcb.10.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pillus L, Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989;58:637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- 49.Reynolds P, Weber S, Prakash L. RAD6 gene of Saccharomyces cerevisiae encodes a protein containing a tract of 13 consecutive aspartates. Proc Natl Acad Sci USA. 1985;82:168–172. doi: 10.1073/pnas.82.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reynolds P, Koken M H K, Hoiejmakers J H J, Prakash S, Prakash L. The rhp6+ gene of Schizosaccharomyces pombe: a structural and functional homolog of the RAD6 gene from the distantly related yeast Saccharomyces cerevisiae. EMBO J. 1990;9:1423–1430. doi: 10.1002/j.1460-2075.1990.tb08258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riggs A D, Pfeiffer G D. X-chromosome inactivation and cell memory. Trends Genet. 1992;8:169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- 52.Rivier D H, Rine J. An origin of DNA replication and a transcriptional silencer require a common element. Science. 1992;256:659–663. doi: 10.1126/science.1585179. [DOI] [PubMed] [Google Scholar]
- 53.Roest H P, Klaveren J V, deWit J, vanGrup C G, Koken M H M, Vermey M, van Roijen J H, Hoogerbrugge J W, Vreeburg J T M, Baarends W M, Bootsma D, Grootegoed J A, Hoeijmakers J H J. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell. 1996;86:799–810. doi: 10.1016/s0092-8674(00)80154-3. [DOI] [PubMed] [Google Scholar]
- 54.Russell P. Gene cloning and expression in fission yeast. In: Nasim A, Young P, Johnson B F, editors. Molecular biology of the fission yeast. New York, N.Y: Academic Press; 1989. pp. 243–271. [Google Scholar]
- 55.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 56.Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoiejmakers J H J, Chambon P, Egly J M. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 57.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh J, Klar A J S. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev. 1992;6:186–196. doi: 10.1101/gad.6.2.186. [DOI] [PubMed] [Google Scholar]
- 59.Singh J, Klar A J S. DNA polymerase α is essential for mating-type switching in fission yeast. Nature. 1993;361:271–273. doi: 10.1038/361271a0. [DOI] [PubMed] [Google Scholar]
- 60.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vivo. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 61.Swerdlow P S, Schuster T, Finley D. A conserved sequence in histone H2A which is a ubiquitination site in higher eukaryotes is not required for growth in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4909–4911. doi: 10.1128/mcb.10.9.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson J S, Hecht A, Grunstein M. Histones and the regulation of heterochromatin in yeast. Cold Spring Harbor Symp Quant Biol. 1994;58:247–256. doi: 10.1101/sqb.1993.058.01.029. [DOI] [PubMed] [Google Scholar]
- 63.Thon G, Klar A J S. The clr 1 locus regulates the expression of cryptic mating-type loci in fission yeast. Genetics. 1992;131:287–296. doi: 10.1093/genetics/131.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thon G, Cohen A, Klar A J S. Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region in Schizosaccharomyces pombe. Genetics. 1994;138:29–38. doi: 10.1093/genetics/138.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Triezenberg S T. Primer extension. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Wiley Interscience; 1994. pp. 4.0.3–4.10.8. [Google Scholar]
- 66.Varshavsky A. The N-end rule. Cold Spring Harbor Symp Quant Biol. 1996;60:461–478. doi: 10.1101/sqb.1995.060.01.051. [DOI] [PubMed] [Google Scholar]
- 67.Wang Z, Svejstrup J Q, Feaver W J, Wu X, Kornberg R D, Friedberg E C. Transcription factor b (TFIIH) is required during nucleotide excision repair in yeast. Nature. 1994;368:74–76. doi: 10.1038/368074a0. [DOI] [PubMed] [Google Scholar]
- 68.Wilson C, Bellen H J, Gehring W J. Position effects on eukaryotic gene expression. Annu Rev Cell Biol. 1990;6:679–714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]
- 69.Wolffe A P. Implications of DNA replication for eukaryotic gene expression. J Cell Sci. 1991;99:201–206. doi: 10.1242/jcs.99.2.201. [DOI] [PubMed] [Google Scholar]
- 70.Wright A, Maundrell K, Heyer W-D, Beach D, Nurse P. Vectors for the construction of gene banks and the integration of cloned genes in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Plasmid. 1986;15:156–158. doi: 10.1016/0147-619x(86)90051-x. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Z, Buchman A R. Identification of a member of a DNA-dependent ATPase family that causes interference with silencing. Mol Cell Biol. 1997;17:5461–5472. doi: 10.1128/mcb.17.9.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhuang Y, Weiner A M. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]