Liu et al. describe a new B cell–intrinsic mechanism underlying increased lupus risk in individuals carrying loss-of-function mutations in NADPH oxidase (NOX2) genes. Mechanistically, reduced NOX2 activity in B cells disrupts endolysosomal trafficking, resulting in amplified Toll-like receptor (TLR) signals and the propensity to humoral autoimmunity.
Abstract
Genome-wide association studies in systemic lupus erythematosus (SLE) have linked loss-of-function mutations in phagocytic NADPH oxidase complex (NOX2) genes, including NCF1 and NCF2, to disease pathogenesis. The prevailing model holds that reduced NOX2 activity promotes SLE via defective efferocytosis, the immunologically silent clearance of apoptotic cells. Here, we describe a parallel B cell–intrinsic mechanism contributing to breaks in tolerance. In keeping with an important role for B cell Toll-like receptor (TLR) pathways in lupus pathogenesis, NOX2-deficient B cells exhibit enhanced signaling downstream of endosomal TLRs, increased humoral responses to nucleic acid-containing antigens, and the propensity toward humoral autoimmunity. Mechanistically, TLR-dependent NOX2 activation promotes LC3-mediated maturation of TLR-containing endosomes, resulting in signal termination. CRISPR-mediated disruption of NCF1 confirmed a direct role for NOX2 in regulating endosomal TLR signaling in primary human B cells. Together, these data highlight a new B cell–specific mechanism contributing to autoimmune risk in NCF1 and NCF2 variant carriers.
Introduction
The phagocyte NADPH oxidase (NOX2) complex facilitates the generation of reactive oxygen species (ROS) in phagosomes. Whereas null mutations in NOX2 proteins result in the immunodeficiency syndrome chronic granulomatous disease (CGD) (Thomas, 2018), common polymorphisms have been linked with the pathogenesis of systemic lupus erythematosus (SLE) and other humoral autoimmune diseases. For example, missense variants in both neutrophil cytosolic factor 1 (NCF1 p.Arg90His) and neutrophil cytosolic factor 2 (NCF2 p.His389Gln) are associated with SLE in diverse ethnic populations (Alarcón-Riquelme et al., 2016; Jacob et al., 2012; Kim-Howard et al., 2014; Yu et al., 2011; Zhao et al., 2017). Functionally, these NCF1 and NCF2 variants result in reduced, but not absent, ROS production (Jacob et al., 2012; Zhao et al., 2017). In keeping with NOX2 enzymatic activity impacting the maintenance of immune tolerance, NCF1 copy number variation is also associated with the development of SLE. Carriers of more than three NCF1 copies are protected from lupus development (odds ratio [OR] = ∼0.5), while those with a single NCF1 allele or mothers of boys with X-linked CGD are at markedly increased lupus risk (OR >3) (Cale et al., 2007; Marciano et al., 2018; Olsson et al., 2017; Zhao et al., 2017). These human genetic data are supported by animal studies showing exacerbated lupus severity following deletion of NOX2 family genes in lupus-prone mice (Campbell et al., 2012; Gordon et al., 2022; Jacob et al., 2017).
Thus, genetic evidence strongly implicates reduced NADPH oxidase activity as a driver of lupus pathogenesis. The predominant mechanistic explanation for this disease association focuses on the roles of NADPH activity in regulating myeloid cell activation. NADPH oxidase deficiency has been shown to disrupt the immunologically silent apoptotic cell clearance by macrophages and to increase Toll-like receptor (TLR)–dependent plasmacytoid dendritic cells (pDC) activation (Geng et al., 2022; Meng et al., 2022; Peng et al., 2007). While these myeloid-intrinsic effects likely contribute to increased risk of humoral autoimmunity, the activation of autoreactive B cells by dual signals downstream of B cell receptors (BCR) and endosomal TLR is a critical step in lupus pathogenesis. After binding self-ligands, BCRs traffic nuclear autoantigens to endosomal compartments, where incorporated RNA and DNA can activate the endosomal receptors TLR7 and TLR9, respectively (Shlomchik, 2009). While both myeloid and B cell lineages express endosomal TLRs, alterations in TLR signaling thresholds, for example TLR7 deletion or overexpression, exert a B cell–intrinsic contribution to lupus development in mouse and man (Brown et al., 2022; Jackson et al., 2014; Walsh et al., 2012).
Given this propensity for TLR-driven humoral autoimmunity, B cell TLR signaling pathways must be tightly regulated. In this context, we recently demonstrated that the αv family of integrins promotes endolysosomal trafficking events through autophagy proteins and thus regulates B cell TLR signaling. Using mice deficient in αVβ3 integrin components, we demonstrated that dysregulated endolysosomal flux results in prolonged TLR-dependent NF-κB activation, enhanced B cell proliferation, a striking increase in TLR-dependent germinal center (GC) responses to viral antigens, and the development of humoral autoimmunity (Acharya et al., 2016, 2020; Raso et al., 2018). Since the phagocytic NADPH oxidase promotes the induction of an equivalent non-canonical autophagy program in macrophages (Martinez et al., 2015), we tested the hypothesis that loss-of-function NCF1 and NCF2 variants promote SLE in a B cell–intrinsic manner via modulation of endosomal TLR signaling thresholds.
Results
B cell–intrinsic deletion of NADPH oxidase complex components enhances humoral responses to virus-like particle (VLP) immunization
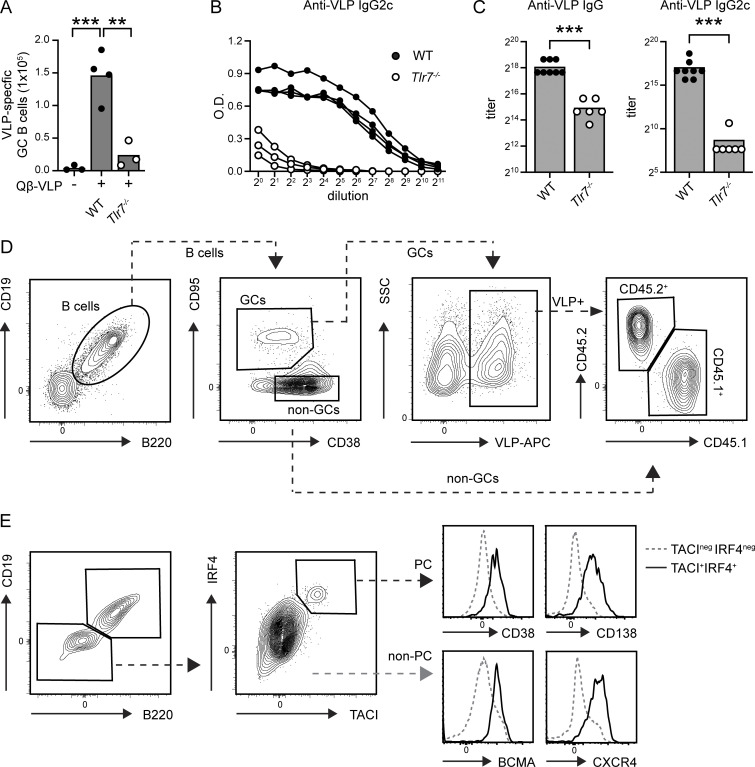
TLR ligands serve as effective vaccine adjuvants, in part by augmenting antigen-specific GC responses (Coffman et al., 2010). Although both myeloid dendritic cells (DC) and B cells express TLRs, DC-specific TLR signals enhance responses to soluble protein antigens chemically linked to a TLR agonist, whereas immunization with VLP, where the nucleic acid (NA) is incorporated within viral capsids, depends on B cell–intrinsic Myd88 activation (Hou et al., 2011). These observations suggest that B cells are “hard-wired” to respond vigorously to viral particles as an immune defense, prompting us to test whether NOX2 impacts TLR-dependent humoral responses. To do this, we immunized wild-type (WT) and Ncf1−/− animals with either nitrophenyl-chicken gamma globulin (NP-CGG) in alum or Qβ-VLP. Preliminary analyses confirmed reduced anti-VLP humoral responses in Tlr7−/− mice (Fig. S1, A–C), in keeping with Qβ-VLP incorporating the TLR7 ligand single-stranded RNA (ssRNA) (Hou et al., 2011). Whereas humoral responses to NP-CGG were equivalent in WT and Ncf1−/− mice (Fig. 1, A–C), immunization with Qβ-VLP promoted a greater expansion of VLP-specific GC B cells and increased IgG and IgG2c anti-VLP titers in NCF1-deficient mice (Fig. 1, D–F). Of note, the IgG2c subclass promotes viral protection by fixing complement and binding to activating Fc receptors (Markine-Goriaynoff and Coutelier, 2002; Nimmerjahn and Ravetch, 2005). Thus, NOX2 complex deletion results in enhanced humoral responses to TLR-dependent viral antigens, but not TLR-independent protein immunization.
Figure S1.
Validation and gating strategies for Qβ-VLP immunization models. (A) Splenic VLP-specific GC B cell counts in WT and Tlr7−/− mice. (B) ELISA dilution curves showing anti-VLP IgG2c titers in WT (solid circles) and Tlr7−/− (open circles) mice. Each line indicates an individual animal. 20 = 1:100 dilution. (C) Anti-VLP IgG (left) and IgG2c (right) titers (by endpoint titer) in WT and Tlr7−/− mice. (A–C) Data were generated using two independent immunization experiments, with representative data from one experiment (A and B) and combined data (C) shown. **, P < 0.01; ***, P < 0.001, by one-way ANOVA, followed by Tukey’s multiple comparison test (A) and two-tailed Mann–Whitney test (C). Animals were immunized with 20 μg Qβ-VLP and analyzed at 12 days after immunization. (D) Gating strategy to identify CD45.1+ versus CD45.2+ VLP-specific GC B cells. (E) Left: Gating strategy to identify murine plasma cells gated as CD19−B220−TACI+IRF4+. Right: Histogram confirming expression of known plasma cell markers (CD38, CD138, BCMA, and CXCR4) on TACI+IRF4+ population (black, solid) versus TACI−IRF4− cells (gray, dotted).
Figure 1.
B cell–intrinsic NADPH oxidase complex deletion promotes TLR-dependent humoral responses. (A) Representative FACs plots showing NP-specific GC B cells in WT and Ncf1−/− animals 14 days after immunization with NP-CGG in alum. Frequencies indicate %FAS+CD38− GC B cells as a percentage of NP+B220+ B cells. (B) Percentage of NP+FAS+CD38− GC B cells of total splenic B cells. (C) Anti-NP30 IgM and IgG titers (normalized to the mean of male WT animals for each experiment). (D) Representative FACs plots showing VLP-specific GC B cells in WT and Ncf1−/− animals 14 days after immunization with Qβ-VLP. Frequencies indicate %FAS+CD38− GC B cells as a percentage of VLP-specific B cells. (E) Percentage of VLP-specific GC B cells of total splenic B cells. (F) Anti-VLP IgG and IgG2c titers in indicated genotypes. (G) Representative FACS plots showing VLP-specific GC B cells in WT:WT and WT:Ncf1−/− animals 14 days after immunization with Qβ-VLP. Cells gated on CD45.2+ B cells from each respective chimera. The number indicates the percentage within the gate. (H) Percentage of VLP-specific FAS+CD38− GC B cells as a proportion of CD45.2+ WT or Ncf1−/− B cells in respective WT:WT and WT:Ncf1−/− chimeras. (I) Selection of VLP-specific CD45.1+ versus CD45.2+ B cells into GC compartment in control (CD45.1+ WT versus CD45.2+ WT) versus NCF1-deficient (CD45.1+ WT versus CD45.2+ Ncf1−/−) competitive chimeras following Qβ-VLP immunization (data normalized to mean WT:WT ratio for each of two independent experiments). (J) Anti-VLP IgG and IgG2c titers from control versus Mb1cre.Cybbfl/fl mice at 14 days after immunization (data normalized to mean titers in male WT animals). (K) Anti-VLP IgG titers at >180 days after immunization (normalized to male WT animals). (L) Representative ELISPOT images of VLP-specific IgG (left) and IgG2c (right) production by BM cells from male and female control and Mb1cre.Cybbfl/fl mice. Data generated at >300 days after Qβ-VLP immunization. (M) ELISPOT spot number per 2 × 106 BM cells from male and female control and Mb1cre.Cybbfl/fl mice. (A–K) Data showed one representative experiment (F, n = 3–10/group), or combined from two (A–E, n = 2–23/group; G–I, n = 4–6/group; K, n = 6–11/group; M, n = 4–10/group) and three (J, n = 11–16/group) independent experiments. Each point represents an individual experimental animal and bars indicate the mean. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 by one-way ANOVA, followed by Tukey’s multiple comparison test (B, C, E, F, H, J, K, and M) or by two-tailed Student’s t test (I).
To test whether NADPH oxidase exerts a B cell–intrinsic impact on TLR-dependent immunity, we first established mixed bone marrow (BM) chimeras by reconstituting sublethally irradiated (500RAD) B cell–deficient (μMT) mice with 50:50 mix of congenically marked CD45.2+ Ncf1−/− or CD45.2+ WT BM and CD45.1+ WT BM. No difference in relative CD45.1+ versus CD45.2+ reconstitution in the B cell compartment was observed between WT:WT or WT:Ncf1−/− chimeras (data not shown). After reconstitution of the peripheral B cell compartment, chimeras where immunized with Qβ-VLP. Notably, a greater proportion of CD45.2+ Ncf1−/− B cells in WT:Ncf1−/− competitive chimeras differentiated into VLP-specific GC B cells than equivalent CD45.2+ WT B cells in control chimeras (Fig. 1, G and H; and Fig. S1 D). In parallel, we calculated VLP+ GC selection (based on the CD45.1:CD45.2 ratio in non-GC and GC B cells) and confirmed that Ncf1−/− B cells exhibit a competitive advantage in GC entry, expansion, and/or selection, relative to CD45.1+ WT control B cells (Fig. 1 I).
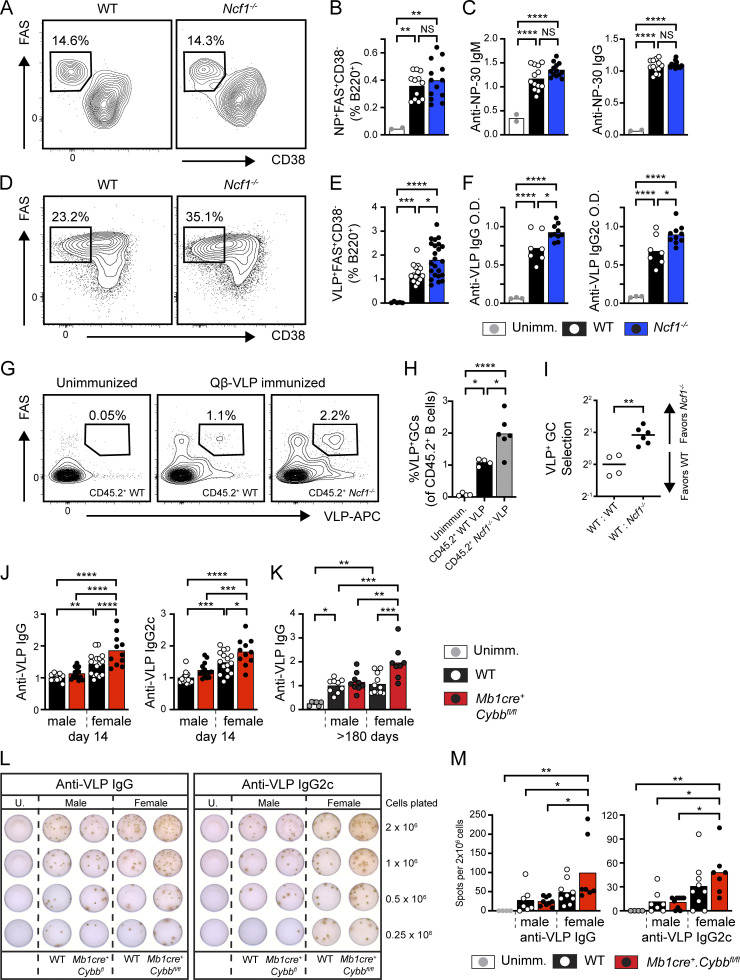
Based on these findings, we tested whether B cell–intrinsic loss of NADPH oxidase complex genes promotes enhanced humoral responses to Qβ-VLP immunization. Since lineage-specific Ncf1−/− mice are not available, we crossed Cybbfl/fl (encoding the NADPH oxidase complex gene Nox2; Pepping et al., 2017) mice with the B cell–specific Mb1cre strain (Hobeika et al., 2006). On day 14 after immunization, Mb1cre.Cybbfl/fl mice exhibited higher anti-VLP titers, in particular IgG2c subclass antibodies. Interestingly, segregating animals by gender uncovered a striking increase in VLP responses in female animals (Fig. 1 J); data are consistent with the observation that human females typically develop higher antibody responses to TLR-dependent viral vaccines, but not adjuvanted protein vaccines (Cook, 2008; Fischinger et al., 2019). Finally, we examined whether the lack of B cell NADPH oxidase expression enhanced the generation of long-lived plasma cells (LLPC) necessary for long-term immunity to previously encountered pathogens. In parallel with increased primary responses, anti-VLP antibody titers remained elevated in female Mb1cre.Cybbfl/fl mice for >180 days after immunization (Fig. 1 K). In addition, ELISPOT quantification of VLP-specific BM plasma cells confirmed increased LLPC in female Mb1cre.Cybbfl/fl mice relative to unimmunized mice, male controls, and male Mb1cre.Cybbfl mice animals, with a trend toward increased LLPC compared with female controls (Fig. 1, L and M). In summary, NADPH oxidase complex gene disruption enhances humoral responses to TLR-containing viral antigens in a B cell–intrinsic manner.
Global and B cell–intrinsic Ncf1 deletion promotes spontaneous GC formation and humoral autoimmunity
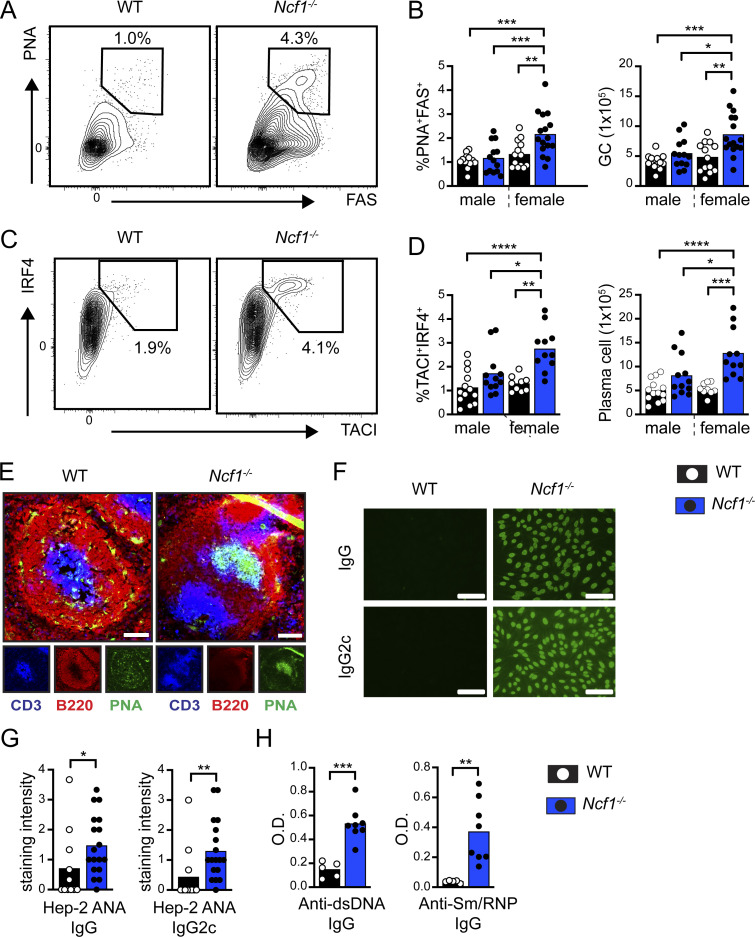
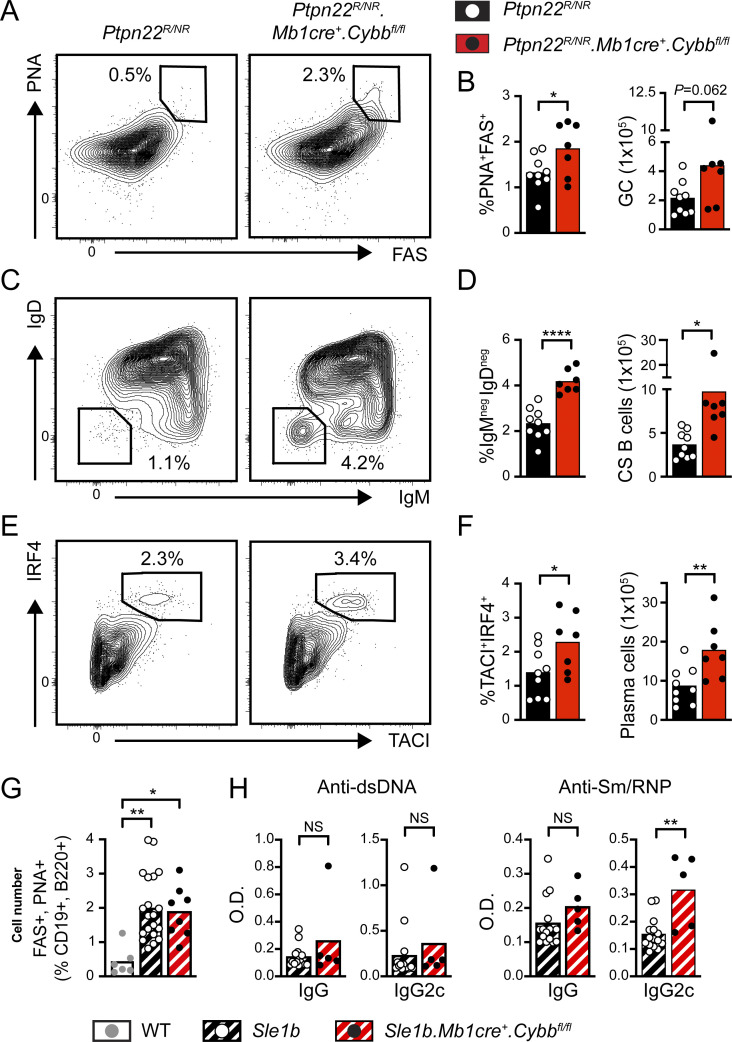
In addition to being the source for pathogenic autoantibodies, B cells initiate an inflammatory cascade leading to the development of SLE. After binding to NA-containing autoantigens, the activation of autoreactive B cells is facilitated by the dual engagement of BCR and TLR signaling pathways. Subsequently, B cells initiate breaks in T cell tolerance and T follicular helper (Tfh) differentiation via B cell–intrinsic MHC class II–dependent antigen presentation, cytokine production, and provision of costimulatory signals (Arkatkar et al., 2017; Chiang et al., 2021; Giles et al., 2015; Jackson et al., 2016). Based on the enhanced VLP responses, we hypothesized that NADPH oxidase complex gene deletion might result in spontaneous humoral autoimmunity by altering B cell TLR signaling thresholds. As predicted, we observed the spontaneous expansion of PNA+FAS+ GC B cells and TACI+IRF4+ plasma cells in the spleens of 1-year-old Ncf1−/− mice (Fig. 2, A–D; and Fig. S1 E). Immunofluorescence staining of splenic sections confirmed the spontaneous formation of PNA+ GCs in Ncf1−/− mice (Fig. 2 E). Importantly, this increase in activated B cell subpopulations only reached statistical significance in female Ncf1−/− animals, in keeping with the known sex bias in human SLE (Amur et al., 2012). Aged female Ncf1−/− mice also exhibited polyclonal autoantibody production, as assessed by Hep2 anti-nuclear antibody (ANA) immunofluorescence staining (Fig. 2, F and G) and specific ELISAs for double-stranded DNA (dsDNA) and Sm/RNP autoantibodies (Fig. 2 H). Thus, in keeping with prior studies using lupus-prone mice (Campbell et al., 2012; Gordon et al., 2022; Jacob et al., 2017), Ncf1 deletion is sufficient to promote the development of female-biased humoral autoimmunity in the non-autoimmune C57BL/6 background.
Figure 2.
Spontaneous humoral autoimmunity in aged Ncf1-deficient mice. (A) Representative FACs plots (gated on CD19+ B cells) showing spontaneous splenic GCs in 1-year-old WT and Ncf1−/− mice. Number: percentage of PNA+FAS+ GC B cells. (B) Percentage and total number of splenic GC B cells in 1-year-old WT and Ncf1−/− mice. (C) Representative FACS plots (gated on B220−CD19− splenocytes) showing the expansion of TACI+IRF4+ plasma cells in Ncf1−/− animals. (D) Percentage and total number of splenic TACI+IRF4+ plasma cells in 1-year-old WT and Ncf1−/− mice. (E) Representative splenic sections stained with B220 (red), PNA (green), and CD3 (blue) showing spontaneous GC formation in Ncf1−/− mice. Scale bars, 100 µm. (F) Hep-2 ANA IgG and IgG2c staining in representative female WT and Ncf1−/− mice. Scale bars, 100 µm. (G) Quantification of Hep-2 ANA IgG and IgG2c staining intensity in female WT and Ncf1−/− mice. ANA staining intensity (range 0–4) was scored by three independent observers blinded to genotype and subsequently averaged. (H) Anti-dsDNA and anti-Sm/RNP IgG titers in 1-year-old female WT and Ncf1−/− mice. (A–H) Data combined from of two (D, n = 9–13/group; H, n = 5–8/group), or three (B, n = 13–16/group; G, n = 11–18/group) independent experimental cohorts. Each point represents an individual experimental animal. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 by one-way ANOVA, followed by Tukey’s multiple comparison test (B and D) or by two-tailed Student’s t test (G and H).
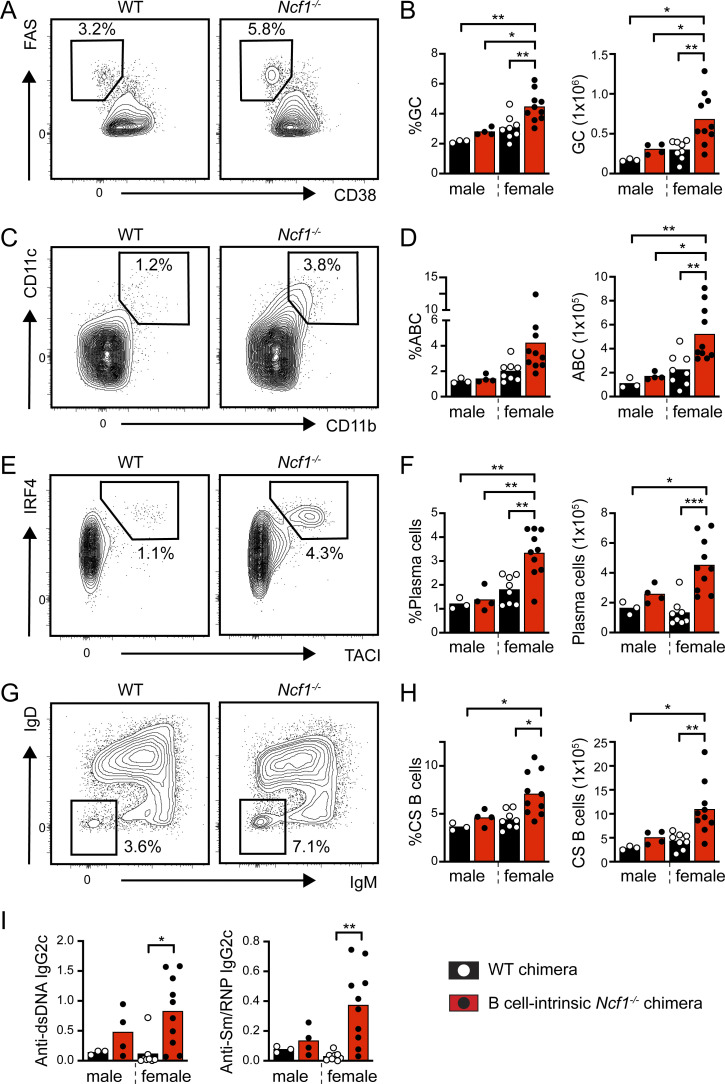
To address the role of B cell–intrinsic NADPH oxidase activity in the maintenance of immune tolerance, we generated mixed BM chimeras in which Ncf1 deletion was limited to the B cell compartment. To do this, we adoptively transferred 20% WT or Ncf1−/− BM together with 80% B cell–deficient (µMT) BM into lethally irradiated µMT recipients. Notably, at 24 wk after transplant, female Ncf1−/− B cell chimeras exhibited spontaneous GC formation and the expansion of CD11b+CD11c+ age/autoimmunity-associated B cells (ABCs; Rubtsov et al., 2011) and splenic plasma cells (Fig. 3, A–F). In addition, loss of B cell Ncf1 expression resulted in an increase in class-switched IgM−IgD− B cells in female chimeras (Fig. 3, G and H). Finally, female B cell–intrinsic Ncf1−/− chimeras exhibited increased anti-dsDNA and anti-Sm/RNP IgG2c titers (Fig. 3 I). Thus, NADPH oxidase activity maintains B cell tolerance and limits the development of humoral autoimmunity in a B cell–intrinsic manner.
Figure 3.
Spontaneous B cell activation in B cell–intrinsic Ncf1-deficient chimeras. (A–H) Flow cytometry analysis of NCF1-deficient chimeras at 24 wk after BM transplant. (A and B) Representative FACS plots (gated on B220+CD19+ splenic B cells) showing spontaneous GCs (A; number: %FAS+CD38− GC B cells in gate), and (B) percentage and total number of splenic GC B cells in indicated chimeras. (C and D) Representative FACS plots (C, gated on B220+CD19+ splenic B cells), and (D) percentage and total number of splenic CD11b+CD11c+ ABCs. (E and F) Representative FACS plots (E, gated on total splenocytes), and (F) percentage and total number of splenic TACI+IRF4+ cells showing the expansion of splenic plasma cells in female B cell–intrinsic Ncf1−/− chimeras. (G and H) Representative FACS plots (gated on B220+CD19+ splenic B cells) showing class-switched B cells (G; number: percentage in IgM−IgD− gate), and (H) percentage and total number of splenic IgM−IgD− class-switched B cells in indicated chimeras. (I) Anti-dsDNA and anti-sm/RNP IgG2c titers at 24-wk after chimera reconstitution. (A–I) Data are combined from three independent chimera cohorts. Each point represents an individual experimental animal sacrificed 24 wk after BM transfer. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 by one-way ANOVA, followed by Tukey’s multiple comparison test.
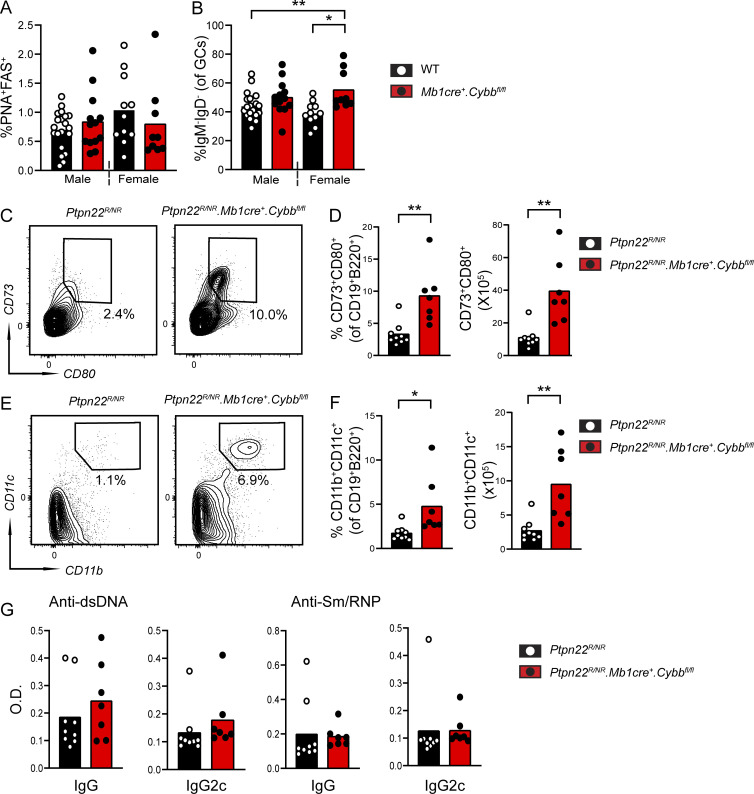
To rule out the possibility that low-level myeloid chimerism contributed to autoimmunity in B cell–intrinsic Ncf1−/− chimeras, we examined whether aged Mb1cre.Cybbfl/fl mice exhibited spontaneous breaks in B cell tolerance. In contrast with B cell Ncf1−/− chimeras, Mb1cre.Cybbfl/fl lacked spontaneous GC formation (Fig. S2 A). However, an increased proportion of GC B cells were class-switched (IgM−IgD−) in 10-mo-old female Mb1cre.Cybbfl/fl mice, supporting a B cell–intrinsic role in GC regulation (Fig. S2 B). We attribute this relative lack of autoimmune B cell activation in Mb1cre.Cybbfl/fl mice versus B cell–intrinsic Ncf1−/− chimeras to elevated serum BAFF and greater exposure to apoptotic cell self-antigens in the context of irradiation-reconstitution. For this reason, we examined whether crossing Mb1cre.Cybbfl/fl animals onto a low-penetrance autoimmune background was sufficient to induce breaks in immune tolerance. A common single-nucleotide polymorphism in PTPN22 (C1858T; R620W) promotes lupus by modulating T cell receptor (TCR) and BCR signaling thresholds (Begovich et al., 2004; Bottini et al., 2004; Kyogoku et al., 2004). A murine knock-in model of the orthologous mouse phosphatase (hereafter Ptpn22RISK/NON-RISK; Ptpn22R/NR) exhibits spontaneous GCs and mild autoimmunity on the mixed C57BL/6J × 129/Sv, but not C57BL/6, background, suggesting that the Ptpn22RISK allele functions as a low-penetrance autoimmune variant (Dai et al., 2013). To test this idea, we crossed Mb1cre.Cybbfl/fl mice and Ptpn22RISK mice. Female 10-mo-old Ptpn22R/NR.Mb1cre.Cybbfl/fl animals exhibited expanded spontaneous GCs, class-switched B cells, and splenic plasma cells compared with Ptpn22R/NR controls (Fig. 4, A–F). In addition, we observed increased CD73+CD80+ memory B cells (Fig. S2, C and D) and ABCs (Fig. S2, E and F), although heterozygous addition of the Ptpn22RISK allele was insufficient to promote autoantibody production (Fig. S2 G).
Figure S2.
B cell–intrinsic Cybb deletion promotes B cell activation in low-penetrance autoimmune strains. (A and B) Percentage of splenic GC B cells (A) and percentage of class-switched B cells (%IgM−IgD− within GC B cell gate) in 10-mo-old WT and Mb1cre.Cybbfl/fl mice. (C) Representative FACS plots showing CD80+CD73+ memory B cells (gated on B220+CD19+ B cells) in Ptpn22R/NR controls and Ptpn22R/NR.Mb1cre.Cybbfl/fl mice. (D) Percentage and total number of CD80+CD73+ memory B cells in indicated strains. (E) Representative FACS plots showing expansion of CD11b+CD11c+ ABCs (gated on B220+CD19+ B cells) in Ptpn22R/NR.Mb1cre.Cybbfl/fl mice relative to Ptpn22R/NR controls. (F) Percentage and total number of CD11b+CD11c+ ABCs in indicated strains. (G) Anti-dsDNA and anti-Sm/RNP IgG and IgG2c titers in 10-mo-old female Ptpn22R/NR controls and Ptpn22R/NR.Mb1cre.Cybbfl/fl mice. (A–G) Data from more than three independent experimental cohorts. Each point represents an individual experimental animal. *, P < 0.05; **, P < 0.01 by one-way ANOVA, followed by Tukey’s multiple comparison test (A and B) or by two-tailed Student’s t test (D, F, and G).
Figure 4.
B cell–intrinsic Cybb deletion facilitates breaks in B cell tolerance on low-penetrance autoimmune backgrounds. (A, C, and E) Representative FACs plots showing expansion of PNA+FAS+ GC B cells (A), IgM−IgD− class-switched B cells (C), and TACI+IRF4+ plasma cells (E) in female Ptpn22R/NR controls and Ptpn22R/NR.Mb1cre.Cybbfl/fl mice. Number equals percentage within the gate. (B, D, and F) Percentage (left) and total number (right) of splenic PNA+FAS+ GC B cells (B), IgM−IgD− class-switched B cells (D), and TACI+IRF4+ plasma cells (F) in female Ptpn22R/NR controls (black) and Ptpn22R/NR.Mb1cre.Cybbfl/fl (red) mice. (G) Percentage of PNA+FAS+ GC B cells in 4-mo-old female WT (gray), Sle1b+/+ (black hashed), and Sle1b+/+.Mb1cre.Cybbfl/fl (red hashed) mice. (H) Anti-dsDNA and anti-Sm/RNP IgG and IgG2c titers from 4-mo-old female Sle1b+/+ (black hashed), and Sle1b+/+.Mb1cre.Cybbfl/fl (red hashed) mice. (A–H) Data are combined from two (H) or three (A–G) independent experimental cohorts. Each point represents an individual experimental animal. *, P < 0.05; **, P < 0.01; and ****, P < 0.0001, by one-way ANOVA, followed by Tukey’s multiple comparison test (G) or by two-tailed Student’s t test (B, D, F, and H).
For this reason, we crossed Mb1cre.Cybbfl/fl mice onto the established lupus-prone Sle1b background (Morel et al., 2000, 2001). Whereas splenic GC B cells were similarly elevated in Sle1b+/+.Mb1cre.Cybbfl/fl mice and Sle1b+/+ controls (Fig. 4 G), loss of B cell NADPH oxidase activity increased anti-Sm/RNP autoantibodies of the pathogenic IgG2c subclass, without impacting anti-dsDNA titers (Fig. 4 H). These findings are reminiscent of previous data showing skewing of the autoantibody repertoire toward RNA-associated autoantigens in Cybb−/− lupus-prone mice (Campbell et al., 2012). In summary, these data indicate that B cell–intrinsic deletion of NADPH oxidase components promotes B cell activation during humoral autoimmunity, lending support for a B cell–specific contribution to autoimmune risk in NCF1 and NCF2 carriers.
Ncf1-deficient B cells exhibit increased endosomal TLR signaling
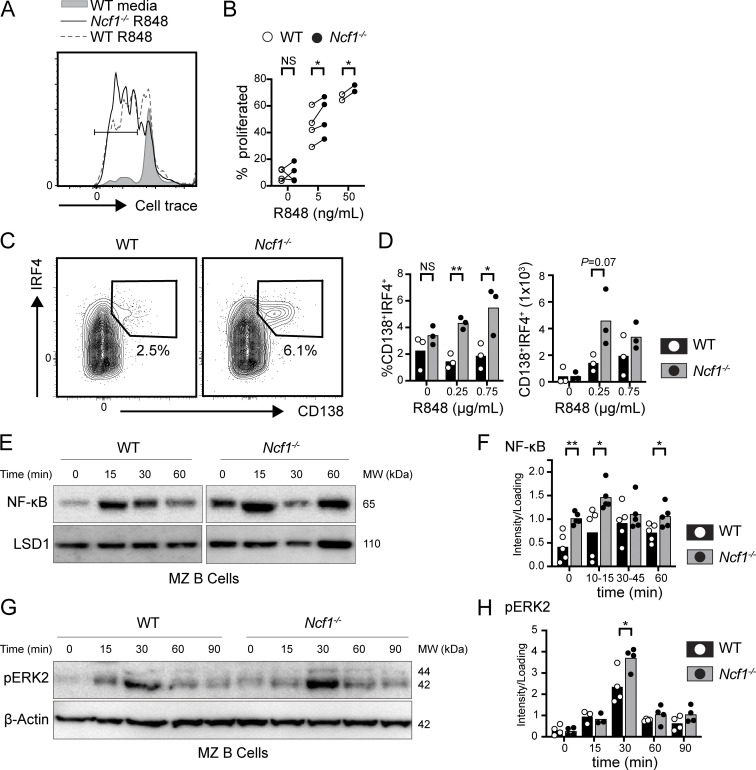
Given known roles for B cell–intrinsic TLR signals in regulating VLP responses and autoimmune pathogenesis, we hypothesized that NADPH oxidase might regulate endosomal TLR signaling in B cells. Consistent with this idea, sorted marginal zone (MZ) B cells from Ncf1−/− mice exhibited enhanced proliferation in vitro following TLR7 agonist R848 stimulation (Fig. 5, A and B). To further characterize these events, we modeled T cell–dependent B cell activation in vitro by culturing splenic B cells from WT and Ncf1−/− mice on CD40L/BAFF-expressing feeder cells together with IL-21 and increasing doses of R848. Notably, R848-stimulated Ncf1−/− B cells exhibited a marked increase in the proportion of antibody-secreting cells (ASCs) following R848 stimulation, with a parallel trend toward higher ASC numbers (Fig. 5, C and D). We observed a similar trend toward increased ASC differentiation in Ncf1−/− B cells following TLR9 agonist CpG stimulation (Fig. S3 A). We also quantified NF-κB p65 nuclear localization following CpG-2395 stimulation of WT and Ncf1−/− MZ B cells. While NF-κB activation in control cells peaked at 15 min and returned to baseline by 60 min, Ncf1−/− B cells exhibited both an increased peak NF-κB response and prolonged NF-κB nuclear localization (Fig. 5, E and F). Interestingly, basal nuclear NF-κB was also elevated in Ncf1−/− MZ B cells, similar to our previous data in αV integrin-deficient B cells (Acharya et al., 2016). ERK phosphorylation downstream of TLR engagement was also increased in Ncf1−/− MZ B cells relative to control cells (Fig. 5, G and H). Thus, disruption of NADPH oxidase activity in B cells alters the kinetics of TLR signaling, resulting in enhanced endosomal TLR signaling and increased TLR-dependent B cell activation.
Figure 5.
Increased endosomal TLR signaling in NCF1-deficient mouse B cells. (A and B) Sorted MZ B cells from female WT and Ncf1−/− mice were stimulated with TLR7 agonist, R848. (A) Histogram depicting CellTrace Violet dilution in WT (R848 0 ng/ml; gray filled), WT (R848 5 ng/ml; gray line), and Ncf1−/− (R848 5 ng/ml; black line) MZ B cells. Gate: percentage proliferated. (B) Proliferation of sorted WT versus Ncf1−/− MZ B cells 48 h after stimulation with indicated R848 dose. Each paired data point represents an independent experiment. (C and D) Splenic B cells cultured for 4 days on CD40L/BAFF-expressing feeder cells plus 10 ng/ml IL-21, and the indicated dose of R848. (C) Representatives FACS plots (gated on live cells) showing in vitro generation of CD138+IRF4+ plasma cells (number equals percentage in gate). (D) Percentage and total number of CD138+IRF4+ plasma cells following stimulation with indicated R848 dose. Each point represents an individual experimental animal. (E) Nuclear translocation of NFκB by western blot in sorted female WT and Ncf1−/− MZ B cells after CpG stimulation at indicated times. Western blot of nuclear LSD1 was used as a protein loading control. MW, molecular weight. (F) Quantification of nuclear NF-κB by densitometry, corrected for loading control. MW, molecular weight. (G and H) Representative western blot of cytosolic pERK2 (G) and pERK2 quantification (H) in sorted female WT and Ncf1−/− MZ B cells after CpG stimulation for indicated times. Actin was used to normalize protein loading. Each point represents an independent experiment. (A–H) Bars indicate mean. Data are combined from two (D), four (B and H), and five (F) independent experiments. *, P < 0.05; and **, P < 0.01 by paired (B) or unpaired Student’s t test (D, F, and H). Source data are available for this figure: SourceData F5.
Figure S3.
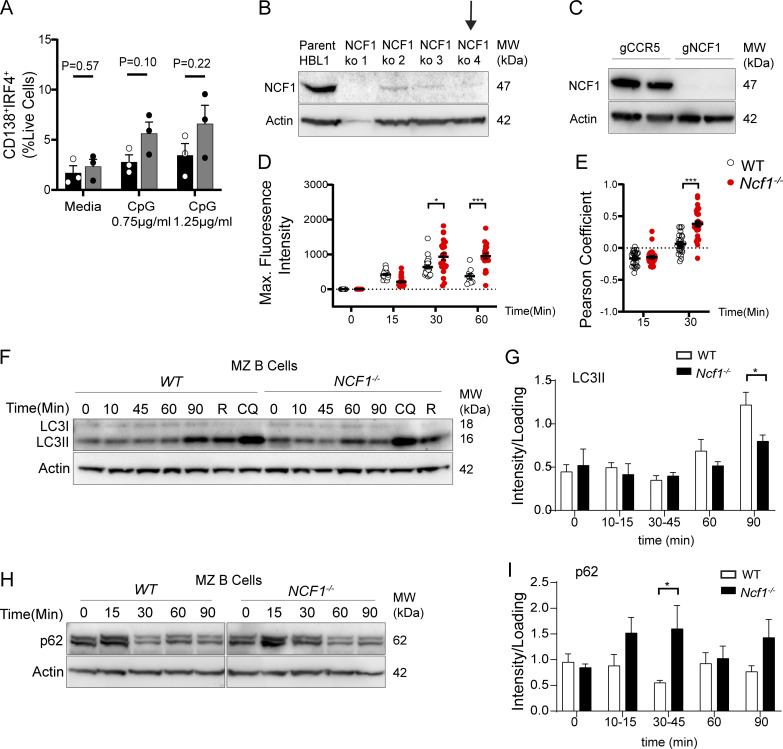
Independent replicates of B cell in vitro stimulation, confirmation of CRISPR-mediated NCF1 deletion, and impact of NCF1 deletion on p62 accumulation. (A) Splenic B cells cultured for 4 days on CD40L/BAFF-expressing feeder cells plus 10 ng/ml IL-21 and indicated dose of CpG. Percentage of CD138+IRF4+ shown. Each point represents the mean of technical replicates for each individual experimental animal. Bars, mean ± SEM. (B) Efficient CRISPR-mediated deletion of NCF1 in HBL1 cells by western blot. Also shown is a western blot for actin to normalized protein loading. Black arrow indicates the clone used for experiments. MW, molecular weight. (C) Western blot showing successful NCF1 deletion in primary human B cells. Actin staining was used to normalize protein loading. MW, molecular weight. (D) Fluorescent CpG maximum signal intensity in control and NCF1−/− HBL1 cells. Each point represents a single cell (n = 9–26 per condition). Independent replicate of data show in Fig. 6 B. (E) Pearson’s coefficient of EEA1 and CpG signal colocalization (above Costes threshold), masked by EEA1 staining after CpG stimulation for 15 and 30 min. Each point represents a single cell (n = 24–37/condition). Independent replicate of data show in Fig. 6 B. (F) Western blot of LC3-II in cytosolic fraction after CpG stimulation for 0–90 min, or after stimulation with rapamycin (R) and chloroquine (CQ) for 90 min to induce autophagy. Actin was used to normalize protein loading. MW, molecular weight. (G) Quantification of cytosolic LC3-II by densitometry (corrected for loading control). (H) Western blot of p62 in cytosolic fraction after stimulation with CpG for 0–90 min. Actin was used to normalize protein loading. MW, molecular weight. (I) Quantification of cytosolic p62 levels by densitometry (corrected for loading control). (B, D, G, and I) Bars show mean ± SEM of each individual experiment. Data were generated from two (A), four (G), five (I) independent experiments. (A, D, F, and H) *, P < 0.05; ***, P < 0.001 by two-tailed Student’s t test. Source data are available for this figure: SourceData FS3.
NCF1 deletion promotes increased TLR signaling via dysregulation of endolysosomal trafficking
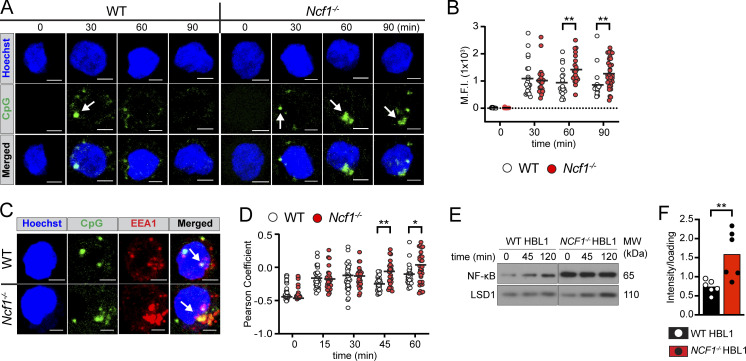
We next sought to understand the mechanism(s) by which Ncf1 deletion facilitates TLR hyperresponsiveness. Prior research has uncovered a critical role for endolysosomal trafficking in regulating the duration and quality of TLR signals (Acharya et al., 2016; Barton and Kagan, 2009). We thus asked whether NCF1 deletion impacts endosomal TLR signaling via modulation of endolysosomal flux. To do this, we used CRISPR/Cas9-mediated gene disruption to generate NCF1−/− HBL1 cells, a cell line derived from a human-activated B cell–like diffuse large B cell lymphoma (ABC-DLBCL) (Fig. S3 B). We next stimulated NCF1+/+ and NCF1−/− HBL1 cells with fluorescently labeled CpG-2395-APC and followed TLR ligand trafficking by confocal microscopy. As predicted, fluorescent CpG was first internalized and then aggregated into intracellular foci by 30 min in NCF1+/+ HBL1 cells. Subsequently, the fluorescent CpG signal was degraded such that few CpG-APC+ puncta were observed at 60 min. In contrast, whereas initial CpG uptake by NCF1−/− HBL1 cells was not affected, a significant amount of CPG-APC+ aggregates persisted for 90 min in the NCF1−/− cells, suggesting that loss of NADPH oxidase activity limited signal degradation (Fig. 6, A and B; replicate experiment in Fig. S3 D).
Figure 6.
NCF1 deletion alters CpG trafficking and degradation in HBL1 cell line. (A) Confocal microscopy showing intracellular trafficking of fluorescence CpG in control versus NCF1-deficient HBL1 cells. Hoechst staining indicates the location of the nucleus. White arrows indicate CpG aggregates. Scale bars, 3 µm. (B) Maximum fluorescence intensity of CpG signal. Z stacks were compressed to a single plane to sum analysis. Each data point represents a single cell (n = 9–32 per condition). Graph shows representative data from one of two independent experiments yielding similar findings. (C) Colocalization of fluorescent CpG and EEA1 at 45 min after CpG stimulation. Hoechst staining indicates the location of the nucleus. White arrows highlight CpG and EEA1 colocalization. (D) Pearson’s coefficient of EEA1 and CpG signal colocalization (above Costes threshold), masked by EEA1 staining after CpG stimulation for 0–60 min. Z stacks were not compressed for this analysis. Each point represents a single cell (n = 25–52/condition). Graph shows representative data from one of two independent experiments yielding similar findings. (E) Nuclear translocation of NF-κB by western blot in control versus NCF1−/− HBL1 cells after CpG stimulation for 0, 45, and 120 min. Nuclear LSD1 was used to normalize protein loading. MW, molecular weight. (F) Quantification of nuclear NF-κB by densitometry at baseline, corrected for loading control. Data are combined from six independent experiments. Each data point represents a single experiment. (B, D, and F) Bars/lines indicate the mean. *, P < 0.05; and **, P < 0.01 by unpaired two-tailed Student’s t test. Source data are available for this figure: SourceData F6.
We previously showed that B cell endosomal TLR signals are tightly regulated by the ordered transition through distinct endolysosomal compartments, with TLR9 persisting in early endosomes in the absence of B cell αV integrin expression (Acharya et al., 2016). To identify the intracellular compartment where CpG accumulated in NCF1−/− HBL1 cells, we performed costaining with the early endosome marker EEA1. Notably, colocalization of EEA1 and CpG-APC at 15 min was similar in NCF1+/+ and NCF1−/− cells, but CpG-APC fluorescence persisted in early endosomes at later timepoints in NCF1-deficient cells (Fig. 6, C and D; replicate experiment in Fig. S3 E). Importantly, TLRs initiate distinct downstream signaling cascades from different endolysosomal compartments, with NF-κB activation occurring in early endosomes (Acharya et al., 2016; Sasai et al., 2010). This compartmentalization of endosomal TLR signaling likely explains the enhanced and prolonged NF-κB activation in CpG-stimulated murine Ncf1−/− MZ B cells. Thus, loss of NCF1 limits CpG degradation, resulting in its accumulation in early endosomal compartments and enhanced TLR signaling.
These data also point to a role for NCF1 in regulating chronic TLR signaling associated with self-antigens. The survival of ABC-DLBCL is facilitated by chronic active BCR signaling that is frequently induced by self-antigen engagement. Indeed, ∼30% ABC-DLBCL express VH4-34 (Young et al., 2015), a VH family member with intrinsic, germline self-reactivity toward multiple self-antigens and enriched in SLE (Cappione et al., 2004; Jenks et al., 2013; Richardson et al., 2013). In addition, ABC-DLBCL frequently carry oncogenic mutations impacting TLR signaling pathways, including the gain-of-function variant in MYD88 (L265P) (Ngo et al., 2011). Since HBL1 cells express both VH4-34 (Young et al., 2015) and MYD88L256P (Ngo et al., 2011), we predicted that recognition of apoptotic debris in culture media is likely to drive dual BCR/TLR9 engagement and constitutive NF-κB activation. Notably, NCF1−/− HBL1 cells exhibited markedly increased nuclear NF-κB at baseline (Fig. 6, E and F). Although additional TLR-independent signals may also contribute to elevated basal NF-κB in NCF1−/− HBL1 cells, these data implicate the NADPH oxidase complex in regulating dual BCR/TLR signaling cascades, given the known roles for a multiprotein supercomplex comprising MYD88, TLR9, and the BCR in the promotion of ABC-DLBCL survival (Phelan et al., 2018).
NCF1 deletion limits LC3 recruitment to early endosomes and delays lysosomal fusion
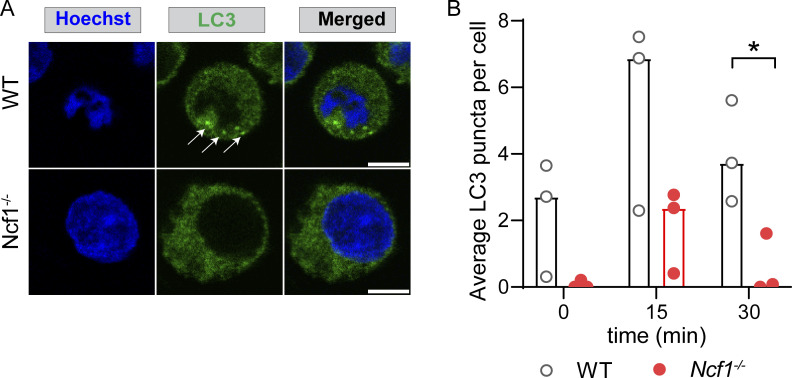
We next sought to address the mechanisms underlying the delay in CpG degradation following NCF1 deletion. Key components of the autophagy machinery are known to regulate endolysosomal trafficking events (Hayashi et al., 2018). Activation of autophagy machinery leads to lipidation of ubiquitin-like protein LC3, which can be conjugated to endosomal membranes. In B cells, TLR-dependent activation of autophagy and recruitment of LC3 to TLR-containing endosomal compartments promotes lysosomal fusion and termination of TLR signaling (Acharya et al., 2016). Based on known roles for NADPH oxidase in the induction of non-canonical autophagy (Martinez et al., 2015), we asked whether the autophagy machinery could be involved in TLR hyperresponsiveness of NCF1−/− B cells. To test this hypothesis, we first assessed the lipidation of LC3 upon TLR stimulation. We sorted MZ B cells from WT and Ncf1−/− mice and quantified CpG-induced LC3-II (lipidated LC3) by western blot. Treatment with rapamycin to induce macroautophagy or hydroxychloroquine, which delays lysosomal degradation, similarly increased levels of LC3-II in Ncf1+/+ and Ncf1−/− MZ B cells, confirming intact autophagy machinery in the absence of NCF1. After CpG stimulation, both Ncf1+/+ and Ncf1−/− MZ B cells exhibited enhanced LC3 lipidation; however, induction of LC3-II was significantly delayed in Ncf1−/− B cells, in particular at 90 min after stimulation (Fig. S4, F and G). As an additional readout of autophagy flux, we quantified levels of p62, an LC3-binding cargo receptor whose levels decrease following lysosomal fusion. Whereas WT p62 levels decreased following CpG stimulation, p62 levels in Ncf1−/− B cells remained elevated in keeping with a role for NADPH oxidase in regulating TLR-induced autophagy flux (Fig. S4, H and I). Moreover, when we assessed LC3 lipidation in response to R848 stimulation by measuring LC3 puncta formation in NCF1+/+ and NCF1−/− HBL1 cells, we found that deletion of NCF1 also impairs LC3 lipidation in response to TLR7 stimulation (Fig. S4).
Figure S4.
NCF1 deletion reduces LC3 lipidation in response to R848 stimulation. (A) Confocal microscopy showing LC3 in control and NCF1-deficient HBL1 cells at 15 min after R848 stimulation. Arrows show LC3 puncta in control cells. Scale bar: 5 µm. (B) Quantification of average LC3 puncta per cell in unstimulated control and NCF1-deficient HBL1 cells through 3D analysis. Every dot represents the average puncta per cell for each individual experiment (n = 3 independent experiments). *, P < 0.05 by two-tailed, paired Student’s t test.
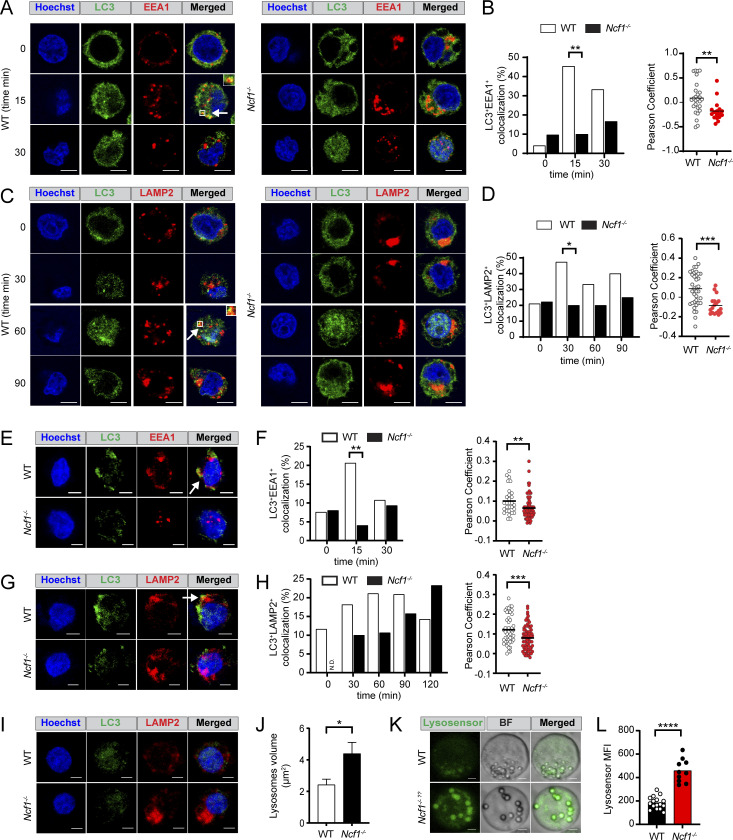
Based on these changes in LC3 lipidation in the Ncf1−/− B cells, we investigated whether NCF1 deletion impacted R848- and CpG-driven recruitment of LC3 to endosomal compartments. Confocal microscopy showed that, whereas rapid EEA1 and LC3 colocalization was observed in R848 stimulated HBL1 cells, NCF1−/− HBL1 cells lacked LC3 recruitment to the EEA1+ early endosomal compartments (Fig. 7, A and B). To extend these findings, we tested whether this defect in LC3 recruitment impacted subsequent endosomal maturation and lysosomal fusion. In control HBL1 cells, colocalization of LC3 and the lysosomal marker LAMP2 peaked at 30 min after R848 stimulation and gradually decreased over time (Fig. 7, C and D). In contrast, LC3 recruitment to LAMP2+ lysosomal compartments was markedly delayed in the absence of NCF1 expression. Similarly, CpG stimulation promoted rapid EEA1 and LC3 colocalization in control HBL1 cells, but this was not seen in NCF1-deficient HBL1 cells (Fig. 7, E and F). Although LC3/LAMP2 colocalization was observed in NCF1−/− HBL1 cells, CpG-induced lysosomal trafficking was markedly delayed in the absence of NCF1 peaking at 60 min in WT control versus 120 min in NCF1−/− HBL1 cells (Fig. 7, G and H). To further validate these findings, we quantified the extent of colocalization between different fluorescent markers using Manders’ overlap coefficient (Fig. S5). This analysis further confirmed that LC3 recruitment to endolysosomal compartments, in response to both TLR7 and TLR9 stimulation, is delayed in the absence of NCF1 expression.
Figure 7.
NADPH oxidase activation promotes LC3-dependent endolysosomal trafficking in TLR-stimulated HBL1 cells. (A) Confocal microscopy showing LC3 and EEA1 colocalization (arrows/insets) in control and NCF1-deficient HBL1 cells at resting conditions (0) and after 15 and 30 min after R848 stimulation. Scale bar, 5 μm. (B) Left: Percentage of control and NCF1-deficient HBL1 cells exhibiting LC3/EEA1 colocalization at 0–30 min after R848 stimulation. Data are combined from two independent experiments yielding similar findings with n = 38–85 cells/condition. Right: Pearson’s coefficient of LC3 and EEA1 signal colocalization (with Costes threshold) at 15 min after R848 stimulation. Each point represents a single cell (n = 17–27 per condition). Graph shows representative data from one of two independent experiments yielding similar findings. (C) Confocal microscopy showing LC3 and LAMP2 colocalization (arrows/insets) in control and NCF1-deficient HBL1 cells at resting condition (0) and 30, 60, 90 min after R848 stimulation. (D) Left: Percentage of control and NCF1-deficient HBL1 cells exhibiting LC3/LAMP2 colocalization at 0–120 min after R848 stimulation. Data are combined from two independent experiments with n = 69–95 cells/condition. Right: Pearson’s coefficient of LC3 and LAMP2 signal colocalization (with Costes threshold) at 60 min after R848 stimulation. Each point represents a single cell (n = 18–37 per condition). Graph shows representative data from one of two independent experiments yielding similar findings. (E) Confocal microscopy showing LC3 and EEA1 colocalization (white arrows) in control and NCF1-deficient HBL1 cells at 15 min after CpG stimulation. Scale bar, 3 μm. (F) Left: Percentage of control and NCF1-deficient HBL1 cells exhibiting LC3/EEA1 colocalization at 0–30 min after CpG stimulation. Data are combined from two independent experiments yielding similar findings with n = 65–123 cells/condition. Right: Pearson’s coefficient of LC3 and EEA1 signal colocalization (no Costes threshold) at 15 min after CpG stimulation. Each point represents a single cell (n = 29–80 per condition). Graph shows representative data from one of two independent experiments yielding similar findings. (G) Confocal microscopy showing LC3 and LAMP2 colocalization (white arrows) in control and NCF1-deficient HBL1 cells at 60 min after CpG stimulation. (H) Left: Percentage of control and NCF1-deficient HBL1 cells exhibiting LC3/LAMP2 colocalization at 0–120 min after CpG stimulation. Data are combined from two independent experiments with n = 20–114 cells/condition. N.D. = not detected. Right: Pearson’s coefficient of LC3 and LAMP2 signal colocalization (no Costes threshold) at 60 min after CpG stimulation. Each point represents a single cell (n = 48–80 per condition). Graph shows representative data from one of two independent experiments yielding similar findings. (I) Representative confocal microscopy images showing control and NCF1-deficient HBL1 cells at baseline without stimulation. (J) Quantification of individual LAMP2+ lysosome volume in unstimulated control and NCF1-deficient HBL1 cells. Bars indicate mean ± SEM. Graph shows representative data from one of three independent experiments yielding similar findings. (K) Representative confocal microscopy images showing Lysosensor staining of control and NCF1-deficient HBL1 cells. (L) Lysosensor mean fluorescence intensity (MFI). (B, D, F, H, J, and L) Each point represents data from an individual control (white) or NCF1-deficient (red) HBL1 cell analyzed. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 by two-tailed Fisher's exact test (B, left; D, left; F, left) and unpaired two-tailed Student’s t test (B, right; D, right; F, right; H, right; J; L).
Figure S5.
NCF1 deletion reduces the rate of LC3 colocalization in early and late endosomes in response to TLR7 and TLR9 ligands. (A) Left: Manders’ coefficient of fraction of early endosomes overlapping with LC3 in resting (0 min) and after 15 and 30 min of R848 stimulation. Graph shows representative data from one of two independent experiments yielding similar findings. Each point represents a cell, 74–89 cells per condition. Right: Graph showing values of the left panel normalized per the average value at time 0 of every condition, graphing the fold change of Manders’ coefficient at every time point. (B) Left: Manders’ coefficient of fraction of early endosomes overlapping with LC3 in resting (0 min) and after 15 and 30 min of CpG stimulation. Graph shows representative data from one of two independent experiments yielding similar findings. Each point represents a cell, 102–149 cells per condition. Right: Graph showing values of the left panel normalized per the average value at time 0 of every condition, graphing the fold change of Manders’ coefficient at every time point. (C) Left: Manders’ coefficient of fraction of lysosomes overlapping with LC3 in resting (0 min) and after 30, 60, and 90 min of R848 stimulation. Graph shows representative data from one of two independent experiments yielding similar findings. Each point represents a cell, 75–119 cells per condition. Right: gGraph showing values of the left panel normalized per the average value at time 0 of every condition, graphing the fold change of Manders’ coefficient at every time point. (D) Left: Manders’ coefficient of fraction of lysosomes overlapping with LC3 in resting (0 min) and after 30, 60, 90, and 120 min of CpG stimulation. Graph shows representative data from one of two independent experiments yielding similar findings. Each point represents a cell, 85–144 cells per condition. Right: Graph showing values of the left panel normalized per the average value at time 0 of every condition, graphing the fold change of Manders’ coefficient at every time point. Bars/lines indicate the mean or mean with SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 by two-way ANOVA, followed by Tukey’s multiple comparison test.
Interestingly, NCF1−/− HBL1 cells also showed significantly enlarged and acidic LAMP2+ lysosomes compared with control HBL1 cells at baseline (Fig. 7, I–L), which was reminiscent of chloroquine-mediated inhibition of autophagosome/lysosome fusion (Mauthe et al., 2018). Thus, NCF1 promotes TLR-dependent recruitment of LC3 to EEA1+ early endosomes, resulting in endosomal maturation and ultimately termination of TLR signals. In the absence of NCF1, LC3 lipidation and recruitment to early endosomes result in delayed endolysosomal fusion, leading to an enhanced threshold of TLR activation responses.
NCF1 deletion enhances TLR-dependent ASC differentiation in primary human B cells
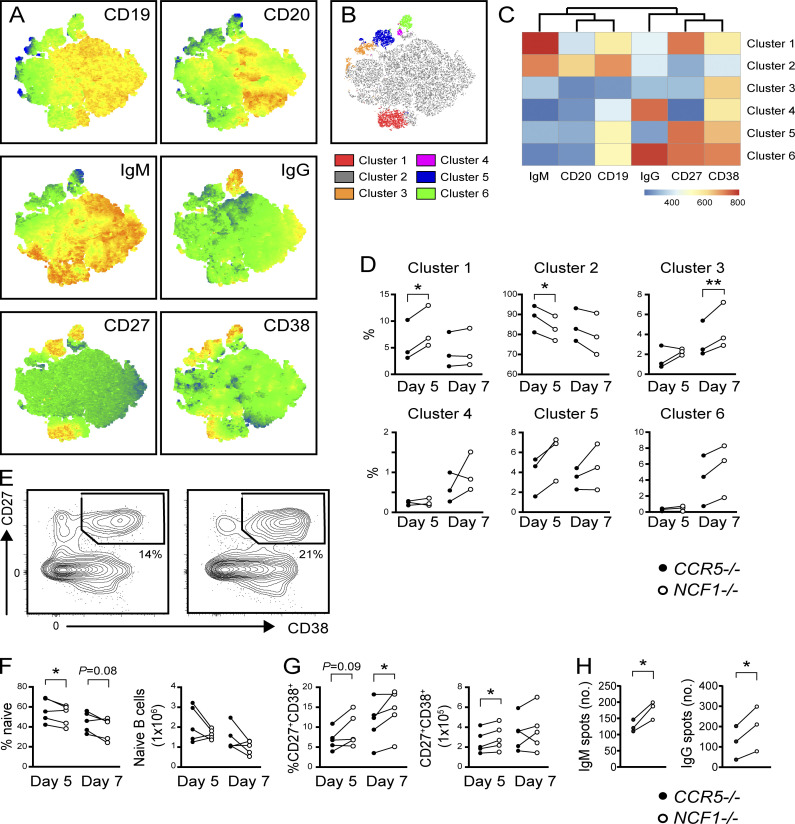
Finally, we assessed whether NCF1 deletion enhanced TLR-driven responses in primary human B cells. To do this, CD19+ B cells from three independent healthy donors were stimulated with a B cell activation cocktail comprising CD40L, cytokines IL-2, IL-10, and IL-15, and the TLR9 agonist CpG2006 (Hung et al., 2018). On day 2, we performed CRISPR/Cas9-mediated gene disruption of NCF1 or the safe harbor locus CCR5, resulting in high-efficiency deletion of NCF1 protein (Fig. S3 C). Cells were analyzed by flow cytometry on days 5 and 7. To aid exploration of these data, we concatenated day 5 and day 7 CCR5−/− and NCF1−/− B cells and generated a t-distributed stochastic neighbor embedding (t-SNE) representation of the data (Fig. 8 A). To uncover differences between the surface phenotypes of CCR5−/− and NCF1−/− B cells, we performed unbiased clustering of the multiparameter data using the FlowSOM algorithm (Van Gassen et al., 2015) (Fig. 8, B and C). On day 5, NCF1−/− B cells exhibited an expansion in cluster 1 (red) corresponding to IgM+ plasmablasts (CD19lowIgM+CD27+CD38+), together with a trend toward an increase in cluster 5 (CD19lowIgM−IgG−CD27+CD38+; blue). This expansion in activated B cells was accompanied by a parallel decrease in CD19+CD20+IgM+IgG−CD38−CD27− “naïve” B cells (cluster 2; gray). By day 7, NCF1−/− B cells exhibited increased CD38+ activated B cells (cluster 3; orange) and a trend toward increased IgM−IgG− and IgG+ CD27+CD38+ plasmablasts (clusters 5 [blue] and 6 [green]) (Fig. 8 D). To validate these findings, we performed an independent experiment using two additional donors followed by manual gating of the combined dataset. Relative to CCR5 controls, NCF1-edited B cells exhibited a relative increase in CD27+CD38+ plasmablasts at day 5 and 7 of culture, which was accompanied by a decrease in naïve B cells (Fig. 8, E–G). Finally, we confirmed that NCF1 deletion increased TLR-dependent generation of IgM− and IgG−producing ASCs by ELISPOT (Fig. 8 H). Together, these data show that NCF1 deletion in primary human B cells results in enhanced in vitro activation following TLR9 ligation, thereby facilitating terminal differentiation into ASCs.
Figure 8.
Increased TLR responses in NCF1-deficient primary human B cells. (A) Heatmaps of marker expression overlaid on composite t-SNE projection from composite in vitro stimulated primary human B cells. (B) t-SNE projection of human B cell clusters identified using FlowSOM. (C) Heatmap of marker MFI in indicated FlowSOM clusters. (D) Percentage of B cells in each FlowSOM cluster in CCR5−/− control (black circles) and NCF1−/− (open circles) human B cells after day 5 and day 7 in vitro stimulation. (E) Representatives FACS plot showing CD27+CD38+ ASCs on day 7. Number indicates percentage in gate. (F) Percentage of naïve and CD27+CD38+ ASCs in CCR5−/− control (black circles) and NCF1−/− (open circles) human B cells after stimulation for 5 and 7 days. (G) Total IgM and IgG producing ASCs by ELISPOT (spots per 1,600 cells plated). (A–G) Data were generated using two independent primary human B cell CRISPR-editing and in vitro stimulation experiments. Each paired data point was generated using a separate PBMC donor. Graphs show combined data (F and G) or representative data from one of two independent experiments (D and H). *, P < 0.05; **, P < 0.01; by two-tailed ratio paired t test.
Discussion
SLE is a severe, multisystem autoimmune disease characterized by polyclonal B cell activation and the production of class-switched autoantibodies. Genome-wide association studies have identified >100 polymorphisms impacting the risk of developing SLE (Deng and Tsao, 2017). However, our understanding of how individual genetic variant promotes autoimmunity remains poor. In addition to the majority of risk polymorphisms mapping to non-coding gene regulatory regions, disease-associated genes frequently exhibit widespread expression in diverse hematopoietic and non-hematopoietic lineages. Given complex interactions between cellular lineages during an autoimmune response, developing a unified framework for how specific genetic variants drive disease pathogenesis is extremely challenging.
In this context, loss-of-function variants of the phagocytic NADPH oxidase (NOX2) complex, NCF1 and NCF2, markedly increase the risk of SLE. This disease association has predominantly been explained by the prediction that reduced NOX2 activity functions to promote pathogenic activation of myeloid lineage cells by self-ligands. For example, BM-derived macrophages (BMDM) from Ncf1Arg90His knock-in mice exhibit defects in the immunologically silent clearance of dead/dying cells, a process termed efferocytosis (Geng et al., 2022; Peng et al., 2007). In addition, Ncf1Arg90His expression was shown to promote pDC activation in SLE via impaired endosomal acidification resulting in enhanced TLR signaling (Meng et al., 2022). In each case, induction of NADPH oxidase served to limit pathogenic activation of endosomal TLRs by self-NA. In the current study, we provide evidence for a parallel B cell–specific mechanism promoting NOX2-dependent lupus risk. Specifically, we show that both murine and human B cells lacking NOX2 complex genes exhibit dysregulated endosomal TLR trafficking, resulting in enhanced TLR signaling, increased TLR-dependent humoral responses, and the propensity toward accelerated lupus-like disease.
Endosomal TLR signals play a critical role in facilitating protective immune responses to invading pathogens, while also inducing tolerance breaks in autoimmunity. To achieve a balance between protective and pathogenic responses, various strategies have evolved to limit TLR activation by self-NA. In addition to sequestering NA-sensing TLRs in endosomal compartments, thereby limiting exposure to circulating self-NA, the induction of intracellular trafficking machinery serves to regulate the nature, amplitude, and duration of TLR signals. In this context, TLR signaling occurs within distinct compartments, such that NF-κB activation in early endosomes occurs before IRF7 induction in late endosomes (Honda et al., 2005). The adaptor protein AP3 has a key role in guiding this ordered transition between endosomal stages, ultimately resulting in endosomal fusion with lysosomes and signal termination (Sasai et al., 2010). Although these processes have been predominantly studied in myeloid lineages, our group has demonstrated that regulation of endolysosomal trafficking similarly impacts TLR signaling thresholds in B cells. Using mice deficient in αVβ3 integrin components, we showed that αV integrin promotes the recruitment of autophagy component LC3 to TLR9-containing endosomes and that this induction of a non-canonical autophagy program facilitates endolysosomal trafficking. In the absence of αVβ3, TLR-dependent endosomal flux in B cells is disturbed, resulting in prolonged NF-κB activation, enhanced B cell proliferation, a striking increase in TLR-dependent GC responses to viral antigens, and the development of humoral autoimmunity (Acharya et al., 2016, 2020; Raso et al., 2018).
The regulation of endolysosomal trafficking in B cells by a non-canonical autophagy program is reminiscent of LC3-associated phagocytosis (LAP) described in macrophages. Consistent with the identification of NOX2 as a key regulator of LAP (Geng et al., 2022; Huang et al., 2009; Ligeon et al., 2021; Martinez et al., 2015; Tam et al., 2014), we report that LC3 recruitment to early endosomes and subsequent endolysosomal trafficking is reduced in NCF1-deficient B cells, resulting in an increased amplitude of TLR signaling. Notably, a recent publication questioned the requirement for NADPH oxidase in LAP (Gordon et al., 2022). While seemingly contradicting our findings, this study quantified LC3 lipidation in BMDM at a single late time point. Here, we show that NADPH oxidase enzymatic activity is not absolutely required for TLR-driven LC3 lipidation, but rather regulates the kinetic of endolysosomal flux resulting in a qualitative difference in TLR signaling. We propose that, by delaying lysosomal fusion and signaling termination, loss-of-function NCF1 and NCF2 variants lower the threshold for TLR activation resulting in a B cell–intrinsic contribution to lupus risk. Thus, our data point to an important role for pathways modulating the strength of TLR signals in regulating B cell activation by self or viral antigens.
Mechanistically, in vitro studies using BMDM have shown that NOX2-dependent ROS inhibits ATG4B, a protease able to delipidate and recycle LC3 from the phagosome surface, resulting in phagosome maturation and accelerated lysosome fusion (Ligeon et al., 2021). In addition, NOX2-derived ROS promotes acidification of phagosomes, such that chemical inhibition of either ROS or the proton channel 1 (Hv1) similarly impacts phagosome maturation (Geng et al., 2022). Additional studies are needed to address whether these pathways also impact TLR-dependent LC3 lipidation and endolysosomal trafficking in B cells.
Importantly, the B cell–intrinsic mechanism we describe here does not preclude a parallel contribution of defective efferocytosis to the pathogenesis of autoimmunity. Rather, we hypothesize that synergistic contributions by myeloid and B cell lineages explain the markedly increased lupus risk in NCF1 and NCF2 variant carriers (OR >2.5 for both genes). In addition, reduced NOX2 activity may also impact other B cell activation pathways beyond endosomal TLRs. For example, BCR ligation promotes NOX-dependent ROS production, although the functional significance remains unclear since BCR-dependent calcium flux is not altered in Ncf1−/− and Cybb−/− B cells (Richards and Clark, 2009; Wheeler and Defranco, 2012). Ultimately, coordination of these dysregulated BCR and TLR signaling pathways in NOX2-deficient B cells is likely to orchestrate breaks in immune tolerance.
Finally, we show that B cell–intrinsic deletion of NOX2 genes specifically enhances humoral responses to immunization with NA-containing VLP. By increasing virus-specific antibody titers and durability, reduced B cell NOX2 activity is predicted to promote long-term protection from pathogen rechallenge. This may explain the observation that, despite markedly increasing the risk of SLE and other humoral autoimmune syndromes, NCF1Arg90His is a common variant with allele frequency ranging from 10 to 20% in Asia (Zhao et al., 2017). By linking the modulation of endolysosomal trafficking to TLR signal strength, our data raise the possibility that NCF1 and NCF2 loss-of-function variants might have been selected in the population in response to evolutionary pressures from viral pathogens.
Materials and methods
Mice
Animal strains include the following: B6 (C57BL/6J; Jax stock #000664), B6 CD45.1 (Jax stock #002014), Mb1cre (Hobeika et al., 2006; Jax stock #020505), Ncf1−/− (Jackson et al., 1995; Jax stock #027331), and Ptpn22RISK (Dai et al., 2013) mice. Cybbfl/fl mice (Pepping et al., 2017) were generously provided by Dr. Annadora Bruce-Keller (Louisiana State University, Baton Rouge, LA, USA), and Sle1b mice (Morel et al., 2000, 2001) were provided by Dr. Ziaur Rahman (Penn State, State College, PA, USA). All mice were maintained on a C57BL/6 background and bred/maintained in a specific pathogen–free facility at Seattle Children’s Research Institute. Animal experiments were conducted according to the protocols approved by the Institutional Animal Care and Use Committee at Seattle Children’s Research Institute.
Immunization models
Mice were intraperitoneally injected with 10 µg ssRNA-containing Qβ-VLP in PBS or 100 µg NP-CGG (Biosearch Technologies) in alum (Imject Alum Adjuvant; 1:1 ratio; Thermo Fisher Scientific). VLP reagents were provided by Baidong Hou (Chinese Academy of Sciences, Beijing, China). Time points for post-immunization analyses are indicated in the manuscript and figure legends.
Antibodies and reagents
Anti-murine antibodies were used in the study and include the following: FAS-BV510 (563646), FAS-BV421(562633), FAS-PE (554258), CD19-PE-Cy7 (1D3, 552854), IgD-BV786 (563618), CD138-APC (558626), B220-PE (RA3-6E2, 553090), and CD24-FITC (M1/69, 553261) were from BD Biosciences. B220-Percp-Cy5.5 (RA3-6B2, 103236), B220-BV650 (103241), B220-PE-Cy7 (RA3-6B2, 103222), CD19-BV605 (115539), CD267 (TACI)-PE (133403), CD21-BV421 (7E9, 123414), CD23-PE-Cy7 (B3B4, 101614), and CD138-APC-Cy7 (281-2, 142529) were from BioLegend. B220-APC-Cy7 (RA3-6B2, 47-0452), CD11b-PE (M1/70, 12-0112), CD11b-APC-eF780(M1/70, 47011282), and IgG (H+L)-HRP (62-6520) were from Thermo Fisher Scientific. IgM- PerCP-eFluor 710 (5016074) was from Thermo Fischer Scientific. IgD-PE (11-26, 1120-09) IgM-HRP (1020-05), IgG-HRP (1030-05), IgG2b-HRP (1090-05), IgG2c-HRP (1079-05), and IgG2c-FITC (1079-02) were from Southern Biotech. CD38-AF700 (90, 56-0381-82), CD38-FITC (11-0381-82), IRF4-PerCP-eFluor 710(46-9858-82), CD11c-APC (17-0114-81), and CD3-AF647(500A2, HM3421) were from Life Technologies. IgG-FITC (F2012), β-actin (A5316-.2MlL), and LC3 (L7543-100 μl) were from Sigma-Aldrich. NF-κB (8242S), Phospho-p44/42 MAPK (Erk1/2) (4376 s), Histone 3 (4499S), and LSD1(2139S) were from Cell Signaling Technology. P62 (03-GP62-C) was from American Research Products. Anti-human antibodies that were used in the study included the following: CD38-PerCP-CY5.5 (HIT2, 551400), CD3-AF700 (SP34-2, 42530), and CD20-FITC (L27, 347673) were from BD Bioscience. IgM-PacBlue (314514) and CD138-BV605 (356519) were from BioLegend. CD27-APC (O323, 17-0279-42) was from Life Technologies. CD19-PE-Cy7 (HIB19, 25-0199) and LAMP2 (MA1-205) were from Thermo Fisher Scientific. IgG F(ab′)-PE (2042-09), IgG (2040-01), IgM (2020-01), IgG-AP (2040-04), and IgM-AP (2020-04) were from Southern Biotech. EEA1 (1G11, ab70521) was from Abcam. NCF1 (p47phox) (4312 s) was from Cell Signaling. Anti-rabbit IgG-HRP (7074S) was from Cell Signaling Technology. Anti-guinea pig Ig-HRP (P0141) was from Dako. FITC anti-mouse peanut agglutinin (PNA) (Fl-1071) was from Vector Labs. AF647 labeled CpG was acquired from Integrated DNA Technology (IDT). Hoechst (H1399) and NHS Ester (Live-dead dye)-AF350 (A10168) were from Thermo Fisher Scientific. DNA sodium salt from the calf thymus (D3664-5X2MG) was from Sigma-Aldrich. Sm-RNP is from Arotec Diagnostics. NP-CGG (N-5055E-5) and NP-BSA ratio 32 (N-5050H-10) were from Biosearch Technology. All VLP reagents were provided by B. Hou (Chinese Academy of Sciences, Beijing, China).
BM chimeras
For aged autoimmune chimera models, BM was harvested from the femora and tibiae of C57BL/6 (WT), Ncf1−/−, and µMT mice. Extracted cells were resuspended in RBC lysis buffer (ACK) and filtered through 40-µm strainers. Single-cell suspensions were depleted for CD138+ cells (130–098-257; Miltenyi Biotec). CD138-depleted WT or NCF1−/− donor BM was mixed with µMT BM at a 20:80 ratio, and 6 × 106 total BM was injected retro-orbitally into lethally irradiated (450 cGy × 2 doses) µMT recipients. Injected recipients were placed on Baytril-medicated water for 2 wk after irradiation. Resulting BM chimeras were bled and sacrificed 24–26 wk after transplantation. For VLP competitive BM chimera, BM was harvested from the femora and tibiae from congenically marked donors of WT (45.2), NCF1−/− (CD45.2), and WT (CD45.1). Extracted cells were resuspended in RBC lysis buffer (ACK) and filtered through 40-µm strainers. Single-cell suspensions were depleted for CD138+ cells (130–098-257; Miltenyi Biotec). CD138-depleted WT (CD45.2) or Ncf1−/− (CD45.2) donor BM was mixed with WT (CD45.1) BM at a 1:1 ratio, and 6 × 106 total BM was injected retro-orbitally into lethally irradiated (450 cGy × 2 doses) µMT recipients. After 6 wk of hematopoietic reconstitution, experimental animals were injected with 2 µg ssRNA-containing Qβ-VLP and sacrificed at 13 days after immunization.
BM chimera data analysis
The gating strategy for the identification of VLP-specific GC B cells is shown in Fig. S1. Briefly, VLP-specific GC B cells were identified based on positive VLP-AF647 staining within the CD95+CD38− GC gate (of total B220+CD19+ B cells). BM donor was identified using CD45.1 and CD45.2 congenic markers. Reconstitution frequency for each animal was determined by the percent of CD45.2+ cells versus the percent of CD45.1+ cells in the non-GC B cell population.
VLP+ GC selection ratio was calculated using the following formula:
Normalization for cohort batch effect was achieved by dividing the VLP+ GC selection ratio for each animal by the average VLP+ GC selection ratio in the WT:WT control chimeras in each cohort (n = 2 independent BM chimera models).
Flow cytometry and cell sorting
Splenocytes and BM cells were harvested into ice-cold complete RPMI 1640 media (with 10% FBS, 1% pen/strep, 1% sodium pyruvate, 1% HEPES, 1% GlutaMAX, and 0.1% β-ME). Cells were incubated with RBC lysis buffer (ACK; Gibco, Thermo Fisher Scientific) for 2 min and filtered through 40-μm cell strainers (Corning). Single-cell suspensions were stained with fluorescence-labeled antibodies for 20 min at 4°C. For the detection of NP or VLP-specific cells, PE-NP and AF647-QβVLP were used to stain the cells together with other surface antibodies. Fixation and permeabilization were performed for intracellular staining (Cytofix/Cytoperm; BD Biosciences) or intranuclear staining (True-Nuclear; BioLegend). Samples were acquired using an LSRII flow cytometer (BD) and analyzed by FlowJo software (Tree Star, Inc.). For sorting of spleen MZ B cells, B cells were enriched with a negative selection cocktail (Stem Cell Technologies) and were labeled with anti-B220-PE, anti-CD21-BV421, anti-CD23-PEcy7, and anti-CD24-FITC antibodies. Cells were sorted with FACs Aria (BD).
In vitro proliferation assay
Sorted MZ cells were resuspended and stained with 1:1,000 diluted CellTrace Violet (C34557; Thermo Fisher Scientific) at 5 × 107 cells/ml. Cells were then incubated with the stain at 37°C and agitated every 5 min for 15 min. 5 ml of prewarmed complete RPMI media was added for 5 min at 37°C to quench the staining. Cells were spun down and incubated with media with or without R848.
ANA quantification
Diluted serum (1:200) was added to fixed Hep-2 ANA slides (30472; Bio-Rad). FITC-conjugated goat anti-mouse IgG or IgG2c (1:200) served as the detection antibody, and slides were counterstained with DAPI. Fluorescence images were obtained using a Leica DM6000B microscope, Leica DFL300 FX camera, and Leica Application Suite Advanced Fluorescence software at ×40 with a constant 5-s exposure.
Spleen immunofluorescence staining
Mouse spleens were embedded in an optimal cutting temperature compound and frozen over dry ice. 10-μm sections were cut on a cryostat, mounted on slides (Superfrost Excell; Thermo Fisher Scientific), and fixed in −20°C acetone for 20 min. After rehydration in staining buffer (PBS, 1% goat serum, 1% BSA, 0.1% Tween 20), slides were stained with anti-mouse B220-PE (1:100 dilution), PNA-FITC (1:750 dilution), and CD3 (1:50 dilution). Images were acquired using a Leica DM6000B microscope, Leica DFL300 FX camera, and Leica Application Suite Advanced Fluorescence software.
Confocal microscopy and image analysis
HBL cells were seeded onto poly-L-lysine–coated glass coverslips (BD BioCoat) in complete IMDM with (10% FBS, 1% pen/step, 0.1% β-ME) and allowed to attach for 1 h at 37°C. Cells were stimulated with 3 µM of CPG (ODN 2395; InvivoGen) or 1.5 µM of AF647-CpG (Integrated DNA technologies) for the indicated time. After stimulation, cells were fixed with 4% paraformaldehyde for 20 min at room temperature and permeabilized with saponin (0.2% saponin in PBS, 0.03 M sucrose, 1% BSA) for 10 min at room temperature. Non-specific binding was prevented by blocking for 1 h at room temperature with a blocking buffer (2% goat serum, 1% BSA, 0.1% cold fish skin gelatin, 0.1% saponin, and 0.05% Tween-20 in PBS). Cells were incubated overnight at 4°C with specific primary antibodies (LC3 1:500 dilution, EEA1 1:200 dilution, and LAMP2 1:600 dilution) in dilution buffer (PBS, 0.05% Tween-20, 1% BSA, 0.1% saponin). Cells were washed three times in dilution buffer (5 min each) and incubated in secondary antibodies (1:250 dilution) in dilution buffer for 1 h at room temperature. Cell nuclei were stained with Hoechst 33342 and the cover slips were mounted with ProLong Antifade (Invitrogen). Cells were imaged on a 63× oil objective (aperture 1.4) and the images were captured on a Leica TCS SP5 confocal microscope. Post-acquisition analysis, such as contrast adjustment, z-slice stacking, and intensity measurement, was performed using FIJI software. For colocalization quantification, Pearson’s correlation and Manders’ colocalization were calculated using Coloc 2 plugin from FIJI software. Lysosome volume and LC3 puncta quantification were calculated using a 3D object counter in FIJI.
Antibody ELISA
For anti-VLP and anti-NP detection, 96-well Nunc-Immuno MaxiSorp plates (Thermo Fisher Scientific) were coated with 1 µg/ml Qβ-VLP or 50 µg/ml NP-CGG overnight at 4°C in PBS. For autoantibody detection, plates were coated with 0.005% Poly-L-lysine (A-005-C; Milipore Sigma-Aldrich) and then 100 µg/ml calf thymus dsDNA (D3664-5X2MG; Sigma-Aldrich) or 5 µg/ml Sm/RNP (ATR01-10; Arotec Diagnostic Limited) at room temperature for 2 h. Plates were blocked for 1 h with 1% BSA in PBS before incubation with serially diluted serum in 1% BSA and 0.05% Tween in PBS overnight at 4°C. Isotype- and subclass-specific antibodies were detected using goat anti-mouse IgM, IgG, or IgG(1, 2b, 2c, 3)-HRP (1:2,000 dilution; SouthernBiotech). Peroxidase reactions were developed using OptEIA TMB substrate reagent set (BD Biosciences), and reactions stopped using sulfuric acid. Absorbance was measured at 450 nm using a SpectraMax i3X microplate reader (Molecular Devices).
ELISpot
For human total antibody ELISpot, MultiScreen 96-well plates (MSHAS4510; Milipore Sigma-Aldrich) were pretreated with 70% ethanol for <1 min and then were incubated with 10 µg/ml of capture antibodies anti-human IgG or IgM overnight at 4°C. Plates were washed three times with culture media, blocked with media at 37°C for 2 h, and then incubated with cells at 37°C overnight. The wells were washed with PBST (PBS+0.1%Tween) and were added with 1 µg/ml detection antibodies anti-human IgG-AP or IgM-AP in PBST+2% BSA at room temperature for 2 h. Wells were washed three times with PBST. Spots were developed using an AP substrate kit (Vector Laboratories) and counted using a Mabtech IRIS reader. For mouse anti-VLP ELISpot, MultiScreen 96-well ELISPOT plates (MAIPS4510; Millipore Sigma) were coated overnight at 4°C with 2 μg/ml VLPs. Plates were blocked with 10% FBS in complete DMEM (Thermo Fisher Scientific) for 1 h at room temperature and then incubated with cells overnight at 37°C. Plates were washed and incubated with goat anti-mouse IgM, IgG, or IgG2c-HRP (1:2,000 dilution). Spots were developed using the AEC substrate kit (Vector Laboratories) and scanned and counted using the CTL ELISPOT reader.
Western blot
For total cell lysate, cells were lysed for 30 min on ice in radioimmunoprecipitation assay (RIPA) buffer with 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS supplemented with sodium orthovanadate and protease inhibitor cocktail. Lysates were centrifuged for 10 min at 4°C at 14,000 g, and supernatant was collected. For nuclear extracts, cells were lysed after stimulation in hypotonic nuclear extraction buffer (1 M Hepes, PH7.5, 5 M NaCl, 0.5 M EDTA PH8, 50% glycerol, 10% Igepal, and 10% TritonX100) with 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS supplemented with sodium orthovanadate and protease inhibitor cocktail for 10 min followed by centrifugation at 1,700 g for 8 min at 4°C to pellet the nuclei. The nuclei were then resuspended in the RIPA buffer described above. Proteins were separated by electrophoresis using NuPage-Bis-Tris gels (Invitrogen) and blotted onto polyvinylidene difluoride membranes. Nonspecific binding was blocked by 5% BSA in TBS-Tween followed by incubation with primary antibody (1:1,000 dilution) overnight at 4°C and secondary antibody HRP-conjugated antibody (1:5,000 dilution) for 1 h at room temperature. Membranes were washed thoroughly with TBS-Tween (0.1%) after antibody incubations and developed using ECL reagents (Millipore). For reprobing, blots were stripped for 30 min at 37°C with Restore PLUS stripping buffer (Thermo Fisher Scientific). ImageJ software was used for band densitometry.
Gene editing
CRISPR crRNAs targeting NCF1 and CCR5 were identified using the CRISPick tool developed by Broad Institute (Doench et al., 2016; Sanson et al., 2018) and synthesized by Integrated DNA Technologies, Inc. (IDT). For HBL1 editing, NCF1 exon 4 crRNA and trans-activating crRNA (tracrRNA; IDT) hybrids were mixed with Cas9 nuclease (IDT) at the ratio of 1.2:1. Cells were transfected by Neon electroporation (MPK1025; Thermo Fisher Scientific). Transfected cells were single-cell sorted, expanded with media, and lysed with the methods described above. Western blot was used to validate knockout on protein level. For human peripheral blood mononuclear cell (PBMC) editing, NCF1 exon 2 and exon 4 crRNA and transactivating crRNA hybrids were mixed with Cas9 nuclease (IDT) at the ratio of 2.4:1. CCR5 crRNA hybrids were mixed with the same molar ratio for the control group. Cells were transfected by Lonza 4D Nucleofector Unit (AAF-1003B and X; Lonza). Transfected cells were lysed with the methods described above, and western blot was used to validate knockout on the protein level. Guide sequences used for CRISPR editing are provided in Table 1.
Table 1.
Guide sequences used for CRISPR editing
| Gene target | Guide sequence (5′–3′) | tracrRNA binding sequence (5′–3′) |
|---|---|---|
| CCR5 | CAAUGUGUCAACUCUUGACA | GUUUUAGAGCUAUGCU |
| NCF1 exon 2 | CACCAGGAACAUGUACACCU | |
| NCF1 exon 4 | GCACCUUGAAGAAGUCGAGG |
Murine in vitro B cell stimulation
Splenic B cells were enriched from mouse spleens using the EasySep Mouse B cells Isolation kit (catalog #19854A; StemCell Tech). BALB/c 3T3 fibroblasts stably transfected with both CD40L and BAFF (40LB cells) (Nojima et al., 2011) were cultured in DMEM (10566016; Thermo Fisher Scientific) with 10% FBS and 1% pen/strep and were inactivated by 8,000 rad irradiations 1 day prior to plating. B cells, enriched from splenocytes by EasySep mouse B cells isolation kit (19854; StemCell Tech), were plated onto 40LB cells cultured in complete IMDM with (10% FBS, 1% pen/step, 0.1% β-ME) and stimulated with 10 ng/ml recombinant murine IL-21 (210-21; Pepro Tech) and/or 0.25 μg/ml, 0.75 μg/ml R848 (tlrl-r848-5; Invivogen). On day 4, cells were analyzed by flow cytometry.
Human B cell plasma cell differentiation
Human B cells were isolated from PBMCs by EasySep Human B cell isolation kit (17964; StemCell Tech) and were cultured in complete IMDM media with (10% FBS and 0.1% β-ME) and stimulated with B cell activation cocktail (Hung et al., 2018) 100 ng/ml MCD40L (ALX-522-100-C010; Enzo Life Sciences), 1 μg/ml CpG ODN2006 (NC9902842; Invivogen), 50 ng/ml IL-2 (200-02; Peprotech), 50 ng/ml IL-10 (200-10; Peprotech), and 10 ng/ml IL-15 (130-095-765; Miltenyi Biotec). Cells were edited with Cas9-RNP described above on day 2, and culture volume was double with the same media on day 3. Cell densities were maintained at 1.5 million cells per ml from days 4 to 7 and analyzed by flow cytometry on days 5 and 7 and by ELISPOT on day 7.
Statistical analysis
Statistical significance was calculated using two-tailed paired, unpaired Student’s t test, two-tailed Fisher's exact test, or ANOVA as indicated in each figure by Prism (GraphPad) software Version 9.0. The P values were considered significant when P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), and P < 0.0001 (****). No statistical tests were used to predetermine the sample size.
Online supplemental material
Fig. S1 shows Qβ-VLP immunization of Tlr7−/− mice, the flow cytometry gating strategy used to analyze the WT:Ncf1−/− competitive chimera model, and the gating strategy and surface phenotype of TACI+IRF4+ mouse plasma cells. Fig. S2 includes additional immunophenotyping data from the Mb1cre.Cybbfl/fl and Ptpn22R/NR.Mb1cre.Cybbfl/fl mouse models. Fig. S3 includes additional data from the murine WT versus Ncf1−/− B cell in vitro stimulation experiment (Fig. S3 A), confirmation of successful CRISPR-mediated disruption of NCF1 in HBL1 cells (Fig. S3 B) and primary human B cell (Fig. S3 C), an independent replicate experiment of the microscopy data shown in Fig. 6 (Fig. S3, D and E), and LC3-II lipidation and p62 degradation by western blot in CpG-stimulated Ncf1+/+ and Ncf1−/− MZ B cells (Fig. S3, F–I). Fig. S4 shows LC3 lipidation in repose to R848 stimulation. Fig. S5 shows EEA1 and LC3 colocalization, and LC3 and LAMP2 colocalization in TLR7 and TLR9-stimulated WT versus NCF1−/− HBL1 cells.
Supplementary Material
contains original blots for Fig. 5.
contains original blots for Fig. 6.
contains original blots for Fig. S3.
Acknowledgments
We thank Karen Sommer and Andrea Repele for laboratory management, Jonathan Woods for technical assistance, and Socheath Jit Khim for animal colony management.
This work was supported by National Institutes of Health grants 5R01AI151167 (to M. Acharya), R01AR073938 (to S.W. Jackson), and R01AR075813 (to S.W. Jackson). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions: S. Liu, M. Acharya, and S.W. Jackson conceptualized the research project and wrote the manuscript. S. Liu, J. Lagos, N.M. Shumlak, A.D. Largent, S.T.E. Lewis, U. Holder, and S.W. Du performed experiments. Y. Liu and B. Hou generated VLP reagents and provided technical assistance regarding VLP immunization models. All authors reviewed and commented on the manuscript.
Data availability
Data are available in the article itself and its supplementary materials. The original data are available from the corresponding author(s) upon reasonable request.
References
- Acharya, M., Raso F., Sagadiev S., Gilbertson E., Kadavy L., Li Q.Z., Yan M., Stuart L.M., Hamerman J.A., and Lacy-Hulbert A.. 2020. B cell αv integrins regulate TLR-driven autoimmunity. J. Immunol. 205:1810–1818. 10.4049/jimmunol.1901056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya, M., Sokolovska A., Tam J.M., Conway K.L., Stefani C., Raso F., Mukhopadhyay S., Feliu M., Paul E., Savill J., et al. 2016. αv Integrins combine with LC3 and atg5 to regulate Toll-like receptor signalling in B cells. Nat. Commun. 7:10917. 10.1038/ncomms10917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón-Riquelme, M.E., Ziegler J.T., Molineros J., Howard T.D., Moreno-Estrada A., Sánchez-Rodríguez E., Ainsworth H.C., Ortiz-Tello P., Comeau M.E., Rasmussen A., et al. 2016. Genome-wide association study in an amerindian ancestry population reveals novel systemic lupus erythematosus risk loci and the role of European admixture. Arthritis Rheumatol. 68:932–943. 10.1002/art.39504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amur, S., Parekh A., and Mummaneni P.. 2012. Sex differences and genomics in autoimmune diseases. J. Autoimmun. 38:J254–J265. 10.1016/j.jaut.2011.12.001 [DOI] [PubMed] [Google Scholar]
- Arkatkar, T., Du S.W., Jacobs H.M., Dam E.M., Hou B., Buckner J.H., Rawlings D.J., and Jackson S.W.. 2017. B cell-derived IL-6 initiates spontaneous germinal center formation during systemic autoimmunity. J. Exp. Med. 214:3207–3217. 10.1084/jem.20170580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, G.M., and Kagan J.C.. 2009. A cell biological view of toll-like receptor function: Regulation through compartmentalization. Nat. Rev. Immunol. 9:535–542. 10.1038/nri2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begovich, A.B., Carlton V.E., Honigberg L.A., Schrodi S.J., Chokkalingam A.P., Alexander H.C., Ardlie K.G., Huang Q., Smith A.M., Spoerke J.M., et al. 2004. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am. J. Hum. Genet. 75:330–337. 10.1086/422827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottini, N., Musumeci L., Alonso A., Rahmouni S., Nika K., Rostamkhani M., MacMurray J., Meloni G.F., Lucarelli P., Pellecchia M., et al. 2004. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat. Genet. 36:337–338. 10.1038/ng1323 [DOI] [PubMed] [Google Scholar]
- Brown, G.J., Cañete P.F., Wang H., Medhavy A., Bones J., Roco J.A., He Y., Qin Y., Cappello J., Ellyard J.I., et al. 2022. TLR7 gain-of-function genetic variation causes human lupus. Nature. 605:349–356. 10.1038/s41586-022-04642-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cale, C.M., Morton L., and Goldblatt D.. 2007. Cutaneous and other lupus-like symptoms in carriers of X-linked chronic granulomatous disease: Incidence and autoimmune serology. Clin. Exp. Immunol. 148:79–84. 10.1111/j.1365-2249.2007.03321.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, A.M., Kashgarian M., and Shlomchik M.J.. 2012. NADPH oxidase inhibits the pathogenesis of systemic lupus erythematosus. Sci. Transl. Med. 4:157ra141. 10.1126/scitranslmed.3004801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappione, A.J., Pugh-Bernard A.E., Anolik J.H., and Sanz I.. 2004. Lupus IgG VH4.34 antibodies bind to a 220-kDa glycoform of CD45/B220 on the surface of human B lymphocytes. J. Immunol. 172:4298–4307. 10.4049/jimmunol.172.7.4298 [DOI] [PubMed] [Google Scholar]
- Chiang, K., Largent A.D., Arkatkar T., Thouvenel C.D., Du S.W., Shumlak N., Woods J., Li Q.Z., Liu Y., Hou B., et al. 2021. Cutting edge: A threshold of B cell costimulatory signals is required for spontaneous germinal center formation in autoimmunity. J. Immunol. 207:2217–2222. 10.4049/jimmunol.2100548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman, R.L., Sher A., and Seder R.A.. 2010. Vaccine adjuvants: Putting innate immunity to work. Immunity. 33:492–503. 10.1016/j.immuni.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, I.F. 2008. Sexual dimorphism of humoral immunity with human vaccines. Vaccine. 26:3551–3555. 10.1016/j.vaccine.2008.04.054 [DOI] [PubMed] [Google Scholar]
- Dai, X., James R.G., Habib T., Singh S., Jackson S., Khim S., Moon R.T., Liggitt D., Wolf-Yadlin A., Buckner J.H., and Rawlings D.J.. 2013. A disease-associated PTPN22 variant promotes systemic autoimmunity in murine models. J. Clin. Invest. 123:2024–2036. 10.1172/JCI66963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y., and Tsao B.P.. 2017. Updates in lupus genetics. Curr. Rheumatol. Rep. 19:68. 10.1007/s11926-017-0695-z [DOI] [PubMed] [Google Scholar]
- Doench, J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R., et al. 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34:184–191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischinger, S., Boudreau C.M., Butler A.L., Streeck H., and Alter G.. 2019. Sex differences in vaccine-induced humoral immunity. Semin. Immunopathol. 41:239–249. 10.1007/s00281-018-0726-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng, L., Zhao J., Deng Y., Molano I., Xu X., Xu L., Ruiz P., Li Q., Feng X., Zhang M., et al. 2022. Human SLE variant NCF1-R90H promotes kidney damage and murine lupus through enhanced Tfh2 responses induced by defective efferocytosis of macrophages. Ann. Rheum. Dis. 81:255–267. 10.1136/annrheumdis-2021-220793 [DOI] [PubMed] [Google Scholar]
- Giles, J.R., Kashgarian M., Koni P.A., and Shlomchik M.J.. 2015. B cell-specific MHC class II deletion reveals multiple nonredundant roles for B cell antigen presentation in murine lupus. J. Immunol. 195:2571–2579. 10.4049/jimmunol.1500792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, R.A., Giannouli C., Raparia C., Bastacky S.I., Marinov A., Hawse W., Cattley R., Tilstra J.S., Campbell A.M., Nickerson K.M., et al. 2022. Rubicon promotes rather than restricts murine lupus and is not required for LC3-associated phagocytosis. JCI Insight. 7:e155537. 10.1172/jci.insight.155537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, K., Taura M., and Iwasaki A.. 2018. The interaction between IKKalpha and LC3 promotes type I interferon production through the TLR9-containing LAPosome. Sci. Signal. 11:eaan4144. 10.1126/scisignal.aan4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobeika, E., Thiemann S., Storch B., Jumaa H., Nielsen P.J., Pelanda R., and Reth M.. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. USA. 103:13789–13794. 10.1073/pnas.0605944103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, K., Ohba Y., Yanai H., Negishi H., Mizutani T., Takaoka A., Taya C., and Taniguchi T.. 2005. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 434:1035–1040. 10.1038/nature03547 [DOI] [PubMed] [Google Scholar]
- Hou, B., Saudan P., Ott G., Wheeler M.L., Ji M., Kuzmich L., Lee L.M., Coffman R.L., Bachmann M.F., and DeFranco A.L.. 2011. Selective utilization of Toll-like receptor and MyD88 signaling in B cells for enhancement of the antiviral germinal center response. Immunity. 34:375–384. 10.1016/j.immuni.2011.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., Canadien V., Lam G.Y., Steinberg B.E., Dinauer M.C., Magalhaes M.A., Glogauer M., Grinstein S., and Brumell J.H.. 2009. Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl. Acad. Sci. USA. 106:6226–6231. 10.1073/pnas.0811045106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, K.L., Meitlis I., Hale M., Chen C.Y., Singh S., Jackson S.W., Miao C.H., Khan I.F., Rawlings D.J., and James R.G.. 2018. Engineering protein-secreting plasma cells by homology-directed repair in primary human B cells. Mol. Ther. 26:456–467. 10.1016/j.ymthe.2017.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S.H., Gallin J.I., and Holland S.M.. 1995. The p47phox mouse knock-out model of chronic granulomatous disease. J. Exp. Med. 182:751–758. 10.1084/jem.182.3.751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S.W., Jacobs H.M., Arkatkar T., Dam E.M., Scharping N.E., Kolhatkar N.S., Hou B., Buckner J.H., and Rawlings D.J.. 2016. B cell IFN-γ receptor signaling promotes autoimmune germinal centers via cell-intrinsic induction of BCL-6. J. Exp. Med. 213:733–750. 10.1084/jem.20151724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S.W., Scharping N.E., Kolhatkar N.S., Khim S., Schwartz M.A., Li Q.Z., Hudkins K.L., Alpers C.E., Liggitt D., and Rawlings D.J.. 2014. Opposing impact of B cell-intrinsic TLR7 and TLR9 signals on autoantibody repertoire and systemic inflammation. J. Immunol. 192:4525–4532. 10.4049/jimmunol.1400098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, C.O., Eisenstein M., Dinauer M.C., Ming W., Liu Q., John S., Quismorio F.P. Jr., Reiff A., Myones B.L., Kaufman K.M., et al. 2012. Lupus-associated causal mutation in neutrophil cytosolic factor 2 (NCF2) brings unique insights to the structure and function of NADPH oxidase. Proc. Natl. Acad. Sci. USA. 109:E59–E67. 10.1073/pnas.1113251108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, C.O., Yu N., Yoo D.G., Perez-Zapata L.J., Barbu E.A., Kaplan M.J., Purmalek M., Pingel J.T., Idol R.A., and Dinauer M.C.. 2017. Haploinsufficiency of NADPH oxidase subunit neutrophil cytosolic factor 2 is sufficient to accelerate full-blown lupus in NZM 2328 mice. Arthritis Rheumatol. 69:1647–1660. 10.1002/art.40141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks, S.A., Palmer E.M., Marin E.Y., Hartson L., Chida A.S., Richardson C., and Sanz I.. 2013. 9G4+ autoantibodies are an important source of apoptotic cell reactivity associated with high levels of disease activity in systemic lupus erythematosus. Arthritis Rheum. 65:3165–3175. 10.1002/art.38138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Howard, X., Sun C., Molineros J.E., Maiti A.K., Chandru H., Adler A., Wiley G.B., Kaufman K.M., Kottyan L., Guthridge J.M., et al. 2014. Allelic heterogeneity in NCF2 associated with systemic lupus erythematosus (SLE) susceptibility across four ethnic populations. Hum. Mol. Genet. 23:1656–1668. 10.1093/hmg/ddt532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyogoku, C., Langefeld C.D., Ortmann W.A., Lee A., Selby S., Carlton V.E., Chang M., Ramos P., Baechler E.C., Batliwalla F.M., et al. 2004. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am. J. Hum. Genet. 75:504–507. 10.1086/423790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligeon, L.A., Pena-Francesch M., Vanoaica L.D., Núñez N.G., Talwar D., Dick T.P., and Münz C.. 2021. Oxidation inhibits autophagy protein deconjugation from phagosomes to sustain MHC class II restricted antigen presentation. Nat. Commun. 12:1508. 10.1038/s41467-021-21829-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciano, B.E., Zerbe C.S., Falcone E.L., Ding L., DeRavin S.S., Daub J., Kreuzburg S., Yockey L., Hunsberger S., Foruraghi L., et al. 2018. X-linked carriers of chronic granulomatous disease: Illness, lyonization, and stability. J. Allergy Clin. Immunol. 141:365–371. 10.1016/j.jaci.2017.04.035 [DOI] [PubMed] [Google Scholar]
- Markine-Goriaynoff, D., and Coutelier J.P.. 2002. Increased efficacy of the immunoglobulin G2a subclass in antibody-mediated protection against lactate dehydrogenase-elevating virus-induced polioencephalomyelitis revealed with switch mutants. J. Virol. 76:432–435. 10.1128/JVI.76.1.432-435.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J., Malireddi R.K., Lu Q., Cunha L.D., Pelletier S., Gingras S., Orchard R., Guan J.L., Tan H., Peng J., et al. 2015. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat. Cell Biol. 17:893–906. 10.1038/ncb3192 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mauthe, M., Orhon I., Rocchi C., Zhou X., Luhr M., Hijlkema K.J., Coppes R.P., Engedal N., Mari M., and Reggiori F.. 2018. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 14:1435–1455. 10.1080/15548627.2018.1474314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Y., Ma J., Yao C., Ye Z., Ding H., Liu C., Li J., Li G., He Y., Li J., et al. 2022. The NCF1 variant p.R90H aggravates autoimmunity by facilitating the activation of plasmacytoid dendritic cells. J. Clin. Invest. 132:e153619. 10.1172/JCI153619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, L., Blenman K.R., Croker B.P., and Wakeland E.K.. 2001. The major murine systemic lupus erythematosus susceptibility locus, Sle1, is a cluster of functionally related genes. Proc. Natl. Acad. Sci. USA. 98:1787–1792. 10.1073/pnas.98.4.1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, L., Croker B.P., Blenman K.R., Mohan C., Huang G., Gilkeson G., and Wakeland E.K.. 2000. Genetic reconstitution of systemic lupus erythematosus immunopathology with polycongenic murine strains. Proc. Natl. Acad. Sci. USA. 97:6670–6675. 10.1073/pnas.97.12.6670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo, V.N., Young R.M., Schmitz R., Jhavar S., Xiao W., Lim K.H., Kohlhammer H., Xu W., Yang Y., Zhao H., et al. 2011. Oncogenically active MYD88 mutations in human lymphoma. Nature. 470:115–119. 10.1038/nature09671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn, F., and Ravetch J.V.. 2005. Divergent immunoglobulin g subclass activity through selective Fc receptor binding. Science. 310:1510–1512. 10.1126/science.1118948 [DOI] [PubMed] [Google Scholar]
- Nojima, T., Haniuda K., Moutai T., Matsudaira M., Mizokawa S., Shiratori I., Azuma T., and Kitamura D.. 2011. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat. Commun. 2:465. 10.1038/ncomms1475 [DOI] [PubMed] [Google Scholar]
- Olsson, L.M., Johansson A.C., Gullstrand B., Jönsen A., Saevarsdottir S., Rönnblom L., Leonard D., Wetterö J., Sjöwall C., Svenungsson E., et al. 2017. A single nucleotide polymorphism in the NCF1 gene leading to reduced oxidative burst is associated with systemic lupus erythematosus. Ann. Rheum. Dis. 76:1607–1613. 10.1136/annrheumdis-2017-211287 [DOI] [PubMed] [Google Scholar]
- Peng, Y., Martin D.A., Kenkel J., Zhang K., Ogden C.A., and Elkon K.B.. 2007. Innate and adaptive immune response to apoptotic cells. J. Autoimmun. 29:303–309. 10.1016/j.jaut.2007.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepping, J.K., Vandanmagsar B., Fernandez-Kim S.O., Zhang J., Mynatt R.L., and Bruce-Keller A.J.. 2017. Myeloid-specific deletion of NOX2 prevents the metabolic and neurologic consequences of high fat diet. PLoS One. 12:e0181500. 10.1371/journal.pone.0181500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan, J.D., Young R.M., Webster D.E., Roulland S., Wright G.W., Kasbekar M., Shaffer A.L. III, Ceribelli M., Wang J.Q., Schmitz R., et al. 2018. A multiprotein supercomplex controlling oncogenic signalling in lymphoma. Nature. 560:387–391. 10.1038/s41586-018-0290-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raso, F., Sagadiev S., Du S., Gage E., Arkatkar T., Metzler G., Stuart L.M., Orr M.T., Rawlings D.J., Jackson S.W., et al. 2018. αv Integrins regulate germinal center B cell responses through noncanonical autophagy. J. Clin. Invest. 128:4163–4178. 10.1172/JCI99597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, S.M., and Clark E.A.. 2009. BCR-induced superoxide negatively regulates B-cell proliferation and T-cell-independent type 2 Ab responses. Eur. J. Immunol. 39:3395–3403. 10.1002/eji.200939587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, C., Chida A.S., Adlowitz D., Silver L., Fox E., Jenks S.A., Palmer E., Wang Y., Heimburg-Molinaro J., Li Q.Z., et al. 2013. Molecular basis of 9G4 B cell autoreactivity in human systemic lupus erythematosus. J. Immunol. 191:4926–4939. 10.4049/jimmunol.1202263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov, A.V., Rubtsova K., Fischer A., Meehan R.T., Gillis J.Z., Kappler J.W., and Marrack P.. 2011. Toll-like receptor 7 (TLR7)-driven accumulation of a novel CD11c⁺ B-cell population is important for the development of autoimmunity. Blood. 118:1305–1315. 10.1182/blood-2011-01-331462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanson, K.R., Hanna R.E., Hegde M., Donovan K.F., Strand C., Sullender M.E., Vaimberg E.W., Goodale A., Root D.E., Piccioni F., and Doench J.G.. 2018. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 9:5416. 10.1038/s41467-018-07901-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai, M., Linehan M.M., and Iwasaki A.. 2010. Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science. 329:1530–1534. 10.1126/science.1187029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomchik, M.J. 2009. Activating systemic autoimmunity: B’s, T’s, and tolls. Curr. Opin. Immunol. 21:626–633. 10.1016/j.coi.2009.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam, J.M., Mansour M.K., Khan N.S., Seward M., Puranam S., Tanne A., Sokolovska A., Becker C.E., Acharya M., Baird M.A., et al. 2014. Dectin-1-dependent LC3 recruitment to phagosomes enhances fungicidal activity in macrophages. J. Infect. Dis. 210:1844–1854. 10.1093/infdis/jiu290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, D.C. 2018. How the phagocyte NADPH oxidase regulates innate immunity. Free Radic. Biol. Med. 125:44–52. 10.1016/j.freeradbiomed.2018.06.011 [DOI] [PubMed] [Google Scholar]
- Van Gassen, S., Callebaut B., Van Helden M.J., Lambrecht B.N., Demeester P., Dhaene T., and Saeys Y.. 2015. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 87:636–645. 10.1002/cyto.a.22625 [DOI] [PubMed] [Google Scholar]
- Walsh, E.R., Pisitkun P., Voynova E., Deane J.A., Scott B.L., Caspi R.R., and Bolland S.. 2012. Dual signaling by innate and adaptive immune receptors is required for TLR7-induced B-cell-mediated autoimmunity. Proc. Natl. Acad. Sci. USA. 109:16276–16281. 10.1073/pnas.1209372109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, M.L., and Defranco A.L.. 2012. Prolonged production of reactive oxygen species in response to B cell receptor stimulation promotes B cell activation and proliferation. J. Immunol. 189:4405–4416. 10.4049/jimmunol.1201433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, R.M., Wu T., Schmitz R., Dawood M., Xiao W., Phelan J.D., Xu W., Menard L., Meffre E., Chan W.C., et al. 2015. Survival of human lymphoma cells requires B-cell receptor engagement by self-antigens. Proc. Natl. Acad. Sci. USA. 112:13447–13454. 10.1073/pnas.1514944112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, B., Chen Y., Wu Q., Li P., Shao Y., Zhang J., Zhong Q., Peng X., Yang H., Hu X., et al. 2011. The association between single-nucleotide polymorphisms of NCF2 and systemic lupus erythematosus in Chinese mainland population. Clin. Rheumatol. 30:521–527. 10.1007/s10067-010-1567-3 [DOI] [PubMed] [Google Scholar]
- Zhao, J., Ma J., Deng Y., Kelly J.A., Kim K., Bang S.Y., Lee H.S., Li Q.Z., Wakeland E.K., Qiu R., et al. 2017. A missense variant in NCF1 is associated with susceptibility to multiple autoimmune diseases. Nat. Genet. 49:433–437. 10.1038/ng.3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains original blots for Fig. 5.
contains original blots for Fig. 6.
contains original blots for Fig. S3.
Data Availability Statement
Data are available in the article itself and its supplementary materials. The original data are available from the corresponding author(s) upon reasonable request.