Abstract
The human HIRA gene has been named after Hir1p and Hir2p, two corepressors which together appear to act on chromatin structure to control gene transcription in Saccharomyces cerevisiae. HIRA homologs are expressed in a regulated fashion during mouse and chicken embryogenesis, and the human gene is a major candidate for the DiGeorge syndrome and related developmental disorders caused by a reduction to single dose of a fragment of chromosome 22q. Western blot analysis and double-immunofluorescence experiments using a specific antiserum revealed a primary nuclear localization of HIRA. Similar to Hir1p, HIRA contains seven amino-terminal WD repeats and probably functions as part of a multiprotein complex. HIRA and core histone H2B were found to physically interact in a yeast double-hybrid protein interaction trap, in GST pull-down assays, and in coimmunoprecipitation experiments performed from cellular extracts. In vitro, HIRA also interacted with core histone H4. H2B- and H4-binding domains were overlapping but distinguishable in the carboxy-terminal region of HIRA, and the region for HIRA interaction was mapped to the amino-terminal tail of H2B and the second α helix of H4. HIRIP3 (HIRA-interacting protein 3) is a novel gene product that was identified from its HIRA-binding properties in the yeast protein interaction trap. In vitro, HIRIP3 directly interacted with HIRA but also with core histones H2B and H3, suggesting that a HIRA-HIRIP3-containing complex could function in some aspects of chromatin and histone metabolism. Insufficient production of HIRA, which we report elsewhere interacts with homeodomain-containing DNA-binding factors during mammalian embryogenesis, could perturb the stoichiometric assembly of multimolecular complexes required for normal embryonic development.
Developmental anomalies are frequently observed in humans in association with deletions affecting the proximal region of the long arm of chromosome 22. These 22q deletion disorders (22DD) include the DiGeorge syndrome (Mendelion inheritance in man [MIM] 188400) and the velocardiofacial syndrome (MIM 192430), whose phenotypes overlap partially. Main clinical features associated with 22DD comprise abnormalities of the face and palate, hypoplastic parathyroid glands, and conotruncal malformations (38), all probably resulting from anomalies of neural crest cells in the embryological region of the pharyngeal arches and pouches (26). Genetically, 90% of all patients have a large (approximately 3-Mb-long) 22q deletion. Although most deletions occur de novo, up to 28% could be inherited (38). In these familial cases whose transmission is autosomal dominant, the phenotypic expression of the same chromosomal defect is largely variable. The additional lack of correlation between the extent of the deletion and the intensity of the phenotype seems to argue against different contiguous genes being each responsible for distinct clinical features. Recently, however, the hypothesis that two causative genes each mapping to the same 22q region may together be responsible for the disorders has been reconsidered (5, 9).
In the absence of any mutation identified in the minority of patients without a confirmed 22q deletion, none of the genes cloned from the large commonly deleted region has been definitely linked to 22DD, making it necessary to study each plausible candidate in detail in order to evaluate its possible implication. This task will be facilitated by recent reports of a few patients whose unusually small 22q deletions with variant proximal and distal chromosomal breakpoints have reduced the critical region to less than 500 kb (6, 15, 28). Of the five genes characterized within the region, neither CTP, which codes for a mitochondrial citrate transport protein (20), CLTD (21), a clathrin heavy chain-like gene, nor the ubiquitously expressed TMVCF gene, which encodes a putative transmembrane protein (41), appears to be a plausible candidate. GSCL, a small (less than 4-kb) gene identified by systematic genomic sequencing, is more intriguing since it contains regions of homology to Goosecoid, a homeodomain gene whose specific expression pattern in the mouse suggests a role in the development of neural tissues (15).
The HIRA gene (27), first reported as TUPLE1 (18) for its partial similarity to the yeast general transcriptional repressor TUP1 (51), also appears to be an interesting candidate. It consists of 25 exons scattered over 100 kb of genomic DNA which is entirely reduced to single copy in 22DD patients (30). In situ hybridization experiments have demonstrated high levels of transcripts in the heart, cranial neural folds, pharyngeal arches, and circumpharyngeal neural crest of murine embryos (53) and in the neuroepithelium, neural crest-derived regions of the head, branchial arches, and pharyngeal pouches of chicken embryos (37). These evolutionarily conserved spatiotemporal expression patterns suggest that haploinsufficiency of HIRA could play an important part in the genesis of 22DD. HIRA was named for its sequence similarity to two yeast proteins, Hir1p and Hir2p (40). In the HIR family, which also comprises HIRA homologs subsequently identified in mice, chickens, and the fish Fugu rubripes, all protein sequences can be aligned over their entire lengths (29, 37, 39, 53). Shared features include basic nuclear localization signals, absence of an identifiable DNA- or RNA-binding motif, and seven characteristic WD repeats conserved at the amino terminus of all family members but Hir2p. WD repeats are ancient motifs that have been detected throughout the eukaryotic kingdom (33). They are present in a set of functionally diverse proteins that are part of macromolecular complexes (33) and in several instances have been shown to provide interfaces for protein interactions (14, 24, 25, 42, 50).
Contrasting with the single HIRA gene present in higher eukaryotes, the two budding yeast genes HIR1 and HIR2 likely have diverged and specialized functionally following an ancestral duplication event. HIR1 and HIR2 had been identified in the course of a genetic screen for Saccharomyces cerevisiae mutants with deregulated core histone gene transcription (34). In a wild-type strain, transcription of core histone genes is repressed outside late G1 and S phases, while in either a hir1 or hir2 mutant, transcription becomes constitutive throughout the cell cycle. To perform their cyclic repressive function, Hir1p and Hir2p, which can be coimmunoprecipitated (43), require the presence of other proteins, including the products of the SPT4, SPT5, and SPT6 genes, which are known for their impact on chromatin (3, 44) and transcription elongation by RNA polymerase II (19, 49). A regulatory protein complex targeted to the negative element identified in the promoter region of regulated histone gene loci by a DNA-binding factor that has been detected but not characterized would provoke transcriptional repression, probably in association with a local remodeling of chromatin structure (43).
In humans, where the number and complexity of core histone genes (1) far exceed those in yeast, it appeared that the main function of HIRA proteins would not necessarily involve the regulation of histone gene transcription. Rather, we hypothesized that during evolution, HIRA proteins could have built upon ancient biochemical properties to acquire the developmental role that is suggested by their regulated expression pattern during vertebrate embryogenesis. To explore the function of HIRA in multicellular species, we have looked for HIRA-interacting proteins (HIRIPs). We here report that human HIRA is a primarily nuclear protein that directly binds core histones H2B and H4, using overlapping but distinguishable domains outside its WD repeat region. HIRIP3, one of the other HIRA interactors identified, is a novel protein which also displays binding to two core histones, H2B and H3, suggesting that HIRA and HIRIP3 can assemble into a protein complex with a role at the level of chromatin components or structure.
MATERIALS AND METHODS
Preparation and affinity purification of anti-HIRA antiserum 1455.
The rabbit antiserum 1455, generated against the carboxy-terminal peptide of human HIRA NH2-CQEQLDILRDK-COOH (amino acids 1007 to 1017), was produced by Eurogentec (Belgium). The serum was affinity purified against a histidine-tagged carboxy-terminal fragment of HIRA (amino acids 930 to 1017) bacterially produced from the pQE32-HIRA3′ construct. After a 3-h induction (37°C) in the presence of 0.5 mM isopropyl-β-d-thiogalactopyranoside, Escherichia coli XL1-Blue harboring the pQE32-HIRA3′ construct was sonicated in buffer A (20 mM HEPES [pH 7.4], 20 mM imidazole, 10% glycerol, 2 mM β-mercaptoethanol, cocktail of protease inhibitors). After centrifugation (30 min, 10,000 rpm), the pellet was resuspended and incubated (3 h, 4°C) in buffer A supplemented with 3 M urea, 500 mM KCl, and 1% Triton X-100. After centrifugation (30 min, 10,000 rpm, 4°C), the supernatant was incubated overnight at 4°C in the presence of nickel-agarose beads (Qiagen). The beads were washed in buffer A supplemented with 3 M urea, 250 mM KCl, and 1% Triton X-100 in the first wash, with 2 M urea, 250 mM KCl, and 0.5% Triton X-100 in the second wash, and with 250 mM KCl in the final wash. The His6-tagged HIRA fragment was eluted by incubating the beads (10 min at 4°C) in 300 mM imidazole–20 mM HEPES (pH 7.4)–100 mM KCl–10% glycerol in the presence of protease inhibitors, subjected to 15% polyacrylamide gel electrophoresis (PAGE), and transferred onto Polyscreen polyvinylidene difluoride (PVDF) membranes (Dupont-NEN). The corresponding membrane fragment, 2 by 120 mm, was cut, saturated (1 h, room temperature) in phosphate-buffered saline (PBS) containing 5% bovine serum albumin and 0.3% Tween 20 (solution 1), and incubated overnight (4°C) with a 500-μl aliquot of serum 1455 diluted 1/10 in solution 1. The membrane fragment was washed once with 50 mM Tris (pH 7.5)–150 mM NaCl and once in PBS (each time for 20 min at room temperature). Affinity-purified antibodies were eluted in 500 μl of 0.2 M glycine (pH 2.8)–1 mM EGTA and neutralized with 1 M Trizma Base.
Western blot analysis.
For preparation of cellular extracts, frozen K562 cells were thawed in 10 mM HEPES (pH 7.4)–1.5 mM MgCl2–0.1 mM EGTA–0.5 mM dithiothreitol–5% glycerol in the presence of a cocktail of antiproteases. The supernatant of a low-speed centrifugation (2,000 rpm, 4°C) was used as the cytosolic extract. Following one wash, the pellet was resuspended in the same buffer adjusted to 350 mM NaCl and incubated on ice for 30 min. The supernatant of a high-speed centrifugation (10,000 rpm, 4°C) was used as the nuclear extract. Protein concentrations were determined by the Bradford assay (Bio-Rad). Polyacrylamide (10%) gel electrophoresis was performed with 30 μg of nuclear or cytosolic extracts deposited in each lane. Transfer was performed onto Polyscreen PVDF membranes. After saturation (1 h at room temperature in solution 1), the membrane was incubated for staining by using affinity-purified rabbit antiserum 1455 diluted 1/250 in 10 ml of solution 1. Three washes were performed in PBS with 0.3% Tween 20 (solution 2) followed by incubation (1 h) with a 1/10,000 dilution of a goat anti-rabbit immunoglobulin G (IgG) antiserum conjugated to peroxidase (Pierce). After three washes in solution 2, the reaction was developed by enhanced chemiluminescence using the Amersham system. Membrane stripping was performed in 62.5 mM Tris (pH 6.2)–2% sodium dodecyl sulfate (SDS)–100 mM β-mercaptoethanol (30 min, 50°C) followed by two washes (20 min, room temperature) in PBS. For staining inhibition assays, 30 μg of the immunizing HIRA peptide was mixed with the undiluted antiserum 1455 (20 min, room temperature) prior to addition of the rabbit antibody to the 10 ml of staining solution. As a control, 30 μg of an irrelevant peptide from the human T-cell leukemia virus type 1 envelope protein was used in place of the HIRA peptide. The final staining was performed with a 1/1,000 dilution of an anti-cyclin B1 monoclonal antibody (Santa Cruz Biotechnology) developed by peroxidase-conjugated goat anti-mouse IgG antibody (Pierce) diluted 1/10,000.
Immunofluorescence analysis.
HeLa and EW11 cells were grown on coverslips and fixed in 3% freshly prepared formaldehyde (15 min, room temperature). All subsequent steps were performed at room temperature. Cells were treated with 10% fetal calf serum (60 min) and permeabilized with 0.5% Triton X-100 (5 min). For K562 cells, fixation and permeabilization were performed in suspension. After a wash in 1× PBS, cells were incubated for 30 min with the appropriate antibodies diluted in PBS. The human anti-lamin B monoclonal antibody, kindly provided by J.-C. Brouet, has been described elsewhere (32). The anti-HIRA rabbit serum was affinity purified as described above. Double staining was performed by simultaneously incubating the two primary antibodies for 1 h, followed by extensive washing with PBS prior to staining development (1 h) with a mixture of species-specific fluoresceinated goat anti-human IgG and Texas red-conjugated goat anti-rabbit IgG antibodies (Jackson Laboratories). Nuclear DNA staining was obtained by incubation (5 min) with 4,6-diamidino-2-phenylindole (DAPI) solution (0.4 μg/ml). All samples were mounted in the presence of the Immumount antifade solution (Shandon Laboratories). An imaging system consisting of a Provis AX70 (Olympus) microscope equipped with a 60× oil immersion objective lens carrying a piezoelectric Z-axis focus device, a charge-coupled device camera (Photometrics), and a set of computer-controlled excitation filters was used to generate optical sections of fluorescently labeled cells. The light haze inherent to fluorescent signals was deblurred mathematically, using the Exhaustive Photon Reassignment software (Scanalytics, Fairfax, Va.) (7).
Two-hybrid screen in yeast.
The screening procedure to identify HIRIPs in a two-hybrid system performed with the budding yeast S. cerevisiae was based on an established protocol (17). The pLexA-HIRA construction used to produce the bait was obtained by insertion of nucleotides 87 to 3115 of the complete HIRA cDNA in the EcoRI-XhoI sites of the pEG202 vector. To generate this HIRA fragment, PCR amplification was performed on DNA from pcDNA3-HIRA (see below), using 5′-CGGAATTCACCAACCACAATGGCAAGC-3′ (forward primer) and 5′-GCGCTCGAGACTTGTCCCTCAGGATG-3′ (reverse primer) (restriction sites are underlined), followed by digestion with EcoRI and XhoI. S. cerevisiae EGY48 was transformed with this construct, resulting in the expression of a fusion between the LexA DNA-binding domain and HIRA (complete except for amino acids 1 to 9). The bait-expressing yeast strain was further transformed with a HeLa cDNA library whose construction in the pJG4-5 vector has been described elsewhere (17), for simultaneous expression of the bait and galactose-inducible proteins fused carboxy-terminal to the bacterial transcriptional activator B42.
Other constructions and plasmids.
For affinity purification of antipeptide rabbit serum 1455, a fragment corresponding to nucleotides 3015 to 3501 in the HIRA cDNA was PCR amplified by using the sense (5′-GCGGATCCAGTCCAGCCACGAGTA-3′) and antisense (5′-TTGGAGGGAGGGATGAGC-3′) oligonucleotides and digested with BamHI and PstI (internal site). The BamHI-PstI 292-bp-long 5′ fragment was purified and introduced in the same sites of the pQE32 vector (Qiagen) to create pQE32-HIRA3′ for expression of a HIRA carboxy-terminal fragment (residues 930 to 1017) tagged with six amino-terminal histidine residues.
For in vitro production of radiolabeled HIRA, a complete coding sequence was assembled in the pcDNA3 vector (Invitrogen). A 5′ 1,729-bp EcoRI-HpaI fragment from the CF18 clone (27) was inserted in place of a similar restriction fragment at the 5′ end of clone 30.25 (27) to produce the pHIRA2 construct. The resulting 3,515-bp EcoRI insert lacked the translation initiation codon of HIRA. An EcoRI-EcoRV 5′ fragment of 1,546 bp containing the translation initiation codon was extracted from the partial HIRA cDNA in the C5 clone (18) and integrated in plasmid pcDNA3 prepared with the same enzymes. The HIRA cDNA 3′ end was introduced at the EcoRV site by using a 2,132-bp EcoRV fragment from pHIRA2. In addition, the resulting construct, pcDNA3-HIRA, was tagged with a carboxy-terminal triple hemagglutinin epitope inserted between the SauI site immediately 5′ of the HIRA stop codon and the 3′ XhoI site from the pcDNA3 multilinker. HIRAΔ1 (HIRA amino acids 1 to 440), HIRAΔ2 (332 to 1017), and HIRAΔ4 (562 to 1017) have been described elsewhere (31a). For production of HIRAΔ3 (amino acids 332 to 737 of HIRA), the HIRAΔ2 construct was transcribed by using T3 RNA polymerase after digestion of the HIRA coding region with BamHI.
For production of HIRA in fusion with glutathione S-transferase (GST), the HIRA-encoding EcoRI-XhoI fragment from pLexA-HIRA was subcloned in the corresponding sites of pGEX-5X1 (Pharmacia) to create pGEX-HIRA. Fusions between GST and fragments of HIRA were all prepared in pGEX-5X1. For HIRA 368-592, the pHIRA2 construct was PCR amplified by using the sense (5′-GAGAATTCGAGGAGGAGAAGAGCCGC-3′) and antisense (5′-AGCCTCGAGCTTTCCACAGCTGTCGG-3′) oligonucleotides. The resulting product was digested by EcoRI-XhoI for insertion in the corresponding sites of pGEX-5X1. HIRA 593-737 was obtained from a BamHI-cut PCR product generated using the sense (5′-CTCGGATCCTAAAAGAGCAGAACCT-3′) and the antisense (5′-AGAGACTCTTCTTTCAC-3′) oligonucleotides and cloned into the BamHI-prepared vector. HIRA 738-826 and HIRA 827-1017 were obtained by inserting BamHI-BglII and BglII-XhoI fragments extracted from the pHIRA2 construct in the BamHI and BamHI-XhoI sites of the vector, respectively.
For GST pull-down experiments, core histones H2A, H2B, H3, and H4 were produced from constructs kindly provided by Nicolas Mermod (Lausanne, Switzerland) and 35S labeled in vitro as fusions with the VP16 viral transactivator as described elsewhere (2). An entire core histone H2B (amino acids 1 to 125) was produced in fusion with GST from pGEX-H2B obtained by introduction in frame in pGEX-5X1 of the EcoRI-XhoI insert—which includes two internal EcoRI sites—that was excised from clone 15 (cl.15). H2B deletion mutants were produced as follows. H2B 1-63 was obtained by deletion of the 3′ EcoRI-XhoI fragment from the pGEX-H2B construct. H2B 1-32 and H2B 1-18 were generated by PCR amplification from pGEX-H2B DNA, using the sense (5′-GCATGGCCTTTGCAGGG-3′) and antisense (5′-TGCTCGAGCTGCGCTTGCGCTTCTTC-3′ and 5′-TGCTCGAGCGCCTTCTTGGAGCCC-3′) primers, respectively, followed by digestion and cloning in the EcoRI-XhoI sites of pGEX-5X1. For expression of GST-H4, the H4-encoding region corresponding to the human H4/a gene (accession no. X60481) was PCR amplified from human genomic DNA by using the sense (5′-GGAATTCTCTGGACGTGGTAAGGGCGGG-3′) and antisense (5′-TAGCTCGAGTGTTTAGTTAGGGCGGC-3′) primers followed by EcoRI-XhoI digestion and cloning in pGEX-5X1. Deletion mutants of H4 have been described elsewhere (48) and were kindly provided by Alain Verreault (Cold Spring Harbor, N.Y.). For in vitro experiments using GST-HIRIP3, the insert in c1.13 was subcloned in the EcoRI-XhoI sites of pGEX-5X1. All constructions were checked as appropriate by restriction enzyme digestion or DNA sequencing using flanking vector sequences or internal oligonucleotides as primers.
In vitro protein interactions.
For production of GST fusion proteins, pGEX constructs were transformed into E. coli XL1-Blue or BL21. Induction of protein expression and immobilization on beads were done following an established protocol (4). 35S-labeled proteins were prepared in vitro from the above-described constructs, using T7 or T3 RNA polymerase and the TnT coupled rabbit reticulocyte lysate system (Promega). Radiolabeled proteins were incubated in the presence of immobilized GST fusion proteins in a final volume of 30 μl of a buffer containing 20 mM Tris (pH 8), 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40 and 200 mg of Pefabloc (Interchim) per ml. After a 1-h incubation at room temperature with gentle rocking, beads were washed five times in 20 mM Tris (pH 8)–150 mM NaCl–0.2% Triton X-100. Bound proteins were released by boiling in gel sample buffer, resolved by SDS-PAGE, and revealed by autoradiography.
Coimmunoprecipitation.
K562 cells were washed twice in PBS and lysed while vortexing (20 min, 4°C) in 50 mM Tris (pH 7.5)–150 mM NaCl–5 mM EDTA–0.5% Nonidet P-40–antiprotease cocktail. The supernatant of a low-speed centrifugation (3,000 rpm, 5 min, 4°C) was subjected to immunoprecipitation (overnight incubation on a rotating wheel at 4°C) using 5 μl of affinity-purified antiserum 1455 or 1 μg of LG11.1, a mouse monoclonal antibody directed to the amino-terminal tail of human H2B that was kindly provided by Sylviane Müller (Strasbourg, France), followed by incubation (2 h, 4°C) with a mixture of 10 μl each of protein A- and protein G-Sepharose (Sigma). After five washes in lysis buffer, the immunoprecipitate was separated by SDS-PAGE, transferred onto a PVDF membrane, and analyzed by Western blotting using either the anti-HIRA rabbit serum 1455 or the anti-H2B monoclonal antibody followed by appropriate peroxidase-conjugated secondary reagents.
HIRIP3 cDNA isolation and DNA sequencing.
The HIRIP3 cl.13 cDNA insert was used to screen a cDNA library prepared from U937 cells differentiated in the presence of tetradecanoyl phorbol acetate (provided by the UK-HGMP Resource Centre, Hinxton, England), yielding three clones, cl.22, cl.17, and cl.31, which were fully sequenced on an ABI 373 instrument (Perkin-Elmer), using flanking vector sequences or internal oligonucleotides as primers.
Database analysis.
The GenBank/EMBL databases were searched for homology by using the set of BLAST programs accessible on-line at the National Center for Biotechnology Information (Bethesda, Md.).
Northern analysis.
Two human multiple-tissue Northern blots were purchased (Clontech) and hybridized according to the manufacturer’s protocol. The final wash was in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at 50°C.
RESULTS
HIRA, a primarily nuclear protein.
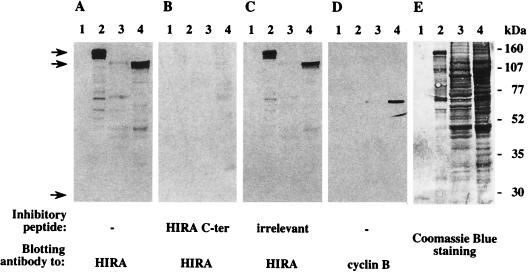
Two putative nuclear localization signals have been conserved between all HIRA proteins in higher eukaryotes, suggesting a nuclear targeting for HIRA molecules. To explore the localization of human HIRA, we generated a rabbit antiserum to a carboxy-terminal peptide of the protein. As seen in Fig. 1A, the affinity-purified serum 1455 recognized the HIRA protein bacterially produced in fusion with GST (calculated molecular mass, 138 kDa [lane 2]) but showed no binding activity to GST alone (26 kDa [lane 1]). The same serum detected a major band whose mobility corresponded to the molecular mass of HIRA (112 kDa) in a nuclear protein extract from the K562 cell line (lane 4), whereas the corresponding cytosolic extract (lane 3) showed little reactivity. To confirm that the 112-kDa signal was HIRA specific, the same blot was sequentially stripped and restained as indicated in Materials and Methods. As seen in Fig. 1B, no signal was obtained when staining with serum 1455 was competed for by preincubation with the immunizing peptide; when restaining was performed in the presence of an irrelevant peptide (Fig. 1C), the GST-HIRA and nuclear HIRA signals observed in Fig. 1A were reproduced. The subsequent detection of cyclin B1 in the nuclear, but not cytosolic, extract (Fig. 1D) provided a control for the nuclear specificity of the signals detected in lane 4. After a final stripping, the blot was stained with Coomassie blue to verify that equivalent cytosolic and nuclear protein amounts had been retained throughout the procedure (Fig. 1E, lanes 3 and 4). Together, these data demonstrated the HIRA specificity of antiserum 1455 and suggested a predominantly nuclear localization of the HIRA protein.
FIG. 1.
Immunoblot analysis using an affinity-purified rabbit antiserum raised to human HIRA. The same blot was subjected to sequential rounds of antibody binding and stripping followed by Coomassie blue staining. Lanes 1 and 2 were loaded with 200 ng of purified GST and GST-HIRA fusion protein, respectively. Lanes 3 and 4 contained 30 μg of cytosolic (lane 3) or nuclear (lane 4) proteins extracted from the human erythroleukemia cell line K562. Staining antibodies were 1455, a rabbit antiserum generated to a carboxy-terminal (C-ter) peptide of HIRA (A to C), and a mouse monoclonal antibody to human cyclin B1 (D) used as a control for nuclear protein extraction. The results shown in panels B and C were obtained by incubating antibody 1455 in the presence of 30 μg of the HIRA immunizing peptide (B) or of an irrelevant peptide (C). Note that the underrepresentation of lower-molecular-weight polypeptides as detected in lanes 3 and 4 in the Coomassie blue staining (E) simply reflects a weaker attachment to the membrane after several rounds of stripping. Initial loads of GST (still weakly detectable in panel E, lane 1) and GST-HIRA (lane 2) were equivalent. Arrows shown left point to migration positions of GST-HIRA, endogenous HIRA, and GST (top to bottom). Migration of molecular weight standards is shown on the right.
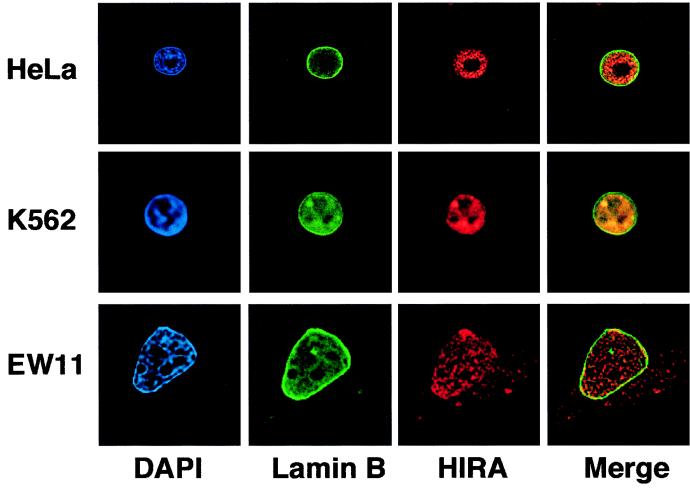
To further analyze the subcellular localization of HIRA, immunofluorescence studies were conducted on three human cell lines, using the anti-HIRA rabbit serum and a monoclonal antibody to lamin B to delineate the nuclear envelope. Conventional fluorescence microscopy images were acquired and deblurred by using a deconvolution process with exhaustive photon reassignment. Representative optical sections from single interphase cells are shown in Fig. 2. In all three cell types, HIRA molecules (red) were detected as numerous dots scattered throughout the nucleus, the nucleoli being excluded from staining. In HeLa and K562 cells, HIRA was largely detected within the limits of the nucleus (green). In EW11 cells, a significant HIRA decoration was also observed beyond the nuclear envelope. Together with the immunoblot analysis, these fluorescence data indicated that the HIRA protein is primarily, though not exclusively, localized in the cell nucleus.
FIG. 2.
Immunofluorescence staining of human cell lines reveals HIRA molecules as primarily nuclear. HeLa, K562, and EW11 cells were stained for HIRA and the nuclear envelope and counterstained for nuclear DNA. Black-and-white fluorescence images were acquired by using a 60× oil immersion objective and treated as described in Materials and Methods. Blue (DAPI; nuclear DNA), green (fluorescein; lamin B), and red (Texas red; HIRA) signals have been pseudo-colored by using Photoshop 4.0. Merge represents an overlay between the green and red images. For each cell line, a single optical section of a representative cell is shown.
Identification in a two-hybrid screen of core histone H2B as a HIRA interactor.
To determine the function of the HIRA protein in multicellular species, we looked for HIRIPs. A yeast two-hybrid screen was undertaken as described in Materials and Methods, using as bait an almost complete version (amino acids 10 to 1017) of human HIRA produced by fusion with the LexA DNA-binding domain. Ten true positives were eventually selected from 2 × 106 transformants plated. As summarized in Table 1, these 10 clones contained four different polyadenylated inserts whose deduced products were named HIRIP1 to HIRIP4. The HIRIP1 and HIRIP2 cDNA inserts (cl.14 [accession no. AJ223352] and cl.15 [accession no. AJ223353]) consisted of 829 and 808 nucleotides, respectively. The two open reading frames, 378 nucleotides in length in each case and 90% identical to each other (data not shown), were each found to encode core histone H2B. Contrasting with the two coding regions, the 5′ ends and 3′ untranslated regions could be aligned neither between each other nor to entries in nucleotide databases, indicating that they represented transcripts from two new H2B loci in the human genome.
TABLE 1.
Summary of confirmed HIRIPs
| HIRIP | Representative clonea | No. of clonesa | cDNA | Homology |
|---|---|---|---|---|
| HIRIP1 | cl.14 | 1 | Complete | Core histone H2B |
| HIRIP2 | cl.15 | 1 | Complete | Core histone H2B |
| HIRIP3 | cl.13 | 7 | Partial | None |
| HIRIP4 | cl.21 | 1 | Partial | DnaJ protein family |
Identified in a two-hybrid screen.
In vitro interaction between HIRA and core histones.
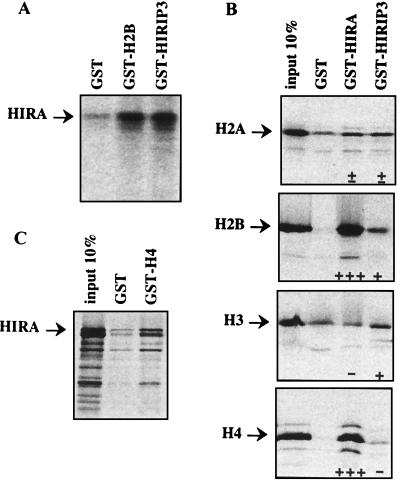
To confirm the HIRA-H2B interaction detected in yeast, an independent assay was used. The HIRIP1 and HIRIP2 H2B products were expressed in fusion with GST, immobilized on glutathione-agarose beads, and tested for interaction with HIRA produced in vitro. HIRA was specifically retained on beads covered with the GST-HIRIP1 (GST-H2B) fusion product compared to background binding to GST alone (Fig. 3A). Identical results were obtained with GST-HIRIP2 (not shown). Therefore, the interaction detected between HIRA and H2B in the yeast environment was verified in the absence of any contaminating yeast protein.
FIG. 3.
In vitro interaction between HIRA, core histones, and HIRIP3. (A) 35S-labeled HIRA binds GST-H2B and GST-HIRIP3 but not GST immobilized on glutathione beads. (B) Each of the four core histones was translated in vitro as a fusion with VP16 and tested for binding to GST, GST-HIRA, and GST-HIRIP3. The input 10% lane was loaded with 1/10 of the volume of radioactively labeled fusion protein used in each interaction assay. Semiquantitative results shown at the bottom of the lanes reflect the amounts of radioactive material retained on beads. +++ (which corresponds to greater than the 10% input material) and +, true positives; ± and −, not significantly different from background binding to GST beads. Note that each protein displays a characteristic background affinity to GST beads that was found to be reproducible between at least three distinct experiments. (C) In vitro-labeled HIRA protein binds core histone H4 immobilized on beads.
We next wished to investigate whether HIRA also had the capacity to interact with H2A, H3, or H4, the other histone components in the core particle. In this set of experiments, the HIRA protein was produced in fusion with GST, bound to glutathione beads, and tested for interaction with each of the four core histones transcribed and translated in vitro. Since cysteine and methionine residues are scarce in core histones, labeled products were generated from previously described constructs (2) that code for fusions between the same amino-terminal fragment of the VP16 protein and each of the four core histones. The results shown in Fig. 3B are from one representative experiment among many that were performed. As expected, H2B (in fusion with VP16) bound GST-HIRA but not GST alone, thus verifying the reciprocal interaction previously observed between GST-H2B and in vitro-produced HIRA. In addition, we found that H4 (VP16-H4) consistently exhibited strong HIRA-binding levels. In contrast, no HIRA-specific binding was observed with VP16-H2A or VP16-H3. Together, these data indicated that binding of H2B and H4 to HIRA did not depend solely on the VP16 moiety, which was identical in all four fusions tested. Finally, the HIRA-H4 interaction was confirmed with in vitro-translated HIRA and immobilized GST-H4 (Fig. 3C).
In vivo interaction between HIRA and core histone H2B.
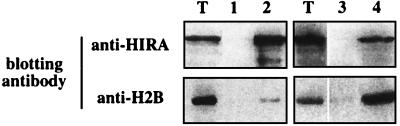
We then attempted to detect the existence of an in vivo interaction between endogenous HIRA and core histone H2B. A total protein extract (500 μg) prepared from the K562 cell line was submitted to immunoprecipitation with the affinity-purified anti-HIRA antiserum 1455. Western blotting using the same anti-HIRA reagent indicated that approximately 30% of HIRA molecules had been precipitated (Fig. 4, top, lane 2; compare with HIRA protein detected in 50 μg [10% input] of total cell lysate as loaded in the lanes marked T). No such band was detected when the same cellular extract was submitted to immunoprecipitation with an irrelevant rabbit antiserum in place of 1455 (lane 1). The lower part of the same blot was stained with LG11.1, a mouse monoclonal antibody that specifically detects core histone H2B in the total lysate (Fig. 4, bottom, lanes T). A small fraction (approximately 1%) of the H2B molecules present in the cellular extract was coimmunoprecipitated with HIRA when 1455 (lane 2), but not the irrelevant antiserum (lane 1), was used as the precipitating antibody. When the converse experiment was conducted with LG11.1, which precipitated H2B with ∼50% efficiency, approximately 3% of HIRA molecules were coimmunoprecipitated with H2B (lane 4), confirming that fractions of the two molecules are physically associated in a cellular context.
FIG. 4.
HIRA and core histone H2B coimmunoprecipitate from cellular extracts. Material was precipitated from 500 μg of a K562 protein extract by using an irrelevant rabbit antiserum (lane 1), 1455 (lane 2), an irrelevant mouse monoclonal antibody (lane 3), or LG11.1 (lane 4), subjected to electrophoresis in a 15% acrylamide gel, and transferred for Western blotting analysis using the anti-HIRA antiserum 1455 (top) or the anti-H2B mouse antibody LG11.1 (bottom) as the staining reagent. Lanes T were loaded with 50 μg (10% input) of total extract. Note that each of the right-hand panels corresponds to a single exposure from the same blot.
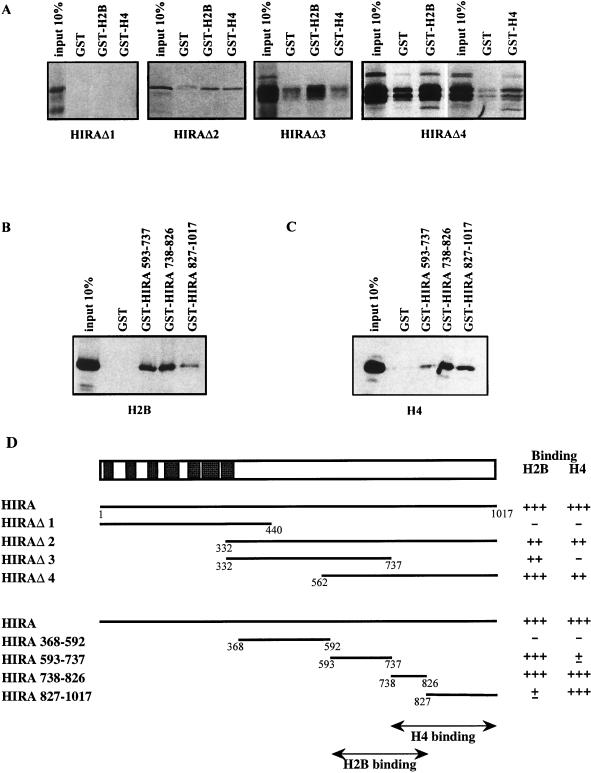
Overlapping but distinguishable H2B- and H4-binding domains in HIRA.
To determine the domains responsible for core histone binding within the HIRA molecule, a series of HIRA deletion mutants (represented in Fig. 5D) were constructed. The amino-terminal fragment (amino acids 1 to 440) that contains the entire WD repeat region of HIRA as well as its evolutionarily conserved glutamine-rich segment immediately carboxy terminal to the seventh WD repeat was produced from HIRAΔ1 and found to lack any histone-binding capacity; in contrast, an overlapping carboxy-terminal fragment (HIRAΔ2, amino acids 332 to 1017) bound GST-H2B and GST-H4 but not GST alone (Fig. 5A). It therefore appeared that the entire WD repeat region of HIRA was neither sufficient nor necessary for interaction with H2B or H4. The large HIRA polypeptide in HIRAΔ2 was further analyzed by using mutants HIRAΔ3 and HIRAΔ4 (amino acids 332 to 737 and 562 to 1017 [Fig. 5A]). The corresponding overlapping fragments displayed different histone-binding activities. Both bound GST-H2B efficiently. In contrast, GST-H4 retained amino acids 562 to 1017 but not 332 to 737 of HIRA (Fig. 5A).
FIG. 5.
Core histone-binding domains in HIRA protein. (A) HIRA deletion mutants indicated below the gels were radioactively labeled in vitro and tested for binding to GST, GST-H2B, and GST-H4 proteins immobilized on glutathione beads. (B and C) Binding of in vitro-translated H2B (B) and H4 (C) to HIRA deletion mutants. (D) The entire HIRA protein is schematized (top), with closed rectangles representing the seven WD repeats. HIRA deletion fragments are shown below with summarized H2B and H4 binding results. Numbers refer to amino acid residues in the HIRA protein sequence.
To further define the H2B- and H4-binding domains within HIRA, subfragments from the carboxy-terminal part of the protein were produced as GST fusions (schematized in Fig. 5D, bottom) and tested for the capacity to retain in vitro-produced core histones. Neither H2B nor H4 bound HIRA amino acids 368 to 592 (data not shown). H2B bound HIRA amino acids 593 to 737 and 738 to 826 strongly but 827 to 1017 weakly (Fig. 5B). In contrast, H4 bound amino acids 738 to 826 and 827 to 1017 strongly but 593 to 737 weakly (Fig. 5C). Together, these data (summarized in Fig. 5D) demonstrated that the H2B- and H4-binding domains reside within fragments 593 to 826 and 738 to 1017 of HIRA, respectively. We therefore conclude that H2B- and H4-binding domains are overlapping but distinguishable in the carboxy-terminal half of the HIRA molecule.
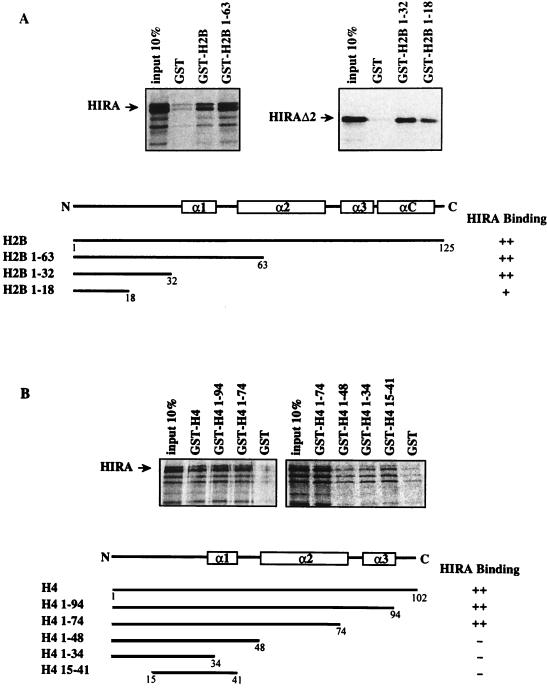
HIRA-binding domains in core histones H2B and H4.
We also wished to delineate which domains were responsible for HIRA binding in both H2B and H4. Deletion mutants were used to produce histone fragments in fusion with GST. Results displayed in Fig. 6A revealed that the amino-terminal half of H2B (amino acids 1 to 63) was sufficient for HIRA binding. Furthermore, an H2B mutant which encoded only amino acids 1 to 32 of H2B retained HIRA as efficiently as the entire core histone, whereas a further deletion allowing expression of amino acids 1 to 18 only significantly reduced the HIRA-binding level. Thus, the entire amino-terminal tail region immediately preceding the first α helix in histone H2B appears to be required for maximal HIRA-binding levels. Similar experiments were conducted with deletion fragments of histone H4. As seen in Fig. 6B, a deletion mutant encoding amino acids 1 to 74 of histone H4 retained as much in vitro-translated HIRA as the entire core histone. In contrast, a substantial decrease in HIRA-binding level was observed when H4 was further deleted, resulting in the absence of the second α helix in the corresponding product (amino acids 1 to 48). Together with the absence of HIRA binding to H4 fragments contributed by amino acids 1 to 34 (the amino-terminal tail) and 15 to 41, which include the first α helix, we conclude that the second α helix of core histone H4 is crucial for HIRA binding (Fig. 6B). Thus, the HIRA protein binds regions of H2B and H4 that are both precisely delineated and structurally different from each other.
FIG. 6.
HIRA-binding domains in core histones H2B and H4. HIRA and HIRAΔ2 were produced in vitro and tested for binding to immobilized core histones H2B (A) and H4 (B) and their deletion fragments, as schematized below the gels, with semiquantitative binding results shown at the right. Also included is a schematic representation of structural elements in the two core histones, with open rectangles corresponding to α helices (31). Note that the slightly higher background binding of HIRA to GST-H4 1-34 and GST-H4 15-41 than to GST-H4 1-48 or GST alone stemmed from higher amounts of corresponding GST fusion proteins being immobilized on glutathione beads, as revealed by Coomassie blue staining (not shown).
Interaction between HIRIP3, a novel HIRA interacting protein, and core histones.
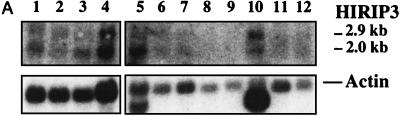
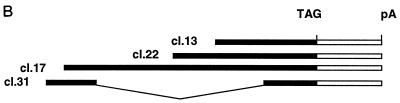
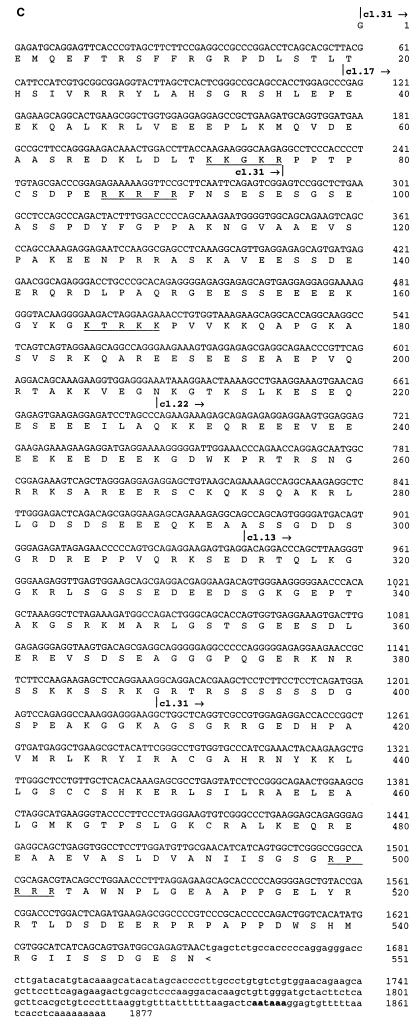
In addition to core histone H2B, the two-hybrid screen described above also identified novel proteins as potential HIRA interactors (Table 1). The cDNA insert in cl.13, one of seven identical HIRIP3 clones, consisted of 931 nucleotides followed by a short poly(A) tail 21 bp downstream of a canonical polyadenylation signal. Northern blot analysis performed on polyadenylated mRNAs of fetal and adult origins (Fig. 7A) revealed two low-level transcripts of approximately 2.0 and 2.9 kb that were similarly expressed in most samples, although there appeared to be a predominance of the larger transcript in adult muscle (Fig. 7A, lane 10). In an effort to isolate full-length cDNAs, iterative library screenings were performed. We isolated three additional clones, cl.17, cl.22, and cl.31, whose nucleotide sequences could be aligned with each other and with c1.13 to extend the HIRIP3 nucleotide sequence in the 5′ direction (Fig. 7B). The largest insert found in cl.17 (accession no. AJ223350) was entirely colinear with those in cl.22 and cl.13, whereas the insert in cl.31 (accession no. AJ223349) lacked an internal 938-bp fragment (Fig. 7B). It thus appeared that the smaller and larger transcripts detected by Northern blot analysis could be represented on the one hand by cl.31 and on the other hand by cl.17, cl.22, and cl.13. A 1,877-bp-long composite HIRIP3 nucleotide sequence was assembled (Fig. 7C; accession no. AJ223351). It consisted of a long reading frame open at its 5′ end that may still lack up to 1 kb of 5′ sequence for assembly of a complete cDNA. The partial HIRIP3 deduced protein, 551 amino acids in length, would be highly charged with 20.1 and 21.1% acidic and basic residues, respectively, including several stretches of lysine and arginine residues that could act as nuclear localization signals (Fig. 7C). No significant homology was detected in protein databases.
FIG. 7.
HIRIP3. (A) Two low-abundance HIRIP3 transcripts are detected by Northern blot analysis. Commercial Northern blots prepared from fetal (lanes 1 to 4) and adult (lanes 5 to 12) tissues (2 μg of polyadenylated mRNA per lane) were sequentially hybridized with the HIRIP3 cl.13 cDNA insert (top; exposure, 5 days) and with an actin probe as control for mRNA loading (bottom; exposure, 3 h). Loaded samples originated from fetal brain (lane 1), lung (lane 2), liver (lane 3), and kidney (lane 4) and from adult heart (lane 5), brain (lane 6), placenta (lane 7), lung (lane 8), liver (lane 9), skeletal muscle (lane 10), kidney (lane 11), and pancreas (lane 12). Sizes of the two HIRIP3 transcripts are indicated. (B) Schematic alignment of isolated HIRIP3 cDNAs. All clones were entirely sequenced and are shown approximately to scale. The open reading frame is shown as a solid box. TAG and pA refer to stop codon and poly(A) tail, respectively. (C) HIRIP3 composite nucleotide sequence (accession no. AJ223351) and deduced translation product. Uppercase, predicted coding region; lowercase, 3′ untranslated region; bold, polyadenylation signal. The 5′ end (and internal limits in the case of clone cl.31) of each cDNA is reported above the nucleotide sequence. Basic pentapeptides (four arginine or lysine amino acids among five consecutive residues) defining putative nuclear localization signals are underlined.
The HIRA-HIRIP3 interaction detected in yeast was reproduced in GST pull-down experiments with in vitro-labeled HIRA binding GST-HIRIP3 immobilized on beads (Fig. 3A). Since HIRIP3 bound HIRA, which we had shown directly interacted with core histones H2B and H4, we next investigated whether HIRIP3 immobilized on beads would be capable of retaining radiolabeled core histones. In the absence of any HIRA molecules, 35S-labeled H2B, and to a lesser extent H3, significantly bound HIRIP3, whereas H2A and H4 did not (Fig. 3B). In summary, both HIRA and its interactor HIRIP3 displayed in vitro binding to two of the four core histones, the former to H2B and H4, the latter to H2B and H3, compatible with the hypothesis of a HIRA-HIRIP3-containing molecular complex playing a role in some aspect(s) of core histone metabolism.
DISCUSSION
In this study, we have started to analyze the properties of the HIRA protein, the product of a major candidate for developmental disorders associated with the loss of a chromosome 22q fragment. Using HIRA as bait in a two-hybrid protein interaction trap, we have identified four novel genes, HIRIP1 to HIRIP4, whose translation products interacted with HIRA, both in vivo and in GST pull-down experiments. HIRIP1 and HIRIP2 contributed two different cDNAs that were both found to encode core histone H2B. The two polyadenylated cDNAs were unprocessed. Their 5′ and 3′ untranslated regions, different from each other, were absent from nucleotide databases, indicating that they represent newly identified H2B genes, thus adding to the accumulating number of histone genes identified in humans (1).
H2B was the only core histone identified from the oligo(dT)-primed cDNA library. As the vast majority of histone transcripts undergo a processing of their 3′ untranslated regions that produces poly(A)-less mRNAs, it appeared likely that histone mRNAs would be underrepresented in this library. We thus decided to test whether in vitro, other core histones would interact with HIRA. Indeed, in GST pull-down experiments, HIRA was found to strongly interact not only with H2B but also with histone H4. The interactions detected between HIRA and H2B or H4 were observed whether it was HIRA or the core histones that were immobilized on beads, with the interacting partner being radioactively labeled in vitro. In the cases where HIRA or fragments thereof were bound on beads, core histones were translated as fusions with a VP16 fragment which contributed most cysteine and methionine residues for 35S labeling. In these interaction assays, however, the VP16 moiety in VP16-H2B or VP16-H4 played no role, as demonstrated by the lack of binding of VP16-H2A or VP16-H3, which contained the same VP16 fragment. A further indication of the physiological relevance of the reported interactions was provided by HIRA and H2B coimmunoprecipitating from cellular extracts. This protein association was demonstrated with either the anti-HIRA antiserum or an anti-H2B monoclonal antibody as the precipitating reagent. Although they did not formally demonstrate that HIRA and core histone H2B are in direct contact in vivo, these coimmunoprecipitation results did indicate that a fraction of HIRA and histone H2B molecules are part of a cellular complex. In the experimental conditions used here, we have not been able to quantitatively precipitate histone H4 along with HIRA and H2B. This could reveal a more labile association of HIRA with H4 than with H2B.
In vitro, the interactions between HIRA and core histones H2B and H4 depended on overlapping domains that were entirely localized to the carboxy-terminal half of HIRA, whereas the WD repeat region contributes the amino-terminal third of the protein. In this respect, HIRA resembles Tup1p, a protein that associates with Ssn6p to assemble a transcriptionally repressive complex in S. cerevisiae (8, 23, 36, 45, 46, 52). Both HIRA and Tup1p are proteins with seven WD repeats and the capacity to interact with two of the four core histones: HIRA with H2B and H4 (this study) and Tup1p with H3 and H4 (11). In both molecules, the histone interaction region lies separate from the seven WD repeats which probably confer each of the two proteins with a β-propellor scaffold structure (14, 24, 42) for protein interactions. Tup1p is known to associate with various transcription factors, including the homeodomain-containing α2 protein that targets transcriptional repression to a-type specific loci (25). Likewise, an interaction has recently been discovered (31a) between HIRA and the homeodomain-containing DNA-binding transcription factor Pax3 (16). Haploinsufficiency of Pax-3 is responsible for the Splotch phenotype in mice (12) and for Waardenburg syndrome in humans (35), disorders that include defects of neural crest derivatives. It will be important to define whether, like its yeast homologs Hir1p and Hir2p, HIRA has a role in transcriptional regulation and more specifically whether it can regulate Pax3-dependent transcription, which could be of direct relevance to the presumed implication of HIRA haploinsufficiency in the genesis of the DiGeorge syndrome and related 22DD.
Initially, the finding that HIRA interacted directly with histone H2B was somewhat surprising because neither Hir1p nor Hir2p had been reported as physically interacting with a histone gene product. However, hints that this may be the case exist since Hir1p and Hir2p appear to act on chromatin structure to repress transcription at core histone gene promoters and other loci (43). Furthermore, the regulatory activity of Hir1p and Hir2p requires the products of the SPT4, SPT5, and SPT6 genes (10), three transcriptional regulators that are known to act on chromatin structure. Indeed, Spt6p directly binds core histone H3 (3), whereas Spt4p and Spt5p and their human homologs have recently been shown to form a complex regulating elongation of gene transcription by RNA polymerase II (19, 49). It will be interesting to test for direct interactions between Hir1p, Hir2p, and core histones and also to investigate whether the two yeast proteins and their homologs in higher eukaryotes fulfill further regulatory functions at the levels of transcription elongation and chromatin metabolism.
Such a chromatin-related function of HIRA may well depend on a HIRA-containing multiprotein complex that could also contain HIRIP3, the other HIRA interactor that we have reported here. The partial 551-amino-acid sequence of HIRIP3 reveals no homolog in databases, nor does it contain a recognizable protein domain. In GST pull-down experiments, HIRIP3 was found to interact with two of the four core histones. These in vitro interactions appeared significant because in contrast to HIRA, which strongly interacted with H2B and H4, HIRIP3 was found to specifically bind H2B and H3. The different biochemical features of HIRA and HIRIP3 would confer complementary histone-binding properties upon a putative HIRA-HIRIP3-containing complex. In the absence of anti-HIRIP3 antibodies, it was not possible to test for the presence of HIRIP3 in the cellular HIRA-histone H2B-containing precipitates. When available, such an anti-HIRIP3 reagent will also allow us to investigate the subcellular localization of the protein.
We have found that the H2B- and H4-binding domains in HIRA are not superimposable, an indication that a single HIRA protein may simultaneously contact two different core histones. The regions of HIRA interaction are structurally different in the two histones. H2B binds HIRA through its N-terminal tail region (amino acids 1 to 32). As seen in the recently described atomic model of the nucleosome, this fragment of H2B consists of an unstructured terminal segment followed by a very basic octapeptide passing between the gyres of the DNA superhelix through a channel formed by aligned minor grooves (31). This HIRA interaction region in H2B would be compatible with a role of HIRA at the level of the nucleosome. In contrast, in histone H4, the major HIRA-binding region corresponds to the second α helix, which is not exposed in the nucleosomal structure. It thus appears unlikely that HIRA would interact with both H2B and H4 when assembled within the nucleosomal octamer. This conclusion is consistent with our inability to detect an interaction between HIRA and a reconstituted nucleosome or a core particle (data not shown).
Other possibilities exist regarding the functional significance of HIRA interacting with nucleosomal components. The immunofluorescence and Western blot experiments that we have performed with a HIRA-specific antiserum have demonstrated that the subcellular location of HIRA is primarily within the nucleus, as predicted from the two putative nuclear localization signals detected in its primary amino acid sequence (27) and from the reported nuclear localization of its yeast homologs Hir1p and Hir2p (40). As exemplified in the EW11 cell line, however, a minor fraction of HIRA molecules were detected in the cytoplasm, suggesting that HIRA could also carry a function outside the nucleus, e.g., as a histone transporter. It is intriguing, in this regard, that the seven WD repeats in HIRA are found highly conserved in the corresponding region of the p60 subunit of the human chromatin assembly factor CAF-1. CAF-1 is a complex of three polypeptides associated with acetylated histones H3 and H4 and implicated in their deposition during DNA replication (22, 47) and nucleotide excision repair (13). Further experiments will be conducted to explore whether HIRA and its associated molecular partners can function in histone transport, assembly, or disassembly or as a transcriptional regulator.
ACKNOWLEDGMENTS
We thank Roger Brent for making kits for protein interaction traps available, Nassos Alevizopoulos, Nicolas Mermod, and Alain Verreault for sharing expression constructs, and Jean-Pierre Brouet and Sylviane Müller for providing antibodies.
S.L. benefited from doctoral fellowships from the Ministère de la Recherche and the Association pour la Recherche contre le Cancer, and J.-P.Q. benefited from a postdoctoral fellowship of the Ligue Nationale contre le Cancer. This work was supported by grants from the Association pour la Recherche contre le Cancer, the Human Frontiers Science Program Organization, and the Ligue Nationale contre le Cancer (G.A. and M.L.) and from the Association Française contre les Myopathies and the Fondation pour la Recherche Médicale (M.L.).
REFERENCES
- 1.Albig W, Kioschis P, Poustka A, Meergans K, Doenecke D. Human histone gene organisation: nonregular arrangement within a large cluster. Genomics. 1997;40:314–322. doi: 10.1006/geno.1996.4592. [DOI] [PubMed] [Google Scholar]
- 2.Alevizopoulos A, Dusserre Y, Tsai-Pflugfelder M, von der Weid T, Wahli W, Mermod N. A proline-rich TGF-β-responsive transcriptional activator interacts with histone H3. Genes Dev. 1995;9:3051–3066. doi: 10.1101/gad.9.24.3051. [DOI] [PubMed] [Google Scholar]
- 3.Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 4.Bours V, Franzoso G, Azarenko V, Park S, Kanno T, Brown K, Siebenlist U. The oncoprotein BCL-3 directly transactivates through kappaB motifs via association with DNA-binding p50β homodimers. Cell. 1993;72:729–739. doi: 10.1016/0092-8674(93)90401-b. [DOI] [PubMed] [Google Scholar]
- 5.Budarf M L, Emanuel B S. Progress in the autosomal segmental aneusomy syndromes (SASs): single or multi-locus disorders? Hum Mol Genet. 1997;6:1657–1665. doi: 10.1093/hmg/6.10.1657. [DOI] [PubMed] [Google Scholar]
- 6.Carlson C, Sirotkin H, Pandita R, Goldberg R, McKie J M, Wadey R, Patanjali S R, Weissman S M, Anyane-Yeboa K, Warburton D, Scambler P, Shprintzen R J, Kucherlapati R, Morrow B E. Molecular definition of 22q11 deletions in 151 velo-cardio-facial syndrome patients. Am J Hum Genet. 1997;61:620–629. doi: 10.1086/515508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter K C, Bowman D G, Carrington W, Fogarty K, McNeil J A, Fay F S, Lawrence J B. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993;259:1330–1335. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- 8.Cooper J P, Roth S Y, Simpson R T. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev. 1994;8:1400–1410. doi: 10.1101/gad.8.12.1400. [DOI] [PubMed] [Google Scholar]
- 9.Dallapiccola B, Pizzuti A, Novelli G. How many breaks do we need to CATCH on 22q11? Am J Hum Genet. 1996;59:7–11. [PMC free article] [PubMed] [Google Scholar]
- 10.DeSilva H, Lee K, Osley M A. Functional dissection of yeast Hir1p, a WD repeat-containing transcriptional corepressor. Genetics. 1998;148:657–667. doi: 10.1093/genetics/148.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edmonson D G, Smith M M, Roth S Y. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 12.Epstein D J, Vekemans M, Gros P. Splotch (Sp2H), a mutation affecting development of the mouse neural tube, shows a deletion within the paired homeodomain of Pax-3. Cell. 1991;67:767–774. doi: 10.1016/0092-8674(91)90071-6. [DOI] [PubMed] [Google Scholar]
- 13.Gaillard P-H L, Martini E M D, Kaufman P D, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: A new role for chromatin assembly factor-I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 14.Gaudet R, Bohm A, Sigler P B. Crystal structure at 2.4 A resolution of the complex of transducin βγ and its regulator, phosducin. Cell. 1996;87:577–588. doi: 10.1016/s0092-8674(00)81376-8. [DOI] [PubMed] [Google Scholar]
- 15.Gottlieb S, Emanuel B S, Driscoll D A, Sellinger B, Wang Z, Roe B, Budarf M L. The DiGeorge syndrome minimal critical region contains a Goosecoid-like (GSCL) homeobox gene that is expressed early in human development. Am J Hum Genet. 1997;60:1194–1201. [PMC free article] [PubMed] [Google Scholar]
- 16.Goulding M D, Chalepakis G, Deutsch U, Erselius J R, Gruss P. Pax-3, a novel murine DNA-binding protein expressed during early neurogenesis. EMBO J. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 18.Halford S, Wadey R, Roberts C, Daw S C M, Whiting J A, O’Donnell H, Dunham I, Bentley D R, Lindsay E A, Baldini A, Francis F, Lehrach H, Williamson R, Wilson D I, Goodship J, Cross I, Burn J, Scambler P J. Isolation of a putative transcriptional regulator from the region of 22q11 deleted in DiGeorge syndrome, Shprintzen syndrome and familial congenital heart disease. Hum Mol Genet. 1993;2:2099–2107. doi: 10.1093/hmg/2.12.2099. [DOI] [PubMed] [Google Scholar]
- 19.Hartzog G A, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heisterkamp N, Mulder M P, Langeveld A, ten Hoeve J, Wang Z, Roe B A, Groffen J. Localization of the human mitochondrial citrate transporter protein gene to chromosome 22q11 in the DiGeorge syndrome critical region. Genomics. 1995;29:451–456. doi: 10.1006/geno.1995.9982. [DOI] [PubMed] [Google Scholar]
- 21.Holmes S E, Riazi M A, Gong W, McDermid H E, Sellinger B T, Hua A, Chen F, Wang Z, Zhang G, Roe B, Gonzalez I, McDonald-McGinn D M, Zackai E, Emanuel B S, Budarf M L. Disruption of the clathrin heavy chain-like gene (CLTCL) associated with features of DGS/VCFS: a balanced (21;22)(p12;q11) translocation. Hum Mol Genet. 1997;6:357–367. doi: 10.1093/hmg/6.3.357. [DOI] [PubMed] [Google Scholar]
- 22.Kaufman P D, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 23.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 24.Komachi K, Johnson A D. Residues in the WD repeats of Tup1 required for interaction with α2. Mol Cell Biol. 1997;17:6023–6028. doi: 10.1128/mcb.17.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komachi K, Redd M J, Johnson A D. The WD repeats of Tup1 interact with the homeo domain protein α2. Genes Dev. 1994;8:2857–2867. doi: 10.1101/gad.8.23.2857. [DOI] [PubMed] [Google Scholar]
- 26.Lammer E J, Opitz J M. The DiGeorge anomaly as a developmental field defect. Am J Med Genet Suppl. 1986;2:113–127. doi: 10.1002/ajmg.1320250615. [DOI] [PubMed] [Google Scholar]
- 27.Lamour V, Lécluse Y, Desmaze C, Spector M, Bodescot M, Aurias A, Osley M A, Lipinski M. A human homolog of the S. cerevisiae HIRI and HIR2 transcriptional repressors cloned from the DiGeorge syndrome critical region. Hum Mol Genet. 1995;4:791–799. doi: 10.1093/hmg/4.5.791. [DOI] [PubMed] [Google Scholar]
- 28.Lévy A, Demczuk S, Aurias A, Depétris D, Mattei M-G, Philip N. Interstitial 22q11 microdeletions excluding the ADU breakpoint in a patient with DiGeorge syndrome. Hum Mol Genet. 1995;4:2417–2419. doi: 10.1093/hmg/4.12.2417. [DOI] [PubMed] [Google Scholar]
- 29.Llevadot, R., X. Estivill, P. Scambler, and M. Pritchard. Isolation and genomic characterization of the TUPLE1/HIRA gene of the pufferfish Fugu rubripes. Gene, in press. [DOI] [PubMed]
- 30.Lorain S, Demczuk S, Lamour V, Toth S, Aurias A, Roe B A, Lipinski M. Structural organization of the WD repeat protein-encoding gene HIRA in the DiGeorge syndrome critical region of human chromosome 22. Genome Res. 1996;6:43–50. doi: 10.1101/gr.6.1.43. [DOI] [PubMed] [Google Scholar]
- 31.Lüger K, Mäder A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 31a.Magnaghi, P., et al. Submitted for publication.
- 32.Mariette X, Brouet J-C, Danon F, Tsapis A, Lassoued K. Nucleotide sequence analysis of the VL and VH domains of the five human IgM directed to lamin B. Evidence for an antigen-driven process in the generation of human autoantibodies to lamin B. Arthritis Rheum. 1993;36:1315–1324. doi: 10.1002/art.1780360916. [DOI] [PubMed] [Google Scholar]
- 33.Neer E J, Schmidt C J, Nambudripad R, Smith T F. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 34.Osley M A, Lycan D. trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol Cell Biol. 1987;7:4204–4210. doi: 10.1128/mcb.7.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Read A P, Newton V E. Waardenburg syndrome. J Med Genet. 1997;34:656–665. doi: 10.1136/jmg.34.8.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redd M J, Arnaud M B, Johnson A D. A complex composed of Tup1 and Ssn6 represses transcription in vitro. Mol Cell Biol. 1997;17:11193–11197. doi: 10.1074/jbc.272.17.11193. [DOI] [PubMed] [Google Scholar]
- 37.Roberts C, Daw S, Halford S, Scambler P J. Cloning and developmental expression analysis of chick Hira (Chira), a candidate gene for DiGeorge syndrome. Hum Mol Genet. 1997;6:237–246. doi: 10.1093/hmg/6.2.237. [DOI] [PubMed] [Google Scholar]
- 38.Ryan A K, Goodship J A, Wilson D I, Philip N, Lévy A, Seidel H, Schuffenhauer S, Oechsler H, Belohradsky B, Prieur M, Aurias A, Raymond F L, Clayton-Smith J, Hatchwell E, McKeon C, Beemer F A, Novelli G, Hurst J A, Ignatius J, Green A J, Winter R M, Brueton L, Brøndum-Nielsen K, Stewart F, Van Essen T, Patton M, Paterson H, Scambler P J. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J Med Genet. 1997;34:798–804. doi: 10.1136/jmg.34.10.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scamps C, Lorain S, Lamour V, Lipinski M. The HIR protein family: isolation and characterization of a complete murine cDNA. Biochim Biophys Acta. 1996;1306:5–8. doi: 10.1016/0167-4781(96)00010-3. [DOI] [PubMed] [Google Scholar]
- 40.Sherwood P W, Tsang S V, Osley M A. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:28–38. doi: 10.1128/mcb.13.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sirotkin H, Morrow B, Saint-Jore B, Puech A, Das Gupta R, Patanjali S R, Skoultchi A I, Weissman S M, Kucherlapati R. Identification, characterization, and precise mapping of a human gene encoding a novel membrane-spanning protein from the 22q11 region deleted in velo-cardio-facial syndrome. Genomics. 1997;42:245–251. doi: 10.1006/geno.1997.4734. [DOI] [PubMed] [Google Scholar]
- 42.Sondek J, Bohm A, Lambright D G, Hamm H E, Sigler P B. Crystal structure of a GA protein βγ dimer at 2.1 A resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 43.Spector M S, Raff A, DeSilva H, Lee K, Osley M A. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol Cell Biol. 1997;17:545–552. doi: 10.1128/mcb.17.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swanson M S, Winston F. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132:325–336. doi: 10.1093/genetics/132.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional corepressor complex. Nature. 1994;369:758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- 46.Varanasi U, Klis M, Mikesell P B, Trumbly R J. The Cyc8 (Ssn6)-Tup1 corepressor complex is composed of one Cyc8 and four Tup1 subunits. Mol Cell Biol. 1996;16:6707–6714. doi: 10.1128/mcb.16.12.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 48.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1997;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 49.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog G A, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wall M A, Coleman D E, Lee E, Iniguez-Lluhi J A, Posner B A, Gilman A G, Sprang S R. The structure of the G protein heterotrimer Giα1β12. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 51.Williams F E, Trumbly R J. Characterization of TUP1, a mediator of glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6500–6511. doi: 10.1128/mcb.10.12.6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams F E, Varanasi U, Trumbly R J. The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiae are associated in a protein complex. Mol Cell Biol. 1991;11:3307–3316. doi: 10.1128/mcb.11.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilming L G, Snoeren C A S, Van Rijswijk A, Grosveld F, Meijers C. The murine homologue of HIRA, a DiGeorge syndrome candidate gene, is expressed in embryonic structures affected in human CATCH22 patients. Hum Mol Genet. 1997;6:247–259. doi: 10.1093/hmg/6.2.247. [DOI] [PubMed] [Google Scholar]