Abstract
A fluent conversation with a virtual assistant, person-tailored news feeds, and deep-fake images created within seconds—all those things that have been unthinkable for a long time are now a part of our everyday lives. What these examples have in common is that they are realized by different means of machine learning (ML), a technology that has fundamentally changed many aspects of the modern world. The possibility to process enormous amount of data in multi-hierarchical, digital constructs has paved the way not only for creating intelligent systems but also for obtaining surprising new insight into many scientific problems. However, in the different areas of biosciences, which typically rely heavily on the collection of time-consuming experimental data, applying ML methods is a bit more challenging: Here, difficulties can arise from small datasets and the inherent, broad variability, and complexity associated with studying biological objects and phenomena. In this Review, we give an overview of commonly used ML algorithms (which are often referred to as “machines”) and learning strategies as well as their applications in different bio-disciplines such as molecular biology, drug development, biophysics, and biomaterials science. We highlight how selected research questions from those fields were successfully translated into machine readable formats, discuss typical problems that can arise in this context, and provide an overview of how to resolve those encountered difficulties.
I. INTRODUCTION
In many areas of medicine and materials science, analyzing complex datasets is a crucial task; those datasets, for instance, consist of images that can be used to identify pathologies or to quantify the progress of diseases1–3 as well as for detecting defects on materials4–6 and monitoring experimental7–9 and production10,11 processes. When performed manually, those tasks require time-consuming expert involvement but, nevertheless, may remain error-prone and biased. This is where computer-based decision processes can help. In the recent decade, machine learning (ML) approaches have gained vastly increased attention and have been successfully applied to different problems. Machine learning is a field of data science that encompasses a variety of algorithms that automatically learn from provided information and then draw conclusions. Such approaches aim at simplifying, extending, or replacing human decision and analysis processes. Examples include object detection12,13 and monitoring,14,15 identification of patterns or correlations between datasets,16,17 as well as data classification,18–20 regression,21,22 or clustering23,24 (Fig. 1).
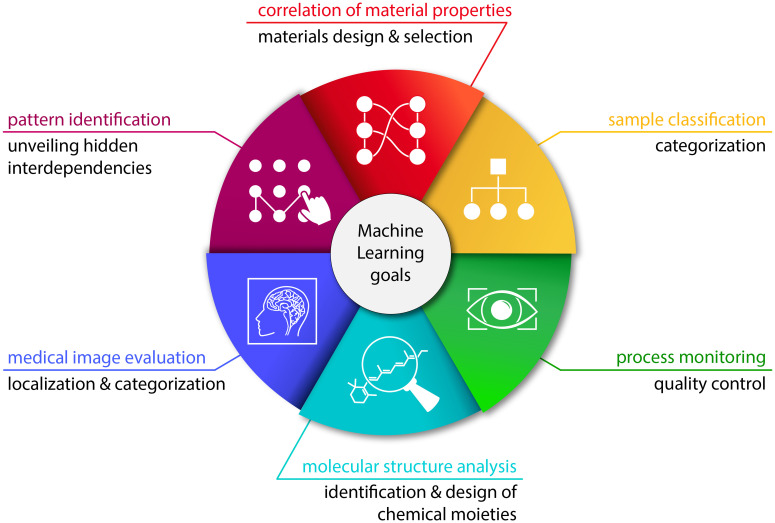
FIG. 1.
Typical objectives of machine learning approaches. A ML-based analysis of data from the biosciences can have different goals. Typical examples include the correlation of material properties, the classification of samples, the identification of patterns, process monitoring, molecular structure analysis, and the evaluation of medical images. Correlating material properties can, for instance, be useful to predict the behavior or certain characteristics of materials to provide guidance for a target-oriented design or selection process. Sample classification finds broad applications in areas where samples need to be assigned to discrete categories, e.g., for the classification of disease patterns based on various biomarkers. ML-based process monitoring can be an essential part of quality control to automatically identify and react to defects or variations in the process flow. Analyzing molecular structures by means of ML allows us to scan large databases to identify or even to design chemical moieties with certain properties. Another growing area of application for ML is the automated evaluation of medical images to, e.g., localize and categorize organs or pathological manifestations in tissues. Finally, one versatile purpose of ML is to identify patterns in databases to unveil hidden dependencies between different characteristics and attributes.
A key task for which machine learning has turned out to be highly helpful is image analysis.25–28 Here, image segmentation and object detection methods can be used to automatically identify and locate the presence of certain objects within an image or video.29–31 By receiving example images as an input, the algorithms learn to find informative regions in the pictures and extract characteristic features such as edges or specific shapes from them.32,33 At the moment, such approaches are extensively applied to face recognition or autonomous driving tasks; yet, this technique offers great potential in other areas as well where decisions are made based on visual impressions: The progression of glaucoma,34–36 dementia,37,38 or cancer39–42 was successfully extracted from medical images, cell nuclei were detected in microscope images,43,44 microtissue-contraction measurements were automatically analyzed in laboratory experiments,45 and additive manufacturing processes of biomaterials were optimized.46,47
In addition to analyzing images, ML algorithms can also handle other data types such as numerical values or text. Instead of an image, the samples then comprise multiple input parameters (commonly referred to as features) and—optionally—an output label or value. In materials science, such data analyses can uncover links among the composition, structure, and characteristics of known materials and extrapolate this knowledge to propose potential new materials with predefined properties.48–50 Here, the algorithms search for patterns and correlations in the dataset, from which conclusions can be drawn.51 With such an approach, it was possible to explore therapeutics that target specific diseases52–55 to study glycan functions,56,57 to enhance single molecule sensing,58 and to improve manufacturing processes such as 3D bioprinting59 or microparticle production.60
By mapping such input data, e.g., experimental findings, onto output labels, predictive algorithms can be established. Depending on the type of possible outputs, one can distinguish between classification and regression attempts. Classification describes the prediction of discrete outputs, i.e., samples are assigned to specific classes. Examples are the categorization of surfaces with regard to their wetting behavior61 or sorting the state of polymer conformations.62 In contrast, regression algorithms predict properties that can be described by continuous values such as interaction affinities,63–65 transcriptional activities of DNA motifs,66 or material parameters describing mechanical responses.67,68 These approaches are especially useful when mathematical equations based on physical models are still unknown.
II. PRINCIPLES, ADVANTAGES, AND LIMITATIONS OF DIFFERENT ML ALGORITHMS
Considering the large variety of available ML algorithms, selecting the most suitable one for a given problem is not always trivial: The best choice depends on the problem statement, the database, the desired output, interpretability, and many other factors. In Sec. II, we give an overview over common learning strategies, we highlight selected ML models (including random ensemble-based, probabilistic, linear, and deep learning methods), and we explain their working principles and characteristics. Although some models can make use of different learning strategies, in the following, each of them is assigned to the most commonly used one. Graphical representations of the algorithms discussed here are depicted in Fig. 2, and an overview of the advantages and disadvantages is given in Table I.
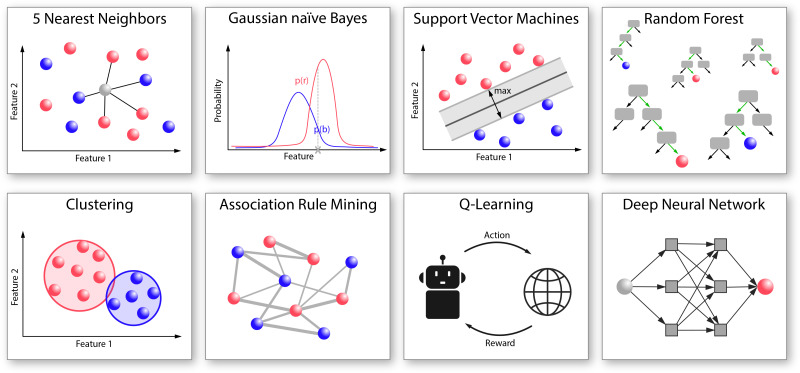
FIG. 2.
Schematic representation of typical ML algorithms used for analyzing problems from the different fields of biosciences. The k nearest neighbor (KNN) algorithm classifies a query sample according to the k samples that are most similar to it, i.e., which have the lowest distance in an n-dimensional hyperspace (here, n corresponds to the number of analyzed features). The Gaussian Naïve Bayes algorithm determines conditional probabilities and classifies samples based on a “most probable” principle. Support vector machines define hyperplanes in the n-dimensional feature space to distinctly separate samples of different classes while maximizing the distance of all samples to this separating hyperplane. The Random Forest classifier combines many randomly generated, uncorrelated decision trees to perform predictions in a popular-vote-like manner. Clustering refers to algorithms that group unlabeled data based on their characteristics. Association rule mining describes the process of finding dependencies that govern correlations and associations between samples. Q-learning assesses the quality of each action available for a given state by rewarding a subset of desired outcomes. Deep neural networks mimic the structure of the human brain by combining activatable units in consecutive, interconnected layers that process information in various manners.
TABLE I.
Overview of the advantages and disadvantages of the different ML algorithms discussed here.
| K nearest neighbors 69–72 | |
| No training phase needed | High dimensionality leads to decreased accuracies |
| Intuitive and simple algorithm | Can become slow for big datasets |
| Easily adapts to new training data | Needs feature scaling |
| Only one hyperparameter to tune | Has problems with imbalanced datasets |
| Missing values are problematic | |
| Naïve Bayes 73–78 | |
| Very fast | The assumption of independent features that equally contribute to the output rarely holds true |
| Needs less training data than most other algorithms | Zero probability problem: If one feature of a sample exhibits a value of zero probability according to the trained model, the class will be assigned a probability of zero. |
| Works well with high-dimensional data | |
| Support vector machines 79–86 | |
| Kernel functions can be used to solve complex problems | Choosing an appropriate kernel can be difficult |
| Effective in high-dimensional spaces even for comparably small sample sizes | Training times can become long with large datasets |
| Memory efficient, as it uses a subset of training points for the decision function | Limited capability to handle noisy or strongly overlapping classes |
| Random forest 87–93 | |
| Robust to outliers, noise, and imbalanced datasets | Long training times for large datasets |
| Lower risk of overfitting | Little control over model formation |
| Runs efficiently with large datasets | Limited ability to extrapolate |
| Easy data preparation | |
| Can handle high dimensionalities | |
| Clustering 94–99 | |
| Can handle unlabeled data | It can be difficult to interpret the sorting decision |
| Algorithms of different complexity are available | Big datasets can lead to long running times |
| Can be used on very small and very large datasets | The criteria to stop clustering or the number of clusters need to be defined |
| Association rule mining 100–104 | |
| Offers an easy way to detect correlations in unsorted datasets | Does not guarantee statistical significance |
| Unveils relationships between elements | Requires nominal variables; continuous values need to be translated |
| Q-Learning 105–109 | |
| After sufficient training, it finds optimal actions | Can be computationally expensive since each state/action pair needs to be evaluated multiple times |
| Can solve problems without explicitly being told how to | Does not include risk assessments into the decision making |
| Can have problems with high dimensionality | |
| Deep neural networks 110–113 | |
| Highly flexible and suitable to approximate complex functions | Requires lots of training data |
| Can be difficult to interpret (black box) | |
| Once trained, the predictions are fast | Training can be computationally expensive |
| There are multiple different network architectures already available | Finding the best network architecture can be challenging |
Overall, data fed into an algorithm can serve three different purposes: First, a “training set” is required to allow the algorithms to develop a model. Second, a “test set” is used for validation, and this set contains data the algorithms are only confronted with once they have established the model. Third, once validation was successful, so-called “query samples” are fed into the algorithm with the aim to get classified or to make predictions for. In all those datasets, input variables that quantify individual measurable characteristics of a data point are referred to as “features,” outputs assigned to training or test samples are called “labels,” and the output of the algorithm (be it continuous or discrete values) created for a query sample is called “prediction.”
A. Supervised learning
In supervised learning, models are developed based on labeled data—similar to how parents teach their children to name objects. The algorithm needs to be provided a training dataset, containing a sufficiently large number of samples; each of them is represented by input data—i.e., information (descriptors) that is likely to characterize the desired output—and corresponding output labels. Such datasets could, for example, comprise histological images of cancerous tissue (input) labeled with the name of the affected organ (output),114 or they could link the composition of a polymeric biomaterial (input) to its mechanical behavior (output).115,116 With such information offered, the ML models aim at identifying relationships between the input and the output and can then perform classification or prediction tasks for new data they were not confronted with before.
1. k nearest neighbor (KNN) algorithms
The simple but powerful k nearest neighbor algorithm follows the assumption that similarity between samples is accompanied by proximity in the data space; in other words, similar samples are expected to come with similar inputs. Instead of developing a generalized model, predictions are made by comparing a query sample to the training data. Then, the k nearest neighbors, i.e., the most similar samples according to their feature values, are identified, and a prediction is made considering the labels of those data points in a popular-vote-like manner. The number of neighbors k can be varied to find a valid compromise between robustness toward outliers (which is achieved for high values of k) and distinctness (which is a typical result for low values of k).117,118
KNN algorithms can be used for multi-class problems,119 and their accuracy can easily be improved by adding more data points to the training set. Providing more input data, however, typically comes at the cost of long computational runtimes.69 Moreover, KNN algorithms have limitations when it comes to handling imbalanced datasets70 (e.g., training data with a dominant class): For predictions to be reliable, a certain amount of data points from all classes is required to achieve a suitable (local) density in the data space. Also, KNN algorithms tend to struggle with large numbers of input features—a phenomenon, which is known as “curse of dimensionality.”71 Finally, as the input features are usually weighted equally when calculating the distance of a query sample to its nearest neighbors, it is important to ensure that the input features have the same scale72 (which is why some preprocessing of the data might be required).
2. Naïve Bayes methods
Naïve Bayes approaches are probabilistic learning methods that are mostly used for classification tasks. Here, the training data are used to determine likelihood distributions (e.g., Gaussian, multinomial, Bernoulli, or categorical distributions120,121) of the feature values representing each class. Then, the probability that a query sample belongs to one of the classes is calculated based on the Naïve assumption that all features are independent and contribute equally to the output. The corresponding mathematical relationship is formulated in Bayes' theorem.122 Although Naïve Bayes approaches typically rely on over-simplified assumptions, those algorithms can outperform even highly sophisticated methods.123
Compared to other algorithms, Naïve Bayes classifiers can be extremely fast73 and require a small amount of training data only.74 Owing to the independent likelihood estimation applied to each feature, those algorithms also perform well when tasked with high-dimensional problems75 (i.e., those, where many input features are considered) and multi-class classifications119—and they can process both, categorical124,125 and continuous input data.126 However, the simplified assumptions made by Naïve Bayes classifiers do not always hold true when real-life problems are studied: Here, only rarely all features of a sample are truly independent;76 similarly, it is not likely that all sample features contribute equally to the output77 and all feature distributions meet the assumed profile. Furthermore, categorical inputs of the query sample that were not present in the training data will lead to an incorrect probability of zero, known as the “zero frequency problem.”78
3. Support vector machines (SVMs)
Support vector machines (SVMs) define hyperplanes in the n-dimensional feature space, which then can be used to either distinctly separate the dataset into single-variety classes (i.e., for classification) or to approximate the training data (i.e., for regression). To allow for handling problems that would otherwise involve complex mathematical operations, kernel functions that transform input data into higher dimensionality can be integrated into those models.127,128
Since only a subset of training points is used for calculating the decision function, support vector methods can handle data spaces of high dimensionality79,80 while remaining efficient regarding memory and runtime.81 However, for large datasets, the training times can increase significantly.82 Due to the large variety of kernel functions that can be selected and specified for creating the decision function,83,84 the algorithms are very versatile and can even be applied to unstructured data. Still, support vector classifiers can have problems with handling very noisy data129 or classes that strongly overlap.85,86
4. Decision trees and random forest (RF) algorithms
Decision trees are flow chart-like representations of hierarchical decision-making models that are created by analyzing a labeled training set. They consist of nodes (i.e., consecutive stages in which distinct decisions are made) and branches that connect these nodes. Starting with a root node, the training data are (based on individual input features) split in a stepwise manner by creating and answering simple true/false questions. A new (=query) sample can then be classified/predicted by running through the tree using the input values of this new sample and the previously established decision rules.
According to the principle of swarm intelligence, the accuracy of such an approach can be improved by combining an ensemble of non-correlating decision trees—a random forest.130 Enforcing this mandatory variation among the trees is mainly achieved by applying two methods known as feature randomization (here, only a random subset of features is provided for splitting the data) and bootstrap aggregation (short: bagging, i.e., randomly eliminating samples of the training set and replacing them with duplicates of the remaining samples).131
Random forest algorithms can achieve very high accuracies even in high-dimensional data spaces.87 These algorithms run efficiently for large datasets,88 and they can handle variable input data types, including binary, categorical, and numerical features.132 They are well suitable for unbalanced data,89 robust toward non-linearity,90 and outliers91 and—when a sufficient number of independent decision trees is used—rather insensitive to overfitting. Moreover, the decision criteria chosen by the decision trees can be extracted and used to rank the importance of individual features for the categorization process.133,134 However, the self-directed formation of the different trees strongly restricts options to influence random forest algorithms. Importantly, random forest models are not able to extrapolate correlations, and this limits them to making predictions within the created knowledge space.92 Finally, even though running efficiently once the model has been established, training can be computationally costly93 since many trees (usually between 100 and 1000) must be created to obtain a robust random forest.
B. Unsupervised learning
When it is not clear yet what the algorithm is supposed to find, or if labeled data are not available, unsupervised machine learning is more suitable. In such a data-driven approach, the algorithm is simply fed with unsorted input data and allowed to draw its own conclusions by either autonomously clustering the samples or by identifying trends, similarities, extreme points, or patterns in the data. With such a strategy, it was possible to quantify the morphological heterogeneity of cells based on a specified set of geometrical parameters135 and to automatically control the quality of electro-spun nanofibers.136
1. Clustering
An important concept in the field of unsupervised learning is clustering; this approach can be used to identify patterns in a set of unlabeled data. Here, a dataset (containing input values only) is analyzed by sorting the samples into subgroups (clusters) by identifying similarities among them. A common subtype of this approach is k-means clustering. Here, the samples are assigned to k clusters in an exclusive manner by iteratively adjusting cluster centroids until the variety of samples within the formed clusters is minimized while the variety between the clusters is maximized. K-means clustering algorithms are simple and fast, which is why they can handle large datasets.94 They can easily adapt to new samples or data, and their sorting result can be influenced by predefining the initial centroids.95,96 Yet, identifying the correct number k of clusters to be formed can be far from trivial and might require preliminary analyses.97,98 Also, as common for distance-based algorithms, high data dimensionalities can cause issues.99 Finally, basic k-means algorithms encounter problems when the created clusters differ in terms of size or density; however, generalization methods can be applied to deal with this particular issue.137
In addition to the rather simple k-means clustering algorithms, there are also other clustering variants that are selected when more complex datasets need to be processed. Mean-shift clustering, for example, searches regions of high data density by sliding pre-defined analysis windows over the data until the windows containing the highest number of data points are identified. There are two main advantages of this algorithm variant: First, the number of final clusters does not need to be pre-defined; second, centroids in close proximity to each other are automatically merged. A very powerful extension of such mean-shift clustering is the DBSCAN method (density-based spatial clustering of applications with noise), which is capable of identifying clusters of any shape and size while detecting and ignoring outliers. In addition, methods that establish clusters of different hierarchies were shown to work efficiently as well.138
2. Association rule mining
Another popular example of an unsupervised ML method is association rule mining. This approach aims at unveiling correlations between variables in a set of unlabeled data. Such association rules can be interpreted as “if–then” statements, where certain variables (antecedents) are linked to correlating ones (consequents). To identify the most important rules, the dataset is first searched for such if-then patterns, which are then ranked using different significance measures. A major drawback of this approach is that calculating those metrics for all identified relations becomes computationally expensive rather soon. The so-called a priori algorithm provides a good solution to this problem: Here, item sets containing variables or subsets with low importance in one metric are quickly eliminated, and this drastically reduces the amount of data that need to be analyzed regarding the other measures. In addition, there is a broad variety of other approaches for association rule mining that allow for handling different datasets and problems of higher complexity.100–102 Yet, in any case, a sufficiently high data density is essential for these algorithms to avoid random correlations from becoming too prominent.
C. Reinforcement learning
A third learning strategy is reinforcement learning—an action-focused training approach. Here, the machine chooses from different possible actions and is punished or rewarded depending on whether or not it made a “correct” choice. Typically, this is implemented by the algorithm trying to optimize a reward function: Here, positive values are assigned when the algorithm chooses the desired outcome, which presents an incentive for the machine to make this choice; consistently, assigning negative values to “wrong” choices serves as a punishment rendering undesired behavior less likely. With this reward/penalty strategy, a machine can, for example, learn to play a simple board game by repeatedly exploring possible actions in a trial-and-error like fashion and trying to maximize the cumulative reward that is granted upon victory. So far, in materials science, reinforcement learning has been applied to a lower extent than supervised or unsupervised learning strategies. Nevertheless, reinforcement-based training strategies were shown to be suitable for controlling the growth of microbial co-cultures in bioreactors139 and for automatically designing RNA sequences with desired secondary structures.140
1. Q-learning
Q-learning is a simple but efficient method to teach an algorithm to automatically act and react in the context of playing a game or to perform certain workflows. By repeatedly (over thousands or even millions of trials) exploring all available actions during the training phase and iteratively assessing their quality based on the final received reward, the algorithm learns to identify the best available action for a given state.
A major advantage of Q-learning is that it does not require an actual model of the environment. The algorithm does not undergo any explicit external teaching step but learns on its own by autonomously exploring the possible options. This allows for gaining competence in areas that might otherwise remain unexplored by humans. Such wide-ranging exploration, however, can easily become computationally expensive. Another drawback is that—in its basic form—Q-learning is only useful for stationary environments; for non-stationary problems, new training is required to adapt the decision values. However, there are several modified versions of Q-learning, where these issues are dealt with.105–107
D. Deep learning
In addition to the learning strategies discussed so far, there are also “deep learning” approaches. Deep learning can be performed in a supervised, unsupervised, or reinforced manner and aims at mimicking the anatomical structure of biological neural networks and the decision-making process of the human brain. Therefore, multi-hierarchical structures of algorithms are established that can handle and analyze data at different levels of abstraction. This approach holds the potential to analyze even highly complex problems but comes at a prize: Owing to the autonomous, multi-stage data processing procedure, such algorithms act as a black-box. In addition to the provided input, only the generated results are accessible: It remains concealed how exactly the algorithm arrived at a particular decision, and this makes it difficult to rationalize the models suggested by deep learning. Nevertheless, deep learning models have demonstrated tremendous success across a plethora of research areas including biomaterials science; for instance, they precisely predicted the skin permeation behavior of drugs released from biopolymeric films,141 supported the design of anti-fouling polymer coatings and materials,142–145 size-tunable poly(lactic-co-glycolic acid) particles,146 or nucleus-targeting polypeptides,147 they successfully detected single molecule activity from patch-clamp electrophysiology trials,148 and they could accurately model biopolymerization processes.149,150
1. Deep neural networks (DNNs)
Deep neural networks (DNNs) denote digital constructs that mimic the architecture and mode of operation of the human brain. Here, the key players are artificial neurons—small, digital units that can be triggered with a (typically) non-linear activation function. Those neurons are structured in subsequent, interconnected layers, and the individual computations made by each neuron are eventually combined into a final output. Each neuron transforms the received input variables and transmits the result to the next layer. Between each input and output layer, there can be a variable number of “hidden” layers comprising different numbers of neurons with distinct activation functions. A basic example of a DNN making use of forward-only data processing is the so-called multi-layer perceptron (MLP). MLPs are suitable for supervised learning problems (both, regression and classification tasks) and are basically able to model any non-linear function, which is why they are also referred to as “universal function approximators.” Recurrent neural networks (RNNs) are extensions of such DNNs and aim at including more complex information into the decision-making process: Different from MLPs, RNNs combine information from preceding and subsequent layers with the goal of not only to analyze single elements but also to consider their context as well.
DNNs are especially suitable for large-scale datasets, for problems that are too complex for other ML algorithms, and when the problem space is not well understood. Their architecture can be flexibly adapted to other problems, applications, learning strategies, or data types. These networks are able to handle data of high dimensionality, can analyze problems at different levels of abstraction, and learn progressively over time. For DNNs to outperform other ML techniques, though, usually a very large amount of data is needed, and this comes with high computational costs. However, once the costly training phase is completed, making predictions on query samples can be very fast. For instance, a deep model that learned to segment and track cells from microscopy images (which involved large experimental and computational costs) was able, after training, to perform segmentation tasks in less than a second.151 Owing to the high complexity of DNNs in combination with the low transparency of their decision-making process, choosing the right approach and interpreting the obtained results or models can be extremely challenging.
2. Convolutional neural networks (CNNs)
When aiming at processing images or videos, convolutional neural networks (CNNs) usually are the method of choice. When given an image as an input, CNNs use trainable weights to assign importance gradings to different aspects of an image or to objects within the image. The networks can then be used to analyze or classify images, or to identify trained objects within an image. For this purpose, CNNs mainly make use of three procedures: convolution, pooling, and flattening. For image convolution, filters are applied to each pixel. This can help the network to identify certain structures such as edges or peaks. Pooling can lower the computational cost by combining pixels from the same region into one, thus reducing the size of the image. After applying (multiple) convolution and pooling steps, the individual pixels of the resulting image matrix are fed into a standard neural network—a process, which is referred to as “flattening.”
III. SELECTED EXAMPLES OF MACHINE LEARNING APPLICATIONS FROM DIFFERENT BIOSCIENCES
For years, ML approaches have been an integral part of many scientific areas and have been used to develop computer vision for autonomous systems,152,153 to design synthetic materials,154,155 or for human behavioral analysis.156–159 Yet, their application in biophysics or biomaterials science has been less frequent. The scientific questions addressed in these bio-disciplines are characterized by a very high complexity that arises from biological variance and, thus, noisy, divergent data. Hence, it can be quite challenging to translate experimental results from those areas into a format that can be well interpreted by ML models and algorithms. However, once this major hurdle is taken, ML approaches can deliver highly valuable insight into bio-based data as well: Implementation of ML was successfully achieved in the fields of biofabrication,160–165 biosensors and -markers,166–174 pharmaceutical science,175–185 pathophysiology,186–198 biomacromolecule science,199–210 gene analysis,211–221 biomaterials,222–231 and process optimization232–240 (Fig. 3; for more details, see Table II). In this section, we discuss selected examples from those areas, and we highlight what type of data was used by the different ML algorithms to obtain predictions or classifications that—using classical data analysis approaches—would have either been way more time consuming to achieve or outright impossible.
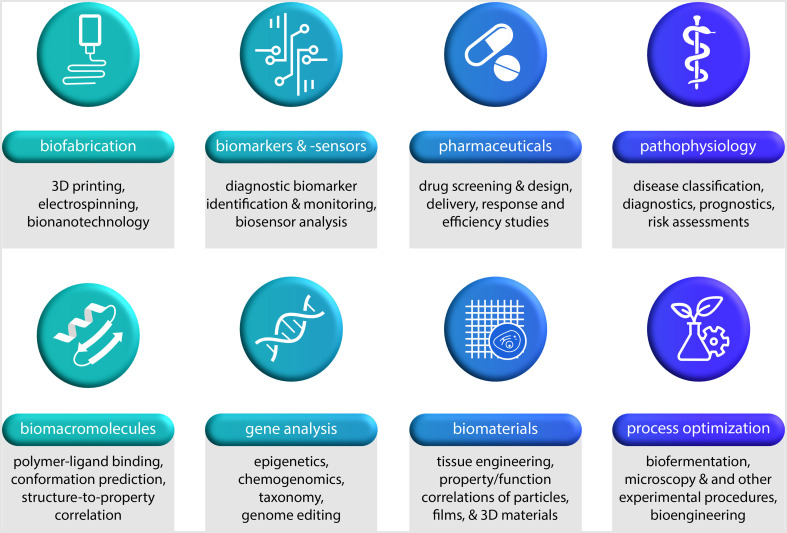
FIG. 3.
Research areas from the biosciences in which machine learning has already been successfully applied. ML approaches were successfully implemented in different fields dealing with biofabrication, biomarkers and sensors, pharmaceuticals, pathophysiology, biomacromolecules, gene analysis, biomaterials, or process optimization. Biofabrication includes various production methods, such as 3D printing or electrospinning; here, ML can be used for process and quality control or for the a priori definition of process parameters. In the context of biomarkers and biosensors, ML can support the identification and the monitoring of diagnostic molecules, and it can assist in the analysis of signals. Pharmaceutical sciences benefit from ML in drug screening and design applications as well as in extensive studies on drug delivery, response, and efficiency. In the context of pathophysiology, ML can help with the classification of diseases as well with diagnostics, prognostics, and the assessment of risk levels. Moreover, a ML-driven analysis of biomacromolecules can help us to investigate polymer-ligand binding, to predict molecule conformations, and to correlate molecular structures with their properties. As part of gene analysis, ML can be employed in the fields of epigenetics, chemogenomics, taxonomy, and genome editing. Biomaterials science and development profit strongly from an ML-driven correlation of properties and functions of different materials including particles, films, or three-dimensional bulk materials. As a final example, process optimization can be achieved by ML-based monitoring and an analysis of microscopy or other experimental procedures/bioengineering processes.
TABLE II.
Overview of studies from various research areas, in which ML was applied.
| Question | Approach | Outcome | Study |
|---|---|---|---|
| Predicting biophysical interactions | |||
| Affinity of protein-peptide interactions across multiple protein families | Hierarchical statistical model | Interaction affinities were successfully predicted based on the amino acid sequences and the inferred structured Hamiltonians (mathematical functions that map the state of a system to its energy). | 16 |
| The model outperformed both, other computational methods293–295 and high-throughput experimental assays developed for the same purpose | |||
| Good performance in high-data and low-data domains | |||
| Protein-ligand binding | SVM, random forest, gradient boosting tree, and a CNN | Successful prediction of protein-ligand binding affinities based on molecular descriptors obtained from topological models | 63 |
| Comparable to or even outperforming other state-of-the-art models296–299 | |||
| Powerful feature engineering | |||
| Compound-protein interactions | Combination of GNNs and CNNs (both supervised); networks were analyzed with neural attention mechanisms | Data-driven representations of compounds (as graphs) and proteins (as sequences of characters) were achieved that proved to be more robust than traditional chemical and biological feature vectors | 64 |
| Competitive or even better performance compared to state-of-the-art models300,301 | |||
| Wettability of a surface based on its topography | KNN, linear regression, Naïve Bayes, random forest, and a DNN | Successful mapping of surface topography parameters to the wetting behavior of the surfaces | 61 |
| Feature elimination was performed to reduce dimensionality and to identify the most influential surface parameters, the choice of which otherwise relies on expert assessment | |||
| The random forest outperformed the other models | |||
| Pathogen attachment to macromolecular coatings | Bayesian regularized artificial neural networks | Successful mapping of individual pathogen attachment to copolymers represented by a set of molecular descriptors | 145 |
| Multiple-pathogen modeling was achieved | |||
| Functional interactions between human genes | Decision tree, logistic regression, Naïve Bayes, random forest | Phylogenetic profiling was performed, and the combination with ML considerably improved the prediction of functional interactions between genes | 217 |
| The random forest outperformed the other models | |||
| Cytotoxicity of nanoparticles (NPs) | Association rule mining | Knowledge about the toxicity of inorganic, organic and carbon-based NPs was extracted from the literature | 257 |
| NPs properties most relevant for their toxicity were identified with a focus on hidden relationships | |||
| Molecular analysis | |||
| Identifying polymer states | DNN | Based on a simulated 3D polymer configuration represented by spatial coordinates, the model can identify different configurational patterns | 62 |
| Phase transition points identified by the model compared well with those obtained from independent specific-heat calculations | |||
| Designing functional protein sequences | Generative model | The model was trained on evolutional protein sequence data and, by this, learned sequence constraints | 202 |
| A diverse library of nanobody sequences was designed that significantly increases the efficiency of discovering stable, functional nanobodies compared to synthetic libraries | |||
| Predicting protein liquid–liquid phase separation | DNN | The ML classifier was trained based on a pre-analysis of datasets comprising proteins of different phase separation tendencies and learned the underlying principles of phase separation behavior with similar accuracy to classifiers using knowledge-based features | 209 |
| Analyzing the structural folding of proteins | Naïve Bayes, SVM, Bayesian generalized linear model | The classifiers accurately predicted mainfolds of proteins based on provided biophysical properties of the amino acids | 252 |
| The Bayesian model outperformed the other two models | |||
| Investigating structures and functions of proteins | Unsupervised language processing (transformer neural networks) | Based on the amino acid character sequences of more than 250 × 106 proteins as an input, knowledge of intrinsic biological properties was developed without supervision | 259 |
| Sensing of single molecules | CNN | A CNN was trained to classify translocation events of single molecules based on time-series signals obtained from nanopore sensors The network was able to automatically extract such information with higher accuracies than previously possible | 58 |
| Disease classification | |||
| Automated detection of glaucoma | Modified CNN (DenseNet), decision trees | Multiple different models were combined to automatically detect glaucoma based on medical images as well as demographic and systemic data | 36 |
| The model shed light onto features that were previously not considered for diagnosis | |||
| Predicting the primary origin of cancer | CNN with an attention model | The model was trained based on labeled images of tumors of known primary origin | 114 |
| The trained model first classified unknown tumors to be either metastatic or primary; then it predicted its site of origin with high accuracy | |||
| Detection of brain tumors | Random forest, SVM, decision trees | Based on geometric features extracted from MRI images, the different models were able to distinguish normal from abnormal brain images | 88 |
| The SVM had the highest sensitivity for detecting brain tumors, whereas the RF had the highest accuracy | |||
| Assessing sepsis through biomarker host response | Naïve Bayes, decision trees | Multiple biomarker measures from plasma samples were used to distinguish septic from healthy cohorts with high accuracies | 168 |
| Naïve Bayes and decision trees performed better than other classifiers—especially regarding the small data size | |||
| COVID-19 detection from x-ray images | Pretrained CNN | Transfer learning (based on a CNN trained on images of general objects) was employed to train a CNN to analyze chest x-ray images | 249 |
| The model successfully distinguished between healthy patients and those suffering from pulmonary diseases; from ill patients, it could identify those with COVID-19 and marked regions of interest in the x-ray images | |||
| Classification of EEG signals in dementia | MLP, logistic regression, SVM | Different feature sets extracted from EEG signals obtained from neurological patients were analyzed and used to make highly accurate predictions of cognitive disorders | 196 |
| The MLP outperformed the other models, and a combination of two different feature sets was shown to entail the most accurate results | |||
| Biomaterials design | |||
| Antifouling polymer brushes | DNN, SVR | A DNN was trained on a benchmark database to rationalize the antifouling properties of existing polymer brushes | 142 |
| A functional group-based SVR was then used to design new antifouling polymer brushes that indeed showed excellent protein resistance properties | |||
| Abiotic nuclear-targeting mini-proteins | Directed, evolution-inspired deep learning | The ML model was provided with data from high-throughput experiments and was then capable of predicting activities of mini-proteins in cells and to decipher sequence-activity predictions for new designs | 147 |
| The ML-designed mini-proteins were more effective than any previously known variant | |||
| Gas-separation polymer membranes | Regression | A rather small set of known polymer membranes (represented by binary fingerprints) and their experimental gas permeability data were used to train the model to predict the gas-separation behavior of a large dataset of polymers that have not been tested for these properties yet | 21 |
| Tested membranes produced from the most promising candidates (based on the prediction) were shown to exhibit excellent gas-separation performance | |||
| Mechanically tough bio-nano-composites | Decision tree and random forest (both as regressors) | Using material compositions linked to the resulting fracture toughness obtained from experimental trials and finite elements analysis, the ML models successfully predicted composition/strength relationships which assist the design of new composites without time-consuming trial-and-error experimentation | 68 |
| Stabilized silver clusters | SVM | The algorithm learned how the sequence of 10 base pair DNA strands correlates to the wavelength of fluorescent light emitted from silver-DNA clusters | 166 |
| With the motifs extracted from the analysis, the model was able to predict the fluorescence color of silver clusters with DNA sequences of variable length | |||
| 3D-printable bioinks | Regression | Different bioink formulations were evaluated regarding their rheological properties and printability, and a general relationship between those properties was established | 59 |
| Cell image analysis | |||
| Extracting biological information from bright field images | Generative adversarial neural network | After being trained on a dataset comprising bright field and fluorescently labeled cell images, the model was able to virtually stain cellular compartments, which eliminates the need for actual (possibly toxic) staining | 278 |
| Quantitative measures of cellular structures were then extracted from the virtually stained images | |||
| Identifying cell morphologies | Image segmentation, principal component analysis, k-means clustering | Cell contours were first identified by image segmentation. After aligning the cell shapes, a principal component analysis was conducted and the cell shape was reconstructed based on the determined eigen-vectors. Finally, different shape modes were identified by k-means clustering. | 135 |
| The protocol is highly automated and very fast in quantifying the cell morphologies | |||
| Predicting osteogenic differentiation | SVM | Based on the cell morphology recorded after 1 day of incubation on nanofiber scaffolds, a pretrained classifier was able to successfully predict the osteogenic differentiation fate of cells | 226 |
| Detecting leukemia | CNN | Characteristic features of white blood cell leukemia were extracted from images and sorted regarding importance | 302 |
| By applying statistics-based feature elimination, the model outperformed several CNN-only based models | |||
| Tracking cell migration | CNN | Stain-free, instance-aware segmentation of cells from phase contrast images was achieved with a CNN and provided unique identifiers for each cell | 237 |
| Based on those identifiers, the same cell could be followed in a series of images taken at different times | |||
| Highly accurate visualization and analysis of cell migration was achieved | |||
| Pharmaceutical development | |||
| Analyzing existing drugs regarding their suitability to target SARS-CoV-2 | Natural language processing with self-attention mechanism | A pre-trained model was used to predict binding affinities between antiviral drugs (represented as strings) and amino acid sequences of the target proteins without providing explicit structural information on the binding epitope | 55 |
| A list of antiviral drugs with good inhibitory potencies against SARS-CoV-2 related proteins was identified | |||
| Identifying self-aggregating drug formulations | Random forest | First, a RF model was used to identify self-aggregating drugs | 177 |
| Then, another RF model precisely predicted the co-aggregation properties of different drugs and excipients and was able to find suitable excipients for a novel drug | |||
| Generation of anticancer molecules | Conditional generative model | A reinforcement learning-based model was trained to design anticancer molecules with specific drug sensitivity and toxicity properties to target individual transcriptomic profiles | 271 |
| Such designed molecules exhibit (in silico) comparable physicochemical properties as existing cancer drugs | |||
| Predicting cancer patient drug responses | Linear regression, ridge regression, support vector regression | Based on transcriptomic data obtained from 3D culture models, different biomarkers were identified that allow for accurate patient/drug response predictions | 273 |
| Identifying drug targets | Naïve Bayes | Multiple different data types were combined to train the model based on a dataset of known molecule/target correlations | 180 |
| Novel drug binding targets were predicted | |||
| Biofabrication | |||
| Predicting the molecular weight of synthesized bio-molecules | MLP, SVM | Biopolymers were synthesized via enzymatic polymerization, and various reaction parameters were tuned to alter the molecular weight of the product | 150 |
| An SVM was shown to be highly suitable to predict the molecular weight despite the small training data size | |||
| Controlling the size of elastin-based particles | K-means clustering | A dataset comprising the properties of elastin-based particles and the corresponding fabrication parameters were analyzed by the clustering algorithm | 60 |
| The influence of the fabrication parameters on the size of the created particles was revealed, and this information was used to fine-tune the fabrication process | |||
| Controlling microbial co-cultures in bioreactors | Q-learning | Process feedback via a trained reinforcement learning model successfully supported maintaining populations at pre-defined target levels | 139 |
| The model was shown to be robust toward variations in the initial states and targets and outperformed standard control approaches | |||
| Identifying high-quality printing configurations | Random forest | With the printing conditions (resulting from the material composition) and the printing parameters as inputs, a classification model could distinguish between “high” and “low” quality prints, and a regression model returned a direct quality metric | 245 |
| The random forest outperformed a simple linear model | |||
| Monitoring anomalies in 3D bioprinting | CNN, SVM | SVM models were trained to predict whether a specific defect is directly visible in the image of a printed object | 274 |
| A CNN was trained to provide information about the applied printing pattern and the occurring printing anomalies | |||
| The combined model accurately detected and recognized anomalies in various different printing patterns | |||
A. Supervised learning approaches
When applying supervised learning strategies, the researchers still have a good level of control over how the algorithms are trained and what type of predictions they try to achieve. For instance, Tourlomousis et al.241 used a supervised SVM algorithm to investigate the mechano-sensing response of cells to electrospun fibrous materials (Fig. 4). Therefore, they compared the morphologies of cells after they were cultivated on different substrate geometries. They correlated cell morphology parameters (e.g., cell area, ellipticity, or number of focal adhesions per cell) obtained from confocal microscopy images with architectural features of the substrate (e.g., fiber diameter, pore size, or degree of uniform fiber alignment). With this ML strategy, it was possible to investigate yet unexplored design spaces to yield specific designs qualified at the single-cell level. The authors demonstrated that certain geometrical characteristics of fiber-based materials can be mapped onto unique aspects of cell morphologies—and this is an important step toward a shape-driven pathway to controlling cellular phenotypes.
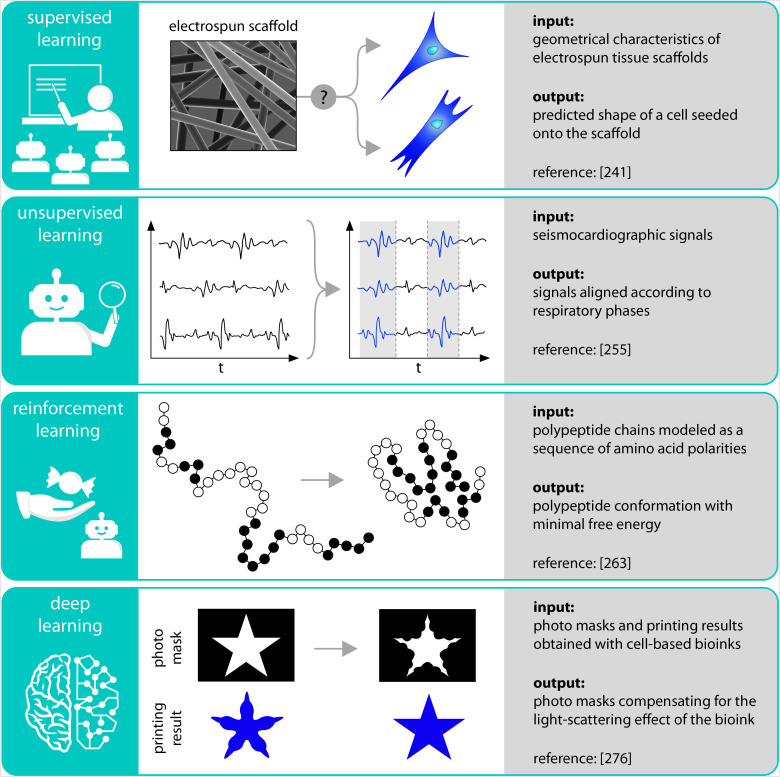
FIG. 4.
Schematic representation of selected examples from the biosciences, where ML algorithms have been successfully applied. By using a supervised approach, the geometrical characteristics of electrospun scaffolds were successfully linked to the resulting shape of cells seeded onto the scaffold. An unsupervised ML algorithm could group seismocardiographic signals according to the respiratory phases during which they were acquired to allow for a more direct signal comparison. Reinforcement learning was employed to find energetically optimal conformations of polypeptides. Finally, deep learning was applied to generate photo masks that compensate for the light-scattering effects of cells present in the used bioink.
Other studies went beyond purely analyzing datasets and used the knowledge generated by ML algorithms to tailor materials for specific applications. For instance, Sujeeun et al.242 utilized multiple supervised learning algorithms for the development of scaffolds for tissue regeneration; such scaffolds are typically used to provide structural support for cell attachment and to enable cell proliferation. Here, the main challenge was to browse through a plethora of available polymeric materials to identify the most suitable candidate that meets specified requirements regarding, e.g., biocompatibility, biodegradability, mechanical strength, porosity, and wound healing behavior. To do so, in vitro cell viability data (obtained from an MTT [3–(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay) were combined with physico-chemical properties (e.g., the dimensions of fibers and pores, Young's modulus, or water contact angles) of different scaffolds to model the material-cell interactions. The established correlations then served for reverse-engineering scaffolds with desired performance. Six basic supervised approaches, including KNN and SVM, were compared, and a random forest classifier achieved the highest accuracy. Moreover, this RF algorithm could provide deeper insight into the identified correlations and demonstrated that two selected material characteristics (the pore and fiber diameter) have the strongest influence on the material-cell interaction. Finally, by performing preliminary in vivo biocompatibility experiments, the authors were able to show that the determined correlations also hold true (at least to a certain extent) when the material is placed into a living organism. The authors mention, however, that an integration of more advanced techniques, such as reinforcement learning or transfer learning (see Chap. 4), should be considered to obtain a more generalized and robust model that is applicable to unknown scaffolds.
In addition to those applications related to tissue engineering, basic supervised ML algorithms were proven to be handy for various other tasks: RF models, for example, can support the design of self-assembling dipeptide hydrogels243 and anti-biofouling surfaces244 and can supervise 3D bioprinting.245 With KNN and SVM algorithms, it is possible to differentiate healthy from apoptotic cells,246 to detect pneumonia247 or COVID-19248,249 by extracting features from x-ray images, to diagnose Parkinson based on recordings of speech disorders,250 and to classify white blood cells.251 Finally, Naïve Bayes models can classify protein folding patterns,252 identify post-transcriptional modifications in RNA sequences,253 and support the detection of brain tumors.254
B. Unsupervised learning approaches
Different from supervised learning approaches, unsupervised algorithms process unlabeled data. For instance, Gamage et al.255 employed a k-means clustering algorithm to group seismocardiographic signals (SCG) according to the patients' different respiratory states (Fig. 4). SCG is a noninvasive technique that monitors heart function by measuring cardiac-related vibrations on the chest surface. Since the measured signals are typically a convolution of respiratory movements and heart contractions, a direct comparison of two different measurements is difficult. By subdividing the obtained signals and using the vibration amplitudes in those subsignals as input features to cluster the generated subsequences based on their similarity, the SCG data were automatically separated into classes of different lung volumes (high or low) or different flow directions (inhaling or exhaling process). Indeed, within those categories, a comparison of the vibration signals to assess cardiac health (and to detect anomalies) is feasible. Hence, an ML-supported analysis of SCG signals may eliminate the necessity of additional (simultaneous, but independent) respiratory measurements.
Another unsupervised clustering approach was reported by Helfrecht et al.256 who aimed at identifying secondary and tertiary structures in proteins and rationalizing their formation. Here, the idea was not to use common structural descriptions of molecules that are based on predefined motifs such as intramolecular hydrogen bonds or distinct dihedral angle patterns (two strategies, which often rely on human intuition/approximations and only cover a predefined subset of molecular motifs) but to develop a more general approach that is readily applicable to various macromolecules. Therefore, the positions of all atoms in a given protein backbone were combined into an input vector whose complexity was reduced into 6–10 features based on a principle component analysis; then, a density-based algorithm was employed to cluster those reduced vectors: Regions in the feature space with high data density were defined as clusters that are separated from each other by low density areas. Even though several of the formed molecule clusters can belong to the same category of secondary structures (e.g., α-helices or β-strands), a similarly good over-all classification could be achieved with this ML-based approach as with traditional methods. Furthermore, the authors compared unsupervised and supervised methods: Their example highlighted that a supervised approach is suitable to adapt existing motif definitions or to test whether the chosen input data sufficiently represent the output. Unsupervised learning, in contrast, turned out to be better suitable for finding new patterns in the feature space.
In addition to clustering, which is certainly one of the most important techniques where unsupervised ML is applied for, unsupervised association rule mining was shown to be a useful tool to highlight hidden correlations between data such as between the material properties and production process of nanoparticles and their cytotoxicity.257 Moreover, unsupervised learning methods were successfully employed for image processing or pattern analysis. For instance, the autonomous detection of characteristic features from abdominal computed tomography (CT) images enabled the reconstruction of CT images captured with low radiation doses.258 Owing to this reduced radiation exposure, the concomitant risk of side effects for patients (such as developing new cancer) is minimized while sufficient image quality is maintained. Furthermore, unsupervised models were able to rationalize and predict selected functional properties (e.g., the biological activity259 or thermostability260) of proteins based on their sequences only, they could unravel the structure of block copolymer micelles,261 and they managed to successfully and automatically recognize the origin tissue of metastatic tumor cells.262
C. Reinforcement learning approaches
Reinforcement learning does not aim at identifying correlations or classifying samples according to given labels (which are typical goals for supervised and unsupervised learning strategies) but makes use of learning procedures to perform certain actions “correctly.” An interesting example for a reinforcement based learning approach was presented by Jafari and Javidi.263 Here, the researchers tried to obtain a complete prediction of the conformation of a polypeptide based on hydrophobic interactions only (Fig. 4). For this purpose, polypeptides were modeled as a sequence of amino acid polarities: For instance, the sequence “HHHPP” would represent a polypeptide with three hydrophobic (H) amino acids followed by two polar (P) ones. Then, the possible conformational space of a polypeptide is given as a bidimensional Cartesian grid with two constraints: First, two consecutive amino acids must be vertical or horizontal neighbors in the grid; second, two amino acids cannot be superimposed. To find the ideal overall conformation, a Q-learning algorithm with a dedicated reward function was employed, which aimed at minimizing the free energy of the polypeptide. In this model, the only actions available to the algorithm are moving a given amino acid from its current position in the grid to a neighboring position. With this approach, conformations of minimal free energy were identified (and found to agree with classical calculations using complex models) without explicitly implementing biophysical knowledge; moreover, it was faster than other state-of-the-art approaches. Remarkably, the “long short-term memory” network (a subtype of recurrent neural networks) used in this study proved to be particularly capable of handling sequential data such as chains of amino acids.
Interestingly, reinforcement learning was also successfully used for target-oriented design tasks such as de novo drug development: Popova et al.264 employed reinforcement learning to combine two independent supervised learning algorithms; here, the first one was capable of creating drug-like molecules, and the second one could predict certain properties of molecular structures. After individual, supervised training phases of both algorithms (in which either learned how to fulfill its particular task), they were jointly re-trained in a reinforcement approach to deliberately bias the creation of new molecules toward variants with desired properties: The first algorithm received a reward only if the properties predicted by the second matched the predefined goal. By adjusting this reward, the created molecule library was successfully tailored to contain drugs with specific physical properties, biological activity, or chemical substructures. Overall, this study impressively demonstrated how reinforcement learning can be used for generating property-optimized chemical libraries of novel compounds.
Overall, reinforcement learning is currently gaining an importance. It was recently used to control and optimize bioprocesses,265 to adapt cold atmospheric plasma conditions to optimally eliminate cancer cells,266 or to identify efficient surgical cardiac ablation strategies for atrial fibrillation.267 Moreover, reinforcement learning was shown to be useful for controlling tumor growth,268 to optimize cancer therapy,269,270 and for the development and dosing of anti-cancer drugs.271–273
D. Deep learning approaches
Deep learning is a special subtype of machine learning, where all types of (supervised, unsupervised, or reinforcement) approaches are solved by algorithms that try to mimic the structure and function of the human brain. These algorithms are often difficult to interpret, but they come with the advantage of high variability and the potential to model even highly complex systems. A process-oriented application of deep learning that recently gained considerable importance addresses 3D bioprinting: Here, deep learning-based algorithms can be used for monitoring the printing procedure to determine optimal process parameters or for detecting anomalies in the printed products.274,275 Moreover, an advanced deep learning approach was demonstrated by Guan et al.276; here, the researchers set out to compensate for cell-induced light scattering effects in light-based bioprinting—a common fabrication technology used for tissue engineering and regenerative medicine purposes (Fig. 4). To obtain the desired structures, a typical approach is to illuminate a reservoir containing the bioink while using a photo mask that only allows curing in predefined regions. However, the light-scattering effect brought about by cells embedded into the bioink impacts the photopolymerization process and entails a reduced printing resolution. To determine the correlation between the used photo mask and the resulting printing pattern, a convolutional neural network was employed: Pairs of graphical representations of the photo mask on the one hand and the printing result on the other hand were processed with several subsequent convolution and deconvolution steps to model the transformation of the former into the latter. With such a trained network, a photo mask was generated that was supposed to compensate the light-scattering effect of this particular bioink sample based on a desired printing output. Indeed, with this approach, a considerable improvement of the printing resolution was achieved; without the help provided by ML, a similar result would have required an extensive and costly trial-and-error style optimization for each individual structure.
Overall, deep learning techniques have been proven to be particularly useful for processing and analyzing images. This includes assessing the damage mechanics of bone tissue based on microCT images,277 extracting quantitative properties of cells from bright-field images,278 or compensating optical errors in microscopy images to obtain reliable images even under difficult conditions.279 Skärberg et al.280 employed a deep learning approach to analyze images of porous polymer films; here, the aim was to obtain a better understanding of how to tune those materials for controlled drug release. Therefore, they collected combined focused ion beam and scanning electron microscopy images of polymer films with different porosities and fed them into a convolutional neural network for segmentation. From the obtained dataset, 100 images (which corresponds to ∼0.4% of the total dataset) were manually segmented and used for training. To increase the dataset size, those images were subdivided, resulting in over 19 × 106 training samples. The trained CNN was then able to automatically identify pores in the images; thus, important information was retrieved that is needed for further sample analysis but that otherwise could only be gathered through expensive expert assessments. In fact, the results received with the CNN were comparable to manual segmentations and better than those previously obtained with a random forest classifier that was trained on scale-space features. Hence, extending the training set by augmenting data (for more information on this particular method, see Sec. IV) was an important step to achieve a robust ML model capable of competing with actual expert judgments.
The potential applications of deep learning approaches are virtually limitless, and many highly sophisticated neural network architectures have been developed and applied to different problem sets. For instance, generative models, such as generative adversarial neural networks, Gaussian mixture models, or hidden Markov models, are unsupervised approaches that can learn patterns from given input data; then, those models can generate new examples that could plausibly stem from the original dataset. Such algorithms were shown to be useful for the design and discovery of drugs,281,282 for the development of complex materials with desired elasticity and porosity283 or tissue engineering-related properties,284 to create synthetic data (e.g., photo-realistic images285 or biomedical signals286) for network training, and for analytical tasks such as identifying cell morphologies typical for cancer.287 Whether for an automated evaluation of tumor spheroid behavior in 3D cultures288 or for identifying cancer based on RNA data,289 for predicting the in vivo fate of nanomaterials based on mass spectrometry,290 to detect the presence of viral DNA sequences from metagenomic contigs,291 or to autonomously detect sleep apnea events from electrocardiogram signals,292 deep learning can be considered the ML equivalent of a Swiss-Army Knife as it can be a helpful tool in many fields of research.
IV. BIGGER IS BETTER BUT HARD TO GET—HOW TO HANDLE SMALL DATA
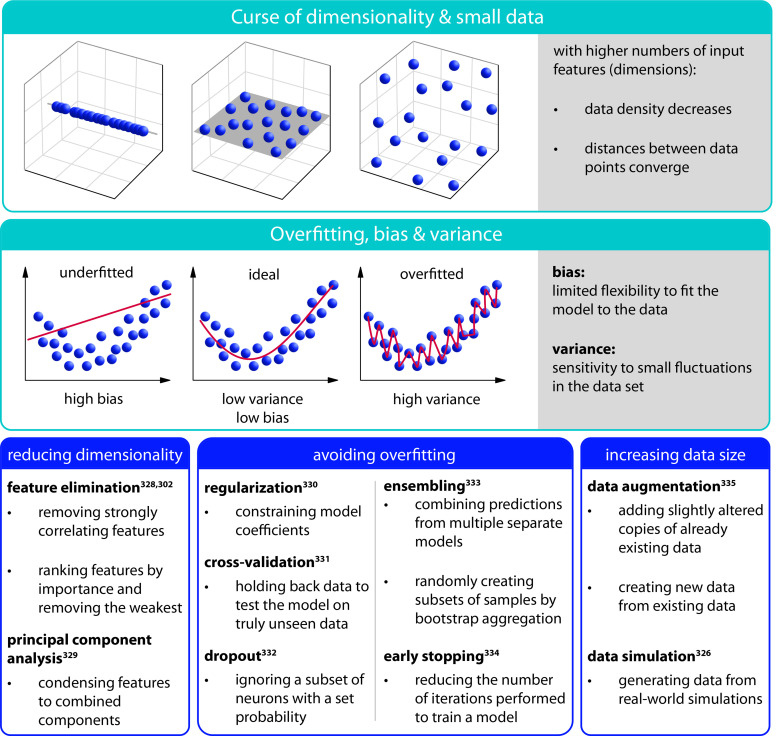
The performance of all ML algorithms critically depends on the amount of existing knowledge, i.e., the size of the database available for training. Whereas “Big Data” are a phrase commonly used in the context of machine learning, generating large volumes of data from experimental trials is often very challenging: The costs and time requirements associated with experimental studies are typically significant. When the training set is too small, commonly encountered problems include overfitting, biased predictions, or a phenomenon known as the “curse of dimensionality” (Fig. 5). Overfitting refers to algorithms that represent the training data in too much detail. Typically, this happens when a model depicts the variations (and, sometimes, even noise) in the training data to such an extent that it negatively impacts the performance of the model when confronted with new data.303 Data bias denotes a type of prejudice or favoritism toward a certain class or a decision that is based on wrong assumptions, which are made based on (non-ideal) training data.304 This can, for instance, occur when the sample set used does not sufficiently represent the whole problem, hence (possibly) neglecting concealed factors or if the model does not properly fit the training data.305,306 Finally, a prominent issue of small datasets occurs with increasing dimensionality (i.e., with increasing numbers of features added): When the total amount of training data stays the same, the density of data points decreases with every dimension added to a multi-dimensional feature space, and low data density can lead to reduced accuracy. Thus, a frequently asked question is: How many data points are actually necessary to establish robust models that provide reliable results? Answering this question is, however, not trivial as several factors need to be taken into account: the complexity of the problem, the chosen algorithm, the number and type of input features, and the noise level in the available data.
FIG. 5.
Typical challenges that can arise when applying ML methods and available remedies to deal with them. High dimensionality, small datasets, overfitting, bias, and variance are common difficulties encountered when using ML. High dimensionality entails a decrease in the data density in the feature space and leads to an equalization of distances between data points. This becomes particularly problematic when datasets are too small to compensate for these effects. Overfitting refers to ML models that approximate the training data too well. Overfitted models show a high sensitivity to small fluctuations in the dataset—a phenomenon which is referred to as “high variance.” In contrast, when the models are not able to sufficiently capture the relationship between input and output, the model is underfitting the training data. Such limited flexibility to fit the model to the data is called “bias.” Those problems, however, can be tackled for the following strategies: Reducing the dimensionality can be achieved by performing a feature elimination302,328 or by condensing the feature space via a principal component analysis.329 Overfitting can be avoided or at least reduced by including regularization,330 early stopping,334 or dropouts332 into the ML models, by using multiple independent predictors (ensembles),333 or by validating the models using cross-validation.331 Finally, the size of a dataset can be increased by simulating326 or augmenting data.335
There are some established rules of thumb that can help researchers to navigate this issue: In a regression problem, the number of training samples should be ten times as high as the number of dimensions of the investigated problem; and at least 1000 images per class should be available for computer vision tasks.307 However, under certain conditions, good prediction accuracies have also been reported for much smaller datasets. For instance, Shaikhina et al.308 successfully established a deep neural network for predicting the compressive strength of human trabecular bones in severe osteoarthritic conditions, and they could achieve this by using data from 35 bone specimens only. Here, the versatile design of DNNs came in handy: The number of hidden layers as well as the number or neurons and their activation functions were iteratively adjusted until the predictive accuracy of the model reached a maximum. Similarly, basic (non-deep) ML models can be optimized with respect to both, the desired problem and the available dataset: Every ML model is characterized by a set of distinct parameters, which are typically referred to as hyperparameters. Examples for such hyperparameters are the number of neighbors considered in a KNN model, the allowed dimensions of the trees in a RF model, or the set amount of penalty for misclassified samples in an SVM; however, also more advanced parameters can be adjusted. With such optimized algorithms, even fewer than 65 samples were shown to be sufficient to train various algorithms including RF or SVM models.309,310
Another important realization in this context is that, even though each research topic is distinct, most questions asked are not entirely unique. Thus, machine learning models that were trained for a certain task can often be used as a starting point for similar problems (this is referred to as transfer learning).311 Then, only few data points of the target problem are needed to transfer models generated from the source task to the target task—a procedure known as few-shot312 or even one-shot learning.313 With this approach, neural networks trained on large-scale image datasets of various macroscopic objects were successfully employed to classify electroencephalogram (EEG) signals obtained from patients diagnosed with delirium,314 or to identify diseases on grape leaves.315
Of course, no algorithm can generate knowledge where no data exist—all models are based on the assumption that the training data cover a suitable and representative subset of the problem at hand. Inter- or extrapolation procedures can (to a certain extent) fill in local gaps, where data are missing, but the machines and models generated by them will only be as reliable as the data fed into them. Even though there might not be a pre-trained algorithm for every research problem, there is a huge amount of data documented in the literature or even stored in readily accessible repositories. From those sources, it is often possible to selectively extract a subset of data to complement one's own dataset, thus increasing the amount of training data. Intriguingly, the collection of such supplemental data is not limited to data already available in a numerical form; especially the extraction of data from texts has been quite successful recently:316 for instance, unsupervised algorithms were—without having been provided with explicit chemical knowledge—able to understand the structure of the periodic table from text-based sources only, and they could recognize complex structure-property relationships of materials for specific applications, such as energy conversion,317 nanomedicine,318 or pharmaceutics,319 even years before they were actually realized.320,321
Gathering data from various sources can, of course, involve considerable effort in terms of retrieving and formatting. Other—possibly less expensive—approaches to extend the training dataset (to improve model generalization and robustness) make use of augmented or synthetic data. Data augmentation refers to a strategy where slightly altered copies of existing data are added to the training set. In the case of images, for example, augmented data can be created by rotating, shifting, splitting, zooming, or flipping the original pixel matrix.322 With these transformations, Liang et al.323 used 48 microscopy images obtained from collagenous tissue to create >300 000 training images; with this augmented dataset, they then successfully trained a CNN to predict non-linear stress-strain responses of the tissue. Importantly, such an approach is not limited to images—also other data types can be augmented, e.g., by superimposing random noise324 or by adding synthetically generated features; examples for the latter include crude estimations of the property-to-predict325 or calculated characteristics derived from empirical models.326 When training samples are created entirely from simulations, this is referred to as synthetic or in silico data. Indeed, by complementing experimental datasets with large amounts of such in silico data, Tulsyan et al.327 were able to develop a reliable ML-based monitoring system for biopharmaceutical manufacturing processes—a task that was previously very difficult due to the lack of data.
V. CONCLUSION AND OUTLOOK
Ongoing challenges encountered in the context of ML include having to deal with insufficient data quality, data scarcity, under- or overfitting of the models on the training data, biased training sets, and high computational costs. Indeed, for a long time, the application of ML techniques for bio-related research questions has been severely restricted by the range of difficulties associated with such problems, i.e., small datasets, complex problem definitions, and biological variability. However, some of those issues can now successfully be dealt with: Once the research questions have been translated into computer-readable formats, various methods can be used to increase the data density and to optimize the models in a way that common problems, such as overfitting and bias, are reduced. Even though the training phase of such algorithms might be computationally and/or experimentally costly, once trained, the models can make predictions very quickly.328–335
The black-box character of most deep learning methods and the increasing complexity of advanced algorithms in combination with the lack of experienced users especially entails a completely new set of hurdles on the path to fully exploiting the potential of ML. ML nowadays includes a diverse spectrum of different algorithms that can be employed for a plethora of different purposes, and the continuous advancement and expansion of the ML portfolio open up an ever-increasing number of possible applications in all kinds of scientific areas. Generative adversarial neural networks, for example, have successfully been employed to mimic any type of data (including images, numerical, or binary data), which then can be used to either increase the training dataset and/or to generate results. As neural networks are automatically developed inspired by human evolution, evolutionary machine learning approaches can decrease the required expert knowledge needed for creating deep ML models. Attention mechanisms are very recent but promising strategies to improve deep model performances by putting a stronger focus on a few, more relevant aspects while paying less attention to the rest. Finally, by integrating statistical properties into variables, Bayesian neural networks are especially suitable for research problems dealing with sparse data. With these improved techniques available now, current ML models are well-equipped to explore the diverse range of structures, effects, and mechanisms of bio-related systems in more detail, and it is clear that we will encounter many more exciting results in the near future.
ACKNOWLEDGMENTS
The authors thank Jochen Mück for helpful discussions regarding terminology. This project was conducted in the framework of the innovation network “ARTEMIS” by the Technical University of Munich.
AUTHOR DECLARATIONS
Conflict of Interest
The authors have no conflicts to disclose.
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1. Patriat R., Niederer J., Kaplan J., Huffmaster S. A., Petrucci M., Eberly L., Harel N., and MacKinnon C., “ Morphological changes in the subthalamic nucleus of people with mild-to-moderate Parkinson's disease: A 7T MRI study,” Sci. Rep. 10(1), 8785 (2020). 10.1038/s41598-020-65752-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang L., Yan Y., Zhang L., Liu Y., Luo R., and Chang Y., “ Substantia nigra neuromelanin magnetic resonance imaging in patients with different subtypes of Parkinson disease,” J. Neural Transm. 128(2), 171–179 (2021). 10.1007/s00702-020-02295-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dorbala S., Cuddy S., and Falk R. H., “ How to image cardiac amyloidosis: A practical approach,” Cardiovasc. Imaging 13(6), 1368–1383 (2020). 10.1016/j.jcmg.2019.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abdelrahman M., Reutzel E. W., Nassar A. R., and Starr T. L., “ Flaw detection in powder bed fusion using optical imaging,” Addit. Manuf. 15, 1–11 (2017). 10.1016/j.addma.2017.02.001 [DOI] [Google Scholar]
- 5. Gobert C., Reutzel E. W., Petrich J., Nassar A. R., and Phoha S., “ Application of supervised machine learning for defect detection during metallic powder bed fusion additive manufacturing using high resolution imaging,” Addit. Manuf. 21, 517–528 (2018). 10.1016/j.addma.2018.04.005 [DOI] [Google Scholar]
- 6. Groschner C. K., Choi C., and Scott M. C., “ Machine learning pipeline for segmentation and defect identification from high-resolution transmission electron microscopy data,” Microsc. Microanal. 27(3), 549–556 (2021). 10.1017/S1431927621000386 [DOI] [PubMed] [Google Scholar]
- 7. Rhoads D. D., “ Computer vision and artificial intelligence are emerging diagnostic tools for the clinical microbiologist,” J. Clin. Microbiol. 58(6), e00511–e00520 (2020). 10.1128/JCM.00511-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhu X., Mohsin A., Zaman W. Q., Liu Z., Wang Z., Yu Z., Tian X., Zhuang Y., Guo M., and Chu J., “ Development of a novel noninvasive quantitative method to monitor Siraitia grosvenorii cell growth and browning degree using an integrated computer‐aided vision technology and machine learning,” Biotechnol. Bioeng. 118(10), 4092–4104 (2021). 10.1002/bit.27886 [DOI] [PubMed] [Google Scholar]
- 9. Fei C., Cao X., Zang D., Hu C., Wu C., Morris E., Tao J., Liu T., and Lampropoulos G., “ Machine learning techniques for real-time UV-Vis spectral analysis to monitor dissolved nutrients in surface water,” in AI and Optical Data Sciences II ( International Society for Optics and Photonics, 2021), Vol. 11703, p. 117031D. [Google Scholar]
- 10. Roach D. J., Rohskopf A., Hamel C. M., Reinholtz W. D., Bernstein R., Qi H. J., and Cook A. W., “ Utilizing computer vision and artificial intelligence algorithms to predict and design the mechanical compression response of direct ink write 3D printed foam replacement structures,” Addit. Manuf. 41, 101950 (2021). 10.1016/j.addma.2021.101950 [DOI] [Google Scholar]
- 11. Martynenko A., “ Computer vision for real-time control in drying,” Food Eng. Rev. 9(2), 91–111 (2017). 10.1007/s12393-017-9159-5 [DOI] [Google Scholar]
- 12. Alam M. M. and Islam M. T., “ Machine learning approach of automatic identification and counting of blood cells,” Healthcare Technol. Lett. 6(4), 103–108 (2019). 10.1049/htl.2018.5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamanishi C., Parigoris E., and Takayama S., “ Kinetic analysis of label-free microscale collagen gel contraction using machine learning-aided image analysis,” Front. Bioeng. Biotechnol. 8, 1–8 (2020). 10.3389/fbioe.2020.582602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Park S., Ahn J. W., Jo Y., Kang H.-Y., Kim H. J., Cheon Y., Kim J. W., Park Y., Lee S., and Park K., “ Label-free tomographic imaging of lipid droplets in foam cells for machine-learning-assisted therapeutic evaluation of targeted nanodrugs,” ACS Nano 14(2), 1856–1865 (2020). 10.1021/acsnano.9b07993 [DOI] [PubMed] [Google Scholar]
- 15. Spanoudaki V., Doloff J. C., Huang W., Norcross S. R., Farah S., Langer R., and Anderson D. G., “ Simultaneous spatiotemporal tracking and oxygen sensing of transient implants in vivo using hot-spot MRI and machine learning,” Proc. Natl. Acad. Sci. 116(11), 4861–4870 (2019). 10.1073/pnas.1815909116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cunningham J. M., Koytiger G., Sorger P. K., and AlQuraishi M., “ Biophysical prediction of protein–peptide interactions and signaling networks using machine learning,” Nat. Methods 17(2), 175–183 (2020). 10.1038/s41592-019-0687-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jones P., Coupette F., Härtel A., and Lee A. A., “ Bayesian unsupervised learning reveals hidden structure in concentrated electrolytes,” J. Chem. Phys. 154(13), 134902 (2021). 10.1063/5.0039617 [DOI] [PubMed] [Google Scholar]
- 18. Clauser J. C., Maas J., Arens J., Schmitz-Rode T., Steinseifer U., and Berkels B., “ Automation of hemocompatibility analysis using image segmentation and supervised classification,” Eng. Appl. Artif. Intell. 97, 104009 (2021). 10.1016/j.engappai.2020.104009 [DOI] [Google Scholar]
- 19. Chu A., Nguyen D., Talathi S. S., Wilson A. C., Ye C., Smith W. L., Kaplan A. D., Duoss E. B., Stolaroff J. K., and Giera B., “ Automated detection and sorting of microencapsulation via machine learning,” Lab Chip 19(10), 1808–1817 (2019). 10.1039/C8LC01394B [DOI] [PubMed] [Google Scholar]
- 20. Madiona R. M., Winkler D. A., Muir B. W., and Pigram P. J., “ Optimal machine learning models for robust materials classification using ToF-SIMS data,” Appl. Surf. Sci. 487, 773–783 (2019). 10.1016/j.apsusc.2019.05.123 [DOI] [Google Scholar]
- 21. Barnett J. W., Bilchak C. R., Wang Y., Benicewicz B. C., Murdock L. A., Bereau T., and Kumar S. K., “ Designing exceptional gas-separation polymer membranes using machine learning,” Sci. Adv. 6(20), eaaz4301 (2020). 10.1126/sciadv.aaz4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liang N., Li B., Jia Z., Wang C., Wu P., Zheng T., Wang Y., Qiu F., Wu Y., and Su J., “ Ultrasensitive detection of circulating tumour DNA via deep methylation sequencing aided by machine learning,” Nat. Biomed. Eng. 5, 586–599 (2021). 10.1038/s41551-021-00746-5 [DOI] [PubMed] [Google Scholar]
- 23. Campano C., Lopez-Exposito P., Gonzalez-Aguilera L., Blanco Á., and Negro C., “ In-depth characterization of the aggregation state of cellulose nanocrystals through analysis of transmission electron microscopy images,” Carbohydr. Polym. 254, 117271 (2021). 10.1016/j.carbpol.2020.117271 [DOI] [PubMed] [Google Scholar]
- 24. Ruggeri F. S., Flagmeier P., Kumita J. R., Meisl G., Chirgadze D. Y., Bongiovanni M. N., Knowles T. P., and Dobson C. M., “ The influence of pathogenic mutations in α-synuclein on biophysical and structural characteristics of amyloid fibrils,” ACS Nano 14(5), 5213–5222 (2020). 10.1021/acsnano.9b09676 [DOI] [PubMed] [Google Scholar]
- 25. Litjens G., Kooi T., Bejnordi B. E., Setio A. A. A., Ciompi F., Ghafoorian M., Van Der Laak J. A., Van Ginneken B., and Sánchez C. I., “ A survey on deep learning in medical image analysis,” Med. Image Anal. 42, 60–88 (2017). 10.1016/j.media.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 26. Yin P., Yuan R., Cheng Y., and Wu Q., “ Deep guidance network for biomedical image segmentation,” IEEE Access 8, 116106–116116 (2020). 10.1109/ACCESS.2020.3002835 [DOI] [Google Scholar]
- 27. LaLonde R., Xu Z., Irmakci I., Jain S., and Bagci U., “ Capsules for biomedical image segmentation,” Med. Image Anal. 68, 101889 (2021). 10.1016/j.media.2020.101889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sekuboyina A., Husseini M. E., Bayat A., Löffler M., Liebl H., Li H., Tetteh G., Kukačka J., Payer C., and Štern D., “ VerSe: A vertebrae labelling and segmentation benchmark for multi-detector CT images,” Med. Image Anal. 73, 102166 (2021). 10.1016/j.media.2021.102166 [DOI] [PubMed] [Google Scholar]
- 29. Berg S., Kutra D., Kroeger T., Straehle C. N., Kausler B. X., Haubold C., Schiegg M., Ales J., Beier T., and Rudy M., “ Ilastik: Interactive machine learning for (bio) image analysis,” Nat. Methods 16(12), 1226–1232 (2019). 10.1038/s41592-019-0582-9 [DOI] [PubMed] [Google Scholar]
- 30. Hesamian M. H., Jia W., He X., and Kennedy P., “ Deep learning techniques for medical image segmentation: Achievements and challenges,” J. Digital Imaging 32(4), 582–596 (2019). 10.1007/s10278-019-00227-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H., Menegaux A., Schmitz‐Koep B., Neubauer A., Bäuerlein F. J., Shit S., Sorg C., Menze B., and Hedderich D., “ Automated claustrum segmentation in human brain MRI using deep learning,” Hum. Brain Mapp. 42(18), 5862–5872 (2021). 10.1002/hbm.25655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiao L., Zhang F., Liu F., Yang S., Li L., Feng Z., and Qu R., “ A survey of deep learning-based object detection,” IEEE Access 7, 128837–128868 (2019). 10.1109/ACCESS.2019.2939201 [DOI] [Google Scholar]
- 33. Zhao Z.-Q., Zheng P., Xu S-t., and Wu X., “ Object detection with deep learning: A review,” IEEE Trans. Neural Networks Learn. Syst. 30(11), 3212–3232 (2019). 10.1109/TNNLS.2018.2876865 [DOI] [PubMed] [Google Scholar]
- 34. Wang X., Chen H., Ran A.-R., Luo L., Chan P. P., Tham C. C., Chang R. T., Mannil S. S., Cheung C. Y., and Heng P.-A., “ Towards multi-center glaucoma OCT image screening with semi-supervised joint structure and function multi-task learning,” Med. Image Anal. 63, 101695 (2020). 10.1016/j.media.2020.101695 [DOI] [PubMed] [Google Scholar]
- 35. An G., Omodaka K., Hashimoto K., Tsuda S., Shiga Y., Takada N., Kikawa T., Yokota H., Akiba M., and Nakazawa T., “ Glaucoma diagnosis with machine learning based on optical coherence tomography and color fundus images,” J. Healthcare Eng. 2019, 4061313. 10.1155/2019/4061313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mehta P., Petersen C. A., Wen J. C., Banitt M. R., Chen P. P., Bojikian K. D., Egan C., Lee S.-I., Balazinska M., and Lee A. Y., “ Automated detection of glaucoma with interpretable machine learning using clinical data and multimodal retinal images,” Am. J. Ophthalmol. 231, 154–169 (2021). 10.1016/j.ajo.2021.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bruun M., Koikkalainen J., Rhodius-Meester H. F., Baroni M., Gjerum L., van Gils M., Soininen H., Remes A. M., Hartikainen P., and Waldemar G., “ Detecting frontotemporal dementia syndromes using MRI biomarkers,” NeuroImage: Clinical 22, 101711 (2019). 10.1016/j.nicl.2019.101711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dolph C. V., Alam M., Shboul Z., Samad M. D., and Iftekharuddin K. M., “ Deep learning of texture and structural features for multiclass Alzheimer's disease classification,” in 2017 International Joint Conference on Neural Networks (IJCNN) ( IEEE, 2017), pp. 2259–2266. [Google Scholar]
- 39. Kasivisvanathan V., Rannikko A. S., Borghi M., Panebianco V., Mynderse L. A., Vaarala M. H., Briganti A., Budäus L., Hellawell G., and Hindley R. G., “ MRI-targeted or standard biopsy for prostate-cancer diagnosis,” New Engl. J. Med. 378(19), 1767–1777 (2018). 10.1056/NEJMoa1801993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Amrane M., Oukid S., Gagaoua I., and Ensari T., “ Breast cancer classification using machine learning,” in 2018 Electric Electronics, Computer Science, Biomedical Engineerings' Meeting (EBBT) ( IEEE, 2018), pp. 1–4. [Google Scholar]
- 41. Goldenberg S. L., Nir G., and Salcudean S. E., “ A new era: Artificial intelligence and machine learning in prostate cancer,” Nat. Rev. Urol. 16(7), 391–403 (2019). 10.1038/s41585-019-0193-3 [DOI] [PubMed] [Google Scholar]
- 42. Houssein E. H., Emam M. M., Ali A. A., and Suganthan P. N., “ Deep and machine learning techniques for medical imaging-based breast cancer: A comprehensive review,” Expert Syst. Appl. 167, 114161 (2021). 10.1016/j.eswa.2020.114161 [DOI] [Google Scholar]
- 43. Caicedo J. C., Roth J., Goodman A., Becker T., Karhohs K. W., Broisin M., Molnar C., McQuin C., Singh S., and Theis F. J., “ Evaluation of deep learning strategies for nucleus segmentation in fluorescence images,” Cytom. Part A 95(9), 952–965 (2019). 10.1002/cyto.a.23863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Englbrecht F., Ruider I. E., and Bausch A. R., “ Automatic image annotation for fluorescent cell nuclei segmentation,” PLoS One 16(4), e0250093 (2021). 10.1371/journal.pone.0250093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gracioso Martins A. M., Wilkins M. D., Ligler F. S., Daniele M. A., and Freytes D. O., “ Microphysiological system for high-throughput computer vision measurement of microtissue contraction,” ACS Sens. 6(3), 985–994 (2021). 10.1021/acssensors.0c02172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ng W. L., Chan A., Ong Y. S., and Chua C. K., “ Deep learning for fabrication and maturation of 3D bioprinted tissues and organs,” Virtual Phys. Prototyping 15(3), 340–358 (2020). 10.1080/17452759.2020.1771741 [DOI] [Google Scholar]
- 47. Radcliffe A. J. and Reklaitis G. V., “ An application of computer vision for optimal sensor placement in drop printing,” in Computer Aided Chemical Engineering ( Elsevier, 2020), Vol. 48, pp. 457–462. [Google Scholar]
- 48. Chen C. T. and Gu G. X., “ Effect of constituent materials on composite performance: Exploring design strategies via machine learning,” Adv. Theory Simul. 2(6), 1900056 (2019). 10.1002/adts.201900056 [DOI] [Google Scholar]
- 49. Hattrick-Simpers J. R., Gregoire J. M., and Kusne A. G., “ Perspective: Composition–structure–property mapping in high-throughput experiments: Turning data into knowledge,” APL Mater. 4(5), 053211 (2016). 10.1063/1.4950995 [DOI] [Google Scholar]
- 50. Butler K. T., Davies D. W., Cartwright H., Isayev O., and Walsh A., “ Machine learning for molecular and materials science,” Nature 559(7715), 547–555 (2018). 10.1038/s41586-018-0337-2 [DOI] [PubMed] [Google Scholar]
- 51. McMillan P. F., Machine Learning Reveals the Complexity of Dense Amorphous Silicon ( Nature Publishing Group, 2021). [DOI] [PubMed] [Google Scholar]
- 52. Zhavoronkov A., Ivanenkov Y. A., Aliper A., Veselov M. S., Aladinskiy V. A., Aladinskaya A. V., Terentiev V. A., Polykovskiy D. A., Kuznetsov M. D., and Asadulaev A., “ Deep learning enables rapid identification of potent DDR1 kinase inhibitors,” Nat. Biotechnol. 37(9), 1038–1040 (2019). 10.1038/s41587-019-0224-x [DOI] [PubMed] [Google Scholar]
- 53. Yamanluirt G., Berns E. J., Xue A., Lee A., Bagheri N., Mrksich M., and Mirkin C. A., “ Exploration of the nanomedicine-design space with high-throughput screening and machine learning,” in Spherical Nucleic Acids ( Jenny Stanford Publishing, 2020), pp. 1687–1716. [Google Scholar]
- 54. Rodriguez S., Hug C., Todorov P., Moret N., Boswell S. A., Evans K., Zhou G., Johnson N. T., Hyman B. T., and Sorger P. K., “ Machine learning identifies candidates for drug repurposing in Alzheimer's disease,” Nat. Commun. 12(1), 1033 (2021). 10.1038/s41467-021-21330-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Beck B. R., Shin B., Choi Y., Park S., and Kang K., “ Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model,” Comput. Struct. Biotechnol. J. 18, 784–790 (2020). 10.1016/j.csbj.2020.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bojar D., Powers R. K., Camacho D. M., and Collins J. J., “ Deep-learning resources for studying glycan-mediated host-microbe interactions,” Cell Host Microbe 29(1), 132–144.e133 (2021). 10.1016/j.chom.2020.10.004 [DOI] [PubMed] [Google Scholar]
- 57. Burkholz R., Quackenbush J., and Bojar D., “ Using graph convolutional neural networks to learn a representation for glycans,” Cell Rep. 35(11), 109251 (2021). 10.1016/j.celrep.2021.109251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Misiunas K., Ermann N., and Keyser U. F., “ QuipuNet: Convolutional neural network for single-molecule nanopore sensing,” Nano Lett. 18(6), 4040–4045 (2018). 10.1021/acs.nanolett.8b01709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee J., Oh S. J., An S. H., Kim W.-D., and Kim S.-H., “ Machine learning-based design strategy for 3D printable bioink: Elastic modulus and yield stress determine printability,” Biofabrication 12(3), 035018 (2020). 10.1088/1758-5090/ab8707 [DOI] [PubMed] [Google Scholar]
- 60. Cobb J. S., Engel A., Seale M. A., and Janorkar A. V., “ Machine learning to determine optimal conditions for controlling the size of elastin-based particles,” Sci. Rep. 11(1), 6343 (2021). 10.1038/s41598-021-85601-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rickert C. A., Hayta E. N., Selle D. M., Kouroudis I., Harth M., Gagliardi A., and Lieleg O., “ Machine learning approach to analyze the surface properties of biological materials,” ACS Biomater. Sci. Eng. 7(9), 4614–4625 (2021). 10.1021/acsbiomaterials.1c00869 [DOI] [PubMed] [Google Scholar]
- 62. Wei Q., Melko R. G., and Chen J. Z., “ Identifying polymer states by machine learning,” Phys. Rev. E 95(3), 032504 (2017). 10.1103/PhysRevE.95.032504 [DOI] [PubMed] [Google Scholar]
- 63. Meng Z. and Xia K., “ Persistent spectral–based machine learning (PerSpect ML) for protein-ligand binding affinity prediction,” Sci. Adv. 7(19), eabc5329 (2021). 10.1126/sciadv.abc5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tsubaki M., Tomii K., and Sese J., “ Compound–protein interaction prediction with end-to-end learning of neural networks for graphs and sequences,” Bioinformatics 35(2), 309–318 (2019). 10.1093/bioinformatics/bty535 [DOI] [PubMed] [Google Scholar]
- 65. Pronobis W., Tkatchenko A., and Müller K.-R., “ Many-body descriptors for predicting molecular properties with machine learning: Analysis of pairwise and three-body interactions in molecules,” J. Chem. Theory Comput. 14(6), 2991–3003 (2018). 10.1021/acs.jctc.8b00110 [DOI] [PubMed] [Google Scholar]
- 66. Huang C. Y., Cassidy C. J., Medrano C., and Kadonaga J. T., “ Identification of the human DPR core promoter element using machine learning,” Nature 585(7825), 459–463 (2020). 10.1038/s41586-020-2689-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang T., Shao M., Guo R., Tao F., Zhang G., Snoussi H., and Tang X., “ Surrogate model via artificial intelligence method for accelerating screening materials and performance prediction,” Adv. Funct. Mater. 31(8), 2006245 (2021). 10.1002/adfm.202006245 [DOI] [Google Scholar]
- 68. Daghigh V., T. E. Lacy, Jr. , Daghigh H., Gu G., Baghaei K. T., Horstemeyer M. F., and C. U. Pittman, Jr. , “ Machine learning predictions on fracture toughness of multiscale bio-nano-composites,” J. Reinf. Plast. Compos. 39(15–16), 587–598 (2020). 10.1177/0731684420915984 [DOI] [Google Scholar]
- 69. Maillo J., Ramírez S., Triguero I., and Herrera F., “ kNN-IS: An iterative Spark-based design of the k-nearest neighbors classifier for big data,” Knowl.-Based Syst. 117, 3–15 (2017). 10.1016/j.knosys.2016.06.012 [DOI] [Google Scholar]
- 70. Mullick S. S., Datta S., and Das S., “ Adaptive learning-based k-nearest neighbor classifiers with resilience to class imbalance,” IEEE Trans. Neural Networks Learn. Syst. 29(11), 5713–5725 (2018). 10.1109/TNNLS.2018.2812279 [DOI] [PubMed] [Google Scholar]
- 71. Bhat A. D., Acharya H. R., and Srikanth H., “ A novel solution to the curse of dimensionality in using KNNs for image classification,” in 2019 2nd International Conference on Intelligent Autonomous Systems (ICoIAS) ( IEEE, 2019), pp. 32–36. [Google Scholar]
- 72. Pandey A. and Jain A., “ Comparative analysis of KNN algorithm using various normalization techniques,” Int. J. Comput. Network Inf. Secur. 9(11), 36 (2017). 10.5815/ijcnis.2017.11.04 [DOI] [Google Scholar]
- 73. Singh A. and Lakshmiganthan R., “ Impact of different data types on classifier performance of random forest, Naive Bayes, and k-nearest neighbors algorithms,” (IJACSA) International Journal of Advanced Computer Science and Applications 8(12), 1–10 (2017). 10.14569/IJACSA.2017.081201 [DOI] [Google Scholar]
- 74. Abdelwahab O., Bahgat M., Lowrance C. J., and Elmaghraby A., “ Effect of training set size on SVM and Naive Bayes for Twitter sentiment analysis,” in 2015 IEEE International Symposium on Signal Processing and Information Technology (ISSPIT) ( IEEE, 2015), pp. 46–51. [Google Scholar]
- 75. Reddy G. T., Reddy M. P. K., Lakshmanna K., Kaluri R., Rajput D. S., Srivastava G., and Baker T., “ Analysis of dimensionality reduction techniques on big data,” IEEE Access 8, 54776–54788 (2020). 10.1109/ACCESS.2020.2980942 [DOI] [Google Scholar]
- 76. Arar Ö. F. and Ayan K., “ A feature dependent Naive Bayes approach and its application to the software defect prediction problem,” Appl. Soft Comput. 59, 197–209 (2017). 10.1016/j.asoc.2017.05.043 [DOI] [Google Scholar]
- 77. Yang J., Ye Z., Zhang X., Liu W., and Jin H., “ Attribute weighted Naive Bayes for remote sensing image classification based on cuckoo search algorithm,” in 2017 International Conference on Security, Pattern Analysis, and Cybernetics (SPAC) ( IEEE, 2017), pp. 169–174. [Google Scholar]
- 78. Kikuchi M., Kawakami K., Watanabe K., Yoshida M., and Umemura K., “ Unified likelihood ratio estimation for high-to zero-frequency N-grams,” IEICE Trans. Fundam. Electron. Commun. Comput. Sci. E104.A(8), 1059–1074 (2021). 10.1587/transfun.2020EAP1088 [DOI] [Google Scholar]
- 79. Bai Y., Sun Z., Zeng B., Long J., Li L., de Oliveira J. V., and Li C., “ A comparison of dimension reduction techniques for support vector machine modeling of multi-parameter manufacturing quality prediction,” J. Intell. Manuf. 30(5), 2245–2256 (2019). 10.1007/s10845-017-1388-1 [DOI] [Google Scholar]
- 80. Adiwijaya W. U., Lisnawati E., Aditsania A., and Kusumo D. S., “ Dimensionality reduction using principal component analysis for cancer detection based on microarray data classification,” J. Comput. Sci. 14(11), 1521–1530 (2018). 10.3844/jcssp.2018.1521.1530 [DOI] [Google Scholar]
- 81. Hossain S., Mou R. M., Hasan M. M., Chakraborty S., and Razzak M. A., “ Recognition and detection of tea leaf's diseases using support vector machine,” in 2018 IEEE 14th International Colloquium on Signal Processing & Its Applications (CSPA) ( IEEE, 2018), pp. 150–154. [Google Scholar]
- 82. Zareapoor M., Shamsolmoali P., Jain D. K., Wang H., and Yang J., “ Kernelized support vector machine with deep learning: An efficient approach for extreme multiclass dataset,” Pattern Recognit. Lett. 115, 4–13 (2018). 10.1016/j.patrec.2017.09.018 [DOI] [Google Scholar]
- 83. Feizizadeh B., Roodposhti M. S., Blaschke T., and Aryal J., “ Comparing GIS-based support vector machine kernel functions for landslide susceptibility mapping,” Arabian J. Geosci. 10(5), 122 (2017). 10.1007/s12517-017-2918-z [DOI] [Google Scholar]
- 84. Achirul Nanda M., Boro Seminar K., Nandika D., and Maddu A., “ A comparison study of kernel functions in the support vector machine and its application for termite detection,” Information 9(1), 5 (2018). 10.3390/info9010005 [DOI] [Google Scholar]
- 85. Lee H. K. and Kim S. B., “ An overlap-sensitive margin classifier for imbalanced and overlapping data,” Expert Syst. Appl. 98, 72–83 (2018). 10.1016/j.eswa.2018.01.008 [DOI] [Google Scholar]
- 86. Wang H., Shao Y., Zhou S., Zhang C., and Xiu N., “ Support vector machine classifier via L0/1 soft-margin loss,” IEEE Trans. Pattern Anal. Mach. Intell. (2021). 10.1109/TPAMI.2021.3092177 [DOI] [PubMed] [Google Scholar]
- 87. Dewi K. C., Murfi H., and Abdullah S., “ Analysis accuracy of random forest model for Big Data—A case study of claim severity prediction in car insurance,” in 2019 5th International Conference on Science in Information Technology (ICSITech) ( IEEE, 2019), pp. 60–65. [Google Scholar]
- 88. Thayumanavan M. and Ramasamy A., “ An efficient approach for brain tumor detection and segmentation in MR brain images using random forest classifier,” Concurrent Eng. 29(3), 266–274 (2021). 10.1177/1063293X211010542 [DOI] [Google Scholar]
- 89. Zhou X., Lu P., Zheng Z., Tolliver D., and Keramati A., “ Accident prediction accuracy assessment for highway-rail grade crossings using random forest algorithm compared with decision tree,” Reliab. Eng. Syst. Saf. 200, 106931 (2020). 10.1016/j.ress.2020.106931 [DOI] [Google Scholar]
- 90. Pu Z., Li Z., Ke R., Hua X., and Wang Y., “ Evaluating the nonlinear correlation between vertical curve features and crash frequency on highways using random forests,” J. Transp. Eng., Part A: Syst. 146(10), 04020115 (2020). 10.1061/JTEPBS.0000410 [DOI] [Google Scholar]
- 91. de Santana F. B., Neto W. B., and Poppi R. J., “ Random forest as one-class classifier and infrared spectroscopy for food adulteration detection,” Food Chem. 293, 323–332 (2019). 10.1016/j.foodchem.2019.04.073 [DOI] [PubMed] [Google Scholar]
- 92. Zhu T., “ Analysis on the applicability of the random forest,” in Journal of Physics: Conference Series ( IOP Publishing, 2020), Vol. 1607, p. 012123. [Google Scholar]
- 93. Aung Y. Y. and Min M. M., “ An analysis of random forest algorithm based network intrusion detection system,” in 2017 18th IEEE/ACIS International Conference on Software Engineering, Artificial Intelligence, Networking and Parallel/Distributed Computing (SNPD) ( IEEE, 2017), pp. 127–132. [Google Scholar]
- 94. Capó M., Pérez A., and Lozano J. A., “ An efficient approximation to the K-means clustering for massive data,” Knowl.-Based Syst. 117, 56–69 (2017). 10.1016/j.knosys.2016.06.031 [DOI] [Google Scholar]
- 95. Motwani M., Arora N., and Gupta A., “ A study on initial centroids selection for partitional clustering algorithms,” in Software Engineering ( Springer, 2019), pp. 211–220. [Google Scholar]
- 96. Nidheesh N., Nazeer K. A., and Ameer P., “ An enhanced deterministic K-means clustering algorithm for cancer subtype prediction from gene expression data,” Comput. Biol. Med. 91, 213–221 (2017). 10.1016/j.compbiomed.2017.10.014 [DOI] [PubMed] [Google Scholar]
- 97. Syakur M., Khotimah B., Rochman E., and Satoto B. D., “ Integration k-means clustering method and elbow method for identification of the best customer profile cluster,” in IOP Conference Series: Materials Science and Engineering ( IOP Publishing, 2018), Vol. 336, p. 012017. [Google Scholar]
- 98. Yuan C. and Yang H., “ Research on K-value selection method of K-means clustering algorithm,” J 2(2), 226–235 (2019). 10.3390/j2020016 [DOI] [Google Scholar]
- 99. Fränti P. and Sieranoja S., “ K-means properties on six clustering benchmark datasets,” Appl. Intell. 48(12), 4743–4759 (2018). 10.1007/s10489-018-1238-7 [DOI] [Google Scholar]
- 100. Rathee S. and Kashyap A., “ Adaptive-miner: An efficient distributed association rule mining algorithm on Spark,” J. Big Data 5(1), 1–17 (2018). 10.1186/s40537-018-0112-0 [DOI] [Google Scholar]
- 101. Abdel-Basset M., Mohamed M., Smarandache F., and Chang V., “ Neutrosophic association rule mining algorithm for big data analysis,” Symmetry 10(4), 106 (2018). 10.3390/sym10040106 [DOI] [Google Scholar]
- 102. Chiclana F., Kumar R., Mittal M., Khari M., Chatterjee J. M., and Baik S. W., “ ARM–AMO: An efficient association rule mining algorithm based on animal migration optimization,” Knowl.-Based Syst. 154, 68–80 (2018). 10.1016/j.knosys.2018.04.038 [DOI] [Google Scholar]
- 103. Kaushik M., Sharma R., Peious S. A., Shahin M., Yahia S. B., and Draheim D., “ A systematic assessment of numerical association rule mining methods,” SN Comput. Sci. 2(5), 1–13 (2021). 10.1007/s42979-021-00725-2 [DOI] [Google Scholar]
- 104. Yazgana P. and Kusakci A. O., “ A literature survey on association rule mining algorithms,” Southeast Eur. J. Soft Comput. 5(1), 5–14 (2016). 10.21533/scjournal.v5i1.102 [DOI] [Google Scholar]
- 105. Majeed S. J. and Hutter M., “ On Q-learning convergence for non-Markov decision processes,” in International Joint Conference on Artificial Intelligence ( AAAI Press, 2018), pp. 2546–2552. [Google Scholar]
- 106. Padakandla S., Prabuchandran K., and Bhatnagar S., “ Reinforcement learning algorithm for non-stationary environments,” Appl. Intell. 50(11), 3590–3606 (2020). 10.1007/s10489-020-01758-5 [DOI] [Google Scholar]
- 107. Malik H. and Almutairi A., “ Modified fuzzy-Q-learning (MFQL)-based mechanical fault diagnosis for direct-drive wind turbines using electrical signals,” IEEE Access 9, 52569–52579 (2021). 10.1109/ACCESS.2021.3070483 [DOI] [Google Scholar]
- 108. Low E. S., Ong P., and Cheah K. C., “ Solving the optimal path planning of a mobile robot using improved Q-learning,” Rob. Auton. Syst. 115, 143–161 (2019). 10.1016/j.robot.2019.02.013 [DOI] [Google Scholar]
- 109. Yang L. and Wang M., “ Sample-optimal parametric q-learning using linearly additive features,” in International Conference on Machine Learning (PMLR, 2019), pp. 6995–7004. [Google Scholar]
- 110. Cichy R. M. and Kaiser D., “ Deep neural networks as scientific models,” Trends Cognit. Sci. 23(4), 305–317 (2019). 10.1016/j.tics.2019.01.009 [DOI] [PubMed] [Google Scholar]
- 111. Miikkulainen R., Liang J., Meyerson E., Rawal A., Fink D., Francon O., Raju B., Shahrzad H., Navruzyan A., and Duffy N., “ Evolving deep neural networks,” in Artificial Intelligence in the Age of Neural Networks and Brain Computing ( Elsevier, 2019), pp. 293–312. [Google Scholar]
- 112. Khaki S. and Wang L., “ Crop yield prediction using deep neural networks,” Front. Plant Sci. 10, 621 (2019). 10.3389/fpls.2019.00621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Samek W., Montavon G., Lapuschkin S., Anders C. J., and Müller K.-R., “ Explaining deep neural networks and beyond: A review of methods and applications,” Proc. IEEE 109(3), 247–278 (2021). 10.1109/JPROC.2021.3060483 [DOI] [Google Scholar]
- 114. Lu M. Y., Chen T. Y., Williamson D. F., Zhao M., Shady M., Lipkova J., and Mahmood F., “ AI-based pathology predicts origins for cancers of unknown primary,” Nature 594(7861), 106–110 (2021). 10.1038/s41586-021-03512-4 [DOI] [PubMed] [Google Scholar]
- 115. Gu G. X., Chen C.-T., and Buehler M. J., “ De novo composite design based on machine learning algorithm,” Extreme Mech. Lett. 18, 19–28 (2018). 10.1016/j.eml.2017.10.001 [DOI] [Google Scholar]
- 116. Ghouli S., Ayatollahi M. R., Bahrami B., and Jamali J., “ In-situ optical approach to predict mixed mode fracture in a polymeric biomaterial,” Theor. Appl. Fract. Mech. 115, 103211 (2021). 10.1016/j.tafmec.2021.103211 [DOI] [Google Scholar]
- 117. Thanh Noi P. and Kappas M., “ Comparison of random forest, k-nearest neighbor, and support vector machine classifiers for land cover classification using Sentinel-2 imagery,” Sensors 18(1), 18 (2018). 10.3390/s18010018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Gao W., Yang B.-B., and Zhou Z.-H., “ On the resistance of nearest neighbor to random noisy labels,” e-print arXiv:1607.07526 (2016).
- 119. Mandal L. and Jana N. D., “ A comparative study of Naive Bayes and k-NN algorithm for multi-class drug molecule classification,” in 2019 IEEE 16th India Council International Conference (INDICON) ( IEEE, 2019), pp. 1–4. [Google Scholar]
- 120. Singh G., Kumar B., Gaur L., and Tyagi A., “ Comparison between multinomial and Bernoulli Naïve Bayes for text classification,” in 2019 International Conference on Automation, Computational and Technology Management (ICACTM) ( IEEE, 2019), pp. 593–596. [Google Scholar]
- 121. Panigrahi R. and Kumar L., “ Application of Naïve Bayes classifiers for refactoring prediction at the method level,” in 2020 International Conference on Computer Science, Engineering and Applications (ICCSEA) ( IEEE, 2020), pp. 1–6. [Google Scholar]
- 122. Berrar D., “ Bayes' theorem and Naive Bayes classifier,” in Encyclopedia of Bioinformatics and Computational Biology: ABC of Bioinformatics ( Elsevier Science Publisher, Amsterdam, 2018), pp. 403–412. [Google Scholar]
- 123. VanderPlas J., Python Data Science Handbook: Essential Tools for Working with Data ( O'Reilly Media, Inc., 2016). [Google Scholar]
- 124. Li Y., Pu Q., Li S., Zhang H., Wang X., Yao H., and Zhao L., “ Machine learning methods for research highlight prediction in biomedical effects of nanomaterial application,” Pattern Recognit. Lett. 117, 111–118 (2019). 10.1016/j.patrec.2018.11.008 [DOI] [Google Scholar]
- 125. Zheng Z., Cai Y., Yang Y., and Li Y., “ Sparse weighted Naive Bayes classifier for efficient classification of categorical data,” in 2018 IEEE Third International Conference on Data Science in Cyberspace (DSC) ( IEEE, 2018), pp. 691–696. [Google Scholar]
- 126. Abass Z. K., Hasan T. M., and Abdullah A. K., “ Brain computer interface enhancement based on stones blind source separation and Naive Bayes classifier,” in International Conference on New Trends in Information and Communications Technology Applications ( Springer, 2020), pp. 17–28. [Google Scholar]
- 127. Padierna L. C., Carpio M., Rojas-Dominguez A., Puga H., and Fraire H., “ A novel formulation of orthogonal polynomial kernel functions for SVM classifiers: The Gegenbauer family,” Pattern Recognit. 84, 211–225 (2018). 10.1016/j.patcog.2018.07.010 [DOI] [Google Scholar]
- 128. Hong H., Pradhan B., Bui D. T., Xu C., Youssef A. M., and Chen W., “ Comparison of four kernel functions used in support vector machines for landslide susceptibility mapping: A case study at Suichuan area (China),” Geomatics Nat. Hazards Risk 8(2), 544–569 (2017). 10.1080/19475705.2016.1250112 [DOI] [Google Scholar]
- 129. Shen X., Niu L., Qi Z., and Tian Y., “ Support vector machine classifier with truncated pinball loss,” Pattern Recognit. 68, 199–210 (2017). 10.1016/j.patcog.2017.03.011 [DOI] [Google Scholar]
- 130. Breiman L., “ Random forests,” Mach. Learn. 45(1), 5–32 (2001). 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 131. Lee T.-H., Ullah A., and Wang R., “ Bootstrap aggregating and random forest,” in Macroeconomic Forecasting in the Era of Big Data ( Springer, 2020), pp. 389–429. [Google Scholar]
- 132. Kirasich K., Smith T., and Sadler B., “ Random forest vs logistic regression: Binary classification for heterogeneous datasets,” SMU Data Sci. Rev. 1(3), 9 (2018). [Google Scholar]
- 133. Fabris F., Doherty A., Palmer D., De Magalhães J. P., and Freitas A. A., “ A new approach for interpreting random forest models and its application to the biology of ageing,” Bioinformatics 34(14), 2449–2456 (2018). 10.1093/bioinformatics/bty087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Gonçalves A. V., Schneider I. J. C., Amaral F. V., Garcia L. P., and de Araújo G. M., “ Feature importance investigation for estimating COVID-19 infection by random forest algorithm,” in International Conference on Data and Information in Online ( Springer, 2021), pp. 272–285. [Google Scholar]
- 135. Phillip J. M., Han K.-S., Chen W.-C., Wirtz D., and Wu P.-H., “ A robust unsupervised machine-learning method to quantify the morphological heterogeneity of cells and nuclei,” Nat. Protoc. 16(2), 754–774 (2021). 10.1038/s41596-020-00432-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Ieracitano C., Paviglianiti A., Campolo M., Hussain A., Pasero E., and Morabito F. C., “ A novel automatic classification system based on hybrid unsupervised and supervised machine learning for electrospun nanofibers,” IEEE/CAA J. Autom. Sin. 8(1), 64–76 (2020). 10.1109/JAS.2020.1003387 [DOI] [Google Scholar]
- 137. Bai L., Liang J., and Guo Y., “ An ensemble clusterer of multiple fuzzy k-means clusterings to recognize arbitrarily shaped clusters,” IEEE Trans. Fuzzy Syst. 26(6), 3524–3533 (2018). 10.1109/TFUZZ.2018.2835774 [DOI] [Google Scholar]
- 138. Gupta N. T., Adams K. D., Briggs A. W., Timberlake S. C., Vigneault F., and Kleinstein S. H., “ Hierarchical clustering can identify B cell clones with high confidence in Ig repertoire sequencing data,” J. Immunol. 198(6), 2489–2499 (2017). 10.4049/jimmunol.1601850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Treloar N. J., Fedorec A. J., Ingalls B., and Barnes C. P., “ Deep reinforcement learning for the control of microbial co-cultures in bioreactors,” PLoS Comput. Biol. 16(4), e1007783 (2020). 10.1371/journal.pcbi.1007783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Eastman P., Shi J., Ramsundar B., and Pande V. S., “ Solving the RNA design problem with reinforcement learning,” PLoS Comput. Biol. 14(6), e1006176 (2018). 10.1371/journal.pcbi.1006176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Salma H., Melha Y. M., Sonia L., Hamza H., and Salim N., “ Efficient prediction of in vitro piroxicam release and diffusion from topical films based on biopolymers using deep learning models and generative adversarial networks,” J. Pharm. Sciences 110(6), 2531–2543 (2021). 10.1016/j.xphs.2021.01.032 [DOI] [PubMed] [Google Scholar]
- 142. Liu Y., Zhang D., Tang Y., Zhang Y., Gong X., Xie S., and Zheng J., “ Machine learning-enabled repurposing and design of antifouling polymer brushes,” Chem. Eng. J. 420, 129872 (2021). 10.1016/j.cej.2021.129872 [DOI] [Google Scholar]
- 143. Le T. C., Penna M., Winkler D. A., and Yarovsky I., “ Quantitative design rules for protein-resistant surface coatings using machine learning,” Sci. Rep. 9(1), 265 (2019). 10.1038/s41598-018-36597-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Echezarreta-López M. and Landin M., “ Using machine learning for improving knowledge on antibacterial effect of bioactive glass,” Int. J. Pharm. 453(2), 641–647 (2013). 10.1016/j.ijpharm.2013.06.036 [DOI] [PubMed] [Google Scholar]
- 145. Mikulskis P., Hook A., Dundas A. A., Irvine D., Sanni O., Anderson D., Langer R., Alexander M. R., Williams P., and Winkler D. A., “ Prediction of broad-spectrum pathogen attachment to coating materials for biomedical devices,” ACS Appl. Mater. Interfaces 10(1), 139–149 (2018). 10.1021/acsami.7b14197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Damiati S. A., Rossi D., Joensson H. N., and Damiati S., “ Artificial intelligence application for rapid fabrication of size-tunable PLGA microparticles in microfluidics,” Sci. Rep. 10(1), 19517 (2020). 10.1038/s41598-020-76477-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Schissel C. K., Mohapatra S., Wolfe J. M., Fadzen C. M., Bellovoda K., Wu C.-L., Wood J. A., Malmberg A. B., Loas A., and Gómez-Bombarelli R., “ Deep learning to design nuclear-targeting abiotic miniproteins,” Nat. Chem. 13(10), 992–1000 (2021). 10.1038/s41557-021-00766-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Celik N., O'Brien F., Brennan S., Rainbow R. D., Dart C., Zheng Y., Coenen F., and Barrett-Jolley R., “ Deep-channel uses deep neural networks to detect single-molecule events from patch-clamp data,” Commun. Biol. 3(1), 3 (2020). 10.1038/s42003-019-0729-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Wong Y. J., Arumugasamy S. K., and Jewaratnam J., “ Performance comparison of feedforward neural network training algorithms in modeling for synthesis of polycaprolactone via biopolymerization,” Clean Technol. Environ. Policy 20(9), 1971–1986 (2018). 10.1007/s10098-018-1577-4 [DOI] [Google Scholar]
- 150. Arumugasamy S. K., Chen Z., Van Khoa L. D., and Pakalapati H., “ Comparison between artificial neural networks and support vector machine modeling for polycaprolactone synthesis via enzyme catalyzed polymerization,” Process Integr. Optim. Sustainability 5(3), 599–607 (2021). 10.1007/s41660-021-00163-w [DOI] [Google Scholar]
- 151. Lugagne J.-B., Lin H., and Dunlop M. J., “ DeLTA: Automated cell segmentation, tracking, and lineage reconstruction using deep learning,” PLoS Comput. Biol. 16(4), e1007673 (2020). 10.1371/journal.pcbi.1007673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Cao Y., Xiao C., Cyr B., Zhou Y., Park W., Rampazzi S., Chen Q. A., Fu K., and Mao Z. M., “ Adversarial sensor attack on lidar-based perception in autonomous driving,” in Proceedings of the 2019 ACM SIGSAC Conference on Computer and Communications Security ( ACM, 2019), pp. 2267–2281. [Google Scholar]
- 153. Janai J., Güney F., Behl A., and Geiger A., “ Computer vision for autonomous vehicles: Problems, datasets and state of the art,” Found. Trends® Comput. Graph. Vision 12(1–3), 1–308 (2020). 10.1561/0600000079 [DOI] [Google Scholar]
- 154. Schmidt J., Marques M. R., Botti S., and Marques M. A., “ Recent advances and applications of machine learning in solid-state materials science,” npj Comput. Mater. 5(1), 83 (2019). 10.1038/s41524-019-0221-0 [DOI] [Google Scholar]
- 155. Podryabinkin E. V., Tikhonov E. V., Shapeev A. V., and Oganov A. R., “ Accelerating crystal structure prediction by machine-learning interatomic potentials with active learning,” Phys. Rev. B 99(6), 064114 (2019). 10.1103/PhysRevB.99.064114 [DOI] [Google Scholar]
- 156. Kashif M., Hussain A., Munir A., Siddiqui A. B., Abbasi A., Aakif M., Malik A. J., Alazemi F. E., and Song O.-Y., “ A machine learning approach for expression detection in healthcare monitoring systems,” Comput. Mater. Continua 67(2), 2123–2139 (2021). 10.32604/cmc.2021.014782 [DOI] [Google Scholar]
- 157. Xu X., Wang J., Peng H., and Wu R., “ Prediction of academic performance associated with internet usage behaviors using machine learning algorithms,” Comput. Hum. Behav. 98, 166–173 (2019). 10.1016/j.chb.2019.04.015 [DOI] [Google Scholar]
- 158. Lv Z., Qiao L., and Singh A. K., “ Advanced machine learning on cognitive computing for human behavior analysis,” in IEEE Transactions on Computational Social Systems ( IEEE, 2020), pp. 1194–1202. [Google Scholar]
- 159. Peterson J. C., Bourgin D. D., Agrawal M., Reichman D., and Griffiths T. L., “ Using large-scale experiments and machine learning to discover theories of human decision-making,” Science 372(6547), 1209–1214 (2021). 10.1126/science.abe2629 [DOI] [PubMed] [Google Scholar]
- 160. Bone J. M., Childs C. M., Menon A., Poczos B., Feinberg A. W., LeDuc P. R., and Washburn N. R., “ Hierarchical machine learning for high-fidelity 3D printed biopolymers,” ACS Biomater. Sci. Eng. 6(12), 7021–7031 (2020). 10.1021/acsbiomaterials.0c00755 [DOI] [PubMed] [Google Scholar]
- 161. Zhu Z., Ng D. W. H., Park H. S., and McAlpine M. C., “ 3D-printed multifunctional materials enabled by artificial-intelligence-assisted fabrication technologies,” Nat. Rev. Mater. 6(1), 27–47 (2021). 10.1038/s41578-020-00235-2 [DOI] [Google Scholar]
- 162. Liu Y., Han F., Li F., Zhao Y., Chen M., Xu Z., Zheng X., Hu H., Yao J., and Guo T., “ Inkjet-printed unclonable quantum dot fluorescent anti-counterfeiting labels with artificial intelligence authentication,” Nat. Commun. 10(1), 2409 (2019). 10.1038/s41467-019-10406-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Toscano J. D., Li Z., Segura L. J., and Sun H., “ A machine learning approach to model the electrospinning process of biocompatible materials,” in International Manufacturing Science and Engineering Conference ( American Society of Mechanical Engineers, 2020), Vol. 84263, p V002T006A031. [Google Scholar]
- 164. Ramzi A. B., Baharum S. N., Bunawan H., and Scrutton N. S., “ Streamlining natural products biomanufacturing with omics and machine learning driven microbial engineering,” Front. Bioeng. Biotechnol. 8, 608918 (2020). 10.3389/fbioe.2020.608918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Oyetunde T., Bao F. S., Chen J.-W., Martin H. G., and Tang Y. J., “ Leveraging knowledge engineering and machine learning for microbial bio-manufacturing,” Biotechnol. Adv. 36(4), 1308–1315 (2018). 10.1016/j.biotechadv.2018.04.008 [DOI] [PubMed] [Google Scholar]
- 166. Copp S. M., Swasey S. M., Gorovits A., Bogdanov P., and Gwinn E. G., “ General approach for machine learning-aided design of DNA-stabilized silver clusters,” Chem. Mater. 32(1), 430–437 (2019). 10.1021/acs.chemmater.9b04040 [DOI] [Google Scholar]
- 167. Becht E., Tolstrup D., Dutertre C.-A., Morawski P. A., Campbell D. J., Ginhoux F., Newell E. W., Gottardo R., and Headley M. B., “ High-throughput single-cell quantification of hundreds of proteins using conventional flow cytometry and machine learning,” Sci. Adv. 7(39), eabg0505 (2021). 10.1126/sciadv.abg0505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Sardesai A. U., Tanak A. S., Krishnan S., Striegel D. A., Schully K. L., Clark D. V., Muthukumar S., and Prasad S., “ An approach to rapidly assess sepsis through multi-biomarker host response using machine learning algorithm,” Sci. Rep. 11(1), 16905 (2021). 10.1038/s41598-021-96081-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Rojas R. F., Huang X., and Ou K.-L., “ A machine learning approach for the identification of a biomarker of human pain using fNIRS,” Sci. Rep. 9(1), 5645 (2019). 10.1038/s41598-019-42098-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Odish O. F., Johnsen K., van Someren P., Roos R. A., and van Dijk J. G., “ EEG may serve as a biomarker in Huntington's disease using machine learning automatic classification,” Sci. Rep. 8(1), 16090 (2018). 10.1038/s41598-018-34269-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Baik S., Lee J., Jeon E. J., Park B-y., Kim D. W., Song J. H., Lee H. J., Han S. Y., Cho S.-W., and Pang C., “ Diving beetle–like miniaturized plungers with reversible, rapid biofluid capturing for machine learning–based care of skin disease,” Sci. Adv. 7(25), eabf5695 (2021). 10.1126/sciadv.abf5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Robison H. M., Chapman C. A., Zhou H., Erskine C. L., Theel E., Peikert T., Lindestam Arlehamn C. S., Sette A., Bushell C., and Welge M., “ Risk assessment of latent tuberculosis infection through a multiplexed cytokine biosensor assay and machine learning feature selection,” Sci. Rep. 11(1), 20544 (2021). 10.1038/s41598-021-99754-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Green E. M., van Mourik R., Wolfus C., Heitner S. B., Dur O., and Semigran M. J., “ Machine learning detection of obstructive hypertrophic cardiomyopathy using a wearable biosensor,” npj Digital Med. 2(1), 57 (2019). 10.1038/s41746-019-0130-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Mi X., Zou B., Zou F., and Hu J., “ Permutation-based identification of important biomarkers for complex diseases via machine learning models,” Nat. Commun. 12(1), 3008 (2021). 10.1038/s41467-021-22756-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Kumar R., Le N., Tan Z., Brown M. E., Jiang S., and Reineke T. M., “ Efficient polymer-mediated delivery of gene-editing ribonucleoprotein payloads through combinatorial design, parallelized experimentation, and machine learning,” ACS Nano 14(12), 17626–17639 (2020). 10.1021/acsnano.0c08549 [DOI] [PubMed] [Google Scholar]
- 176. Tréguier J., Bugnicourt L., Gay G., Diallo M., Islam S. T., Toro A., David L., Théodoly O., Sudre G., and Mignot T., “ Chitosan films for microfluidic studies of single bacteria and perspectives for antibiotic susceptibility testing,” mBio 10(4), e01375-19 (2019). 10.1128/mBio.01375-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Reker D., Rybakova Y., Kirtane A. R., Cao R., Yang J. W., Navamajiti N., Gardner A., Zhang R. M., Esfandiary T., and L'Heureux J., “ Computationally guided high-throughput design of self-assembling drug nanoparticles,” Nat. Nanotechnol. 16(6), 725–733 (2021). 10.1038/s41565-021-00870-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Golriz Khatami S., Mubeen S., Bharadhwaj V. S., Kodamullil A. T., Hofmann-Apitius M., and Domingo-Fernández D., “ Using predictive machine learning models for drug response simulation by calibrating patient-specific pathway signatures,” npj Syst. Biol. Appl. 7(1), 40 (2021). 10.1038/s41540-021-00199-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Piazza I., Beaton N., Bruderer R., Knobloch T., Barbisan C., Chandat L., Sudau A., Siepe I., Rinner O., and de Souza N., “ A machine learning-based chemoproteomic approach to identify drug targets and binding sites in complex proteomes,” Nat. Commun. 11(1), 4200 (2020). 10.1038/s41467-020-18071-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Madhukar N. S., Khade P. K., Huang L., Gayvert K., Galletti G., Stogniew M., Allen J. E., Giannakakou P., and Elemento O., “ A Bayesian machine learning approach for drug target identification using diverse data types,” Nat. Commun. 10(1), 5221 (2019). 10.1038/s41467-019-12928-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Kobayashi H., Lei C., Wu Y., Mao A., Jiang Y., Guo B., Ozeki Y., and Goda K., “ Label-free detection of cellular drug responses by high-throughput bright-field imaging and machine learning,” Sci. Rep. 7(1), 12454 (2017). 10.1038/s41598-017-12378-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Sarmadi M., Behrens A. M., McHugh K. J., Contreras H. T., Tochka Z. L., Lu X., Langer R., and Jaklenec A., “ Modeling, design, and machine learning-based framework for optimal injectability of microparticle-based drug formulations,” Sci. Adv. 6(28), eabb6594 (2020). 10.1126/sciadv.abb6594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Theodoris C. V., Zhou P., Liu L., Zhang Y., Nishino T., Huang Y., Kostina A., Ranade S. S., Gifford C. A., Uspenskiy V., and Malashicheva A., “ Network-based screen in iPSC-derived cells reveals therapeutic candidate for heart valve disease,” Science 371(6530), eabd0724 (2021). 10.1126/science.abd0724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Morris A., McCorkindale W., Drayman N., Chodera J. D., Tay S., London N., and Consortium C. M., “ Discovery of SARS-CoV-2 main protease inhibitors using a synthesis-directed de novo design model,” Chem. Commun. 57, 5909–5912 (2021). 10.1039/D1CC00050K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Chodera J., Lee A. A., London N., and von Delft F., “ Crowdsourcing drug discovery for pandemics,” Nat. Chem. 12(7), 581–581 (2020). 10.1038/s41557-020-0496-2 [DOI] [PubMed] [Google Scholar]
- 186. Wood D. E., White J. R., Georgiadis A., Van Emburgh B., Parpart-Li S., Mitchell J., Anagnostou V., Niknafs N., Karchin R., and Papp E., “ A machine learning approach for somatic mutation discovery,” Sci. Transl. Med. 10(457), eaar7939 (2018). 10.1126/scitranslmed.aar7939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Huda A., Castaño A., Niyogi A., Schumacher J., Stewart M., Bruno M., Hu M., Ahmad F. S., Deo R. C., and Shah S. J., “ A machine learning model for identifying patients at risk for wild-type transthyretin amyloid cardiomyopathy,” Nat. Commun. 12(1), 2725 (2021). 10.1038/s41467-021-22876-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Kim S. H., Jeon E.-T., Yu S., Kyungmi O., Kim C. K., Song T.-J., Kim Y.-J., Heo S. H., Park K.-Y., and Kim J.-M., “ Interpretable machine learning for early neurological deterioration prediction in atrial fibrillation-related stroke,” Sci. Rep. 11, 20610 (2021). 10.1038/s41598-021-99920-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Ricciardi C., Edmunds K. J., Recenti M., Sigurdsson S., Gudnason V., Carraro U., and Gargiulo P., “ Assessing cardiovascular risks from a mid-thigh CT image: A tree-based machine learning approach using radiodensitometric distributions,” Sci. Rep. 10(1), 2863 (2020). 10.1038/s41598-020-59873-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Jurmeister P., Bockmayr M., Seegerer P., Bockmayr T., Treue D., Montavon G., Vollbrecht C., Arnold A., Teichmann D., and Bressem K., “ Machine learning analysis of DNA methylation profiles distinguishes primary lung squamous cell carcinomas from head and neck metastases,” Sci. Transl. Med. 11(509), eaaw8513 (2019). 10.1126/scitranslmed.aaw8513 [DOI] [PubMed] [Google Scholar]
- 191. Chen S., Jiang L., Gao F., Zhang E., Wang T., Zhang N., Wang X., and Zheng J., “ Machine learning-based pathomics signature could act as a novel prognostic marker for patients with clear cell renal cell carcinoma,” Br. J. Cancer 126, 771–777 (2021). 10.1038/s41416-021-01640-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Qiu S., Joshi P. S., Miller M. I., Xue C., Zhou X., Karjadi C., Chang G. H., Joshi A. S., Dwyer B., and Zhu S., “ Development and validation of an interpretable deep learning framework for Alzheimer's disease classification,” Brain 143(6), 1920–1933 (2020). 10.1093/brain/awaa137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Yu J., Zhou Y., Yang Q., Liu X., Huang L., Yu P., and Chu S., “ Machine learning models for screening carotid atherosclerosis in asymptomatic adults,” Sci. Rep. 11(1), 22236 (2021). 10.1038/s41598-021-01456-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Chiu Y.-C., Zheng S., Wang L.-J., Iskra B. S., Rao M. K., Houghton P. J., Huang Y., and Chen Y., “ Predicting and characterizing a cancer dependency map of tumors with deep learning,” Sci. Adv. 7(34), eabh1275 (2021). 10.1126/sciadv.abh1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Gemein L. A., Schirrmeister R. T., Chrabąszcz P., Wilson D., Boedecker J., Schulze-Bonhage A., Hutter F., and Ball T., “ Machine-learning-based diagnostics of EEG pathology,” NeuroImage 220, 117021 (2020). 10.1016/j.neuroimage.2020.117021 [DOI] [PubMed] [Google Scholar]
- 196. Ieracitano C., Mammone N., Hussain A., and Morabito F. C., “ A novel multi-modal machine learning based approach for automatic classification of EEG recordings in dementia,” Neural Networks 123, 176–190 (2020). 10.1016/j.neunet.2019.12.006 [DOI] [PubMed] [Google Scholar]
- 197. An J.-Y., Lin K., Zhu L., Werling D. M., Dong S., Brand H., Wang H. Z., Zhao X., Schwartz G. B., and Collins R. L., “ Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder,” Science 362(6420), eaat6576 (2018). 10.1126/science.aat6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Brückner D. B., Arlt N., Fink A., Ronceray P., Rädler J. O., and Broedersz C. P., “ Learning the dynamics of cell–cell interactions in confined cell migration,” Proc. Natl. Acad. Sci. 118(7), e2016602118 (2021). 10.1073/pnas.2016602118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Méndez-Lucio O., Ahmad M., del Rio-Chanona E. A., and Wegner J. K., “ A geometric deep learning approach to predict binding conformations of bioactive molecules,” Nat. Mach. Intell. 3, 1033–1039 (2021). 10.1038/s42256-021-00409-9 [DOI] [Google Scholar]
- 200. Webb M. A., Jackson N. E., Gil P. S., and de Pablo J. J., “ Targeted sequence design within the coarse-grained polymer genome,” Sci. Adv. 6(43), eabc6216 (2020). 10.1126/sciadv.abc6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Uesawa Y., “ Quantitative structure–activity relationship analysis using deep learning based on a novel molecular image input technique,” Bioorg. Med. Chem. Lett. 28(20), 3400–3403 (2018). 10.1016/j.bmcl.2018.08.032 [DOI] [PubMed] [Google Scholar]
- 202. Shin J.-E., Riesselman A. J., Kollasch A. W., McMahon C., Simon E., Sander C., Manglik A., Kruse A. C., and Marks D. S., “ Protein design and variant prediction using autoregressive generative models,” Nat. Commun. 12(1), 2403 (2021). 10.1038/s41467-021-22732-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Matsunaga Y. and Sugita Y., “ Linking time-series of single-molecule experiments with molecular dynamics simulations by machine learning,” Elife 7, e32668 (2018). 10.7554/eLife.32668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Wang J., Ferguson A., and Team J. W., “ Machine learning of protein folding funnels from experimentally measurable observables,” in APS March Meeting Abstracts, 2018. [Google Scholar]
- 205. Chen X., Yang B., and Lin Z., “ A random forest learning assisted “divide and conquer” approach for peptide conformation search,” Sci. Rep. 8(1), 8796 (2018). 10.1038/s41598-018-27167-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Moman E., Grishina M. A., and Potemkin V. A., “ Nonparametric chemical descriptors for the calculation of ligand-biopolymer affinities with machine-learning scoring functions,” J. Comput.-Aided Mol. Des. 33(11), 943–953 (2019). 10.1007/s10822-019-00248-2 [DOI] [PubMed] [Google Scholar]
- 207. Moebel E., Martinez-Sanchez A., Lamm L., Righetto R., Wietrzynski W., Albert S., Lariviere D., Fourmentin E., Pfeffer S., and Ortiz J., “ Deep learning improves macromolecule identification in 3D cellular cryo-electron tomograms,” Nat. Methods 18, 1386–1394 (2021). 10.1038/s41592-021-01275-4 [DOI] [PubMed] [Google Scholar]
- 208. Bandyopadhyay S. and Mondal J., “ A deep autoencoder framework for discovery of metastable ensembles in biomacromolecules,” e-print arXiv:2106.00724 (2021). [DOI] [PubMed]
- 209. Saar K. L., Morgunov A. S., Qi R., Arter W. E., Krainer G., and Knowles T. P., “ Learning the molecular grammar of protein condensates from sequence determinants and embeddings,” Proc. Natl. Acad. Sci. 118(15), e2019053118 (2021). 10.1073/pnas.2019053118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Yang Q., Bassyouni A., Butler C. R., Hou X., Jenkinson S., and Price D. A., “ Ligand biological activity predicted by cleaning positive and negative chemical correlations,” Proc. Natl. Acad. Sci. 116(9), 3373–3378 (2019). 10.1073/pnas.1810847116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Sharifi S., Pakdel A., Ebrahimi M., Reecy J. M., Fazeli Farsani S., and Ebrahimie E., “ Integration of machine learning and meta-analysis identifies the transcriptomic bio-signature of mastitis disease in cattle,” PLoS one 13(2), e0191227 (2018). 10.1371/journal.pone.0191227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Fu C., Zhang X., Veri A. O., Iyer K. R., Lash E., Xue A., Yan H., Revie N. M., Wong C., and Lin Z.-Y., “ Leveraging machine learning essentiality predictions and chemogenomic interactions to identify antifungal targets,” Nat. Commun. 12(1), 6497 (2021). 10.1038/s41467-021-26850-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Jiang B., Mu Q., Qiu F., Li X., Xu W., Yu J., Fu W., Cao Y., and Wang J., “ Machine learning of genomic features in organotropic metastases stratifies progression risk of primary tumors,” Nat. Commun. 12(1), 6692 (2021). 10.1038/s41467-021-27017-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Kim W., Kim T. H., Oh S. J., Kim H. J., Kim J. H., Kim H.-A., Jung J.-Y., Choi I. A., and Lee K. E., “ Association of TLR 9 gene polymorphisms with remission in patients with rheumatoid arthritis receiving TNF-α inhibitors and development of machine learning models,” Sci. Rep. 11(1), 20169 (2021). 10.1038/s41598-021-99625-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Huang Y., Sun X., Jiang H., Yu S., Robins C., Armstrong M. J., Li R., Mei Z., Shi X., and Gerasimov E. S., “ A machine learning approach to brain epigenetic analysis reveals kinases associated with Alzheimer's disease,” Nat. Commun. 12(1), 4472 (2021). 10.1038/s41467-021-24710-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Scott M. A., Woolums A. R., Swiderski C. E., Perkins A. D., and Nanduri B., “ Genes and regulatory mechanisms associated with experimentally-induced bovine respiratory disease identified using supervised machine learning methodology,” Sci. Rep. 11(1), 22916 (2021). 10.1038/s41598-021-02343-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Stupp D., Sharon E., Bloch I., Zitnik M., Zuk O., and Tabach Y., “ Co-evolution based machine-learning for predicting functional interactions between human genes,” Nat. Commun. 12(1), 6454 (2021). 10.1038/s41467-021-26792-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Gussow A. B., Park A. E., Borges A. L., Shmakov S. A., Makarova K. S., Wolf Y. I., Bondy-Denomy J., and Koonin E. V., “ Machine-learning approach expands the repertoire of anti-CRISPR protein families,” Nat. Commun. 11(1), 3784 (2020). 10.1038/s41467-020-17652-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Trofimov A. A., Pawlicki A. A., Borodinov N., Mandal S., Mathews T. J., Hildebrand M., Ziatdinov M. A., Hausladen K. A., Urbanowicz P. K., and Steed C. A., “ Deep data analytics for genetic engineering of diatoms linking genotype to phenotype via machine learning,” npj Comput. Mater. 5(1), 67 (2019). 10.1038/s41524-019-0202-3 [DOI] [Google Scholar]
- 220. Cheng C.-Y., Li Y., Varala K., Bubert J., Huang J., Kim G. J., Halim J., Arp J., Shih H.-J. S., and Levinson G., “ Evolutionarily informed machine learning enhances the power of predictive gene-to-phenotype relationships,” Nat. Commun. 12(1), 5627 (2021). 10.1038/s41467-021-25893-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Zhang J. X., Yordanov B., Gaunt A., Wang M. X., Dai P., Chen Y.-J., Zhang K., Fang J. Z., Dalchau N., and Li J., “ A deep learning model for predicting next-generation sequencing depth from DNA sequence,” Nat. Commun. 12(1), 4387 (2021). 10.1038/s41467-021-24497-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Leng Y., Tac V., Calve S., and Tepole A. B., “ Predicting the mechanical properties of biopolymer gels using neural networks trained on discrete fiber network data,” Comput. Methods Appl. Mech. Eng. 387, 114160 (2021). 10.1016/j.cma.2021.114160 [DOI] [Google Scholar]
- 223. Entekhabi E., Nazarpak M. H., Sedighi M., and Kazemzadeh A., “ Predicting degradation rate of genipin cross-linked gelatin scaffolds with machine learning,” Mater. Sci. Eng.: C 107, 110362 (2020). 10.1016/j.msec.2019.110362 [DOI] [PubMed] [Google Scholar]
- 224. Özkan M., Borghei M., Karakoç A., Rojas O. J., and Paltakari J., “ Films based on crosslinked TEMPO-oxidized cellulose and predictive analysis via machine learning,” Sci. Rep. 8(1), 4748 (2018). 10.1038/s41598-018-23114-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Röding M., Fager C., Olsson A., von Corswant C., Olsson E., and Lorén N., “ Three‐dimensional reconstruction of porous polymer films from FIB‐SEM nanotomography data using random forests,” J. Microsc. 281(1), 76–86 (2021). 10.1111/jmi.12950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Chen D., Dunkers J. P., Losert W., and Sarkar S., “ Early time-point cell morphology classifiers successfully predict human bone marrow stromal cell differentiation modulated by fiber density in nanofiber scaffolds,” Biomaterials 274, 120812 (2021). 10.1016/j.biomaterials.2021.120812 [DOI] [PubMed] [Google Scholar]
- 227. Robles-Bykbaev Y., Naya S., Díaz-Prado S., Calle-López D., Robles-Bykbaev V., Garzón L., Sanjurjo-Rodríguez C., and Tarrío-Saavedra J., “ An artificial-vision-and statistical-learning-based method for studying the biodegradation of type I collagen scaffolds in bone regeneration systems,” PeerJ 7, e7233 (2019). 10.7717/peerj.7233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Liu Z., Shi Y., Chen H., Qin T., Zhou X., Huo J., Dong H., Yang X., Zhu X., and Chen X., “ Machine learning on properties of multiscale multisource hydroxyapatite nanoparticles datasets with different morphologies and sizes,” npj Comput. Mater. 7(1), 142 (2021). 10.1038/s41524-021-00618-1 [DOI] [Google Scholar]
- 229. Cao Y., Karimi M., Kamrani E., Nourani P., Manesh A. M., Momenieskandari H., and Anqi A. E., “ Machine learning methods help accurate estimation of the hydrogen solubility in biomaterials,” Int. J. Hydrogen Energy 47(6), 3611–3624 (2022). 10.1016/j.ijhydene.2021.10.259 [DOI] [Google Scholar]
- 230. Daghigh V., T. E. Lacy, Jr. , Daghigh H., Gu G., Baghaei K. T., Horstemeyer M. F., and C. U. Pittman, Jr. , “ Heat deflection temperatures of bio-nano-composites using experiments and machine learning predictions,” Mater. Today Commun. 22, 100789 (2020). 10.1016/j.mtcomm.2019.100789 [DOI] [Google Scholar]
- 231. Mitterwallner B. G., Schreiber C., Daldrop J. O., Rädler J. O., and Netz R. R., “ Non-Markovian data-driven modeling of single-cell motility,” Phys. Rev. E 101(3), 032408 (2020). 10.1103/PhysRevE.101.032408 [DOI] [PubMed] [Google Scholar]
- 232. Zhang J., Petersen S. D., Radivojevic T., Ramirez A., Pérez-Manríquez A., Abeliuk E., Sánchez B. J., Costello Z., Chen Y., and Fero M. J., “ Combining mechanistic and machine learning models for predictive engineering and optimization of tryptophan metabolism,” Nat. Commun. 11(1), 4880 (2020). 10.1038/s41467-020-17910-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Li C., Wang Y., Sha S., Yin H., Zhang H., Wang Y., Zhao B., and Song F., “ Analysis of the tendency for the electronic conductivity to change during alcoholic fermentation,” Sci. Rep. 9(1), 5512 (2019). 10.1038/s41598-019-41225-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Hlangwani E., Doorsamy W., Adebiyi J. A., Fajimi L. I., and Adebo O. A., “ A modeling method for the development of a bioprocess to optimally produce umqombothi (a South African traditional beer),” Sci. Rep. 11(1), 20626 (2021). 10.1038/s41598-021-00097-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Durand A., Wiesner T., Gardner M.-A., Robitaille L.-É., Bilodeau A., Gagné C., De Koninck P., and Lavoie-Cardinal F., “ A machine learning approach for online automated optimization of super-resolution optical microscopy,” Nat. Commun. 9(1), 5247 (2018). 10.1038/s41467-018-07668-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Kistenev Y. V., Vrazhnov D., Nikolaev V., Sandykova E., and Krivova N., “ Analysis of collagen spatial structure using multiphoton microscopy and machine learning methods,” Biochemistry 84(1), S108–S123 (2019). 10.1134/S0006297919140074 [DOI] [PubMed] [Google Scholar]
- 237. Tsai H.-F., Gajda J., Sloan T. F., Rares A., and Shen A. Q., “ Usiigaci: Instance-aware cell tracking in stain-free phase contrast microscopy enabled by machine learning,” SoftwareX 9, 230–237 (2019). 10.1016/j.softx.2019.02.007 [DOI] [Google Scholar]
- 238. Anderson T. I., Vega B., and Kovscek A. R., “ Multimodal imaging and machine learning to enhance microscope images of shale,” Comput. Geosci. 145, 104593 (2020). 10.1016/j.cageo.2020.104593 [DOI] [Google Scholar]
- 239. He Y., Xu W., Zhi Y., Tyagi R., Hu Z., and Cao G., “ Rapid bacteria identification using structured illumination microscopy and machine learning,” J. Innovative Opt. Health Sci. 11(1), 1850007 (2018). 10.1142/S1793545818500074 [DOI] [Google Scholar]
- 240. Mazurenko S., Prokop Z., and Damborsky J., “ Machine learning in enzyme engineering,” ACS Catal. 10(2), 1210–1223 (2019). 10.1021/acscatal.9b04321 [DOI] [Google Scholar]
- 241. Tourlomousis F., Jia C., Karydis T., Mershin A., Wang H., Kalyon D. M., and Chang R. C., “ Machine learning metrology of cell confinement in melt electrowritten three-dimensional biomaterial substrates,” Microsyst. Nanoeng. 5(1), 15 (2019). 10.1038/s41378-019-0055-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Sujeeun L. Y., Goonoo N., Ramphul H., Chummun I., Gimié F., Baichoo S., and Bhaw-Luximon A., “ Correlating in vitro performance with physico-chemical characteristics of nanofibrous scaffolds for skin tissue engineering using supervised machine learning algorithms,” R. Soc. Open Sci. 7(12), 201293 (2020). 10.1098/rsos.201293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Li F., Han J., Cao T., Lam W., Fan B., Tang W., Chen S., Fok K. L., and Li L., “ Design of self-assembly dipeptide hydrogels and machine learning via their chemical features,” Proc. Natl. Acad. Sci. 116(23), 11259–11264 (2019). 10.1073/pnas.1903376116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Liu Y., Zhang D., Tang Y., Zhang Y., Chang Y., and Zheng J., “ Machine learning-enabled design and prediction of protein resistance on self-assembled monolayers and beyond,” ACS Appl. Mater. Interfaces 13(9), 11306–11319 (2021). 10.1021/acsami.1c00642 [DOI] [PubMed] [Google Scholar]
- 245. Conev A., Litsa E. E., Perez M. R., Diba M., Mikos A. G., and Kavraki L. E., “ Machine learning-guided three-dimensional printing of tissue engineering scaffolds,” Tissue Eng. Part A 26(23–24), 1359–1368 (2020). 10.1089/ten.tea.2020.0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Li Y., Nowak C. M., Pham U., Nguyen K., and Bleris L., “ Cell morphology-based machine learning models for human cell state classification,” npj Syst. Biol. Appl. 7(1), 23 (2021). 10.1038/s41540-021-00180-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Masad I. S., Alqudah A., Alqudah A. M., and Almashaqbeh S., “ A hybrid deep learning approach towards building an intelligent system for pneumonia detection in chest X-ray images,” Int. J. Electr. Comput. Eng. 11(6), 5530–5540 (2021). [Google Scholar]
- 248. Tuncer T., Dogan S., and Ozyurt F., “ An automated residual exemplar local binary pattern and iterative ReliefF based COVID-19 detection method using chest X-ray image,” Chemom. Intell. Lab. Syst. 203, 104054 (2020). 10.1016/j.chemolab.2020.104054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Brunese L., Mercaldo F., Reginelli A., and Santone A., “ Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays,” Comput. Methods Programs Biomed. 196, 105608 (2020). 10.1016/j.cmpb.2020.105608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Lahmiri S., Dawson D. A., and Shmuel A., “ Performance of machine learning methods in diagnosing Parkinson's disease based on dysphonia measures,” Biomed. Eng. Lett. 8(1), 29–39 (2018). 10.1007/s13534-017-0051-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Malkawi A., Al-Assi R., Salameh T., Alquran H., and Alqudah A. M., “ White blood cells classification using convolutional neural network hybrid system,” in 2020 IEEE 5th Middle East and Africa Conference on Biomedical Engineering (MECBME) ( IEEE, 2020), pp. 1–5. [Google Scholar]
- 252. Rajendran S. and Jothi A., “ Sequentially distant but structurally similar proteins exhibit fold specific patterns based on their biophysical properties,” Comput. Biol. Chem. 75, 143–153 (2018). 10.1016/j.compbiolchem.2018.05.009 [DOI] [PubMed] [Google Scholar]
- 253. Dou L., Li X., Ding H., Xu L., and Xiang H., “ iRNA-m5C_NB: A novel predictor to identify RNA 5-methylcytosine sites based on the Naive Bayes classifier,” IEEE Access 8, 84906–84917 (2020). 10.1109/ACCESS.2020.2991477 [DOI] [Google Scholar]
- 254. Zaw H. T., Maneerat N., and Win K. Y., “ Brain tumor detection based on Naïve Bayes Classification,” in 2019 5th International Conference on Engineering, Applied Sciences and Technology (ICEAST) ( IEEE, 2019), pp. 1–4. [Google Scholar]
- 255. Gamage P. T., Azad M. K., Taebi A., Sandler R. H., and Mansy H. A., “ Clustering seismocardiographic events using unsupervised machine learning,” in 2018 IEEE Signal Processing in Medicine and Biology Symposium (SPMB) ( IEEE, 2018), pp. 1–5. [Google Scholar]
- 256. Helfrecht B. A., Gasparotto P., Giberti F., and Ceriotti M., “ Atomic motif recognition in (bio) polymers: Benchmarks from the protein data bank,” Front. Mol. Biosci. 6, 24 (2019). 10.3389/fmolb.2019.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Gul G., Yildirim R., and Ileri-Ercan N., “ Cytotoxicity analysis of nanoparticles by association rule mining,” Environ. Sci.: Nano 8(4), 937–949 (2021). 10.1039/D0EN01240H [DOI] [Google Scholar]
- 258. Kuanar S., Athitsos V., Mahapatra D., Rao K., Akhtar Z., and Dasgupta D., “ Low dose abdominal CT image reconstruction: An unsupervised learning based approach,” in 2019 IEEE International Conference on Image Processing (ICIP) ( IEEE, 2019), pp. 1351–1355. [Google Scholar]
- 259. Rives A., Meier J., Sercu T., Goyal S., Lin Z., Liu J., Guo D., Ott M., Zitnick C. L., and Ma J., “ Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences,” Proc. Natl. Acad. Sci. 118(15), e2016239118 (2021). 10.1073/pnas.2016239118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Yoshida K., Kawai S., Fujitani M., Koikeda S., Kato R., and Ema T., “ Enhancement of protein thermostability by three consecutive mutations using loop-walking method and machine learning,” Sci. Rep. 11(1), 11883 (2021). 10.1038/s41598-021-91339-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Ziolek R. M., Smith P., Pink D. L., Dreiss C. A., and Lorenz C. D., “ Unsupervised learning unravels the structure of four-arm and linear block copolymer micelles,” Macromolecules 54(8), 3755–3768 (2021). 10.1021/acs.macromol.0c02523 [DOI] [Google Scholar]
- 262. Bushnell G. G., Hardas T. P., Hartfield R. M., Zhang Y., Oakes R. S., Ronquist S., Chen H., Rajapakse I., Wicha M. S., and Jeruss J. S., “ Biomaterial scaffolds recruit an aggressive population of metastatic tumor cells in vivo,” Cancer Res. 79(8), 2042–2053 (2019). 10.1158/0008-5472.CAN-18-2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Jafari R. and Javidi M. M., “ Solving the protein folding problem in hydrophobic-polar model using deep reinforcement learning,” SN Appl. Sci. 2(2), 259 (2020). 10.1007/s42452-020-2012-0 [DOI] [Google Scholar]
- 264. Popova M., Isayev O., and Tropsha A., “ Deep reinforcement learning for de novo drug design,” Sci. Adv. 4(7), eaap7885 (2018). 10.1126/sciadv.aap7885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Petsagkourakis P., Sandoval I. O., Bradford E., Zhang D., and del Rio-Chanona E. A., “ Reinforcement learning for batch bioprocess optimization,” Comput. Chem. Eng. 133, 106649 (2020). 10.1016/j.compchemeng.2019.106649 [DOI] [Google Scholar]
- 266. Hou Z., Lee T., and Keidar M., “ Reinforcement learning with safe exploration for adaptive plasma cancer treatment,” IEEE Trans. Radiat. Plasma Med. Sci. 6(4), 482–492 (2022). 10.1109/TRPMS.2021.3094874 [DOI] [Google Scholar]
- 267. Seno H., Yamazaki M., Shibata N., Sakuma I., and Tomii N., “ In-silico deep reinforcement learning for effective cardiac ablation strategy,” J. Med. Biol. Eng. 41, 935–965 (2021). 10.1007/s40846-021-00664-6 [DOI] [Google Scholar]
- 268. Yazdjerdi P., Meskin N., Al-Naemi M., Al Moustafa A.-E., and Kovács L., “ Reinforcement learning-based control of tumor growth under anti-angiogenic therapy,” Comput. Methods Programs Biomed. 173, 15–26 (2019). 10.1016/j.cmpb.2019.03.004 [DOI] [PubMed] [Google Scholar]
- 269. Tseng H. H., Luo Y., Cui S., Chien J. T., Ten Haken R. K., and Naqa I. E., “ Deep reinforcement learning for automated radiation adaptation in lung cancer,” Med. Phys. 44(12), 6690–6705 (2017). 10.1002/mp.12625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Padmanabhan R., Meskin N., and Haddad W. M., “ Reinforcement learning-based control of drug dosing for cancer chemotherapy treatment,” Math. Biosci. 293, 11–20 (2017). 10.1016/j.mbs.2017.08.004 [DOI] [PubMed] [Google Scholar]
- 271. Born J., Manica M., Oskooei A., Cadow J., Markert G., and Martínez M. R., “ PaccMannRL: De novo generation of hit-like anticancer molecules from transcriptomic data via reinforcement learning,” Iscience 24(4), 102269 (2021). 10.1016/j.isci.2021.102269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Eckardt J.-N., Wendt K., Bornhäuser M., and Middeke J. M., “ Reinforcement learning for precision oncology,” Cancers 13(18), 4624 (2021). 10.3390/cancers13184624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Kong J., Lee H., Kim D., Han S. K., Ha D., Shin K., and Kim S., “ Network-based machine learning in colorectal and bladder organoid models predicts anti-cancer drug efficacy in patients,” Nat. Commun. 11(1), 5485 (2020). 10.1038/s41467-020-19313-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Jin Z., Zhang Z., Shao X., and Gu G. X., “ Monitoring anomalies in 3D bioprinting with deep neural networks,” ACS Biomater. Sci. Eng. (2021). 10.1021/acsbiomaterials.0c01761 [DOI] [PubMed] [Google Scholar]
- 275. Yu C. and Jiang J., “ A perspective on using machine learning in 3D bioprinting,” Int. J. Bioprint. 6(1), 253 (2020). 10.18063/ijb.v6i1.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Guan J., You S., Xiang Y., Schimelman J., Alido J., Ma X., Tang M., and Chen S., “ Compensating the cell-induced light scattering effect in light-based bioprinting using deep learning,” Biofabrication 14(1), 015011 (2021). 10.1088/1758-5090/ac3b92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Shen S. C., Fernández M. P., Tozzi G., and Buehler M. J., “ Deep learning approach to assess damage mechanics of bone tissue,” J. Mech. Behav. Biomed. Mater. 123, 104761 (2021). 10.1016/j.jmbbm.2021.104761 [DOI] [PubMed] [Google Scholar]
- 278. Helgadottir S., Midtvedt B., Pineda J., Sabirsh A., Adiels C. B., Romeo S., Midtvedt D., and Volpe G., “ Extracting quantitative biological information from bright-field cell images using deep learning,” Biophys. Rev. 2(3), 031401 (2021). 10.1063/5.0044782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Xin L., Xiao W., Cao R., Wu X., Ferraro P., and Pan F., “ Automatic compensation of phase aberration in digital holographic microscopy with deep neural networks for monitoring the morphological response of bone cells under fluid shear stress,” in Optical Methods for Inspection, Characterization, and Imaging of Biomaterials V ( International Society for Optics and Photonics, 2021), Vol. 11786, p. 117860O. [Google Scholar]
- 280. Skärberg F., Fager C., Mendoza‐Lara F., Josefson M., Olsson E., Lorén N., and Röding M., “ Convolutional neural networks for segmentation of FIB‐SEM nanotomography data from porous polymer films for controlled drug release,” J. Microsc. 283(1), 51–63 (2021). 10.1111/jmi.13007 [DOI] [PubMed] [Google Scholar]
- 281. Lin E., Lin C.-H., and Lane H.-Y., “ Relevant applications of generative adversarial networks in drug design and discovery: Molecular de novo design, dimensionality reduction, and de novo peptide and protein design,” Molecules 25(14), 3250 (2020). 10.3390/molecules25143250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Bian Y. and Xie X.-Q., “ Generative chemistry: Drug discovery with deep learning generative models,” J. Mol. Model. 27(3), 1–18 (2021). 10.1007/s00894-021-04674-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Mao Y., He Q., and Zhao X., “ Designing complex architectured materials with generative adversarial networks,” Sci. Adv. 6(17), eaaz4169 (2020). 10.1126/sciadv.aaz4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Zhang H., Yang L., Li C., Wu B., and Wang W., “ Scaffoldgan: Synthesis of scaffold materials based on generative adversarial networks,” Comput.-Aided Des. 138, 103041 (2021). 10.1016/j.cad.2021.103041 [DOI] [Google Scholar]
- 285. Calimeri F., Marzullo A., Stamile C., and Terracina G., “ Biomedical data augmentation using generative adversarial neural networks,” in International Conference on Artificial Neural Networks ( Springer, 2017), pp. 626–634. [Google Scholar]
- 286. Hazra D. and Byun Y.-C., “ SynSigGAN: Generative adversarial networks for synthetic biomedical signal generation,” Biology 9(12), 441 (2020). 10.3390/biology9120441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Aida S., Okugawa J., Fujisaka S., Kasai T., Kameda H., and Sugiyama T., “ Deep learning of cancer stem cell morphology using conditional generative adversarial networks,” Biomolecules 10(6), 931 (2020). 10.3390/biom10060931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Chen Z., Ma N., Sun X., Li Q., Zeng Y., Chen F., Sun S., Xu J., Zhang J., and Ye H., “ Automated evaluation of tumor spheroid behavior in 3D culture using deep learning-based recognition,” Biomaterials 272, 120770 (2021). 10.1016/j.biomaterials.2021.120770 [DOI] [PubMed] [Google Scholar]
- 289. Xiao Y., Wu J., Lin Z., and Zhao X., “ A semi-supervised deep learning method based on stacked sparse auto-encoder for cancer prediction using RNA-seq data,” Comput. Methods Programs Biomed. 166, 99–105 (2018). 10.1016/j.cmpb.2018.10.004 [DOI] [PubMed] [Google Scholar]
- 290. Lazarovits J., Sindhwani S., Tavares A. J., Zhang Y., Song F., Audet J., Krieger J. R., Syed A. M., Stordy B., and Chan W. C., “ Supervised learning and mass spectrometry predicts the in vivo fate of nanomaterials,” ACS Nano 13(7), 8023–8034 (2019). 10.1021/acsnano.9b02774 [DOI] [PubMed] [Google Scholar]
- 291. Tampuu A., Bzhalava Z., Dillner J., and Vicente R., “ ViraMiner: Deep learning on raw DNA sequences for identifying viral genomes in human samples,” PLoS One 14(9), e0222271 (2019). 10.1371/journal.pone.0222271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Erdenebayar U., Kim Y. J., Park J.-U., Joo E. Y., and Lee K.-J., “ Deep learning approaches for automatic detection of sleep apnea events from an electrocardiogram,” Comput. Methods Programs Biomed. 180, 105001 (2019). 10.1016/j.cmpb.2019.105001 [DOI] [PubMed] [Google Scholar]
- 293. Kundu K., Mann M., Costa F., and Backofen R., “ MoDPepInt: An interactive web server for prediction of modular domain–peptide interactions,” Bioinformatics 30(18), 2668–2669 (2014). 10.1093/bioinformatics/btu350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Stormo G. D., Schneider T. D., Gold L., and Ehrenfeucht A., “ Use of the ‘Perceptron’ algorithm to distinguish translational initiation sites in E. coli,” Nucl. Acids Res. 10(9), 2997–3011 (1982). 10.1093/nar/10.9.2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Miller M. L., Jensen L. J., Diella F., Jørgensen C., Tinti M., Li L., Hsiung M., Parker S. A., Bordeaux J., and Sicheritz-Ponten T., “ Linear motif atlas for phosphorylation-dependent signaling,” Sci. Signal. 1(35), ra2 (2008). 10.1126/scisignal.1159433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Wójcikowski M., Kukiełka M., Stepniewska-Dziubinska M. M., and Siedlecki P., “ Development of a protein–ligand extended connectivity (PLEC) fingerprint and its application for binding affinity predictions,” Bioinformatics 35(8), 1334–1341 (2019). 10.1093/bioinformatics/bty757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Jiménez J., Skalic M., Martinez-Rosell G., and De Fabritiis G., “ K DEEP: Protein-ligand absolute binding affinity prediction via 3D-convolutional neural networks,” J. Chem. Inf. Model. 58(2), 287–296 (2018). 10.1021/acs.jcim.7b00650 [DOI] [PubMed] [Google Scholar]
- 298. Stepniewska-Dziubinska M. M., Zielenkiewicz P., and Siedlecki P., “ Development and evaluation of a deep learning model for protein–ligand binding affinity prediction,” Bioinformatics 34(21), 3666–3674 (2018). 10.1093/bioinformatics/bty374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Boyles F., Deane C. M., and Morris G. M., “ Learning from the ligand: Using ligand-based features to improve binding affinity prediction,” Bioinformatics 36(3), 758–764 (2020). 10.1093/bioinformatics/btz665 [DOI] [PubMed] [Google Scholar]
- 300. Wallach I., Dzamba M., and Heifets A., “ AtomNet: A deep convolutional neural network for bioactivity prediction in structure-based drug discovery,” e-print arXiv:1510.02855 (2015).
- 301. Ragoza M., Hochuli J., Idrobo E., Sunseri J., and Koes D. R., “ Protein-ligand scoring with convolutional neural networks,” J. Chem. Inf. Model. 57(4), 942–957 (2017). 10.1021/acs.jcim.6b00740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Sahlol A. T., Kollmannsberger P., and Ewees A. A., “ Efficient classification of white blood cell leukemia with improved swarm optimization of deep features,” Sci. Rep. 10(1), 2536 (2020). 10.1038/s41598-020-59215-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Ying X., “ An overview of overfitting and its solutions,” in Journal of Physics: Conference Series ( IOP Publishing, 2019), Vol. 1168, p. 022022. [Google Scholar]
- 304. Mehrabi N., Morstatter F., Saxena N., Lerman K., and Galstyan A., “ A survey on bias and fairness in machine learning,” ACM Comput. Surv. 54(6), 1–35 (2021). 10.1145/3457607 [DOI] [Google Scholar]
- 305. Vokinger K. N., Feuerriegel S., and Kesselheim A. S., “ Mitigating bias in machine learning for medicine,” Commun. Med. 1(1), 25 (2021). 10.1038/s43856-021-00028-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Obermeyer Z., Powers B., Vogeli C., and Mullainathan S., “ Dissecting racial bias in an algorithm used to manage the health of populations,” Science 366(6464), 447–453 (2019). 10.1126/science.aax2342 [DOI] [PubMed] [Google Scholar]
- 307. van Smeden M., Moons K. G., de Groot J. A., Collins G. S., Altman D. G., Eijkemans M. J., and Reitsma J. B., “ Sample size for binary logistic prediction models: Beyond events per variable criteria,” Stat. Methods Med. Res. 28(8), 2455–2474 (2019). 10.1177/0962280218784726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Shaikhina T., Lowe D., Daga S., Briggs D., Higgins R., and Khovanova N., “ Machine learning for predictive modelling based on small data in biomedical engineering,” IFAC-PapersOnLine 48(20), 469–474 (2015). 10.1016/j.ifacol.2015.10.185 [DOI] [Google Scholar]
- 309. Rahmati O., Tahmasebipour N., Haghizadeh A., Pourghasemi H. R., and Feizizadeh B., “ Evaluation of different machine learning models for predicting and mapping the susceptibility of gully erosion,” Geomorphology 298, 118–137 (2017). 10.1016/j.geomorph.2017.09.006 [DOI] [Google Scholar]
- 310. Perry G. L. and Dickson M. E., “ Using machine learning to predict geomorphic disturbance: The effects of sample size, sample prevalence, and sampling strategy,” J. Geophys. Res.: Earth Surf. 123(11), 2954–2970, 10.1029/2018JF004640 (2018). [DOI] [Google Scholar]
- 311. Rogers A. W., Vega‐Ramon F., Yan J., del Río‐Chanona E. A., Jing K., and Zhang D., “ A transfer learning approach for predictive modelling of bioprocesses using small data,” Biotechnol. Bioeng. 119(2), 411–422 (2021). 10.1002/bit.27980 [DOI] [PubMed] [Google Scholar]
- 312. Qi G.-J. and Luo J., “ Small data challenges in big data era: A survey of recent progress on unsupervised and semi-supervised methods,” in IEEE Transactions on Pattern Analysis and Machine Intelligence ( IEEE, 2020), pp. 2168–2187. [DOI] [PubMed] [Google Scholar]
- 313. Altae-Tran H., Ramsundar B., Pappu A. S., and Pande V., “ Low data drug discovery with one-shot learning,” ACS Cent. Sci. 3(4), 283–293 (2017). 10.1021/acscentsci.6b00367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Bahador N. and Kortelainen J., “ Deep learning-based classification of multichannel bio-signals using directedness transfer learning,” Biomed. Signal Process. Control 72, 103300 (2022). 10.1016/j.bspc.2021.103300 [DOI] [Google Scholar]
- 315. Aravind K., Raja P., Aniirudh R., Mukesh K., Ashiwin R., and Vikas G., “ Grape crop disease classification using transfer learning approach,” in International Conference on ISMAC in Computational Vision and Bio-Engineering ( Springer, 2018), pp. 1623–1633. [Google Scholar]
- 316. Hakimi O., Krallinger M., and Ginebra M.-P., “ Time to kick-start text mining for biomaterials,” Nat. Rev. Mater. 5(8), 553–556 (2020). 10.1038/s41578-020-0215-z [DOI] [Google Scholar]
- 317. Court C. J., Jain A., and Cole J. M., “ Inverse design of materials that exhibit the magnetocaloric effect by text-mining of the scientific literature and generative deep learning,” Chem. Mater. 33(18), 7217–7231 (2021). 10.1021/acs.chemmater.1c01368 [DOI] [Google Scholar]
- 318. Ye J., Xu B., Fan B., Zhang J., Yuan F., Chen Y., Sun Z., Yan X., Song Y., and Song S., “ Discovery of selenocysteine as a potential nanomedicine promotes cartilage regeneration with enhanced immune response by text mining and biomedical databases,” Front. Pharmacol. 11, 1138 (2020). 10.3389/fphar.2020.01138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Rincón-López J., Almanza-Arjona Y. C., Riascos A. P., and Rojas-Aguirre Y., “ When cyclodextrins met data science: Unveiling their pharmaceutical applications through network science and text-mining,” Pharmaceutics 13(8), 1297 (2021). 10.3390/pharmaceutics13081297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Tshitoyan V., Dagdelen J., Weston L., Dunn A., Rong Z., Kononova O., Persson K. A., Ceder G., and Jain A., “ Unsupervised word embeddings capture latent knowledge from materials science literature,” Nature 571(7763), 95–98 (2019). 10.1038/s41586-019-1335-8 [DOI] [PubMed] [Google Scholar]
- 321. Kim E., Huang K., Saunders A., McCallum A., Ceder G., and Olivetti E., “ Materials synthesis insights from scientific literature via text extraction and machine learning,” Chem. Mater. 29(21), 9436–9444 (2017). 10.1021/acs.chemmater.7b03500 [DOI] [Google Scholar]
- 322. Mongkhonthanaphon S. and Limpiyakorn Y., “ A deep neural network for pixel-wise classification of titanium microstructure,” Int. J. Mach. Learn. Comput. 10(1), 128–133 (2020). 10.18178/ijmlc.2020.10.1.909 [DOI] [Google Scholar]
- 323. Liang L., Liu M., and Sun W., “ A deep learning approach to estimate chemically-treated collagenous tissue nonlinear anisotropic stress-strain responses from microscopy images,” Acta Biomater. 63, 227–235 (2017). 10.1016/j.actbio.2017.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Korpela J., Suzuki H., Matsumoto S., Mizutani Y., Samejima M., Maekawa T., Nakai J., and Yoda K., “ Machine learning enables improved runtime and precision for bio-loggers on seabirds,” Commun. Biol. 3(1), 633 (2020). 10.1038/s42003-020-01356-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Zhang Y. and Ling C., “ A strategy to apply machine learning to small datasets in materials science,” npj Comput. Mater. 4(1), 25 (2018). 10.1038/s41524-018-0081-z [DOI] [Google Scholar]
- 326. Oyetunde T., Liu D., Martin H. G., and Tang Y. J., “ Machine learning framework for assessment of microbial factory performance,” PLoS One 14(1), e0210558 (2019). 10.1371/journal.pone.0210558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Tulsyan A., Garvin C., and Ündey C., “ Advances in industrial biopharmaceutical batch process monitoring: Machine‐learning methods for small data problems,” Biotechnol. Bioeng. 115(8), 1915–1924 (2018). 10.1002/bit.26605 [DOI] [PubMed] [Google Scholar]
- 328. Nair M., Bica I., Best S. M., and Cameron R. E., “ Feature importance in multi-dimensional tissue-engineering datasets: Random forest assisted optimization of experimental variables for collagen scaffolds,” Appl. Phys. Rev. 8(4), 041403 (2021). 10.1063/5.0059724 [DOI] [Google Scholar]
- 329. Yang W., Si Y., Wang D., and Guo B., “ Automatic recognition of arrhythmia based on principal component analysis network and linear support vector machine,” Comput. Biol. Med. 101, 22–32 (2018). 10.1016/j.compbiomed.2018.08.003 [DOI] [PubMed] [Google Scholar]
- 330. Tian Y. and Zhang Y., “ A comprehensive survey on regularization strategies in machine learning,” Inf. Fusion 80, 146–166 (2021). 10.1016/j.inffus.2021.11.005 [DOI] [Google Scholar]
- 331. Xiong Z., Cui Y., Liu Z., Zhao Y., Hu M., and Hu J., “ Evaluating explorative prediction power of machine learning algorithms for materials discovery using k-fold forward cross-validation,” Comput. Mater. Sci. 171, 109203 (2020). 10.1016/j.commatsci.2019.109203 [DOI] [Google Scholar]
- 332. Zhang Y.-D., Pan C., Sun J., and Tang C., “ Multiple sclerosis identification by convolutional neural network with dropout and parametric ReLU,” J. Comput. Sci. 28, 1–10 (2018). 10.1016/j.jocs.2018.07.003 [DOI] [Google Scholar]
- 333. Hyun J. C., Kavvas E. S., Monk J. M., and Palsson B. O., “ Machine learning with random subspace ensembles identifies antimicrobial resistance determinants from pan-genomes of three pathogens,” PLoS Comput. Biol. 16(3), e1007608 (2020). 10.1371/journal.pcbi.1007608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Lippeveld M., Knill C., Ladlow E., Fuller A., Michaelis L. J., Saeys Y., Filby A., and Peralta D., “ Classification of human white blood cells using machine learning for stain‐free imaging flow cytometry,” Cytom. Part A 97(3), 308–319 (2020). 10.1002/cyto.a.23920 [DOI] [PubMed] [Google Scholar]
- 335. Loey M., Manogaran G., and Khalifa N. E. M., “ A deep transfer learning model with classical data augmentation and CGAN to detect COVID-19 from chest CT radiography digital images,” Neural Comput. Appl. 2020, 1–13. 10.1007/s00521-020-05437-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.