Visual Abstract
Keywords: metabolism, pathology, renal function
Abstract
Key Points
d-Alanine affects the circadian clock to regulate gluconeogenesis in the kidney.
d-Alanine itself has a clear intrinsic circadian rhythm, which is regulated by urinary excretion, and acts on the circadian rhythm.
d-Alanine is a signal activator for circadian rhythm and gluconeogenesis through circadian transcriptional network.
Background
The aberrant glucose circadian rhythm is associated with the pathogenesis of diabetes. Similar to glucose metabolism in the kidney and liver, d-alanine, a rare enantiomer of alanine, shows circadian alteration, although the effect of d-alanine on glucose metabolism has not been explored. Here, we show that d-alanine acts on the circadian clock and affects glucose metabolism in the kidney.
Methods
The blood and urinary levels of d-alanine in mice were measured using two-dimensional high-performance liquid chromatography system. Metabolic effects of d-alanine were analyzed in mice and in primary culture of kidney proximal tubular cells from mice. Behavioral and gene expression analyses of circadian rhythm were performed using mice bred under constant darkness.
Results
d-Alanine levels in blood exhibited a clear intrinsic circadian rhythm. Since this rhythm was regulated by the kidney through urinary excretion, we examined the effect of d-alanine on the kidney. In the kidney, d-alanine induced the expressions of genes involved in gluconeogenesis and circadian rhythm. Treatment of d-alanine mediated glucose production in mice. Ex vivo glucose production assay demonstrated that the treatment of d-alanine induced glucose production in primary culture of kidney proximal tubular cells, where d-amino acids are known to be reabsorbed, but not in that of liver cells. Gluconeogenetic effect of d-alanine has an intraday variation, and this effect was in part mediated through circadian transcriptional network. Under constant darkness, treatment of d-alanine normalized the circadian cycle of behavior and kidney gene expressions.
Conclusions
d-Alanine induces gluconeogenesis in the kidney and adjusts the period of the circadian clock. Normalization of circadian cycle by d-alanine may provide the therapeutic options for life style–related diseases and shift workers.
Introduction
The circadian clock plays a key role in glucose metabolism.1–3 Worsened glucose metabolism and aberrant circadian rhythm are associated with type 2 diabetes pathogenesis,4 suggesting a potential role for nutrient levels in linking circadian rhythm and pathogenesis.5 Gluconeogenesis is a part of glucose metabolism that results in the glucose production from the intermediate metabolites, such as pyruvate and amino acids. Gluconeogenesis occurs mainly in the liver and kidney.6–8 In the kidney, gluconeogenesis occurs in the proximal tubules, where gluconeogenetic enzymes, such as glucose-6-phosphatase, are abundantly present.6,9 The glucose produced through gluconeogenesis is essential for maintaining blood glucose levels, particularly under fasting conditions. Gluconeogenesis in the kidney is activated on taking exercises10 or under pathological conditions, such as diabetes or metabolic acidosis.11,12 On the basis of the comparable tissue blood flow and enzymatic abundance, the kidney is considered to share the importance with the liver in gluconeogenesis.6,9 The enzymatic activities of gluconeogenesis have the circadian rhythm in the kidney and liver,13,14 although it remains unclear how gluconeogenesis in the kidney is connected with the circadian clock.
d-Alanine, a rare enantiomer of alanine, may link glucose metabolism and the circadian clock. Naturally occurring proteins consist entirely of l-amino acids, whereas d-amino acids are rare enantiomers whose presence in animals have only recently been demonstrated.15,16 Among d-amino acids, d-alanine is known to have a circadian rhythm in its blood and urine levels.17 Moreover, d-alanine is found in glucose metabolism-related tissues, such as the pancreatic islet, adrenal gland, and pituitary gland,18 suggesting a potential role for d-alanine levels in linking circadian rhythm and pathogenesis. The kidney regulates the levels of d-alanine and other d-amino acids through the balance between reabsorption at proximal tubules and urinary excretion.19–21 d-Amino acids have recently been shown to have several physiological functions,22–26 but the role of d-alanine and its regulation by the kidney in the context of glucose metabolism remains unclear.
In this study, we investigated how d-alanine affects glucose metabolism and identified the mode of action of d-alanine in gluconeogenesis in the kidney via modulation of the circadian clock.
Methods
Animals
C57BL/6 mice were purchased from SLC Inc. (Tokyo, Japan). d-amino acids oxidase (Dao)–deficient mice were described previously.27
For the analysis of circadian oscillations in d-alanine, male mice (age 7–10 weeks) were individually caged and housed in light–tight, ventilated closets within a temperature-controlled facility and humidity-controlled facility. Mice were entrained on a 12-hour light: 12-hour dark (LD) cycle for 2 weeks, and blood samples were collected from trunk after decapitation. Urine samples were collected through bladder puncture. For sleep deprivation, mice were gently touched with a soft swab to make them awake under light.28
For the intraperitoneal injection (ip), mice starved for 6 hours and then anesthetized with pentobarbital sodium (75 mg/kg ip; Kyoritsu Seiyaku, Tokyo, Japan) were administered with d-alanine (2801, Peptide Institute, Ibaraki, Japan) at the dose of 12.5 μmol/g. Preliminary, mice were administered with 12.5 μmol/g of d-alanine for three times with 12 hours of interval and confirmed the sufficient increase in the blood d-alanine level. Blood samples were collected transcardially, and kidneys were harvested after infusion of saline. For measuring blood levels of glucose and insulin, starved mice were administered with d-alanine without anesthesia, and blood samples were collected from tail veins.
All animal experiments were conducted in compliance with the Guidelines for Japanese Animal Protection and Management Law and performed under protocols approved by the Animal Research Committee of National Institutes of Biomedical Innovation, Health and Nutrition (NIBIOHN), Osaka University, and Kyoto University.
Sample Preparation for Two-Dimensional High-Performance Liquid Chromatography
Sample preparation for two-dimensional high-performance liquid chromatography (2D-HPLC) was performed, as previously described.18,29 In brief, 20-fold volumes of methanol were added to the sample, and 10 μl of the supernatant obtained from the methanol homogenate was used for 4-fluoro-7-nitro-2,1,3-benzoxadiazole (NBD) derivatization (0.5 μl of the plasma was used for the reaction). After drying the solution, 20 μl of 200 mM sodium borate buffer (pH, 8.0) and 5 μl of fluorescence labeling reagent (40 mM 4-fluoro-7-nitro-2,1,3-benzoxadiazole in anhydrous acetonitrile [MeCN]) were added and then heated at 60°C for 2 minutes. An aqueous 0.1% (v/v) trifluoroacetic acid (TFA) solution (75 μl) was added, and 2 μl of the reaction mixture was subjected to 2D-HPLC.
Determination of Alanine Enantiomers by 2D-HPLC
The enantiomers of alanine were quantified using the 2D-HPLC platform, as previously described,18,29 with the shape-fitting algorithm.30 In brief, the NBD derivatives of the amino acids were separated from numerous intrinsic substances using a reversed-phase (RP) column (Singularity RP column, 1.0 mm i.d.×50 mm; provided by KAGAMI Inc., Ibaraki, Japan) with the gradient elution using aqueous mobile phases containing MeCN and formic acid. To separately determine the d and l forms of alanine, the fraction of alanine was automatically collected using a multiloop valve, and transferred to enantioselective column (Singularity CSP-001S, 1.5 mm i.d.×75 mm; KAGAMI Inc.). The mobile phases are the mixed solution of methanol-MeCN containing formic acid, and the fluorescence detection of the NBD-amino acids was performed at 530 nm with excitation at 470 nm. The fluorescence detector uses two photomultiplier tubes to cover high and low ranges and enables simultaneous and accurate measurement of both abundant l-alanine and trace d-alanine in human samples. d-Alanine ratio was calculated as plasma d-alanine levels divided by the sum of l-alanine and d-alanine levels.
Calculation of Fractional Excretion
Fractional excretion (FE, %) was calculated from the clearance of substrate divided by creatinine clearance, as follows:
where Us and Ps represent urine and plasma levels of substrate, respectively. FE is the ratio of a substrate filtered by the kidney glomeruli that is excreted in the urine. Serum and urine creatinine were measured enzymatically, and d-alanine/l-alanine was measured as beforementioned using the same sample. Low and high FE indicates the dominance of tubular reabsorption and excretion, respectively.
RNA Sequencing
RNA was extracted using TRIzol (15596018, Thermo Fisher Scientific, Waltham). Library preparation was performed using a TruSeq stranded mRNA sample prep kit (Illumina, San Diego, CA) or NEBNext UltraDirectional RNA Library Prep Kit for Illumina according to the manufacturer's instructions. Sequencing was performed on an Illumina HiSeq 2500 platform in a 75-base single-end or on a NovaSeq6000 in a 150-base pair-end mode. Illumina Casava1.8.2 software was used for base calling. The sequenced reads were mapped to the mouse reference genome sequences (mm10) using TopHat version 2.0.13 in combination with Bowtie2 version 2.2.3 and SAMtools version 0.1.19. The fragments per kilobase of exon per million mapped fragments were calculated using Cufflinks version 2.2.1. Raw sequencing data are available from Gene Expression Omnibus under accession numbers GSE227752 and GSE227755.
Enrichment Analysis of Biologic Pathways, Gene Ontology Terms, and Transcription Factors
To interpret the RNA sequencing data and elucidate the pathways involved, we used TargetMine31 and Metascape, an integrated warehouse of human and mouse biologic data from data sources, such as Reactome and Kyoto Encyclopedia of Genes and Genomes. The genes upregulated in d-alanine–treated mice compared with vehicle-treated mice with a 1.5-fold cut-off change in average values were uploaded to TargetMine and Metascape. Enrichment analysis was performed based on the Kyoto Encyclopedia of Genes and Genomes functional hierarchy. Enrichment of the Reactome pathway and gene ontology terms32 was estimated by hypergeometric distribution and the inferred P values, which were further adjusted for multiple test corrections to control the false discovery rate using the Benjamini and Hochberg procedure. Visualization was performed using R (version 4.1.0). Thereafter, an enrichment analysis was performed to evaluate the effects of transcription factors on their binding target genes.33 We used a data mining platform, chromatin immunoprecipitation (ChIP)-Atlas,34 for visualization of genomic data on integrative genomics viewer and utilization of publicly available ChIP sequencing (ChIP-seq) data.
Extraction of Intergenic Interactions by Random Forest Method
Intergenic interactions were extracted using an iterative random forest.35 In this method, differences in gene expression patterns between the two groups were first learned by a random forest. The random forest method is a learning method called ensemble learning consisting of a majority decision of multiple decision trees.36 The decision tree is a hierarchical stack of multiple discriminants on the basis of gene expression levels that enables nonlinear learning. Then, genes and their interactions important for distinguishing between groups were extracted from the learned random forest through the statistical processing of the decision pathways of multiple decision trees. The iterative random forests were set as follows: 20 decision trees, two branches from the parent node to the child node, and three longest path. The results are shown as stability scores, whose higher level reflects a central role in intergenic interactions.
Insulin Clamp Experiment
The experiment with the insulin clamp was performed under conscious and unrestrained conditions, as described previously.37 In brief, after the mice had fasted starting at 6 am, the catheters were inserted into the mice and were connected to PE20 tubes (Instech, Plymouth Meeting, PA) at 8:30 am. The mice were returned to a cage where their free movement was possible. Thereafter, we started bolus priming of 2.5 μCi, followed by a 0.05 μCi/min infusion of 3-[3H]-glucose (Catalog No. ART0124; Muromachi Yakuhin, Tokyo, Japan); this was maintained for 2 hours as the basal period at 9 am. A pancreatic insulin clamp with priming of 15 mU/kg, followed by infusion of 1.5 mU/kg per minute human insulin (Catalog No. I9278; Sigma-Aldrich, St. Louis, MO) mixed with 3 μg/kg per minute somatostatin (Catalog No. H1490; Bachem, Bubendorf, Switzerland) was then initiated at 9 am that lasted for 2 hours. During this clamping period, a 20% glucose solution was infused at a variable rate to maintain the plasma glucose concentrations within 80–110 mg/dl. The glucose infusion rate was fixed at 0.33 mg/min for all mice during the first 30 minutes and then modified as needed thereafter. The blood glucose concentrations were measured 120, 60, 30, 15, and 0 minutes before the insulin clamping was started and at 30 minutes and after a 10-minute interval until 2 hours. Blood samples were obtained at t=−30, −15, 0, 80, 90, 100, 110, and 120 minutes and centrifuged at 3000 rpm for 1 minute for determining the 3-[3H]-glucose–specific activity. The plasma was then stored, deproteinized with Ba(OH)2 and ZnSO4, and dried. Red blood cells were returned to the mice to prevent anemia. At t=120 minutes, hematocrit was confirmed to be >40%, and the mice were euthanized by isoflurane inhalation and cervical subluxation.
Ex Vivo Glucose Production Assay
Proximal tubular epithelial cells (PTECs) were isolated from 6-week-old male mice, as described previously.38 After anesthetization and perfusion with saline, the kidneys from mice were immediately removed and placed in cold PBS (4°C). After the renal capsule was removed, the kidney was cut sagittally, and the cortex was finely minced and transferred to Hanks' Balanced Salt Solution containing 0.25 mg/ml Liberase (Roche, Penzberg, Germany) for 30 minutes at 37°C with gentle shaking. After digestion, suspended cells were washed in glucose-free HEPES buffer consisted of 5.4 mM KCl, 143 mM NaCl, 1.8 mM CaCl2, 0.9 mM NaH2PO4, 0.8 mM MgSO4, and 5 mM HEPES buffer, pH 7.4, for 30 minutes at 37°C with gentle shaking. Then, PTECs were incubated with glucose-free HEPES buffer containing either l-alanine or d-alanine at 1 mM or indicated levels in the legend for 1 hour. Usually, the doses of approximately 10 mM of d-amino acids were selected in cell culture experiments to eliminate the effect of l-amino acids, which are present abundantly.39,40 In this experiment, we reduced the dose of d-alanine as we performed in l-amino acid-free medium. Glucose formation was calculated from the glucose levels in the medium measured by the glucose oxidase method using the Amplex Red and horseradish peroxidase (A22189 Invitrogen, Manassas, CA).
Measurement of Pyruvate
Samples were prepared from PTECs cultured in glucose-free HEPES buffer containing either l-alanine or d-alanine. Supernatants or cell lysates harvested in PBS were analyzed. Separately, we measured the concentration of pyruvate produced from PBS containing 2.5 units/ml purified DAO (A5222, Sigma), 100 µM flavin adenine dinucleotide (Nacalai Tesque, Japan) and several concentrations of d-alanine (1, 10, 100, 1000 µM) after 1-hour incubation at 37°C.
For the measurement of pyruvate, 10 μl of the sample was mixed with 5 μl of water and 85 μl of MeCN and then centrifuged at 800 g at 4°C for 10 minutes. The obtained supernatant (10 μl) was mixed with 20 μl of an MeCN solution containing 5 mM 4-nitro-7-piperazino-2,1,3-benzoxadiazole and 10 mM 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride. After the mixture was incubated at 25°C for 60 minutes, an aqueous TFA solution (0.1% [v/v]; 220 μl) was added and 100 μl of the reaction mixture was subjected to high-performance liquid chromatography for fluorescence detection. Pyruvate was separated as an NBD derivative using the tandemly connected reversed-phase columns (Singularity RP18, 1.0 mm i.d.×250 mm) at 40°C. For the mobile phase, an aqueous solution containing 15% ethanol and 0.05% TFA was used, and the flow rate was 25 µl/min. Fluorescence detection was performed at 530 nm with excitation at 470 nm. After confirming the pyruvate production, pyruvate assay kit (2390092, AAT Bioquest, CA) was used.
Assessment of Glucose-6-Phosphatase and dao Activity
Glucose-6-phosphatase activity was assessed, as described previously with some modifications.41 In brief, the kidney cortex was homogenized in ice–cold saline, and the supernatant was collected after centrifugation for 10 minutes at 3000 g. Ten microliter of the supernatants was mixed with 150 μl of 0.1 M malate buffer containing 200 mM glucose-6-phosphate and was incubated at 37°C for 10 minutes. After centrifugation for 10 minutes at 4500 rpm, 50 μl of Taussky-Shorr Color Reagent (ammonium molybdate solution with ferrous sulfate) was applied. The amount of inorganic phosphate liberated from glucose-6-phosphate in the reaction solution was measured by spectroscopy with the absorbance at 660 nm.
The DAO activity was assessed, as described previously with some modifications.42 In brief, the kidney cortex was homogenized in 10 mM boric acid buffer, and the supernatant was collected after centrifugation at 4°C for 10 minutes at 5000 g. The supernatant was ultra-filtrated at 4°C for 40 minutes at 5000 g. The supernatant was reacted in 50 mM boric acid buffer containing 0.1 mM flavin adenine dinucleotide, 0.005 units horseradish peroxidase, 0.2 mM 10-acetyl-3,7-dihydroxyphenoxazine, and 4 mM d-alanine for 30 minutes. Reaction solutions were measured by absorbance at 570 nm. We diluted DAO (0.001, 0.005, 0.01, 0.05, 0.1, 0.5, and 1 U/ml, A5222, Sigma) for the standard curve.
Glucose and Insulin Tolerance Test
To mice that had been fasted for 6 hours, either glucose (2 g/kg body weight, Nacalai Tesque, 16806-25) or insulin (0.83 U/kg body weight, Humulin R; Lilly, Indianapolis, IN) was intraperitoneally injected for the glucose or insulin tolerance test, respectively. Blood samples were collected from the tail vein before administering glucose or insulin and after 30, 60, and 120 minutes. Levels of glucose in whole blood and insulin in plasma were determined using a blood glucose monitoring unit (Glutest Sensor Neo, Sanwa Kagaku Kenkyusho, Aichi, Japan) and insulin ELISA kit (M1104, Morinaga, Kanagawa, Japan), respectively.
Circadian Rhythmicity Determination
To identify the rhythmic transcripts in time series data, we used the following two common algorithms: ARSER43 and JTK_CYCLE.44 The phase of the rhythmic features was determined as an output measure. These analyses were performed using the MetaCycle platform of the R software package.45
Quantitative PCR (qPCR) and Western blot
qPCR and Western blot analyses were performed, as described previously.46 Data were normalized with mRNA level of β-actin. The sequences of the primers used were as follows: Pck1-F, 5′-gatgggcatatctgtgctgg-3´; Pck1-R, 5′-cagccacccttcctccttag-3´; Fbp1-F, 5′-caccgcgatcaaagccatct-3´; Fbp1-R, 5′-aggtagcgtaggacgacttca-3´; G6pc-F, 5′-tttccccaccaggtcgtggct-3´; G6pc-R, 5′-cccattctggccgctcacacc-3´; G6pc3-F, 5′-agtaccagctacggctccaa-3´; G6pc3-R, 5′-tagggtgcaagtcgaggaac-3´; Arntl-F, 5′-cctaatctccagggcagcagat-3´; Arntl-R, 5′-tccagtcttggcatcaatgagt-3´; Cry1-F, 5′-cgaatgaatgcaaactccct-3´; Cry1-R, 5′-aaaaatttcacgccacaggag-3´; Per1-F, 5′-cccagctttacctgcagaag-3´; Per1-R, 5′-agctggggcagtttcctatt-3´; Per2-F, 5′-gcggatgctcgtggaatctt-3´; Per2-R, 5′-gctccttcagggtccttatc-3´; β-actin-F, 5′-tgacaggatgcagaaggaga-3´; β-actin-R, 5′-acatctgctggaaggtggac-3′.
ChIP-qPCR
ChIP was performed using the ChIP Kit (ab500, Abcam), as described previously.47 In brief, the kidney sample was fixed in 1.5% paraformaldehyde solution at room temperature for 15 minutes, and fixation was stopped using 0.125 M glycine. The sample was added to ice–cooled PBS and ground for mild strokes. After centrifugation, the ground sample was resuspended in an FA lysis buffer (50 mM HEPES-KOH, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, and protease inhibitors) and sonicated for 10–20 cycles using the Bioruptor (UCD-250, Cosmo Bio Co., Ltd., Tokyo, Japan) on ice–cool condition to shear DNA to an average fragment size of 200–1000 bp. After diluting each sample with ChIP buffer, we added the primary antibody to the samples and incubated them under rotational agitation overnight and adsorbed protein G magnetic beads. Finally, immunoprecipitated chromatin was analyzed using a genomic DNA purification kit and subjected to qPCR analysis. The sequences of the primers used were as follows: G6pc3-F, 5′-aaggcaagtttctcgcacat-3´; G6pc3-R, 5′-tggaccatttgggaggaagg-3′.
Antibodies and Reagents
Antibodies against anti-Bmal (ab93806) were from Abcam (Cambridge, the United Kingdom). Reagents used were insulin (I9278-5ML, Sigma-Aldrich), l-lactate (129-02666, Fujifilm Wako Chemicals Corporation, Osaka, Japan), and l-alanine (2701, Peptide Institute).
Constant Dark Experiment and Behavior Analysis
For the constant dark experiments, individually caged adult male mice (age 7–10 weeks) were entrained on a LD cycle for at least 2 weeks and transferred to the constant darkness (DD) facility. For behavioral activity monitoring, the diet and water were replaced with an alanine-free diet generated using the FR-2 formula (Oriental Kobo, Tokyo, Japan) and water 1 week before transfer to the DD. Two weeks later, the water was replaced with one dissolved with 0.5% d-alanine. Locomotor activity was detected with passive (pyroelectric) infrared sensors (FA-05 F5B; Omron), and data were analyzed using ClockLab software (Actimetrics) developed on MATLAB (Mathworks).48
Statistics
Sample size was selected based on previously published data or preliminary experiments. Animals in the same litter were randomly assigned to different treatment groups. No animals or data were excluded from the analysis. Continuous variables are presented as means SEM, or as medians and ranges when indicated. Numbers of experiments are provided in the figure legends. Either two-tailed t test (paired or unpaired) or one-way or two-way ANOVA with Dunnett's post hoc test was used. Correlation was assessed using Pearson's correlation coefficient. Data were analyzed using GraphPad Prism or STATA. Statistical significance was defined as P < 0.05.
Results
Blood Level of d-Alanine Forms a Clear Circadian Rhythm
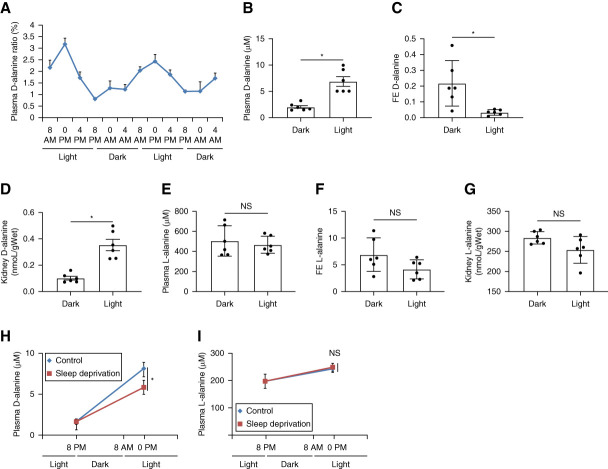
To investigate intrinsic circadian oscillations in d-alanine, we analyzed d-alanine levels in blood from mice bred under a LD cycle condition (L, 8 am–8 pm; D, 8 pm–8 am) (Figure 1A and Supplemental Figure 1, A and B). Mice are known to be asleep in the light and awake in the dark. d-Alanine levels in blood clearly show an intrinsic circadian rhythm, similarly to rats.17 The mean blood d-alanine ratios, calculated as d-alanine levels per total alanine level, at 0 pm and 8 pm were 3.18% and 0.80%, and the mean blood levels of d-alanine were 9.2 and 2.6 µM, respectively. On the other hand, the blood level of l-alanine did not show the clear circadian rhythm. Alanine has the chiral-specific circadian rhythm.
Figure 1.
d-Alanine has an intrinsic circadian rhythm, which is affected by urinary excretion and sleep. (A) Plasma d-alanine ratio of mice bred under a LD cycle condition (L, 8 am–8 pm; D, 8 pm–8 am) n=4. (B) Plasma levels, (C) urinary FE, and (D) levels in kidney of d-alanine in mice in the light (0 pm) or dark (8 pm). (E) Plasma levels, (F) urinary FE, and (G) levels in kidney of l-alanine in mice in the light (0 pm) or dark (8 pm). (H) Levels of d-alanine and (I) l-alanine in plasma from mice that were either asleep or deprived for sleep from 8 am to 0 pm Data, means±SEM. Statistic (B–I) two-tailed unpaired t test. *P < 0.05. FE, fractional excretion; LD, 12-hour light:12-hour dark; NS, not significant.
Kidney and Sleep Regulate the Circadian Rhythm of d-Alanine
The kidney is the key regulator of blood d-alanine levels due to urinary excretion.19,20,49,50 Generally, d-amino acids in blood are filtered through the glomeruli of the kidney, followed by the reabsorption into the proximal tubules, and the rest are excreted. The urinary FE ratio has a clear chiral selectivity. In case of serine, 85% of d-serine is excreted into urine, while <1% of l-serine is urinary excreted.19 Because of this high FE, the blood d-serine level is determined by the kidney and reflects the glomerular filtration rate. To determine whether the kidney contributes to d-alanine circadian oscillations, we measured the urinary FE of d-alanine. When mice were in the light (0 pm), the blood d-alanine levels increased (Figure 1B), whereas the FE of d-alanine decreased (Figure 1C). Similarly, d-alanine levels in the kidney increased due to higher reabsorption at 0 pm (Figure 1D). These behaviors of d-alanine were opposite when mice in the dark (8 pm; Figure 1, B–D). By contrast, levels of l-alanine did not show the circadian rhythm (Figure 1, E–G).
To verify that the circadian clock affects blood d-alanine levels, we deprived sleep from mice (Figure 1H). d-Alanine levels were low in the dark (8 pm), and this level increased in the light (0 pm). Compared with control mice, d-alanine levels did not increase in sleep-deprived mice, suggesting that sleep restores d-alanine levels. Sleep deprivation did not affect the blood level of l-alanine (Figure 1L). Overall, urinary excretion from the kidney and sleep contributes in forming the intrinsic circadian rhythm of d-alanine.
d-Alanine Activates Signals of Gluconeogenesis and Circadian Rhythm
Since kidney uptakes d-alanine with the circadian rhythm, we investigated the physiological functions of d-alanine in the kidney. To eliminate the effect of circadian variability of intrinsic d-alanine concentrations, we used d-alanine at relatively high dose to induce the sufficiently higher blood level. We injected d-alanine to mice intraperitoneally at the dose of 12.5 μmol/g, after confirming the sufficient increase in the blood d-alanine level (Supplemental Figure 2). To identify the early signals that d-alanine mediates, we obtained gene expression profiles of the kidney from mice 2 hours after administration of d-alanine once. Pathway enrichment analysis identified that d-alanine is involved in a number of biologic processes. These include developmental, metabolic, rhythmic, immunologic process, longevity regulating pathway, necroptosis, bladder cancer, infection, and cellular signals (Figure 2A). Deep learning analysis using an iterative random forest algorithm identified a subset of genes with a higher stability score, reflecting their central roles in intergenic interactions in the kidneys of d-alanine treated mice (Figure 2B). The top gene we identified was G6pc3, a subset of gluconeogenic genes.51 Indeed, d-alanine specifically affects metabolic pathways, such as type II diabetes mellitus and fructose metabolism (Figure 2A). In addition, three genes, G6pc3, Mbnl2, and Cpeb1, are associated with the circadian clock.52,53 Volcano plots indicated that d-alanine upregulated both gluconeogenic genes (Pck1 and G6pc) and circadian genes (Per1 and Bhlhe40) (Figure 2C). Gene ontology analysis found that d-alanine affected rhythmic process (Supplemental Figure 3). Transcription factor enrichment analysis identified Cry2, a key circadian regulator, as being responsible for the gene network induced by d-alanine (Figure 2D). Overall, d-alanine possesses the potential to activate gluconeogenesis and modulate circadian rhythm signals.
Figure 2.
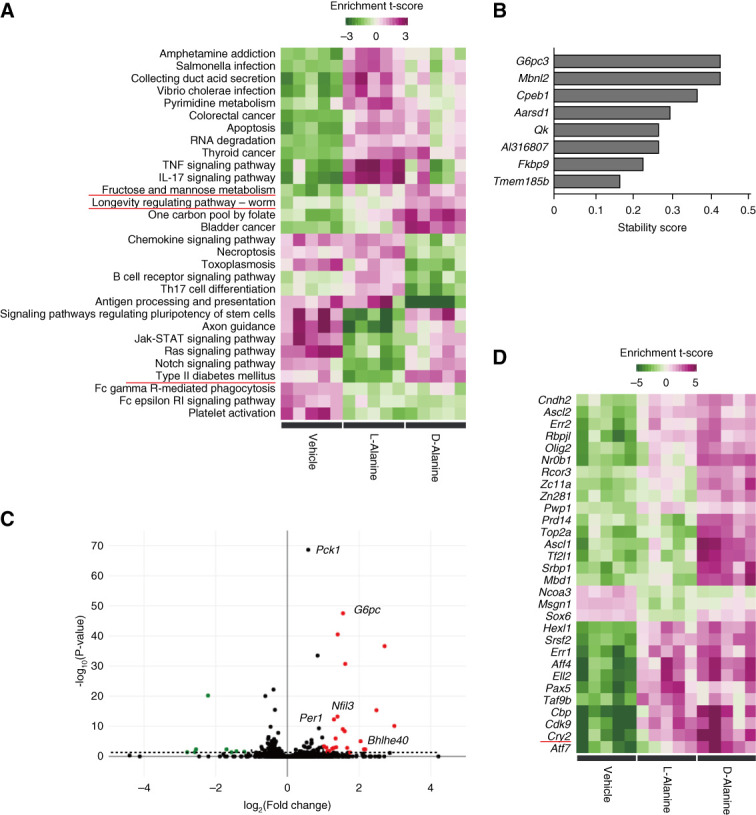
d-Alanine activates the signals of gluconeogenesis and circadian rhythm in the kidney. (A) Pathway enrichment analysis of the kidney from mice that were intraperitoneally treated with 12.5 μmol/g of l-alanine and d-alanine. Kidneys were harvested 2 hours after treatment. (B) Stability score of genes whose intergenic interactions were dominant in mice treated with d-alanine versus vehicle-treated mice, learned using an iterative random forest algorithm. (C) Volcano plot. Red and green dots indicate increase (>0) and decrease (<0) in expression with statistical significance. The symbols of genes involved in gluconeogenesis pathway and circadian rhythm are listed. (D) Gene sets of transcriptional factor enrichment analysis. Estimated effects of transcriptional factors are presented in heatmaps as enrichment t score.
d-Alanine Induces Gluconeogenesis in the Kidney
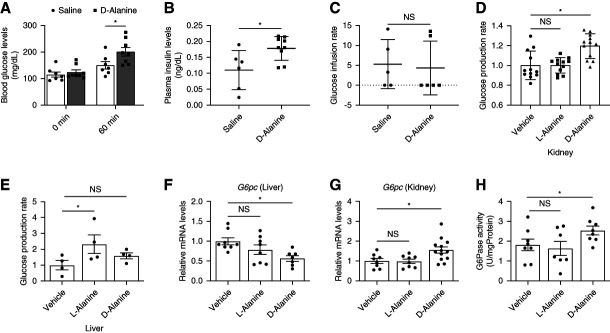
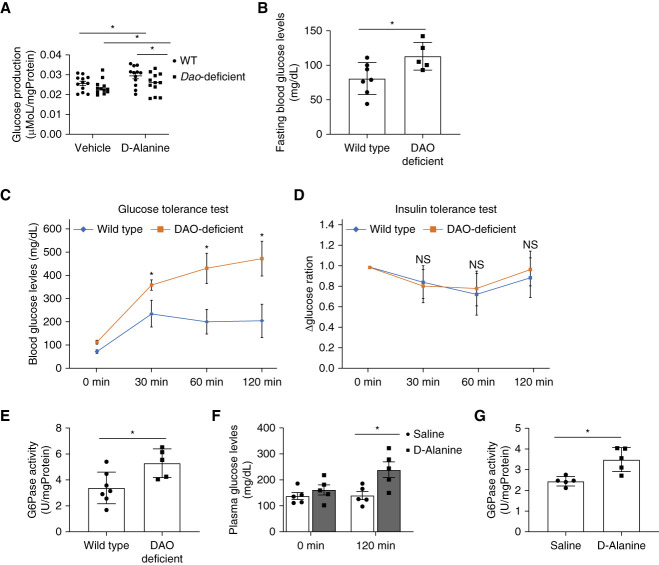
We investigated the gluconeogenetic capacity of d-alanine. d-Alanine injection increased the levels of glucose and insulin in blood (Figure 3, A and B). To monitor insulin resistance, we performed an insulin clamp test under physiological conditions. d-Alanine did not affect insulin resistance (Figure 3C), suggesting that d-alanine increases the blood glucose level by increasing glucose production, leading to the secretion of insulin.
Figure 3.
d-Alanine induces glucose production in the kidney. Levels of (A) glucose and (B) insulin in plasma from mice intraperitoneally administered with saline or d-alanine at the dose of 12.5 μmol/g. Levels of insulin were measured 60 minutes after injection, n=7–9. (C) Glucose infusion rate during the euglycemic–hyperinsulinemic clamp test (1.5 mU/kg body weight/min) in mice treated with intraperitoneally injected saline and d-alanine, n=5–6. (D and E) Glucose production rates of (D) isolated proximal tubular or (E) liver cells cultured in glucose-free medium containing 1 mM of l-alanine or d-alanine for 1 hour. Data are expressed as fold change relative to mean value of the vehicle. (F and G) Relative G6pc expression in the (F) liver and (G) kidney and (H) G6pc enzymatic activity in kidney harvested from mice at 1 hour after intraperitoneal injection of l-alanine and d-alanine, n=8–11. Data, means±SEM. Statistic (A) two-way ANOVA and (B and C) two-tailed unpaired t test and (D–H) one-way ANOVA. *P < 0.05.
Gluconeogenesis occurs in the kidney and liver.6–8 We directly measured the glucose production rate by d-alanine in the kidney and liver. Since the analysis using cell lines is inappropriate as they lack the key gluconeogenetic enzymes,9 we established an ex vivo method to monitor the glucose production rate using kidney tubular cells collected from the kidney or liver cells. Ex vivo analysis indicated chiral selective induction of glucose production in both the kidney and liver (Figure 3, D and E); d-Alanine induced glucose production exclusively in the kidney, whereas l-alanine did so in the liver. d-Alanine induces the expression of G6pc mRNA, which encodes the key gluconeogenesis enzyme, within the kidney, but not in the liver (Figure 3, F and G). In the kidney, d-alanine activated G6pc activity (Figure 3H). Therefore, d-alanine has the capacity to induce gluconeogenesis in the kidney through gene expression.
d-Alanine also Serves as a Substrate for Gluconeogenesis
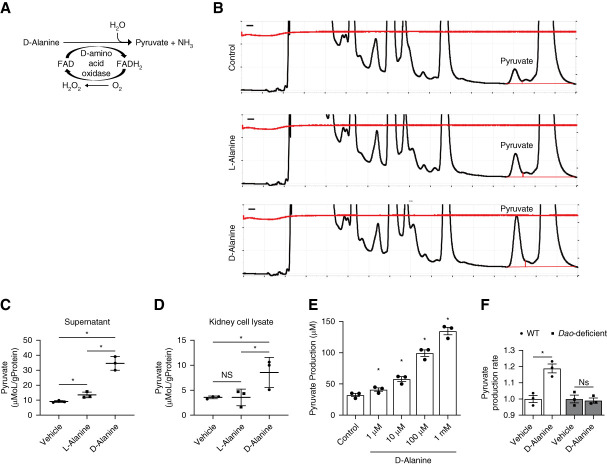
d-Amino acids are known to be metabolized by DAO, which is expressed mainly in the proximal tubules of the kidney.54 On oxidation, d-amino acids are converted to imino acids. We postulated that pyruvate, a gluconeogenesis substrate, is produced by oxidation of d-alanine on the basis of the structure (Figure 4A). We monitored pyruvate levels by detecting fluorescent-labeled pyruvate using high-performance liquid chromatography (Figure 4B). d-Alanine treatment increased the levels of pyruvate in both supernatant and cell lysates from ex vivo cultured kidney (Figure 4, C and D). Pyruvate production is dependent on DAO since purified DAO can produce pyruvate (Figure 4E). In line with this, d-alanine did not induce pyruvate production in kidneys from Dao-deficient mice (Figure 4F).
Figure 4.
d-Alanine is metabolized to pyruvate by dao. (A) Enzymatic oxidation of d-amino acids by DAO. (B) Representative chromatograms of pyruvate in cell lysate of isolated proximal tubular cells with glucose-free medium containing 200 µM l-alanine or d-alanine for 1 hour, obtained by a HPLC system. Pyruvate production in (C) supernatant or (D) cell lysate of isolated proximal tubular cells in glucose-free medium containing 200 µM l-alanine or d-alanine for 1 hour. Data were normalized with weights of cell lysate protein. (E) Pyruvate production from d-alanine incubated with purified dao for 1 hour, n=3. (F) Pyruvate production rates of proximal tubular cells from wild-type mice and Dao deficiency mice cultured in glucose-free medium containing 1 mM d-alanine for 1 hour. Pyruvate was measured using kit in (E and F). Data, means±SEM. Statistic (C and D) two-tailed unpaired t test and (E and F) one-way ANOVA. *P < 0.05. DAO, d-amino acid oxidase; FAD, flavin adenine dinucleotide; FADH2, dihydroflavine-adenine dinucleotide; HPLC, high-performance liquid chromatography; WT, wild type.
d-Alanine Activates Gluconeogenesis as a Mediator in Dao-Deficient Mice
We investigated whether the effects of d-alanine as a substrate is sufficient to induce gluconeogenesis. For this purpose, we used Dao-deficient mice. We treated d-alanine to ex vivo cultured kidney from Dao-deficient mice. Gluconeogenesis was also activated in Dao-deficient mice after d-alanine treatment albeit at lower levels than in wild-type kidney (Figure 5A). Therefore, d-alanine as a substrate is insufficient to induce gluconeogenesis, and d-alanine also serves as a mediator in Dao-deficient mice.
Figure 5.
d-Alanine induces glucose production in the kidney of Dao deficiency mouse. (A) Glucose production rates of proximal tubular cells from wild-type mice and Dao deficiency mice cultured in glucose-free medium containing 1 mM d-alanine for 1 hour. (B) The level of plasma glucose in wild-type mice or Dao deficiency mice after fasting for 6 hours. (C) Glucose and (D) insulin tolerance test and (E) G6pc enzymatic activity in wild-type or Dao deficiency mice, n=5–7. (F) Levels of glucose in plasma and (G) G6pc enzymatic activity in the kidney from Dao deficiency mice that were received intraperitoneal injection of saline or d-alanine, n=5. Data, means±SEM. Statistic (A, B, and E–G) two-tailed unpaired t test and (C and D) two-way ANOVA. *P < 0.05.
While we intended to confirm this finding in vivo, we detected the abnormal glucose metabolism in Dao-deficient mice. Both the fasting glucose level and glucose tolerance of Dao-deficient mice were impaired, while their insulin sensitivity was reserved (Figure 5, B–D). G6pc activity in Dao-deficient mice was also elevated (Figure 5E). The glucose metabolism of Dao-deficient mice was largely impaired. Increased level of d-alanine due to the lack of oxidation18 may affect the glucose metabolism in Dao-deficient mice.
We treated d-alanine to Dao-deficient mice. Treatment of d-alanine increased the blood level of glucose (Figure 5F). G6pc activity of kidney from d-alanine-treated Dao-deficient mice was further elevated when compared with that from vehicle-treated Dao-deficient mice (Figure 5G). d-Alanine still has the capacity to induce gluconeogenesis of kidney in Dao-deficient mice.
Intraday Variations of the Effects of d-Alanine on Gluconeogenesis in the Kidney
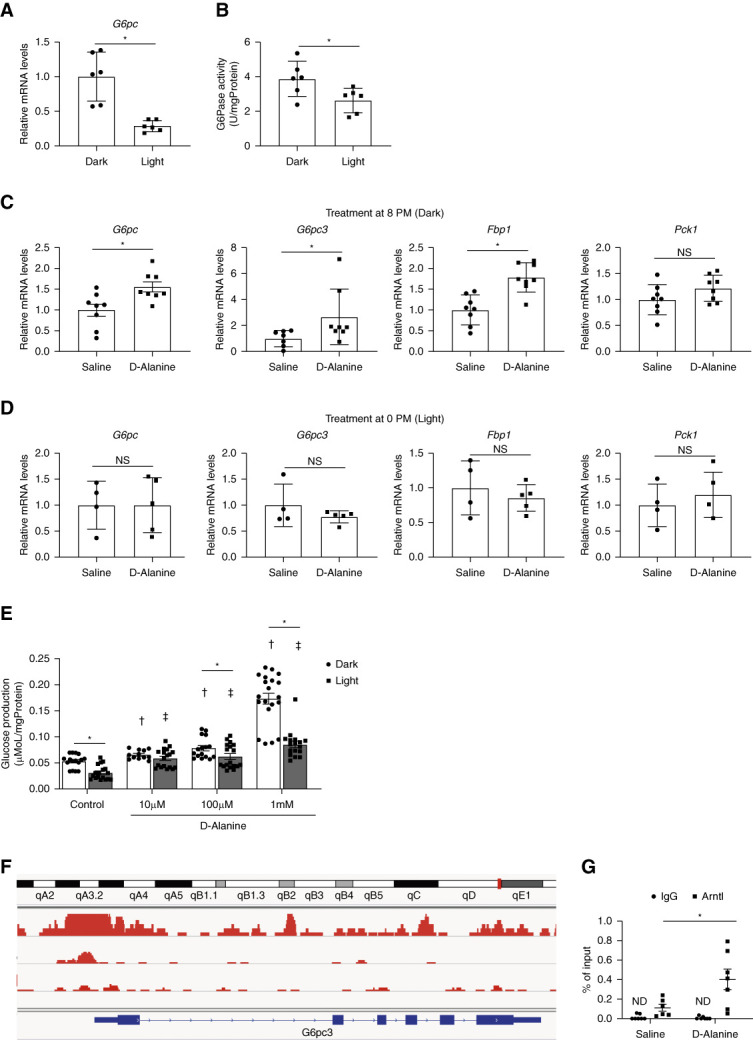
d-Alanine has a clear intrinsic circadian rhythm, whereas it activates gluconeogenesis. Since gluconeogenesis in the kidney has a circadian rhythm,13 we wondered whether metabolic effects of d-alanine in the kidney could be sensitive to the circadian rhythm. We investigated whether intraday variations of d-alanine treatment affect the gluconeogenesis in the kidney and glucose metabolism. The mRNA level of G6pc was higher in mice under darkness at 8 pm than that in mice under light at 0 pm (Figure 6A). In line with this, the kidney's gluconeogenic capacity was higher at 8 pm in waking mice, as indicated by G6pc activity (Figure 6B).13 Kidneys of mice under darkness are likely to be d-alanine-sensitive and d-alanine treatment at 8 pm, when blood d-alanine level is low, but not at 0 pm, when blood d-alanine level is high, augmented the mRNA expression of G6pc (Figure 6, C and D). On treatment at darkness, d-alanine induces G6pc expression.
Figure 6.
d-Alanine mediates circadian transcriptional factors for gluconeogenesis. (A) G6pc expression and (B) glucose-6-phosphatase (G6Pase) activity in the kidney from mice in the dark or light. (C and D) Expression levels of gluconeogenetic genes (G6pc, G6pc3, Fbp1, and Pck1) in the kidney from mice intraperitoneally treated with d-alanine at either (C) 8 pm or (D) 0 pm (E) glucose production assay from d-alanine in isolated proximal tubular cells from mice in the dark or light. (F) Arntl-binding promoter regions of G6pc3 gene in mice kidney. The genomic data were visualized on the IGV browser platform using ChIP-Atlas. (G) ChIP-qPCR analysis of kidneys from mice treated with d-alanine at 8 pm, n=4–6. Data, means±SEM. Statistic (A−D and G) two-tailed unpaired t test and (E) two-way ANOVA. *P < 0.05 versus treatment-matched vehicle control or dark group; †P < 0.05 versus vehicle-treated dark group; ‡P < 0.05 versus vehicle-treated light group. ChIP, chromatin immunoprecipitation; IGV, integrative genomics viewer; ND, not detected.
To further examine the intraday variations directly, we measured the glucose production rate by d-alanine ex vivo (Figure 6E). In line with Figure 6B, the basal gluconeogenetic activity was lower in kidneys from mice under light than those from mice under dark. In the kidneys from mice under light, the activation of gluconeogenesis by d-alanine was confirmed in its low dose. In the kidneys from mice under dark, gluconeogenesis was also activated by d-alanine in its low dose, whereas it was further enhanced in its high dose. The effects of d-alanine show clear intraday variations.
d-Alanine Enriches Circadian Transcriptional Factor on Promoter Regions of G6pc
We investigated the mode of intraday regulation of G6pc expression by d-alanine. Since Cry2 was identified as the key transcriptional factor in the gene network induced by d-alanine (Figure 2D), we performed the transcriptional analysis of circadian genes on G6pc gene. Analysis of public ChIP-seq data using ChIP-Atlas revealed the enrichment of Arntl, an upstream regulator of Cry2 (Figure 2D), in the promoter region of G6pc3 (Figure 6F). ChIP-qPCR analysis confirmed that d-alanine induced the enrichment of Arntl at the G6pc3 promoter region when it was administered at 8 pm, when gluconeogenesis was active (Figure 6G). d-Alanine was suggested to regulate the gluconeogenic genes through the circadian transcriptional factors (Figure 2D).
d-Alanine Acts on the Circadian Rhythm
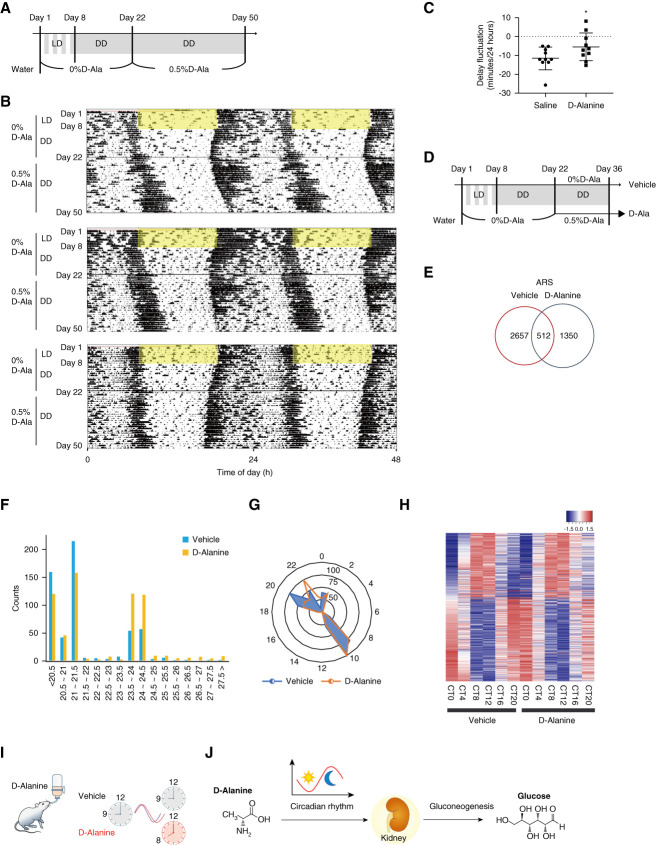
d-Alanine can affect circadian rhythm at the level of gene expression (Figure 2, A–D, Figure 6G, and Supplemental Figure 3). We investigated the effects of d-alanine on the circadian response in vivo. For this purpose, we determined the behavioral effect of d-alanine. Mice were fed an alanine-free water and diet for 21 days and then placed with water containing 0.5% d-alanine, afterward (Figure 7A). At day 8, mice were kept in DD. The blood level of d-alanine was confirmed to surpass its intraday variation sufficiently through the oral treatment (Supplemental Figure 4, A and B). After d-alanine administration, we observed a daily delay in activity time (Figure 7B). As animals were kept in DD, the onset of daily behavioral activity shifted forward approximately 10 minutes every circadian cycle, but the magnitude of this shift was reduced by d-alanine treatment (Figure 7C and Supplemental Figure 4C). Treatment of d-alanine normalized the circadian cycle in DD.
Figure 7.
d-Alanine normalizes circadian cycles in DD. (A) Experimental protocol. Mice were entrained on a LD cycle for at least 2 weeks and then transferred to DD. One week before behavioral measurement (day 1), mice began to be fed an alanine-free diet and water. Two weeks after transferring to DD, water was replaced with a solution of 0.5% d-alanine in water. (B) Representative actograms from mice subjected to the experimental protocol in (A). Data are shown in double-plotted format. Each horizontal line represents 48 hours; the second 24-hour period is plotted to the right and below the first. The periods of light-on are indicated by yellow. (C) Delay fluctuation per 24 hours was calculated from actograms in (B), n=10. (D) Experimental protocol. Mice were treated as in (A) except the duration of treatment with 0.5% d-alanine in water was shortened to 14 days. (E) Venn diagram representing rhythmic genes of the kidney from saline or d-alanine–treated mice. Rhythmic genes were identified using ARS algorithm, n=4. (F) The number of common rhythmic genes identified using ARS algorithm in kidney from vehicle-treated and d-alanine–treated mice. (G) Radar plots and (H) heatmap depicting the phase analysis of common rhythmic genes identified using ARS algorithm in the kidney from vehicle-treated and d-alanine–treated mice. In radar plots, the numbers of rhythmic genes were denoted, and phases were adjusted for 24 hours. (I) Schematic summary representing the effect of d-alanine on circadian rhythm. (J) Schematic summary of this study. d-Alanine affects circadian rhythm and induces gluconeogenesis at the transcriptional level in the kidney. d-Alanine is a key metabolite that connects gluconeogenesis and circadian rhythm. Data, means±SEM. Statistic, two-tailed unpaired t test. *P < 0.05. D, dark; DD, constant darkness; L, light.
We investigated whether d-alanine can normalize the circadian rhythm in gene expression levels. Mice were fed an alanine-free water and diet for 21 days and then placed with either water containing 0.5% d-alanine or vehicle (Figure 7D). At day 8, mice were kept in DD. The kidneys were harvested 14 days after treatment with d-alanine, and the potential effects of d-alanine treatment on the kidney circadian gene expression profiles were examined. By using ARSER, an algorithm for detecting periodic oscillations,43 we identified 512 periodic transcripts common to both d-alanine–treated mice and vehicle-treated control mice that had an altered period in d-alanine–treated mice (Figure 7, E–H). Figure 7E shows the number of common rhythmic genes identified using ARSES algorithm in the kidney from vehicle-treated and d-alanine–treated mice. Genes are tended to show the longer periodic oscillations in d-alanine–treated mice (Figure 7F). Radar plots and heatmap of rhythmic genes confirmed the shift of periodic oscillations by the treatment with d-alanine (Figure 7, G and H). The periods were longer in mice treated with d-alanine (21.30 hours for vehicle versus 22.05 hours for d-alanine, Figure 7G). Analysis using another rhythm-detecting algorithm, JTK,44 yielded similar results (Supplemental Figure 4, D–F). d-Alanine is therefore likely to have a period-lengthening effect on both circadian locomotor activity rhythm and the kidney gene mRNA expression rhythm (Figure 7I).
Discussion
In this study, we demonstrated that d-alanine links gluconeogenesis in the kidney and the circadian clock (Figure 7J). d-Alanine activates gluconeogenesis through circadian transcriptional network. Exogenous d-alanine modifies organismal circadian behavior and kidney gene expression, whereas the d-alanine level in the blood is regulated by urinary excretion from the kidney. d-Alanine, a molecule with a clear circadian rhythm, maintains glucose metabolism by regulating gluconeogenesis and circadian rhythm.
The level of d-alanine in the blood has a circadian rhythm with a clear sign curve. To our knowledge, this rhythm is a fairly pronounced variation among known metabolites. For example, melatonin shows a circadian rhythm with an on–off patterns.55 The clear circadian rhythm of d-alanine enables the identification of daily peaks and bottoms. d-Alanine is a natural nutrient that originates from daily food or the intestinal microbiota.25 The kidney regulates the circadian rhythm of d-alanine through tubular reabsorption. After glomerular filtration, approximately 70%–80% of d-alanine is reabsorbed in the proximal tubules as assessed by creatinine-based FE, whereas remainder is excreted in the urine.19 Since d-alanine accumulates in the blood of patients with a reduced glomerular filtration rate, the d-alanine level in the blood serves as a kidney biomarker.16,19 In mice, d-alanine levels in the blood decrease at night when they are active, whereas they increase during day when they are inactive or fall asleep.17 We found that sleep increases d-alanine levels in the blood, potentially by suppressing urinary excretion. A nocturnal shift in the urinary excretion of sodium is reportedly associated with variations in BP.56 Similar to the handling of electrocytes, the kidney modulates the nocturnal urinary excretion of d-alanine to form a clear circadian rhythm.
d-Alanine activates gluconeogenesis as a signal mediator through circadian network. d-Alanine induces the expressions of circadian clock genes. d-Alanine also induces expressions of gluconeogenetic genes through the activation of circadian transcriptional factors. The clear circadian rhythm of d-alanine may also take part in this regulation. Oxidation of d-alanine to pyruvate could contribute in gluconeogenesis. This multifacet interaction between d-alanine and circadian rhythm controls the gluconeogenesis in the kidney.
This d-alanine circadian interaction maintains the circadian cycle in vivo. d-Alanine normalizes the aberrant circadian rhythm by shifting the phases and correcting the shortening of behavioral activity and kidney gene expression rhythm in constant dark. Intrinsic circadian rhythm of d-alanine may also prevent the onset of circadian-related diseases, such as diabetes and cardiovascular diseases. Moreover, treatment with d-alanine is a potential therapeutic option for the life style–related diseases or shift workers through the correction of behavioral rhythm.
The effect of d-alanine on gluconeogenesis has a clear intraday variation. Treatment of d-alanine enhanced glucose production in mice kidneys from both dark and light groups, whereas the sensitivity to d-alanine was different in dose dependency (Figure 6E). Although speculative, d-alanine is likely involved in a multifacet regulatory system of gluconeogenesis in the kidney. Under darkness, d-alanine may potentially induce more glucose production. In this condition, G6pc activity is high despite the low blood d-alanine level (Figure 6B). The reduction of blood d-alanine level was mediated by urinary excretion to potentially suppress further induction of gluconeogenesis when gluconeogenetic activity is high. d-Alanine may play the complementary role in the regulation of gluconeogenesis.
Besides glucose metabolism, d-alanine is involved in a broad range of biologic processes. These include circadian-related processes, such as immunologic, metabolic processes, and cancer. d-Alanine may link circadian rhythm and associated biologic processes through the circadian gene network as seen in this study of gluconeogenesis. Recently, d-alanine is found to be protective against viral infections and inflammatory bowel disease,24,57 and these effects may be explained by d-alanine-circadian interaction. d-Alanine and its related signals are likely involved in a wide range of diseases. The study of d-alanine, whose existence in human body was not even known until recently, will unravel the pathophysiology and lead to the therapeutic options for the metabolic and immunologic diseases.
This study has some limitations. Since the results of this study solely depend on rodent studies, it is necessary to examine the effects of d-alanine in human for further applications. We identified the key role of d-alanine in transcriptional network, and further studies are necessary to identify the detailed molecular mechanisms. We confirmed that d-alanine activates gluconeogenesis in ex vivo kidney even at low dose; however, it was difficult to examine gluconeogenetic effect at lower dose in vivo because of the presence of intrinsic d-alanine and strong DAO activity in the kidney. Despite these, the effect of d-alanine as a circadian modulator suggests a strong potential for the future clinical implementations.
In conclusion, d-alanine is a metabolite that connects multiple biologic processes with circadian rhythm. d-Alanine induces gluconeogenesis and maintains circadian rhythm through the regulation of circadian transcriptional network. With a clear intrinsic circadian rhythm, d-alanine forms the circadian–gluconeogenesis axis in the kidney. With its association with a wide range of biologic processes, the study of d-alanine will provide key pathophysiologic concepts in broad range of diseases.
Supplementary Material
Acknowledgments
We thank Hiroshi Imoto, Eiichi Negishi, Shoto Ishigo (KAGAMI Inc.), and Y. Tanaka (NIBIOHN) for technical support.
Footnotes
See related article, “D-Amino Acids in Kidney Diseases” on pages 173–174.
Present address: Mitsuaki Yoshino, MBX Techno, Osaka, Japan.
Disclosures
A. Hesaka reports the following: Ownership Interest: KAGAMI Inc. Y. Isaka reports the following: Consultancy: Kirin Co. Ltd. and Sanwa Kagaku Kenkyusyo Co. Ltd.; Research Funding: Kirin Co. Ltd.; Advisory or Leadership Role: Kirin Co. Ltd. and Sanwa Kagaku Kenkyusyo Co. Ltd.; and Speakers Bureau: Astellas Pharma Inc., AstraZeneca plc, Kirin Co. Ltd., Kissei Pharmaceutical Co. Ltd., Mitsubishi Tanabe Pharma, Otsuka Pharmaceutical Co. Ltd., and Sanwa Kagaku Kenkyusyo Co. Ltd. E. Kawakami reports the following: Consultancy: The Phage; Ownership Interest: BIC Camera, McDonald's Holdings Company (Japan), Ltd., Mitsubishi Paper Mills Limited, and Takara Tomy; and Honoraria: Chugai Pharmaceutical Co., Ltd. and Lundbeck. T. Kimura reports the following: Ownership Interest: KAGAMI Inc.; Research Funding: KAGAMI Inc. and Kyowa Hakko Kirin Co., Ltd; and Patents or Royalties: KAGAMI Inc. M. Mita reports the following: Ownership Interest: KAGAMI Inc.; Patents or Royalties: KAGAMI Inc.; Advisory or Leadership Role: KAGAMI Inc.; and Other Interests or Relationships: JSN. M. Nakane reports the following: Ownership Interest: KAGAMI Inc.; and Patents or Royalties: KAGAMI Inc. Y. Tsukamoto reports the following: Ownership Interest: KAGAMI Inc. M. Yoshino reports the following: Ownership Interest: MBX Co., Ltd; and Advisory or Leadership Role: MBX Co., Ltd. All remaining authors have nothing to disclose.
Funding
T. Kimura: Japan Agency of Medical Research and Development (JP21gm5010001), Manpei Suzuki Diabetes Foundation (NA), and Japan Society for the Promotion of Science (22K19414). Y. Isaka: Japan Society for the Promotion of Science (21H02935).
Author Contributions
Conceptualization: Tomonori Kimura, Shinsuke Sakai.
Data curation: Tomonori Kimura, Shinsuke Sakai, Youichi Tanaka.
Formal analysis: Masao Doi, Eiryo Kawakami, Tomonori Kimura, Shinsuke Sakai, Youichi Tanaka, Mitsuaki Yoshino.
Funding acquisition: Yoshitaka Isaka, Tomonori Kimura.
Investigation: Masao Doi, Atsuko Fukushima, Kenji Hamase, Atsushi Hesaka, Chin-Ling Hsieh, Yoshitaka Isaka, Tomonori Kimura, Shihoko Kimura-Ohba, Hiroyo Matsumura, Masashi Mita, Maiko Nakane, Daisuke Okuzaki, Hiraku Ono, Shinsuke Sakai, Youichi Tanaka, Yusuke Tsukamoto, Kotaro Yokote.
Methodology: Masao Doi, Kenji Hamase, Eiryo Kawakami, Tomonori Kimura, Hiraku Ono, Shinsuke Sakai, Youichi Tanaka.
Project administration: Masao Doi, Yoshitaka Isaka, Tomonori Kimura, Kotaro Yokote.
Supervision: Masao Doi, Kenji Hamase, Yoshitaka Isaka, Tomonori Kimura.
Validation: Masao Doi, Kenji Hamase, Yoshitaka Isaka, Tomonori Kimura, Shinsuke Sakai.
Visualization: Masao Doi, Tomonori Kimura, Shinsuke Sakai, Youichi Tanaka.
Writing – original draft: Tomonori Kimura, Shinsuke Sakai.
Data Sharing Statement
Original data created for the study are or will be available in a persistent repository upon publication. All data is included in the manuscript and/or supporting information. Raw Data/Source Data. Gene Expression Omnibus (GEO). Sequencing data were deposited into GEO under the accession no. GSE227752 and GSE227755.
Supplemental Material
This article contains the following supplemental material online at http://links.lww.com/KN9/A422.
Supplemental Figure 1. d-Alanine has an intrinsic circadian rhythm.
Supplemental Figure 2. Plasma level of d-alanine in mice.
Supplemental Figure 3. d-Alanine modifies transcripts of circadian and gluconeogenic genes in kidney.
Supplemental Figure 4. d-Alanine normalizes circadian cycles in constant darkness.
References
- 1.Van Cauter E, Blackman JD, Roland D, Spire JP, Refetoff S, Polonsky KS. Modulation of glucose regulation and insulin secretion by circadian rhythmicity and sleep. J Clin Invest. 1991;88(3):934–942. doi: 10.1172/JCI115396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134(5):728–742. doi: 10.1016/j.cell.2008.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asher G, Sassone-Corsi P. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161(1):84–92. doi: 10.1016/j.cell.2015.03.015 [DOI] [PubMed] [Google Scholar]
- 4.Qian J, Scheer F. Circadian system and glucose metabolism: implications for physiology and disease. Trends Endocrinol Metab. 2016;27(5):282–293. doi: 10.1016/j.tem.2016.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330(6009):1349–1354. doi: 10.1126/science.1195027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerich JE, Meyer C, Woerle HJ, Stumvoll M. Renal gluconeogenesis: its importance in human glucose homeostasis. Diabetes Care. 2001;24(2):382–391. doi: 10.2337/diacare.24.2.382 [DOI] [PubMed] [Google Scholar]
- 7.Owen OE, Felig P, Morgan AP, Wahren J, Cahill GF. Liver and kidney metabolism during prolonged starvation. J Clin Invest. 1969;48(3):574–583. doi: 10.1172/JCI106016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stumvoll M, Meyer C, Perriello G, Kreider M, Welle S, Gerich J. Human kidney and liver gluconeogenesis: evidence for organ substrate selectivity. Am J Physiol. 1998;274(5):E817–E826. doi: 10.1152/ajpendo.1998.274.5.E817 [DOI] [PubMed] [Google Scholar]
- 9.Krebs H, Bennett D, Gasquet P, Gascoyne T, Yoshida T, Yoshida T. Renal Gluconeogenesis: the effect of diet on the gluconeogeneic capacity of rat-kidney-cortex slice. Biochem J. 1963;86(1):22–27. doi: 10.1042/bj0860022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogers SA, Karl IE, Hammerman MR. Growth hormone directly stimulates gluconeogenesis in canine renal proximal tubule. Am J Physiol. 1989;257(5 Pt 1):E751–E756. doi: 10.1152/ajpendo.1989.257.5.E751 [DOI] [PubMed] [Google Scholar]
- 11.Jenssen T, Nurjhan N, Consoli A, Gerich JE. Failure of substrate-induced gluconeogenesis to increase overall glucose appearance in normal humans: demonstration of hepatic autoregulation without a change in plasma glucose concentration. J Clin Invest. 1990;86(2):489–497. doi: 10.1172/JCI114735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longshaw ID, Pogson CI. The effect of steroids and ammonium chloride acidosis on phosphoenolpyruvate carboxykinase in rat kidney cortex. I. Differentiation of the inductive process and characterization of enzyme activities. J Clin Invest. 1972;51(9):2277–2283. doi: 10.1172/JCI107037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagai K, Suda M, Yamagishi O, Toyama Y, Nakagawa H. Studies on the circadian rhythm of phosphoenolpyruvate carboxykinase: III. circadian rhythm in the kidney. J Biochem. 1975;77(6):1249–1254. doi: 10.1093/oxfordjournals.jbchem.a130322 [DOI] [PubMed] [Google Scholar]
- 14.Reinke H, Asher G. Circadian clock control of liver metabolic functions. Gastroenterology. 2016;150(3):574–580. doi: 10.1053/j.gastro.2015.11.043 [DOI] [PubMed] [Google Scholar]
- 15.Nagata Y. Involvement of D-amino acid oxidase in elimination of D-serine in mouse brain. Experientia. 1992;48(8):753–755. doi: 10.1007/BF02124295 [DOI] [PubMed] [Google Scholar]
- 16.Kimura T Hamase K Miyoshi Y, et al. Chiral amino acid metabolomics for novel biomarker screening in the prognosis of chronic kidney disease. Sci Rep. 2016;6:26137. doi: 10.1038/srep26137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morikawa A, Hamase K, Miyoshi Y, Koyanagi S, Ohdo S, Zaitsu K. Circadian changes of d-alanine and related compounds in rats and the effect of restricted feeding on their amounts. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;875(1):168–173. doi: 10.1016/j.jchromb.2008.04.004 [DOI] [PubMed] [Google Scholar]
- 18.Miyoshi Y, Hamase K, Tojo Y, Mita M, Konno R, Zaitsu K. Determination of d-serine and d-alanine in the tissues and physiological fluids of mice with various d-amino-acid oxidase activities using two-dimensional high-performance liquid chromatography with fluorescence detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(24):2506–2512. doi: 10.1016/j.jchromb.2009.06.028 [DOI] [PubMed] [Google Scholar]
- 19.Hesaka A Sakai S Hamase K, et al. D-Serine reflects kidney function and diseases. Sci Rep. 2019;9(1):5104–5108. doi: 10.1038/s41598-019-41608-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawamura M Hesaka A Taniguchi A, et al. Measurement of glomerular fi ltration rate using endogenous D -serine clearance in living kidney transplant donors and recipients. EClinicalMedicine. 2022;43:101223. doi: 10.1016/j.eclinm.2021.101223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taniguchi A Kawamura M Sakai S, et al. D-asparagine is an ideal endogenous molecule for measuring the glomerular filtration rate. Kidney Int Rep. 2023;8(6):1192–1200. doi: 10.1016/j.ekir.2023.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kartvelishvily E, Shleper M, Balan L, Dumin E, Wolosker H. Neuron-derived D-serine release provides a novel means to activate N-methyl-D-aspartate receptors. J Biol Chem. 2006;281(20):14151–14162. doi: 10.1074/jbc.M512927200 [DOI] [PubMed] [Google Scholar]
- 23.Hesaka A Tsukamoto Y Nada S, et al. D-serine mediates cellular proliferation for kidney remodeling. Kidney360. 2021;2(10):1611–1624. doi: 10.34067/KID.0000832021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura-Ohba S Asaka MN Utsumi D, et al. D-Alanine as a biomarker and a therapeutic option for severe influenza virus infection and COVID-19. Biochim Biophys Acta Mol Basis Dis. 2023;1869(1):166584. doi: 10.1016/j.bbadis.2022.166584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasabe J Miyoshi Y Rakoff-Nahoum S, et al. Interplay between microbial d-amino acids and host d-amino acid oxidase modifies murine mucosal defence and gut microbiota. Nat Microbiol. 2016;1(10):16125. doi: 10.1038/nmicrobiol.2016.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakade Y Iwata Y Furuichi K, et al. Gut microbiota – derived D-serine protects against acute kidney injury. JCI Insight. 2018;3(20):e97957. doi: 10.1172/jci.insight.97957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konno R, Yasumura Y. Mouse mutant deficient in D-amino acid oxidase activity. Genetics. 1983;103(2):277–285. doi: 10.1093/genetics/103.2.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuji S Brace CS Yao R, et al. Sleep–wake patterns are altered with age, Prdm13 signaling in the DMH, and diet restriction in mice. Life Sci Alliance. 2023;6(6):e202301992. doi: 10.26508/lsa.202301992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamase K Miyoshi Y Ueno K, et al. Simultaneous determination of hydrophilic amino acid enantiomers in mammalian tissues and physiological fluids applying a fully automated micro-two-dimensional high-performance liquid chromatographic concept. J Chromatogr A. 2010;1217(7):1056–1062. doi: 10.1016/j.chroma.2009.09.002 [DOI] [PubMed] [Google Scholar]
- 30.Hamase K Ikeda T Ishii C, et al. Determination of trace amounts of chiral amino acids in complicated biological samples using two-dimensional high-performance liquid chromatography with an innovative “shape-fitting” peak identification/quantification method. Chromatography. 2018;39(3):147–152. doi: 10.15583/jpchrom.2018.019 [DOI] [Google Scholar]
- 31.Chen YA, Tripathi LP, Mizuguchi K. TargetMine, an integrated data warehouse for candidate gene prioritisation and target discovery. PLoS One. 2011;6(3):e17844. doi: 10.1371/journal.pone.0017844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashburner M Ball C Blake J, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawakami E, Nakaoka S, Ohta T, Kitano H. Weighted enrichment method for prediction of transcription regulators from transcriptome and global chromatin immunoprecipitation data. Nucleic Acids Res. 2016;44(11):5010–5021. doi: 10.1093/nar/gkw355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oki S Ohta T Shioi G, et al. ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 2018;19(12):e46255. doi: 10.15252/embr.201846255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basu S, Kumbier K, Brown JB, Yu B. Iterative random forests to discover predictive and stable high-order interactions. Proc Natl Acad Sci U S A. 2018;115(8):1943–1948. doi: 10.1073/pnas.1711236115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breiman L. Random forests. Machine Learn. 2001;45(1):5–32. doi: 10.1023/a:1010933404324 [DOI] [Google Scholar]
- 37.Sakai G Inoue I Suzuki T, et al. Effects of the activation of three major hepatic Akt substrates on glucose metabolism in male mice. Endocrinology. 2017;158(8):2659–2671. doi: 10.1210/en.2016-1969 [DOI] [PubMed] [Google Scholar]
- 38.Kimura T Takabatake Y Takahashi A, et al. Autophagy protects the proximal tubule from degeneration and acute ischemic injury. J Am Soc Nephrol. 2011;22(5):902–913. doi: 10.1681/ASN.2010070705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki M Sujino T Chiba S, et al. Host-microbe cross-talk governs amino acid chirality to regulate survival and differentiation of B cells. Sci Adv. 2021;7(10):eabd6480. doi: 10.1126/sciadv.abd6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okada A Nangaku M Jao TM, et al. D-serine, a novel uremic toxin, induces senescence in human renal tubular cells via GCN2 activation. Sci Rep. 2017;7(1):11168. doi: 10.1038/s41598-017-11049-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swanson MA, Swanson A. Phosphatases of liver. J Biol Chem. 1950;184(2):647–659. doi: 10.1016/S0021-9258(19)50999-1 [DOI] [PubMed] [Google Scholar]
- 42.Rosini E, Caldinelli L, Piubelli L. Assays of D-amino acid oxidase activity. Front Mol Biosci. 2017;4:102. doi: 10.3389/fmolb.2017.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang R, Su Z. Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. Bioinformatics. 2010;26(12):168–174. doi: 10.1093/bioinformatics/btq189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient non-parametric algorithm for detecting rhythmic components in genome-scale datasets. J Biol Rhythms. 2010;25(5):372–380. doi: 10.1177/0748730410379711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB. MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics. 2016;32(21):3351–3353. doi: 10.1093/bioinformatics/btw405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakai S Yamamoto T Takabatake Y, et al. Proximal tubule autophagy differs in type 1 and 2 diabetes. J Am Soc Nephrol. 2019;30(6):929–945. doi: 10.1681/ASN.2018100983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Meter M Kashyap M Rezazadeh S, et al. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun. 2014;5:5011. doi: 10.1038/ncomms6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doi M Shimatani H Atobe Y, et al. Non-coding cis-element of Period2 is essential for maintaining organismal circadian behaviour and body temperature rhythmicity. Nat Commun. 2019;10(1):2563. doi: 10.1038/s41467-019-10532-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimura T, Hesaka A, Isaka Y. d-amino acids and kidney diseases. Clin Exp Nephrol. 2020;24(5):404–410. doi: 10.1007/s10157-020-01862-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakade Y Iwata Y Sakai N, et al. Increased levels of oral Streptococcus-derived d-alanine in patients with chronic kidney disease and diabetes mellitus. Sci Rep. 2022;12(1):21773. doi: 10.1038/s41598-022-26175-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guionie O, Clottes E, Stafford K, Burchell A. Identification and characterisation of a new human glucose-6-phosphatase isoform. FEBS Lett. 2003;551(1-3):159–164. doi: 10.1016/s0014-5793(03)00903-7 [DOI] [PubMed] [Google Scholar]
- 52.Kim JS Coon SL Weller JL, et al. Muscleblind-like 2: circadian expression in the mammalian pineal gland is controlled by an adrenergic-cAMP mechanism. J Neurochem. 2009;110(2):756–764. doi: 10.1111/j.1471-4159.2009.06184.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kojima S, Sher-Chen EL, Green CB. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 2012;26(24):2724–2736. doi: 10.1101/gad.208306.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koga R, Miyoshi Y, Sakaue H, Hamase K, Konno R. Mouse d-amino-acid oxidase: distribution and physiological substrates. Front Mol Biosci. 2017;4:82. doi: 10.3389/fmolb.2017.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGeer EG, McGeer PL. Circadian rhythm in pineal tyrosine hydroxylase. Science. 1966;153(3731):73–74. doi: 10.1126/science.153.3731.73 [DOI] [PubMed] [Google Scholar]
- 56.Fujii T Uzu T Nishimura M, et al. Circadian rhythm of natriuresis is disturbed in nondipper type of essential hypertension. Am J Kidney Dis. 1999;33(1):29–35. doi: 10.1016/S0272-6386(99)70254-4 [DOI] [PubMed] [Google Scholar]
- 57.Umeda S Sujino T Miyamoto K, et al. D-amino acids ameliorate experimental colitis and cholangitis by inhibiting growth of proteobacteria: potential therapeutic role in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2023;16(6):1011–1031. doi: 10.1016/j.jcmgh.2023.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data created for the study are or will be available in a persistent repository upon publication. All data is included in the manuscript and/or supporting information. Raw Data/Source Data. Gene Expression Omnibus (GEO). Sequencing data were deposited into GEO under the accession no. GSE227752 and GSE227755.