Abstract
Brf is the TFIIB-related component of Saccharomyces cerevisiae RNA polymerase III transcription initiation factor IIIB (TFIIIB). An extensive set of Brf fragments has been examined for the abilities to assemble the TFIIIB-DNA complex and recruit RNA polymerase III to accurately initiate transcription. The principal TFIIIB-assembly function of Brf was found to be contributed by a C-proximal segment spanning amino acids 435 to 545, while the principal transcription-directing function was contributed by a segment of its N-proximal, TFIIB-homologous half. The diverse activities of Brf were also reconstituted from combined fragments. The fragments spanning amino acids 1 to 282 and 284 to 596 were found to assemble into TFIIIB-DNA and TFIIIC-TFIIIB-DNA complexes that were very stable, transcriptionally highly active, and indistinguishable (by in vitro footprinting) from complexes formed with intact Brf. The proximities of the individual halves of split Brf to DNA were extensively mapped by photochemical cross-linking of the TFIIIB-DNA complex. We also identified sites of interaction of Brf fragments with TATA-binding protein (TBP), taking advantage of a recently completed mutational analysis of the TBP surface. The constraints established by these analyses specify a global model of the functional segments of Brf and how they fit into the structure of the TFIIIB-DNA complex.
Eukaryotic nuclear and archaeal transcription apparatuses share an essential common feature: the RNA polymerase (pol) is brought to the transcriptional start site by a DNA-bound assembly of transcription initiation proteins. The promoter-marking assembly of archaeal RNA polymerases and the core promoter-marking assembly of eukaryotic pol II each consist of two proteins: TATA-binding protein (TBP) and the phylogenetically related transcription factor B (TFB) or TFIIB (for archaeal or pol II transcription systems, respectively) (43).
The situation differs only slightly for transcription by pol III, whose promoter-marking assembly, TFIIIB, is composed of three subunits: TBP, Brf (named for its relatedness to TFIIB) (7, 11, 33), and B" (18, 24). In Saccharomyces cerevisiae, the three components of TFIIIB are held together by a more stable interaction between Brf and TBP (which together make up the B′ component of TFIIIB) and by a weaker interaction between TBP-DNA-bound Brf and B" (17, 22); a weaker direct B"-TBP interaction has also been noted (12, 37). Each of these TFIIIB components is required for all pol III transcription in yeast. Homologs of Brf and B" serve as components of the pol III system of humans (42, 44) but are not ubiquitously required. It appears instead that specialized alternative promoter-marking assemblies participate in different classes of human pol III promoters (16, 34, 42, 48).
The homology relationship of Brf with TFIIB is confined to the N-terminal half of the much larger Brf; similarities between the C-proximal half of Brf and proteins of the pol II transcription initiation apparatus have not been discerned, at least at the level of the primary amino acid sequence (8, 11, 44).
We and others are undertaking molecular genetic dissections of TFIIIB (12, 21, 30, 37, 38, 40; see also references 10 and 27). The experiments presented below analyze the TFIIIB assembly properties and transcriptional capabilities of a large set of Brf fragments. Functional Brf can be assembled from selected pairs of these fragments, effectively splitting the protein at diverse sites and generating internal deletions without the constraint of joining together domains or regions of Brf that might normally be well separated in space. The interactions of the individual fragments of split Brf with TBP and with serial locations along the DNA-binding site of TFIIIB have also been mapped. The results of these analyses suffice for the specification of a global map of protein and DNA interactions in the TFIIIB-promoter complex.
MATERIALS AND METHODS
DNA.
Plasmids pU6LboxB and pTZ1 have been described elsewhere (26, 47). The 75-bp TA-30-B6 DNA probe for electrophoretic mobility shift assay (EMSA) was generated by annealing the two synthetic oligonucleotides shown in Fig. 1a, one of which was 5′ end labeled with T4 polynucleotide kinase and [γ-32P]ATP. The 240-bp TA-30 variant SUP4 tRNA gene probe for EMSA and methidiumpropyl-EDTA (MPE)-Fe(II) footprinting was generated by PCR as described previously (19, 30). The 59-bp photochemical cross-linking probes were generated essentially as described elsewhere (21) with the primers shown in Fig. 4a.
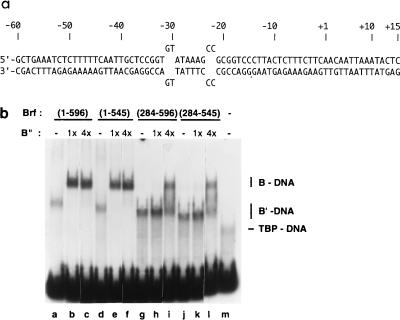
FIG. 1.
Effects of N- and C-terminal truncations in Brf on the TFIIIC-independent assembly of B′ (Brf plus TBP)-DNA and TFIIIB-DNA complexes. (a) The 75-bp TA-30-B6 probe for EMSA for TFIIIC-independent assembly of TBP, Brf, and B". (b) EMSA for TFIIIB-DNA complex formation. Ten femtomoles of the TA-30-B6 probe was incubated with 10 fmol of TBP, 50 fmol of full-length Brf or the indicated truncated Brf, and B" (1× = 50 fmol), as indicated above each lane. Reaction conditions were otherwise as specified in Materials and Methods, and electrophoresis was done in Mg2+-containing native gels. Control reactions (not shown) demonstrated that Brf-DNA complexes did not form in the absence of TBP. TBP-DNA, B′-DNA, and TFIIIB-DNA complexes are identified at the right.
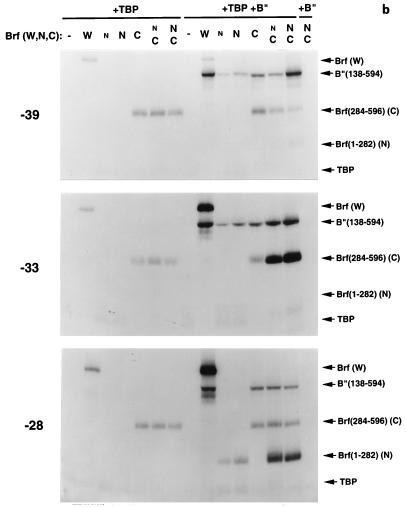
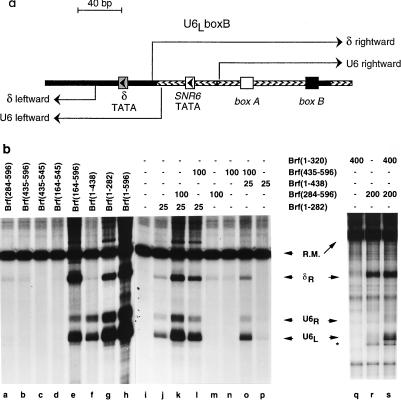
FIG. 4.
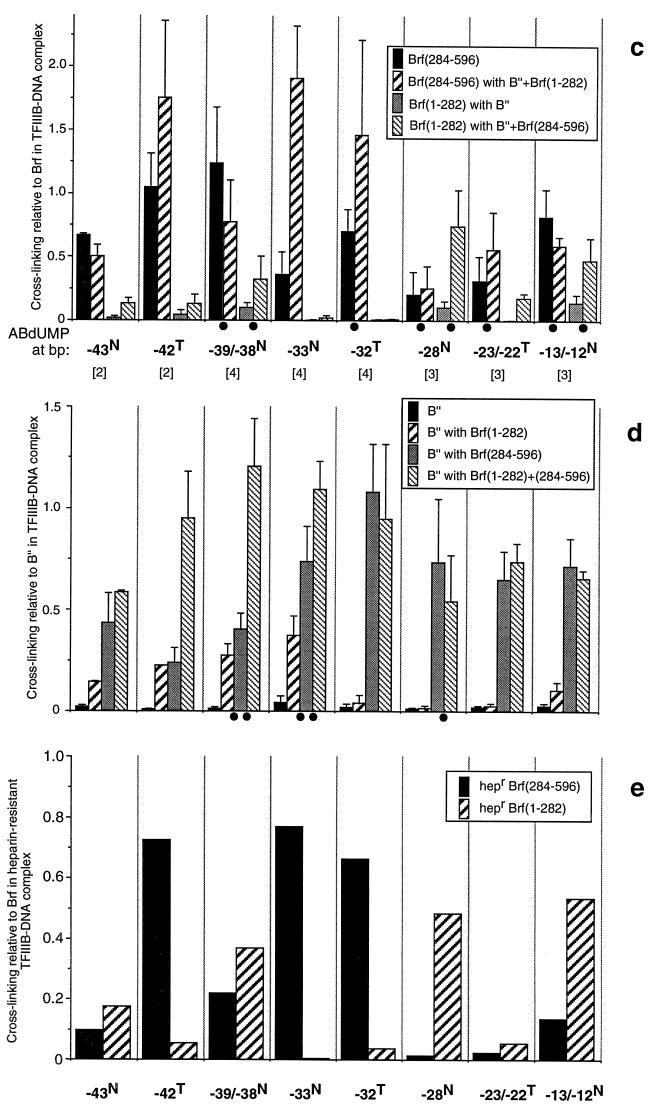
Mapping the disposition of Brf fragments in transcription factor complexes by photochemical cross-linking: Brf split at amino acid 283. (a) DNA probes. The sequence of the SNR6 gene probe with a TGT mutant TATA box is shown. Locations at which dTMP was replaced by the photoactive ABdUMP are circled. The primers used to generate these probes are indicated in capital letters above and below the sequence. These primers were extended by incorporating ABdUMP(u) and the indicated radioactive nucleotide (lowercase letters, underlined) and subsequently extended to full length. Probes are designated by the location of ABdUMP and by the photoactive strand (N, nontranscribed; T, transcribed). (b) Analysis of cross-linking to bp −39/−38, −33, and −28 in the nontranscribed strand. The proteins present with DNA are indicated above the lanes, and cross-linked proteins are identified at the right. The use of TBPm3 in conjunction with the TGT-mutated TATA box allows unidirectional and TFIIIC-independent assembly of TFIIIB on this DNA. Samples contained 400 fmol of TBPm3, 700 fmol of intact Brf (designated W), 150 or 300 fmol Brf(1-282) (designated n or N, respectively), 150 fmol Brf(284-596) (designated C), and 100 fmol B"(138-594), as indicated. (c to e) Quantitative analysis of cross-linking efficiencies along the DNA-binding site. The eight probes indicated between panels c and d were analyzed two, three, or four times, as indicated in brackets. Cross-linking was quantified, normalized to the level of the reference protein indicated on the ordinate of each panel, and averaged. Error bars indicate the average deviation (for experiments repeated twice) or the standard error of the mean (for experiments repeated three or four times). Because the normalization contributed substantially to these variations, rank orders that were consistently observed are also designated (• between columns) below the abscissas of panels c and d (for experiments repeated three or four times only and when not otherwise obvious). (c) Cross-linking efficiencies of Brf fragments separately and in combination relative to those of intact Brf in TFIIIB-DNA complexes; (d) cross-linking efficiencies of B" in TFIIIB-DNA complexes assembled with Brf fragments separately or in combination, normalized to that of B" in the TFIIIB-DNA complex with intact Brf; (e) cross-linking efficiencies of Brf fragments relative to that of intact Brf in heparin-challenged TFIIIB-DNA complexes reconstituted with Brf(1-282) and Brf(284-596).
Proteins.
TFIIIC, pol III, recombinant TBP, recombinant TBPm3, Brf(317-596) (the Brf fragment spanning amino acids 317 to 596), recombinant wild-type B" (native purification), and recombinant B"(137-594) were purified and assayed as described elsewhere (21, 25, 30, 47). Full-length Brf containing hexahistidine tags at both N and C termini was generated and purified as follows. The expression vector pET21b was modified by inserting TATG(CAC)6CCCATG between the NdeI and BamHI sites (top strand shown; the bottom strand provided the appropriate overhangs), generating pET21b-H6-Nco with an NcoI site now overlapping the BamHI site. The C-terminally His6-tagged Brf gene contained on an NcoI/BlpI fragment from pSH360 (11) was then ligated into the equivalent sites of pET21b-H6-Nco and subsequently transformed into Escherichia coli BL21(DE3)pLysS. Induced cells (33 g) were lysed under native conditions as described for B"(138-594) (30). The lysate was sedimented at 22,000 × g for 1 h, the pellet was resuspended and solubilized in buffer A (50 mM Tris-Cl [pH 8.0], 0.1% [wt/vol] Tween 20, 7 mM β-mercaptoethanol, 10% [vol/vol] glycerol, 6 M guanidine HCl, 0.5 mM phenylmethylsulfonyl fluoride, 1 μg each of leupeptin and pepstatin per ml) and then sedimented at 22,000 × g for 30 min to remove insoluble material, and the supernatant fluid was applied to a 16-ml Ni-nitrilotriacetic acid (NTA) agarose column. The column was developed sequentially with buffers containing 100 mM NaxHxPO4, 7 M deionized urea, β-mercaptoethanol, and protease inhibitors as in buffer A (32 ml of each) at pHs 6.7, 6.3, 5.9, 5.7, 5.5, 5.1, and 4.7, respectively. The major contaminants [Brf(165-596) and other singly hexahistidine-tagged truncated forms of Brf] eluted at pH 5.9 and 5.7. Doubly hexahistidine-tagged Brf, which eluted at pH 5.5 and 5.1, was adjusted to pH 7.8 with Tris base and then dialyzed for 6 to 12 h sequentially against buffer D500 (30) with 10 μM ZnSO4 also containing 3, 1.5, 0.75, and 0 M urea.
N- and C-terminal deletions of Brf were generated by PCR amplification from pSH360 with a 1:1 mixture of Pfu and Taq DNA polymerases and oligonucleotides (sequences available upon request) that introduced NcoI sites at the N termini and XhoI sites at the C termini for insertion into pET21b-H6-Nco. N-terminal amino acid sequences of full-length Brf (doubly hexahistidine tagged), Brf(1-n), and Brf(164-n) were MHHHHHHP; Brf(284-n) and Brf(435-n) were preceded by MHHHHHHPM, and Brf(320-n) was preceded by MHHHHHHPME. Brf C termini at amino acids 165, 282, 320, 438, 545, and 596 were followed by an UAA termination codon; full-length Brf had six histidine residues added at its C terminus. The resulting plasmids were introduced into E. coli BL21(DE3) containing the argU expression plasmid pUBS520 (32, 39), generously provided by I. Willis (Albert Einstein College of Medicine) and R. Mattes (Max-Planck-Institut für Züchtungsforschung, Cologne, Germany). Brf fragments for most EMSA and transcription experiments were purified from lysates of induced 50-ml cultures on Ni-NTA agarose under native conditions, as described for B"(137-596) (30). Three Brf fragments were also purified under denaturing conditions from cells lysed by resuspension in buffer A (specified above) also containing 7 M guanidine–HCl, removal of insoluble debris, and Ni-NTA agarose chromatography, as described for doubly His-tagged Brf except that the pH 5.9 and 5.7 elution steps were omitted. The fraction eluting at pH 5.5 was neutralized and chromatographed on Superose 12 in buffer D500 containing 7 M urea, and Brf was renatured by stepwise dialysis out of urea, as described above for doubly His-tagged Brf. Renatured Brf(1-282) and Brf(284-596) were used for photochemical cross-linking analysis and renatured Brf(1-282), Brf(284-596), and Brf(1-438) were used for some transcription assays.
Quantities of TFIIIC are specified as femtomoles of DNA-binding activity, and quantities of pol III are specified as femtomoles of enzyme active for specific transcription (30). B", B"(137-594), TBP, TBPm3, and Brf proteins are specified as femtomoles of protein (relative to a standard curve for Coomassie blue-stained bovine serum albumin [BSA] on sodium dodecyl sulfate [SDS]-polyacrylamide gels or determined by the predicted extinction coefficient of the protein [for B" and TBP]). B", B"(137-594), and TBP were estimated to be nearly 100% active, and full-length Brf was estimated to be 20% active, in the formation of heparin-resistant TFIIIB-DNA complexes (25).
Assays.
Protein-DNA complexes for EMSA, transcription, and photochemical cross-linking were formed in 16- to 20-μl volumes containing 40 mM Tris-Cl (pH 8.0), 7 mM MgCl2, 3 mM dithiothreitol (or 3 mM 2-mercaptoethanol for photochemical cross-linking), 4 to 6% (vol/vol) glycerol, 100 μg of BSA per ml (60 μg/ml for photochemical cross-linking), and 50 to 70 mM NaCl for TFIIIC-independent, or 70 to 90 mM NaCl for TFIIIC-dependent, DNA complex formation and transcription. For EMSA with the TA-30-B6 probe (10 to 20 fmol), reaction mixtures were incubated for 60 min at 20°C and contained 100 ng of poly(dG-dC) · poly(dG-dC), 10 to 100 fmol of TBP, 50 to 100 fmol of Brf or Brf fragment, and 50 to 300 fmol of B", when present (ranges in amounts referring to titrations). For EMSA with the SUP4 TA-30 probe (9 fmol), reaction mixtures were incubated for 40 min at 20°C and contained the quantities of TFIIIC, TBP, Brf or Brf fragment, and B" noted in the appropriate figure legend.
Protein-DNA complexes were analyzed on 4% polyacrylamide gels containing 20 mM Tris-Cl (pH 8.0), 2 mM EDTA, and 4% (vol/vol) glycerol or containing 20 mM Tris-Cl (pH 8.0), 0.5 mM EDTA, 2.5 mM MgCl2, 0.5 mM dithiothreitol, and 4% (vol/vol) glycerol. The electrode buffers lacked glycerol and dithiothreitol and were recirculated during electrophoresis.
Reaction mixtures for transcription contained 200 ng of pU6LboxB or pTZ1 supercoiled plasmid DNA as the template, 8 to 16 fmol of pol III, 40 to 80 fmol of TFIIIC (with pTZ1 only), 50 to 200 fmol of TBP, 150 to 300 fmol of B", and 50 to 400 fmol of Brf or its truncated variants (the ranges for TBP and Brf refer to titrations). Following complex formation for 40 or 60 min at 20°C, 5 μl of a nucleotide mixture providing 200 μM ATP, 100 μM CTP, 100 μM GTP, and 25 μM [α-32P]UTP (10,000 cpm/pmol) was added for 30 min of transcription. Samples were processed for electrophoresis on 10% polyacrylamide gels containing 8 M urea as described previously (30). The quantified transcription reaction mixtures represented in Tables 1 and 2 and Fig. 2 contained 50 fmol of pU6LboxB, 100 fmol of TBP, 150 fmol of B", 10 fmol of pol III, 300 to 350 fmol of Brf (or its truncated variants), and 50 mM NaCl (120 mM NaCl with full-length Brf). Transcription was quantified, relative to that in control reactions with intact Brf included in each experiment, by phosphorimaging.
TABLE 1.
TFIIIC-independent transcription with Brf fragments
| Brf fragment | pU6LboxB transcript yielda
|
||
|---|---|---|---|
| U6 leftward | U6 rightward | δ rightward | |
| 1-165 | — | — | — |
| 1-282 | 68 | 60 | 71 |
| 1-320 | 2 | 2 | 1 |
| 1-438 | 12 | 18 | 3 |
| 1-545 | 29 | 54 | 37 |
| 164-596 | 16 | 10 | 65 |
| 284-596 | 1 | — | 6 |
| 317-596 | 1 | — | 7 |
| 435-596 | <1 | — | 4 |
| 284-545 | — | — | — |
| 164-545 | <1 | — | 3 |
| 320-545 | — | — | — |
| 435-545 | — | — | — |
| 164-438 | — | — | — |
| 164-320 | — | — | — |
Transcript yield relative to that of intact Brf (set to 100). —, not detectable.
TABLE 2.
Complementation for TFIIIC-independent transcription with Brf fragmentsa
| Brf fragment and transcript | Complementation with indicated Brf fragment
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1-165
|
1-282
|
1-438
|
164-320
|
164-438
|
||||||
| Txn | Stim | Txn | Stimb | Txn | Stim | Txn | Stim | Txn | Stim | |
| 164-596 | ||||||||||
| δ | 65 | — | ||||||||
| r | 23 | √ | ||||||||
| l | 21 | √ | ||||||||
| 284-596 | ||||||||||
| δ | 10 | — | 96 | 30 | 109 | 13 | 159 | 16 | ||
| r | — | 73 | 16 | 114 | 4 | 11 | 22c | |||
| l | 1 | — | 66 | 30 | 131 | 8 | 16 | 21 | ||
| 317-596 | ||||||||||
| δ | 90 | 24 | 89 | 14 | ||||||
| r | 69 | 15 | 117 | 4 | ||||||
| l | 66 | 28 | 126 | 7 | ||||||
| 435-596 | ||||||||||
| δ | 10 | — | 81 | 25 | 94 | 20 | 98 | 18 | 92 | 17 |
| r | — | 60 | 16 | 125 | 5 | 3 | 7c | 5 | 13c | |
| l | <1 | — | 57 | 29 | 132 | 8 | 6 | 14 | 23 | 56 |
| 164-545 | ||||||||||
| δ | 12 | 5 | ||||||||
| r | 13 | 9 | ||||||||
| l | 11 | 8 | ||||||||
| 284-545 | ||||||||||
| δ | 64 | 12 | ||||||||
| r | 54 | 9 | ||||||||
| l | 54 | 15 | ||||||||
| 435-545 | ||||||||||
| δ | 83 | 11 | 29 | 8 | ||||||
| r | 60 | 7 | 65 | 3 | ||||||
| l | 65 | 16 | 76 | 5 | ||||||
Txn, maximum yield of the specified U6LboxB transcript (δ rightward [δ], U6 rightward [r], and U6 leftward [l]), relative to that of intact Brf (set to 100). Stim, maximum stimulation (fold) relative to the individual Brf fragments. —, stimulation not detected; √, stimulation detected but less than twofold.
Measured in transcription buffer containing 100 mM (instead of 50 mM) NaCl, reducing Brf(1-282)-mediated transcription to 3 to 5% of that obtained with intact Brf.
Gross underestimate, reflecting stimulation over residual background.
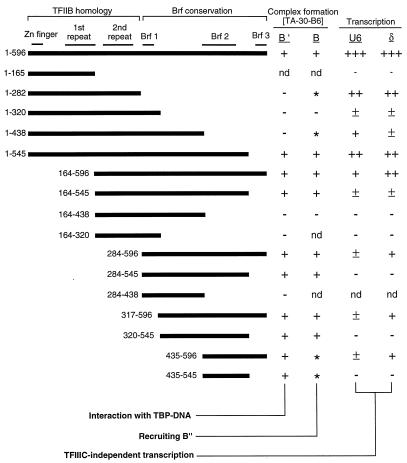
FIG. 2.
Transcription activities and interactions of Brf fragments. Formation of TFIIIB-DNA and B′ (TBP-Brf)-DNA complexes with the TA-30-B6 DNA probe (Fig. 1a) was analyzed by gel retardation. Transcription was measured with the SNR6 gene-related pU6LboxB construct (Fig. 3a). The locations of landmark features of Brf—the TFIIB-homologous putative Zn-binding domain, the two TFIIB-related sequence repeats, and the three segments of conserved sequence among fungal Brfs—are indicated at the top, and the N- and C-terminal Brf deletion mutants are designated at the left. The four columns at the right summarize the ability of each truncated Brf to allow formation of B′-DNA and TFIIIB-DNA complexes (nd, not determined; ∗, unstable to gel electrophoresis but detected by DNA-protein photochemical cross-linking) and to allow TFIIIC-independent transcription directed by the SNR6 (U6) and δ TATA boxes present on plasmid pU6LboxB (+++, 100%; ++, 30 to 80%; +, 5 to 20%; ±, <1 to 4%; −, not reliably detected).
Reaction mixtures for photochemical cross-linking contained 10 fmol of photoreactive DNA probe, 100 ng of poly(dG-dC) · poly(dG-dC), 400 fmol of TBPm3, 100 fmol of B"(137-594) when present, and 150 to 300 fmol of the specified Brf fragments or 350 to 700 fmol of full-length Brf. UV irradiation and nuclease treatment prior to electrophoresis on SDS–10 or 15% polyacrylamide gels was carried out as described elsewhere (3). Cross-linking was quantified by phosphorimage analysis and normalized to cross-linking of intact Brf and B"(138-594) within the TFIIIB-DNA complex that served as the control for each experiment.
Reaction mixtures for footprinting with MPE-Fe(II) contained, in 15 μl of buffer (20 mM Tris-Cl [pH 8], 95 mM NaCl, 3.5 mM MgCl2, 2.6 mM dithiothreitol, 100 μg of BSA per ml), 55 fmol of TA-30 variant SUP4 gene probe, 100 ng of pGEM (carrier) DNA, 250 fmol of TBP, 500 fmol of Brf or 500 fmol each of Brf(1-282) and Brf(284-596), as specified, 500 fmol of B", as specified, and 62 fmol of TFIIIC. DNA cutting at 21°C was started by adding MPE-Fe(II) to 2 μM and sodium ascorbate to 1 mM and stopped by adding 2 μl of 50 μM bathophenanthroline and 2 μl of 40% (vol/vol) glycerol. Reaction mixtures were resolved by electrophoresis on nondenaturing 4% acrylamide gels as specified above and exposed without drying to an image plate to locate the desired protein-DNA complex. The latter was cut out of the gel and eluted into 20 mM Tris-Cl (pH 8)–0.2% (wt/vol) SDS–0.5 mM EDTA–1 M LiCl. DNA was extracted from the eluate with phenol-chloroform, precipitated with ethanol, redissolved, denatured, and resolved by electrophoresis on a DNA-sequencing (10% acrylamide) gel. Dried gels were analyzed on a phosphorimager as described previously (30).
RESULTS
A Brf sequence that is required for stable interaction with the other components of the TFIIIB-DNA complex.
In preceding work (21), we analyzed three N-terminal deletion variants of Brf and showed that removing 164 N-proximal TFIIB-homologous amino acids does not abolish the ability to direct TFIIIC-independent TFIIIB-DNA complex assembly or transcription. In fact, the C-terminal half of Brf retains some ability to interact with TBP and with the B" component of TFIIIB. We have now combined nested series of C-terminal and N-terminal deletions in order to define the role of individual parts of Brf in assembling TFIIIB-DNA complexes.
The first series of experiments examines the ability of truncated Brf to interact with TBP and B" at a TATA box. The DNA that was used for most of the analysis has a 6-bp TATA box, TATAAA, with 4-nucleotide loops at sites at which bound TBP kinks DNA (14, 15, 20, 28, 29). Because it forms a more stable complex with wild-type TBP than does the corresponding perfect duplex DNA, this loop-containing DNA (designated TA-30-B6 [Fig. 1a]) facilitates analysis of B′ (i.e., TBP plus Brf)-DNA complexes by gel electrophoresis. At the same time, the loop-containing TA-30-B6 DNA probe retains specificity of interactions, in the sense that DNA binding of Brf required TBP and B" binding required Brf.
The analysis (examples of experiments are shown in Fig. 1b; the collected data are summarized in Fig. 2) showed that amino acids 435 to 545 (principally comprising Brf homology region 2) are essential and sufficient for stable attachment of Brf to the TBP-DNA complex: B′-DNA complexes with Brf fragments encompassing Brf homology region 2 were readily detected, but the corresponding complex with Brf(1-282) was not (Fig. 1b and 2). Conversely, Brf fragments lacking Brf homology region 2—Brf(1-282), Brf(1-438), and Brf(164-438)—did not form TFIIIB-DNA complexes that were stable to gel electrophoresis (Fig. 2), although a weak stabilization was noted for Brf(1-282) at elevated concentrations of B" (data not shown). Further analysis also showed that the C-terminal 52 amino acids of Brf, which include Brf homology region 3, were not required for interaction with TBP or for recruitment of B" into TFIIIB-DNA complexes (Fig. 1b, lanes d to f and j to l). Photochemical cross-linking experiments (see below) showed that the amino acid 435–545 segment of Brf, containing homology region 2, is also necessary and sufficient for B" recruitment to a stable DNA complex.
The preceding experiments specify requirements for formation of protein-DNA complexes that are sufficiently stable to survive gel electrophoresis. This is a relatively stringent criterion. A weaker capacity for interaction with TBP and B" resides in the N-terminal, TFIIB-related half of Brf and was detected by other means, as shown below.
Transcription.
The next experiments examined effects of Brf truncations on TFIIIC-independent transcription of pU6LboxB, a construct related to the U6 snRNA gene (SNR6) with two TATA boxes, TATAAATA, in different surrounding sequence; each TATA box generates a pair of divergent transcripts (Fig. 3a) (21). As reported previously (21), Brf(165-596) is able to direct transcription of the pU6L construct, albeit less efficiently than does intact Brf. Under the conditions of the analysis, divergent transcription directed by the SNR6 TATA box was considerably diminished (>6-fold), but rightward transcription directed by the upstream δ TATA box was much less affected (Table 1; Fig. 3b [compare lanes e and h]).
FIG. 3.
TFIIIC-independent transcription. (a) The pU6LboxB construct. The start sites and termination sites of leftward and rightward transcription directed by the SNR6 (U6) TATA box and by the δ TATA box are indicated. Rightward transcription initiating to the right of the SNR6 TATA box can be directed by TFIIIC binding to boxes B and A. (b) TFIIIC-independent transcription of pU6LboxB with Brf fragments individually and in combination. Transcripts are identified at the side [R.M., recovery marker; ∗, downstream-shifted initiation with Brf(284-596)], and Brf fragments are indicated above the lanes. Transcription was performed with 100 fmol of pU6LboxB DNA, 16 fmol of pol III, 100 fmol of TBP, 100 fmol of B", and 300 fmol of Brf fragment or intact Brf (lanes a to h) or as specified above lanes i to s. The weaker complementation of Brf(1-320) with Brf(284-596) was analyzed in lanes q to s with additional TBP (200 fmol) and B" (300 fmol).
More extensive N-terminal deletions [Brf(284-596), Brf(317-596), and Brf(435-596)] nearly abolished TFIIIC-independent transcription directed by the U6 TATA box, with only barely detectable levels of leftward transcription remaining (Fig. 3b, lanes a, b, and r; Table 1). The transcriptional start site of this residual activity appeared to shift downstream by ∼4 bp (Fig. 3b, lane r). Rightward transcription directed by the upstream TATA element (δ) was also greatly diminished, but it was surprising to see retention of some transcription activity in a Brf fragment entirely lacking its TFIIB-related component.
C-terminal deletions (Table 1; Fig. 3b) showed transcription only moderately reduced upon removal of amino acids 545 to 596 (Table 1). To our surprise, removal of the entire Brf-specific, C-proximal half of Brf (amino acids 283 to 596) allowed nearly full retention of transcription directed both from the upstream (δ) and U6 TATA boxes (Fig. 3b, lane g; Table 1), but this transcription required higher concentrations of B" than did full-length Brf (data not shown). In contrast, Brf(1-320) retained very little transcription activity (Table 1; Fig. 3b, lane q). Experiments described below show that Brf(1-320) also was only weakly active in complementation assays, suggesting that it may have contained predominantly incorrectly folded material. Brf(1-438) also was less active than Brf(1-282) (Table 1; Fig. 3b [compare lanes f and g]), but it is not likely that this was due solely to a low fraction of correctly folded molecules, because Brf(1-438) was active in complementation assays described below. It is also noteworthy that rightward transcription mediated by the δ TATA box was preferentially lost when amino acids 439 to 596 were removed (Table 1; Fig. 3b, lane f). Combinations of N- and C-terminal deletions also yielded proteins that either remained or became transcriptionally inactive: whereas Brf(1-545) was highly active and Brf(164-596) retained high activity for transcription directed by the δ TATA box, Brf(164-545) was essentially inert (lane d), with rightward transcription mediated by the δ TATA box only barely detectable (in the original data; lost in reproduction).
Split Brf.
The availability of many inactive and only partially active Brf fragments made it possible to test for the reconstitution of Brf activity by complementation. Examples of complementation are shown in Fig. 3b (lanes i to s), and the outcome of a more extensive survey is presented in Table 2. Complementation was assessed by two criteria: (i) the maximum transcriptional activity achieved with a combination of two Brf fragments relative to intact Brf; and (ii) the maximum fold stimulation generated by combining two fragments relative to the sum of residual activities of each fragment. The first criterion was satisfied by using relatively high concentrations of all transcription components and assaying at low ionic strength. The second criterion is an ideal (near-infinite stimulation) that was achieved for complementation with Brf(1-282) or Brf(1-438) by lowering the concentration of transcription factors and/or elevating the ionic strength. The same reaction conditions were retained for maximal stimulation and maximal transcription, as long as that allowed an at least 20-fold increase in one of the pU6LboxB transcripts. Combining Brf(1-282) with Brf(284-596) generated strong complementation according to either criterion, with 6- to 9-fold stimulation at relatively low fragment concentrations (Fig. 3b, lanes j, k, and m) and 16- to 30-fold stimulation at elevated ionic strength (Table 2). Complementation was also generated when Brf(1-438) and Brf(435-596) were combined (Fig. 3b, lanes n to p; Table 2), and weak complementation was detected when Brf(1-320) and Brf(284-596) were combined (Fig. 3b, lanes q to s). Evidence for complementation of fragments generated by cleavage at amino acid 164 was principally provided by combining Brf(1-165) with Brf(164-545) (Table 2). Brf(164-596) had a relatively high activity of its own (reference 21 and Table 1), and the generally poor activity of the Brf(1-165) fragment (conceivably reflecting improper refolding of this poorly expressed protein) generated only equivocal evidence for complementation of Brf(1-165) and Brf(164-596). The very weak activity of Brf(164-545) alone (Table 1) made it easier to detect the complementation.
It also proved possible to remove large internal segments, reconstituting U6 RNA synthesis with Brf(1-282) and Brf(435-596), for example (Fig. 3b, lanes j, l, and n). Complementation of Brf(1-282) with Brf(435-545), removing Brf homology regions 1 and 3 by combining a large internal deletion with a smaller C-terminal deletion, was also highly effective (Table 2). It was even possible to reconstitute activity with large polypeptide duplications, combining Brf(1-438) or Brf(1-320) with Brf(284-596) and combining Brf(1-438) with Brf(317-596), for example (Table 2; Fig. 3b, lanes q to s). Removing the N-terminal 163 amino acids (including the putative Zn finger and the first TFIIB-related repeat) still allowed Brf to be split at amino acids 435 and 283 (the latter in conjunction with a doubling up of amino acids 284 to 319), with retention of some activity. All complementing pairs (Table 2) included Brf homology region 2, pointing once again to its importance in assembly and function of the transcription initiation complex.
These results show clearly that Brf can be split at amino acids 283 and 435 and, with retention of partial activity, at amino acid 320. They imply the existence of at least three independent folding domains in Brf: amino acids 1 to 282, 283 to 434, and 435 to 596.
Mapping protein and DNA interactions with split Brf.
The ability to split Brf offers the opportunity to map the interactions of the resulting fragments with each other and with DNA by photochemical cross-linking. DNA with the photoactive nucleotide 5-[N′-(p-azidobenzoyl)-3-aminoallyl]dUMP (ABdUMP) (1, 3) and a radioactive nucleotide inserted at specific sites of an SNR6 gene-related DNA probe (Fig. 4a) was constructed as described previously (21). This DNA was designed to generate unidirectional TFIIIC-independent assembly of TFIIIB-DNA complexes by virtue of its variant 8-bp TATA box, TGTAAATA, to which mutant TBPm3 (41) binds in a unique orientation (47). (Uniqueness of assembly is, of course, crucial for unambiguous interpretation of cross-linking experiments.)
Complexes of Brf fragments and combinations of fragments with and without B" were built up on DNA with variously placed ABdUMP residues, in the presence of excess TBPm3. Sample cross-linking experiments with Brf reconstituted from its 1-282 and 284-596 fragments are shown in Fig. 4b. Results were analyzed by comparing the cross-linking efficiencies of the split Brf fragments with that of full-length Brf within a TFIIIB-DNA complex. The efficiency of B" cross-linking in complexes with Brf fragments and with intact Brf was quantified in the same way. In making comparisons in this manner, it is necessary to bear in mind that the efficiency of cross-linking of full-length Brf and B" varies between DNA sites, that is, between probes (Table 3). The N-terminally truncated B"(137-594) was used for this analysis for technical reasons, but the DNA-binding, transcription, and photochemical cross-linking properties of full-length B" and B"(137-594) are indistinguishable (reference 30 and data not shown). Results of the analysis for Brf split at amino acid 283 are summarized in Fig. 4c to e. The following are the salient observations.
TABLE 3.
Variations in efficiency of cross-linking along the DNA-binding site of the TFIIIB-DNA complex
| Probe | Relative cross-linking efficiencya
|
|
|---|---|---|
| Brf | B"(137–594) | |
| −43N | 7–9 | 2–4 |
| −42T | 2–6 | 2–8 |
| −33N | 70–110 | 10–20 |
| −32T | 7–20 | 1–2 |
| −28N | 40–70 | 5 |
| −23/−22T | 40–70 | 5–10 |
| −13/−12N | 100–110 | 14–25 |
Relative to cross-linking at −39/−38N (set to 100), corrected for specific activity differences between probes. The range indicates the experiment-to-experiment variation.
(i) Cross-linking of intact Brf to DNA was closely reflected in the properties of its C-terminal half. When Brf(284-596) was brought to a TBP-DNA complex by itself, cross-linking was seen between bp −43 and −12 (Fig. 4c) (cross-linking efficiency is expressed relative to that of intact Brf in the TFIIIB-DNA complex, but it should be noted [Table 3] that intact Brf is relatively inefficiently cross-linked at bp −43, −42, and −32). The level of Brf(284-596) cross-linking did not significantly change upon separately adding Brf(1-282) or B" (data not shown). The most striking feature of the cross-linking pattern of Brf(284-596) by itself (Fig. 4c), with Brf(1-282), or with B" (data not shown) was the deficit of cross-linking at bp −33 (in the nontranscribed strand); bp −33 was the preferred site of cross-linking of full-length Brf in the context of the TFIIIB-DNA complex (Table 3). Cross-linking of Brf(284-596) at −28N was also markedly low relative to that of intact Brf (Fig. 4b, middle panel). Forming the complete TFIIIB-DNA complex with split Brf greatly increased cross-linking of Brf(284-596) at −33N but had little or only modest effect elsewhere (in particular, no effect was seen at −28N and cross-linking at −39N reproducibly decreased). The cross-linking efficiency of full-length Brf was also lower at −33N in the absence of B" than in its presence, indicating that efficient cross-linking at this site is also B" dependent (data not shown).
(ii) Cross-linking of Brf(1-282) was not detectable at any site in the absence of B" (e.g., Fig. 4b). EMSA also failed to provide evidence that a complete B′ complex formed with Brf split at amino acid 283 (data not shown). The absence of an effect of Brf(1-282) on the cross-linking of Brf(284-596) in the absence of B" (Fig. 4c) further indicated a failure to form this complex; Brf(1-282) cross-linked to DNA at −39/−38N, −28N, and −13/−12N only when B" was also provided (Fig. 4c and data not shown), but with comparably low efficiencies (∼10% relative to that of full-length Brf). Addition of B" and Brf(284-596) to form the TFIIIB-DNA complex with split Brf allowed relatively inefficient cross-linking of Brf(1-282) to be detected at −43N, −42T, and −23/−22T, as well as enhanced cross-linking at −39/−38N, −28N, and −13N; there was no cross-linking at −33N and −32T. It is noteworthy that the efficiency of Brf(1-282) cross-linking at −28N preferentially increased and approached that of full-length Brf. With one exception (−42T), sites of stronger Brf(1-282) cross-linking relative to that of full-length Brf correlated with weaker cross-linking of Brf(284-596).
The trend to a neatly partitioned occupancy of DNA sites between the N- and C-proximal halves of Brf in Fig. 4c and d is partially obscured by two factors: first, Brf(284-596) formed a stable complex with TBP-DNA even in the absence of the Brf(1-282) segment and of B"; second, assembly of Brf(1-282) into the TFIIIB-DNA complex was limiting (data not shown, but see Fig. 4b) so that some of the cross-linked TFIIIB-DNA complexes lacked Brf(1-282). Treatment with the polyanion heparin disrupts partial complexes, and thereby simplifies the analysis, but was not included in the preceding analysis because it also generates its own complication: heparin binds to sites within the TFIIIB-DNA complex that might otherwise be accessible to the photoactive nucleotide (Fig. 3 of reference 18 and reference 2). While keeping this complication in mind, it is nevertheless useful to examine the results of a subsidiary photochemical cross-linking analysis of heparin-resistant TFIIIB-DNA complexes with Brf split at amino acid 283 (Fig. 4e): the Brf(284-596) segment accounted for 100% of the cross-linking of split Brf at −33N and −32T, for 93% of cross-linking at −42T, and for 37, 20, and 3% of cross-linking at −39/−38N, −13/−12N, and −28N, respectively.
(iii) B" cross-linked very weakly to DNA in the presence of TBP and in the absence of Brf or Brf fragments (Fig. 4d). This may reflect the previously noted direct B"-TBP interaction (37).
(iv) B" was brought into the TFIIIB-DNA complex by either half of Brf but not with the same disposition relative to DNA (Fig. 4d; note Table 3). In the complex with Brf(1-282), B" was within cross-linking reach of DNA upstream, at −43N, −42T, −39/−38N, and −33N, but not further downstream, at −32T, −28N, and −23/−22T, and only barely at −13/−12N. In the complex with Brf(284-596), B" was cross-linked at all sites: with nearly full efficiency to full efficiency at −33N and further downstream, and at somewhat lower efficiency upstream of −33. Addition of the Brf(1-282) fragment increased the efficiency of B" cross-linking upstream at −43N, −42T, −39/−38N, and, to some extent, −33N but not further downstream. We conclude that Brf(284-596) holds all of B" tightly to the TBP-DNA complex and that Brf(1-282) holds only the part of B" that is in vicinity of DNA upstream of bp −33.
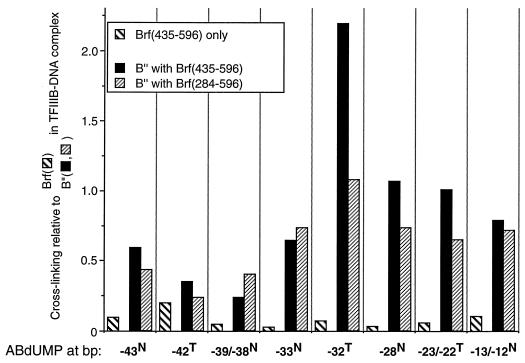
Similar experiments were done with Brf reconstituted from its 1-282 and 435-596 fragments, that is, with the segment from amino acids 283 to 434 missing (Fig. 5). Brf(435-596) cross-linked poorly at all sites relative to full-length Brf (5 to 20% relative cross-linking efficiency). The presence of Brf(1-282) and/or B" had little or no effect on the efficiency of cross-linking of Brf(435-596) (data not shown). Nevertheless, Brf(435-596) by itself proved as competent as Brf(284-596) for recruitment and efficient cross-linking of B". Brf(435-596) increased the cross-linking efficiency of Brf(1-282) in the presence of B", similar to what had been seen with Brf(284-596) (Fig. 4) but with only one-half of the quantitative effect (data not shown). Brf(435-545) and Brf(435-596) were comparably competent in recruitment of B" for cross-linking to the −39/38N probe, but Brf(435-545) was itself cross-linked poorly (data not shown, but see below).
FIG. 5.
Analysis of the assembly of DNA complexes with Brf(1-282) and Brf(435-596) by photochemical cross-linking. The cross-linking efficiency of Brf(435-596) in the B′-DNA complex is shown relative to that of intact Brf in the TFIIIB-DNA complex. Recruitment of B" to DNA by Brf(435-596) and Brf(284-596) is compared and normalized to the cross-linking efficiencies of B" in TFIIIB-DNA complexes formed with intact Brf.
The most straightforward interpretation of these observations is that amino acids 435 to 545 of Brf, containing Brf homology region 2, serve as the dominant link between TBP and Brf and also between B" and Brf. The segment of the C-terminal half of Brf that closely approaches DNA is contained between amino acids 284 and 434. The N-terminal, TFIIB-related half of Brf likewise interacts with both TBP and B" but less stably. Further insight into sites of interaction of each half of Brf with TBP are provided by the next experiment.
A set of 91 sequence substitutions on the surface of human TBPm3 (hTBPm3; mutated so as to recognize TGTAAATA) and an analogous subset of mutations in yeast TBP were examined recently for ability to promote B′-DNA and TFIIIB-DNA complex formation with yeast Brf and B" on the SNR6 (U6 gene) TATA box and for ability to promote pol III-directed SNR6 transcription (6, 12, 40). This analysis defined an extended interaction surface on TBP, where radical sequence substitutions significantly reduce the affinity of the TBP-DNA complex for full-length Brf and for the C-terminal half of Brf [e.g., Brf(317-596)], implying that most of the affinity for TBP lies in the C-terminal half of Brf. The Brf interaction epitope of TBP is positively charged and lies on the top (DNA-distal) surface and side of that half of TBP that comprises its N-proximal pseudo-repeat sequence. The Brf interaction surface of TBP is far removed from its TFIIB-binding surface.
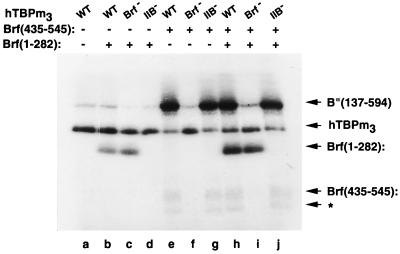
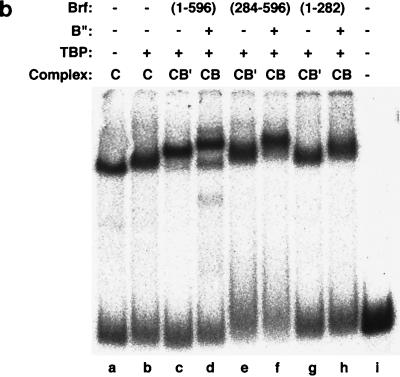
The just-described DNA cross-linking experiments implicate sequences within amino acids 435 to 545 of Brf in interaction with these epitopes on TBP. A triple mutation on the top surface of hTBPm3 combining three Brf-binding-defective substitutions (R231E, R235E, and R239S) was found to nearly abolish TFIIIB-DNA complex formation and SNR6 transcription (40). The expectation that this construct, hTBPm3(R231E+R235E+R239S), should be defective in the assembly and photochemical cross-linking of Brf(435-545) but not of Brf(1-282) was confirmed [compare lanes h and i in Fig. 6; since photochemical cross-linking of Brf(435-545) at bp −28 is quite weak, B" cross-linking provides a simple and more sensitive measure for Brf(435-545) assembly (compare lanes e to g with a)]. A second triple mutation in hTBPm3 had also been constructed at three residues (E284, E286, and L287) that are critical for TFIIB interaction but have no discernible effect on Brf binding or SNR6 transcription (40). As expected, the E284R-E286R-L287E triple mutation had no effect on the assembly and cross-linking of Brf(435-545), but Brf(1-282) was now unable to assemble and be cross-linked to DNA at bp −28 (compare lanes h and j). This result strongly indicated that the TFIIB-related N-terminal half of Brf interacts with TBP in a region that overlaps or lies near the TFIIB-binding domain. (Librizzi and coworkers [32] previously interpreted a Brf-generated quantitative footprint change to suggest that Brf might contact TBP in a TFIIB-like manner.) This interaction between the N-terminal half of Brf and TBP does not appear to play a significant role in the TFIIIC-independent transcriptional activity of Brf since SNR6 transcription with hTBPm3(E284R+E286R+L287E) is unimpaired (40). TFIIIC-dependent transcription on the SUP4 tRNA gene, but not TFIIIB-SUP4 DNA complex formation, was, however, defective (data not shown).
FIG. 6.
The N- and C-proximal halves of Brf interact with opposite ends of TBP: analysis by photochemical cross-linking. TFIIIB-DNA complexes were formed with 200 fmol of hTBPm3 (WT [wild type]), of the Brf interaction-defective hTBPm3(R231E+R235E+R239S) (Brf−), or of the TFIIB interaction-defective hTBPm3(E284R+E286R+L287E) (IIB−), 300 fmol each of Brf(1-282) and Brf(435-545) as indicated above each lane, 200 fmol of B"(137-594), and 8 fmol of −28N DNA probe. Cross-linked proteins are indicated at the right. The asterisk marks a fragment of Brf(435-545) that appears to be competent to assemble TFIIIB-DNA complexes.
In a recent analysis of the yeast TFIIIB-DNA complex, Colbert et al. (12) examined the minimum extent of additional DNA that is required to assemble Brf-TBP-DNA complexes that are stable to gel electrophoresis. They found extensive additional DNA required downstream of the TATA box but very little, if any, additional DNA required upstream of the TATA box. In contrast, stable TFIIIB complexes required a DNA extension upstream of the TATA box and less DNA downstream than for the Brf-TBP-DNA complex. One might conclude that these findings imply placement of B" and Brf in a TFIIIB-DNA complex upstream and downstream, respectively, of the TATA box. The experiments presented here expose a more feature-rich landscape of Brf and B" cross-linking to DNA upstream as well as downstream of the TATA box.
Nevertheless, the results of these two analyses are complementary rather than conflicting, for the following reasons. First, a determination of minimum DNA size (judged in terms of a particular criterion that is implicit in the specific conditions of a gel electrophoresis assay) does not preclude additional contributions to protein-DNA affinity originating outside that minimum domain. Second, the photochemical cross-linking experiments with ABdUMP-containing DNA examine close protein proximity, at up to 8 to 9 Å from the pyrimidine ring, rather than direct, in-the-groove protein-DNA contact. Third, our findings of close Brf proximity to DNA upstream of the TATA box (Fig. 4 and 5) refer to a TFIIIB-DNA complex. Cross-linking of Brf at bp −33, for example, largely depends on assembly of the TFIIIB-DNA complex (Fig. 4b). This is the result that one would anticipate if B" binding brought upstream DNA and Brf closer together, either by further folding the DNA or by generating a conformational change in Brf.
Interactions with TFIIIC: assembly of transcription factor complexes and transcription on the SUP4 tRNA gene.
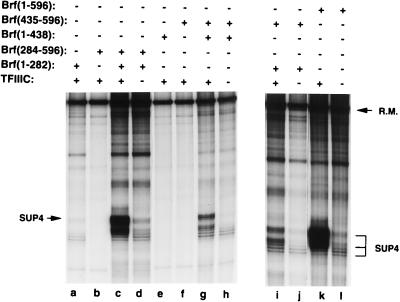
When TFIIIC directs the assembly of the TFIIIB-DNA complex (as it probably does on all yeast pol III genes, including the U6 snRNA gene, in vivo [5, 13]), the assembly-initiating interaction involves Brf instead of TBP (24). The TFIIIC-dependent pathway to transcription places stricter demands on Brf. Of all of the individual fragments tested, only Brf(1-545) retained a small fraction of transcriptional activity (∼5 to 10% of that of intact Brf), while the residual activity of Brf(1-282) was only barely detectable (<1%) and the distribution of transcriptional start sites on the SUP4 gene was altered (Fig. 7; compare lane a with lane k). These low residual activities of Brf fragments made it possible to test with high sensitivity for reconstitution of activity in TFIIIC-dependent transcription from combinations of fragments. Brf split at amino acids 283 and 437 was highly active (Fig. 7; compare lanes a to d and e to h with lanes k and l), generating 45 and 25% of the SUP4 transcription activity, respectively, of intact Brf. Even the combination of Brf(1-282) with Brf(435-596) generated readily detectable transcription activity (10% of the SUP4 transcription activity of intact Brf) above a low background of TFIIIC-independent transcription (lanes i and j).
FIG. 7.
TFIIIC-dependent transcription of the SUP4 gene with Brf split at amino acids 282 and 435. Components (400 fmol of intact Brf or the indicated fragments, 100 fmol of TBP, 300 fmol of B", 76 fmol of TFIIIC, as indicated, 16 fmol of pol III, and 100 fmol of the SUP4 gene-containing plasmid pTZ1) are specified above each lane. SUP4 transcripts and the recovery marker (R.M.) are identified at the sides.
The TFIIIC-dependent assembly of DNA-complexes containing Brf fragments 1-282 and 284-596, separately and together, was also examined by gel retardation, and the internal structures of complexes were probed by two-dimensional footprinting with MPE-Fe(II) (9). We used the TA-30 variant of the SUP4 gene for this analysis. In the TA-30 gene, the AT-rich upstream segment of the SUP4 gene is replaced by TATAAA centered 30 bp upstream of the transcriptional start site, surrounded by GC-rich sequence (Fig. 1a). The TA-30 SUP4 gene restricts initiation of transcription predominantly to a single site in vitro (in contrast to the wild-type SUP4 gene [Fig. 7, lane k]) because the TATAAA box restricts TFIIIB assembly more uniformly to a single upstream site while still retaining nearly complete TFIIIC dependence for transcription (19).
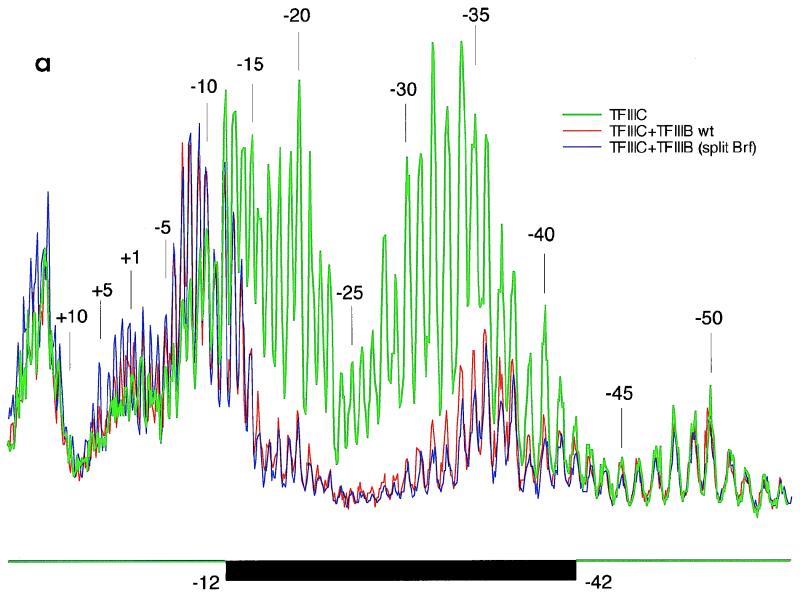
TFIIIC-B-DNA complexes formed on the TA-30 gene with Brf split at amino acid 283 were indistinguishable from complexes formed with intact Brf by the criterion of MPE-Fe(II) footprinting over the entire length of DNA covered by the assembled proteins (Fig. 8a). It appeared that all interactions affecting the disposition of protein around DNA [i.e., Brf-TBP, Brf-B", and Brf-TFIIIC(τ120)] are quite precisely matched when this split Brf and intact Brf are used to assemble the TFIIIC-B-DNA complex.
FIG. 8.
TFIIIC interaction. (a) MPE-Fe(II) footprints of TFIIIC-B-DNA complexes assembled on TA-30 DNA with intact Brf and with Brf split at amino acid 283. Reaction mixtures were assembled as described below and subjected to reaction with MPE-Fe(II) as specified in Materials and Methods. The resulting material was separated electrophoretically, and the gel was exposed without drying to a phosphorimaging screen as shown in panel b. Gel slices corresponding to the appropriate bands were excised; DNA was extracted, processed, separated on a sequencing gel as specified in Materials and Methods, and analyzed on a phosphorimaging detector. The density traces shown have been normalized at 5′ unprotected sequence. Green trace, TFIIIC-DNA complex; red trace, TFIIIC-TFIIIB-DNA complex assembled with intact Brf; blue trace, TFIIIC-TFIIIB-DNA complex assembled with Brf(1-282) and Brf(284-596). (b) EMSA for assembly and stability of TFIIIC-B′-DNA and TFIIIC-TFIIIB-DNA complexes with Brf(1-282) and Brf(284-596). Nine femtomoles of TA-30 DNA probe was incubated with 31 fmol of TFIIIC, 200 fmol of TBP, 300 fmol of B", and 200 fmol of intact Brf (as marked above lanes a to d) or with 31 fmol of TFIIIC, 500 fmol of TBP, 500 fmol of Brf fragment, and 300 fmol of B" (as indicated above lanes e to h). Electrophoresis was in Mg2+-containing gel. The result shown was generated in a footprinting experiment: DNA-protein mixtures were reacted with MPE-Fe(II) before electrophoresis, and the exposure shown was made without drying the gel.
Each half of Brf was found to interact with TFIIIC, but the evidence indicated much weaker interactions. TFIIIC-B[Brf(284-596)]-DNA complexes were sufficiently stable to be detected as a well-defined band in EMSA (done in the presence of Mg2+) (Fig. 8b, lane f). A separate experiment showed that assembly of this complex on the TA-30 gene was TFIIIC dependent even at the elevated concentrations of components that were required to drive association with Brf(284-596): the interaction with the TA-30 DNA probe and its 6-bp TATAAA box was unstable on the gel in the absence of TFIIIC (data not shown). However, the MPE-Fe(II) footprint of the TFIIIC-B[Brf(284-596)]-DNA complex, analyzed by cutting the corresponding band of protein-DNA complex from the gel (Fig. 8b, lane f), was grossly aberrant; for example, it lacked the footprint segment upstream of the TATAAA box (bp −33 to −42) that is conferred by B" recruitment into the TFIIIC-B-DNA complex (data not shown). Formation of a TFIIIC-B[Brf(1-282)]-DNA complex was also detected by EMSA, as judged by a small mobility shift (Fig. 8b, lane h), and weak TFIIIC-dependent photochemical cross-linking of Brf(1-282) was detected (data not shown). The observed competence for transcription, although extremely low, also implied the ability to form such a complex. The data presented here made very clear that the two halves of Brf strongly reinforce each other’s interactions with TFIIIC, TBP, B", and DNA.
DISCUSSION
Each half of Brf interacts with DNA-bound TBP: the C-proximal half of Brf interacts with the N-proximal pseudo-repeat lobe of TBP, and the N-proximal, TFIIB-related half of Brf interacts with the C-proximal pseudo-repeat lobe of TBP (Fig. 6). A direct interaction between the C-proximal halves of human and yeast Brf with TBP has been noted previously, but tests for an interaction with the N-proximal half of Brf yielded an inconsistent outcome or weak interaction (10, 27, 44). The interaction of the C-proximal half of Brf is the more stable one, yielding a B′-DNA complex that is detected by gel electrophoresis (Fig. 1b and 2) and also by photochemical cross-linking (Fig. 4). Brf also has multiple sites of interaction with B", so that TFIIIB complexes can be formed by each half of Brf separately (Fig. 1b, 2, and 4). The TBP- and B"-interacting domains of the C-terminal half of Brf both reside within the amino acid 435-545 segment, which contains Brf homology region 2 (Fig. 5 and 6; Table 2). A temperature sensitivity-conferring mutation in this region (L→S at amino acid 462) has been found to yield a Brf protein that is defective in binding to TBP-DNA (12) either as a result of defective folding or through loss of a specific protein-protein contact. Brf homology region 2 has a substantial degree of conservation among homologs from a worm and the human. We anticipate that Brf homology region 2 will also turn out to be the principal anchor for human Brf on TBP, as has been suggested on the basis of sequence conservation (34).
Each half of Brf also has some competence to generate transcription, but the N-terminal half plays the major role in pol III recruitment (Table 1; Fig. 3b); the relatively high transcriptional activity of Brf(1-282) and Brf(164-596) in δ-directed transcription points more specifically to the importance of amino acids 164 to 282, which encompass the second TFIIB-related sequence repeat (Fig. 2), in pol III recruitment. It is likely that the corresponding segment in human Brf will play a similar role in pol III recruitment and/or initiation of transcription, since the human pol III RPC39 subunit is homologous to the yeast pol III rpc34 subunit and likewise interacts with human Brf (human TFIIIB90) (45).
Some of the truncated Brf proteins differentially retain TFIIIC-independent transcription activity on our two test promoters. For example, Brf(1-438) was seen preferentially to lose, and Brf(164-596) was seen preferentially to retain, the capacity for transcription initiating at the δ promoter. This is consistent with two interpretations: (i) formation of a TFIIIB-DNA complex with these Brf fragments may require interaction with DNA sequence lying outside the TATA box that is not rate limiting for the transcriptionally more active full length Brf; and (ii) loss of protein-protein contacts mediated by the missing Brf segments may place additional requirements on the flexibility of TATA box-flanking DNA for securing required interaction with B" and pol III.
Each half of Brf also interacts with TFIIIC. A small shift of electrophoretic mobility suggested that Brf(1-282) was brought into a TFIIIB-DNA complex under the direction of TFIIIC (Fig. 8b), but its TFIIIC-dependent assembly was only weakly detected by photochemical cross-linking and its transcriptional activity was extremely low. Brf(284-596) formed a more stable but structurally aberrant and transcriptionally inactive TFIIIC-B-DNA assembly (Fig. 8b). It is surprising to find both halves of Brf required to properly integrate B" into the preinitiation complex in the presence of TFIIIC, whereas each half of Brf has the ability to do this in the absence of TFIIIC. Evidently, the role of TFIIIC as the assembly factor for the TFIIIB-DNA preinitiation complex (23) is more subtle than one might have imagined: it brings Brf and TBP to a DNA-binding site but must also be steered away (by Brf?) from interfering with entry of B" into the preinitiation complex.
The low transcriptional activity of various N-terminally and C-terminally deleted Brf fragments on the SNR6 gene construct and inactivity of these fragments in TFIIIC-dependent transcription made it possible to test for the retention of transcription activity when Brf is split at internal sites and when internal overlaps and gaps are generated. Brf split down the middle (at amino acid 283) was found to function well for TFIIIC-independent and TFIIIC-requiring transcription (Table 2; Fig. 7), form a TFIIIC-B-DNA complex that was indistinguishable from its intact-Brf counterpart in MPE-Fe(II) footprinting (Fig. 8a), and assemble a heparin-stable TFIIIB-DNA complex (Fig. 4f). Brf split at amino acid 437 was also transcriptionally active (Table 2; Fig. 3 and 7). TFIIIC-independent transcription was seen to be relatively permissive for extensive internal deletions in Brf. However, while Brf homology regions 1 and 3 could be removed, all transcriptionally effective complementing pairs of Brf fragments included Brf homology region 2 (Table 2; Fig. 3). (We also encountered some discrepancies [Table 2]. For example, Brf split between amino acids 317 and 320 [with small duplications] was much less active than was Brf internally split so as to entirely lack amino acids 283 to 434 [data not shown]. This and similar inconsistencies may reflect variable ability to properly fold and/or refold different overproduced Brf fragments.)
The close correspondence between the properties of intact Brf and Brf split at amino acid 283 allowed us to examine the disposition of the two halves of Brf in the TFIIIB-DNA complex. Eight DNA sites, covering 11 bp, were probed in this analysis (Fig. 4c to e). At five of these sites, cross-linking in the complete TFIIIB-DNA complex was predominantly to one or the other half of Brf (Fig. 4e). Comparison of the efficiencies of cross-linking of Brf(284-596) and Brf(435-596) (Fig. 4c and 5) suggests that proximity of Brf to DNA at bp −33 is primarily contributed by the nonessential amino acid 284-434 segment and is not critical to assembly of TFIIIB on DNA.
Further information for constructing a model of the disposition of Brf in the TFIIIB-DNA complex comes from a recently completed analysis of TBP mutations that interfere with the TBP-Brf interaction. TBP mutants interfering with binding of intact Brf also failed to interact with Brf(317-596) (40), with Brf(352-596) (12), or with Brf(435-545) (Fig. 6). Conversely, a triple mutation in TBP that prevents TFIIB binding also prevented cross-linking of Brf(1-282) to a DNA site at the upstream end of the U6 TATA box in a TFIIIB-DNA complex assembled with Brf(1-282) and Brf(435-545) (Fig. 6). This result identifies a site of interaction on the C-proximal lobe of TBP for the N-proximal TFIIB-related half of Brf and a site on the N-proximal lobe of TBP for the C-proximal Brf homology domain 2. It is the latter interaction that contributes the major part of the affinity of Brf for TBP.
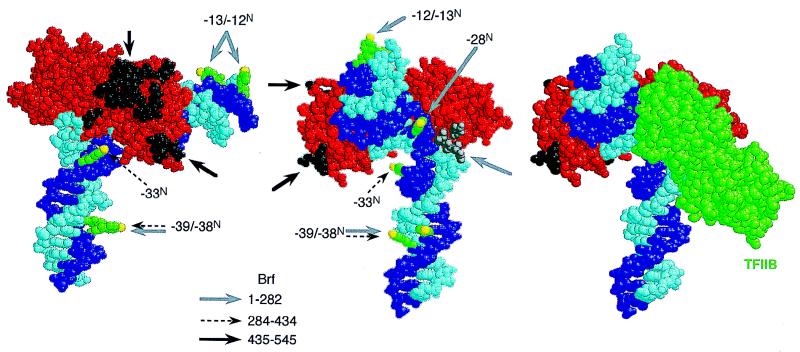
These constraints specify a global model of how Brf assembles into a TFIIIB-DNA complex. Figure 9 shows the TBP-DNA complex backbone of this model and identifies six widely separated reference points that are contacted by Brf. Four points of close vicinity to DNA, extending from upstream of the TATA box to the underside and further downstream, are represented by the N atoms (in yellow) at the tips of ABdUMP side chains (in green) projecting out of the DNA major groove. These sites span more than two and one-half turns of DNA helix but are brought closer together by the TBP-imposed DNA bend. Intact Brf cross-links with comparable efficiency to these four sites in a TFIIIB-DNA complex (Table 3). Two surfaces for protein-protein contact, one on each lobe of TBP, are represented in black and gray, respectively.
FIG. 9.
Model of the disposition of Brf in the TFIIIB-DNA complex. Two views of the TBP-DNA complex are shown (derived from reference 35). The existing DNA sequence was changed to place T at −33N, −32T, −23T, and −22T, and DNA was also extended at each end with undistorted B helix (corresponding to SNR6 DNA sequence). DNA strands are in dark (transcribed) and light (nontranscribed) blue; core TBP (Arabidopsis thaliana residues 12 to 198) is in red. The photoactive side chains of ABdUMP at bp −39, −38, −33, −28, −13, and −12 are shown, fully extended and projecting out of the major groove, in green, with their nitrene nitrogens in yellow. The TBP residues corresponding to the surface required for stable Brf binding (40) are shown in black. The corresponding TBP epitope required for cross-linking of Brf(1-282) and for TFIIB binding is shown in gray. Gray, dashed, and black arrows point to sites of proximity and interaction of Brf fragments 1-282, 284-434, and 435-545, respectively. At the right is shown the TFIIB-TBP-DNA complex (35). The structure comprises amino acids 113 to 316 of TFIIB (in green), homologous to amino acids 79 to 289 of yeast Brf.
In order to reach all of these sites, Brf has to intertwine with a TBP-DNA complex, reaching the underside of the TATA box as well as the lateral and DNA-distal sides at each end of TBP. Three domains of Brf make distinctive contacts with these six sites.
Brf(1-282), the TFIIB-homologous segment of Brf, occupies a TFIIB-related space in the complex. It contacts DNA at the downstream side (−13/−12N) and under the TBP-imposed bend at −28N but is also within cross-linking reach of the upstream −39/−38N site. The space filled by Brf(1-282) may be less extended than indicated here because DNA is further bent in the TFIIIB-DNA complex (4, 15a, 31). The similarity in disposition of TFIIB and its Brf(1-282) homolog in the pol II and pol III preinitiation complexes is certainly compatible with a previous proposal that the relationship between the direction of transcription and the orientation of TBP is the same for yeast pol II and pol III (47).
Brf(284-434) contributes the primary DNA proximity of the C-proximal half of Brf, primarily at −33N but also at −39/−38N (Fig. 4f), but this is not essential for Brf function (Table 2; Fig. 7).
Brf(435-545) is the site of tightest TBP binding, mapped in detail by Shen et al. (40); the contact surface on TBP is shown in Fig. 9 in black. This Brf segment is also the site of a tight linkage to B" (Fig. 4).
The similarity in disposition of TFIIB and of its homologous Brf segment in space, and the existence of a direct Brf-pol III interaction (27, 46), raises an interesting question. Why is B" absolutely required for all yeast pol III transcription, in DNA as well as in chromatin, in supercoiled as well as linear DNA, in TFIIIC-dependent as well as TFIIIC-independent transcription, and with yeast-derived biochemically purified components as well as with E. coli-synthesized recombinant proteins? Why do Brf and TBP, the B′ components of TFIIIB, not suffice for at least some basal transcriptional activity? It is known that B" drastically changes the stability of association of the other two components of TFIIIB with promoters. B" probably also further bends the Brf-TBP-DNA complex. Are these further structural constraints essential for transcriptional initiation by pol III? If they are, why is that the case?
It may be helpful to look to bacterial transcription for assessing these questions and their possible answers. Initiation of transcription by E. coli RNA polymerase requires successive steps of promoter attachment (recruitment), isomerization of the closed promoter-polymerase complex, DNA strand separation around the transcriptional start site, and promoter clearance. The activities of bacterial promoters can be limited or activated at each of these steps (36). We propose that the requirement for B" points to a role for TFIIIB in transcription by pol III that extends beyond simply and passively marking the promoter for polymerase attachment.
ACKNOWLEDGMENTS
We thank C. J. Brent and G. A. Letts for assistance with experiments, H. M. Vu for generous help with construction of one of the figures, Y. Shen and A. J. Berk for generously supplying key materials in advance of publication, and C. A. P. Joazeiro for helpful comments on the manuscript.
Support of our research by the NIGMS is gratefully acknowledged.
REFERENCES
- 1.Bartholomew B, Kassavetis G A, Braun B R, Geiduschek E P. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 1990;9:2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartholomew B, Kassavetis G A, Geiduschek E P. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol Cell Biol. 1991;11:5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartholomew B, Tinker R L, Kassavetis G A, Geiduschek E P. Photochemical cross-linking assay for DNA tracking by replication proteins. Methods Enzymol. 1995;262:476–494. doi: 10.1016/0076-6879(95)62039-7. [DOI] [PubMed] [Google Scholar]
- 4.Braun B R, Kassavetis G A, Geiduschek E P. Bending of the Saccharomyces cerevisiae 5S rRNA gene in transcription factor complexes. J Biol Chem. 1992;267:22562–22569. [PubMed] [Google Scholar]
- 5.Brow D A, Guthrie C. Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes Dev. 1990;4:1345–1356. doi: 10.1101/gad.4.8.1345. [DOI] [PubMed] [Google Scholar]
- 6.Bryant G O, Martel L S, Burley S K, Berk A J. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes Dev. 1996;10:2491–2504. doi: 10.1101/gad.10.19.2491. [DOI] [PubMed] [Google Scholar]
- 7.Buratowski S, Zhou H. Functional domains of transcription factor TFIIB. Proc Natl Acad Sci USA. 1993;90:5633–5637. doi: 10.1073/pnas.90.12.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buratowski S, Zhou H. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:221–230. doi: 10.1016/0092-8674(92)90351-c. [DOI] [PubMed] [Google Scholar]
- 9.Cartwright I L, Elgin S C. Nonenzymatic cleavage of chromatin. Methods Enzymol. 1989;170:359–369. doi: 10.1016/0076-6879(89)70056-2. [DOI] [PubMed] [Google Scholar]
- 10.Chaussivert N, Conesa C, Shaaban S, Sentenac A. Complex interactions between yeast TFIIIB and TFIIIC. J Biol Chem. 1995;270:15353–15358. doi: 10.1074/jbc.270.25.15353. [DOI] [PubMed] [Google Scholar]
- 11.Colbert T, Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 1992;6:1940–1949. doi: 10.1101/gad.6.10.1940. [DOI] [PubMed] [Google Scholar]
- 12.Colbert T, Lee S, Schimmack G, Hahn S. Architecture of protein and DNA contacts within the TFIIIB-DNA complex. Mol Cell Biol. 1998;18:1682–1691. doi: 10.1128/mcb.18.3.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eschenlauer J B, Kaiser M W, Gerlach V L, Brow D A. Architecture of a yeast U6 RNA gene promoter. Mol Cell Biol. 1993;13:3015–3026. doi: 10.1128/mcb.13.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grove, A., A. Galeone, E. Yu, L. Mayol, and E. P. Geiduschek. Affinity and polarity of binding of the TATA binding protein governed by flexure at the TATA box. J. Mol. Biol., in press. [DOI] [PubMed]
- 15.Grove A, Geiduschek E P. Localizing flexibility within the target site of DNA-bending proteins. In: Marshak D, editor. Techniques in protein chemistry VIII. San Diego, Calif: Academic Press; 1997. pp. 585–592. [Google Scholar]
- 15a.Grove, A., and G. A. Kassavetis. Unpublished data.
- 16.Henry R W, Sadowski C L, Kobayashi R, Hernandez N. A TBP-TAF complex required for transcription of human snRNA genes by RNA polymerase II and III. Nature. 1995;374:653–656. doi: 10.1038/374653a0. [DOI] [PubMed] [Google Scholar]
- 17.Huet J, Conesa C, Manaud N, Chaussivert N, Sentenac A. Interactions between yeast TFIIIB components. Nucleic Acids Res. 1994;22:3433–3439. doi: 10.1093/nar/22.16.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joazeiro C A, Kassavetis G A, Geiduschek E P. Identical components of yeast transcription factor IIIB are required and sufficient for transcription of TATA box-containing and TATA-less genes. Mol Cell Biol. 1994;14:2798–2808. doi: 10.1128/mcb.14.4.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joazeiro C A P, Kassavetis G A, Geiduschek E P. Alternative outcomes in assembly of promoter complexes: the roles of TBP and a flexible linker in placing TFIIIB on tRNA genes. Genes Dev. 1996;10:725–739. doi: 10.1101/gad.10.6.725. [DOI] [PubMed] [Google Scholar]
- 20.Juo Z S, Chiu T K, Leiberman P M, Baikalov I, Berk A J, Dickerson R E. How proteins recognize the TATA box. J Mol Biol. 1996;261:239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- 21.Kassavetis G A, Bardeleben C, Kumar A, Ramirez E, Geiduschek E P. Domains of the Brf component of RNA polymerase III transcription factor IIIB (TFIIIB): functions in assembly of TFIIIB-DNA complexes and recruitment of RNA polymerase to the promoter. Mol Cell Biol. 1997;17:5299–5306. doi: 10.1128/mcb.17.9.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kassavetis G A, Bartholomew B, Blanco J A, Johnson T E, Geiduschek E P. Two essential components of the Saccharomyces cerevisiae transcription factor IIIB: transcription and DNA-binding properties. Proc Natl Acad Sci USA. 1991;88:7308–7312. doi: 10.1073/pnas.88.16.7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kassavetis G A, Braun B R, Nguyen L H, Geiduschek E P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 24.Kassavetis G A, Joazeiro C A, Pisano M, Geiduschek E P, Colbert T, Hahn S, Blanco J A. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992;71:1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- 25.Kassavetis G A, Nguyen S T, Kobayashi R, Kumar A, Geiduschek E P, Pisano M. Cloning, expression, and function of TFC5, the gene encoding the B" component of the Saccharomyces cerevisiae RNA polymerase III transcription factor TFIIIB. Proc Natl Acad Sci USA. 1995;92:9786–9790. doi: 10.1073/pnas.92.21.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kassavetis G A, Riggs D L, Negri R, Nguyen L H, Geiduschek E P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989;9:2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khoo B, Brophy B, Jackson S P. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994;8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 28.Kim J L, Nikolov D B, Burley S K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 29.Kim Y, Geiger J H, Hahn S, Sigler P B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 30.Kumar A, Kassavetis G A, Geiduschek E P, Hambalko M, Brent C J. Functional dissection of the B" component of RNA polymerase III transcription factor (TF)IIIB: a scaffolding protein with multiple roles in assembly and initiation of transcription. Mol Cell Biol. 1997;17:1868–1880. doi: 10.1128/mcb.17.4.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leveillard T, Kassavetis G A, Geiduschek E P. Saccharomyces cerevisiae transcription factors IIIB and IIIC bend the DNA of a tRNA(Gln) gene. J Biol Chem. 1991;266:5162–5168. [PubMed] [Google Scholar]
- 32.Librizzi M D, Moir R D, Brenowitz M, Willis I M. Expression and purification of the RNA polymerase III transcription specificity factor IIIB70 from Saccharomyces cerevisiae and its cooperative binding with TATA-binding protein. J Biol Chem. 1996;271:32695–32701. doi: 10.1074/jbc.271.51.32695. [DOI] [PubMed] [Google Scholar]
- 33.López-De-León A, Librizzi M, Puglia K, Willis I M. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:211–220. doi: 10.1016/0092-8674(92)90350-l. [DOI] [PubMed] [Google Scholar]
- 34.Mital R, Kobayashi R, Hernandez N. RNA polymerase III transcription from the human U6 and adenovirus type 2 VAI promoters has different requirements for human BRF, a subunit of human TFIIIB. Mol Cell Biol. 1996;16:7031–7042. doi: 10.1128/mcb.16.12.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 36.Record M T, Jr, Reznikoff W S, Craig M L, McQuade K L, Schlax P J. Escherichia coli RNA polymerase (Eς70), promoters, and the kinetics of the steps of transcription initiation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 792–820. [Google Scholar]
- 37.Roberts S, Miller S J, Lane W S, Lee S, Hahn S. Cloning and functional characterization of the gene encoding the TFIIIB90 subunit of RNA polymerase III transcription factor TFIIIB. J Biol Chem. 1996;271:14903–14909. doi: 10.1074/jbc.271.25.14903. [DOI] [PubMed] [Google Scholar]
- 38.Rüth J, Conesa C, Dieci G, Lefèbvre O, Dusterhoft A, Ottonello S, Sentenac A. A suppressor of mutations in the class III transcription system encodes a component of yeast TFIIIB. EMBO J. 1996;15:1941–1949. [PMC free article] [PubMed] [Google Scholar]
- 39.Schenk P M, Baumann S, Mattes R, Steinbiss H H. Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. BioTechniques. 1995;19:196–200. [PubMed] [Google Scholar]
- 40.Shen Y, Kassavetis G, Bryant G O, Berk A J. Polymerase (Pol) III TATA box-binding protein (TBP)-associated factor Brf binds to a surface on TBP also required for activated Pol II transcription. Mol Cell Biol. 1998;18:1692–1700. doi: 10.1128/mcb.18.3.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strubin M, Struhl K. Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell. 1992;68:721–730. doi: 10.1016/0092-8674(92)90147-5. [DOI] [PubMed] [Google Scholar]
- 42.Teichmann M, Dieci G, Huet J, Rüth J, Sentenac A, Seifart K H. Functional interchangeability of TFIIIB components from yeast and human cells in vitro. EMBO J. 1997;16:4708–4716. doi: 10.1093/emboj/16.15.4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomm M. Archaeal transcription factors and their role in transcription initiation. FEMS Microbiol Rev. 1996;18:159–171. doi: 10.1111/j.1574-6976.1996.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, Roeder R G. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc Natl Acad Sci USA. 1995;92:7026–7030. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Roeder R G. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 46.Werner M, Chaussivert N, Willis I M, Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J Biol Chem. 1993;268:20721–20724. [PubMed] [Google Scholar]
- 47.Whitehall S K, Kassavetis G A, Geiduschek E P. The symmetry of the yeast U6 RNA gene’s TATA box and the orientation of the TATA-binding protein in yeast TFIIIB. Genes Dev. 1995;9:2974–2985. doi: 10.1101/gad.9.23.2974. [DOI] [PubMed] [Google Scholar]
- 48.Yoon J B, Murphy S, Bai L, Wang Z, Roeder R G. Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol Cell Biol. 1995;15:2019–2027. doi: 10.1128/mcb.15.4.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]