Dear Editor,
In development, after cells make commitment to their fates, they undergo a continuous and adaptive maturation process to eventually reach their terminal states. Cell maturity often decays during aging and pathogenesis. The immature phenotypes of stem cell-differentiated cells deposit a major bottleneck in regenerative medicine. Thus, cell maturation studies have recently become a new frontier in developmental biology (Alvarez-Dominguez and Melton, 2022).
In cardiac maturation, cardiomyocytes undergo morphology and gene expression changes. Structurally, cardiomyocyte maturation is characterized by the expansion of myofibrils, the biogenesis of mitochondria, and the formation of transverse tubules (T-tubule), which are invaginations of plasma membranes that facilitate electrophysiology. Cardiomyocytes also exhibit tremendous transcription changes (Li et al., 2022), such as myofibril isoform switching and the upregulation of fatty acid oxidation and oxidative phosphorylation genes. These changes collectively establish the robust cardiac function that can be sustained throughout human life.
We recently discovered serum response factor (SRF) as a central transcription factor that regulates cardiomyocyte maturation (Guo et al., 2018). The activity of SRF is tightly regulated by its myocardin-family co-factors including MYOCD (myocardin), MRTFA (myocardin-related transcription factor-A) and MRTFB (myocardin-related transcription factor-B) (Pipes et al., 2006). MYOCD and MRTFA/B are thought to participate in distinct signal pathways due to their distinct responses to actin dynamics (Pipes et al., 2006). The RPEL domain of MRTFA/B binds monomeric actin, which masks nuclear localization signals to reduce the nuclear localization of MRTFA/B, but the RPEL domain of MYOCD exhibits weak binding to monomeric actin.
The actin-MRTF-SRF axis was initially discovered in non-muscle cell types. Whether cardiomyocytes use the same mechanism to modulate gene expression remains poorly studied. We recently showed that cardiac α-actin (ACTC1) interacts with MRTFA and perturbs its nuclear localization (Guo et al., 2021). Strikingly, we found that the actin cytoskeleton was naturally in a highly polymeric state in the heart. Therefore, MRTFA was constitutively in the nuclei of cardiomyocytes in the wild-type, physiological context (Guo et al., 2021). These data suggested the hypothesis that MRTFA, MRTFB and MYOCD could be all constitutively active in cardiomyocyte nuclei and therefore redundantly co-activate transcription through SRF.
We previously injected AAV9-Tnnt2-Cre (AAV-Cre) vector into Mrtfa−/−;MrtfbF/F;RosaCas9-GFP mice to study the role of MRTFA/B in cardiomyocyte maturation (Guo et al., 2021). The RosaCas9-GFP allele could be further harnessed to inactivate Myocd in order to inactivate all three myocardin-family members. For this purpose, we designed a pair of sgRNAs to target a coding exon shared by all Myocd splicing variants (Figure S1A). AAV9-U6-Myocd-sgRNA-Tnnt2-Cre (AAV-Myocd-sgRNA) vectors that express these sgRNAs were administered at 3×109 vg/g to postnatal day 1 (P1) RosaCas9-GFP mice, and heart tissues were collected at P7. Reverse transcription-PCR (RT-PCR) validated genomic deletion between the sgRNA-targeted sites in AAV-Myocd-sgRNA-treated hearts (Figure S1B). Real-time quantitative PCR (RT-qPCR) confirmed 70% reduction of Myocd mRNA in AAV-Myocd-sgRNA-treated hearts (Figure S1C). Thus, a CRISPR/Cas9-based system to deplete Myocd in the heart was successfully established.
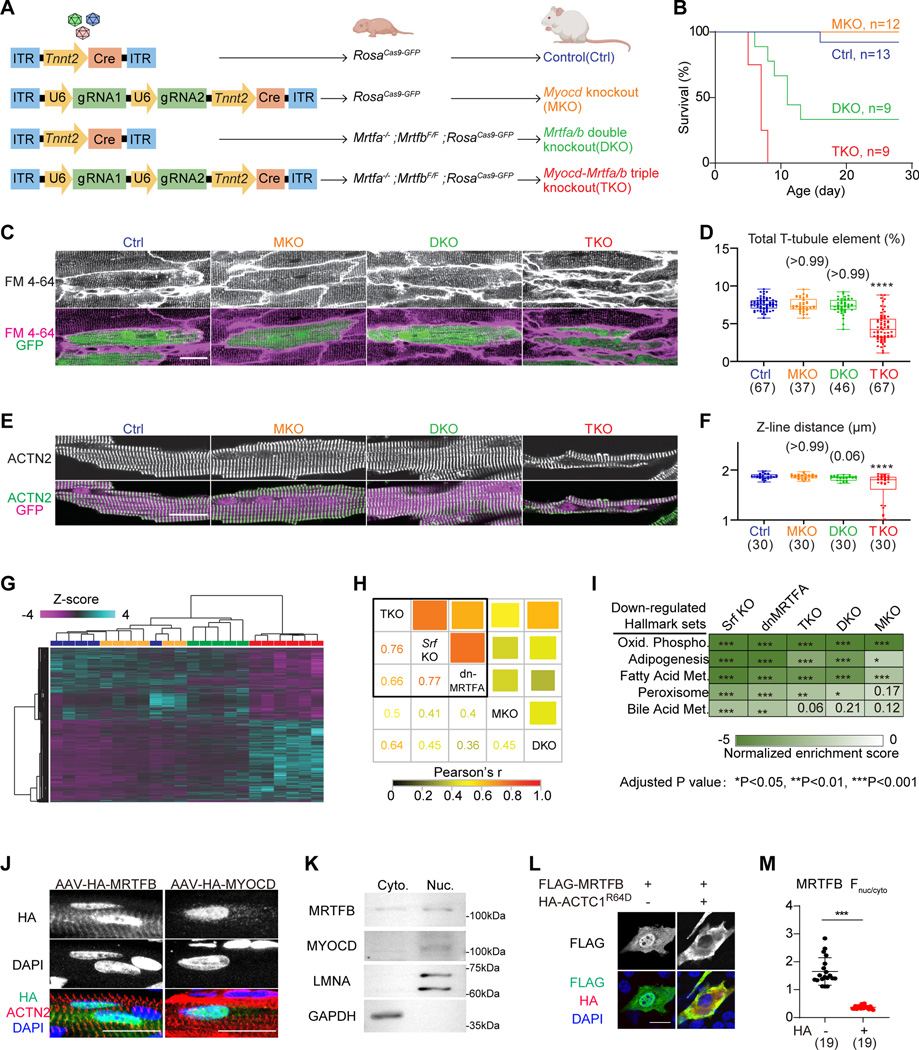
We next combined CRISPR/Cas9 and Cre-loxP systems to obtain Mrtfa/Mrtfb/Myocd triple knockouts (TKO), Mrtfa/Mrtfb double knockouts (DKO), and Myocd knockouts (MKO) as shown in Figure A. AAV- Cre-treated RosaCas9-GFP mice served as non-mutant controls (Ctrl). We firstly treated P1 animals with 3×109 vg/g AAV, a high dose that empirically infected more than 70% cardiomyocytes in the heart. After AAV administration, TKO mice all died before P10 (Figure B). About 67% of the DKO mice died by one month, while all MKO mice survived (Figure B). Echocardiographic analysis showed significantly reduced fractional shortening (FS), and increased systolic left ventricular internal diameter (LVID;s) in the TKO group at P7, while less severe cardiac dysfunction was observed in DKO or MKO groups (Figure S2A).
Figure: A shared role of the myocardin-family transcriptional coactivators in cardiomyocyte maturation.
(A) A schematic diagram showing the production of mice with different mutations of the myocardin-family genes. (B) Survival curves of mice that were injected with 3×109 AAV particles at P1. (C) In situ confocal imaging of hearts that were perfused with the cell membrane dye FM4–64. (D) T-tubule organization was quantified by AutoTT software. (E) Fluorescence confocal imaging of isolated cardiomyocytes immunolabelled with ACTN2 antibody to detect sarcomere Z-lines. (F) Distance between adjacent Z-lines was quantified. (G) Hierarchical clustering analysis of differentially expressed genes identified by RNA-Seq. (H) Pair-wise correlation among MKO, DKO, TKO, the published Srf KO and dnMRTFA data for the union of all differentially expressed genes. Both color and square size represents Pearson’s r. (I) Heatmap of top down-regulated hallmark gene sets.
(J) Confocal images of P14 immunostained heart sections that were treated with AAV-MRTFB or AAV-MYOCD at P1. (K) Western blot analysis of endogenous MRTFB and MYOCD in cytoplasmic (Cyto.) versus nuclear (Nuc.) fractions of a wild-type heart. (L) Immunofluorescent staining of NIH3T3 cells that were treated with the indicated plasmids. (M) Quantification of nuclear versus cytoplasmic fluorescence intensity. Scale bars indicate 20 μm in all relevant panels. Box-and-whisker plots: horizontal lines indicate 25th, 50th and 75th quantiles; whiskers extend to maximum or minimum values. Kruskal-Wallis test with Dunn’s multiple comparison correction: *Padj (adjusted P-value)<0.05, ***Padj (adjusted P value)<0.001, ****Padj (adjusted P value)<0.0001, non-significant Padj value in parenthesis above the whiskers. The number of measured cells were indicated in parenthesis below the plots.
Next, we treated the P1 animals with a lower dose of AAV (3×108 vg/g) to generate genetic mosaics, which circumvented animal death and allowed investigation of the cell-autonomous functions of myocardin family members. One month after AAV injection, in situ confocal imaging of hearts loaded with the plasma membrane dye FM 4–64 revealed severe defects in T-tubule organization in the TKO group as was quantified by the AutoTT software (Figure C–D). Immunostaining of ACTN2, a marker for sarcomere Z-lines, revealed disrupted sarcomere organization and reduced sarcomere length in TKO cardiomyocytes (Figure E–F). TKO cardiomyocytes also exhibited reduced cardiomyocyte size (Figure S2B). These morphological phenotypes were less severe in MKO or DKO groups (Figure C–F&S2B ).
Next, we purified AAV-transduced, GFP+ cardiomyocytes by fluorescence-activated cell sorting to perform low-input RNA sequencing analysis by SMART-Seq2. Principal component analysis (PCA) on 5–6 biological replicates per group demonstrated clear separation between TKO and the other three groups (Figure S3A). Hierarchical clustering revealed TKO as the major group exhibiting gene expression changes (Figure G). Pair-wise correlation analysis showed robust correlation between the TKO differential expression data and previously published Srf knockout (Srf KO, GSE109425) (Guo et al., 2018) or dnMRTF overexpression (dnMRTF-OE, GSE136096) (Guo et al., 2021) data, while DKO and MKO data exhibited weaker correlation with the other groups (Figure H).
Gene set enrichment analysis (GSEA) showed that all five genetic manipulations (Srf KO, dnMRTFA-OE, TKO, DKO, and MKO) downregulated oxidative phosphorylation and fatty acid metabolism gene expression programs (Figure I) whose upregulation are hallmarks of cardiomyocyte maturation. We also assessed key gene sets relevant to sarcomere contraction, mitochondrial respiration, cardiac electrophysiology, and calcium handling. Changes in these gene sets clustered the TKO, Srf KO, and dnMRTFA-OE groups (Figure S3B–C). Gene expression changes in DKO also showed fairly strong correlation, whereas correlation was weakest for MKO. These data showed that the three myocardin-family cofactors play an overlapping role in determining SRF activity during cardiomyocyte maturation.
We previously reported that the monomeric actin level was low in postnatal cardiomyocytes in vivo, so MRTFA was constitutively localized in cell nuclei (Guo et al., 2021). To test whether MRTFB and MYOCD behave following the same mechanism in cardiomyocytes, we constructed AAV vectors to deliver HA-tagged MRTFB or MYOCD to P1 hearts and performed immunostaining analysis of the HA tag in cardiac sections at P14. This analysis showed that MRTFB partially localized in the cell nucleus and additionally colocalized with ACTN2 on Z-lines (Figure J). By contrast, MYOCD was exclusively positioned in the cell nucleus (Figure J). Subcellular fractionation analysis of the heart further confirmed the nuclear localization of endogenous MRTFB and MYOCD (Figure K).
We wondered if ACTC1 could regulate the subcellular localization of MRTFB and MYOCD. Thus we expressed FLAG-tagged MRTFB or MYOCD in NIH3T3 cells in the presence or absence of ACTC1R64D, a mutant with reduced actin polymerization(Guo et al., 2021). Quantification of nuclear and cytoplasmic FLAG signals showed significantly reduced nuclear MRTFB after ACTC1R64D treatment (Figure L–M). By contrast, the localization of MYOCD was unaltered by ACTC1R64D (Figure S4). Thus, ACTC1 can selectively regulate the subcellular localization of MRTFB but not MYOCD.
Myocardin-family transcription cofactors are key transcriptional regulators of cardiac development and homeostasis, but the relative contribution of MYOCD versus MRTFA/B was unknown. Due to their different responses to monomeric actin, which we confirmed in this study, MYOCD was thought to function differently from MRTFA/B. Here we combined CRISPR/Cas9 and Cre-LoxP systems and clearly showed that these three factors function redundantly in postnatal cardiomyocyte maturation and function. Because the monomeric actin level is low in cardiomyocytes (Guo et al., 2021), MRTFA and MRTFB physiologically localize in the cell nucleus and share function with MYOCD to interact with SRF and activate downstream genes. This insight provides guidance to enhance actin polymerization as a new avenue to manipulate cardiomyocyte maturation.
Myocardin-family cofactors lack DNA binding domains and therefore interact with the genome indirectly. Due to their low endogenous expression levels and the lack of good antibodies for chromatin immunoprecipitation (ChIP) in heart tissues, this study was not able to acquire quality data to analyze the genome occupancy of these cofactors in vivo. Future advancement in ChIP-Seq techniques is necessary to identify the direct binding sites of each myocardin family members and distinguish their shared versus unique downstream gene targets.
Supplementary Material
Acknowledgments
We appreciate technical support by the Dana-Farber Flow Cytometry Core, the HMS Biopolymers Facility, and the animal facilities at Boston Children’s Hospital.
Funding information
This work was funded by the National Key R&D Program of China (2022YFA1104800 to Y.G.), the National Natural Science Foundation of China (82222006, 32100660 and 82170367), Beijing Nova Program (Z211100002121003 and 20220484205) and Beijing Municipal Natural Science Foundation (7232094). W.T.P. was supported by National Institute of Health (R01HL146634 and UM1HL098166). E.D.D. was supported by the Natural Science Foundation of China (82070235 and 92168113), the CAMS Innovation Fund for Medical Sciences (2021-I2M-5-003) and Haihe Laboratory of Cell Ecosystem Innovation Fund (HH22KYZX0047).
Footnotes
Competing Interest Statement
The authors declare no competing interests.
Reference
- Alvarez-Dominguez JR and Melton DA (2022) ‘Cell maturation: Hallmarks, triggers, and manipulation’, Cell 185(2): 235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Cao Y, Jardin BD, Sethi I, Ma Q, Moghadaszadeh B, Troiano EC, Mazumdar N, Trembley MA, Small EM et al. (2021) ‘Sarcomeres regulate murine cardiomyocyte maturation through MRTF-SRF signaling’, Proc Natl Acad Sci U S A 118(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Jardin BD, Zhou P, Sethi I, Akerberg BN, Toepfer CN, Ai Y, Li Y, Ma Q, Guatimosim S. et al. (2018) ‘Hierarchical and stage-specific regulation of murine cardiomyocyte maturation by serum response factor’, Nat Commun 9(1): 3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yao F, Yu P, Li D, Zhang M, Mao L, Shen X, Ren Z, Wang L. and Zhou B. (2022) ‘Postnatal state transition of cardiomyocyte as a primary step in heart maturation’, Protein Cell 13(11): 842–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipes GC, Creemers EE and Olson EN (2006) ‘The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis’, Genes Dev 20(12): 1545–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.