Abstract
Regulation of the mRNA cap binding protein (eIF4E) is critical to the control of cellular proliferation since this protein is the rate-limiting factor in translation initiation and transforms fibroblasts and since eIF4E mutants arrest budding yeast in the G1 phase of the cell cycle (cdc33). We previously demonstrated regulation of eIF4E by altered transcription of its mRNA in serum-stimulated fibroblasts and in response to c-myc. To identify additional factors regulating eIF4E transcription, we used linker-scanning constructs to characterize sites in the promoter of the eIF4E gene required for its expression. Promoter activity was dependent on sites at −5, −25, −45, and −75; the site at −75 included a previously described myc box. Electrophoretic mobility shift assays identified DNA-protein interactions at −25 and revealed a binding site (TTACCCCCCCTT) that is unique to the eIF4E promoter. Proteins of 68 and 97 kDa bound this site in UV cross-linking and Southwestern experiments. Levels of 4E regulatory factor activities correlated with c-Myc levels, eIF4E expression levels, and protein synthesis in differentiating U937 and HL60 cells, suggesting that these activities may function to regulate protein synthesis rates during differentiation. Since the eIF4E promoter lacked typical TATA and initiator elements, further studies of this novel initiator-homologous element should provide insights into mechanisms of transcription initiation and growth regulation.
The mRNA cap binding protein eIF4E, which recognizes the m7GpppN cap structure on all mRNA molecules, is a critical regulator of cell growth and protein synthesis (65). In binding the cap structure, it also participates in pleiotropic functions including regulation of mRNA export from the nucleus, splicing, and mRNA stability (27, 47). eIF4E is especially important in regulating new protein synthesis because its low abundance makes it rate limiting for translation initiation (13, 27). eIF4E mutants alter G1 cell cycle regulation (cdc33) and eIF4E overexpression in mammalian cells is both mitogenic and tumorigenic (2, 66), indicating that levels of eIF4E are critical for cell division as well as protein synthesis control.
eIF4E abundance varies in response to growth stimulation (13, 56). In addition, various signaling pathways regulate its activity by phosphorylation of critical residues (especially serine 209 [18–20, 28, 36, 52, 71]) and 4E binding proteins inhibit its function (50). Nevertheless, its overall abundance is of key importance to its regulation, since simple overexpression of its mRNA is sufficient to transform cells (10, 42). Reasoning that the regulation of translation initiation factors may be particularly important at points in the cell cycle where protein synthesis is rate limiting for growth, we found that levels of eIF4E mRNA peaked at the restriction point in late G1 in growth factor-stimulated fibroblasts (56).
Recent studies have focused renewed attention on mechanisms that regulate the transcription of gene products governing protein synthesis rates (3, 38, 69). Yeast ribosomal protein levels are regulated by several transcription factors (32, 38, 46). In mammalian cells, a transcriptional element in the promoter of the gene encoding eukaryotic initiation factor 2α (eIF2α) is shared among several promoters of genes encoding mitochondrial proteins (4) and promoters of several cell cycle regulators (21, 68), thereby coordinating protein synthesis, energy metabolism, and cell cycle control. A newly described transcription factor, nuclear regulatory factor 1 (NRF-1), regulates eIF2α through that site, and its sequence is homologous to those of a family of developmental regulators in Drosophila (14, 69). In addition, the retinoblastoma tumor suppressor gene (Rb) inhibits RNA polymerase I and III transcription and is thought to inhibit protein synthesis by decreasing ribosomal biogenesis (3, 7, 41, 72). Taken together, the examples of yeast ribosomal biogenesis, NRF-1, and Rb provide important illustrations of transcriptional regulation of factors regulating protein synthesis.
The c-myc oncogene is a key regulator of cell proliferation and differentiation (16, 43). Despite intense scrutiny of its biochemical functions, less is known about the c-myc target genes that mediate its functions in growth control (23). We previously observed that the peak levels of eIF4E mRNA at the restriction point in late G1 coincided with peak levels of c-Myc (56). Consequently, we evaluated the transcriptional response of eIF4E mRNA to c-myc by using cells expressing estradiol-regulated c-myc fusion constructs (myc-er cells) (56). Transcriptional increases of eIF4E mRNA in myc-er cells coincided with elevated protein synthesis that preceded the start of DNA synthesis (15, 54, 56). These experiments led us to clone the proximal promoter of the eIF4E gene (35), which revealed myc box motifs at −77 and −232 that were essential for promoter function. Surprisingly, this promoter lacked both TATA and initiator (INR) elements, which are usually necessary to guide the formation of RNA polymerase II transcription complexes (22, 70). To understand the function of myc, direct interactions between Myc binding in candidate target genes and transactivation or repression of basic promoter elements in the same genes must eventually be identified (61). The absence of the usual promoter elements in the eIF4E promoter that are required for RNA polymerase II function led us to search for alternative sequences that might function in initiating its transcription.
In this study, we identified transcription elements required for eIF4E expression by using reporter constructs containing linker-scanning mutations of the eIF4E promoter. Although coordination of growth and division is often studied at the restriction point when cell proliferation is induced, we sought to determine whether this coordination is also seen when cells exit the cell cycle. Among its many functions, c-myc regulates cell cycle arrest and differentiation in hematopoietic cells (25, 31, 73). Since c-myc plays a prominent role in arresting DNA synthesis during differentiation of HL60 and U937 cells, its target genes should be similarly down-regulated and studies of this regulation should provide additional insights into mechanisms coordinating protein synthesis and cell division.
MATERIALS AND METHODS
Phages, plasmids, and nucleic acids.
Standard manipulations of Escherichia coli, nucleic acids, and tissue culture cells were performed essentially as described previously (60). With peIF4ECAT as a starting plasmid [p4ECAT(403) from reference 35), a series of eIF4E-CAT linker scanner constructs were made by the PCR-based overlap extension technique (29), as modified by Datta (8). Briefly, sense oligonucleotides encoding the designed 10-base substitution (but otherwise complementary to eIF4E promoter sequences; typically 15 to 20 bases on either end of the 10-base linker) were used in PCRs with peIF4ECAT as a template, along with an antisense primer complementary to sequences flanking the 3′ end of the eIF4E promoter sequences. The PCR products were purified by agarose gel electrophoresis and used in a second PCR with peIF4ECAT as a template and a sense primer complementary to sequences flanking the 5′ end of the eIF4E promoter sequences. The PCR products were then digested with PstI and XhoI and subcloned into the same sites of peIF4ECAT. DNA sequencing of the entire eIF4E promoter region verified all the introduced substitutions. These constructs successively replaced every 10 bases with the sequence ACTCTAGACT, which contained no known transcription-activating elements.
A mouse genomic DNA library (strain 129SVJ) cloned in λ FIX II phage (Stratagene, La Jolla, Calif.) was screened with a Klenow-labeled fragment containing human eIF4E genomic sequences from an FspI site at −110 to a BssHII site at +103. Positive phages were plaque purified. Five phages were mapped and found to represent two independent inserts. Mouse genomic sequences between an XbaI site at −1045 and an ApaI site at +235 were subcloned into plasmid vectors for sequencing. Sequences were obtained with an automated sequencer through the Massachusetts General Hospital core sequencing facility. The mouse genomic sequences were compared to equivalent human sequences by using Genetics Computer Group software from the University of Wisconsin package.
Unless otherwise designated, oligonucleotides used in electrophoretic mobility shift assay (EMSA) experiments were custom synthesized by Gibco-BRL and are summarized in Fig. 1.
FIG. 1.
Oligonucleotides used in the EMSA experiments.
Cells, transfections, and CAT assays.
HeLa and HL60 cells were obtained from the American Type Culture Collection, Manassas, Va.; rat embryo fibroblasts transfected with c-myc (REF-myc) or the neomycin resistance gene (REF-neo) were the generous gift of R. A. Weinberg. U937 cells were the generous gift of Ben Kreskel and Alan Ezekowitz. Adherent cells were routinely grown in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum; HL60 and U937 cells were routinely grown in RPMI with 10% fetal bovine serum (FCS).
Transfections were accomplished by standard calcium phosphate coprecipitation (60). For the linker-scanning constructs, HeLa cells were transfected with 10 μg of eIF4E-CAT reporter constructs and 3 μg of pSVtkhGH, which is not regulated by c-myc (37). We determined human growth hormone levels in supernatant media by using a commercial radioimmunoassay kit (Allegro Inc.) to normalize for transfection efficiency. Chloramphenicol acetyltransferase (CAT) assays were performed by thin-layer chromatography, using standard methods. All CAT assays were performed with duplicate points, and each experiment was repeated three times.
DNA binding assays.
Nuclear extracts were prepared from 108 of the indicated cells during logarithmic growth for EMSA by a modification of the Dignam method (11, 12, 48). The modified extraction buffer used (0.5% deoxycholate, 1% octyl-β-glucoside, 20 mM HEPES [pH 7.9], 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol [DTT], 25% glycerol) results in improved yields of c-Myc in the lysates. Binding reaction mixtures included 10 μg of the indicated extracts. We evaluated gel shift activity in standard EMSA binding buffer (10 mM Tris [pH 7.5], 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 5 mM MgCl2, 5% glycerol [1]) with poly(dI-dC) (0.1 μg/μl) as a nonspecific competitor. Complexes formed in binding buffer were resolved on 6% nondenaturing polyacrylamide gels containing 1× TBE (0.090 M Tris-borate [pH 8.0], 0.002 M EDTA).
Oligonucleotides were labeled with polynucleotide kinase and [γ-32P]dATP if they contained blunt ends or by Klenow reactions with [α-32P]dCTP if they contained 5′ overhangs. Each binding-reaction mixture contained 0.1 to 0.5 ng of labeled oligonucleotide. Competition experiments were performed with the indicated molar excess of unlabeled oligonucleotides.
UV cross-linking and Southwestern analyses.
The LS3 trimer oligonucleotide, AAGGGGGGG TAAGAGGAAGAAGGGGGGG TAAGAG GAAGAAG G GGGGGTAAGAGGAAACTCTAGACT, was annealed with AGTCTAGAGTTT. These oligonucleotides were then radiolabeled with 5-bromo-2′-dUTP (50 μM) and α32-P-labeled dCTP (5 μM) by using standard Klenow reaction mixtures together with cold dATP (50 μM) and dGTP (50 μM) (6). The 66-bp trimeric oligonucleotide was then purified by electroelution by using polyacrylamide gel electrophoresis and size markers to identify the full-length product. The full-length cross-linking probe was then incubated with 25 μg of nuclear lysates from HeLa cells, 1 mg of bovine serum albumin per ml, and 20 μg of poly(dI-dC) under standard EMSA conditions (6). Each reaction mixture was irradiated from 5 cm directly above by inverting a UV transilluminator of 305 nm and intensity 7,000 μW/cm2 for a period experimentally determined to optimize cross-linking (20 min). After incubation with DNase I (2 μg per reaction), the samples were analyzed on 10% denaturing polyacrylamide gels to identify proteins which bound the eIF4E promoter sites.
Southwestern analyses were performed essentially as described previously (26, 63, 67, 68). Nuclear extracts (50 μg) from the indicated cell types were run on 10% denaturing polyacrylamide gels, electroblotted to nitrocellulose for 12 h, and allowed to dry. The filters were then immersed for 10 min in denaturation-renaturation buffer containing 6 M guanidine hydrochloride. Partial renaturation of immobilized proteins was performed by five successive incubations of the filters in buffer containing progressive (twofold) dilutions of guanidine hydrochloride and finally in buffer lacking the denaturant. The filters were blocked with 5% nonfat dry milk in binding buffer (50 mM Tris [pH 7.5], 50 mM NaCl, 1 mM EDTA, 1 mM DTT) and were then washed twice with 0.25% nonfat dry milk in binding buffer. The filters were hybridized in binding buffer containing Klenow-labeled LS3 trimer oligonucleotide probe, 0.25% nonfat dry milk, and 1 μg sonicated salmon sperm DNA per ml for 60 min at room temperature. The filters were then washed three times with binding buffer containing 0.25% nonfat dry milk alone and dried.
Expression analysis of U937 and HL60 cells.
HL60 and U937 cells were initially seeded at 105 cells per 150-mm plate for all determinations. Differentiation was induced with 5 nM 12-O-tetradecanoylphorbol-13-acetate (TPA). Cells from individual plates were harvested at the indicated time points for RNA and protein analyses.
Levels of expression of eIF4E, c-myc, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNAs were analyzed with total cellular RNA (5) from U937 and HL60 cells that was size fractionated (10 μg/lane) on formaldehyde-agarose gels, transferred to Hybond-N nylon matrices, and cross-linked with UV light. Filters were hybridized in a rapid hybridization solution (Rapidhybe; Amersham) at 65°C with eIF4E, c-myc, or GAPDH cDNA fragments α32-P labeled by the Klenow reaction with random priming.
Levels of expression of eIF4E, c-Myc, and actin proteins were compared in U937 and HL60 cells by using immunoblots containing 50 μg of total protein harvested in Laemmli loading buffer as described previously (56, 60). Briefly, 50 μg of cell lysate was analyzed on a standard 10% denaturing polyacrylamide gel. Proteins were electroblotted onto a nylon membrane (Immobilon; Millipore) and blocked overnight in 5% dry milk. The membrane was cut according to the molecular weights of the proteins to be identified, and the identical blot was incubated with anti-c-Myc (9E10; Santa Cruz), anti-actin (N-350; Boehringer), or anti-eIF4E (rabbit polyclonal; gift of Nahum Sonenberg) antibodies. The secondary antibodies used were those included in an enhanced chemiluminescence detection kit (Amersham) and were chosen on the basis of the species used for the primary antibodies.
Protein and DNA synthesis rates in HL60 and U937 cells were determined by adding 20 μCi of [35S]methionine (Dupont-NEN) to each plate along with 20 μCi of [3H]thymidine for 3 h. The labeled cells were lysed, and incorporated counts were determined by trichloroacetic acid precipitation as described previously (9). Identical plates were also harvested at the end of the 3-h incubations in 250 μl of Laemmli loading buffer; 50-μl samples of this lysate were compared on standard protein electrophoresis gels at each time point. The pattern of proteins displayed on these one-dimensional gels has been used to evaluate the synthesis rates of the most abundant individual proteins in cells (9).
Nucleotide sequence accession number.
The nucleotide sequence of the mouse eIF4E promoter has been deposited in GenBank (accession no. AF079156).
RESULTS
Linker-scanning reporter constructs of the eIF4E promoter reveal novel sites necessary for its transcription at nucleotides −5, −25, and −45.
Our previous analysis of the eIF4E promoter revealed a unique transcriptional start site, although TATA and initiator elements were absent; classic c-myc and CCAAT box motifs were also identified (35). To identify additional elements required for transcriptional regulation of eIF4E, we constructed a series of linker-scanning constructs within the eIF4E proximal promoter. Linker-scanning constructs were made by site-directed mutagenesis in which 10 successive nucleotides were replaced with the sequence ACTCTAGACT (Fig. 2). This sequence was chosen because it contained no homology to any known transcription factor binding sites and was designed to include an XbaI site. These constructs were transfected into HeLa cells, and promoter activity was determined by standard CAT assays (Fig. 2). Mutations at four sites markedly decreased promoter activity compared to that of the unaltered eIF4E-CAT construct. The loss of the c-Myc binding site in the reporter construct containing a linker spanning −71 to −80 (LS8) resulted in a 10-fold decrease in promoter activity. Reporter activity was also markedly decreased in the constructs containing linkers spanning −1 to −10 (LS1), −21 to −30 (LS3), and −41 to −50 (LS5). These sites were located downstream of the CCAAT box (position −59), in a region of the promoter that typically directs transcriptional initiation.
FIG. 2.
Identification of sites within the eIF4E promoter necessary for promoter activity. (Top) General scheme of linker-scanning mutations. (Bottom) The indicated eIF4E-CAT linker-scanner constructs were transfected into HeLa cells and analyzed for CAT activity. The effect of the mutation on CAT expression compared with that of the unaltered eIF4E-CAT construct is presented as the mean and standard error. The mean and standard error are based on three transfections, each performed in duplicate (n = 6 for each determination).
Critical transcriptional elements are often conserved among different species. To help us identify regions of the eIF4E promoter that might specify conserved promoter elements, we screened a murine genomic library for phages containing genomic eIF4E sequences. Probes made with human promoter sequences between FspI and BssHII sites (−110 to −122) detected a single band on murine Southern blots (data not shown). We used this probe to screen a murine genomic library and identified three independent phages carrying eIF4E promoter sequences. We subcloned sequences between an XbaI site at −1043 and an ApaI site at +240 for use in automated sequencing. A comparison of the human and mouse eIF4E promoters revealed extensive homology between the two species throughout the whole promoter region (Fig. 3). The distal c-Myc binding site at −232 was conserved, and its flanking sequences were similar to those of the human site. The proximal c-Myc site at −77 and its flanking sequences were identical between the two species. Additionally, the three proximal sites identified by the linker-scanning mutations LS1, LS3, and LS5 were highly conserved. TATA and consensus binding sites for initiator regions were again absent from the mouse promoter. A canonical CCAAT box was present in both species at position −59.
FIG. 3.
Comparison of sequences from the mouse and human eIF4E promoter. The sequences of the mouse and human promoter regions extending 5′ to the PstI site used to make peIF4E-CAT are compared. The mouse and human promoters are markedly similar in the regions of the human promoter used in these studies. MB1 and MB2 identify the two myc boxes previously identified in this promoter (35). Exon 1 is shaded. The CCAAT box and the three linker sites critical to promoter functions are boxed and indicated.
EMSAs reveal a novel polypyrimidine transcription element between −17 and −28 in the eIF4E promoter.
Although the linker-scanning mutations identified sites at −5, −25, and −45 that were critical to eIF4E promoter activity, none of the sites corresponded to known targets for DNA binding proteins. Consequently, we used electrophoretic mobility gel shifts to define candidate DNA binding activities interacting with the eIF4E promoter. To identify DNA binding activities, we generated three sets of probes containing eIF4E promoter sequences between −59 and +3. These sets included insertion mutations, 5′ deletions, and 3′ deletions (Fig. 4A, B, and C, respectively). The probes containing sequential insertion mutations (Fig. 4A) were generated by digesting LS1 through LS5 with MscI and XhoI, resulting in a series of constructs with the sequence ACTCTAGACT successively replacing 10 nucleotides at a time. Unaltered eIF4ECAT was similarly digested to provide a wild-type, control probe. When the constructs were analyzed in standard EMSAs, it was found that the insertion of sequences between −11 and −30 abolished gel shift activity (probes MX2 and MX3 [Fig. 4A, lanes 1, 4, and 5]. Activity was also decreased by the insertion of a linker between −41 and −50 (probe MX5 [lane 2]), although this effect was less marked. These data identified an activity that bound DNA sequences between −11 and −30 which was critical for eIF4E promoter activity and provides independent confirmation of these sequences compared to the previous reporter construct experiments.
FIG. 4.
EMSA experiments identify a unique protein binding region within the eIF4E proximal promoter sequences. (A) LS1 through LS5 digested with MscI and XhoI generated a series of insertions across the promoter. These oligonucleotides and the corresponding sequences from unaltered eIF4E-CAT were radiolabeled and analyzed by standard EMSA. The indicated cold competitor oligonucleotide (1000× Cold) contained the wild-type eIF4E oligonucleotide to evaluate specificity. The sites where linker sequences replace endogenous sequence are written in lowercase and are underlined for emphasis throughout this figure. (B) LS2 through LS7 were digested with XbaI and XhoI, generating a series of 5′ deletion mutants. These oligonucleotides were radiolabeled and EMSAs were performed as above. The indicated cold competitor oligonucleotide (1000× Cold) contained the wild-type eIF4E oligonucleotide. (C) LS1 through LS5 were digested with MscI and XbaI, generating a series of 3′ deletion mutants. These oligonucleotides and the corresponding sequences from unaltered eIF4E-CAT were radiolabeled and EMSAs were performed as above. The indicated cold competitor oligonucleotide (1000× Cold) contained wild-type eIF4E.
We further confirmed the importance of the −11 to −30 site by using sequential 5′ and 3′ deletions (Fig. 4B and C, respectively). The 5′ deletions were generated by cutting LS2 through LS7 with XbaI and XhoI; the reporter construct containing wild-type eIF4E sequences was identically digested (Fig. 4B). Gel shift activity was lost when sequences between −21 and −30 were deleted (XX3 [Fig. 4B, lanes 1, 5, and 6]), precisely matching our results obtained with the insertion constructs. To generate nested 3′ deletions, LS1 through LS5 were digested with MscI and XbaI (Fig. 4C). Deletion of sequences between −1 and −10 actually increased binding compared to a wild-type eIF4E probe (MB1 [Fig. 4C, lanes 1 and 2]). In contrast, deletion of sequences between −11 and −20 resulted in a loss of gel shift activity (MB2 [lanes 1 and 3]). Our results suggested that DNA binding activity was critically dependent upon intact sequences between −11 and −30 and that flanking sequences centered at nucleotides −5 and −45 modulated the activity.
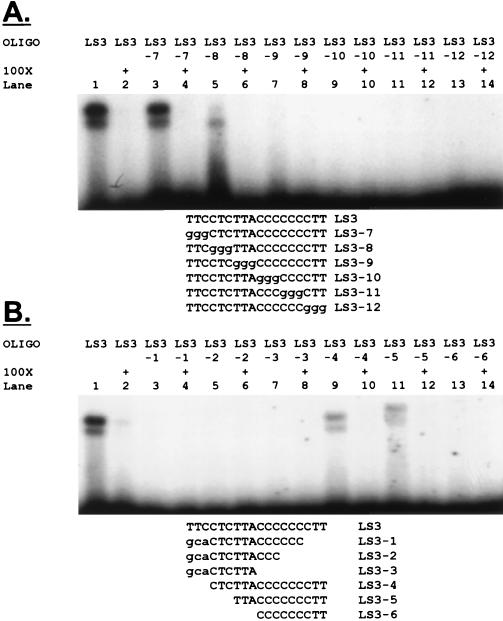
To determine the minimum sequence necessary for binding activity, we made a series of 3-nucleotide insertion and deletion mutations spanning nucleotides −15 to −30. The first series of insertion construct oligonucleotides, in which every 3 nucleotides was sequentially replaced by the sequence GGG, revealed the core sequence necessary for binding activity as TTACCCCCCCTT (Fig. 5A). Addition of the 5′-flanking CTC sequence resulted in maximum activity (compare lanes 3 and 5). This binding sequence was confirmed by using oligonucleotides in which 3 nucleotides were successively deleted from either end of the sequence (Fig. 4B), although the 5′ terminal TTC also appeared to contribute to binding in this experiment (compare lanes 1 and 9). Again, we identified the TTACCCCCCCTT core as necessary for binding activity, with the 5′-flanking TTCCTC being needed to maximize activity. The core DNA binding motif apparently extends from nucleotides −28 to −17 and is contained primarily within LS3.
FIG. 5.
A core 12-nucleotide element is sufficient for binding activity. (A) A series of LS3 oligonucleotides was generated in which every 3 nucleotides were sequentially replaced by GGG. The indicated oligonucleotides were analyzed by EMSA. Cold competitor oligonucleotides contained the same sequences as the probes to demonstrate specificity. (B) Another series of mutant LS3 oligonucleotides was generated in which deletions of three nucleotides were made sequentially from both ends. The indicated oligonucleotides were analyzed by EMSA. Cold competitor oligonucleotides contained the same sequences as the probes.
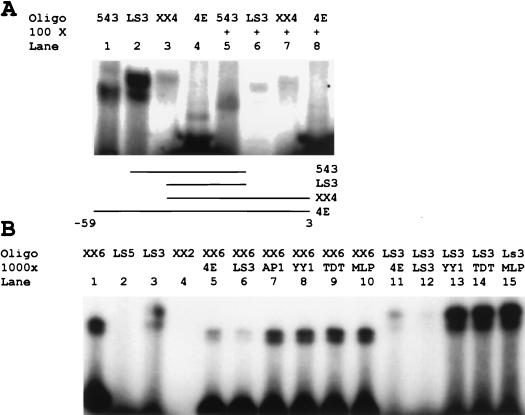
We further explored the apparent modulation of binding activity by the flanking sites at nucleotides −5 and −45 by generating a series of oligonucleotides containing various combinations of the sites centered at nucleotides −5, −22, and −45. Oligonucleotide 543 contained the sites centered at −22 and −45 but lacked the site at nucleotide −5. Oligonucleotide LS3 contained only the core −22 binding site, flanked by the CTC motif to generate maximum binding activity. XbaI and XhoI digests of LS4 produced the XX4 oligonucleotide. This oligonucleotide contained the sites centered at −5 and −22 but lacked the site centered at −45. Oligonucleotide 4E contained all three sites. We found maximum binding with the LS3 oligonucleotide (Fig. 6A). Activity was partially decreased in the presence of the site centered at nucleotide −45 (543) and decreased significantly with addition of the site at nucleotide −5 (XX4). Indeed, the two activities which bind to the 4E nucleotide are barely seen in Fig. 6A compared with Fig. 4 because of the difference in the exposure time of gels needed to accommodate the strong binding of the LS3 oligonucleotide. This suggests that flanking sites in both LS1 and LS5 affect binding to the core LS3 activity. The site centered in LS1 markedly inhibited binding, and the site at −45 curbed this effect somewhat, although it was inhibitory on its own.
FIG. 6.
Core binding activity at the eIF4E binding site differs from known initiator elements in cross-competition experiments. (A) Core binding seen at a unique binding site in the eIF4E promoter is modified by interactions at flanking sites. The LS543 oligonucleotide and the LS3 oligonucleotide were directly radiolabeled. LS4 and eIF4E-CAT were digested with XbaI and XhoI and radiolabeled. The resulting oligonucleotides were subjected to EMSA as described in the text. Cold competitor oligonucleotide (row 100×) was included to demonstrate specificity. Lines in the schematic below the gel show what portion of the whole region between −59 and +3 is included in each of the indicated oligonucleotides. (B) Core binding activity at the eIF4E binding site differs from known initiator elements. The LS5 and LS3 oligonucleotides were directly radiolabeled. LS2 and LS6 were digested with XbaI and XhoI and radiolabeled. The resulting oligonucleotides were subjected to EMSA as described above. Cold competitor oligonucleotides (row 1000×) were used to determine specificity (see Fig. 1 for identification of competitors).
The TTACCCCCCCTT sequence was not homologous to any known DNA element or the binding site of any transcription factor in current databases. The absence of a TATA sequence led us to consider possible similarities between the eIF4E polypyrimidine element and initiator sequences. Although the novel element appeared similar to classic initiator sites because of its high pyrimidine content, it was not located at the site of transcription initiation as expected of typical initiator sites (64). To evaluate this apparent similarity in sequences, we used cross-competition experiments with previously described binding sites for various initiator elements (Fig. 6B). We performed cross-competition experiments with both the core LS3 site and an oligonucleotide containing the entire proximal promoter region. Cold competitor oligonucleotides containing the Yin-Yang 1 (YY1) binding site, the initiator region of the terminal deoxynucleotidyltransferase promoter (TDT), and the initiator region of the adenovirus major late promoter (MLP) were compared. An AP-1 binding-site oligonucleotide unrelated to these factors was used as a control. Binding activity was unaffected by any competitor; no cross-reactivity was observed with the −22 site alone or in the presence of the flanking activities at sites −5 and −45. Competition with the homologous cold oligonucleotides confirmed the specificity of the binding activity. This result suggested that the binding activity seen at the site at −25 is due to a novel polypyrimidine DNA binding site which does not correspond to known initiator elements.
Identification of proteins which bind the novel −22 polypyrimidine element by using UV cross-linking.
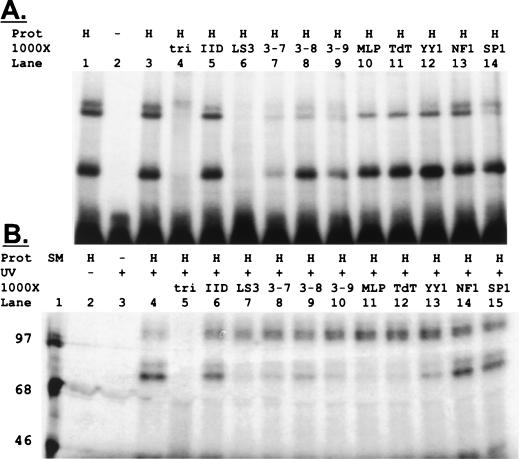
Our data suggested that a novel polypyrimidine site centered at nucleotide −22 was responsible for binding activity. To identify proteins that bound this site, we used UV cross-linking studies. We first compared multimeric LS3 site probes and found that a trimeric probe resulted in the highest binding affinity (data not shown). This oligonucleotide, the LS3 trimer (tri), was radiolabeled with [α-32P]dCTP and with 5-bromo-2′-deoxyuridine along with excess cold dGTP and dATP. Full-length, double-stranded probes were purified for these studies. The trimeric probe bound three retarded complexes in EMSAs: one doublet of slow-migrating complexes and a single fast-migrating complex (Fig. 7A). Cross-competition with cold trimeric probe (tri) confirmed binding specificity. We then used cold oligonucleotides containing the indicated sites in cross-competitions. The lower band in this EMSA appeared more specific, competing only with the LS3 monomer, the LS3-7 oligonucleotide, and trimer oligonucleotides. The upper doublet appeared less specific. No competition was observed with TATA, NF-1, or SP-1.
FIG. 7.
Novel 97- and 68-kDa proteins bind the LS3 site in UV cross-linking experiments. (A) A trimeric form of the LS3 oligonucleotide, AAGGGGGGGTAAGAGGAAGAAGGGGGGGTAAGAGGAAGAAGGGGGGGTAAGAGGAAACTCTAGACT, was primed with AGTCTAGAGT and labeled with 5-bromo-2′-dUTP and [α-32P]dCTP, and an excess of cold dGTP and dATP. A full length, double-stranded, trimeric probe containing all 66 bp was purified by polyacrylamide gel electrophoresis. This oligonucleotide (lane tri) was analyzed by EMSA as described above (Fig. 4). The cold competitor oligonucleotides used to demonstrate specificity included TATA (lane IID), the LS3 monomer (lane LS3), the LS3-7, LS3-8, and LS3-9 mutant oligonucleotides, the initiator region binding sites in the adenovirus major late promoter (lane MLP), the initiator region of the terminal deoxynucleotidyltransferase promoter (lane TDT), the Yin-Yang 1 (lane YY1) binding site, and unrelated NF-1 and SP-1 sites. (B) The full-length, double stranded LS3 trimer oligonucleotide was radiolabeled with 5-bromo-2′-dUTP and [α-32P]dCTP along with cold dATP and dGTP in standard Klenow reactions. The 66-bp probe was then incubated with 25 μg of nuclear lysates from HeLa cells as described in Materials and Methods. Each reaction mixture was irradiated with a UV transilluminator for a period experimentally determined to optimize cross-linking. After incubation with DNase I, the samples were analyzed on a 10% denaturing polyacrylamide gel. The indicated cold competitor oligonucleotides were used to demonstrate specificity (see Fig. 1 for identification of competitors). Size markers are indicated (SM).
We then used UV cross-linking to identify proteins binding at the LS3 site. The affinity-labeled, full-length, double-stranded LS3 trimer oligonucleotide was incubated with nuclear lysates from HeLa cells under standard EMSA conditions and then subjected to UV irradiation to cross-link DNA binding proteins to the radioactive oligonucleotide. After incubation with DNase I, the lysates were resolved on 10% denaturing polyacrylamide gels (Fig. 7B). Two specific proteins, of 97 and 68 kDa, bound the −22 site. Cross-competition reactions with the TFII-D, LS3 trimer, LS3 monomer, LS3-7, LS3-8, LS3-9, MLP, TDT, YY1, NF-1, and SP-1 oligonucleotides were also performed in the cross-linking studies. We found that the 97-kDa protein corresponded to the more specific lower band on the EMSA, since only the cold LS3 trimer oligonucleotide itself competed for binding. Binding to the 97-kDa protein was not competed by any other polypyrimidine elements used, nor did it compete with the unrelated control factors, NF-1 and SP-1. The 68-kDa protein appeared less specific. It did not cross-react with TFII-D or with the NF-1 or SP-1 oligonucleotides but was competed by all polypyrimidine oligonucleotides as seen with the upper band in the EMSA analysis.
Levels of the novel 97- and 68-kDa proteins identified in Southwestern blots correlated with expression of c-Myc and with down-regulation of protein synthesis during differentiation.
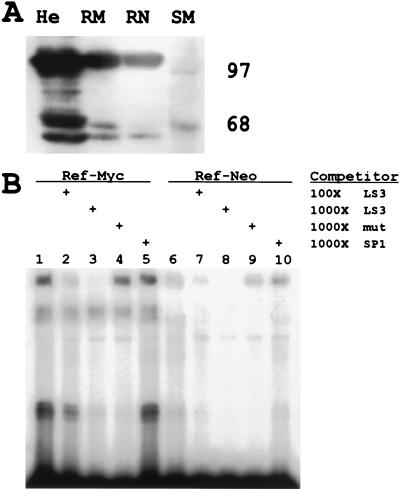
We also identified the 97- and 68-kDa proteins by using the same trimeric probe in Southwestern analyses of three cell lines (HeLa, REF-myc, and REF-neo cells) expressing different levels of eIF4E and c-Myc (Fig. 8A). Renatured filters were probed with the radiolabeled LS3 trimer oligonucleotide. The amounts of the 97- and 68-kDa LS3 binding activity corresponded to the amounts of eIF4E and c-Myc present in the cells. HeLa cells express high levels of c-Myc, rat embryo fibroblasts stably transfected with c-myc (REF-myc) express moderate levels of c-Myc, and rat embryo fibroblasts stably transfected with the neomycin resistance gene (REF-neo) express low levels of the protein (35, 56).
FIG. 8.
Levels of LS3-interacting proteins correlate with c-Myc in fibroblast cell lines. (A) Nuclear extracts (50 μg) from HeLa (lane He), REF-myc (lane RM), and REF-neo (lane RN) cells blotted on nitrocellulose filters were renatured through guanidine hydrochloride and then hybridized in Southwestern binding buffer containing the α-32P-labeled LS3 trimer oligonucleotide probe. c-Myc levels decrease from 90,000 copies of c-Myc protein in HeLa cells (49) to 30% of that level in REF-myc cells and 10% in REF-neo cells (35). Size markers (lane SM) are indicated. (B) Binding of the α-32P-labeled LS3 trimeric probe to REF-myc and REF-neo nuclear extracts (10 μg) was compared in EMSA experiments. Binding conditions were identical to those used for experiments in previous figures. The indicated amounts of cold-competitor oligonucleotides (LS3 and SP1) were added in the designated lanes to evaluate the specificity of binding. A trimeric version of mutant oligonucleotide LS3-8 (mut) is included in lanes 4 and 9.
To confirm this finding, we performed an EMSA with nuclear extracts from REF-myc and REF-neo cells together with the radiolabeled LS3 trimer oligonucleotide (Fig. 8B). Again, we found that LS3 trimeric binding activity corresponded to relative c-Myc levels in cells.
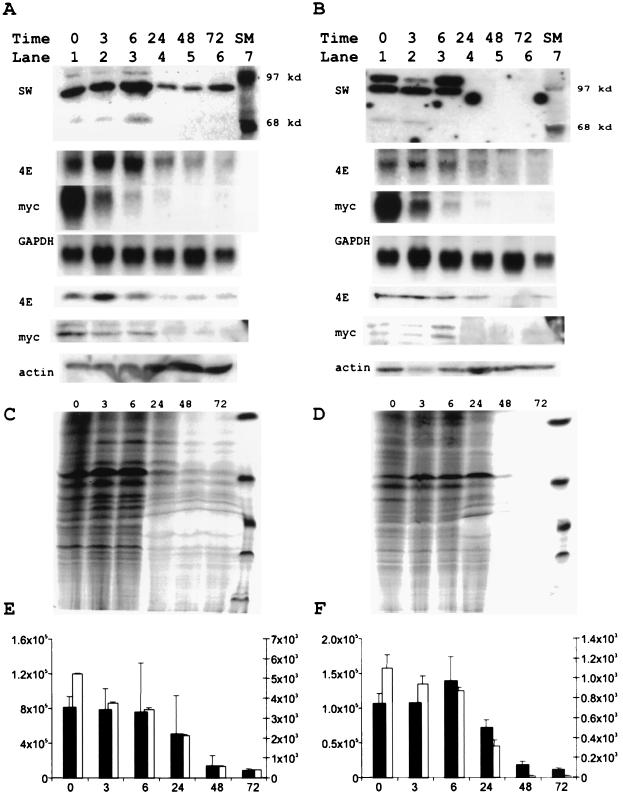
We assessed potential functional roles for the 97- and 68-kDa proteins in the regulation of both eIF4E and net protein synthesis by evaluating their expression patterns during differentiation of U937 and HL60 model cell lines. We induced differentiation and analyzed LS3 trimer binding activity at various time points after differentiation by using Southwestern blots (Fig. 9A and B). We found that levels of the 97- and 68-kDa proteins decreased markedly at 24 h and remained low through 72 h. Although c-myc Northern blots revealed immediate decreases in c-myc mRNA levels, the level of c-Myc protein did not decrease until 24 h in Western blots. The levels of eIF4E mRNA likewise fell at 24 h, corresponding to the decreased c-Myc protein and LS3 binding activity, and this decrease was accompanied by decreased eIF4E protein levels. All of these decreases corresponded with matching changes in both protein and DNA synthesis (Fig. 9C through F). These correlations were seen in both U937 and HL60 cells.
FIG. 9.
Decreased eIF4E expression during myeloid differentiation correlates with decreased levels of the 4E regulatory factors, c-Myc and protein synthesis. (A and B) For Southwestern analyses, protein lysates from U937 (A) and HL60 (B) cells were prepared with Laemmli buffer at 0, 3, 6, 24, 48, and 72 h after the addition of TPA. The lysates (50 μg) at the indicated time points were analyzed with the radioactively labeled LS3 trimer oligonucleotide in a Southwestern assay (SW panels). Size markers (lane SM) are indicated. For Northern blots, total cellular RNA was harvested from U937 (second through fourth panels in panel A) and HL60 (second through fourth panels in panel B) cells after the addition of TPA. RNA was size fractionated, run on formaldehyde-agarose gels, blotted, and hybridized with eIF4E, c-myc, or GAPDH plasmid fragments (4E, myc, and GAPDH, respectively). The protein lysates used for the Southwestern analyses were additionally run on 10% denaturing polyacrylamide gels, blotted, and probed with anti-eIF4E, anti-c-Myc, and anti-actin antibodies for the U937 (fifth through seventh panels [4E, myc, and actin] in panel A) and HL60 (fifth through seventh panels [4E, myc, and actin] in panel B) cells. (C through F) U937 (C and E) and HL60 (D and F) cells were pulse-labeled for 3 h with [35S]methionine and [3H]thymidine at the indicated time points. Aliquots of protein lysates at each time point were harvested directly in Laemmli buffer and run on 10% denaturing polyacrylamide gels to simultaneously evaluate protein synthesis rates of multiple individual proteins (C and D). Counts incorporated during pulse labeling were further evaluated by trichloroacetic acid precipitation of cell lysates (E and F). [35S]methionine (solid bars; y axis on right) and [3H]thymidine (open bars; y axis on left) incorporation is displayed as the mean and standard deviation of four determinations at each time point to evaluate the regulation of net protein synthesis (35S) and DNA synthesis (3H) during differentiation.
DISCUSSION
Translation initiation factor eIF4E levels must be regulated within a narrow concentration range because as little as a threefold increase in the level of eIF4E transforms cells while inactivation of eIF4E arrests cell growth (2, 66). Tight regulation of eIF4E is especially important since the abundance of critical translation factors differentially affects both translation of specific mRNA molecules and global protein synthesis (17, 39, 45, 51, 55, 57). In contrast to the narrow range within which eIF4E is regulated in non-differentiating tissues, we observed a fifty-fold decrease in eIF4E mRNA during growth arrest in differentiating myeloblasts (Fig. 9). Thus, although eIF4E is typically regulated within a narrow range of concentrations in model fibroblast cells, transcriptional controls of eIF4E expression can also respond over a much wider range in more complex tissues. These contrasting regulatory requirements have apparently selected for tight conservation of eIF4E promoter sequences between diverse species, since the murine and human promoters revealed a high overall degree of sequence conservation (Fig. 3). Although most previous efforts to understand eIF4E regulation focused on its phosphorylation (18–20, 28, 36, 52, 71) or its interactions with inhibitory proteins (50), our results emphasized the need for additional investigation of its transcriptional regulation.
We used linker-scanning mutagenesis of the eIF4E promoter to explore mechanisms regulating eIF4E expression and identified sites necessary for its transcription. Using this approach, we recognized the same proximal c-Myc box at nucleotide −75 that we had previously described (35). Linker-scanning mutations further identified a novel pyrimidine-rich site, TTACCCCCCCTT, which was also critical for promoter function. This sequence motif has not been previously identified as a transcriptional target in any other gene (74). Intriguingly, its position 25 nucleotides upstream of the unique transcription initiation site in eIF4E places it in the normal location for a TATA box (30, 53, 75). Nevertheless, its sequence did not fit any known TATA-binding motif (40), and the molecular masses of the eIF4E regulatory factors (Fig. 7 to 9) clearly distinguished them from the TATA-binding protein (38 kDa).
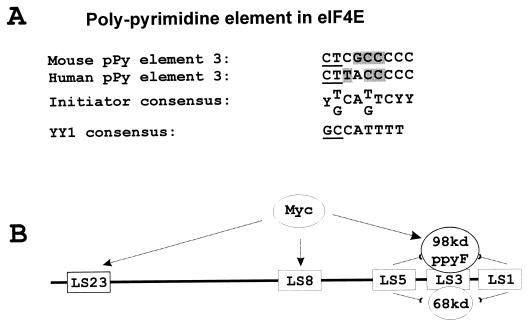
The absence of TATA or INR elements from the 4E promoter therefore prompted us to investigate possible sequence similarities between the eIF4E −25 element and INR motifs (Fig. 10) (34, 40). Our comparison between the TTACCCCCCCTT sequence in the eIF4E promoter and published initiator consensus sites revealed similarities in the pyrimidine-rich sequences flanking a conserved adenosine (Fig. 10, pPy element 3). In contrast, typical INR sites usually contain thymidine residues 3′ to the adenosine where the eIF4E element contained cytosines. The eIF4E polypyrimidine element also contained CTT 5′ to the conserved adenosine where YY1 consensus sites usually contain GCC. These sequence differences may explain the absence of cross-competition between the eIF4E element and either YY1 or INR oligonucleotides (Fig. 6). Moreover, the molecular masses of the 4E regulatory factor differed from those of any of the proteins known to bind initiator sites including TFII-I, YY1, and USF (58, 59, 62, 64). These data, taken together, strongly suggest that these eIF4E regulatory factors have not been previously recognized.
FIG. 10.
The polypyrimidine element in eIF4E is related to other initiator regions but contains significant differences. (A) Sequence is conserved at polypyrimidine (pPy) element 3 between mouse and human sequences. This sequence is further compared with the consensus sequence for an initiator region and for the YY1 element. Underlining indicates nucleotides required for YY1 binding which differ in the eIF4E element. Shaded boxes indicate nucleotides normally required for initiator binding which differ in the eIF4E element. (B) Model for potential direct and indirect effects of c-myc on the eIF4E promoter. c-myc may activate the eIF4E promoter by directly interacting with either or both of its myc boxes (LS8 and LS23). Alternatively, c-myc may indirectly regulate eIF4E through regulation of proteins binding at the LS3 site.
The dependence of eIF4E transcription on a gene-specific regulatory factor is similar to the situation seen for the eIF2α promoter. The eIF2α gene also lacks a TATA site and requires the binding of a unique transcription factor, NRF-1, at −21 for expression (4, 14, 33, 69). In general, transcription initiation of most genes is accomplished through the common basal transcription apparatus, which is then regulated by nearby activating sequences that achieve specificity through combinatorial interactions. More rarely, promoters achieve regulatory specificity via gene-specific activators (24). The transcriptional mechanisms regulating translation initiation factors eIF4E and eIF2α are therefore quite interesting. The binding of eIF4E regulatory factors (4ERFs) at the typical location of a TATA box and the similarity of their binding site to initiator sequences suggest that they might play a role in formation of the preinitiation complex. The unique specificity of the 4ERF binding sites, their high degree of regulation during differentiation, their molecular masses, and their unique functions in transcriptional control suggest that the eIF4E regulatory factors are novel and may reveal new mechanisms of transcriptional control.
Whether oncogenic proteins involved in transcriptional activation exert their effects by targeting single genes and pathways or through global effects on transcription of many or all genes remains an open question (44). Despite the many biological functions that have been suggested for c-myc, the mechanism(s) by which it transactivates its targets and the identities of myc-regulated genes continue to challenge investigators. The translation factor eIF4E is an appealing candidate because of its functions in growth control and the presence of the CACGTG myc boxes in its promoter. We were therefore surprised to find that levels of the 4ERFs were increased in cells expressing higher levels of c-Myc (Fig. 8 and 9), providing a second, indirect mechanism by which c-myc might regulate eIF4E (Fig. 10B).
Cell growth is required before S phase starts in both yeast and mammalian cells. Translational control of G1 cyclin synthesis responds to the ribosomal content of the yeast cell, thereby coupling cell growth and division in yeast (51). In contrast to yeast, recent findings that the Rb gene product controls transcription of cell cycle regulators and ribosomal biogenesis suggest that shared regulatory molecules may coordinate cell growth with cell division in mammalian cells (3, 72). Our findings suggest a potentially similar role for c-myc in the coordination of cell growth and division by regulating the cell cycle regulator CDC25a (21) and by regulating eIF4E and eIF2α (56). The 4ERFs may also be important in these control mechanisms, since they may themselves be c-myc targets and function to coordinate cell growth and division.
ACKNOWLEDGMENTS
This work, Kelly Johnston, Michael Polymenis, John Branda, and Emmett Schmidt were supported by PHS grant RO1-CA63117. Shanping Wang was supported by departmental funds of The Pediatric Service, Massachusetts General Hospital.
We thank R. Weinberg, A. Ezekowitz, and B. Kreskel for providing cell lines.
REFERENCES
- 1.Bernards R. N-myc disrupts protein kinase C-mediated signal transduction in neuroblastoma. EMBO J. 1991;10:1119–1125. doi: 10.1002/j.1460-2075.1991.tb08052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner C, Nakayama N, Goebl M, Tanaka K, Toh-e A, Matsumoto K. CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:3556–3559. doi: 10.1128/mcb.8.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavanaugh A H, Hempel W M, Taylor L J, Rogalsky V, Todorov G, Rothblum L I. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 4.Chau C M, Evans M J, Scarpulla R C. Nuclear respiratory factor 1 activation sites in genes encoding the gamma-subunit of ATP synthase, eukaryotic initiation factor 2 alpha, and tyrosine aminotransferase. Specific interaction of purified NRF-1 with multiple target genes. J Biol Chem. 1992;267:6999–7006. [PubMed] [Google Scholar]
- 5.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease activity. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 6.Chodosh L A, Carthew R W, Sharp P A. A single polypeptide possesses the binding and transcription activities of the adenovirus major late transcription factor. Mol Cell Biol. 1986;6:4723–4733. doi: 10.1128/mcb.6.12.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu W M, Wang Z, Roeder R G, Schmid C W. RNA polymerase III transcription repressed by Rb through its interactions with TFIIIB and TFIIIC2. J Biol Chem. 1997;272:14755–14761. doi: 10.1074/jbc.272.23.14755. [DOI] [PubMed] [Google Scholar]
- 8.Datta A K. Efficient amplification using “megaprimer” by asymmetric polymerase chain reaction. Nucleic Acids Res. 1995;23:4530–4531. doi: 10.1093/nar/23.21.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Benedetti A, Joshi-Barve S, Rinker-Schaeffer C, Rhoads R E. Expression of antisense RNA against initiation factor eIF-4E mRNA in HeLa cells results in lengthened cell division times, diminished translation rates, and reduced levels of both eIF-4E and the p220 component of eIF-4F. Mol Cell Biol. 1991;11:5435–5445. doi: 10.1128/mcb.11.11.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Benedetti A, Rhoads R E. Overexpression of eukaryotic protein synthesis initiation factor 4E in HeLa cells results in aberrant growth and morphology. Proc Natl Acad Sci USA. 1990;87:8212–8216. doi: 10.1073/pnas.87.21.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 13.Duncan R, Hershey J W. Regulation of initiation factors during translational repression caused by serum depletion. Abundance, synthesis, and turnover rates. J Biol Chem. 1985;260:5486–5492. [PubMed] [Google Scholar]
- 14.Efiok B J, Chiorini J A, Safer B. A key transcription factor for eukaryotic initiation factor-2 alpha is strongly homologous to developmental transcription factors and may link metabolic genes to cellular growth and development. J Biol Chem. 1994;269:18921–18930. [PubMed] [Google Scholar]
- 15.Eilers M, Picard D, Yamamoto K R, Bishop J M. Chimaeras of myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature. 1989;340:66–68. doi: 10.1038/340066a0. [DOI] [PubMed] [Google Scholar]
- 16.Evan G I, Littlewood T D. The role of c-myc in cell growth. Curr Opin Genet Dev. 1993;3:44–49. doi: 10.1016/s0959-437x(05)80339-9. [DOI] [PubMed] [Google Scholar]
- 17.Fagan R J, Lazaris-Karatzas A, Sonenberg N, Rozen R. Translational control of ornithine aminotransferase. Modulation by initiation factor eIF-4E. J Biol Chem. 1991;266:16518–16523. [PubMed] [Google Scholar]
- 18.Flynn A, Proud C G. Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J Biol Chem. 1995;270:21684–21688. doi: 10.1074/jbc.270.37.21684. [DOI] [PubMed] [Google Scholar]
- 19.Frederickson R M, Montine K S, Sonenberg N. Phosphorylation of eukaryotic translation initiation factor 4E is increased in Src-transformed cell lines. Mol Cell Biol. 1991;11:2896–2900. doi: 10.1128/mcb.11.5.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frederickson R M, Sonenberg N. Signal transduction and regulation of translation initiation. Semin Cell Biol. 1992;3:107–115. doi: 10.1016/s1043-4682(10)80020-0. [DOI] [PubMed] [Google Scholar]
- 21.Galaktionov K, Chen X, Beach D. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 22.Gao M, Rychlik W, Rhoads R E. Cloning and characterization of human eIF4E genes. J Biol Chem. 1998;273:4622–4628. doi: 10.1074/jbc.273.8.4622. [DOI] [PubMed] [Google Scholar]
- 23.Grandori C, Eisenman R N. Myc target genes. Trends Biochem Sci. 1997;22:177–181. doi: 10.1016/s0968-0004(97)01025-6. [DOI] [PubMed] [Google Scholar]
- 24.Hansen S K, Takada S, Jacobson R H, Lis J T, Tjian R. Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell. 1997;91:71–83. doi: 10.1016/s0092-8674(01)80010-6. [DOI] [PubMed] [Google Scholar]
- 25.Heikkila R, Schwab G, Wickstrom E, Loke S L, Pluznik D H, Watt R, Neckers L M. A c-myc antisense oligodeoxynucleotide inhibits entry into S phase but not progress from G0 to G1. Nature. 1987;328:445–449. doi: 10.1038/328445a0. [DOI] [PubMed] [Google Scholar]
- 26.Hendrickson W, Schleif R. A dimer of AraC protein contacts three adjacent major groove regions of the aral DNA site. Proc Natl Acad Sci USA. 1985;82:3129–3133. doi: 10.1073/pnas.82.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hershey J W. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- 28.Hiremath L S, Hiremath S T, Rychlik W, Joshi S, Domier L L, Rhoads R E. In vitro synthesis, phosphorylation, and localization on 48 S initiation complexes of human protein synthesis initiation factor 4E. J Biol Chem. 1989;264:1132–1138. [PubMed] [Google Scholar]
- 29.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann A, Oelgeschlager T, Roeder R G. Considerations of transcriptional control mechanisms: do TFIID-core promoter complexes recapitulate nucleosome-like functions? Proc Natl Acad Sci USA. 1997;94:8928–8935. doi: 10.1073/pnas.94.17.8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holt J T, Redner R L, Nienhuis A W. An oligomer complementary to c-myc mRNA inhibits proliferation of HL-60 promyelocytic cells and induces differentiation. Mol Cell Biol. 1988;8:963–973. doi: 10.1128/mcb.8.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huet J, Cottrelle P, Cool M, Vignais M L, Thiele D, Marck C, Buhler J M, Sentenac A, Fromageot P. A general upstream binding factor for genes of the yeast translational apparatus. EMBO J. 1985;4:3539–3547. doi: 10.1002/j.1460-2075.1985.tb04114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Humbelin M, Safer B, Chiorini J A, Hershey J W, Cohen R B. Isolation and characterization of the promoter and flanking regions of the gene encoding the human protein-synthesis-initiation factor 2 alpha. Gene. 1989;81:315–324. doi: 10.1016/0378-1119(89)90192-3. [DOI] [PubMed] [Google Scholar]
- 34.Hyde-DeRuyscher R P, Jennings E, Shenk T. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 1995;23:4457–4465. doi: 10.1093/nar/23.21.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones R M, Branda J, Johnston K A, Polymenis M, Gadd M, Rustgi A, Callanan L, Schmidt E V. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol Cell Biol. 1996;16:4754–4764. doi: 10.1128/mcb.16.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joshi B, Cai A L, Keiper B D, Minich W B, Mendez R, Beach C M, Stepinski J, Stolarski R, Darzynkiewicz E, Rhoads R E. Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. J Biol Chem. 1995;270:14597–14603. doi: 10.1074/jbc.270.24.14597. [DOI] [PubMed] [Google Scholar]
- 37.Kaddurah-Daouk R, Greene J M, Baldwin A S, Jr, Kingston R E. Activation and repression of mammalian gene expression by the c-myc protein. Genes Dev. 1987;1:347–357. doi: 10.1101/gad.1.4.347. [DOI] [PubMed] [Google Scholar]
- 38.Klein C, Struhl K. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol Cell Biol. 1994;14:1920–1928. doi: 10.1128/mcb.14.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koromilas A E, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIf-4e. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kraus R J, Murray E E, Wiley S R, Zink N M, Loritz K, Gelembiuk G W, Mertz J E. Experimentally determined weight matrix definitions of the initiator and TBP binding site elements of promoters. Nucleic Acids Res. 1996;24:1531–1539. doi: 10.1093/nar/24.8.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larminie C G, Cairns C A, Mital R, Martin K, Kouzarides T, Jackson S P, White R J. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J. 1997;16:2061–2071. doi: 10.1093/emboj/16.8.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lazaris-Karatzas A, Montine K S, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 43.Lemaitre J M, Buckle R S, Mechali M. c-Myc in the control of cell proliferation and embryonic development. Adv Cancer Res. 1996;70:95–144. doi: 10.1016/s0065-230x(08)60873-8. [DOI] [PubMed] [Google Scholar]
- 44.Lewin B. Oncogenic conversion by regulatory changes in transcription factors. Cell. 1991;64:303–312. doi: 10.1016/0092-8674(91)90640-k. [DOI] [PubMed] [Google Scholar]
- 45.Lodish H F. Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature. 1974;251:385–388. doi: 10.1038/251385a0. [DOI] [PubMed] [Google Scholar]
- 46.Mager W H, Planta R J. Multifunctional DNA-binding proteins mediate concerted transcription activation of yeast ribosomal protein genes. Biochim Biophys Acta. 1990;1050:351–355. doi: 10.1016/0167-4781(90)90193-6. [DOI] [PubMed] [Google Scholar]
- 47.Merrick W C. Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miltenberger R J, Sukow K A, Farnham P J. An E-box-mediated increase in cad transcription at the G1/S-phase boundary is suppressed by inhibitory c-Myc mutants. Mol Cell Biol. 1995;15:2527–2535. doi: 10.1128/mcb.15.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore J P, Hancock D C, Littlewood T D, Evan G I. A sensitive and quantitative enzyme-linked immunosorbance assay for the c-myc and N-myc oncoproteins. Oncogene Res. 1987;2:65–80. [PubMed] [Google Scholar]
- 50.Pause A, Belsham G J, Gingras A C, Donze O, Lin T A, Lawrence J C, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 51.Polymenis M, Schmidt E V. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rinker-Schaeffer C W, Austin V, Zimmer S, Rhoads R E. Ras transformation of cloned rat embryo fibroblasts results in increased rates of protein synthesis and phosphorylation of eukaryotic initiation factor 4E. J Biol Chem. 1992;267:10659–10664. [PubMed] [Google Scholar]
- 53.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 54.Rosenwald I B. Upregulated expression of the genes encoding translation initiation factors eIF-4E and eIF-2alpha in transformed cells. Cancer Lett. 1996;102:113–123. doi: 10.1016/0304-3835(96)04171-7. [DOI] [PubMed] [Google Scholar]
- 55.Rosenwald I B, Lazaris-Karatzas A, Sonenberg N, Schmidt E V. Elevated levels of cyclin D1 protein in response to increased expression of eukaryotic initiation factor 4E. Mol Cell Biol. 1993;13:7358–7363. doi: 10.1128/mcb.13.12.7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosenwald I B, Rhoads D B, Callanan L D, Isselbacher K J, Schmidt E V. Increased expression of eukaryotic translation initiation factors eIF-4E and eIF-2 alpha in response to growth induction by c-myc. Proc Natl Acad Sci USA. 1993;90:6175–6178. doi: 10.1073/pnas.90.13.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci USA. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roy A L, Malik S, Meisterernst M, Roeder R G. An alternative pathway for transcription initiation involving TFII-I. Nature. 1993;365:355–359. doi: 10.1038/365355a0. [DOI] [PubMed] [Google Scholar]
- 59.Roy A L, Meisterernst M, Pognonec P, Roeder R G. Cooperative interaction of an initiator-binding transcription initiation factor and the helix-loop-helix activator USF. Nature. 1991;354:245–248. doi: 10.1038/354245a0. [DOI] [PubMed] [Google Scholar]
- 60.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 61.Schmidt E V. MYC family ties. Nat Genet. 1996;14:8–10. doi: 10.1038/ng0996-8. [DOI] [PubMed] [Google Scholar]
- 62.Shi Y, Seto E, Chang L S, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 63.Singh H, LeBowitz J H, Baldwin A S, Jr, Sharp P A. Molecular cloning of an enhancer binding protein: isolation by screening of an expression library with a recognition site DNA. Cell. 1988;52:415–423. doi: 10.1016/s0092-8674(88)80034-5. [DOI] [PubMed] [Google Scholar]
- 64.Smale S T, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 65.Sonenberg N. Cap-binding proteins of eukaryotic messenger RNA: functions in initiation and control of translation. Prog Nucleic Acid Res Mol Biol. 1988;35:173–207. doi: 10.1016/s0079-6603(08)60614-5. [DOI] [PubMed] [Google Scholar]
- 66.Sonenberg N. Regulation of translation and cell growth by eIF-4E. Biochimie. 1994;76:839–846. doi: 10.1016/0300-9084(94)90185-6. [DOI] [PubMed] [Google Scholar]
- 67.Swick A G, Lane M D. Identification of a transcriptional repressor down-regulated during preadipocyte differentiation. Proc Natl Acad Sci USA. 1992;89:7895–7899. doi: 10.1073/pnas.89.17.7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vinson C R, LaMarco K L, Johnson P F, Landschulz W H, McKnight S L. In situ detection of sequence-specific DNA binding activity specified by a recombinant bacteriophage. Genes Dev. 1988;2:801–806. doi: 10.1101/gad.2.7.801. [DOI] [PubMed] [Google Scholar]
- 69.Virbasius C A, Virbasius J V, Scarpulla R C. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 1993;7:2431–2445. doi: 10.1101/gad.7.12a.2431. [DOI] [PubMed] [Google Scholar]
- 70.Weis L, Reinberg D. Transcription by RNA polymerase II: initiator-directed formation of transcription-competent complexes. FASEB J. 1992;6:3300–3309. doi: 10.1096/fasebj.6.14.1426767. [DOI] [PubMed] [Google Scholar]
- 71.Whalen S G, Gingras A C, Amankwa L, Mader S, Branton P E, Aebersold R, Sonenberg N. Phosphorylation of eIF-4E on serine 209 by protein kinase C is inhibited by the translational repressors, 4E-binding proteins. J Biol Chem. 1996;271:11831–11837. doi: 10.1074/jbc.271.20.11831. [DOI] [PubMed] [Google Scholar]
- 72.White R J, Trouche D, Martin K, Jackson S P, Kouzarides T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996;382:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- 73.Wickstrom E L, Bacon T A, Gonzalez A, Lyman G H, Wickstrom E. Anti-c-myc DNA increases differentiation and decreases colony formation by HL-60 cells. In Vitro Cell Dev Biol Anim. 1989;25:297–302. doi: 10.1007/BF02628470. [DOI] [PubMed] [Google Scholar]
- 74.Wingender E, Kel A E, Kel O V, Karas H, Heinemeyer T, Dietze P, Knueppel R, Romaschenko A G, Kolchanov N A. Towards a federated database system on transcriptional regulation. Nucleic Acids Res. 1997;25:265–268. doi: 10.1093/nar/25.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]