Abstract
The Hox gene cluster is an iconic example of evolutionary conservation between divergent animal lineages, providing evidence for ancient similarities in the genetic control of embryonic development. However, there are differences between taxa in gene order, gene number and genomic organisation implying conservation is not absolute. There are also examples of radical functional change of Hox genes; for example, the ftz, zen and bcd genes in insects play roles in segmentation, extraembryonic membrane formation and body polarity, rather than specification of anteroposterior position. There have been detailed descriptions of Hox genes and Hox gene clusters in several insect species, including important model systems, but a large-scale overview has been lacking. Here we extend these studies using the publicly-available complete genome sequences of 243 insect species from 13 orders. We show that the insect Hox cluster is characterised by large intergenic distances, consistently extreme in Odonata, Orthoptera, Hemiptera and Trichoptera, and always larger between the ‘posterior’ Hox genes. We find duplications of ftz and zen in many species and multiple independent cluster breaks, although certain modules of neighbouring genes are rarely broken apart suggesting some organisational constraints. As more high-quality genomes are obtained, a challenge will be to relate structural genomic changes to phenotypic change across insect phylogeny.
Keywords: Homeobox, Hexapoda, Gene cluster, Shx
1. Introduction
Insects display an astounding range of developmental and morphological diversity. Comprising over half of all described animal species, insect diversity has been attributed to high rates of speciation and adaptive radiation in association with flowering plant diversification, underpinned by dynamic rates of gene and genome evolution. Together with the orders Protura, Diplura and Collembola, insects make up the Hexapoda, a clade within Arthropoda consisting of six-legged, mostly terrestrial species. Within Hexapoda there have been several major evolutionary transitions associated with novel phenotypic traits. The evolution of insect wings is one such event which resulted in diversification of body forms within the clade Pterygota [1]. A later event was the emergence of complete metamorphosis in the holometabolous insects, thought to have permitted rapid diversification. Indeed, the most diverse and speciose insect orders are found within the holometabolous pterygotes (Hymenoptera, Coleoptera, Diptera and Lepidoptera). While the insect body plan is generally well conserved, a myriad of morphological novelties have emerged through insect radiation, ranging from pronotal horns on some beetles, sucking mouthparts in Hemiptera and (most) Lepidoptera, stings in bees and wasps, and halteres in Diptera and Strepsiptera.
Changes in developmental processes underlie morphological diversity, and ultimately these developmental changes must be underpinned by inherited genetic changes. Identifying these genetic changes is one of the goals of evolutionary developmental biology (evo-devo) although this is a difficult task when the morphological transitions occurred tens or hundreds of millions of years ago. One place to start is with the genes shared between taxa, and with key roles in development: a set of genes sometimes called the developmental toolkit. The Hox genes are examples of such core developmental genes, encoding position along the anteroposterior axis of most animal embryos. Furthermore, Hox genes code for transcription factors that activate and repress cascades of downstream genes to sculpt the morphology appropriate to that position. Later in development, Hox genes also orchestrate cell differentiation decisions, primarily though not exclusively within their original embryonic expression domains [2]. Changes in the content, order and expression domains of these genes have been implicated in a huge range of morphological novelties in the arthropod body plan [2].
The insect Hox cluster is thought to have consisted ancestrally of 10 genes: labial (lab), proboscipedia (pb), zerknüllt (zen), Deformed (Dfd), Sex combs reduced (Scr), fushi tarazu (ftz), Antennapedia (Antp), Ultrabithorax (Ubx), abdominal-A (abdA), and Abdominal-B (AbdB), similar to that of the bilaterian ancestor [3], [4]. Of these, zen and ftz have ‘altered’ roles, having switched from their ancestral roles in anteroposterior position specification to extraembryonic membrane patterning (zen) and segmentation (ftz). Until recently, we have lacked a thorough knowledge of the multiple evolutionary paths that have been taken from this ancestral state across the diversity of insect orders. Here, we provide an updated view of the evolution of the Hox gene cluster across the largest sample of insect genomes sampled to date. We focus on the evolution of Hox cluster organisation, and do not discuss recent work on Hox gene regulation or the changing downstream functions of Hox genes, such as co-option of Hox genes to accessory roles in different orders (e.g. [5], [6], [7], [8], [9]). Furthermore, we focus on protein-coding loci within Hox clusters, and do not cover non-coding RNAs since these cannot be predicted reliably from genome sequences. There is certainly good evidence for antisense lncRNAs produced from within the Hox cluster of insects and other arthropods, as well as annelids, but comparative data are sparse (for review see [10]). We do not report new sequence data here: these analyses are based on publicly-available complete genome sequences, interpreted in the light of previous analyses.
2. Hox genes in a new era of insect genomics
In the pre-genomic era of molecular biology, from the early 1980s to around 2005, Hox gene clusters were analysed either by painstaking positional cloning of mutants or by cross- hybridisation of probes to genomic libraries followed by laborious genomic walking and clone-by-clone sequencing, sometimes coupled with in situ hybridization to chromosome spreads [11], [12]. These heroic efforts were limited to a few species, but they started to generate a picture of comparative stasis of insect Hox gene clusters. For example, in the fruitfly Drosophila melanogaster, the insect in which Hox genes were first studied, it was clear that the zen gene had undergone tandem duplication to give three genes: zen, z2 and bcd. Otherwise, there were no examples of Hox gene duplication in that evolutionary lineage. Similarly, in the red flour beetle Tribolium castaneum, there is a zen duplication to give two genes, but otherwise the cluster is unaltered [13], [14]. It was, of course, clear from the earliest days that splitting of the cluster was possible, as evidenced by the cluster split in Drosophila melanogaster with a 9.6 Mb gap between Ubx and Antp. It was also clear that intergenic distances could be very large, as in Drosophila melanogaster and the locust Schistocerca gregaria [11]. Nonetheless, these are relatively minor changes compared to whole Hox gene cluster duplication in vertebrates and extensive posterior Hox gene duplication in vertebrates and amphioxus [15], [16].
From the mid-2000s onwards, complete genome sequencing began to be applied to single species, or small sets of related species, and the evolutionary picture was refined. First, it became clear that splitting of the cluster was not a unique event in Drosophila melanogaster, as there have been independent splits at different positions in some other Drosophila species [17]. Second, lability of the zen gene, in terms of propensity to duplication, was reinforced by a remarkable discovery in the genome of the silk moth Bombyx mori [18]. This analysis revealed extensive tandem gene duplication, generating at least 13 copies of zen: one locus with an amino acid sequence similar to the ancestral zen gene, and 12 that have diverged extensively and given the name Shx (Special homeobox) genes. Later analysis of a refined genome assembly suggested the number may be even greater: zen plus 15 Shx loci, although not all can encode functional proteins [19]. Genome sequences of two butterflies (Heliconius melpomene and the Monarch Danaus plexippus) revealed presence of four Shx genes [20], [21], as did low coverage genome skims of the Comma butterfly Polygonia c-album, Speckled Wood butterfly Pararge aegeria, Scarlet Tiger moth Callimorpha dominula and Horse Chestnut leaf-miner Cameraria ohridella [19].
The above historical perspective is one of a gradually unfolding picture emerging in a piecemeal manner as each additional genome is sequenced or analysed. But the landscape is now changing rapidly. In 2018, the Earth BioGenome Project was announced, as a bold vision to determine the complete genome sequence of all living eukaryote species [22]. This vision has galvanised action from over 40 affiliated projects, each attempting to determine high quality genome sequences at scale [23]. Among these, the project that has generated the largest number of high quality insect genomes to date is the Wellcome Trust-funded ‘Darwin Tree of Life’ (DToL), focussed on species living in Britain and Ireland [24]. Since 2019, the DToL project has generated 381 complete genome assemblies with over 2000 more species in the genome sequencing pipeline (data as of July 2022: https://portal.darwintreeoflife.org/tracking). These genome assemblies have been determined using long-read DNA sequencing technology (primarily PacBio HiFi) and scaffolding to chromosome-level using Hi-C. As such, they surpass in quality the large majority of genome assemblies previously available. In particular, the large contig sizes scaffolded to chromosomes provides opportunity to determine gene order and distances. Importantly, all data from the DToL project are released openly.
In reviewing the evolution of Hox gene clusters, we consider that the landscape of the field has changed so remarkably in the past two years that we cannot draw conclusions solely from previously published analyses. Instead, we supplement previous findings with analyses of the openly released data from DToL and other genome sequencing projects. We do not report new experimental data here, but rather draw new conclusions from available data. We use these data to summarise patterns of Hox gene duplication and the changes to genomic organisation across insects, using genomic data from 243 species representing 13 insect orders, plus one order of non-insect hexapod as an outgroup (Table 1). We show that insects continue to be an important model for understanding Hox gene evolution and, with the development of further methods and models for genetic manipulation from a phylogenetically diverse set of orders, will be vital for progress in the field of evolutionary developmental biology [25].
Table 1.
Order, species and genomes used in this study.
| Order | Species | Genome |
|---|---|---|
| Trichoptera | Limnephilus lunatus | GCA_917563855.1_iiLimLuna2.1_genomic |
| Trichoptera | Limnephilus marmoratus | GCA_917880885.1_iiLimMarm1.1_genomic |
| Trichoptera | Limnephilus rhombicus | GCA_929108145.1_iiLimRhom1.1_genomic |
| Trichoptera | Glyphotaelius pellucidus | GCA_936435175.1_iiGlyPell1.1_genomic |
| Trichoptera | Eubasilissa regina | GCA_022840565_Eubasilissa_regina |
| Phasmatodea | Timema cristinae | GCA_002926335_ipTimCris1 |
| Plecoptera | Nemoura dubitans | GCA_921293005.1_ipNemDubi1.1_genomic |
| Plecoptera | Nemurella pictetii | GCA_921293315.1_ipNemPict2.1_genomic |
| Plecoptera | Brachyptera putata | GCA_907164805.1_ipBraPut3m.1_genomic |
| Coleoptera | Pyrochroa serraticornis | GCA_905333025.1_icPyrSerr1.1_genomic |
| Coleoptera | Rhagonycha fulva | GCA_905340355.1_icRhaFulv1.1_genomic |
| Coleoptera | Coccinella septempunctata | GCA_907165205.1_icCocSept1.1_genomic |
| Coleoptera | Malachius bipustulatus | GCA_910589415.1_icMalBipu1.1_genomic |
| Coleoptera | Adalia bipunctata | GCA_910592335.1_icAdaBipu1.1_genomic |
| Coleoptera | Ocypus olens | GCA_910593695.1_icOcyOlen1.1_genomic |
| Coleoptera | Cantharis rustica | GCA_911387805.1_icCanRust1.1_genomic |
| Coleoptera | Harmonia axyridis | GCA_914767665.1_icHarAxyr1.1_genomic |
| Coleoptera | Apoderus coryli | GCA_911728435.1_icApoCory1.1_genomic |
| Coleoptera | Pterostichus madidus | GCA_911728475.1_icPteMadi1.1_genomic |
| Coleoptera | Agrypnus murinus | GCA_929113105.1_icAgrMuri1.1_genomic |
| Coleoptera | Podabrus alpinus | GCA_932274525.1_icPodAlpi1.1_genomic |
| Coleoptera | Philonthus cognatus | GCA_932526585.1_icPhiCogn1.1_genomic |
| Coleoptera | Leistus spinibarbis | GCA_933228885.1_icLeiSpin1.1_genomic |
| Coleoptera | Polydrusus cervinus | GCA_935413205.1_icPolCerv1.1_genomic |
| Coleoptera | Melolontha melolontha | GCA_935421215.1_icMelMelo1.1_genomic |
| Coleoptera | Rutpela maculata | GCA_936432065.1_icLepMacu1.1_genomic |
| Coleoptera | Halyzia sedecimguttata | GCA_937662695.1_icHalSede1.1_genomic |
| Coleoptera | Ophonus ardosiacus | GCA_943142095.1_icOphArdo1.1_genomic |
| Lepidoptera | Micropterix aruncella | GCA_944548615.1_ilMicArun2.1_genomic |
| Lepidoptera | Autographa gamma | GCA_905146925.1_ilAutGamm1.1_genomic |
| Lepidoptera | Laspeyria flexula | GCA_905147015.1_ilLasFlex1.1_genomic |
| Lepidoptera | Inachis io | GCA_905147045.1_ilAglIoxx1.1_genomic |
| Lepidoptera | Pieris brassicae | GCA_905147105.1_ilPieBrab1.1_genomic |
| Lepidoptera | Blastobasis lacticolella | GCA_905147135.1_ilBlaLact1.1_genomic |
| Lepidoptera | Nymphalis urticae | GCA_905147175.1_ilAglUrti1.1_genomic |
| Lepidoptera | Euproctis similis | GCA_905147225.1_ilEupSimi1.1_genomic |
| Lepidoptera | Erynnis tages | GCA_905147235.1_ilEryTage1.1_genomic |
| Lepidoptera | Hypena proboscidalis | GCA_905147285.1_ilHypProb1.1_genomic |
| Lepidoptera | Mythimna impura | GCA_905147345.1_ilMytImpu1.1_genomic |
| Lepidoptera | Apotomis turbidana | GCA_905147355.1_ilApoTurb1.1_genomic |
| Lepidoptera | Aricia agestis | GCA_905147365.1_ilAriAges1.1_genomic |
| Lepidoptera | Hylaea fasciaria | GCA_905147375.1_ilHylFasc1.1_genomic |
| Lepidoptera | Limenitis camilla | GCA_905147385.1_ilLimCami1.1_genomic |
| Lepidoptera | Xestia xanthographa | GCA_905147715.1_ilXesXant1.1_genomic |
| Lepidoptera | Phlogophora meticulosa | GCA_905147745.1_ilPhlMeti2.1_genomic |
| Lepidoptera | Thyatira batis | GCA_905147785.1_ilThyBati1.1_genomic |
| Lepidoptera | Pieris rapae | GCA_905147795.1_ilPieRapa1.1_genomic |
| Lepidoptera | Phalera bucephala | GCA_905147815.1_ilPhaBuce1.1_genomic |
| Lepidoptera | Endotricha flammealis | GCA_905163395.1_ilEndFlam1.1_genomic |
| Lepidoptera | Noctua fimbriata | GCA_905163415.1_ilNocFimb1.1_genomic |
| Lepidoptera | Mamestra brassicae | GCA_905163435.1_ilMamBras1.1_genomic |
| Lepidoptera | Pararge aegeria | GCA_905163445.1_ilParAegt1.1_genomic |
| Lepidoptera | Craniophora ligustri | GCA_905163465.1_ilCraLigu1.1_genomic |
| Lepidoptera | Cosmia trapezina | GCA_905163495.1_ilCosTrap1.1_genomic |
| Lepidoptera | Lymantria monacha | GCA_905163515.1_ilLymMona1.1_genomic |
| Lepidoptera | Notocelia uddmanniana | GCA_905163555.1_ilNotUddm1.1_genomic |
| Lepidoptera | Celastrina argiolus | GCA_905187575.1_ilCelArgi3.1_genomic |
| Lepidoptera | Cyaniris semiargus | GCA_905187585.1_ilCyaSemi1.1_genomic |
| Lepidoptera | Colias croceus | GCA_905220415.1_ilColCroc2.1_genomic |
| Lepidoptera | Amphipyra tragopoginis | GCA_905220435.1_ilAmpTrag2.1_genomic |
| Lepidoptera | Deilephila porcellus | GCA_905220455.1_ilDeiPorc1.1_genomic |
| Lepidoptera | Ennomos fuscantarius | GCA_905220475.1_ilEnnFusc2.1_genomic |
| Lepidoptera | Laothoe populi | GCA_905220505.1_ilLaoPopu1.1_genomic |
| Lepidoptera | Lysandra coridon | GCA_905220515.1_ilLysCori1.1_genomic |
| Lepidoptera | Mellicta athalia | GCA_905220545.1_ilMelAtha1.1_genomic |
| Lepidoptera | Melitaea cinxia | GCA_905220565.1_ilMelCinx1.1_genomic |
| Lepidoptera | Nymphalis polychloros | GCA_905220585.1_ilNymPoly1.1_genomic |
| Lepidoptera | Spilosoma lubricipeda | GCA_905220595.1_ilSpiLubr1.1_genomic |
| Lepidoptera | Tinea trinotella | GCA_905220615.1_ilTinTrin1.1_genomic |
| Lepidoptera | Boloria selene | GCA_905231865.2_ilBolSele5.2_genomic |
| Lepidoptera | Pieris napi | GCA_905231885.1_ilPieNapi4.1_genomic |
| Lepidoptera | Vanessa atalanta | GCA_905147765.1_ilVanAtal1.1_genomic |
| Lepidoptera | Notodonta dromedarius | GCA_905147325.1_ilNotDrom1.1_genomic |
| Lepidoptera | Vanessa cardui | GCA_905220365.1_ilVanCard2.1_genomic |
| Lepidoptera | Hecatera dysodea | GCA_905332915.1_ilHecDyso1.1_genomic |
| Lepidoptera | Mimas tiliae | GCA_905332985.1_ilMimTili1.1_genomic |
| Lepidoptera | Lycaena phlaeas | GCA_905333005.1_ilLycPhla1.1_genomic |
| Lepidoptera | Lysandra bellargus | GCA_905333045.1_ilLysBell1.1_genomic |
| Lepidoptera | Maniola jurtina | GCA_905333055.1_ilManJurt1.1_genomic |
| Lepidoptera | Pheosia tremula | GCA_905333125.1_ilPheTrem1.1_genomic |
| Lepidoptera | Abrostola tripartita | GCA_905340225.1_ilAbrTrip1.1_genomic |
| Lepidoptera | Noctua pronuba | GCA_905220335.1_ilNocPron1.1_genomic |
| Lepidoptera | Atethmia centrago | GCA_905333075.2_ilAteCent1.2_genomic |
| Lepidoptera | Glaucopsyche alexis | GCA_905404095.1_ilGlaAlex1.1_genomic |
| Lepidoptera | Pheosia gnoma | GCA_905404115.1_ilPheGnom1.1_genomic |
| Lepidoptera | Hesperia comma | GCA_905404135.1_ilHesComm1.1_genomic |
| Lepidoptera | Biston betularia | GCA_905404145.1_ilBisBetu1.1_genomic |
| Lepidoptera | Plebejus argus | GCA_905404155.1_ilPleArgu1.1_genomic |
| Lepidoptera | Anthocharis cardamines | GCA_905404175.1_ilAntCard3.1_genomic |
| Lepidoptera | Fabriciana adippe | GCA_905404265.1_ilFabAdip1.1_genomic |
| Lepidoptera | Hedya salicella | GCA_905404275.1_ilHedSali1.1_genomic |
| Lepidoptera | Erannis defoliaria | GCA_905404285.1_ilEraDefo1.1_genomic |
| Lepidoptera | Ochlodes sylvanus | GCA_905404295.1_ilOchSylv3.1_genomic |
| Lepidoptera | Leptidea sinapis | GCA_905404315.1_ilLepSina1.1_genomic |
| Lepidoptera | Autographa pulchrina | GCA_905475315.1_ilAutPulc1.1_genomic |
| Lepidoptera | Clostera curtula | GCA_905475355.1_ilCloCurt1.1_genomic |
| Lepidoptera | Schrankia costaestrigalis | GCA_905475405.1_ilSchCost1.1_genomic |
| Lepidoptera | Ochropleura plecta | GCA_905475445.1_ilOchPlec1.1_genomic |
| Lepidoptera | Zeuzera pyrina | GCA_907165235.1_ilZeuPyri1.1_genomic |
| Lepidoptera | Habrosyne pyritoides | GCA_907165245.1_ilHabPyri1.1_genomic |
| Lepidoptera | Zygaena filipendulae | GCA_907165275.1_ilZygFili1.1_genomic |
| Lepidoptera | Crocallis elinguaria | GCA_907269065.1_ilCroElin1.1_genomic |
| Lepidoptera | Idaea aversata | GCA_907269075.1_ilIdaAver1.1_genomic |
| Lepidoptera | Blastobasis adustella | GCA_907269095.1_ilBlaAdus2.1_genomic |
| Lepidoptera | Mythimna ferrago | GCA_910589285.1_ilMytFerr1.1_genomic |
| Lepidoptera | Noctua janthe | GCA_910589295.1_ilNocJant1.1_genomic |
| Lepidoptera | Bembecia ichneumoniformis | GCA_910589475.1_ilBemIchn1.1_genomic |
| Lepidoptera | Ennomos quercinarius | GCA_910589525.1_ilEnnQuei1.1_genomic |
| Lepidoptera | Carcina quercana | GCA_910589575.1_ilCarQuer1.1_genomic |
| Lepidoptera | Chrysoteuchia culmella | GCA_910589605.1_ilChrCulm1.1_genomic |
| Lepidoptera | Tinea semifulvella | GCA_910589645.1_ilTinSemi1.1_genomic |
| Lepidoptera | Acronicta aceris | GCA_910591435.1_ilAcrAcer1.1_genomic |
| Lepidoptera | Cydia splendana | GCA_910591565.1_ilCydSple1.1_genomic |
| Lepidoptera | Ypsolopha scabrella | GCA_910592155.1_ilYpsScab1.1_genomic |
| Lepidoptera | Amphipyra berbera | GCA_910594945.1_ilAmpBerb1.1_genomic |
| Lepidoptera | Parapoynx stratiotata | GCA_910589355.1_ilParStra1.1_genomic |
| Lepidoptera | Pyrgus malvae | GCA_911387765.1_ilPyrMalv3.1_genomic |
| Lepidoptera | Thymelicus sylvestris | GCA_911387775.1_ilThySylv1.1_genomic |
| Lepidoptera | Apamea monoglypha | GCA_911387795.1_ilApaMono1.1_genomic |
| Lepidoptera | Neomicropteryx cornuta | GCA_020383195.1_ilNeoCorn1.1_genomic |
| Lepidoptera | Hemaris fuciformis | GCA_907164795.1_ilHemFuc1.1_genomic |
| Lepidoptera | Papilio machaon | GCA_912999745.1_ilPapMach1.1_genomic |
| Lepidoptera | Sesia apiformis | GCA_914767545.1_ilSesApif2.1_genomic |
| Lepidoptera | Hydraecia micacea | GCA_914767645.1_ilHydMica1.1_genomic |
| Lepidoptera | Ptilodon capucinus | GCA_914767695.1_ilPtiCapc1.1_genomic |
| Lepidoptera | Agrochola circellaris | GCA_914767755.1_ilAgrCirc1.1_genomic |
| Lepidoptera | Eupsilia transversa | GCA_914767815.1_ilEupTran1.1_genomic |
| Lepidoptera | Agriopis aurantiaria | GCA_914767915.1_ilAgrAura1.1_genomic |
| Lepidoptera | Eilema depressum | GCA_914767945.1_ilEilDepe1.1_genomic |
| Lepidoptera | Eilema sororculum | GCA_914829495.1_ilEilSoro1.1_genomic |
| Lepidoptera | Spilarctia lutea | GCA_916048165.1_ilSpiLutu1.1_genomic |
| Lepidoptera | Griposia aprilina | GCA_916610205.1_ilGriApri1.1_genomic |
| Lepidoptera | Omphaloscelis lunosa | GCA_916610215.1_ilOmpLuno1.1_genomic |
| Lepidoptera | Mesoligia furuncula | GCA_916614155.1_ilMesFuru1.1_genomic |
| Lepidoptera | Xestia c-nigrum | GCA_916618015.1_ilXesCnig1.1_genomic |
| Lepidoptera | Emmelina monodactyla | GCA_916618145.1_ilEmmMono1.1_genomic |
| Lepidoptera | Agrochola macilenta | GCA_916701695.1_ilAgrMaci1.1_genomic |
| Lepidoptera | Orgyia antiqua | GCA_916999025.1_ilOrgAnti1.1_genomic |
| Lepidoptera | Erebia ligea | GCA_917051295.1_ilEreLige1.1_genomic |
| Lepidoptera | Dryobotodes eremita | GCA_917490735.1_ilDryErem1.1_genomic |
| Lepidoptera | Selenia dentaria | GCA_917880725.1_ilSelDent1.1_genomic |
| Lepidoptera | Synanthedon vespiformis | GCA_918317495.1_ilSynVesp1.1_genomic |
| Lepidoptera | Notodonta ziczac | GCA_918843915.1_ilNotZicz1.1_genomic |
| Lepidoptera | Eulithis prunata | GCA_918843925.1_ilEulPrun1.1_genomic |
| Lepidoptera | Philereme vetulata | GCA_918857605.1_ilPhiVetu1.1_genomic |
| Lepidoptera | Melanargia galathea | GCA_920104075.1_ilMelGala2.1_genomic |
| Lepidoptera | Furcula furcula | GCA_911728495.1_ilFurFurc1.1_genomic |
| Lepidoptera | Peribatodes rhomboidaria | GCA_911728515.1_ilPerRhom1.1_genomic |
| Lepidoptera | Pammene fasciana | GCA_911728535.1_ilPamFasc1.1_genomic |
| Lepidoptera | Aporia crataegi | GCA_912999735.1_ilApoCrat1.1_genomic |
| Lepidoptera | Hydriomena furcata | GCA_912999785.1_ilHydFurc1.1_genomic |
| Lepidoptera | Campaea margaritaria | GCA_912999815.1_ilCamMarg1.1_genomic |
| Thysanoptera | Thrips palmi | GCF_012932325_itThrPalm1 |
| Collembola | Folsomia candida | GCF_002217175_hcFolCand1 |
| Hymenoptera | Bombus hortorum | GCA_905332935.1_iyBomHort1.1_genomic |
| Hymenoptera | Bombus pascuorum | GCA_905332965.1_iyBomPasc1.1_genomic |
| Hymenoptera | Bombus campestris | GCA_905333015.1_iyBomCamp1.1_genomic |
| Hymenoptera | Vespula germanica | GCA_905340365.1_iyVesGerm1.1_genomic |
| Hymenoptera | Vespula vulgaris | GCA_905475345.1_iyVesVulg1.1_genomic |
| Hymenoptera | Nomada fabriciana | GCA_907165295.1_iyNomFabr1.1_genomic |
| Hymenoptera | Vespa crabro | GCA_910589235.1_iyVesCrab1.1_genomic |
| Hymenoptera | Cerceris rybyensis | GCA_910591515.1_iyCerRyby1.1_genomic |
| Hymenoptera | Nysson spinosus | GCA_910591585.1_iyNysSpin1.1_genomic |
| Hymenoptera | Ectemnius continuus | GCA_910591665.1_iyEctCont1.1_genomic |
| Hymenoptera | Bombus terrestris | GCA_910591885.1_iyBomTerr1.1_genomic |
| Hymenoptera | Andrena haemorrhoa | GCA_910592295.1_iyAndHaem1.1_genomic |
| Hymenoptera | Ectemnius lituratus | GCA_910593735.1_iyEctLitu1.1_genomic |
| Hymenoptera | Dolichovespula media | GCA_911387685.1_iyDolMedi1.1_genomic |
| Hymenoptera | Bombus hypnorum | GCA_911387925.1_iyBomHypn1.1_genomic |
| Hymenoptera | Dolichovespula saxonica | GCA_911387935.1_iyDolSaxo1.1_genomic |
| Hymenoptera | Osmia bicornis | GCA_907164935.1_iyOsmBic2.1_genomic |
| Hymenoptera | Vespa velutina | GCA_912470025.1_iyVesVel2.1_genomic |
| Hymenoptera | Seladonia tumulorum | GCA_913789895.1_iySelTumu1.1_genomic |
| Hymenoptera | Sphecodes monilicornis | GCA_913789915.1_iySphMoni1.1_genomic |
| Hymenoptera | Tenthredo notha | GCA_914767705.1_iyTenNoth1.1_genomic |
| Hymenoptera | Anoplius nigerrimus | GCA_914767735.1_iyAnoNige1.1_genomic |
| Hymenoptera | Ancistrocerus nigricornis | GCA_916049575.1_iyAncNigr1.1_genomic |
| Hymenoptera | Macropis europaea | GCA_916610135.1_iyMacEuro1.1_genomic |
| Hymenoptera | Lasioglossum morio | GCA_916610235.1_iyLasMori1.1_genomic |
| Hymenoptera | Lasioglossum lativentre | GCA_916610255.1_iyLasLatv2.1_genomic |
| Hymenoptera | Athalia rosae | GCA_917208135.1_iyAthRosa1.1_genomic |
| Hymenoptera | Mimumesa dahlbomi | GCA_917499265.1_iyMimDahl1.1_genomic |
| Hymenoptera | Ichneumon xanthorius | GCA_917499995.1_iyIchXant1.1_genomic |
| Hymenoptera | Dolichovespula sylvestris | GCA_918808275.1_iyDolSylv1.1_genomic |
| Hymenoptera | Bombus sylvestris | GCA_911622165.1_iyBomSyle1.1_genomic |
| Hymenoptera | Andrena dorsata | GCA_929108735.1_iyAndDors1.1_genomic |
| Hymenoptera | Andrena minutula | GCA_929113495.1_iyAndMinu1.1_genomic |
| Hymenoptera | Bombus pratorum | GCA_930367275.1_iyBomPrat1.1_genomic |
| Diptera | Scaeva pyrastri | GCA_905146935.1_idScaPyra1.1_genomic |
| Diptera | Syritta pipiens | GCA_905187475.1_idSyrPipi1.1_genomic |
| Diptera | Tachina fera | GCA_905220375.1_idTacFera2.1_genomic |
| Diptera | Xylota sylvarum | GCA_905220385.1_idXylSylv2.1_genomic |
| Diptera | Eristalis tenax | GCA_905231855.1_idEriTena2.1_genomic |
| Diptera | Volucella inanis | GCA_907269105.1_idVolInan1.1_genomic |
| Diptera | Eristalis pertinax | GCA_907269125.1_idEriPert2.1_genomic |
| Diptera | Bibio marci | GCA_910594885.1_idBibMarc1.1_genomic |
| Diptera | Xanthogramma pedissequum | GCA_910595825.1_idXanPedi1.1_genomic |
| Diptera | Chrysotoxum bicinctum | GCA_911387755.1_idChrBici1.1_genomic |
| Diptera | Melanostoma mellinum | GCA_914767635.1_idMelMell2.1_genomic |
| Diptera | Coremacera marginata | GCA_914767935.1_idCorMarg1.1_genomic |
| Diptera | Thecocarcelia acutangulata | GCA_914767995.1_idTheAcut1.1_genomic |
| Diptera | Bellardia pandia | GCA_916048285.1_idBelPand1.1_genomic |
| Diptera | Platycheirus albimanus | GCA_916050605.1_idPlaAlba1.1_genomic |
| Diptera | Cheilosia vulpina | GCA_916610125.1_idCheVulp2.1_genomic |
| Diptera | Eristalis arbustorum | GCA_916610145.1_idEriArbu1.1_genomic |
| Diptera | Gymnosoma rotundatum | GCA_916610165.1_idGymRotn1.1_genomic |
| Diptera | Criorhina berberina | GCA_917880715.1_idCriBerb1.1_genomic |
| Diptera | Eupeodes latifasciatus | GCA_920104205.1_idEupLati1.1_genomic |
| Diptera | Clusia tigrina | GCA_920105625.1_idCluTigr1.1_genomic |
| Diptera | Sicus ferrugineus | GCA_922984085.1_idSicFerr1.1_genomic |
| Diptera | Sarcophaga caerulescens | GCA_927399465.1_idSarCaer1.1_genomic |
| Diptera | Volucella inflata | GCA_928272305.1_idVolInfl1.1_genomic |
| Diptera | Epistrophe grossulariae | GCA_929447395.1_idEpiGros1.1_genomic |
| Diptera | Myathropa florea | GCA_930367185.1_idMyaFlor2.1_genomic |
| Diptera | Pollenia angustigena | GCA_930367215.1_idPolAngu1.1_genomic |
| Diptera | Sarcophaga rosellei | GCA_930367235.1_idSarRose1.1_genomic |
| Diptera | Sarcophaga variegata | GCA_932273835.1_idSarVari1.1_genomic |
| Diptera | Leucozona laternaria | GCA_932273885.1_idLeuLate1.1_genomic |
| Diptera | Protocalliphora azurea | GCA_932274085.1_idProAzur1.1_genomic |
| Diptera | Nephrotoma flavescens | GCA_932526605.1_idNepFlae1.1_genomic |
| Diptera | Epicampocera succincta | GCA_932526305.1_idEpiSucc1.1_genomic |
| Diptera | Bombylius major | GCA_932526495.1_idBomMajo1.1_genomic |
| Diptera | Rhingia campestris | GCA_932526625.1_idRhiCamp1.1_genomic |
| Diptera | Stomorhina lunata | GCA_933228675.1_idStoLuna1.1_genomic |
| Diptera | Machimus atricapillus | GCA_933228815.1_idMacAtri3.1_genomic |
| Diptera | Cheilosia pagana | GCA_936431705.1_idChePaga1.1_genomic |
| Diptera | Nowickia ferox | GCA_936439885.1_idNowFero1.1_genomic |
| Diptera | Sarcophaga subvicina | GCA_936449025.1_idSarSubv1.1_genomic |
| Diptera | Thecophora atra | GCA_937620795.1_idTheAtra2.1_genomic |
| Diptera | Cistogaster globosa | GCA_937654795.1_idCisGlob1.1_genomic |
| Diptera | Bombylius discolor | GCA_939192795.1_idBomDisc1.1_genomic |
| Diptera | Phyto melanocephala | GCA_941918925.1_idPhyMeln1.1_genomic |
| Diptera | Calliphora vomitoria | GCA_942486065.1_idCalVomi1.1_genomic |
| Odonata | Ischnura elegans | GCA_921293095.1_ioIscEleg1.1_genomic |
| Odonata | Platycnemis pennipes | GCA_933228895.1_ioPlaPenn1.1_genomic |
| Odonata | Pantala flavescens | GCA_020796165_Panflav1_CAAS_Pfla_1.0 |
| Psocodea | Liposcelis brunnea | GCA_023512825_ipLipBrun1 |
| Orthoptera | Schistocerca piceifrons | GCA_021461385_ioSchPice1 |
| Orthoptera | Schistocerca gregaria | GCA_023897955_ioSchGreg1 |
| Orthoptera | Schistocerca americana | GCA_021461395_ioSchAmer1 |
| Hemiptera | Aelia acuminata | GCA_911387785.1_ihAelAcum1.1_genomic |
| Hemiptera | Acanthosoma haemorrhoidale | GCA_930367205.1_ihAcaHaem1.1_genomic |
| Neuroptera | Chrysoperla carnea | GCA_905475395.1_inChrCarn1.1_genomic |
| Neuroptera | Chrysopa pallens | GCA_020423425_inChrPall1 |
3. Insect Hox gene clusters
3.1. Gene loss in insect Hox clusters
There are no clear examples of Hox gene loss within insects, at least for the ‘canonical’ Hox genes that play roles in specifying anteroposterior position. All eight of the expected canonical Hox genes - pb, lab, Dfd, Scr, Antp, Ubx, abdA, AbdB - are present in all insects (Fig. 1). This contrasts to some other arthropod lineages (see [26]). For example, within Crustacea the abdA gene is proposed to be missing in three barnacles that have been studied (Elminius modestus, Trypetesa lampas and Sacculina carcini) and within Chelicerata the same gene has not been found in two mites (Archegozetes longisetosus and Tetranychus urticae) and a pycnogonid (Endeis spinosa), although not all these surveys were based on high quality genome assemblies (see [26]).
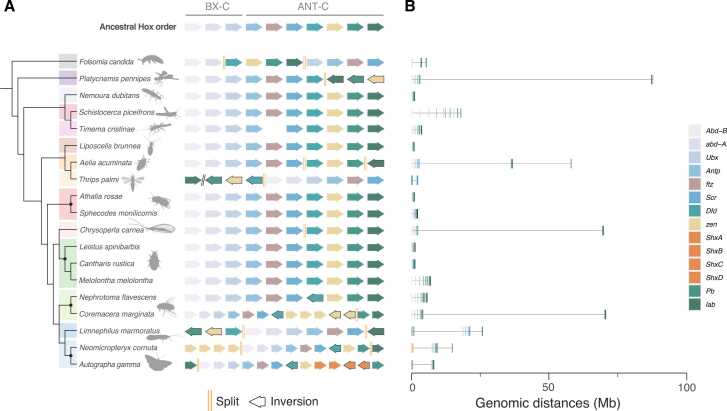
Fig. 1.
Genomic organisation and gene orientation across insect Hox clusters (A) Left shows the phylogeny for subset of species analysed. Hexapod orders, from top to bottom are: Collembola (grey), Odonata (dark purple), Plecoptera (light purple), Orthoptera (dark pink), Phasmatodea (light pink), Psocodea (brown), Hemiptera (dark orange), Thysanoptera (light orange), Hymenoptera (dark red), Neuroptera (light red), Coleoptera (dark green), Diptera (light green), Trichoptera (dark blue), and Lepidoptera (light blue).Dots on the phylogeny represent hexapod orders for which data are shown from more than one species. Right shows the order and transcriptional orientation of Hox genes (coloured as per the legend) in each species. Splits within the Hox cluster are denoted by double orange lines, inversions are annotated with a black border around the gene. Slanted double black lines represent translocation to a separate scaffold. (B) Structure of the Hox cluster per species shown using actual genomic distances. Each line represents a Hox gene as it occurs in the genome, coloured as per the legend. Genomic distances are shown in Megabases.
The ftz gene, which evolved a role in segmental patterning in insects rather than specification of position, seems to be absent in the genome of the stick insect Timema cristina (order Phasmatodea; [27]; assembly tcristinae_2.1). However, since this is a finding from analysis of a single genome assembly, verification is needed. The other Hox gene with a changed function in insects, the paralogy group 3 gene zen, is present in most insects. Interestingly, zen appears to be lost completely from the genomes of two related flies, Epicampocera succincta and Thecocarcelia acutangulata, which are both within the dipteran family Tachinidae. Similar loss of zen may have occurred within some Chelicerata, where this gene is reported absent from the genomes of the mites Tetranychus urticae [28] and Metaseiulus occidentalis [29]. Other cases of gene loss affect more recent duplicates. For example, zen has undergone tandem duplication in several lineages of insects and in some cases there has been secondary loss of derived zen duplicates (see Section 3.4). This includes a shared loss of the zen-derived ShxD gene in all Lycaenidae butterflies.
3.2. Splits, rearrangements and inversions in the insect Hox cluster
Even before the molecular cloning of Hox genes, it was clear that the mutant loci giving homeotic phenotypes in Drosophila melanogaster were located in two distinct complexes on chromosome 3: the ANT-C and the BX-C [30], [31]. Cloning revealed the ANT-C contains from lab to Antp (of the ancestral gene order), whereas BX-C contains the genes from Ubx to AbdB, with a 9.6 Mb gap between them. Splits have also occurred, at different positions, in other Drosophila species [17]. In the mosquito Anopheles gambiae, the cluster is not split. The clear implication is that an unbroken Hox cluster is ancestral for this clade of Diptera and by implication (assuming a split cluster cannot reform into a complete cluster) ancestral for all insects, as it is for the Bilateria. In surveying the structure of Hox clusters across insects, therefore, we are not asking whether a split cluster is ancestral. Instead, we can ask whether there are particular intergenic regions where splits are more frequent evolutionarily, and conversely whether particular sets of Hox genes always stay together in evolution. Here we define intergenic regions as the genomic content between the homeobox sequences of the Hox genes, as current data do not allow us to identify the ends of every transcription unit.
Examination of 243 insect genomes reveals Hox cluster splits in many species (Fig. 1). For example, these include splits in the Hox clusters of Platycnemis pennipes (Odonata), Aelia acuminata (Hemiptera), Chrysoperla carnea (Neuroptera), Coremacera marginata (Diptera), Limnephilus marmoratus (Trichoptera) and in all Lepidoptera. In some cases, these splits lead to dramatic expansion in the overall size of the Hox cluster. For example, the lab, pb and zen genes in Platycnemis pennipes are located ∼84 Mb from the rest of the cluster. Similarly, splits between Scr and Dfd and between pb and lab in Aelia acuminata, resulted in genomic distances of ∼33 Mb and ∼21 Mb between these genes, respectively. In Chrysoperla carnea the split occurs between Scr and Dfd and results in a distance of ∼67 Mb, and in Coremacera marginata a distance of ∼66 Mb separates zen and pb. The cluster split in Lepidoptera lies between lab and the rest of the cluster, and is present in every lepidopteran species analysed (124 species) including two representatives of the most basal family, Micropterygidae (Neomicropteryx cornuta and Micropterix aruncella).
The lab gene is found distal to the ‘posterior’ end of the cluster in most Lepidoptera (represented in Fig. 1 by the Silver-Y moth Autographa gamma). This repositioning is clearly a secondary event following ‘escape’ of the Hox gene from tight linkage to other Hox genes, since in Neomicropteryx cornuta (in the basal family Micropterygidae) the split has occurred but the repositioning has not. The finding that the lab gene is also distant from the rest of the Hox cluster in Trichoptera (caddisfly) genomes suggests this split probably occurred prior to the common ancestor of Lepidoptera+Trichoptera (Amphiesmenoptera). Interestingly, relocation of lab to a different chromosome was also found in two mosquito species, Aedes aegypti and Culex quinquefasciatus [32].
Similar cases of translocation or inversion have occurred in Odonata, Thysanoptera and Trichoptera, after splitting of the Hox cluster. This is the case for zen, pb and lab split from the rest of the cluster in Platycnemis pennipes (Odonata), Thrips palmi (Thysanoptera) and Limnephilus marmoratus (Trichoptera). Other inversions of genes occur unrelated to splits, for example, Dfd is inverted in Coremacera marginata and Nephrotoma flavescens (Diptera) and Neomicropteryx cornuta and Autographa gamma (Lepidoptera) (Fig. 1).
While splits have occurred frequently in insect evolution, the overall genomic order of Hox genes in insects is comparable to that seen for the homologous genes in vertebrates. This represents the colinear correspondence between gene order and the body position where each gene is expressed and functional during early embryonic development, for those Hox genes that still play this role (Fig. 2). Although we do not find clear cases of shuffling this order when the genes are together in a single intact cluster, there are cases of rearrangement caused by cluster breakage, in some cases involving inversions. Interestingly, these changes are almost always associated with paralogy groups 1–4: lab, pb, zen and Dfd (Fig. 2). The rearrangements found affect these four Hox genes in different ways. In each case of gene, or gene block rearrangement, there is a link between splits in the Hox cluster and subsequent rearrangement events within insect orders.
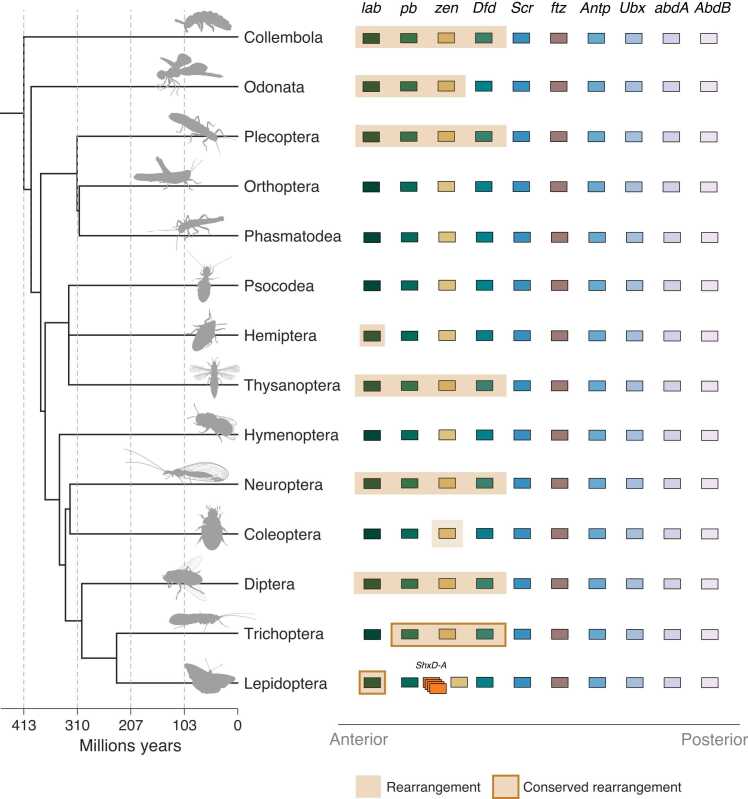
Fig. 2.
Hox genes prone to rearrangement in insect Hox clusters Left shows a time calibrated species tree of insect Orders analysed in this study. Right shows the composition of the Hox gene cluster, in their ancestral order. Shaded orange regions represent genes that have undergone rearrangement from the ancestral order of Hox genes. Those conserved across species sampled have a border around the box. Splits in the Hox cluster are not depicted.
In some insect orders, there are rearrangements in all species sampled; for example, five Trichoptera (caddisfly) species have pb, zen and Dfd in derived positions. In all four species in the family Limnephilidae, pb, zen and Dfd are located at the ‘posterior’ (AbdB) end of the cluster, with an inversion in two species, Limnephilus marmoratus and Limnephilus rhombicus. In Eubasilissa regina (family Phryganeidae), pb, zen and Dfd are found outside the cluster, upstream of lab. Similarly, in all Lepidoptera (butterflies and moths; 124 species) lab is found away from the rest of the Hox cluster. The rearrangement of lab away from the rest of the Hox cluster was noted previously in the silk moth Bombyx mori [18], [33]; the higher quality genome assemblies now available confirm that the lab gene is usually located at the ‘posterior’ end of the lepidopteran Hox cluster, separated by a large distance (from 1.4 Mb in Tinea semifulvella to 24 Mb in Phalera bucephala) containing numerous non-Hox genes.
In two of the three Odonata (dragonfly and damselfly) species analysed, lab, pb and zen are rearranged, but Dfd is in its ancestral position in the cluster. In the white-legged damselfly Platycnemis pennipes there has been an inversion that switched the order and transcriptional orientation of these genes as a block, and in the blue-tailed damselfly Ischnura elegans there has been an inversion plus a translocation to the other end of the cluster. In Plecoptera (stoneflies), Thysanoptera (thrips) and Neuroptera (lacewings and allies) various rearrangements are found. In certain species in these groups lab, pb, zen and Dfd have all been translocated to the ‘posterior’ end of the cluster, nearer to AbdB, with a subsequent inversion of this gene cassette in the plecopteran species Nemurella pictetii. In the only thysanopteran species in our dataset (Thrips palmi), an additional rearrangement resulted in lab positioned on a separate scaffold to the rest of the cluster and pb, zen and Dfd translocated to the posterior end. In Hemiptera, lab is located at the ‘posterior’ end of the cluster in Acanthosoma haemorrhoidale, although larger rearrangements affecting lab, pb, zen and Dfd in another hemipteran species (Diaphorina citri) has been observed [34]. Diptera displays the largest number of rearrangements, with at least five different rearrangement events occurring across the tree, resulting in translocations of one or more of the lab, pb, zen and Dfd genes to the opposite end of the gene cluster. In Coleoptera, three species show translocation of zen copies outside of the Hox cluster, resulting from independent lineage-specific events.
3.3. Hox cluster size across insects
While splits and rearrangements in the Hox cluster occur frequently across insects, there are certain genes which have rarely been split apart in the insect genomes studied to date. For example, the three genes found in the Bithorax complex of Drosophila (AbdB, abdA and Ubx) are found in the same order, in all 243 insect genomes studied, although we note that a cluster split between Ubx and abdA occurred in a clade of Drosophila [17]. Within the set of genes corresponding to the ANT-C of Drosophila (Fig. 1, Fig. 2), the genes Antp, ftz and Scr are most conserved in their organisation and orientation. To our knowledge, there are no known cases of split between these genes, indicating there may be a selective pressure to maintain their linkage. Indeed, overall there are relatively few cluster splits between AbdB and Scr.
When the intergenic distances between each pair of genes (measured as the distance between homeobox sequences of the Hox genes) are compared between insect orders, a very intriguing pattern emerges (Fig. 3A). Excluding the first three Hox genes located at the ‘anterior’ end of the cluster (lab, pb and zen), which underwent significant rearrangements in many different species, we see that the intergenic distances between the next four genes (Antp, ftz, Scr and Dfd) are consistently small. These four ‘tightly linked’ genes are all orthologues of the ANT-C genes of Drosophila melanogaster. In contrast, the intergenic distances between the three orthologues of the BX-C genes (AbdB, abdA and Ubx) are consistently larger. This trend is most easily seen when the distances are compared within an insect order, and is seen regardless of whether the insect order has more or less ‘relaxed’ organisation of the Hox cluster (Fig. 3A). This may imply that there is a deep and fundamental difference between Hox gene organisation between ANT-C and BX-C genes, dating to long before the homeotic complex split in Drosophila. Interestingly, the intergenic distance between Ubx and Antp in most insects (the position of the BX-C/ANT-C split in Drosophila melanogaster) falls into the range of the BX-C intergenic distances, even in gene clusters that are not split.
Fig. 3.
Hox cluster size across insects. (A) Distribution of intergenic regions per Hox gene in different insect orders. Only shows orders where the Hox genes are conserved in the ‘normal’ order. (B) Distribution of core Hox cluster (AbdB to Scr) size for each order, ordered by phylogeny. (C) Correlation between core Hox cluster size and genome size. Each dot represents a species, which are colour by order as per the figure legend. Dots are sized by the total number of Hox genes present in each species. In (A) and (B) boxplots have a rectangle between the 25th and 75th percentiles of the range, with the median as a dark line, whiskers reach the largest and smallest values within 1.5x Interquartile range, and outliers are points beyond 1.5 × Interquartile range (plotted using geom_boxplot in ggplot2).
The relative conservation in gene order and organisation from AbdB to Scr across all orders provides a useful opportunity to compare the evolution of the Hox cluster size across insects. The size of this conserved core region of the Hox cluster ranges from 0.57 Mb (Common Plume moth Emmelina monodactyla; Lepidoptera) to 5.8 Mb (Tachina fera; Diptera). These genomic distances are much larger than the same region in vertebrates where whole Hox clusters are only ∼0.1 Mb [35], [36]. Odonata, Hemiptera and Trichoptera have consistently large sizes for this core cluster, reflecting large intergenic distances, while Coleoptera and Diptera each show great variation in cluster size across the order (Fig. 3B). For most insect orders, the size of the core part of the Hox cluster correlates with genome size (Fig. 3C). However, Diptera and Lepidoptera both show low correlation values (r = 0.37 and 0.27, respectively), suggesting that there are other factors driving the size of the Hox cluster other than genome size in these groups of insects.
The size of the Hox cluster in Schistocerca piceifrons (Orthoptera; assembly iqSchPice1.1) is significantly expanded with larger distances between all genes, relative to most other insects, suggesting relaxation of the constraints acting on the whole cluster (Fig. 1). When we compare cluster size across other Schistocerca species (Schistocerca americana; iqSchAmer2.1 and Schistocerca gregaria; iqSchGreg1.1), the total size of the Hox cluster ranges from 16 Mb to 17.8 Mb, and the ‘core’ Hox cluster size (AbdB to Scr) ranges from 10.8 Mb to 12.2 Mb, significantly larger than any other insect species analysed in this study (Fig. 3B). This contrasts with earlier (pre-genomic) analysis in Schistocerca gregaria, where the total cluster size was determined using chromosomal in situ hybridization and estimated to be at least 700Kb in length, and no longer that 2 Mb in total [11]. Although linkage in the Hox cluster in this genus has relaxed significantly, there are no rearrangements found in the order of the Hox genes within the genome.
3.4. Tandem duplication of insect Hox genes: Zerknüllt and fushi tarazu
Tandem duplication within a Hox gene cluster is rare, with some of the clearest examples being the initial expansion of the Hox cluster in early bilaterian evolution [37], [38] and expansions at the ‘posterior’ end of the cluster in vertebrates, amphioxus and echinoderms [15], [39]. In analysing publicly-available insect genomes, we find only two cases of putative tandem duplication of a canonical Hox gene (Fig. 4): two copies of Dfd present in Acronicta aceris (Sycamore Moth) and two copies of pb present in Micropterix aruncella. While genomic position and gene trees provide support for these putative Hox duplicates as real events, since they are present in a single species each of the findings needs further verification. Indeed, it is expected that tandem duplications of canonical Hox genes would be deleterious since they could disrupt the spatial regulation of these genes, and thereby disrupt anteroposterior body patterning. In contrast, the two Hox genes that have derived roles, ftz and zen, might be expected to have less constraint against duplication. This is because the zen gene lost its ancestral homeotic function in early insect evolution, acquiring a novel role in the formation of extraembryonic tissues, while ftz has a new role in segmentation.
Fig. 4.
Copy number of Hox genes across insects. Left shows the phylogeny of all species analysed, with block colours signifying the Order to which they belong. Heatmap shows the copy number of all Hox genes per species. Barchart of the right shows the total number of Hox genes annotated per species.
As noted in Section 3.1, a putative loss of ftz is observed in one insect, and conversely there are two ftz copies in Spilarctia lutea (Buff Ermine Moth), as well as two closely related wasps: Vespula germanica and Vespula vulgaris (Fig. 4). Finding the duplication in related species gives stronger support to this observation. Duplications of zen in insects have been known about and intensively studied for many years. First, there is a well-studied duplication of zen in Tribolium castaneum, which gave zen and zen2 [13]. Recent work has shown that this duplication is shared by three closely related Tribolium species, and that the gene products interact in a negative feedback loop that may confer precision of temporal expression [40]. Second, it was shown over 20 years ago that the dipteran bcd gene is a derived tandem duplicate of zen [41]. This duplication was followed by extensive sequence divergence in the locus which became bcd, in a classic case of ‘asymmetric sequence divergence’ where one daughter gene undergoes far more sequence change than the other [42], [43]. Key amino acid changes in the homeodomain include a mutation from glutamine to lysine at position 50 (Q50K) and a switch from methionine to arginine at position 54 (M54R); these substitutions contributed to changing downstream targets and altered biological role [44], [45]. Third, a further duplication of zen within the Drosophila genus produced zen2. Fourth, the most dramatic cases of zen duplication have been reported in Lepidoptera. In ditrysian lepidopterans, the zen gene duplicated to give four additional fast-evolving copies named ShxA, ShxB, ShxC and ShxD [19]. These highly derived Shx genes are expressed in the developing serosa, not the embryo itself, and may pre-pattern this extraembryonic tissue, as judged by the striking pattern of maternal RNA localisation [19]. These genes duplicated even further in Bombyx mori resulting in at least 12, possibly 15, tandem Shx gene copies [18], [19]. Furthermore, recent work has shown that the extreme duplication in Bombyx is not unique: at least 18 other lineages of Lepidoptera have highly expanded sets of Shx genes, in some cases reaching over 100 homeobox copies (Fig. 4). There has also been occasional loss of specific Shx genes; for example, ShxD was lost in butterflies of the family Lycaenidae (‘blues’ and their allies) and fritillary butterflies of the genus Melitaea [46]).
With the availability of many high-quality insect genomes, it is now possible to ask if there are additional cases of zen duplication, in addition to those mentioned above (Fig. 4). Within the coleopteran species for which genomes are available, multiple duplications of zen occur in the Cucujiformia infraorder and range from 2 copies in Polydrusus cervinus (weevil) to 17 copies in Harmonia axyridis (harlequin ladybird) and 19 copies in Pyrochroa serraticornis (cardinal beetle). This is in addition to the well-studied duplication in Tribolium. In Diptera, copy number of zen ranges from a single copy in the early diverging lineages to 118 in Tachina fera and 93 in Sarcophaga variegata. Even within a family of flies for which there is a large number of species sampled, Syrphidae, there is significant variation in zen copy number between these related species (Fig. 4). The number of copies and the branching patterns within the gene tree (Fig. 4) suggest that large tandem duplication events occurred multiple times independently in this lineage.
It is striking that duplication of zen, and its progenitors (Shx in Lepidoptera) occur only in the highly speciose orders Diptera, Lepidoptera and Coleoptera, within the holometabolous insects. As described above, zen lost its homeotic function early on in insect evolution, and in many insect species is involved in development of extraembryonic membranes. In insects these membranes consist of two distinct layers: the amnion and serosa (these form a single epithelium known as the amnioserosa in higher flies) [47], [48]. The amnion is the inner membrane which surrounds the ventral side of the developing embryo, while the serosa is an outer membrane which lies just inside the chorion and envelops the embryo, amnion and yolk [49], [50], [51]. This structure is hypothesised to be involved in a wide range of functions unrelated to development of body form, such as a general protective role including structural stability, water regulation and desiccation resistance [52], [53], [54], and innate immune response [55], [56], [57]. Interestingly, while this dual structure is present in most pterygote insects, derived hymenopterans (Apocrita) usually lack an amnion, or have a temporary amniotic-like structure which covers the yolk [58], [59]. It is intriguing to consider whether the highly dynamic copy numbers of zen along with its functions in the extraembryonic tissues may have played a role in facilitating speciation and adaptation to diverse habitats in Diptera, Lepidoptera and Coleoptera. Indeed, it is striking that copy number variation of zen is particularly variable in highly speciose families such as Syrphidae and Coccinellidae (Fig. 4). However, whether these large expansions in gene number are functional, or even expressed during early development, requires further analyses. Furthermore, the neutral theory posits that increases in copy number may not always have adaptive significance, and may instead result from mutational processes within the genome, affected by intragenomic variation in copying fidelity and the effects of transposable element accumulation.
4. Conclusions
We are entering a new era of genomics, as new technologies are facilitating the imminent sequencing and assembly of thousands of eukaryotic species. At the time of writing, there are more than 200 high quality, complete insect genomes available for analysis, and although this number is expected to rise very rapidly, now is an excellent time to pause and take stock of the lessons that can be learned. This is an opportune time for two reasons. First, the available high quality genomes span a wide phylogenetic diversity of insects, including representatives of at least 13 orders (Odonata, Plecoptera, Orthoptera, Phasmatodea, Psocodea, Hemiptera, Thysanoptera, Hymenoptera, Neuroptera, Coleoptera, Diptera, Trichoptera, Lepidoptera). Second, within some taxa (notably Lepidoptera and Diptera) ‘deep dives’ have been undertaken, yielding genomes from closely related species, thereby permitting insights into genomic change on shorter time frames. Here we have used these data, in combination with previously published analyses, to compare Hox gene cluster organisation across insects. We have searched for patterns of evolutionary conservation or general trends across insects, examples of convergent evolution, and anomalies.
First, we examine gene loss and conclude that canonical Hox genes have not been lost in insect evolution: pb, lab, Dfd, Scr, Antp, Ubx, abdA, AbdB are present in all insects studied. The two ‘non-canonical’ Hox genes, zen and ftz, are lost rarely. We find two closely related insect species putatively lacking zen, possibly a shared loss inherited from a common ancestor, and one example of a putative loss of ftz. The rarity of these losses highlights that further verification is needed. However, the finding that canonical Hox genes are never lost in insects has a biological implication. We suggest that each Hox gene has remained indispensable through insect radiation because segment number and tagmatization, has remained consistent, giving no opportunity for gene redundancy and loss.
Second, we find many independent cases of splitting of the insect Hox gene cluster, in an analogous fashion to the separation of ANT-C and BX-C in Drosophila melanogaster. Although these splits can occur in several different places in the Hox cluster, they are most commonly seen affecting the first four paralogy groups (PG1 to PG4): lab, pb, zen, and Dfd. There are cases where just lab (PG1) is split away (Lepidoptera and Trichoptera), one dipteran in which lab plus pb are separated away, Odonata with lab, pb and zen split away, and many insects with a split between Dfd and Scr (separating PG1 to PG4 from the rest). We do not find cases of complete ‘atomisation’ of the Hox cluster, as seen in larvacean chordates and predatory mites for example. It would be interesting to compare patterns of Hox cluster breakage and rearrangement with the overall genome-wide recombination and inversion rates for each taxa, to test if Hox cluster rearrangements reflect general genomic properties. From the pattern of splitting observed, we suggest that insect Hox genes are not generally regulated as a whole cluster, but there are selective pressures acting to prevent many rearrangements. These selective pressures could include shared regulation of neighbouring games, interdigitated control (enhancers for one Hox gene located beyond the neighbouring gene) or simply a high density of regulatory elements. We suggest these constraints are lowest around paralogy groups 1–4. We speculate that shared and interdigitated control may have evolved around ‘posterior’ insect Hox genes to fine-tune expression within overlapping domains in the abdomen.
Third, it has long been known that insect Hox gene clusters have much larger intergenic distances than in vertebrates. We find that intergenic distances in the Hox cluster vary greatly across insects, with particularly large genomic distances in Orthoptera, Odonata, Hemiptera and Trichoptera, and highly variable intergenic distances in Coleoptera and Diptera. Intergenic lengths correlate with genome size in most, but not all, insect orders. We note a striking and puzzling trend in intergenic distance within insect Hox clusters: the distances between ‘posterior’ genes are consistently greater than distances between each pair of ‘central’ or ‘anterior’ genes. Specifically, intergenic distances from AbdB to Antp are greater than intergenic distances across the rest of the cluster. We do not know the biological basis for this observation. Counterintuitively, the region with the largest intergenic distances is also the region least prone to genomic rearrangement in evolution. We suggest that fundamental mechanisms of gene regulation may be different at the two ends of the insect Hox gene cluster.
Fourth, we examine gene duplication and conclude that insect Hox genes are rarely duplicated, with the exception of zen. We do find putative cases of Dfd duplication and pb duplication, but these are seen in single genomes and require further verification. A ftz duplication is seen in genome assemblies for two wasps and can be treated as more definitive. The zen gene, in contrast, has undergone tandem duplication many times independently, undergoing dramatic copy number expansion in some insect lineages. The most striking examples of zen duplication are seen in genomes from the highly speciose orders, Coleoptera, Diptera and Lepidoptera, where over 100 zen-derived homeobox sequences can be present in some species. It is unclear why such dramatic copy number changes have occurred, and indeed whether retention of extra genes was selectively advantageous through subfunctionalization, neofunctionalization or simply dosage effects. The fact that zen genes play roles in extraembryonic patterning, rather than position-specific cell fate in the embryo, may underpin why tandem duplications are not instantly deleterious, but this does not seem to explain the preponderance of zen gene arrays observed. Further work is required to determine if the locus is particularly prone to unequal crossover at meiosis, and therefore a hotspot of mutation, and/or whether duplicated zen genes were repeatedly recruited to novel roles in extraembryonic membrane patterning as insects adapted to their multitude of ecological niches.
Declarations of interest
None.
Acknowledgements
We acknowledge the contributions of the many laboratories and research groups that have made insect genome sequences openly available for analysis, including the Darwin Tree of Life project coordinated by the Wellcome Sanger Institute. This research was funded by the Wellcome Trust Darwin Tree of Life Discretionary Award [218328] and the John Fell OUP Research Fund. For the purpose of open access, the authors have applied a CC BY public copyright licence.
Contributor Information
Peter O. Mulhair, Email: peter.mulhair@biology.ox.ac.uk.
Peter W.H. Holland, Email: peter.holland@biology.ox.ac.uk.
References
- 1.Dudley R. Princeton University Press; 2002. The Biomechanics of Insect Flight: Form, Function, Evolution. [Google Scholar]
- 2.Hughes C.L., Kaufman T.C. Hox genes and the evolution of the arthropod body plan. Evol. Dev. 2002;4:459–499. doi: 10.1046/j.1525-142x.2002.02034.x. [DOI] [PubMed] [Google Scholar]
- 3.de Rosa R., Grenier J.K., Andreeva T., Cook C.E., Adoutte A., Akam M., Carroll S.B., Balavoine G. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399:772–776. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- 4.Cook C.E., Smith M.L., Telford M.J., Bastianello A., Akam M. Hox genes and the phylogeny of the arthropods. Curr. Biol. 2001;11:759–763. doi: 10.1016/s0960-9822(01)00222-6. [DOI] [PubMed] [Google Scholar]
- 5.Crumière A.J.J., Khila A. Hox genes mediate the escalation of sexually antagonistic traits in water striders. Biol. Lett. 2019;15 doi: 10.1098/rsbl.2018.0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian L., Rahman S.R., Ezray B.D., Franzini L., Strange J.P., Lhomme P., Hines H.M. A homeotic shift late in development drives mimetic color variation in a bumble bee. Proc. Natl. Acad. Sci. USA. 2019;116:11857–11865. doi: 10.1073/pnas.1900365116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rafiqi A.M., Rajakumar A., Abouheif E. Origin and elaboration of a major evolutionary transition in individuality. Nature. 2020;585:239–244. doi: 10.1038/s41586-020-2653-6. [DOI] [PubMed] [Google Scholar]
- 8.Liu B.-P., Ding G., Miao Y., Hua B.-Z. The Hox gene Abdominal-B regulates the appendage development during the embryogenesis of scorpionflies. Insect Mol. Biol. 2022 doi: 10.1111/imb.12790. [DOI] [PubMed] [Google Scholar]
- 9.Murugesan S.N., Connahs H., Matsuoka Y., Das Gupta M., Tiong G.J.L., Huq M., Gowri V., Monroe S., Deem K.D., Werner T., Tomoyasu Y., Monteiro A. Butterfly eyespots evolved via cooption of an ancestral gene-regulatory network that also patterns antennae, legs, and wings. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2108661119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novikova E.L., Kulakova M.A. There and back again: hox clusters use both DNA strands. J. Dev. Biol. 2021;9 doi: 10.3390/jdb9030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrier D.E., Akam M. Organization of the Hox gene cluster in the grasshopper, Schistocerca gregaria. Proc. Natl. Acad. Sci. USA. 1996;93:13024–13029. doi: 10.1073/pnas.93.23.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holland, The fall and rise of evolutionary developmental biology, Syst. Assoc. Spec. Vol. (2004). 〈https://books.google.com/books?hl=en&lr=&id=qnUhL9z9QqcC&oi=fnd&pg=PA261&dq=The+Fall+and+Rise+of+Evolutionary+Developmental+Biology&ots=ss4mShxI2l&sig=lOQvpEl35SP065UFXJF2k4x2lqs〉.
- 13.Brown S.J., Fellers J.P., Shippy T.D., Richardson E.A., Maxwell M., Stuart J.J., Denell R.E. Sequence of the Tribolium castaneum homeotic complex: the region corresponding to the Drosophila melanogaster antennapedia complex. Genetics. 2002;160:1067–1074. doi: 10.1093/genetics/160.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Zee M., Berns N., Roth S. Distinct functions of the tribolium zerknu¨llt genes in serosa specification and dorsal closure. Curr. Biol. 2005;15:624–636. doi: 10.1016/j.cub.2005.02.057. [DOI] [PubMed] [Google Scholar]
- 15.Ferrier D.E., Minguillón C., Holland P.W., Garcia-Fernàndez J. The amphioxus Hox cluster: deuterostome posterior flexibility and Hox14. Evol. Dev. 2000;2:284–293. doi: 10.1046/j.1525-142x.2000.00070.x. [DOI] [PubMed] [Google Scholar]
- 16.Pascual-Anaya J., D’Aniello S., Kuratani S., Garcia-Fernàndez J. Evolution of Hox gene clusters in deuterostomes. BMC Dev. Biol. 2013;13:26. doi: 10.1186/1471-213X-13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Negre B., Ruiz A. HOM-C evolution in Drosophila: is there a need for Hox gene clustering? Trends Genet. 2007;23:55–59. doi: 10.1016/j.tig.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Chai C.-L., Zhang Z., Huang F.-F., Wang X.-Y., Yu Q.-Y., Liu B.-B., Tian T., Xia Q.-Y., Lu C., Xiang Z.-H. A genomewide survey of homeobox genes and identification of novel structure of the Hox cluster in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2008;38:1111–1120. doi: 10.1016/j.ibmb.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Ferguson L., Marlétaz F., Carter J.-M., Taylor W.R., Gibbs M., Breuker C.J., Holland P.W.H. Ancient expansion of the hox cluster in lepidoptera generated four homeobox genes implicated in extra-embryonic tissue formation. PLOS Genet. 2014;10 doi: 10.1371/journal.pgen.1004698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhan S., Merlin C., Boore J.L., Reppert S.M. The monarch butterfly genome yields insights into long-distance migration. Cell. 2011;147:1171–1185. doi: 10.1016/j.cell.2011.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heliconius Genome Consortium Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewin H.A., Robinson G.E., Kress W.J., Baker W.J., Coddington J., Crandall K.A., Durbin R., Edwards S.V., Forest F., Gilbert M.T.P., Goldstein M.M., Grigoriev I.V., Hackett K.J., Haussler D., Jarvis E.D., Johnson W.E., Patrinos A., Richards S., Castilla-Rubio J.C., van Sluys M.-A., Soltis P.S., Xu X., Yang H., Zhang G. Earth biogenome project: sequencing life for the future of life. Proc. Natl. Acad. Sci. USA. 2018;115:4325–4333. doi: 10.1073/pnas.1720115115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewin H.A., Richards S., Lieberman Aiden E., Allende M.L., Archibald J.M., Bálint M., Barker K.B., Baumgartner B., Belov K., Bertorelle G., Blaxter M.L., Cai J., Caperello N.D., Carlson K., Castilla-Rubio J.C., Chaw S.-M., Chen L., Childers A.K., Coddington J.A., Conde D.A., Corominas M., Crandall K.A., Crawford A.J., DiPalma F., Durbin R., Ebenezer T.E., Edwards S.V., Fedrigo O., Flicek P., Formenti G., Gibbs R.A., Gilbert M.T.P., Goldstein M.M., Graves J.M., Greely H.T., Grigoriev I.V., Hackett K.J., Hall N., Haussler D., Helgen K.M., Hogg C.J., Isobe S., Jakobsen K.S., Janke A., Jarvis E.D., Johnson W.E., Jones S.J.M., Karlsson E.K., Kersey P.J., Kim J.-H., Kress W.J., Kuraku S., Lawniczak M.K.N., Leebens-Mack J.H., Li X., Lindblad-Toh K., Liu X., Lopez J.V., Marques-Bonet T., Mazard S., Mazet J.A.K., Mazzoni C.J., Myers E.W., O’Neill R.J., Paez S., Park H., Robinson G.E., Roquet C., Ryder O.A., Sabir J.S.M., Shaffer H.B., Shank T.M., Sherkow J.S., Soltis P.S., Tang B., Tedersoo L., Uliano-Silva M., Wang K., Wei X., Wetzer R., Wilson J.L., Xu X., Yang H., Yoder A.D., Zhang G. The earth biogenome project 2020: starting the clock. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2115635118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The Darwin Tree of Life Project Consortium Sequence locally, think globally: the Darwin tree of life project. Proc. Natl. Acad. Sci. USA. 2022 doi: 10.1073/pnas.2115642118. 〈https://www.pnas.org/content/pnas/119/4/e2115642118〉 accessed March 25, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heffer A., Pick L. Conservation and variation in Hox genes: how insect models pioneered the evo-devo field. Annu. Rev. Entomol. 2013;58:161–179. doi: 10.1146/annurev-ento-120811-153601. [DOI] [PubMed] [Google Scholar]
- 26.Pace R.M., Grbić M., Nagy L.M. Composition and genomic organization of arthropod Hox clusters. Evodevo. 2016;7:11. doi: 10.1186/s13227-016-0048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soria-Carrasco V., Gompert Z., Comeault A.A., Farkas T.E., Parchman T.L., Johnston J.S., Buerkle C.A., Feder J.L., Bast J., Schwander T., Egan S.P., Crespi B.J., Nosil P. Stick insect genomes reveal natural selection’s role in parallel speciation. Science. 2014;344:738–742. doi: 10.1126/science.1252136. [DOI] [PubMed] [Google Scholar]
- 28.Grbić M., Van Leeuwen T., Clark R.M., Rombauts S., Rouzé P., Grbić V., Osborne E.J., Dermauw W., Ngoc P.C.T., Ortego F., Hernández-Crespo P., Diaz I., Martinez M., Navajas M., Sucena É., Magalhães S., Nagy L., Pace R.M., Djuranović S., Smagghe G., Iga M., Christiaens O., Veenstra J.A., Ewer J., Villalobos R.M., Hutter J.L., Hudson S.D., Velez M., Yi S.V., Zeng J., Pires-daSilva A., Roch F., Cazaux M., Navarro M., Zhurov V., Acevedo G., Bjelica A., Fawcett J.A., Bonnet E., Martens C., Baele G., Wissler L., Sanchez-Rodriguez A., Tirry L., Blais C., Demeestere K., Henz S.R., Gregory T.R., Mathieu J., Verdon L., Farinelli L., Schmutz J., Lindquist E., Feyereisen R., Van de Peer Y. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479:487–492. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoy M.A., Waterhouse R.M., Wu K., Estep A.S., Ioannidis P., Palmer W.J., Pomerantz A.F., Simão F.A., Thomas J., Jiggins F.M., Murphy T.D., Pritham E.J., Robertson H.M., Zdobnov E.M., Gibbs R.A., Richards S. Genome sequencing of the phytoseiid predatory mite metaseiulus occidentalis reveals completely atomized Hox genes and superdynamic intron evolution. Genome Biol. Evol. 2016;8:1762–1775. doi: 10.1093/gbe/evw048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis E.B. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 31.Kaufman T.C., Lewis R., Wakimoto B. Cytogenetic analysis of chromosome 3 in drosophila melanogaster: the homoeotic gene complex in polytene chromosome interval 84a-B. Genetics. 1980;94:115–133. doi: 10.1093/genetics/94.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthews B.J., Dudchenko O., Kingan S.B., Koren S., Antoshechkin I., Crawford J.E., Glassford W.J., Herre M., Redmond S.N., Rose N.H., Weedall G.D., Wu Y., Batra S.S., Brito-Sierra C.A., Buckingham S.D., Campbell C.L., Chan S., Cox E., Evans B.R., Fansiri T., Filipović I., Fontaine A., Gloria-Soria A., Hall R., Joardar V.S., Jones A.K., Kay R.G.G., Kodali V.K., Lee J., Lycett G.J., Mitchell S.N., Muehling J., Murphy M.R., Omer A.D., Partridge F.A., Peluso P., Aiden A.P., Ramasamy V., Rašić G., Roy S., Saavedra-Rodriguez K., Sharan S., Sharma A., Smith M.L., Turner J., Weakley A.M., Zhao Z., Akbari O.S., Black W.C., 4th, Cao H., Darby A.C., Hill C.A., Johnston J.S., Murphy T.D., Raikhel A.S., Sattelle D.B., Sharakhov I.V., White B.J., Zhao L., Aiden E.L., Mann R.S., Lambrechts L., Powell J.R., Sharakhova M.V., Tu Z., Robertson H.M., McBride C.S., Hastie A.R., Korlach J., Neafsey D.E., Phillippy A.M., Vosshall L.B. Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature. 2018;563:501–507. doi: 10.1038/s41586-018-0692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasukochi Y., Ashakumary L.A., Wu C., Yoshido A., Nohata J., Mita K., Sahara K. Organization of the Hox gene cluster of the silkworm, Bombyx mori: a split of the Hox cluster in a non-Drosophila insect. Dev. Genes Evol. 2004;214:606–614. doi: 10.1007/s00427-004-0441-1. [DOI] [PubMed] [Google Scholar]
- 34.Shippy T.D., Hosmani P.S., Flores-Gonzalez M., Mueller L.A., Hunter W.B., Brown S.J., D’Elia T., Saha S. Annotation of Hox cluster and Hox cofactor genes in the Asian citrus psyllid, Diaphorina citri, reveals novel features. Gigabyte. 2022;2022:1–18. doi: 10.46471/gigabyte.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duboule D. The rise and fall of Hox gene clusters. Development. 2007;134:2549–2560. doi: 10.1242/dev.001065. [DOI] [PubMed] [Google Scholar]
- 36.Darbellay F., Bochaton C., Lopez-Delisle L., Mascrez B., Tschopp P., Delpretti S., Zakany J., Duboule D. The constrained architecture of mammalian Hox gene clusters. Proc. Natl. Acad. Sci. USA. 2019;116:13424–13433. doi: 10.1073/pnas.1904602116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holland P.W.H. Evolution of homeobox genes. Wiley Interdiscip. Rev. Dev. Biol. 2013;2:31–45. doi: 10.1002/wdev.78. [DOI] [PubMed] [Google Scholar]
- 38.Nong W., Cao J., Li Y., Qu Z., Sun J., Swale T., Yip H.Y., Qian P.Y., Qiu J.-W., Kwan H.S., Bendena W., Tobe S., Chan T.F., Yip K.Y., Chu K.H., Ngai S.M., Tsim K.Y., Holland P.W.H., Hui J.H.L. Jellyfish genomes reveal distinct homeobox gene clusters and conservation of small RNA processing. Nat. Commun. 2020;11:3051. doi: 10.1038/s41467-020-16801-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szabó R., Ferrier D.E.K. Two more Posterior Hox genes and Hox cluster dispersal in echinoderms. BMC Evol. Biol. 2018;18:203. doi: 10.1186/s12862-018-1307-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gurska D., Vargas Jentzsch I.M., Panfilio K.A. Unexpected mutual regulation underlies paralogue functional diversification and promotes epithelial tissue maturation in Tribolium. Commun. Biol. 2020;3:552. doi: 10.1038/s42003-020-01250-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stauber M., Jäckle H., Schmidt-Ott U. The anterior determinant bicoid of Drosophila is a derived Hox class 3 gene. Proc. Natl. Acad. Sci. USA. 1999;96:3786–3789. doi: 10.1073/pnas.96.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holland P.W.H., Marlétaz F., Maeso I., Dunwell T.L., Paps J. New genes from old: asymmetric divergence of gene duplicates and the evolution of development. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017;372 doi: 10.1098/rstb.2015.0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bao R., Dia S.E., Issa H.A., Alhusein D., Friedrich M. Comparative evidence of an exceptional impact of gene duplication on the developmental evolution of drosophila and the higher diptera. Front. Ecol. Evol. 2018;6 doi: 10.3389/fevo.2018.00063. [DOI] [Google Scholar]
- 44.Liu Q., Onal P., Datta R.R., Rogers J.M., Schmidt-Ott U., Bulyk M.L., Small S., Thornton J.W. Ancient mechanisms for the evolution of the bicoid homeodomain’s function in fly development. eLife. 2018;7 doi: 10.7554/eLife.34594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Onal P., Gunasinghe H.I., Umezawa K.Y., Zheng M., Ling J., Azeez L., Dalmeus A., Tazin T., Small S. Suboptimal intermediates underlie evolution of the bicoid homeodomain. Mol. Biol. Evol. 2021;38:2179–2190. doi: 10.1093/molbev/msab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahola V., Lehtonen R., Somervuo P., Salmela L., Koskinen P., Rastas P., Välimäki N., Paulin L., Kvist J., Wahlberg N., Tanskanen J., Hornett E.A., Ferguson L.C., Luo S., Cao Z., de Jong M.A., Duplouy A., Smolander O.-P., Vogel H., McCoy R.C., Qian K., Chong W.S., Zhang Q., Ahmad F., Haukka J.K., Joshi A., Salojärvi J., Wheat C.W., Grosse-Wilde E., Hughes D., Katainen R., Pitkänen E., Ylinen J., Waterhouse R.M., Turunen M., Vähärautio A., Ojanen S.P., Schulman A.H., Taipale M., Lawson D., Ukkonen E., Mäkinen V., Goldsmith M.R., Holm L., Auvinen P., Frilander M.J., Hanski I. The Glanville fritillary genome retains an ancient karyotype and reveals selective chromosomal fusions in Lepidoptera. Nat. Commun. 2014;5:4737. doi: 10.1038/ncomms5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt-Ott U. The amnioserosa is an apomorphic character of cyclorrhaphan flies. Dev. Genes Evol. 2000;210:373–376. doi: 10.1007/s004270000068. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt-Ott U., Kwan C.W. How two extraembryonic epithelia became one: serosa and amnion features and functions of Drosophila’s amnioserosa. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2022;377 doi: 10.1098/rstb.2021.0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panfilio K.A. Extraembryonic development in insects and the acrobatics of blastokinesis. Dev. Biol. 2008;313:471–491. doi: 10.1016/j.ydbio.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt-Ott U., Kwan C.W. Morphogenetic functions of extraembryonic membranes in insects. Curr. Opin. Insect Sci. 2016;13:86–92. doi: 10.1016/j.cois.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 51.Panfilio K.A., Chuva de Sousa Lopes S.M. The extended analogy of extraembryonic development in insects and amniotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2022;377 doi: 10.1098/rstb.2021.0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goltsev Y., Rezende G.L., Vranizan K., Lanzaro G., Valle D., Levine M. Developmental and evolutionary basis for drought tolerance of the Anopheles gambiae embryo. Dev. Biol. 2009;330:462–470. doi: 10.1016/j.ydbio.2009.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jacobs C.G.C., Rezende G.L., Lamers G.E.M., van der Zee M. The extraembryonic serosa protects the insect egg against desiccation. Proc. Biol. Sci. 2013;280 doi: 10.1098/rspb.2013.1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farnesi L.C., Menna-Barreto R.F.S., Martins A.J., Valle D., Rezende G.L. Physical features and chitin content of eggs from the mosquito vectors Aedes aegypti, Anopheles aquasalis and Culex quinquefasciatus: connection with distinct levels of resistance to desiccation. J. Insect Physiol. 2015;83:43–52. doi: 10.1016/j.jinsphys.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 55.Gorman M.J., Kankanala P., Kanost M.R. Bacterial challenge stimulates innate immune responses in extra-embryonic tissues of tobacco hornworm eggs. Insect Mol. Biol. 2004;13:19–24. doi: 10.1111/j.1365-2583.2004.00454.x. [DOI] [PubMed] [Google Scholar]
- 56.Jacobs C.G.C., Spaink H.P., van der Zee M. The extraembryonic serosa is a frontier epithelium providing the insect egg with a full-range innate immune response. eLife. 2014;3 doi: 10.7554/eLife.04111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacobs C.G.C., van der Hulst R., Chen Y.-T., Williamson R.P., Roth S., van der Zee M. Immune function of the serosa in hemimetabolous insect eggs. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2022;377 doi: 10.1098/rstb.2021.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bull A.L. Stages of living embryos in the jewel wasp Mormoniella (nasonia) vitripennis (walker) (hymenoptera: pteromalidae) Int. J. Insect Morphol. Embryol. 1982;11:1–23. [Google Scholar]
- 59.Fleig R., Sander K. Honeybee morphogenesis: embryonic cell movements that shape the larval body. Development. 1988;103:525–534. [Google Scholar]