Abstract
PBREM, the phenobarbital-responsive enhancer module of the cytochrome P-450 Cyp2b10 gene, contains two potential nuclear receptor binding sites, NR1 and NR2. Consistent with the finding that anti-retinoid X receptor (RXR) could supershift the NR1-nuclear protein complex, DNA affinity chromatography with NR1 oligonucleotides enriched the nuclear orphan receptor RXR from the hepatic nuclear extracts of phenobarbital-treated mice. In addition to RXR, the nuclear orphan receptor CAR was present in the same enriched fraction. In the phenobarbital-treated mice, the binding of both CAR and RXR was rapidly increased before the induction of CYP2B10 mRNA. In vitro-translated CAR bound to NR1, but only in the presence of similarly prepared RXR. PBREM was synergistically activated by transfection of CAR and RXR in HepG2 and HEK293 cells when the NR1 site was functional. A CAR-RXR heterodimer has thus been characterized as a trans-acting factor for the phenobarbital-inducible Cyp2b10 gene.
Cytochromes P-450 (CYPs) comprise a superfamily of heme-thiolate proteins. They function as monooxygenases that are activated by accepting electrons from NADPH-CYP reductase (19). The CYP enzymes display diverse functions, from the synthesis and degradation of biological signaling molecules such as steroid hormones and fatty acid derivatives to the metabolism of xenobiotic chemicals including pharmaceutical drugs and environmental contaminants and carcinogens. Phenobarbital (PB) is the prototype of a large group of structurally diverse xenobiotic chemicals that induce the subset of the CYP genes within the CYP2A, CYP2B, CYP2C, and CYP3A subfamilies, with the CYP2B genes being the most effectively induced (3, 4, 8, 14, 24). PB-type inducers regulate mainly at the transcription level. Compared with the well-known Ah receptor-mediated regulation of the CYP1A1 gene (4, 6), the mechanism by which PB induces transcription of the CYP2B genes has been elusive.
Recently, PB-responsive enhancer activity has been associated with DNA sequences found approximately -2.3 kbp upstream of the initiation site of the rat CYP2B2 and mouse Cyp2b10 genes (8, 9, 18, 23). The core enhancer sequence is the 51-bp DNA sequence located at bp -2339 to -2289 of the Cyp2b10 gene and was designated phenobarbital-responsive enhancer module (PBREM) (8–10). PBREM seems to be a general PB-responsive enhancer since it responds to numerous PB-type inducers including 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP), polychlorinated biphenyls, chlorinated pesticides, organic solvents, and some plant products such as camphor (10). The PBREM sequences are conserved and functional in the PB-inducible rat CYP2B genes but are mutated and nonfunctional in the noninducible mouse Cyp2b9 gene (8). The nuclear factors that regulate the PBREM activity have not been identified.
PBREM contains putative nuclear receptor binding sites, NR1 and NR2, that flank a nuclear factor 1 (NF1) binding site. Specific mutations of these NR sites resulted in a complete loss of the responsiveness of PBREM to PB-type inducers (8, 10). In this study, we have identified the nuclear orphan receptors CAR and retinoid X receptor (RXR), which bind to the NR1 site of PBREM in response to PB induction. Additionally, we have demonstrated an activation of PBREM by these orphan receptors in transformed cell lines. CAR is known to activate a subset of retinoic acid-responsive elements (1), and the Cyp2b10 gene appears to be the first identified target gene for this orphan receptor.
MATERIALS AND METHODS
Reagents.
Antibodies against mouse isoforms of TRα1 (sc-772X), RARα/β/γ (sc-773X), RXRα (sc-553X), RXRβ (sc-831X), RXRγ (sc-555X), and c-jun (sc-044X) were purchased from Santa Cruz Biotechnology. Anti-COUP-TFII and anti-HNF4 were kind gifts from Ming-Jer Tsai and James Darnell, Jr., respectively. Anit-hCAR was raised in rabbits against the bacterial recombinant protein. Another antibody to mCAR was also raised by immunizing rabbits by the peptide (CALFSPDRPGVTQREEIDQLQE) containing 21 residues from 276 to 296 of murine CAR (mCAR). The following expression plasmids were kindly provided: RXR by Ronald Evans, TRα1 by Leslie DeGroot, COUP-TFI by Ming-Jer Tsai, FXR by Cary Weinberger, pCMV-LXR by David Mangelsdorf, pCMV-HNF4 by James Darnell, Jr., and pCMV-MB67 by David Moore. [α-32P]dATP (>6,000 Ci/mmol), [14C]dichloroacetylchloramphenicol (56 mCi/mmol), and l-[14C]leucine (>300 mCi/mmol) were purchased from Amersham.
Transient transfection.
The primary hepatocytes were prepared from 2-month-old C57BL/KS/J male mice (Jackson Laboratory) by two-step collagenase perfusion (7). These hepatocytes were electroporated with 30 μg each of individual enhancer-pBLCAT2 reporter plasmids (15) and 10 μg of pSVβgal control plasmid (Promega). Transfected cells on dishes (2 × 106 to 3 × 106 cells) were cultured for 24 h in the absence or presence of inducers under the previous conditions. HepG2 and HEK293 cells were cultured in minimal essential medium supplemented with 10% fetal bovine serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml and transfected by the calcium phosphate method (CellPhect kit; Pharmacia). As described in our previous papers (7, 8), the cell extracts from either transfected hepatocytes or the transformed cell lines were assayed for protein or β-galactosidase, heat treated for 20 min, and assayed for chloramphenicol acetyltransferase (CAT) activity. All the experiments were done in triplicate, and the data were normalized for β-galactosidase activity.
DNA affinity chromatography.
Adult Crl:CDS-1(ICR)BR mice (Charles River Breeding Co.) were treated with PB (100 mg/kg of body weight, injected intraperitoneally). Between 3 and 5 h after PB treatment, the mouse liver nuclear extracts were prepared and were applied to a heparin-agarose column (22, 25). The fractions which exhibited NR1 binding activity were subsequently incubated with NR1-conjugated Dynabeads under the conditions previously described (22, 25). Pooled fractions from the agarose column were dialyzed against buffer A (25 mM Tris-HCl [pH 7.5] buffer containing 0.5 mM EDTA, 0.5 mM dithiothreitol, 10% glycerol, and 0.05% Nonidet P-40 [NP-40]) and incubated for 30 min at 4°C with 20 μg of herring sperm DNA per ml, 10 μg of poly(dI · dC) per ml, and 25 μg of NR1* oligonucleotides per ml. Following incubation for 30 min at 4°C, the beads were washed three times with buffer A that contained increasing concentrations of NaCl (0.1, 0.2, 0.3, and 0.5 M).
Recombinant proteins.
The CARs were bacterially expressed. For mCAR, the coding sequence was amplified from mouse liver cDNAs with 5′AGTCTCGGATCCATGACAGCTATGCTAACACT3′ and 5′AGAGTCCTCGAGTCAACTGCAAATCTCCCCGA3′ as primers (GenBank no. AF009327) and the amplified mCAR DNA fragment was cloned into pGEX-4T.3 (Pharmacia) at the BamHI and XhoI sites. By using the MB67 (human CAR [hCAR])-bearing pT7HisMyc plasmid (1), the recombinant hCAR was expressed in inclusion bodies and purified by metal affinity chromatography on His-Bind Resin (Novagen Inc.) in the presence of 6 M guanidine. The radiolabeled RXRα, hCAR, and mCAR were synthesized by using one-step in vitro transcription and translation (TNT coupled reticulocyte lysate system; Promega).
Gel shift assays and Western blot analysis.
Gel shift assays were done in 10 μl of 10 mM HEPES (pH 7.6)–0.5 mM dithiothreitol–15% glycerol–2 μg of poly(dl · dC)–0.05% NP-40–50 mM NaCl with approximately 30,000 cpm of 32P-end-labeled oligonucleotide probe. In competition experiments, mixtures of unlabeled oligonucleotides in 20- or 50-fold excess were added before the start of the binding reaction initiated by nuclear extract. In antibody supershift experiments nuclear extracts were preincubated at 25°C with 1 μg of preimmune immunoglobulin G (IgG) or specific IgG for 15 min before being subjected to electrophoresis on a 5% acrylamide gel. For Western blot analysis, the proteins resolved on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel (10% polyacrylamide) were transferred to a polyvinylidene difluoride membrane that was incubated with anti-RXR or anti-hCAR (anti-recombinant hCAR and anti-hCAR peptide). After the secondary anti-rabbit IgG-horseradish peroxidase (1:5,000 dilution), the immunoreactive bands were visualized with enhanced chemiluminescence reagents (Amersham Life Science Inc.).
RESULTS
Functional analysis of the NR sites of PBREM.
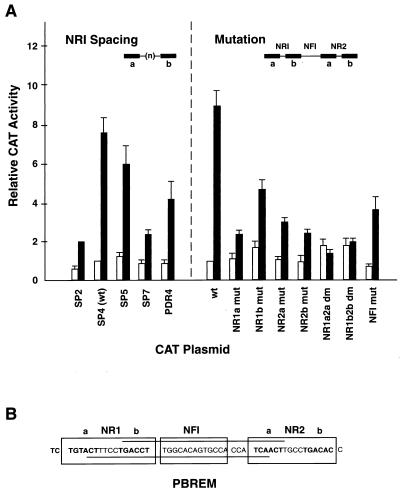
Spacing and mutation variants of the NR1 site were introduced into the context of the wild-type 51-bp PBREM sequence. The mutated elements were ligated to thymidine kinase (tk)-CAT reporter plasmids and transfected into mouse primary hepatocytes to test their responsiveness (Fig. 1). As expected, the wild-type PBREM displayed the highest enhancer activity, showing 7- to 10-fold induction of CAT expression. We altered the number of nucleotides in the spacer of the wild-type NR1 site of PBREM from 4 to 2, 5, or 7 (SP2-CAT, SP5-CAT, or SP7-CAT). The responsiveness of the altered PBREMs was decreased dramatically to only threefold induction by the 2- or 7-bp spacer. Alteration of the NR1 sequence to the perfect AGGTCA repeats (PDR4-CAT) also resulted in a significant loss of PBREM activity. Additionally, the responsiveness of the SP5-CAT plasmid was relatively high, about 75% of the wild-type value. Thus, NR1 responded most efficiently when it retained the wild-type characteristics of DR4, although it responded well as a DR5 motif.
FIG. 1.
Functional assays to define the NR1 site as a DR4 motif. (A) The 51-bp enhancer–tk-CAT plasmids of the spacing and mutation variations of the 51-bp enhancer element were placed in front of the tk-CAT plasmids and were transfected into mouse primary hepatocytes. The CAT activity of the transfected cell extracts in the absence or the presence of 50 nM TCPOBOP is shown by open and solid bars, respectively. The data shown are means and standard deviations from three or four independent transfections, relative to the activity of the wild-type 51-bp enhancer element (wt = 100). (B) The PBREM sequence and motifs are depicted. The half-sites (a and b) of the putative NR binding sites are shown in boldface type. The bipartite NF1 binding motif is boxed.
The roles of each half-site in responsiveness were also examined (Fig. 1). For this purpose, the individual half-sites were mutated by a 4-bp central CTGG substitution within the context of the PBREM–TK-CAT plasmids. Each of these half-site mutations decreased the responsiveness of the PBREM to less than threefold induction, whereas double mutations of both NR1 and NR2 half-sites (NR1a2a dm and NR1b2b dm) completely abolished activity. Mutation of the NF1 site decreased the basal activity by 20% and reduced the responsiveness to 4.6-fold induction. While the NF1 mutation was as effective as a single half-site mutation in decreasing the PBREM activity, it did not abolish the activity. These results showed that both NR sites play major roles in the activation of PBREM and that the individual NR half-sites are essential for full responsiveness.
Binding analysis of the NR1 site.
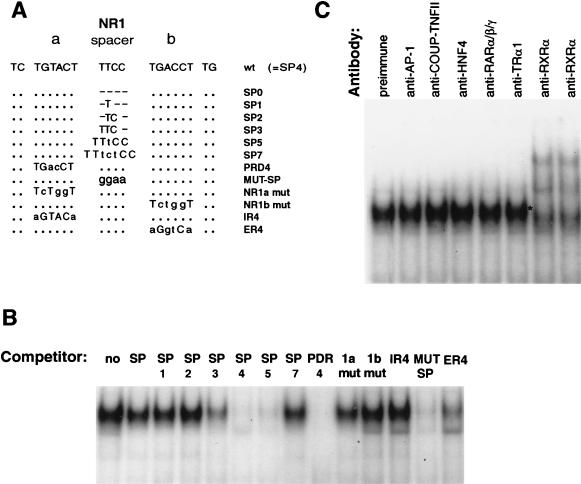
The functional analysis was followed by a search for nuclear proteins that could bind to the NR1 site. Since the binding of a nuclear orphan receptor is dictated by the sequence, orientation, and spacing of the half-sites (16, 17), several oligonucleotides containing altered spacing and substitution were designed as NR1 binding competitors (Fig. 2A). The binding of NR1 probe to the PB-treated liver nuclear extracts was completely abolished by oligonucleotide SP4 and effectively abolished by SP5 but was decreased only slightly by SP3 (Fig. 2B). Neither PDR4 nor MUT-SP competed with NR1 for binding to nuclear proteins: the 4-bp TTCC spacer was changed to GGAA in MUT-SP. The competition was decreased when the four central nucleotides of either NR1a or NR1b was mutated to CTGG (NR1a mut and NR1b mut) or when the direct repeat was changed to an inverted or everted repeat (IR4 and ER4). Thus, the characteristics of binding of NR1 to the nuclear protein were consistent with the transcription activity studies, indicating that NR1 is most active in the original imperfect DR4 motif.
FIG. 2.
Gel shift assay to define the NR1 site as a DR4 motif and RXR binding. (A) Sequences of indicated NR1 mutants used as competitors for NR1 binding are compared to the wild-type NR1 (SP4 = wt). Spacer mutations are denoted by SP with numbers of bases. In MUT-SP, four bases of spacer are mutated. In addition to a random mutation in either the NR1a or NR1b site (NR1a mut and NR1b mut), these sites are mutated to create a perfect direct repeat with different orientations (PDR4, IR4, and ER4). The dots and hyphens indicate no change and deletions, respectively. The different nucleotides are shown in lowercase type. (B) Competition for NR1 binding was done with a 20-fold excess of indicated oligonucleotides. (C) Supershift assays were done by incubating preimmune IgG (1 μg) or indicated antibodies (1.5 μg) with liver nuclear extract from PB-treated mice. The results are representative of three independent experiments.
Supershift assays performed with antibodies of various DR4 motif binding receptors such as COUP-TF, thyroid hormone receptor (TR), retinoic acid receptor (RAR) and RXR indicated that only anti-RXRα created a new, slower-migrating complex (Fig. 2C). Under the gel shift conditions, the major NR1 binding complex of the PB-treated liver nuclear extracts appeared to contain RXR. Anti-HNF4 and anti-AP-1 were used as controls, since HNF4 and AP-1 are not DR4 binding proteins. As expected, these antibodies did not supershift the NR1 complex. The results strongly suggested that the NR1-nuclear protein complex contains RXR.
RXRα, RXRβ, and mCAR as NR1 binding proteins.
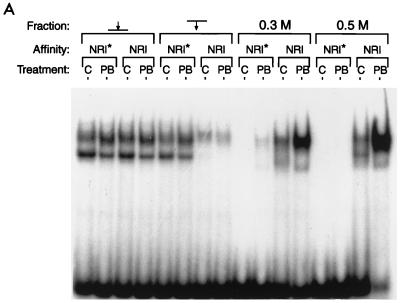
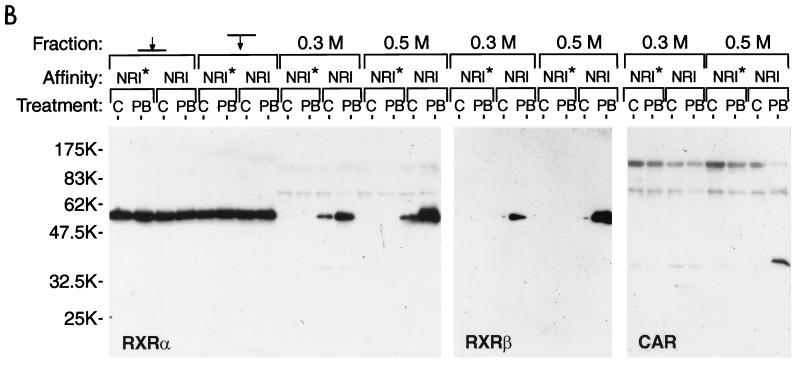
A related CYP2B gene, the mouse Cyp2b9 gene, is not PB inducible. The sequence of this gene corresponding to PBREM had diverged, resulting in a nonfunctional PBREM element (8, 9). Moreover, the mutated NR1 sequence (NR1*) of the Cyp2b9 gene did not compete with the functional NR1 for binding to nuclear proteins. Using DNA-affinity beads of the NR1 or NR1* oligonucleotides, we performed chromatography on the hepatic nuclear extracts from the PB-treated (for 3 to 5 h) and untreated mice. Gel shift assays showed that the NR1 binding proteins were recovered only from the NR1 affinity beads and were enriched mainly in the 0.5 M NaCl eluates from the samples from the PB-treated mice (Fig. 3A, right-hand lane). As shown in Fig. 3B, the subsequent Western blot analysis with anti-RXRα and anti-RXRβ detected a single band that was similar in its apparent molecular mass to RXR (60 kDa). Since anti-RXRγ did not react with the band (data not shown), the NR1 binding proteins with molecular masses near 60 kDa were RXRα and RXRβ. Consistent with the results of the gel shift assays, RXRs were purified only from the samples from the PB-treated mice. Since the RXRs in the various fractions applied to the column exhibited the same levels, PB appeared to induce an RXR complex that could bind to NR1 probe. The RXR binding thus seems to be specific to the NR1 site and to PB induction.
FIG. 3.
DNA affinity chromatography of RXRs and CAR. Pooled fractions of the liver nuclear extracts from a heparin-agarose column were incubated with either NR1 oligonucleotide-conjugated or NR1* oligonucleotide-conjugated magnetic beads. C and PB denote the extracts isolated from the untreated and the PB-treated (for 3 to 5 h) mice. The proteins were eluted from the beads with 0.3 and then 0.5 M NaCl. (A) The eluted proteins were subjected to a gel shift assay with NR1 as the probe. Nuclear extracts (1 μg of protein) and approximately 0.1 to 0.05 μg of the eluted proteins were used for gel shift assay. (B) For Western blots, 10 μg of nuclear extracts and 0.05 μg of the eluted proteins were resolved on an SDS–10% polyacrylamide gel and immunoblotted with anti-RXRα or anti-RXRβ IgG (1:3,000 dilution) or anti-hCAR serum (1:250 dilution). The arrows pointing to and from the bars indicate the applied and path-through fractions, respectively. Prestained Protein Marker Broad Range (New England Biolabs) was used as the molecular mass maker (shown in thousands). The results are representative of at least two independent purifications.
Given the facts that induction by PB of the Cyp2b10 gene was liver specific (7), that NR1 binding protein preferred the DR4 and DR5 motif, and that RXR was present in the NR1 binding complex, we assumed that the NR1 binding protein was a RXR heterodimer with a liver-enriched orphan receptor. Since the orphan nuclear receptor CAR met these criteria, its association with NR1 was examined. Antibodies (anti-hCAR) were raised against the bacterially expressed hCAR and the 21-residue peptide of mCAR in rabbits. Both antibodies cross-reacted with the bacterially expressed mCAR (data not shown). These anti-CAR antibodies could not shift the NR1-nuclear protein complex in the gel shift assay, because their antigenic sites may be masked by protein-protein and/or protein-DNA interactions of the complex. We therefore used these antibodies in a Western blot analysis to detect mCAR. Anti-hCAR visualized a specific band on a Western blot of the 0.5 M NaCl eluate of the PB-treated extracts from the NR1-affinity beads (Fig. 3B right-hand lane). The apparent molecular mass of this immunoreactive band was 40 kDa, which agreed with the molecular mass (40,893 Da) calculated from the deduced amino acid sequence of mCAR1 (2). This indicated that the NR1 binding complex contains both mCAR and RXR, presumably as a heterodimer.
Time-dependent binding of mCAR1 and RXR after PB treatment.
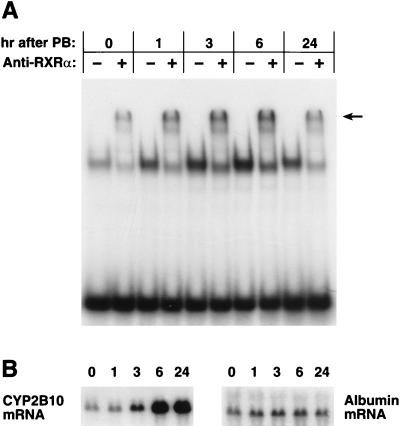
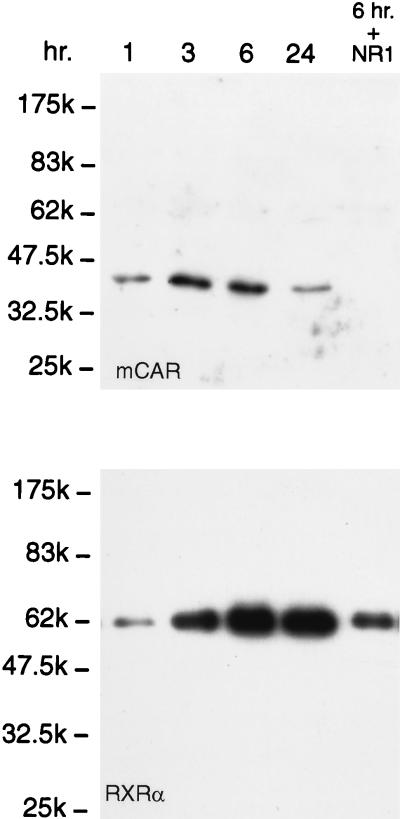
Supershift assays with anti-RXRα and the hepatic nuclear extracts prepared at various times after PB treatment showed that the level of the NR1-RXR complex had an initial sharp increase that peaked at 3 h and then began to decrease at 24 h (Fig. 4). Importantly, this increase in the level of the complex preceded the rise in the CYP2B10 mRNA level. To obtain direct evidence of increasing binding of the nuclear orphan receptors CAR and RXR to NR1, we precipitated the NR1-nuclear protein complex by using NR1 affinity beads and analyzed it by Western blotting (Fig. 5). Anti-hCAR detected a single band at the molecular mass corresponding to mCAR, and this band was not recovered in the presence of the competitive NR1 oligonucleotide. In accordance with the increase in the level of the supershift band and preceding the mRNA induction, mCAR dramatically increased its binding to NR1, with the level of the complex peaking at 3 h after PB treatment. RXRα (or RXRβ [data not shown]) also displayed a rapid increase in its binding to NR1 after PB treatment (Fig. 5). The high binding level of RXR, however, remained at 24 h, at which time the level of mCAR was significantly decreased, suggesting that mCAR was not the only orphan receptor that bound to NR1 as an RXR heterodimer in response to PB induction. Since the time course of the mCAR binding was more consistent with those of the anti-RXR supershifts and of the CYP2B10 mRNA level, the CAR-RXR heterodimer was implicated as the regulatory factor for the PB-inducible transcription. Because of their later appearance after PB treatment, other possible RXR heterodimers may display low-affinity binding to NR1 and may not regulate the activation of PBREM.
FIG. 4.
Supershift displaying the PB-dependent increase in RXR binding to NR1. (A) Liver nuclear extracts were prepared from 20 PB-treated mice injected at each time point. Three independent nuclear extracts were prepared from each time point, and 1 μg of the nuclear proteins per line was incubated with NR1 oligoprobe as described in Materials and Methods and resolved on a 5% polyacrylamide gel. The arrow indicates a band shifted by anti-RXRα. (B) Total RNA (6 μg per lane) prepared from the same liver pool at each time point was subjected to Northern hybridization with the CYP2B10 and albumin cDNA probes. The number above each lane indicates the time (in hours) after PB treatment. These results are representative of two or more independent experiments.
FIG. 5.
Western blot displaying the PB-dependent binding of mCAR1 and RXR to NR1. Liver nuclear extracts (1 mg of proteins in 1 ml of the incubation mixture) from the PB-treated mice at each time point (1 to 24 h) were incubated with NR1 affinity beads. In the right-hand lane (6 hr. + NR1), an excess amount of NR1 oligonucleotide (10 μg) was included as a competitor during the incubation. A 30-μl volume of the 100-μl eluates with 0.5 M NaCl was subjected to Western blot analysis with anti-RXRα IgG (1:3,000 dilution) and anti-CAR (peptide) serum (1:250 dilution). The results are representative of five or more independent experiments.
In vitro binding to NR1 and synergistic activation of PBREM.
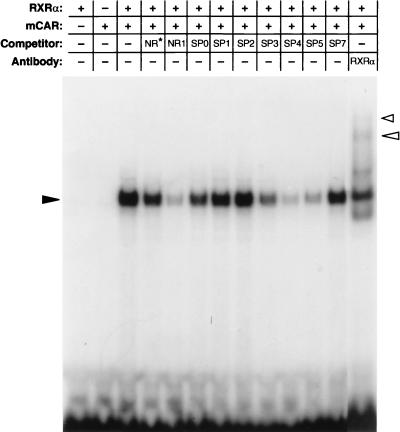
Since the key NR1-nuclear protein complex appeared to be the mCAR-RXR heterodimer, in vitro-translated RXR and CAR (either mCAR or hCAR) were used for the gel shift assay to verify their binding to NR1 (Fig. 6). Neither CAR nor RXRα alone was able to bind to NR1, but their mixture displayed specific binding to NR1. The binding was effectively competed with NR1 oligonucleotide but not with NR1* oligonucleotide. With respect to the specificity of spacing variations (Fig. 2B), the binding of the in vitro-translated mCAR-RXR heterodimer to NR1 was competed most effectively by SP4, followed by SP5 and then by SP3. The SP0, SP1, SP2, and SP7 oligonucleotides did not affect the NR1 binding to the in vitro-translated mCAR-RXR heterodimer.
FIG. 6.
Binding of in vitro-translated CAR and RXR to NR1 probe. Gel shifts were performed using in vitro-translated CARs (1.25 μl each) and RXR (1.25 μl) and 32P-labeled NR1 probe. For competitions, a 50-fold molar excess of each cold oligonucleotide was added to the reaction mixtures. The figure was generated from an experiment with mCAR since hCAR bound to the NR1 probe with the same specificity as did mCAR. The solid arrowhead indicates a gel shift band formed with RXR, mCAR, and NR1, while the open arrowheads point to supershift bands by anti-RXR. The results are representative of two or more independent experiments.
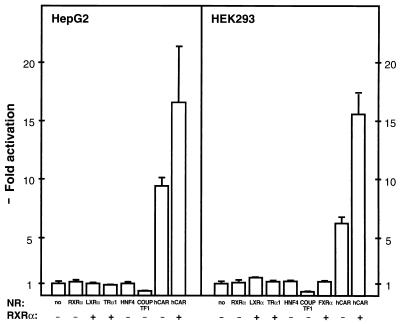
Transfection of RXR alone did not activate PBREM in HepG2 or HEK293 cells (Fig. 7). Transfection of hCAR, on the other hand, resulted in an approximately 10-fold increase of the PBREM activity without the presence of a PB-type inducer. Moreover, a synergistic increase was observed when RXR was transfected in addition to hCAR. Endogenously expressed RXR in the transfected cells might have contributed the 10-fold increase by hCAR and the relatively small increase by further transfection of RXR. Other nuclear orphan receptors including LXRα, TRα1, and HNF4 failed to induce transcription from the PBREM-linked reporter gene (Fig. 7). The activation was also specific to the NR1 sequence, since space alterations (SP2 and SP7) and half-site mutations (NR1a mut and NR1b mut) decreased the activation to approximately 2-fold from the original 10-fold.
FIG. 7.
Activation of PBREM by CAR and RXR in the transformed cell lines. The HepG2 (or HEK293) cells were transfected at 30 to 40% confluence with 0.5 μg (or 0.4 μg) of the appropriate CAT reporter plasmid, 0.5 μg (or 0.125 μg) of β-galactosidase plasmid, and 0.15 μg (or 0.075 μg) of expression vector for individual nuclear receptors. At 48 h after transfection, cell extracts were assayed for β-galactosidase and CAT activities. The fold inductions of the normalized CAT activities are compared with the activity from the PBREM–tk-CAT alone (set to 100). Standard deviations were calculated from at least three independent experiments.
DISCUSSION
Inducible gene transcription as a result of exposure to xenobiotic chemicals is characteristic of the xenobiotic-metabolizing enzymes. PB alone can induce the genes encoding CYP, NADPH-CYP reductase, and different transferases involved in conjugation reactions, in addition to many other genes (9, 24). We have now identified the nuclear orphan receptors CAR and RXR as factors regulating the transcription of the PB-inducible Cyp2b10 genes. Presumably as the RXR heterodimer, CAR regulates the enhancer activity of the 51-bp PBREM through its binding to the NR1 site. The Cyp2b10 gene is the first target gene identified for the orphan receptor CAR.
hCAR (originally called MB67) was initially cloned by screening a human liver cDNA library with degenerate oligonucleotides corresponding to the DNA binding domain (P box) of the RAR/TR orphan receptor subfamily (1). CAR is a liver-enriched nuclear receptor and, as a heterodimer with RXR, can activate a subset of retinoic acid response elements (RAREs) consisting of direct repeats related to the hexamer AGGTCA (1, 2). In the light of the fact that the CAR-RXR heterodimer can regulate the PBREM activity in the transformed cell lines, does CAR bind to NR1 in vivo in response to PB induction so as to regulate the PB-inducible transcription of the Cyp2b10 gene? Indeed, CAR binding to NR1, presumably as a heterodimer with RXR, increased sharply after PB treatment. The PB-induced increase in CAR binding occurred in accordance with that of the supershift band by anti-RXR and preceded the accumulation of CYP2B10 mRNA. Moreover, the mRNA level decreased as the level of CAR decreased at 24 h after PB induction. These results therefore provide compelling evidence that the CAR-RXR heterodimer is a trans-acting factor responsible for NR1-mediated transcription activity from PBREM in the liver. PBREM is a composite regulatory element consisting of two NR sites with different sequences and NF1 site. It responds to the numerous structurally unrelated PB-type inducers (10). Our present studies therefore do not limit orphan receptors involved in the induction of the CYP2B gene by PB only to CAR.
Since none of the transformed cell lines, including HepG2 cells, responds to PB by inducing the CYP2B genes, it is not possible at present to investigate directly whether and how PB can activate the binding of mCAR to PBREM. The constitutive (or intrinsic) activation by mCAR in cell lines in the absence of PB may have several explanations. mCAR may be activated by a potential ligand present in the cell culture medium. Alternatively, an unknown repressor of mCAR in the liver may not have been present in the cell lines. The repressors can be proteins, such as SHP, which represses the activation of RAREs by various orphan receptors including mCAR (20), or they can be metabolites, such as geranylgeraniol, that repress the nuclear orphan receptor LXRα (5). In addition, a role for protein phosphorylation and dephosphorylation in the PB induction of CYP2B genes is evident (11, 21). CAR may be constitutively activated by the lack of proper signaling pathways in the transformed cell lines. However, each of these possible explanations may be a clue to uncovering the induction mechanism in the liver.
With respect to the scores of the PB-type inducers, many of the members of this immense group of structurally unrelated chemicals can activate transcription from the PBREM-CAT reporter gene in the mouse primary hepatocytes (10). It would be interesting to see whether the CAR-RXR heterodimer can mediate most of these activations. Expression of the PB-inducible P-450 genes can also be affected by endocrine factors, such as glucocorticoid, sex, and thyroid hormones (reference 9 and references therein). Since these endocrine factors often control their target genes through nuclear steroid hormone receptors, these factors may interfere with PB induction signals by inducing cross talk between nuclear receptors. The CYP-dependent metabolism can, on the other hand, produce a practically unlimited number of potential ligands (both endogenous hormones and exogenous chemicals) for the nuclear receptors. It appears that the induction of the CYP genes may depend on a tight interaction between members of the two superfamilies, those of the nuclear receptor and the CYP genes themselves. Our present finding that the CAR-RXR heterodimer is a PB-responsive trans-acting factor strengthens this view, and as an additional example, the CYP4A gene is activated by exposure to peroxisome proliferators through binding of the PPAR-RXR heterodimer (12, 13). The PXR-RXR heterodimer may activate transcription of CYP3A gene by synthetic glucocorticoids such as pregnenolone 16α-carbonitrile and dexamethasone t-butylacetate (13a). Finally, a large group of the nuclear orphan receptors may provide cells with the capability to induce the various CYP genes and other genes responsive to unlimited numbers of xenobiotic chemicals.
ACKNOWLEDGMENTS
We thank Cary Weinberger and Anton Jetten (both at National Institute of Environmental Health Sciences) for helpful discussion and comments on the manuscript.
P.H. and I.Z. contributed equally to this work.
REFERENCES
- 1.Baes M, Gulick T, Choi H, Martinoli M, Simha D, Moore D D. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol. 1994;14:1544–1552. doi: 10.1128/mcb.14.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi H-S, Chung M, Tzameli I, Simha D, Lee Y-K, Seol W, Moore D D. Differential transactivation by two isoforms of the orphan nuclear hormone receptor CAR. J Biol Chem. 1997;272:23565–23571. doi: 10.1074/jbc.272.38.23565. [DOI] [PubMed] [Google Scholar]
- 3.Conney A H. Pharmacological implications of microsomal enzyme induction. Pharmacol Rev. 1967;19:317–366. [PubMed] [Google Scholar]
- 4.Denison M S, Whitlock J P., Jr Xenobiotic-inducible transcription of cytochrome P450 genes. J Biol Chem. 1995;270:18175–18178. doi: 10.1074/jbc.270.31.18175. [DOI] [PubMed] [Google Scholar]
- 5.Forman B M, Ruan B, Chen J, Scroepfer G J, Jr, Evans R M. The orphan nuclear receptor LXRα is positively and negatively regulated by distinct products of mevalonate metabolism. Proc Natl Acad Sci. 1997;94:10588–10593. doi: 10.1073/pnas.94.20.10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 7.Honkakoski P, Moore R, Gynther J, Negishi M. Characterization of phenobarbital-inducible mouse Cyp2b10 gene transcription in primary hepatocytes. J Biol Chem. 1996;271:9746–9753. doi: 10.1074/jbc.271.16.9746. [DOI] [PubMed] [Google Scholar]
- 8.Honkakoski P, Negishi M. Characterization of a phenobarbital-responsive enhancer module in mouse P450 Cyp2b10 gene. J Biol Chem. 1997;272:14943–14949. doi: 10.1074/jbc.272.23.14943. [DOI] [PubMed] [Google Scholar]
- 9.Honkakoski P, Negishi M. Regulatory DNA elements of phenobarbital-responsive cytochrome P450 CYP2B genes. J Biochem Mol Toxicol. 1998;12:3–9. doi: 10.1002/(sici)1099-0461(1998)12:1<3::aid-jbt2>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 10.Honkakoski P, Moore R, Washburn K, Negishi M. Activation by diverse xenochemicals of the 51-bp phenobarbital-responsive enhancer module of the Cyp2b10 gene. Mol Pharmacol. 1998;53:597–601. doi: 10.1124/mol.53.4.597. [DOI] [PubMed] [Google Scholar]
- 11.Honkakoski P, Negishi M. Protein serine/threonine phosphatase inhibitors suppress phenobarbital-induced Cyp2b10 gene transcription in mouse primary hepatocytes. Biochem J. 1998;330:889–895. doi: 10.1042/bj3300889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Issemann I, Green S. Activation of a member of the steroid hormone receptor by peroxisome proliferators. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 13.Johnson E J, Palmer C A, Griffin K J, Hsu M-H. Role of the peroxisome proliferator-activating receptor in cytochrome P450 4A gene regulation. FASEB J. 1996;10:1241–1248. doi: 10.1096/fasebj.10.11.8836037. [DOI] [PubMed] [Google Scholar]
- 13a.Kliwere S A, Moore J T, Wada L, Staudinger J L, Watson M A, Jones S A, McKee D D, Oliver B B, Wilson T M, Zetterstrom R H, Permann T, Lehmann J M. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 14.Lubet R A, Dragnev K H, Chauhan D P, Nims R W, Diwan B A, Ward J M, Jones C R, Rice J M, Miller M S. A pleiotropic response to phenobarbital-type enzyme inducers in the F344/NCr rat: effects of chemicals of varied structure. Biochem Pharmacol. 1992;43:1067–1078. doi: 10.1016/0006-2952(92)90614-o. [DOI] [PubMed] [Google Scholar]
- 15.Luckow B, Schütz G. CAT construction with multiple unique restriction sites for the functional analysis of eucaryotic promoters and regulatory elements. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 18.Park Y, Li H, Kemper B. Phenobarbital induction mediated by a distal CYP2B2 sequence in rat liver transiently transfected in situ. J Biol Chem. 1996;271:23725–23728. doi: 10.1074/jbc.271.39.23725. [DOI] [PubMed] [Google Scholar]
- 19.Porter T D, Coon M J. Cytochrome P-450: multiplicity of isoforms, substrates, and catalytic and regulatory mechanism. J Biol Chem. 1991;266:13469–13472. [PubMed] [Google Scholar]
- 20.Seol W, Choi H-S, Moore D D. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science. 1996;272:1336–1339. doi: 10.1126/science.272.5266.1336. [DOI] [PubMed] [Google Scholar]
- 21.Sidhu J S, Omiecinski C J. cAMP-associated inhibition of phenobarbital-inducible cytochrome P450 gene expression in primary hepatocyte cultures. J Biol Chem. 1995;270:12762–12773. doi: 10.1074/jbc.270.21.12762. [DOI] [PubMed] [Google Scholar]
- 22.Sueyoshi T, Kobayashi R, Nishio K, Aida K, Moore R, Wada T, Handa H, Negishi M. A nuclear factor (NF2d9) that binds to the male-specific P-450 (Cyp2d-9) in mouse liver. Mol Cell Biol. 1995;15:4158–4166. doi: 10.1128/mcb.15.8.4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trottier E, Belzil A, Stoltz C, Anderson A. Localization of a phenobarbital-responsive element (PBRE) in the 5′-flanking region of the rat CYP2B2 gene. Gene. 1995;158:263–268. doi: 10.1016/0378-1119(94)00916-g. [DOI] [PubMed] [Google Scholar]
- 24.Waxman D J, Azaroff L. Phenobarbital induction of cytochrome P-450 gene expression. Biochem J. 1992;281:577–592. doi: 10.1042/bj2810577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokomori N, Kobayashi R, Moore R, Sueyoshi T, Negishi M. DNA methylation site in the male-specific P-450 (Cyp2d-9) promoter and the heteromeric transcription factor GABP. Mol Cell Biol. 1995;15:5355–5362. doi: 10.1128/mcb.15.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]