Abstract
Transparent immunodeficient animal models not only enhance in vivo imaging investigations of visceral organ development but also facilitate in vivo tracking of transplanted tumor cells. However, at present, transparent and immunodeficient animal models are confined to zebrafish, presenting substantial challenges for real-time, in vivo imaging studies addressing specific biological inquiries. Here, we employed a mitf−/−/prkdc−/−/il2rg−/− triple-knockout strategy to establish a colorless and immunodeficient amphibian model of Xenopus tropicalis. By disrupting the mitf gene, we observed the loss of melanophores, xanthophores, and granular glands in Xenopus tropicalis. Through the endogenous mitf promoter to drive BRAFV600E expression, we confirmed mitf expression in melanophores, xanthophores and granular glands. Moreover, the reconstruction of the disrupted site effectively reinstated melanophores, xanthophores, and granular glands, further highlighting the crucial role of mitf as a regulator in their development. By crossing mitf−/− frogs with prkdc−/−/il2rg−/− frogs, we generated a mitf−/−/prkdc−/−/il2rg−/− Xenopus tropicalis line, providing a colorless and immunodeficient amphibian model. Utilizing this model, we successfully observed intravital metastases of allotransplanted xanthophoromas and migrations of allotransplanted melanomas. Overall, colorless and immunodeficient Xenopus tropicalis holds great promise as a valuable platform for tumorous and developmental biology research.
Subject terms: Disease model, Cancer models
Xenopus tropicalis with triple knockout of mitf, prkdc, and il2rg is a colorless immunodeficient animal model. The mitf−/− frog further underscores the vital role of the mitf in the development of melanophores, xanthophores, and granular glands.
Introduction
Transparent animal models are invaluable for conducting precise in vivo imaging investigations in the realms of developmental biology and tumor studies1. Immunodeficient animal models, in particular, prove beneficial for addressing various biological inquiries, including the in vivo tracking of transplanted tumor cells1,2. Transparent immunodeficient animal models, beyond their facilitation of in vivo imaging studies pertaining to organ development, also provide essential support for the dynamic tracking and investigation of transplanted tumor cells3,4. However, to the best of our knowledge, the current scope of transparent immunodeficient animal models is confined to the zebrafish model. The use of zebrafish introduces substantial challenges for addressing biological inquiries such as real-time, in vivo imaging studies of lung development, orthotopic transplantation of lung cancer, and the progression of lung cancer, in addition to issues like genetic redundancy5–8. Xenopus tropicalis is a well-established, diploid amphibian animal model that has been extensively utilized in developmental genetics and tumor biology fields9,10. Despite its utility, to the best of our knowledge, reports of transparent and immunodeficient Xenopus tropicalis have yet to be published11–14. Hence, the transparent glassfrog serves as an inspiration for us to develop another transparent and immunodeficient amphibian model in Xenopus tropicalis, thereby diversifying the range of transparent immunodeficient animal models. This will enable future utilization of in vivo imaging in transparent immunodeficient frogs to address questions that may not be easily answered in the zebrafish model15.
Presently, pigment cells identified in mammalian skin exclusively comprise melanocytes. With the exception of the retinal pigment epithelium, pigment cells in vertebrates derive from neural crest cells, demonstrating a substantial degree of developmental conservation16,17. In contrast, the skin of fish and amphibians typically encompasses three primary types of pigment cells: melanophores (named melanocytes in mammals), xanthophores, and iridophores18. These cutaneous pigment cells exert a profound influence on skin transparency12,13,19. In casper zebrafish, transparent skin arises from the absence of both light-absorbing melanophores and reflective iridophores, which is associated with the double mutant of mitfa and mpv174,20,21. MPV17 is a human gene encoding a mitochondrial inner membrane protein (MPV17), and its malfunction or loss can cause rare autosomal recessive disorders termed mitochondrial DNA depletion syndromes22. Adult mice with homozygous mutations in the MPV17 gene developed nephrotic syndrome and chronic renal failure, with a survival rate of less than 10% at six months23. In zebrafish, the orthologous gene of human MPV17 is transparent (tra), also known as mpv17, and its mutations can lead to complete loss or significant reduction of iridophores in zebrafish21,24. Nonetheless, the precise contribution of mpv17 towards the transparent skin phenotype in casper zebrafish still requires further investigation25. In Xenopus tropicalis, mutants were engineered through tyr knockout, slc2a7 knockout, or a triple knockout involving tyr, slc2a7, and hps6, aiming to develop a transparent frog. Nonetheless, challenges arise from the impact of granule glands on skin transparency and heightened mortality rates in the offspring14. Considering these factors, and to ensure optimal survival rates and minimal impact on health in the final construction of transparent immunodeficient frogs, we have chosen to initiate model development by attempting to knock out the mitf gene in Xenopus tropicalis.
Mutated mitfa has been unequivocally found to cause a depletion of light-absorbing melanophores in casper zebrafish, which is a crucial aspect leading to the skin transparency4,20. Mitf is the mammalian ortholog of zebrafish mitfa and has homologs across species, ranging from primitive metazoans to higher-order mammals. Mitf encodes a transcription factor, microphthalmia-associated transcription factor (Mitf), which belongs to the MiT family and contains a basic helix-loop-helix leucine zipper (bHLH-Zip) DNA binding and dimerization domain26. In mammals, a wide variety of Mitf isoforms have been identified, and express widely or in a tissue-specific manner26. In casper zebrafish, the mutant mitfa113stop is instrumental in rendering the skin transparent by disrupting Mitf’s regulatory role in melanophore development, thus preventing skin from melanin pigmentation20. Of note, mitfa is also expressed in zebrafish xanthophores, yet its function in xanthophore development remains uncertain27,28. While there are no reports investigating mitf in Xenopus tropicalis, limited data from Xenopus laevis suggests that this gene could act as a master regulator of melanophore development in Xenopus tropicalis29–32. Therefore, disrupting mitf presents an attractive opportunity to establish transparent Xenopus tropicalis.
Up till now, intravital imaging of engrafted tumor cells and therapy response studies have established that transparent and immunodeficient zebrafish is a first-class animal model3,5. In zebrafish, an ample level of immunodeficiency for tumor transplantation can be attained by disrupting prkdc to impair T and B lymphocyte development, and mutating il2rg to eliminate mature natural killer (NK) cells3. PRKDC is an essential gene for the DNA-dependent protein kinase (DNA-PK) complex, which consists of Ku80 (encoded by XRCC5), Ku70 (encoded by XRCC6), and the catalytic subunit of DNK-PKcs (encoded by PRKDC)33. This complex plays a crucial role in repairing double-strand breaks in genomic DNA and facilitating V(D)J recombination in the T and B lymphocytes of mammals and amphibians alike34,35. IL2RG encodes IL2RG (CD132), which is a shared receptor subunit for six cytokines (IL-4, IL-7, IL-9, IL-15 and IL-21), plays a vital role in lymphoid development by activating JAK3 and is expressed on the surface of T, NK, NKT and dendritic cells36. In Xenopus tropicalis, tumor transplantations are feasible in thymectomized or sub-lethally gamma-irradiated or rag2−/− individuals that serve as recipients11,37. Other immunodeficient Xenopus tropicalis models have been developed via the knockout of XNC10.1 or foxn1, but their suitability for tumor transplantation is yet to be validated38,39. Therefore, we envisioned the generation of a transparent and immunodeficient frog through the dual disruption of prkdc and il2rg, building upon the foundation of mitf null Xenopus tropicalis.
Eventually, through the founder generations of mitf−/− and prkdc−/−/il2rg−/− frogs, the mitf−/−/prkdc−/−/il2rg−/− triple-knockout Xenopus tropicalis line was established. Actually, this frog line resulted in a colorless and immunodeficient amphibian animal model of Xenopus tropicalis, making it a suitable recipient for transplantation of xenogeneic skin, allogeneic xanthophoroma, and melanoma. Moreover, knocking out mitf in Xenopus tropicalis revealed previously unknown functions of mitf in the development of granular glands and xanthophores. Collectively, our mitf−/−/prkdc−/−/il2rg−/− Xenopus tropicalis line presents a valuable amphibian tool for intravital investigations of tumorous and developmental biology.
Results
Biallelic mitf disruption creates colorless Xenopus tropicalis
Due to the transparency of skin in Xenopus tropicalis is influenced by light-absorbing melanophores, necessitating the targeting of mitf to impede melanophore development. In recognition of the intricate transcriptional regulation of the mitf locus26, a set of guide RNAs were tailored to specifically target exons of mitf to knockout mitf (Supplementary Fig. 1 and Supplementary Fig. 2a). All guide RNAs effectively induced damage to their respective targets, however, melanophore loss was only observed in the mosaic founder generations that were targeted by gRNA T5 or gRNA T7 (Supplementary Fig. 2b, c, Supplementary Fig. 3a). As shown in Supplementary Fig. 1, the target site for gRNA T5 was located upstream of the core domain of Mitf, while the target site for gRNA T7 was located in the helix2 domain of Mitf. Therefore, G0 founder tadpoles (79/108) generated by gRNA T7, which exhibited a noticeable lack of melanophores, were selected for further experimentation (Fig. 1a). To verify the genotype-phenotype specificity of SpCas9 mitf site of gRNA T7, genomic material from ten randomly assigned melanophore-phenotypic tadpoles was extracted and performed off-target analyses. Using CRISPOR40, we determined the genome-wide potential off-target sites and selected those with no more than five base mismatches (Supplementary Data 1). Upon further inspection, we found that none of the off-target sites containing four or fewer mismatches exhibited any detectable mutations (Supplementary Fig. 3b and Supplementary Data 2). Consequently, these findings suggested off-target effects were infrequent in the tadpoles examined.
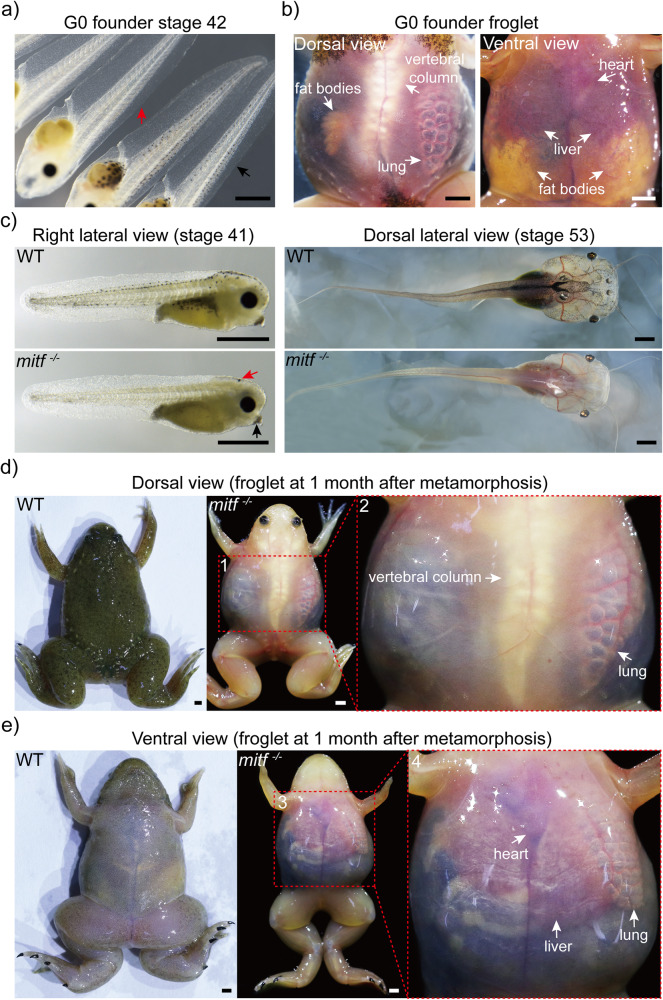
Fig. 1. Biallelic mitf disruption makes Xenopus tropicalis transparent within two months post-metamorphosis.
a After using the CRISPR/Cas9 system to knock out mitf via gRNA T7, founder generation (G0) mosaic tadpoles at stage 42 exhibited a loss of melanophores (indicated by red arrow), compared to wild type (WT) tadpoles from the same batch (indicated by black arrow). 79 mosaic tadpoles and 20 wild type tadpoles were observed. b One month after metamorphosis, the transparent skin of the mosaic froglet allowed for external visibility of internal organs such as the fat bodies, lung, liver, heart, and vertebral column. Here was shown one representative froglet, out of a total of 11 mosaic froglets. c The F1 mitf−/− Xenopus tropicalis tadpole exhibited melanophores loss throughout the entire body at stages 41 and 53 compared to wild type tadpoles from the same batch. Notably, at stage 41, the melanin pigmentation in the eyes of mitf−/− Xenopus tropicalis was generated by retinal pigment epithelium (RPE) cells, and redistribution of melanin in oocytes resulted in some melanin pigment in the head (indicated by red arrow) and cement gland (indicated by black arrow). However, the melanin pigment in the head and cement gland disappeared in later development stages, such as stage 53. Representative photographs were exhibited from 35 F1 mitf−/− Xenopus tropicalis tadpoles at stage 41, 15 F1 mitf−/− Xenopus tropicalis tadpoles at stage 53, as well as from 10 wild type tadpoles each at stage 41 and 53. d, e One month after metamorphosis, the F1 mitf−/− Xenopus tropicalis froglet exhibited transparent skin from both dorsal (d) and ventral (e) views. Red dashed boxes 2 and 4 corresponded to magnified views of red dashed boxes 1 and 3, respectively. The transparent skin allowed for external visibility of internal organs including the lung, liver, heart, and vertebral column within one month after metamorphosis as shown. Ten F1 mitf−/− Xenopus tropicalis froglets and ten wild-type Xenopus tropicalis froglets were used to provide representative photographs as shown. Each scale bar is 1 mm.
Mosaic tadpoles generated via gRNA T7 exhibited a surprising transparency of the skin and made certain internal organs externally visible beyond one-month postmetamorphosis (Fig. 1b). The mosaic frogs (G0 founders) were subsequently raised to sexual maturity and their offspring (F1 generations) obtained through reciprocal breeding of G0 founders. Of the F1 tadpoles, 46% displayed normal melanophore development without any observable differences, while the remainder showed melanophore disappearance (Supplementary Fig. 4a). From this F1 generations, ten tadpoles exhibiting normal melanophores or fifteen offspring with melanophore loss were randomly selected and subjected to genotyping. Seven offspring with melanophore-present exhibited heterozygous genotypes, while the remaining melanophore-present offspring were of the wild type (Supplementary Fig. 4b). All 15 melanophore-loss F1 offspring exhibited compound mutations of biallelic mitf, indicating a complete loss of Mitf function in these melanophore-loss offspring (Supplementary Fig. 4c). Individuals from the F1 generation carrying both alleles of the mitf disruptive mutation, which can cause frameshift mutations, were selected for further breeding to obtain the F2 generation (Supplementary Fig. 4d). Hereinafter, mutants displaying a functional null of biallelic mitf were referred to as mitf−/− individuals, and those exhibiting a functional null of monoallelic mitf as mitf+/− individuals.
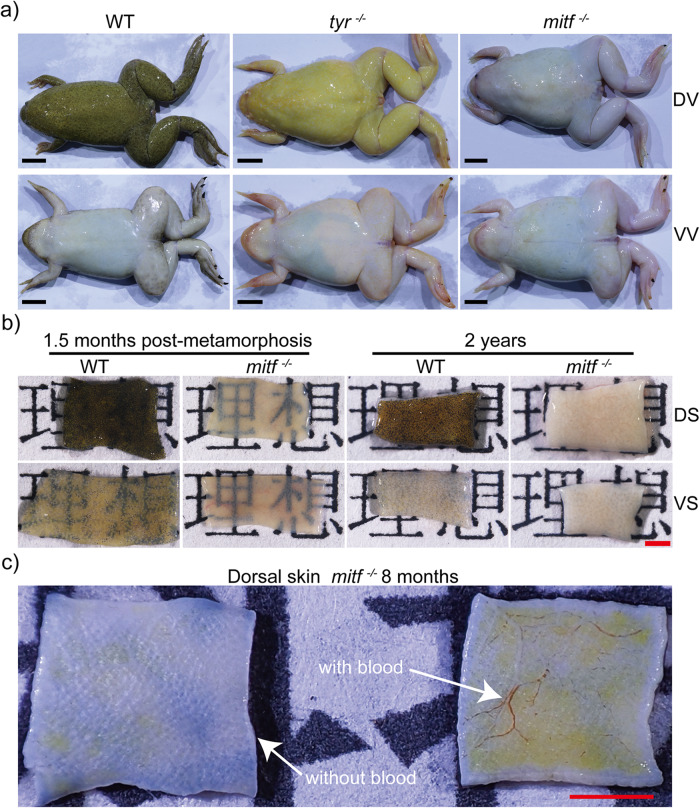
The F1 mitf−/− Xenopus tropicalis tadpoles solely possessed melanin pigmentation in the eyes, with a lack of pigmentation observed in the other part of body, resulting in the transparent skin, in addition to some residual reflective cells in the peritoneum visible post-stage 48 (Fig. 1c and Supplementary Fig. 5). After metamorphosis, the F1 mitf−/− frog skin became transparent within a two-month period, enabling clear visualization of internal organs such as the liver and lungs from an external perspective (Fig. 1d, e). Beyond this period, however, the F1 mitf−/− frog skin turned colorless and opaque, impeding external visualization of the internal organs (Fig. 2a). To compare skin transparency among adult wild type, mitf−/−, and tyr−/− Xenopus tropicalis41, their skin transparency was evaluated for light transmission. Figure 2b clearly indicated that the skin of mitf−/− Xenopus tropicalis exhibited the highest degree of light transmission, enabling a clear view of the Chinese characters “理想” behind it. Given the potential impact of blood on the transparency of amphibian skin15, we examined the influence of blood on the transparency of adult mitf−/− Xenopus tropicalis skin. The results revealed that the exclusion of blood had a negligible effect on the skin transparency of Xenopus tropicalis (Fig. 2c). Moreover, we conducted a comprehensive examination of melanophores throughout the entire body of adult mitf−/− Xenopus tropicalis. We found a lack of black pigmentation in most internal organs, with the exception of melanomacrophage centers in the lungs42, black claws resulting from modified corneocytes of the corneous layer43, and black eyes due to retinal pigment epithelium cells32 (Supplementary Fig. 6). Importantly, F1 mitf−/− Xenopus tropicalis exhibited normal growth and reproductive behavior, and we easily generated the F2 generation through germline transmission of F1 frogs. Furthermore, a noticeable difference in skin color was observed between mitf−/− and tyr−/− Xenopus tropicalis, which lacked melanin pigmentation due to the inability to biosynthesize melanin41,44, indicating possible alterations in other pigment cell types in mitf−/− Xenopus tropicalis (Fig. 2a).
Fig. 2. The skin of mitf−/− adult frogs turns colorless and opaque.
a The representative photographs of adult wild type (WT), tyr−/−, and mitf−/− Xenopus tropicalis were displayed to showcase their dorsal view (DV) and ventral view (VV). The photographs were obtained from a sample of 15 adult frogs of each genotype, all of which were 1 year old. b Skin samples approximately 2 mm×5 mm in size were obtained from both dorsal skin (DS) and ventral skin (VS) of wild-type and mitf−/− Xenopus tropicalis aged 2 years or 1.5 months postmetamorphosis. These samples were utilized in transillumination experiments to assess their translucency properties. Specifically, the experiment involved positioning each sample on a sheet of paper with the word “理想“ and photographing them under consistent lighting and camera settings to assess the visibility of the word. Three frogs were used for each genotype, and one sample of DS and VS skin was collected per frog. Representative photographs of each genotype were presented to illustrate the experimental findings. c Skin samples were obtained with blood or after exlusion of blood from dorsal regions of the same mitf−/− Xenopus tropicalis aged 8 months. Three frogs were used for this experiment. The representative photograph was presented. The black scale bar in a is 1 cm. The red scale bar in b and c is 1 mm.
Together, by functionally nullifying biallelic mitf mutations, we’ve successfully created a colorless line of Xenopus tropicalis. This novel line displays transparent skin and visible internal organs within two months postmetamorphosis, and lacks cutaneous melanophores throughout its lifespan. This newly developed amphibian model holds great potential for use in tumorous and developmental studies3,4.
Crucial role of mitf in melanophore, xanthophore, and granular gland developments in Xenopus tropicalis
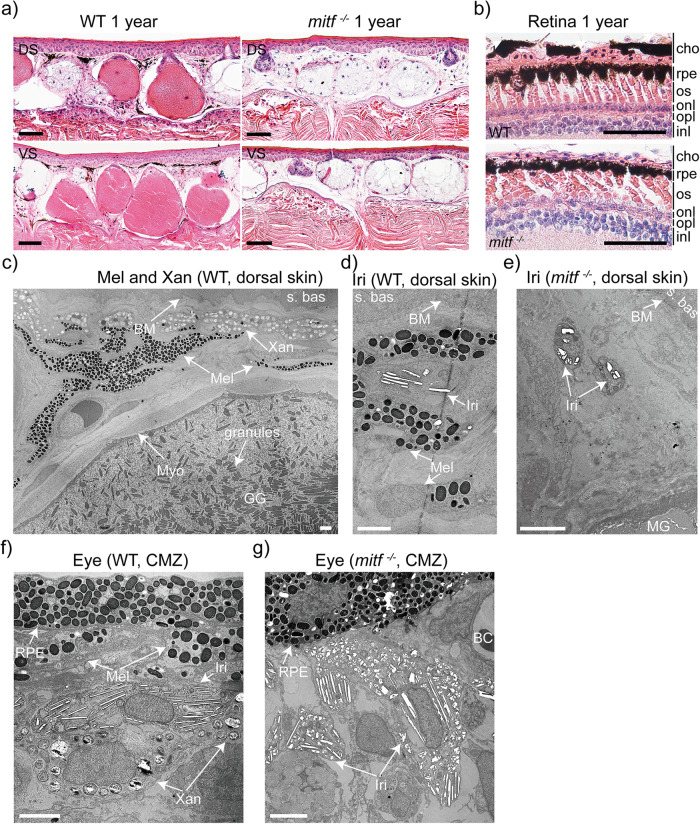
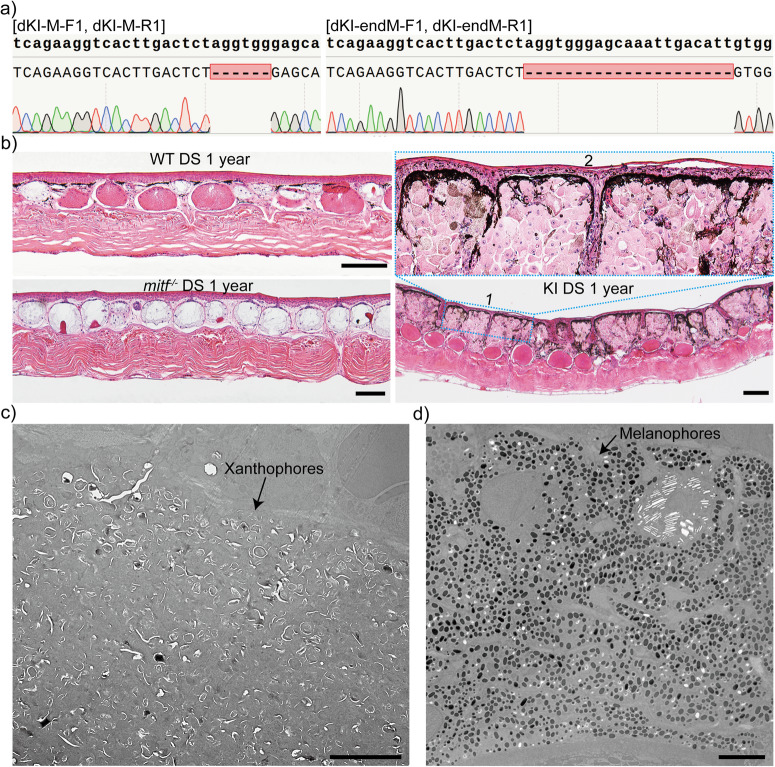
Mitf plays a crucial role as a master regulator in the development of melanocytes across mammals and fish26. Loss or malfunction of Mitf results in the absence of melanocytes in these species. To confirm the absence of melanophores in mitf−/− frogs, skin and eye samples of mitf−/− and wild type Xenopus tropicalis were subjected to histopathological observation. Unexpectedly, the skin of mitf−/− Xenopus tropicalis exhibited a concurrent absence of melanophores and granular glands (Fig. 3a). The loss of melanophores was also confirmed in the choroid of mitf−/− Xenopus tropicalis (Fig. 3b). Moreover, the absence of both granular glands and melanophores was confirmed through transmission electron microscopy (TEM) analysis of skin and eye samples of mitf−/− and wild-type Xenopus tropicalis (Fig. 3c–h and Supplementary Fig. 7). Furthermore, through the characterization of the granular gland development process in both wild type and mitf−/− Xenopus tropicalis, we observed that gland rudiments in the skin of mitf−/− Xenopus tropicalis may undergo normal development at stage 58, but mature granular glands fail to emerge at stage 60 (Supplementary Fig. 8). Notably, throughout the complete developmental stages of mitf−/− Xenopus tropicalis, xanthophores were notably absent, alongside the absence of granular glands in the entire organism (Fig. 3c-h and Supplementary Fig. 9).
Fig. 3. Melanophores, xanthophores, and granular glands were absent in mitf−/−Xenopus tropicalis.
a Histological structure representative of the dorsal (up line) and ventral (bottom line) skin of 1-year-old WT and mitf−/− Xenopus tropicalis was displayed. b The histological structure representative of the retina in eyes of 1-year-old WT and mitf−/− Xenopus tropicalis was shown. cho, choroid; rpe, retinal pigmented epithelium; pl, photoreceptor layer of rods and cones; onl, outer nuclear layer; opl, outer plexiform layer; inl, inner nuclear layer; ipl, inner plexiform layer; gcl, ganglion cell layer. Five frogs of each genotype underwent H&E staining. Two dorsal skin samples, two ventral skin samples, and both eyes were collected from each frog. Histological examination of each collected sample was conducted on 10 paraffin sections. c, d The transmission electron microscopy (TEM) examination of the dorsal skin of 1-year-old WT Xenopus tropicalis was exhibited. The results revealed the presence of melanophores and xanthophores in the dermis (c), and the coexistence of iridophores and melanophores (d). e The TEM examination of the dorsal skin of 1-year-old mitf−/− Xenopus tropicalis was exhibited. f, g The TEM examination of the ciliary marginal zone in retina of 1-year-old WT and mitf−/− Xenopus tropicalis was exhibited. The results revealed the presence of melanophores, xanthophores, and iridophores in the WT ciliary marginal zone (f), while melanophores and xanthophores were absent in the mitf−/− ciliary marginal zone (g). TEM analysis was performed on three frogs of each genotype. For each frog, three 1 mm×1 mm skin samples were collected from distinct regions on the dorsal skin, and one eye sample was randomly selected. Five ultrathin sections were generated from each sample for generating the representative results presented here. Mel, melanophore; Xan, xanthophore; Iri, iridophore; BM, basement membrane; MG, mucous gland; GG, granular gland; Myo, myoepithelial cell; s.bas, stratum basale; s.spi, stratum spisosum; s.gra, stratum granulosum; s.cor, stratum corneum; BC, blood cell; CMZ, ciliary marginal zone. The black scale bar in a–b is 50 μm. The scale bar in c–g is 2 μm.
As regards the granular glands of Xenopus tropicalis, they share numerous structural and functional similarities with mammalian exocrine sweat glands45, including similar tissue architecture enveloped by myoepithelial cells46, expression of the classical aquaporin Aqp5 that regulates water homeostasis and other physiological processes47, and secretion of antimicrobial peptides, such as Nmb and Pgq, for safeguarding the skin against pathogenic microorganisms48,49 (Supplementary Fig. 10). Nonetheless, to the best of our knowledge, the involvement of Mitf in the development of granular or exocrine sweat glands has yet to be reported45,46. Hence, it is imperative to conduct a meticulous examination of the impact of mutated MITF on the formation of sweat glands in mice and humans, in light of the findings from mitf−/− Xenopus tropicalis. In terms of xanthophores in Xenopus tropicalis, their gene regulation network and developmental pathways have not been extensively investigated in comparison to their zebrafish counterparts13,50. Additionally, there are no reports on the role of mitf in xanthophore development in any species to date26,51. Nevertheless, given the intricate transcriptional regulation and functional diversity of the mitf locus26, it is plausible that mitf plays a crucial role in the development of xanthophores and granular glands in Xenopus tropicalis, as indicated by the phenotype of mitf−/− Xenopus tropicalis. Therefore, subsequent studies were necessary to firmly establish the genotype-phenotype specificity of mitf in relation to xanthophores and granular glands in Xenopus tropicalis.
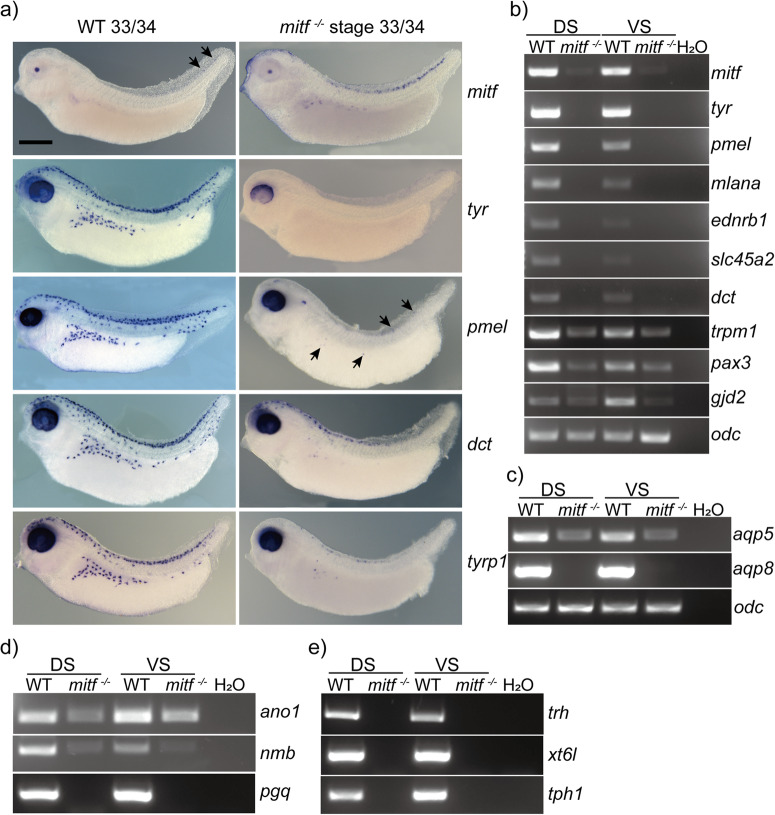
To uncover the disruption of melanophore development in mitf−/− Xenopus tropicalis, we analyzed the expression of mitf and melanophore markers (tyr, pmel, dct, and tyrp1) using whole mount in situ hybridization26. Our results showed that the observed changes in marker gene expression in early embryonic melanocytes of Mitf-mutated mice and zebrafish were also apparent in mitf−/− Xenopus tropicalis20. Specifically, melanoblasts labeled by dct, pmel, and tyrp1 were detected, but in a substantially reduced number (Fig. 4a and Supplementary Fig. 11–20). Notably, we observed a persistent absence of mature melanophore marker gene (tyr)-labeled cells in mitf−/− Xenopus tropicalis (Fig. 4a and Supplementary Fig. 14), underscoring the essential role of mitf in the maintenance and survival of melanoblasts in Xenopus tropicalis, as in mammals and fish26,52. Concomitantly, reverse transcription-polymerase chain reaction (RT-PCR) analysis further confirmed the evident reduction or absence of melanophore markers (tyr, pmel, mlana, ednrb1, slc45a2, and dct) in the skin of mitf−/− individuals compared to that of wild-type Xenopus tropicalis (Fig. 4b). Therefore, the whole-mount in situ hybridization and RT-PCR results suggest a total absence of mature melanophores throughout the body of mitf−/− Xenopus tropicalis. This is likely attributed to the insufficient Mitf activity in melanoblasts, posing challenges to their survival.
Fig. 4. The expression of crucial genes associated with melanophores, xanthophores, and granular glands was reduced or lost in mitf−/−Xenopus tropicalis.
a Whole-mount in situ hybridization analysis of stage 33/34 Xenopus tropicalis embryos demonstrated a reduction or absence of mRNA hybridization signals in mitf−/− embryos compared to the WT. The black arrows indicated some weaker mRNA hybridization signals. At least five embryos were used for in situ hybridization of each gene in every genotype embryo. b–e Representative results of RT-PCR analysis for several melanophores-related genes, including mitf, tyr, pmel, mlana, ednrb1, slc45a2, and dct, as well as xanthophores-related genes, trpm1, pax3, and gid2, were presented in b. The expression of aquaporin-related genes, such as aqp5 and aqp8, as well as Ca2+-activated chloride channel anoctamin 1 (ano1), and antimicrobial peptides-related genes, including nmb and pgq, in the granular glands were displayed in c and d, respectively. Several hormone-related genes of granular glands, including trh, xt6l, and tph1, were also evaluated (e). The representative results of this analysis were presented in panels c–e. odc was used as the RNA loading control and c–e shared the RNA loading control in c. For RT-PCR analysis, each gene was subjected to three repetitions using three replicates of total RNA samples, while each total RNA sample was extracted from the skin of three individual frogs. The original data for the mentioned RT-PCR can be found in Supplementary Fig. 32 and Supplementary Fig. 33. Additionally, the original data for the RT-PCR in Supplementary Fig. 25 can be found in Supplementary Fig. 34. DS, dorsal skin; VS, ventral skin. The black scale bar in a is 0.5 mm.
Given the absence of xanthophores and granular glands in mitf−/− Xenopus tropicalis, we investigated whether there were changes in gene expression related to these features. As there were limited literatures on xanthophore-related genes in this species, we used Xenopus tropicalis homologs of zebrafish xanthophore-related genes (trpm1, pax3, and gjd2) for RT-PCR analysis53. The expression of these genes in the skin of mitf−/− Xenopus tropicalis was evidently lower compared to the wild type, suggesting an abnormality in xanthophore development in these frogs (Fig. 4b). To corroborate the loss of granular gland phenotype in mitf−/− Xenopus tropicalis, genes pertinent to granular gland function, including aquaporins (aqp5 and aqp8)47, Ca2+-activated chloride channel anoctamin 1 (ano1)45,54, antimicrobial peptides (nmb and pgq)48,49, and hormones (trh, xt6l, and tph1)55, were selected and subjected to RT-PCR analysis. Notably, Aqp5 and Ano1 are established markers for exocrine sweat glands in some mammals. As expected, the skin of mitf−/− Xenopus tropicalis exhibited evidently reduced expression of all these genes (Fig. 4c–e). Together, the molecular evidence provided here also suggests abnormal developments of xanthophores and granular glands (Fig. 4).
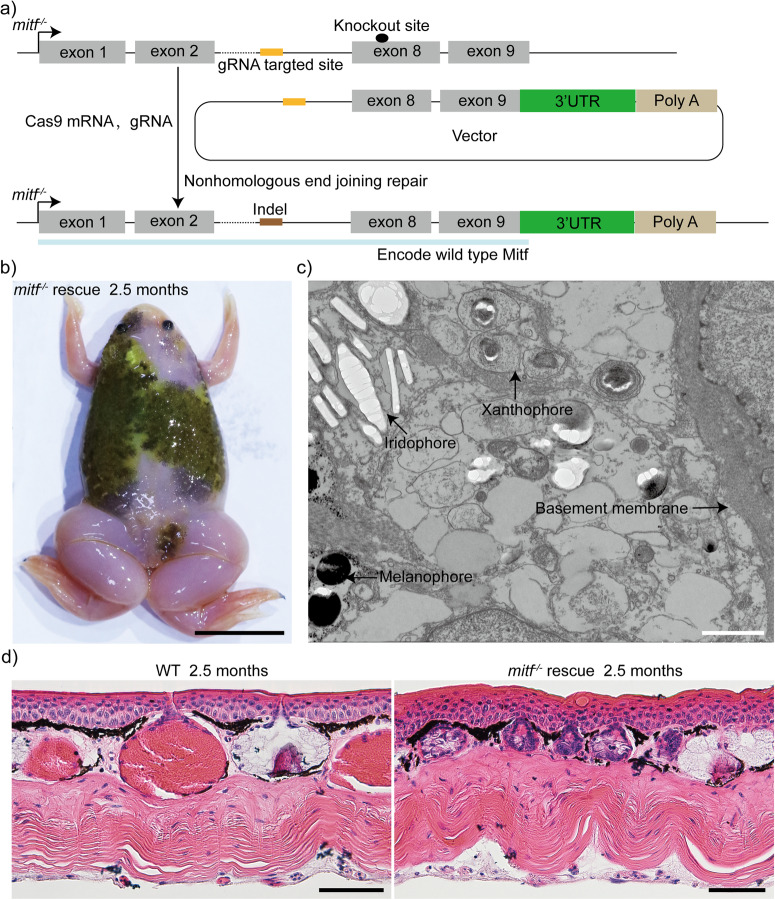
To strengthen the genotype-phenotype specificity of mitf−/− Xenopus tropicalis, we conducted a rescue experiment by integrating a foreign DNA fragment into the disrupted mitf locus to repair the damaged site (Fig. 5a). Concretely, we inserted a foreign DNA fragment that included the final two exons and the 3′UTR region of the mitf locus. This successfully repaired the damaged mitf site in mitf−/− Xenopus tropicalis, as confirmed by the diverse degrees of melanophore rescue phenotypes observed in the mosaic Xenopus tropicalis (Fig. 5b-c). In addition, xanthophores were also restored in the melanocytes-bearing skin of the mosaic frog (Fig. 5c). Although we didn’t find mature granular glands in the rescued skin of mosaic frogs, early-stage granular glands56 were detected in the rescued skin (Fig. 5d). The lack of developed mature granular glands in the rescued skin of mosaic frogs is likely closely related to their mosaic nature. Consequently, these findings further corroborated the genotype-phenotype specificity of the melanophores, xanthophores, and granular glands loss caused by mitf knockout in Xenopus tropicalis.
Fig. 5. Restoration of the damaged mitf site in mitf−/−Xenopus tropicalis results in the rescue of the phenotype of melanophores, xanthophores, and granular glands.
a The precise integration of the DNA fragment encompassing the final two exons and the 3′UTR region of mitf into the mitf−/− genomic locus via CRISPR/Cas9-mediated non-homologous end joining repair has been demonstrated. b A rescued phenotype was demonstrated in a representative 2.5-month-old froglet of the mosaic founders. Approximately 300 embryos of mitf−/− Xenopus tropicalis were injected, and eventually, six tadpoles were found to have a rescued phenotype of melaphores. c TEM examination of the dorsal skin of the rescued froglet depicted in b revealed a significant resurgence of both xanthophores and melanophores. We sampled three distinct regions of the dorsal skin. Six ultrathin sections were subsequently generated from each of the three samples to yield the representative results presented here. d The histological structure of the dorsal skin from 2.5-month-old WT froglets and rescued froglets was presented. Three 2.5-month-old WT froglets and three rescued froglets depicted in b were subjected to H&E staining, and two dorsal skin samples with 3 mm×3 mm were obtained from each frog. Histological examination was performed on ten paraffin sections for each collected sample to generate the representative results presented here. Scale bar in b, 1 cm; Scale bar in c, 1 μm; Scale bar in d, 50 μm.
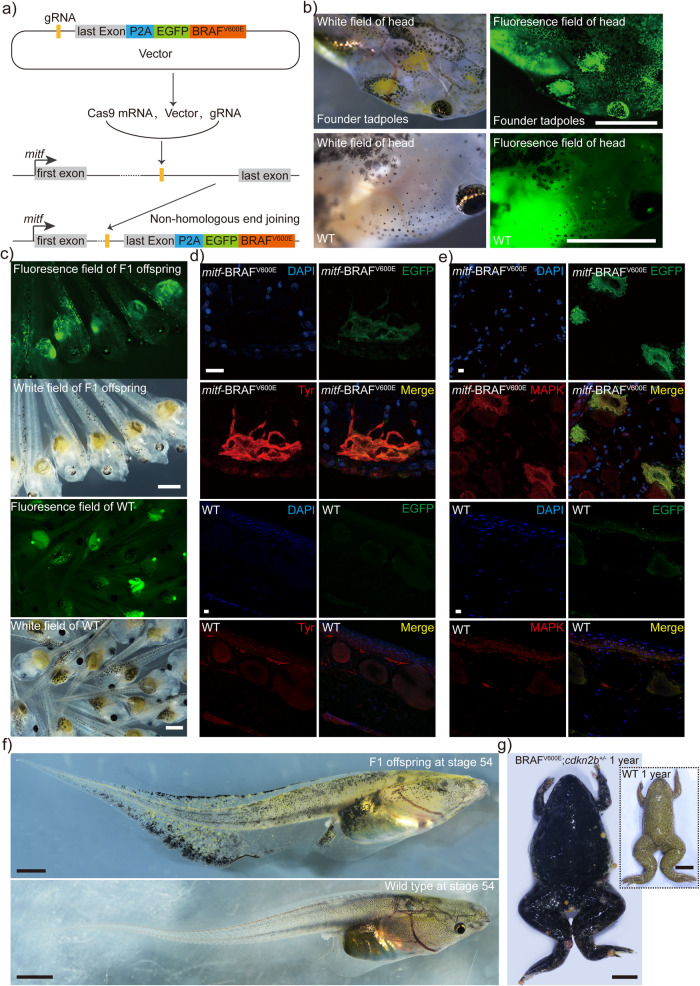
Coincidentally, as shown in Fig. 6a, we were also utilizing the strategy of the specific integration of human BRAFT1799A into the mitf locus to establish a Xenopus tropicalis melanoma model. This model happened to address the issue of the lack of specific and reliable Mitf antibodies for detecting mitf expression in Xenopus tropicalis. BRAFT1799A is a frequently occurring point mutation in the BRAF oncogene, associated with the development of human nevus and melanoma57. This mutation encodes BRAFV600E, which substitutes valine with glutamic acid at amino acid 600 of the kinase domain, thereby transforming the protein from an inactive to an active form. The resultant BRAFV600E protein continuously activates the MAPK oncogenic signaling cascade and induces the formation of melanocytic nevus and melanoma in mice and zebrafish when expressed in melanocytes57. In this Xenopus tropicalis melanoma model, the BRAFV600E expression was under endogenous mitf promoter control in theory. Thus, a similar phenotype of aberrant melanophore proliferation observed in xanthophores and granular glands would indicate the presence of mitf expression in xanthophores and granular glands. Mosaic G0 founders of targeted integration of BRAFT1799A into the mitf locus, designated as mitf-BRAFV600E, were generated by injecting a mixture of SpCas9 mRNA, gRNA, and donor vector plasmids into zygotes at the 1-cell stage. This resulted in mosaic G0 founders displaying abnormal proliferations of xanthophores and melanophores, as expected (Fig. 6b). The F1 generation of mitf-BRAFV600E Xenopus tropicalis was obtained through outcrossing the mosaic G0 founders with wild-type individuals, as shown in Fig. 6c. The expression of the fusion protein EGFP-BRAFV600E in Tyr-positive melanophores led to the activation of the MAPK signaling pathway, as demonstrated in Fig. 6d, e. These results indicated that not only did BRAFT1799A integrate correctly as expected, but it also expressed BRAFV600E under the control of the endogenous mitf promoter. Furthermore, the expressed fusion protein exhibited sustained kinase activation function. As expected, melanocytic nevi-like lesions were firstly apparent throughout the bodies of the F1 generations of mitf-BRAFV600E Xenopus tropicalis, and corresponding tadpoles at stage 57/58 exhibited abnormal proliferation of xanthophores and melanophores (Fig. 6b, c, f). Adult mitf-BRAFV600E;cdkn2b+/− Xenopus tropicalis spontaneously develop nevi caused by abnormal proliferation of melanophores and xanthophoromas caused by abnormal proliferation of xanthophores58 (Fig. 6g) (A manuscript focusing on the tumor formations, eye degeneration, nevi, and bone dysplasia in this line is currently under preparation). Moreover, after genotyping, we confirmed the correct integration of BRAFT1799A in the target lines (Fig. 7a and Supplementary Fig. 21). Histopathological assessments revealed abnormal proliferations of melanophores and xanthophores, along with the presence of abundant granular glands in this line (Fig. 7b–d). Consequently, these findings were indicative of the expression of mitf in xanthophores and granular glands.
Fig. 6. The integration of the BRAFT1799A within the mitf locus elicits spontaneously formed melanophores-related nevi and xanthophores-like moles in Xenopus tropicalis.
a The strategy of precisely integrating the human BRAFT1799A mutation into the mitf genomic locus through non-homologous end joining repair mediated by CRISPR/Cas9 has been demonstrated. This involves placing the mutation downstream of the last exon of mitf, thereby utilizing the endogenous promoter of mitf to drive BRAFV600E expression. b At stage 56, following precise integration of human BRAFT1799A into the mitf genomic locus, the founder generation tadpoles exhibited a mosaic phenotype characterized by aberrant proliferation of melanophores and xanthophores. Additionally, a significant proliferation of xanthophores was observed using spontaneous green fluorescence. The control group exhibited the absence of this phenotype in WT tadpoles. A total of eight such tadpoles were observed at this stage. c Compared to the control group of WT tadpoles, the F1 generation mitf-BRAFV600E tadpoles at stage 46 were observed and found to exhibit EGFP signals. d, e Immunofluorescence analysis was performed on three tail samples obtained from three stage 56 mitf-BRAFV600E tadpoles from the F1 generation. Every sample was fixed, embedded, and sectioned into six paraffin slices for the analysis. The results demonstrated co-localization of Tyr and EGFP (d), as well as co-localization of EGFP and MAPK signaling pathway activation (e). The WT control group showed an absence of EGFP signals, and cells with activated MAPK pathway were also scarce. The activation of the MAPK signaling pathway was evidenced by positive fluorescence signal of the antibody-ERK1 + ERK2. The images presented here were representative of the observed results. f A typical photograph of F1 mitf-BRAFV600E tadpoles at stage 54 and the corresponding batch of WT tadpoles was presented. g Compared to a 1-year-old WT frog, a representative image of a 1-year-old cdkn2b+/−;mitf-BRAFV600E frog showed spontaneously formed nevi and xanthophoromas. Scale bar in b, 2 mm; Scale bar in c, 1 mm; Scale bar in d and e, 10 μm; Scale bar in f, 5 mm; Scale bar in g, 1 cm.
Fig. 7. The integration of the BRAFT1799A within the mitf locus results in a substantial proliferation of melanophores and xanthophores, as well as a significant increase in the number of granular glands.
a The Sanger sequencing of the integrated upstream and downstream repair site confirmed the correct integration of BRAFT1799A within the design target. For a detailed explanation of the detection principle, please refer to Supplementary Fig. 18. Ten mitf-BRAFV600E tadpoles were subjected to genotyping. b The histological structure of the dorsal skin (DS) from 1-year-old WT, F2 mitf−/−, and F1 mitf-BRAFV600E Xenopus tropicalis specimens was depicted. Blue dashed box 2 corresponded to the magnified view of blue dashed box 1. Five frogs of each genotype underwent H&E staining. Two dorsal skin samples were collected from each frog. Histological examination of each collected sample was conducted on 10 paraffin sections for generating the representative results presented here. c, d Upon TEM examination of the dorsal skin of 1-year-old F1 mitf-BRAFV600E Xenopus tropicalis, a notable proliferation of xanthophores (c) and melanophores (d) was observed. We conducted TEM analysis on three frogs of mitf-BRAFV600E Xenopus tropicalis, collecting three samples from distinct regions on the dorsal skin for each frog. Six ultrathin sections were generated from each sample to generate the representative results presented here. The black scale bar in b is 100 μm. The scale bar in c, d is 5 μm.
Collectively, the findings reported above demonstrated that abrogating mitf via the gRNA T7 resulted in the disappearance of light-absorbing melanophores, granular glands and light-absorbing/reflecting xanthophores throughout the entirety of Xenopus tropicalis’ body, revealing that mitf functioned not only as a master regulator of melanophores but also as a key factor in orchestrating the development of xanthophores and granular glands. Still, the exact molecular mechanisms by which mitf regulates the development of xanthophores and granular glands remain to be deciphered.
A colorless and immunodeficient Xenopus tropicalis line is established through the mitf/prkdc/il2rg triple-knockout
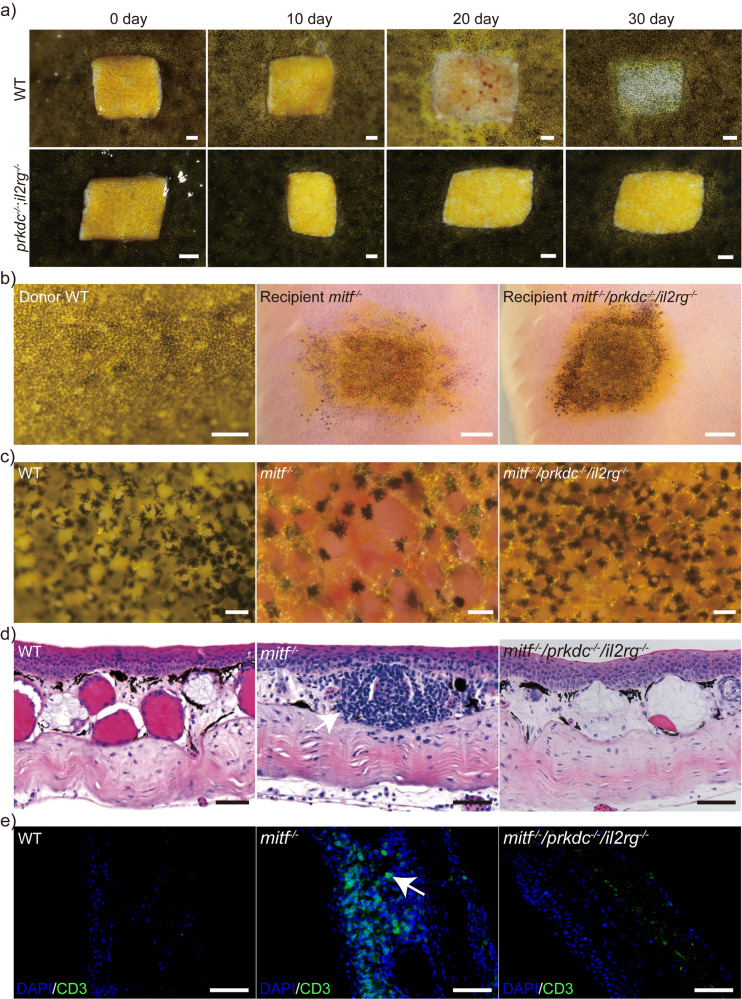
To generate immunodeficient Xenopus tropicalis for tumor transplantation, we employed the CRISPR/Cas9 system to simultaneously knock out prkdc and il2rg genes, which are essential for T cell, B cell, and NK cell development3. The dual knockout strategy was depicted in Supplementary Fig. 22a, where SpCas9 mRNA and gRNAs were co-injected into fertilized Xenopus tropicalis eggs at the 1-cell stage, resulting in mosaic G0 founders with a 60% efficiency of knockout for prkdc and il2rg individually, thereby implying a 36% efficiency for prkdc/il2rg double-knockout (Supplementary Fig. 22b). Of these G0 founders, 53% (72/135) exhibited significant thymic abnormalities at stage 56, as shown in Supplementary Fig. 22c. Subsequently, we raised these tadpoles to sexual maturity and carried out assortative mating to produce offspring with various genotypes, selecting mutants with Indels of prkdc and il2rg able to cause frameshift mutations. Among these mutants, individuals with 4 bp and 22 bp deletions in il2rg alleles, and 11 bp deletions in both prkdc alleles, were denoted as prkdc−/−/il2rg−/− and comprised 20% of the animals (26/128). The thymus and spleen of prkdc−/−/il2rg−/− Xenopus tropicalis were stunted and structurally abnormal, with a significant downregulation of T-cell, B-cell, and NK-cell marker genes in the spleen, liver, lung, and blood, indicating a deficiency in the development of T cells, B cells, and NK cells (Supplementary Fig. 23 and Supplementary Fig. 24). The significant reduction in il2 expression in the spleen of prkdc−/−/il2rg−/− Xenopus tropicalis further indicates abnormal T-cell development in these frogs (Supplementary Fig. 25). Additionally, we performed skin transplantation using Xenopus tropicalis of the wild type and prkdc−/−/il2rg−/− mutants as an indicator of immune deficiency59. The results showed that wild type Xenopus tropicalis exhibited immunological rejection to allogeneic or xenogeneic skin grafts (The skin transplants from Xenopus tropicalis, Xenopus laevis, and zebrafish, all exhibited a certain immune rejection response with a 3/3 rate), while the prkdc−/−/il2rg−/− mutants accepted skin grafts from different species with varying degrees of rejection depending on evolutionary distance (3/3 Xenopus tropicalis skin transplants showed no rejection after one year, while 3/3 Xenopus laevis had mild rejection, and 3/3 zebrafish exhibited strong rejection after 14 days), for instance, completely accepting skin grafts from allogeneic Xenopus tropicalis yet showing some immunological rejection to skin grafts from xenogeneic zebrafish (Fig. 8a and Supplementary Fig. 26). Overall, these findings indicated that prkdc−/−/il2rg−/− Xenopus tropicalis mutants were promising subjects for tumor cell transplantation studies across species while indicating their potential utility in tumor immunology research.
Fig. 8. The mitf/prkdc/il2rg triple-knockout results in the creation of a line of colorless and immunodeficient Xenopus tropicalis.
a Three WT and three prkdc−/−/il2rg−/− Xenopus tropicalis aged 6 months were used as transplant recipients. The donors were three tyr−/− Xenopus tropicalis aged 2 years, and their dorsal skins were grafted onto the recipients’ dorsal skin. The grafting outcomes were monitored for a 30-day period post-transplantation. The representative results were presented here and the transplanted donor skins were approximately 5 mm × 5 mm in size. b–e Three mitf−/− and three mitf−/−/prkdc−/−/il2rg−/− Xenopus tropicalis froglets were selected as transplant recipients at the age of two months. The donor skins were obtained from three WT Xenopus tropicalis at the same age and were grafted onto the dorsal skin of the recipients. The transplanted donor skins were approximately 3 mm×3 mm in size, and the WT skin and representative photos of the transplanted donor skins were taken on the 52nd day post-transplantation (b). The magnified view of the corresponding skin in b was shown in c. d The histological structure of the corresponding skin in c was presented through histological examination of 18 paraffin sections from the collected sample, which provided the representative results presented here. e T cell infiltration following skin allografts in mitf−/− and mitf−/−/prkdc−/−/il2rg−/− Xenopus tropicalis. T cells were labeled with the CD3 antibody, and cell nuclei were stained with DAPI. The sample preparation method was the same as for d. Scale bar in a and b, 1 mm; Scale bar in c, 100 μm; Scale bar in d and e, 50 μm.
Next, we investigated whether Xenopus tropicalis with a triple-knockout of mitf/prkdc/il2rg was both colorless and immunodeficient. To generate this line, mitf−/− mutants were crossbred with prkdc−/−/il2rg−/− individuals, resulting in F2 generations that showed colorlessness and the mitf/prkdc/il2rg triple-knockout genotype (mitf−/−/prkdc−/−/il2rg−/−) in a frequency of 22/1086, following Mendelian inheritance principles (Supplementary Fig. 27). To assess the immunodeficiency of this strain, we performed allotransplantation of wild-type skin to both mitf−/− and mitf−/−/prkdc−/−/il2rg−/− Xenopus tropicalis, and observed immune rejection responses and T cell infiltration. Skin allograft transplantation experiments conducted in thymectomized and wild-type Xenopus laevis have provided evidence that CD4 and CD8 positive T lymphocytes play a crucial role in the rejection response of skin grafts in Xenopus frogs59. Results showed that recipients of mitf−/− individuals experienced immune rejection with an elevated number of T cells (CD3+ cells) in the transplanted skin, whereas recipients of mitf−/−/prkdc−/−/il2rg−/− individuals did not experience immune rejection and the transplanted skin developed similarly to skin from wild type frogs without transplantation (Fig. 8b–e). Thus, these results demonstrated that mitf−/−/prkdc−/−/il2rg−/− Xenopus tropicalis was a colorless and immunodeficient line.
Visualizing allografted tumor cell metastasis in mitf−/−/prkdc−/−/il2rg−/−Xenopus tropicalis
To ascertain the suitability of mitf−/−/prkdc−/−/il2rg−/− Xenopus tropicalis for tumor allotransplantation, we conducted allotransplantations of xanthophoromas and melanomas in these amphibians. The xanthophoroma tissues used for allotransplantation were sourced from the skin of cdkn2b−/−;mitf-BRAFV600E Xenopus tropicalis. When these donors of xanthophoroma tissues were transplanted to wild type and mitf−/−/prkdc−/−/il2rg−/− Xenopus tropicalis recipients, no visible metastasis was observed within a month for both recipients (Supplementary Fig. 28a–e). Xanthophoromas metastasis was observed in the mitf−/−/prkdc−/−/il2rg−/− recipients for up to two months post-transplantation, while no signs of metastasis were seen in the wild type recipients during the same period (Supplementary Fig. 28f-g). At five months post-transplantation, systemic metastasis of xanthophoromas was evident in the mitf−/−/prkdc−/−/il2rg−/− recipients, but not in the wild type recipients (Fig. 9a). Furthermore, through a comparative analysis of the green fluorescence signal in the dorsal thigh skin of the transplanted mitf−/−/prkdc−/−/il2rg−/− recipient with that of the mitf−/− frog, we discovered that mitf−/−/prkdc−/−/il2rg−/− Xenopus tropicalis was suitable for intravital imaging of dynamic changes in tumor cells using green fluorescence signals post-transplantation (Fig. 9b). Conversely, wild type Xenopus tropicalis was deemed unsuitable due to the presence of intense autofluorescence. Remarkably, xanthophoromas were observed to metastasize to the entire body of mitf−/−/prkdc−/−/il2rg−/− recipients about one year after transplantation, resulting in grave lesions necessitating the sacrifice of the recipients (Supplementary Fig. 28h). Conversely, the transplanted xanthophoromas in wild type recipients stayed at the site of transplantation, as opposed to metastasizing or being eliminated by the host immune system (Supplementary Fig. 28i). These findings suggested that the use of mitf−/−/prkdc−/−/il2rg−/− Xenopus tropicalis was a viable method for tumor allogeneic transplantation and intravital imaging of transplanted tumor cells. Furthermore, the observed mutual restriction mechanism between the wild type recipients and transplanted xanthophoromas may have implications for the development of tumor immunotherapy.
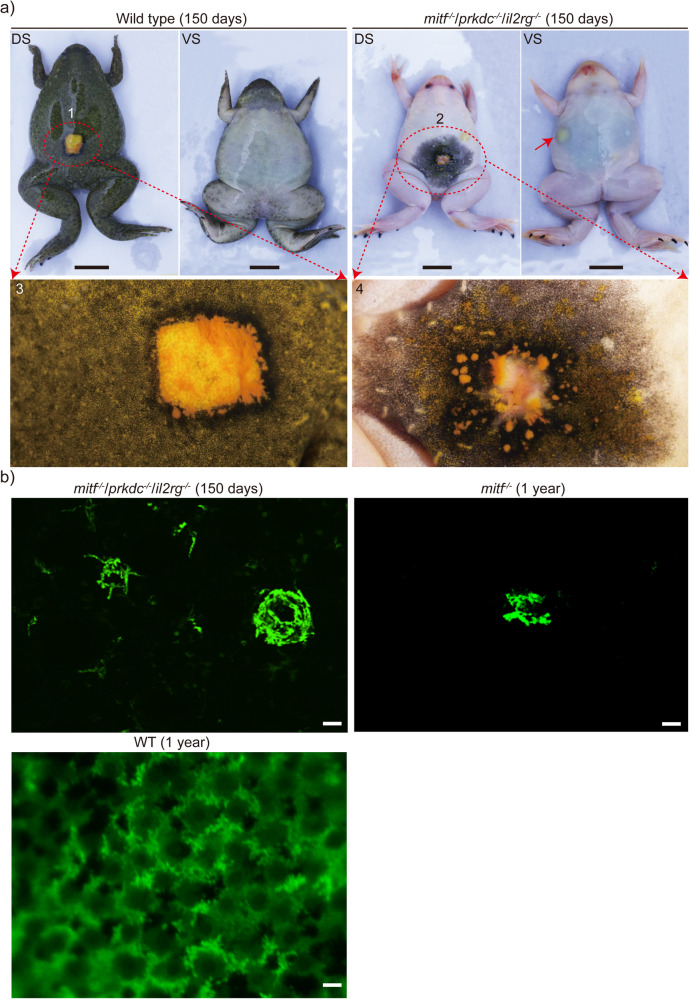
Fig. 9. Allogeneic transplantations of xanthophoromas were conducted in mitf−/−/prkdc−/−/il2rg−/−Xenopus tropicalis, resulting in the observed systemic metastasis of transplanted xanthophoromas in the recipient animals.
a At 150 days post-transplantation, xanthophoromas were observed intravitally in both dorsal skin (DS) and ventral skin (VS), and the enlarged views of transplanted xanthophoromas in ellipses 1 and 2 were depicted in 3 and 4, respectively. Xanthophoromas from eight stage 58 cdkn2b−/−;mitf-BRAFV600E tadpoles were transplanted onto the dorsal skin of four WT frogs and four mitf−/−/prkdc−/−/il2rg−/− frogs, which served as the transplant recipients. The transplants were monitored for up to one year. The presented data represented representative results on day 150 post-transplantation, and additional representative observation data are provided in Supplementary Fig. 28. b Metastasized xanthophoroma cells were intravitally observed under fluorescence field conditions in the right thigh of the recipient depicted in panel a. To provide a representative control, intravital observations were also conducted on the right thigh of three 1-year-old mitf−/− frogs that had not undergone xanthophoroma transplantation, using fluorescence field conditions. Control group WT exhibits spontaneous green fluorescence on the dorsal skin of one-year-old Xenopus tropicalis. The scale bar in a is 1 cm. The scale bar in b is 10 μm.
Melanoma tissues from tp53△7/△7 Xenopus tropicalis donors were utilized for melanoma allotransplantations60. As previously reported, the melanoma developed similarly to human melanoma60. Due to limited nevi and melanoma samples, four dysplastic nevi and four metastatic melanomas were selected and transplanted into mitf−/−/prkdc−/−/il2rg−/− recipients (experimental group) and mitf−/− recipients (control group), respectively, as demonstrated in Supplementary Fig. 29a. Unfortunately, one dysplastic nevus and one metastatic melanoma that were transplanted into their respective groups became detached accidentally on the fourth day after transplantation, leaving only one transplanted frog per group to continue with the experiment. Thirty days post-transplantation of dysplastic nevi, migration of donor melanophores was observed exclusively in the recipient skin of mitf−/−/prkdc−/−/il2rg−/− Xenopus tropicalis, but not in that of the mitf−/− frog (Supplementary Fig. 29b-g). Notably, even after 40 days of transplantation in the skin of the mitf−/− frog, the migration of donor melanophores remained undetectable (Fig. 10a, c). However, the migration of donor melanophores was observed in the mitf−/−/prkdc−/−/il2rg−/− recipient within the same time frame, and the morphology of these donor melanophores was indistinguishable from that of differentiated Xenopus tropicalis melanophores12 (Fig. 10b, d). As for metastatic melanoma tissues allotransplantations, migration of melanoma cells was also exclusively observed in the mitf−/−/prkdc−/−/il2rg−/− recipient (Supplementary Fig. 23j–m and Fig. 10e–h). Unlike dysplastic nevi transplantation results, the morphology of migrating melanomas exhibited an undifferentiated state after 40 days of transplantation (Fig. 10e–h). To supplement the inventory of melanoma transplants, allogeneic transplantation experiments were conducted utilizing spontaneously occurring dysplastic nevi from cdkn2b−/−/tp53−/− Xenopus tropicalis. The outcomes revealed that mitf−/− recipients manifested immune rejection, contrasting with the absence of such rejection in mitf−/−/prkdc−/−/il2rg−/− recipients (Supplementary Fig. 30). Dysplastic nevi transplanted onto both mitf−/− and mitf−/−/prkdc−/−/il2rg−/− recipients did not exhibit notable melanophore migration, potentially attributed to the advanced age of the recipients. Taken together, the transplantation of dysplastic nevi and metastatic melanomas in mitf−/− Xenopus tropicalis recipients was observed to result in a lack of migration and clear signs of clearance by the host’s immune system (Fig. 10d, h). Interestingly, xanthophoromas and melanomas transplanted into immunocyte-normal recipients (wild type and mitf−/− Xenopus tropicalis) demonstrated contrastive behaviors: persistent in situ growth and immune rejection, respectively. These discrepancies could be attributed to either variable oncogenic potential of donor tumors or idiosyncratic interactions between tumorous cells and the recipient’s immune system.
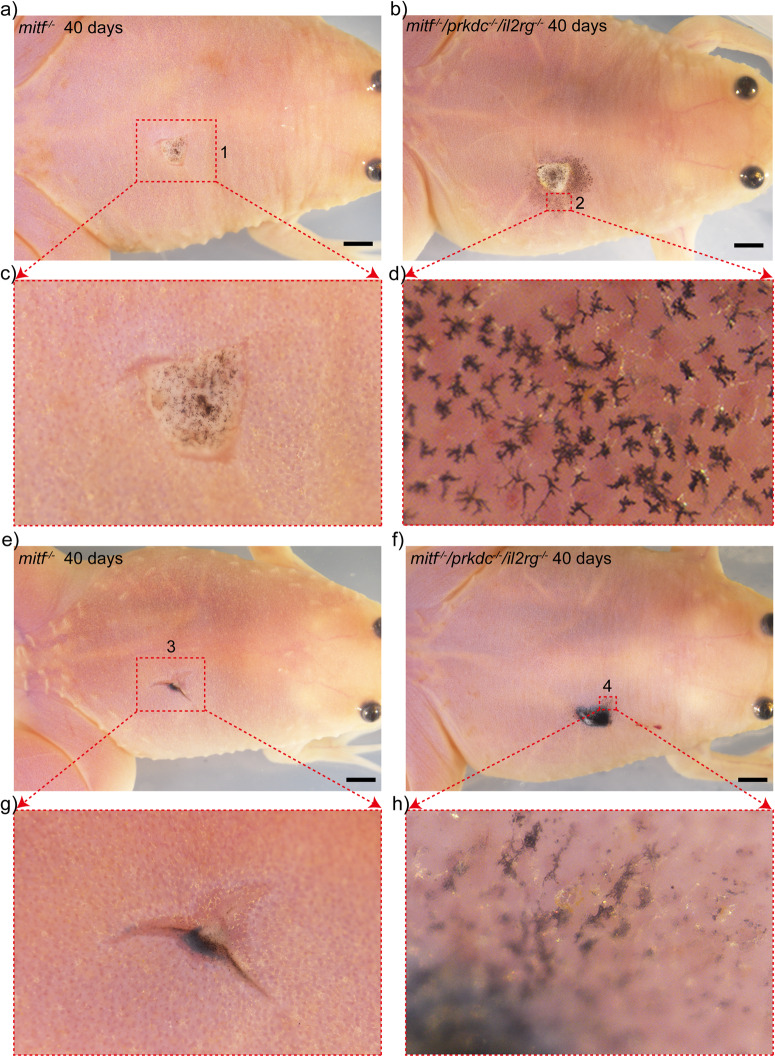
Fig. 10. Allogeneic transplantations of melanomas were conducted in mitf−/−/prkdc−/−/il2rg−/−Xenopus tropicalis, resulting in the observed migrations of transplanted melanomas in the recipient animals.
a–h At day 40 post-transplantation, the transplanted nevi and metastatic melanoma were examined in mitf−/− (a, c, e, g) and mitf−/−/prkdc−/−/il2rg−/− (b, d, f, h) recipients. The enlarged views of the transplanted nevi and metastatic melanoma were portrayed in panels c and d (red dashed boxes 1 and 2) and g and h (red dashed boxes 3 and 4), respectively. Each scale bar is 1 mm.
Collectively, as shown in Fig. 11, our studies suggested that mitf−/−/prkdc−/−/il2rg−/− Xenopus tropicalis was a valuable tool for transplanting tumors and intravitally observing the initial stages of metastasis, particularly the migration of melanomas. Furthermore, our results also implied that a comprehensive examination of tumor response following transplantation of xanthophoromas and melanomas in Xenopus tropicalis could hold promise for advancing the development of tumor immunotherapy.
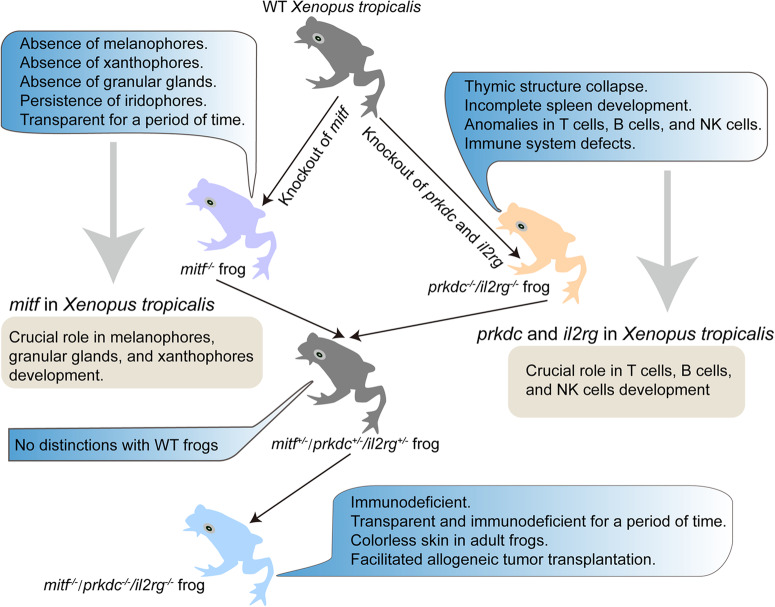
Fig. 11. Schematic of mitf−/−/prkdc−/−/il2rg−/−Xenopus tropicalis construction.
The process revealed the crucial role of mitf in the development of melanophores, xanthophores, and granular glands. The resulting mitf−/−/prkdc−/−/il2rg−/− Xenopus tropicalis is suitable for research on allograft tumor transplantation.
Discussion
In this study, we developed an animal model of tumor allograft recipient utilizing a genetically engineered amphibian, the colorless and immunodeficient Xenopus tropicalis line generated by mitf/prkdc/il2rg triple-knockout, enabling intravital observations of transplanted xanthophoromas’ and melanomas’ migration and metastasis. Concomitantly, we discovered that mitf−/− Xenopus tropicalis lacked not only melanophores but also xanthophores and granular glands. By integrating BRAFT1799A into the mitf locus and conducting rescue experiments of mitf−/− damage site targeted repair, we demonstrated that mitf was also expressed in xanthophores and granular glands, and played a crucial role in their development, revealing novel functions for the mitf locus in the development of xanthophores and granular glands.
In melanocyte development, several signaling pathways, including MC1R, PI3K, MAPK, and WNT, intricately regulate the process through the mediation of MITF26,61. MITF’s regulatory influence manifests through the modulation of its expression levels. For instance, the collaborative orchestration of the PI3K, MAPK, and WNT pathways regulates MITF expression by manipulating the transcription factor BRN2 (POU3F2), thus influencing MITF’s function26. Additionally, MITF’s functionality is also subject to control through the regulation of its phosphorylation status. The phosphorylation at S73, S69, S298, and the three C-terminal sites are modulated by PI3K, MAPK, and WNT signaling pathways, impacting MITF’s activity62–64. Research on the regulation of Mitf in melanocyte development in Xenopus tropicalis is limited. Nevertheless, considering the conservation of melanocyte development in vertebrates, the regulation of Mitf protein stability, nuclear translocation, and protein expression levels plays a crucial role in controlling melanocyte development. While the role of Mitf in regulating melanocytes and melanophores is well-established, its involvement in xanthophores has not been extensively studied26. Mutated zebrafish carrying mitfaC417T/C417T, which produces a stop codon substituting glutamine 113, display an increase in iridophores but an absence in melanophores and a slight reduction in xanthophore pigmentation20. Knockout of mitf in tilapia resulted in the loss of melanophores, increased iridophores and erythrophores, and enlarged xanthophores65. The underlying mechanisms behind the altered xanthophores in zebrafish and tilapia are still unclear, and knockout of mitf in fish leading to the loss of xanthophores has not been reported. In zebrafish, xanthophores and melanophores can be derived from a common Tuba8l3-positive progenitor cell, and the developmental fate of these progenitor cells is determined by the expression of mitfa and kita for melanophores while csf1ra for xanthophores51. Recent studies using single-cell RNA sequencing in zebrafish have confirmed that mitfa expression occurs in xanthophore progenitor cells27. Additionally, in vitro studies have shown that amphibian xanthophores could transdifferentiate into melanophores66. It is thus plausible that Xenopus tropicalis utilizes a similar mechanism wherein both melanophores and xanthophores stem from a common progenitor cell, and mitf expression may be crucial for the development of these cells. A potential consequence of mitf loss-of-function in Xenopus tropicalis would therefore be the absence of both xanthophores and melanophores.
The granular glands of Xenopus tropicalis exhibit strong similarities in cellular composition and gene expression in comparison to the exocrine sweat glands of humans and mice45–48,67. Nevertheless, no studies have been conducted on the role of MITF in sweat gland development in either of these species46. Here, our study demonstrated the crucial role of Mitf in the development of granular glands in Xenopus tropicalis through evidence supporting the genotype-phenotype specificity of granular gland loss caused by mitf knockout. While Mitf mutant studies in mice primarily focused on melanocyte-related diseases, the potential sweat gland-related phenotype has not been reported, possibly because sweat glands are relatively small and located in the palmar-plantar region26,46. Given the complexity of the transcriptional regulatory network of the mitf locus26,68, it was possible that the granular glands specific transcript of mitf may lose its function due to the disrupted site in mitf−/− Xenopus tropicalis, leading to a loss of granular glands in these frogs. However, specific mitf transcripts in the oocytes and RPE of mitf−/− Xenopus tropicalis may remain unaffected by the mutants, thus allowing for normal melanin deposition in oocytes and RPE development in eyes (Supplementary Fig. 31), similar to some Mitf/mitf mutated mice and zebrafish, which exhibit normal RPE-specific mitf transcripts26,68. Therefore, exploring the molecular mechanism underlying granular gland loss in mitf−/− Xenopus tropicalis represents an interesting issue to address, and a thorough examination of Mitf functions in mouse sweat glands is warranted.
In humans, the MITF locus also plays a crucial role in in the development and function of various immunocytes, such as dendritic cells, B cells, and mast cells69. The mitf locus in Xenopus tropicalis is hypothesized to share similarities with the Mitf locus in mammals, allowing for the transcription of multiple isoforms, each exhibiting distinct expression patterns and functions. In mammals, it is well-established that Mitf is also expressed in mast cells, and various mast cell defects have been observed in several Mitf mutant mouse strains69. Mitf plays a pivotal role in guiding the differentiation of pre-BMPs (pre-basophil and mast cell progenitors) into mast cells69. Given the essential role of mast cells in the inflammatory process, the impact of mitf knockout on immune rejection defects in Xenopus tropicalis is anticipated to be relatively modest. This perspective is reinforced by the notable infiltration of CD3+ cells in mitf−/− Xenopus tropicalis following allogeneic skin transplantation (Fig. 8). Additionally, the loss of granular glands in mitf−/− Xenopus tropicalis results in a weakened immune response due to the absence of antibacterial substances secreted by these glands. Despite this, allograft transplantation in mitf−/− recipients results in an increased number of immunocytes and subsequent immune rejection, ultimately leading to allograft destruction. Thus, in the colorless and immunodeficient Xenopus tropicalis line of mitf/prkdc/il2rg triple-knockout, further exploration is needed to determine the contribution of the mitf knockout to the immune deficiency of this organism.
In conclusion, the mitf−/−/prkdc−/−/il2rg−/− Xenopus tropicalis provide a valuable recipient model for studying the molecular mechanisms of tumor metastasis in this species and a possibility for establishing xenograft models in amphibians in the future.
Methods
Xenopus tropicalis and Xenopus laevis maintenance
Adult Xenopus tropicalis and Xenopus laevis were procured from Nasco (Fort Atkinson, WI, USA; http://www.enasco.com) and were maintained at the laboratory facility in compliance with approved standards for husbandry and management. Specifically, for mitf−/−/prkdc−/−/il2rg−/− or prkdc−/−/il2rg−/− Xenopus tropicalis, despite being maintained under the same husbandry conditions as WT Xenopus tropicalis, it is essential to keep the rearing water clean and promptly remove food residues and excreta. All the experimental procedures involving Xenopus tropicalis and Xenopus laevis were conducted under the approval of the Institutional Animal Care and Use Committee at the Southern University of Science and Technology. We have complied with all relevant ethical regulations for animal use.
Polymerase chain reaction (PCR), reverse transcription-polymerase chain reaction (RT-PCR) analysis, and quantitative real-time PCR (qPCR)
To conduct PCR, we employed TransTaq® DNA Polymerase High Fidelity (HiFi) (AP131-11, Transgen Biotech) as the manual. To perform RT-PCR analyses, we employed the TransZol Up Plus RNA Kit (ER501-01, Transgen Biotech) to extract total RNA, followed by cDNA synthesis using the TransScript® Uni One-Step gDNA Removal and cDNA Synthesis SuperMix (AU311-03, Transgen Biotech). The TransTaq® DNA Polymerase High Fidelity (HiFi) (AP131-11, Transgen Biotech) was also used in RT-PCR and qPCR analyses. The specific PCR, RT-PCR and qPCR primer sequences utilized in this study were listed in Supplementary Data 3.
Preparation of Cas9 mRNA, guide RNA (gRNA) and vectors, embryo microinjection, genotyping and off-target analysis
Preparations of Cas9 mRNA and guide RNA were performed according to the procedure in our previously published methodology60. Specifically, the linearization of pCS2-SpCas9 plasmid was facilitated via the Not I endonuclease and ensued in vitro transcription through the mMessage mMachine SP6 Kit (Ambion, Austin, TX, USA). The gRNA target sequences for mitf, prkdc, and il2rg were compiled in Supplementary Data 3. The gRNA template was obtained by PCR amplification utilizing the pUC57-T7-gRNA scaffold vector, along with the Trac-reverse primer and a gRNA forward primer harboring a T7 promoter and the specific gRNA target sequence. The primer sequences were listed in Supplementary Data 3. The gRNA in vitro transcription was conducted using the Transcript Aid T7 High Yield Transcription Kit (Thermo Fisher Scientific, Rockford, IL, USA). For purification of Cas9 mRNA and gRNA, the RNeasy Mini Kit and miRNeasy Mini Kit (Qiagen, Hilden, Germany) were employed, respectively.
To create the pBS-mitf.le-P2A-CreERT2-P2A-EGFP-BRAFT1799A-SV40 poly(A)-ela-GFP-SV40 poly(A) donor vector for the mitf locus, a Xenopus tropicalis genomic fragment (named mitf.le) containing the target site and last exon was amplified by PCR and inserted into the pBluescript II SK+ plasmid (Agilent Technologies, Santa Clara, CA, USA). BRAFT1799A was obtained through homologous recombination with a PCR product generated from a PCR-amplified human genomic fragment from Hela Cells, which was a kind gift from Chunhui Hou Lab (Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, China). The pEASY®-Basic Seamless Cloning and Assembly Kit (CU201-02, Transgen Biotech) was used for the homologous recombination. A Not I-digested ela-GFP fragment from pElastase-GFP, containing a 203 bp rat pancreas-specific Elastase I promoter, GFP coding sequence, and an SV40 polyadenylation signal, was inserted as a reporter. A pBS-mitf.le(8-9)-3′UTR donor vector for repairing the mitf disrupted site of mitf−/− Xenopus tropicalis was also created by cloning a PCR-amplified Xenopus tropicalis genomic fragment (named mitf.le(8-9)) containing the integration target site and the last two exons into the pBluescript II SK+ plasmid. The FastDigest Enzyme including BamHI (FD0054, Thermo Fisher Scientific, Rockford, IL, USA), EcoRI (FD0274, Thermo Fisher Scientific, Rockford, IL, USA), HindIII (FD0504, Thermo Fisher Scientific, Rockford, IL, USA), and NdeI (FD0584, Thermo Fisher Scientific, Rockford, IL, USA) and T4 DNA Ligase (FL101-01, Transgen Biotech) was also used to construct donor vectors. The constructed vectors sequence information was shown in Supplementary Data 4.
Xenopus tropicalis fertilized eggs were acquired via hormone-induced mating after pre-priming adult male and female frogs with 20 units of human chorionic gonadotropin (HCG) one day in advance and a full dose of 145 units of HCG 2-3 h before collecting the embryos. To induce gene disruption or integration, we injected a mixture of Cas9 mRNA (300 picograms per egg) and gRNA (200 picograms per egg), or Cas9 mRNA (300 picograms per egg), gRNA (200 picograms per egg), and a donor vector (20 picograms per egg) into the animal pole of fertilized eggs at a volume of 2 nL per egg.
To assess the targeting efficiency, we collected 5–8 healthy embryos at random, 24 h post-microinjection, and proceeded to extract and amplify genomic DNA of the targeted region via PCR. Subsequently, we performed TA cloning by inserting the purified PCR products into the pMD18-T vector (Takara, Katsu, Japan). Sanger DNA sequencing analysis was then conducted on randomly selected single colonies to identify any Indel mutations. Genotyping of adult frogs was performed using small pieces of nails, and the primer sequences used for the PCR genotyping were listed in Supplementary Data 3.
The CRISPOR tool was employed for off-target prediction analysis40. To detect off-target effects, we focused on potential sites with a maximum of four mismatches, as detailed in Supplementary Data 1 and Supplementary Data 2. The resulting potential off-target sites were identified and genotyped by Sanger sequencing, and corresponding PCR primers were provided in Supplementary Data 2.
Hematoxylin-Eosin (H&E) staining, immunofluorescence, whole-mount in situ hybridization (WISH) and transmission electron microscopy (TEM)
The H&E staining procedure was carried out in accordance with our previously published methodology60. Detailedly, Xenopus tropicalis tissues were excised and fixed in FAS eyeball fixative (Servicebio, Wuhan, China) at room temperature for 24 h. Following fixation, the samples underwent a series of dehydration steps in ascending concentrations of ethanol (75%, 85%, 95%, and 100%) for 45 min each, and were then subjected to two rounds of xylene treatment, for 1 and 3 h respectively, before being embedded in paraffin wax. The paraffin-embedded samples were sliced into 6 μm thick sections and underwent dewaxing in xylene thrice for 5 min each, and subsequently rehydrated with 100%, 95%, and 70% ethanol for 5 min each, prior to H&E staining with the H&E staining Kit (Baso, Zhuhai, China). For immunofluorescence, the rehydrated sections underwent destaining with a solution containing 0.45% H2O2 and 1X Tris/Tricine/SDS (T1165, Sigma-Aldrich®) for 15 min at 80 °C to remove melanin pigmentations. The sections were then washed twice with 1X PBS for 5 min each to remove the destaining solution. Antigen retrieval of the destaining sections was performed using citrate antigen retrieval solution (8.1 mM trisodium citrate dihydrate and 1.9 mM citric acid) for 12 h at 60 °C, followed by washing with 1 X PBS solution twice for 5 min each to remove the citrate antigen retrieval solution. The following primary antibodies were then employed: anti-Tyrosinase (ab180753, Abcam), anti-EGFP (ab184601, Abcam), anti-CD31 (ab5690, Abcam), and anti-ERK1 + ERK2 (184699, Abcam). Additionally, two secondary antibodies were utilized: Goat Anti-Mouse IgG H&L (Alexa Fluor® 488) (ab150113, Abcam) and Goat Anti-Rabbit IgG H&L (Alexa Fluor® 594) (ab150080, Abcam). Mounting Medium With DAPI – Aqueous & Fluoroshield (ab104139, Abcam) was employed to stain the cell nucleus. Fluorescence imaging was conducted using a Leica SP8 confocal microscope (TCS SP8, Leica), while H&E-stained sections were visualized with an Olympus bx53 upright microscope (Olympus, Japan).
Regarding WISH, Xenopus tropicalis embryos were fixed in MEMFA solution (0.1 M MOPS pH 7.4, 2 mM EDTA, 3.7% Formaldehyde) for 1 h at room temperature and preserved in ethanol at −20 °C. The digoxigenin-labeled antisense probe was synthesized by employing digoxigenin RNA-labeling mix (Roche) and T7 RNA polymerase (M0251S, New England Biolabs) based on a PCR-amplified template containing the T7 promoter. The procedure of WISH was conducted following the established protocol of Harland70, with certain alterations documented in Hollemann et al.71. The relevant PCR primers for the probes were listed in Supplementary Data 3.
As for TEM, a 1 mm × 1 mm section of skin tissue was extracted from frogs and immersed in 2% glutaraldehyde solution for 12 h at 4 °C to fix the sample. Subsequently, the tissue was washed four times with 10 mM PBS for 10 min each time, followed by fixation in 1% osmium tetroxide solution at room temperature for 3 h. The sample was again washed twice with 10 mM PBS for 10 min each time and then rinsed twice with distilled water for 10 min each time. The sample was then treated with 2% uranyl acetate solution for 2 h at room temperature or overnight at 4 °C, and washed with distilled water until the solution was clear. To prepare the tissue for embedding, a 30%, 50%, and 75% acetone solution was prepared with distilled water. The sample was sequentially treated with 30% acetone once, 50% acetone once, 75% acetone once, and 100% acetone twice, each for 10 min at room temperature. In addition, the 100% resin was prepared in advance. The method of preparation of 100% resin was to add Epoxy resin 9.8 g, DDSA 5.6 g, NMH 4.6 g, and DMP-30 0.28 mL in order, and then rotate the mixture at room temperature on a rotary mixer for at least 4 h. The 100% resin was diluted with acetone to prepare 25%, 50%, and 75% resin solutions, and the sample was sequentially treated with 25% resin-acetone once, 50% resin-acetone once, 75% resin-acetone once, and 100% resin once, each for 1.5–2 h at room temperature. Finally, the sample was treated with fresh 100% resin overnight at 4 °C, placed in an embedding box, and embedded at 60 °C for 2 days to complete the embedding. To observe and photograph the sample, 70 nm thin slices were cut using an ultramicrotome (EM UC7, Leica) and placed on copper grids (AGH100, Electron Microscopy China). The sample was imaged using a 120KV biology transmission electron microscope (HT7700 Exalens, Hitachi, Japan).
Statistics and reproducibility
Data were analyzed utilizing GraphPad Prism version 9 (GraphPad Prism Software, Inc., CA, USA) and expressed as means ± SD. Unpaired two-tailed t-tests were employed to compare two groups, with statistical significance defined at P < 0.05. Reproducibility was maintained through a minimum of three replicates. The accompanying figure legend provides comprehensive information on the data processing methodology.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of additional supplementary files
Acknowledgements
We thank Longjian Niu, Qinnan Yan and Jiarong Wu for their technical supports and Dong Liu Lab for their zebrafish and Pingyuan Zou for frog husbandry. This work was supported by Shenzhen Science and Technology Program (JCYJ20210324120205015), Guangdong Provincial Key Laboratory of Cell Microenvironment and Disease Research (2017B030301018), and Shenzhen Key Laboratory of Cell Microenvironment (ZDSYS20140509142721429). Additionally, the authors would like to dedicate this paper in honor of the forthcoming wedding of Dr. Rensen Ran and Miss Hongjiao Wang.
Author contributions
R.R. and Y.C. conceived the project. R.R. and L.L. performed the experiments and analyzed the data together with T.X., J.H. and H.H. R.R. and Y.C. wrote the manuscript with input from all the authors. All authors read and approved the final manuscript.
Peer review
Peer review information
Communications Biology thanks Takashi Kato, Avery Angell Swearer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Christina Karlsson Rosenthal. A peer review file is available.
Data availability
All study data are included in the article and/or supplemental information.
Code availability
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Rensen Ran, Lanxin Li.
Contributor Information
Rensen Ran, Email: sankeshumy@163.com.
Yonglong Chen, Email: chenyl@sustech.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-05967-3.
References
- 1.Ye W, Chen Q. Potential Applications and Perspectives of Humanized Mouse Models. Ann. Rev. Animal Biosci. 2022;10:395–417. doi: 10.1146/annurev-animal-020420-033029. [DOI] [PubMed] [Google Scholar]
- 2.Pittet MJ, Weissleder R. Intravital imaging. Cell. 2011;147:983–991. doi: 10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan C, et al. Visualizing Engrafted Human Cancer and Therapy Responses in Immunodeficient Zebrafish. Cell. 2019;177:1903–1914 e1914. doi: 10.1016/j.cell.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White RM, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osmani N, Goetz JG. Multiscale Imaging of Metastasis in Zebrafish. Trends Cancer. 2019;5:766–778. doi: 10.1016/j.trecan.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Wenner M. The most transparent research. Nat. Med. 2009;15:1106–1109. doi: 10.1038/nm1009-1106. [DOI] [PubMed] [Google Scholar]
- 7.Renshaw S, Loynes C, Elworthy S, Ingham P, Whyte M. Modeling inflammation in the zebrafish: how a fish can help us understand lung disease. Exp. Lung Res. 2007;33:549–554. doi: 10.1080/01902140701756778. [DOI] [PubMed] [Google Scholar]
- 8.Wu, X., Hua, X., Xu, K., Song, Y. & Lv, T. Zebrafish in Lung Cancer Research. Cancers15, 4721 (2023). [DOI] [PMC free article] [PubMed]
- 9.Hellsten U, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Robertis, E. & Gurdon, J. XenopusA Brief History of in Biology. Cold Spring Harb. Protoc.2021, 469–472 (2021). [DOI] [PubMed]
- 11.Tulkens, D. et al. Engraftment of Allotransplanted Tumor Cells in Adult rag2 Mutant Xenopus tropicalis. Cancers (Basel)14, 4560 (2022). [DOI] [PMC free article] [PubMed]
- 12.Nakayama T, et al. no privacy, a Xenopus tropicalis mutant, is a model of human Hermansky-Pudlak Syndrome and allows visualization of internal organogenesis during tadpole development. Dev. Biol. 2017;426:472–486. doi: 10.1016/j.ydbio.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 13.Nakajima K, Shimamura M, Furuno N. Generation of no-yellow-pigment Xenopus tropicalis by slc2a7 gene knockout. Dev. Dyn. 2021;250:1420–1431. doi: 10.1002/dvdy.334. [DOI] [PubMed] [Google Scholar]
- 14.Nakajima K, Tazawa I, Furuno N. Generation of translucent Xenopus tropicalis through triple knockout of pigmentation genes. Dev. Growth Diff. 2023;65:591–598. doi: 10.1111/dgd.12891. [DOI] [PubMed] [Google Scholar]
- 15.Taboada C, et al. Glassfrogs conceal blood in their liver to maintain transparency. Science (New York, N.Y.) 2022;378:1315–1320. doi: 10.1126/science.abl6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centeno P, Pavet V, Marais R. The journey from melanocytes to melanoma. Nat. Rev. Cancer. 2023;23:372–390. doi: 10.1038/s41568-023-00565-7. [DOI] [PubMed] [Google Scholar]
- 17.Lin J, Fisher D. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–850. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- 18.Schartl M, et al. What is a vertebrate pigment cell? Pigment Cell Melanoma Res. 2016;29:8–14. doi: 10.1111/pcmr.12409. [DOI] [PubMed] [Google Scholar]
- 19.Sumida M, et al. The first see-through frog created by breeding: description, inheritance patterns, and dermal chromatophore structure. Sci. Rep. 2016;6:24431. doi: 10.1038/srep24431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lister J, Robertson C, Lepage T, Johnson S, Raible D. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Dev. (Cambridge, England) 1999;126:3757–3767. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- 21.D’Agati G, et al. A defect in the mitochondrial protein Mpv17 underlies the transparent casper zebrafish. Dev. Biol. 2017;430:11–17. doi: 10.1016/j.ydbio.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spinazzola A, et al. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat. Genet. 2006;38:570–575. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- 23.Weiher H, Noda T, Gray DA, Sharpe AH, Jaenisch R. Transgenic mouse model of kidney disease: Insertional inactivation of ubiquitously expressed gene leads to nephrotic syndrome. Cell. 1990;62:425–434. doi: 10.1016/0092-8674(90)90008-3. [DOI] [PubMed] [Google Scholar]
- 24.Martorano, L. et al. The zebrafish orthologue of the human hepatocerebral disease gene MPV17 plays pleiotropic roles in mitochondria. Dis. Model Mech.12, dmm037226 (2019). [DOI] [PMC free article] [PubMed]
- 25.Bian, C. et al. Genome and Transcriptome Sequencing of casper and roy Zebrafish Mutants Provides Novel Genetic Clues for Iridophore Loss. Int. J. Mol. Sci.21, 2385 (2020). [DOI] [PMC free article] [PubMed]
- 26.Goding CR, Arnheiter H. MITF-the first 25 years. Genes Dev. 2019;33:983–1007. doi: 10.1101/gad.324657.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brombin A, et al. Tfap2b specifies an embryonic melanocyte stem cell that retains adult multifate potential. Cell Rep. 2022;38:110234. doi: 10.1016/j.celrep.2021.110234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parichy D, Ransom D, Paw B, Zon L, Johnson S. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Dev. (Cambridge, England) 2000;127:3031–3044. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- 29.Fukuzawa T. A wide variety of Mitf transcript variants are expressed in the Xenopus laevis periodic albino mutant. Genes Cells. 2018 doi: 10.1111/gtc.12606. [DOI] [PubMed] [Google Scholar]
- 30.Kawasaki A, et al. Mitf contributes to melanosome distribution and melanophore dendricity. Pigment. Cell Melanoma Res. 2008;21:56–62. doi: 10.1111/j.1755-148X.2007.00420.x. [DOI] [PubMed] [Google Scholar]
- 31.Kumasaka M, Sato H, Sato S, Yajima I, Yamamoto H. Isolation and developmental expression of Mitf in Xenopus laevis. Dev. Dyn. 2004;230:107–113. doi: 10.1002/dvdy.20019. [DOI] [PubMed] [Google Scholar]
- 32.Kumasaka M, Sato S, Yajima I, Goding CR, Yamamoto H. Regulation of melanoblast and retinal pigment epithelium development by Xenopus laevis Mitf. Dev. Dyn. 2005;234:523–534. doi: 10.1002/dvdy.20505. [DOI] [PubMed] [Google Scholar]
- 33.Hnizda A, Blundell TL. Multicomponent assemblies in DNA-double-strand break repair by NHEJ. Curr. Opin. Struct. Biol. 2019;55:154–160. doi: 10.1016/j.sbi.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 34.Daza P, et al. Mechanisms of nonhomologous DNA end-joining in frogs, mice and men. Biol. Chem. 1996;377:775–786. doi: 10.1515/bchm3.1996.377.12.775. [DOI] [PubMed] [Google Scholar]
- 35.Meek K, Gupta S, Ramsden D, Lees-Miller S. The DNA-dependent protein kinase: the director at the end. Immunol. Rev. 2004;200:132–141. doi: 10.1111/j.0105-2896.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 36.Spolski R, Gromer D, Leonard W. The γ family of cytokines: fine-tuning signals from IL-2 and IL-21 in the regulation of the immune response. F1000Research. 2017;6:1872. doi: 10.12688/f1000research.12202.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banach M, Robert J. Tumor immunology viewed from alternative animal models-the Xenopus story. Curr. Pathobiol. Rep. 2017;5:49–56. doi: 10.1007/s40139-017-0125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banach M, Edholm ES, Robert J. Exploring the functions of nonclassical MHC class Ib genes in Xenopus laevis by the CRISPR/Cas9 system. Dev. Biol. 2017;426:261–269. doi: 10.1016/j.ydbio.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakai Y, Nakajima K, Robert J, Yaoita Y. Ouro proteins are not essential to tail regression during Xenopus tropicalis metamorphosis. Genes Cells. 2016;21:275–286. doi: 10.1111/gtc.12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haeussler M, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo X, et al. Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development. 2014;141:707–714. doi: 10.1242/dev.099853. [DOI] [PubMed] [Google Scholar]
- 42.Zuasti A, Jiménez-Cervantes C, García-Borrón J, Ferrer C. The melanogenic system of Xenopus laevis. Arch. Histol. Cytol. 1998;61:305–316. doi: 10.1679/aohc.61.305. [DOI] [PubMed] [Google Scholar]
- 43.Maddin HC, Eckhart L, Jaeger K, Russell AP, Ghannadan M. The anatomy and development of the claws of Xenopus laevis (Lissamphibia: Anura) reveal alternate pathways of structural evolution in the integument of tetrapods. J. Anat. 2009;214:607–619. doi: 10.1111/j.1469-7580.2009.01052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hearing VJ. Determination of melanin synthetic pathways. J. Invest. Dermatol. 2011;131:E8–E11. doi: 10.1038/skinbio.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui CY, Schlessinger D. Eccrine sweat gland development and sweat secretion. Exp. Dermatol. 2015;24:644–650. doi: 10.1111/exd.12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu C, Fuchs E. Sweat gland progenitors in development, homeostasis, and wound repair. Cold Spring Harb. Perspect. Med. 2014;4:a015222. doi: 10.1101/cshperspect.a015222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shibata Y, et al. Gene expression and localization of two types of AQP5 in Xenopus tropicalis under hydration and dehydration. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014;307:R44–56,. doi: 10.1152/ajpregu.00186.2013. [DOI] [PubMed] [Google Scholar]
- 48.Simmaco M, Mignogna G, Barra D. Antimicrobial peptides from amphibian skin: what do they tell us? Biopolymers. 1998;47:435–450. doi: 10.1002/(SICI)1097-0282(1998)47:6<435::AID-BIP3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 49.Hirooka, A. et al. The gastrin-releasing peptide/bombesin system revisited by a reverse-evolutionary study considering Xenopus. Sci. Rep.11, 13315 (2021). [DOI] [PMC free article] [PubMed]
- 50.Yasutomi M, Hama T. Electron microscopic study on the xanthophore differentiation in Xenopus laevis, with special reference to their pterinosomes. J. Ultrastructure Res. 1972;38:421–432. doi: 10.1016/0022-5320(72)90080-9. [DOI] [PubMed] [Google Scholar]
- 51.Patterson LB, Parichy DM. Zebrafish Pigment Pattern Formation: Insights into the Development and Evolution of Adult Form. Annu. Rev. Genet. 2019;53:505–530. doi: 10.1146/annurev-genet-112618-043741. [DOI] [PubMed] [Google Scholar]
- 52.Nakayama A, et al. Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mech. Dev. 1998;70:155–166. doi: 10.1016/S0925-4773(97)00188-3. [DOI] [PubMed] [Google Scholar]
- 53.Lencer, E., Prekeris, R. & Artinger, K. Single-cell RNA analysis identifies pre-migratory neural crest cells expressing markers of differentiated derivatives. eLife10, e66078 (2021). [DOI] [PMC free article] [PubMed]
- 54.Huanosta-Gutierrez A, et al. TMEM16A alternative splicing isoforms in Xenopus tropicalis: distribution and functional properties. Biochem. Biophys. Res. Commun. 2014;446:1096–1101. doi: 10.1016/j.bbrc.2014.03.057. [DOI] [PubMed] [Google Scholar]
- 55.Bennett G, Marsden C, Clothier R, Waters A, Balls M. Co-existence of thyrotrophin releasing hormone and 5-hydroxytryptamine in the skin of Xenopus laevis. Comparat Biochem. Physiol. C: Comparat. Pharmacol. 1982;72:257–261. doi: 10.1016/0306-4492(82)90092-2. [DOI] [PubMed] [Google Scholar]
- 56.Flucher BE, Lenglachner-Bachinger C, Pohlhammer K, Adam H, Mollay C. Skin peptides in Xenopus laevis: morphological requirements for precursor processing in developing and regenerating granular skin glands. J. Cell Biol. 1986;103:2299–2309. doi: 10.1083/jcb.103.6.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patton EE, et al. Melanoma models for the next generation of therapies. Cancer Cell. 2021;39:610–631. doi: 10.1016/j.ccell.2021.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schartl M, et al. A mutated EGFR is sufficient to induce malignant melanoma with genetic background-dependent histopathologies. J. Invest. Dermatol. 2010;130:249–258. doi: 10.1038/jid.2009.213. [DOI] [PubMed] [Google Scholar]
- 59.Nedelkovska, H. & Robert, J. Comparative Study of Skin Graft Tolerance and Rejection in the Frog Xenopus Laevis. In Skin Grafts - Indications, Applications and Current Research. 175–196 (2011).
- 60.Ran R, et al. Disruption of tp53 leads to cutaneous nevus and melanoma formation in Xenopus tropicalis. Mol. Oncol. 2022;16:3554–3567. doi: 10.1002/1878-0261.13301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Albrecht LV, Tejeda-Munoz N, De Robertis EM. Cell Biology of Canonical Wnt Signaling. Annu. Rev. Cell Dev. Biol. 2021;37:369–389. doi: 10.1146/annurev-cellbio-120319-023657. [DOI] [PubMed] [Google Scholar]
- 62.Ngeow KC, et al. BRAF/MAPK and GSK3 signaling converges to control MITF nuclear export. Proc. Natl. Acad. Sci. USA. 2018;115:E8668–E8677. doi: 10.1073/pnas.1810498115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ploper D, et al. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc. Natl. Acad. Sci. USA. 2015;112:E420–E429. doi: 10.1073/pnas.1424576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takeda K, et al. Ser298 of MITF, a mutation site in Waardenburg syndrome type 2, is a phosphorylation site with functional significance. Human Mol. Genet. 2000;9:125–132. doi: 10.1093/hmg/9.1.125. [DOI] [PubMed] [Google Scholar]
- 65.Wang C, et al. Knockout of microphthalmia-associated transcription factor (mitf) confers a red and yellow tilapia with few pigmented melanophores. Aquaculture. 2023;565:739151. doi: 10.1016/j.aquaculture.2022.739151. [DOI] [Google Scholar]
- 66.Ide H. Transdifferentiation of amphibian chromatophores. Curr. Top Dev. Biol. 1986;20:79–87. doi: 10.1016/S0070-2153(08)60655-9. [DOI] [PubMed] [Google Scholar]
- 67.Dockray G, Hopkins C. Caerulein secretion by dermal glands in Xenopus laevis. J. Cell Biol. 1975;64:724–733. doi: 10.1083/jcb.64.3.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- 69.Kim S, Song HS, Yu J, Kim YM. MiT Family Transcriptional Factors in Immune Cell Functions. Mol Cells. 2021;44:342–355. doi: 10.14348/molcells.2021.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harland R. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/S0091-679X(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 71.Hollemann, T., Panitz, F. & Pieler, T. In situ hybridization techniques with Xenopus embryos. In A Comparative Methods Approach to the Study of Oocytes and Embryos, (Ed Richter, J. D.) 279–290 (Oxford University Press, 1999).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary files
Data Availability Statement
All study data are included in the article and/or supplemental information.
Not applicable.