Abstract
Survival from ovarian cancer depends on the resection status after primary surgery. We performed genome-wide association analyses for resection status of 7705 ovarian cancer patients, including 4954 with high-grade serous carcinoma (HGSOC), to identify variants associated with residual disease. The most significant association with resection status was observed for rs72845444, upstream of MGMT, in HGSOC (p = 3.9 × 10−8). In gene-based analyses, PPP2R5C was the most strongly associated gene in HGSOC after stage adjustment. In an independent set of 378 ovarian tumours from the AGO-OVAR 11 study, variants near MGMT and PPP2R5C correlated with methylation and transcript levels, and PPP2R5C mRNA levels predicted progression-free survival in patients with residual disease. MGMT encodes a DNA repair enzyme, and PPP2R5C encodes the B56γ subunit of the PP2A tumour suppressor. Our results link heritable variation at these two loci with resection status in HGSOC.
Subject terms: Ovarian cancer, Predictive markers
Introduction
Epithelial ovarian cancer (EOC) is a leading cause of cancer death in women1. Most patients with EOC cannot be cured as more than 70% of patients are diagnosed with advanced disease (stage III or IV)2 and because tumours develop resistance against systemic therapy3. Quality of treatment is an independent prognostic parameter in patients with EOC4. Maximal-effort cytoreductive surgery represents a major therapeutic cornerstone and improved surgical techniques have resulted in higher rates of total macroscopic tumour debulking5,6. Several analyses have shown that residual disease following primary debulking surgery is strongly associated with survival7. For example, the overall survival of patients with FIGO IIIC EOC increases from 34 months in patients with incomplete resection, to 81 months in those with complete resection8. The incorporation of extended surgical techniques in the upper abdomen such as diaphragmatic peritoneal stripping or splenectomy has been shown to further increase rates of complete tumour resection9,10, and consequently of progression-free and overall survival11,12.
Despite this progress, there are several reasons why complete cytoreduction cannot be achieved in all patients with EOC. Even in specialised centres, ~30% of patients have macroscopic residual disease after surgery13. The main reason for residual disease is disseminated miliary carcinomatosis scattered over the viscera and the meso of the small bowel14. Such residual disease is apparently due to local tumour spread, which might be influenced by biological features, and some have proposed gene expression variations associated with residual disease15,16. While the focus on biological factors influencing residual disease in EOC has been on factors originating from the tumour, there are also interactions between ovarian cancer cells and other cell types such as the connective tissue or the mesothelium17. In addition, there is evidence from the Ovarian Cancer Association Consortium (OCAC) that women who were using hormone therapy at the time of diagnosis of EOC less frequently have residual disease after surgery18. However, whether and how inherited genetic factors influence residual disease is not known. Compared to transcriptomic or proteomic approaches, a genome-wide association study (GWAS) is independent of tissue-specific differences and may be helpful to identify heritable factors for residual disease.
We hypothesised that residual disease in EOC is partially dependent on inherited factors and therefore performed GWAS analyses for risk of residual disease in a large case series of patients with ovarian cancer. The methylation and expression of candidate genes resulting from these analyses were then tested in an independent series of ovarian tumour samples with known debulking status (residual disease (RD) = 0 cm vs > 0 cm) and patient survival.
Results
Genome-wide association study for residual disease identifies rs72845444 at the MGMT locus and three further candidate genes
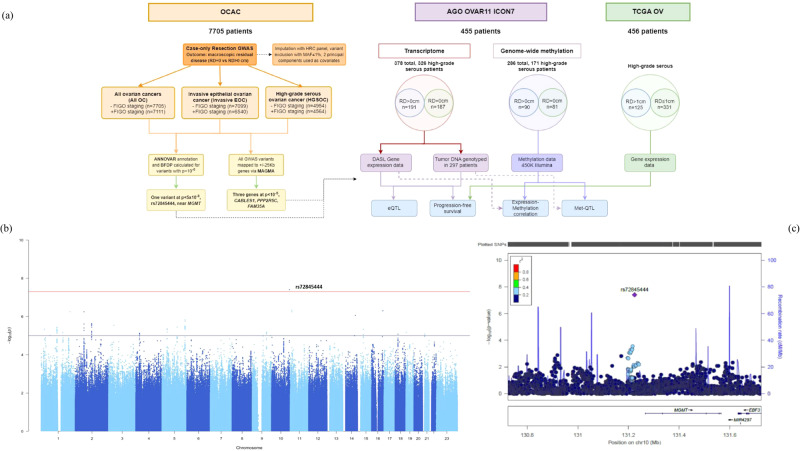
We applied a two-stage approach to identify potential genetic variants associated with resection status (Fig. 1a). First, we undertook a GWAS of resection status in a large dataset from the Ovarian Cancer Association Consortium using complete resection vs any residual disease. Second, we tested the identified variants for an effect on gene transcript levels and on gene methylation in an independent tumour bank and clinical dataset from the AGO-OVAR 11 study. We also evaluated their correlation with progression-free survival in patients with no macroscopic residual disease (RD = 0) vs patients with residual disease (RD > 0). The validity of these results was additionally tested in the TCGA data set (Fig. 1a).
Fig. 1. Workflow of the GWAS and follow-up study.
a Study workflow combining three analyses of OCAC GWAS data for overall, invasive-only and high-grade serous ovarian cancer (left) with AGO-OVAR 11 and TCGA gene expression and clinical datasets. b Manhattan plot depicting GWAS results in high-grade serous ovarian cancer (unadjusted for stage) with rs72845444 as the top hit. Blue line: p = 1 × 10−5, red line: p = 5 × 10−8. c Locus Zoom regional association plot for variant rs72845444, close to MGMT.
We extracted the OncoArray and COGS genotyping data from the Ovarian Cancer Association Consortium database19 for 7705 patients with information on resection status to perform a GWAS in a case-only design, with macroscopic residual disease (yes/no) as the binary outcome variable. Age-adjusted logistic regression analyses were performed for all OC, invasive EOC, or HGSOCs, with or without adjusting for stage, and the sample numbers for each GWAS analysis are listed in Supplementary Table 1. We identified one variant, rs72845444 (chr10:131224242:A:G; EAF = 0.02; OR = 2.11, 95%CI = 1.61–2.74; p = 3.9 × 10−8) that was strongly associated with debulking status in the high-grade serous ovarian cancer group (Fig. 1b, Table 1). This variant is located about 40 kbp upstream of the MGMT gene on chromosome 10q26.3 (Fig. 1c). When we repeated the analyses with an adjustment for FIGO stage, this association was only modestly reduced (OR = 2.11, 95% CI = 1.58–2.82, p = 4.9 × 10−7). Two further variants, rs72859096 and rs12292762 at the PARVA locus, were also highly associated (p = 6.5 × 10−8 and p = 7.4 × 10−8, respectively) but this potential association disappeared after adjustment for stage, suggesting that these variants may be linked with tumour stage. No further strong associations were found after adjustment for stage. A list of variants at p < 10−6 in all GWAS analysis, along with their allele frequencies and additionally calculated Bayesian False Discovery Probability (BFDP) scores are provided in Supplementary Table 2.
Table 1.
Outcomes from GWAS and MAGMA analyses with and without adjustment for stage
| Study Characteristics | Study Name | GIF (lambda) | SNPs above GWS | Genes from MAGMA at p < 10−5 (25Kb window) |
|---|---|---|---|---|
| Age-adjusted (invasive and non-invasive) | All OC - FIGO | 0.995 | ||
| Age-adjusted (invasive) | Invasive EOC - FIGO | 1.001 | ||
| Age-adjusted (high-grade serous) | HGSOC - FIGO | 1.011 | rs72845444 (10:131224242:A:G, p = 3.89x10−8) near MGMT | CABLES1 (p = 2.63 × 10−6) |
| Age and stage adjusted (invasive and non-invasive) | All OC + FIGO | 0.981 | FAM35A (p = 6.36 × 10−6) | |
| Age and stage-adjusted (invasive) | Invasive EOC + FIGO | 0.986 | ||
| Age and stage adjusted (high-grade serous) | HGSOC + FIGO | 1.004 | PPP2R5C (p = 4.78 × 10−6) |
Most significant variants and genes for the GWAS and MAGMA analyses in all OC (invasive and non-invasive), invasive OC and high-grade serous-only ovarian cancer (HGSOC) patient groups are noted. The analyses were run just age-adjusted or age and stage adjusted, respectively. Associations at p < 5 × 10−8 were considered significant, and MAGMA p < 10−5 was considered noteworthy for follow-up.
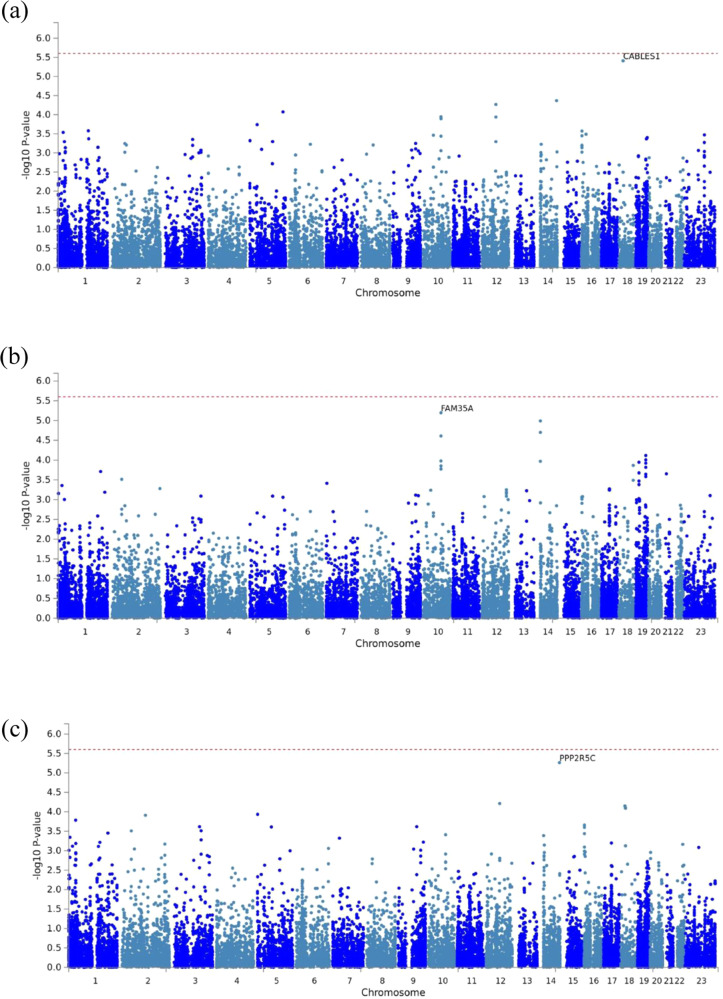
We then performed gene-based MAGMA analyses on all the GWAS datasets, to identify cumulative effects of SNPs within and around single genes (up to 25 kbp distance) (Fig. 2a–c, Supplementary Table 3). No gene passed the genome-wide significance threshold of 2.5 × 10−6 in these analyses, but three genes were identified at p < 10−5, with CABLES1 and PPP2R5C in high-grade serous ovarian cancers, without or with adjustment for stage, respectively, and FAM35A in all ovarian cancers after adjustment for stage (Table 1). That PPP2R5C and FAM35A were only associated after adjustment for stage suggested they could represent independent predictors of residual disease. The GWAS summary statistics of the most significant SNPs underlying these MAGMA gene-associations are in Supplementary Table 2.
Fig. 2. MAGMA gene-based association analyses.
Manhattan plots for the MAGMA gene-based association analyses in high-grade serous ovarian cancer without or with adjustment for stage (a, c) and in overall ovarian cancers after adjustment for stage (b). Indicated are the top genes CABLES1 (a), FAM35A (b), and PPP2R5C (c), respectively.
Association of PPP2R5C risk alleles with mRNA levels in ovarian tumours
We investigated transcript levels in ovarian cancer tissue stratified by debulking status in the genes identified as associated with residual disease. We analysed the log2-fold change in mRNA levels for seven transcripts (four from PPP2R5C and one from each of the genes MGMT, CABLES1, and FAM35A, see Supplementary Table 4 for Illumina Probe IDs) in a series of 378 tumour tissues from the AGO-OVAR 11 study20,21. In a comparison between patients undergoing complete resection vs patients with residual disease, the mRNA levels for none of these genes were statistically associated with residual disease, neither in all EOC nor in high-grade serous tumours (Supplementary Table 5). We then investigated whether the GWAS-identified variants may be expression quantitative trait loci (eQTLs) and whether their effect may be dependent on the resection status. We therefore directly genotyped the tumour samples of the AGO-OVAR 11 study for the most strongly associated variants in CABLES1 (rs77770767, rs28589524, rs6507532, rs4281829), PPP2R5C (rs2448233, rs59784377, rs3783362, rs79999043), FAM35A (rs11492866) as well as for the MGMT variant rs72845444 (Supplementary Table 6). While there was no association between variants near CABLES1 or MGMT with the mRNA levels of their respective genes, rs11492866 showed a borderline association with FAM35A levels (pTrend = 0.04) (Supplementary Fig. 1i). Furthermore, the genotypes for rs2448233, rs3783362, and rs59784377 were associated with PPP2R5C mRNA levels in an allelic dose-dependent manner, or when rare allele carriers were combined in a dominant model, respectively (Table 2, further data in Supplementary Fig. 2(viii, xii) and Supplementary Fig. 3ii, iv, xii, xvi). These associations were observed mainly for the major isoform, transcript variant 1, for which the rare alleles were associated with lower PPP2R5C mRNA levels. Additional associations were observed with minor isoforms when stratified by resection status (Table 2, Supplementary Fig. 4(vii, xvi) and Supplementary Fig. 5(ii, v, viii, ix, xii).
Table 2.
Association of PPP2R5C variants with mRNA levels of PPP2R5C isoforms
| Model | Variant ID | ILMN_1780913 | ILMN_1789283 | ILMN_2364971 | ILMN_1795846 |
|---|---|---|---|---|---|
| Isoform 1 | Isoforms 1,2,3 | Isoforms 1,2,3 | Isoform 3 | ||
| Overall allelic | rs2448233 | 0.072 | 0.200 | 0.076 | 0.569 |
| rs3783362 | 0.032 | 0.519 | 0.437 | 0.150 | |
| rs59784377 | 0.003 | 0.552 | 0.529 | 0.827 | |
| rs79999043 | 0.890 | 0.400 | 0.976 | 0.113 | |
| Overall dominant | rs2448233 | 0.041 | 0.072 | 0.024 | 0.728 |
| rs3783362 | 0.203 | 0.858 | 0.248 | 0.283 | |
| rs59784377 | 0.014 | 0.706 | 0.291 | 0.564 | |
| rs79999043 | 0.632 | 0.187 | 0.909 | 0.037 | |
| No residual disease (RD = 0) dominant | rs2448233 | 0.448 | 0.623 | 0.132 | 0.810 |
| rs3783362 | 0.635 | 0.304 | 0.643 | 0.025 | |
| rs59784377 | 0.352 | 0.063 | 0.795 | 0.435 | |
| rs79999043 | 0.978 | 0.471 | 0.381 | 0.071 | |
| Any residual disease (RD > 0) dominant | rs2448233 | 0.073 | 0.057 | 0.123 | 0.427 |
| rs3783362 | 0.003 | 0.125 | 0.543 | 0.899 | |
| rs59784377 | 0.007 | 0.104 | 0.053 | 0.806 | |
| rs79999043 | 0.544 | 0.040 | 0.580 | 0.038 |
Four PPP2R5C variant genotypes were tested for association with mRNA levels of PPP2R5C isoforms (measured by four illumine probes as indicated) in the AGO-OVAR 11 study under an allelic or dominant model, respectively. Samples were further stratified into complete resection vs residual disease (RD = 0 vs RD > 0). Transcript isoforms indicated represent NM_002719 (1), NM_178586 (2) and NM_178587 (3) in the NCBI Genbank, respectively. Probe ID ILMN_1780913 captured PPP2R5C isoform 1, ILMN_1789283 mapped onto isoforms 1, 2, and 3, ILMN_2364971 matched isoforms 1, 2, and 3, and ILMN_1795846 matched isoform 3. Bold values indicate p < 0.05.
Association of risk alleles at MGMT and PPP2R5C with gene methylation in ovarian tumours
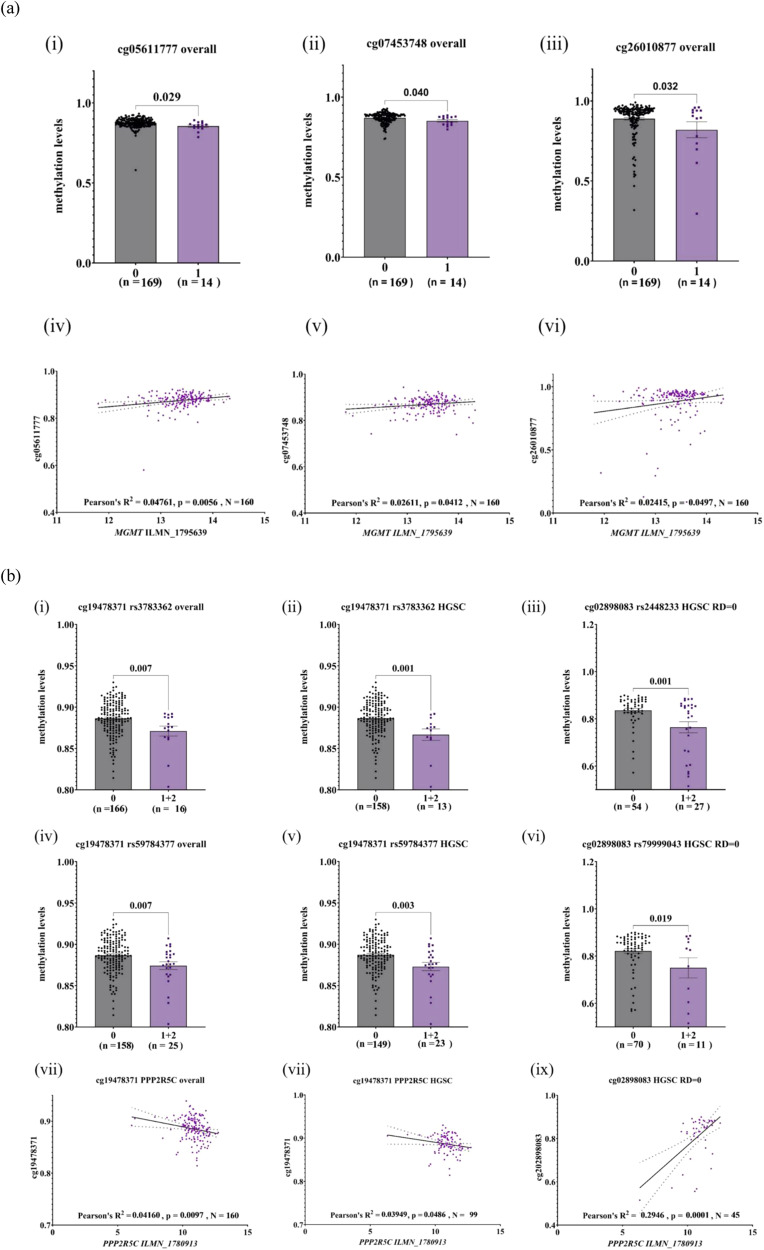
We then tested for met-QTLs in the vicinity of the genes MGMT, PPP2R5C, CABLES1 and FAM35A and found multiple potential SNP-methylation and gene transcript-methylation associations (Supplementary Table 7b). For MGMT, the risk allele of GWAS variant rs72845444 was associated with hypomethylation at three methylation sites that also correlated with lower MGMT mRNA levels (cg05611777 p = 0.006; cg07453748 p = 0.04; cg26010877 p = 0.05; N = 160) (Fig. 3a). In HGSOC samples, only cg26010877 correlated with both rs7284544 (p = 0.04) and MGMT expression (p = 0.03; N = 99). The detected effects were not due to outliers as their removal generally improved the associations. At the PPP2R5C gene, the rare alleles of variants rs3783362 and rs59784377 were associated with hypomethylation of cg19478371 overall and in HGSOC samples (Fig. 3b), and cg19478371 inversely correlated with PPP2R5C transcript levels overall (p = 0.01; N = 160) and weakly in HGSOC (p = 0.05; N = 99) (Fig. 3b). In HGSOC samples from patients with no residual disease, cg02898083 was associated with rs2448233 and rs7999043 (p = 0.001 and p = 0.02, respectively (Fig. 3b)), as well as with PPP2R5C transcript levels (p = 0.0001; N = 45) (Supplementary Table 7e and Fig. 3b). Taken together, these analyses supported a functional role for the MGMT single variant rs72845444 and suggest a more complex pattern of regulation for PPP2R5C.
Fig. 3. Methylation-QTL and correlation with expression.
CpG sites that were nominally significant met-QTLs and also correlated with gene expression at (a) MGMT and (b) PPP2R5C in overall, high-grade serous (HGSOC), HGSOC optimal or sub-optimal groups. Plotted are methylation intensity levels (y-axis) vs genotype of the specified SNP (x-axis), or log2 normalised gene expression levels of the corresponding gene transcript (x-axis). The CpG sites are indicated by Illumina cg-Probe IDs, and the Illumina probe per gene is specified by ILMN IDs. p values are indicated after unpaired t tests between two groups, or following Pearson’s correlation R2 values and number of samples (N).
Association of MGMT and PPP2R5C with progression-free survival in patients with residual disease
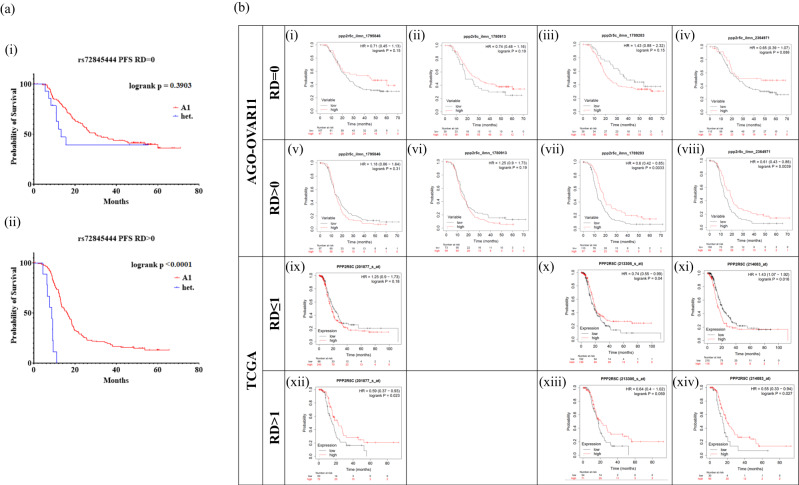
We also determined whether the GWAS-identified variants and candidate gene transcript levels were associated with progression-free survival (PFS) in the 378 patients of the AGO-OVAR 11 trial (number ISRCTN91273375). The rare allele of rs72845444 (MGMT) was associated with a worse PFS especially in patients with residual disease (logrank p < 0.001), although carrier numbers were small (10/96 with complete resection, 6/115 with residual disease; Fig. 4a(ii)). None of the other single variants were associated with PFS at p < 0.05. We then examined the impact of tumour mRNA levels of CABLES1, FAM35A, MGMT and PPP2R5C on PFS in the AGO-OVAR 11 study, and in the publicly available TCGA datasets. We found no evidence of association for CABLES1, FAM35A or MGMT. However, PPP2R5C mRNA levels positively correlated with PFS in patients with residual disease in the AGO-OVAR 11 dataset with respect to the probes that detect all three major PPP2R5C isoforms (HR 0.60, p = 0.003, and HR 0.61, p = 0.004 (Fig. 4b(vii, viii)), and this was supported by the TCGA data (HR 0.64, p = 0.059, and HR 0.55, p = 0.027) (Fig. 4b(xiii, xiv)).
Fig. 4. Progression-free survival analysis stratified by debulking status.
a Kaplan-Meier plots for patients in AGO-OVAR 11 with optimal (RD = 0, top) or suboptimal (RD > 0, bottom) debulking stratified by rs72845444 genotype. b Kaplan-Meier plots for high-grade serous patients with optimal (top) or suboptimal (bottom) debulking stratified by PPP2R5C mRNA levels in the AGO-OVAR 11 (top panel, RD = 0 vs RD > 0) and the TCGA cohort (bottom panel, RD < = 1 cm vs RD > 1 cm). PPP2R5C mRNA levels were measured by four different probes per study as indicated within the figures (Illumina IDs from the AGO-OVAR 11 dataset or specific probe set from the TCGA data accessed via KM-Plotter). For the TCGA dataset, patients were split by auto-selected best cutoff, and high-grade serous patients were chosen, followed by further selection of debulking status. Probe ID ILMN_1780913 captured PPP2R5C isoform 1, ILMN_1789283 mapped onto isoforms 1, 2, and 3, ILMN_2364971 matched isoforms 1, 2, and 3, and ILMN_1795846 matched isoform 3. Transcript isoforms indicated represent NM_002719 (1), NM_178586 (2) and NM_178587 (3) in the NCBI Genbank, respectively.
Discussion
Although some evidence has been obtained for modulation of therapeutic response by the genomic background of the patient, our knowledge about the prognostic role of genetic factors remains incomplete22–24. We have identified candidate loci associated with residual disease after primary debulking surgery in patients with advanced EOC, providing further insight into the pathophysiology of this devastating condition. Our study indicates that inherited factors are also involved in the complex scenery of residual disease after debulking surgery. All molecular-pathologic studies conducted on residual disease have so far focused on the tumour itself as the object of interest15,16, including studies that proposed underlying genetic signatures25,26. However, the prediction of resection status after primary debulking surgery in patients with EOC has proven challenging by means of existing gene expression analyses from tumour tissue20,27,28. Earlier studies of potential associations between BRCA1 or BRCA2 germline mutations and residual disease in advanced EOC revealed conflicting results, as some authors have found significant associations29, while others did not30. Our present study assumed that genome-wide association analyses could help to pinpoint potential inherited predictors of resection status.
The most significant single variant from our GWAS was rs72845444, located upstream of the MGMT gene. MGMT encodes O6-methylguanine DNA methyltransferase which repairs the mutagenic DNA lesion O6-methylguanine back to guanine and prevents mismatch and errors during DNA replication and transcription. This role may be consistent with a progressive accumulation of mutations in EOC. In our experiments, we were not able to find a significant association of rs72845444 with MGMT mRNA in the AGO-OVAR 11 samples and therefore the direction of effect could not easily be fixed at the transcript level. This could be due to the relatively low minor allele frequency of rs72845444 which limits the power of eQTL analyses. However, we obtained evidence that rs72845444 predicted gene methylation and at least one of the CpG sites near MGMT also correlated with MGMT mRNA levels. Thus, if rs72845444 exerts its effect through MGMT, it may partly occur through an effect on gene methylation to regulate gene expression and, consequently, cellular sensitivity to alkylating agents. Interestingly, promoter methylation of MGMT is a known biomarker in predicting the prognosis of patients with glioblastoma multiforme31,32. Although MGMT methylation and gross total resection have been reported as independent prognostic factors33, others found MGMT methylation to be associated with the extent of resection in this common brain tumour34.
We further investigated cumulative effects of variants using a gene-wide approach and identified three candidate genes in different analyses: CABLES1 and PPP2R5C in high-grade serous ovarian cancers without and with adjustment for FIGO stage, respectively, and FAM35A in the overall EOC analysis. In our subsequent analysis of the AGO-OVAR 11 tumour samples, the mRNA levels for PPP2R5C correlated with resection status, whereas no strong evidence was obtained for CABLES1 or FAM35A. Furthermore, we have shown associations of specific GWAS-derived genetic variants in PPP2R5C with the levels of its transcript, and this was partly dependent on the resection status. The genetic variants at PPP2R5C exhibited their association independent of each other as they are virtually unlinked (highest r2 is 0.24 for rs2448233 and rs59784377). Beyond the prediction of residual disease, we analysed the potential of PPP2R5C levels specifically to predict progression-free survival in patients with suboptimal debulking, and this was seen in both the AGO-OVAR 11 and TCGA data sets. Taken together, these results provide convergent evidence for a consistent association between germline variants in PPP2R5C, its methylation and mRNA levels, the resection status and progression-free survival.
PPP2R5C encodes the serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, B56γ, that regulates the activity of the PP2A enzyme and can direct it to cancer-specific targets of dephosphorylation, including TP53. The activation of TP53 through PP2A(B56γ) is dependent on DNA damage-induced activation of ATM which then phosphorylates TP53 as well as B56γ35. However, PPP2R5C was identified after stratification for high-grade serous cancer in our study, and most high-grade serous ovarian tumours harbour a mutation in TP53, making this pathway an unlikely explanation. More recently, a complex of PP2A/B56 with BRCA2 has been described to be required for DNA repair by homologous recombination36. This connection of PP2A/B56γ with homology-directed repair (HDR) as a positive regulator of BRCA2 function may be important for the results in our study. We found that rare alleles of GWAS variants were associated with lower PPP2R5C mRNA levels in patients with high-grade serous tumours and residual disease, suggesting lower levels of PP2A activation. It is conceivable that in such incompletely debulked tumours, impairment of HDR may have contributed to the resection status and to worse survival. PP2A is a druggable tumour suppressor that has been proposed for targeted anticancer therapy, most recently also for ovarian cancer37–39. Our data are consistent with recent observations that PP2A genes essential for cellular transformation (B56α, B56γ and PR72) are heterozygously lost in the majority of HGSC and their loss correlates with worse overall patient survival which could be antagonised by stabilisation of PP2A expressed from the remaining allele39.
This study used the screening approach of GWAS to identify inherited factors responsible for residual disease in patients undergoing primary debulking surgery. We had a large patient series from the OCAC available to identify genomic variants and genes associated with resection status. A limitation here was that we did not stratify for neoadjuvant chemotherapy due to insufficient data. Furthermore, although MGMT and PPP2R5C were supported by both genetic association and eQTL/mQTL evidence, the results were below genome-wide significance and therefore will need to be replicated in subsequent studies. We then used a well-described patient cohort from the ICON7 trial (number ISRCTN91273375) to analyse the potential impact of the identified variants on gene methylation, gene expression and progression-free survival. Although this analysis uncovered associations with both methylation and expression, the study size was limited and the role of the identified variants will warrant further investigation. Additionally, we did not have information on copy number variants or specific gene mutations in this patient set, in order to perform an adjusted mQTL analysis. Finally, different definitions of optimal vs suboptimal disease in the AGO-OVAR 11 data vs TCGA data may also have limited the comparability of stratified groups in these data sets. From a clinical point of view, it is important to point out that the results generated here should not be used to minimise surgical resection or to reduce attempts to further increase complete resection rates in each unit. Nevertheless, our study provides evidence that there are biologic reasons for residual disease, despite maximal surgical effort. As we included all stages in the GWAS, some of the genomic variants (such as those in PARVA) may act through their effect on stage. However, in the stage-adjusted analyses, the associations with MGMT and PPP2R5C variants still stood out.
In summary, our GWAS provided strong evidence for candidate genomic loci associated with resection status in patients with EOC undergoing primary debulking surgery and identified a potential role for inherited variants at two genes involved in DNA repair, MGMT and PPP2R5C, in modulating gene expression, debulking outcome and progression-free survival. Future prospective studies should test genomic markers at these genes as predictive factors for resection status and prognostic factors for survival in patients with epithelial ovarian cancer.
Methods
Patients
The studies in the Ovarian Cancer Association Consortium that contributed to the GWAS meta-analyses have been described previously (Supplementary Table 1a)19. A total of 7705 female individuals had information on residual disease (RD) after primary surgery and were included in our case-only logistic regression analysis for resection status, comparing macroscopic complete resection vs any RD. Of those, 7111 individuals had information on FIGO stage and could be included in an analysis adjusted for stage. RD was defined as the maximum dimension of disease remaining following primary debulking surgery. The actual size of residual tumour was extracted from surgery reports at each participating site and recorded in centimeters. Samples stratified by country, debulking status, FIGO stage, age, and histotype are shown in Supplementary Table 1a. For the analysis, we defined macroscopic complete resection as no residual disease (RD = 0 cm). Researchers were not blinded to resection status, and randomisation of groups was not necessary for this study. The OCAC study was approved by the Duke University Health System Institutional Review Board (IRB) under two separate protocols, one for the collection of the data (IRB Protocol #: Pro00013555), and a second for the analysis and distribution of the data (IRB Protocol #: Pro00013556). This study was conducted in accordance with the Helsinki Declaration and all participants provided signed consent. All participants were of European descent.
Ovarian tumour tissues were derived from 455 female patients of the AGO-OVAR 11 trial, the German contribution to the ICON7 multicenter phase III trial (Supplementary Table 1b)40. The median age at diagnosis for this cohort was 58.5 years (ranging from 19 to 81 years). 425 of the 455 tumour samples had been tested for genome-wide methylation, and transcriptome-wide gene expression data was available for 378 of the 455. Of the latter, 279 tumour DNA samples were available for genotyping in the present study. Patients with gene expression data (n = 378) were divided based on RD into 187 patients (49.4%) with complete resection (RD = 0), and 191 (50.6%) patients having had residual disease (RD > 0). 326/378 patients (86.2%) had high-grade serous histology, of whom 154 underwent complete resection and 172 had residual disease (Fig. 1a). PFS was calculated from the date of randomisation to the date of the first indication of disease progression or death, whichever occurred first. Disease progression was defined according to the Response Evaluation Criteria in Solid Tumors (RECIST) guidelines on the basis of radiologic, clinical, or symptomatic indicators of progression41.
GWAS analyses
The dataset from genotyped samples was imputed using the Haplotype Reference Consortium panel. We excluded variants with MAF ≤ 1% and performed age-adjusted logistic regression analyses with residual disease (yes/no) as the binary outcome variable. An initial logistic regression of residual disease by age was significant (log OR = 0.023, SE = 0.019, z = 11.93). Therefore, age was included to improve power slightly. Three initial analyses were performed: for all ovarian cancers, all invasive EOC, and limited to high-grade serous ovarian histology.
Being a case-only analysis, we tested how many principal components (PCs) should be included in the GWAS analyses. Our initial logistic regression analysis to test the association between multiple PCs and the outcome variable (residual disease status) showed that only the first two PCs contributed to the outcome (p < 0.05), whereas none of the further PCs (3–9) were significantly associated (p > 0.05). This suggested that adding any further PCs would not improve the accuracy. Therefore we only included the first two PCs for each panel (Oncoarray and COGS).
Summary statistics were visualised via Manhattan and QQ plots generated using the qqman R package42 and the genomic inflation factor (λ) was calculated via R 3.6.2. Our study was estimated to have >80% power to detect effect sizes >1.2 for variants with minor allele frequencies larger than 0.14 at a genome-wide significance level α = 5 × 10−8.
Adding stage information to the logistic regression analysis with age was highly significant (log OR = 0.26, se = 0.08, z-score = 32.8) with the pseudo-R2 going up from 0.0136 (age only) to 0.158 with age and FIGO stage. We therefore performed three further logistic regression analyses with adjustment for FIGO stage.
We also calculated the Bayes False Discovery Probability (BFDP) for all the variants with MAF > 0.01 and p < 10−6 using the genetic analysis package GAP43 in R v3.6.2. Priors of 1:1000, 1:10,000 and 1:100,000 were tested for odds ratios of 1.5 or 2, with similar outcomes (Supplementary Table 2b). A BFDP < 20% was considered strong evidence for an association.
For a gene-wide analysis of cumulative SNP effects, summary statistics were uploaded into FUMA v1.3.6144, and MAGMA v1.07245 was used to perform gene-based testing. SNPs were mapped within a 25 kb window from the transcription start site (TSS) of genes, and a genome-wide gene-based testing was performed to identify significant genes within each GWAS analysis (i.e. all OC, invasive EOC or just HGSOC, ±FIGO stage). MAGMA genome-wide significance threshold was calculated to be p = 2.5 × 10−6 after mapping variants to 20,016 protein-coding genes.
Gene expression and survival analyses
Log2 normalised gene expression data from cDNA-mediated annealing, selection, extension and ligation (DASL) assays was available for 378 patients from the AGO-OVAR 11 trial, as described previously21, along with clinical variables. For the top genes from the SNP-based ANNOVAR and MAGMA predictions, gene expression data was converted into Z scores. These data were then stratified based on resection (complete resection (RD = 0, n = 187) or residual disease (RD > 0, n = 191)) as well as by histology or grade, and t-tests were performed using R 3.6.2. to identify targets with differential expression among the tested cohorts. For in silico annotation of variants in terms of eQTL effects we used SNiPA v.3.446.
For the top genes from the SNP-based ANNOVAR and MAGMA predictions, gene expression data available from the AGO-OVAR 11 trial was used to plot gene-based progression-free survival curves for this patient cohort after stratification based on resection status (complete resection (RD = 0, n = 187) or residual disease (RD > 0, n = 191)). Gene expression data was converted into Z scores and divided into quartiles, and survival curves were plotted with custom plotting in KM plotter. In parallel, progression-free survival curves were plotted in the TCGA ovarian cancer dataset using KM plotter47 using auto-select best cutoff for a total of 456 patients with serous histology and high-grade cancer, with RD ≤ 1 cm (n = 331) and RD > 1 cm (n = 125) defined as optimal/suboptimal in the available dataset.
Variant genotyping and eQTL analysis
Tumour DNA was still available for 297 out of 378 patients from the AGO-OVAR 11 trial, from which patients with high-grade serous histology (n = 211) and complete resection (n = 96) or residual disease (n = 115) were selected for variant genotyping via SNPtype assays (Fluidigm). Assays were designed for 10 variants of interest with allele-specific primers (Fluidigm; Supplementary Table 6) and allele-specific PCR products were detected with FAM or HEX-labelled universal probes (Fluidigm). Variant genotype was then tested for association with log2 normalised DASL gene expression data for variant-gene pairs of interest under an allelic model via GraphPad Prism v9.0 using Student’s t test to compare two groups or ANOVA between three groups. A linear test for trend was performed after ANOVA as well as after a linear regression analysis to check whether the genotype was associated with the transcript levels under an allelic model. Sequences for the selected Illumina Human HT-12 WG-DASL V4.0 R2 expression bead chip assays are provided in Supplementary Table 4. eQTL analysis was also performed for patients after stratification by debulking. Variant genotypes were further tested as predictors of progression-free survival in survival analysis via GraphPad Prism v9.0.
Methylation analysis
425 of 455 FFPE tumour samples from the AGO-OVAR 11 study were bisulphite converted and run on the Illumina 450 K Infinium Methylation Beadchip. The Infinium HD FFPE Quality Check assay was performed to remove samples failing (<95% CpG detection) as part of the 450 K ICON7 project. After a rigorous quality check (see Supplementary File 2), methylation data on 286 samples remained. The methylation data, after covariable adjustment, was combined with the available genotype data, as well as the DASL expression data for the corresponding transcripts, and met-QTL and methylation-gene expression correlation analysis (using Pearson’s R) was carried out for 286 patient samples. For methylation-QTL analysis, an association of methylation with SNP genotype was tested using Student’s t test for two groups and ANOVA between three groups. From these 286 samples, 172 had high-grade serous histology, from which 1 patient did not undergo surgery, 81 had RD > 0 and 90 had RD = 0. CpG probes ± 25 kbp from the four genes of interest were analysed (189 probes for MGMT, 83 probes for PPP2R5C, 30 probes for CABLES1 and 14 probes for FAM35A). Tested probe Illumina cg IDs and SNPs per gene are mentioned in Supplementary Table 7a.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank all the study participants who contributed to this study and all the researchers, clinicians, technical and administrative staff who have made this work possible. Acknowledgements for individual studies: AUS: The AOCS also acknowledges the cooperation of the participating institutions in Australia, and the contribution of the study nurses, research assistants and all clinical and scientific collaborators. The complete AOCS Study Group can be found at www.aocstudy.org. We would like to thank all of the women who participated in this research programme; BEL: We would like to thank Gilian Peuteman, Thomas Van Brussel, Annick Van den Broeck and Joke De Roover for technical assistance; MOF: the Total Cancer Care™ Protocol and the Collaborative Data Services and Tissue Core Facilities at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center (P30-CA076292), Merck Pharmaceuticals and the state of Florida; OPL: Members of the OPAL Study Group (http://opalstudy.qimrberghofer.edu.au/); SRO: To thank all members of Scottish Gynaecological Clinical Trails group and SCOTROC1 investigators; UHN: Princess Margaret Cancer Centre Foundation-Bridge for the Cure; VAN: BC Cancer Foundation, VGH & UBC Hospital Foundation; WMH: We thank the Gynaecological Oncology Biobank at Westmead, a member of the Australasian Biospecimen Network-Oncology group. The Ovarian Cancer Association Consortium is funded by generous contributions from its research investigators and through anonymous donations. OCAC was funded by a grant from the Ovarian Cancer Research Fund (OCRF). The OCAC OncoArray genotyping project was funded through grants from the U.S. National Institutes of Health (CA1X01HG007491-01 (C.I.A.), U19-CA148112 (T.A.S.), R01-CA149429 (C.M.P.) and R01-CA058598 (M.T.G.); Canadian Institutes of Health Research (MOP-86727 (L.E.K.) and the Ovarian Cancer Research Fund (A.B.). The COGS project was funded through a European Commission’s Seventh Framework Programme grant (agreement number 223175 - HEALTH-F2-2009-223175) and in part by the US National Cancer Institute GAME-ON Post-GWAS Initiative (U19-CA148112). This study made use of data generated by the Wellcome Trust Case Control consortium that was funded by the Wellcome Trust under award 076113. The results published are in part based upon data generated by The Cancer Genome Atlas Pilot Project established by the National Cancer Institute and National Human Genome Research Institute (dbGap accession number phs000178.v8.p7). Funding for individual studies: AUS: The Australian Ovarian Cancer Study (AOCS) was supported by the U.S. Army Medical Research and Materiel Command (DAMD17-01-1-0729), National Health & Medical Research Council of Australia (199600, 400413 and 400281), Cancer Councils of New South Wales, Victoria, Queensland, South Australia and Tasmania and Cancer Foundation of Western Australia (Multi-State Applications 191, 211 and 182). AOCS gratefully acknowledges additional support from Ovarian Cancer Australia and the Peter MacCallum Foundation; BAV: ELAN Funds of the University of Erlangen-Nuremberg; BEL: National Kankerplan; CNI: Instituto de Salud Carlos III (PI 19/01730); Ministerio de Economía y Competitividad (SAF2012); HAW: U.S. National Institutes of Health (R01-CA58598, N01-CN-55424 and N01-PC-67001); HOP: University of Pittsburgh School of Medicine Dean’s Faculty Advancement Award (F. Modugno), Department of Defense (DAMD17-02-1-0669, OC20085) and United States National Cancer Institute (R21-CA267050, K07-CA080668, R01-CA95023, MO1-RR000056); LAX: American Cancer Society Early Detection Professorship (SIOP-06-258-01-COUN) and the National Center for Advancing Translational Sciences (NCATS), Grant UL1TR000124; MAC: National Institutes of Health (R01-CA2482288, P30-CA15083, P50-CA136393); Mayo Foundation; Minnesota Ovarian Cancer Alliance; Fred C. and Katherine B. Andersen Foundation; Fraternal Order of Eagles; MAL: Funding for this study was provided by research grant R01- CA61107 from the National Cancer Institute, Bethesda, MD, research grant 94 222 52 from the Danish Cancer Society, Copenhagen, Denmark, the Mermaid I project; and the Mermaid III project; MAY: National Institutes of Health (R01-CA248288, P30-CA15083, P50-CA136393); Mayo Foundation; Minnesota Ovarian Cancer Alliance; Fred C. and Katherine B. Andersen Foundation; MOF: Moffitt Cancer Center, Merck Pharmaceuticals, the state of Florida, Hillsborough County, and the city of Tampa; NCO: National Institutes of Health (R01-CA76016) and the Department of Defense (DAMD17-02-1-0666); NEC: National Institutes of Health R01-CA54419 and P50-CA105009 and Department of Defense W81XWH-10-1-02802; NOR: Helse Vest, The Norwegian Cancer Society, The Research Council of Norway; OPL: National Health and Medical Research Council (NHMRC) of Australia (APP1025142, APP1120431) and Brisbane Women’s Club; ORE: Sherie Hildreth Ovarian Cancer (SHOC) Foundation; PVD: Canadian Cancer Society and Cancer Research Society GRePEC Program; SRO: Cancer Research UK (C536/A13086, C536/A6689) and Imperial Experimental Cancer Research Centre (C1312/A15589); UHN: Princess Margaret Cancer Centre Foundation-Bridge for the Cure; VAN: BC Cancer Foundation, VGH & UBC Hospital Foundation; VTL: NIH K05-CA154337; WMH: National Health and Medical Research Council of Australia, Enabling Grants ID 310670 & ID 628903. Cancer Institute NSW Grants 12/RIG/1-17 & 15/RIG/1-16. The AGO-OVAR 11 study was funded by Roche Pharma AG.
Author contributions
The study was conceptualised by F.H., T.D., P.D.P., G.C-T., S.J.R., and A.B. GWAS analysis was performed by J.P.T. Further genotyping, data analysis and post-GWAS analysis were performed by D.R., P.Sc., and L-M.Spei. RNA sequencing and methylation data were generated by S.Ko., B.Wi. and J.Pf. OCAC database was maintained and coordinated by M.J.R. Authors on behalf of OCAC contributed samples and clinical data to OCAC. Authors on behalf of the AGO contributed samples and clinical data to the AGO-OVAR 11 study. The manuscript was drafted by D.R., F.H. and T.D., with critical input from members of the writing group (J.T., A.DF., M.J.R., A.B., P.D.P, G.C-T., S.J.R.). All authors were involved in the further editing of the manuscript and agreed with this version of the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
Summary statistics from the six GWASs in this study will be available at GWAS Catalogue (accession GCP ID: GCP000727; GCST IDs for each GWAS: All_OC_FIGO GCST90292521, All_OC_no_FIGO GCST90292522, HGSOC_FIGO GCST90292523, HGSOC_no_FIGO GCST90292524, Invasive_EOC_FIGO GCST90292525, Invasive_EOC_no_FIGO GCST90292526). OCAC summary results are available from the combined iCOGS, Oncoarray, GWAS meta-analyses and can be looked up at the OCAC website https://ocac.ccge.medschl.cam.ac.uk/data-projects/results-lookup-by-region/. Individual-level genotyping data generated in this study are not publicly available due to patient privacy requirements but can be applied for through established OCAC procedures. Derived data supporting the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
U.C. received honoraria for lectures from Lilly and AstraZeneca and is on the advisory board of AstraZeneca. J.P. received honoraria from Roche Pharma AG, AstraZeneca, Amgen, Clovis Oncology, MSD Oncology, GSK, Chugai Pharma, Teva, Medupdate, SAI MedPartners, Decision Resources, Simon-Kucher and partners, Juniper, Bionest partners, Vox Bio, Axiom healthcare strategies, Prosapient, iMed Institut, Lilly, and is a consultant/advisor for AstraZeneca, Roche, Pharma AG, Tesaro, Clovis Oncology, MSD Oncology. P.A.F. conducts research funded by Amgen, Novartis and Pfizer and received Honoraria from Roche, Novartis and Pfizer. None of these sponsors had any role in the design, data acquisition or interpretation of results in the present study.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lists of authors and their affiliations appear at the end of the paper.
Contributor Information
Thilo Dörk, Email: doerk.thilo@mh-hannover.de.
Florian Heitz, Email: F.Heitz@kem-med.com, Email: florian.heitz@gmx.net.
AOCS Group:
David Bowtell, Sian Fereday, Nadia Traficante, and Jillian Hung
OPAL Study Group:
Michael Friedlander, Andreas Obermair, Peter Grant, Vanessa Beesley, Penelope Blomfield, Alison Brand, Alison Davis, Yee Leung, James Nicklin, Michael Quinn, Karen Livingstone, Helen O’Neill, and Merran Williams
Supplementary information
The online version contains supplementary material available at 10.1038/s41525-024-00395-y.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Jelovac D, Armstrong DK. Recent progress in the diagnosis and treatment of ovarian cancer. CA Cancer J. Clin. 2011;61:183–203. doi: 10.3322/caac.20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freimund AE, Beach JA, Christie EL, Bowtell DDL. Mechanisms of drug resistance in high-grade serous ovarian cancer. Hematol. Oncol. Clin. North Am. 2018;32:983–996. doi: 10.1016/j.hoc.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Rochon J, du Bois A. Clinical research in epithelial ovarian cancer and patients’ outcome. Ann. Oncol. 2011;22:vii16–vii19. doi: 10.1093/annonc/mdr421. [DOI] [PubMed] [Google Scholar]
- 5.Tseng JH, et al. Continuous improvement in primary Debulking surgery for advanced ovarian cancer: do increased complete gross resection rates independently lead to increased progression-free and overall survival? Gynecol. Oncol. 2018;151:24–31. doi: 10.1016/j.ygyno.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norppa N, Staff S, Helminen M, Auranen A, Saarelainen S. Improved survival after implementation of ultra-radical surgery in advanced epithelial ovarian cancer: Results from a tertiary referral center. Gynecol. Oncol. 2022;165:478–485. doi: 10.1016/j.ygyno.2022.03.023. [DOI] [PubMed] [Google Scholar]
- 7.Chang S-J, Bristow RE. Evolution of surgical treatment paradigms for advanced-stage ovarian cancer: Redefining ‘optimal’ residual disease. Gynecol. Oncol. 2012;125:483–492. doi: 10.1016/j.ygyno.2012.02.024. [DOI] [PubMed] [Google Scholar]
- 8.du Bois A, et al. Role of surgical outcome as prognostic factor in advanced epithelial ovarian cancer: a combined exploratory analysis of 3 prospectively randomized phase 3 multicenter trials. Cancer. 2009;115:1234–1244. doi: 10.1002/cncr.24149. [DOI] [PubMed] [Google Scholar]
- 9.Kommoss S, et al. Prognostic impact of additional extended surgical procedures in advanced-stage primary ovarian cancer. Ann. Surg. Oncol. 2010;17:279–286. doi: 10.1245/s10434-009-0787-8. [DOI] [PubMed] [Google Scholar]
- 10.Eisenhauer EL, et al. The addition of extensive upper abdominal surgery to achieve optimal cytoreduction improves survival in patients with stages IIIC–IV epithelial ovarian cancer. Gynecol. Oncol. 2006;103:1083–1090. doi: 10.1016/j.ygyno.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 11.Chi DS, et al. Improved progression-free and overall survival in advanced ovarian cancer as a result of a change in surgical paradigm. Gynecol. Oncol. 2009;114:26–31. doi: 10.1016/j.ygyno.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Harter P, et al. Impact of a structured quality management program on surgical outcome in primary advanced ovarian cancer. Gynecol. Oncol. 2011;121:615–619. doi: 10.1016/j.ygyno.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Manning-Geist BL, et al. A novel classification of residual disease after interval debulking surgery for advanced-stage ovarian cancer to better distinguish oncologic outcome. Am. J. Obstet. Gynecol. 2019;221:326.e1–326.e7. doi: 10.1016/j.ajog.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Heitz F, et al. Pattern of and reason for postoperative residual disease in patients with advanced ovarian cancer following upfront radical debulking surgery. Gynecol. Oncol. 2016;141:264–270. doi: 10.1016/j.ygyno.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 15.Riester M, et al. Risk prediction for late-stage ovarian cancer by meta-analysis of 1525 patient samples. JNCI J. Natl Cancer Inst. 2014;106:1–12. doi: 10.1093/jnci/dju048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z, et al. Suboptimal cytoreduction in ovarian carcinoma is associated with molecular pathways characteristic of increased stromal activation. Gynecol. Oncol. 2015;139:394–400. doi: 10.1016/j.ygyno.2015.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenny HA, et al. Mesothelial cells promote early Ovarian cancer metastasis through fibronectin secretion. J. Clin. Investig. 2014;124:4614–4628. doi: 10.1172/JCI74778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brieger KK, et al. Menopausal hormone therapy prior to the diagnosis of ovarian cancer is associated with improved survival. Gynecol. Oncol. 2020;158:702–709. doi: 10.1016/j.ygyno.2020.06.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phelan CM, et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat. Genet. 2017;49:680–691. doi: 10.1038/ng.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heitz F, et al. Dilution of molecular-pathologic gene signatures by medically associated factors might prevent prediction of resection status after debulking surgery in patients with advanced ovarian cancer. Clin. Cancer Res. 2020;26:213–219. doi: 10.1158/1078-0432.CCR-19-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kommoss S, et al. Bevacizumab may differentially improve ovarian cancer outcome in patients with proliferative and mesenchymal molecular subtypes. Clin. Cancer Res. 2017;23:3794–3801. doi: 10.1158/1078-0432.CCR-16-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glubb DM, et al. Analyses of germline variants associated with ovarian cancer survival identify functional candidates at the 1q22 and 19p12 outcome loci. Oncotarget. 2017;8:64670–64684. doi: 10.18632/oncotarget.18501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talhouk A, et al. Development and validation of the gene expression predictor of high-grade serous ovarian carcinoma molecular SubTYPE (PrOTYPE) Clin. Cancer Res. 2020;26:5411–5423. doi: 10.1158/1078-0432.CCR-20-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinn MCJ, et al. Identification of a locus near ULK1 associated with progression-free survival in ovarian cancer. Cancer Epidemiol. Biomark. Prev. 2021;30:1669–1680. doi: 10.1158/1055-9965.EPI-20-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonome T, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–5486. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tucker SL, et al. Molecular biomarkers of residual disease after surgical debulking of high-grade serous ovarian cancer. Clin. Cancer Res. 2014;20:3280–3288. doi: 10.1158/1078-0432.CCR-14-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berchuck A, et al. Prediction of optimal versus suboptimal cytoreduction of advanced-stage serous ovarian cancer with the use of microarrays. Am. J. Obstet. Gynecol. 2004;190:910–923. doi: 10.1016/j.ajog.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Borley J, Wilhelm-Benartzi C, Brown R, Ghaem-Maghami S. Does tumour biology determine surgical success in the treatment of epithelial ovarian cancer? A systematic literature review. Br. J. Cancer. 2012;107:1069–1074. doi: 10.1038/bjc.2012.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SR, et al. Maximizing cancer prevention through genetic navigation for Lynch syndrome detection in women with newly diagnosed endometrial and nonserous/nonmucinous epithelial ovarian cancer. Cancer. 2021;127:3082–3091. doi: 10.1002/cncr.33625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ataseven B, et al. Clinical outcome in patients with primary epithelial ovarian cancer and germline BRCA1/2-mutation—real life data. Gynecol. Oncol. 2021;163:569–577. doi: 10.1016/j.ygyno.2021.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Hegi ME, et al. Clinical trial substantiates the predictive value of O-6-Methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin. Cancer Res. 2004;10:1871–1874. doi: 10.1158/1078-0432.CCR-03-0384. [DOI] [PubMed] [Google Scholar]
- 32.Della Monica R, et al. MGMT and whole‐genome DNA methylation impacts on diagnosis, prognosis and therapy of glioblastoma multiforme. Int. J. Mol. Sci. 2022;23:7148. doi: 10.3390/ijms23137148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gessler F, et al. Surgery for glioblastoma in light of molecular markers: impact of resection and MGMT promoter methylation in newly diagnosed IDH-1 wild-type glioblastomas. Neurosurgery. 2019;84:190–197. doi: 10.1093/neuros/nyy049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Incekara F, et al. The association between the extent of glioblastoma resection and survival in light of MGMT promoter methylation in 326 patients with newly diagnosed IDH-wildtype glioblastoma. Front. Oncol. 2020;10:1–8. doi: 10.3389/fonc.2020.01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shouse GP, Nobumori Y, Panowicz MJ, Liu X. ATM-mediated phosphorylation activates the tumor-suppressive function of B56γ–PP2A. Oncogene. 2011;30:3755–3765. doi: 10.1038/onc.2011.95. [DOI] [PubMed] [Google Scholar]
- 36.Ambjørn SM, et al. A complex of BRCA2 and PP2A-B56 is required for DNA repair by homologous recombination. Nat. Commun. 2021;12:5748. doi: 10.1038/s41467-021-26079-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perrotti D, Neviani P. Protein phosphatase 2A: a target for anticancer therapy. Lancet Oncol. 2013;14:e229–e238. doi: 10.1016/S1470-2045(12)70558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruvolo PP. The broken “Off” switch in cancer signaling: PP2A as a regulator of tumorigenesis, drug resistance, and immune surveillance. BBA Clin. 2016;6:87–99. doi: 10.1016/j.bbacli.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avelar, R. A. et al. Small molecule-mediated stabilization of PP2A modulates the Homologous Recombination pathway and potentiates DNA damage-induced cell death. Mol. Cancer Ther. 10.1158/1535-7163.MCT-21-0880 (2023). [DOI] [PMC free article] [PubMed]
- 40.Perren TJ, et al. A phase 3 trial of bevacizumab in ovarian cancer. N. Engl. J. Med. 2011;365:2484–2496. doi: 10.1056/NEJMoa1103799. [DOI] [PubMed] [Google Scholar]
- 41.Therasse P, et al. New guidelines to evaluate the response to treatment in solid tumors. J. Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 42.D. Turner S. qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. J. Open Source Softw. 2018;3:731. doi: 10.21105/joss.00731. [DOI] [Google Scholar]
- 43.Zhao JH. Gap: genetic analysis package. J. Stat. Softw. 2007;23:1–18. doi: 10.18637/jss.v023.i08. [DOI] [Google Scholar]
- 44.Watanabe K, Taskesen E, Van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1–10. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11:1–19. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arnold M, Raffler J, Pfeufer A, Suhre K, Kastenmüller G. SNiPA: An interactive, genetic variant-centered annotation browser. Bioinformatics. 2015;31:1334–1336. doi: 10.1093/bioinformatics/btu779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lánczky A, Győrffy B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J. Med. Internet Res. 2021;23:e27633. doi: 10.2196/27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics from the six GWASs in this study will be available at GWAS Catalogue (accession GCP ID: GCP000727; GCST IDs for each GWAS: All_OC_FIGO GCST90292521, All_OC_no_FIGO GCST90292522, HGSOC_FIGO GCST90292523, HGSOC_no_FIGO GCST90292524, Invasive_EOC_FIGO GCST90292525, Invasive_EOC_no_FIGO GCST90292526). OCAC summary results are available from the combined iCOGS, Oncoarray, GWAS meta-analyses and can be looked up at the OCAC website https://ocac.ccge.medschl.cam.ac.uk/data-projects/results-lookup-by-region/. Individual-level genotyping data generated in this study are not publicly available due to patient privacy requirements but can be applied for through established OCAC procedures. Derived data supporting the findings of this study are available from the corresponding author upon reasonable request.