Abstract
The yeast transcriptional adapter Gcn5p serves as a histone acetyltransferase, directly linking chromatin modification to transcriptional regulation. Two human homologs of Gcn5p have been reported previously, hsGCN5 and hsP/CAF (p300/CREB binding protein [CBP]-associated factor). While hsGCN5 was predicted to be close to the size of the yeast acetyltransferase, hsP/CAF contained an additional 356 amino-terminal residues of unknown function. Surprisingly, we have found that in mouse, both the GCN5 and the P/CAF genes encode proteins containing this extended amino-terminal domain. Moreover, while a shorter version of GCN5 might be generated upon alternative or incomplete splicing of a longer transcript, mRNAs encoding the longer protein are much more prevalent in both mouse and human cells, and larger proteins are detected by GCN5-specific antisera in both mouse and human cell extracts. Mouse GCN5 (mmGCN5) and mmP/CAF genes are ubiquitously expressed, but maximum expression levels are found in different, complementary sets of tissues. Both mmP/CAF and mmGCN5 interact with CBP/p300. Interestingly, mmGCN5 maps to chromosome 11 and cosegregates with BRCA1, and mmP/CAF maps to a central region of chromosome 17. As expected, recombinant mmGCN5 and mmP/CAF both exhibit histone acetyltransferase activity in vitro with similar substrate specificities. However, in contrast to yeast Gcn5p and the previously reported shorter form of hsGCN5, mmGCN5 readily acetylates nucleosomal substrates as well as free core histones. Thus, the unique amino-terminal domains of mammalian P/CAF and GCN5 may provide additional functions important to recognition of chromatin substrates and the regulation of gene expression.
Transcription is a complex process requiring the coordinate action of multiple basal and transactivating proteins. In eukaryotic cells, this process is complicated further by the packaging of DNA into chromatin. Nucleosomes provide the fundamental repeat unit of chromatin, consisting of two molecules of each of the four core histones (H2A, H2B, H3, and H4) and ∼146 bp of DNA wound in almost two turns around the exterior of the histone octamer (37). Individual nucleosomes as well as more highly folded structures are generally inhibitory to the initiation of transcription. Alterations in nucleosomal structure and in chromatin packing often accompany transcriptional activation (12).
Posttranslational acetylation of the histones has long been correlated with transcriptional activation (36, 39, 40). Acetylation neutralizes the charge associated with epsilon amino groups of lysine residues, thereby loosening contacts between the histones and the negatively charged DNA. Histone acetylation also influences compaction of nucleosomal arrays, yielding less condensed chromatin structures (16). Both of these effects can increase transactivator binding to nucleosomal DNA, facilitating transcriptional activation.
A molecular basis for the linkage between histone acetylation and gene activation was provided by the discovery that the yeast transcriptional adapter Gcn5p serves as the catalytic subunit of a histone acetyltransferase type A activity (5). Gcn5p is associated with two multisubunit complexes in yeast, which include Ada proteins (Ada2p, Ada3p, and Ada5p) and/or certain Spt proteins (6, 14, 18, 24, 25, 31). These complexes are required for transcriptional activation by particular transactivators, including heterologous VP16 derivatives and endogenous Gcn4p (3, 13, 24, 34). Components of the Gcn5p-Adap complex contact both transactivator proteins and basal transcription proteins, thus providing an adapter or coactivator function, in addition to histone acetyltransferase activity (2, 34). Association with both Ada2p and acetyltransferase activity is required for Gcn5p function in vivo (8, 38).
Human homologs of GCN5 have been cloned based on sequence and functional similarities of their predicted products to the yeast protein. A cDNA predicted to encode a protein of similar size and with overall homology to yeast Gcn5p has been described (7, 41). A human ADA2 gene has also been cloned, indicating a conservation of adapter and histone acetyltransferase functions across species (7). In addition, a cDNA encoding a second, larger Gcn5-related protein that possesses unique sequences in its amino-terminal half has been identified. This protein, P/CAF (p300/CREB binding protein [CBP]-associated factor), associates with two highly related proteins, p300 and CBP, that have a region of homology with ADA2 (41). Interestingly, p300 and CBP are also histone acetyltransferases (1, 29). Interactions between P/CAF and p300 or CBP are disrupted by the viral E1A oncogene product, and this disruption is required for cellular transformation by E1A (41). Proper association of these histone acetyltransferase activities, then, is extremely important for normal cell growth (32).
In order to further study the functions of histone acetyltransferases in the growth and development of mammalian cells, we endeavored to isolate sequences encoding mouse GCN5 (mmGCN5) and mmP/CAF. To our surprise, although our mmGCN5 exhibited 98% identity with the reported human GCN5 (hsGCN5) sequence, the mouse cDNA encoded an extended amino-terminal domain with high similarity to a corresponding domain in P/CAF. Upon further examination, we found that the reported hsGCN5 cDNA (41) may result from an incompletely spliced transcript, and that a more prevalent transcript exists that potentially encodes a longer hsGCN5 protein similar to that encoded by the mouse cDNA that we isolated. Moreover, in contrast to previous reports that yeast and human GCN5 proteins acetylate only free core histones, the full-length recombinant mmGCN5 protein containing this extended amino-terminal region acetylates both free and nucleosomal histones H3 and H4. These results suggest that this additional domain in the mammalian GCN5 acetyltransferase facilitates chromatin recognition. Interestingly, P/CAF and GCN5 are expressed in inverse ratios in many mouse tissues, indicating that these proteins may serve tissue-specific functions.
MATERIALS AND METHODS
cDNA library screening.
Nested PCR with degenerate oligomers and a mouse embryonic cDNA library (13.5 days postcoitum [dpc]) as the template was performed to generate a fragment of the mmGCN5 cDNA. Oligomers were chosen from regions of sequence conserved between yeast and Tetrahymena, which correspond to amino acids 131 to 244 of the yeast protein sequence. A single band of 123 bp was generated and cloned into pBluescript (Stratagene). Sequencing revealed 80% nucleotide identity and 94% identity at the amino acid level to the reported hsGCN5. This PCR product and human EST clones (IMAGE clone no. 243927) with similarity to GCN5 were used together to screen a cDNA library under conditions of low stringency as previously described (11). Clones were plaque purified and rescued as per the manufacturer’s protocol. Sequencing revealed two types of clones, some with similarity to hsGCN5 and some with similarity to hsP/CAF. All of the P/CAF clones contained only a short piece of P/CAF, and rescreening of the library failed to isolate any longer clones. Therefore, an oligomer corresponding to the 5′-most sequence of mmP/CAF was used to screen a 10.5-dpc embryonic mouse plasmid library with GeneTrapper technology (Gibco BRL). Additional clones, corresponding to full-length P/CAF sequences, were isolated according to the manufacturer’s protocol.
Genomic library screening.
A mouse genomic library, Lambda FIXII (Stratagene), was screened by using a mixture of a 5′ fragment of the mmGCN5 cDNA and a 5′ fragment of the mmP/CAF cDNA. Positive plaques were picked and subjected to secondary screening. Phage DNA was prepared from positive plaques by standard procedures. Genomic inserts were released from phage DNA by NotI digestion and subsequently subcloned into Bluescript KS(+) (Stratagene).
Sequencing analysis.
DNA sequencing was performed by using the Thermo-Sequenase radiolabeled terminator cycle sequencing kit (Amersham Life Science). Sequencing amplification conditions were 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min for 40 cycles. Alternatively, automated sequencing was carried out by the Sequencing Core Facility at the M. D. Anderson Cancer Center.
Sequence alignment.
Published sequences were obtained by searching the GenBank, PIR-Protein, and SWISS-PROT databases. Sequence alignment was carried out with the Genetics Computer Group (GCG) (Wisconsin Package version 9.1; GCG, Madison, Wis.) Pileup program. Percent identity between two proteins was calculated by using the GCG Bestfit program.
Linkage analysis mapping.
Restriction fragment length polymorphisms for mmGCN5 or mmP/CAF in C57BL/6J and SPRET/Ei subspecies were determined by using genomic DNA purchased from the Jackson Laboratory. The Jackson Laboratory interspecific backcross panel (C57BL/6JEi × SPRET/Ei)F1 × SPRET/Ei, known as Jackson BSS (33), was then used to map the chromosomal locations of the mmGCN5 and mmP/CAF genes. Predigested panels (BglII digestion for P/CAF or XbaI digestion for GCN5) were analyzed by Southern blotting with a GCN5 or P/CAF intronic probe. Typing results were processed via the Jackson Laboratory database analysis (see http://www.jax.org/resources/documents/cmdata for raw data).
RT-PCR.
Isolation of total RNA from various mouse tissues was performed as described previously (10). RNA was digested with RNase A-free DNase I (Ambion) for 30 min at 37°C. Reverse transcriptase PCR (RT-PCR) was performed with an RT-PCR kit (Perkin-Elmer) according to manufacturer’s protocols. Reverse transcription was carried out at 42°C for 15 min, followed by heating at 95°C for 5 min. PCRs were carried out at 95°C for 60 s and 60°C for 60 s for 35 cycles as suggested by the manufacturer. Primer A (see Fig. 3B for sequence location) for RT-PCR is CTGGTGCCTGAGAAGAGGAC; primer B (see Fig. 3B) is CTCCGAAGGTGGCATGGTGAAG.
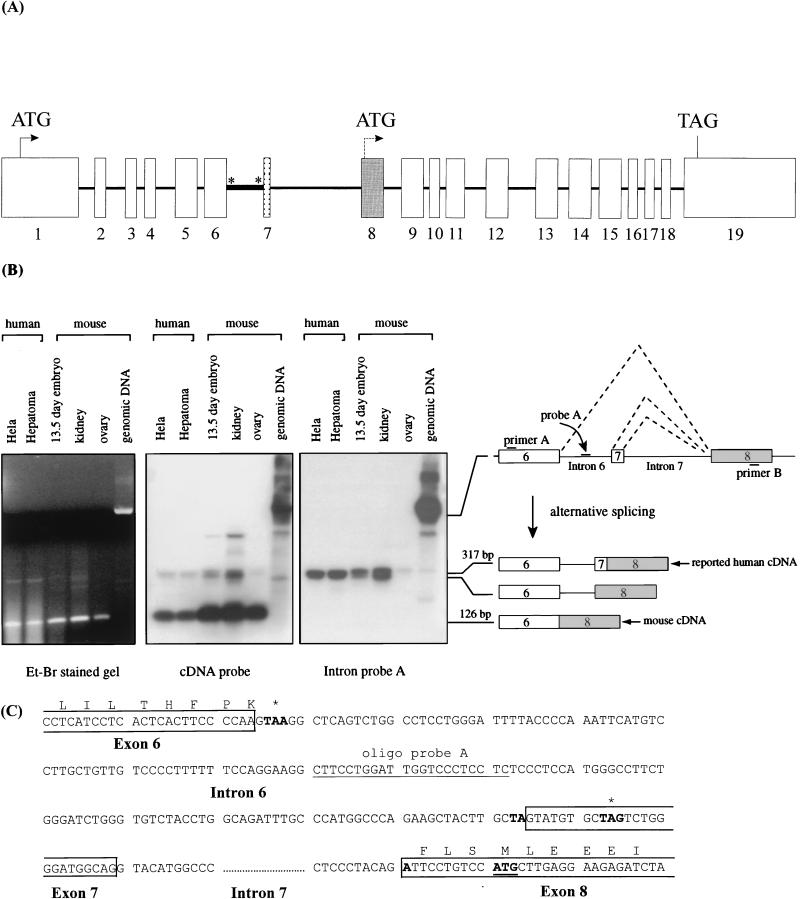
FIG. 3.
(A) Genomic structure of mmGCN5. Exons in the mmGCN5 gene are shown in boxes, while introns are represented by the intervening lines. The thick line indicates the intron that is retained in some alternatively spliced variants and which is homologous with sequences found in the previously reported 5′ untranslated region of the hsGCN5 cDNA. The asterisks indicate in-frame stop codons that would prevent translation of the full-length protein. Exon 7 is the exon that is skipped in the mouse cDNA. Exon 8 is the first coding exon in the reported hsGCN5 cDNA. The positions of the translation start codon (ATG) and the termination codon (TAG) are indicated. (B) Coexistence of multiple forms of mammalian GCN5 transcripts. Left panel, DNase I-treated RNAs from mouse kidney, ovary, and embryo and human HeLa and hepatoma cell lines were RT-PCR amplified by using primers A and B shown in the diagram on the right. RT-PCR products were resolved on an ethidium bromide (Et-Br)-stained agarose gel. Mouse genomic DNA was also amplified under the same conditions. A prevalent product corresponding to the size of the mouse cDNA (without introns 6 and 7) was amplified from all of the RNA samples, while other, larger products corresponding to the size of the reported human cDNA (containing intron 6 but not intron 7) were barely detected. Middle panel, the RT-PCR products were transferred to nylon membrane and hybridized with mmGCN5 cDNA sequences. Right panel, the same blot from the middle panel was stripped and rehybridized with a probe (probe A) specific to the conserved intron 6. (C) The RT-PCR products described above were gel purified and then amplified by PCR with a nested pair of primers. The PCR products were then subjected to DNA sequencing. The nucleotide sequence of the 317-bp fragment is shown. The smaller (292-bp) fragment has the same sequence as the larger fragment except that it lacks exon 7 sequences. Exons are boxed, and introns are numbered underneath. In-frame stop codons are in boldface and marked by asterisks. The mmGCN5 reading frame separated by intron 6 and exon 7 is shown. The initiator methionine codon for the reported hsGCN5 (41) protein is in boldface and underlined.
RNA analysis.
Total RNA from adult mouse tissues or whole embryos (13.5 dpc) was extracted as described previously (10). RNAs were electrophoresed on a 1.1% agarose gel containing formaldehyde, along with RNA molecular size markers (Gibco BRL). RNA was transferred to a GeneScreen Plus membrane (NEN Life Science) in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Hybridization was carried out with mmGCN5- and mmP/CAF-specific probes.
GCN5 protein analysis.
Mouse embryos (12.5 dpc) were homogenized in radioimmunoprecipitation assay buffer (1× phosphate-buffered saline, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 100 μg of phenylmethylsulfonyl fluoride per ml, 1 μg of aprotinin per ml) and then centrifuged at 15,000 × g for 20 min at 4°C. Supernatant was collected for Western blotting, and GCN5 was immunoprecipitated with the polyclonal hsGCN5 antibody (generously provided by Shelley Berger, Wistar Institute) by the protocol of Santa Cruz Biotech, Inc. HeLa cell nuclear extract was kindly provided by Warren Liao and Yongsheng Ren (M.D. Anderson Cancer Center).
Cloning and expression of full-length mmGCN5 and mmP/CAF.
Comparison of the mmGCN5 genomic and cDNA clones revealed that the isolated cDNA lacks the sequences encoding the first 74 amino acids. These sequences (which lack introns) were excised from the GCN5 genomic clone by NcoI and BssHII digestion and inserted into the appropriate position of the cDNA clone to generate a full-length mmGCN5 cDNA, as verified by DNA sequencing. Full-length mmGCN5 was subcloned into the NcoI and HindIII sites of the pRSETB vectors (Invitrogen), such that an N-terminal His6 tag was fused in frame with the coding region. Similarly, full-length mmP/CAF was subcloned into the BamHI and KpnI sites of the pRSETB vector. His6-tagged proteins were induced in BL21-DE3 bacterial cells by addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Recombinant protein was purified by using nickel-nitrilotriacetic acid resin (Qiagen) according to the manufacturer’s protocol. Purified recombinant proteins were verified by Western blot analysis with an antibody specific to the His6 tag (Clontech).
Acetyltransferase assays.
Acetyltransferase assays were performed as previously described (4, 5). HeLa cell mononucleosomes or core histones were the kind gift of Jerry Workman, and the cysteine-linked peptides (corresponding to amino acids 1 to 20 of H3 or to this same region with substitution of acetyl-lysine at positions 9 and 14) were the gift of C. David Allis. Calf thymus histones were purchased from Worthington Biochemical Corporation (Freehold, N.J.). Acetylation assays were performed in 10- to 30-μl volumes with either 10 μg of histones or the indicated amount of synthetic peptide. Following incubation at 30°C for 30 min, an aliquot of each reaction mixture was processed for liquid scintillation counting (P81 filter assay) as described by Brownell et al. (5), and when appropriate, another aliquot was electrophoresed on an SDS–22% polyacrylamide gel and histones were visualized by Coomassie blue staining and autoradiography.
GST fusion protein interaction assays.
Glutathione S-transferase (GST)–CBP/p300 interaction assays were performed as described by Yang et al. (41) except that crude bacterial lysates containing His-tagged recombinant P/CAF, GCN5, or HIRA were used and the interactions were detected by Western blotting with the His tag antibody.
RESULTS
Cloning of mmGCN5.
In order to study the function of acetyltransferases in a mammalian system, we endeavored to clone mouse GCN5 homologs. First we generated a fragment of the mmGCN5 cDNA using a nested PCR strategy employing degenerate primers homologous to conserved regions of the yeast GCN5 and the Tetrahymena p55 genes. To further enhance the probability of identifying GCN5-related sequences, this fragment was used together with a human GCN5 EST to screen a 13.5-dpc mouse embryonic cDNA library under conditions of low stringency (11). Multiple positive clones were identified, and upon sequencing, these were found to contain open reading frames predicted to encode proteins with significant homology to either hsGCN5 or hsP/CAF (Fig. 1).
FIG. 1.
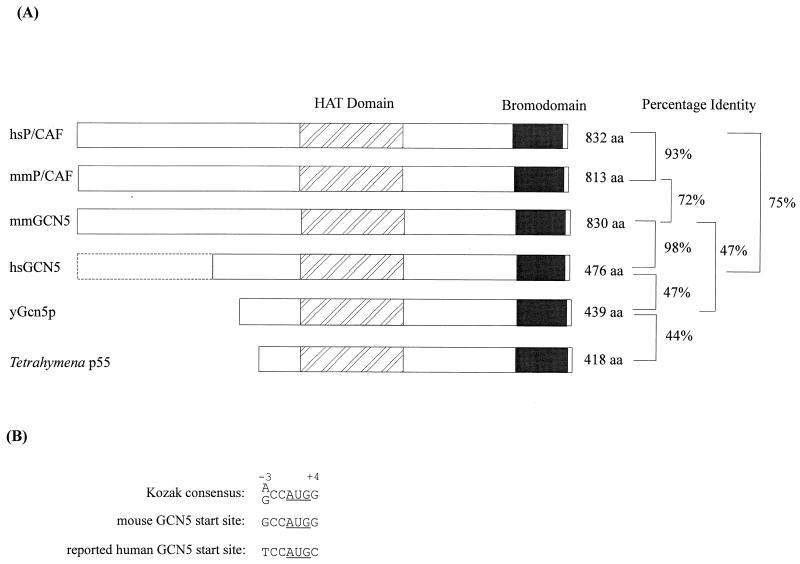
Alignment of the mmGCN5, mmP/CAF, and reported hsGCN5 amino acid sequences. Identical amino acids are shaded. Amino acid deletions are indicated with a dotted line. The locations of the histone acetyltransferase (HAT)/acetyl coenzyme A binding regions (20, 38) and the bromodomain motif (17) are indicated. The full bromodomain likely encompasses amino acids 363 to 472 of hsGCN5 (19).
One cDNA clone contained an open reading frame encoding 756 amino acids, and the C-terminal portion of this predicted amino acid sequence exhibited 98% identity with the reported hsGCN5 sequence, but only 71% homology to the hsP/CAF sequence, over the length of the predicted proteins (Fig. 2A). We tentatively concluded that this cDNA clone likely contains the mmGCN5 gene, as confirmed below.
FIG. 2.
Comparison of GCN5 and P/CAF sequences across species. (A) Schematic comparisons of mmGCN5 and mmP/CAF sequences with GCN5 and P/CAF proteins from other species. Published sequences were obtained by searching the PIR-Protein and SWISS-PROT databases. Positions of the putative catalytic domains and the bromodomains are indicated above the diagram. Percent identities between proteins are indicated on the right. yGcn5p, yeast Gcn5p; HAT, histone acetyltransferase; aa, amino acids. The dotted box indicates the existence of a predicted extended amino-terminal region in the hsGCN5 protein. (B) Comparison of the Kozak consensus sequence for translation start sites, the predicted mmGCN5 translation start sites, and the previously reported hsGCN5 translation start site (41). The underlined AUG is the codon for the initiator methionine. The A of the AUG is designated +1. In the consensus sequence, the nucleotides at positions −3 and +4 have the greatest impact on translation efficiency; 97% of vertebrate mRNAs have a purine (A or G, preferably A) in position −3, and 46% have a G in position +4 (21, 22).
We next used a fragment from the 5′ end of this clone to screen a library of mouse genomic sequences. Three different clones were isolated, and restriction analysis and sequencing indicated that all three clones harbored the entire mmGCN5 gene. Comparison of the genomic and cDNA clones of mmGCN5 revealed that the cDNA clone isolated as described above actually lacked the first 74 amino-terminal codons and that the mmGCN5 gene is divided into 19 exons and contains relatively small (85-bp to 1-kb) introns (Fig. 3A). We inserted sequences from the genomic clone containing the missing amino-terminal codons into the cDNA clone to generate a full-length (encoding 830 amino acids) recombinant mmGCN5 cDNA.
The two previously reported hsGCN5 sequences differ in the position of the initiating methionine, such that one reported sequence contains 49 additional amino-terminal amino acids relative to the other (7, 41). The mmGCN5 open reading frame also encodes these additional amino acids, but the open reading frame is further extended for some distance upstream of these sequences, potentially encoding 356 additional amino acids. The context of the predicted translation initiation site in this extended region of mmGCN5 matches well the Kozak consensus sequence (Fig. 2B) (21, 22). Moreover, the amino acids in this amino-terminal extension exhibit more than 66% identity to sequences in the corresponding regions of both mouse (see below) and human P/CAF, and the length of this extended region is similar to that of the P/CAF proteins. These data indicate that mmGCN5 encodes a protein that is very homologous to yeast Gcn5p and is almost identical to the previously reported hsGCN5 but that contains an extended N-terminal domain homologous to P/CAF in both size and sequence.
Incomplete splicing might yield a shorter GCN5 protein in mouse and human cells.
We were interested in determining the basis of the incongruity in size between mmGCN5 and the reported human cDNA. Inspection of the mmGCN5 genomic sequence revealed the presence of an intron (intron 6 in Fig. 3A) 10 bp upstream of the previously reported upstream-most hsGCN5 translation initiation site (41). Sequences highly similar (91% identical) to these intronic sequences are also present in the predicted 5′ untranslated region of the reported hsGCN5 cDNA but are absent in the mouse cDNA we isolated as described above. These comparisons suggest either that the mouse and human GCN5 genes are subject to differential splicing events, in which this intron is either removed (mouse) or retained (human), or that the previously identified human cDNA sequence is incomplete. Interestingly, a conserved, in-frame stop codon is found near the beginning of intron 6, and retention of this intron would prevent translation of the larger protein in both mouse and human cells, perhaps yielding a smaller protein with a size corresponding to that previously predicted for hsGCN5.
To investigate the possibility of alternative (or incomplete) splicing of mouse and human GCN5 transcripts, we performed RT-PCR on total RNA isolated from human HeLa cells, human hepatoma cells, mouse kidney, mouse ovary, and a 13.5-dpc mouse embryo. All RNAs were treated with RNase-free DNase I before RT-PCR to remove any genomic DNA from the samples. An mmGCN5 genomic DNA clone was used in a separate reaction, as a positive control for the presence of the intron sequences. Two primers corresponding to conserved sequences in exons 6 and 8, which flank introns 6 and 7 (Fig. 3A and B), were used for the amplification. The RT-PCR products were separated on an agarose gel, transferred to a membrane, and then probed sequentially with mmGCN5 cDNA sequences or intron 6 sequences.
A predominant RT-PCR product of a size corresponding to the spliced cDNA (lacking the intron) was amplified from mouse embryonic, kidney, and ovarian RNAs (lower band in Fig. 3B). As expected, this product was significantly smaller (126 bp) than the amplification product from the genomic DNA (about 1 kb), which contains introns 6 and 7. This small product hybridized to the mmGCN5 cDNA sequences but not to the intron 6 probe, consistent with the removal of these intronic sequences by splicing. In contrast, two less abundant, closely spaced bands were detected by both the cDNA and the intron 6 probes. An intron 7 probe hybridized only to the genomic DNA but failed to detect any of the RT-PCR products (data not shown), suggesting that intron 7 had been removed in all of the transcripts. Sequencing of the larger, closely spaced RT-PCR products revealed that they represent two alternatively spliced variants of mmGCN5 (Fig. 3C). Both of these variants retained intron 6, but one also contained a novel 25-bp exon (exon 7) located between introns 6 and 7. Intron 7 was removed from both of these alternatively spliced products, bringing the stop codons in intron 6 to a position just upstream of the ATG sequence corresponding to the previously predicted translation start site of hsGCN5. Together these data indicate that the predominant form of the mouse cDNA is completely spliced, lacks these stop codons, and therefore is predicted to encode the longer version of GCN5. However, the two minor RT-PCR products that we observed might encode shorter GCN5 proteins, consisting of the amino-terminal, P/CAF-like domain in isolation or of the C-terminal domain, which is most similar to yeast GCN5.
RT-PCR of total RNA from human cells revealed a similar mixture of completely and incompletely spliced RNAs. For example, two RT-PCR products were generated from the human HeLa cell and hepatoma cell RNAs. The size of the more abundant, smaller product again is consistent with a spliced cDNA lacking sequences homologous to the mouse intron 6 and exon 7, and this product hybridizes only to cDNA sequences. The less abundant, larger product hybridizes to both intron and exon sequences (Fig. 3B, middle panel). We suggest that the longer product likely corresponds to the hsGCN5 cDNA sequences previously reported, whereas the more prevalent, shorter form represents a spliced product predicted to encode a longer protein analogous to that encoded by the mouse cDNA isolated as described above.
Long GCN5 proteins are present in both human and mouse cells.
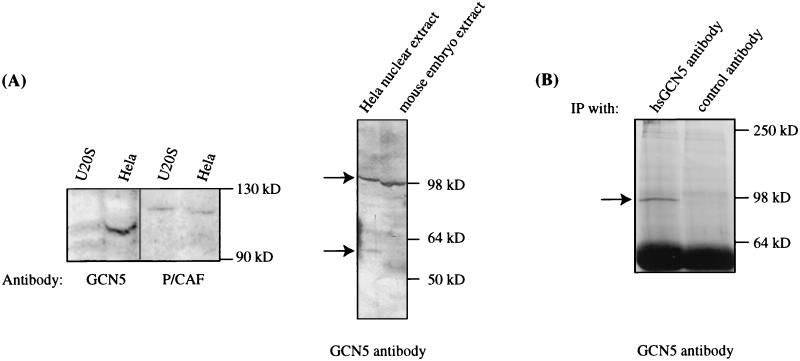
To identify the size of the native mammalian GCN5 protein(s), total cell extracts prepared from a 12.5-dpc mouse embryo or human HeLa cells were probed with a polyclonal serum raised against the previously described hsGCN5 (generously provided by Shelley Berger, Wistar Institute). The hsGCN5-specific antiserum detected a 98-kDa protein in the HeLa cell nuclear extracts, consistent with the predicted size of the full-length GCN5 protein containing the extended amino-terminal region (Fig. 4A, left panel). To ensure that this band corresponded to mmGCN5 and that the hsGCN5 antibody did not cross-react with P/CAF, we compared the relative signals obtained with the hsGCN5 antibody and a P/CAF antibody (generously provided by Yoshihiro Nakatani, National Institutes of Health) with extracts from U2OS cells or HeLa cells. The P/CAF antibody recognized a single band in the U20S extract, consistent with previous reports that P/CAF is well expressed in these cells (41), and in the HeLa cell nuclear extract. The hsGCN5 antibody, however, did not recognize any proteins of a similar size in either extract but did recognize a prominent band of ∼98 kDa in the HeLa cell nuclear extract. Therefore, the hsGCN5 antibody does not appear to cross-react significantly with PCAF, and we conclude that the 98-kDa protein recognized by this antibody in HeLa cell extracts is GCN5.
FIG. 4.
Detection of both long and short GCN5 proteins. (A) Left panel, protein extracts were prepared from U2OS cells or HeLa cell nuclei and probed with polyclonal antibodies to hsP/CAF or hsGCN5, as indicated. Right panel, protein extracts were prepared from 12.5-dpc mouse embryos or HeLa cells and probed with the hsGCN5 antibody. (B) GCN5 proteins were immunoprecipitated (IP) from 12.5-dpc mouse embryos and then probed with hsGCN5 antibody. An unrelated HIRA polyclonal antibody was used as a negative control.
The hsGCN5 antibody also recognized a faint 60-kDa band (lower arrow in right panel of Fig. 4A) in the HeLa cell extracts, close to the predicted size of the shorter GCN5 protein described previously (38) and above. Thus, both the long and short forms of GCN5 appear to be expressed in these cells, but the longer form appears to be predominant. Interestingly, the long form of GCN5 was the only form detected in mouse embryo extracts. The expression of GCN5 protein in the embryonic extracts is consistent with high levels of GCN5-specific RNA detected in these tissues (see Fig. 5). Moreover, since only very low levels of P/CAF RNA were detected at this (or any) stage of mouse embryogenesis (data not shown and see Fig. 5), these data further support our conclusion that the hsGCN5 antibody recognizes mmGCN5 rather than mmP/CAF. Neither the long nor the short form of GCN5 was detected by control, preimmune serum in either the mouse or human extracts (data not shown).
FIG. 5.
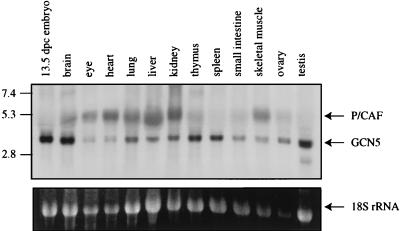
Ubiquitous and complementary expression of mmGCN5 and mmP/CAF. (Top panel) Total RNA was isolated from various mouse tissues and embryos as indicated. Northern blot hybridization was performed with a mixture of mmGCN5 and mmP/CAF cDNA probes. Two transcripts were detected and are indicated by arrows on the right. The identities of the transcripts were confirmed by Northern blot hybridization with single GCN5 or P/CAF probes (data not shown). Positions of RNA molecular size standards (in kilobases) are shown on the left. (Bottom panel) Ethidium bromide-stained gel showing 18S rRNAs.
We also used the anti-hsGCN5 serum to immunoprecipitate GCN5 proteins from the mouse embryo extract. Precipitated proteins were then detected by Western blotting with the same serum. Again, a 98-kDa protein was detected by the hsGCN5 antibody but not by a control rabbit serum (Fig. 4B). Unfortunately, the shorter form of GCN5, if it was present, would comigrate with the immunoglobulin G band and thus could not be detected by this approach. Nevertheless, these experiments confirm the presence of the longer GCN5 protein in mouse embryos.
Cloning of mmP/CAF.
A second GCN5-related cDNA clone that contained a high degree of similarity to hsP/CAF was isolated in our screen of the mouse cDNA library. Since all initial clones appeared to be incomplete, containing an 867-bp fragment of the cDNA (relative to the human sequence), a second library was screened by using GeneTrapper technology. Multiple full-length cDNAs containing an open reading frame predicted to encode 813 amino acids were obtained. This open reading frame exhibited 93% identity to the hsP/CAF cDNA sequence but only 75% identity to the reported hsGCN5 cDNA sequence (41). We therefore designated this clone mmP/CAF. Both the mmGCN5 and the mmP/CAF sequences possess predicted catalytic domains and bromodomains identified in a number of recently identified histone acetyltransferases, including several highly conserved amino acids near the putative catalytic center (Fig. 1).
Using a fragment from the 5′ region of the mmP/CAF cDNA as a probe, we identified multiple clones from a library of mouse genomic sequences that contained P/CAF sequences. Four of these contained different portions of the cDNA sequence. These clones indicate that in contrast to the mmGCN5 gene, which contains small introns (a few hundred base pairs each), the mmP/CAF gene contains very large introns (16 to 20 kb). Because of these large introns, we have not completed cloning of mmP/CAF genomic sequences.
Interestingly, several clones identified in our genomic screens apparently contain a P/CAF pseudogene. No intronic sequences are present in these clones, and several base substitutions, relative to the cDNA sequence, are scattered throughout the predicted coding region of the pseudogene. RT-PCR analysis indicates that the pseudogene is not expressed in several mouse tissues examined, including brain, eye, heart, lung, liver, kidney, thymus, spleen, fat, diaphragm, small intestine, ovary, testis, or a 13.5-dpc embryo (data not shown).
Ubiquitous but complementary expression of mmGCN5 and mmP/CAF.
To examine and compare the expression of mmGCN5 and mmP/CAF, total RNA was extracted from various mouse tissues, subjected to denaturing electrophoresis, transferred to a membrane, and then probed with mmGCN5- or mmP/CAF-specific sequences.
A single transcript of 3.3 kb was detected in all tissues with the GCN5 probe, consistent with size of the cDNA clone we isolated. Similarly, a single, ubiquitous transcript was detected with the P/CAF probe, and the size of this RNA, 4.4 kb, is similar to that of the P/CAF cDNA that we isolated. Interestingly, the P/CAF RNA always exhibited a broader banding pattern than did the GCN5 RNA. These two RNAs were clearly distinguished from one another when probed on the same blot, and a differential pattern of expression was detected (Fig. 5). For example, the ratio of mmGCN5 to mmP/CAF expression is higher in brain, thymus, spleen, testis, and 13.5-dpc embryonic tissue, while this ratio is much lower in heart, liver, kidney, and skeletal muscle. Western blot analysis of GCN5 protein levels (with the polyclonal antiserum to hsGCN5 described above) in various mouse tissues confirmed the general pattern of expression indicated by this RNA analysis (data not shown).
Chromosomal locations of the mmGCN5 and mmP/CAF genes.
The chromosomal location of the mmGCN5 gene was mapped by standard linkage analysis with the Jackson Laboratory interspecific backcross panel (C57BL/6Jei × SPRET/Ei)F1 × SPRET/Ei, also known as Jackson BSS (33). mmGCN5 mapped cleanly to a distal region on chromosome 11 and cosegregated tightly with BRCA1, as well as with a number of other genes previously mapped to that locus (data not shown, but raw data from the Jackson Laboratory are available at http://www.jax.org/resources/documents/cmdata). Interestingly, the hsGCN5 gene was recently mapped by fluorescent in situ hybridization analysis to a syntenic region of human chromosome 17 (9) and was also found to cosegregate with human BRCA1.
The location of mmP/CAF was mapped in a similar fashion, using the same backcross panel. In this case we used a probe specific for intronic sequences to ensure that we mapped the authentic mmP/CAF gene and not the P/CAF pseudogene. This analysis indicated that mmP/CAF is located 32 centimorgans from the centromere of mouse chromosome 17 and that it cosegregates with the DNA marker D17Bir8 (see www address above).
mmGCN5 encodes a histone acetyltransferase with substrate specificity similar to that of P/CAF.
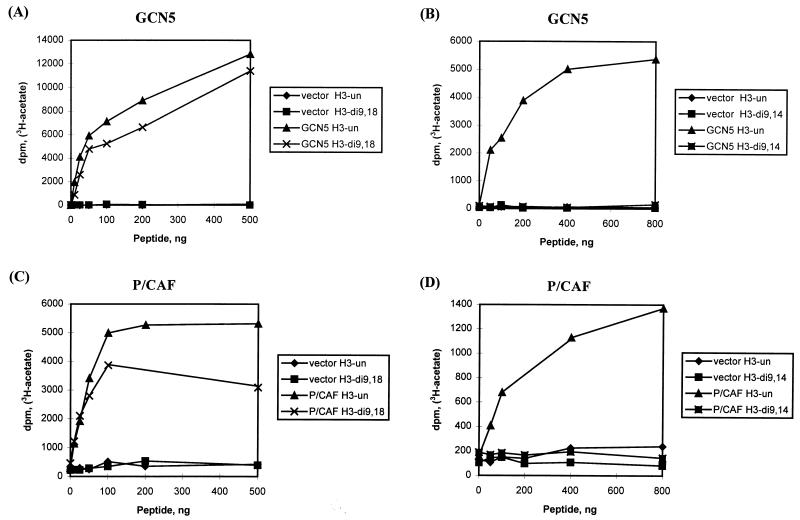
The high degree of homology between the mouse, human, and yeast GCN5 proteins strongly predicts that mmGCN5 and mmP/CAF will exhibit histone acetyltransferase activity. We confirmed this initially by examining the activities of the isolated, conserved acetylase domains of mmGCN5 and mmP/CAF, expressed as recombinant proteins in Escherichia coli. As expected, this domain of mmGCN5 was quite active as a histone acetylase, and it preferentially acetylated free (nonnucleosomal) histone H3, and to a lesser degree H4, as does yeast Gcn5p (23) and the previously reported form of the hsGCN5 protein (41). Full-length mmGCN5 and mmP/CAF recombinant proteins (also expressed in bacteria) exhibited this same substrate specificity towards free histones (Fig. 6 and data not shown).
FIG. 6.
Acetylation of histone H3 synthetic peptides by mmGCN5 and mmP/CAF. (A and B) Results of acetyltransferase assays with recombinant full-length mmGCN5 and synthetic peptides corresponding to the amino-terminal tail of histone H3. (C and D) Results of peptide assays with recombinant mmP/CAF. Peptides were either unacetylated (un) or synthesized with acetyl-lysine residues at either K9 and K18 (di9,18) or K9 and K14 (di9,14) in H3. Vector indicates assay of a control extract, made from bacteria transformed by the recombinant vector without an acetyltransferase insert, subjected to the His tag purification procedure.
To determine which residues of histone H3 were acetylated by mmGCN5, we performed assays with synthetic peptides corresponding to the amino-terminal tail of this histone. As expected, we found that the full-length GCN5 protein efficiently acetylated peptides corresponding to the first 20 amino acids of histone H3 (Fig. 6A and B). This domain alone, then, is sufficient for binding to the enzyme and subsequent catalysis. However, mmGCN5 could not acetylate a peptide that contained acetyl-lysine moieties at positions 9 and 14 (Fig. 6B), suggesting that one or both of these lysines may be a target site for mmGCN5. In contrast, mmGCN5 readily acetylated a peptide containing acetyl-lysine moieties at positions 9 and 18 (Fig. 6A). Taken together, these data suggest that K14 is the preferred acetylation site in H3 for mmGCN5. Similar assays performed with H4 peptides indicate that K8 is the preferred site of acetylation in H4 (data not shown). These results are consistent with the site specificity determined for recombinant yeast Gcn5p, which was confirmed by protein sequencing of acetylated histones (23). Importantly, these results indicate that the extended amino-terminal domain of mmGCN5 does not change the histone or lysine residue specificity of the enzyme.
The specificity of mmP/CAF was also tested with the peptide substrates. In all respects, mmP/CAF exhibited a substrate specificity identical to that of mmGCN5 (Fig. 6C and D).
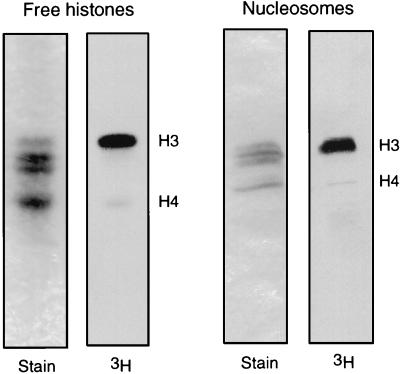
One striking difference between the previously reported, shorter form of recombinant hsGCN5 (or yeast Gcn5p) and recombinant hsP/CAF was the ability of P/CAF to acetylate nucleosomal substrates (23, 41). Given the homology between the amino-terminal portions of P/CAF and mmGCN5, we asked whether the full-length recombinant mmGCN5 could also acetylate histones within a nucleosome. We found that mmGCN5, like hsP/CAF, can acetylate nucleosomal H3 and, to a lesser degree, H4 (Fig. 7). In agreement with previously reported results (23, 41), we also found that the short form of mmGCN5 or yeast Gcn5p was unable to acetylate nucleosomes (data not shown). These results suggest that one function of the amino-terminal domains of mammalian GCN5 and P/CAF may be to facilitate the recognition of chromatin templates.
FIG. 7.
Acetylation of nucleosomal histones by mmGCN5 and mmP/CAF. Acetyltransferase assays were performed with HeLa cell mononucleosomes or free histones as indicated, and an aliquot of each assay mixture was resolved on an SDS–22% polyacrylamide gel. Coomassie blue-stained gels and corresponding autoradiographs are shown. In both assays, histones H3 and H4 were acetylated by the recombinant full-length mmGCN5.
mmGCN5 and mmP/CAF both interact with CBP and p300.
hsP/CAF interacts with CBP and p300 (41). Given the similarity between mmGCN5 and mmP/CAF, we examined the abilities of both of these proteins to bind to CBP or p300 in vitro (Fig. 8).
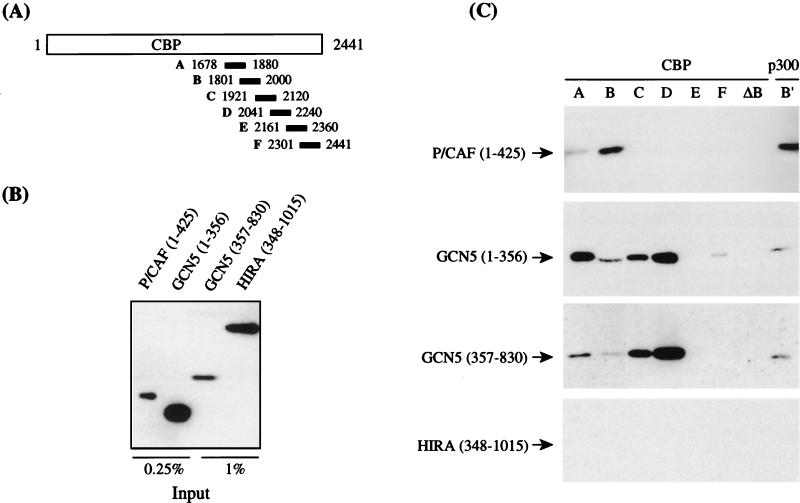
FIG. 8.
mmGCN5 and mmP/CAF both interact with CBP and p300 in vitro. (A) Fragments of CBP fused to GST that were used for the interaction assays in panel C. These fragments span the region of homology to ADA2 and extend into the transactivation domain of CBP (41). (B) Recombinant, His-tagged proteins used in the interaction assays of panel C were resolved by SDS-polyacrylamide gel electrophoresis and probed with an antibody specific for the His tag. The amounts shown represent 0.25 or 1% of the protein used in the assay, as indicated. (C) GST-CBP or -p300 fusion proteins were mixed with crude bacterial lysates containing P/CAF, GCN5, or HIRA protein. Interacting proteins were recovered by using glutathione-Sepharose and detected on Western blots with the anti-His tag antibody. The p300 B′ fragment is homologous to the CBP B fragment, and the CBP ΔB fragment is missing residues 1801 to 1851 (41).
Whole-cell lysates from bacteria expressing fragments of CBP fused to GST (fusion constructs were kindly provided by Y. Nakatani, National Institutes of Health) (41) were mixed with lysates from cells expressing the amino-terminal domain of mmP/CAF, the amino-terminal domain of mmGCN5, or the C-terminal domain of mmGCN5. The CBP fragments (A to F) spanned the ADA2 homology domain and extended into the transcriptional activation domain (41). A fragment of p300 (B′) homologous to the B fragment of CBP was also tested. GST fusion proteins were purified together with any interacting proteins by using glutathione-Sepharose, and the interacting proteins were identified by Western blotting with an antiserum specific for the six-histidine tag present in the recombinant mmP/CAF or mmGCN5 protein.
The amino-terminal domain of mmP/CAF selectively bound to fragments A and B of CBP and the corresponding B′ fragment of p300. In some experiments, we also observed binding to the D fragment, but we never observed binding to fragment C, E, or F. A deletion within the B fragment (ΔB) of CBP that removed residues 1801 to 1851 eliminated binding. This pattern of binding to the CBP/p300 fragments is extremely similar to that previously reported for hsP/CAF (41), as expected.
A recombinant form of hsGCN5, which lacked the amino-terminal domain reported here for mmGCN5, failed to bind CBP or p300 in previous experiments by Yang et al. (41). This form of hsGCN5 corresponds to the C-terminal region of mmGCN5. We therefore compared binding of the amino-terminal and the C-terminal halves of mmGCN5 to the GST-CBP and -p300 fragments. Surprisingly, we found that both of these mmGCN5 domains bound to CBP fragments A to D, with little or no binding to fragment E, fragment F, or the ΔB fragment. Both the amino-terminal and C-terminal regions of mmGCN5 also bound to the p300 B′ fragment. The amount of the GST fusion proteins recovered from the GST columns that did not exhibit binding to the GCN5 fragments was greater than or equal to that of the GST fusions that did exhibit binding (data not shown), so the absence of binding was not due to reduced amounts of the E, F, or ΔB fragment. In addition, the selective binding of the mmGCN5 peptides to CBP fragments A to D indicates that these interactions are not nonspecifically mediated by the GST moiety, since this moiety is also present in fragments E, F, and ΔB. The specificity of the interactions was further tested by using an unrelated protein, HIRA, which failed to bind to any of the GST-CBP or -p300 fragments. Thus, CBP fragments A to D do not exhibit general, nonspecific binding to random proteins. We conclude that mmGCN5 contains two distinct CBP/p300 interaction domains and that these domains interact with a broader region in CBP than does P/CAF. Importantly, our finding that mmGCN5 and mmP/CAF can both interact with CBP/p300 indicates that these proteins are very similar in function as well as in structure.
DISCUSSION
The recent identification of nuclear histone acetyltransferases has directly linked chromatin modification with transcriptional regulation (1, 5, 26, 29). We report here the cloning of mmGCN5 and mmP/CAF sequences. We find that mmGCN5 differs from yeast GCN5 and the previously reported hsGCN5 sequences in that it encodes a large N-terminal domain similar to that found in P/CAF. Our data indicate that hsGCN5 contains this extra domain as well. While this domain does not appear to affect the histone specificity of the acetyltransferase, it does afford the enzyme the ability to modify nucleosomal substrates in vitro.
In vivo, both the yeast and mammalian enzymes must interact with and modify nucleosomal histones. In yeast, this is accomplished by association of Gcn5p into high-molecular-weight protein complexes that can modify nucleosomes and that recognize additional histones (14). At least some of the Gcn5p-associated proteins are conserved in higher eukaryotes, and Gcn5-Ada complexes have been identified in human cells (7), further indicating that these enzymes serve similar functions across species. We scanned the yeast genome database to determine whether a protein homologous to the amino-terminal domains of mmGCN5 or mmP/CAF might exist that could be a component of the Gcn5p-containing complexes. However, we found no such homologs.
Interestingly, a single GCN5-related gene has been identified in Drosophila. This gene exhibits high similarity to mammalian P/CAF (34a) and encodes the extended N-terminal domain. Although this domain is apparently not needed in yeast, its functions are not restricted to mammals.
Our work indicates that multiple differentially spliced forms of GCN5 transcripts coexist in both mouse and human cells, which may generate different isoforms of GCN5 proteins. Of course, we cannot rule out the possibility that the less abundant products represent incompletely spliced RNAs, but it is interesting that intron 6 and the stop codons therein are conserved between human and mouse. Since we detected transcripts containing intron 6 in both mouse and human tissues, we are intrigued by the possibility that shortened GCN5 proteins, containing either the N-terminal domain alone or the C-terminal domain alone, may provide an additional level of regulation of GCN5 functions. For example, we detected the full-length GCN5 protein in mouse embryo extracts and some human cells, but we detected a shorter, less abundant protein in HeLa cells in addition to the full-length protein. It will be especially interesting to determine whether various forms of GCN5 proteins are differentially regulated in different cell types or at different developmental stages.
The long form of mmGCN5 is very similar to mmP/CAF in structure, in acetyltransferase activity and substrate specificity, and in interactions with CBP/p300. Additional experiments are needed to determine whether these two proteins are functionally redundant in vivo. Even if GCN5 and P/CAF perform the same functions, they might be utilized at different developmental stages or in different cell types or tissues. The similarity between these proteins is somewhat reminiscent of that between CBP and p300. These two proteins also appear to be functionally equivalent in vitro, but mutations in p300 and CBP cause different phenotypes (27, 30), indicating that the proteins are not functionally redundant in vivo. It will be interesting to determine whether the same is true for mmGCN5 and mmP/CAF.
Several histone acetyltransferases, including p300/CBP and P/CAF, have been implicated in growth control and tumorigenesis (30, 32, 41). p300/CBP physically interacts with the tumor suppressor p53 and potentiates sequence-specific DNA binding and transactivation by p53 through acetylation of its C-terminal domain (15). Moreover, mutations in p300 have been found in certain colorectal and gastric cancers (27). CBP mutations are also involved in the etiology of certain acute myeloid leukemias and Rubinstein-Taybi syndrome (30), a developmental disorder with a high incidence of neoplasms. In addition, P/CAF counteracts the transforming activity elicited by oncoprotein E1A, and overexpression of P/CAF has been shown to inhibit cell cycle progression (41). Therefore, histone acetyltransferases have been postulated to be negative regulators of cell growth and, possibly, tumor suppressors. We (this study) and others (9) have found that GCN5 cosegregates with the tumor suppressor BRCA1 gene (1-centimorgan interval) in a highly syntenic region in mouse (chromosome 11) and human (chromosome 17). Interestingly, loss of heterozygosity on human chromosome 17 is a frequent genetic alteration in sporadic breast and ovarian cancers, where mutations in BRCA1 and BRCA2 are rarely found (28, 35). Indeed, a novel tumor suppressor gene involved in these cancers has been postulated to be located adjacent to the BRCA1 locus (28, 35). GCN5 may provide an attractive candidate for this novel tumor suppressor. The isolation and characterization of mmGCN5 and mmP/CAF reported here should facilitate further study of the roles of these genes and of histone acetylation in normal mammalian development, as well as in abnormal events leading to tumorigenesis.
ACKNOWLEDGMENTS
We thank Jerry Workman and Patrick Grant for the kind gift of HeLa cell mononucleosomes and histones. We thank David Allis for the gift of H3 amino-terminal peptides, and we thank E. Smith and D. Allis for the gift of oligomers for PCR and for sharing results prior to publication. We also thank Shelley Berger for antiserum specific for hsGCN5, Yongshen Ren for the gift of HeLa cell nuclear extracts, and Yoshihiro Nakatani for the GST-CBP and GST-p300 fusion constructs and hsP/CAF antibodies. Some DNA sequencing was performed by the UTMDACC Sequencing Core Facility. We are grateful to Lucy Rowe and Mary Barter at the Jackson Laboratory for their assistance in the mouse chromosome mapping analysis. We thank Karen Hensley for help in preparation of some graphics and Aurora Diaz for help in preparing the manuscript.
W.X. is supported by a Rosalie B. Hite Fellowship, and D.G.E. is supported by a Theodore Law UCF Scientific Fund Fellowship. This work was supported by grants to S.Y.R. from the Robert A. Welch Foundation, the USARMC, and the Breast Cancer Research Center at UTMDACC.
The first two authors contributed equally to this work.
REFERENCES
- 1.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 2.Barlev N, Candau R, Wang L, Darpino P, Silverman N, Berger S. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA binding protein. J Biol Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- 3.Berger S L, Pina B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 4.Brownell J E, Allis C D. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 6.Candau R, Berger S L. Structural and functional analysis of yeast putative adaptors: evidence for an adaptor complex in vivo. J Biol Chem. 1996;271:5237–5243. doi: 10.1074/jbc.271.9.5237. [DOI] [PubMed] [Google Scholar]
- 7.Candau R, Moore P A, Wang L, Barlev N, Ying C Y, Rosen C A, Berger S L. Identification of functionally conserved human homologs of the yeast adapters ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Candau R, Zhou J, Allis C D, Berger S L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter K C, Wang L, Shell B K, Zamir I, Berger S L, Moore P A. The human transcriptional adaptor genes TADA2L and GCN5L2 colocalize to chromosome 17q12-q21 and display a similar tissue expression pattern. Genomics. 1997;40:497–500. doi: 10.1006/geno.1996.4605. [DOI] [PubMed] [Google Scholar]
- 10.Chomcznski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.Edmondson D G, Olson E N. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 1989;3:628–640. doi: 10.1101/gad.3.5.628. [DOI] [PubMed] [Google Scholar]
- 12.Edmondson D G, Roth S Y. Chromatin and transcription. FASEB J. 1996;10:1173–1182. doi: 10.1096/fasebj.10.10.8751719. [DOI] [PubMed] [Google Scholar]
- 13.Georgakopoulos T, Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant P A, Duggan L, Cote J, Roberts S, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast GCN5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an ADA complex and the SAGA (SPT/ADA) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 15.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 16.Hansen J C. The core histone amino-termini: combinatorial interaction domains that link chromatin structure with function. Chem Tracts Biochem Mol Biol. 1997;10:56–69. [Google Scholar]
- 17.Haynes S R, Dollard C, Winston F, Beck S, Trowsdale J, Dawid I B. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horiuchi J, Silverman N, Marcus G A, Guarente L. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol Cell Biol. 1995;15:1203–1209. doi: 10.1128/mcb.15.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeanmougin F, Wurtz J-M, Le Douarin B, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 20.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T-M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for co-activators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 21.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–904. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 23.Kuo M H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmondson D G, Roth S Y, Allis C D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 24.Marcus G, Silverman N, Berger S, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2, putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcus G A, Horiuchi J, Silverman N, Guarente L. ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol Cell Biol. 1996;16:3197–3205. doi: 10.1128/mcb.16.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 27.Muraoka M, Konishi M, Kikuchi-Yanoshita R, Tanaka K, Shitara N, Chong J, Iwama T, Miyaki M. p300 gene alterations in colorectal and gastric carcinomas. Oncogene. 1996;12:1565–1569. [PubMed] [Google Scholar]
- 28.Niederacher D, Picard F, Roeyen C V, An H, Bender H G, Beckmann M W. Patterns of allelic loss on chromosome 17 in sporadic beast carcinomas detected by fluorescent-labeled microsatellite analysis. Genes Chromosomes Cancer. 1997;18:181–192. doi: 10.1002/(sici)1098-2264(199703)18:3<181::aid-gcc5>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 29.Ogryzko V V, Schlitz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 30.Petrij F, Giles R H, Dauwerse H G, Saris J J, Hennekam R C M, Masuno M, Tommerup N, van Ommen G-J B, Goodman R H, Peters D J M, Breuning M H. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 31.Roberts S M, Winston F. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3206–3213. doi: 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roth S Y. Something about silencing. Nat Genet. 1996;14:3–4. doi: 10.1038/ng0996-3. [DOI] [PubMed] [Google Scholar]
- 33.Rowe L B, Nadeau J H, Turner R, Frankel W N, Letts V A, Eppig J T, Ko M S H, Thurston S J, Birkenmeier E H. Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mamm Genome. 1994;5:253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- 34.Silverman N, Agapite J, Guarente L. Yeast ADA2 binds to the VP16 protein activation domain and activates transcription. Proc Natl Acad Sci USA. 1994;91:11665–11668. doi: 10.1073/pnas.91.24.11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Smith, E., and D. Allis. Personal communication.
- 35.Tangir J, Muto M G, Berkowitz R S, Welch W R, Bell D A, Mok S C. A 400 kb novel deletion unit centromeric to the BRCA1 gene in sporadic epithelial ovarian cancer. Oncogene. 1996;12:735–740. [PubMed] [Google Scholar]
- 36.Turner B M. Histone acetylation and control of gene expression. J Cell Sci. 1991;99:13–20. doi: 10.1242/jcs.99.1.13. [DOI] [PubMed] [Google Scholar]
- 37.van Holde K E. Chromatin. New York, N.Y: Springer-Verlag; 1989. [Google Scholar]
- 38.Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, Allis C D, Berger S L. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol Cell Biol. 1997;17:519–527. doi: 10.1128/mcb.17.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolffe A. Chromatin structure and function. London, United Kingdom: Academic Press; 1992. [Google Scholar]
- 40.Wolffe A P, Pruss D. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 41.Yang X-J, Ogryzko V V, Nishikawa J-I, Howard B H, Nakatani Y. A p300/CBP associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]