Abstract
This study was targeted at investigating the biological functions of E74-like ETS transcription factor 1 (ELF1) in pancreatic cancer (PC) and its underlying mechanism. ELF1 expression in PC tissues was detected by quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) and immunohistochemistry. Cell counting kit-8 (CCK-8) method, EdU method and flow cytometry were used to detect the cell proliferation and apoptosis of PC cell lines after transfection. A subcutaneous tumorigenesis model was constructed to validate the oncogenic role of ELF1 in vivo. PROMO database was used to predict the binding site of ELF1 on the promoter region of doublecortin-like kinase 1 (DCLK1). Dual-luciferase reporter gene assay, chromatin immunoprecipitation-quantitative polymerase chain reaction (ChIP-qPCR) assay and quantitative real-time PCR were performed to detect the binding of ELF1 to the promoter region of DCLK1. The effect of ELF1 on DCLK1 expression was detected by Western blot assay. It was found that ELF1 expression in PC tissues and cells was up-regulated. ELF1 overexpression promoted the proliferation and inhibited the apoptosis of PC cells, while knocking down ELF1 had the opposite effects. ELF1 could bind to the promoter region of DCLK1 and ELF1 overexpression promoted the expression of DCLK1. Bioinformatics analysis suggested that Janus kinase (JAK) - signal transducer and activator of transcription (STAT) signaling pathway was associated to DCLK1 expression, and overexpression of ELF1 promoted the expression of Janus kinase 2 (JAK2) and signal transducer and activator of transcription 3 (STAT3). In conclusion, ELF1 promoted the malignant progression of PC via regulating DCLK1/ JAK/STAT signaling pathway.
Keywords: ELF1, DCLK1, pancreatic cancer, proliferation, apoptosis
Introduction
Pancreatic cancer (PC) is a tumor of digestive system with extremely high malignancy [1], and ranks the seventh leading cause of cancer-related deaths [2,3]. Early symptoms of PC are insidious, so most patients already have distant metastases once they are diagnosed [4,5]. In recent years, the prognosis of patients with PC has been significantly improved as the progress of comprehensive treatment. However, the 5-year survival rate of PC patients is still less than 10% [6-8]. Therefore, it is necessary to further clarify the molecular mechanism of PC progression.
Transcription factor is a class of DNA-binding proteins modulating expressions of multiple genes via binding to the specific region of the promoter of a gene [9]. Transcription factors, currently accounting for about 20% of all oncogenes, are associated with the pathogenesis and progression of multiple human diseases, including cancer [10]. For example, ETV4 activates ANXA2 transcription by binding to ANXA2 promoter region, which in turn activates the Wnt/β-catenin pathway and accelerates the progression of HBV-related liver cancer [11]; E2F1 binds to the promoter region of TINCR and activates the transcription of TINCR, thus facilitating the proliferation and inhibiting the apoptosis of gastric cancer cells [12]. NR5A2 transcription is activated by BRD4, subsequently promotes PC cell growth, migration and invasion [13]. In addition, the role of E74-like ETS transcription factor 1 (ELF1) has been reported in many diseases. For example, ELF1 is up-regulated in osteosarcoma cells, and ELF1 up-regulates the expression of ZCCHC3 by activating FOXD3-AS1, thereby promoting the aggressiveness and epithelial-mesenchymal transition of osteosarcoma cells [14]. In this study, bioinformatics analysis showed that ELF1 expression in PC was up-regulated and correlated with shorter overall survival time of patients. However, how ELF1 functions in PC deserves further clarification.
In addition, doublecortin like kinase 1 (DCLK1) is up-regulated in PC and the overexpression of DCLK1 promotes the proliferation, migration and invasion of PC cells [15]. Bioinformatics analysis showed that ELF1 could bind to DCLK1 promoter region, and their expression levels were positively correlated. Therefore, this study focused on exploring the biological function of ELF1 in PC and its regulatory effects on DCLK1 transcription.
Materials and methods
Tissue sample collection
PC tissues and non-cancerous pancreatic tissues were obtained during surgery from Department of Hepato-Pancreato-Biliary Surgery, the Second Affiliated Hospital of Zhejiang University School of Medicine. All subjects did not receive chemotherapy, radiotherapy, and other anti-cancer treatments before the surgery. This study was conducted with the approval and under the guidance of the Ethics Committee of the Second Affiliated Hospital of Zhejiang University School of Medicine (approval number: 2017ZJDX036).
Immunohistochemistry
PC tissues (n=50) and unpaired non-cancerous pancreatic tissues (n=20) were used for immunohistochemistry. After the removal, the tissues were washed with normal saline and then fixed by formalin. After the tissues were embedded in paraffin, tissue sections were prepared. Next, the sections were dewaxed in xylene and rehydrated by gradient ethanol. Then the sections were incubated in 3% H2O2 for 30 min to block the activity of endogenous peroxidase. Next, the sections were heated in boiling sodium citrate buffer (0.01 M, pH 6.0) for 10 min, to repair the antigens. Next, the tissues were blocked with goat serum for 30 min, and then incubated with anti-ELF1 antibody (ab64937, 1:100, Abcam Inc, Cambridge, UK) in a wet box overnight at room temperature. The tissues were then incubated at room temperature for 1 h with the secondary antibody (Proteintech, Wuhan, China). After rinsing with phosphate buffer saline (PBS), DAB was added for color developing. PBS was used as a blank control instead of the primary antibody. The staining intensity of ELF1 was scored by two experienced pathologists, and clarified into “high expression”, “low expression” and “negative”. The Human Protein Atlas (www.proteinatlas.org) was also searched to check the immunohistochemical staining of ELF1 in PC tissues and non-cancerous tissues.
Cell culture
PC cell lines (PANC-1, BxPc-3, Capan-2) and human normal pancreatic ductal epithelial cell line hTERT-HPNE were available from the American Type Culture Collection (Manassas, VA, USA). The cells were cultured in Roswell Park Memorial Institute-1640 medium (RPMI-1640, Gibco, Carlsbad, CA, USA) or Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Carlsbad, CA, USA) and placed in an incubator containing 5% CO2 at 37°C. The medium contained 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA), 100 U/mL penicillin (Gibco, Carlsbad, CA, USA) and 0.1 mg/mL streptomycin (Gibco, Carlsbad, CA, USA).
Cell transfection
Empty plasmid (negative control, NC), ELF1 overexpression plasmid (ELF1), DCLK1 overexpression plasmid (DCLK1), small interfering RNA (siRNA) targeting ELF1 (si-ELF1) (si-ELF1-1: sense, 5’-CACUUCAAAUAGGAAUCAAC-3’; anti-sense, 5’-GUUGAUUCCUAUUUGAAGUG-3’; si-ELF1-2: sense, 5’-UCCGACCGAGUCGUCCAUGUA-3’; anti-sense, 5’-UACAUGGACGACUCGGUCGGA-3’), siRNA targeting DCLK1 (si-DCLK1; sense, 5’-GAUCGAUACUUCAAAGGGA-3’; anti-sense, 5’-UCCCUUUGAAGUAUCGAUC-3’), and negative control (si-NC; sense, 5’-UUCUCCGAACGUGUCACGUTT-3’; anti-sense, 5’-ACGUGACACGUUCGGAGAATT-3’) were purchased from GenePharma (Shanghai, China). When the cells reached 60-80% confluence, the above plasmids or siRNAs were transfected into Capan-2 and PANC-1 cells with LipofectamineTM 2000 kit (Invitrogen, Carlsbad, CA, USA). 48 h later, the transfection efficiency was detected by quantitative Real-time PCR (qRT-PCR) and the cells were collected for subsequent analyses.
qRT-PCR
Forty-four pairs of PC tissue samples and adjacent normal samples were collected for qRT-PCR. They were obtained from the surgically resected tumor tissues and adjacent non-cancerous tissues, and the samples were subsequently frozen in liquid nitrogen until RNA extraction. Total RNA was extracted by a TRIzol kit (Invitrogen, Carlsbad, CA, USA) and reversely transcribed into cDNA by a Miscript Reverse Transcription Kit (Qiagen GmbH, Hilden, Germany). Then, the PCR reaction was carried out with a miScript SYBR Green PCR Kit (Qiagen GmbH, Hilden, Germany) on a Rotorgene 3000 series PCR machine (Qiagen GmbH, Hilden, Germany). Ultimately, quantitative analysis was performed with the Rotor Gene software. The 2-ΔΔCt method was used to calculate the relative expression of ELF1 mRNA and DCLK1 mRNA, with GAPDH as the internal reference. The primer sequences: ELF1 Forward: 5’-TGTGTGATAGGTCTGCGAAAA-3’, ELF1 Reverse: 5’-ATAAGGGCAAGGACATT-3’; DCLK1 Forward: 5’-CAGCGCCATCAAATACCTGC-3’, DCLK1 Reverse: 5’-TGGTCATCACCACTTCCACG-3’; GAPDH Forward: 5’-GAAGGTGAAGGTCGGAGTC-3’, GAPDH Reverse: 5’-GAAGATGGTGATGGGATTTC-3’.
Cell counting kit-8 (CCK-8) assay
The viability of Capan-2 and PANC-1 cells was detected by a CCK-8 kit (Dojindo, Shanghai, China). These cells were inoculated on 96-well plates at a density of 1 × 103 cells/well. After 24, 48 or 72 h, 10 μl of CCK-8 reagent was loaded into each well and the cells were incubated at 37°C in 5% CO2 for 2 h. After that, the optical density (OD) values at 450 nm per well were probed by a microplate reader (Molecular Devices, Sunnyvale, CA, USA).
EdU assay
Capan-2 and PANC-1 cells were respectively inoculated into a 24-well plate. A BeyoClick™ EdU-488 cell proliferation Kit (Beyotime, Shanghai, China) was used for the EdU assay. Each well was added with 200 μL of 5 μmol/L EdU working solution for 2 h. The cells were followingly fixed with paraformaldehyde for 10 min and stained with Apollo for 30 min. Besides, the cells were incubated with 1 × Hoechst 33342 DNA staining solution (Dojin, Tokyo, Japan) in darkness for 20 min at ambient temperature. After the cells were washed, the cells were observed under a fluorescence microscope and the results were analyzed.
Flow cytometry assay
The apoptosis of PC cells was detected by the Apoptosis Detection Kit. The PC cells were cultured for 24 h, trypsinized with 0.25% trypsin and then collected after centrifugation. After being washed twice with PBS containing 5% BSA, the cells were mixed with 5 µl of Annexin V-FITC solution (YEASEN Biotech Co., Ltd., Shanghai, China) and 10 µl of propidium iodide (PI) solution (YEASEN Biotech Co., Ltd., Shanghai, China) and incubated for 20 min. After the cells were washed with PBS, the apoptotic rate was detected by a flow cytometer (BD Biosciences, San Jose, CA, USA).
Dual luciferase report gene assay
Through the PROMO database, the binding sites between ELF1 and DCLK1 promoter regions were predicted, and the DCLK1 promoter sequence fragment was amplified by PCR and cloned into a pGL3-Basic vector (Promega, Madison, WI, USA) to construct a pGL3-DCLK1-wild type (DCLK1-WT) and a pGL3-DCLK1-mutant (DCLK1-MUT) reporter vector. The vectors and the ELF1 overexpression plasmid and si-ELF1 were then co-transfected into Capan-2 and PANC-1 cells by LipofectamineTM 2000 kit. 48 h later, the luciferase activity was detected by a dual-luciferase reporter assay system (Promega, Madison, WI, USA).
Chromatin immunoprecipitation (ChIP) assay
ChIP experiment was performed with the EZ-ChIPTM kit (Millipore, Billerica, MA, USA). Capan-2 and PANC-1 cells were fixed with formaldehyde and terminated with glycine after incubation. The cells were scraped to get cell precipitation, and then the cell lysis buffer containing phenylmethanesulfonyl fluoride (PMSF) was added and the nuclear precipitate was obtained. The DNA was cut by ultrasound in an ice bath. 10% of the supernatant of the nuclear lysate cut by ultrasound was used as the control, and the remaining 90% of the lysate and magnetic beads coupled with anit-ELF1 antibody were mixed, shaken, incubated and centrifuged, and the DNA bound to ELF1 was eluted with fresh elution buffer. Next, the DNA was purified by DNA purification kit and then detected by qRT-PCR.
Western blot
The cells were collected and incubated with pre-cooled RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) for 20 min on ice and then centrifuged for 20 min (13000 r/min, 4°C). Next, the supernatant was collected as the sample, with the protein quantification performed by the bicinchoninic acid (BCA) protein quantification kit (Beyotime Biotechnology, Shanghai, China). After the concentration of the protein sample was adjusted, the protein was denatured by boiling for 5 min and protein samples were loaded on a 12% sodium dodecyl sulfate-polyacrylamide gel (Sigma Aldrich, St. Louis, MO, USA) and electrophoresed for 2 h and transferred to polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). Then, the membranes were washed with tris buffered saline tween (TBST), and incubated overnight at 4°C with primary antibodies including anti-ELF1 (ab64937, 1:1000, Abcam Inc., Cambridge, UK), anti-DCLK1 (ab106635, 1:500, Abcam Inc., Cambridge, UK), anti-Janus kinase 2 (JAK2) (ab108596, 1:500, Abcam Inc., Cambridge, UK), anti-signal transducer and activator of transcription 3 (STAT3) (ab109085, 1:500, Abcam Inc., Cambridge, UK) and anti-GAPDH (ab181602, 1:500, Abcam Inc., Cambridge, UK). After rinsing with TBST again, the PVDF membranes were incubated with secondary antibody Goat Anti-Rabbit IgG H&L (ab205718, 1:1000, Abcam Inc, Cambridge, UK) at room temperature for 2 h and then immersed in TBST again. Finally, the protein bands were visualized with an ECL luminescence kit (Santa Cruz, CA, USA), with GAPDH as the internal reference.
Animal experiments
The processes of the animal experiments of the present study were performed after the approval of the Animal Research Ethics Committee of the Second Affiliated Hospital of Zhejiang University School of Medicine. The mice (4 weeks old, BALB/c, male, bought from Hio-bio, Hangzhou, China) were divided into two groups (ELF1 overexpression group and the control group, and 3 mice per group). Capan-2 cells were then subcutaneously inoculated into the back of the nude mice (2 × 107 cells per mouse, suspended in sterile PBS). The mice were fed for 15 days, and then were sacrificed, and the formed tumors were isolated, and the size was compared.
Statistical analysis
SPSS 24.0 statistical software (SPSS Inc., Chicago, IL, USA) was applied to analyze the experimental data. Student’s t-test was adopted for the comparison between two groups, and one-way ANOVA was used for comparisons among multiple groups. P < 0.05 indicates statistically significant differences.
Results
The expression characteristics of ELF1 in PC tissues
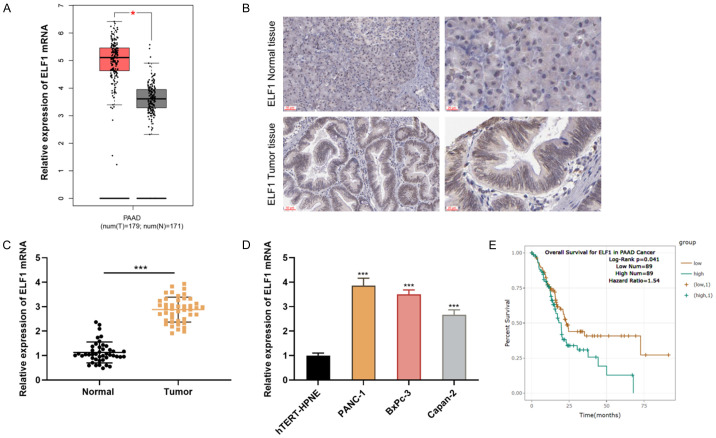
GEPIA database (http://gepia.cancer-pku.cn/) showed that ELF1 expression in PC tissue samples was significantly up-regulated (Figure 1A). Next, immunohistochemistry results obtained from the Human Protein Atlas database (https://www.proteinatlas.org/) showed that ELF1 had a higher expression in PC tissues than that in normal pancreatic tissues; and ELF1 protein expression in PC tissues (n=50) and non-cancerous pancreatic tissues (n=20) was also evaluated by immunohistochemistry, and it showed that ELF1 expression was up-regulated in PC tissues (Chi-square value =13.693; P value =0.001) (Figure 1B; Table 1). qRT-PCR showed that, ELF1 mRNA expression in PC tissues was up-regulated as against the adjacent tissues (Figure 1C). In addition, ELF1 expression was up-regulated in PC cells compared with the immortalized pancreatic cell line (Figure 1D). In addition, StarBase database showed a negative correlation between ELF1 expression and the overall survival of patients, suggesting its oncogenic role (Figure 1E).
Figure 1.
ELF1 is up-regulated in PC, and it is a potential prognostic factor. A. ELF1 expression characteristics in PC tissues and non-cancerous tissues were analyzed using GEPIA database. B. Representative IHC staining of ELF1 in PC tissues and non-cancerous tissues were provided (scale bars =50 μm and 20 μm, respectively). C. The expression of ELF1 mRNA in PC tissues (n=44) and normal tissues (n=44) was determined by qRT-PCR. D. The expression of ELF1 mRNA in PC cell lines (PANC-1, BxPc-3, Capan-2) and hTERT-HPNE cells was determined by qRT-PCR. E. The association between the prognosis of PC patients and the expression of ELF1 was analyzed using StarBase database. *P < 0.05 and ***P < 0.001
Table 1.
The statistical analysis of the results of immunohistochemistry
| PC tissues | Non-cancerous tissues | Chi-square value | P value | |
|---|---|---|---|---|
| ELF1 high | 15 | 4 | 13.693 | 0.001* |
| ELF1 low | 30 | 6 | ||
| ELF1 negative | 5 | 10 |
Pearson’s chi-squared test.
Effects of ELF1 on proliferation and apoptosis of PC cells
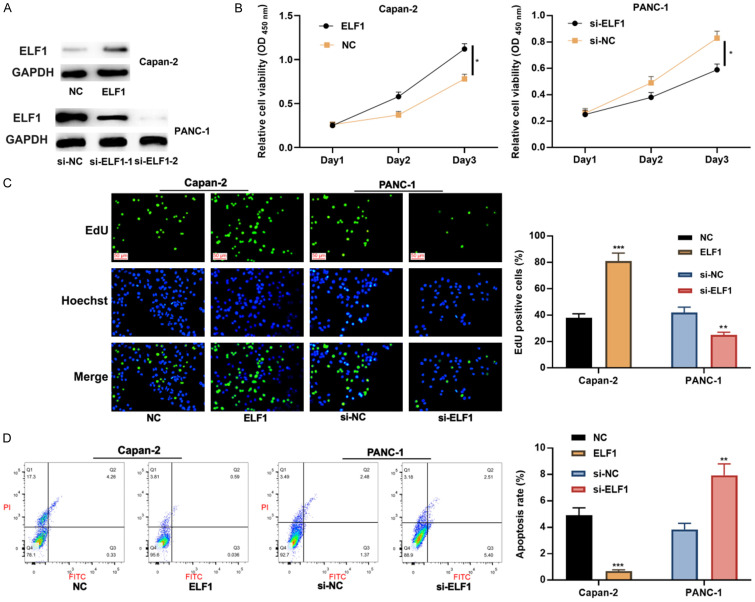
To clarify the biological functions of ELF1 in Capan-2 and PANC-1 cells, Capan-2 cells were transfected with ELF1 overexpression plasmids, and PANC-1 cells were transfected with si-ELF1, and Western blot was used to verify the success of transfection (Figure 2A). Considering the si-ELF1-2 had high knockdown efficiency, it was selected for subsequent experiments. CCK-8 assay, EdU assay and flow cytometry showed that compared with the control group, the up-regulation of ELF1 promoted the proliferation and restrained the apoptosis of PC cells, while knocking down ELF1 functioned oppositely (Figure 2B-D).
Figure 2.
Effects of ELF1 on the proliferation and apoptosis of PC cells. A. The transfection efficiency of ELF1 overexpression plasmid and si-ELF1 was detected by Western blot. B. The effects of ELF1 overexpression or knockdown on the viability of Capan-2 and PANC-1 cells were detected by CCK-8 assay. C. The effects of ELF1 overexpression or knockdown on the proliferation of Capan-2 and PANC-1 cells were detected by EdU assay (scale bars =50 μm). D. The effects of ELF1 overexpression or knockdown on the apoptosis of Capan-2 and PANC-1 cells were detected by flow cytometry. *P < 0.05, **P < 0.01, and ***P < 0.001.
ELF1 promotes the transcription of DCLK1
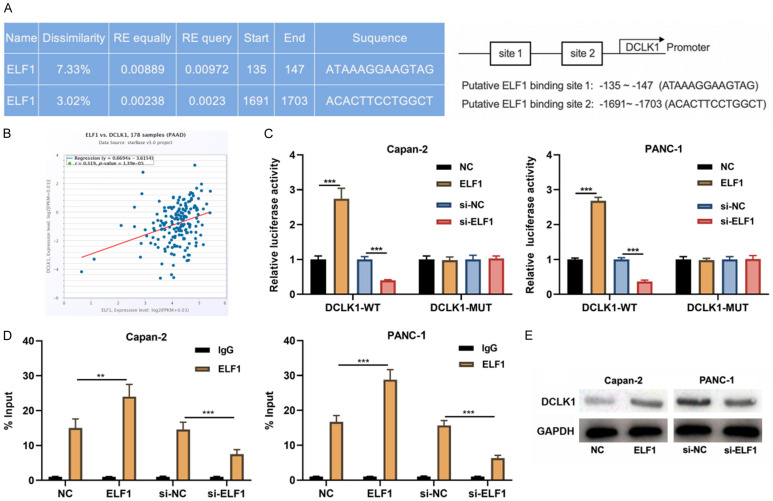
To explore the downstream mechanism of ELF1, PROMO database was searched and two binding sites were found between ELF1 and the promoter region of DCLK1 (Figure 3A). Previous studies have shown DCLK1 facilitates the malignant biological behaviors of PC cells [15-17]. Consistently, we also observed that DCLK1 overexpression promoted the proliferation and inhibited the apoptosis of PANC-1 cells (Supplementary Figure 1). Additionally, StarBase database showed a positive correlation between ELF1 expression and DCLK1 expression in PC tissues (Figure 3B). It was supposed that ELF1 could promote the transcription of DCLK1. As expected, dual luciferase reporter gene assay showed that in Capan-2 and PANC-1 cells, overexpression of ELF1 increased the luciferase activity of DCLK WT reporter, while knocking down of ELF1 worked oppositely; overexpression or knockdown of ELF1 did not significantly change the luciferase activity of DCLK1-MUT (Figure 3C). ChIP-qPCR showed that overexpression of ELF1 enhanced the binding between ELF1 and DCLK1 promoter in Capan-2 and PANC-1 cells, while knocking down E2F1 had the opposite effects (Figure 3D). In addition, Western blot showed that ELF1 overexpression promoted DCLK1 expression, and knockdown ELF1 worked oppositely (Figure 3E).
Figure 3.
ELF1 transcriptionally promotes DCLK1 expression. A. PROMO database was used to predict the binding sites between ELF1 and DCLK1 promoter region. B. The correlation between ELF1 expression and DCLK1 expression in PC samples was analyzed by the StarBase database. C. Dual-luciferase reporter gene assay was performed to detect the effects of ELF1 overexpression and knockdown on the luciferase activities of DCLK1-WT and DCLK1-MUT. D. ChIP-qPCR assay was applied to detect the binding of ELF1 to the DCLK1 promoter region. E. Western blot was used to detect the effect of ELF1 overexpression or knock-down on the expression of DCLK1 in Capan-2 and PANC-1 cells. **P < 0.01 and ***P < 0.001.
Effects of ELF1 and DCLK1 on proliferation and apoptosis of PC cells
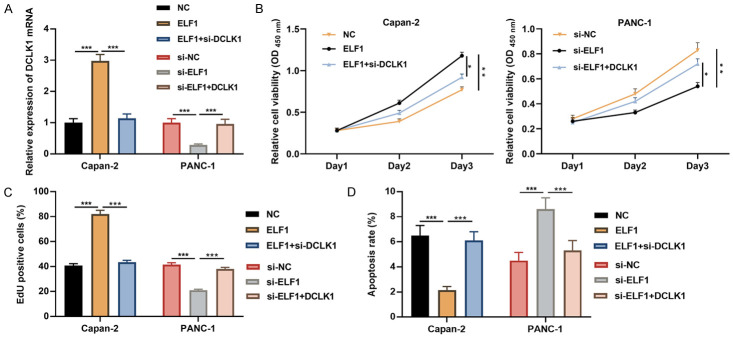
To further confirm whether ELF1 promotes the progression of PC by regulating DCLK1, we co-transfected the ELF1 overexpression plasmid and si-DCLK1 into Capan-2 cells and the si-ELF1 and DCLK1 overexpression plasmids into PANC-1 cells, and qRT-PCR suggested that the transfection is successful (Figure 4A). CCK-8 assay, EdU assay and flow cytometry showed that overexpression of ELF1 accelerated cell growth and inhibited cell apoptosis, while knockdown of DCLK1 weakened this effect; knocking down ELF1 inhibited cell proliferation and promoted apoptosis, while the DCLK1 overexpression reversed this effect (Figure 4B-D). These data suggested the biological functions of ELF1 were partially dependent on DCLK1.
Figure 4.
ELF1 interferes with cell proliferation and apoptosis by targeting DCLK1. The ELF1 overexpression plasmid and si-DCLK1 were co-transfected into Capan-2 cells, and the si-ELF1 and DCLC1 overexpression plasmids were co-transfected into PANC-1 cells. A. DCLK1 mRNA expression was probed by qRT-PCR after transfection. B, C. The viability of Capan-2 and PANC-1 cells was detected by CCK-8 and EdU assays. D. The apoptosis of Capan-2 and PANC-1 cells was detected by flow cytometry. *P < 0.05, **P < 0.01, and ***P < 0.001.
DCLK1 activates the JAK-STAT signaling pathway
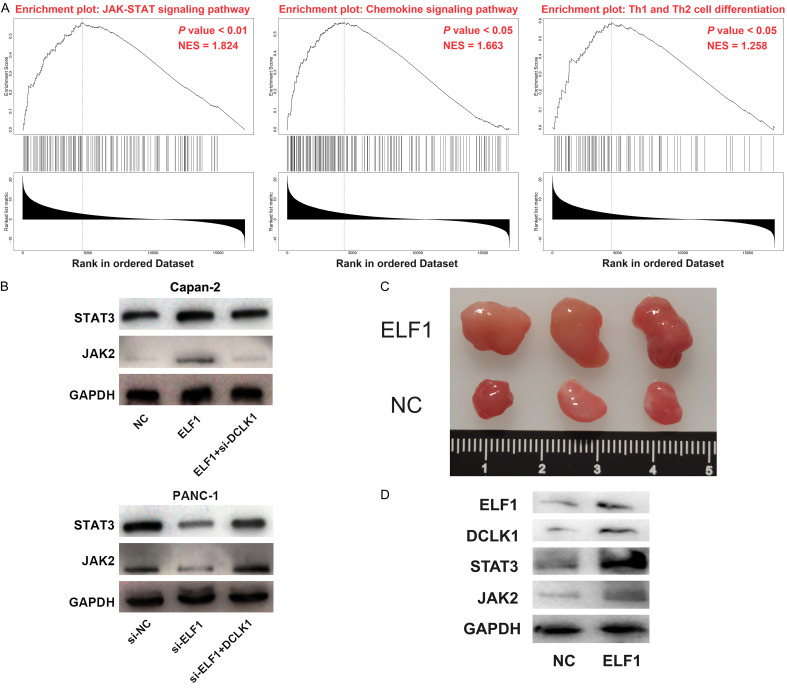
To further elaborate on the downstream mechanism of DCLK1 in regulating the biological function of PC cells, we used LinkedOmics database (http://www.linkedomics.org) to conduct gene set enrichment analysis, and proved that DCLK1 expression was correlated with JAK-STAT signaling pathway (Figure 5A). Janus kinase 2 (JAK2) is a transcriptional co-activator of the JAK-STAT signaling pathway located at the upstream of STAT3 signaling pathway and is the major activator of STAT3 [18]. EdU assay, flow cytometry and Western blot showed that DCLK1 overexpression promoted the proliferation of PANC-1 cells, and repressed the apoptosis, accompanied with up-regulated JAK2 expression and STAT3 expression in PANC-1 cells, while the treatment of Ganoderic acid A (a JAK-STAT pathway inhibitor) totally reversed these effects (Supplementary Figure 1). These results supported that DCLK1 promoted the malignant biological behaviors of PC cells via modulating JAK/STAT pathway. Western blot showed that overexpression of ELF1 promoted the expression of STAT3 and JAK2, while knocking down DCLK1 weakened these effects; knockdown of ELF1 inhibited the expression of STAT3 and JAK2, while overexpression of DCLK1 worked oppositely (Figure 5B). These data supported that ELF1 potentially modulate DCLK1/JAK/STAT axis. To further validate the oncogenic role of ELF1 in PC, a tumorigenesis model in nude mice was constructed, and the results showed that Capan-2 cells with ELF1 overexpression could form larger tumor in nude mice compared with the control cells (Figure 5C). Additionally, the expression levels of ELF1, DCLK1, STAT3 and JAK2 in the tumor tissues of ELF1 overexpression group were obviously higher than those in the control group (Figure 5D), which further supported that ELF1 promoted PC progression via DCLK1/JAK/STAT axis.
Figure 5.
ELF1 modulates JAK-STAT signaling pathway via DCLK1. A. Gene set enrichment analysis was adopted to analyze the signaling pathway related to DCLK1. B. The ELF1 overexpression plasmid and si-DCLK1 were co-transfected into Capan-2 cells, and the si-ELF1 and DCLK1 overexpression plasmids were co-transfected into PANC-1 cells. JAK2 expression and STAT3 expression were analyzed by Western blot assay. C. After ELF1 overexpression plasmids were transfected into Capan-2 cells, the Capan-2 cells with ELF1 overexpression and the control Capan-2 cells were subcutaneously transplanted into nude mice, and the tumor size of the two groups (3 mice per group) was compared. D. The expression levels of ELF1, DCLK1, STAT3 and JAK2 in the tumor tissues of the two groups of mice were detected by Western blot and compared.
Discussion
Due to insidious early symptoms and rapid disease progression, as well as the low surgical resection rate and insensitivity to chemotherapy, the prognosis of PC is extremely poor, with 90% of patients surviving for less than one year. In this context, it is crucial to seek new treatment strategies [19]. Here we found that the expression of ELF1 in PC tissues and cell lines was up-regulated and interrelated with a short survival time of patients, indicating that it could be a prognostic marker and therapeutic target for PC.
E26 transformation-specific transcription factor family is a large and important family of evolutionarily conservative transcription factors [20-22]. Among them, the gene of ELF1, as one of its important family members, is located in the 13q13.3-13q14.11 region, and it is mainly expressed in lymphocytes. Moreover, ELF1 acts as an enhancer to regulate the transcription of various genes, thus participates in the progression of certain tumors [23-25]. For example, highly expressed ELF1 is associated with poor prognosis of the patients with endometrial cancer and ovarian cancer [26,27]. ELF1 is upregulated in gliomas and ELF1 promotes the malignant progression of gliomas by activating MEIS1 and regulating the GFI1/FBW7 axis [28]. ELF1 is also up-regulated in oral squamous cell carcinoma, and as a transcription factor of CTNB1, ELF1 promotes the growth of oral squamous carcinoma cells by facilitating the expression of CTNB1 [29]. However, the biological function of ELF1 in PC has not been fully clarified yet. Here we found up-regulating ELF1 promotes the proliferation and inhibits the apoptosis of PC cells; knocking down ELF1 inhibits the proliferation and induces the apoptosis of PC cells. Based on our findings, we conclude that ELF1 plays a role as an oncoprotein in PC.
DCLK1 is a transmembrane microtubule-associated protein kinase with a DCX domain and a C-terminal serine/threonine protein kinase domain, which is considered to be a crucial modulator of cell movement [30]. Previous studies have shown that DCLK1 is highly expressed in a variety of cancers and has been identified as a potential oncogene associated with cancer progression, in intestinal tumor, non-small cell lung cancer and nasopharyngeal carcinoma [31-34]. In addition, there are a large number of reports about the role of DCLC1 in PC. For example, DCLC1 is overexpressed in PC and is interrelated to the poor prognosis; depletion of DCLK1 inhibits epithelial-mesenchymal transition by down-regulating Bmi-1, which in turn represses the migration and invasion of PC cells [35]. Up-regulation of DCLK1 activates KRAS, which enhances PI3K/AKT/mTOR signaling and facilitates the migration and invasion of PC cells [36]; another study reports that DCLK1 is up-regulated in PC, and miR-195 directly targets DCLK1 to inhibit the proliferation, migration and aggressiveness of PC cells [37]. In this study, we found that ELF1 could bind to the promoter region of DCLK1, thus activating transcription of DCLK1. Our data give a reasonable explanation for DCLK1 dysregulation in PC.
Janus kinase/signal transducer and activator of transcription (JAK/STAT) signal pathway is a classical pathway involved in tumorigenesis and development. For example, in PC, IL28RA activates JAK-STAT pathway to accelerate the cell proliferation and invasion [38]. In the present work, with in vitro and in vivo data, we found overexpression of ELF1 activates the JAK-STAT signaling pathway by targeting DCLK1. Collectively, the ELF1/DCLK1/JAK-STAT signaling pathway may accelerate the progression of PC.
Conclusion
In conclusion, to our best knowledge, this is the first study that provides direct evidence for the role of ELF1 in PC progression. Mechanistically, ELF1 activates the JAK-STAT signaling pathway by targeting DCLK1, thus promoting the malignant progression of PC. This study potentially offers novel treatment targets for PC patients. Nonetheless, there are some limitations to this work. Firstly, this study is still limited to single-center samples. In the future, tissue sample size is necessary to be enlarged to explore the prognostic prediction value of ELF1 in depth. Additionally, the regulatory effects of ELF1 and DCLK1 on the other members of JAK-STAT signaling still remain to be investigated.
Acknowledgements
This work was supported by General Project of National Natural Science Foundation of China (No. 72074188).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Lee SH, Hwang HK, Lee WJ, Kang CM. MCT4 as a potential therapeutic target to augment gemcitabine chemosensitivity in resected pancreatic cancer. Cell Oncol (Dordr) 2021;44:1363–1371. doi: 10.1007/s13402-021-00643-8. [DOI] [PubMed] [Google Scholar]
- 2.Arslan C, Yalcin S. Current and future systemic treatment options in metastatic pancreatic cancer. J Gastrointest Oncol. 2014;5:280–295. doi: 10.3978/j.issn.2078-6891.2014.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Zhang S, Zhao P, Zeng H, Zou X, He J. Annual report on status of cancer in China, 2010. Chin J Cancer Res. 2014;26:48–58. doi: 10.3978/j.issn.1000-9604.2014.01.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen S, Hao Y, Huang S, Leng D, Ma Y. Resection of rectal metastasis after previous radical surgery for pancreatic cancer: case report and literature review. Medicine (Baltimore) 2023;102:e36365. doi: 10.1097/MD.0000000000036365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Z, Chen Z, Peng M, Zhang Z, Luo W, Shi R, Wang L, Hong Y. MicroRNA MiR-490-5p suppresses pancreatic cancer through regulating epithelial-mesenchymal transition via targeting MAGI2 antisense RNA 3. Bioengineered. 2022;13:2673–2685. doi: 10.1080/21655979.2021.2024653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu H, Li T, Du Y, Li M. Pancreatic cancer: challenges and opportunities. BMC Med. 2018;16:214. doi: 10.1186/s12916-018-1215-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rombouts SJ, Vogel JA, van Santvoort HC, van Lienden KP, van Hillegersberg R, Busch OR, Besselink MG, Molenaar IQ. Systematic review of innovative ablative therapies for the treatment of locally advanced pancreatic cancer. Br J Surg. 2015;102:182–193. doi: 10.1002/bjs.9716. [DOI] [PubMed] [Google Scholar]
- 8.Huang X, Wang Z, Hou S, Yue C, Li Z, Hu W, Lu H. Long non-coding RNA DSCAM-AS1 promotes pancreatic cancer progression via regulating the miR-136-5p/PBX3 axis. Bioengineered. 2022;13:4153–4165. doi: 10.1080/21655979.2021.2016326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sunkel BD, Stanton BZ. Pioneer factors in development and cancer. iScience. 2021;24:103132. doi: 10.1016/j.isci.2021.103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambert M, Jambon S, Depauw S, David-Cordonnier MH. Targeting transcription factors for cancer treatment. Molecules. 2018;23:1479. doi: 10.3390/molecules23061479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun T, Zhang J. ETV4 mediates the Wnt/β-catenin pathway through transcriptional activation of ANXA2 to promote hepatitis B virus-associated liver hepatocellular carcinoma progression. J Biochem. 2021;170:663–673. doi: 10.1093/jb/mvab088. [DOI] [PubMed] [Google Scholar]
- 12.Xu TP, Wang YF, Xiong WL, Ma P, Wang WY, Chen WM, Huang MD, Xia R, Wang R, Zhang EB, Liu YW, De W, Shu YQ. E2F1 induces TINCR transcriptional activity and accelerates gastric cancer progression via activation of TINCR/STAU1/CDKN2B signaling axis. Cell Death Dis. 2017;8:e2837. doi: 10.1038/cddis.2017.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue Y, Wu T, Sheng Y, Zhong Y, Hu B, Bao C. MicroRNA-1252-5p, regulated by Myb, inhibits invasion and epithelial-mesenchymal transition of pancreatic cancer cells by targeting NEDD9. Aging (Albany NY) 2021;13:18924–18945. doi: 10.18632/aging.203344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L. ELF1-activated FOXD3-AS1 promotes the migration, invasion and EMT of osteosarcoma cells via sponging miR-296-5p to upregulate ZCCHC3. J Bone Oncol. 2020;26:100335. doi: 10.1016/j.jbo.2020.100335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou B, Sun C, Hu X, Zhan H, Zou H, Feng Y, Qiu F, Zhang S, Wu L, Zhang B. MicroRNA-195 suppresses the progression of pancreatic cancer by targeting DCLK1. Cell Physiol Biochem. 2017;44:1867–1881. doi: 10.1159/000485876. [DOI] [PubMed] [Google Scholar]
- 16.Ge Y, Liu H, Zhang Y, Liu J, Yan R, Xiao Z, Fan X, Huang X, An G. Inhibition of DCLK1 kinase reverses epithelial-mesenchymal transition and restores T-cell activity in pancreatic ductal adenocarcinoma. Transl Oncol. 2022;17:101317. doi: 10.1016/j.tranon.2021.101317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito H, Tanaka S, Akiyama Y, Shimada S, Adikrisna R, Matsumura S, Aihara A, Mitsunori Y, Ban D, Ochiai T, Kudo A, Arii S, Yamaoka S, Tanabe M. Dominant expression of DCLK1 in human pancreatic cancer stem cells accelerates tumor invasion and metastasis. PLoS One. 2016;11:e0146564. doi: 10.1371/journal.pone.0146564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin XM, Chen H, Zhan XL. MiR-203 regulates JAK-STAT pathway in affecting pancreatic cancer cells proliferation and apoptosis by targeting SOCS3. Eur Rev Med Pharmacol Sci. 2019;23:6906–6913. doi: 10.26355/eurrev_201908_18730. [DOI] [PubMed] [Google Scholar]
- 19.Zhao X, Zhang X, Zhang X, Jiang T, Zhai J, Wang H, Huang M, Lang R, He Q. MiR-374b-5p inhibits KDM5B-induced epithelial-mesenchymal transition in pancreatic cancer. Am J Cancer Res. 2021;11:3907–3920. [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang W, Xu Y, Chen X, Pan S, Zhu X. E26 transformation-specific variant 4 as a tumor promotor in human cancers through specific molecular mechanisms. Mol Ther Oncolytics. 2021;22:518–527. doi: 10.1016/j.omto.2021.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sizemore GM, Pitarresi JR, Balakrishnan S, Ostrowski MC. The ETS family of oncogenic transcription factors in solid tumours. Nat Rev Cancer. 2017;17:337–351. doi: 10.1038/nrc.2017.20. [DOI] [PubMed] [Google Scholar]
- 22.Oh S, Shin S, Janknecht R. ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta. 2012;1826:1–12. doi: 10.1016/j.bbcan.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang M, Yang C, Liu X, Zheng J, Xue Y, Ruan X, Shen S, Wang D, Li Z, Cai H, Liu Y. An upstream open reading frame regulates vasculogenic mimicry of glioma via ZNRD1-AS1/miR-499a-5p/ELF1/EMI1 pathway. J Cell Mol Med. 2020;24:6120–6136. doi: 10.1111/jcmm.15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang X, Brown C, Ni W, Maynard E, Rigby AC, Oettgen P. Critical role for the Ets transcription factor ELF-1 in the development of tumor angiogenesis. Blood. 2006;107:3153–60. doi: 10.1182/blood-2005-08-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tzenov YR, Andrews PG, Voisey K, Popadiuk P, Xiong J, Popadiuk C, Kao KR. Human papilloma virus (HPV) E7-mediated attenuation of retinoblastoma (Rb) induces hPygopus2 expression via Elf-1 in cervical cancer. Mol Cancer Res. 2013;11:19–30. doi: 10.1158/1541-7786.MCR-12-0510. [DOI] [PubMed] [Google Scholar]
- 26.Gerloff A, Dittmer A, Oerlecke I, Holzhausen HJ, Dittmer J. Protein expression of the Ets transcription factor Elf-1 in breast cancer cells is negatively correlated with histological grading, but not with clinical outcome. Oncol Rep. 2011;26:1121–1125. doi: 10.3892/or.2011.1409. [DOI] [PubMed] [Google Scholar]
- 27.Takai N, Miyazaki T, Nishida M, Shang S, Nasu K, Miyakawa I. Clinical relevance of Elf-1 overexpression in endometrial carcinoma. Gynecol Oncol. 2003;89:408–13. doi: 10.1016/s0090-8258(03)00131-8. [DOI] [PubMed] [Google Scholar]
- 28.Cheng M, Zeng Y, Zhang T, Xu M, Li Z, Wu Y. Transcription factor ELF1 activates MEIS1 transcription and then regulates the GFI1/FBW7 axis to promote the development of glioma. Mol Ther Nucleic Acids. 2020;23:418–430. doi: 10.1016/j.omtn.2020.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao C, Qiao T, Yang S, Liu L, Zheng M. SNHG17/miR-384/ELF1 axis promotes cell growth by transcriptional regulation of CTNNB1 to activate Wnt/β-catenin pathway in oral squamous cell carcinoma. Cancer Gene Ther. 2022;29:122–132. doi: 10.1038/s41417-021-00294-9. [DOI] [PubMed] [Google Scholar]
- 30.Gagliardi G, Moroz K, Bellows CF. Immunolocalization of DCAMKL-1, a putative intestinal stem cell marker, in normal colonic tissue. Pathol Res Pract. 2012;208:475–479. doi: 10.1016/j.prp.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 31.Ferguson FM, Nabet B, Raghavan S, Liu Y, Leggett AL, Kuljanin M, Kalekar RL, Yang A, He S, Wang J, Ng RWS, Sulahian R, Li L, Poulin EJ, Huang L, Koren J, Dieguez-Martinez N, Espinosa S, Zeng Z, Corona CR, Vasta JD, Ohi R, Sim T, Kim ND, Harshbarger W, Lizcano JM, Robers MB, Muthaswamy S, Lin CY, Look AT, Haigis KM, Mancias JD, Wolpin BM, Aguirre AJ, Hahn WC, Westover KD, Gray NS. Discovery of a selective inhibitor of doublecortin like kinase 1. Nat Chem Biol. 2020;16:635–643. doi: 10.1038/s41589-020-0506-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandrakesan P, Yao J, Qu D, May R, Weygant N, Ge Y, Ali N, Sureban SM, Gude M, Vega K, Bannerman-Menson E, Xia L, Bronze M, An G, Houchen CW. Dclk1, a tumor stem cell marker, regulates pro-survival signaling and self-renewal of intestinal tumor cells. Mol Cancer. 2017;16:30. doi: 10.1186/s12943-017-0594-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Zhan Y, Abuduwaili K, Wang X, Shen Y, Nuerlan S, Liu C. Knockdown of long non-coding RNA HOTAIR suppresses cisplatin resistance, cell proliferation, migration and invasion of DDP-resistant NSCLC cells by targeting miR-149-5p/doublecortin-like kinase 1 axis. Cancer Manag Res. 2020;12:7725–7737. doi: 10.2147/CMAR.S246299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, Li H, You J, Xue H, Tan X, Chao C. MicroRNA-223-5p suppresses the progression of nasopharyngeal carcinoma by targeting DCLK1. Oncol Lett. 2021;21:396. doi: 10.3892/ol.2021.12657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Wang Y, Ge J, Li W, Yin L, Zhao Z, Liu S, Qin H, Yang J, Wang L, Ni B, Liu Y, Wang H. Doublecortin-like kinase 1 (DCLK1) regulates B cell-specific moloney murine leukemia virus insertion site 1 (Bmi-1) and is associated with metastasis and prognosis in pancreatic cancer. Cell Physiol Biochem. 2018;51:262–277. doi: 10.1159/000495228. [DOI] [PubMed] [Google Scholar]
- 36.Qu D, Weygant N, Yao J, Chandrakesan P, Berry WL, May R, Pitts K, Husain S, Lightfoot S, Li M, Wang TC, An G, Clendenin C, Stanger BZ, Houchen CW. Overexpression of DCLK1-AL increases tumor cell invasion, drug resistance, and KRAS activation and can be targeted to inhibit tumorigenesis in pancreatic cancer. J Oncol. 2019;2019:6402925. doi: 10.1155/2019/6402925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon MS, Chung HK, Xiao L, Yu TX, Wang SR, Piao JJ, Rao JN, Gorospe M, Wang JY. MicroRNA-195 regulates Tuft cell function in the intestinal epithelium by altering translation of DCLK1. Am J Physiol Cell Physiol. 2021;320:C1042–C1054. doi: 10.1152/ajpcell.00597.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang L, Wei WC, Meng XN, Gao J, Guo N, Wu FT, Zeng WW. Significance of IL28RA in diagnosis of early pancreatic cancer and its regulation to pancreatic cancer cells by JAK/STAT signaling pathway - effects of IL28RA on pancreatic cancer. Eur Rev Med Pharmacol Sci. 2019;23:9863–9870. doi: 10.26355/eurrev_201911_19550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.