Abstract
Increasing evidence indicates that long noncoding RNAs (lncRNAs) are therapeutic targets and key regulators of tumors development and progression, including melanoma. Long intergenic non-protein-coding RNA 511 (LINC00511) has been demonstrated as an oncogenic molecule in breast, stomach, colorectal, and lung cancers. However, the precise role and functional mechanisms of LINC00511 in melanoma remain unknown. This study confirmed that LINC00511 was highly expressed in melanoma cells (A375 and SK-Mel-28 cells) and tissues, knockdown of LINC00511 could inhibit melanoma cell migration and invasion, as well as the growth of subcutaneous tumor xenografts in vivo. By using Chromatin immunoprecipitation (ChIP) assay, it was demonstrated that the transcription factor Yin Yang 1 (YY1) is capable of binding to the LINC00511 promoter and enhancing its expression in cis. Further mechanistic investigation showed that LINC00511 was mainly enriched in the cytoplasm of melanoma cells and interacted directly with microRNA-150-5p (miR-150-5p). Consistently, the knockdown of miR-150-5p could recover the effects of LINC00511 knockdown on melanoma cells. Furthermore, ADAM metallopeptidase domain expression 19 (ADAM19) was identified as a downstream target of miR-150-5p, and overexpression of ADAM19 could promote melanoma cell proliferation. Rescue assays indicated that LINC00511 acted as a competing endogenous RNA (ceRNA) to sponge miR-150-5p and increase the expression of ADAM19, thereby activating the PI3K/AKT pathway. In summary, we identified LINC00511 as an oncogenic lncRNA in melanoma and defined the LINC00511/miR-150-5p/ADAM19 axis, which might be considered a potential therapeutic target and novel molecular mechanism the treatment of patients with melanoma.
Keywords: LINC00511, melanoma, ADAM19, ceRNA, PI3K/AKT
Introduction
Melanoma is a high-grade malignant tumor originating from melanocytes in the neural crest, and its morbidity rate has been increasing annually [1,2]. Despite advances in treatment, the five-year survival rate of patients with melanoma remains low due to its highly metastatic nature and resistance to treatment [3,4]. The development of melanoma is multifactorial with an interplay between environmental exposure and genetic susceptibility, with ultraviolet exposure being the most critical risk factor [5-7]. Studies have demonstrated that long noncoding RNAs (lncRNAs) regulate melanoma progression through various epigenetic mechanisms, including chromatin modification and remodeling and gene transcription regulation, and act as competing endogenous RNAs (ceRNAs) [8]. Accordingly, it is essential to investigate the molecular regulatory mechanisms implicated in melanoma is essential for the effective treatment of this disease.
LncRNAs are RNA molecules longer than 200 nucleotides that lack an open reading frame [9]. They perform critical regulatory functions in diverse biological processes by acting as guides, scaffolds, decoys, signaling molecules, or sponges to modulate microRNA (miRNA) activity. Numerous studies have demonstrated that lncRNAs play a key role in cancer progression at multiple levels, including epigenetic, transcriptional, and post-transcriptional regulation [10]. For instance, the lncRNA CASC15 facilitates melanoma proliferation and migration through the epigenetic regulation of PDCD4 [11]. The lncRNA THOR promotes melanoma cell invasion and metastasis by interacting with the RNA-binding protein, IGF2BP1, to stabilize the IGF2 and CD44 genes [12]. The lncRNA MALAT1 acts as a molecular sponge for miR-22, modulating the expression of its downstream targets, MMP14 and Snail, and facilitating melanoma cell growth and metastasis [13]. In contrast, the lncRNA-HOXA11-AS can prevent the proliferation and metastasis of melanoma cells while promoting apoptosis by regulating the miR-152-3p/ITGA9 axis [14]. Additionally, lncRNAs MEG3 [15], CASC2 [16], LINC00518 [17], and SAMMSON [18] have been identified as potential biomarkers and therapeutic targets for melanoma. Given these findings, it is essential to investigate the precise molecular mechanisms underlying lncRNA function in melanoma to develop effective treatment strategies.
The LINC00511 RNA gene is located on chromosome 17q24.3, spanning a transcriptional length of 2265 bp [19]. It encodes the lncRNA LINC00511, which plays a crucial role in the development and progression of various tumors by regulating tumor cell migration, invasion, proliferation, and other malignant phenotypes. For example, LINC00511 regulates cell movement in glioblastoma cells by functioning as a molecular sponge for miR-126-5p, which indicates its potential as a prognostic marker for glioblastoma [20]. LINC00511 stimulates the growth and invasion of non-small cell lung cancer (NSCLC) cells by targeting miR-625-5p/GSPT1 [21]. In bladder cancer, LINC00511 suppresses miR-143-3p expression and promotes PCMT1 production, thereby preventing the proliferation and invasion of bladder cancer cells [22]. Moreover, LINC00511 inhibits the occurrence and progression of colorectal cancer through the suppression of the miR-625/WEE1 and miR-29c-3p/NFIA axes [23,24]. However, the role of LINC00511 in melanoma remains unclear.
Several studies have established that lncRNAs participate in cancer development by regulating the ceRNA network balance [25]. For instance, the lncRNA SNHG16 promotes cervical cancer progression by acting as an endogenous ‘sponge’ that interacts with the miR-216A-5p/ZEB1 axis [26]. The lncRNA SNHG6, promotes the malignant phenotype of colorectal cancer by upregulating EZH2 through sponge-like activity on miR-26a/b and miR-214 [27]. The lncRNA ARNILA functions as a ceRNA of miR-204 and upregulates its downstream target gene, SOX4, thus contributing to the development of triple-negative breast cancer and facilitating tumor cell invasion and metastasis [28]. Additionally, the lncRNA DSCAM-AS1 promotes AKT3 overexpression in colorectal cancer by acting as a molecular sponge for miR-384 and promoting the progression of NSCLC by targeting the miR-577/HMGB1 axis [29,30]. Hence, clarifying the specific mechanisms underlying the regulatory network of ceRNAs will improve our understanding of the molecular mechanisms underlying cancer development.
Consistently, this study aimed to elucidate the biological function of LINC00511 and explore its ceRNA regulatory mechanism in melanoma progression. These findings will potentially help in the identification of novel therapeutic targets and the development of diagnostic biomarkers.
Materials and methods
Cell culture
The human melanoma cell lines (A375 and SK-Mel-28) and human normal melanocytes (PIG1) used in this study were procured from Pricella (Wuhan, China). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with fetal bovine serum (FBS).
siRNA transfection and lentiviral infection
According to the manufacturer’s manual, transfection was carried out using polyplus jetPRIME® reagent (Illkirch, France). The siRNAs targeting LINC00511 and ADAM19, negative control (NC) si-RNA, miR-150-5p inhibitor, miR-150-5p mimics, and negative control miR-150-5p NC were all purchased from GenePharma (Shanghai, China). The overexpression vectors for LINC00511 and ADAM19, as well as the controls, were purchased from Genechem Co., Ltd. (Shanghai, China). Stable cell lines were established using lentiviral vectors from Genechem Co., Ltd. for the siRNA-mediated knockdown of LINC00511 and ADAM19. Prior to lentiviral transduction, A375 cells were seeded at a density of 5 × 104 cells/well in a 6-well plate. Cells were transduced with lentiviral vectors and 10 μg/mL polybrene (Sigma, Aldrich, St. Louis, MO, USA) at a multiplicity of infection of 10. Medium was changed 12 h post-transduction. To select for stably transduced cells, cells were cultured with a medium containing 2 μg/mL puromycin (Sigma-Aldrich) for 48 h. Stable transduced cells were cultured in medium containing 0.5 μg/mL puromycin. Transduction efficiency was assessed 72 h post-transduction using fluorescence microscopy and further confirmed using qRT-PCR and western blotting. The small interference sequences are listed in Table S1.
Quantitative reverse transcription-PCR (qRT-PCR)
The TRIzol method (Takara, Otsu, Shiga, Japan) was used for the isolation and extraction of total RNA. Nanodrop one spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to assess the quantity and quality of the RNA. The primers used in this study are listed in Table S2.
Western blot analysis
To analyze the relevant proteins, the cells were lysed in the RIPA lysis buffer (Elabscience, USA). The BCA assay kit was used to calculate the protein concentration (Thermo Fisher Scientific, Waltham, MA, USA). Proteins separated after electrophoresis were transferred onto polyvinylidene difluoride membranes, blocked, and overnight incubated with specific primary antibodies against ADAM19, AKT, p-AKT, PI3K, p-PI3K, GAPDH, Bax, Bcl-2, MMP9, and PCNA followed by one-hour incubation using secondary antibodies at room temperature. The blots were analyzed using the ImageJ software (NIH, Version 1.53). The source files of western blotting are provided in Table S3.
Tissue samples
Melanoma tissue microarray (TMA) (IWLT-N-47ML61) was obtained from Shanghai Outdo Biotech Company (China). This tissue microarray contained 37 carcinoma tissue samples and 10 normal tissue samples. Details of each sample are listed in Table S4.
Xenograft tumor assay
We selected immunodeficient male nude mice (5-6 weeks old, Beijing Experimental Animal Center, Beijing, China), which were randomly divided into experimental and control groups, with 6 mice in each group. Subcutaneous injections were performed according to the grouping, with a cell count of approximately 1 × 107/0.1 ml. All mice had ad libitum access to food and water. Sterile food, drinking water, and padding were changed every 3 days. Moreover, tumor volume was monitored to assess tumor growth. Approximately 4 weeks later, the mice were euthanized to obtain tumor tissue for weighing and measurement analysis. Tumor volume was calculated using the following formula: Tumor volume = ½LW2, where L is the maximum length and W is the minimum length. All procedures in these studies complied with the guidelines for the use of laboratory animals and the care guidelines of the National Research Council. This study was approved by the Ethics Committee of Inner Mongolia University (IMU-mouse-2022-051).
Cell proliferation assay
A certain number of cells were plated following the guidelines of the Cell Counting Kit-8 (CCK-8) (Everbright America, Suzhou, China). Ten microliters of CCK-8 solution was added per well after the cells had adhered, followed by 1 h incubation at 37°C. The absorbance was measured at 450 nm at designated time points to generate proliferation curves, and the experiments were conducted in triplicate.
The 5-ethynyl-2’-deoxyuridine (EdU) proliferation assay was performed using the Beyoclick™ EdU Cell Proliferation Kit (Beyotime Biotechnology, Shanghai, China). Fluorescence microscopy (Nikon) was employed to capture images, and EdU-positive and total cells were counted in each field.
Apoptosis analysis
The TransDetect Annexin V-FITC/PI Apoptosis Detection Kit and TUNEL Apoptosis Assay Kit (Promega, Madison, WI, USA) were used to conduct apoptosis assays. Flow cytometry was performed to assess the cell apoptosis status, and the cell fluorescence intensities were measured using a confocal microscope (Leica, Wetzlar, Germany).
Immunohistochemistry analysis (IHC)
Tumor tissue was subjected to various treatments, including dewaxing in xylene and hydration in ethanol. Endogenous peroxidase activity was blocked by incubating the sections in a 3% H2O2 solution. Next, antigen retrieval was carried out by boiling the samples in a citric acid solution (pH = 6.0). Non-specific binding was blocked by applying host serum before incubating with anti-ADAM19 and anti-Ki-67 primary antibodies overnight at 4°C. A biotinylated secondary antibody was subsequently applied, followed by incubation with HRP-conjugated streptavidin. The tissue was counterstained with hematoxylin, dehydrated, and mounted after incubation in a 3,3’-diaminobenzidine solution. Finally, the images were obtained using an inverted microscope (Nikon). Analysis of the findings was conducted employing ImageJ software.
Fluorescence in situ hybridization (FISH) assay
The FISH kit (GenePharma, Shanghai, China) was used to determine the subcellular localization of LINC00511. Correlative fluorescence images were obtained using a laser confocal scanning microscope (Leica, Wetzlar, Germany).
Chromatin immunoprecipitation (ChIP) assay
The ChIP experiment was performed using the EZ-CHIP KIT (Millipore, Billerica, MA, USA). First, A375 and SK-Mel-28 cells (1 × 107 cells/well in a 6-well plate) were fixed with 1% formaldehyde at room temperature for 10 min, followed by termination with glycine and collection in lysis buffer. After centrifugation at 13000 rpm for 5 min, the supernatant was aspirated. The lysates were then sonicated, washed, and subjected to magnetic bead pretreatment. The precleared chromatin was then incubated with 5 μg of anti-YY1 ChIP-grade antibody or IgG antibody at 4°C with slow rotation for 12-16 h. Subsequently, 20 μL of protein A/G beads were added, and the mixture was rotated at room temperature for 30 min. This was followed by washing and elution with elution buffer. After reverse cross-linking, RNase A and Proteinase K treatment, the purified immunoprecipitated DNA was analyzed using qRT-PCR. The primers sequences used in the ChIP experiment are listed in Table S2.
RNA pull-down assay
For the biotinylated miRNA pull-down assay, A375 and SK-Mel-28 cells (1 × 107 cells/well in a 6-well plate) were prepared for detection of the binding of biotinylated miR-150-5p to LINC00511/ADAM19. Biotinylated miR-150-5p and biotinylated miR-NC were transiently transfected into A375 and SK-Mel-28 cells. After 48 h, whole cell lysates were prepared and mixed with streptavidin magnetic beads (Invitrogen, Waltham, MA, USA) and incubated at 4°C with rotation for 16 h. The beads were then collected and thoroughly washed, and the RNA was finally eluted and purified for further analysis. The levels of specific RNA were detected using qRT-PCR.
RNA immunoprecipitation assay (RIP)
The RIP kit (GENESEED, Guangzhou, China) was used for the RIP assay. A375 and SK-Mel-28 cells (1 × 107 cells/well in a 6-well plate) were washed twice with cold phosphate-buffered saline and then centrifuged at 1500 rpm at 4°C for 10 min to collect the cells. After the supernatant was discarded, lysis buffer was added to the cell pellet, which was then incubated on ice for 10 min. The lysate was then centrifuged at 4°C at 14,000 rpm for 10 min. Subsequently, 100 μL of the supernatant was aliquoted as the input control, and 900 μL of the supernatant was placed in an RNase-free tube, which was incubated with pretreated beads 4°C with gentle rotation for 30 min. Ago-2 or IgG antibody was added and incubated overnight at 4°C with gentle rotation. After 5-10 washes, the purified immunoprecipitated RNA was used for analysis. Enriched RNA was further analyzed using qRT-PCR.
Luciferase reporter assay
One day before transfection, 5 × 103 cells were plated onto a 96-well plate. After 24 h incubation, the cells were co-transfected with the pmirGLO reporter vector encoding either the LINC00511 Mut or Wt gene, the ADAM19 Mut or Wt gene, and miR-mimics or mimics-NC, and incubated for another 48 h post-transfection. Luciferase activities were detected using the Dual-luciferase Reporter System (Promega, USA).
Bioinformatics analysis
LINC00511 expression data were obtained from the GSE183878 dataset of the Gene Expression Omnibus (GEO). Additionally, the expression data for LINC00511 and ADAM19 were derived using the GEPIA platform. For the prediction of the upstream transcription factor, PROMO, hTFtarget, and JASPAR databases were employed. The interaction between lncRNA and miRNA was predicted using Starbase. A subcellular localization database (lncLocator) was used to predict the cellular location of lncRNA. We used multiple online databases, such as miRDB, TargetScan, ENCORI, and miR walk, for identifying target mRNAs that directly interact with miR-150-5p and predicted the possible interactions between the relevant lncRNAs and miRNAs. The common target mRNAs were examined, and visually represented using the Draw Venn Diagram Website Tool. All website URLs used in the bioinformatic analysis are listed in Table S5.
Statistical analysis
The data were statistically evaluated using the SPSS software version 22 (SPSS; USA) and GraphPad Prism 8 (San Diego, California, USA). Differences in experimental groups were evaluated using Student’s t-test (two-tailed) or ANOVA. P < 0.05 indicated statistically significant difference in the experimental results.
Results
LINC00511 is overexpressed in melanoma cells
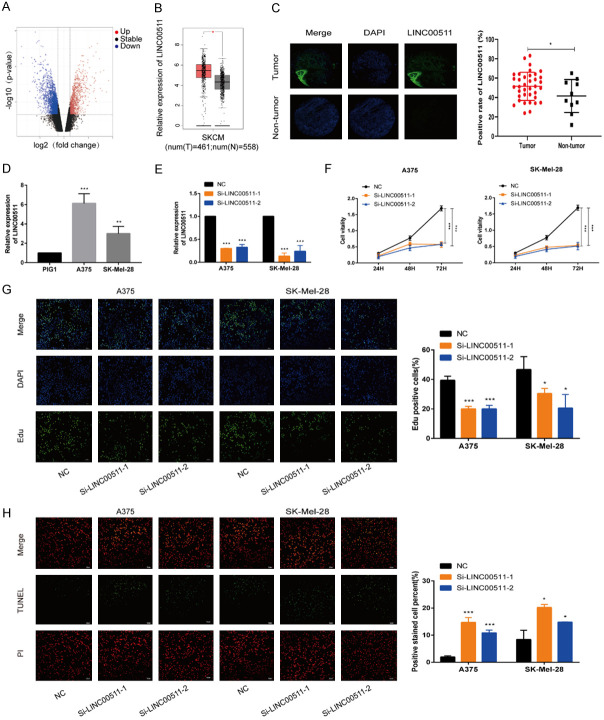
To investigate the expression of LINC00511 in melanoma, we performed in-silico data mining using high-throughput sequencing data from the GEO dataset GSE183878. The volcano plot displays the differentially expressed genes (Figure 1A), and the heatmap highlights a significantly higher expression of LINC00511 in melanoma samples compared to healthy tissues (Figure S1). These finding were further supported by data from The Cancer Genome Atlas database on Skin Cutaneous Melanoma (TCGA-SKCM), confirming the upregulation of LINC00511 in melanoma tissues (Figure 1B). Immunofluorescence analysis of melanoma tissue microarray demonstrated a significant increase in LINC00511 expression in melanoma compared to that in normal samples. Additionally, high LINC00511 expression was associated with tumor metastasis (Figure 1C and Table 1). Moreover, qRT-PCR revealed that LINC00511 was overexpressed in the melanoma cell lines A375 and SK-Mel-28 compared to that in the non-transformed, normal melanocyte cell line PIG1 (Figure 1D).
Figure 1.
LINC00511 was over-expressed in melanoma cells. A. The results from the GEO database showed that LINC00511 expression was significantly upregulated in melanoma tissues compared with normal tissues. The fold change (FC) of genes was assessed by log transformation. |log FC| > 2 and adjusted P < 0.05 were defined as the screened threshold. B. Based on the TCGA dataset, LINC00511 expression was found to be increased in melanoma tissues. C. The expression of LINC00511 in melanoma tissues and nor-tumor tissues by ISH (scale bars, 500 µm). D. Melanoma cells exhibited elevated levels of LINC00511 expression. E. Relative LINC00511 levels were in melanoma cells transfected with si-NC and si-LINC00511-1, -2. F, G. The proliferation of transfected melanoma cells was assessed using CCK8 and EdU experiments. H. The effect of transfection with si-NC, si-LINC00511-1, and -2 on the rate of apoptosis was evaluated using TUNEL analysis. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, not significant.
Table 1.
Correlation between LINC00511 expression and clinicopathologic features in melanoma
| Characteristics | Cases | LINC00511 expression | P-value | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Gender | ||||
| Male | 20 | 10 | 10 | 0.591 |
| Female | 17 | 7 | 10 | |
| Age | ||||
| > 65 | 16 | 6 | 10 | 0.368 |
| ≤ 65 | 21 | 11 | 10 | |
| Tumor site | ||||
| Nasopharynx | 19 | 6 | 13 | 0.072 |
| Other sites | 18 | 11 | 7 | |
| Tumor size | ||||
| > 1.9 cm | 17 | 10 | 7 | 0.549 |
| ≤ 1.9 cm | 20 | 10 | 10 | |
| LN Metastasis | ||||
| Yes | 8 | 1 | 7 | 0.032* |
| No | 29 | 16 | 13 | |
| Pigmentation | ||||
| Yes | 19 | 8 | 11 | 0.630 |
| No | 18 | 9 | 9 | |
| Histological type | ||||
| Epithelium | 20 | 8 | 12 | 0.431 |
| Other types | 17 | 9 | 8 | |
P < 0.05 was considered statistically significant.
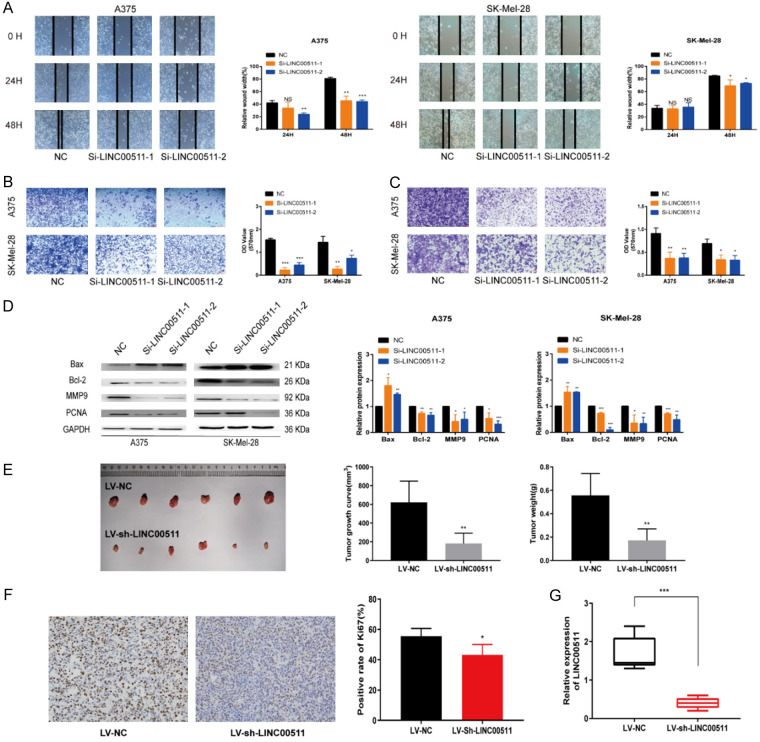
We conducted in vitro and in vivo experiments to explore the functional role of LINC00511 in melanoma. The knockdown of LINC00511 in melanoma cells was confirmed by qRT-PCR (Figure 1E). Partial silencing of LINC00511 resulted in the inhibition of melanoma cell proliferation, as demonstrated by the CCK-8 and EdU assays (Figure 1F, 1G). Moreover, LINC00511 knockdown increased the rate of apoptosis (Figure 1H) and attenuated melanoma cell migration, and invasion (Figure 2A-C). Western blot analysis confirmed that LINC00511 depletion directly affected the expression of apoptosis and migration markers (Figure 2D). In contrast, LINC00511 overexpression promoted melanoma cell proliferation, migration, and infiltration (Figure S2A-G) and altered the expression of markers related to cell apoptosis and migration (Figure S2H). These findings highlight the critical role of LINC00511 in melanoma progression in vitro.
Figure 2.
LINC00511 was over-expressed in melanoma cells. A. Metastasis ability was measured by wound-healing assay. B, C. The migration and invasion abilities of A375 and SK-Mel-28 cells were detected (scale bars, 100 µm). D. Relative expression levels of protein markers were observed in A375 and SK-Mel-28 cells transfected with si-NC, si-LINC00511-1, and -2 by western blot. E. The tumor’s morphology, growth curve, volume, and weight were observed. F. The expression of Ki-67 was assessed by IHC (scale bars, 200 µm). G. The levels of LINC00511 were measured in xenograft tissues by qRT-PCR. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, not significant.
To evaluate the effect of LINC00511 on tumor progression in vivo, the LINC00511 knockdown cells were established and transfection efficiency was confirmed by fluorescence and qRT-PCR analysis (Figure S2I). We injected A375 cells with stable downregulation of LINC00511 (LV-sh-LINC00511) or control cells into the subcutaneous tissues of nude mice. LINC00511 knockdown considerably inhibited tumor growth, as evidenced by decreased tumor volume and weight (Figure 2E) and decreased Ki67 expression (Figure 2F). Further, qRT-PCR analysis confirmed the effective inhibition of LINC00511 expression by LV-sh-LINC00511 (Figure 2G). Collectively, these findings further support the role of LINC00511 in promoting melanoma progression both in vitro and in vivo.
Transcription factor Yin Yang 1 (YY1) mediates the upregulation of LINC00511
The transcription factor YY1 induces the activation of various lncRNAs at the transcriptional level [31]. We identified overlapping transcription factors (YY1, AR) through the PROMO, hTFtarget, and JASPAR websites (Figure 3A). As the regulatory mechanism between YY1 and LINC00511 in melanoma remains unclear, we chose YY1 as a candidate transcription factor that regulates LINC00511. We predicted its binding motif using the JASPAR website (Figure 3B). qRT-PCR and western blot analysis showed that LINC00511 knockdown inhibited YY1 expression in melanoma cells (Figure 3C, 3D). YY1 mRNA and protein levels decreased to varying degrees following YY1 knockdown (Figure 3E, 3F), and YY1 knockdown reduced LINC00511 expression (Figure 3G). The ChIP assay confirmed the binding of YY1 with the LINC00511 promoter region (Figure 3H). Additionally, a luciferase reporter assay demonstrated a reduction in the luciferase activity of the LINC00511 promoter-wt upon YY1 knockdown (Figure 3I). Collectively, these findings suggest that YY1 enhances the expression of LINC00511 at the transcriptional level in melanoma cells.
Figure 3.
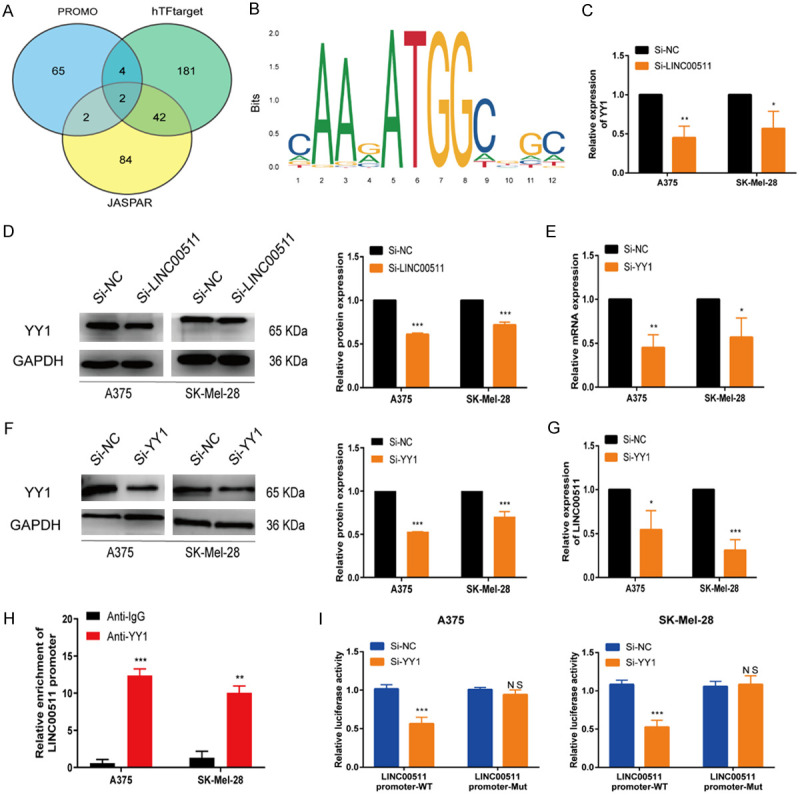
Transcription factor YY1 mediates the upregulation of LINC00511. A. The intersection of PROMO, hTFtarget, and JASPAR databases was utilized to identify transcription factors upstream of LINC00511. B. The DNA motif of YY1 on the promoter LINC00511. C, D. The expression of YY1 in A375 and SK-Mel-28 cells after LINC00511 knockdown was detected by qRT-PCR and western blot. E, F. The efficiency of YY1 knockdown in A375 and SK-Mel-28 cells was determined by qRT-PCR and western blot. G. Adopted qRT-PCR assay to detect the relative expression of LINC00511 by YY1 depletion. H. The ChIP assay confirmed the binding affinity between YY1 and the LINC00511 promoter. I. The luciferase activity of the wild/mutant LINC00511 promoter was detected upon YY1 depletion. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, not significant.
LINC00511 acts as a sponge for miR-150-5p
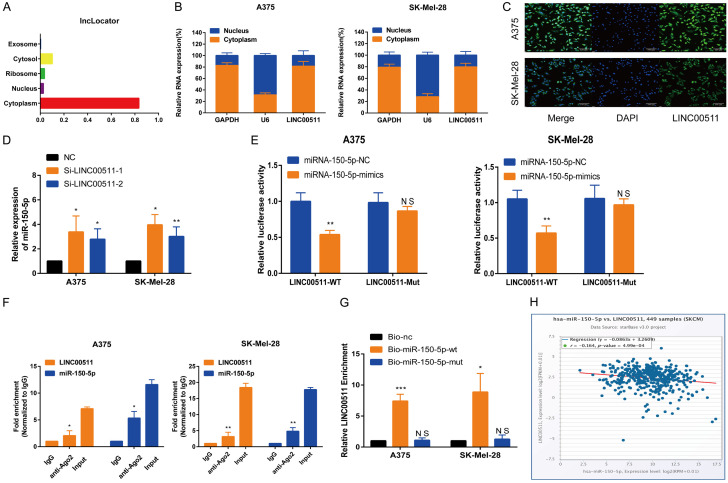
The biological functions of lncRNAs are highly dependent on their subcellular location [32]; therefore, to investigate the regulatory and biological functions of LINC00511 in melanoma, we examined its subcellular localization. The lncLocator database predicted that LINC00511 was predominantly located in the cytoplasm (Figure 4A), which was confirmed by nuclear-cytoplasmic separation and fluorescence in-situ hybridization (FISH) assays (Figure 4B, 4C). These results indicate that LINC00511 exerts its regulatory effects in the cytoplasm. In cancer, ceRNA-mediated posttranscriptional regulatory mechanisms are commonly implicated, where in lncRNAs act as ceRNAs and target miRNAs to influence the development and progression of cancer [33]. We used the starBase database for screening miRNAs and to investigate the involvement of LINC00511 in ceRNA regulation. We identified ten putative miRNAs that could interact with LINC00511 (Table S6). Based on our previous research and functional analysis [34], we hypothesized that miR-150-5p is a downstream target of LINC00511. Consistently, LINC00511 knockdown was found to increase the expression of miR-150-5p (Figure 4D). In addition, luciferase assays confirmed the direct binding of LINC00511 to miR-150-5p at endogenous levels (Figure 4E). Additionally, the RIP assay demonstrated significant enrichment of both miR-150-5p and LINC00511 in the anti-Ago2 fraction (Figure 4F). Furthermore, RNA pull-down analysis indicated that the wild-type LINC00511 was enriched in miR-150-5p vector-transfected cells compared to that in cells transfected with the empty vector (Figure 4G). Pearson’s correlation analysis revealed a negative correlation between the expression of LINC00511 and miR-150-5p in melanoma (Figure 4H). Collectively, these results support the role of LINC00511 as a ceRNA that targets miR-150-5p in melanoma cells.
Figure 4.
LINC00511 acted as a sponge for miR-150-5p. A. The localization of LINC00511 was predicted using the lncRNA subcellular localization predictor, lncLocator. B. The LINC00511 molecule was mostly located in the cytoplasm. C. FISH assay was conducted to determine the subcellular localization of LINC00511 in A375 and SK-Mel-28 cells. D. The expression levels of miR-150-5p were identified using qRT-PCR in A375 and SK-Mel-28 cells with LINC00511 knockdown. E. Luciferase activity was measured in A375 and SK-Mel-28 cells co-transfected with miR-NC and miR-150-mimics containing LINC00511-wt or LINC00511-mut. F. The RIP assay revealed that both LINC00511 and miR-150-5p expressions were enhanced in the mixture immunoprecipitated by anti-Ago2. G. The miRNA pull-down assays confirmed the binding ability between LINC00511 and miR-150-5p. H. Scatter-plots show a negative correlation between LINC00511 and miR-150-5p at the mRNA level in 499 SKCM tissues. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, not significant.
miR-150-5p directly targets ADAM19 in melanoma
Online databases (miRDB, TargetScan, ENCORI, and miRWalk) were used to predict the target mRNAs of miR-150-5p and identify their molecular role in melanoma progression. Based on this analysis, we identified ten potential target genes-ADIPOR2, CHD2, ADAM19, HILPDA, TADA1, PAPPA, BASP1, MTCH2, DCAF6, and ENSA. Among these genes, ADAM19 was overexpressed in melanoma tissues and cell lines (Figure 5A-C). Furthermore, an analysis of TCGA data revealed significantly higher levels of ADAM19 in samples of patients with melanoma than in those of healthy controls (Figure S3). Increased ADAM19 expression was associated with decreased overall survival, advanced histopathological stage, and accelerated disease progression (Figures 5D, S4A, S4B). Consistently, ADAM19 expression was elevated in melanoma tissues, and its expression was correlated with tumor metastasis (Figure 5E, 5F and Table 2). Additionally, the luciferase reporter assay confirmed that ADAM19 directly binds to miR-150-5p (Figure 5G), which was also validated by the RIP and miRNA pull-down assays (Figure 5H, 5I). Collectively, these findings confirm that ADAM19 is a downstream target of miR-150-5p in melanoma.
Figure 5.
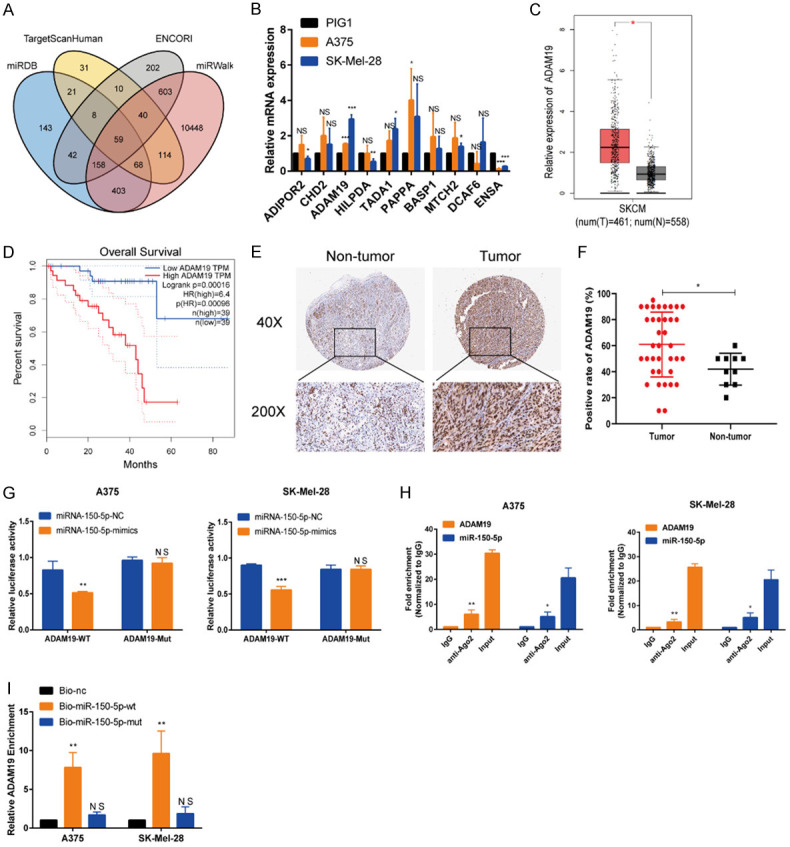
miR-150-5p directly targeted ADAM19 in melanoma. A. The intersection of miRDB, Target Scan Human, ENCORI, and miRWalk databases was used to identify ADAM19 as the downstream target of miR-150-5p. B. The top ten genes were selected for qRT-PCR analysis of their expression in A375 and SK-Mel-28 cells. C. Based on the TCGA dataset, ADAM19 expression is found to be increased in melanoma tissues. D. Overall survival analysis based on the TCGA dataset. E, F. The expression of ADAM19 in melanoma tissues and nor-tumor tissues by IHC (scale bars, 500 µm; scale bars, 100 µm). G. Luciferase activity assay was used to confirm luciferase reporter. H. RIP assays were carried out to demonstrate the coexistence of miR-150-5p and ADAM19 in RISCs. I. ADAM19 was pulled down by the biotin-miR-150-5p probe, but not by the biotin-miR-150-NC probe. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, not significant.
Table 2.
Correlation between ADAM19 expression and clinicopathologic features in melanoma
| Characteristics | Cases | ADAM19 expression | P-value | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Gender | ||||
| Male | 20 | 13 | 7 | 0.531 |
| Female | 17 | 7 | 10 | |
| Age | ||||
| > 65 | 16 | 9 | 7 | 0.424 |
| ≤ 65 | 21 | 11 | 10 | |
| Tumor site | ||||
| Nasopharynx | 19 | 10 | 9 | 0.637 |
| Other sites | 18 | 10 | 8 | |
| Tumor size | ||||
| > 1.9 cm | 17 | 9 | 8 | 1.000 |
| ≤ 1.9 cm | 20 | 11 | 9 | |
| LN Metastasis | ||||
| Yes | 8 | 4 | 4 | 0.007* |
| No | 29 | 16 | 13 | |
| Pigmentation | ||||
| Yes | 19 | 12 | 7 | 0.669 |
| No | 18 | 8 | 10 | |
| Histological type | ||||
| Epithelium | 20 | 11 | 9 | 0.491 |
| Other types | 17 | 9 | 8 | |
P < 0.05 was considered statistically significant.
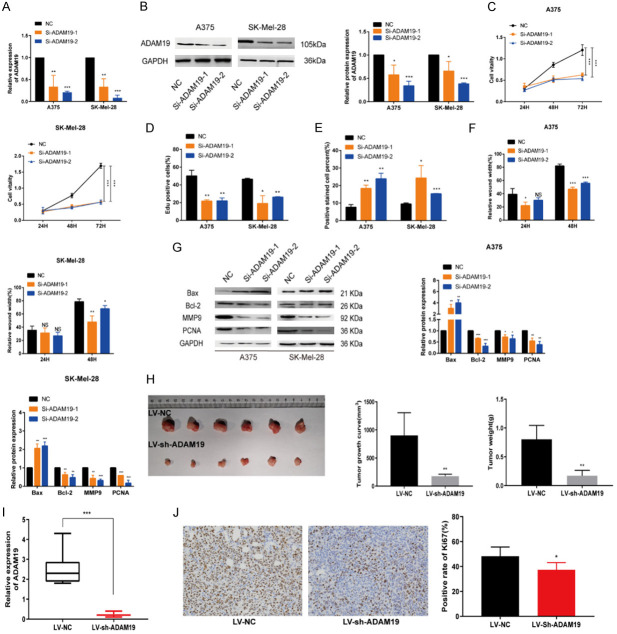
Silencing ADAM19 suppresses melanoma progression in vitro and in vivo
To specifically target ADAM19 in melanoma cells, we used si-RNAs (si-ADAM19-1 and -2), and a NC siRNA (si-NC) was used for comparison (Figure 6A, 6B). Functional assays showed that ADAM19 knockdown significantly inhibited melanoma cell proliferation, suppressed antiapoptotic properties, and impaired migration and invasion (Figures 6C-F and S5A-E). In contrast, ADAM19 overexpression promoted melanoma cell proliferation (Figure S6A-C), suppressed cell apoptosis (Figure S6D), and increased cell motility (Figure S6E-G). Western blot analysis revealed that depletion of ADAM19 altered the expression of markers associated with cell apoptosis and decreased the expression of migration markers (Figure 6G), whereas its overexpression exerted the opposite effects (Figure S6H). Additionally, we established ADAM19 knockdown cells, and fluorescence, qRT-PCR, and western blotting confirmed successful transfection (Figure S6I, S6J). We conducted in vivo experiments by injecting A375 cells with stable ADAM19 downregulation into the groin of nude mice. The results demonstrated significant suppression of tumor growth (Figure 6H). qRT-PCR analysis confirmed that ADAM19 expression was lower in tissues transfected with LV-sh-ADAM19 than in the control tissues (Figure 6I). IHC revealed decreased Ki-67 expression in the experimental group compared to that in the control group (Figure 6J). These findings suggest that ADAM19 promotes melanoma progression both in vitro and in vivo.
Figure 6.
Silencing ADAM19 suppressed melanoma progression in vitro and in vivo. A. The efficiency of si-ADAM19 knockdown was determined by qRT-PCR. B. The protein level of ADAM19 was illustrated by Western blot assay after knocking down ADAM19 in A375 and SK-Mel-28 cells. C, D. CCK8 and EdU experiments were used to evaluate the proliferation rate of transfected melanoma cells. E. The influence of ADAM19 knockdown on the rate of apoptosis was assessed by TUNEL analysis. F. Metastatic ability was assessed using the wound-healing assay. G. Relative expression levels of Bax, Bcl-2, MMP9, and PCNA were observed in A375 and SK-Mel-28 cells with ADAM19 knockdown. H. The tumor’s morphology, growth curve, volume, and weight were observed. I. The levels of ADAM19 in xenograft tissues were measured using qRT-PCR. J. The expression of Ki-67 was examined through IHC (scale bars, 200 µm). ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, not significant.
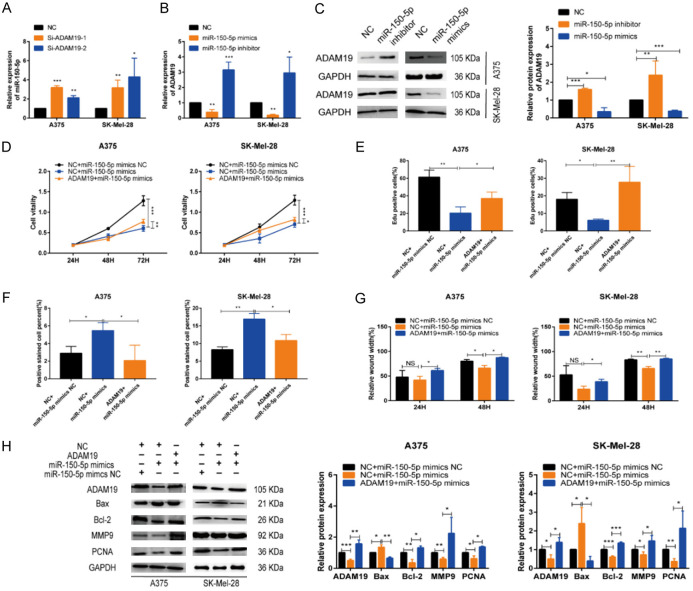
ADAM19 reverses the impact of miR-150-5p on melanoma cells
To validate whether the observed miR-150-5p-mediated effects in melanoma cells can be reversed by restoring ADAM19 expression, we conducted rescue experiments. The results confirmed the reciprocal relationship between the expression of ADAM19 and miR-150-5p (Figure 7A-C), which was restored via transfection with ADAM19 or miR-150-5p (Figures S7A, S8A). Results of various biological assays, including cell proliferation, apoptosis, cell scratch, and Transwell assays, demonstrated that ADAM19 partially counteracted the biological impact of miR-150-5p on melanoma cells (Figures 7D-G, S7B-F, S8B-G). Furthermore, western blot analysis revealed that the inhibitory effect of miR-150-5p on melanoma cells was partially attenuated by ADAM19 overexpression (Figure 7H). These findings indicate that miR-150-5p facilitates melanoma growth by negatively modulating ADAM19.
Figure 7.
ADAM19 reversed the impact of miR-150-5p on melanoma cells. A. Expression levels of miR-150-5p were identified in melanoma cells after ADAM19 overexpression. B, C. Relative expression levels of ADAM19 are observed in A375 and SK-Mel-28 cells transfected with miR-150-5p. D, E. CCK8 and EdU assays to proliferation rate of transfected cells. F. The influence of ADAM19 and miR-150-5p mimics on the rate of apoptosis was evaluated through TUNEL analysis. G. Scratch wound healing assay is used to evaluate migration capacity. H. Relative expression levels of ADAM19, Bax, Bcl-2, MMP9, PCNA were observed in A375 and SK-Mel-28 cells transfected with ADAM19 and miR-150-5p mimics. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, not significant.
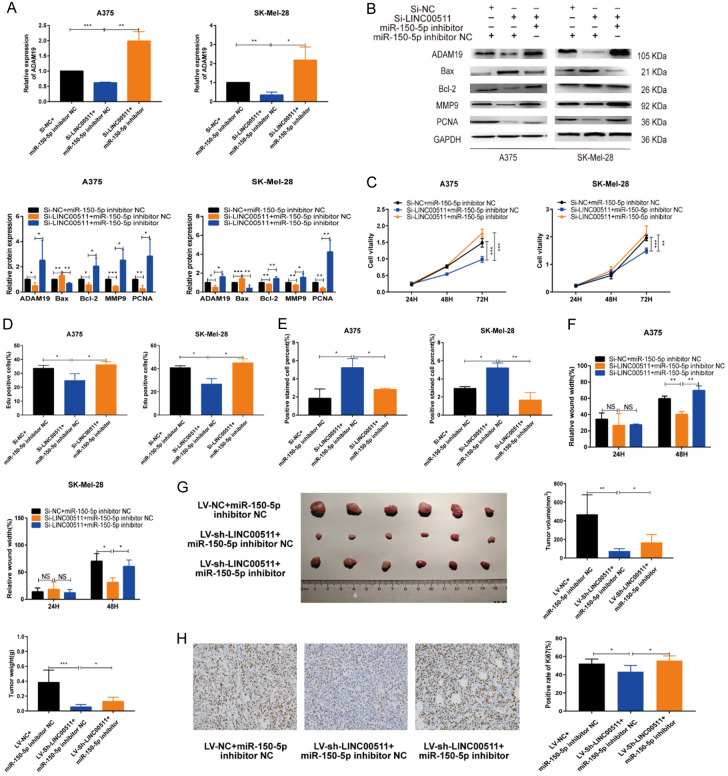
LINC00511 acts as a sponge for miR-150-5p to upregulate ADAM19
qRT-PCR analysis revealed that ADAM19 was downregulated upon LINC00511 knockdown, and this effect was reversed by miR-150-5p inhibition (Figure 8A). Furthermore, miR-150-5p inhibition reversed the effect of LINC00511 knockdown on the expression of ADAM19, BAX, Bcl-2, MMP9, and PCNA (Figure 8B). Results of cell proliferation, wound healing, apoptosis, and transwell assays demonstrated that the differential inhibitory and promoting effects of LINC00511 on melanoma cells were reversed by miR-150-5p inhibitors or mimics, respectively (Figures 8C-F, S9A-E, S10A-F). Moreover, in vivo experiments demonstrated that mice in the LINC00511 knockdown group showed markedly decreased tumor size and tumor weight compared to that in the control group, while miR-150-5p inhibition promoted LINC00511 knockdown-induced tumor progression (Figure 8G). Additionally, IHC staining showed that knockdown of LINC00511 inhibited the expression of Ki67 expression, which was rescued by the knockdown of miR-150-5p (Figure 8H). qRT-PCR analysis showed that silencing LINC00511 inhibited ADAM19 expression in melanoma cells, which was reversed by ADAM19 overexpression (Figure S11A). Functional experimental results demonstrated that ADAM19 overexpression reversed the impact of LINC00511 silencing on melanoma cells by inhibiting cell migration, proliferation, and invasion and increasing the rate of cell apoptosis (Figure S11B-G). Moreover, ADAM19 overexpression reversed the effect of LINC00511 knockdown on the expression of apoptosis- and migration-related proteins (Figure S11H). In addition, in vivo experiments have shown that overexpression of ADAM19 promotes tumor progression induced by the knockdown of LINC00511 (Figure S12A). IHC demonstrated that knockdown of LINC00511 inhibits Ki67 expression, while overexpression of ADAM19 restored Ki67 expression (Figure S12B). These findings suggest that LINC00511 functions as a sponge for miR-150-5p to regulate ADAM19 expression.
Figure 8.
LINC00511 acted as a sponge for miR-150-5p to up-regulate ADAM19 expression. A. The expression levels of ADAM19 in A375 and SK-Mel-28 cells transfected with si-LINC00511 and miR-150-5p inhibitor were identified using qRT-PCR. B. Relative expression levels of ADAM19, Bax, Bcl-2, MMP9, PCNA were observed in A375 and SK-Mel-28 cells transfected with si-LINC00511 and miR-150-5p inhibitor. C, D. CCK8 and EdU assays to proliferation rate of transfected cells. E. The influence of si-LINC00511 and miR-150-5p inhibitor on the rate of apoptosis was evaluated through TUNEL analysis. F. Metastasis ability was measured using wound-healing assay in melanoma cells transfected with si-LINC00511and miR-150-5p inhibitor. G. Gross appearance, tumor volume, and tumor weight of each group. H. IHC staining results (Ki-67). ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, not significant.
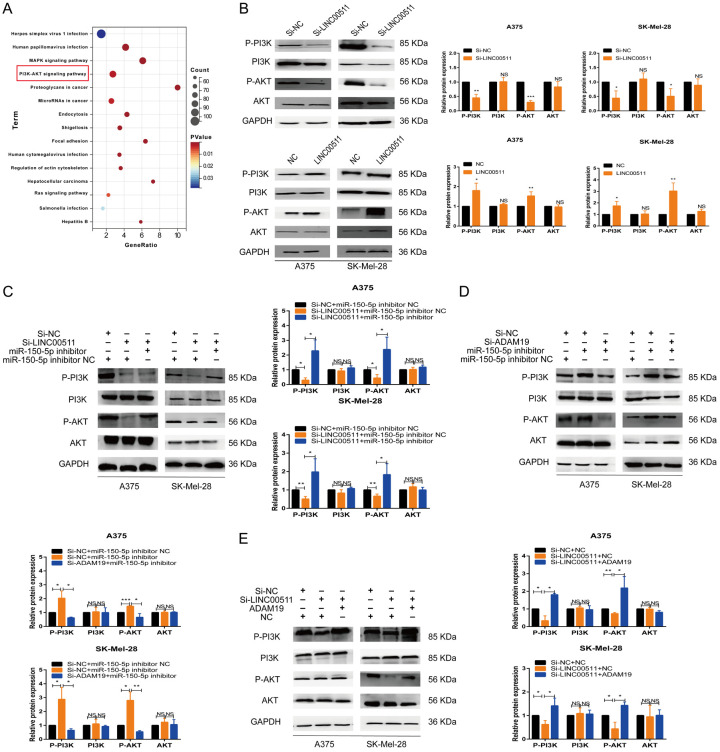
LINC00511 activates the PI3K/AKT pathway in melanoma cells through the miR-150-5p/ADAM19 axis
PI3K/AKT signaling plays a critical role in tumor initiation, progression, and therapeutic response. Previous studies have reported the key role of the PI3K-AKT pathway in melanoma progression, which is consistent with the findings of KEGG enrichment analysis findings (Figure 9A) [35-37]. Silencing LINC00511 significantly reduced the levels of phosphorylated PI3K (p-PI3K) and phosphorylated AKT (p-AKT) compared to those in the control group. However, there were no significant differences in the total PI3K and AKT levels. Conversely, LINC00511 overexpression increased the phosphorylation of these proteins but did not affect the levels of total PI3K and AKT (Figure 9B). The rescue experiment further demonstrated that the miR-150-5p inhibitor prevented the effects of LINC00511 and ADAM19 knockdown on p-PI3K and p-AKT and that ADAM19 overexpression partially restored the phosphorylation of these proteins altered by LINC00511 depletion (Figure 9C-E). Additionally, LINC00511 partially reversed the effects of the PI3K inhibitor (LY294002) and activator (740Y-P) on melanoma cell proliferation (Figure S13). These findings suggest that LINCC0511 affects melanoma progression by activating the PI3K/AKT pathway via the miR-150-5p/ADAM19 axis.
Figure 9.
LINC00511 activated the PI3K/AKT pathway in melanoma cell through the miR-150-5p/ADAM19 axis. A. KEGG enrichment analysis. B. The effects of the LINC00511 gene on PI3K, p-PI3K, p-Akt and Akt expression in A375 and SK-Mel-28 cells were detected by western blot analysis. C-E. The relative protein expression of PI3K, p-PI3K, p-Akt and Akt was analyzed in A375 and SK-Mel-28 cells transfected with si-NC/si-LINC00511+inhibitor NC/miR-150-5p inhibitor, si-NC/si-ADAM19/+inhibitor NC/miR-150-5p inhibitor, si-NC/si-LINC00511+NC/ADAM19 respectively. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, not significant.
Discussion
Melanoma is a highly aggressive skin cancer associated with a poor prognosis and a high metastatic rate, resulting in a significant annual mortality rate [38,39]. Despite recent advances in treatment modalities such as surgery, radiotherapy, immunotherapy, and targeted therapy, the prognosis of patients with melanoma remains unfavorable [40]. Accumulating evidence supports the involvement of lncRNAs in melanoma initiation, development, and metastasis, as they influence crucial cellular processes [41]. Hence, in-depth investigations of the underlying mechanisms and biological functions of lncRNAs can reveal potential new therapeutic targets for melanoma.
Our findings highlight the significant role of LINC00511 in promoting melanoma migration and invasion while inhibiting the growth of xenografted tumors in vivo. LINC00511 is a cancer-promoting factor in melanoma and has been implicated in several types of cancers, showing promise as a potential biomarker for cancer diagnosis, prognosis, and treatment strategies. For instance, LINC00511 overexpression has been linked to cell invasion and migration in cervical cancer, which suggesting its potential as a novel biomarker [42]. In hepatocellular carcinoma, LINC00511 was found to be upregulated, and its overexpression induced vascular invasion and lymph node metastasis [43]. A meta-analysis revealed a close relationship between LINC00511 overexpression and poor prognosis (overall, progression-free, or relapse-free survival) in lung cancer, pancreatic ductal adenocarcinoma, and breast cancer [44]. Additionally, inhibition of LINC00511 reportedly suppresses cell growth and metastasis in glioma [20], NSCLC [21], and melanoma [45]. These studies, combined with our findings, suggest that targeting LINC00511 could be an effective therapeutic strategy for melanoma and suggest its potential role as a key regulator of multiple tumor types.
Upstream regulatory factors, such as transcription factors, can contribute to the dysregulation of lncRNA expression in cancers [46]. In the present study, the transcription factor YY1 was identified as an upstream transcriptional regulator of LINC00511 using database analysis. YY1 is a member of the GLI-Kruppel class of zinc finger proteins and is involved in cell proliferation, invasion, and metastasis, among other biological processes [47]. In addition, YY1 acts as a master modulator of the regulatory epigenetic network and modulates the expression of downstream target genes, thereby affecting tumorigenesis in pancreatic [48], lung [49], colorectal [50], and breast cancers [51]. Moreover, YY1 has been found to participate in several biological processes by activating the expression of various lncRNAs, including ZFPM2-AS12 [52], PKMYT1AR [53], SNHG17 [54], and LINC00673 [55]. Our study revealed direct binding of YY1 to the promoter region of LINC00511 and consistently, we found that downregulation of YY1 led to decreased expression of LINC00511. Previous studies have demonstrated that elevated YY1 expression in melanoma promotes cell proliferation and migration [56]. In melanoma, YY1 has been implicated in the regulation of autophagy and lysosomal biogenesis in conjunction with the TFEB transcription [57] and the regulation of the miR-9/RYBP axis [58]. A recent study reported the role of the YY1-induced lncRNA SNHG8 in the promotion of melanoma tumorigenesis via the microRNA-656-3p/SERBP1 axis [59]. Collectively, our findings reveal a novel regulatory mechanism in melanoma, wherein the YY1-mediated upregulation of LINC00511 promotes melanoma development.
Mechanisms underlying the functions of ceRNA have garnered significant attention in cancer biology; endogenous lncRNAs contain miRNA-responsive elements and modulate mRNAs by acting as molecular sponges and binding to specific miRNAs [60]. For instance, LINC00612 functions as a ceRNA of miR-214-5p, indirectly upregulating SOX4 and promoting the proliferation, invasion, and epithelial-mesenchymal transition of osteosarcoma cells [61]. The lncRNA SNHG8 can bind to miR-152 and increase c-MET expression in endometrial cancer [62]. The ceRNA activity of LINC00665, which acts as a molecular sponge for miR-98, has been implicated in the activation of the AKR1B10-ERK signaling pathway and the progression of lung adenocarcinoma [63]. The biological function of lncRNAs is predominantly determined by their specific subcellular localization [32]. LINC00511 is primarily localized in the cytoplasm of melanoma cells, which suggests that it may regulate critical gene expression through the ceRNA network. Previous studies have demonstrated that LINC00511 promotes gastric cancer progression by regulating NFIX expression and targeting miR-625 [64]. In hepatocellular carcinoma, LINC00511 acts as a ceRNA to regulate the miR-195/EYA1 axis and promotes the malignant behavior of hepatocellular carcinoma [65]. Moreover, LINC00511 can affect cancer development, progression, and prognosis by sponging various miRNAs, including miRNA-29b-3p [66], miRNA-29c [67], miRNA-124-3p [68], miRNA-185 [69], miRNA-424 [70], miRNA-618 [71], and miRNA-765 [72]. Bioinformatics analysis revealed that LINC00511 has potential miR-150-5p-binding sites, suggesting its role as a ceRNA for miR-150-5p. This miRNA has been identified as a tumor suppressor in several cancers, including thyroid cancer [73], colorectal cancer [74], NSCLC [75], and melanoma [76]. In the present study, FISH, RIP, and RNA pull-down assays confirmed that LINC00511 acts as a ceRNA by sponging miR-150-5p in melanoma cells. Zhang et al. found that LINC00511 acts as a sponge for miR-150-5p to regulate cell proliferation and apoptosis in osteoarthritis [77]. Wu et al. reported that LINC00511 suppresses lung squamous cell carcinoma migration and proliferation by suppressing miR-150-5p and activating TADA1 [78]. Consistent with these findings, our study indicates that the tumor-promoting effect of LINC00511 in melanoma may be largely attributable to its regulation of miR-150-5p activity. Therefore, an in-depth exploration of the mechanisms underlying the functions of ceRNAs can assist researchers in gaining a better understanding of the structure and function of gene regulatory networks in tumor regulatory mechanisms. Further, these findings can contribute to the implementation of precision therapy and further advance clinical diagnosis, and the development of treatment modalities.
As a crucial member of the ADAM family, ADAM19 plays a role in cell adhesion, proteolysis, and phenotypic alteration [79]. Abnormal ADAM19 expression has been reported in various malignancies, with high expression being associated with increased cancer cell invasion and migration in NSCLC and nasopharyngeal carcinoma [80,81]. Conversely, ADAM19 has been found to exert a tumor-suppressive effect on prostate cancer, attenuating the proliferation and motility of prostate cancer cells upon overexpression [82]. ADAM19 is also a target of multiple miRNAs, including miR-145 [83], miR-30c [84], miR-361-3p [85], and miR-144-3p [86]. The present study validated ADAM19 as a direct downstream target of miR-150-5p and proposed that this relation could be determinant for its biological effects in melanoma cells. Numerous studies have suggested that lncRNAs regulate the expression of miRNA target genes by sequestering miRNAs and inhibiting their interactions with protein-coding transcripts [48]. Herein, we demonstrated that LINC00511 enhances the expression of ADAM19 by targeting miR-150-5p in melanoma cells. Furthermore, the inhibitory effects of LINC00511 on melanoma cell migration and invasion can be counteracted by the inhibition of miR-150-5p or ADAM19 overexpression. Overall, our findings indicate that the LINC00511/miR-150-5p/ADAM19 axis plays a crucial role in regulating melanoma progression, thereby supporting the involvement of LINC00511 in the ceRNA regulatory system in melanoma cells.
The PI3K/AKT signaling pathway promotes the growth, survival, and invasion of cancer cells. Aberrant expression of lncRNAs can activate the PI3K/AKT pathway and contributes to the progression of different cancers, including melanoma [35,36]. Downregulation of the lncRNA MIAT inhibits the activation of PI3K/AKT signaling pathway, thereby reducing melanoma cell invasion and proliferation [87]. The lncRNA DBH-AS1 induces melanoma progression through the miR-233-3p/IGF-1R/PI3K/AKT pathway [88]. Our data revealed that LINC00511 promotes melanoma progression by modulating the miR-150-5p/ADAM19 axis to enhance the activity of the PIK3/AKT pathway. A recent study demonstrated that LINC00511 promotes gastric cancer progression by inhibiting PTEN to activate the PI3K/AKT pathway [89]. These findings suggest that LINC00511 functions as an oncogene, promoting cancer occurrence and development by driving the activation of the PI3K/AKT signaling pathway.
Conclusion
In conclusion, our study provides valuable insights into the complex molecular landscape of melanoma. The activation of LINC00511 by YY1, its ceRNA activity, and its role in regulating ADAM19 and the PI3K/AKT signaling pathway contribute to our understanding of melanoma. These findings pave the way for the development of targeted and personalized therapies for patients with melanoma, ultimately improving their clinical outcomes (Figure 10). However, the present study has some limitations. Firstly, the findings of this study need to be verified using clinical samples from patients with melanoma. Secondly, a metastasis model should be established to explore the impact of the LINC00511-miR-150-5p-ADAM19 axis on melanoma cell metastasis. This is also a future research direction.
Figure 10.
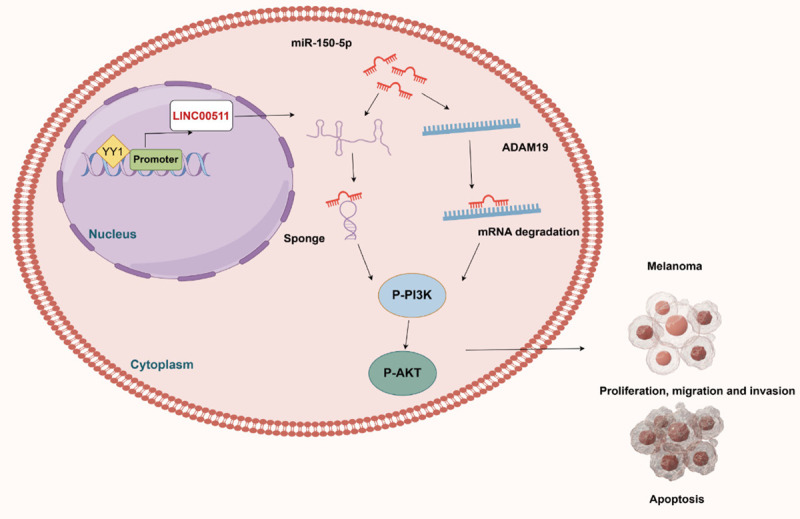
A proposed model in which YY1-induced LncRNA00511 promotes melanoma progression by regulating ADAM19 expression and PI3K/AKT signaling through competitive binding with miR-150-5p.
Acknowledgements
This study was supported by grants from the Key Technology Research Plan Project of Inner Mongolia Autonomous Region (2021GG0153), and the National Natural Science Foundation of China (31760333).
Disclosure of conflict of interest
None.
Abbreviations
- ceRNAs
Competing endogenous RNAs
- GEO
gene expression omnibus
- lncRNAs
long noncoding RNAs
- LINC00511
long intergenic non-protein-coding RNA 511
- miR-150-5p
microRNA-150-5p
- YY1
Yin Yang 1
Supporting Information
References
- 1.Puyana C, Denyer S, Burch T, Bhimani AD, McGuire LS, Patel AS, Mehta AI. Primary malignant melanoma of the brain: a populationbased study. World Neurosurg. 2019;130:e1091–e1097. doi: 10.1016/j.wneu.2019.07.095. [DOI] [PubMed] [Google Scholar]
- 2.Gaweł-Bęben K, Kukula-Koch W, Hoian U, Czop M. Characterization of cistus × incanus L. and cistus ladanifer L. extracts as potential multifunctional antioxidant ingredients for skin protecting cosmetics. Antioxidants (Basel) 2020;9:202. doi: 10.3390/antiox9030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rinck-Junior JA, Torricelli C, Gomez GVB, Oliveira C, Moraes AM, Lourenço GJ, Lima CSP. Influence of functional variants Asp312Asn and Lys751Gln of xeroderma pigmentosum group D (XPD) and glutathione S-transferase Mu 1 (GSTM1) and theta 1 (GSTT1) genes on cutaneous melanoma susceptibility and prognosis. Exp Dermatol. 2019;28:631–635. doi: 10.1111/exd.13914. [DOI] [PubMed] [Google Scholar]
- 4.Huang YL, Xu Q, Wang X. Long noncoding RNA DSCAM-AS1 is associated with poor clinical prognosis and contributes to melanoma development by sponging miR-136. Eur Rev Med Pharmacol Sci. 2019;23:2888–2897. doi: 10.26355/eurrev_201904_17567. [DOI] [PubMed] [Google Scholar]
- 5.Allen KJH, Jiao R, Malo ME, Frank C, Fisher DR, Rickles D, Dadachova E. Comparative radioimmunotherapy of experimental melanoma with novel humanized antibody to melanin labeled with 213Bismuth and 177Lutetium. Pharmaceutics. 2019;11:348. doi: 10.3390/pharmaceutics11070348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shannan B, Perego M, Somasundaram R, Herlyn M. Heterogeneity in melanoma. Cancer Treat Res. 2016;167:1–15. doi: 10.1007/978-3-319-22539-5_1. [DOI] [PubMed] [Google Scholar]
- 7.Ribero S, Glass D, Bataille V. Genetic epidemiology of melanoma. Eur J Dermatol. 2016;26:335–339. doi: 10.1684/ejd.2016.2787. [DOI] [PubMed] [Google Scholar]
- 8.Sarkar D, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Epigenetic regulation in human melanoma: past and future. Epigenetics. 2015;10:103–121. doi: 10.1080/15592294.2014.1003746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun CC, Zhu W, Li SJ, Hu W, Zhang J, Zhuo Y, Zhang H, Wang J, Zhang Y, Huang SX, He QQ, Li DJ. FOXC1-mediated LINC00301 facilitates tumor progression and triggers an immune-suppressing microenvironment in nonsmall cell lung cancer by regulating the HIF1α pathway. Genome Med. 2020;12:77. doi: 10.1186/s13073-020-00773-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin Y, Zhao B, Li D, Yin G. Long non-coding RNA CASC15 promotes melanoma progression by epigenetically regulating PDCD4. Cell Biosci. 2018;8:42. doi: 10.1186/s13578-018-0240-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Hosono Y, Niknafs YS, Prensner JR, Iyer MK, Dhanasekaran SM, Mehra R, Pitchiaya S, Tien J, Escara-Wilke J, Poliakov A, Chu SC, Saleh S, Sankar K, Su F, Guo S, Qiao Y, Freier SM, Bui HH, Cao X, Malik R, Johnson TM, Beer DG, Feng FY, Zhou W, Chinnaiyan AM. Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA. Cell. 2017;171:1559–1572. e20. doi: 10.1016/j.cell.2017.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luan W, Li L, Shi Y, Bu X, Xia Y, Wang J, Djangmah HS, Liu X, You Y, Xu B. Long non-coding RNA MALAT1 acts as a competing endogenous RNA to promote malignant melanoma growth and metastasis by sponging miR-22. Oncotarget. 2016;7:63901–63912. doi: 10.18632/oncotarget.11564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, Zhang J, Zhang Q, Xu H, Liu L. Long non-coding RNA HOXA11-as modulates proliferation, apoptosis, metastasis and EMT in cutaneous melanoma cells partly via miR-152-3p/ITGA9 axis. Cancer Manag Res. 2021;13:925–939. doi: 10.2147/CMAR.S281920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L, Zhu L, Li Y, Zheng Z, Lin X, Yang C. LncRNA MEG3 promotes melanoma growth, metastasis and formation through modulating miR-21/E-cadherin axis. Cancer Cell Int. 2020;20:12. doi: 10.1186/s12935-019-1087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Qian W, Feng F, Cao Q, Li Y, Hou Y, Zhang L, Fan J. Upregulated lncRNA CASC2 may inhibit malignant melanoma development through regulating miR-18a-5p/RUNX1. Oncol Res. 2019;27:371–377. doi: 10.3727/096504018X15178740729367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luan W, Ding Y, Ma S, Ruan H, Wang J, Lu F. Long noncoding RNA LINC00518 acts as a competing endogenous RNA to promote the metastasis of malignant melanoma via miR-204-5p/AP1S2 axis. Cell Death Dis. 2019;10:855. doi: 10.1038/s41419-019-2090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, Radaelli E, Eyckerman S, Leonelli C, Vanderheyden K, Rogiers A, Hermans E, Baatsen P, Aerts S, Amant F, Van Aelst S, Van den Oord J, de Strooper B, Davidson I, Lafontaine DL, Gevaert K, Vandesompele J, Mestdagh P, Marine JC. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531:518–522. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 19.Ghafouri-Fard S, Safarzadeh A, Hussen BM, Taheri M, Ayatollahi SA. A review on the role of LINC00511 in cancer. Front Genet. 2023;14:1116445. doi: 10.3389/fgene.2023.1116445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Y, Tian M, Liu J, Wang K. LINC00511 facilitates temozolomide resistance of glioblastoma cells via sponging miR-126-5p and activating Wnt/β-catenin signaling. J Biochem Mol Toxicol. 2021;35:e22848. doi: 10.1002/jbt.22848. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y, Wang S, Mu X. Long non-coding RNA LINC00511 promotes proliferation, invasion, and migration of non-small cell lung cancer cells by targeting miR-625-5p/GSPT1. Transl Cancer Res. 2021;10:5159–5173. doi: 10.21037/tcr-21-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong LM, Zhang XL, Mao MH, Li YP, Zhang XY, Xue DW, Liu YL. LINC00511/miRNA-143-3p modulates apoptosis and malignant phenotype of bladder carcinoma cells via PCMT1. Front Cell Dev Biol. 2021;9:650999. doi: 10.3389/fcell.2021.650999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian X, Jiang C, Zhu Z, Han G, Xu N, Ye J, Wang R. Long non-coding RNA LINC00511 facilitates colon cancer development through regulating microRNA-625-5p to target WEE1. Cell Death Discov. 2022;8:233. doi: 10.1038/s41420-021-00790-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y, Zhang Y, Ding M, Xu R. LncRNA LINC00511 acts as an oncogene in colorectal cancer via sponging miR-29c-3p to upregulate NFIA. Onco Targets Ther. 2021;13:13413–13424. doi: 10.2147/OTT.S250377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wozniak M, Czyz M. The functional role of long non-coding RNAs in melanoma. Cancers (Basel) 2021;13:4848. doi: 10.3390/cancers13194848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu H, Zeng Y, Zhou CC, Ye W. SNHG16/miR-216-5p/ZEB1 signal pathway contributes to the tumorigenesis of cervical cancer cells. Arch Biochem Biophys. 2018;637:1–8. doi: 10.1016/j.abb.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Xu M, Chen X, Lin K, Zeng K, Liu X, Xu X, Pan B, Xu T, Sun L, He B, Pan Y, Sun H, Wang S. lncRNA SNHG6 regulates EZH2 expression by sponging miR-26a/b and miR-214 in colorectal cancer. J Hematol Oncol. 2019;12:3. doi: 10.1186/s13045-018-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang F, Shen Y, Zhang W, Jin J, Huang D, Fang H, Ji W, Shi Y, Tang L, Chen W, Zhou G, Guan X. An androgen receptor negatively induced long non-coding RNA ARNILA binding to miR-204 promotes the invasion and metastasis of triple-negative breast cancer. Cell Death Differ. 2018;25:2209–2220. doi: 10.1038/s41418-018-0123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Sun H, Zhang J. LncRNA DSCAM-AS1 promotes colorectal cancer progression by acting as a molecular sponge of miR-384 to modulate AKT3 expression. Aging (Albany NY) 2020;12:9781–9792. doi: 10.18632/aging.103243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu Z, Pan XX, You DY. LncRNA DSCAM-AS1 promotes non-small cell lung cancer progression via regulating miR-577/HMGB1 axis. Neoplasma. 2020;67:871–879. doi: 10.4149/neo_2020_190826N821. [DOI] [PubMed] [Google Scholar]
- 31.Wang XL, Li J, Cao YH. Crosstalk between YY1 and lncRNAs in cancer: a review. Medicine (Baltimore) 2022;101:e31990. doi: 10.1097/MD.0000000000031990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Zhou L, Lu C, Shen Q, Su Y, Zhi Z, Wu F, Zhang H, Wen Z, Chen G, Li H, Xia Y, Tang W. Long non-coding RNA FAL1 functions as a ceRNA to antagonize the effect of miR-637 on the down-regulation of AKT1 in Hirschsprung’s disease. Cell Prolif. 2018;51:e12489. doi: 10.1111/cpr.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun W, Yang Y, Xu C, Guo J. Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet. 2017;216-217:105–110. doi: 10.1016/j.cancergen.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Han P, Gopalakrishnan C, Yu H, Wang E. Gene regulatory network rewiring in the immune cells associated with cancer. Genes (Basel) 2017;8:308. doi: 10.3390/genes8110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahmani M, Aust MM, Benson EC, Wallace L, Friedberg J, Grant S. PI3K/mTOR inhibition markedly potentiates HDAC inhibitor activity in NHL cells through BIM- and MCL-1-dependent mechanisms in vitro and in vivo. Clin Cancer Res. 2014;20:4849–4860. doi: 10.1158/1078-0432.CCR-14-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kircher DA, Trombetti KA, Silvis MR, Parkman GL, Fischer GM, Angel SN, Stehn CM, Strain SC, Grossmann AH, Duffy KL, Boucher KM, McMahon M, Davies MA, Mendoza MC, VanBrocklin MW, Holmen SL. AKT1E17K activates focal adhesion kinase and promotes melanoma brain metastasis. Mol Cancer Res. 2019;17:1787–1800. doi: 10.1158/1541-7786.MCR-18-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maddodi N, Setaluri V. Role of UV in cutaneous melanoma. Photochem Photobiol. 2008;84:528–536. doi: 10.1111/j.1751-1097.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- 38.de Oliveira Filho RS, de Oliveira DA, Nisimoto MM, Marti LC. A Review of advanced cutaneous melanoma therapies and their mechanisms, from immunotherapies to lysine histone methyl transferase inhibitors. Cancers (Basel) 2023;15:5751. doi: 10.3390/cancers15245751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Jia Y, Zhang P, Yang H, Cong X, An L, Xiao C. Celastrol self-stabilized nanoparticles for effective treatment of melanoma. Int J Nanomedicine. 2020;15:1205–1214. doi: 10.2147/IJN.S232603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou W, Xu X, Cen Y, Chen J. The role of lncRNAs in the tumor microenvironment and immunotherapy of melanoma. Front Immunol. 2022;13:1085766. doi: 10.3389/fimmu.2022.1085766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nie L, Wu HJ, Hsu JM, Chang SS, Labaff AM, Li CW, Wang Y, Hsu JL, Hung MC. Long noncoding RNAs: versatile master regulators of gene expression and crucial players in cancer. Am J Transl Res. 2012;4:127–150. [PMC free article] [PubMed] [Google Scholar]
- 42.Lu M, Gao Q, Wang Y, Ren J, Zhang T. LINC00511 promotes cervical cancer progression by regulating the miR-497-5p/MAPK1 axis. Apoptosis. 2022;27:800–811. doi: 10.1007/s10495-022-01768-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu P, Cui H, Lei T, Li S, Mai E, Jia F. Linc00511 indicates a poor prognosis of liver hepatocellular carcinoma. Onco Targets Ther. 2019;12:9367–9376. doi: 10.2147/OTT.S228231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agbana YL, Abi ME, Ni Y, Xiong G, Chen J, Yun F, Yi Z, Zhang Q, Yang Z, Kuang Y, Zhu Y. LINC00511 as a prognostic biomarker for human cancers: a systematic review and metaanalysis. BMC cancer. 2020;20:682. doi: 10.1186/s12885-020-07188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang G, Wang Z, Liu J, Feng S, Ji S, Ai D. LINC00511 promotes melanoma progression by targeting miR-610/NUCB2. Open Med (Wars) 2023;18:20230628. doi: 10.1515/med-2023-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Z, Chen X, Lei T, Gu Y, Gu J, Huang J, Lu B, Yuan L, Sun M, Wang Z. Integrative analysis of NSCLC identifies LINC01234 as an oncogenic lncRNA that interacts with HNRNPA2B1 and regulates miR-106b biogenesis. Mol Ther. 2020;28:1479–1493. doi: 10.1016/j.ymthe.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khachigian LM. The Yin and Yang of YY1 in tumor growth and suppression. Int J Cancer. 2018;143:460–465. doi: 10.1002/ijc.31255. [DOI] [PubMed] [Google Scholar]
- 48.Huang X, Pan L, Zuo Z, Li M, Zeng L, Li R, Ye Y, Zhang J, Wu G, Bai R, Zhuang L, Wei L, Zheng Y, Su J, Deng J, Deng S, Zhang S, Zhu S, Che X, Wang C, Wu C, Chen R, Lin D, Zheng J. LINC00842 inactivates transcription co-regulator PGC-1α to promote pancreatic cancer malignancy through metabolic remodelling. Nat Commun. 2021;12:3830. doi: 10.1038/s41467-021-23904-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang T, Wang G, Yang L, Peng B, Wen Y, Ding G, Wang Z. Transcription factor YY1 modulates lung cancer progression by activating lncRNA-PVT1. DNA Cell Biol. 2017;36:947–958. doi: 10.1089/dna.2017.3857. [DOI] [PubMed] [Google Scholar]
- 50.Ye Y, Gu B, Wang Y, Shen S, Huang W. YY1-induced upregulation of long noncoding RNA ARAP1-AS1 promotes cell migration and invasion in colorectal cancer through the Wnt/β-catenin signaling pathway. Cancer Biother Radiopharm. 2019;34:519–528. doi: 10.1089/cbr.2018.2745. [DOI] [PubMed] [Google Scholar]
- 51.Shen B, Li Y, Ye Q, Qin Y. YY1-mediated long non-coding RNA Kcnq1ot1 promotes the tumor progression by regulating PTEN via DNMT1 in triple negative breast cancer. Cancer Gene Ther. 2021;28:1099–1112. doi: 10.1038/s41417-020-00254-9. [DOI] [PubMed] [Google Scholar]
- 52.Yan Z, Yang Q, Xue M, Wang S, Hong W, Gao X. YY1-induced lncRNA ZFPM2-AS1 facilitates cell proliferation and invasion in small cell lung cancer via upregulating of TRAF4. Cancer Cell Int. 2020;20:108. doi: 10.1186/s12935-020-1157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He Y, Jiang X, Duan L, Xiong Q, Yuan Y, Liu P, Jiang L, Shen Q, Zhao S, Yang C, Chen Y. LncRNA PKMYT1AR promotes cancer stem cell maintenance in non-small cell lung cancer via activating Wnt signaling pathway. Mol Cancer. 2021;20:156. doi: 10.1186/s12943-021-01469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Li T, Huang D, Zhang P. Long noncoding RNA SNHG17 induced by YY1 facilitates the glioma progression through targeting miR-506-3p/CTNNB1 axis to activate Wnt/β-catenin signaling pathway. Cancer Cell Int. 2020;20:29. doi: 10.1186/s12935-019-1088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiao K, Ning S, Wan L, Wu H, Wang Q, Zhang X, Xu S, Pang D. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J Exp Clin Cancer Res. 2019;38:418. doi: 10.1186/s13046-019-1421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou S, Li P, Qin L, Huang S, Dang N. Transcription factor YY1 contributes to human melanoma cell growth through modulating the p53 signalling pathway. Exp Dermatol. 2022;31:1563–1578. doi: 10.1111/exd.14628. [DOI] [PubMed] [Google Scholar]
- 57.Du J, Ren W, Yao F, Wang H, Zhang K, Luo M, Shang Y, O’Connell D, Bei Z, Wang H, Xiong R, Yang Y. YY1 cooperates with TFEB to regulate autophagy and lysosomal biogenesis in melanoma. Mol Carcinog. 2019;58:2149–2160. doi: 10.1002/mc.23105. [DOI] [PubMed] [Google Scholar]
- 58.Zhao G, Li Q, Wang A, Jiao J. YY1 regulates melanoma tumorigenesis through a miR-9~RYBP axis. J Exp Clin Cancer Res. 2015;34:66. doi: 10.1186/s13046-015-0177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shan B, Qu S, Lv S, Fan D, Wang S. YY1-induced long non-coding RNA small nucleolar RNA host gene 8 promotes the tumorigenesis of melanoma via the microRNA-656-3p/SERPINE1 mRNA binding protein 1 axis. Bioengineered. 2022;13:4832–4843. doi: 10.1080/21655979.2022.2034586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou Y, Li X, Yang H. LINC00612 functions as a ceRNA for miR-214-5p to promote the proliferation and invasion of osteosarcoma in vitro and in vivo. Exp Cell Res. 2020;392:112012. doi: 10.1016/j.yexcr.2020.112012. [DOI] [PubMed] [Google Scholar]
- 62.Yang CH, Zhang XY, Zhou LN, Wan Y, Song LL, Gu WL, Liu R, Ma YN, Meng HR, Tian YL, Zhang Y. LncRNA SNHG8 participates in the development of endometrial carcinoma through regulating c-MET expression by miR-152. Eur Rev Med Pharmacol Sci. 2018;22:1629–1637. doi: 10.26355/eurrev_201803_14698. [DOI] [PubMed] [Google Scholar]
- 63.Cong Z, Diao Y, Xu Y, Li X, Jiang Z, Shao C, Ji S, Shen Y, De W, Qiang Y. Long non-coding RNA linc00665 promotes lung adenocarcinoma progression and functions as ceRNA to regulate AKR1B10-ERK signaling by sponging miR-98. Cell Death Dis. 2019;10:84. doi: 10.1038/s41419-019-1361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deng H, Huang C, Wang Y, Jiang H, Peng S, Zhao X. LINC00511 promotes the malignant phenotype of clear cell renal cell carcinoma by sponging microRNA-625 and thereby increasing cyclin D1 expression. Aging (Albany NY) 2019;11:5975–5991. doi: 10.18632/aging.102156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu WY, Wei HY, Li KM, Wang RB, Xu XQ, Feng R. LINC00511 as a ceRNA promotes cell malignant behaviors and correlates with prognosis of hepatocellular carcinoma patients by modulating miR-195/EYA1 axis. Biomed Pharmacother. 2020;121:109642. doi: 10.1016/j.biopha.2019.109642. [DOI] [PubMed] [Google Scholar]
- 66.Quan X, Zhao M, Yang X, Zhu Y, Tian X. AP2γ mediated downregulation of lncRNA LINC00511 as a ceRNA suppresses trophoblast invasion by regulating miR-29b-3p/Cyr61 axis. Biomed Pharmacother. 2019;120:109269. doi: 10.1016/j.biopha.2019.109269. [DOI] [PubMed] [Google Scholar]
- 67.Zhang H, Zhao B, Wang X, Zhang F, Yu W. LINC00511 knockdown enhances paclitaxel cytotoxicity in breast cancer via regulating miR-29c/CDK6 axis. Life Sci. 2019;228:135–144. doi: 10.1016/j.lfs.2019.04.063. [DOI] [PubMed] [Google Scholar]
- 68.Li C, Liu H, Yang J, Yang J, Yang L, Wang Y, Yan Z, Sun Y, Sun X, Jiao B. Long noncoding RNA LINC00511 induced by SP1 accelerates the glioma progression through targeting miR-124-3p/CCND2 axis. J Cell Mol Med. 2019;23:4386–4394. doi: 10.1111/jcmm.14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu G, Li Y, Ma Y, Lu J, Chen Y, Jiang Q, Qin Q, Zhao L, Huang Q, Luo Z, Huang S, Wei Z. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res. 2018;37:289. doi: 10.1186/s13046-018-0945-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang RP, Jiang J, Jiang T, Wang Y, Chen LX. Increased long noncoding RNA LINC00511 is correlated with poor prognosis and contributes to cell proliferation and metastasis by modulating miR-424 in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23:3291–3301. doi: 10.26355/eurrev_201904_17691. [DOI] [PubMed] [Google Scholar]
- 71.Guo W, Yu Q, Zhang M, Li F, Liu Y, Jiang W, Jiang H, Li H. Long intergenic non-protein coding RNA 511 promotes the progression of osteosarcoma cells through sponging microRNA 618 to upregulate the expression of maelstrom. Aging (Albany NY) 2019;11:5351–5367. doi: 10.18632/aging.102109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan L, Wu X, Liu Y, Xian W. LncRNA Linc00511 promotes osteosarcoma cell proliferation and migration through sponging miR-765. J Cell Biochem. 2019;120:7248–7256. doi: 10.1002/jcb.27999. [DOI] [PubMed] [Google Scholar]
- 73.Guo K, Qian K, Shi Y, Sun T, Wang Z. LncRNA-MIAT promotes thyroid cancer progression and function as ceRNA to target EZH2 by sponging miR-150-5p. Cell Death Dis. 2021;12:1097. doi: 10.1038/s41419-021-04386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meng X, Sun W, Yu J, Zhou Y, Gu Y, Han J, Zhou L, Jiang X, Wang C. LINC00460-miR-149-5p/miR-150-5p-mutant p53 feedback loop promotes oxaliplatin resistance in colorectal cancer. Mol Ther Nucleic Acids. 2020;22:1004–1015. doi: 10.1016/j.omtn.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H, Kong J, Ding K, Shen HM, Wu H, Xia D, Wu Y. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer. 2017;16:118. doi: 10.1186/s12943-017-0685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liang L, Zhang Z, Qin X, Gao Y, Zhao P, Liu J, Zeng W. Long noncoding RNA ZFAS1 promotes tumorigenesis through regulation of miR-150-5p/RAB9A in melanoma. Melanoma Res. 2019;29:569–581. doi: 10.1097/CMR.0000000000000595. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Y, Dong Q, Sun X. Positive feedback loop LINC00511/miR-150-5p/SP1 modulates chondrocyte apoptosis and proliferation in osteoarthritis. DNA Cell Biol. 2020;39:1506–1512. doi: 10.1089/dna.2020.5718. [DOI] [PubMed] [Google Scholar]
- 78.Wu Y, Li L, Wang Q, Zhang L, He C, Wang X, Liu H. LINC00511 promotes lung squamous cell carcinoma proliferation and migration via inhibiting miR-150-5p and activating TADA1. Transl Lung Cancer Res. 2020;9:1138–1148. doi: 10.21037/tlcr-19-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mochizuki S, Okada Y. ADAMs in cancer cell proliferation and progression. Cancer Sci. 2007;98:621–628. doi: 10.1111/j.1349-7006.2007.00434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shan N, Shen L, Wang J, He D, Duan C. MiR-153 inhibits migration and invasion of human non-small-cell lung cancer by targeting ADAM19. Biochem Biophys Res Commun. 2015;456:385–391. doi: 10.1016/j.bbrc.2014.11.093. [DOI] [PubMed] [Google Scholar]
- 81.Feng X, Xue H, Guo S, Chen Y, Zhang X, Tang X. MiR-874-3p suppresses cell proliferation and invasion by targeting ADAM19 in nasopharyngeal carcinoma. Panminerva Med. 2021;63:238–239. doi: 10.23736/S0031-0808.19.03682-6. [DOI] [PubMed] [Google Scholar]
- 82.Hoyne G, Rudnicka C, Sang QX, Roycik M, Howarth S, Leedman P, Schlaich M, Candy P, Matthews V. Genetic and cellular studies highlight that A disintegrin and metalloproteinase 19 is a protective biomarker in human prostate cancer. BMC Cancer. 2016;16:151. doi: 10.1186/s12885-016-2178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang X, Wang E, Cao J, Xiong F, Yang Y, Liu H. MiR-145 inhibits the epithelial-to-mesenchymal transition via targeting ADAM19 in human glioblastoma. Oncotarget. 2017;8:92545–92554. doi: 10.18632/oncotarget.21442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Q, Yu L, Qin D, Huang R, Jiang X, Zou C, Tang Q, Chen Y, Wang G, Wang X, Gao X. Role of microRNA-30c targeting ADAM19 in colorectal cancer. PLoS One. 2015;10:e0120698. doi: 10.1371/journal.pone.0120698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang Y, Xiao F, Wang L, Wang T, Chen L. Circular RNA has_circ_0000034 accelerates retinoblastoma advancement through the miR-361-3p/ADAM19 axis. Mol Cell Biochem. 2021;476:69–80. doi: 10.1007/s11010-020-03886-5. [DOI] [PubMed] [Google Scholar]
- 86.Shi L, Hong X, Ba L, He X, Xiong Y, Ding Q, Yang S, Peng G. Long non-coding RNA ZNFX1-AS1 promotes the tumor progression and metastasis of colorectal cancer by acting as a competing endogenous RNA of miR-144 to regulate EZH2 expression. Cell Death Dis. 2019;10:150. doi: 10.1038/s41419-019-1332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang Y, Zhang Z, Wu Z, Lin W, Yu M. Downregulation of the expression of the lncRNA MIAT inhibits melanoma migration and invasion through the PI3K/AKT signaling pathway. Cancer Biomark. 2019;24:203–211. doi: 10.3233/CBM-181869. [DOI] [PubMed] [Google Scholar]
- 88.Chen XX, Zhang N, Fu XF, Jiang Y, Wang MY. LncRNA DBH-AS1 facilitates the tumorigenesis of melanoma by targeting miR-233-3p via IGF-1R/Akt signaling. Eur Rev Med Pharmacol Sci. 2020;24:7698–7708. doi: 10.26355/eurrev_202007_22272. [DOI] [PubMed] [Google Scholar]
- 89.Wang Q, Mao X, Luo F, Wang J. LINC00511 promotes gastric cancer progression by regulating SOX4 and epigenetically repressing PTEN to activate PI3K/AKT pathway. J Cell Mol Med. 2021;25:9112–9127. doi: 10.1111/jcmm.16656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.