Abstract
The inflammation-related tumor microenvironment (TME) is one of the major driving forces of hepatocarcinogenesis. We aimed to investigate cell-to-cell communication among Hepatocellular Carcinoma (HCC) through re-analyzing HCC single-cell RNA-seq data, and to confirm such cellular interaction through in vitro and in vivo study. We found a subset of Regulatory B cells with PD-L1 expression (PD-L1+ Bregs), mainly located in adjacent HCC tissues. In co-localization with PD-L1+ Bregs, a subset of Tumor Associated Macrophages with high expression of CXCL12 (CXCL12+ TAMs) was also mainly located in adjacent HCC tissues. Moreover, CXCL12+ TAMs can be stimulated in vitro using an HCC conditional medium. Using CellChat analysis and Multiplex Immunohistochemistry staining (mIHC), CXCL12+ TAMs were found to be first recruited by Cancer-Associated Fibroblasts (CAFs) through a CD74/macrophage migration inhibitory factor (MIF) pattern, and further differentiated into TGF-β-enriched tissues. Furthermore, CXCL12+ TAMs recruited PD-L1+ Bregs via the CXCL12/CXCR4 axis, and CXCR4 expression was significantly positively correlated to PD-L1 expression in PD-L1+ Bregs. At last, we confirmed the communications among CAFs, Macrophages and B cells and their tumor-promoting effects by using an orthotopic mouse model of HCC. Immunosuppressive HCC TME involving cell-to-cell communications comprised MIF-secreting CAFs, CXCL12-secreting TAMs, and PD-L1-producing Bregs, and their regulation could be promising therapeutic targets in future immunotherapy for human HCC.
Keywords: Regulatory B cells, CXCL12, CXCR4, hepatocellular carcinoma, tumor-associated macrophages, cancer-associated fibroblasts
Introduction
Hepatocellular carcinoma (HCC), the most common type of primary liver cancer, is among the leading causes of cancer-related death globally [1]. It has been well established that the inflammation-related tumor microenvironment (TME) is an essential factor in the initiation and progression of HCC. In addition, the immunosuppressive TME has become the most important target for HCC prevention and treatment, especially for novel HCC gene therapy [2].
Tumor-promoting TMEs usually comprise various cells, including tumor cells, hepatic stellate cells, cancer-associated fibroblasts, endothelial cells, neuroendocrine cells, tumor-infiltrating immunocytes, bone marrow-derived cells, and the extracellular matrix [3]. Among all these components, tumor-infiltrating immunocytes play a dominant role in determining the anti- or pro-tumor properties of the TME. Tumor-infiltrating lymphocytes (TILs) are the primary immune components of solid tumors and involve a host-anti-tumor reaction [4]. Recent research has shown that regulatory B cells (Bregs) in the TME can suppress diverse cell subtypes, including TILs, through the secretion of anti-inflammatory mediators such as interleukin-10 (IL-10) and can facilitate the conversion of T cells to Regulatory T cells, thereby attenuating anti-tumor immune responses [5]. However, our knowledge of the recruitment and differentiation of B cell in human HCC was still insufficient.
Bregs are a subset of B cells that play crucial roles in various conditions, including infections, allergies, autoimmune diseases, transplantation, and tumors [6]. The term of “regulatory B cells” was first coined in 2002 by Mizoguchi et al. They demonstrated that IL-10-producing CD1d+ Bregs are induced in a chronic inflammatory environment and dampen the progression of intestinal inflammation [7]. In the TME, Bregs can suppress effector T cells and target other tumor-infiltrating immune cells, such as myeloid-derived suppressor cells, natural killer cells, and macrophages, to hamper anti-tumor immunity [6]. In a mouse model of HCC, Shalapour et al. identified an IgA+ population of plasma cells, a type of Bregs, accumulating in the liver, which suppressed CD8+ T-cell responses through PD-L1 expression. The PD-L1 blockage significantly decreased HCC tumor size, which correlated with increased T-cell activation and decreased IgA+ plasma cell accumulation in the liver [8].
Although accumulating evidences suggested that specific signals control Breg generation in different settings, the extrinsic and intrinsic factors involved in Breg cell regulation are yet to be elucidated [9]. It has been reported that myeloid-derived cells producing IL-6 (in the mesenteric lymph nodes) and both IL-6 and IL-1β (in the spleen) are responsible for Breg induction in arthritis [10], and IL-21-producing CD4+ T cells located in the spleen are responsible for Breg induction in experimental autoimmune encephalitis [11].
Cancer-associated fibroblasts (CAFs) can be summarized as key cellular components within the tumor microenvironment, playing pivotal roles in the development and metastasis of various cancers through mechanisms like cytokine secretion, extracellular matrix remodeling, and angiogenesis [12]. Distinct from normal fibroblasts, CAFs exhibit unique biological characteristics and functional diversity [13]. Their potential as therapeutic targets has gained significant attention in recent years, underscoring their importance in understanding tumor biology and developing novel cancer treatment strategies [14].
Tumor-associated macrophages (TAMs), one of the most abundant stromal cell types in the HCC TME, not only suppress anti-tumor immunity but also secrete various inflammatory mediators to promote tumor progression through inducing extracellular matrix remodeling, angiogenesis, metastasis, and therapeutic resistance [15]. Our research has elucidated that CAFs play a pivotal role in HCC progression, engaging in a reciprocal signaling relationship with TAMs to foster a metastasis-prone environment. The discovery of a previously unrecognized dialogue between CXCL12+ TAMs and PD-L1+ regulatory B cells, facilitated by the CXCL12/CXCR4 axis, adds a layer of complexity to the immune evasion tactics within the TME. Moreover, the collaborative role of TAMs and CAFs in cultivating an immunosuppressive TME underscores an urgent need for therapeutic interventions aimed at dismantling this alliance, intending to restore an immune contexture conducive to tumor eradication.
Materials and methods
Clinical samples
Paraffin-embedded tumors and paired adjacent tissues were collected from 35 patients with HCC who underwent surgical resection between January 2019 and October 2020 at the Affiliated Drum Tower Hospital of Nanjing University Medical School (Nanjing, Jiangsu, China). All the tissues were combined into a single tissue array. None of the patients with HCC had undergone anticancer therapy prior to surgery, and individuals with concurrent autoimmune disease, HIV infection, or syphilis were excluded. The patients’ clinical characteristics were classified according to the Union for International Cancer Control (TNM staging) guidelines. All experiments were performed in compliance with government policies and the principles of the Declaration of Helsinki. The individuals were informed about the study and provided consent prior to specimen collection. This study was approved by the Ethics Committee of the Affiliated Drum Tower Hospital of Nanjing University Medical School (2021-304). The clinical features of all patients involved in this study are presented in Table 1.
Table 1.
The clinicopathological of 35 HCC patients
| Number | |
|---|---|
| Gender | |
| Female | 10 (29.4%) |
| Male | 25 (70.6%) |
| Age | |
| ≥50 | 28 (88.2%) |
| <50 | 7 (11.8%) |
| Capsule | |
| No | 24 (82.4%) |
| Yes | 11 (17.6%) |
| Vessel | |
| No | 29 (76.5%) |
| Yes | 6 (23.5%) |
| Child-Pugh stage | |
| classA | 27 (70.6%) |
| classB | 8 (29.4%) |
| HbsAg | |
| Negative | 10 (23.5%) |
| Positive | 25 (76.5%) |
| Cirrhosis | |
| S0 | 1 (5.9%) |
| S1 | 12 (29.4%) |
| S2 | 8 (29.4%) |
| S3 | 5 (11.8%) |
| S4 | 9 (23.5%) |
| AFP | |
| Abnormal | 15 (41.2%) |
| Normal | 20 (58.8%) |
| TNM | |
| I | 15 (29.4%) |
| II | 15 (52.9%) |
| III | 5 (17.6%) |
Data source
The single-cell transcriptome file of GSE149614 was downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/).
Processing of scRNA-seq data
Processing of scRNA-seq data was performed using the Seurat package (V.4.0.3) in R [16]. Low-quality cells with maximum gene expression ≥4000, minimum gene expression ≤500, or a proportion of mitochondrial gene expression ≥20 were discarded.
Clustering analysis
FindVariableFeatures in the Seurat package was used to identify highly variable genes and then principal component analysis was performed with the top 2000 variable genes. Clusters were partitioned using FindClusters in the Seurat package. SingleR package (V.1.6.1) was then used and the SingleR method was applied to identify each cluster using the Human Primary Cell Atlas database as a reference library [17].
Re-clustering analysis
B cell and macrophage clusters were selected for re-clustering analysis using the Garnett package (V.0.2.17) in R [18]. We populated a marker file that included each of the expected cell types using literature-based markers [18]. The Garnett package enables the use of marker information provided to select cells, which we then used to train a regression-based classifier. After the classifier was trained, it was applied to the single-cell dataset for re-clustering analysis and cell identification. The cells were then projected into a two-dimensional space with a uniform manifold approximation and projection (UMAP).
Deferentially expressed gene (DEG) analysis
The limma package (V.3.48.1) was used for DEGs [19]. DEG analysis was performed between CXCL12+ TAM and other macrophages and between PDL1+ Breg and other B cells. DEGs were determined using an adjusted p-value of <0.05 as the screening criteria. Anti- and pro-inflammatory genes were selected from the DEGs.
Gene set variation analysis (GSVA)
We used the GSVA package (V.1.40.1) for GSVA involving 87 basal metabolic pathway genes [20]. GSVA was performed to compare CXCL12+ TAM and other macrophages.
Cellular interaction analysis
Cell-to-cell interaction analysis was conducted using the CellChat package (V.1.1.3), which can visualize and analyze intercellular communication from scRNA-seq data [21,22]. We followed the official workflow, loaded the normalized counts into CellChat, and applied the standard preprocessing steps, including the functions identifyOverExpressedGenes, identifyOverExpressedInteractions, and projectData with a standard parameter set. We then calculated the potential ligand-receptor interactions between all macrophage clusters and B-cell clusters, based on the functions computeCommunProb, computeCommunProbPathway, and aggregateNet, using standard parameters.
Multiple-colored immunofluorescence (mIHC)
All tissue specimens were evaluated by two independent experienced pathologists. Paraffin-embedded human HCC tissues were cut into 5-μm sections to undergo tissue microarray (TMA) staining. The sections were stained with antibodies against human Tim3 (1:200; Signalway Antibody LLC, #28301), CD8 (1:200, Cell Signaling Technology, C8/144B), CD163 (1:200, Cell Signaling Technology, D6U1J), CD38 (1:200, Proteintech, 25284-1-AP), IgA (1:200, Proteintech, 11449-1-AP), CXCL12 (1:200, Proteintech, 17402-1-AP), CXCR4 (1:50, Abcam, 60042-1-Ig), PD-L1 (1:40, Abcam, ab58810), HRP-conjugated anti-mouse or anti-rabbit IgG (PANOVUE), and PPD 520/540/570/620/650/690/DAPI (PANOVUE). Positive cells were detected using a Vectra 3 automated quantitative pathology imaging system (Vectra 3, Akoya Biosciences) and analyzed using an inForm (PerkinElmer).
In vitro macrophage stimulation and RNA-seq
The human monocyte cell line THP1 was maintained in RPMI 1640 supplemented with 10% heat-inactivated FBS at 37°C and 5% CO2. All experiments were performed using mycoplasma-free cells. For macrophage stimulation, THP1 was treated with 5 ng/mL PMA (Sigma) for 48 h. For TAM stimulation, THP1-derived macrophages were treated in sample tissue lysate-derived conditional medium (adjusted to a final protein concentration of 50 μg/mL) for 48 h. The macrophages were then collected, and RNA library sequencing was performed on Illumina HiseqTM 2500/4000 using Gene Denovo Biotechnology Co., Ltd. (Guangzhou, China).
Tumor infiltrating leukocytes extraction and flow cytometry
Tumor infiltrating leukocytes were extracted according to the methods described previously [23]. Briefly, Fatty, connective, and necrotic tissue was removed. Tissues were minced into 1-2 mm pieces in RPMI 1640, transferred into 15 or 50 mL conical tubes, and incubated with triple enzyme digestion medium containing DNase (30 U/mL), hyaluronidase (0.1 mg/mL), and collagenase (1 mg/mL) for 2 hours at room temperature with gentle shaking. Tissues were resuspended in 10 mL RPMI 1640 and filtered through a 70-μm cell strainer (BD Pharmingen). Tissue trapped by the strainer was placed into individual wells containing 1 mL of T-cell growth medium in a 24-well plate for isolation or detection by flow cytometry.
Flow cytometry and flow cytometry sorting after isolation cells were washed and resuspended in FACS buffer (1× PBS, 2% iFCS, 2 mM EDTA) for counting using a Neubauer chamber in Trypan blue. 2-3^106 cells were used for FACS staining. Single cell suspensions were incubated with viability dye (1:400) in 1× PBS for 20 min. After washing with FACS buffer, cells were incubated with FcR-block (1:100) for 10 min and sequentially stained with the antibody listed in Table S1. After washing, samples were fixed stained for 10 min with streptavidin, washed and fixed with 4% PFA for 30 min and washed with FACS buffer before analysis using an LSRII Flow Cytometer (BD Bioscience). Data was analyzed using FlowJo software (Tree Star Inc.).
An orthotopic mouse model of hepatocellular carcinoma
In investigating the regulation of MIF positive CAF on the production of CXCL12 in infiltrating macrophage, the C57BL/6 (Gemphamatec, Nanjing, China) mice and underwent the were first underwent the treatment of administration of CCl4 20% (vol/vol) three times per week for 6 weeks controlled by olive oil. Both fibrosis and non-fibrosis mouse were used for orthotopic HCC model, which was setup by using mouse HCC cell line Hep1-6 according to previous publication (step 18-42) [24], during the tumor growth, inhibitor of MIF, MIF098 (Aobious, Cat. No. AOB33430), in the dose of 20 mg/kg was injected into mice via tail vein every two weeks. Eight weeks after the orthotopic transplantation, the mice were sacrificed for further investigation including tumor volume measurement and mIHC staining.
For the investigation for the regulation of macrophage derived CXCL12 on the production of PD-L1 in tumor infiltrating B cells, same orthotopic mouse model of HCC was also setup in C57BL/6 mice, 6^106 bone marrow derived mouse macrophage transduced with or without lentiviral derived mouse CXCL12 were adoptively transferred into mice by tail vein injection every two weeks, the inhibitor of CXCR4 AMD3100 (Cat. No. S8030, Selleckchem, TX) was injected into mice treated with CXCL12+ macrophage in the dose of 5 mg/kg every other day, and the production of lentivirus were based on previous publication [25].
Data analysis
Data are presented as mean ± standard error of mean. The χ2 test and Student’s t-test analysis of variance were performed to evaluate the differences in demographic and clinical characteristics. Pearson’s correlation analysis was performed to analyze the relationships between associated factors. Statistical analyses were performed using R and presented using R (version 4.1.0) or GraphPad Prism (CA, USA). In all cases, statistical significance was set at a p-value of <0.05.
Results
High PD-L1 expression IgA-secreting Bregs were mainly distributed in adjacent human HCC tissue
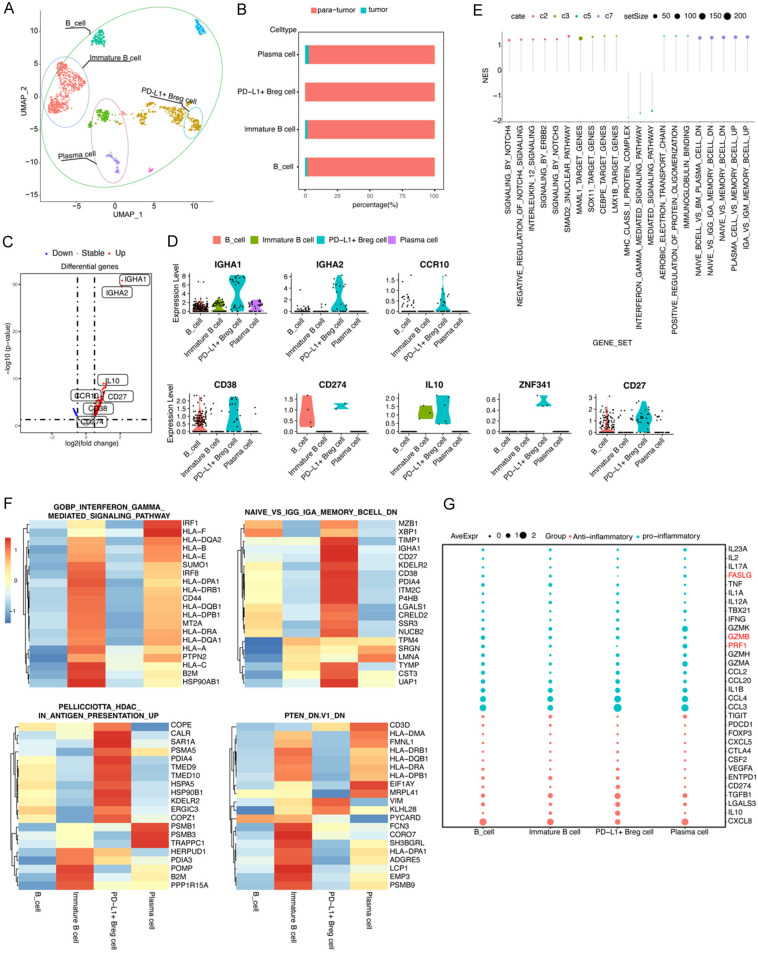
To investigate B cells related to TME in human HCC, we re-analyzed an HCC single-cell dataset with GEO number GSE149614. After discarding low-quality cells, 5979 cells were used for further analysis. We defined all clusters into eight broad cellular lineages, including endothelial cells, hepatocytes, macrophages, T cells, monocytes, smooth muscle cells, NK cells, and B cells, using singleR package (Figure S1). Among these eight cellular lineages, we first focused on the B-cell lineage. After re-clustering the B cells using the Garnett package, different subclusters were visualized using UMAP (Figure 1A). Four B-cell clusters were observed within human HCC TME, namely, PD-L1+ Bregs, immature B cells, B cell, and plasma cell. These four B-cell clusters, especially PD-L1+ Bregs, were mainly distributed in para-tumor tissues (Figure 1B). To determine the unique characteristics of PD-L1+ Bregs, DEG analysis was performed. Compared with the other three B-cell clusters, PD-L1+ Bregs expressed a significantly higher number of marker genes (Table S2), such as IGHA1, IGHA2, CCR10, CD38, CD274, IL10, ZNF341, and CD27 (Figure 1C, 1D). Subsequently, to identify detailed characters of PD-L1+ Breg cells, Gene Set Enrichment Analysis (GSEA) was performed for all B-cell lineage clusters using gene sets from the Molecular Signatures Database (MSigDB, ver.7.4), including curated gene sets (C2), regulatory target gene sets (C3), ontology gene sets (C5), and immunologic signature gene sets (C7). The results indicated that PD-L1+ Bregs were cluster cells with activation of Notch, IL-12, and ERBB2 signaling, with DEGs regulated by transcriptional factors such as MAML1, SOX11, LMX1B, and CEBPE, and demonstrated a cell lineage with “IgA expression memory B cells” (Figure 1E, 1F). This cluster’s secretory gene expression was evaluated, and we found that PD-L1+ Bregs expressed anti-inflammatory genes, such as IL-10, TGF-β, and CD274, and low expression of pro-inflammatory genes, such as GZMB, PRF1, and FASLG (Figure 1G).
Figure 1.
High PD-L1 expression Bregs were spotted as a subset of infiltrated B cells in human HCC indicated through a single-cell RNA-seq. A. The UMAP plot of single B cells obtained from one patient, representing four distinct clusters including PD-L1+ Breg cells. Each dot corresponds to one single cell, and circles are colored according to cell clusters. B. Relative abundance of four types of B cells in scRNA-seq analysis of human liver samples obtained from tumor and para-tumor liver sites. C. A volcano plot of differentially expressed genes (DEGs) is compared between PD-L1+ Breg cells and other B cells. Each colored dot denotes an individual gene with an adjusted p-value of <0.05. D. Violin plots of significant markers in four types of B cells. E. GSEA analysis using C2, C3, C5, and C7 gene sets. F. A heatmap of the top 20 genes expressed in four pathways. G. A dot plot of the expression of pro-inflammatory and anti-inflammatory genes in four types of B cells.
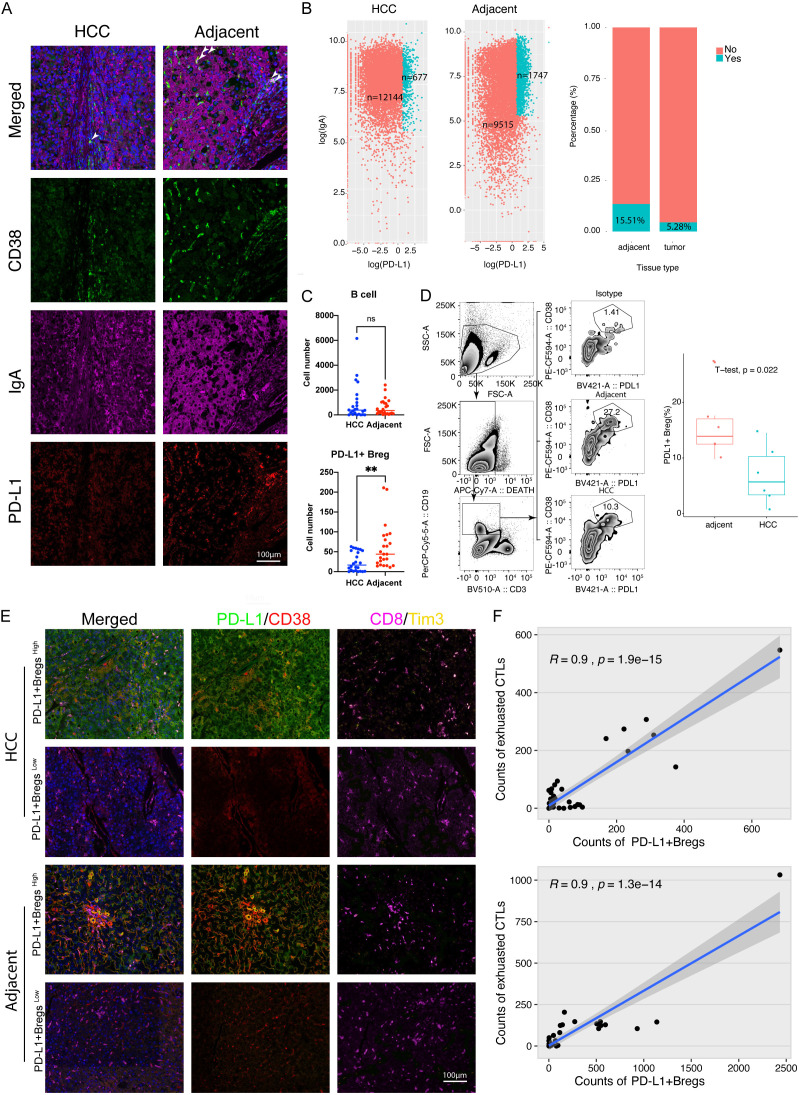
To verify the existence of PD-L1+ Bregs, an mIHC staining assay was performed using an HCC tissue array containing 35 paired tumors and adjacent tumor tissues. PD-L1+ Breg was defined using CD38, PD-L1, and IgA (Figure 2A). Similar to the results of single-cell sequencing, the number of PD-L1+ Bregs (CD38+PD-L1+IgA+) were found to be mainly distributed in adjacent tissues, whereas there was no difference in B cell numbers between tumor and adjacent tumor tissues (Figure 2B, 2C). Moreover, the chi-square test revealed that the number of PD-L1+ Bregs, but not total B cells, was significantly different among varied stages of liver cirrhosis (Tables 2 and 3). To further evaluate the function of PD-L1+ Bregs, we found that the number of PD-L1+ Bregs (PD-L1+CD38+) was significantly correlated with the number of exhausted CTLs (CD8+Tim3+), not only in the tumor but also in adjacent tumor tissues (Figure 2D-F).
Figure 2.
High PD-L1 expression Bregs were mainly located in adjacent HCC tissues and associated with exhausted CTLs in human HCC. A. Representative figures (×200 magnification) of multiple-color staining for CD38, PD-L1, and IgA expression in human HCC and adjacent tissue. B. Plots showing the number (left panel) and percentage (right panel) of IgA+PD-L1+ Breg cells in human HCC and adjacent tissue. C. A quantitative comparison of B cells and IgA+ PD-L1+ B cells in HCC and adjacent tissue using multiple-color staining. Data are presented as means ± SEM and were analyzed with Student’s t-test (**, P<0.01; ns, no significance, P>0.05). D. Comparison and gating strategy of CD19(+) CD38(+) PD-L1(+) Breg cell in TILs of human HCC analyzed by flow cytometry. E. Representative figures (×200 magnification) of multiple-color staining for PD-L1, CD38, CD8, and Tim3 expression in human HCC and adjacent tissues. F. Liner correlation studies were performed between the number of PD-L1+ B cells and exhausted T cells in HCC and adjacent tissue.
Table 2.
The clinicopathological relevance analysis of B cell number in HCC patients
| High (N=17) | Low (N=18) | P-value* | |
|---|---|---|---|
| Gender | |||
| Female | 5 (29.4%) | 5 (27.8%) | 1 |
| Male | 12 (70.6%) | 13 (72.2%) | |
| Age | |||
| ≥50 | 15 (88.2%) | 13 (72.2%) | 0.447 |
| <50 | 2 (11.8%) | 5 (27.8%) | |
| Capsule | |||
| No | 14 (82.4%) | 10 (55.6%) | 0.179 |
| Yes | 3 (17.6%) | 8 (44.4%) | |
| Vessel | |||
| No | 13 (76.5%) | 16 (88.9%) | 0.599 |
| Yes | 4 (23.5%) | 2 (11.1%) | |
| Child-Pugh stage | |||
| classA | 12 (70.6%) | 15 (83.3%) | 0.621 |
| classB | 5 (29.4%) | 3 (16.7%) | |
| HbsAg | |||
| Negative | 4 (23.5%) | 6 (33.3%) | 0.789 |
| Positive | 13 (76.5%) | 12 (66.7%) | |
| Cirrhosis | |||
| S0 | 1 (5.9%) | 0 (0%) | 0.714 |
| S1 | 5 (29.4%) | 7 (38.9%) | |
| S2 | 5 (29.4%) | 3 (16.7%) | |
| S3 | 2 (11.8%) | 3 (16.7%) | |
| S4 | 4 (23.5%) | 5 (27.8%) | |
| AFP | |||
| Abnormal | 7 (41.2%) | 8 (44.4%) | 1 |
| Normal | 10 (58.8%) | 10 (55.6%) | |
| TNM | |||
| I | 5 (29.4%) | 10 (55.6%) | 0.295 |
| II | 9 (52.9%) | 6 (33.3%) | |
| III | 3 (17.6%) | 2 (11.1%) |
Data was analyzed by chi-squared test.
P value in bold indicated statistically significant.
Table 3.
The clinicopathological relevance analysis of PD-L1+Bregs cell number in HCC patients
| High (N=17) | Low (N=18) | P-value* | |
|---|---|---|---|
| Gender | |||
| Female | 2 (11.8%) | 8 (44.4%) | 0.0776 |
| Male | 15 (88.2%) | 10 (55.6%) | |
| Age | |||
| ≥50 | 14 (82.4%) | 14 (77.8%) | 1 |
| <50 | 3 (17.6%) | 4 (22.2%) | |
| Capsule | |||
| No | 11 (64.7%) | 13 (72.2%) | 0.909 |
| Yes | 6 (35.3%) | 5 (27.8%) | |
| Vessel | |||
| No | 14 (82.4%) | 15 (83.3%) | 1 |
| Yes | 3 (17.6%) | 3 (16.7%) | |
| Child-Pugh stage | |||
| classA | 11 (64.7%) | 16 (88.9%) | 0.194 |
| classB | 6 (35.3%) | 2 (11.1%) | |
| HbsAg | |||
| Negative | 6 (35.3%) | 4 (22.2%) | 0.63 |
| Positive | 11 (64.7%) | 14 (77.8%) | |
| Cirrhosis | |||
| S0 | 1 (5.9%) | 0 (0%) | 0.032 |
| S1 | 3 (17.6%) | 9 (50.0%) | |
| S2 | 2 (11.8%) | 6 (33.3%) | |
| S3 | 4 (23.5%) | 1 (5.55%) | |
| S4 | 7 (41.2%) | 2 (11.1%) | |
| AFP | |||
| Abnormal | 6 (35.3%) | 9 (50.0%) | 0.591 |
| Normal | 11 (64.7%) | 9 (50.0%) | |
| TNM | |||
| I | 2 (11.8%) | 13 (72.2%) | 0.0012 |
| II | 9 (52.9%) | 6 (33.3%) | |
| III | 5 (29.4%) | 0 (0%) |
Data was analyzed by chi-squared test.
P value in bold indicated statistically significant.
High-expression CXCL12 TAMs are also mainly located in adjacent tissues in human HCC
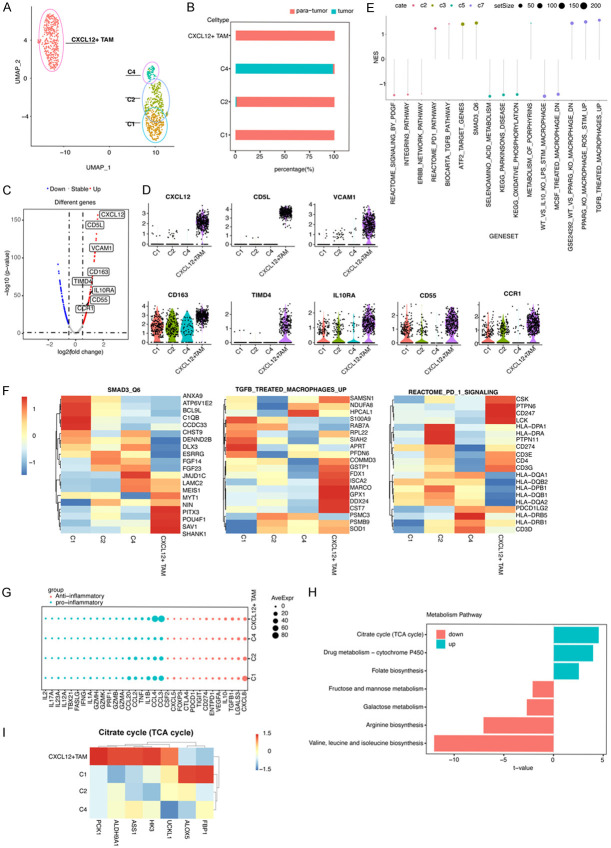
In addition to B cells, we analyzed tumor-infiltrating macrophages using the same single-cell seq dataset and identified another cluster, namely, CXCL12+ TAM, which was distinguished from the other three clusters (Figure 3A). We found that CXCL12+ TAM was mainly distributed in adjacent tissues, similar to the C1 and C2 clusters, and that it differed from the C4 cluster (Figure 3B). Using DEG analysis, CXCL12+ TAMs were found to have hallmark genes, including CXCL12, CD5L, VCAM1, CD163, TIMD4, IL10RA, CD55, and CCR1 (Figure 3C, 3D). Using GSEA with geneset MSigDB C2, C3, C5, and C7, we found that CXCL12+ TAM was inactivated in the PDGF, TGF-β, and ERBB signaling, and activated in PD-1 and TGFB1 signaling, with DEGs regulated by TFs including ATF2 and SMAD3 (Figure 3E, 3F). In addition, our findings also indicated that CXCL12+ TAMs highly expressed pro-inflammatory genes CCL3 and CCL4, anti-inflammatory genes CD274, IL-10, and TGF-β (Figure 3G). Finally, we analyzed the metabolism features of CXCL12+ TAMs using GSVA analysis, compared with other clusters, and we found that CXCL12+ TAM had increased TCA cycles, cytochrome P450 activity, and folate biosynthesis, and decreased metabolism of mannose, fructose, and galactose, biosynthesis of arginine, valine, leucine, and isoleucine (Figure 3H, 3I).
Figure 3.
High expression CXCL12+ tumor-associated macrophages (TMAs) were a subset of infiltrating macrophage in human HCC. (A) The UMAP plot of single macrophages obtained from one patient, representing four distinct clusters including CXCL12+ TMAs. Each dot corresponds to one single cell, with circles colored according to cell clusters. (B) Relative abundance of four types of macrophages in scRNA-seq analysis of human liver samples obtained from tumor and para-tumor liver sites. (C) A volcano plot of differentially expressed genes (DEGs) compared between CXCL12+ TMAs and other macrophages. Each colored dot denotes an individual gene with an adjusted p-value of <0.05. (D) Violin plots of significant markers in four types of macrophage. (E) GSEA analysis using C2, C3, C5, and C7 gene sets. (F) A heatmap of the top 20 genes expressed in three pathways indicated in the figure. (G) A dot plot of the expression of pro-inflammatory and anti-inflammatory genes in four types of macrophages. (H) A bar plot showing the difference between CXCL12+ TMA and other macrophages in the enrichment of metabolic pathways. (I) A heatmap of the expression levels of key enzymes in the metabolic pathway in (H) in four types of macrophages.
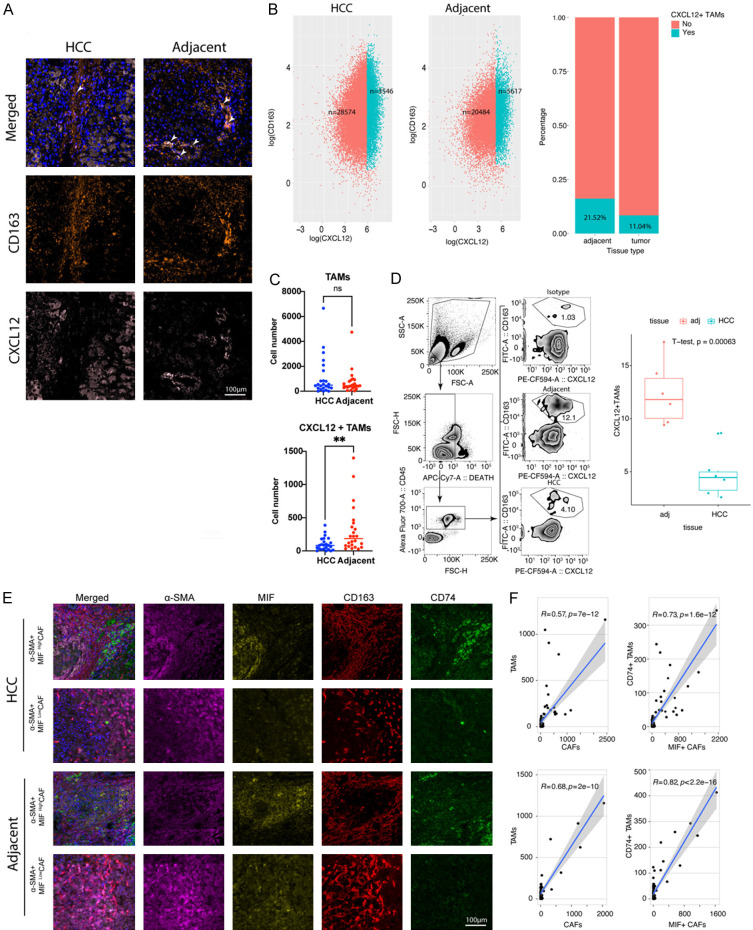
mIHC was then used to verify the existence of CXCL12+ TAMs in the human HCC tissue array (Figure 4A), with CXCL12+ TAMs detected in both HCC and adjacent tissues. However, the amount of CXCL12+ TAM was significantly higher in adjacent tissues than in the intratumor area (Figure 4B, 4C). The chi-square test revealed that the number of CXCL12+ TAMs, but not total TAMs, was significantly different within varied stages of liver cirrhosis (Tables 4 and 5). CellChat analysis suggested a potential interaction between a subset of inflammatory cancer-associated fibroblasts (iCAFs) and CXCL12+ TAMs via the MIF-CD74 axis, as illustrated in Figure S2A-C. To verify this regulation, we used mIHC to analyze the correlation between the number of MIF-producing CAFs and CD74+ TAMs, and found that these two subsets were closely co-located (Figure 4D-F).
Figure 4.
CXCL12+ high-expression tumor-associated macrophages (TAMs) were co-localized with MIF+CAFs in human HCC. A. Representative figures (×200 magnification) of multiple-color staining for CD163, PD-L1, and CXCL12 expression in human HCC and adjacent samples. B. Plots showing the number (left panel) and percentage (right panel) of CXCL12+ TAMs cells in human HCC and adjacent tissue. C. The comparison of TAMs and CXCL12+ TAMs in HCC and adjacent tissues using multiple-color staining. Data are presented as mean ± SEM and were analyzed using Student’s t-test (**, P<0.01; ns, no significance, P>0.05). D. Comparison and gating strategy of CD45(+) CD163(+) CXCL12(+) TAMs in TILs of human HCC analyzed by flow cytometry. E. Representative figures (×200 magnification) for multiple-color staining for α-SMA, MIF, CD163, and CD74 expression in human HCC and adjacent samples. F. Liner correlation studies were performed for TAMs vs CAFs and CD74+TAMs vs MIF+CAFs in HCC and adjacent tissue.
Table 4.
The clinicopathological relevance analysis of TAMs number in HCC patients
| High (N=17) | Low (N=18) | P-value* | |
|---|---|---|---|
| Gender | |||
| Female | 4 (23.5%) | 6 (33.3%) | 0.789 |
| Male | 13 (76.5%) | 12 (66.7%) | |
| Age | |||
| ≥50 | 16 (94.1%) | 12 (66.7%) | 0.108 |
| <50 | 1 (5.9%) | 6 (33.3%) | |
| Capsule | |||
| No | 13 (76.5%) | 11 (61.1%) | 0.539 |
| Yes | 4 (23.5%) | 7 (38.9%) | |
| Vessel | |||
| No | 11 (64.7%) | 18 (100%) | 0.0203 |
| Yes | 6 (35.3%) | 0 (0%) | |
| Child-Pugh stage | |||
| classA | 12 (70.6%) | 15 (83.3%) | 0.621 |
| classB | 5 (29.4%) | 3 (16.7%) | |
| HbsAg | |||
| Negative | 6 (35.3%) | 4 (22.2%) | 0.63 |
| Positive | 11 (64.7%) | 14 (77.8%) | |
| Cirrhosis | |||
| S0 | 0 (0%) | 1 (5.6%) | 0.614 |
| S1 | 6 (35.3%) | 6 (33.3%) | |
| S2 | 3 (17.6%) | 5 (27.8%) | |
| S3 | 2 (11.8%) | 3 (16.7%) | |
| S4 | 6 (35.3%) | 3 (16.7%) | |
| AFP | |||
| Abnormal | 9 (52.9%) | 6 (33.3%) | 0.407 |
| Normal | 8 (47.1%) | 12 (66.7%) | |
| TNM | |||
| I | 5 (29.4%) | 10 (55.6%) | 0.295 |
| II | 9 (52.9%) | 6 (33.3%) | |
| III | 3 (17.6%) | 2 (11.1%) |
Data was analyzed by chi-squared test.
P value in bold indicated statistically significant.
Table 5.
The clinicopathological relevance analysis of CXCL12+ TAMs number in HCC patients
| High (N=17) | Low (N=18) | P-value* | |
|---|---|---|---|
| Gender | |||
| Female | 4 (23.5%) | 6 (33.3%) | 0.789 |
| Male | 13 (76.5%) | 12 (66.7%) | |
| Age | |||
| ≥50 | 15 (88.2%) | 13 (72.2%) | 0.447 |
| <50 | 2 (11.8%) | 5 (27.8%) | |
| Capsule | |||
| No | 12 (70.6%) | 12 (66.7%) | 1 |
| Yes | 5 (29.4%) | 6 (33.3%) | |
| Vessel | |||
| No | 12 (70.6%) | 17 (94.4%) | 0.155 |
| Yes | 5 (29.4%) | 1 (5.6%) | |
| Child-Pugh stage | |||
| classA | 12 (70.6%) | 15 (83.3%) | 0.621 |
| classB | 5 (29.4%) | 3 (16.7%) | |
| HbsAg | |||
| Negative | 5 (29.4%) | 5 (27.8%) | 1 |
| Positive | 12 (70.6%) | 13 (72.2%) | |
| Cirrhosis | |||
| S0 | 0 (0%) | 8 (44.4%) | 0.0035 |
| S1 | 3 (17.6%) | 9 (50.0%) | |
| S2 | 4 (23.5%) | 4 (22.2%) | |
| S3 | 2 (11.7%) | 3 (16.7%) | |
| S4 | 8 (47.1%) | 1 (5.6%) | |
| AFP | |||
| Abnormal | 9 (52.9%) | 6 (33.3%) | 0.407 |
| Normal | 8 (47.1%) | 12 (66.7%) | |
| TNM | |||
| I | 3 (17.6%) | 12 (66.7%) | 0.012 |
| II | 10 (58.8%) | 5 (27.8%) | |
| III | 4 (23.5%) | 1 (5.6%) |
Data was analyzed by chi-squared test.
P value in bold indicated statistically significant.
CXCL12+ TAMs might be a subset of SMAD3-activated TAMs with a poor glycolysis capacity
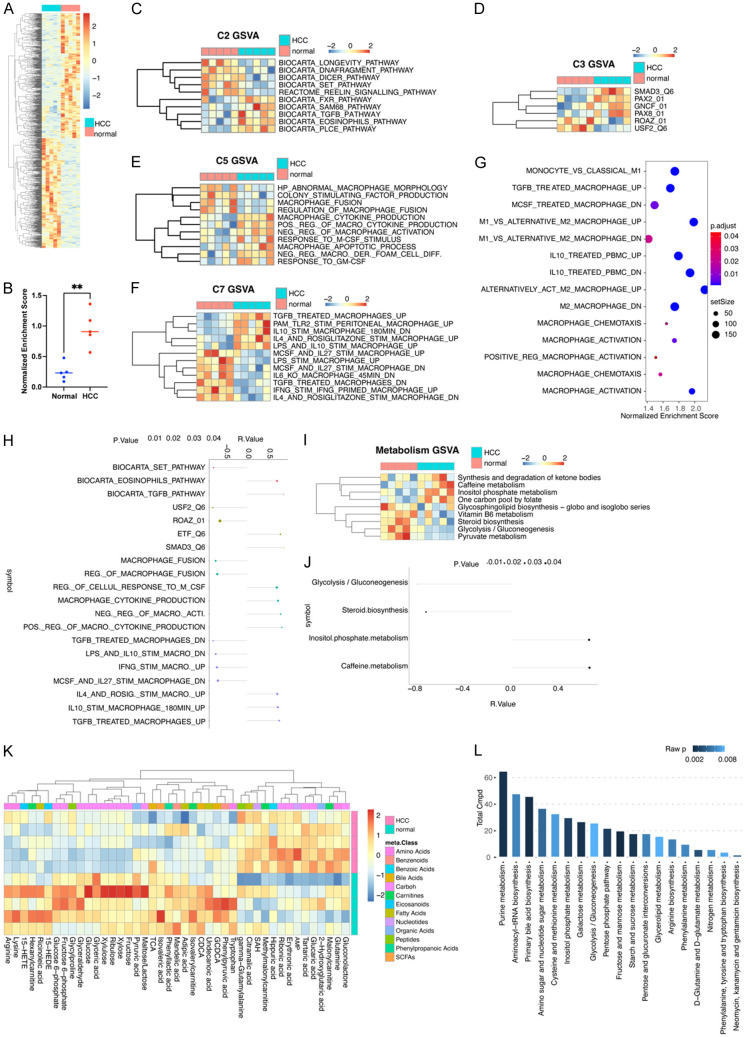
To explore the characters of CXCL12+ TAMs, we used conditional medium (CM) derived from the lysis of normal and HCC tissues to generate TAMs from PMA-treated THP1 cells. Using RNA-seq, we identified 931 DEGs, including 435 upregulated and 496 downregulated (Figure 5A). We then examined the CXCL12+ TAMs differentiation by using GSVA on a customized gene set comprised DEGs of CXCL12+ TAMs in single-cell sequencing. After stimulation with HCC CM, the percentage of CXCL12+ TAMs increased significantly (Figures 5B, S3). Next, to explore the potential regulation of CXCL12+ TAM development, we compared different gene sets from MSigDB (ver.7.4) also using GSVA, including curated gene sets (C2), regulatory target gene sets (C3), ontology gene sets (C5), and immunologic signature gene sets (C7). The most different gene sets are shown in (Figure 5C-F). We noted that the results for HCC CM-stimulated TAMs shared some similarities with the results obtained from sc-seq, especially in relation to TGF-β-related gene sets. For C2 gene sets, we found that either CXCL12+ TAMs or HCC CM-derived TAMs showed an increased NES for the “BIOCARTA_TGFB_PATHWAY” gene set in GSVA. In C3 gene set analysis, it was found that they shared an increased “SMAD3_Q6” gene set, whereas in C7 gene sets, they shared an increased “TGFB_TREATED_MACROPHAGES_UP” gene set. We also analyzed RNA-seq data using GSEA on C7 gene sets, and found that HCC CM medium-derived TAMs had significantly increased NES for the “TGFB_TRE ATED_MACROPHAGE_UP” gene set (Figure 5G). We then performed a multiple correlation analysis between the NES of CXCL12+ TAMs gene sets and the NES of different expression gene sets in C2, C3, C5, and C7, and the results indicated that “BIOCARTA_TGFB_PATHWAY”, “SMAD3_Q6”, and “TGFB_TREATED_MACROPHAGES_UP” all positively correlated with CXCL12+ TAMs (Figure 5H). These findings appear to indicate that the genesis of CXCL12+ TAMs is associated with the TGF-β-related TME within HCC. We used mIHC staining to verify this hypothesis and found that the number of CXCL12+ TAMs (CXCL12+CD163+) and CXCL12+ expression within TAMs were positively associated with TGF-β levels within HCC (Figure S4A, S4B).
Figure 5.
CXCL12+ TAMs are a subset of inducible SMAD3-activated tumor-associated macrophages. A. A heatmap showing differential gene expression between HCC and normal liver tissue. B. A dot plot for GSEA enrichment of the CXCL12+ TAMs gene set. The red color represents HCC (n=5) and blue represents normal liver tissue (n=5). Data are presented as mean ± SEM and were analyzed using Student’s t-test (**P<0.01). C-F. A heatmap showing different GSVA enrichment scores for C2, C3, C5, and C7 gene sets from MSigDB (ver.7.4) between HCC and normal liver tissue. G. A bubble plot showing normalized enrichment scores and a p-value of GSEA using macrophage-related gene sets from MSigDB (ver.7.4). H. A lollipop chart showing the correlation between the GSVA scores for CXCL12+ TAMs and the GSVA scores of differential gene sets between macrophages induced due to HCC and a normal conditional medium. I. A heatmap showing different GSVA enrichment scores for metabolic gene sets from TCGA between HCC and normal liver tissues. J. A lollipop chart showing the correlation between the GSVA scores of CXCL12+ TAMs and GSVA scores of differential metabolism-related gene sets between macrophages induced due to HCC and a normal conditional medium. K. A heatmap showing metabolite differences between HCC and normal liver tissues. L. A bar graph showing enrichment of metabolites and raw p-values.
We also investigated metabolic differences between normal and HCC CM-induced macrophages using GSVA and found that, compared with normal CM-derived macrophages, HCC TAMs showed increased gene signatures of synthesis and degradation of ketone bodies and one carbon pool by folate, and decreased gene signatures of glycolysis/gluconeogenesis and pyruvate metabolism (Figure 5I). After a multiple correlation analysis between differential expression metabolism gene sets and CXCL12+ TAMs gene sets, we found that glycolysis/gluconeogenesis was negatively correlated with CXCL12+ TAM gene sets (Figure 5J). To verify such metabolism change, we used metabolomics detection for normal and HCC CM-stimulated macrophages (Figure 5K), with pathway enrichment for different metabolites. Our observations suggested that CXCL12+ TAMs in HCC exhibit a reduced capacity for glycolysis and gluconeogenesis, as evidenced by our metabolomics analysis (Figure 5K, 5L). In light of this, CXCL12+ TAMs may tentatively be characterized as a potentially inducible group of TAMs with a diminished glycolytic function, possibly associated with SMAD3 activation.
CXCL12-secreting TAMs regulate PD-L1+ Bregs through the CXCL12/CXCR4 axis
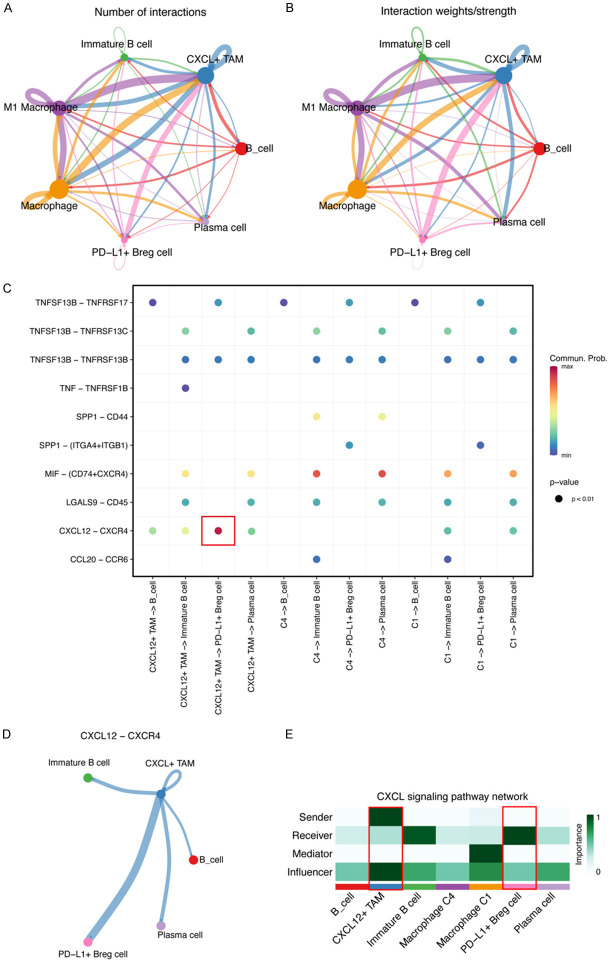
To further determine the interaction between TAMs and B cells, we used CellChat to analyze the number and strength of interactions among the subsets of B cells and macrophage populations. In addition to regulation among different subsets of macrophages, we found strong cell-to-cell communication between CXCL12+ TAMs and IgA+ PD-L1 B cells (Figure 6A, 6B). After fully exploring the different ligand-receptor interaction patterns, CXCL12/CXCR4 was found to have the highest communication probability in cross-talk between CXCL12+ TAMs and PD-L1+ B cells (Figure 6C). We found that cross-talk via CXCL12/CXCR4 may exist between CXCL12+ TAMs and all other B-cell subsets, but the possibility was highest in PD-L1+ B cells (Figure 6D, 6E).
Figure 6.
The CXCL12/CXCR4 axis was a potential cell-to-cell communication manner between CXCL12+ TAMs and PD-L1+ Bregs. A. A circle plot of the number of cellular communications in six identified types of cell populations in liver tissue. The width represents the number of cell interactions. B. A circle plot of the weights/strength of the cellular communications in six identified types of cell populations in liver tissue. The width represents the weights/strength of the interaction. C. Depiction of all the significant ligand-receptor pairs that contribute to the signaling sending from three macrophage types to four B cell types. The dot color and size represents the calculated communication probability and p-values. The p-values were computed using a one-sided permutation test. D. Circle plots of the CXCL12-CXCR4 signaling pathway network between six identified types of cell populations. The width represents the weights/strength of the interaction. E. A heatmap showing the relative importance of each cell group based on the computed four network centrality measures of the CXCL12 signaling pathway network.
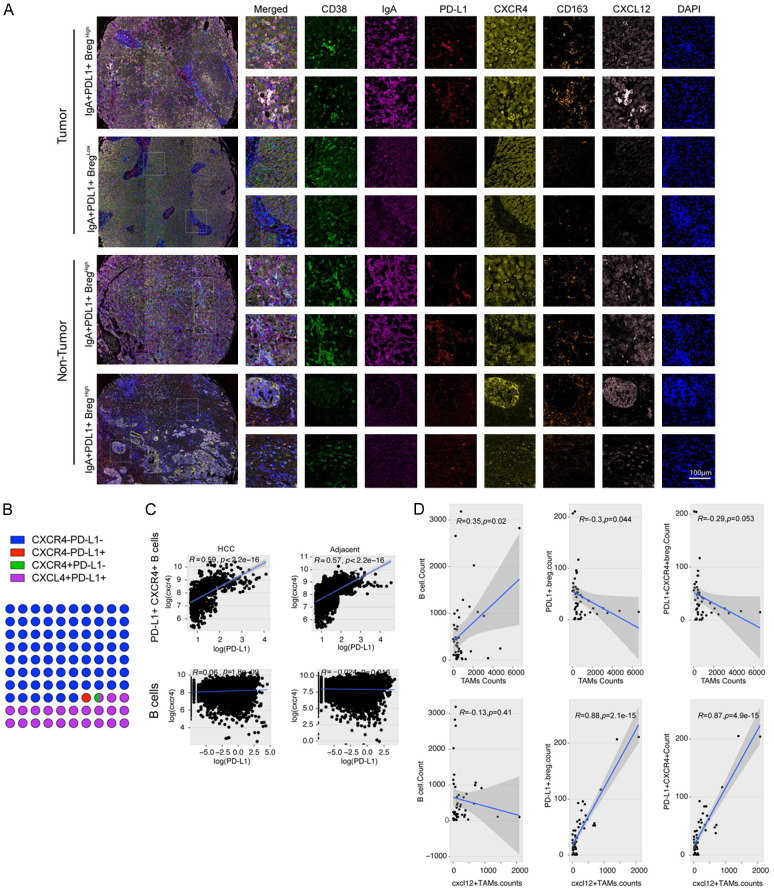
To verify this bioinformatic prediction, we performed mIHC staining for six indices in an HCC tissue array, including CD38, IgA, PD-L1, CXCR4, CXCL12, and CD163 (Figure 7A). After analysis, we found 29947 CD38+ cells (4.00% of total cells) with 86.90% IgA positivity. Among the IgA+ B cells, we found that 76.09% of the cells were PD-L1-/CXCR4- and 21.72% were PD-L1+/CXCR4+, and almost all PD-L1+ B cells 98.87% were CXCR4 positive, which implied that CXCR4 was important for PD-L1 expression in B cells (Figure 7B). Moreover, we found that only CXCR4+ PD-L+ B cells showed a significant positive correlation between CXCR4 and PD-L1 expression in HCC as well as in adjacent tissues (Figure 7C). Next, we evaluated the correlation between cell counts of macrophages and B cells, and found that the number of CXCL12+ TAMs was positively correlated with the number of IgA+ PD-L1+ B cells and IgA+ PD-L1+ CXCR4+ B cells (Figure 7D). Western blot analysis and quantitative real-time PCR revealed that stimulation with CXCL12 up-regulated PD-L1 expression in B cells isolated from mouse spleens, while co-treatment with CXCR4 antibody attenuated this expression (Figure S5A, S5B). These results imply that TAMs are related to the generation of PD-L1+ B cells via the CXCL12/CXCR4 axis.
Figure 7.
CXCL12-secreting TAMs regulate PD-L1+ Bregs through the CXCL12/CXCR4 axis. A. Representative figures for multiple-color staining of CD38, IgA, PD-L1, CXCR4, CD163, and CXCL12 expression in human HCC and adjacent samples. B. Dot plots showing the proportions of four types of cells. Different colors represent different cells. C. Liner correlation studies were performed for CXCR4 expression vs PD-L1 expression in PD-L+ CXCR4+ B cells and B cells in HCC and adjacent tissues. D. Linear correlation studies were performed between a count of TAMs and CXCL12+ TAMs in clinical samples with a count of B cells, PD-L1+ Breg cells, and PD-L1+ CXCR4+ B cells.
The regulation axis of MIF+CAF-CXCL12+macrophage-CXCR4+PDL1+B cell promote tumor growth in vivo
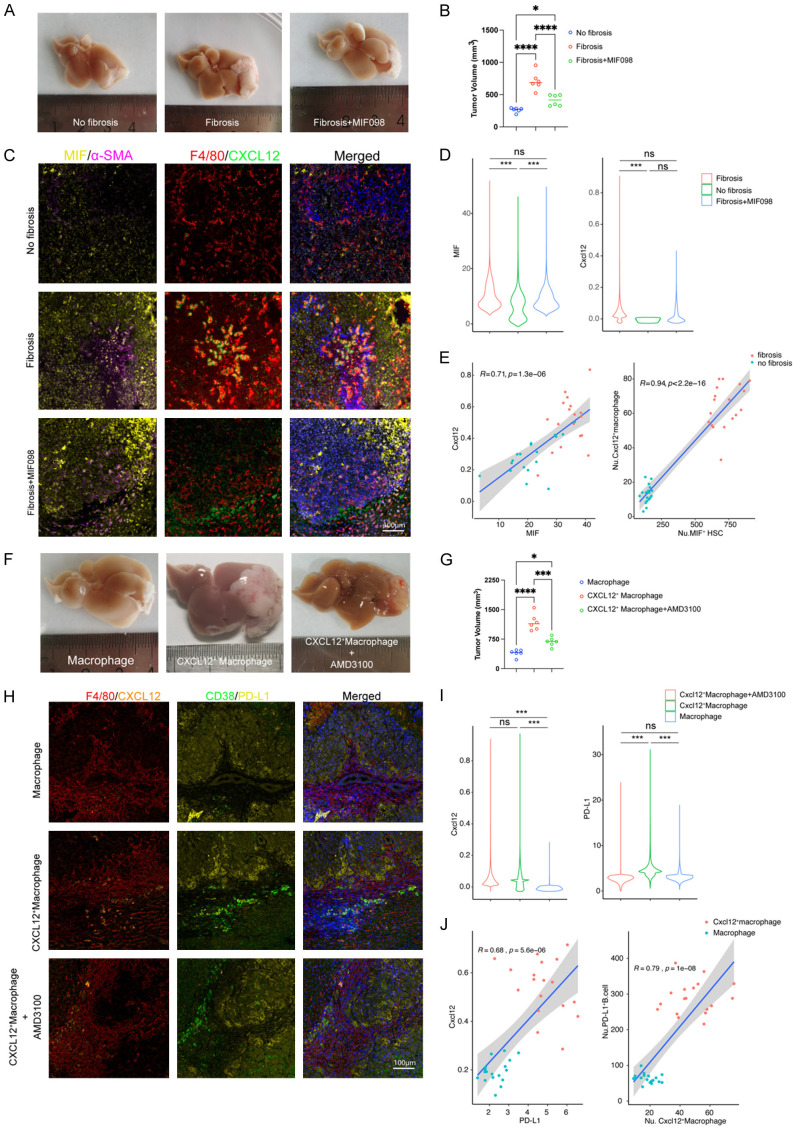
To verify the pro-tumor characters of these cellular interactions, we performed an orthotopic mouse model of hepatocellular carcinoma (Figure S6). For the intercellular interaction between CAFs and macrophages, we first setup a CCL4-induced mice fibrosis model (Figure S7A, S7B), and we found that liver fibrosis can promote tumor growth, macrophage infiltration and CXCL12 expression in macrophage, but when the effect of MIF was blocked by its inhibitor MIF098, tumor growth, macrophage infiltration, and macrophage CXCL12 expression were all restored (Figure 8A-D). In addition, we found that MIF expression in CAFs was positively correlated to CXCL12 expression in macrophages, and the number of MIF+ CAFs was also positively correlated to the number of CXCL12+ macrophages (Figure 8E). These results confirmed both chemotaxis and tumor promoting effect of MIF+CAF-CXCL12+ Macrophage axis. Next, an orthotopic mouse HCC model along with macrophage adoptive transfer assay were performed to verify the effects of tumor promoting and B cell PD-L1 expression regulation of CXCL12+ macrophage/PD-L1+ B cell interaction. We confirmed the expression of CXCL12 in transduced macrophage (Figure S7C, S7D), it can promote tumor growth and PD-L1 expression in tumor infiltrating B cells. However, the promoting effect by CXCL12+ macrophage can be fully restored by using CXCR4 inhibitor AMD3100 (Figure 8F-I). Furthermore, we also found that CXCL12 expression in macrophages was positively linearly correlated to PD-L1 expression in B cell, and the number of CXCL12+ macrophages was positively correlated to the number of PD-L1+ B cells in mice HCC (Figure 8J).
Figure 8.
The regulation axis of MIF+CAF-CXCL12+macrophage-CXCR4+PDL1+B cell promote tumor growth in vivo. A. Representative figures of tumor growth in orthotopic mouse HCC model treated differently indicated in the figure (n=6); B. Dot plots represent a comparison of tumor volumes for different group (***, P≤0.001; **, P≤0.01; *, P≤0.05); C. Representative figures for multiple-color staining for MIF, α-SMA, F4/80 and CXCL12 expression of different group; D. The violin plot of the expression of TGF-β and CXCL12 in the three groups; E. Liner correlation studies were performed for the expression of Cxcl12 vs TGF-β and the number of Cxcl12+ macrophage vs TGF-β+ HSC in different group; F. Representative figure of tumor growth in orthotopic mouse HCC model treated differently indicated in the figure (n=6). G. Dot plots represent a comparison of tumor volumes in different group (***, P≤0.001; **, P≤0.01; *, P≤0.05); H. Representative figures for multiple-color staining for F4/80, CXCL12, CD38 and PD-L1 expression of different groups; I. The violin plot of the expression of Cxcl12 and PD-L1 in the three model; J. Liner correlation studies were performed for the expression of Cxcl12 vs PD-L1 and the number of PD-L1+ B cells vs Cxcl12+ macrophage in the model Cxcl12+ macrophage and Macrophage.
Discussion
In the present study, through a re-analysis of HCC single-cell sequencing data, we found a novel crosstalk between CXCL12-secreting TAMs and PD-L1 expression in HCC-infiltrated IgA+ B cells. We found that CXCL12+ TAMs promoted PD-L1 expression in HCC-infiltrating B cells via the CXCL12/CXCR4 axis. Studies concerning PD-L1-expressing B cells have mainly focused on malignant B cells, including certain aggressive B-cell lymphomas as well as virus- and immunodeficiency-associated malignancies associated with an ineffective T-cell immune response expressing PD-L1 on tumor cells [26]. These malignant B cells expressing PD-L1 can bind to their cognate receptor PD-1 and inhibit the proliferation of activated T cells in peripheral tissues, leading to T cell exhaustion. In malignant B cells, PD-L1 expression has also been observed in normal B cells, especially in tumor-infiltrating B cells. For example, in HCC, Shalapour et al. identified an IgA+ population of plasma cells, a type of regulatory B cell, accumulating in the liver that suppressed CD8+ T-cell responses through PD-L1 expression [8]. PD-L1-producing B cells have also been observed in other malignancies, and PD-L1-producing Breg cells have been reported to induce tumor immunosuppression through inhibition of T-cell proliferation and exhaustion in human breast cancer and metastatic melanoma [27,28]. We also identified a subset of PD-L1-producing IgA+ B cells with a high expression of CXCR4, which is essential for PD-L1 expression triggered by CXCL12 and secreted by TAMs.
Evidence concerning regulation in relation to the genesis and maintenance of PD-L1 production in B cells is limited; however, some studies have indicated that the Breg generation is associated with dendritic cells (DCs) and mast cells in autoimmune diseases. Boldison et al. showed that DCs can stimulate Bregs to produce IL-10 and maintain a tolerogenic state to prevent type 1 diabetes [29]. Plasmacytoid dendritic cells (pDCs) control the fate of B cells as CD19+CD24hiCD38hi Breg cells and plasmablasts through the release of IFN-α and CD40 [30]. Mast cells are another regulator of Bregs; they enhance the population of IL-10-producing Bregs via the CD40L/CD40 axis in vitro and in vivo [31]. Moreover, IL-5 secreted from mast cells is important for maintaining the population of IL-10+ Bregs in peripheral lymphoid tissues [32]. In our study, we identified CXCL12-secreting TAMs as a new cell type that regulates Bregs through the CXCL12/CXCR4 axis. The CXCL12-secreting TAMs that we identified were a subset of TAMs, which were one of the most abundant stromal cell types in the HCC TME, and had mainly accumulated in the para-tumor tissues.
Based on the mIHC results, we found that CXCL12 was not only abundant, but also originated from many cell types within the HCC TME. The CXCL12/CXCR4 axis could be a plausible regulatory mechanism between other component cells and infiltrating B cells. Many studies have indicated that monocytes/macrophages are the main sources of CXCL12. In peripheral blood, monocytes secrete CXCL12 chemokines and express CXCR4 and CXCR7 receptors to form an autocrine/paracrine loop, which helps them differentiate into macrophages with a reduced function to stimulate antigen-specific T-lymphocyte responses [33]. Therefore, CXCL12 is a key factor that recruits immunosuppressive myeloid cells through interaction with CXCR4 [34]. As one of the receptors of CXCL12, CXCR4 triggers downstream signaling pathways, including PI3K/Akt and Ras/Raf/MAPK, promoting tumor growth and metastasis [35-37]. Several recent studies have increased understanding of the CXCL12/CXCR4 axis and PD-L1 expression, and a combination therapy involving a blockage of CXCL12-CXCR4 and PD-1-PD-L1 simultaneously has been shown to improve the therapeutic effect of single anti-PD1/PD-L1 therapy in liver, ovarian [38-40], and pancreatic cancer. We found co-localization of CXCL12+ TAMs and CXCR4+ PD-L1+ Bregs in both tumor and adjacent tissues of human HCC, which might imply that TAMs can recruit PD-L1 expression in Bregs through the CXCL12/CXCR4 axis. Moreover, the expression level of CXCR4 was positively correlated with PD-L1 expression in Bregs. We postulated that downstream signaling pathways, such as PI3K/AKT or RAS/RAF/c-jun, regulate the transcription of PD-L1 mRNA, which has been addressed in other studies [41-45].
Using cell-to-cell interaction analysis, we also identified another cell-to-cell regulation within the HCC TME, which was composed of macrophage MIF-expressing cancer-associated fibroblasts (CAFs) and CD74-expressing TAMs. Additionally, we found that CXCL12+ TAM differentiation was determined through TGF-β-induced SMAD2/3 activation. MIF/CD74 is a well-known pattern that promotes leukocyte recruitment to sites of inflammation [46-48]. MIF-secreting CAFs have been shown to promote the accumulation CXCL12+ TAMs both in human and mice liver cancer. Additionally, both scRNA-seq and macrophage RNA-seq data implied that differentiation of CXCL12+ TAMs was determined through SMAD3 activation. However, the relationship between TGF-β and CXCL12 remains unclear [49-51].
Taken together, we found that CAFs can enhance the recruitment of TAMs from peripheral blood through MIF/CD74-mediated chemotaxis, and differentiate into CXCL12+ TAMs through SMAD3 activation. CXCL12+ TAMs further recruited B cells through CXCL12/CXCR4-mediated chemotaxis and enhanced the exhaustion of CTL cells via CXCR4-related PD-L1 expression.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation (Grant Number: 82173100 to R.J.).
Disclosure of conflict of interest
None.
Abbreviations
- Bregs
Regulatory B cells
- CAFs
Cancer Associated Fibroblasts
- CM
Conditional medium
- CXCL12
C-X-C Motif Chemokine Ligand 12
- CXCR4
C-X-C Motif Chemokine Receptor 4
- HCC
Hepatocellular Carcinoma
- MIF
Macrophage Migration Inhibitory Factor
- PD-L1
Programmed Cell Death 1 Ligand 1
- TAMs
Tumor Associated Macrophages
- TIL
Tumor Infiltrating Lymphocytes
- TME
Tumor Microenvironment
Supporting Information
References
- 1.Global Burden of Disease Liver Cancer Collaboration. Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu MA, Allen C, Al-Raddadi R, Alvis-Guzman N, Amoako Y, Artaman A, Ayele TA, Barac A, Bensenor I, Berhane A, Bhutta Z, Castillo-Rivas J, Chitheer A, Choi JY, Cowie B, Dandona L, Dandona R, Dey S, Dicker D, Phuc H, Ekwueme DU, Zaki MS, Fischer F, Fürst T, Hancock J, Hay SI, Hotez P, Jee SH, Kasaeian A, Khader Y, Khang YH, Kumar A, Kutz M, Larson H, Lopez A, Lunevicius R, Malekzadeh R, McAlinden C, Meier T, Mendoza W, Mokdad A, Moradi-Lakeh M, Nagel G, Nguyen Q, Nguyen G, Ogbo F, Patton G, Pereira DM, Pourmalek F, Qorbani M, Radfar A, Roshandel G, Salomon JA, Sanabria J, Sartorius B, Satpathy M, Sawhney M, Sepanlou S, Shackelford K, Shore H, Sun J, Mengistu DT, Topór-Mądry R, Tran B, Ukwaja KN, Vlassov V, Vollset SE, Vos T, Wakayo T, Weiderpass E, Werdecker A, Yonemoto N, Younis M, Yu C, Zaidi Z, Zhu L, Murray CJL, Naghavi M, Fitzmaurice C. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol. 2017;3:1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hou PP, Luo LJ, Chen HZ, Chen QT, Bian XL, Wu SF, Zhou JX, Zhao WX, Liu JM, Wang XM, Zhang ZY, Yao LM, Chen Q, Zhou D, Wu Q. Ectosomal PKM2 promotes HCC by inducing macrophage differentiation and remodeling the tumor microenvironment. Mol Cell. 2020;78:1192–1206. e1110. doi: 10.1016/j.molcel.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Hernandez-Gea V, Toffanin S, Friedman SL, Llovet JM. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology. 2013;144:512–527. doi: 10.1053/j.gastro.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin LX. Inflammatory immune responses in tumor microenvironment and metastasis of hepatocellular carcinoma. Cancer Microenviron. 2012;5:203–209. doi: 10.1007/s12307-012-0111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarvaria A, Madrigal JA, Saudemont A. B cell regulation in cancer and anti-tumor immunity. Cell Mol Immunol. 2017;14:662–674. doi: 10.1038/cmi.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shang J, Zha H, Sun Y. Phenotypes, functions, and clinical relevance of regulatory B cells in cancer. Front Immunol. 2020;11:582657. doi: 10.3389/fimmu.2020.582657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizoguchi A, Mizoguchi E, Takedatsu H, Blumberg RS, Bhan AK. Chronic intestinal inflammatory condition generates IL-10-producing regulatory B cell subset characterized by CD1d upregulation. Immunity. 2002;16:219–230. doi: 10.1016/s1074-7613(02)00274-1. [DOI] [PubMed] [Google Scholar]
- 8.Shalapour S, Lin XJ, Bastian IN, Brain J, Burt AD, Aksenov AA, Vrbanac AF, Li W, Perkins A, Matsutani T, Zhong Z, Dhar D, Navas-Molina JA, Xu J, Loomba R, Downes M, Yu RT, Evans RM, Dorrestein PC, Knight R, Benner C, Anstee QM, Karin M. Inflammation-induced IgA+ cells dismantle anti-liver cancer immunity. Nature. 2017;551:340–345. doi: 10.1038/nature24302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baba Y, Saito Y, Kotetsu Y. Heterogeneous subsets of B-lineage regulatory cells (Breg cells) Int Immunol. 2020;32:155–162. doi: 10.1093/intimm/dxz068. [DOI] [PubMed] [Google Scholar]
- 10.Rosser EC, Oleinika K, Tonon S, Doyle R, Bosma A, Carter NA, Harris KA, Jones SA, Klein N, Mauri C. Regulatory B cells are induced by gut microbiota-driven interleukin-1β and interleukin-6 production. Nat Med. 2014;20:1334–1339. doi: 10.1038/nm.3680. [DOI] [PubMed] [Google Scholar]
- 11.Yoshizaki A, Miyagaki T, DiLillo DJ, Matsushita T, Horikawa M, Kountikov EI, Spolski R, Poe JC, Leonard WJ, Tedder TF. Regulatory B cells control T-cell autoimmunity through IL-21-dependent cognate interactions. Nature. 2012;491:264–268. doi: 10.1038/nature11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 13.Öhlund D, Elyada E, Tuveson D. Fibroblast heterogeneity in the cancer wound. J Exp Med. 2014;211:1503–1523. doi: 10.1084/jem.20140692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa A, Kieffer Y, Scholer-Dahirel A, Pelon F, Bourachot B, Cardon M, Sirven P, Magagna I, Fuhrmann L, Bernard C, Bonneau C, Kondratova M, Kuperstein I, Zinovyev A, Givel AM, Parrini MC, Soumelis V, Vincent-Salomon A, Mechta-Grigoriou F. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33:463–479. e410. doi: 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587. e3529. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, Butte AJ, Bhattacharya M. Referencebased analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pliner HA, Shendure J, Trapnell C. Supervised classification enables rapid annotation of cell atlases. Nat Methods. 2019;16:983–986. doi: 10.1038/s41592-019-0535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou R, Denisenko E, Ong HT, Ramilowski JA, Forrest ARR. Predicting cell-to-cell communication networks using NATMI. Nat Commun. 2020;11:5011. doi: 10.1038/s41467-020-18873-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park JE, Stephenson E, Polański K, Goncalves A, Gardner L, Holmqvist S, Henriksson J, Zou A, Sharkey AM, Millar B, Innes B, Wood L, Wilbrey-Clark A, Payne RP, Ivarsson MA, Lisgo S, Filby A, Rowitch DH, Bulmer JN, Wright GJ, Stubbington MJT, Haniffa M, Moffett A, Teichmann SA. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuang DM, Xiao X, Zhao Q, Chen MM, Li XF, Liu RX, Wei Y, Ouyang FZ, Chen DP, Wu Y, Lao XM, Deng H, Zheng L. B7-H1-expressing antigen-presenting cells mediate polarization of protumorigenic Th22 subsets. J Clin Invest. 2014;124:4657–4667. doi: 10.1172/JCI74381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reiberger T, Chen Y, Ramjiawan RR, Hato T, Fan C, Samuel R, Roberge S, Huang P, Lauwers GY, Zhu AX, Bardeesy N, Jain RK, Duda DG. An orthotopic mouse model of hepatocellular carcinoma with underlying liver cirrhosis. Nat Protoc. 2015;10:1264–1274. doi: 10.1038/nprot.2015.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner M, Koester H, Deffge C, Weinert S, Lauf J, Francke A, Lee J, Braun-Dullaeus R, Herold J. Isolation and intravenous injection of murine bone marrow derived monocytes. J Vis Exp. 2014:52347. doi: 10.3791/52347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen BJ, Chapuy B, Ouyang J, Sun HH, Roemer MG, Xu ML, Yu H, Fletcher CD, Freeman GJ, Shipp MA, Rodig SJ. PD-L1 expression is characteristic of a subset of aggressive B-cell lymphomas and virus-associated malignancies. Clin Cancer Res. 2013;19:3462–3473. doi: 10.1158/1078-0432.CCR-13-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen M, Wang J, Yu W, Zhang C, Liu M, Wang K, Yang L, Wei F, Wang SE, Sun Q, Ren X. A novel MDSC-induced PD-1-PD-L1+ B-cell subset in breast tumor microenvironment possesses immuno-suppressive properties. Oncoimmunology. 2018;7:e1413520. doi: 10.1080/2162402X.2017.1413520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu H, Xia L, Jia D, Zou H, Jin G, Qian W, Xu H, Li T. PD-L1+ regulatory B cells act as a T cell suppressor in a PD-L1-dependent manner in melanoma patients with bone metastasis. Mol Immunol. 2020;119:83–91. doi: 10.1016/j.molimm.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 29.Boldison J, Da Rosa LC, Davies J, Wen L, Wong FS. Dendritic cells license regulatory B cells to produce IL-10 and mediate suppression of antigen-specific CD8 T cells. Cell Mol Immunol. 2020;17:843–855. doi: 10.1038/s41423-019-0324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menon M, Blair PA, Isenberg DA, Mauri C. A regulatory feedback between plasmacytoid dendritic cells and regulatory B cells is aberrant in systemic lupus erythematosus. Immunity. 2016;44:683–697. doi: 10.1016/j.immuni.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mion F, D’Incà F, Danelli L, Toffoletto B, Guarnotta C, Frossi B, Burocchi A, Rigoni A, Gerdes N, Lutgens E, Tripodo C, Colombo MP, Rivera J, Vitale G, Pucillo CE. Mast cells control the expansion and differentiation of IL-10-competent B cells. J Immunol. 2014;193:4568–4579. doi: 10.4049/jimmunol.1302593. [DOI] [PubMed] [Google Scholar]
- 32.Kim HS, Lee MB, Lee D, Min KY, Koo J, Kim HW, Park YH, Kim SJ, Ikutani M, Takaki S, Kim YM, Choi WS. The regulatory B cellmediated peripheral tolerance maintained by mast cell IL-5 suppresses oxazolone-induced contact hypersensitivity. Sci Adv. 2019;5:eaav8152. doi: 10.1126/sciadv.aav8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sánchez-Martín L, Estecha A, Samaniego R, Sánchez-Ramón S, Vega MÁ, Sánchez-Mateos P. The chemokine CXCL12 regulates monocyte-macrophage differentiation and RUNX3 expression. Blood. 2011;117:88–97. doi: 10.1182/blood-2009-12-258186. [DOI] [PubMed] [Google Scholar]
- 34.Kryczek I, Wei S, Keller E, Liu R, Zou W. Stroma-derived factor (SDF-1/CXCL12) and human tumor pathogenesis. Am J Physiol Cell Physiol. 2007;292:C987–C995. doi: 10.1152/ajpcell.00406.2006. [DOI] [PubMed] [Google Scholar]
- 35.Jin F, Brockmeier U, Otterbach F, Metzen E. New insight into the SDF-1/CXCR4 axis in a breast carcinoma model: hypoxia-induced endothelial SDF-1 and tumor cell CXCR4 are required for tumor cell intravasation. Mol Cancer Res. 2012;10:1021–1031. doi: 10.1158/1541-7786.MCR-11-0498. [DOI] [PubMed] [Google Scholar]
- 36.Guo F, Wang Y, Liu J, Mok SC, Xue F, Zhang W. CXCL12/CXCR4: a symbiotic bridge linking cancer cells and their stromal neighbors in oncogenic communication networks. Oncogene. 2016;35:816–826. doi: 10.1038/onc.2015.139. [DOI] [PubMed] [Google Scholar]
- 37.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 38.Zeng Y, Li B, Liang Y, Reeves PM, Qu X, Ran C, Liu Q, Callahan MV, Sluder AE, Gelfand JA, Chen H, Poznansky MC. Dual blockade of CXCL12-CXCR4 and PD-1-PD-L1 pathways prolongs survival of ovarian tumor-bearing mice by prevention of immunosuppression in the tumor microenvironment. FASEB J. 2019;33:6596–6608. doi: 10.1096/fj.201802067RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Semaan A, Dietrich D, Bergheim D, Dietrich J, Kalff JC, Branchi V, Matthaei H, Kristiansen G, Fischer HP, Goltz D. CXCL12 expression and PD-L1 expression serve as prognostic biomarkers in HCC and are induced by hypoxia. Virchows Arch. 2017;470:185–196. doi: 10.1007/s00428-016-2051-5. [DOI] [PubMed] [Google Scholar]
- 40.Feig C, Jones JO, Kraman M, Wells RJ, Deonarine A, Chan DS, Connell CM, Roberts EW, Zhao Q, Caballero OL, Teichmann SA, Janowitz T, Jodrell DI, Tuveson DA, Fearon DT. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110:20212–20217. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Y, Yang J, Cai Y, Fu S, Zhang N, Fu X, Li L. IFN-γ-mediated inhibition of lung cancer correlates with PD-L1 expression and is regulated by PI3K-AKT signaling. Int J Cancer. 2018;143:931–943. doi: 10.1002/ijc.31357. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Zeng Y, Qu Q, Zhu J, Liu Z, Ning W, Zeng H, Zhang N, Du W, Chen C, Huang JA. PD-L1 induced by IFN-γ from tumor-associated macrophages via the JAK/STAT3 and PI3K/AKT signaling pathways promoted progression of lung cancer. Int J Clin Oncol. 2017;22:1026–1033. doi: 10.1007/s10147-017-1161-7. [DOI] [PubMed] [Google Scholar]
- 43.Lastwika KJ, Wilson W 3rd, Li QK, Norris J, Xu H, Ghazarian SR, Kitagawa H, Kawabata S, Taube JM, Yao S, Liu LN, Gills JJ, Dennis PA. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. 2016;76:227–238. doi: 10.1158/0008-5472.CAN-14-3362. [DOI] [PubMed] [Google Scholar]
- 44.Green MR, Rodig S, Juszczynski P, Ouyang J, Sinha P, O’Donnell E, Neuberg D, Shipp MA. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res. 2012;18:1611–1618. doi: 10.1158/1078-0432.CCR-11-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai TF, Lin JF, Lin YC, Chou KY, Chen HE, Ho CY, Chen PC, Hwang TI. Cisplatin contributes to programmed death-ligand 1 expression in bladder cancer through ERK1/2-AP-1 signaling pathway. Biosci Rep. 2019;39:BSR20190362. doi: 10.1042/BSR20190362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan H, Hall P, Santos LL, Gregory JL, Fingerle-Rowson G, Bucala R, Morand EF, Hickey MJ. Macrophage migration inhibitory factor and CD74 regulate macrophage chemotactic responses via MAPK and Rho GTPase. J Immunol. 2011;186:4915–4924. doi: 10.4049/jimmunol.1003713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi K, Koga K, Linge HM, Zhang Y, Lin X, Metz CN, Al-Abed Y, Ojamaa K, Miller EJ. Macrophage CD74 contributes to MIF-induced pulmonary inflammation. Respir Res. 2009;10:33. doi: 10.1186/1465-9921-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregory JL, Morand EF, McKeown SJ, Ralph JA, Hall P, Yang YH, McColl SR, Hickey MJ. Macrophage migration inhibitory factor induces macrophage recruitment via CC chemokine ligand 2. J Immunol. 2006;177:8072–8079. doi: 10.4049/jimmunol.177.11.8072. [DOI] [PubMed] [Google Scholar]
- 49.Strickland J, Garrison D, Copple BL. Hypoxia upregulates Cxcl12 in hepatocytes by a complex mechanism involving hypoxia-inducible factors and transforming growth factor-β. Cytokine. 2020;127:154986. doi: 10.1016/j.cyto.2020.154986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Z, Gao Z, Xia L, Wang X, Lu L, Wu X. Dysregulation of DPP4-CXCL12 balance by TGF-β1/SMAD pathway promotes CXCR4+ inflammatory cell infiltration in keloid scars. J Inflamm Res. 2021;14:4169–4180. doi: 10.2147/JIR.S326385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu PF, Huang Y, Xu CL, Lin LY, Han YY, Sun WH, Hu GH, Rabson AB, Wang Y, Shi YF. Downregulation of CXCL12 in mesenchymal stromal cells by TGFβ promotes breast cancer metastasis. Oncogene. 2017;36:840–849. doi: 10.1038/onc.2016.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.