Abstract
The stability of the p53 tumor suppressor protein is regulated by interaction with Mdm2, the product of a p53-inducible gene. Mdm2-targeted degradation of p53 depends on the interaction between the two proteins and is mediated by the proteasome. We show here that in addition to the N-terminal Mdm2 binding domain, the C terminus of p53 participates in the ability of p53 to be degraded by Mdm2. In contrast, alterations in the central DNA binding domain of p53, which change the conformation of the p53 protein, do not abrogate the sensitivity of the protein to Mdm2-mediated degradation. The importance of the C-terminal oligomerization domain to Mdm2-targeted degradation of p53 is likely to reflect the importance of oligomerization of the full-length p53 protein for interaction with Mdm2, as previously shown in vitro. Interestingly, the extreme C-terminal region of p53, outside the oligomerization domain, was also shown to be necessary for efficient degradation, and deletion of this region stabilized the protein without abrogating its ability to bind to Mdm2. Mdm2-resistant p53 mutants were not further stabilized following DNA damage, supporting a role for Mdm2 as the principal regulator of p53 stability in cells. The extreme C terminus of the p53 protein has previously been shown to contain several regulatory elements, raising the possibility that either allosteric regulation of p53 by this domain or interaction between this region and a third protein plays a role in determining the sensitivity of p53 to Mdm2-directed degradation.
The p53 tumor suppressor gene product plays an important role in the prevention of malignancies, and the function of this protein is lost in most human cancers (18). Mice lacking p53 are viable, although some show evidence of developmental defects (2, 15), indicating that p53 function is not essential for normal cell growth. p53 activity is strongly stimulated in response to genotoxic stress, such as DNA damage, and leads to the inhibition of cell growth, either by institution of a cell cycle arrest or activation of programmed cell death (apoptosis) (5). Either of these responses prevents the replication of cells with damaged DNA, and loss of this protective function is proposed to allow the outgrowth of cells harboring potentially oncogenic mutations (34). Many activities have been ascribed to p53, the most clearly understood being the ability of p53 to function as a transcription factor (59). Many p53-inducible cell genes have been described, and it is clear that activated cell cycle arrest genes (such as the cyclin-dependent kinase inhibitor p21Waf1/Cip1) or apoptotic genes (such as Bax) are important as mediators of some of the functions of p53 (7, 14, 43, 61, 63). Nevertheless, there is substantial evidence for transcriptionally independent activities of p53, particularly in the activation of the apoptotic response (8, 22, 60).
The p53 protein is maintained in normal cells as an unstable protein at very low levels, and activation of a p53 response leads to rapid accumulation of the p53 protein through posttranscriptional mechanisms (16, 30, 37). Increased levels of p53 are thought to result principally from a dramatic increase in the half-life of the protein, although enhanced rates of protein synthesis also play a role (17). The p53 protein has been shown to be degraded through ubiquitin-dependent proteolysis (36), and recent studies have shown that interaction with the Mdm2 protein can target p53 for degradation (6, 21, 32). Mdm2 is itself a transcriptional target of p53 and binds to a domain in the N terminus of the p53 protein (4, 10, 45, 47). Simple binding of Mdm2 to p53 can inhibit the transcriptional activity and G1 arrest function of p53 (9), probably by obscuring the very closely linked trans-activation domain (47), but there is evidence that the interaction between Mdm2 and p53 is not sufficient to inhibit the transcriptionally independent apoptotic functions of p53 (20). Degradation of p53 targeted by Mdm2 would abrogate all p53 functions, and the transcriptional activation of Mdm2 by p53 provides a regulatory loop in normal cells to prevent activation of a p53 response (62). The importance of Mdm2 in regulating p53 function during normal growth and development is illustrated by the observation that deletion of Mdm2 in mice results in very early embryonic lethality, which is rescued by simultaneous deletion of p53 (29, 46). The stabilization of p53 in response to DNA damage indicates that mechanisms must exist by which p53 can become resistant to degradation by Mdm2. Recent studies have suggested that phosphorylation of either p53 or Mdm2 by DNA-PK can decrease the binding between the two proteins (42, 54). Since binding is necessary for degradation, this provides a mechanism by which p53 may become stabilized despite the presence of Mdm2. However, there is now also evidence that regulation of degradation may also occur through mechanisms other than the inhibition of binding between the two proteins. The p14ARF protein has recently been shown to inhibit Mdm2-mediated degradation of p53 by binding to Mdm2 at a region distinct from the p53 binding site, and interaction with p14ARF does not prevent Mdm2 binding to p53 (49, 55, 64).
The p53 protein contains several well-characterized domains (Fig. 1); the N-terminal trans-activation domain, a proline-rich domain, a central sequence-specific DNA binding domain and the C-terminal region which contains the oligomerization domain, nuclear localization signals, and single-stranded or damaged DNA binding activity (31). The extreme C terminus of the protein has been shown to regulate sequence-specific DNA binding, and the full-length protein is maintained in a non-DNA binding latent form in vitro. This regulation of DNA binding has been attributed to an allosteric mechanism, in which the C terminus of p53 directly binds and occludes the central DNA binding domain (25, 27), although interaction of the C terminus with DNA can also inhibit specific DNA binding (1). The latent form of p53 can be activated to bind DNA by several mechanisms, including modification of the C terminus by phosphorylation (26), glycosylation (53), acetylation (19), mutation (26, 41), or interaction with single-stranded DNA (28). The exact contribution of these mechanisms to the regulation of p53 in vivo is not yet clear, but there is evidence that activation of p53 function and stabilization of the protein are separable steps in the initiation of a p53 response (27, 35, 57).
FIG. 1.
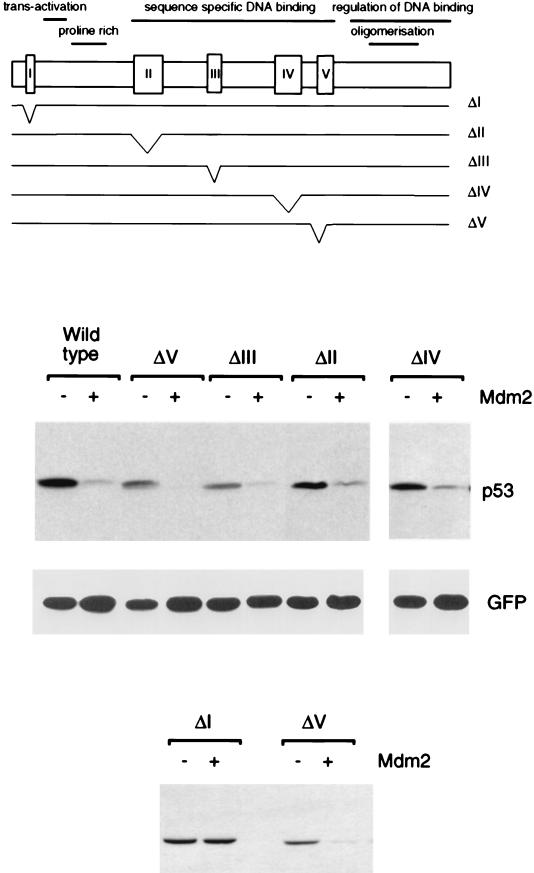
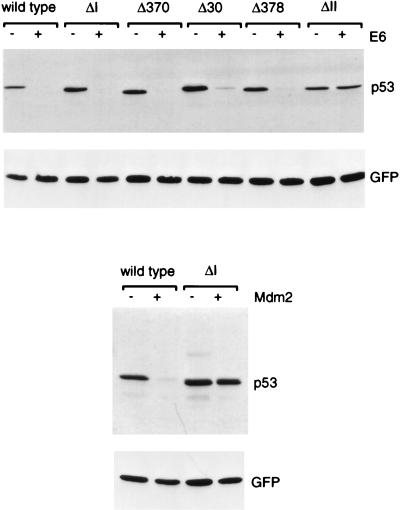
Degradation of p53 deletion mutants by Mdm2. The position of each mutation in full-length p53 is shown. Levels of each p53 protein following transient transfection into Saos-2 cells in the presence or absence of exogenous Mdm2, as determined by Western blotting, are shown below. Equal transfection efficiency is demonstrated by expression of cotransfected green fluorescent protein.
We previously showed that mutational alteration of the N-terminal Mdm2 binding region of p53 rendered the protein resistant to degradation by Mdm2 (32), indicating that this activity of Mdm2 is dependent on an interaction with p53. In this study we examine the contribution of other regions of the p53 protein to sensitivity to Mdm2-targeted degradation.
MATERIALS AND METHODS
Plasmids and antibodies.
Plasmids encoding wild-type and mutant p53 under the control of the cytomegalovirus promoter have been reported previously (11, 39–41). Plasmids encoding for mouse wild-type Mdm2 (pCOC Mdm2 X2) (20), the human mutant Mdm2Δ222-437 (pCHDMΔ222-437) (9), and HPV16 E6 (11) have also been described previously. The p53 ΔII-Δ370 plasmid was constructed by replacing a StuI/BamHI fragment in p53ΔII (39) with the corresponding fragment from p53Δ370. p53ΔI-ΔII has been described previously (3). The GFP expression plasmid pEGFP-N1 was purchased from Clontech (Palo Alto, Calif.).
p53-specific monoclonal antibodies PAb1801 and DO-1 and the Mdm2-specific antibody IF2 were purchased from Oncogene Science (Cambridge, Mass.). The polyclonal rabbit serum CM-1 was from Novocastra (Burlingham, Calif.). Anti-GFP monoclonal antibodies were purchased from Clontech, the anti-actin antibody was purchased from Chemicon (Temecula, Calif.), and the fluorescein isothiocyanate-conjugated rabbit anti-mouse antibody was obtained from Dako (Carpenteria, Calif.).
Cells and transfections.
p53 null Saos-2 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and transiently transfected by the calcium phosphate precipitation method. Unless otherwise indicated, 3 μg of wild-type p53 or mutant p53 plasmid was cotransfected with 9 μg of wild-type mouse Mdm2 encoding plasmid or HPV16 E6 encoding plasmid per 100-mm diameter dish, and cells were harvested 24 h after transfection. For association assays, p53 was cotransfected with the binding competent, degradation-deficient human Mdm2 mutant Δ222-437 (32). In all cases, transfection efficiency was monitored by cotransfection of 1 μg of pEGFP-N1, and equal expression of GFP was verified by Western blotting. To generate MCF-7 cell lines stably expressing p53 mutant proteins, cells were selected after transfection with G418 for 3 weeks. Total cell lysates of pooled transfected cells were analyzed for p53 expression. Where indicated, the cells were first treated with 5 nM actinomycin-D for 16 h.
Protein analysis.
Western blotting and immunoprecipitation were carried out as previously described (40). To analyze association between p53 and Mdm2, transiently transfected cells were divided for Western blotting to determine total p53 expression and immunoprecipitated with the anti-Mdm2 antibody IF2. Immunoprecipitated proteins were then Western blotted, and coprecipitated p53 was detected with rabbit polyclonal antiserum CM-1.
The half-life of p53 protein in transfected cells was determined by radioactive pulse labeling of cells for 30 min and chase in unlabeled medium for 0, 0.5, and 4.5 h. The p53 protein was then immunoprecipitated with the monoclonal antibody PAb1801. Quantification of labeled p53 protein was carried out with the STORM PhosphorImager model 860 (Molecular Dynamics).
For immunofluorescence studies, Saos-2 cells were seeded on coverslips, transiently transfected, and fixed 24 h after transfection in ice-cold methanol. The p53 protein was detected with antibody PAb1801 and a fluorescein isothiocyanate-conjugated secondary antibody. Subcellular localization of p53 was analyzed by confocal microscopy.
RESULTS
Contribution of the p53 conserved regions to degradation by Mdm2.
We utilized a series of previously described p53 deletion mutants (39) to analyze the contribution of the conserved domains of p53 to sensitivity to degradation by Mdm2 (Fig. 1). p53ΔI carries a deletion of the first conserved box in p53 and is unable to bind Mdm2, although it retains wild-type transcriptional activity. p53ΔII, III, IV, and V carry deletions of conserved boxes II to V and fail to bind DNA. These mutants have lost p53 transcriptional activity (39) and adopt a conformation associated with tumor-derived p53 mutants (40). Each of the p53 mutants was transfected in Saos-2 cells, a p53 null human cell line, with or without wild-type Mdm2 (Fig. 1). As shown previously (32), p53ΔI, which fails to bind Mdm2, is resistant to Mdm2-targeted degradation. In contrast, p53 proteins with deletion of conserved regions II to V remained sensitive to degradation by Mdm2, despite loss of function and wild-type conformation. These results are consistent with our previous observations that tumor-derived point mutants within the DNA binding domain also remain sensitive to Mdm2-targeted degradation (32).
Contribution of p53 oligomerization to degradation by Mdm2.
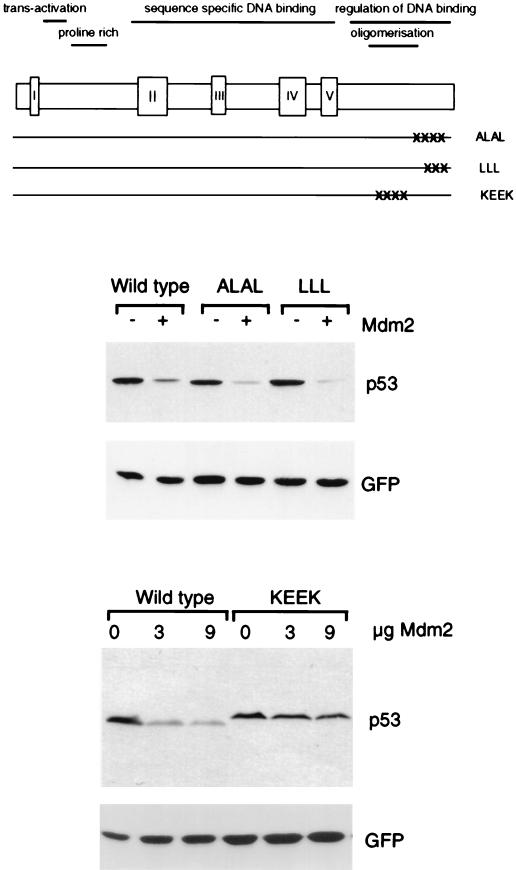
Previous in vitro studies pointed to a contribution of oligomerization of p53 to Mdm2 binding in the context of the full-length p53 protein (40), although the binding site for Mdm2 has been shown to reside within the N terminus of p53 (10, 48). We therefore examined the sensitivity of oligomerization-defective p53 mutants to degradation by Mdm2 after coexpression in Saos-2 cells. These included two-point mutants which have been described as forming dimers, but not tetramers (p53ALAL and p53LLL), and a quadruple-point mutant which is maintained in the monomeric form (p53KEEK) (Fig. 2) (56, 58). The two dimerization-competent mutants both showed sensitivity to degradation by Mdm2 at levels comparable to the wild-type protein (Fig. 2), consistent with observations that these p53 mutants retain the ability to interact with Mdm2 in vitro (40). The oligomerization-defective mutant, however, showed clear resistance to Mdm2-mediated degradation (Fig. 2), although a small reduction in p53 levels was seen with increasing Mdm2, suggesting that oligomerization was not absolutely necessary for sensitivity. Similarly, a large C-terminal deletion which removed the oligomerization domain (p53Δ327) also rendered the protein resistant to Mdm2-targeted degradation (data not shown).
FIG. 2.
Degradation of p53 oligomerization mutants by Mdm2. The position of each mutation in full-length p53 is shown. Levels of each p53 protein following transient transfection into Saos-2 cells in the presence or absence of exogenous Mdm2, as determined by Western blotting, are shown below. Equal transfection efficiency is demonstrated by expression of cotransfected green fluorescent protein.
Contribution of the C terminus of p53 to degradation by Mdm2.
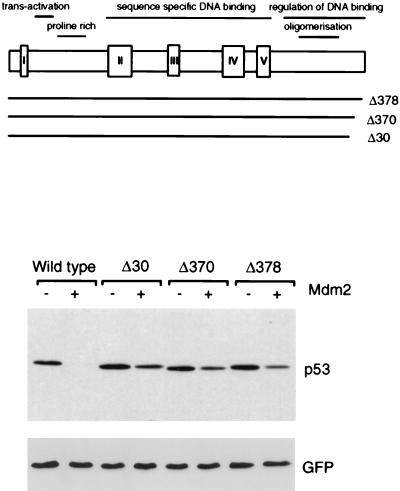
The extreme C terminus of the p53 protein has been shown to play an important role in the regulation of p53 activity, particularly in the maintenance of the protein in a latent, non-DNA binding conformation. We therefore analyzed a series of C-terminal truncation mutants of p53 for their sensitivity to degradation by Mdm2 (Fig. 3). Deletion of the C-terminal 30 or 24 amino acids in p53Δ30 and p53Δ370 has been shown to constitutively activate p53 DNA binding function, although this is not achieved following deletion of the last 16 amino acids in p53Δ378 (41). All of these C-terminal deletion mutants were significantly less-well degraded than the wild-type protein, although comparison with p53ΔI, which is entirely resistant to degradation, showed that each of the C-terminal mutants retained some sensitivity to Mdm2. This sensitivity to Mdm2 was most marked in the least-extensive deletion, p53Δ378 (Fig. 3).
FIG. 3.
Degradation of p53 C-terminal truncation mutants by Mdm2. The position of each mutation in full-length p53 is shown. Levels of each p53 protein following transient transfection into Saos-2 cells in the presence or absence of exogenous Mdm2, as determined by Western blotting, are shown below. Equal transfection efficiency is demonstrated by expression of cotransfected green fluorescent protein.
The resistance of extreme C-terminal p53 mutants to Mdm2-mediated degradation was intriguing, and we examined the contribution of the C-terminal lysine residues, which represent potential targets for ubiquitination. Analysis of p53I381/382/386 (in which the last three lysine residues were mutated to isoleucine) and p53I370/372/373 (in which the penultimate three lysine residues were substituted by isoleucine) showed that neither of these mutants displayed enhanced resistance to Mdm2-mediated degradation (data not shown).
Deletion of the C terminus constitutively stabilizes p53 in the absence of DNA damage.
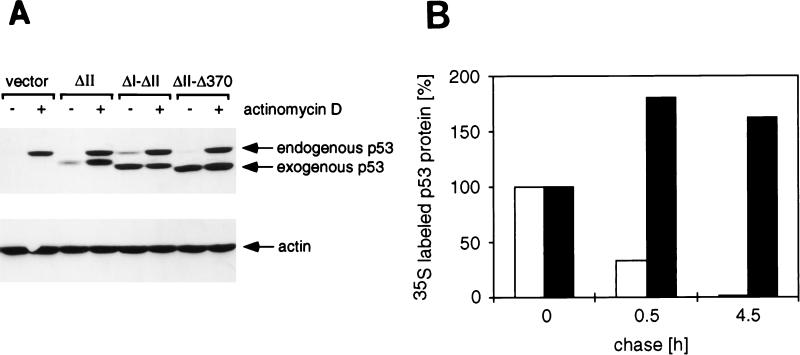
We have identified p53 mutants which are resistant to Mdm2-mediated degradation either due to loss of Mdm2 binding (such as p53ΔI) or through mechanisms other than loss of binding (such as p53Δ370). In order to confirm that these mutants exhibit a longer half-life in cells where p53 stability is normally regulated by endogenous p53, we turned to MCF-7 cells, a human breast carcinoma cell line that expresses low levels of wild-type p53. Expression of exogenous p53 with wild-type activities in these cells leads to cell cycle arrest, and so it is not possible to establish stably expressing cells with these mutants. We therefore made use of our observation that deletion of the conserved regions in the DNA binding domain of p53, which render p53 unable to inhibit cell growth (12), did not prevent sensitivity to Mdm2-mediated degradation. We generated p53 mutants containing the N- and C-terminal deletions in the context of the box II (ΔII) deletion, and MCF-7 lines stably expressing each of the exogenous p53 mutants were generated (Fig. 4). This system to study p53 stability has been previously described (44), making use of a point mutant to inactivate p53 growth-suppressive function. In this analysis the exogenous and endogenous proteins can be distinguished by their sizes. The p53ΔII mutant is expressed at slightly higher levels than the endogenous protein, which is not detectable in the absence of DNA damage (Fig. 4A), probably because the exogenous protein is expressed from the cytomegalovirus promoter rather than the endogenous p53 promoter. In contrast, deletion of the Mdm2 binding region in p53ΔI-ΔII or deletion of the C terminus in p53ΔII-Δ370 resulted in a protein with much-higher steady-state expression levels in these cells (Fig. 4A). Similar results with the Δ30 deletion have been previously published (44). The endogenous p53 protein in these cells can be stabilized by DNA damage and is clearly detected after 16 h of treatment with actinomycin-D. Similarly, the exogenous p53ΔII protein levels increase following treatment. However, the already high levels of p53ΔI-ΔII and p53ΔII-Δ370 are not detectably increased following DNA damage. In order to confirm that the elevated protein levels seen with p53ΔII-Δ370 in undamaged cells are due to increased stability, as was previously shown for p53ΔI (32), the half-lives of p53ΔII and p53ΔII-Δ370 were assessed by radioactive pulse-labeling of the cells (Fig. 4B). These results show that while the half-life of p53ΔII is around 30 min, consistent with the half-life of the endogenous wild-type p53 (35), the half-life of the p53ΔII-Δ370 mutant is in excess of 4.5 h.
FIG. 4.
Stable expression of p53 mutants in MCF-7 cells. (A) Western blot analysis of MCF-7 cells transfected with vector alone, p53ΔII, p53ΔI-ΔII, or p53ΔII-Δ370 as indicated. Cells were either untreated or treated with 5 nM actinomycin-D for 16 h to induce a DNA damage response and stabilize p53. The migration of the endogenous p53 protein and the exogenous mutant p53 proteins is indicated. Equal protein loading is demonstrated by actin expression. (B) Stability of the p53ΔII (white bars) and p53ΔII-Δ370 (black bars) proteins in MCF-7 cells as determined by metabolic pulse-chase labeling. The amount of labeled immunoprecipitated p53 protein remaining at 0.5 and 4.5 h postlabeling was quantified and expressed as a percentage of labeled protein present at the start of the chase.
Degradation of p53 by HPV16 E6.
The p53 protein is also a target for degradation mediated by interaction with the E6 protein encoded by the high-risk genital human papillomaviruses (13, 52). In vitro analyses have shown that deletion of conserved region I or C-terminal truncation of p53 does not impair degradation by E6, although sensitivity to E6 is dependent on p53 protein conformation (38–40). Since the C-terminal region appeared to be important for degradation of p53 by Mdm2 in vivo, we carried out similar analyses of the sensitivity of several p53 mutants to E6 following coexpression in cells (Fig. 5). These analyses confirmed the in vitro results, showing that the N- and C-terminal deletion mutants retain wild-type sensitivity to E6-mediated degradation, while deletion of conserved box V renders p53 resistant to E6.
FIG. 5.
Degradation of p53 deletion and truncation mutants by HPV16 E6. The position of each mutation in full-length p53 is shown in Fig. 1 and 2. Levels of each p53 protein in the presence or absence of HPV16 E6 as determined by Western blotting are shown. Levels of wild-type p53 and p53ΔI in the presence and absence of exogenous Mdm2 from the same experiment are shown for comparison.
Interaction between p53 mutants and Mdm2 in cells.
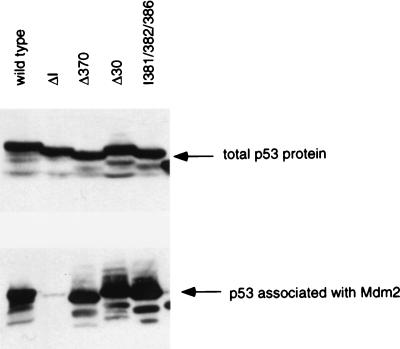
Analysis of the interaction of these p53 mutants with Mdm2 has been predominantly carried out in vitro, and we were concerned that the resistance of some of the C-terminal p53 truncation mutants might reflect a defect in in vivo binding not apparent in the in vitro assays. We therefore carried out coprecipitations of Mdm2 and p53 from cells cotransfected with the p53 mutants and an Mdm2 mutant which retains the ability to bind p53 but cannot mediate its degradation (32) (Fig. 6). The Mdm2 mutant was used to allow stable interaction of all p53 proteins without loss of association due to degradation. Western blot analysis confirmed equal expression of each p53 protein, and coprecipitation through the Mdm2 mutant showed that each of the C-terminal truncations (p53Δ370 and p53Δ30) retained wild-type ability to interact with Mdm2. As expected, the N-terminal deletion p53ΔI failed to bind Mdm2. Interestingly, a more extensive C-terminal deletion which is unable to oligomerize (p53Δ327) retained some binding activity, although this was clearly reduced in comparison to that of the wild-type protein (data not shown), supporting the suggestion that oligomerization contributes to, but is not absolutely essential for, the interaction of full-length p53 with Mdm2.
FIG. 6.
Interaction between p53 mutants and Mdm2 in vivo. The indicated p53 mutants were cotransfected with a human Mdm2 mutant (Δ222-437) which retains the ability to interact with p53 but fails to mediate its degradation. Approximately equal levels of expression of each p53 protein were determined by Western blotting (above). Binding of p53 to Mdm2 was determined by immunoprecipitation of the Mdm2 protein followed by Western blot analysis of the coprecipitated p53 (below).
Nuclear localization of p53 mutants.
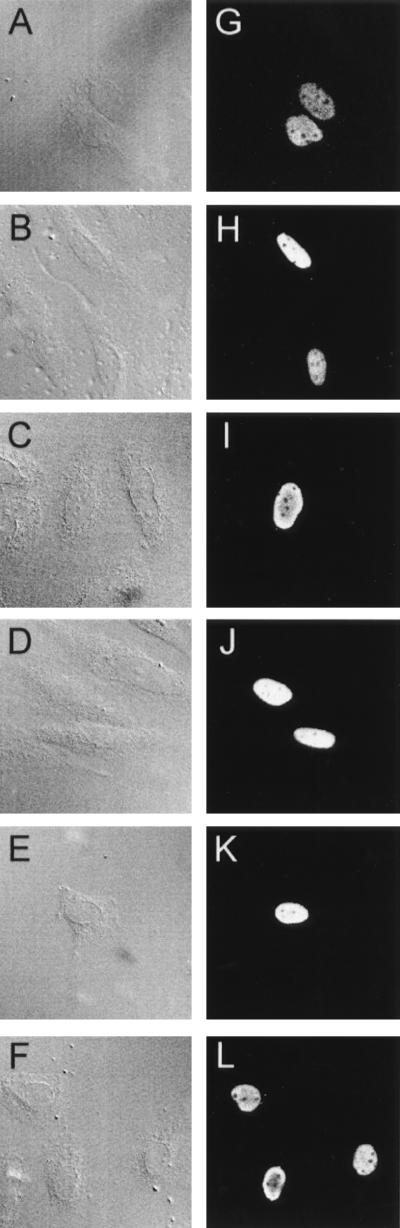
Although the C-terminal p53 deletion mutants clearly retain the ability to interact with Mdm2, both in vitro and in cell extracts, the possibility remained that degradation was not occurring in cells because the p53 mutants were unable to efficiently localize to the nucleus and were therefore unable to interact with Mdm2 in intact cells. All of the C-terminal truncation mutants retain wild-type transcriptional activation activity (41), making it unlikely that they are completely excluded from the nucleus. Immunofluorescence studies of transfected Saos-2 cells confirmed that each of the mutants studied here showed almost exclusive nuclear localization, indistinguishable from that seen with wild-type p53 (Fig. 7). Similar nuclear localization was seen for p53ΔII, p53ΔI-ΔII, and p53ΔII-Δ370 stably expressed in MCF-7 cells as described above (data not shown).
FIG. 7.
Nuclear localization of p53 proteins following transient transfection into Saos-2 cells. Normarski/DIC images (A, B, C, D, E, and F) and immunofluorescent analysis of p53 in transfected cells (G, H, I, J, K, and L) show nuclear localization of wild-type p53 (A and G), p53ΔI (B and H), p53KEEK (C and I), p53Δ30 (D and J), p53Δ370 (E and K), and p53Δ378 (F and L).
DISCUSSION
The ability of Mdm2 to target p53 for degradation likely represents a key mechanism controlling the activity of p53 during cell growth. In this study we have analyzed the regions of p53 which are necessary for sensitivity to Mdm2-mediated degradation and found a contribution of both N- and C-terminal regions of p53. As described previously, interaction between p53 and Mdm2 is necessary for degradation (32), and we found that deletion of either the N-terminal Mdm2 binding domain or the C-terminal oligomerization domain results in loss of sensitivity to degradation, which can be related to defects in binding to Mdm2. These observations support in vitro data suggesting a role for oligomerization of p53 in binding to Mdm2 (40). However, analysis of p53 peptides demonstrates that this activity is not essential for the interaction between the two proteins (48) and most likely only enhances the binding activity of full-length p53. Crystallographic analysis of the p53-Mdm2 interacting domains has shown the N-terminal p53 sequences fitting into a deep hydrophobic cleft formed in Mdm2 (33), and it is not clear how oligomerization of p53 contributes to this interaction. It is possible that the conformation of oligomerized p53 allows access of Mdm2 to the N terminus of the tumor suppressor protein which may be masked in the full-length monomer.
In addition to the requirement for interaction between p53 and Mdm2, our data demonstrated a dissociation between binding and degradation. Mutants of p53 lacking extreme C-terminal sequences show resistance to Mdm2-mediated degradation despite oligomerizing and binding to Mdm2 like wild-type p53. This resistance to Mdm2-mediated degradation is reflected in an extended half-life of a p53 mutant lacking this C-terminal region in stably expressing MCF-7 cells, similar to that seen when the Mdm2 binding site itself is deleted. Interestingly, these constitutively stable mutants could not be further stabilized following DNA damage (Fig. 4A), strongly suggesting that inhibition of the Mdm2-mediated degradation pathway is the principal mechanism by which p53 stability is regulated. Deletion of the C terminus of p53 has previously been shown to result in the activation of constitutive DNA binding activity, probably locking the protein into a DNA binding conformation. However, analysis of several C-terminal mutants suggested that the sensitivity to Mdm2-mediated degradation did not show a simple correlation with the maintenance of the p53 protein in a non-DNA binding state. The smallest C-terminal truncation mutant, p53Δ378, is not constitutively activated for DNA binding (41), but shows enhanced resistance to Mdm2-targeted degradation (although this is less dramatic than that seen with slightly larger C-terminal deletions such as p53Δ370 and p53Δ30). Conversely, the point mutants p53ALAL and p53 I370/372/373 both show some degree of constitutive DNA binding activity (41) but remain sensitive to Mdm2-mediated degradation. These two-point mutants are clearly less efficiently activated for DNA binding than deletion mutants such as p53Δ370 (41), and it remains possible that subtle differences in conformation of these various proteins contribute to their relative sensitivities to Mdm2.
In targeting p53 for ubiquitin-dependent degradation, Mdm2 shows striking functional similarity to the E6 protein encoded by the high-risk genital human papillomavirus types, although there is no clear structural similarity between Mdm2 and E6. However, analysis of the interaction between p53 mutant proteins and E6 or Mdm2 has illustrated a virtual complete discordance between regions of p53 important for binding to the viral and cellular proteins (39, 40). The interaction of p53 with E6 is dependent on maintenance of the wild-type conformation of the p53 protein, mutations within the central DNA binding region resulting in loss of binding and resistance of p53 to E6-mediated degradation. Alterations in the N or C terminus of p53 fail to impede binding or degradation by E6, and monomeric p53 is a target for E6-mediated degradation. By contrast, we show here that degradation by Mdm2 is not prevented by deletions or alterations within the central DNA binding domain of p53, but both N- and C-terminal alterations in the p53 protein render it resistant to Mdm2-mediated degradation. Despite these differences, there may be similarities between the mechanisms of Mdm2- and E6-mediated degradation. Of particular interest is the observation that E6-targeted degradation of p53 involves a third protein, E6-AP (24). The E6-E6-AP complex functions as a ubiquitin ligase (51), forming a trimeric complex with p53 and conjugating ubiquitin to the p53 protein, a prerequisite for targeting to and degradation through the proteasome.
Recently it has been shown that Mdm2 can function as a ubiquitin ligase (23) in vitro. However, efficient degradation of p53 by Mdm2 in vitro could not be detected in conditions under which p53 is degraded by E6 (data not shown), indicating that one or more components of the p53/Mdm2 degradation pathway is missing in this assay. We would like to suggest that a third protein is involved in the Mdm2-targeted degradation of p53. Analysis of Mdm2 has shown that regions in the central and C-terminal part of the protein are important for the degradation of p53, although they do not contribute to binding (32; data not shown). Therefore, as shown here for p53, the binding and degradation activities of Mdm2 are separable. We propose that a third protein necessary for degradation forms a trimeric complex with Mdm2 and p53, contacting domains in both Mdm2 and p53. Deletion of the binding domain in p53, such as in the p53Δ370 and p53Δ30 mutants, results in a much-reduced degradation rate. Many cellular proteins have been reported to bind to the C terminus of p53, including general transcription factors such as TBP and TFIIH and replication and repair proteins such as RPA, XPB, XPD, and CSB (31). Whether these or other proteins can contribute to the degradation of p53 by Mdm2 is currently under investigation.
Another possible explanation for the resistance of the C-terminal p53 mutants is that they impair normal nucleocytoplasmic shuttling of the p53-Mdm2 complex. Mutations of the nuclear export signal in Mdm2 have been shown to impede degradation of p53, suggesting that Mdm2 shuttles p53 to the cytoplasm for degradation (50). Although we have been unable to see difference in the subcellular localization of wild-type and mutant p53 proteins in either transiently transfected cells or stable expression in MCF-7 cells, it is possible that deleting the C terminus of p53 prevents export from the nucleus and so prevents degradation, despite the maintenance of the interaction with Mdm2. This would imply that p53 sequences, as well as the Mdm2 nuclear export sequence, contribute to the subcellular localization of the p53-Mdm2 complex.
The data described here are entirely consistent with a recent study examining the relative stability of mutant p53 proteins expressed in MCF-7 cells (44). Although functionally inactive p53 mutants were maintained at low levels in these cells, alterations in either the N terminus or the C terminus resulted in elevated p53 protein expression. Our results indicate that the stability of these p53 mutants is the consequence of loss of sensitivity to Mdm2-mediated degradation.
ACKNOWLEDGMENTS
We are extremely grateful to Moshe Oren and Arnold Levine for the Mdm2 plasmids and to Stewart Bates for reading the manuscript. We also thank Jim Resau for help with the confocal microscopy.
This work was supported by the National Cancer Institute under contract with ABL.
REFERENCES
- 1.Anderson M E, Woelker B, Reed M, Wang P, Tegtmeyer P. Reciprocal interference between the sequence-specific core and nonspecific C-terminal DNA binding domains of p53: implications for regulation. Mol Cell Biol. 1997;17:6255–6264. doi: 10.1128/mcb.17.11.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong J F, Kaufman M H, Harrison D J, Clarke A R. High-frequency developmental abnormalities in p53-deficient mice. Curr Biol. 1995;5:931–936. doi: 10.1016/s0960-9822(95)00183-7. [DOI] [PubMed] [Google Scholar]
- 3.Ashcroft, M., and K. H. Vousden. Regulation of p53 function and stability by phosphorylation. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 4.Barak Y, Juven T, Haffner R, Oren M. mdm-2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates S, Vousden K H. p53 in signalling checkpoint arrest or apoptosis. Curr Opin Genet Dev. 1996;6:1–7. doi: 10.1016/s0959-437x(96)90004-0. [DOI] [PubMed] [Google Scholar]
- 6.Böttger A, Böttger V, Sparks A, W.-L. L, Howard S F, Lane D P. Design of a synthetic Mdm-2 binding mini protein that activates the p53 response in vivo. Curr Biol. 1997;7:860–869. doi: 10.1016/s0960-9822(06)00374-5. [DOI] [PubMed] [Google Scholar]
- 7.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–556. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 8.Caelles C, Helmberg A, Karin M. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature. 1994;370:220–223. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Wu X, Lin J, Levine A J. mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol Cell Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J D, Marechal V, Levine A J. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crook T, Ludwig R L, Marston N J, Willkomm D, Vousden K H. Sensitivity of p53 lysine mutants to ubiquitin-directed degradation targeted by human papillomavirus E6. Virology. 1996;217:285–292. doi: 10.1006/viro.1996.0115. [DOI] [PubMed] [Google Scholar]
- 12.Crook T, Marston N J, Sara E A, Vousden K H. Transcriptional activation by p53 correlates with suppression of growth but not transformation. Cell. 1994;79:817–827. doi: 10.1016/0092-8674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 13.Crook T, Tidy J A, Vousden K H. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and transactivation. Cell. 1991;67:547–556. doi: 10.1016/0092-8674(91)90529-8. [DOI] [PubMed] [Google Scholar]
- 14.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 15.Donehower L A. The p53-deficient mouse: a model for basic and applied cancer studies. Semin Cancer Biol. 1996;7:269–278. doi: 10.1006/scbi.1996.0035. [DOI] [PubMed] [Google Scholar]
- 16.Fritsche M, Haessler C, Brandner G. Induction of nuclear accumulation of the tumor-suppressor protein p53 by DNA-damaging agents. Oncogene. 1993;8:307–318. [PubMed] [Google Scholar]
- 17.Fu L, Benchimol S. Participation of the human p53 3′UTR in translational repression and activation following gamma-irradiation. EMBO J. 1997;16:4117–4127. doi: 10.1093/emboj/16.13.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 19.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 20.Haupt Y, Barak Y, Oren M. Cell type-specific inhibition of p53-mediated apoptosis by mdm2. EMBO J. 1996;15:1596–1606. [PMC free article] [PubMed] [Google Scholar]
- 21.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 22.Haupt Y, Rowan S, Shaulian E, Vousden K H, Oren M. Induction of apoptosis in HeLa cells by trans-activation deficient p53. Genes Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 23.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 24.Huibregtse J M, Scheffner M, Howley P M. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hupp T R, Lane D P. Regulation of the cryptic sequence-specific DNA-binding function of p53 by protein kinases. Cold Spring Harbor Symp Quant Biol. 1994;59:195–206. doi: 10.1101/sqb.1994.059.01.024. [DOI] [PubMed] [Google Scholar]
- 26.Hupp T R, Meek D W, Midgley C A, Lane D P. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 27.Hupp T R, Sparks A, Lane D P. Small peptides activate the latent sequence-specific DNA binding function of p53. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 28.Jayaraman L, Prives C. Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell. 1995;81:1021–1029. doi: 10.1016/s0092-8674(05)80007-8. [DOI] [PubMed] [Google Scholar]
- 29.Jones S N, Roe A E, Donehower L A, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 30.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 31.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 32.Kubbutat M H G, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 33.Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 34.Lane D P. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 35.Lu X, Burbridge S A, Griffin S, Smith H M. Discordance between accumulated p53 protein levels and its transcriptional activity in response to U.V. radiation. Oncogene. 1997;13:413–418. [PubMed] [Google Scholar]
- 36.Maki C G, Huibregtse J, Howley P M. In vivo ubiquitination and proteosome-mediated degradation of p53. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 37.Maltzman W, Czyzyk L. UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mansur C P, Marcus B, Dalal S, Androphy E J. The domain of p53 required for binding HPV16 E6 is separable from the degradation domain. Oncogene. 1995;10:457–465. [PubMed] [Google Scholar]
- 39.Marston N J, Crook T, Vousden K H. Interaction of p53 with MDM2 is independent of E6 and does not mediate wild type transformation suppressor function. Oncogene. 1994;9:2707–2716. [PubMed] [Google Scholar]
- 40.Marston N J, Jenkins J R, Vousden K H. Oligomerisation of full length p53 contributes to the interaction with mdm2 but not HPV E6. Oncogene. 1995;10:1709–1715. [PubMed] [Google Scholar]
- 41.Marston N J, Ludwig R L, Vousden K H. Activation of p53 DNA binding activity by point mutation. 1998. Oncogene, in press. [DOI] [PubMed] [Google Scholar]
- 42.Mayo L D, Turchi J J, Berberich S J. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res. 1997;57:5013–5016. [PubMed] [Google Scholar]
- 43.McCurrach M E, Connor T M F, Knudson M C, Korsmeyer S J, Lowe S W. bax-deficiency promotes drug resistance and oncogenic transformation by attenuating p53-dependent apoptosis. Proc Natl Acad Sci USA. 1997;94:2345–2349. doi: 10.1073/pnas.94.6.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Midgley C A, Lane D P. p53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene. 1997;15:1179–1189. doi: 10.1038/sj.onc.1201459. [DOI] [PubMed] [Google Scholar]
- 45.Momand J, Zambetti G P, George D L, Levine A J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 46.Montes de Oca Luna R, Wagner D S, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 47.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 48.Picksley S M, Vojtesek B, Sparks A, Lane D P. Immunochemical analysis of the interaction of p53 with MDM2—fine mapping of the MDM2 binding site on p53 using synthetic peptides. Oncogene. 1994;9:2523–2529. [PubMed] [Google Scholar]
- 49.Pomerantz J, Schreiber-Agus N, Liégeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, Cordon-Cardo C, DePinho R A. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 50.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 52.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 53.Shaw P, Freeman J, Bovey R, Iggo R. Regulation of specific DNA binding by p53: evidence for a role of O-glycosylation and charged residues at the carboxy-terminus. Oncogene. 1996;12:921–930. [PubMed] [Google Scholar]
- 54.Shieh S-Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 55.Stott F, Bates S A, James M, McConnell B B, Starborg M, Brookes S, Palmero I, Hara E, Ryan K M, Vousden K H, Peters G. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. 1998. EMBO J., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stürzbecher H W, Brain R, Addison C, Rudge K, Remm M, Grimaldi M, Keenan E, Jenkins J R. A C-terminal alpha-helix plus basic region motif is the major structural determinant of p53 tetramerization. Oncogene. 1992;7:1513–1523. [PubMed] [Google Scholar]
- 57.Sun Y, Bian J, Wang Y, Jacobs C. Activation of p53 transcriptional activity by 1,10-phenanthroline, a metal chelator and redox sensitive compound. Oncogene. 1997;14:385–393. doi: 10.1038/sj.onc.1200834. [DOI] [PubMed] [Google Scholar]
- 58.Tarunina M, Jenkins J R. Human p53 binds DNA as a protein homodimer but monomeric variants retain full transcription transactivation activity. Oncogene. 1993;7:3165–3173. [PubMed] [Google Scholar]
- 59.Vogelstein B, Kinzler K W. p53 function and dysfunction. Cell. 1992;70:523–526. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- 60.Wagner A J, Kokontis J M, Hay N. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- 61.Waldman T, Kinzler K W, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 62.Wu X W, Bayle J H, Olson D, Levine A J. The p53 mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 63.Yin C, Knudson C M, Korsmeyer S J, Van Dyke T. Bax suppresses tumorigenesis and stimulates apoptosis in vivo. Nature. 1997;385:637–640. doi: 10.1038/385637a0. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y, Xiong Y, Yarbrough W G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]