Abstract
Using a new inducible form of phosphatidylinositol 3-kinase (PI 3-kinase) we have found that PI 3-kinase activation has the following effects on cell growth and proliferation. (i) Activation of PI 3-kinase was sufficient to promote entry into S phase of the cell cycle within several hours. This was shown by activation of cyclin-dependent kinase 4 (Cdk4) and Cdk2 and by the induction of DNA synthesis. (ii) PI 3-kinase activation alone was not, however, sufficient to provide for progression through the entire cell cycle. Instead, prolonged activation of PI 3-kinase in the absence of serum stimulation resulted in apoptosis. It is possible that the cells undergo apoptosis because the PI 3-kinase-induced entry into the cell cycle is abnormal. For example, we found that the cyclin E-Cdk2 complex, which normally disappears after entry into S phase of the cell cycle, fails to be downregulated following induction by PI 3-kinase. (iii) Finally, we found that prolonged activation of PI 3-kinase in the presence of serum resulted in cellular changes that resemble those associated with oncogenic transformation. The cells reached high densities, were irregular and refractile in appearance, and formed colonies in soft agar. In contrast, neither PI 3-kinase nor serum stimulation alone could induce these changes. Our results suggest that activation of PI 3-kinase promotes anchorage-independent cell growth and entry into the cell cycle but does not abrogate the growth factor requirement for cell proliferation.
Phosphatidylinositol (PI) 3-kinase has been shown to mediate signaling induced by numerous growth factors and tumor antigens. The intracellular levels of the phospholipid products of PI 3-kinase increase in response to stimulation with growth factors or after oncogenic transformation (for reviews, see references 10, 11, 33, 76, 80). PI 3-kinase signaling appears to be required for a number of mitogens during the G1-to-S-phase transition of the cell cycle (63). Recently, it was demonstrated that PI 3-kinase regulates cell survival in response to various apoptotic stimuli (21, 49).
PI 3-kinase is a heterodimeric complex consisting of an 85-kDa regulatory subunit, p85, and a 110-kDa catalytic subunit, p110 (11, 33). The p85 subunit contains two Src homology 2 (SH2) domains, which bind to tyrosine-phosphorylated receptors after stimulation of cells with growth factors and in this manner recruit the p85-p110 complex to the cell membrane. The region between the two SH2 domains of p85, the iSH2 region, mediates the association with p110, and this interaction is required for the enzymatic activity of p110 (37). Based on this observation we generated a chimeric molecule, p110*, in which the iSH2 region of p85 was covalently linked to its binding site at the p110 N terminus by using a flexible hinge region (30). p110* is a constitutively active PI 3-kinase which can activate signaling pathways independent of growth factor stimulation.
The generation of constitutively active PI 3-kinase molecules has greatly facilitated the analysis of signaling events regulated by PI 3-kinase (18, 30, 40, 64). Constitutively active PI 3-kinases allow the identification and study of responses specifically induced by PI 3-kinase. This approach enables the direct study of PI 3-kinase function without prior growth factor activation. It also eliminates the use of growth factor receptor mutants or PI 3-kinase inhibitors, the specificities of which are controversial. By using constitutively active forms of PI 3-kinase it is possible to test whether PI 3-kinase activation alone is sufficient to induce a certain signaling response.
Since the original description of a constitutively active PI 3-kinase, p110*, which has a high level of specific activity, additional forms of constitutively active PI 3-kinases have been described (18, 40, 61, 64). A second form of constitutively active PI 3-kinase was generated by fusing p110 with a membrane localization signal. This approach targets p110 to the location of its lipid substrates. Membrane-localized versions of p110 are able to induce signaling when overexpressed in a transient system. However, these versions have limited efficacy and do not induce the entire spectrum of PI 3-kinase-mediated responses since they depend on the interaction with endogenous p85 for enzymatic function (18, 40, 61). The most potent constitutively active PI 3-kinase, M · p110*, has a high level of enzymatic activity and is localized to the membrane (40). Transient expression of constitutively active PI 3-kinases has indicated that activation of PI 3-kinase was sufficient to induce a variety of cellular responses. These responses include the regulation of gene expression (16, 30) and the activation of signaling kinases which function in different pathways (18, 40, 83), as well as membrane ruffling (51), endocytosis (46), glucose transport, and DNA synthesis (25, 50, 79). Furthermore, p110* expression was able to rescue cells from undergoing apoptosis in response to various apoptotic stimuli (34, 35, 42, 59).
By using various forms of p110* either in vivo or in a cell-free system it was shown that the PI 3-kinase-produced phospholipids mediate PI 3-kinase signaling (39, 40). One of the products generated by purified p110* protein, PI 3,4-P2, was able to increase the kinase activity of PI 3-kinase effector Akt (also known as Rac protein kinase or protein kinase B) in vitro approximately 10-fold (22, 24, 39). PI 3-kinase-mediated activation of Akt is also controlled at the level of protein kinases, which by themselves are stimulated in the presence of PI 3,4-P2 and PI 3,4,5-P3 (1, 2, 41, 75, 77). Signaling intermediates which bind phospholipids via pleckstrin homology domains such as G-protein exchange factors and GTPase-activating proteins are also candidates for being regulated by the products of PI 3-kinase (27, 58).
Activated forms of Akt stimulate pp70 S6 kinase and are associated with cellular transformation (2, 5, 8). pp70 S6 kinase, an additional downstream effector of PI 3-kinase (13, 14, 54, 83), is required for S-phase transition (45, 62). The immunosuppressant rapamycin interferes with pp70 S6 kinase activation by inhibiting the mammalian TOR homolog Raft/FRAP (7, 9, 43, 66, 67). Rapamycin treatment of cells selectively blocks the pp70 S6 kinase pathway downstream of PI 3-kinase by causing dephosphorylation and inactivation of pp70 S6 kinase (6, 15, 31).
Experiments using constitutively active PI 3-kinase molecules suggest that PI 3-kinase activation is sufficient for the induction of a number of signaling pathways known to promote cell proliferation. However, transient expression systems are overexpression systems and in addition cannot entirely exclude the induction of an autocrine loop. Furthermore, transient approaches do not allow for the determination of the timely order of events. To study the role of PI 3-kinase in the regulation of proliferation and oncogenic transformation more rigorously, we expressed constitutively active PI 3-kinase molecules in an inducible fashion. This approach allows for an unbiased analysis of downstream events and facilitates time course studies to identify the order of responses.
We investigated the effect of PI 3-kinase stimulation on cell division and on processes that regulate cell cycle progression. Cyclin D–cyclin-dependent kinase 4 (Cdk4) (or cyclin D-Cdk6) and cyclin E-Cdk2 complexes regulate the G1-to-S-phase transition during the cell cycle (55, 56, 72, 73). In contrast to the Cdk protein levels, which remain constant during the cell cycle, the levels of the regulatory cyclin components oscillate. Cyclin D-Cdk4 activity is first detected in mid-G1 phase after quiescent cells have been stimulated to enter the cell cycle (52, 53). Next, the cyclin E-Cdk2 complex appears transiently during the G1-to-S-phase transition. Cyclin E is rapidly degraded once the cells enter S phase, and Cdk2 subsequently associates with cyclin A. Our results indicate that PI 3-kinase activation is sufficient for promoting the entry of quiescent cells into the cell cycle by activating G1- and G1/S-phase cyclin-Cdk complexes and for inducing DNA synthesis. Prolonged activation of PI 3-kinase in the absence of other stimuli results in apoptosis, indicating that PI 3-kinase activation is not sufficient for progression through the entire cell cycle. However, in combination with serum treatment, chronic PI 3-kinase activation resulted in cellular changes which are characteristic of cellular transformation.
MATERIALS AND METHODS
Cell culture.
3Y1 rat embryo fibroblasts (36) were cultured at 37°C in Dulbecco’s modified Eagle medium (DMEM) containing 10% bovine calf serum (CS), penicillin (50 μg/ml), and streptomycin (50 μg/ml). Stable cell lines were established by selection in either G418 (800 μg/ml), puromycin (1.5 μg/ml), or hygromycin B (200 μg/ml) after cotransfection of plasmids encoding the respective selectable markers. Transfections were carried out in 10-cm-diameter plates (at 30 to 50% confluency) by using Lipofectamine (Gibco BRL) or FuGene 6 (Boehringer Mannheim) according to the manufacturer’s instructions. COS 7 cells were transiently transfected in 10-cm-diameter plates (50 to 70% confluency) by the DEAE-dextran method (26).
Antibodies.
Murine monoclonal anti-p110 antibody U3A has been described previously (30, 37). Polyclonal anti-Akt antibodies (C-20-G), anti-pp70 S6 kinase antibodies (C-18), anti-estrogen receptor (ER) antibodies (MC-20), anti-Cdk2 antibodies (M2-G), anti-Cdk4 antibodies (H22-G for immunoprecipitation, M22-G for immunoblotting), anti-cyclin E antibodies (M-20), and anti-Jun N-terminal kinase (JNK) antibodies (C-17-G for immunoprecipitation, monoclonal F-3 for immunoblotting) were from Santa Cruz Biotechnology.
Reagents.
4-Hydroxytamoxifen (4-OHT) was purchased from Sigma. Histone H1 and histone H2B were from Boehringer Mannheim. Glutathione S-transferase (GST)-Rb and GST-c-Jun (1-79) were obtained from Santa Cruz Biotechnology.
Plasmids.
The sequence encoding the hormone binding domain (HBD; amino acids 281 to 599 [47]) of the mouse estrogen receptor (mER) was amplified by PCR from a mouse uterus cDNA library (Clontech) with primer mER-s- (5′ AT GGC GCC GGC CGA AAT GAA ATG GGT GCT TCA G 3′) overlapping nucleotides 838 to 862 of the coding strand (A of the start codon is designated nucleotide 1; nucleotides that are changed with respect to the wild-type sequence are underlined) and primer mER-α- (5′ AT GGA TCC GGT ACC TCA GAT CGT GTT GGG GAA GCC CTC TGC 3′) overlapping nucleotides 1774 to 1797. This extended the mER HBD fragment coding sequence at one end by a sequence encoding amino acids GAG as a hinge region (overlapping restriction sites for KasI/NarI/EheI and NaeI/NgoMI) and by a stop codon preceding restriction sites for KpnI/Asp718 and BamHI at the coding sequence for the C-terminal end. The point mutation that changes amino acid 525 of the mER from G to R was introduced by PCR with primer mER GR525-s-(5′ AGT AAC AAA CGC ATG GAG CAT CTC TAC AAC ATG AAA 3′) in combination with primer mER GR525-α-(5′ GAG ATG CTC CAT GCG TTT GTT ACT CAT GTG CCG GAT 3′). The Myc-tagged C-terminal end of myristoylated p110* (M · p110*) (40) was replaced by the mER HBD with the GR525 mutation by using EheI and BamHI in a mammalian expression vector that directs expression from the SRα promoter (78). To modify the C terminus of Akt with the mER GR525 sequence, the Akt1 cDNA encoding a C-terminal fragment was amplified with primer Akt BbrPI-s- (5′ TGG CAG CAC GTG TAC GAG 3′), consisting of nucleotides 1237 to 1254 of the coding strand overlapping a BbrPI site, and primer Akt C-term-α- (5′ T GGA TCC TCA TTA GGC GCC GGC CGT GCT GCT GGC CGA GTA 3′), which overlaps nucleotides 1420 to 1440 of the noncoding strand, restriction sites for KasI/NarI/EheI and NaeI/NgoMI, a stop codon, and a BamHI restriction site. The BbrPI-BamHI fragment encoding the C terminus of myristoylated Akt (M · Akt) (42) was replaced by the sequence encoding the modified C terminus in which the stop codon is preceded by restriction sites. This allowed the fusion of the Akt coding region to the sequence encoding the mER HBD with the GR525 mutation by insertion of the EheI-BamHI fragment described above. The correct sequences of the DNA fragments modified by PCR were confirmed by DNA sequence analysis. The sequence encoding mER HBD GR525 was further used to modify the coding regions for p110* and M · p110* (40) at the coding sequences for their respective C-terminal ends by using the restriction sites described above for M · p110*. Expression of the ER-tagged molecules was analyzed after transient expression in COS 7 cells. A mammalian expression vector for 4-OHT-responsive Myc · ER was generously provided by Catherine Tribouley (Chiron Corporation). Vectors with selectable markers that were used for cotransfections, pL1-3neo (4), pL1-3hyg (38), and pBabe-Puro (57), have been described previously.
Preparation of cell extracts and immunoblotting.
Stably transfected cells in 10-cm-diameter plates were starved for at least 30 h in medium containing 0.5% dialyzed CS and then stimulated with 10% CS–200 nM 4-OHT in dimethylsulfoxide (DMSO) or with DMSO at 37°C for the indicated times. In experiments in which the effect of rapamycin was analyzed, the reagent was added in DMSO to the cells at 20 ng/ml just before stimulation. Cells were washed twice with cold phosphate-buffered saline (PBS) and lysed at 4°C in lysis buffer containing 20 mM Tris (pH 7.5), 137 mM NaCl, 15% (vol/vol) glycerol, 1% (vol/vol) Nonidet P-40 (NP-40), 2 mM phenylmethylsulfonyl fluoride, 10 mg of aprotinin per ml, 20 mM leupeptin, 2 mM benzamidine, 1 mM sodium vanadate, 25 mM β-glycerolphosphate, 50 mM NaF, and 10 mM Na-pyrophosphate. Lysates were cleared by centrifugation at 14,000 × g for 5 min, and aliquots of the lysates were analyzed for protein expression and enzyme activity (see below). Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (44) and transferred to nitrocellulose filters (Schleicher & Schuell). Filters were blocked in TBST buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% [vol/vol] Tween 20, 0.5% [wt/vol] sodium azide) containing 5% (wt/vol) dried milk. The respective antibodies were added in TBST at appropriate dilutions. Bound antibody was detected with anti-mouse-, anti-goat, or anti-rabbit-conjugated alkaline phosphatase (Santa Cruz Biotechnology) in TBST, washed, and developed with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (Promega). Alternatively, horseradish peroxidase-conjugated secondary antibodies were used and developed by enhanced chemiluminescence (Amersham).
In vitro protein kinase assays.
Cell extracts were incubated with the indicated antibodies for 2 h at 4°C. Protein A-Sepharose (Sigma) or protein A/G-agarose beads (Santa Cruz Biotechnology) were used to precipitate the immune complexes. The beads were washed once with 50 mM Tris-HCl (pH 7.5)–0.5 M LiCl–0.5% (vol/vol) NP-40, twice with PBS, and once with 10 mM Tris-HCl (pH 7.5)–10 mM MgCl2, all containing 0.1 mM sodium vanadate, 25 mM β-glycerolphosphate, and 1 mM dithiothreitol (DTT).
For analyzing the kinase activity of Akt, one-third of the immunobeads were subjected to an in vitro kinase reaction and two-thirds were analyzed for the amount of Akt protein by immunoblotting. Akt activity was measured by using histone H2B as a substrate (23) according to the reaction conditions described previously (32). Briefly, the reactions were carried out in 30 μl containing 30 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 5 mM MnCl2, 2 mM DTT, 50 μM ATP, 10 mM β-glycerolphosphate, 1 μM protein kinase A inhibitor peptide (PKA-I), 1 μM protein kinase C inhibitor peptide (PKC-I), and 2 μg of histone H2B in the presence of 5 μCi of [γ-32P]ATP. The reaction mixtures were incubated at 22°C for 20 min, and then the reactions were stopped by the addition of 8 μl of Laemmli sample buffer (44). Half of each reaction mixture was separated by SDS–16% PAGE. The relative amounts of incorporated radioactivity were determined by autoradiography.
For the analysis of JNK activity, half of the immunoprecipitates were subjected to an in vitro kinase reaction and the other half were analyzed for JNK protein levels by immunoblotting. JNK activity was determined by using GST–Jun(1-79) as a substrate as described in reference 17. Briefly, reactions were carried out in 30 μl containing 30 mM HEPES (pH 7.2), 20 mM MgCl2, 2 mM DTT, 20 μM ATP, 20 mM β-glycerolphosphate, 1 mM sodium vanadate, 1 μM PKA-I, 1 μM PKC-I, and 1.5 μg of GST–Jun(1-79) in the presence of 5 μCi of [γ-32P]ATP. The reaction mixtures were incubated at 22°C for 20 min, and then the reactions were stopped by the addition of 8 μl of Laemmli sample buffer. Half of each reaction mixture was separated by SDS–12% PAGE. The relative amounts of incorporated radioactivity were determined by autoradiography.
To measure Cdk2 activity, one half of the anti-CDK2 or anti-cyclin E immunobeads were subjected to an in vitro Cdk2 assay using histone H1 as a substrate (19) and the other half were analyzed for relative Cdk2 protein levels by immunoblotting. Briefly, the in vitro kinase reactions were carried out in 30 μl containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 2 mM DTT, 50 μM ATP, 10 mM β-glycerolphosphate, 1 μM PKA-I, 1 μM PKC-I, and 1.5 μg of histone H1 in the presence of 5 μCi of [γ-32P]ATP. The reaction mixtures were incubated at 22°C for 15 min, and then the reactions were stopped by the addition of sample buffer. Half of each reaction mixture was separated by SDS–16% PAGE. The relative amounts of incorporated radioactivity were determined by autoradiography.
For measuring Cdk4 activity, anti-Cdk4 precipitates were analyzed in an in vitro kinase reaction with GST-Rb as a substrate (52, 53). A typical reaction mixture of 30 μl contained 50 mM HEPES (pH 7.2), 10 mM MgCl2, 2 mM DTT, 25 μM ATP, 10 mM β-glycerolphosphate, 2 μM PKA-I, 2 μM PKC-I, 1 μg of GST-Rb, and 5 μCi of [γ-32P]ATP. The reaction mixtures were incubated at 22°C for 25 min, and then the reactions were stopped by the addition of sample buffer. Half of each reaction mixture was separated by SDS–12% PAGE. Phosphorylated GST-Rb was detected by autoradiography. Half of each anti-Cdk4 precipitate was analyzed for the Cdk4 protein level.
Analysis of intracellular levels of 3′-phosphorylated phosphoinositides.
The generation of PI 3-phosphoinositides in vivo was determined as described previously (40, 69). Briefly, stably transfected cells were starved for 24 h in a medium containing 0.5% serum. The cells were metabolically labeled in phosphate-free medium containing 0.3% dialyzed CS for 12 h by using 1 mCi of [32P]orthophosphate (8,500 to 9,120 Ci/mmol; New England Nuclear) per 10-cm-diameter dish. After stimulation with 200 nM 4-OHT for the times indicated in Fig. 3 or with 2 nM platelet-derived growth factor (PDGF-BB) for 20 min, the phospholipids were extracted by the addition of 750 μl of 1:1 (vol/vol) methanol–1 N HCl, collected into Eppendorf tubes, and mixed with 380 μl of chloroform. After centrifugation, the lower chloroform phase was collected, the interface material was reextracted with chloroform, and the chloroform phases were combined. For analysis of PI 3,4-P2 production, the extracts were directly deacylated and subjected to anion-exchange high-pressure liquid chromatography (HPLC) analysis (69). For analysis of PI 3,4,5-P3 production, the extracts were purified by thin-layer chromatography (TLC) (81) and then processed as described above for HPLC. Peak fractions containing glycerophosphoinositides derived from PI 3,4-P2 or PI 3,4,5-P3 were identified by cochromatography with deacylated 32P-labeled standards produced by p110* in an in vitro PI 3-kinase reaction (40).
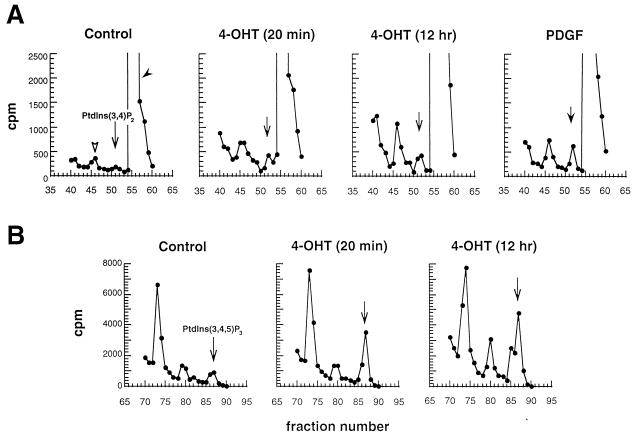
FIG. 3.
Prolonged stimulation of M · p110* · ER results in persistent upregulation of 3′-phosphorylated phosphoinositides. Stable M · p110* · ER-expressing cell lines were metabolically labeled with 32Pi and stimulated with 4-OHT for the indicated times or with PDGF for 20 min as a positive control. Phospholipids were extracted, deacylated, and analyzed by HPLC or a combination of TLC and HPLC as described in Materials and Methods. An identical fraction of each extract from one 10-cm-diameter plate was analyzed. Three independent experiments using different M · p110* cell lines were performed. The results of a representative experiment are shown. (A) PI 3,4-P2 production in cells stimulated with vehicle, 4-OHT, or PDGF. (B) PI 3,4,5-P3 production after treatment with vehicle or 4-OHT. Results are presented as counts per minute per column fraction, with background subtracted. The positions of the standards for deacylated glycerophosphoinositides PI 3,4-P2 [Ptdins(3,4)P2] (A) and PI 3,4,5-P3 [Ptdins(3,4,5)P3] (B) generated with purified recombinant p110* are indicated. In panel A, a solid arrowhead indicates the position of PI 4,5-P2; an open arrowhead indicates the presumed position of PI 3,5-P2 (69).
Determination of the rate of DNA synthesis by incorporation of radiolabeled thymidine.
Cells plated in triplicate samples in 24-well plates were starved in DMEM containing 10 mM HEPES (pH 7.2) and 0.3% dialyzed CS for 48 h. After stimulation with 200 nM 4-OHT or 10% CS for the time indicated in the legends for Fig. 4 and 5 the rate of DNA synthesis was measured by pulse-labeling the cells for 1 h with 0.5 μCi of [3H]thymidine (50 Ci/mmol) in 500 μl of sample per well. The reaction was stopped by precipitation in 5% trichloroacetic acid (TCA). After two additional TCA washes to remove unincorporated radiolabel, precipitated nucleic acids were solubilized in 0.1% SDS–0.25 N NaOH. The samples were neutralized with HCl, and incorporated [3H]thymidine was measured in a scintillation counter.
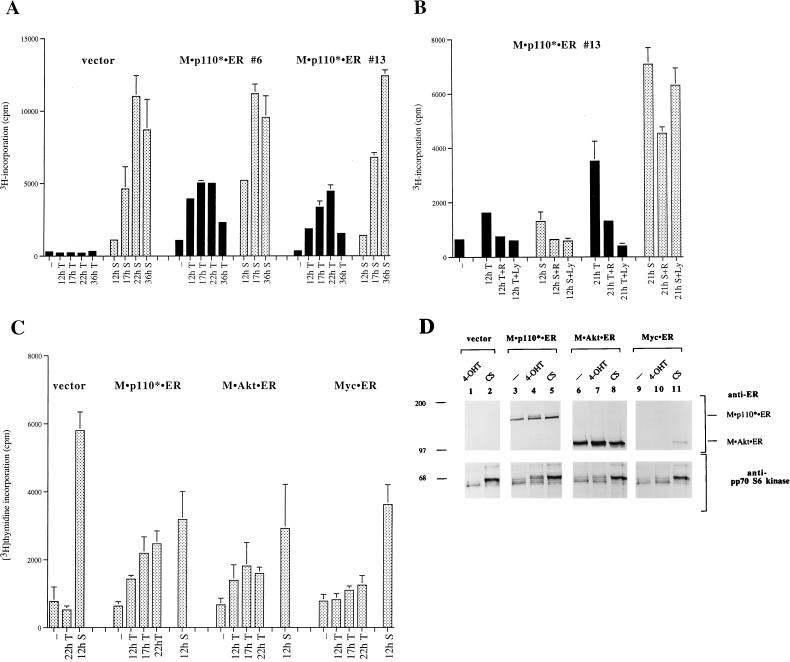
FIG. 4.
Induction of PI 3-kinase is sufficient to induce DNA synthesis in cells expressing M · p110* · ER. (A) 4-OHT treatment results in an increased rate of [3H]thymidine incorporation in M · p110* · ER transfectants. Vector-transfected cells were analyzed in parallel with two cell lines (no. 6 and 13) expressing M · p110* · ER. Cells grown in 24-well plates were starved in 0.5% serum for 48 h and subsequently stimulated with 200 nM 4-OHT (T; black bars) or 10% CS (S; stippled bars) for the indicated times. The rate of DNA synthesis was determined after pulse-labeling with [3H]thymidine for 1 h. Each bar represents the mean of triplicate samples ± the standard deviation. (B) PI 3-kinase activation is required for the initial phase of DNA synthesis. Cells were treated with 4-OHT or CS in the presence or absence of 20 ng of rapamycin (R) per ml or 10 μM LY294002 (LY) for 12 or 21 h. The rate of DNA synthesis was measured by determining [3H]thymidine incorporation after labeling cells for 1 h. Each bar represents the mean of triplicate samples ± the standard deviation. The expression levels of M · p110* · ER in the cell lines used for the experiments shown in panels A and B were comparable to that of endogenous p110 as assessed by anti-p110 immunoblotting (data not shown). (C) Comparison of DNA synthesis rates induced after 4-OHT stimulation of M · p110* · ER, M · Akt · ER, and Myc · ER. Pools of 50 to 100 transfectants each were treated with 4-OHT or CS as indicated. [3H]thymidine incorporation was determined after labeling the cells for 1 h. Each bar represents the mean of triplicate samples ± the standard deviation of two experiments. Each experiment was carried out with pools of independently transfected cells. (D) Expression levels of the 4-OHT-inducible molecules and their potential to activate pp70 S6 kinase. Cell extracts of the transfected populations analyzed in panel C were separated by SDS-PAGE and immunoblotted with anti-ER antibody (upper portion) or anti-pp70 S6 kinase antibody (lower portion). Myc · ER, which is in the nucleus, was not detected after cell lysis by NP-40. Similar results were obtained when single-cell clones were analyzed (data not shown).
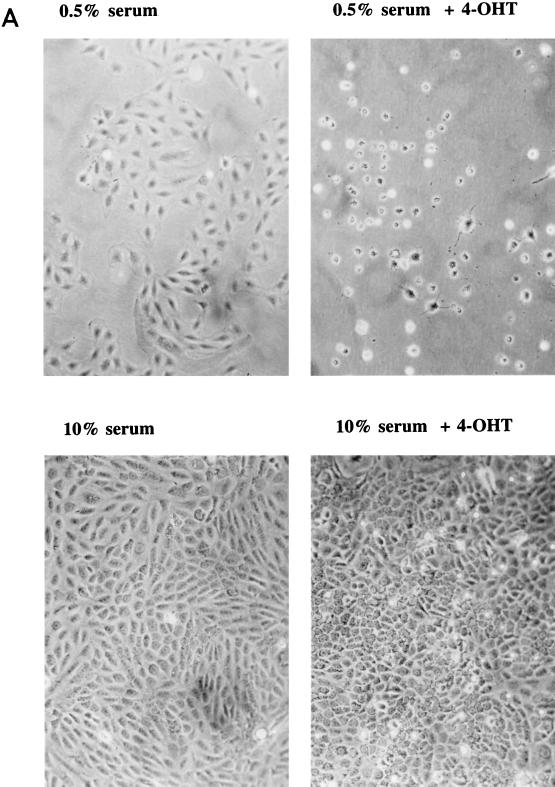
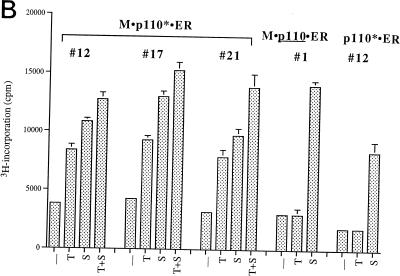
FIG. 5.
Effect of prolonged PI 3-kinase activation on cell growth and cell morphology. (A) Comparison of PI 3-kinase activation by 4-OHT with activation by serum stimulation and activation by 4-OHT treatment in combination with serum. Subconfluent cells expressing M · p110* · ER were growth arrested in 0.5% serum for 36 h and then either treated with vehicle (DMSO) or stimulated in the presence of 200 nM 4-OHT, 10% CS, or a combination of 4-OHT and serum as shown. The cells were photographed 48 h later at a magnification of ×10. The same changes were observed when several independent M · p110* · ER transfectants were analyzed. Control cells that expressed M · p110 · ER showed no obvious morphological changes in response to 4-OHT stimulation (data not shown). (B) Comparison of the effects of 4-OHT-mediated induction of M · p110* · ER, serum stimulation, and 4-OHT treatment plus serum on the rate of DNA synthesis. Various cell lines (indicated by numbers) stably expressing M · p110* · ER, M · p110 · ER, or p110* · ER were synchronized in 0.5% serum and subsequently stimulated with 4-OHT (T), serum (S), or 4-OHT in the presence of serum (T+S) for 15 h. Cells labeled − were left untreated. The rate of DNA synthesis was determined by measuring [3H]thymidine incorporation after 1 h of labeling.
Soft-agar colony formation.
A total of 3 × 105 cells per 60-mm-diameter dish were embedded into 0.5% soft agar (Agar Noble [Difco] in DMEM) in the presence of 10% CS, 10% CS plus 200 nM 4-OHT, or 0.5% CS plus 200 nM 4-OHT. The cells were refed every week by overlaying them with DMEM containing CS and/or 4-OHT. Anchorage-independent cell growth was monitored over a 4-week period.
TUNEL assay and DNA content analysis by flow cytometry.
Cells were plated at 1 × 105 to 2 × 105 cells per 10-cm-diameter plate and arrested in G0 by 0.5% serum treatment for 24 h. The cells were treated with vehicle (DMSO) or stimulated with 200 nM 4-OHT in the presence of 20 ng of rapamycin per ml or in the absence of rapamycin. After treatment with 4-OHT for 40 h or longer the cells exhibited a rounded morphology and began to detach from the plate. Attached cells were harvested by trypsinization and combined with the floating cell population after centrifugation for 3 min at 400 × g. Cells combined from three plates per sample were fixed in 4% formaldehyde in PBS (freshly prepared from paraformaldehyde) and permeabilized in 0.1% Triton X-100–0.1% sodium citrate. After a series of washes in PBS, DNA fragmentation in apoptotic cells was determined by measuring terminal deoxynucleotidyltransferase activity by the terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling, (TUNEL) assay (Boehringer Mannheim). The reaction was stopped after 1 h by washing the cells in PBS, and the cells were resuspended in propidium iodide (10 μg/ml) solution containing RNase (Coulter) for analysis of the DNA content in parallel. Data were then collected on a Becton Dickinson FACScan, 20,000 events per sample, by using Cellquest software. The analysis of the percentage of TUNEL-positive cells was performed with Cellquest software by gating on all populations with the exception of multicellular clumps. DNA content analysis was performed with Verity ModFit software for the Macintosh computer.
RESULTS
Activation of a regulatable p110* efficiently induces cell signaling.
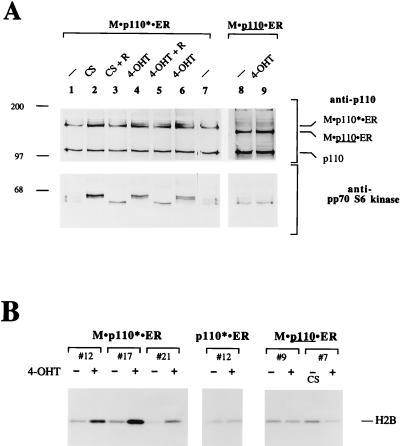
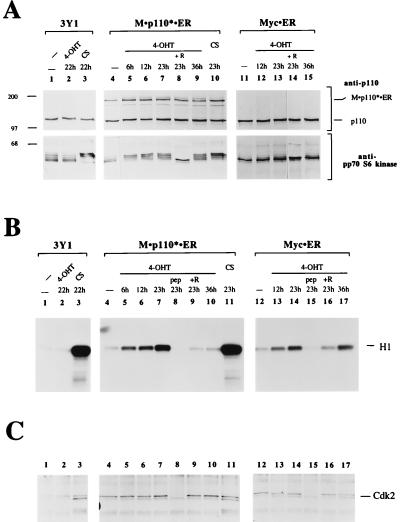
We have previously shown that transient expression of a constitutively active mutant of PI 3-kinase, p110*, is sufficient to induce signaling events such as activation of the fos promoter, pp70 S6 kinase, Akt, and JNK (30, 40, 83). The observation that p110* can activate pathways known to be involved in the regulation of cell proliferation prompted us to test its effect on cell growth and mitogenesis. To this end we generated inducible PI 3-kinase molecules by fusing p110 and p110* to the HBD of a mutant mER. Proteins that are fused to this mutant ER domain are inactive until the addition of 4-OHT (47). We created fusion proteins between the ER mutant and p110* (p110* · ER), a myristoylated version of p110 (M · p110 · ER), and a myristoylated version of p110* (M · p110* · ER) (Fig. 1). We have previously shown that p110*, M · p110, and M · p110* function as constitutively active PI 3-kinases when overexpressed in a transient system. We established stable rat 3Y1 cell lines that express the chimeric molecules. First, we tested whether induction of p110* · ER, M · p110 · ER, or M · p110* · ER could activate pp70 S6 kinase and Akt, both well-established effectors of PI 3-kinase (Fig. 2A and B). The cells were arrested in 0.5% serum and stimulated with 4-OHT. Cell extracts were separated by SDS-PAGE and analyzed for activation of pp70 S6 kinase by Western blotting. The activation of pp70 S6 kinase by phosphorylation can be monitored by its decreased mobility on protein gels (15, 54, 83). Cell lines expressing M · p110* · ER showed activation of pp70 S6 kinase after 15 min of 4-OHT stimulation (Fig. 2A). Control cells stimulated with serum also activated pp70 S6 kinase after 15 min. Both responses were abrogated by pretreatment of the cells with rapamycin.
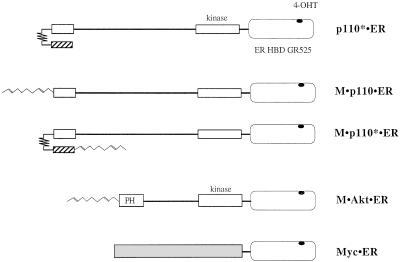
FIG. 1.
Schematic representation of 4-OHT-regulatable molecules used in this study. Constitutively active PI 3-kinase (p110*), which has a high level of specific activity (30), myristoylated p110 (M · p110), which is localized to the membrane (40), and myristoylated p110* (M · p110*) were fused at their respective C termini to the HBD of mER. The ER portion contains a point mutation, GR525, that renders it specific for responding to 4-OHT (47). Myristoylated, constitutively active Akt (M · Akt) and Myc were similarly fused to the ER domain. The p110 and Akt regions with homology to the catalytic domains of protein kinases are depicted by boxes labeled kinase. The domain responsible for the interaction with the inter-SH2 (iSH2) domain of the p85 subunit is shown as a small box at the p110 N terminus. p110* is a chimeric molecule that contains the iSH2 domain of p85 (hatched bar) fused to the N terminus of p110 by a flexible “glycine kinker” (30). M · p110 · ER, M · p110* · ER, and M · Akt · ER contain the myristoylation signal of pp60 c-Src at their respective N-terminal ends (68). The pleckstin homology (PH) domain at the N terminus of Akt is indicated.
FIG. 2.
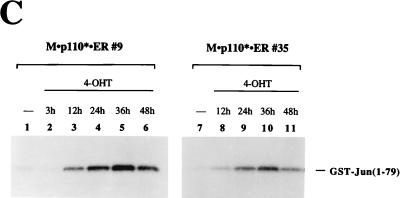
4-OHT efficiently stimulates activation of PI 3-kinase-mediated events in cells expressing M · p110* · ER. (A) Activation of pp70 S6 kinase after induction of M · p110* · ER by 4-OHT. In lanes 2 to 5 and 9, cells stably expressing M · p110* · ER or M · p110 · ER were arrested in 0.5% serum and then treated with 10% CS, 200 nM 4-OHT, or rapamycin (R) (20 ng/ml) as indicated for 15 min. In lane 6 the cells were treated with 4-OHT for 25 h. Cells in lanes 1, 7, and 8 were left untreated. Cell extracts were separated by SDS–8% PAGE and analyzed by Western blotting using anti-p110 antibody (upper portion) and anti-pp70 S6 kinase antibody (lower portion). The positions of M · p110* · ER, M · p110 · ER, and endogenous p110 are indicated on the right. Positions of molecular size markers (in kilodaltons) are shown on the left. (B) Activation of Akt kinase activity in response to 4-OHT treatment in cells expressing M · p110* · ER. Various cell lines stably expressing M · p110* · ER, p110* · ER, or M · p110 · ER were treated with 4-OHT (+) or 10% CS for 15 min or were not treated (−), as shown above each sample. Different cell lines are represented by their clone numbers. Anti-Akt immune precipitates were prepared from cell extracts and analyzed in an in-vitro-kinase assay using histone H2B as a substrate. The reaction mixtures were then separated by SDS–16% PAGE, and the phosphorylation of histone H2B was monitored by autoradiography. The position of histone H2B is indicated on the right. Precipitates from stimulated and unstimulated cells contained comparable amounts of Akt protein (data not shown). (C) Activation of JNK after stimulation of M · p110* · ER. Cell lines stably transfected with M · p110* · ER were treated with 4-OHT for the indicated times; serum stimulation was performed in parallel (data not shown). Anti-JNK precipitates were analyzed for kinase activity in vitro by using GST-Jun(1-79) as a substrate and for the presence of comparable amounts of JNK protein (data not shown). The reactions were analyzed by SDS–12% PAGE, and phosphorylation of the substrate was detected by autoradiography. The position of GST-Jun(1-79) is indicated on the right. The results obtained with two cell lines are shown.
p110* · ER- or M · p110 · ER-expressing cells were not able to activate either S6 kinase or Akt in response to 4-OHT treatment (Fig. 2A and B). When expressed at levels comparable to that of wild-type p110, only M · p110* · ER was capable of activating signaling pathways. This is in agreement with our previous data demonstrating that M · p110* is the most potent constitutively active PI 3-kinase (40). Further, this result is consistent with the hypothesis that an activated form of PI 3-kinase needs to be localized at the cell membrane in order to induce downstream events. Therefore, for the following studies we focused on the analysis of cell lines expressing M · p110* · ER.
M · p110* · ER cells also showed induction of JNK activity after 4-OHT treatment (Fig. 2C), confirming our previous results obtained by transient overexpression (40). JNK activity was maximal after 24 to 36 h and subsequently declined. A similar response was observed when the cells were stimulated with serum (data not shown). These data indicate that activation of JNK is a late response compared to the activation of pp70 S6 kinase or Akt, which occurs within minutes after induction of M · p110* · ER or serum stimulation (Fig. 2A and B).
We have previously suggested that the 3′-phosphorylated phosphoinositides produced by p110* are mediators of PI 3-kinase signaling (39, 40). To analyze the generation of phospholipids after activation of M · p110* · ER, cells were metabolically labeled with 32Pi and stimulated with 4-OHT for the times indicated in Fig. 3. Phospholipids were extracted, deacylated, partially purified by TLC, and subjected to HPLC analysis (69). Levels of PI 3-kinase products PI 3,4-P2 and PI 3,4,5-P3 were increased after 20 min of 4-OHT stimulation; PDGF stimulation served as a positive control (Fig. 3A and B). The phospholipid levels induced after activation of M · p110* · ER were comparable to the levels generated by endogenous PI 3-kinase in response to stimulation with PDGF (Fig. 3A and data not shown). Interestingly, the phospholipid levels induced by M · p110* · ER remained high after prolonged stimulation. This is in contrast to growth factor-mediated induction of the PI 3-kinase-specific phospholipids, the levels of which increase rapidly but only transiently (3). The data show that chronic activation of M · p110* · ER results in a sustained upregulation of 3′-phosphorylated phosphoinositides.
Induction of the PI 3-kinase pathway is sufficient to induce DNA synthesis.
Since induction of PI 3-kinase is sufficient to activate pp70 S6 kinase and since pp70 S6 kinase is required for S-phase transition (45, 62), we investigated whether the activation of PI 3-kinase can induce DNA synthesis. Cell lines expressing M · p110* · ER were serum starved and subsequently stimulated with 4-OHT or serum. DNA synthesis was detected by measuring [3H]thymidine incorporation into newly synthesized DNA. Cell lines expressing M · p110* · ER showed detectable increases in DNA synthesis 12 h after stimulation with 4-OHT (Fig. 4A). The rate of DNA synthesis increased over 22 h and declined after 36 h. In contrast, the rate of DNA synthesis after cells were stimulated with serum remained high at 36 h. Vector-transfected cells and cells expressing either p110* · ER or M · p110 · ER did not show any detectable increase in DNA synthesis after 4-OHT treatment even though they did respond normally to serum stimulation (Fig. 4A and Fig. 5B).
In M · p110* · ER cells the initial rates of DNA synthesis after treatment with serum or 4-OHT were comparable (Fig. 4A). To test whether the initial rate of DNA synthesis in response to serum was dependent on PI 3-kinase, we compared the effects of rapamycin and the PI 3-kinase inhibitor LY294002 (82) on 4-OHT- or serum-stimulated cells (Fig. 4B). At 12 h after 4-OHT or serum stimulation the rate of DNA synthesis was diminished by rapamycin and LY294002. At 21 h the 4-OHT response remained sensitive to rapamycin and LY294002, while the serum response was only moderately affected. These results suggest that activation of PI 3-kinase might primarily regulate the initial phase of DNA synthesis after stimulation of cells with serum growth factors, whereas the later phases of DNA synthesis are regulated by other pathways. This is consistent with the observation that prolonged activation of PI 3-kinase causes a decrease in the rate of DNA synthesis (Fig. 4A).
We next compared the effect of an activated PI 3-kinase on the induction of DNA synthesis with those of activated Akt (M · Akt · ER) and Myc · ER (Fig. 1). Akt can be converted into a constitutively active form by fusion with a membrane localization signal (2, 5, 8). The 4-OHT-inducible form of Myc has been studied extensively (28, 47, 65) and can induce DNA synthesis. Stable rat 3Y1 cell lines were analyzed for the induction of DNA synthesis in response to 4-OHT stimulation. We determined that M · p110* · ER caused a more robust induction of DNA synthesis than M · Akt · ER despite the fact that M · Akt · ER was expressed at substantially higher levels (Fig. 4C and D). Both molecules induced a shift in pp70 S6 kinase mobility. Myc · ER showed the weakest effect on DNA synthesis and did not activate pp70 S6 kinase.
Prolonged activation of PI 3-kinase leads to apoptosis.
We investigated the effects of prolonged activation of PI 3-kinase on cells. Cells that expressed M · p110* · ER were quiesced in 0.5% serum and then stimulated with 4-OHT for 42 h. We observed that the cell morphology changed from flat to round and that the cells detached from the plate (Fig. 5A). The majority of the control cells remained flat and attached to the plate.
In order to determine whether prolonged activation of PI 3-kinase caused cells to undergo apoptosis, we analyzed their fragmented DNA content by the TUNEL assay. We analyzed Myc · ER-expressing control cells in parallel, since Myc has been shown to induce cell death by apoptosis under low-concentration serum conditions (28, 29). Activation of M · p110* · ER by 4-OHT caused a significant proportion of cells to stain TUNEL positive (Table 1). A majority of the same cell population had furthermore shifted into S phase, as determined by flow cytometry after costaining the cells with propidium iodide. Treatment of cells with rapamycin reduced the number of apoptotic cells and prevented entry into S phase. Apoptosis induced by activated Myc · ER affected more cells and was not inhibited by rapamycin treatment (Table 1). Our data suggest that the selective activation of PI 3-kinase is sufficient to induce S-phase entry but that the cells subsequently undergo apoptosis. Interestingly, cells were rescued from p110*-induced apoptosis in the presence of serum (Fig. 5A; also, see below).
TABLE 1.
Effect of prolonged activation of PI 3-kinase on cell viability and DNA content
| Fusion protein/cell line and treatment | % of cells that werea:
|
% of cells inb:
|
||
|---|---|---|---|---|
| TUNEL negative | TUNEL positive | G0/G1 phase | S phase | |
| M · p110* · ER/7 | ||||
| None | 91 | 4 | 60 | 38 |
| 4-OHT | 70 | 23 | 33 | 60 |
| 4-OHT + Rc | 85 | 10 | 50 | 45 |
| M · p110* · ER/13 | ||||
| None | 86 | 3 | 59 | 36 |
| 4-OHT | 63 | 27 | 27 | 66 |
| 4-OHT + R | 73 | 16 | 71 | 28 |
| Myc · ER/1 | ||||
| None | 86 | 7 | ndd | nd |
| 4-OHT | 52 | 42 | nd | nd |
| 4-OHT + R | 55 | 38 | nd | nd |
DNA fragmentation was analyzed by flow cytometry after TUNEL labeling with fluorescein-dUTP.
DNA content analysis was performed by flow cytometry after propidium iodide costaining.
R, rapamycin.
nd, not determined.
PI 3-kinase activation promotes anchorage-independent cell growth in the presence of serum.
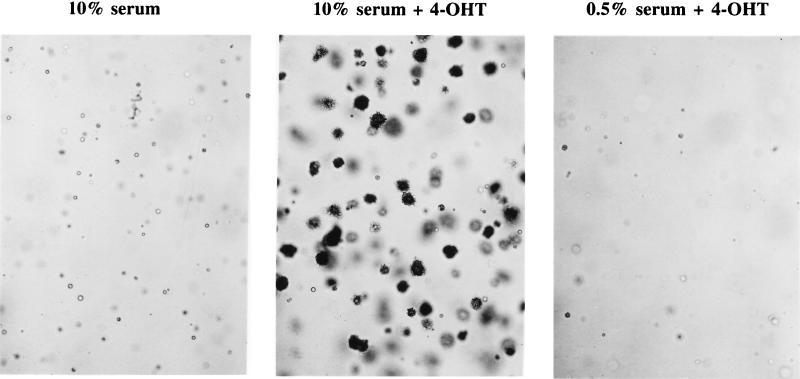
When M · p110* · ER-expressing cells were subjected to continuous treatment with 4-OHT in the presence of 10% serum, the cells no longer detached from the plate (Fig. 5A). Instead, these cells reached high cell densities and became rounded and refractile in appearance, unlike cells cultured in serum alone. The cells also showed an increased rate of DNA synthesis compared to cells treated with either 4-OHT or serum alone (Fig. 5B). These observations prompted us to test whether the chronic activation of PI 3-kinase in combination with serum stimulation leads to additional cellular changes characteristic of transformation. When plated in soft agar containing 4-OHT and serum, cells expressing M · p110* · ER efficiently formed colonies (Fig. 6). Neither treatment with 4-OHT alone nor treatment with serum alone induced the cells to form colonies in soft agar. This shows that chronic activation of the PI 3-kinase pathway in combination with additional stimuli provided by serum leads to anchorage-independent cell growth and possible cellular transformation.
FIG. 6.
Stimulation of PI 3-kinase promotes anchorage-independent cell growth in the presence of serum. 3Y1 cells stably expressing M · p110* · ER were plated in soft agar in the presence of 10% serum, 10% serum plus 200 nM 4-OHT, or 0.5% serum plus 200 nM 4-OHT. Photographs were taken after 4 weeks at a magnification of ×10. The experiment was reproduced with several independent M · p110* · ER transfectants as well as with pools of transfectants. The parental cell line and cells expressing M · p110 · ER or p110* · ER were not able to form colonies in soft agar (data not shown).
PI 3-kinase activation is sufficient to activate Cdks.
The selective activation of the PI 3-kinase pathway was sufficient for the induction of DNA synthesis. In order to verify that cells were entering S phase, we determined whether Cdks were activated. Cells were arrested in G0 by serum starvation and then stimulated with 4-OHT. Cells stimulated with serum served as a positive control. At various times after induction cell extracts were prepared and M · p110* · ER protein levels and responsiveness to 4-OHT were analyzed (Fig. 7A). Cdk2 was precipitated from the cell extracts, and half of each precipitate was subjected to a Cdk2 activity assay using histone H1 as a substrate (19, 71) (Fig. 7B). The other half of the precipitate was analyzed for Cdk2 protein levels (Fig. 7C). PI 3-kinase activation resulted in a substantial increase in Cdk2 activity that could be detected as early as 6 h after stimulation. The activity increased for 22 h after stimulation and decreased at later time points (Fig. 7B, lane 10). The specificity of the immunoprecipitation was confirmed by using the peptide against which the antibody was raised as a competitor (lane 8). Rapamycin treatment reduced Cdk2 activity to background levels. Control cells that did not express M · p110* · ER did not show any Cdk2 activation after 4-OHT treatment. However, Cdk2 activity in these cells was stimulated after serum stimulation. Myc-induced activation of Cdk2 also increased over time but remained high even after 36 h. The results indicate that PI 3-kinase activation is sufficient for the induction of Cdk2 activity. The time course of the Cdk2 activation correlated well with the time course observed for DNA synthesis.
FIG. 7.
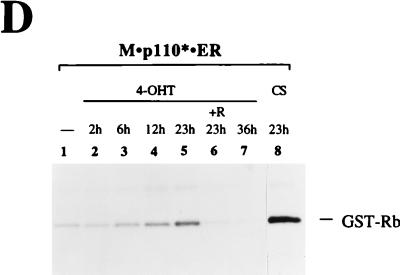
PI 3-kinase stimulation results in increased Cdk activity. (A) Time course study of expression levels and responsiveness to 4-OHT. Parental 3Y1 cells and M · p110* · ER and Myc · ER transfectants were stimulated with 200 nM 4-OHT or 10% CS for the indicated times; the addition of 20 ng of rapamycin per ml is indicated (+R). Cell extracts were analyzed by immunoblotting using anti-p110 (upper portion) or anti-pp70 S6 kinase (lower portion) antibodies. The positions of M · p110* · ER and endogenous p110 are indicated on the right. Positions of molecular size markers (in kilodaltons) are shown on the left. The nuclear localization of Myc · ER precluded its extraction during cell lysis, since the conditions used had to preserve Cdk activities. (B) Cdk2 activity assay. Cdk2 was precipitated from the cell extracts with an anti-Cdk2 antibody, and its kinase activity was analyzed in an in vitro kinase reaction, with histone H1 as a substrate. The reaction mixtures were separated by SDS–16% PAGE, and the incorporation of radiolabeled phosphate into histone H1 was monitored by autoradiography. In lanes 8 and 15 the antigenic peptide (pep) was used as a competitor at 400 ng/ml. The position of histone H1 is indicated on the right. (C) Relative amounts of Cdk2 in the precipitates. Half of the complexes analyzed in panel B were tested for Cdk2 protein levels by anti-Cdk2 immunoblotting. The numbers above each lane correspond to the numbers of the reactions shown in panel B. The position of Cdk2 is indicated on the right. (D) Cdk4 activity assay. M · p110* · ER-expressing cells were treated as described for panel A. Cdk4 was precipitated from cell extracts, and its kinase activity was analyzed with GST-Rb as a substrate. The reaction mixtures were separated by SDS–12% PAGE, and the phosphorylation of GST-Rb was monitored by autoradiography. The position of GST-Rb is indicated on the right. Half of the immunocomplexes were analyzed for Cdk4 protein levels, which were comparable in all samples (data not shown). Five experiments using different M · p110* · ER cell lines were carried out. A representative experiment is shown.
We also investigated whether PI 3-kinase activation can induce the activation of Cdk4. Cdk4 was precipitated from cell lysates, and Cdk4 activity assays were performed with GST-Rb as a substrate (52, 53). Cdk4 activation showed a time course similar to that of Cdk2 activation following 4-OHT treatment. Maximal activation was observed after 22 h of 4-OHT stimulation, and Cdk4 activation was blocked by rapamycin treatment (Fig. 7D). Prolonged activation of PI 3-kinase resulted in a decrease in Cdk4 activity, as observed for Cdk2 above.
The time course of Cdk activation is abnormal after PI 3-kinase activation.
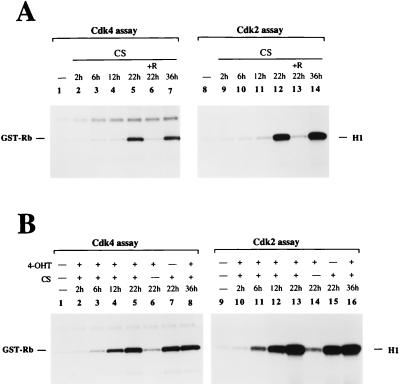
We compared the time courses for the activation of Cdk4 and Cdk2 induced by PI 3-kinase with those induced by serum stimulation (Fig. 8A). The onset of serum-induced Cdk4 and Cdk2 activation (between 12 and 22 h) appeared delayed compared to that induced by 4-OHT (6 to 12 h; Fig. 7). However, serum caused a much greater increase in the Cdk activities, which peaked around 22 h. Cdk4 and Cdk2 activation by serum was also rapamycin sensitive. In contrast to the PI 3-kinase-mediated response, which had disappeared at 36 h, the serum response remained high at 36 h.
FIG. 8.
Cdk4 and Cdk2 activation in cells stably expressing M · p110* · ER after serum stimulation and after stimulation with serum plus 4-OHT. (A) Time course of Cdk4 and Cdk2 activation in response to stimulation with serum. Cells were stimulated with 10% CS for the indicated times, and Cdk activation was analyzed as described in the legend for Fig. 7. R, rapamycin. (B) Time course of Cdk4 and Cdk2 activation in response to treatment with a combination of 4-OHT and serum. Cells were stimulated in the presence of 200 nM 4-OHT and 10% CS at various time points; in lanes 6 and 14 only 4-OHT was added, and in lanes 7 and 15 only CS was added. Cdk activity was analyzed as described above. The presence of comparable protein levels in the anti-Cdk4 and anti-Cdk2 precipitates was monitored by Western blotting (data not shown). Two experiments using different M · p110* · ER cell lines were performed. A representative experiment is shown.
Finally, activation of M · p110* · ER in combination with serum stimulation caused an early onset of Cdk activation (6 to 12 h), which remained high even after 36 h (Fig. 8B). The combination of 4-OHT and serum stimulation appeared to induce a greater response than that observed with either stimulus alone. These data are consistent with the observation that the activation of PI 3-kinase in combination with serum stimulation leads to cellular changes that resemble those of transformation.
The amount of the cyclin E-Cdk2 complex does not oscillate after PI 3-kinase stimulation.
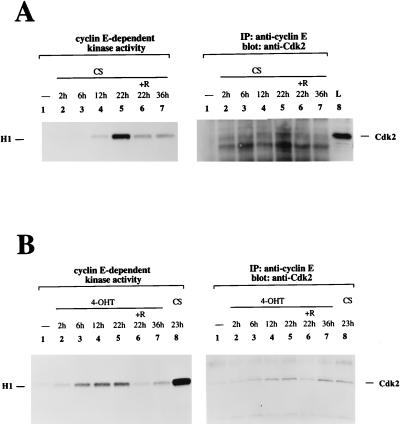
Our results have shown that stimulation of PI 3-kinase is sufficient for transition into S phase but not for progression through the entire cell cycle. The cells subsequently die by undergoing apoptosis. It is possible that the cells undergo apoptosis because they cannot exit from S phase. The transient formation of cyclin E-Cdk2 complexes can serve as a marker for S-phase progression: the cyclin E-Cdk2 complex appears during the transition from G1 to S phase; then, after S-phase entry, cyclin E is rapidly degraded (71). Therefore, we examined cyclin E-dependent kinase activity in cells treated with 4-OHT or with serum (Fig. 9). Cyclin E-dependent kinase activity decreased after its peak at 22 h of serum stimulation, and the Cdk2 protein levels detected in the anti-cyclin E precipitate also decreased after peaking at 22 h (Fig. 9A), presumably because cyclin E is degraded after cells enter S phase. Cyclin E-dependent kinase activity was increased at 6 h after activation of M · p110* · ER, with a peak at 22 h, and had returned to background levels at 36 h (Fig. 9B). However, after 36 h of stimulation with 4-OHT the Cdk2 protein levels in the anti-cyclin E precipitates remained high. Also, 4-OHT-induced cyclin E-dependent kinase activation stayed submaximal compared to the serum-induced activation, although the protein levels were comparable after 22 h of stimulation (Fig. 9B). The results show that the cyclin E-dependent kinase response, which oscillates after growth factor activation, is abnormal when PI 3-kinase is the only stimulus. The finding that the cyclin E-Cdk2 complex was not downregulated indicates that activation of PI 3-kinase alone cannot provide for further progression through the cell cycle after S-phase entry.
FIG. 9.
Comparison between cyclin E-dependent kinase activity induced by serum and that induced by PI 3-kinase activation. (A) Cyclin E-dependent kinase activity and the level of the cyclin E-Cdk2 complex oscillate in response to serum stimulation. Cells were stimulated in the presence of 10% CS for the indicated times. Cyclin E complexes were precipitated from cell extracts and subjected to an in vitro protein kinase assay using histone H1 as a substrate. The reaction mixtures were separated by SDS-PAGE, and histone H1 phosphorylation was detected by autoradiography (left portion). The position of histone H1 is shown on the left. Half of the anti-cyclin E complexes were analyzed for the presence of Cdk2 by immunoblotting (right portion). Lane L shows a control sample for which cell lysate was used to identify the position of Cdk2, which is indicated on the right. R, rapamycin; IP, immunoprecipitate. (B) Selective activation of PI 3-kinase induces an early onset of cyclin E-dependent kinase activity and does not provide for a downregulation of the cyclin E-Cdk2 complex. Cells were stimulated with 4-OHT for the indicated times, and control samples were treated with CS. Cell extracts were analyzed in parallel for cyclin E-dependent kinase activity and for Cdk2 levels in the precipitates as described for panel A. Two experiments using different M · p110* · ER cell lines were performed. A representative experiment is shown.
DISCUSSION
In this study, we describe the effects of activating PI 3-kinase independent of other signaling pathways on cell signaling and proliferation. We found that PI 3-kinase induces immediate-early responses such as the activation of the signaling kinases Akt and pp70 S6 kinase (Fig. 2). The activation of JNK, however, occurred hours later. Activation of PI 3-kinase was also sufficient for the induction of later responses which are associated with entry into the cell cycle, such as activation of Cdk4 and Cdk2 (Fig. 7) and the induction of DNA synthesis (Fig. 4). However, prolonged stimulation of PI 3-kinase resulted in a decrease in both Cdk activities and the rate of DNA synthesis. Cells responded to chronic activation of PI 3-kinase by undergoing apoptosis (Fig. 5 and Table 1). Our data demonstrate that activation of PI 3-kinase is sufficient to promote entry into the cell cycle. However, activation of PI 3-kinase appears to be insufficient to promote progression through the entire cell cycle. Additional signals are required to complement PI 3-kinase function, since serum stimulation rescues the cells from death. It is also possible that the chronic induction of PI 3-kinase is incompatible with normal cell growth and that PI 3-kinase needs to be inactivated for cells to progress through the cell cycle. Indeed, the accumulation of phospholipid products of PI 3-kinase is transient following growth factor stimulation (3), whereas prolonged activation of M · p110* · ER resulted in a persistent upregulation of 3′-phosphorylated phosphoinositides (Fig. 3). We have not been able to reverse the stimulatory effect of 4-OHT on M · p110* · ER in order to test this possibility.
Serum stimulation of M · p110* · ER-expressing cells rescued the apoptotic effect of chronic PI 3-kinase activation (Fig. 5A) and, in combination with stimulation by 4-OHT, induced cellular changes characteristic of oncogenic transformation: (i) cells stimulated with both serum and 4-OHT reached high cell densities and were irregular and refractile in appearance compared to cells cultured in serum alone (Fig. 5A); (ii) these cells had an increased rate of DNA synthesis compared to cells stimulated with either 4-OHT or serum alone (Fig. 5B); and (iii) they were able to form colonies in soft agar (Fig. 6). Furthermore, after serum and 4-OHT treatment, Cdk2 and Cdk4 activities both increased hours earlier than they did after treatment with serum alone (Fig. 8). The Cdk activities remained high after prolonged treatment, which is in contrast to what was found for 4-OHT treatment alone, where Cdk activities returned to background levels (Fig. 7 and 8). Our results suggest that activation of PI 3-kinase can contribute to the transformed phenotype but is not sufficient to transform cells by itself. In contrast, expression of chicken p110 has been reported to be sufficient to transform chicken embryo fibroblasts and to cause hemangiosarcomas in chickens (12). In our system membrane-localized p110 was not potent enough to induce any signaling response when expressed at levels comparable to that of endogenous p110; only membrane-localized p110* was able to induce signaling (Fig. 2A and B and 5B). It is possible that chicken fibroblasts are more permissive for transformation than the rat fibroblasts used here. In our system M · p110* · ER appears to abrogate the requirement for anchorage-dependent growth but not the requirement for growth factors for a complete progression through the cell cycle. Therefore, PI 3-kinase activation could be sufficient to induce a mitogenic response in a certain cellular context.
When PI 3-kinase was selectively activated, the timing of regulatory events appeared to be changed. PI 3-kinase stimulation induced the activation of Cdk4 and Cdk2 earlier than serum stimulation (Fig. 7 to 9). In addition, the Cdk activities decreased after prolonged activation of PI 3-kinase, whereas the serum-induced activation remained high. Also, the level of the cyclin E-Cdk2 complex, which normally oscillates during G1-to-S-phase transition, failed to be downregulated following induction by PI 3-kinase (Fig. 9). These results suggest that the S phase induced by PI 3-kinase may not be normal, and perhaps this causes the cells to undergo apoptosis. Normally, once cells enter S phase they are committed to proceed through the cell cycle (55, 72). After passing a “restriction checkpoint”, no further growth factor-mediated signals are required in order to finish the program. This restriction point is controlled by Cdk4-cyclin D (or Cdk6-cyclin D) and Cdk2-cyclin E complexes in G1. After S-phase entry, the cells have already advanced past the checkpoint and are committed for mitosis. In contrast, cells which entered S phase in response to PI 3-kinase activation were not able to progress through the cell cycle. This suggests that the S-phase transition induced by PI 3-kinase might be partial and therefore not productive. Consistent with this idea is the observation that the activation of Cdks and the rate of DNA synthesis induced by PI 3-kinase remained only partial compared to the same responses induced by serum. It is possible that induction of PI 3-kinase can only stimulate the formation of the cyclin-Cdk complexes and does not provide for their full activation by phosphorylation. This is suggested by the observation that the characteristic faster-migrating form of activated Cdk2 (71) was detectable in serum-, but not in 4-OHT-stimulated samples (Fig. 7C). Furthermore, DNA synthesis induced by PI 3-kinase was only comparable in efficiency to serum stimulation during the initial phase (Fig. 4A and B). The finding that a PI 3-kinase inhibitor blocked DNA synthesis induced by short-term serum stimulation but not by long-term stimulation (Fig. 4B) suggests that PI 3-kinase may be important in early S phase but not in the completion of S phase. Our data are in agreement with results described by Roche et al. (63) demonstrating that PI 3-kinase is required during the G1-to-S-phase transition for DNA synthesis induced by several growth factors.
It is unlikely that JNK activation mediates the apoptotic effect after prolonged activation of M · p110* · ER, since the cells showed a time course for JNK activity in response to serum stimulation similar to that in response to 4-OHT stimulation (Fig. 2C and data not shown). In addition, JNK activation decreased after 36 h of stimulation, before the onset of any detectable apoptosis.
Since the activation of PI 3-kinase in combination with serum stimulation caused cellular changes characteristic of transformation, it is possible that chronic activation of the PI 3-kinase pathway facilitates progression through the G1 phase by reducing the growth factor requirements as suggested for a number of oncogenes (48, 74). M · p110* · ER appeared to be more potent than the inducible forms of activated Akt (M · Akt · ER) or Myc (Myc · ER) in stimulating DNA synthesis (Fig. 4C). Despite its lower expression level M · p110* · ER was more potent in activating pp70 S6 kinase than M · Akt · ER (Fig. 4D), most likely because PI 3-kinase regulates S6 kinase activity by activating at least two pathways, one of which is Akt (1a, 23, 60, 83). Similarly, PI 3-kinase, but not Akt, mediates the invasiveness regulated by integrins (70). Myc on the other hand, appears to signal via pathways different from the ones regulated by PI 3-kinase. Myc · ER induced DNA synthesis, Cdk activation, and apoptosis (28, 47, 65) (Fig. 4 and 7; Table 1) but did not activate pp70 S6 kinase as did M · p110* · ER and M · Akt · ER (Fig. 4D). Further, Myc-induced apoptosis was not affected by rapamycin treatment, which interfered with PI 3-kinase-induced apoptosis (Table 1). This is in agreement with studies showing that Myc can induce apoptosis independent of the phase of the cell cycle (20, 28). Myc-induced apoptosis was more synchronized and more efficient than PI 3-kinase-induced apoptosis.
Recently, it has been suggested that activated PI 3-kinase promotes cell survival in response to several apoptotic stimuli (34, 35, 42, 59). However, here we demonstrated that prolonged activation of PI 3-kinase in the absence of serum results in apoptosis. Activated forms of PI 3-kinase were found to rescue cells from apoptosis following induction of signals by Myc overexpression or UV treatment. PI 3-kinase might be able to function as a survival factor if the pathways which are regulated by its activity can complement signaling responses induced by the other stimuli. Alternatively, a survival function for PI 3-kinase which was found by using transient expression systems might be a temporary response which does not reflect the effect of prolonged activation of PI 3-kinase.
The system described here, which allows for the selective activation of PI 3 kinase function, will enable us to determine which specific pathways cooperate with activated PI 3-kinase for cell cycle progression and to dissect the balance between proliferative responses and cell death. For example, it is possible that the induction of a pathway which promotes cell survival after prolonged activation of PI 3-kinase results in transformation. These and other experiments will help to elucidate the role of PI 3-kinase in regulating proliferation, oncogenic transformation, tumor metastasis, and cell survival.
ACKNOWLEDGMENTS
We thank Catherine Tribouley for generously providing the Myc · ER expression vector. We thank Laurie Goda for the speedy synthesis of oligonucleotides and Moijgan Amir-Ebrahimi and Jeff Tucker for DNA sequence analysis. We are grateful to Kang Dai, Bert Pronk, Christoph Reinhard, Kelly Smith, Anne Roulston, and Ning Lee for sharing their expertise on cell cycle regulation and apoptosis. We thank Kang Dai, Bert Pronk, Christoph Reinhard, Steve Harrison, A. B. Jefferson, Nicholas Marini, and especially Kelly Smith, Kevin Ramer, and Lisa Molz for many helpful comments on the manuscript.
REFERENCES
- 1.Alessi D R, James S R, Downes C P, Holmes A B, Gaffney P R, Reese C B, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 1a.Alessi D R, Kozlowski M T, Weng Q-P, Morrice N, Avruch J. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Cum Biol. 1998;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- 2.Andjelkovic M, Alessi D R, Meier R, Fernandez A, Lamb N J, Frech M, Cron P, Cohen P, Lucocq J M, Hemmings B A. Role of translocation in the activation and function of protein kinase B. J Biol Chem. 1997;272:31515–31524. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- 3.Auger K R, Serunian L A, Soltoff S P, Libby P, Cantley L C. PDGF-dependent tyrosine phosphorylation stimulates production of novel polyphosphoinositides in intact cells. Cell. 1989;57:167–175. doi: 10.1016/0092-8674(89)90182-7. [DOI] [PubMed] [Google Scholar]
- 4.Baim S B, Labow M A, Levine A J, Shenk T. A chimeric mammalian transactivator based on the lac repressor that is regulated by temperature and isopropyl beta-d-thiogalactopyranoside. Proc Natl Acad Sci USA. 1991;88:5072–5076. doi: 10.1073/pnas.88.12.5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellacosa A, Testa J R, Staal S P, Tsichlis P N. A retroviral oncogene, akt, encoding a serine-threonine kinase containing an SH2-like region. Science. 1991;254:274–277. doi: 10.1126/science.254.5029.274. [DOI] [PubMed] [Google Scholar]
- 6.Beretta L, Gingras A C, Svitkin Y V, Hall M N, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 7.Brown E J, Albers M W, Shin T B, Ichikawa K, Keith C T, Lane W S, Schreiber S L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 8.Burgering B M, Coffer P J. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 9.Cafferkey R, Young P R, McLaughlin M M, Bergsma D J, Koltin Y, Sathe G M, Faucette L, Eng W K, Johnson R K, Livi G P. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol Cell Biol. 1993;13:6012–6023. doi: 10.1128/mcb.13.10.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantley L C, Auger K R, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S. Oncogenes and signal transduction. Cell. 1991;64:281–302. doi: 10.1016/0092-8674(91)90639-g. . (Erratum, 65:914, 1991.) [DOI] [PubMed] [Google Scholar]
- 11.Carpenter C L, Cantley L C. Phosphoinositide kinases. Curr Opin Cell Biol. 1996;8:153–158. doi: 10.1016/s0955-0674(96)80060-3. [DOI] [PubMed] [Google Scholar]
- 12.Chang H W, Aoki M, Fruman D, Auger K R, Bellacosa A, Tsichlis P N, Cantley L C, Roberts T M, Vogt P K. Transformation of chicken cells by the gene encoding the catalytic subunit of PI 3-kinase. Science. 1997;276:1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 13.Cheatham B, Vlahos C J, Cheatham L, Wang L, Blenis J, Kahn C R. Phosphatidylinositol 3-kinase activation is required for insulin stimulation of pp70 S6 kinase, DNA synthesis, and glucose transporter translocation. Mol Cell Biol. 1994;14:4902–4911. doi: 10.1128/mcb.14.7.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung J, Grammer T C, Lemon K P, Kazlauskas A, Blenis J. PDGF- and insulin-dependent pp70S6k activation mediated by phosphatidylinositol-3-OH kinase. Nature. 1994;370:71–75. doi: 10.1038/370071a0. [DOI] [PubMed] [Google Scholar]
- 15.Chung J, Kuo C J, Crabtree G R, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 16.Cichy S, Uddin S, Danilkovich A, Guo S, Klippel A, Unterman T G. Protein kinase B/Akt mediates the effects of insulin and phosphatidylinositol 3-kinase on basal hepatic IGFBP-1 gene expression through a conserved insulin response sequence. J Biol Chem. 1998;273:6482–6487. doi: 10.1074/jbc.273.11.6482. [DOI] [PubMed] [Google Scholar]
- 17.Derijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 18.Didichenko S A, Tilton B, Hemmings B A, Ballmer-Hofer K, Thelen M. Constitutive activation of protein kinase B and phosphorylation of p47phox by a membrane-targeted phosphoinositide 3-kinase. Curr Biol. 1996;6:1271–1278. doi: 10.1016/s0960-9822(02)70713-6. [DOI] [PubMed] [Google Scholar]
- 19.Dulic V, Lees E, Reed S I. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 20.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 21.Franke T F, Kaplan D R, Cantley L C. PI3K: downstream AKTion blocks apoptosis. Cell. 1997;88:435–437. doi: 10.1016/s0092-8674(00)81883-8. [DOI] [PubMed] [Google Scholar]
- 22.Franke T F, Kaplan D R, Cantley L C, Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 23.Franke T F, Yang S I, Chan T O, Datta K, Kazlauskas A, Morrison D K, Kaplan D R, Tsichlis P N. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 24.Frech M, Andjelkovic M, Ingley E, Reddy K K, Falck J R, Hemmings B A. High affinity binding of inositol phosphates and phosphoinositides to the pleckstrin homology domain of RAC/protein kinase B and their influence on kinase activity. J Biol Chem. 1997;272:8474–8481. doi: 10.1074/jbc.272.13.8474. [DOI] [PubMed] [Google Scholar]
- 25.Frevert E U, Kahn B B. Differential effects of constitutively active phosphatidylinositol 3-kinase on glucose transport, glycogen synthase activity, and DNA synthesis in 3T3-L1 adipocytes. Mol Cell Biol. 1997;17:190–198. doi: 10.1128/mcb.17.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorman C. High efficiency gene transfer into mammalian cells. In: Glover D M, editor. DNA cloning, a practical approach. II. Oxford, United Kingdom: IRL Press; 1985. pp. 143–190. [Google Scholar]
- 27.Han J, Luby-Phelps K, Das B, Shu X, Xia Y, Mosteller R D, Krishna U M, Falck J R, White M A, Broek D. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science. 1998;279:558–560. doi: 10.1126/science.279.5350.558. [DOI] [PubMed] [Google Scholar]
- 28.Harrington E A, Bennett M R, Fanidi A, Evan G I. c-Myc-induced apoptosis in fibroblasts is inhibited by specific cytokines. EMBO J. 1994;13:3286–3295. doi: 10.1002/j.1460-2075.1994.tb06630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoang A T, Cohen K J, Barrett J F, Bergstrom D A, Dang C V. Participation of cyclin A in Myc-induced apoptosis. Proc Natl Acad Sci USA. 1994;91:6875–6879. doi: 10.1073/pnas.91.15.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Q, Klippel A, Muslin A J, Fantl W J, Williams L T. Ras-dependent induction of cellular responses by constitutively active phosphatidylinositol-3 kinase. Science. 1995;268:100–102. doi: 10.1126/science.7701328. [DOI] [PubMed] [Google Scholar]
- 31.Jefferies H B, Fumagalli S, Dennis P B, Reinhard C, Pearson R B, Thomas G. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones P F, Jakubowicz T, Pitossi F J, Maurer F, Hemmings B A. Molecular cloning and identification of a serine/threonine protein kinase of the second-messenger subfamily. Proc Natl Acad Sci USA. 1991;88:4171–4175. doi: 10.1073/pnas.88.10.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kapeller R, Cantley L C. Phosphatidylinositol 3-kinase. Bioessays. 1994;16:565–576. doi: 10.1002/bies.950160810. [DOI] [PubMed] [Google Scholar]
- 34.Kauffmann-Zeh A, Rodriguez-Viciana P, Ulrich E, Gilbert C, Coffer P, Downward J, Evan G. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature. 1997;385:544–548. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- 35.Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne P H, Downward J. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura G, Itagaki A, Summers J. Rat cell line 3y1 and its virogenic polyoma- and sv40-transformed derivatives. Int J Cancer. 1975;15:694–706. doi: 10.1002/ijc.2910150419. [DOI] [PubMed] [Google Scholar]
- 37.Klippel A, Escobedo J A, Hirano M, Williams L T. The interaction of small domains between the subunits of phosphatidylinositol 3-kinase determines enzyme activity. Mol Cell Biol. 1994;14:2675–2685. doi: 10.1128/mcb.14.4.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klippel A, Escobedo J A, Hu Q, Williams L T. A region of the 85-kilodalton (kDa) subunit of phosphatidylinositol 3-kinase binds the 110-kDa catalytic subunit in vivo. Mol Cell Biol. 1993;13:5560–5566. doi: 10.1128/mcb.13.9.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klippel A, Kavanaugh W M, Pot D, Williams L T. A specific product of phosphatidylinositol 3-kinase directly activates the protein kinase Akt through its pleckstrin homology domain. Mol Cell Biol. 1997;17:338–344. doi: 10.1128/mcb.17.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klippel A, Reinhard C, Kavanaugh W M, Apell G, Escobedo M-A, Williams L T. Membrane localization of phosphatidylinositol 3-kinase is sufficient to activate multiple signal-transducing kinase pathways. Mol Cell Biol. 1996;16:4117–4127. doi: 10.1128/mcb.16.8.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohn A D, Takeuchi F, Roth R A. Akt, a pleckstrin homology domain containing kinase, is activated primarily by phosphorylation. J Biol Chem. 1996;271:21920–21926. doi: 10.1074/jbc.271.36.21920. [DOI] [PubMed] [Google Scholar]
- 42.Kulik G, Klippel A, Weber M J. Antiapoptotic signalling by the insulin-like growth factor I receptor, phosphatidylinositol 3-kinase, and Akt. Mol Cell Biol. 1997;17:1595–1606. doi: 10.1128/mcb.17.3.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva N R, Hall M N. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 44.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 45.Lane H A, Fernandez A, Lamb N J, Thomas G. p70s6k function is essential for G1 progression. Nature. 1993;363:170–172. doi: 10.1038/363170a0. [DOI] [PubMed] [Google Scholar]
- 46.Li G, D’Souza-Schorey C, Barbieri M A, Roberts R L, Klippel A, Williams L T, Stahl P D. Evidence for phosphatidylinositol 3-kinase as a regulator of endocytosis via activation of Rab5. Proc Natl Acad Sci USA. 1995;92:10207–10211. doi: 10.1073/pnas.92.22.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Littlewood T D, Hancock D C, Danielian P S, Parker M G, Evan G I. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J J, Chao J R, Jiang M C, Ng S Y, Yen J J, Yang-Yen H F. Ras transformation results in an elevated level of cyclin D1 and acceleration of G1 progression in NIH 3T3 cells. Mol Cell Biol. 1995;15:3654–3663. doi: 10.1128/mcb.15.7.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marte B M, Downward J. PKB/Akt: connecting phosphoinositide 3-kinase to cell survival and beyond. Trends Biochem Sci. 1997;22:355–358. doi: 10.1016/s0968-0004(97)01097-9. [DOI] [PubMed] [Google Scholar]
- 50.Martin S S, Haruta T, Morris A J, Klippel A, Williams L T, Olefsky J M. Activated phosphatidylinositol 3-kinase is sufficient to mediate actin rearrangement and GLUT4 translocation in 3T3-L1 adipocytes. J Biol Chem. 1996;271:17605–17608. doi: 10.1074/jbc.271.30.17605. [DOI] [PubMed] [Google Scholar]
- 51.Martin S S, Rose D W, Saltiel A R, Klippel A, Williams L T, Olefsky J M. Phosphatidylinositol 3-kinase is necessary and sufficient for insulin-stimulated stress fiber breakdown. Endocrinology. 1996;137:5045–5054. doi: 10.1210/endo.137.11.8895379. [DOI] [PubMed] [Google Scholar]
- 52.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J Y. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol. 1994;14:2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ming X F, Burgering B M, Wennstrom S, Claesson-Welsh L, Heldin C H, Bos J L, Kozma S C, Thomas G. Activation of p70/p85 S6 kinase by a pathway independent of p21ras. Nature. 1994;371:426–429. doi: 10.1038/371426a0. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 55.Morgan D O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 56.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 57.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nimnual A S, Yatsula B A, Bar-Sagi D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- 59.Philpott K L, McCarthy M J, Klippel A, Rubin L L. Activated phosphatidylinositol 3-kinase and Akt kinase promote survival of superior cervical neurons. J Cell Biol. 1997;139:809–815. doi: 10.1083/jcb.139.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pullen N, Dennis P B, Andjelkovic M, Dufner A, Kozma S C, Hemmings B A, Thomas G. Phosphorylation and activation of p70s6k by PDK1. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 61.Reif K, Nobes C D, Thomas G, Hall A, Cantrell D A. Phosphatidylinositol 3-kinase signals activate a selective subset of Rac/Rho-dependent effector pathways. Curr Biol. 1996;6:1445–1455. doi: 10.1016/s0960-9822(96)00749-x. [DOI] [PubMed] [Google Scholar]
- 62.Reinhard C, Fernandez A, Lamb N J, Thomas G. Nuclear localization of p85s6k: functional requirement for entry into S phase. EMBO J. 1994;13:1557–1565. doi: 10.1002/j.1460-2075.1994.tb06418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roche S, Koegl M, Courtneidge S A. The phosphatidylinositol 3-kinase alpha is required for DNA synthesis induced by some, but not all, growth factors. Proc Natl Acad Sci USA. 1994;91:9185–9189. doi: 10.1073/pnas.91.19.9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodriguez-Viciana P, Warne P H, Vanhasenbroeck B, Waterfield M D, Downward J. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J. 1996;15:2442–2451. [PMC free article] [PubMed] [Google Scholar]
- 65.Rudolph B, Saffrich R, Zwicker J, Henglein B, Muller R, Ansorge W, Eilers M. Activation of cyclin-dependent kinases by Myc mediates induction of cyclin A, but not apoptosis. EMBO J. 1996;15:3065–3076. [PMC free article] [PubMed] [Google Scholar]
- 66.Sabatini D M, Erdjument-Bromage H, Lui M, Tempst P, Snyder S H. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 67.Sabers C J, Martin M M, Brunn G J, Williams J M, Dumont F J, Wiederrecht G, Abraham R T. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 68.Schultz A M, Henderson L E, Oroszlan S, Garber E A, Hanafusa H. Amino terminal myristoylation of the protein kinase p60src, a retroviral transforming protein. Science. 1985;227:427–429. doi: 10.1126/science.3917576. [DOI] [PubMed] [Google Scholar]
- 69.Serunian L A, Auger K R, Cantley L C. Identification and quantification of polyphosphoinositides produced in response to platelet-derived growth factor stimulation. Methods Enzymol. 1991;198:78–87. doi: 10.1016/0076-6879(91)98010-4. [DOI] [PubMed] [Google Scholar]
- 70.Shaw L M, Rabinovitz I, Wang H H, Toker A, Mercurio A M. Activation of phosphoinositide 3-OH kinase by the α6β4 integrin promotes carcinoma invasion. Cell. 1997;91:949–60. doi: 10.1016/s0092-8674(00)80486-9. [DOI] [PubMed] [Google Scholar]
- 71.Sheaff R J. Regulation of mammalian cyclin-dependent kinase 2. Methods Enzymol. 1997;283:173–193. doi: 10.1016/s0076-6879(97)83015-7. [DOI] [PubMed] [Google Scholar]
- 72.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 73.Sherr C J. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 74.Steiner P, Philipp A, Lukas J, Godden-Kent D, Pagano M, Mittnacht S, Bartek J, Eilers M. Identification of a Myc-dependent step during the formation of active G1 cyclin-cdk complexes. EMBO J. 1995;14:4814–4826. doi: 10.1002/j.1460-2075.1995.tb00163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stephens L, Anderson K, Stokoe D, Erdjument-Bromage H, Painter G F, Holmes A B, Gaffney P R, Reese C B, McCormick F, Tempst P, Coadwell J, Hawkins P T. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science. 1998;279:710–714. doi: 10.1126/science.279.5351.710. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 76.Stephens L R, Jackson T R, Hawkins P T. Agonist-stimulated synthesis of phosphatidylinositol(3,4,5)-trisphosphate: a new intracellular signalling system? Biochim Biophys Acta. 1993;1179:27–75. doi: 10.1016/0167-4889(93)90072-w. [DOI] [PubMed] [Google Scholar]
- 77.Stokoe D, Stephens L R, Copeland T, Gaffney P R, Reese C B, Painter G F, Holmes A B, McCormick F, Hawkins P T. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 78.Takebe Y, Seiki M, Fujisawa J-I, Hoy P, Yokota K, Arai K-I, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanti J F, Gremeaux T, Grillo S, Calleja V, Klippel A, Williams L T, Van Obberghen E, Le Marchand-Brustel Y. Overexpression of a constitutively active form of phosphatidylinositol 3-kinase is sufficient to promote Glut 4 translocation in adipocytes. J Biol Chem. 1996;271:25227–25232. doi: 10.1074/jbc.271.41.25227. [DOI] [PubMed] [Google Scholar]
- 80.Toker A, Cantley L C. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature. 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- 81.Traynor-Kaplan A E, Harris A L, Thompson B L, Taylor P, Sklar L A. An inositol tetrakisphosphate-containing phospholipid in activated neutrophils. Nature. 1988;334:353–356. doi: 10.1038/334353a0. [DOI] [PubMed] [Google Scholar]
- 82.Vlahos C J, Matter W F, Hui K Y, Brown R F. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 83.Weng Q P, Andrabi K, Klippel A, Kozlowski M T, Williams L T, Avruch J. Phosphatidylinositol 3-kinase signals activation of p70 S6 kinase in situ through site-specific p70 phosphorylation. Proc Natl Acad Sci USA. 1995;92:5744–5748. doi: 10.1073/pnas.92.12.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]