Summary
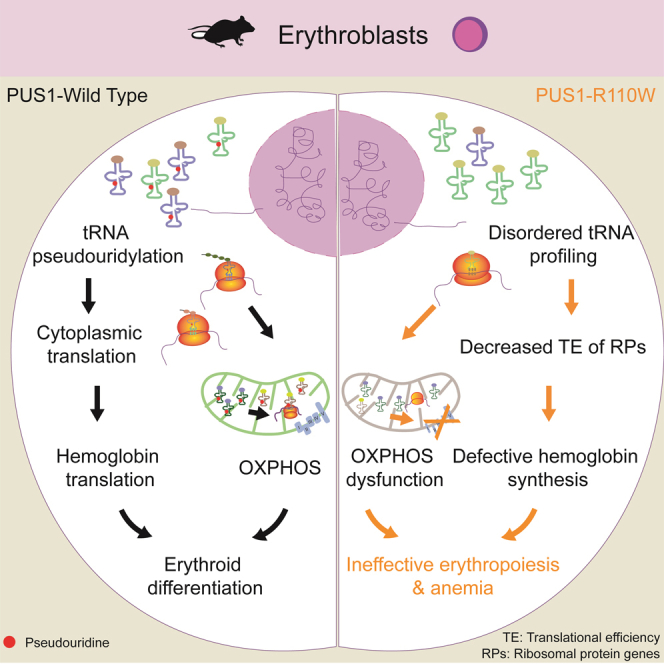
Pseudouridylation plays a regulatory role in various physiological and pathological processes. A prime example is the mitochondrial myopathy, lactic acidosis, and sideroblastic anemia syndrome (MLASA), characterized by defective pseudouridylation resulting from genetic mutations in pseudouridine synthase 1 (PUS1). However, the roles and mechanisms of pseudouridylation in normal erythropoiesis and MLASA-related anemia remain elusive. We established a mouse model carrying a point mutation (R110W) in the enzymatic domain of PUS1, mimicking the common mutation in human MLASA. Pus1-mutant mice exhibited anemia at 4 weeks old. Impaired mitochondrial oxidative phosphorylation was also observed in mutant erythroblasts. Mechanistically, mutant erythroblasts showed defective pseudouridylation of targeted tRNAs, altered tRNA profiles, decreased translation efficiency of ribosomal protein genes, and reduced globin synthesis, culminating in ineffective erythropoiesis. Our study thus provided direct evidence that pseudouridylation participates in erythropoiesis in vivo. We demonstrated the critical role of pseudouridylation in regulating tRNA homeostasis, cytoplasmic translation, and erythropoiesis.
Subject areas: Molecular biology, Cell biology
Graphical abstract
Highlights
-
•
PUS1R110W mutant mice (R110W) exhibited anemia and OXPHOS defects
-
•
R110W phenocopied the hematologic and mitochondrial features of patients with MLASA
-
•
Pseudouridine defects alter the expression of PUS1-targeted mt-tRNATyr and mt-tRNAIle
-
•
Decreased TE of RPs and globin synthesis in R110W lead to futile erythropoiesis
Molecular biology; Cell biology
Introduction
Pseudouridine (Ψ), also known as the fifth nucleotide, is the most abundant RNA modification.1,2 It was initially discovered in transfer RNAs (tRNAs) and plays an essential role in maintaining their stability, thereby regulating translation.3,4 The modification of Ψ in tRNA fragments has been shown to determine cell fate, and its dysregulation has been implicated in leukemogenesis.4,5 More recently, Ψ modification has also been identified in mRNA and long non-coding RNA,6,7,8 indicating its involvement in regulating gene expression through various mechanisms, including influencing alternative pre-mRNA processing9,10 and ensuring the fidelity of translation in ribosomal RNA (rRNA).11,12 Despite these significant findings, the roles of Ψ in a variety of biological processes remain to be fully understood.
The isomerization of uridine to pseudouridine can be catalyzed through either a small nucleolar RNA (snoRNAs)-dependent mechanism requiring the box H/ACA ribonucleoproteins,13 or through an RNA-independent mechanism involving the pseudouridine synthase (PUS) enzymes.14 PUS enzymes are classified into six families: RluA, RsuA, TruA, TruB, TruD, and Pus10.14,15,16 Mutations in pseudouridine synthases have been associated with various human diseases.17 Among these, human mitochondrial myopathy, lactic acidosis, and sideroblastic anemia (MLASA) was reported to be associated with mutations in PUS1,18 a member of TruA family. Previous studies have shown that PUS1 is responsible for pseudouridylation at various positions in several mitochondrial-tRNA and cytoplasmic-tRNA in mammals,6,19,20,21 some of which are involved in tRNA stability.22,23,24,25 To date, several PUS1 mutations have been reported in patients with MLASA, with the R144W mutation-where the arginine residue is replaced with tryptophan-being the most common.18,19 This missense mutation affects a highly conserved domain (RTDKGV) that contains the catalytic aspartate residue as the active site for the conversion of U-to-Ψ. In vitro assays have confirmed that the R144W mutant significantly reduced its pseudouridylation activity, probably by sterically blocking the active site,26 indicating that defective pseudouridylation of PUS1 targets may underlie the molecular mechanism of MLASA.
In addition to PUS1, two other genes implicated in MLASA are mitochondrial tyrosyl-tRNA synthetase (YARS2)27 and mitochondrial-DNA encoded ATP synthase membrane subunit 6 (MT-ATP6).28,29 Patients carrying pathogenic mutations in PUS1, YARS2, and MT-ATP6 exhibit similar symptoms, with mitochondrial dysfunction being a shared clinical manifestation in all patients with MLASA, suggesting that the disruption of mitochondrial function may be a common mechanism underlying MLASA. Mitochondria are not only crucial for energy production but also act as integration centers for various metabolic and biogenesis pathways. Interestingly, genes associated with congenital sideroblastic anemia (CSA), including MLASA, are mainly involved in mitochondrial metabolism, including heme biosynthesis, iron-sulfur cluster biogenesis, and mitochondrial respiratory chain biosynthesis.30 While the roles of heme and iron-sulfur cluster biosynthesis in iron metabolism and erythropoiesis are well-established, the exact mechanisms by which dysfunction in mitochondrial respiratory chain biosynthesis leads to CSA are incompletely understood.
Erythropoiesis is a tightly regulated and intricate process that begins in the bone marrow, arising from a multipotent stem cell and culminating in the formation of mature erythrocytes. Hematopoietic stem cells (HSCs) serve as the starting point in this process, giving rise to common myeloid progenitors (CMPs), which subsequently differentiate into megakaryocyte-erythroid progenitors (MEPs). MEPs subsequently generate erythroid-committed precursors (burst-forming unit-erythroid, BFU-E, and colony-forming unit-erythroid, CFU-E) and mature red blood cells (RBCs). The maturation process from BFU-E and CFU-E to mature RBCs is termed terminal erythropoiesis.31 Proerythroblasts undergo morphological changes, produce erythroid-specific proteins such as hemoglobin, and exhibit reduced proliferative capacity as they sequentially differentiate into basophilic, polychromatophilic, and orthochromatic erythroblasts to produce mature RBCs.32
RBCs need to respond rapidly and synthesize sufficient hemoglobin to meet environmental changes.32,33 Fine-tuning of translation control is essential to ensure that protein synthesis provides the necessary proteins for the physiological function of erythrocytes without excessive production. Thus, erythropoiesis evolves a comprehensive and multi-faceted regulatory network that involves mitochondrial biogenesis, ribosome abundance, translation initiation, and the balanced synthesis of heme and globin chains, and so forth.33 However, it remains unclear whether pseudouridine deficiency disrupts the homeostasis of globin chains and heme synthesis by altering the tRNA stability or abundance, ultimately leading to anemia.
In this study, we generated a mouse model carrying the PUS1 R110W mutation, which is designed to mimic the R144W mutation in MLASA patient. We found that R110W-mutant PUS1 protein maintained comparable levels and cellular distribution to the wild-type (WT) PUS1 protein, making it an ideal model for dissecting the enzymatic-dependent role of PUS1 in pseudouridylation and erythropoiesis. Our findings revealed that PUS1 R110W mutant mice exhibited defective pseudouridylation of specific tRNAs, impaired mitochondrial oxidative phosphorylation (OXPHOS), reduced cytoplasmic translation, and consequently developed anemia.
Results
Pseudouridine synthase 1 R110W mutant mice developed anemia at 4 weeks old
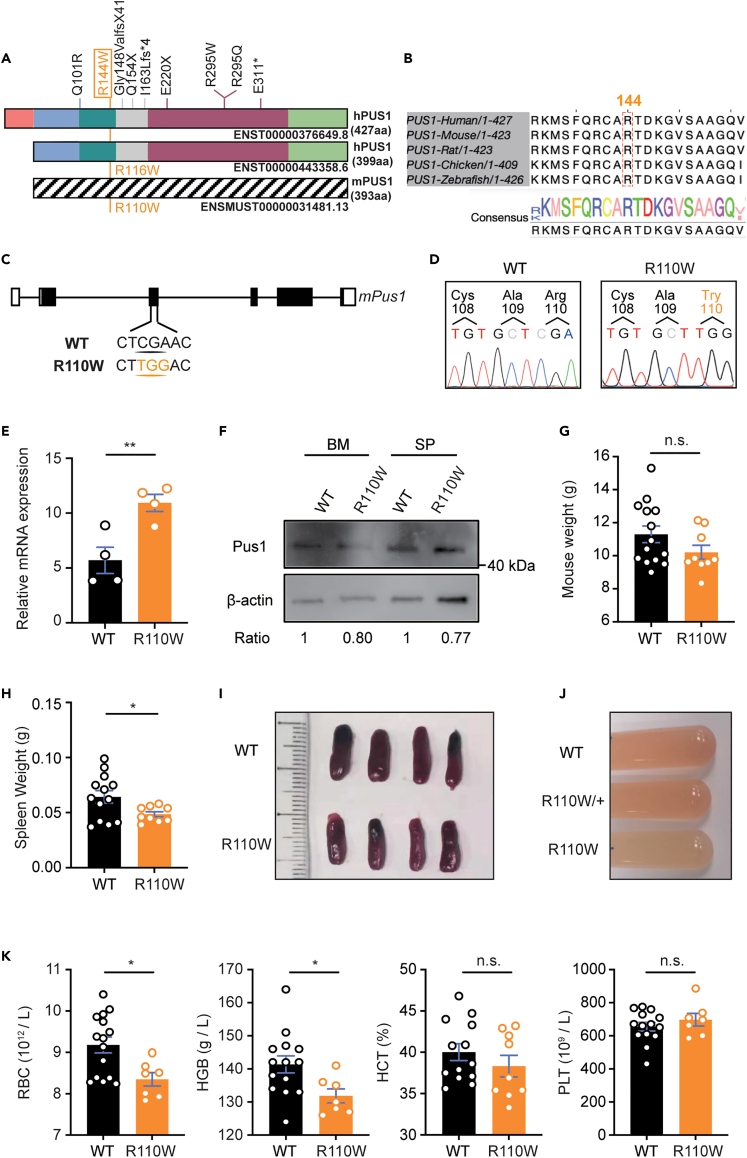
There are two isoforms of PUS1 protein in both humans and mice, with the shorter one located in the nucleus, whereas the longer one, which contains a mitochondrial targeting signal in mitochondria.34 The R144W mutation in human PUS1-427 (hPUS1-427aa) (corresponding to R116W in the 399aa short form of hPUS1-399aa) is the most frequent mutation observed in patients with MLASA (Figure 1A). The arginine residue (R144) in the enzymatic domain, which has an equivalent mutation in mice known as R110W (Figure 1A), is highly conserved across vertebrates (Figure 1B). To investigate the pathogenesis of the R144W mutation in MLASA, we generated Pus1-mutant mice by introducing a CGA to TGG substitution at position 444–446 in the Pus1 gene by CRISPR-Cas9 (referred to as R110W mice hereafter) (Figure 1C), and confirmed it by Sanger sequencing (Figure 1D). The RNA level of Pus1 in BM cells from R110W mice was higher than that in controls (Figure 1E), whereas the protein level of PUS1 in both BM and spleen cells was comparable between these two groups (Figure 1F). Further, immunofluorescence staining revealed similar co-localization of PUS1 with DAPI-labeled nuclei and Mitotracker-labeled mitochondria in both mutant and wild type (WT) cells, consistent with a previous report,19 suggesting that the cellular distribution of PUS1 was not affected by the R110W mutation (Figure S1A). Phenotypic analysis revealed that the body weights of 4-week-old R110W mice were slightly lower than controls (Figure 1G), while the weights and sizes of the spleen decreased significantly in R110W mice (Figures 1H and 1I). Additionally, the flushed BM cells from the lower limbs appeared paler in R110W mice than in controls (Figure 1J).
Figure 1.
R110W mice developed anemia at 4 weeks old
(A) Illustration of the two human PUS1 isoforms and the mutations reported in patients with MLASA. The hPUS1-427aa contains a mitochondrial targeting signal and is localized within mitochondria, while the hPUS1-399aa isoform lacks a mitochondrial targeting signal and is localized in the nucleus. The mutation R144W in hPUS1-427aa corresponds to R116W in hPUS1-399aa and R110W in mPUS1.
(B) The amino acid Arginine 144 (144R) in the enzymatic region of PUS1 is highly conserved across vertebrates. The alignment was performed using T-Coffee (https://tcoffee.crg.eu).
(C) Construction strategy of R110W mice using CRISPR-Cas9 technology with CGA replaced with TGG.
(D) Confirmation of the Pus1 mutation by the Sanger sequence of mouse genomic DNA. The location of this mutation is denoted in orange text. Primers used are listed in Table S1.
(E) Relative expression of Pus1 in whole BM cells of R110W mice and WT littermates normalized to 18s; n = 4.
(F) The protein level of mutant Pus1 in the bone marrow (BM) and spleen of mutant and wild-type (WT) littermates was analyzed by Western blotting. The ratio represents the fold change in expression between the two groups as measured by densitometric analysis.
(G) Body weights of the WT and R110W male mice at 4 weeks old (n = 14 for WT; n = 9 for R110W).
(H) Spleen weights of the WT and R110W male mice at 4 weeks old (n = 13 for WT; n = 9 for R110W).
(I) Representative images of the spleen in the WT and R110W male mice (n = 4).
(J) Representative images of BM cells flushed from WT littermates, heterozygous and homozygous mice of R110W.
(K) Complete blood count analysis of peripheral blood from 4-week-old R110W and WT male mice (n = 15 for WT; n = 7 for R110W). RBC, Red blood cells; HGB, Hemoglobin; HCT, Hematocrit; PLT, Platelets. Data are represented as Mean ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S1.
Complete blood count analysis revealed that R110W mice had lower levels of RBCs and hemoglobin compared with controls at 4 weeks old, while no obvious differences were observed in platelet numbers (Figures 1K and S1B). The proportions of peripheral blood (PB) cells were similar between R110W and control mice (Figure S1C), and the serum iron levels were also comparable (Figure S1D). These findings indicate that R110W mutation in PUS1 causes anemia in mice without affecting other hematopoietic lineages in a steady state.
Mutation in pseudouridine synthase 1 caused impaired erythropoiesis in an intrinsic manner
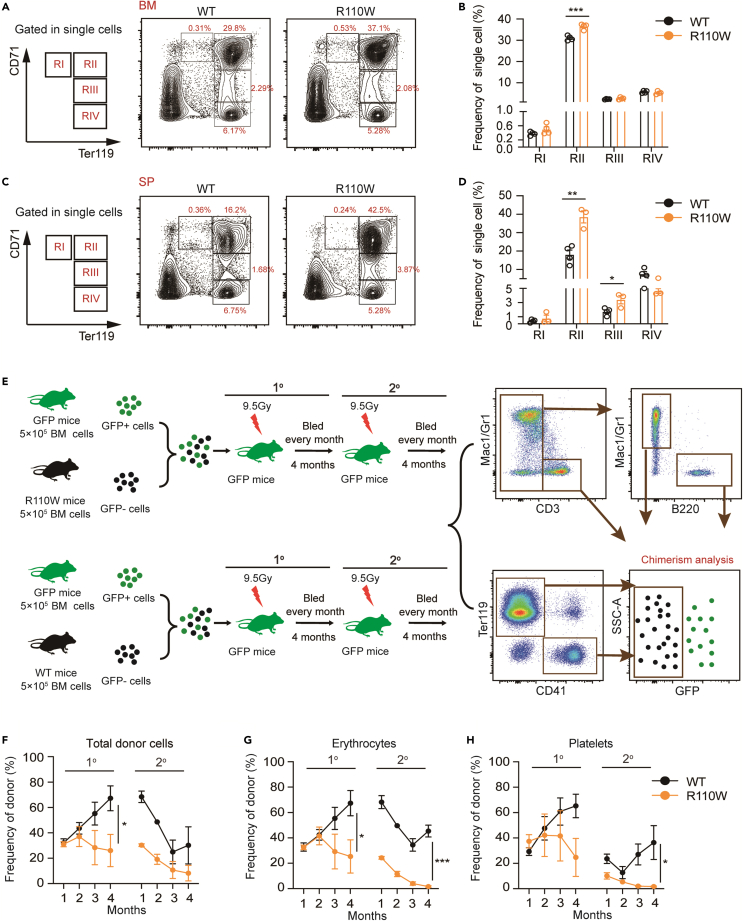
To explore whether anemia is caused by impaired erythropoiesis in R110W mice, we analyzed the frequencies of immature nucleated erythroblasts in the BM and spleen with the expression of CD71 and Ter119. CD71 (transferrin receptor) mediates the uptake of transferrin-iron complexes,35 and its expression increased from the proerythroblast stage, peaks in basophilic and polychromatophilic erythroblasts, decreases in orthochromatophilic cells, and is absent in mature erythrocytes.36 Although no ring sideroblasts were observed in R110W BM erythroblasts (Figure S1E), an increase in the proportion of basophilic erythroblasts and higher numbers of proerythroblasts and basophilic erythroblasts in R110W BM cells were observed (Figures 2A, 2B, and S2A). Similarly, elevated proportions of basophilic and chromatophilic erythroblasts were found in the R110W spleen (Figures 2C, 2D, and S2B). Furthermore, we found the mean fluorescence intensity (MFI) of CD71 in R110W mice was significantly higher than that of the controls (Figures S2C and S2D). To rule out the possibility that the increased CD71 level was due to ferroptosis,37,38 we also analyzed lipid peroxidation, another marker of ferroptosis.39 Lipid peroxidation levels were similar in both groups of erythroblasts (Figures S2E and S2F), suggesting the increased CD71 level was mainly caused by defective erythropoiesis rather than increased ferroptosis.
Figure 2.
Mutation of Pus1 impaired erythroid lineage cell commitment and differentiation in mice
(A) Flow cytometric gating strategy of erythroblasts in BM by CD71 and Ter119 staining. Region I (CD71highTer119intermediate) represents proerythroblasts, region II (CD71highTer119+) are basophilic erythroblasts, region III (CD71intermediateTer119+) reflects late basophilic and chromatophilic erythroblasts and region IV (CD71-Ter119+) are orthochromatophilic erythroblasts.41.
(B) The proportion of erythroblasts in BM at different stages during terminal erythropoiesis (n = 4, male mice).
(C) Flow cytometric gating strategy of erythroblasts in the spleen.
(D) The proportion of erythroblasts in the spleen at different stages during terminal erythropoiesis. (n = 4, male mice).
(E) Schematic diagram for competitive serial transplantation assay. 5×105 BM cells from R110W or control mice, together with 5×105 BM cells from actin-EGFP mice as protective cells, were transplanted into lethally irradiated actin-EGFP mice to evaluate the intrinsic regulation of PUS1 on hematopoiesis. Secondary transplantation was performed 4 months later.
(F) The total frequencies of donor cells derived erythrocytes (Ter119+) and platelets (CD41+) in the PB of recipient male mice at indicated time point during primary and secondary transplantation (n = 4).
(G‒H) The frequencies of donor cells derived mature erythrocytes (Ter119+) (G) and platelets (CD41+) (H) in the PB of recipient male mice during primary and secondary transplantation (n = 4). Data are represented as Mean ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figures S1–S4.
To further investigate whether impaired erythropoiesis originated at the stem cell level, we analyzed the proportion of hematopoietic stem and progenitor cells (HSPC) in Pus1-mutant mice. We found the frequencies of LSK+(Lin-Sca-1+c-Kit+) cells, LKS- (Lin-c-Kit+Sca-1-) cells, long-term HSCs, short-term HSCs, and multipotent progenitor cells were comparable between the two groups (Figures S3A‒S3D). The proportion of MEP cells decreased in the BM of R110W mice, while CMP and granulocyte-macrophage progenitor (GMP) remained unchanged between the two groups (Figures S3E and S3F). We further evaluated the functional BFU-E and CFU-E by colony-forming assay, and observed similar colony morphology and numbers of BFU-E (Figures S3G and S3H) and CFU-E (Figures S3I and S3J) in both groups. Taken together, these findings suggest that impaired erythropoiesis in R110W mice does not appear to originate from abnormalities at the stem cell level but rather occurs at later stages of erythropoiesis in a steady state.
To determine whether the defect in erythropoiesis is intrinsic or extrinsic, we employed transgenic GFP-positive mice as the recipients and the source of protective BM cells, which carried an EGFP transgene under the control of the Actin promoter, resulting in the labeling of all blood cell lineages, including erythrocytes, platelets, granulocytes, monocytes, and lymphocytes with EGFP. Under the transplantation conditions, the cells differentiated from protective cells were GFP-positive, while blood cells from BM cells of wild-type or mutant donor mice were GFP-negative, allowing us to quantify the percentage and absolute number of GFP-negative cells in the bone marrow and peripheral blood of recipient mice to evaluate the degree of chimerism. Briefly, we performed serial transplantation assays with actin-EGFP transgenic mice as donor mice, in which the hematopoietic cells were labeled as GFP+ (Figure S4A).40 We transplanted 5×105 BM cells from R110W or control mice together with 5×105 BM cells from actin-EGFP mice as protective cells into lethally irradiated actin-EGFP mice to evaluate the intrinsic regulation of PUS1 on hematopoiesis, particularly erythropoiesis. Secondary transplantation was performed 4 months later (Figure 2E). During the first transplantation, the total RBCs and platelet chimerism derived from R110W cells in the PB of recipients was extremely lower than those of controls (Figures 2F and S4B). Interestingly, this diminution of RBCs and platelets individually upon reconstitution was even more conspicuous during serial transplantation (Figures 2G and 2H). Furthermore, a significant decrease in the proportion of donor-derived erythroblasts was also observed in the BM of R110W recipients during secondary transplantation (Figure S4C). Surprisingly, lower reconstitution of myeloid cells during the first transplantation and T/B cells during secondary transplantation derived from R110W were observed (Figure S4D). These differences were not evident under steady-state conditions (Figure S1C), suggesting that the PUS1 R110W mutation specifically affects the multilineage differentiation potential of HSPCs under transplantation pressure. Taken together, these results indicate that the PUS1 R110W mutation not only impairs the commitment and differentiation of RBCs and platelets but also attenuates the self-renewal capacity and multilineage differentiation potential of HSCs under transplantation pressure.
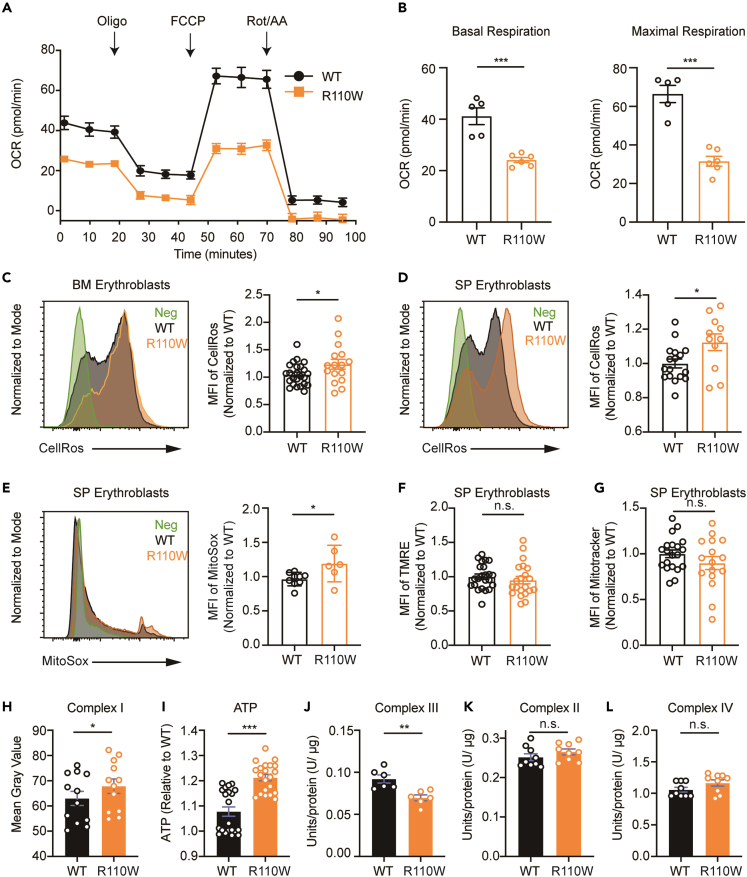
Pseudouridine synthase 1 mutation impaired the mitochondrial oxidative phosphorylation of bone marrow erythroblasts
To assess the impact of the R110W mutation on mitochondrial function, we measured mitochondrial OXPHOS levels using a Seahorse XF24 Extracellular Flux Analyzer. The results showed that the oxygen consumption rate of R110W BM erythroblasts, particularly the basal and maximum oxygen consumption rates, was significantly decreased, compared with controls (Figures 3A and 3B). We further examined the mitochondrial membrane potential (MMP), mitochondrial mass, mitochondrial superoxide (MitoSox), and cellular reactive oxygen species (ROS) levels in BM and spleen erythroblasts of the two groups. Erythroblasts from Pus1-mutant BM and spleen exhibited markedly elevated cellular ROS levels (Figures 3C and 3D). The MitoSox level was also increased in mutant spleen erythroblasts but not in BM erythroblasts (Figures 3E and S4E). Moreover, no significant differences in MMP and mitochondrial mass in erythroblasts were observed between the two groups (Figures 3F, 3G, S4F, and S4G).
Figure 3.
Pus1 mutation resulted in mitochondrial respiratory chain dysfunction
(A) Mitochondrial respiration in R110W and control BM erythroblasts measured by cellular OCR at baseline and after various inhibitors treatment. The OCR prior to the addition of oligomycin represents basal respiration. Mitochondrial ATP synthesis, proton translocation, and O2 uptake were measured after the addition of oligomycin (Oligo). Addition of carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP) defines maximal respiratory capacity, while rotenone plus antimycin A (Rot/AA) describes spare respiratory capacity (n = 6, male mice).
(B) OCR of basal respiration (left) and maximal respiratory capacity (right) from (A) (n = 6, male mice).
(C and D) Mean fluorescence intensity (MFI) peak (left) and values (right) of CellROS in BM (C) or SP (D) erythroblasts between WT and mutant male mice (n = 17–24 for WT; n = 11–17 for R110W).
(E) MFI peak (left) and values (right) of MitoSox in spleen erythroblasts between WT and mutant male mice (n = 9 for WT; n = 6 for R110W).
(F‒G) MFI values of TMRE (F) and mitotracker (G) in spleen erythroblasts between WT and mutant male mice (n = 20–24 for WT; n = 16–20 for R110W).
(H) Complex I activity of BM cells in the two groups, see the detailed protocol in the STAR methods (n = 12, male mice).
(I) ATP level of BM cells in the two groups, see the detailed protocol in the STAR methods (n = 6, each sample includes three or four replicates, male mice).
(J) Complex III activity of spleen cells in the two groups, see the detailed protocol in the STAR methods (n = 6, male mice).
(K‒L) Complex II (K) and complex IV (L) activity of BM cells in the two groups, see the detailed protocol in the STAR methods (n = 3, each sample includes three replicates, male mice). Data are represented as Mean ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S4.
To elucidate the underlying causes of abnormal mitochondrial OXPHOS in R110W mice, we assessed the enzyme activities of mitochondrial complex I-IV subunits and cellular ATP levels in BM or spleen cells. Compared with controls, complex I activity (Figure 3H) and ATP levels (Figure 3I) were increased in R110W mice. Conversely, complex III activity was decreased (Figure 3J). The enzymatic activities of complex II and IV remained unchanged (Figures 3K and 3L). These findings indicate that the R110W mutation in PUS1 specifically affects the function of certain mitochondrial OXPHOS complexes, which consequently contribute to the impaired mitochondrial oxygen consumption and elevated ROS levels observed in R110W mice.
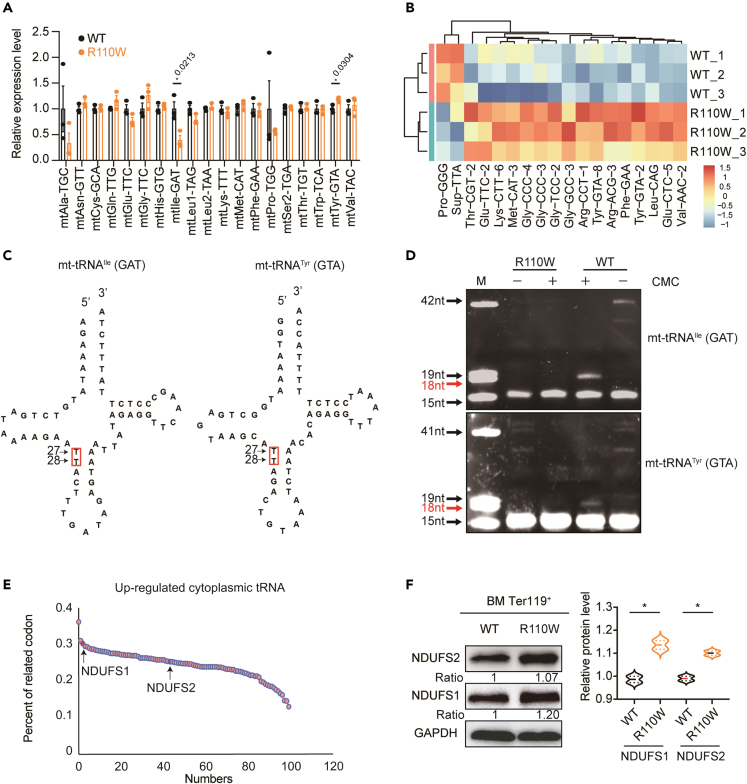
Pseudouridine synthase 1 R110W mutation altered mitochondrial and cytoplasmic transfer RNAs profiles
To gain further insights into the mechanisms underlying blocked erythropoiesis and mitochondrial dysfunction caused by the Pus1 mutation, we conducted an analysis of the overall tRNA profile with a tRNA PCR array, which allowed us to identify specific tRNAs that may be affected by Pus1 mutation. In the mitochondrial tRNA PCR array analysis, we observed decreased levels of mt-tRNAIle (GAT) and increased levels of mt-tRNATyr (GTA) in R110W BM erythroblasts in comparison with controls, while all other mitochondrial tRNAs remained unchanged (Figure 4A; Table S2). In contrast to the mitochondrial tRNAs, the levels of a large number of cytoplasmic tRNAs were changed. Among the 18 differentially expressed cytoplasmic tRNAs identified in the BM erythroblasts, 16 cytoplasmic tRNAs were significantly upregulated in R110W mice, while only two tRNAs were downregulated. Between these two decreased tRNAs, only tRNAPro (GGG) was involved in amino acid transport, while the other one was a suppressor tRNA (tRNA-Sup-TTA) (Figures 4B; Table S2).
Figure 4.
Pus1-mutant mice presented defective pseudouridylation of targeted tRNAs
(A) Expression levels of mitochondrial tRNA in BM erythroblasts of the two groups, all of the detected mitochondrial tRNAs were listed in the panel. (n = 3, female mice).
(B) Heatmap showing the expression level of cytoplasmic tRNAs with significant differences between the two groups (n = 3, female mice).
(C) Sequences and secondary structures of the mitochondrial tRNA with differential expression in R110W BM erythroblasts. Red boxes indicate putative Pus1 target sites that we examined by the CMC primer extension assay.
(D) CMC primer extension assay of mitochondrial tRNAIle (GAT) and tRNATyr (GTA). M, marker, a pool of indicated length of RNAs. Red arrows indicate sites that contain pseudouridine modifications in WT but are not found in the R110W cells. The detailed sequences used are shown in the supplementary materials.
(E) Nuclear-encoded mitochondrial OXPHOS genes ranked by the frequencies containing codons of the up-regulated cytosolic tRNAs in R110W cells. The arrows indicate the ranking of NDUFS1 and NDUFS2, respectively.
(F) Expression changes of nuclear-encoded mitochondrial respiratory chain-related proteins in R110W BM erythroblasts by Western blotting. Data are represented as Mean ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S5 and Table S2.
PUS1 modifies uridines at positions 27 and 28 in most cytoplasmic and mitochondrial tRNAs.42,43 To investigate whether the differentially expressed tRNAs were pseudouridine-modified by PUS1, we performed a CMC primer extension assay targeting positions 27 and 28 of two differentially expressed mitochondrial tRNAs (Figure 4C). Interestingly, both the upregulated mt-tRNATyr (GTA) and the downregulated mt-tRNAIle (GAT) contained Ψ at position 28 in WT cells, while it was absent in Pus1-mutant cells (Figure 4D). Besides, mt-tRNAHis(GTG)44 has been reported as a target of PUS1 in mice at positions 27 and 28, while no changes in expression were observed in mutant mice, suggesting a differential effect of PUS1 on the expression of tRNAs undergoing pseudouridylation.
Since the differentially expressed tRNAs may influence the translation of proteins that rely on their cognate codons, we calculated the cumulative proportion of codon usages corresponding to the changed mitochondrial or cytoplasmic tRNAs. Based on these values, we ranked the mitochondrial genome-encoded or nuclear-encoded OXPHOS proteins (Figures 4E and S5A). Despite the high codon usage of differentially expressed mt-tRNAs for mt-CYTB, no significant differences were observed in the protein level of mt-CYTB. Similarly, the protein levels of mt-ND3, mt-CO1, and mt-CO2 were comparable between R110W BM erythroblasts and controls (Figure S5B). This could be attributed to the combined effect of the upregulated and downregulated codon usages of tRNAs on the translation of the same protein.
In R110W BM erythroblasts, we observed elevated levels of nuclear-encoded complex I proteins, specifically NDUFS1 (ranking 3%) and NDUFS2 (ranking 43%) (Figure 4F). However, no significant changes were seen in Ter119- non-erythroblasts (Figure S5B). These findings suggest that the PUS1 mutation specifically affects the expression of nuclear-encoded complex I subunits in erythroblasts, which may contribute to the increased activity of complex I observed in our study (Figure 3H).
Overall, the Pus1 mutation disrupts mitochondrial and cytoplasmic tRNA profiles, leading to altered translational levels of specific genes involved in mitochondrial function.
R110W bone marrow erythroblasts exhibited decreased translation efficiency of ribosomal protein genes
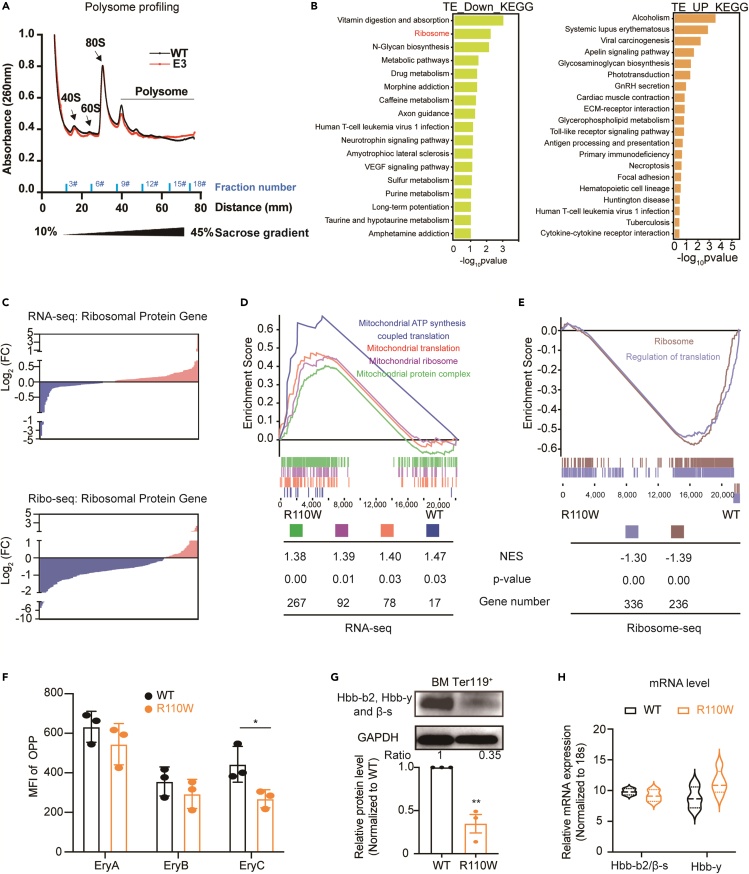
Since a considerable number of cytoplasmic tRNAs were altered in R110W, we speculated that the global translation of cytoplasmic protein may be affected. To characterize the translation dynamics resulting from the PUS1 mutation, polysome profiles of erythroblasts from WT and R110W mice were performed. The data showed that the PUS1 R110W mutation resulted in a slight decrease of 40S, 60S subunits and polysomes (Figure 5A), suggesting a mild defect of ribosome formation in R110W. The lower level of pre-ribosomal subunits (40S and 60S) can occur by different mechanisms, including impaired synthesis of ribosomal proteins, defective of rRNA transcription and the rRNA processing pathway, or disruptions of the ribosome assembly process.
Figure 5.
BM erythroblasts of Pus1-mutant mice showed decreased translation efficiency of ribosomal protein
(A) Polysome profiles were performed by fractionation on a 10 to 45% sucrose gradient. Fractions were collected and corresponding tube numbers were indicated, and the optical density at 260 nm (A260) was measured. The positions of the 40S and 60S native subunits and the 80S monosomes are indicated (n = 3, female mice).
(B) KEGG enrichment analysis of down-regulated translational efficiency genes (log2FC < −1, p < 0.05, left) and up-regulated translational efficiency genes (log2FC > 1, p < 0.05, right).
(C) RNA expression changes of ribosomal protein genes by RNA-seq (upper) and translational expression changes of ribosomal protein gene by Ribo-seq (bottom). Blue, downregulated genes; Pink, upregulated genes. Related genes are listed in Table S4.
(D) GSEA performed on all genes detected by RNA-seq. NES: normalized enrichment score.
(E) GSEA performed on all genes detected by Ribo-seq. NES: normalized enrichment score.
(F) MFI values of OPP incorporation in BM erythroblasts (n = 3, female mice). The gating strategy is shown in Figure S7B.
(G) Expression of Hbb-b2, Hbb-y, and β-s protein in BM erythroblasts (upper) by Western blotting and quantification (bottom).
(H) Relative mRNA level of Hbb-b2/β-s and Hbb-y (n = 3). Primers used are listed in Table S1. Data are represented as Mean ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figures S6 and S7.
To further elucidate the potential impact of the R110W on rRNA processing, fractions containing RNAs with varying sedimentation were collected into 18 tubes, as depicted in Figure 5A. The expression levels of mature and intermediate products of rRNA in the initial 6 tubes were analyzed with RT-qPCR, and no significant differences were observed in the expression of either mature or intermediate rRNA products compared to the control group (Figure S6A), suggesting the R110W mutation may have no effect on rRNA processing.
To further assess the impact of the PUS1 mutation on ribosomal protein translation, we performed RNA sequencing (RNA-seq) and ribosome profiling (Ribo-seq) on BM erythroblasts from R110W mice and controls (Figure S6B). We calculated the translational efficiency (TE) for each transcript and found that the number of genes with decreased TE was higher than that with increased TE in R110W (Figure S6C; Table S3). Further KEGG enrichment analysis revealed that the transcripts with decreased TE in R110W mice were enriched in the ribosome and metabolic-related pathways (Figure 5B). We further analyzed the differential TE of ribosomal protein genes using RNA-seq and Ribo-seq and determined whether the differential TE of ribosomal protein genes was influenced by transcription, translation, or both. We found that while the number of upregulated and downregulated ribosomal protein genes at the RNA level was comparable, a significant portion of these genes exhibited decreased translational levels (Figures 5C; Table S4), suggesting that the decreased TE of ribosomal protein genes was mainly caused by reduced translation. Gene Set Enrichment Analysis (GSEA) further supported these findings, showing the enrichment of mitochondrial translation, mitochondrial ribosome, and mitochondrial protein complex enriched in R110W erythroblasts in RNA-seq (Figure 5D). Conversely, translation and ribosome-related signaling pathways, such as the ribosome and regulation of translation, were inversely correlated with the R110W group in ribosome-seq (Figure 5E), confirming that the TE of ribosome-related proteins was downregulated in R110W mice.
O-propargyl-puromycin (OPP, an analog of puromycin) incorporation assay was further used to determine the cytoplasmic translation between R110W and control groups. The results showed that the MFI of OPP in LSK+ cells were increased in R110W mice, while it was comparable in Ter119+ erythroblasts between the two groups (Figure S7A). The OPP incorporation in the immature nucleated erythroblasts at different stages was further identified, and decreased OPP incorporation was observed in BM orthochromatic erythroblasts of R110W mice (Figures 5F and S7B). These findings suggest that the effect of PUS1 mutation on cytoplasmic translation varied during the developmental stages of hematopoietic cells, and with decreased protein synthesis in the late-stage erythroblasts of mutant mice. Given the high synthesis activity of hemoglobin proteins during erythropoiesis, which represents approximately 98% of the proteome,45 we investigated the protein levels of Hbb-b2, Hbb-y, and β-s, which are crucial components of hemoglobin. We found a striking decrease in hemoglobin protein levels in R110W erythroblasts (Figure 5G), while the transcriptional levels of these genes remained unchanged (Figure 5H).
Overall, these results suggest that the impact of the PUS1 mutation on translation may not be attributed to its effect on the rRNA maturation of ribosome biogenesis. The overall decrease in translational efficiency of ribosomal protein genes and the absence of tRNA pseudouridylation may be the reasons for the translation impairment caused by the PUS1 mutation. The widespread translational changes in R110W BM erythroblasts, with a particular impact on ribosomal protein synthesis, lead to reduced protein synthesis in the late-stage erythroblasts and impaired hemoglobin translation, which likely contributes to the impaired erythropoiesis observed in R110W mice.
Disturbances of oxidative phosphorylation and cytoplasmic translation collectively contribute to the pathogenesis of mitochondrial myopathy, lactic acidosis, and sideroblastic anemia syndrome
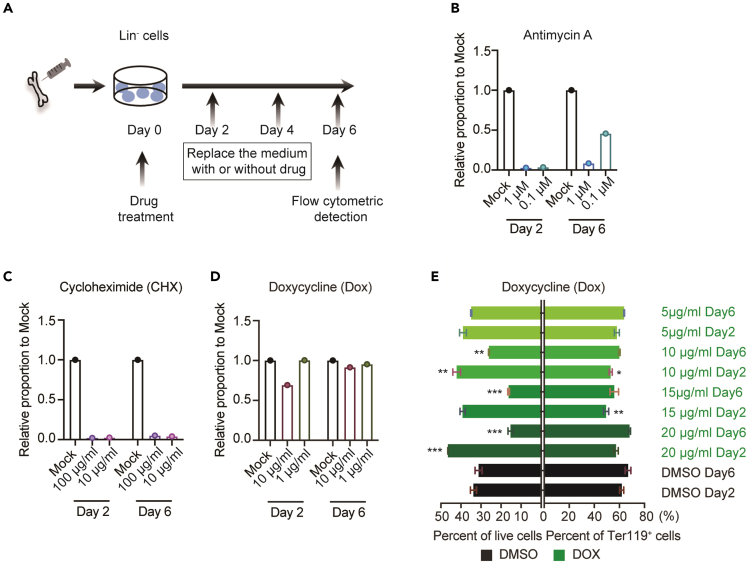
The findings of mitochondrial OXPHOS and mitochondrial and cytoplasmic translation abnormalities during erythroid differentiation in R110W mutant mice prompted us to further dissect which process may account for the defective erythropoiesis. BM Lin- cells from WT mice were isolated and cultured in an erythroid differentiation system with varying concentrations of different inhibitors of mitochondrial oxidative respiration for 48 h, and the ratio of differentiated erythrocytes (Ter119+) was measured on the second and sixth day of the differentiation process (Figures 6A and S8A). We found a concentration-dependent inhibition of erythroid differentiation by complex III inhibitor Antimycin A. Interestingly, the inhibitory effect of a low concentration of Antimycin A (0.1 μM) could be partially relieved by day 6 (Figures 6B and S8B), suggesting a reversible nature of the inhibition. These results indicate that the impairment of OXPHOS, specifically through the inhibition of complex III, plays a role in the defective erythropoiesis in R110W.
Figure 6.
Inhibition of the mitochondrial electron transport chain and cytoplasmic translation exerted inhibitory effects on erythropoiesis
(A) Strategy of in vitro erythropoiesis differentiation.
(B) Quantification of erythroid cells that were differentiated from sorted Lin- cells of WT mice with erythropoiesis differentiation culture medium in the presence or absence of antimycin A. Inhibitors were both incubated with cells for 48 h, and then the drugs were withdrawn.
(C and D) Quantification of erythroid cells that were differentiated from Lin- cells of WT mice cultured in liquid medium in the presence or absence of cycloheximide (C) or doxycycline (D), inhibitors both incubate with cells for 48 h.
(E) Quantification of live cells or Ter119+ erythroid cells at day 6 after differentiation by the treatment with doxycycline for 2 days or 6 days. Data are represented as Mean ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figures S8 and S9.
To assess the impact of translation inhibitors on erythroid differentiation, we treated Lin- cells during erythropoiesis with two different inhibitors for 48 h: cycloheximide (CHX), which inhibits cytoplasmic translation, and doxycycline (DOX), which inhibits mitochondrial translation. The results showed that both low and high concentrations of CHX significantly inhibited the differentiation of erythroid cells (Figures 6C and S8B). As for Dox, a high concentration exhibited a modest inhibitory effect on early differentiation (Day 2), but the proportion of Ter119+ cells recovered to a comparable level to that of DMSO control by day 6 (Figures 6D and S8B), suggesting a transient effect of mitochondrial translation inhibitor on erythropoiesis. We further increased the concentration range of DOX and prolonged the drug incubation for six days to examine the effect of DOX on erythropoiesis. We found that higher concentrations and longer incubation of DOX significantly reduced cell viability, but did not affect the erythroid differentiation (Figure 6E). These results suggest that cytoplasmic translation plays an important role in regulating erythropoiesis, whereas mitochondrial translation may have less of an effect. Taken together, our results highlight the importance of OXPHOS and cytoplasmic translation in erythroid differentiation and suggest that disturbances in these processes, rather than mitochondrial translation, may contribute to the defective erythropoiesis observed in MLASA.
It was reported that the targets of PUS1 also included mRNA and lncRNA.46,47 We referred to previous studies on PUS1 targeting mRNAs, highlighting genes related to OXPHOS and cytoplasmic translation in Figure S9A, to see whether the expression of these genes was changed in R110W groups. We analyzed the TE of these genes using RNA-seq and Ribo-seq data and found that only RPS8 exhibited significant differences between the two groups (Figure S9B). We further assessed the relevant protein levels of some selected genes. The results indicated that EIF4E2, EIF3H, EEF1A1, RPS8, and MT-CO1 showed no significant differences in protein levels between the two groups (Figure S9C).
These data suggest that despite the loss of PUS1 enzymatic activity, there were no notable alterations in the protein levels of PUS1 mRNA targets. Therefore, the reduced protein translation observed in PUS1 R110W erythropoietic cells may be not related to PUS1 targeted mRNA coding genes involved in the OXPHOS pathway or cytosolic protein regulation.
Discussion
In this study, we have presented compelling evidence demonstrating the involvement of pseudouridylation in erythropoiesis in vivo through the establishment of an MLASA mouse model harboring the most common enzymatic inactivating PUS1 mutation associated with patients with MLASA. Our findings in R110W mice, characterized by mitochondrial dysfunction and anemia, closely resemble the primary clinical manifestations observed in patients with MLASA. Importantly, we have identified that loss of tRNA pseudouridylation affects the TE of specific genes by modulating tRNA abundance, and reduces hemoglobin production, thereby leading to ineffective hematopoiesis. Furthermore, our study has revealed that the impairment of OXPHOS and cytoplasmic translation collectively contribute to the pathogenesis of MLASA.
RNA modifications play a critical role in controlling RNA functions and gene expression. Although some studies have suggested non-enzymatic role(s) of PUSs,3 most studies have utilized knockout or knockdown systems, which do not allow clear differentiation between the enzymatic-dependent or -independent functions of PUSs. In this study, we have established a unique mouse model carrying the enzymatically inactive R110W-mutant PUS1, which exhibits comparable protein level and cellular distribution to WT PUS1, thus providing an ideal model to dissect the enzymatic role of PUS1 in biological processes. Similar to patients with MLASA, the R110W mice displayed mild growth retardation but survive and manifested a specific defect in erythropoiesis, underscoring the relatively specific role of PUS1 mediated pseudouridine in erythropoiesis.
As a critical component of both mitochondrial and cytoplasmic translation machinery, tRNA are tightly regulated via various post-transcriptional modifications. Among these modifications, pseudouridine has been implicated in influencing tRNA structure, stability, and the translation efficiency.48 Interestingly, in our study, we made an intriguing observation that two newly identified PUS1-targeted mt-tRNAs mt-tRNATyr (GTA) and mt-tRNAIle (GAT) showed opposite expression patterns with depleted pseudouridine in R110W erythroblasts, suggesting the complex regulatory mechanism involving pseudouridine or other post-transcriptional modifications in regulating tRNA stability. Given the differential expression of these two tRNAs, it is not surprising that no obvious changes were observed in the level of mt-CYTB, which utilizes these two tRNAs in a high ratio of as cognate codons. Apart from its mitochondrial localization, PUS1 is also present in the nucleus and converts uridine into pseudouridine in several cytosolic tRNAs. In our study, the levels of numerous cytosolic tRNAs were altered by R110W-mutant PUS1. Despite the upregulation of two nuclear encoded mito-proteins, NDUFS1 and NDUFS2, the overall translation efficiency was reduced at the late stage of erythropoiesis, resulting in ineffective erythropoiesis in PUS1 mutant cells.
OXPHOS is a key energy producing process that involves coupling electron transport chain, while producing many essential intermediates for other metabolic pathways such as the tricarboxylic acid (TCA) cycle.49 OXPHOS dysfunction is a common characteristic observed in patients with MLASA and has been recapitulated in our MLASA mouse model and PUS1 knockout mice.44 However, the signaling link between OXPHOS and erythropoiesis remains elusive. Impaired OXPHOS causes disturbed energy production and increased ROS production due to electron transport disruption.50 It may also perturb related metabolites,51 such as succinyl-CoA, a substrate involved in iron metabolism and heme biosynthesis, thereby affecting erythropoiesis. We observed decreased OXPHOS and increased ROS levels in the erythroblasts of R110W mice, suggesting that oxidative stress, as an unfavorable environment cue, may be involved in the regulation of erythropoiesis. Using an in vitro erythroid differentiation system, we further demonstrated that OXPHOS inhibition significantly impeded the progression of erythroid differentiation, providing direct evidence for the impact of OXPHOS disruption on erythropoiesis.
During the late stage of erythropoiesis, translation becomes a pivotal biological process that is tightly regulated by various internal and external signals. For instance, when cellular levels of ROS accumulate or iron availability is limited, the rate of protein synthesis rate is attenuated. Ribosomal proteins play a crucial role in translation, and perturbations in these proteins, as observed in Diamond–Blackfan anemia (DBA), can lead to decreased protein synthesis.52,53 In our study, we also observed a reduction in cytoplasmic protein synthesis in R110W-mutant orthochromatic erythroblasts, resulting in decreased hemoglobin levels. This impaired protein synthesis may be the primary cause of the blockage in erythroid maturation, ultimately leading to anemia.
In summary, our study has uncovered the critical role of PUS1-mediated pseudouridylation in maintaining mitochondrial function during erythropoiesis. We have provided evidence that the catalytically inactivated R110W mutation in PUS1 leads to defective tRNA pseudouridylation and alters the abundance of both cytoplasmic and mitochondrial tRNAs. The disruption of cyto- and mito-tRNAs collectively results in impaired OXPHOS, reduced TE of ribosomal protein genes, and diminished hemoglobin translation in BM erythroblasts, all of which contribute to defective erythropoiesis. Our study suggests that modulating mitochondrial homeostasis and protein translation may hold promise as potential therapeutic strategies for congenital anemias and mitochondria-related anemia syndromes.
Limitations of the study
Although we provide direct evidence that pseudouridylation plays a role in erythropoiesis in vivo, further investigation into this mechanism in human samples would be valuable. Given the interspecies differences in iron metabolism, erythrocyte development, and mitochondrial homeostasis between humans and animal models, additional experiments are warranted to confirm the relevance of these findings to patients.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD71 (Transferrin Receptor) Monoclonal Antibody, clone R17217 (RI7 217.1.4), APC | eBioscience | Cat# 17-0711-82; RRID: AB_1834355 |
| TER-119 Monoclonal Antibody, clone TER-119, PE-Cyanine7 | eBioscience | Cat# 25-5921-82; RRID: AB_469661 |
| CD3e Monoclonal Antibody, clone 145-2C11, PE-Cyanine7 | eBioscience | Cat# 25-0031-82; RRID: AB_469572 |
| anti-mouse/human CD11b Antibody, clone M1/70, PE |
Biolegend | Cat# 101207; RRID: AB_312790 |
| Ly-6G/Ly-6C Monoclonal Antibody, clone RB6-8C5, PE | eBioscience | Cat# 12-5931-82; RRID: AB_466045 |
| CD45R (B220) Monoclonal Antibody, clone RA3-6B2, APC | eBioscience | Cat# 17-0452-82; RRID: AB_469395 |
| Rat Anti-Mouse CD41 Antibody, clone MWReg30, PE | BD Pharmingen | Cat# 558040; RRID: AB_397004 |
| anti-mouse CD4 Antibody, clone GK1.5, APC/Cyanine7 | BioLegend | Cat# 100414; RRID: AB_312699 |
| CD8a Monoclonal Antibody, clone 53-6.7, APC-eFluor 780 | eBioscience | Cat# 47-0081-82; RRID: AB_1272185 |
| CD117 (c-Kit) Monoclonal Antibody, clone 2B8, APC-eFluor 780 | eBioscience | Cat# 47-1171-82; RRID: AB_1272177 |
| Ly-6A/E (Sca-1) Monoclonal Antibody, clone D7, PE-Cyanine7 | eBioscience | Cat# 25-5981-82; RRID: AB_469669 |
| Rat Anti-Mouse CD34, clone RAM34, BV421 | BD Horizon | Cat# 562608; RRID: AB_11154576 |
| anti-mouse CD135 Antibody, clone A2F10, PE | BioLegend | Cat# 135305; RRID: AB_1877218 |
| CD16/CD32 Monoclonal Antibody, clone 93, PerCP-Cyanine5.5 | eBioscience | Cat# 45-0161-82, RRID: AB_996659 |
| CellROX™ Green Reagent, for oxidative stress detection | Invitrogen | Cat# C10444 |
| MitoSOX Mitochondrial Superoxide Indicators | Invitrogen | Cat# M36008 |
| MitoTracker™ Green FM | Invitrogen | Cat# M7514 |
| TMRE | Invitrogen | Cat# T669 |
| Anti-Ter-119 MicroBeads, mouse | Miltenyi biotech | Cat# 130-049-901; RRID: AB_2936424 |
| Anti-PUS1 (A-4) antibody | Santa Cruz biotechnology | Cat# sc-390043 |
| Anti-Ndufs1 antibody | Abcam | Cat# ab169540; RRID: AB_2687932 |
| Recombinant Anti-NDUFS2 antibody | Abcam | Cat# ab192022; RRID: AB_2895019 |
| Rabbit Anti-GAPDH Monoclonal Antibody | Cell Signaling Technology | Cat# 2118; RRID: AB_561053 |
| Anti-Hemoglobin β/γ/δ/ε antibody | Santa Cruz biotechnology | Cat# sc-390668 |
| MT-ND3 Rabbit pAb | ABclonal | Cat# A17969; RRID: AB_2861771 |
| MT-ND6 Rabbit pAb | ABclonal | Cat# A17991; RRID: AB_2861788 |
| MTCO1 Rabbit pAb | ABclonal | Cat# A17889; RRID: AB_2861744 |
| MTCO2 antibody | Proteintech | Cat# 55070-1-AP; RRID: AB_10859832 |
| CYTB antibody | Proteintech | Cat# 55090-1-AP; RRID: AB_2881266 |
| β-Actin (8H10D10) Mouse mAb | Cell Signaling Technology | Cat# 3700; RRID: AB_2242334 |
| EIF3H Rabbit pAb | ABclonal | Cat# A18026; RRID: AB_2861822 |
| EIF4E2 Polyclonal Antibody | ABclonal | Cat# A4305; RRID: AB_2765612 |
| eEF1A1 Rabbit mAb | ABclonal | Cat# A23515 |
| RPS8 Rabbit mAb | ABclonal | Cat# A21124 |
| Bacterial and virus strains | ||
| Trans1-T1 Phage Resistant Chemically Competent Cell | TransGen Biotech | Cat# CD501-03 |
| Chemicals, peptides, and recombinant proteins | ||
| N-cyclohexyl-N′-(2- morpholinoethyl) carbodiimide | Sigma | Cat# C106402 |
| Glycogen, RNA grade | Thermo Scientific | Cat# R0551 |
| TRIzol™ Reagent | Invitrogen | Cat# 15596018 |
| EDTA (0.5 M), pH 8.0, RNase-free | Invitrogen | Cat# AM9260G |
| Fast SYBR Green Master Mix | Clontech | Cat# 639676 |
| RNase H (5 U/μL) | Thermo Scientific | Cat# EN0202 |
| Holo-Transferrin, powder, BioReagent, suitable for cell culture, ≥97% | Sigma | Cat# T0665 |
| Bovine Serum Albumin, heat shock fraction, protease free, low endotoxin, suitable for cell culture, pH 7, ≥98% | Sigma | Cat# A4919 |
| Insulin-Transferrin-Selenium-Ethanolamine (ITS -X) (100X) | Gibco | Cat# 51500056 |
| Glutamax supplement | Gibco | Cat# 3505061 |
| Epo | 3SBIO INC | |
| Dexamethasone | Sigma | Cat# D2915 |
| Penicillin-Streptomycin | HyClone | Cat# SV30010 |
| Recombinant Murine SCF | peprotech | Cat# 250-03 |
| Antimycin A | Sigma | Cat# A8674 |
| Cycloheximide | Sigma | Cat# S7418 |
| Doxycycline monohydrate | Selleck | Cat# S6068 |
| FCCP | Selleck | Cat# S8276 |
| MethoCult™ SF M3436 | STEMCELL Technologies | Cat# 03436 |
| MethoCult™ M3334 | STEMCELL Technologies | Cat# 03334 |
| Critical commercial assays | ||
| DNeasy Blood & Tissue Kit | Qiagen | Cat# 69506 |
| Lineage Cell Depletion Kit | Miltenyi Biotec | Cat# 130-110-470 |
| PrimeScript™ II 1st Strand cDNA Synthesis Kit | Takara | Cat# 6210B |
| Evo M-MLV Mix Kit with gDNA Clean for qPCR | Accurate Biology | Cat# AG11728 |
| Pierce™ BCA Protein Assay Kit | Thermo Scientific | Cat# 23225 |
| Mitochondria Isolation Kit | Thermo Scientific | Cat# 89874 |
| Image-iT® Lipid Peroxidation Kit | Invitrogen | Cat# D3861 |
| Seahorse XF Cell Mito Stress Test Kit | Agilent | Cat# 103015-100 |
| Click-iT® Plus OPP Protein Synthesis Assay Kits | Invitrogen | Cat# C10457 |
| Iron Assay Kit | Sigma | Cat# MAK025 |
| Complex I Enzyme Activity Dipstick Assay Kit | Abcam | Cat# ab109720 |
| Succinate Dehydrogenase Assay Kit | Abcam | Cat# ab228560 |
| Mitochondrial Complex III Activity Assay Kit | Abcam | Cat# ab287844 |
| Cytochrome c Oxidase Assay Kit | Abcam | Cat# ab239711 |
| CellTiter-Glo® 2.0 Cell Viability Assay kit | Promega | Cat# G9241 |
| NucleoBond Xtra Midi Plus kit | MACHEREY-NAGEL | Cat# 740410.5 |
| Deposited data | ||
| nrStar™ Mouse tRNA PCR Array | This paper | Table S2 |
| RNA sequencing | This paper | GSA: CRA009788 |
| Ribosome profiling | This paper | GSA: CRA009788 |
| Experimental models: Organisms/strains | ||
| Mouse: Pus1R110W/R110W mice | This paper | N/A |
| Mouse: β-actin-GFP transgenic reporter mice (actin-EGFP mice) in a C57BL/6-Ly5.2 background | Tao Cheng’s lab | Dong F et al.54 |
| Oligonucleotides | ||
| PCR, qPCR and CMCT primer extension assay primers see Table S1 | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/www.graphpad.com/scientific-software/prism/; RRID: SCR_002798 |
| FlowJo | BD Biosciences | https://www.flowjo.com/solutions/flowjo/downloads; RRID: SCR_008520 |
| fastp (version 0.18.0) | Chen et al.55 | https://github.com/OpenGene/fastp; RRID: SCR_016962 |
| HISAT2 | Kim et al.56 | http://daehwankimlab.github.io/hisat2/; RRID: SCR_015530 |
| StringTie v1.3.1 | Pertea et al.57 | https://ccb.jhu.edu/software/stringtie/#install; RRID: SCR_016323 |
| DESeq2 | Love MI et al.58 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html; RRID: SCR_015687 |
| RiboWaltz | Lauria F et al.59 | https://github.com/LabTranslationalArchitectomics/riboWaltz; RRID: SCR_016948 |
| RSEM | Li et al.60 | https://github.com/deweylab/RSEM; RRID: SCR_013027 |
| Fiji | Schindelin, J et al.61 | https://imagej.net/software/fiji/downloads; RRID: SCR_002285 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yajing Chu (chuyajing@ihcams.ac.cn).
Materials availability
Mice line and all unique/stable reagents generated in this study are available on a reasonable request from the lead contact, Yajing Chu (chuyajing@ihcams.ac.cn).
Data and code availability
-
•
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA009788) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. These accession numbers are also listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Mice
CRISPR-Cas9 system was used to generate Pus1R110W/+ mice by creating a missense mutation in exon 3. Pus1R110W/+ mice were crossed to generate Pus1R110W/R110W (R110W). Genotyping of mice were determined with genomic DNA PCR followed by Sanger sequencing (BGI tech company). Primer sequences are listed in Table S1. Age- and gender-matched littermates were used as control. β-actin-GFP transgenic reporter mice (actin-EGFP mice) in a C57BL/6-Ly5.2 background were presented by Tao Cheng’s lab.54 8–12 weeks old actin-EGFP mice were served as donors and recipients after confirming GFP-positive cell rate in PB. All animals were bred in a specific-pathogen-free facility. All experiments were conducted under the institutional guidelines of the Institutional Animal Care and Use Committee of SKLEH. Mice used here were on C57BL/6-Ly5.2 (Ly5.2, CD45.2) genetic background.
Method details
tRNA PCR array
nrStar Mouse tRNA PCR Array was performed to determine the tRNA abundance by Aksomics company. Pellet enriched Ter119+ cells by centrifugation. Lyse cells in Trizol Reagent by repetitive pipetting. Then RNA extraction, RNA demethylation, first strand cDNA synthesis, real-time PCR were carried out to obtain the raw data of tRNA expression. The raw data was analyzed by ΔΔCt method and showed in Table S2.
RNA sequencing
Total RNA was extracted using Trizol reagent kit (Invitrogen) according to the manufacturer’s protocol. RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies) and checked by RNase-free agarose gel electrophoresis. mRNA was enriched by Oligo(dT) beads. The enriched mRNA was then fragmented into short fragments using fragmentation buffer and reversely transcribed into cDNA by using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB). The purified double-stranded cDNA fragments were end repaired, A-tailed, and ligated to Illumina sequencing adapters. The resulting cDNA library was sequenced using Illumina Novaseq6000 by Gene Denovo Biotechnology Co. (Guangzhou, China).
Reads were further filtered by fastp (version 0.18.0).55 After removing the rRNA mapped reads, paired-end clean reads were mapped to the reference genome using HISAT2.56 The mapped reads of each sample were assembled by StringTie57 v1.3.1 in a reference-based approach. For each transcription region, an FPKM (fragment per kilobase of transcript per million mapped reads) value was calculated to quantify its expression abundance. RNAs differential expression analysis was performed by DESeq2.58
Preparation and processing of Ribo-seq data
1 mL of pre-cooled PBS (containing 0.1 mg/mL cycloheximide) was added to the Ter119+ erythroblasts and mixed well, followed by the centrifugation. After repeated treatment with 1 mL of pre-cooled PBS (containing 0.1 mg/mL cycloheximide), the precipitates were frozen in liquid nitrogen and stored at −80°C.
Low-quality reads are filtered using fastp, followed by STAR for genome-wide alignments. Then, according to the position of the 5’end of the reads, the remaining ribosome footprints (RF) were aligned to different regions of the genome (5′ UTR, CDS, 3′UTR and intron). The R package “RiboWaltz” was used to calculate optimal P-site offsets.59 The counts of reads in the open reading frame of the coding gene were calculated using RSEM software60 and standardized using FPKM. Translational efficiency (TE) for each transcript was calculated by dividing normalized ribosome-protected fragment reads (as assessed by Ribo-seq) by the normalized RNA-seq reads. TE levels of Differentially expressed genes by Ribo-seq and RNA-seq are showed in Table S3.
BM competitive transplantation
For continuous competitive reconstitution assays, 5 × 105 BM cells from R110W mutant mice and their littermate controls were mixed with 5×105 GFP+ BM competitor cells and injected intravenously into lethally-irradiated EGFP recipient mice. Donor-derived chimerism (including RBC, Platelet, T cells, B cells and myeloid cells) of peripheral blood samples from recipient mice was analyzed at 4, 8, 12, 16 weeks after transplantation according to a previous study.62 Secondary transplantation was performed by transferring 1 × 106 BM cells of the primary recipient mice into lethally irradiated actin-EGFP mice. After 4 months, all recipient mice were sacrificed for donor-derived erythropoiesis and HSPC analysis.
Blood routine examination
Blood samples are harvested at different developmental stages of mice. The procedure for blood routine examination has been previously described.63,64 Briefly, 20 μL PB was collected from tail vein of mice into 1.5 mL tube containing 120 μL PBE (PBS with 2 mM EDTA and 2% FBS). Then the 1:7 diluted blood were measured and analyzed by XN-1000 V hematology analyzer (Sysmex).
RNA extraction
Total RNA was extracted using Trizol reagent (Invitrogen) following the manufacturer’s protocol, with Phasemaker tube (Invitrogen) to accommodate RNA extraction.
CMC labeling
Total RNA was CMC (N-cyclohexyl-N′-(2- morpholinoethyl) carbodiimide) labeled as previously described.65 Briefly, 3 - 10 μg total RNA in 12 μL RNase-free water was added to 24 μL of 1× TEU buffer (50 mM Tris-HCl (pH 8.3), 4 mM EDTA, 7 M urea), and 4 μL TEU buffer with (CMC+) or without (CMC-) 1 M CMC. The incubation was performed at 30°C for 16 h. After incubation, 160 μL of 50 mM KOAc (pH = 7), 200 mM KCl, 3 μL of 5 μg/μL glycogen, and 550 μL of ethanol were mixed with the samples, followed by > 2 h incubation at −80°C and centrifugation at 14 krpm for 30 min to remove excess CMC. After removing the supernatant, 500 μL of 75% ethanol was added to the pellet, kept at at −80°C for >2 h, and centrifuged at 14 krpm for 30 min. The precipitation needed to repeated once more. RNA pellets recovered from the CMC+ and CMC- samples were then separately dissolved in 40 μL of 50 mM Na2CO3, 2 mM EDTA (pH = 10.4) and incubated at 37°C for 6 h. The precipitation and washing procedure was then repeated.
Reverse transcription
After CMC labeling, RNA samples were dissolved in 8–15 μL Nuclease-free water, and the RNA concentration was determined by Nanodrop. Reverse transcription was carried out using PrimeScript II 1st Strand cDNA Synthesis Kit (Takara) following the manufacturer’s protocol. More than 1 μg of RNA was used as input for reverse transcription. Targeted tRNA primers were modified at the 5′ end with Cy3 (Sangon Biotech). When reverse transcription was complete, 1 μL of RNase H was added and incubated at 37°C for 20 min to remove RNA. Then, 75% ethanol was used to precipitate the cDNA.
Gel electrophoresis
The cDNA pellet was dissolved in 7 μL of TE buffer, mixed with 7 μL of 2 × TBE-Urea loading buffer (denaturating urea gel) (Real-Times Biotechnology), and incubated at 70°C for 5 min. The entire mixture was loaded onto a pre-run 22% denaturing urea gel containing 1 × TBE. The bands were determined using the Tanon imaging system at 605 nm.
In vitro erythropoiesis assay
This in vitro erythropoiesis assay was modified from a previously published article.66 Briefly, Lin− cells were enriched and plated on fibronectin-coated (2 g/cm2) Costar 24-well Clear TC-treated Multiple Well Plates (Corning) at a cell density of 1 × 105/mL in erythropoietic medium with drug or DMSO treatment. The erythropoietic medium consisted of Iscove’s modified Dulbecco’s medium (IMDM, Gibco) containing 15% FBS, 1% detoxified BSA (low endotoxin), 200 μg/mL holotransferrin, 10 μg/mL recombinant human insulin, 2 mM L-glutamine, 10−4 M (0.1 mM) β-mercaptoethanol, 50 units/ml penicillin G and 50 g/mL streptomycin, 10 units/ml Epo, 10 ng/mL SCF (R & D Systems, Minneapolis, MN), 10 μM dexamethasone (Sigma). After 48 h incubation, the erythropoietic medium was replaced with maintenance medium for 48 h including Iscove’s modified Dulbecco’s medium (IMDM, Gibco) supplemented with 20% FBS, 2 mM L-glutamine, 0.1 mM β-mercaptoethanol, 50 units/ml penicillin G and 50 g/mL streptomycin. Then, half of the medium was discarded and replaced with fresh maintenance medium. The drugs used are listed below, Antimycin A (Sigma, A8674), Cycloheximide (NSC-185) (Sigma, S7418), Doxycycline monohydrate (Selleck, S6068).
Immunoblotting
The Immunoblotting was performed as documented previously and underwent minor modifications.67 Enriched Ter119+ cell lysates were harvested by RIPA Buffer (Cell Signaling Technology), incubated on ice for 30 min, and 1×SDS-PAGE loading buffer was added. The samples were subjected to electrophoresis on 12.5% SDS-PAGE gels after heating them at 100°C for 10 min. Then, proteins were transferred to polyvinylidene difluoride (PVDF) membrane (Millipore) by wet-based transfer experiment (Bio-Rad). Immobilon Western Chemiluminescent HRP Substrate (Millipore) was used to detect HRP-labeled antibodies and the protein bands were detected with a ChemiDoc Imaging Systems machine (Bio-Rad) and quantified with ImageJ software. The antibodies used in this study are listed below. Anti-PUS1 (A-4) antibody (Santa Cruz biotechnology, sc-390043), Anti-Ndufs1 antibody (abcam, ab169540), Anti-Ndufs2 antibody (abcam, ab192022), Anti-GAPDH antibody (Cell Signaling Technology, 2118), Anti-Hemoglobin β/γ/δ/ε antibody (Santa Cruz biotechnology, sc-390668), Anti-ND3 antibody (abclonal, A17969), Anti-ND6 antibody (abclonal, A17991), Anti-CO1 antibody (abclonal, A17889), Anti-CO2 antibody (Proteintech, 55070-1-AP), Anti-CYTB antibody (proteintech, 55090-1-AP), Anti-β-Actin antibody (Cell Signaling Technology, 3700). The relative density was quantified using Fiji.61
Measurements of oxygen consumption rates
Oxygen consumption rates (OCRs) were measured using a Seahorse Bioscience Extracellular Flux Analyzer (XFe24) according to manufacturer’s instructions. Freshly isolated whole bone marrow cells of mice were enriched for the Ter119+ cells using magnetic cell sorting and 800,000 cells/well in 100 μL were immobilized to XFe24 plate (Agilent) pre-coated with Cell-Tak (Corning). The plate was incubated in an incubator at 37°C without CO2 for 40 min to equilibrate and 400 μL assay medium was added subsequently. Oxygen consumption measurements, taken every 10 min, were measured at baseline and after sequential addition of 1 μM oligomycin, 2 μM FCCP, and 1 μM rotenone/antimycin A.
Flow cytometry analysis and cell enrichment
Staining and washing procedures for flow cytometry have been previously described.68 The lineage cocktail markers used include CD3, CD4, CD8, B220, Mac1, Gr1 and Ter119. Cells were analyzed using an LSR II or Canto II instrument (BD Biosciences). Lin− cells and Ter119+ cells were enriched with Lineage microbeads and Ter119 microbeads (Miltenyi Biotec), respectively, according to the manufacturer’s instructions.
Mitochondrial mass and membrane potential measurement
Mitochondrial mass and mitochondrial membrane potential (MMP) were determined using flow cytometry as documented previously.69 Briefly, 1 × 106 BM or SP cells from mice were resuspended with 1mL pre-warmed PBS, incubated with cell surface antibody and 50 nM MitoTracker Green (Invitrogen) for 30 min at 37°C, and measured with 488 nm excitation. MMP was determined by TMRE staining. Cells were centrifuged to obtain a cell pellet and the supernatant was aspirated. Then, cells were gently resuspended in prewarmed (37°C) PBS. 200 nM TMRE (Invitrogen) and cell surface markers were added to cell suspension. For FCCP (carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone) control, 1 μL 50 μM FCCP (Selleck) was added to cell suspension for 15 min at 37°C followed by 200 nM TMRE staining. Cells were then washed, resuspended and analyzed on a BD LSR II or Canto II.
ROS and MitoSox detection
Mitochondrial superoxide and cellular ROS levels were assessed by staining with MitoSOX, CellROX (Invitrogen) and H2DCFDA (Invitrogen), respectively. For MitoSOX measurement, 1 × 106 BM or SP cells were resuspended in 500 μL PBS and incubated with cell surface markers and 0.5 μL 5 mM MitoSOX reagent stock solution. Incubations were performed in the dark for 20 min at RT, then cells were analyzed by flow cytometry. For CellROX and H2DCFDA measurement, cells were resuspended with 1 mL pre-warmed PBS and incubated with CellROX (2.5 μM final concentration) and H2DCFDA (10 μM final concentration) for 30 min at 37°C. Then cells were washed, resuspended and analyzed on BD LSR II or Canto II.
Lipid peroxidation measurement
Lipid peroxidation were determined using the Image-iT Lipid Peroxidation Kit (Thermo Fisher). Ter119+ cells were isolated from BM and SP cells of mice by magnetic-activated cell sorting (MACS). The Image-iT Lipid Peroxidation Sensor (Component A) was added to the cells at a final concentration of 10 μM. Cells were then incubated for 30 min at 37°C. For the positive control, cumene hydroperoxide (Component B) was added to the cells at a final concentration of 100 μM and incubated at 37°C for 2 h. Then, the Image-iT Lipid Peroxidation Sensor (Component A) was added at a final concentration of 10 μM for the last 30 min of incubation. Lipid peroxidation was determined by quantifying the fluorescence intensities in the FITC channel on a BD LSR II or Canto II instrument.
Cell separation by magnetic-activated cell sorting
BM and SP cells were collected and suspended in 3 mL PBE. After centrifugation at 1500 rpm for 8 min, cell pellets were harvested and resuspend in 90 μL of buffer per 107 total cells. 10 μL of anti Ter-119 MicroBeads (Miltenyl Biotec) per 107 total cells were added to the cell suspension and incubated for 15 min at 4 − 8°C. Cells were washed by adding 1–2 mL of PBS per 107 cells and centrifuged at 300 × g for 10 min. Cells were resuspended at 108/500 μL in PBS for magnetic separation by MS column (Miltenyl Biotec) or LS column (Miltenyl Biotec). Ter119+ and Ter119- Cells were harvested and collected for next experiment.
Iron detection
This protocol refers to the manufacturer’s documentation. Serum iron was analyzed using Iron Assay Kit from Sigma. Standards of 0, 2, 4, 6, 8, and 10 nmol/well were prepared as described in the protocol. Then, proper volumes of blood serum from mice were used to detected serum iron at the range of standards. Ferrous (Fe2+) iron and total iron were detected using a microplate reader by measuring absorbance at 593 nm. The level of ferric (Fe3+) iron was obtained by subtracting the ferrous iron from the total iron.
Colony formation assay
For BFU-E assay, 1 × 105 BM cells were plated in 24 - well plates containing 500 μL Methocult methylcellulose (Stem Cell Technologies, MethoCult SF M3436). The number of colonies formed on each plate was counted after culturing 10–12 days which was incubated at 37°C, in 5% CO2.
For CFU-E assay, 4 × 103 BM cells were plated in 24 - well plates containing 500 μL Methocult methylcellulose (Stem Cell Technologies, MethoCult M3334). The number of colonies formed on each plate was counted after 3–4 days culturing which was cultured at 37°C in a humidified atmosphere with 5% CO2.
Enzyme activity measurements
Activities of Complex I (NADH dehydrogenase), Complex II (succinate dehydrogenase), Complex III (ubiquinol-cytochrome c oxidoreductase complex), Complex IV (cytochrome c oxidase) were determined using Abcam’s Complex I Enzyme Activity Dipstick Assay Kit (abcam, ab109720), Succinate Dehydrogenase Assay Kit (abcam, ab228560), Mitochondrial Complex III Activity Assay Kit (abcam, ab287844), Cytochrome c Oxidase Assay Kit (ab239711) according to the manufacturer’s instructions, respectively.
ATP production determination
The CellTiter-Glo 2.0 Cell Viability Assay kit (Promega, G9241) was used for ATP determination according to the manufacturer’s instructions. Briefly, the reagents of this assay were equilibrated to room temperature before the experiment. The 100 μL detection reagent was added to 100 μL medium containing 1 - 2 × 106 cells, and mixed on the orbital shaker for 2 min to induce cell lysis. After incubation at room temperature for 10 min, the luminescence was read on a microplate reader (Syneregy H4, Bio-Tek). Different concentrations of ATP (Sigma-Aldrich, A7699) standards were prepared with the medium and determined together with the samples.
OPP protein synthesis assay
Nascent protein synthesis in gated cells was determined by the Click-iT Plus OPP Protein Synthesis Assay Kits (ThermoFisher). Briefly, 5 × 106 BM or SP cells were centrifuged and resuspended in 500 μL IMDM containing 10% fetal bovine serum (FBS). Click-iT OPP (Component A) was added to a final concentration of 20 μM along with FACS antibodies. Cells were incubated at 37°C for 60 min. Fixation was carried out by 500 μL 4% PFA at room temperature for 15 min. Cells were washed once with pre-warmed PBS. Permeabilization was performed using 500 μL 0.5% Triton X-100 in PBS at room temperature for 15 min. After washing cells with PBS, the Click-iT Plus OPP reaction cocktail was added to cells following the manufacturer’s protocol. Once the incubation was completed, cells were washed and resuspended in PBS, and analyzed using FACS canto II.
Quantification and statistical analysis
All data were analyzed using GraphPad Prism 8 and presented as Mean ± SEM. One-way ANOVA and unpaired Student's t-test were used for two groups and multiple-group comparisons, respectively. p < 0.05 was considered significant (asterisks indicate ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001).
Acknowledgments
This work was supported by funds from the Ministry of Science and Technology of China (2022YFA1103301 to WPY and YJC, 2020YFE0203000 to YJC); the National Natural Science Foundation of China (82150710556 and 82170135 to WPY, 82170117 to YJC, 82270145 to SJ); Natural Science Foundation of Tianjin City (21JCYBJC01170 to YJC); the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences, CIFMS (2021-I2M-1-040 to WPY, 2021-I2M-1-073 to SJ) and Haihe Laboratory of Cell Ecosystem Innovation Fund (22HHXBSS00037 to WPY and YJC).
We also appreciate the kind giving of β-actin-GFP transgenic reporter mice (actin-EGFP mice) from Tao Cheng’s laboratory. We also would like to thank all our colleagues at the State Key Laboratory of Experimental Hematology, Institute of Hematology and Blood Diseases Hospital, CAMS/PUMC.
Author contributions
YJC, JS, and WPY conceived the project, designed the study, and revised the article. DYS, BCW, and HYL performed the experiments, wrote the article. YL, QYM, MTC, and TL assisted with experiments and data analysis, YWM and LS contributed to the research design and article discussion. All authors reviewed the article.
Declaration of interests
The authors declare no competing interests.
Published: February 17, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109265.
Contributor Information
Weiping Yuan, Email: wpyuan@ihcams.ac.cn.
Jun Shi, Email: shijun@ihcams.ac.cn.
Yajing Chu, Email: chuyajing@ihcams.ac.cn.
Supplemental information
References
- 1.Cohn W.E. Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: isolation, structure, and chemical characteristics. J. Biol. Chem. 1960;235:1488–1498. [PubMed] [Google Scholar]
- 2.Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M., et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song J., Zhuang Y., Zhu C., Meng H., Lu B., Xie B., Peng J., Li M., Yi C. Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat. Chem. Biol. 2020;16:160–169. doi: 10.1038/s41589-019-0420-5. [DOI] [PubMed] [Google Scholar]
- 4.Guzzi N., Muthukumar S., Cieśla M., Todisco G., Ngoc P.C.T., Madej M., Munita R., Fazio S., Ekström S., Mortera-Blanco T., et al. Pseudouridine-modified tRNA fragments repress aberrant protein synthesis and predict leukaemic progression in myelodysplastic syndrome. Nat. Cell Biol. 2022;24:299–306. doi: 10.1038/s41556-022-00852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guzzi N., Cieśla M., Ngoc P.C.T., Lang S., Arora S., Dimitriou M., Pimková K., Sommarin M.N.E., Munita R., Lubas M., et al. Pseudouridylation of tRNA-Derived Fragments Steers Translational Control in Stem Cells. Cell. 2018;173:1204–1216.e26. doi: 10.1016/j.cell.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., Gilbert W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., León-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S., et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao B.S., He C. Pseudouridine in a new era of RNA modifications. Cell Res. 2015;25:153–154. doi: 10.1038/cr.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez N.M., Su A., Burns M.C., Nussbacher J.K., Schaening C., Sathe S., Yeo G.W., Gilbert W.V. Pseudouridine synthases modify human pre-mRNA co-transcriptionally and affect pre-mRNA processing. Mol. Cell. 2022;82:645–659.e9. doi: 10.1016/j.molcel.2021.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge J., Yu Y.T. RNA pseudouridylation: new insights into an old modification. Trends Biochem. Sci. 2013;38:210–218. doi: 10.1016/j.tibs.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma S., Entian K.D. Chemical Modifications of Ribosomal RNA. Methods Mol. Biol. 2022;2533:149–166. doi: 10.1007/978-1-0716-2501-9_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decatur W.A., Fournier M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 13.Czekay D.P., Kothe U. H/ACA Small Ribonucleoproteins: Structural and Functional Comparison Between Archaea and Eukaryotes. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.654370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rintala-Dempsey A.C., Kothe U. Eukaryotic stand-alone pseudouridine synthases - RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 2017;14:1185–1196. doi: 10.1080/15476286.2016.1276150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spenkuch F., Motorin Y., Helm M. Pseudouridine: still mysterious, but never a fake (uridine) RNA Biol. 2014;11:1540–1554. doi: 10.4161/15476286.2014.992278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamma T., Ferré-D'Amaré A.R. Pseudouridine synthases. Chem. Biol. 2006;13:1125–1135. doi: 10.1016/j.chembiol.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Shaheen R., Han L., Faqeih E., Ewida N., Alobeid E., Phizicky E.M., Alkuraya F.S. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum. Genet. 2016;135:707–713. doi: 10.1007/s00439-016-1665-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bykhovskaya Y., Casas K., Mengesha E., Inbal A., Fischel-Ghodsian N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA) Am. J. Hum. Genet. 2004;74:1303–1308. doi: 10.1086/421530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patton J.R., Bykhovskaya Y., Mengesha E., Bertolotto C., Fischel-Ghodsian N. Mitochondrial myopathy and sideroblastic anemia (MLASA): missense mutation in the pseudouridine synthase 1 (PUS1) gene is associated with the loss of tRNA pseudouridylation. J. Biol. Chem. 2005;280:19823–19828. doi: 10.1074/jbc.M500216200. [DOI] [PubMed] [Google Scholar]
- 20.Bohnsack M.T., Sloan K.E. The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease. Cell. Mol. Life Sci. 2018;75:241–260. doi: 10.1007/s00018-017-2598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borchardt E.K., Martinez N.M., Gilbert W.V. Regulation and Function of RNA Pseudouridylation in Human Cells. Annu. Rev. Genet. 2020;54:309–336. doi: 10.1146/annurev-genet-112618-043830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lecointe F., Namy O., Hatin I., Simos G., Rousset J.P., Grosjean H. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J. Biol. Chem. 2002;277:30445–30453. doi: 10.1074/jbc.M203456200. [DOI] [PubMed] [Google Scholar]
- 23.Alexandrov A., Chernyakov I., Gu W., Hiley S.L., Hughes T.R., Grayhack E.J., Phizicky E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 24.Tomita K., Ueda T., Watanabe K. The presence of pseudouridine in the anticodon alters the genetic code: a possible mechanism for assignment of the AAA lysine codon as asparagine in echinoderm mitochondria. Nucleic Acids Res. 1999;27:1683–1689. doi: 10.1093/nar/27.7.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adachi H., De Zoysa M.D., Yu Y.T. Post-transcriptional pseudouridylation in mRNA as well as in some major types of noncoding RNAs. Biochim. Biophys. Acta. Gene Regul. Mech. 2019;1862:230–239. doi: 10.1016/j.bbagrm.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sibert B.S., Fischel-Ghodsian N., Patton J.R. Partial activity is seen with many substitutions of highly conserved active site residues in human Pseudouridine synthase 1. RNA. 2008;14:1895–1906. doi: 10.1261/rna.984508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riley L.G., Cooper S., Hickey P., Rudinger-Thirion J., McKenzie M., Compton A., Lim S.C., Thorburn D., Ryan M.T., Giegé R., et al. Mutation of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia--MLASA syndrome. Am. J. Hum. Genet. 2010;87:52–59. doi: 10.1016/j.ajhg.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ardissone A., Lamantea E., Quartararo J., Dallabona C., Carrara F., Moroni I., Donnini C., Garavaglia B., Zeviani M., Uziel G. A Novel Homozygous YARS2 Mutation in Two Italian Siblings and a Review of Literature. JIMD Rep. 2015;20:95–101. doi: 10.1007/8904_2014_397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burrage L.C., Tang S., Wang J., Donti T.R., Walkiewicz M., Luchak J.M., Chen L.C., Schmitt E.S., Niu Z., Erana R., et al. Mitochondrial myopathy, lactic acidosis, and sideroblastic anemia (MLASA) plus associated with a novel de novo mutation (m.8969G>A) in the mitochondrial encoded ATP6 gene. Mol. Genet. Metab. 2014;113:207–212. doi: 10.1016/j.ymgme.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossmann M.P., Hoi K., Chan V., Abraham B.J., Yang S., Mullahoo J., Papanastasiou M., Wang Y., Elia I., Perlin J.R., et al. Cell-specific transcriptional control of mitochondrial metabolism by TIF1γ drives erythropoiesis. Science (New York, N.Y.) 2021;372:716–721. doi: 10.1126/science.aaz2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moras M., Lefevre S.D., Ostuni M.A. From Erythroblasts to Mature Red Blood Cells: Organelle Clearance in Mammals. Front. Physiol. 2017;8:1076. doi: 10.3389/fphys.2017.01076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caulier A.L., Sankaran V.G. Molecular and cellular mechanisms that regulate human erythropoiesis. Blood. 2022;139:2450–2459. doi: 10.1182/blood.2021011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vatikioti A., Karkoulia E., Ioannou M., Strouboulis J. Translational regulation and deregulation in erythropoiesis. Exp. Hematol. 2019;75:11–20. doi: 10.1016/j.exphem.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Fernandez-Vizarra E., Berardinelli A., Valente L., Tiranti V., Zeviani M. Nonsense mutation in pseudouridylate synthase 1 (PUS1) in two brothers affected by myopathy, lactic acidosis and sideroblastic anaemia (MLASA) J. Med. Genet. 2007;44:173–180. doi: 10.1136/jmg.2006.045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsee D.K., Pinkus G.S., Yu H. CD71 (transferrin receptor): an effective marker for erythroid precursors in bone marrow biopsy specimens. Am. J. Clin. Pathol. 2010;134:429–435. doi: 10.1309/ajcpcrk3moaoj6at. [DOI] [PubMed] [Google Scholar]
- 36.Grzywa T.M., Nowis D., Golab J. The role of CD71(+) erythroid cells in the regulation of the immune response. Pharmacol. Ther. 2021;228 doi: 10.1016/j.pharmthera.2021.107927. [DOI] [PubMed] [Google Scholar]
- 37.Feng H., Schorpp K., Jin J., Yozwiak C.E., Hoffstrom B.G., Decker A.M., Rajbhandari P., Stokes M.E., Bender H.G., Csuka J.M., et al. Transferrin Receptor Is a Specific Ferroptosis Marker. Cell Rep. 2020;30:3411–3423.e7. doi: 10.1016/j.celrep.2020.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X., Comish P.B., Tang D., Kang R. Characteristics and Biomarkers of Ferroptosis. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.637162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H., An P., Xie E., Wu Q., Fang X., Gao H., Zhang Z., Li Y., Wang X., Zhang J., et al. Characterization of ferroptosis in murine models of hemochromatosis. Hepatology. 2017;66:449–465. doi: 10.1002/hep.29117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan X.W., Liao H., Sun L., Okabe M., Xiao Z.C., Dawe G.S. Fetal microchimerism in the maternal mouse brain: a novel population of fetal progenitor or stem cells able to cross the blood-brain barrier? Stem Cell. 2005;23:1443–1452. doi: 10.1634/stemcells.2004-0169. [DOI] [PubMed] [Google Scholar]
- 41.Socolovsky M., Nam H., Fleming M.D., Haase V.H., Brugnara C., Lodish H.F. Ineffective erythropoiesis in Stat5a(-/-)5b(-/-) mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- 42.Czudnochowski N., Wang A.L., Finer-Moore J., Stroud R.M. In human pseudouridine synthase 1 (hPus1), a C-terminal helical insert blocks tRNA from binding in the same orientation as in the Pus1 bacterial homologue TruA, consistent with their different target selectivities. J. Mol. Biol. 2013;425:3875–3887. doi: 10.1016/j.jmb.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Behm-Ansmant I., Massenet S., Immel F., Patton J.R., Motorin Y., Branlant C. A previously unidentified activity of yeast and mouse RNA:pseudouridine synthases 1 (Pus1p) on tRNAs. RNA. 2006;12:1583–1593. doi: 10.1261/rna.100806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mangum J.E., Hardee J.P., Fix D.K., Puppa M.J., Elkes J., Altomare D., Bykhovskaya Y., Campagna D.R., Schmidt P.J., Sendamarai A.K., et al. Pseudouridine synthase 1 deficient mice, a model for Mitochondrial Myopathy with Sideroblastic Anemia, exhibit muscle morphology and physiology alterations. Sci. Rep. 2016;6 doi: 10.1038/srep26202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathangasinghe Y., Fauvet B., Jane S.M., Goloubinoff P., Nillegoda N.B. The Hsp70 chaperone system: distinct roles in erythrocyte formation and maintenance. Haematologica. 2021;106:1519–1534. doi: 10.3324/haematol.2019.233056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X., Zhu P., Ma S., Song J., Bai J., Sun F., Yi C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015;11:592–597. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- 47.Zhang M., Jiang Z., Ma Y., Liu W., Zhuang Y., Lu B., Li K., Peng J., Yi C. Quantitative profiling of pseudouridylation landscape in the human transcriptome. Nat. Chem. Biol. 2023;19:1185–1195. doi: 10.1038/s41589-023-01304-7. [DOI] [PubMed] [Google Scholar]
- 48.Shi H., Chai P., Jia R., Fan X. Novel insight into the regulatory roles of diverse RNA modifications: Re-defining the bridge between transcription and translation. Mol. Cancer. 2020;19:78. doi: 10.1186/s12943-020-01194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang J.X., Thompson K., Taylor R.W., Oláhová M. Mitochondrial OXPHOS Biogenesis: Co-Regulation of Protein Synthesis, Import, and Assembly Pathways. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21113820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zorov D.B., Juhaszova M., Sollott S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014;94:909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martínez-Reyes I., Chandel N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020;11:102. doi: 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choesmel V., Bacqueville D., Rouquette J., Noaillac-Depeyre J., Fribourg S., Crétien A., Leblanc T., Tchernia G., Da Costa L., Gleizes P.E. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109:1275–1283. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang B., Wang C., Wan Y., Gao J., Ma Y., Zhang Y., Tong J., Zhang Y., Liu J., Chang L., et al. Decoding the pathogenesis of Diamond-Blackfan anemia using single-cell RNA-seq. Cell Discov. 2022;8:41. doi: 10.1038/s41421-022-00389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dong F., Hao S., Zhang S., Zhu C., Cheng H., Yang Z., Hamey F.K., Wang X., Gao A., Wang F., et al. Differentiation of transplanted haematopoietic stem cells tracked by single-cell transcriptomic analysis. Nat. Cell Biol. 2020;22:630–639. doi: 10.1038/s41556-020-0512-1. [DOI] [PubMed] [Google Scholar]
- 55.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lauria F., Tebaldi T., Bernabò P., Groen E.J.N., Gillingwater T.H., Viero G. riboWaltz: Optimization of ribosome P-site positioning in ribosome profiling data. PLoS Comput. Biol. 2018;14 doi: 10.1371/journal.pcbi.1006169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamamoto R., Morita Y., Ooehara J., Hamanaka S., Onodera M., Rudolph K.L., Ema H., Nakauchi H. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154:1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 63.Li M., Qiu C., Bian Y., Shi D., Wang B., Ma Q., Wang X., Shi J., Zhang L., Ma Y., et al. SETD5 modulates homeostasis of hematopoietic stem cells by mediating RNA Polymerase II pausing in cooperation with HCF-1. Leukemia. 2022;36:1111–1122. doi: 10.1038/s41375-021-01481-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang B., Xia M., Chen T., Li M., Shi D., Wang X., Pang A., Zhou J., Yuan W., Chu Y. Loss of Tet2 affects platelet function but not coagulation in mice. Blood Sci. 2020;2:129–136. doi: 10.1097/bs9.0000000000000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang W., Eckwahl M.J., Zhou K.I., Pan T. Sensitive and quantitative probing of pseudouridine modification in mRNA and long noncoding RNA. Rna. 2019;25:1218–1225. doi: 10.1261/rna.072124.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schneider R.K., Schenone M., Ferreira M.V., Kramann R., Joyce C.E., Hartigan C., Beier F., Brümmendorf T.H., Germing U., Platzbecker U., et al. Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat. Med. 2016;22:288–297. doi: 10.1038/nm.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chu Y., Chen Y., Guo H., Li M., Wang B., Shi D., Cheng X., Guan J., Wang X., Xue C., et al. SUV39H1 regulates the progression of MLL-AF9-induced acute myeloid leukemia. Oncogene. 2020;39:7239–7252. doi: 10.1038/s41388-020-01495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang L., Cui Z., Shi D., Chu Y., Wang B., Wan Y., Ma Q., Zhang R., Li H., Cheng X., et al. Polyclonal evolution of Fanconi anemia to MDS and AML revealed at single cell resolution. Exp. Hematol. Oncol. 2022;11:64. doi: 10.1186/s40164-022-00319-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X., Zhang Y., Ni M., Cao H., Signer R.A.J., Li D., Li M., Gu Z., Hu Z., Dickerson K.E., et al. Regulation of mitochondrial biogenesis in erythropoiesis by mTORC1-mediated protein translation. Nat. Cell Biol. 2017;19:626–638. doi: 10.1038/ncb3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA009788) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. These accession numbers are also listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.