In the crystal structure of methyl 4-[4-(difluoromethoxy)phenyl]-2,7,7-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (I), molecules are linked by N—H⋯O and C—H⋯O interactions, forming a tri-periodic network, while molecules of isopropyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (II) and tert-butyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (III) are linked by N—H⋯O, C—H⋯F and C—H⋯π interactions, forming layers parallel to (002).
Keywords: crystal structure; 1,4-dihydropyridine ring; cyclohexene ring; quinoline ring system; disorder; van der Waals interactions; Hirshfeld surface analysis
Abstract
The crystal structures and Hirshfeld surface analyses of three similar compounds are reported. Methyl 4-[4-(difluoromethoxy)phenyl]-2,7,7-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate, (C21H23F2NO4), (I), crystallizes in the monoclinic space group C2/c with Z = 8, while isopropyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate, (C23H27F2NO4), (II) and tert-butyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate, (C24H29F2NO4), (III) crystallize in the orthorhombic space group Pbca with Z = 8. In the crystal structure of (I), molecules are linked by N—H⋯O and C—H⋯O interactions, forming a tri-periodic network, while molecules of (II) and (III) are linked by N—H⋯O, C—H⋯F and C—H⋯π interactions, forming layers parallel to (002). The cohesion of the molecular packing is ensured by van der Waals forces between these layers. In (I), the atoms of the 4-difluoromethoxyphenyl group are disordered over two sets of sites in a 0.647 (3): 0.353 (3) ratio. In (III), the atoms of the dimethyl group attached to the cyclohexane ring, and the two carbon atoms of the cyclohexane ring are disordered over two sets of sites in a 0.646 (3):0.354 (3) ratio.
1. Chemical context
Inflammation is a defense tool developed by the immune system to eliminate abnormal conditions resulting from harmful stimuli caused by pathogens, damaged cells, toxic compounds and traumatic cells. Inflammatory processes are important in terms of providing hemostasis of the body. Inflammatory mediators such as cytokines, chemokines and leukocytes secreted by the immune system during inflammation regulate the vital functions of the cell such as survival, growth and proliferation. In some cases, persistent and uncontrolled acute inflammatory responses cause chronic inflammation (Chen et al., 2018 ▸; Aqdas & Sung, 2023 ▸).
Cancer is a dangerous disease with a high incidence all over the world. Although chemotherapy, radiotherapy and surgical interventions are among the current treatment methods, there are cases where these methods are insufficient. In addition, cancer is a disease that progresses rapidly and can recur even after treatment. Therefore, there is an urgent need for new treatments and new therapeutic agents (Shaheen et al., 2020 ▸). Tumor tissues are formed by the abnormal and damaged proliferation of cancer cells. Inflammation mediators multiply uncontrollably by immune cells in the microenvironment of tumor tissue (Aqdas & Sung, 2023 ▸). This uncontrolled development of inflammation is the root cause of many chronic diseases and cancers. Therefore, it is very important to develop new anti-inflammatory treatments (Wu et al., 2022 ▸).
1,4-DHPs and their condensed derivatives are heterocyclic compounds with many pharmacological and biological activities. These compounds were described in the literature for the first time with their calcium channel modulator activities, and then various activities such as anticancer and anti-ischemic were discovered (Bryzgalov et al., 2023 ▸). Lerkadipine, which is a calcium channel blocker in the pharmaceutical market, has also been shown by in vivo studies to be effective in melanoma and non-small-cell lung cancer. Based on this information, new compounds with anti-inflammatory effects have been obtained with modifications made on 1,4-DHPs and their activities have been proven (Pan et al., 2022 ▸) (Fig. 1 ▸). Hexahydroquinolines are heterocyclic rings obtained by the condensation of 1,4-DHPs with the cyclohexane ring. In recent years, it has been seen that hexahydroquinoline derivatives have many biological activities such as analgesic, anticancer, antibacterial, antituberculosis, antimalarial, antioxidant, anti-inflammatory, anti-Alzheimer’s. Therefore, the hexahydroquinoline ring system is a very well-established motif for medicinal chemistry and has been the subject of many studies in recent years (Ranjbar et al., 2019 ▸).

Figure 1.
Structure of lercanidipine
2. Structural commentary
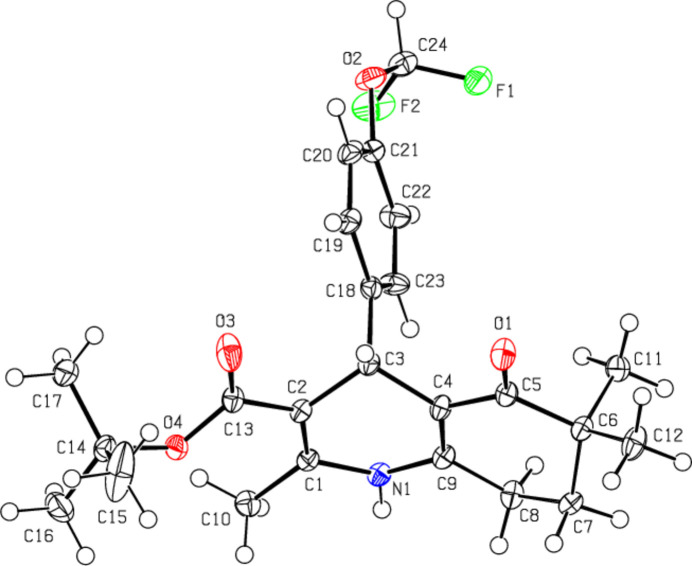
The 1,4-dihydropyridine ring (N1/C1/C6–C9) of compound (I) (Fig. 2 ▸) adopts a distorted boat conformation [puckering parameters (Cremer & Pople, 1975 ▸) are Q T = 0.196 (3) Å, θ = 72.2 (9)° and φ = 185.8 (8)°], while the cyclohexene ring (C1–C6) has a distorted half-chair conformation [puckering parameters are Q T = 0.466 (3) Å, θ = 123.1 (4)° and φ = 295.3 (4)°]. The atoms of the 4-difluoromethoxyphenyl group in (I) are disordered over two sets of sites with refined occupancy factors of 0.647 (3) and 0.353 (3). The major (C15–C20) and minor (C15A–C20A) disorder components of the 4-[4-(difluoromethoxy]phenyl ring make dihedral angles of 80.84 (15) and 85.81 (27)°, respectively, with the mean plane of the quinoline ring system [N1/C1–C9; maximum deviation = 0.382 (2) Å for C3].
Figure 2.
The molecular structure of (I) with displacement ellipsoids drawn at the 30% probability level. Only the major component of disorder is shown for clarity.
In (II) (Fig. 3 ▸), the 1,4-dihydropyridine ring (N1/C1/C6–C9) and the cyclohexene ring (C1–C6) both have distorted boat conformations [puckering parameters are Q T = 0.3187 (9) Å, θ = 105.86 (16)° and φ = 359.72 (17)° for the 1,4-dihydropyridine ring, and Q T = 0.4332 (11) Å, θ = 131.14 (13)° and φ = 301.37 (17)° for the cyclohexene ring]. The 4-[4-(difluoromethoxy]phenyl ring (C17–C22) makes a dihedral angle of 86.39 (4)° with the mean plane of the quinoline ring system [N1/C1–C9; maximum deviation = 0.421 (1) Å for C3].
Figure 3.
The molecular structure of (II) with displacement ellipsoids drawn at the 50% probability level.
In (III) (Fig. 4 ▸), the 1,4-dihydropyridine ring (N1/C1–C4/C9) and the cyclohexene ring (C4–C9) both have distorted boat conformations [puckering parameters are Q T = 0.3403 (14) Å, θ = 73.4 (2)° and φ = 180.4 (3)° for the 1,4-dihydropyridine ring, and Q T = 0.420 (5) Å, θ = 131.7 (6)° and φ = 356.2 (10)° for the cyclohexene ring]. The two carbon atoms (C7/C7A and C8/C8A) in the cyclohexane ring of the quinoline ring system are disordered over two sets of sites in a 0.646 (3):0.354 (3) ratio. The 4-[4-(difluoromethoxy]phenyl ring (C18–C23) makes dihedral angles of 84.47 (4) and 88.71 (5)°, respectively, with the mean planes of the major and minor disorder components of the quinoline ring system [N1/C1–C9; maximum deviation = −0.427 (3) Å for C7 in the major component and N1/C1–C6/C7A/C8A/C9; maximum deviation = 0.392 (3) Å for C3 in the minor component].
Figure 4.
The molecular structure of (III) with displacement ellipsoids drawn at the 50% probability level. Only the major component of disorder is shown for clarity.
Bond lengths and angles in all compounds are in agreement with those reported for the related compounds discussed in the Database survey section.
3. Supramolecular features and Hirshfeld surface analysis
In the crystal structure of (I), molecules are linked by N—H⋯O and C—H⋯O interactions, forming a tri-periodic network (Table 1 ▸; Figs. 5 ▸, 6 ▸ and 7 ▸), while molecules of (II) and (III) are linked by N—H⋯O, C—H⋯F and C—H⋯π interactions, forming layers parallel to (002) [Table 2 ▸, Figs. 8 ▸, 9 ▸, 10 ▸ and 11 ▸; C3—H3B⋯Cg3
a
: H3B⋯Cg3
a
= 3.6716 (14) Å, C3—H3B⋯Cg3
a
= 158°; symmetry code: (a) 1 − x,
 + y,
+ y,
 − z; Cg3 is the centroid of the 4-difluoromethoxyphenyl ring (C17–C22) for (II), and Table 3 ▸, Figs. 12 ▸, 13 ▸, 14 ▸ and 15 ▸; C7—H7B⋯Cg4
b
: H7B⋯Cg4
b
= 3.687 (2) Å, C7—H7B⋯Cg4
b
= 158°; symmetry code: (b) 1 − x, −
− z; Cg3 is the centroid of the 4-difluoromethoxyphenyl ring (C17–C22) for (II), and Table 3 ▸, Figs. 12 ▸, 13 ▸, 14 ▸ and 15 ▸; C7—H7B⋯Cg4
b
: H7B⋯Cg4
b
= 3.687 (2) Å, C7—H7B⋯Cg4
b
= 158°; symmetry code: (b) 1 − x, −
 + y,
+ y,
 − z; Cg4 is the centroid of the 4-difluoromethoxy-phenyl ring (C18–C23) for (III)]. The cohesion of the molecular packing is ensured by van der Waals forces between these layers.
− z; Cg4 is the centroid of the 4-difluoromethoxy-phenyl ring (C18–C23) for (III)]. The cohesion of the molecular packing is ensured by van der Waals forces between these layers.
Table 1. Hydrogen-bond geometry (Å, °) for (I) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O1i | 0.90 (3) | 1.93 (4) | 2.834 (3) | 174 (3) |
| C12—H12A⋯O2 | 0.98 | 2.32 | 2.831 (4) | 111 |
| C12—H12C⋯F2ii | 0.98 | 2.63 | 3.449 (5) | 141 |
| C12—H12C⋯F1A ii | 0.98 | 2.41 | 3.291 (7) | 150 |
| C14—H14C⋯O4A iii | 0.98 | 2.66 | 3.551 (6) | 152 |
| C17—H17A⋯F1 | 0.95 | 2.43 | 2.975 (4) | 117 |
| C17—H17A⋯F1iv | 0.95 | 2.56 | 3.488 (4) | 165 |
| C21—H21A⋯O2v | 1.00 | 2.44 | 3.155 (5) | 128 |
| C21A—H21B⋯O2v | 1.00 | 2.50 | 3.062 (7) | 115 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 ; (iv)
; (iv)
 ; (v)
; (v)
 .
.
Figure 5.
The N—H⋯O and C—H⋯O contacts (solid lines) of (I), shown along the a-axis. Only the major component of disorder is shown for clarity.
Figure 6.
The N—H⋯O and C—H⋯O contacts (solid lines) of (I), shown along the b-axis.
Figure 7.
The N—H⋯O and C—H⋯O contacts (solid lines) of (I), shown along the c-axis.
Table 2. Hydrogen-bond geometry (Å, °) for (II) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O1i | 0.863 (16) | 1.967 (16) | 2.8258 (12) | 173.0 (14) |
| C2—H2A⋯F2ii | 0.99 | 2.40 | 3.1626 (13) | 133 |
| C12—H12A⋯O3 | 0.98 | 2.18 | 2.7991 (14) | 120 |
| C19—H19A⋯F2 | 0.95 | 2.37 | 2.9106 (14) | 116 |
| C23—H23A⋯F1iii | 1.00 | 2.63 | 3.3972 (14) | 133 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
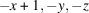 .
.
Figure 8.
The N—H⋯O and C—H⋯F contacts (solid lines) of (II), shown along the a-axis.
Figure 9.
The N—H⋯O and C—H⋯F contacts (solid lines) of (II), shown along the b-axis.
Figure 10.
The N—H⋯O and C—H⋯F contacts (solid lines) of (II), shown along the c-axis.
Figure 11.
The C—H⋯π contacts (solid lines) of (II), shown along the a-axis.
Table 3. Hydrogen-bond geometry (Å, °) for (III) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O1i | 0.88 (2) | 1.97 (2) | 2.8418 (16) | 171.2 (19) |
| C8A—H8A⋯F2ii | 0.99 | 2.53 | 3.168 (19) | 130 |
| C8A—H8AB⋯F2ii | 0.99 | 2.48 | 3.168 (19) | 126 |
| C10—H10A⋯O4 | 0.98 | 2.27 | 2.7834 (18) | 112 |
| C15—H15A⋯O3 | 0.98 | 2.47 | 3.038 (3) | 116 |
| C16—H16C⋯F1iii | 0.98 | 2.62 | 3.573 (2) | 164 |
| C17—H17B⋯O3 | 0.98 | 2.41 | 2.969 (2) | 116 |
| C22—H22A⋯F2 | 0.95 | 2.37 | 2.9091 (19) | 116 |
| C24—H24A⋯O4iv | 1.00 | 2.65 | 3.4638 (18) | 139 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
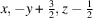 ; (iv)
; (iv)
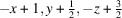 .
.
Figure 12.
The N—H⋯O and C—H⋯F contacts (solid lines) of (III), shown along the a-axis. Only the major component of disorder is shown for clarity.
Figure 13.
The N—H⋯O and C—H⋯F contacts (solid lines) of (III), shown along the b-axis.
Figure 14.
The N—H⋯O and C—H⋯F contacts (solid lines) of (III), shown along the c-axis.
Figure 15.
The C—H⋯π contacts (solid lines) of (III), shown along the a-axis.
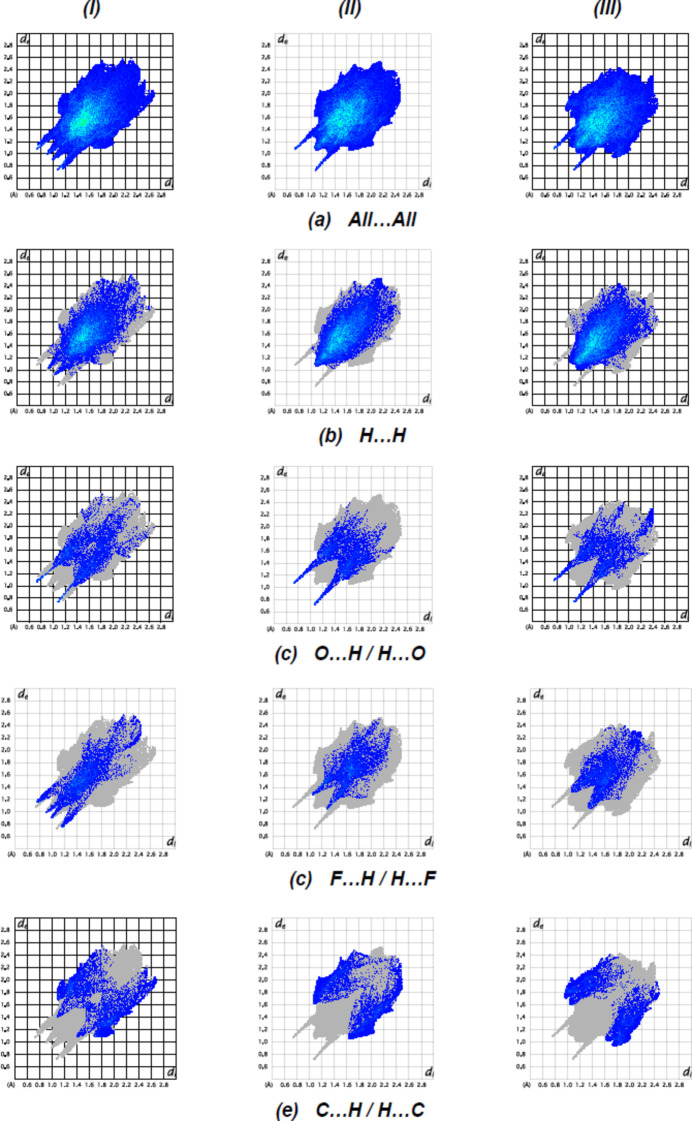
To quantify the intermolecular interactions between the molecules of (I), (II) and (III) in their respective crystal structures, the Hirshfeld surfaces and their corresponding two-dimensional fingerprint plots were calculated using the software package Crystal Explorer 17.5 (Spackman et al., 2021 ▸). The two-dimensional fingerprint plots are shown in Fig. 16 ▸. The dominant interactions of all compounds are H⋯H [(I): 49.1%, (II): 55.5% and (III): 58.9%], O⋯H/H⋯O [(I): 17.5%, (II): 14.9% and (III): 12.7%], F⋯H/H⋯F [(I): 16.2%, (II): 14.1% and (III): 12.9%] and C⋯H/H⋯C [(I) 11.7%, (II): 14.5% and (III): 12.0% ]. The percentage contributions of interatomic contacts calculated for each compound are given in Table 4 ▸. These interactions play a crucial role in the overall consolidation of the crystal packing. The presence of different functional groups in the compounds leads to some differences in the remaining weak interactions.
Figure 16.
Two-dimensional fingerprint graphs showing the H⋯H, O⋯H/H⋯O, F⋯H/H⋯F and C⋯H/H⋯C interactions of (I), (II) and (III).
Table 4. Percentage contributions of interatomic contacts to the Hirshfeld surface for the compounds.
| Contact | Percentage contribution | ||
|---|---|---|---|
| (I) | (II) | (III) | |
| H⋯H | 49.1 | 55.5 | 58.9 |
| O⋯H/H⋯O | 17.5 | 14.9 | 12.7 |
| F⋯H/H⋯F | 16.2 | 14.1 | 12.9 |
| C⋯H/H⋯C | 11.7 | 14.5 | 12.0 |
| F⋯F | 1.8 | – | 0.2 |
| O⋯C/C⋯O | – | 1.2 | 1.0 |
| F⋯O/O⋯F | 0.8 | – | 0.2 |
| N⋯H/H⋯N | 0.5 | 0.2 | 0.2 |
| F⋯C/C⋯F | 0.5 | 1.5 | 1.4 |
| O⋯N/N⋯O | 0.3 | 0.5 | 0.4 |
| O⋯O | 0.1 | – | – |
| C⋯C | 0.1 | 0.4 | 0.1 |
4. Database survey
A search of the Cambridge Structural Database (CSD, Version 5.42, update of September 2021; Groom et al., 2016 ▸) for similar structures with the 1,4,5,6,7,8-hexahydroquinoline group showed that the nine results most closely related to the title compound are LIMYUF (Pehlivanlar et al., 2023 ▸), WEZJUK (Yıldırım et al., 2023 ▸), ECUCUE (Yıldırım et al., 2022 ▸), LOQCAX (Steiger et al., 2014 ▸), NEQMON (Öztürk Yildirim, et al., 2013 ▸), PECPUK (Gündüz et al., 2012 ▸), IMEJOA (Linden et al., 2011 ▸), PUGCIE (Mookiah et al., 2009 ▸), UCOLOO (Linden et al., 2006 ▸) and DAYJET (Linden et al., 2005 ▸). In all these compounds, molecules are linked by N—H⋯O hydrogen bonds. Furthermore, C—H⋯F hydrogen bonds in LIMYUF, C—H⋯O hydrogen bonds in WEZJUK, ECUCUE, NEQMON, IMEJOA and PUGCIE and C—H⋯π interactions in LIMYUF, WEZJUK and ECUCUE were also observed.
5. Synthesis and crystallization
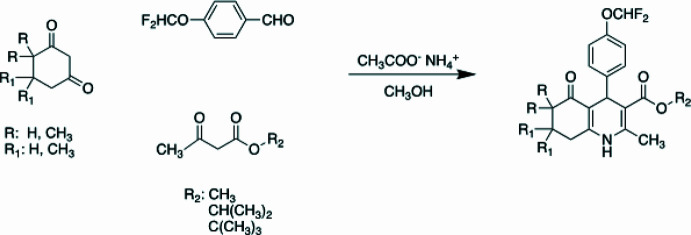
The target compounds were synthesized by 5,5-dimethylcyclohexane-1,3-dione/4,4-dimethylcyclohexane-1,3-dione (1 mmol), 4-difluoromethoxybenzaldehyde (1 mmol), methyl acetoacetate/isopropyl acetoacetate/tert-butyl acetoacetate (1 mmol), and ammonium acetate (5 mmol), which were refluxed for 8 h in absolute methanol (10 ml). The progress of the reactions were monitored by TLC and after the reactions were seen to be complete, they were cooled to room temperature. The obtained precipitates were filtered and recrystallized from methanol for further purification. The synthetic route is shown in Fig. 17 ▸. The structures of the compounds were elucidated by IR, 1H-NMR, 13C-NMR and HRMS analysis.
Figure 17.
Synthetic scheme
Methyl 4-[4-(difluoromethoxy)phenyl]-2,7,7-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (I): Yield: 59%; Yellow solid; mp: 478–479 K; IR (ν, cm−1) 3208 (N—H stretching); 3076 (C—H stretching, aromatic); 2956 (C—H stretching, aliphatic); 1700 (C=O stretching, ester); 1649 (C=O stretching, ketone). 1H NMR (500 MHz, DMSO-d 6, ppm): δ 0.84 (3H; s; 7-CH3), 1.00 (3H; s; 7-CH3), 1.98 (1H; d; J = 16,05; kinolin H8a), 2.17 (1H; d; J = 16.05 Hz; quinoline H8b), 2.29 (3H; s; 2-CH3), 2.29 (1H; d; J = 16.05 Hz quinoline H6a), 2.30 (2H; d; J = 16.05 Hz; quinoline H6b), 3.53 (3H; s; COOCH3), 4.86 (H; s; quinoline H4), 6.99 (2H; d; J = 8.6 Hz; Ar-H3, Ar-H5), 7.13 (1H; t; J = 74.4 Hz; OCHF2), 7.17 (2H; d; J = 8.6 Hz; Ar-H2, Ar-H6), 9.14 (1H; s; NH). 13C NMR (125 MHz, DMSO-d 6, ppm): δ 18.8 (2-CH3), 26.9 (7-CH3), 29.5 (C-7), 32.6 (C-8), 35.6 (C-4), 50.6 (C-6), 51.1 (COOCH3), 103.4 (C-3), 110.2 (C-4a), 114.8 (C3′), 116.9, 118.6, 118.9 (OCHF2), 129.2 (C2′), 145.0 (C1′), 145.9 (C-2), 149.4 (C-8a), 150.06 (C4′), 167.6 (COOCH3), 194.7 (C-5). HRMS (ESI/Q-TOF): m/z calculated for C21H23F2NO4 [M + H]+, 392,1673; found 392.1825.
Isopropyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (II): Yield: 37%; White solid; mp: 486–487 K; IR (ν, cm−1) 3194 (N—H stretching); 2970 (C—H stretching, aromatic); 2939 (C—H stretching, aliphatic); 1674 (C=O stretching, ester). 1H NMR (400 MHz, DMSO-d 6, ppm): δ 0.86 (3H; s; 6-CH3), 0.96 (3H; s; 6-CH3), 1.0 [3H; d; J = 6.4 Hz; COOCH(CH3)2a], 1.15 [3H; d; J=6.4 Hz; COOCH(CH3)2b], 1.67–1.70 (2H; m; quinoline H7), 2.44 (3H; m; quinoline H8), 2.24 (3H; s; 2-CH3), 4.77–4.82 [1H; m; COOCH(CH3)2], 4.81 (1H; s; quinoline H4), 6.95 (2H; d; J = 8 Hz; Ar-H3) 7.09 (1H; t; J = 74.4 Hz; OCHF2), 7.14 (2H; d; J = 8 Hz; Ar-H2), Ar-H6, 9.01 (1H; s; NH). 13C NMR (100 MHz, DMSO-d 6, ppm): δ 18.2 (2-CH3), 21.5 [COOCH(CH3)2a], 21.8 [COOCH(CH3)2b], 22.8 (C-8), 24.0 (6-CH3), 25.0 (C-7), 34.0 (C-4), 35.5 (C-6), 66.0 [COOCH(CH3)2] 103.3 (C-3), 108.9 (C-4a), 113.8 (C3′), 116.6, 118.0, 118.9 (OCHF2), 128.8 (C2′), 144.7 (C1′), 144.9 (C-2), 149.3 (C-8a), 149.7 (C4′), 166.2 [COOCH(CH3)2], 199.3 (C-5). HRMS (ESI/Q-TOF): m/z calculated for C23H27F2NO4 [M + H]+, 420.1986; found 420.2150.
tert-Butyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (III): Yield: 20%; White solid; mp: 456–457 K; IR (ν, cm−1) 3194 (N—H stretching); 2962 (C—H stretching, aromatic); 2931 (C—H stretching, aliphatic); 1674 (C=O stretching, ester). 1H NMR (400 MHz, DMSO-d 6, ppm): δ 0.86 (3H; s; 6-CH3), 0.95 (3H; s; 6-CH3), 1.30 [9H; s; COOC(CH3)3], 1.65–1.69 (2H; m; quinoline H7), 2.20 (3H; s; 2-CH3), 2.44–2.47 (2H; m; quinoline H8), 4.76 (1H; s; quinoline H4), 6.96 (2H; d; J = 8.4 Hz; Ar-H3, Ar-H5), 7.10 (1H; t; J = 74.4 Hz; OCHF2), 7.13 (2H; d; J = 8 Hz; Ar-H2, Ar-H6), 8.95 (1H; s; NH). 13C NMR (100 MHz, DMSO-d 6, ppm): δ 18.1 (2-CH3), 22.8 (C-8), 24.0 (6-CH3), 25.0 (C-7), 27.8 [COOC(CH3)3], 34.0 (C-4), 35.7 (C-6), 78.7 [COOC(CH3)3], 104.4 (C-3), 108.7 (C-4a), 113.8 (C3′), 116.3, 118.0, 118.9 (OCHF2), 128.7 (C2′), 143.9 (C1’), 144.9 (C-2), 148.7 (C-8a), 149.7 (C4′), 166.3 (COOC(CH3)3), 199.2 (C-5). HRMS (ESI/Q-TOF): m/z calculated for C24H29F2NO4 [M + H]+, 434.2143; found 434.2321.
6. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 5 ▸. In (I), (II) and (III), the N-bound H atom was located in a difference Fourier map and refined freely [N1—H1N = 0.90 (3) Å for (I), N1—H1N = 0.863 (16) Å for (II) and N1—H1N = 0.88 (2) Å for (III)]. The C-bound H atoms of all compounds were positioned geometrically [C—H = 0.95–1.00 Å] and refined using a riding model withU iso(H) = 1.2 or 1.5U eq(C). In (I), the atoms of the 4-difluoromethoxy-phenyl group are disordered over two sets of sites with refined occupancy factors of 0.647 (3):0.353 (3). In (III), the carbon atoms (C10, C13–C24) of the methyl and tert-butyl formate group attached to the 1,4-dihydropyridine ring were refined isotropically for a stable structure. The atoms (C11/C11A and C12/C12A) of the dimethyl group attached to the cyclohexane ring, and the two carbon atoms (C7/C7A and C8/C8A) in the anticlockwise direction after the carbon atom to which the dimethyl group of the cyclohexane ring is attached, were refined as disordered over two sets of sites in a 0.646 (3):0.354 (3) ratio.
Table 5. Experimental details.
| (I) | (II) | (III) | |
|---|---|---|---|
| Crystal data | |||
| Chemical formula | C21H23F2NO4 | C23H27F2NO4 | C24H29F2NO4 |
| M r | 391.40 | 419.45 | 433.48 |
| Crystal system, space group | Monoclinic, C2/c | Orthorhombic, P b c a | Orthorhombic, P b c a |
| Temperature (K) | 100 | 100 | 100 |
| a, b, c (Å) | 19.705 (3), 15.389 (2), 14.1279 (19) | 12.255 (3), 15.694 (3), 21.903 (4) | 12.4094 (8), 15.9871 (12), 21.9629 (15) |
| α, β, γ (°) | 90, 113.801 (4), 90 | 90, 90, 90 | 90, 90, 90 |
| V (Å3) | 3919.7 (9) | 4212.3 (14) | 4357.2 (5) |
| Z | 8 | 8 | 8 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα |
| μ (mm−1) | 0.10 | 0.10 | 0.10 |
| Crystal size (mm) | 0.30 × 0.25 × 0.17 | 0.31 × 0.23 × 0.08 | 0.31 × 0.27 × 0.09 |
| Data collection | |||
| Diffractometer | Bruker D8 Quest with Photon 2 detector | Bruker D8 Quest with Photon 2 detector | Bruker APEXII CCD |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) | Multi-scan (SADABS; Krause et al., 2015 ▸) | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.603, 0.746 | 0.684, 0.747 | 0.374, 0.746 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 47858, 4871, 3288 | 102650, 8537, 6743 | 56620, 6654, 4732 |
| R int | 0.082 | 0.073 | 0.142 |
| (sin θ/λ)max (Å−1) | 0.667 | 0.788 | 0.715 |
| Refinement | |||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.073, 0.186, 1.05 | 0.050, 0.128, 1.03 | 0.059, 0.163, 1.05 |
| No. of reflections | 4871 | 8537 | 6654 |
| No. of parameters | 332 | 280 | 329 |
| No. of restraints | 361 | 0 | 68 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.58, −0.58 | 0.58, −0.42 | 0.37, −0.31 |
Supplementary Material
Crystal structure: contains datablock(s) I, II, III. DOI: 10.1107/S2056989024001233/jy2044sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989024001233/jy2044Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989024001233/jy2044IIsup3.hkl
Structure factors: contains datablock(s) III. DOI: 10.1107/S2056989024001233/jy2044IIIsup4.hkl
Supporting information file. DOI: 10.1107/S2056989024001233/jy2044Isup5.cml
Supporting information file. DOI: 10.1107/S2056989024001233/jy2044IIsup6.cml
Supporting information file. DOI: 10.1107/S2056989024001233/jy2044IIIsup7.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
Authors’ contributions are as follows. Conceptualization, RS and SÖY; methodology, RS and GÇ; investigation, RS and SÖY; writing (original draft), GÇ and MA; writing (review and editing of the manuscript), RS and SÖY; crystal data production and validation, RJB and SÖY; visualization, MA; funding acquisition, RJB; resources, AB, RJB and RS.
supplementary crystallographic information
Methyl 4-[4-(difluoromethoxy)phenyl]-2,7,7-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (I). Crystal data
| C21H23F2NO4 | F(000) = 1648 |
| Mr = 391.40 | Dx = 1.326 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 19.705 (3) Å | Cell parameters from 9994 reflections |
| b = 15.389 (2) Å | θ = 2.6–29.2° |
| c = 14.1279 (19) Å | µ = 0.10 mm−1 |
| β = 113.801 (4)° | T = 100 K |
| V = 3919.7 (9) Å3 | Prism, colorless |
| Z = 8 | 0.30 × 0.25 × 0.17 mm |
Methyl 4-[4-(difluoromethoxy)phenyl]-2,7,7-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (I). Data collection
| Bruker D8 Quest with Photon 2 detector diffractometer | 3288 reflections with I > 2σ(I) |
| φ and ω scans | Rint = 0.082 |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | θmax = 28.3°, θmin = 1.7° |
| Tmin = 0.603, Tmax = 0.746 | h = −26→24 |
| 47858 measured reflections | k = −20→20 |
| 4871 independent reflections | l = −18→18 |
Methyl 4-[4-(difluoromethoxy)phenyl]-2,7,7-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (I). Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.073 | Hydrogen site location: mixed |
| wR(F2) = 0.186 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.043P)2 + 11.081P] where P = (Fo2 + 2Fc2)/3 |
| 4871 reflections | (Δ/σ)max = 0.001 |
| 332 parameters | Δρmax = 0.58 e Å−3 |
| 361 restraints | Δρmin = −0.58 e Å−3 |
Methyl 4-[4-(difluoromethoxy)phenyl]-2,7,7-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (I). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Methyl 4-[4-(difluoromethoxy)phenyl]-2,7,7-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (I). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O1 | 0.09716 (12) | 0.54160 (14) | 0.69583 (14) | 0.0510 (5) | |
| O2 | 0.15731 (12) | 0.17128 (13) | 0.53819 (18) | 0.0594 (6) | |
| O3 | 0.15902 (12) | 0.23777 (13) | 0.68037 (16) | 0.0553 (6) | |
| N1 | 0.12023 (13) | 0.42587 (15) | 0.40402 (16) | 0.0392 (5) | |
| C1 | 0.10494 (13) | 0.49275 (16) | 0.45557 (17) | 0.0320 (5) | |
| C2 | 0.07419 (16) | 0.57420 (17) | 0.39524 (18) | 0.0432 (7) | |
| H2A | 0.019785 | 0.568190 | 0.357785 | 0.052* | |
| H2B | 0.095506 | 0.581955 | 0.343212 | 0.052* | |
| C3 | 0.09123 (15) | 0.65493 (18) | 0.4646 (2) | 0.0432 (7) | |
| C4 | 0.06672 (14) | 0.63670 (17) | 0.5521 (2) | 0.0392 (6) | |
| H4A | 0.084333 | 0.684614 | 0.603066 | 0.047* | |
| H4B | 0.011840 | 0.636679 | 0.523487 | 0.047* | |
| C5 | 0.09443 (13) | 0.55203 (17) | 0.60759 (18) | 0.0336 (5) | |
| C6 | 0.11516 (12) | 0.48321 (15) | 0.55589 (16) | 0.0292 (5) | |
| C7 | 0.14879 (14) | 0.40124 (16) | 0.61533 (18) | 0.0332 (5) | |
| C8 | 0.14482 (13) | 0.32712 (16) | 0.54191 (19) | 0.0342 (5) | |
| C9 | 0.13559 (14) | 0.34270 (17) | 0.4431 (2) | 0.0377 (6) | |
| C10 | 0.0482 (2) | 0.7326 (2) | 0.4006 (3) | 0.0720 (12) | |
| H10A | −0.005015 | 0.719741 | 0.371558 | 0.108* | |
| H10B | 0.064070 | 0.743656 | 0.344258 | 0.108* | |
| H10C | 0.058084 | 0.784075 | 0.444977 | 0.108* | |
| C11 | 0.17443 (16) | 0.67588 (19) | 0.5086 (2) | 0.0457 (7) | |
| H11A | 0.184624 | 0.725991 | 0.555128 | 0.068* | |
| H11B | 0.189114 | 0.689477 | 0.451748 | 0.068* | |
| H11C | 0.202669 | 0.625559 | 0.547039 | 0.068* | |
| C12 | 0.13988 (19) | 0.2777 (2) | 0.3665 (2) | 0.0547 (8) | |
| H12A | 0.177848 | 0.234171 | 0.402577 | 0.082* | |
| H12B | 0.152879 | 0.307481 | 0.314772 | 0.082* | |
| H12C | 0.091700 | 0.248966 | 0.332042 | 0.082* | |
| C13 | 0.15455 (14) | 0.23808 (18) | 0.5819 (2) | 0.0429 (6) | |
| C14 | 0.1635 (2) | 0.1524 (2) | 0.7254 (3) | 0.0746 (11) | |
| H14A | 0.167604 | 0.158165 | 0.796548 | 0.112* | |
| H14B | 0.207276 | 0.121931 | 0.725548 | 0.112* | |
| H14C | 0.118808 | 0.119180 | 0.684523 | 0.112* | |
| C15 | 0.22622 (13) | 0.4139 (4) | 0.6988 (2) | 0.0329 (10) | 0.647 (3) |
| C16 | 0.28551 (17) | 0.4229 (3) | 0.67009 (18) | 0.0388 (11) | 0.647 (3) |
| H16A | 0.276995 | 0.423772 | 0.598944 | 0.047* | 0.647 (3) |
| C17 | 0.35727 (14) | 0.4308 (2) | 0.7454 (3) | 0.0430 (10) | 0.647 (3) |
| H17A | 0.397791 | 0.436936 | 0.725802 | 0.052* | 0.647 (3) |
| C18 | 0.36973 (14) | 0.4295 (2) | 0.8495 (2) | 0.0426 (11) | 0.647 (3) |
| C19 | 0.31044 (19) | 0.4205 (3) | 0.87829 (18) | 0.0449 (11) | 0.647 (3) |
| H19A | 0.318964 | 0.419649 | 0.949438 | 0.054* | 0.647 (3) |
| C20 | 0.23869 (16) | 0.4127 (3) | 0.8029 (3) | 0.0414 (11) | 0.647 (3) |
| H20A | 0.198167 | 0.406484 | 0.822581 | 0.050* | 0.647 (3) |
| O4 | 0.44015 (17) | 0.4369 (2) | 0.9302 (2) | 0.0575 (10) | 0.647 (3) |
| C21 | 0.5006 (3) | 0.4146 (5) | 0.9161 (5) | 0.081 (2) | 0.647 (3) |
| H21A | 0.542852 | 0.412128 | 0.985748 | 0.097* | 0.647 (3) |
| F1 | 0.51546 (16) | 0.4803 (3) | 0.8608 (3) | 0.0920 (13) | 0.647 (3) |
| F2 | 0.4900 (2) | 0.3321 (2) | 0.8754 (4) | 0.0751 (13) | 0.647 (3) |
| C15A | 0.2335 (2) | 0.4232 (7) | 0.6827 (5) | 0.0348 (18) | 0.353 (3) |
| C16A | 0.2824 (3) | 0.4423 (5) | 0.6369 (4) | 0.0375 (18) | 0.353 (3) |
| H16B | 0.264737 | 0.445770 | 0.563751 | 0.045* | 0.353 (3) |
| C17A | 0.3570 (3) | 0.4564 (4) | 0.6983 (4) | 0.0342 (15) | 0.353 (3) |
| H17B | 0.390378 | 0.469487 | 0.667015 | 0.041* | 0.353 (3) |
| C18A | 0.3828 (2) | 0.4514 (4) | 0.8053 (4) | 0.0373 (15) | 0.353 (3) |
| C19A | 0.3339 (3) | 0.4323 (5) | 0.8511 (4) | 0.0391 (16) | 0.353 (3) |
| H19B | 0.351570 | 0.428885 | 0.924261 | 0.047* | 0.353 (3) |
| C20A | 0.2593 (3) | 0.4182 (6) | 0.7897 (5) | 0.0387 (17) | 0.353 (3) |
| H20B | 0.225928 | 0.405167 | 0.820999 | 0.046* | 0.353 (3) |
| O4A | 0.4570 (2) | 0.4712 (3) | 0.8666 (4) | 0.0458 (14) | 0.353 (3) |
| C21A | 0.5040 (4) | 0.4078 (6) | 0.9164 (6) | 0.068 (3) | 0.353 (3) |
| H21B | 0.554500 | 0.433982 | 0.951241 | 0.081* | 0.353 (3) |
| F1A | 0.5034 (3) | 0.3581 (5) | 0.8348 (4) | 0.0512 (16) | 0.353 (3) |
| F2A | 0.4849 (3) | 0.3731 (4) | 0.9915 (4) | 0.0766 (19) | 0.353 (3) |
| H1N | 0.1163 (19) | 0.437 (2) | 0.339 (3) | 0.062 (10)* | |
| H7 | 0.1208 (15) | 0.3857 (17) | 0.655 (2) | 0.032 (7)* |
Methyl 4-[4-(difluoromethoxy)phenyl]-2,7,7-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (I). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0691 (14) | 0.0580 (13) | 0.0330 (10) | 0.0189 (10) | 0.0279 (10) | 0.0037 (9) |
| O2 | 0.0485 (12) | 0.0372 (11) | 0.0726 (15) | −0.0057 (9) | 0.0037 (11) | −0.0120 (10) |
| O3 | 0.0576 (13) | 0.0419 (11) | 0.0525 (12) | 0.0102 (9) | 0.0079 (10) | 0.0168 (9) |
| N1 | 0.0505 (13) | 0.0445 (12) | 0.0249 (10) | −0.0165 (10) | 0.0177 (9) | −0.0080 (9) |
| C1 | 0.0326 (12) | 0.0362 (12) | 0.0232 (11) | −0.0098 (10) | 0.0071 (9) | −0.0013 (9) |
| C2 | 0.0513 (16) | 0.0430 (14) | 0.0220 (11) | −0.0171 (12) | 0.0008 (11) | 0.0029 (10) |
| C3 | 0.0431 (15) | 0.0394 (14) | 0.0312 (13) | −0.0079 (11) | −0.0017 (11) | 0.0056 (10) |
| C4 | 0.0329 (13) | 0.0366 (13) | 0.0387 (13) | 0.0039 (10) | 0.0047 (10) | 0.0011 (11) |
| C5 | 0.0293 (12) | 0.0429 (14) | 0.0268 (11) | 0.0042 (10) | 0.0094 (9) | 0.0015 (10) |
| C6 | 0.0266 (11) | 0.0351 (12) | 0.0223 (10) | 0.0006 (9) | 0.0061 (9) | 0.0006 (9) |
| C7 | 0.0314 (12) | 0.0382 (13) | 0.0266 (11) | 0.0062 (10) | 0.0083 (10) | 0.0019 (10) |
| C8 | 0.0256 (11) | 0.0342 (12) | 0.0382 (13) | −0.0008 (9) | 0.0081 (10) | −0.0040 (10) |
| C9 | 0.0348 (13) | 0.0401 (14) | 0.0387 (13) | −0.0116 (11) | 0.0154 (11) | −0.0121 (11) |
| C10 | 0.072 (2) | 0.0413 (17) | 0.062 (2) | −0.0130 (15) | −0.0159 (17) | 0.0200 (15) |
| C11 | 0.0473 (16) | 0.0479 (16) | 0.0353 (13) | −0.0163 (13) | 0.0101 (12) | −0.0057 (12) |
| C12 | 0.0607 (19) | 0.0528 (18) | 0.0557 (18) | −0.0183 (15) | 0.0288 (15) | −0.0275 (15) |
| C13 | 0.0257 (12) | 0.0411 (15) | 0.0479 (15) | −0.0017 (10) | 0.0002 (11) | 0.0012 (12) |
| C14 | 0.070 (2) | 0.051 (2) | 0.080 (2) | 0.0077 (17) | 0.0066 (19) | 0.0312 (18) |
| C15 | 0.0309 (19) | 0.034 (2) | 0.028 (2) | 0.0096 (16) | 0.0063 (15) | 0.0006 (17) |
| C16 | 0.0326 (19) | 0.050 (3) | 0.0262 (19) | 0.0093 (18) | 0.0040 (16) | −0.0102 (19) |
| C17 | 0.0315 (19) | 0.053 (2) | 0.034 (2) | 0.0103 (17) | 0.0024 (17) | −0.0150 (19) |
| C18 | 0.040 (2) | 0.037 (2) | 0.0295 (19) | 0.0117 (18) | −0.0084 (18) | −0.0116 (16) |
| C19 | 0.059 (3) | 0.043 (2) | 0.0213 (18) | 0.013 (2) | 0.0046 (17) | −0.0017 (16) |
| C20 | 0.047 (2) | 0.045 (2) | 0.0258 (18) | 0.013 (2) | 0.0087 (17) | 0.0010 (16) |
| O4 | 0.0514 (17) | 0.0443 (17) | 0.0397 (16) | 0.0138 (14) | −0.0202 (13) | −0.0107 (13) |
| C21 | 0.047 (3) | 0.075 (3) | 0.076 (3) | 0.025 (3) | −0.021 (3) | −0.036 (3) |
| F1 | 0.0350 (16) | 0.141 (3) | 0.077 (2) | 0.0084 (18) | −0.0005 (15) | −0.059 (2) |
| F2 | 0.0411 (19) | 0.065 (2) | 0.093 (3) | 0.0140 (15) | −0.0006 (19) | −0.028 (2) |
| C15A | 0.039 (3) | 0.034 (3) | 0.022 (3) | 0.007 (3) | 0.002 (3) | −0.005 (3) |
| C16A | 0.031 (3) | 0.037 (4) | 0.034 (3) | 0.004 (3) | 0.002 (3) | −0.002 (3) |
| C17A | 0.032 (3) | 0.032 (3) | 0.032 (3) | 0.001 (2) | 0.006 (3) | −0.005 (3) |
| C18A | 0.033 (3) | 0.036 (3) | 0.031 (3) | 0.004 (2) | 0.001 (2) | −0.004 (3) |
| C19A | 0.035 (3) | 0.043 (3) | 0.027 (3) | −0.003 (3) | −0.001 (3) | 0.000 (3) |
| C20A | 0.035 (3) | 0.044 (3) | 0.031 (3) | 0.001 (3) | 0.007 (3) | −0.002 (3) |
| O4A | 0.034 (2) | 0.041 (3) | 0.043 (3) | 0.002 (2) | −0.004 (2) | 0.004 (2) |
| C21A | 0.045 (4) | 0.060 (4) | 0.065 (5) | 0.013 (4) | −0.012 (4) | −0.011 (4) |
| F1A | 0.033 (3) | 0.080 (4) | 0.038 (3) | 0.013 (3) | 0.011 (2) | −0.020 (3) |
| F2A | 0.055 (3) | 0.110 (5) | 0.062 (3) | 0.005 (3) | 0.021 (3) | 0.046 (3) |
Methyl 4-[4-(difluoromethoxy)phenyl]-2,7,7-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (I). Geometric parameters (Å, º)
| O1—C5 | 1.236 (3) | C12—H12C | 0.9800 |
| O2—C13 | 1.212 (3) | C14—H14A | 0.9800 |
| O3—C13 | 1.358 (4) | C14—H14B | 0.9800 |
| O3—C14 | 1.447 (4) | C14—H14C | 0.9800 |
| N1—C1 | 1.363 (3) | C15—C16 | 1.3900 |
| N1—C9 | 1.378 (3) | C15—C20 | 1.3900 |
| N1—H1N | 0.90 (3) | C16—C17 | 1.3900 |
| C1—C6 | 1.357 (3) | C16—H16A | 0.9500 |
| C1—C2 | 1.499 (3) | C17—C18 | 1.3900 |
| C2—C3 | 1.533 (4) | C17—H17A | 0.9500 |
| C2—H2A | 0.9900 | C18—C19 | 1.3900 |
| C2—H2B | 0.9900 | C18—O4 | 1.400 (3) |
| C3—C4 | 1.524 (4) | C19—C20 | 1.3900 |
| C3—C10 | 1.532 (4) | C19—H19A | 0.9500 |
| C3—C11 | 1.535 (4) | C20—H20A | 0.9500 |
| C4—C5 | 1.505 (3) | O4—C21 | 1.330 (6) |
| C4—H4A | 0.9900 | C21—F2 | 1.375 (7) |
| C4—H4B | 0.9900 | C21—F1 | 1.379 (8) |
| C5—C6 | 1.436 (3) | C21—H21A | 1.0000 |
| C6—C7 | 1.512 (3) | C15A—C16A | 1.3900 |
| C7—C15 | 1.518 (3) | C15A—C20A | 1.3900 |
| C7—C8 | 1.522 (3) | C16A—C17A | 1.3900 |
| C7—C15A | 1.589 (4) | C16A—H16B | 0.9500 |
| C7—H7 | 0.97 (3) | C17A—C18A | 1.3900 |
| C8—C9 | 1.354 (4) | C17A—H17B | 0.9500 |
| C8—C13 | 1.465 (4) | C18A—C19A | 1.3900 |
| C9—C12 | 1.501 (4) | C18A—O4A | 1.399 (4) |
| C10—H10A | 0.9800 | C19A—C20A | 1.3900 |
| C10—H10B | 0.9800 | C19A—H19B | 0.9500 |
| C10—H10C | 0.9800 | C20A—H20B | 0.9500 |
| C11—H11A | 0.9800 | O4A—C21A | 1.334 (7) |
| C11—H11B | 0.9800 | C21A—F2A | 1.370 (7) |
| C11—H11C | 0.9800 | C21A—F1A | 1.379 (9) |
| C12—H12A | 0.9800 | C21A—H21B | 1.0000 |
| C12—H12B | 0.9800 | ||
| C13—O3—C14 | 114.9 (3) | H12A—C12—H12C | 109.5 |
| C1—N1—C9 | 123.2 (2) | H12B—C12—H12C | 109.5 |
| C1—N1—H1N | 117 (2) | O2—C13—O3 | 121.4 (3) |
| C9—N1—H1N | 119 (2) | O2—C13—C8 | 128.5 (3) |
| C6—C1—N1 | 120.3 (2) | O3—C13—C8 | 110.0 (2) |
| C6—C1—C2 | 122.7 (2) | O3—C14—H14A | 109.5 |
| N1—C1—C2 | 116.9 (2) | O3—C14—H14B | 109.5 |
| C1—C2—C3 | 112.34 (19) | H14A—C14—H14B | 109.5 |
| C1—C2—H2A | 109.1 | O3—C14—H14C | 109.5 |
| C3—C2—H2A | 109.1 | H14A—C14—H14C | 109.5 |
| C1—C2—H2B | 109.1 | H14B—C14—H14C | 109.5 |
| C3—C2—H2B | 109.1 | C16—C15—C20 | 120.0 |
| H2A—C2—H2B | 107.9 | C16—C15—C7 | 119.0 (2) |
| C4—C3—C10 | 110.0 (3) | C20—C15—C7 | 120.9 (2) |
| C4—C3—C2 | 108.1 (2) | C15—C16—C17 | 120.0 |
| C10—C3—C2 | 109.2 (2) | C15—C16—H16A | 120.0 |
| C4—C3—C11 | 110.3 (2) | C17—C16—H16A | 120.0 |
| C10—C3—C11 | 108.7 (2) | C16—C17—C18 | 120.0 |
| C2—C3—C11 | 110.6 (3) | C16—C17—H17A | 120.0 |
| C5—C4—C3 | 114.5 (2) | C18—C17—H17A | 120.0 |
| C5—C4—H4A | 108.6 | C19—C18—C17 | 120.0 |
| C3—C4—H4A | 108.6 | C19—C18—O4 | 116.4 (3) |
| C5—C4—H4B | 108.6 | C17—C18—O4 | 123.6 (3) |
| C3—C4—H4B | 108.6 | C18—C19—C20 | 120.0 |
| H4A—C4—H4B | 107.6 | C18—C19—H19A | 120.0 |
| O1—C5—C6 | 120.8 (2) | C20—C19—H19A | 120.0 |
| O1—C5—C4 | 119.8 (2) | C19—C20—C15 | 120.0 |
| C6—C5—C4 | 119.5 (2) | C19—C20—H20A | 120.0 |
| C1—C6—C5 | 119.9 (2) | C15—C20—H20A | 120.0 |
| C1—C6—C7 | 121.2 (2) | C21—O4—C18 | 120.7 (3) |
| C5—C6—C7 | 118.9 (2) | O4—C21—F2 | 107.8 (6) |
| C6—C7—C15 | 113.7 (3) | O4—C21—F1 | 107.6 (5) |
| C6—C7—C8 | 110.86 (19) | F2—C21—F1 | 118.1 (5) |
| C15—C7—C8 | 112.6 (3) | O4—C21—H21A | 107.6 |
| C6—C7—C15A | 105.9 (4) | F2—C21—H21A | 107.6 |
| C8—C7—C15A | 108.7 (4) | F1—C21—H21A | 107.6 |
| C6—C7—H7 | 107.8 (15) | C16A—C15A—C20A | 120.0 |
| C15—C7—H7 | 102.1 (15) | C16A—C15A—C7 | 121.6 (4) |
| C8—C7—H7 | 109.2 (15) | C20A—C15A—C7 | 118.3 (4) |
| C15A—C7—H7 | 114.3 (16) | C17A—C16A—C15A | 120.0 |
| C9—C8—C13 | 120.4 (2) | C17A—C16A—H16B | 120.0 |
| C9—C8—C7 | 121.2 (2) | C15A—C16A—H16B | 120.0 |
| C13—C8—C7 | 118.4 (2) | C16A—C17A—C18A | 120.0 |
| C8—C9—N1 | 119.6 (2) | C16A—C17A—H17B | 120.0 |
| C8—C9—C12 | 127.0 (3) | C18A—C17A—H17B | 120.0 |
| N1—C9—C12 | 113.4 (2) | C19A—C18A—C17A | 120.0 |
| C3—C10—H10A | 109.5 | C19A—C18A—O4A | 120.3 (4) |
| C3—C10—H10B | 109.5 | C17A—C18A—O4A | 119.5 (4) |
| H10A—C10—H10B | 109.5 | C18A—C19A—C20A | 120.0 |
| C3—C10—H10C | 109.5 | C18A—C19A—H19B | 120.0 |
| H10A—C10—H10C | 109.5 | C20A—C19A—H19B | 120.0 |
| H10B—C10—H10C | 109.5 | C19A—C20A—C15A | 120.0 |
| C3—C11—H11A | 109.5 | C19A—C20A—H20B | 120.0 |
| C3—C11—H11B | 109.5 | C15A—C20A—H20B | 120.0 |
| H11A—C11—H11B | 109.5 | C21A—O4A—C18A | 119.8 (5) |
| C3—C11—H11C | 109.5 | O4A—C21A—F2A | 110.1 (7) |
| H11A—C11—H11C | 109.5 | O4A—C21A—F1A | 101.2 (6) |
| H11B—C11—H11C | 109.5 | F2A—C21A—F1A | 121.3 (7) |
| C9—C12—H12A | 109.5 | O4A—C21A—H21B | 107.8 |
| C9—C12—H12B | 109.5 | F2A—C21A—H21B | 107.8 |
| H12A—C12—H12B | 109.5 | F1A—C21A—H21B | 107.8 |
| C9—C12—H12C | 109.5 | ||
| C9—N1—C1—C6 | 10.0 (4) | C9—C8—C13—O2 | 1.7 (4) |
| C9—N1—C1—C2 | −167.7 (2) | C7—C8—C13—O2 | −175.6 (3) |
| C6—C1—C2—C3 | 28.0 (4) | C9—C8—C13—O3 | −177.0 (2) |
| N1—C1—C2—C3 | −154.2 (2) | C7—C8—C13—O3 | 5.7 (3) |
| C1—C2—C3—C4 | −51.7 (3) | C6—C7—C15—C16 | −74.8 (3) |
| C1—C2—C3—C10 | −171.3 (3) | C8—C7—C15—C16 | 52.4 (4) |
| C1—C2—C3—C11 | 69.1 (3) | C6—C7—C15—C20 | 108.0 (3) |
| C10—C3—C4—C5 | 169.0 (2) | C8—C7—C15—C20 | −124.8 (3) |
| C2—C3—C4—C5 | 49.9 (3) | C20—C15—C16—C17 | 0.0 |
| C11—C3—C4—C5 | −71.1 (3) | C7—C15—C16—C17 | −177.2 (4) |
| C3—C4—C5—O1 | 158.9 (2) | C15—C16—C17—C18 | 0.0 |
| C3—C4—C5—C6 | −23.1 (3) | C16—C17—C18—C19 | 0.0 |
| N1—C1—C6—C5 | −176.0 (2) | C16—C17—C18—O4 | 179.9 (4) |
| C2—C1—C6—C5 | 1.6 (4) | C17—C18—C19—C20 | 0.0 |
| N1—C1—C6—C7 | 4.5 (4) | O4—C18—C19—C20 | −179.9 (3) |
| C2—C1—C6—C7 | −177.9 (2) | C18—C19—C20—C15 | 0.0 |
| O1—C5—C6—C1 | 173.5 (2) | C16—C15—C20—C19 | 0.0 |
| C4—C5—C6—C1 | −4.5 (3) | C7—C15—C20—C19 | 177.1 (4) |
| O1—C5—C6—C7 | −7.0 (4) | C19—C18—O4—C21 | 155.4 (5) |
| C4—C5—C6—C7 | 175.0 (2) | C17—C18—O4—C21 | −24.5 (6) |
| C1—C6—C7—C15 | 110.1 (3) | C18—O4—C21—F2 | −51.8 (7) |
| C5—C6—C7—C15 | −69.4 (3) | C18—O4—C21—F1 | 76.6 (6) |
| C1—C6—C7—C8 | −18.1 (3) | C6—C7—C15A—C16A | −64.9 (6) |
| C5—C6—C7—C8 | 162.4 (2) | C8—C7—C15A—C16A | 54.3 (6) |
| C1—C6—C7—C15A | 99.6 (4) | C6—C7—C15A—C20A | 118.7 (4) |
| C5—C6—C7—C15A | −79.9 (4) | C8—C7—C15A—C20A | −122.1 (4) |
| C6—C7—C8—C9 | 20.0 (3) | C20A—C15A—C16A—C17A | 0.0 |
| C15—C7—C8—C9 | −108.7 (3) | C7—C15A—C16A—C17A | −176.4 (8) |
| C15A—C7—C8—C9 | −96.0 (4) | C15A—C16A—C17A—C18A | 0.0 |
| C6—C7—C8—C13 | −162.7 (2) | C16A—C17A—C18A—C19A | 0.0 |
| C15—C7—C8—C13 | 68.6 (3) | C16A—C17A—C18A—O4A | −175.9 (6) |
| C15A—C7—C8—C13 | 81.3 (4) | C17A—C18A—C19A—C20A | 0.0 |
| C13—C8—C9—N1 | 174.5 (2) | O4A—C18A—C19A—C20A | 175.9 (6) |
| C7—C8—C9—N1 | −8.2 (4) | C18A—C19A—C20A—C15A | 0.0 |
| C13—C8—C9—C12 | −5.2 (4) | C16A—C15A—C20A—C19A | 0.0 |
| C7—C8—C9—C12 | 172.0 (2) | C7—C15A—C20A—C19A | 176.5 (8) |
| C1—N1—C9—C8 | −8.1 (4) | C19A—C18A—O4A—C21A | 75.5 (8) |
| C1—N1—C9—C12 | 171.7 (2) | C17A—C18A—O4A—C21A | −108.6 (7) |
| C14—O3—C13—O2 | −2.9 (4) | C18A—O4A—C21A—F2A | −66.7 (9) |
| C14—O3—C13—C8 | 175.9 (2) | C18A—O4A—C21A—F1A | 62.8 (8) |
Methyl 4-[4-(difluoromethoxy)phenyl]-2,7,7-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (I). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1i | 0.90 (3) | 1.93 (4) | 2.834 (3) | 174 (3) |
| C12—H12A···O2 | 0.98 | 2.32 | 2.831 (4) | 111 |
| C12—H12C···F2ii | 0.98 | 2.63 | 3.449 (5) | 141 |
| C12—H12C···F1Aii | 0.98 | 2.41 | 3.291 (7) | 150 |
| C14—H14C···O4Aiii | 0.98 | 2.66 | 3.551 (6) | 152 |
| C17—H17A···F1 | 0.95 | 2.43 | 2.975 (4) | 117 |
| C17—H17A···F1iv | 0.95 | 2.56 | 3.488 (4) | 165 |
| C21—H21A···O2v | 1.00 | 2.44 | 3.155 (5) | 128 |
| C21A—H21B···O2v | 1.00 | 2.50 | 3.062 (7) | 115 |
Symmetry codes: (i) x, −y+1, z−1/2; (ii) x−1/2, −y+1/2, z−1/2; (iii) −x+1/2, y−1/2, −z+3/2; (iv) −x+1, y, −z+3/2; (v) x+1/2, −y+1/2, z+1/2.
Isopropyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (II). Crystal data
| C23H27F2NO4 | Dx = 1.323 Mg m−3 |
| Mr = 419.45 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pbca | Cell parameters from 9744 reflections |
| a = 12.255 (3) Å | θ = 2.3–34.0° |
| b = 15.694 (3) Å | µ = 0.10 mm−1 |
| c = 21.903 (4) Å | T = 100 K |
| V = 4212.3 (14) Å3 | Plate, colorless |
| Z = 8 | 0.31 × 0.23 × 0.08 mm |
| F(000) = 1776 |
Isopropyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (II). Data collection
| Bruker D8 Quest with Photon 2 detector diffractometer | 6743 reflections with I > 2σ(I) |
| φ and ω scans | Rint = 0.073 |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | θmax = 34.1°, θmin = 2.3° |
| Tmin = 0.684, Tmax = 0.747 | h = −15→19 |
| 102650 measured reflections | k = −24→24 |
| 8537 independent reflections | l = −32→34 |
Isopropyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (II). Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.050 | Hydrogen site location: mixed |
| wR(F2) = 0.128 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0602P)2 + 1.7687P] where P = (Fo2 + 2Fc2)/3 |
| 8537 reflections | (Δ/σ)max < 0.001 |
| 280 parameters | Δρmax = 0.58 e Å−3 |
| 0 restraints | Δρmin = −0.42 e Å−3 |
Isopropyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (II). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Isopropyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (II). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| F1 | 0.58006 (8) | 0.07640 (5) | 0.03801 (3) | 0.0364 (2) | |
| F2 | 0.64472 (7) | 0.00608 (5) | 0.11460 (4) | 0.0349 (2) | |
| O1 | 0.34068 (6) | 0.45116 (5) | 0.16296 (3) | 0.01817 (15) | |
| O2 | 0.29601 (6) | 0.27319 (5) | 0.34751 (4) | 0.02216 (16) | |
| O3 | 0.41830 (6) | 0.29307 (5) | 0.42284 (3) | 0.01679 (14) | |
| O4 | 0.46707 (7) | 0.03988 (5) | 0.11352 (4) | 0.02388 (17) | |
| N1 | 0.63612 (7) | 0.39398 (5) | 0.29383 (4) | 0.01310 (14) | |
| C1 | 0.59145 (7) | 0.43524 (5) | 0.24486 (4) | 0.01197 (15) | |
| C2 | 0.66452 (8) | 0.49617 (6) | 0.21159 (5) | 0.01519 (17) | |
| H2A | 0.708624 | 0.464675 | 0.181174 | 0.018* | |
| H2B | 0.715096 | 0.523476 | 0.240904 | 0.018* | |
| C3 | 0.59723 (8) | 0.56435 (6) | 0.17941 (5) | 0.01630 (17) | |
| H3A | 0.645890 | 0.598521 | 0.152912 | 0.020* | |
| H3B | 0.565594 | 0.603050 | 0.210406 | 0.020* | |
| C4 | 0.50494 (8) | 0.52715 (6) | 0.14056 (4) | 0.01366 (16) | |
| C5 | 0.43666 (8) | 0.46417 (6) | 0.17775 (4) | 0.01252 (16) | |
| C6 | 0.48713 (7) | 0.41800 (5) | 0.22754 (4) | 0.01160 (15) | |
| C7 | 0.42689 (7) | 0.34420 (5) | 0.25648 (4) | 0.01133 (15) | |
| H7A | 0.347627 | 0.359096 | 0.258478 | 0.014* | |
| C8 | 0.46808 (7) | 0.33050 (6) | 0.32152 (4) | 0.01173 (15) | |
| C9 | 0.57283 (7) | 0.35044 (6) | 0.33615 (4) | 0.01219 (15) | |
| C10 | 0.43280 (9) | 0.59982 (7) | 0.11768 (6) | 0.0233 (2) | |
| H10A | 0.402326 | 0.630702 | 0.152635 | 0.035* | |
| H10B | 0.373259 | 0.576488 | 0.092926 | 0.035* | |
| H10C | 0.476524 | 0.638908 | 0.092797 | 0.035* | |
| C11 | 0.55042 (10) | 0.47676 (8) | 0.08592 (5) | 0.0232 (2) | |
| H11A | 0.599642 | 0.513276 | 0.062280 | 0.035* | |
| H11B | 0.489998 | 0.457985 | 0.059883 | 0.035* | |
| H11C | 0.590601 | 0.426920 | 0.100808 | 0.035* | |
| C12 | 0.63489 (8) | 0.33279 (7) | 0.39391 (4) | 0.01612 (17) | |
| H12A | 0.595564 | 0.290247 | 0.418173 | 0.024* | |
| H12B | 0.641987 | 0.385556 | 0.417509 | 0.024* | |
| H12C | 0.707624 | 0.311051 | 0.383743 | 0.024* | |
| C13 | 0.38614 (8) | 0.29573 (6) | 0.36382 (4) | 0.01362 (16) | |
| C14 | 0.34202 (9) | 0.25787 (7) | 0.46746 (5) | 0.0208 (2) | |
| H14A | 0.299229 | 0.210294 | 0.448771 | 0.025* | |
| C15 | 0.26612 (14) | 0.32793 (11) | 0.48827 (7) | 0.0461 (4) | |
| H15A | 0.226327 | 0.350637 | 0.453038 | 0.069* | |
| H15B | 0.308701 | 0.373654 | 0.507282 | 0.069* | |
| H15C | 0.214105 | 0.304965 | 0.518006 | 0.069* | |
| C16 | 0.41149 (11) | 0.22458 (8) | 0.51914 (5) | 0.0268 (2) | |
| H16A | 0.462584 | 0.181853 | 0.503408 | 0.040* | |
| H16B | 0.364532 | 0.198499 | 0.550169 | 0.040* | |
| H16C | 0.452451 | 0.271766 | 0.537407 | 0.040* | |
| C17 | 0.43931 (7) | 0.26381 (6) | 0.21739 (4) | 0.01218 (15) | |
| C18 | 0.54172 (8) | 0.23882 (6) | 0.19612 (5) | 0.01650 (17) | |
| H18A | 0.603453 | 0.272969 | 0.205585 | 0.020* | |
| C19 | 0.55622 (8) | 0.16531 (7) | 0.16141 (5) | 0.01900 (19) | |
| H19A | 0.626783 | 0.148978 | 0.147783 | 0.023* | |
| C20 | 0.46524 (8) | 0.11639 (6) | 0.14715 (4) | 0.01634 (17) | |
| C21 | 0.36233 (8) | 0.14000 (6) | 0.16660 (4) | 0.01612 (17) | |
| H21A | 0.300604 | 0.106507 | 0.155987 | 0.019* | |
| C22 | 0.34957 (8) | 0.21340 (6) | 0.20194 (4) | 0.01432 (16) | |
| H22A | 0.278885 | 0.229241 | 0.215665 | 0.017* | |
| C23 | 0.55424 (9) | 0.01649 (7) | 0.08023 (5) | 0.01990 (19) | |
| H23A | 0.537493 | −0.038295 | 0.058878 | 0.024* | |
| H1N | 0.7011 (13) | 0.4077 (9) | 0.3055 (7) | 0.020 (3)* |
Isopropyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (II). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| F1 | 0.0592 (6) | 0.0287 (4) | 0.0212 (3) | −0.0035 (4) | 0.0095 (3) | 0.0009 (3) |
| F2 | 0.0279 (4) | 0.0283 (4) | 0.0485 (5) | 0.0093 (3) | −0.0177 (3) | −0.0111 (3) |
| O1 | 0.0114 (3) | 0.0238 (3) | 0.0193 (3) | −0.0028 (3) | −0.0020 (3) | 0.0071 (3) |
| O2 | 0.0142 (3) | 0.0315 (4) | 0.0207 (3) | −0.0073 (3) | −0.0003 (3) | 0.0069 (3) |
| O3 | 0.0158 (3) | 0.0214 (3) | 0.0132 (3) | −0.0012 (3) | 0.0028 (2) | 0.0032 (2) |
| O4 | 0.0220 (4) | 0.0201 (3) | 0.0295 (4) | −0.0038 (3) | 0.0025 (3) | −0.0103 (3) |
| N1 | 0.0088 (3) | 0.0158 (3) | 0.0146 (3) | −0.0017 (3) | −0.0007 (3) | 0.0018 (3) |
| C1 | 0.0103 (3) | 0.0120 (3) | 0.0136 (4) | −0.0007 (3) | 0.0009 (3) | 0.0002 (3) |
| C2 | 0.0106 (4) | 0.0165 (4) | 0.0185 (4) | −0.0031 (3) | 0.0004 (3) | 0.0036 (3) |
| C3 | 0.0162 (4) | 0.0141 (4) | 0.0187 (4) | −0.0040 (3) | −0.0003 (3) | 0.0028 (3) |
| C4 | 0.0126 (4) | 0.0143 (4) | 0.0140 (4) | −0.0014 (3) | 0.0010 (3) | 0.0032 (3) |
| C5 | 0.0116 (4) | 0.0129 (3) | 0.0130 (4) | −0.0002 (3) | 0.0013 (3) | 0.0009 (3) |
| C6 | 0.0098 (3) | 0.0118 (3) | 0.0132 (4) | −0.0011 (3) | 0.0008 (3) | 0.0015 (3) |
| C7 | 0.0089 (3) | 0.0122 (3) | 0.0129 (4) | −0.0013 (3) | 0.0004 (3) | 0.0014 (3) |
| C8 | 0.0106 (3) | 0.0124 (3) | 0.0122 (4) | −0.0004 (3) | 0.0008 (3) | 0.0016 (3) |
| C9 | 0.0116 (4) | 0.0122 (3) | 0.0128 (4) | 0.0005 (3) | 0.0003 (3) | 0.0006 (3) |
| C10 | 0.0192 (5) | 0.0204 (4) | 0.0303 (5) | −0.0010 (4) | −0.0028 (4) | 0.0115 (4) |
| C11 | 0.0246 (5) | 0.0276 (5) | 0.0173 (4) | −0.0048 (4) | 0.0053 (4) | −0.0025 (4) |
| C12 | 0.0138 (4) | 0.0200 (4) | 0.0146 (4) | 0.0000 (3) | −0.0025 (3) | 0.0018 (3) |
| C13 | 0.0130 (4) | 0.0132 (3) | 0.0146 (4) | 0.0004 (3) | 0.0017 (3) | 0.0026 (3) |
| C14 | 0.0191 (5) | 0.0250 (5) | 0.0182 (4) | 0.0016 (4) | 0.0068 (4) | 0.0083 (4) |
| C15 | 0.0432 (8) | 0.0545 (9) | 0.0405 (7) | 0.0299 (7) | 0.0263 (7) | 0.0257 (7) |
| C16 | 0.0316 (6) | 0.0323 (6) | 0.0167 (4) | 0.0083 (5) | 0.0048 (4) | 0.0073 (4) |
| C17 | 0.0115 (4) | 0.0127 (3) | 0.0123 (4) | −0.0015 (3) | −0.0006 (3) | 0.0018 (3) |
| C18 | 0.0117 (4) | 0.0167 (4) | 0.0211 (4) | −0.0031 (3) | 0.0006 (3) | −0.0029 (3) |
| C19 | 0.0144 (4) | 0.0187 (4) | 0.0239 (5) | −0.0022 (3) | 0.0024 (4) | −0.0052 (4) |
| C20 | 0.0176 (4) | 0.0152 (4) | 0.0162 (4) | −0.0016 (3) | −0.0004 (3) | −0.0025 (3) |
| C21 | 0.0142 (4) | 0.0173 (4) | 0.0169 (4) | −0.0038 (3) | −0.0020 (3) | −0.0009 (3) |
| C22 | 0.0113 (4) | 0.0159 (4) | 0.0158 (4) | −0.0017 (3) | −0.0013 (3) | 0.0008 (3) |
| C23 | 0.0222 (5) | 0.0189 (4) | 0.0186 (4) | 0.0022 (4) | −0.0028 (4) | −0.0025 (3) |
Isopropyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (II). Geometric parameters (Å, º)
| F1—C23 | 1.3561 (13) | C10—H10A | 0.9800 |
| F2—C23 | 1.3501 (13) | C10—H10B | 0.9800 |
| O1—C5 | 1.2369 (12) | C10—H10C | 0.9800 |
| O2—C13 | 1.2136 (12) | C11—H11A | 0.9800 |
| O3—C13 | 1.3521 (12) | C11—H11B | 0.9800 |
| O3—C14 | 1.4608 (12) | C11—H11C | 0.9800 |
| O4—C23 | 1.3445 (14) | C12—H12A | 0.9800 |
| O4—C20 | 1.4089 (12) | C12—H12B | 0.9800 |
| N1—C1 | 1.3673 (12) | C12—H12C | 0.9800 |
| N1—C9 | 1.3884 (12) | C14—C16 | 1.5098 (16) |
| N1—H1N | 0.863 (16) | C14—C15 | 1.5106 (18) |
| C1—C6 | 1.3606 (13) | C14—H14A | 1.0000 |
| C1—C2 | 1.4991 (13) | C15—H15A | 0.9800 |
| C2—C3 | 1.5236 (14) | C15—H15B | 0.9800 |
| C2—H2A | 0.9900 | C15—H15C | 0.9800 |
| C2—H2B | 0.9900 | C16—H16A | 0.9800 |
| C3—C4 | 1.5311 (14) | C16—H16B | 0.9800 |
| C3—H3A | 0.9900 | C16—H16C | 0.9800 |
| C3—H3B | 0.9900 | C17—C18 | 1.3950 (13) |
| C4—C10 | 1.5276 (14) | C17—C22 | 1.3963 (13) |
| C4—C5 | 1.5297 (13) | C18—C19 | 1.3930 (14) |
| C4—C11 | 1.5390 (15) | C18—H18A | 0.9500 |
| C5—C6 | 1.4481 (13) | C19—C20 | 1.3892 (14) |
| C6—C7 | 1.5127 (12) | C19—H19A | 0.9500 |
| C7—C8 | 1.5265 (13) | C20—C21 | 1.3817 (14) |
| C7—C17 | 1.5323 (13) | C21—C22 | 1.3966 (14) |
| C7—H7A | 1.0000 | C21—H21A | 0.9500 |
| C8—C9 | 1.3595 (13) | C22—H22A | 0.9500 |
| C8—C13 | 1.4712 (13) | C23—H23A | 1.0000 |
| C9—C12 | 1.5019 (13) | ||
| C13—O3—C14 | 117.69 (8) | H11A—C11—H11C | 109.5 |
| C23—O4—C20 | 121.93 (9) | H11B—C11—H11C | 109.5 |
| C1—N1—C9 | 122.21 (8) | C9—C12—H12A | 109.5 |
| C1—N1—H1N | 118.8 (10) | C9—C12—H12B | 109.5 |
| C9—N1—H1N | 116.2 (10) | H12A—C12—H12B | 109.5 |
| C6—C1—N1 | 120.05 (8) | C9—C12—H12C | 109.5 |
| C6—C1—C2 | 123.54 (8) | H12A—C12—H12C | 109.5 |
| N1—C1—C2 | 116.37 (8) | H12B—C12—H12C | 109.5 |
| C1—C2—C3 | 110.46 (8) | O2—C13—O3 | 122.53 (9) |
| C1—C2—H2A | 109.6 | O2—C13—C8 | 122.95 (9) |
| C3—C2—H2A | 109.6 | O3—C13—C8 | 114.49 (8) |
| C1—C2—H2B | 109.6 | O3—C14—C16 | 105.76 (9) |
| C3—C2—H2B | 109.6 | O3—C14—C15 | 108.70 (9) |
| H2A—C2—H2B | 108.1 | C16—C14—C15 | 111.89 (11) |
| C2—C3—C4 | 112.90 (8) | O3—C14—H14A | 110.1 |
| C2—C3—H3A | 109.0 | C16—C14—H14A | 110.1 |
| C4—C3—H3A | 109.0 | C15—C14—H14A | 110.1 |
| C2—C3—H3B | 109.0 | C14—C15—H15A | 109.5 |
| C4—C3—H3B | 109.0 | C14—C15—H15B | 109.5 |
| H3A—C3—H3B | 107.8 | H15A—C15—H15B | 109.5 |
| C10—C4—C5 | 109.91 (8) | C14—C15—H15C | 109.5 |
| C10—C4—C3 | 108.98 (8) | H15A—C15—H15C | 109.5 |
| C5—C4—C3 | 110.77 (8) | H15B—C15—H15C | 109.5 |
| C10—C4—C11 | 109.76 (9) | C14—C16—H16A | 109.5 |
| C5—C4—C11 | 106.27 (8) | C14—C16—H16B | 109.5 |
| C3—C4—C11 | 111.13 (8) | H16A—C16—H16B | 109.5 |
| O1—C5—C6 | 121.38 (8) | C14—C16—H16C | 109.5 |
| O1—C5—C4 | 119.16 (8) | H16A—C16—H16C | 109.5 |
| C6—C5—C4 | 119.38 (8) | H16B—C16—H16C | 109.5 |
| C1—C6—C5 | 120.78 (8) | C18—C17—C22 | 117.93 (9) |
| C1—C6—C7 | 119.60 (8) | C18—C17—C7 | 120.50 (8) |
| C5—C6—C7 | 119.36 (8) | C22—C17—C7 | 121.58 (8) |
| C6—C7—C8 | 109.72 (7) | C19—C18—C17 | 121.99 (9) |
| C6—C7—C17 | 110.35 (7) | C19—C18—H18A | 119.0 |
| C8—C7—C17 | 111.88 (7) | C17—C18—H18A | 119.0 |
| C6—C7—H7A | 108.3 | C20—C19—C18 | 118.55 (9) |
| C8—C7—H7A | 108.3 | C20—C19—H19A | 120.7 |
| C17—C7—H7A | 108.3 | C18—C19—H19A | 120.7 |
| C9—C8—C13 | 125.54 (8) | C21—C20—C19 | 121.00 (9) |
| C9—C8—C7 | 119.98 (8) | C21—C20—O4 | 113.85 (9) |
| C13—C8—C7 | 114.47 (8) | C19—C20—O4 | 125.15 (9) |
| C8—C9—N1 | 118.91 (8) | C20—C21—C22 | 119.62 (9) |
| C8—C9—C12 | 129.36 (8) | C20—C21—H21A | 120.2 |
| N1—C9—C12 | 111.73 (8) | C22—C21—H21A | 120.2 |
| C4—C10—H10A | 109.5 | C17—C22—C21 | 120.89 (9) |
| C4—C10—H10B | 109.5 | C17—C22—H22A | 119.6 |
| H10A—C10—H10B | 109.5 | C21—C22—H22A | 119.6 |
| C4—C10—H10C | 109.5 | O4—C23—F2 | 112.53 (9) |
| H10A—C10—H10C | 109.5 | O4—C23—F1 | 111.46 (9) |
| H10B—C10—H10C | 109.5 | F2—C23—F1 | 105.81 (10) |
| C4—C11—H11A | 109.5 | O4—C23—H23A | 109.0 |
| C4—C11—H11B | 109.5 | F2—C23—H23A | 109.0 |
| H11A—C11—H11B | 109.5 | F1—C23—H23A | 109.0 |
| C4—C11—H11C | 109.5 | ||
| C9—N1—C1—C6 | −16.13 (13) | C7—C8—C9—N1 | 8.88 (13) |
| C9—N1—C1—C2 | 165.96 (8) | C13—C8—C9—C12 | 8.63 (16) |
| C6—C1—C2—C3 | 27.78 (13) | C7—C8—C9—C12 | −171.62 (9) |
| N1—C1—C2—C3 | −154.38 (8) | C1—N1—C9—C8 | 16.09 (13) |
| C1—C2—C3—C4 | −50.39 (11) | C1—N1—C9—C12 | −163.49 (8) |
| C2—C3—C4—C10 | 172.05 (8) | C14—O3—C13—O2 | 2.82 (14) |
| C2—C3—C4—C5 | 51.01 (11) | C14—O3—C13—C8 | −178.91 (8) |
| C2—C3—C4—C11 | −66.87 (11) | C9—C8—C13—O2 | −174.48 (10) |
| C10—C4—C5—O1 | 34.21 (12) | C7—C8—C13—O2 | 5.75 (13) |
| C3—C4—C5—O1 | 154.69 (9) | C9—C8—C13—O3 | 7.26 (13) |
| C11—C4—C5—O1 | −84.49 (11) | C7—C8—C13—O3 | −172.51 (8) |
| C10—C4—C5—C6 | −148.80 (9) | C13—O3—C14—C16 | 153.97 (9) |
| C3—C4—C5—C6 | −28.32 (12) | C13—O3—C14—C15 | −85.73 (13) |
| C11—C4—C5—C6 | 92.50 (10) | C6—C7—C17—C18 | 48.03 (11) |
| N1—C1—C6—C5 | 176.85 (8) | C8—C7—C17—C18 | −74.44 (11) |
| C2—C1—C6—C5 | −5.39 (14) | C6—C7—C17—C22 | −132.01 (9) |
| N1—C1—C6—C7 | −9.05 (13) | C8—C7—C17—C22 | 105.53 (10) |
| C2—C1—C6—C7 | 168.71 (8) | C22—C17—C18—C19 | −1.13 (15) |
| O1—C5—C6—C1 | −177.45 (9) | C7—C17—C18—C19 | 178.83 (9) |
| C4—C5—C6—C1 | 5.63 (13) | C17—C18—C19—C20 | 0.81 (16) |
| O1—C5—C6—C7 | 8.44 (13) | C18—C19—C20—C21 | 0.29 (16) |
| C4—C5—C6—C7 | −168.48 (8) | C18—C19—C20—O4 | −178.78 (10) |
| C1—C6—C7—C8 | 29.90 (11) | C23—O4—C20—C21 | 165.58 (10) |
| C5—C6—C7—C8 | −155.92 (8) | C23—O4—C20—C19 | −15.29 (16) |
| C1—C6—C7—C17 | −93.83 (10) | C19—C20—C21—C22 | −1.02 (15) |
| C5—C6—C7—C17 | 80.35 (10) | O4—C20—C21—C22 | 178.15 (9) |
| C6—C7—C8—C9 | −29.89 (11) | C18—C17—C22—C21 | 0.38 (14) |
| C17—C7—C8—C9 | 92.94 (10) | C7—C17—C22—C21 | −179.59 (8) |
| C6—C7—C8—C13 | 149.89 (8) | C20—C21—C22—C17 | 0.68 (15) |
| C17—C7—C8—C13 | −87.29 (9) | C20—O4—C23—F2 | 61.18 (13) |
| C13—C8—C9—N1 | −170.87 (8) | C20—O4—C23—F1 | −57.50 (13) |
Isopropyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (II). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1i | 0.863 (16) | 1.967 (16) | 2.8258 (12) | 173.0 (14) |
| C2—H2A···F2ii | 0.99 | 2.40 | 3.1626 (13) | 133 |
| C12—H12A···O3 | 0.98 | 2.18 | 2.7991 (14) | 120 |
| C19—H19A···F2 | 0.95 | 2.37 | 2.9106 (14) | 116 |
| C23—H23A···F1iii | 1.00 | 2.63 | 3.3972 (14) | 133 |
Symmetry codes: (i) x+1/2, y, −z+1/2; (ii) −x+3/2, y+1/2, z; (iii) −x+1, −y, −z.
tert-Butyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (III) . Crystal data
| C24H29F2NO4 | Dx = 1.322 Mg m−3 |
| Mr = 433.48 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pbca | Cell parameters from 8996 reflections |
| a = 12.4094 (8) Å | θ = 2.3–30.3° |
| b = 15.9871 (12) Å | µ = 0.10 mm−1 |
| c = 21.9629 (15) Å | T = 100 K |
| V = 4357.2 (5) Å3 | Plate, colorless |
| Z = 8 | 0.31 × 0.27 × 0.09 mm |
| F(000) = 1840 |
tert-Butyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (III) . Data collection
| Bruker APEXII CCD diffractometer | 4732 reflections with I > 2σ(I) |
| φ and ω scans | Rint = 0.142 |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | θmax = 30.6°, θmin = 1.9° |
| Tmin = 0.374, Tmax = 0.746 | h = −17→17 |
| 56620 measured reflections | k = −22→22 |
| 6654 independent reflections | l = −31→31 |
tert-Butyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (III) . Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.059 | Hydrogen site location: mixed |
| wR(F2) = 0.163 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0685P)2 + 0.6147P] where P = (Fo2 + 2Fc2)/3 |
| 6654 reflections | (Δ/σ)max = 0.001 |
| 329 parameters | Δρmax = 0.37 e Å−3 |
| 68 restraints | Δρmin = −0.31 e Å−3 |
tert-Butyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (III) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
tert-Butyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (III) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| F1 | 0.57517 (9) | 0.94793 (6) | 0.95792 (5) | 0.0321 (2) | |
| F2 | 0.64034 (9) | 1.01340 (7) | 0.88027 (6) | 0.0401 (3) | |
| O1 | 0.32590 (9) | 0.56945 (7) | 0.83355 (5) | 0.0243 (2) | |
| O2 | 0.46372 (9) | 0.98316 (7) | 0.88197 (5) | 0.0257 (3) | |
| O3 | 0.28837 (10) | 0.73942 (9) | 0.65077 (6) | 0.0347 (3) | |
| O4 | 0.41461 (9) | 0.72089 (6) | 0.57815 (5) | 0.0202 (2) | |
| N1 | 0.62614 (10) | 0.63107 (7) | 0.71106 (6) | 0.0164 (2) | |
| C1 | 0.56462 (11) | 0.67209 (8) | 0.66740 (6) | 0.0153 (3) | |
| C2 | 0.46038 (11) | 0.69097 (8) | 0.68021 (6) | 0.0161 (3) | |
| C3 | 0.41866 (11) | 0.67977 (8) | 0.74513 (6) | 0.0161 (3) | |
| H3A | 0.340091 | 0.666165 | 0.743048 | 0.019* | |
| C4 | 0.47622 (11) | 0.60720 (8) | 0.77510 (6) | 0.0168 (3) | |
| C5 | 0.42280 (11) | 0.55954 (8) | 0.82239 (7) | 0.0174 (3) | |
| C6 | 0.48853 (12) | 0.49691 (8) | 0.85970 (7) | 0.0185 (3) | |
| C7 | 0.58499 (18) | 0.46128 (13) | 0.82234 (11) | 0.0190 (5) | 0.646 (3) |
| H7A | 0.632708 | 0.429090 | 0.849834 | 0.023* | 0.646 (3) |
| H7B | 0.557191 | 0.422424 | 0.790953 | 0.023* | 0.646 (3) |
| C8 | 0.6512 (9) | 0.5311 (7) | 0.7910 (4) | 0.0193 (9) | 0.646 (3) |
| H8A | 0.694593 | 0.560970 | 0.821974 | 0.023* | 0.646 (3) |
| H8B | 0.701396 | 0.505677 | 0.761262 | 0.023* | 0.646 (3) |
| C11 | 0.5333 (2) | 0.54966 (15) | 0.91386 (11) | 0.0229 (5) | 0.646 (3) |
| H11A | 0.571754 | 0.598568 | 0.897985 | 0.034* | 0.646 (3) |
| H11B | 0.582927 | 0.515342 | 0.937978 | 0.034* | 0.646 (3) |
| H11C | 0.473433 | 0.568179 | 0.939625 | 0.034* | 0.646 (3) |
| C12 | 0.4219 (2) | 0.42518 (15) | 0.88388 (12) | 0.0263 (6) | 0.646 (3) |
| H12A | 0.361872 | 0.447214 | 0.907996 | 0.039* | 0.646 (3) |
| H12B | 0.467089 | 0.389375 | 0.909586 | 0.039* | 0.646 (3) |
| H12C | 0.393637 | 0.392387 | 0.849735 | 0.039* | 0.646 (3) |
| C7A | 0.6013 (3) | 0.5198 (3) | 0.86174 (19) | 0.0187 (8) | 0.354 (3) |
| H7AA | 0.641688 | 0.476110 | 0.884062 | 0.022* | 0.354 (3) |
| H7AB | 0.609014 | 0.572916 | 0.884527 | 0.022* | 0.354 (3) |
| C8A | 0.6504 (16) | 0.5304 (12) | 0.7986 (7) | 0.0181 (13) | 0.354 (3) |
| H8AA | 0.654909 | 0.475285 | 0.778147 | 0.022* | 0.354 (3) |
| H8AB | 0.724400 | 0.553046 | 0.802297 | 0.022* | 0.354 (3) |
| C11A | 0.4353 (4) | 0.4888 (3) | 0.9231 (2) | 0.0263 (10) | 0.354 (3) |
| H11D | 0.449832 | 0.539399 | 0.946901 | 0.039* | 0.354 (3) |
| H11E | 0.464945 | 0.440054 | 0.944246 | 0.039* | 0.354 (3) |
| H11F | 0.357278 | 0.481932 | 0.918181 | 0.039* | 0.354 (3) |
| C12A | 0.4654 (4) | 0.4132 (3) | 0.8248 (2) | 0.0243 (9) | 0.354 (3) |
| H12D | 0.387682 | 0.402116 | 0.824822 | 0.036* | 0.354 (3) |
| H12E | 0.503045 | 0.367006 | 0.845007 | 0.036* | 0.354 (3) |
| H12F | 0.490895 | 0.418052 | 0.782704 | 0.036* | 0.354 (3) |
| C9 | 0.58058 (11) | 0.59079 (8) | 0.75968 (6) | 0.0167 (3) | |
| C10 | 0.62756 (12) | 0.68867 (9) | 0.61031 (7) | 0.0207 (3) | |
| H10A | 0.595418 | 0.735945 | 0.588470 | 0.031* | |
| H10B | 0.626017 | 0.638871 | 0.584301 | 0.031* | |
| H10C | 0.702331 | 0.702011 | 0.620876 | 0.031* | |
| C13 | 0.37937 (12) | 0.72026 (9) | 0.63625 (7) | 0.0186 (3) | |
| C14 | 0.34411 (13) | 0.74324 (9) | 0.52654 (7) | 0.0214 (3) | |
| C15 | 0.2548 (2) | 0.67935 (14) | 0.52089 (12) | 0.0622 (8) | |
| H15A | 0.207089 | 0.683389 | 0.556308 | 0.093* | |
| H15B | 0.286095 | 0.623115 | 0.519003 | 0.093* | |
| H15C | 0.213484 | 0.690132 | 0.483698 | 0.093* | |
| C16 | 0.4207 (2) | 0.73950 (15) | 0.47310 (8) | 0.0477 (6) | |
| H16A | 0.480326 | 0.778580 | 0.479785 | 0.072* | |
| H16B | 0.382166 | 0.754898 | 0.435784 | 0.072* | |
| H16C | 0.449097 | 0.682580 | 0.469071 | 0.072* | |
| C17 | 0.30051 (15) | 0.83103 (10) | 0.53339 (7) | 0.0292 (4) | |
| H17A | 0.360098 | 0.869678 | 0.541516 | 0.044* | |
| H17B | 0.249301 | 0.832725 | 0.567338 | 0.044* | |
| H17C | 0.263853 | 0.847613 | 0.495754 | 0.044* | |
| C18 | 0.43219 (11) | 0.75974 (9) | 0.78268 (6) | 0.0168 (3) | |
| C19 | 0.34560 (12) | 0.81251 (9) | 0.79479 (7) | 0.0192 (3) | |
| H19A | 0.276094 | 0.798378 | 0.779861 | 0.023* | |
| C20 | 0.35903 (12) | 0.88595 (9) | 0.82852 (7) | 0.0213 (3) | |
| H20A | 0.299016 | 0.920984 | 0.836923 | 0.026* | |
| C21 | 0.46028 (12) | 0.90708 (9) | 0.84950 (7) | 0.0192 (3) | |
| C22 | 0.54853 (13) | 0.85619 (10) | 0.83835 (7) | 0.0247 (3) | |
| H22A | 0.618017 | 0.871002 | 0.852956 | 0.030* | |
| C23 | 0.53307 (12) | 0.78277 (10) | 0.80522 (7) | 0.0229 (3) | |
| H23A | 0.593071 | 0.747316 | 0.797753 | 0.028* | |
| C24 | 0.55049 (13) | 1.00616 (9) | 0.91461 (7) | 0.0233 (3) | |
| H24A | 0.535474 | 1.060914 | 0.934921 | 0.028* | |
| H1N | 0.6911 (17) | 0.6159 (12) | 0.6999 (9) | 0.030 (5)* |
tert-Butyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (III) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| F1 | 0.0440 (6) | 0.0243 (5) | 0.0281 (5) | 0.0072 (4) | −0.0078 (5) | −0.0008 (4) |
| F2 | 0.0317 (6) | 0.0286 (5) | 0.0601 (7) | −0.0069 (4) | 0.0200 (5) | −0.0048 (5) |
| O1 | 0.0156 (5) | 0.0303 (6) | 0.0270 (6) | 0.0012 (4) | 0.0020 (5) | 0.0096 (4) |
| O2 | 0.0261 (6) | 0.0196 (5) | 0.0313 (6) | 0.0068 (4) | −0.0035 (5) | −0.0056 (4) |
| O3 | 0.0185 (6) | 0.0584 (8) | 0.0272 (6) | 0.0125 (5) | 0.0026 (5) | 0.0157 (6) |
| O4 | 0.0201 (5) | 0.0228 (5) | 0.0176 (5) | 0.0039 (4) | −0.0023 (4) | 0.0010 (4) |
| N1 | 0.0112 (5) | 0.0181 (5) | 0.0199 (6) | 0.0011 (4) | 0.0002 (5) | 0.0010 (4) |
| C1 | 0.0147 (6) | 0.0137 (6) | 0.0175 (6) | −0.0006 (5) | −0.0017 (5) | −0.0001 (5) |
| C2 | 0.0155 (6) | 0.0143 (6) | 0.0186 (6) | −0.0001 (5) | −0.0007 (5) | 0.0019 (5) |
| C3 | 0.0121 (6) | 0.0179 (6) | 0.0184 (6) | 0.0020 (5) | −0.0003 (5) | 0.0022 (5) |
| C4 | 0.0136 (6) | 0.0175 (6) | 0.0194 (6) | 0.0003 (5) | −0.0028 (6) | 0.0035 (5) |
| C5 | 0.0154 (6) | 0.0176 (6) | 0.0193 (6) | −0.0006 (5) | −0.0023 (6) | 0.0019 (5) |
| C6 | 0.0193 (7) | 0.0162 (6) | 0.0201 (7) | 0.0014 (5) | −0.0006 (6) | 0.0034 (5) |
| C7 | 0.0182 (10) | 0.0158 (9) | 0.0231 (10) | 0.0033 (7) | −0.0011 (9) | 0.0010 (7) |
| C8 | 0.0145 (14) | 0.0217 (14) | 0.022 (2) | −0.0021 (12) | −0.0006 (15) | 0.0053 (15) |
| C11 | 0.0241 (12) | 0.0249 (11) | 0.0195 (10) | 0.0039 (9) | −0.0038 (10) | 0.0001 (8) |
| C12 | 0.0198 (11) | 0.0246 (11) | 0.0345 (13) | −0.0014 (9) | 0.0000 (10) | 0.0132 (10) |
| C7A | 0.0147 (15) | 0.0200 (15) | 0.0213 (16) | 0.0008 (13) | −0.0055 (14) | 0.0033 (13) |
| C8A | 0.013 (2) | 0.018 (2) | 0.023 (3) | 0.007 (2) | −0.003 (2) | 0.008 (2) |
| C11A | 0.024 (2) | 0.032 (2) | 0.023 (2) | 0.0093 (17) | 0.0002 (18) | 0.0048 (17) |
| C12A | 0.024 (2) | 0.0181 (18) | 0.031 (2) | −0.0044 (15) | 0.0053 (18) | −0.0015 (16) |
| C9 | 0.0146 (6) | 0.0153 (6) | 0.0203 (7) | −0.0006 (5) | −0.0022 (6) | 0.0015 (5) |
| C10 | 0.0172 (7) | 0.0236 (7) | 0.0212 (7) | −0.0008 (5) | 0.0019 (6) | 0.0012 (5) |
| C13 | 0.0162 (6) | 0.0203 (6) | 0.0192 (7) | −0.0002 (5) | −0.0008 (6) | 0.0039 (5) |
| C14 | 0.0269 (8) | 0.0177 (6) | 0.0196 (7) | 0.0011 (6) | −0.0087 (6) | 0.0010 (5) |
| C15 | 0.0774 (17) | 0.0428 (11) | 0.0665 (15) | −0.0359 (12) | −0.0534 (14) | 0.0279 (11) |
| C16 | 0.0590 (14) | 0.0649 (13) | 0.0190 (8) | 0.0378 (11) | −0.0010 (9) | −0.0003 (8) |
| C17 | 0.0353 (9) | 0.0287 (8) | 0.0236 (7) | 0.0132 (7) | −0.0059 (7) | −0.0002 (6) |
| C18 | 0.0151 (7) | 0.0198 (6) | 0.0156 (6) | 0.0018 (5) | 0.0014 (5) | 0.0022 (5) |
| C19 | 0.0141 (6) | 0.0197 (6) | 0.0239 (7) | 0.0018 (5) | 0.0007 (6) | 0.0038 (5) |
| C20 | 0.0183 (7) | 0.0193 (6) | 0.0262 (7) | 0.0055 (5) | 0.0034 (6) | 0.0026 (5) |
| C21 | 0.0218 (7) | 0.0177 (6) | 0.0181 (6) | 0.0029 (5) | 0.0011 (6) | 0.0005 (5) |
| C22 | 0.0172 (7) | 0.0293 (8) | 0.0274 (8) | 0.0046 (6) | −0.0044 (6) | −0.0068 (6) |
| C23 | 0.0164 (7) | 0.0269 (7) | 0.0255 (7) | 0.0063 (6) | −0.0009 (6) | −0.0073 (6) |
| C24 | 0.0223 (7) | 0.0178 (6) | 0.0299 (8) | 0.0001 (5) | 0.0027 (7) | −0.0004 (6) |
tert-Butyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (III) . Geometric parameters (Å, º)
| F1—C24 | 1.3658 (18) | C7A—C8A | 1.525 (16) |
| F2—C24 | 1.3510 (19) | C7A—H7AA | 0.9900 |
| O1—C5 | 1.2374 (18) | C7A—H7AB | 0.9900 |
| O2—C24 | 1.3449 (19) | C8A—C9 | 1.553 (18) |
| O2—C21 | 1.4107 (17) | C8A—H8AA | 0.9900 |
| O3—C13 | 1.2127 (19) | C8A—H8AB | 0.9900 |
| O4—C13 | 1.3490 (18) | C11A—H11D | 0.9800 |
| O4—C14 | 1.4757 (17) | C11A—H11E | 0.9800 |
| N1—C9 | 1.3692 (18) | C11A—H11F | 0.9800 |
| N1—C1 | 1.3902 (18) | C12A—H12D | 0.9800 |
| N1—H1N | 0.88 (2) | C12A—H12E | 0.9800 |
| C1—C2 | 1.3578 (19) | C12A—H12F | 0.9800 |
| C1—C10 | 1.501 (2) | C10—H10A | 0.9800 |
| C2—C13 | 1.470 (2) | C10—H10B | 0.9800 |
| C2—C3 | 1.5273 (19) | C10—H10C | 0.9800 |
| C3—C4 | 1.5132 (18) | C14—C16 | 1.511 (3) |
| C3—C18 | 1.531 (2) | C14—C17 | 1.512 (2) |
| C3—H3A | 1.0000 | C14—C15 | 1.512 (2) |
| C4—C9 | 1.364 (2) | C15—H15A | 0.9800 |
| C4—C5 | 1.449 (2) | C15—H15B | 0.9800 |
| C5—C6 | 1.5295 (19) | C15—H15C | 0.9800 |
| C6—C7A | 1.447 (4) | C16—H16A | 0.9800 |
| C6—C12 | 1.510 (3) | C16—H16B | 0.9800 |
| C6—C11A | 1.546 (5) | C16—H16C | 0.9800 |
| C6—C7 | 1.559 (3) | C17—H17A | 0.9800 |
| C6—C11 | 1.560 (3) | C17—H17B | 0.9800 |
| C6—C12A | 1.570 (4) | C17—H17C | 0.9800 |
| C7—C8 | 1.547 (10) | C18—C19 | 1.3918 (19) |
| C7—H7A | 0.9900 | C18—C23 | 1.396 (2) |
| C7—H7B | 0.9900 | C19—C20 | 1.398 (2) |
| C8—C9 | 1.468 (11) | C19—H19A | 0.9500 |
| C8—H8A | 0.9900 | C20—C21 | 1.380 (2) |
| C8—H8B | 0.9900 | C20—H20A | 0.9500 |
| C11—H11A | 0.9800 | C21—C22 | 1.386 (2) |
| C11—H11B | 0.9800 | C22—C23 | 1.394 (2) |
| C11—H11C | 0.9800 | C22—H22A | 0.9500 |
| C12—H12A | 0.9800 | C23—H23A | 0.9500 |
| C12—H12B | 0.9800 | C24—H24A | 1.0000 |
| C12—H12C | 0.9800 | ||
| C24—O2—C21 | 121.96 (12) | H11D—C11A—H11E | 109.5 |
| C13—O4—C14 | 122.42 (12) | C6—C11A—H11F | 109.5 |
| C9—N1—C1 | 122.21 (12) | H11D—C11A—H11F | 109.5 |
| C9—N1—H1N | 117.9 (13) | H11E—C11A—H11F | 109.5 |
| C1—N1—H1N | 116.2 (13) | C6—C12A—H12D | 109.5 |
| C2—C1—N1 | 119.02 (13) | C6—C12A—H12E | 109.5 |
| C2—C1—C10 | 129.03 (13) | H12D—C12A—H12E | 109.5 |
| N1—C1—C10 | 111.95 (12) | C6—C12A—H12F | 109.5 |
| C1—C2—C13 | 125.87 (13) | H12D—C12A—H12F | 109.5 |
| C1—C2—C3 | 119.37 (12) | H12E—C12A—H12F | 109.5 |
| C13—C2—C3 | 114.74 (12) | C4—C9—N1 | 119.68 (13) |
| C4—C3—C2 | 109.63 (11) | C4—C9—C8 | 125.1 (4) |
| C4—C3—C18 | 110.75 (11) | N1—C9—C8 | 115.2 (4) |
| C2—C3—C18 | 111.59 (11) | C4—C9—C8A | 120.9 (7) |
| C4—C3—H3A | 108.3 | N1—C9—C8A | 119.4 (7) |
| C2—C3—H3A | 108.3 | C1—C10—H10A | 109.5 |
| C18—C3—H3A | 108.3 | C1—C10—H10B | 109.5 |
| C9—C4—C5 | 120.75 (13) | H10A—C10—H10B | 109.5 |
| C9—C4—C3 | 119.19 (12) | C1—C10—H10C | 109.5 |
| C5—C4—C3 | 119.94 (12) | H10A—C10—H10C | 109.5 |
| O1—C5—C4 | 121.30 (13) | H10B—C10—H10C | 109.5 |
| O1—C5—C6 | 119.73 (13) | O3—C13—O4 | 123.28 (14) |
| C4—C5—C6 | 118.97 (12) | O3—C13—C2 | 122.98 (14) |
| C7A—C6—C5 | 111.51 (19) | O4—C13—C2 | 113.70 (12) |
| C12—C6—C5 | 113.18 (14) | O4—C14—C16 | 102.37 (13) |
| C7A—C6—C11A | 114.0 (3) | O4—C14—C17 | 111.14 (12) |
| C5—C6—C11A | 107.99 (19) | C16—C14—C17 | 109.82 (14) |
| C12—C6—C7 | 109.13 (15) | O4—C14—C15 | 109.50 (13) |
| C5—C6—C7 | 111.51 (13) | C16—C14—C15 | 111.71 (19) |
| C12—C6—C11 | 109.71 (17) | C17—C14—C15 | 111.91 (17) |
| C5—C6—C11 | 104.16 (13) | C14—C15—H15A | 109.5 |
| C7—C6—C11 | 108.97 (16) | C14—C15—H15B | 109.5 |
| C7A—C6—C12A | 114.1 (3) | H15A—C15—H15B | 109.5 |
| C5—C6—C12A | 101.5 (2) | C14—C15—H15C | 109.5 |
| C11A—C6—C12A | 106.9 (3) | H15A—C15—H15C | 109.5 |
| C8—C7—C6 | 112.2 (4) | H15B—C15—H15C | 109.5 |
| C8—C7—H7A | 109.2 | C14—C16—H16A | 109.5 |
| C6—C7—H7A | 109.2 | C14—C16—H16B | 109.5 |
| C8—C7—H7B | 109.2 | H16A—C16—H16B | 109.5 |
| C6—C7—H7B | 109.2 | C14—C16—H16C | 109.5 |
| H7A—C7—H7B | 107.9 | H16A—C16—H16C | 109.5 |
| C9—C8—C7 | 111.1 (7) | H16B—C16—H16C | 109.5 |
| C9—C8—H8A | 109.4 | C14—C17—H17A | 109.5 |
| C7—C8—H8A | 109.4 | C14—C17—H17B | 109.5 |
| C9—C8—H8B | 109.4 | H17A—C17—H17B | 109.5 |
| C7—C8—H8B | 109.4 | C14—C17—H17C | 109.5 |
| H8A—C8—H8B | 108.0 | H17A—C17—H17C | 109.5 |
| C6—C11—H11A | 109.5 | H17B—C17—H17C | 109.5 |
| C6—C11—H11B | 109.5 | C19—C18—C23 | 117.70 (13) |
| H11A—C11—H11B | 109.5 | C19—C18—C3 | 121.64 (13) |
| C6—C11—H11C | 109.5 | C23—C18—C3 | 120.65 (12) |
| H11A—C11—H11C | 109.5 | C18—C19—C20 | 121.21 (14) |
| H11B—C11—H11C | 109.5 | C18—C19—H19A | 119.4 |
| C6—C12—H12A | 109.5 | C20—C19—H19A | 119.4 |
| C6—C12—H12B | 109.5 | C21—C20—C19 | 119.40 (13) |
| H12A—C12—H12B | 109.5 | C21—C20—H20A | 120.3 |
| C6—C12—H12C | 109.5 | C19—C20—H20A | 120.3 |
| H12A—C12—H12C | 109.5 | C20—C21—C22 | 121.11 (14) |
| H12B—C12—H12C | 109.5 | C20—C21—O2 | 114.03 (13) |
| C6—C7A—C8A | 112.7 (8) | C22—C21—O2 | 124.86 (14) |
| C6—C7A—H7AA | 109.0 | C21—C22—C23 | 118.53 (14) |
| C8A—C7A—H7AA | 109.0 | C21—C22—H22A | 120.7 |
| C6—C7A—H7AB | 109.0 | C23—C22—H22A | 120.7 |
| C8A—C7A—H7AB | 109.0 | C22—C23—C18 | 122.05 (14) |
| H7AA—C7A—H7AB | 107.8 | C22—C23—H23A | 119.0 |
| C7A—C8A—C9 | 110.2 (11) | C18—C23—H23A | 119.0 |
| C7A—C8A—H8AA | 109.6 | O2—C24—F2 | 112.74 (14) |
| C9—C8A—H8AA | 109.6 | O2—C24—F1 | 111.38 (12) |
| C7A—C8A—H8AB | 109.6 | F2—C24—F1 | 105.20 (13) |
| C9—C8A—H8AB | 109.6 | O2—C24—H24A | 109.1 |
| H8AA—C8A—H8AB | 108.1 | F2—C24—H24A | 109.1 |
| C6—C11A—H11D | 109.5 | F1—C24—H24A | 109.1 |
| C6—C11A—H11E | 109.5 | ||
| C9—N1—C1—C2 | 16.6 (2) | C3—C4—C9—N1 | −10.2 (2) |
| C9—N1—C1—C10 | −162.96 (12) | C5—C4—C9—C8 | −5.5 (5) |
| N1—C1—C2—C13 | −168.52 (13) | C3—C4—C9—C8 | 170.5 (5) |
| C10—C1—C2—C13 | 10.9 (2) | C5—C4—C9—C8A | −9.4 (8) |
| N1—C1—C2—C3 | 9.82 (19) | C3—C4—C9—C8A | 166.6 (8) |
| C10—C1—C2—C3 | −170.73 (13) | C1—N1—C9—C4 | −16.5 (2) |
| C1—C2—C3—C4 | −32.19 (17) | C1—N1—C9—C8 | 162.9 (4) |
| C13—C2—C3—C4 | 146.33 (12) | C1—N1—C9—C8A | 166.7 (8) |
| C1—C2—C3—C18 | 90.88 (15) | C7—C8—C9—C4 | 26.3 (8) |
| C13—C2—C3—C18 | −90.60 (14) | C7—C8—C9—N1 | −153.0 (4) |
| C2—C3—C4—C9 | 32.34 (18) | C7A—C8A—C9—C4 | −19.0 (15) |
| C18—C3—C4—C9 | −91.22 (16) | C7A—C8A—C9—N1 | 157.8 (7) |
| C2—C3—C4—C5 | −151.65 (13) | C14—O4—C13—O3 | −1.5 (2) |
| C18—C3—C4—C5 | 84.79 (16) | C14—O4—C13—C2 | 176.23 (12) |
| C9—C4—C5—O1 | −174.39 (14) | C1—C2—C13—O3 | −176.94 (15) |
| C3—C4—C5—O1 | 9.7 (2) | C3—C2—C13—O3 | 4.6 (2) |
| C9—C4—C5—C6 | 6.6 (2) | C1—C2—C13—O4 | 5.3 (2) |
| C3—C4—C5—C6 | −169.33 (12) | C3—C2—C13—O4 | −173.09 (12) |
| O1—C5—C6—C7A | −152.9 (2) | C13—O4—C14—C16 | 176.95 (15) |
| C4—C5—C6—C7A | 26.1 (2) | C13—O4—C14—C17 | 59.76 (18) |
| O1—C5—C6—C12 | 28.8 (2) | C13—O4—C14—C15 | −64.4 (2) |
| C4—C5—C6—C12 | −152.19 (17) | C4—C3—C18—C19 | −134.72 (14) |
| O1—C5—C6—C11A | −26.9 (3) | C2—C3—C18—C19 | 102.85 (15) |
| C4—C5—C6—C11A | 152.1 (2) | C4—C3—C18—C23 | 46.27 (18) |
| O1—C5—C6—C7 | 152.31 (15) | C2—C3—C18—C23 | −76.16 (16) |
| C4—C5—C6—C7 | −28.68 (19) | C23—C18—C19—C20 | −0.2 (2) |
| O1—C5—C6—C11 | −90.31 (18) | C3—C18—C19—C20 | −179.23 (13) |
| C4—C5—C6—C11 | 88.70 (17) | C18—C19—C20—C21 | 0.9 (2) |
| O1—C5—C6—C12A | 85.2 (2) | C19—C20—C21—C22 | −0.8 (2) |
| C4—C5—C6—C12A | −95.7 (2) | C19—C20—C21—O2 | 178.96 (13) |
| C12—C6—C7—C8 | 174.7 (4) | C24—O2—C21—C20 | 168.08 (14) |
| C5—C6—C7—C8 | 48.9 (4) | C24—O2—C21—C22 | −12.1 (2) |
| C11—C6—C7—C8 | −65.5 (4) | C20—C21—C22—C23 | 0.1 (2) |
| C6—C7—C8—C9 | −47.3 (6) | O2—C21—C22—C23 | −179.67 (14) |
| C5—C6—C7A—C8A | −55.3 (8) | C21—C22—C23—C18 | 0.6 (2) |
| C11A—C6—C7A—C8A | −177.9 (8) | C19—C18—C23—C22 | −0.6 (2) |
| C12A—C6—C7A—C8A | 58.9 (9) | C3—C18—C23—C22 | 178.48 (14) |
| C6—C7A—C8A—C9 | 52.0 (13) | C21—O2—C24—F2 | 60.27 (18) |
| C5—C4—C9—N1 | 173.81 (13) | C21—O2—C24—F1 | −57.73 (18) |
tert-Butyl 4-[4-(difluoromethoxy)phenyl]-2,6,6-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (III) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1i | 0.88 (2) | 1.97 (2) | 2.8418 (16) | 171.2 (19) |
| C8A—H8A···F2ii | 0.99 | 2.53 | 3.168 (19) | 130 |
| C8AA—H8AB···F2ii | 0.99 | 2.48 | 3.168 (19) | 126 |
| C10—H10A···O4 | 0.98 | 2.27 | 2.7834 (18) | 112 |
| C15—H15A···O3 | 0.98 | 2.47 | 3.038 (3) | 116 |
| C16—H16C···F1iii | 0.98 | 2.62 | 3.573 (2) | 164 |
| C17—H17B···O3 | 0.98 | 2.41 | 2.969 (2) | 116 |
| C22—H22A···F2 | 0.95 | 2.37 | 2.9091 (19) | 116 |
| C24—H24A···O4iv | 1.00 | 2.65 | 3.4638 (18) | 139 |
Symmetry codes: (i) x+1/2, y, −z+3/2; (ii) −x+3/2, y−1/2, z; (iii) x, −y+3/2, z−1/2; (iv) −x+1, y+1/2, −z+3/2.
References
- Aqdas, M. & Sung, M. H. (2023). Trends Immunol. 44, 32–43. [DOI] [PMC free article] [PubMed]
- Bruker (2018). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bryzgalov, A., Tolstikova, T., Koshcheev, B. & Maksimov, A. (2023). Results Chem. 5, 100705.
- Chen, L., Deng, H., Cui, H., Fang, J., Zuo, Z., Deng, J., Li, Y., Wang, X. & Zhao, L. (2018). Oncotarget, 9, 7204–7218. [DOI] [PMC free article] [PubMed]
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Gündüz, M. G., Butcher, R. J., Öztürk Yildirim, S., El-Khouly, A., Şafak, C. & Şimşek, R. (2012). Acta Cryst. E68, o3404–o3405. [DOI] [PMC free article] [PubMed]
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Linden, A., Gündüz, M. G., Şimşek, R. & Şafak, C. (2006). Acta Cryst. C62, o227–o230. [DOI] [PubMed]
- Linden, A., Şafak, C., Şimşek, R. & Gündüz, M. G. (2011). Acta Cryst. C67, o80–o84. [DOI] [PubMed]
- Linden, A., Şimşek, R., Gündüz, M. & Şafak, C. (2005). Acta Cryst. C61, o731–o734. [DOI] [PubMed]
- Mookiah, P., Rajesh, K., Narasimhamurthy, T., Vijayakumar, V. & Srinivasan, N. (2009). Acta Cryst. E65, o2664. [DOI] [PMC free article] [PubMed]
- Öztürk Yildirim, S., Butcher, R. J., Gündüz, M. G., El-Khouly, A., Şimşek, R. & Şafak, C. (2013). Acta Cryst. E69, o40–o41. [DOI] [PMC free article] [PubMed]
- Pan, C., Luo, M., Lu, Y., Pan, X., Chen, X., Ding, L., Che, J., He, Q. & Dong, X. (2022). Bioorg. Chem. 125, 105820. [DOI] [PubMed]
- Pehlivanlar, E., Yıldırım, S. Ö., Şimşek, R., Akkurt, M., Butcher, R. J. & Bhattarai, A. (2023). Acta Cryst. E79, 664–668. [DOI] [PMC free article] [PubMed]
- Ranjbar, S., Edraki, N., Firuzi, O., Khoshneviszadeh, M. & Miri, R. (2019). Mol. Divers. 23, 471–508. [DOI] [PubMed]
- Shaheen, M. A., El-Emam, A. A. & El-Gohary, N. S. (2020). Bioorg. Chem. 105, 104274. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spackman, P. R., Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Jayatilaka, D. & Spackman, M. A. (2021). J. Appl. Cryst. 54, 1006–1011. [DOI] [PMC free article] [PubMed]
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Steiger, S. A., Monacelli, A. J., Li, C., Hunting, J. L. & Natale, N. R. (2014). Acta Cryst. C70, 790–795. [DOI] [PMC free article] [PubMed]
- Wu, B., Sodji, Q. H. & Oyelere, A. K. (2022). Cancers, 14, 552. [DOI] [PMC free article] [PubMed]
- Yıldırım, S. Ö., Akkurt, M., Çetin, G., Şimşek, R., Butcher, R. J. & Bhattarai, A. (2022). Acta Cryst. E78, 798–803. [DOI] [PMC free article] [PubMed]
- Yıldırım, S. Ö., Akkurt, M., Çetin, G., Şimşek, R., Butcher, R. J. & Bhattarai, A. (2023). Acta Cryst. E79, 187–191. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, II, III. DOI: 10.1107/S2056989024001233/jy2044sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989024001233/jy2044Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989024001233/jy2044IIsup3.hkl
Structure factors: contains datablock(s) III. DOI: 10.1107/S2056989024001233/jy2044IIIsup4.hkl
Supporting information file. DOI: 10.1107/S2056989024001233/jy2044Isup5.cml
Supporting information file. DOI: 10.1107/S2056989024001233/jy2044IIsup6.cml
Supporting information file. DOI: 10.1107/S2056989024001233/jy2044IIIsup7.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report