The crystal structure of the tetraethylammonium salt of the non-steroidal anti-inflammatory drug nimesulide (polymorph II), C8H20N+·C13H11N2O5S−, was determined using single-crystal X-ray diffraction. There are differences in the geometry of both the nimesulide anion and the tetraethylammonium cation in polymorphs I and II of the title compound.
Keywords: nimesulide, N-(4-nitro-2-phenoxyphenyl)methanesulfonamide, tetraethylammonium salt, API, crystal structure, polymorphism
Abstract
The crystal structure of the tetraethylammonium salt of the non-steroidal anti-inflammatory drug nimesulide (polymorph II) (systematic name: tetraethylammonium N-methanesulfonyl-4-nitro-2-phenoxyanilinide), C8H20N+·C13H11N2O5S−, was determined using single-crystal X-ray diffraction. The title compound crystallizes in the monoclinic space group P21/c with one tetraethylammonium cation and one nimesulide anion in the asymmetric unit. In the crystal, the ions are linked by C—H⋯N and C—H⋯O hydrogen bonds and C—H⋯π interactions. There are differences in the geometry of both the nimesulide anion and the tetraethylammonium cation in polymorphs I [Rybczyńska & Sikorski (2023 ▸). Sci. Rep. 13, 17268] and II of the title compound.
1. Chemical context
Nimesulide [systematic name: N-(4-nitro-2-phenoxyphenyl)methanesulfonamide] is an active pharmaceutical ingredient (API) categorized among non-steroidal anti-inflammatory drugs (NSAIDs). This is a drug that effectively manages acute pain and primary dysmenorrhea as a result of its antipyretic, analgesic, and anti-inflammatory properties (Kress et al., 2016 ▸; Vane & Botting, 1998 ▸). Similar to other NSAIDs, its action involves inhibiting cyclooxygenase – an enzyme crucial in prostaglandin synthesis within cell membranes (Bennett & Villa, 2000 ▸).

The crystal structure of nimesulide is known – it exists in the form of two polymorphs (Dupont et al., 1995 ▸; Sanphui et al., 2011 ▸; Banti et al., 2016 ▸). However, only a few structures of multi-component crystals containing nimesulide have been described in the literature, such as co-crystals (Wang et al., 2020 ▸) and metal complexes (Banti et al., 2016 ▸), but only two, previously examined by us, structures of organic salts of nimesulide (Rybczyńska & Sikorski, 2023 ▸) are known. One of these salts is the tetraethylammonium salt of nimesulide (polymorph I). We became interested in it because the quaternary tetraethylammonium cation has interesting biological activities: it is a ganglionic blocker and inhibitor at nicotinic acetylcholine (Kleinhaus & Prichard, 1977 ▸; Akk & Steinbach, 2003 ▸), and is a common organic structure-directing agent (OSDA) (Schmidt et al., 2016 ▸).
In this research communication, as a continuation of our recent study on the tetraalkylammonium salts of nimesulide (Rybczyńska & Sikorski, 2023 ▸), we report on the crystal structure, conformational analysis of ions and analysis of intermolecular interactions in the crystal of tetraethylammonium salt of nimesulide (polymorph II).
2. Structural commentary
The title compound crystallizes in the monoclinic P21/c space group with one tetraethylammonium cation and one nimesulide anion in the asymmetric unit (Table 1 ▸, Fig. 1 ▸). For comparison, polymorph I crystallizes in the monoclinic P21/n space group with one ion pair in the asymmetric unit.
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 and Cg2 are the centroids of the C1–C6 and C13–C18 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C11—H11A⋯O20i | 0.96 | 2.65 | 3.360 (4) | 131 |
| C11—H11C⋯O20ii | 0.96 | 2.66 | 3.555 (4) | 156 |
| C17—H17A⋯N7iii | 0.93 | 2.70 | 3.478 (4) | 142 |
| C24—H24C⋯O10 | 0.96 | 2.47 | 3.415 (4) | 168 |
| C25—H25A⋯O9iv | 0.97 | 2.47 | 3.155 (5) | 128 |
| C26—H26C⋯O9v | 0.96 | 2.58 | 3.541 (4) | 175 |
| C27—H27B⋯O9v | 0.97 | 2.52 | 3.267 (4) | 134 |
| C14—H14A⋯Cg1ii | 0.93 | 3.07 | 3.951 (5) | 158 |
| C24—H24A⋯Cg2v | 0.96 | 2.88 | 3.608 (5) | 134 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 ; (iv)
; (iv)
 ; (v)
; (v)
 .
.
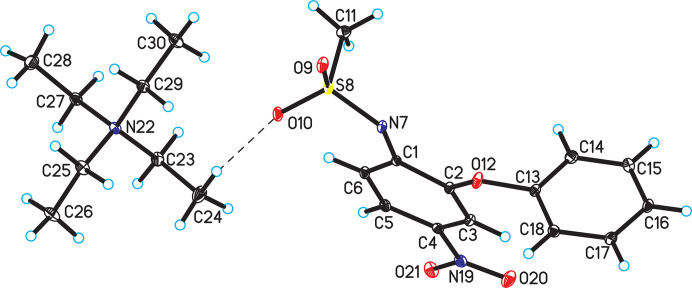
Figure 1.
Crystal structure of title compound with the atom-labeling scheme (displacement ellipsoids are drawn at the 25% probability level; hydrogen bonds are represented by dashed lines).
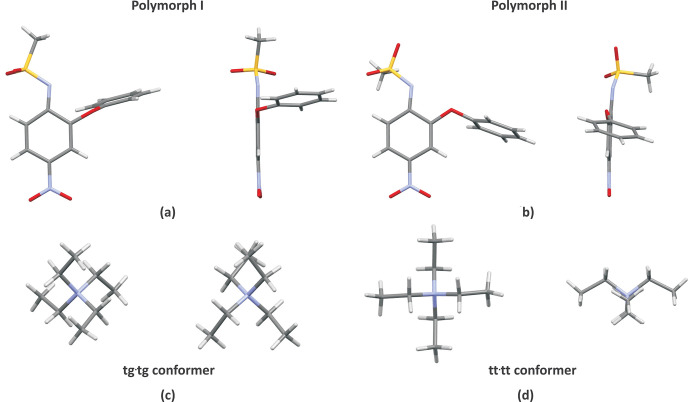
In the crystal structure of the title compound, nimesulide occurs in an ionized form, which is confirmed by the C1—N7 [d(C—N) = 1.365 (4) Å] and N7—S8 [d(N—S) = 1.584 (2) Å] bond lengths and the value of the C1—N7—S8 angle [∠(C—N—S) = 122.7 (2)°] in the sulfonamide group. Similar d(N—S) values are also observed in the crystal structure of polymorph I [1.589 (2) Å], but the d(C—N) distance is slightly shorter and the ∠(C—N—S) angle is smaller for polymorph I [1.345 (3) Å and 119.2 (2)°, respectively]. There are also differences in the arrangement of the methyl group from the sulfonamide moiety and the phenoxy group within the nimesulide anion (Fig. 2 ▸). In the crystal of polymorph I, the methyl group lies almost in the plane of the phenyl ring of the nimesulide anion [with torsion angle ∠(C1—N7—S8—C11) = −174.7 (2)°], while in the crystal of polymorph II it is almost perpendicular [torsion angle ∠(C1—N7—S8—C11) = −74.0 (3)°]. In turn, in the crystal of polymorph I, the phenoxy group is tilted and twisted relative to the benzene ring of nimesulide, with a torsion angle of ∠ (C3—C2—O12—C13) = 88.5 (2)° and an interplanar angle of 84.8 (2)°, while in the crystal of polymorph II the values of these angles are 20.9(4 and 78.3 (2)°, respectively.
Figure 2.
Comparison of the geometries of the nimesulide anion (a) and (b) and the tetraethylammonium cation (c) and (d) in the crystals of the two polymorphs of the tetraethylammonium salt of nimesulide.
Differences in the geometry of the tetraethylammonium cation in the crystals of the two polymorphs of the title compound are also observed (Fig. 2 ▸). In the case of polymorph I, the cation adopts the geometry of a tg·tg conformer, while in the crystal of polymorph II it exists in a tt·tt conformer (Ikuno et al., 2015 ▸; Schmidt et al., 2016 ▸; Takekiyo & Yoshimura, 2006 ▸). Both conformers of the tetraethylammonium cation are also observed in other tetraethylammonium salts (e.g. de Arriba et al., 2011 ▸; Evans et al., 1990 ▸; Warnke et al., 2010 ▸; Lutz et al., 2014 ▸; Brahim et al., 2018 ▸). It is interesting that the distribution of conformers of the tetraethylammonium cation in tetraethylammonium hydroxide solution is temperature dependent (the tt·tt conformer dominates at lower temperatures), and higher concentrations lead to a greater proportion of the tg·tg conformer (Ikuno et al., 2015 ▸; Schmidt et al., 2016 ▸; Takekiyo & Yoshimura, 2006 ▸). This may explain why only a few single crystals of polymorph II were obtained as a result of the synthesis of the title compound carried out under specific conditions (see: Synthesis and crystallization section).
The changes in the conformation of both the nimesulide anion and the tetraethylammonium cation results in an increase in the volume of the unit cell from 2300.6 (2) Å3 (polymorph I) to 2330.0 (4) Å3 (polymorph II). Moreover, the crystal density decreases (1.292 and 1.272 g cm−3 for polymorph I and II, respectively), as well as the Kitaigorodskii packing index (with the percentage of filled space equal to 66.7 and 66.0% for polymorphs I and II, respectively). This indicates a more favorable molecular packing in the crystal of polymorph I.
3. Supramolecular features
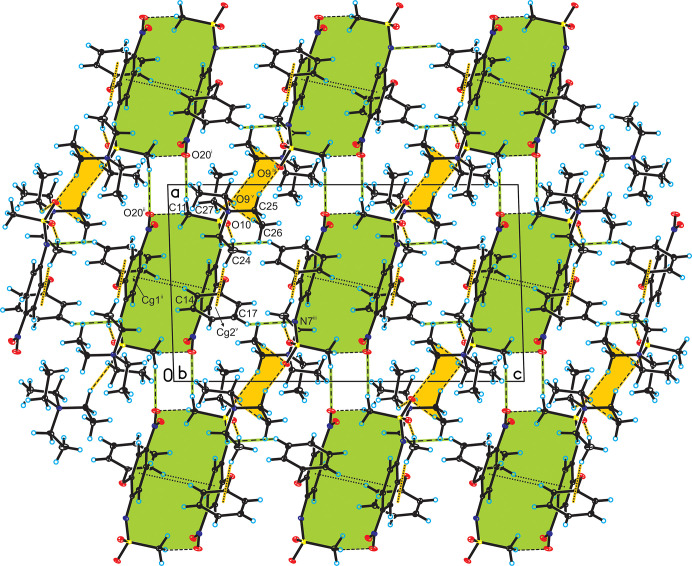
In the crystal of the title compound, neighboring nimesulide anions are linked by C14—H14A⋯π interactions [d(H⋯Cg) = 3.07 Å; Fig. 3 ▸, Table 1 ▸], forming a homodimer. Adjacent homodimers are linked through Cphenoxy—H⋯N− and Cmethyl—H⋯Onitro hydrogen bonds, building porous organic frameworks along the b-axis (Fig. 3 ▸, Table 1 ▸). The tetraethylammonium cations are located in the voids of these networks and linked with the nimesulide anions via Cmethyl—H⋯Osulfo hydrogen bonds and C24—H24A⋯πphenoxy interactions [d(H⋯Cg) = 2.88 Å; Fig. 3 ▸, Table 1 ▸].
Figure 3.
Crystal packing of the title compound viewed along the b axis (interactions between nimesulide anions are highlighted in green, whereas interactions between the nimesulide anion and tetraethylammonium cation are highlighted in orange).
4. Database survey
In the Cambridge Structural Database (CSD version 5.43, update of 03/2023; Groom et al., 2016 ▸) there are only 13 structures involving a nimesulide molecule or ion, viz., the crystal structures of two polymorphs of nimesulide [refcodes WINWUL (Dupont et al., 1995 ▸), WINWUL01, WINWUL02 (Sanphui et al., 2011 ▸), and WINWUL03 (Banti et al., 2016 ▸)], five structures of nimesulide–silver complexes (refcodes EXEZUE, EXIBAQ, EXIBAU, EXIBIY, EXIBOE; Banti et al., 2016 ▸), the crystal structures of tetramethylammonium and tetraethylammonium salts of nimesulide (polymorph I; CCDC 2281374 and CCDC 2281375; Rybczyńska & Sikorski, 2023 ▸), and four structures of co-crystals of nimesulide with pyridine derivatives (refcodes LAKLOC, LAKLUI, LAKMAP, and LAKMET; Wang et al., 2021 ▸). In the CSD, there are also 5062 structures of tetraethylammonium salts: 728 of them are structures of organic compounds involving the tetraethylammonium cation, including three structures of sulfonamide salts (refcodes RALGOC, RALGUI, and RALHAP; de Arriba et al., 2011 ▸).
5. Synthesis and crystallization
All chemicals were purchased from Sigma-Aldrich and used without any further purification. Nimesulide (0.05 g, 0.162 mmol) was dissolved in 0.12 ml of tetraethylammonium hydroxide (20 wt.% in H2O, d = 1.01 g cm−3 in 293 K, 0.162 mmol) and 5 cm3 of ethanol. The solution was mixed and heated until boiling. The solution was allowed to evaporate in place without sunlight for a few days, giving yellow crystals of polymorph I and a small amount of yellow crystals of polymorph II (m.p. = 388 K). The mixture of polymorphs was separated by mechanical means.
6. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All H atoms were placed geometrically and refined using a riding model with C—H = 0.93–0.97 Å and U iso(H) = 1.2U eq(C) [C—H = 0.96 Å and U iso(H) = 1.5U eq(C) for the methyl groups]. The most disagreeable reflections (621) and (589) with an error/s.u. of more than 10 were omitted using the OMIT instruction in SHELXL (Sheldrick, 2015b ▸).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C8H20N+·C13H11N2O5S− |
| M r | 437.55 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 291 |
| a, b, c (Å) | 11.0276 (10), 10.7661 (8), 19.635 (2) |
| β (°) | 91.792 (9) |
| V (Å3) | 2330.0 (4) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.17 |
| Crystal size (mm) | 0.42 × 0.20 × 0.09 |
| Data collection | |
| Diffractometer | Oxford Diffraction Ruby CCD |
| Absorption correction | Multi-scan (CrysAlis RED; Oxford Diffraction, 2008 ▸). |
| T min, T max | 0.966, 0.998 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 15531, 4094, 2549 |
| R int | 0.073 |
| (sin θ/λ)max (Å−1) | 0.595 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.066, 0.128, 1.10 |
| No. of reflections | 4094 |
| No. of parameters | 276 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.16, −0.23 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989024001300/dx2059sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989024001300/dx2059Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989024001300/dx2059Isup3.mol
Supporting information file. DOI: 10.1107/S2056989024001300/dx2059Isup4.cml
CCDC reference: 2332021
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The title compound is protected by European patent application No. EP22208962 submitted by the authors of this paper.
supplementary crystallographic information
Crystal data
| C8H20N+·C13H11N2O5S− | Dx = 1.247 Mg m−3 |
| Mr = 437.55 | Melting point: 388 K |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 11.0276 (10) Å | Cell parameters from 15531 reflections |
| b = 10.7661 (8) Å | θ = 3.3–25.0° |
| c = 19.635 (2) Å | µ = 0.17 mm−1 |
| β = 91.792 (9)° | T = 291 K |
| V = 2330.0 (4) Å3 | Plate, yellow |
| Z = 4 | 0.42 × 0.20 × 0.09 mm |
| F(000) = 936 |
Data collection
| Oxford Diffraction Ruby CCD diffractometer | 4094 independent reflections |
| Radiation source: fine-focus sealed tube | 2549 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.073 |
| Detector resolution: 10.4002 pixels mm-1 | θmax = 25.0°, θmin = 3.3° |
| ω scans | h = −13→12 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2008). | k = −12→11 |
| Tmin = 0.966, Tmax = 0.998 | l = −23→23 |
| 15531 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.066 | H-atom parameters constrained |
| wR(F2) = 0.128 | w = 1/[σ2(Fo2) + (0.0316P)2 + 0.0136P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.10 | (Δ/σ)max = 0.001 |
| 4094 reflections | Δρmax = 0.16 e Å−3 |
| 276 parameters | Δρmin = −0.23 e Å−3 |
| 0 restraints |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N22 | 0.8646 (2) | 0.6786 (2) | 0.66998 (12) | 0.0466 (7) | |
| C23 | 0.7364 (3) | 0.6589 (3) | 0.6432 (2) | 0.0679 (10) | |
| H23A | 0.738581 | 0.638164 | 0.595204 | 0.082* | |
| H23B | 0.692237 | 0.736376 | 0.647115 | 0.082* | |
| C24 | 0.6669 (4) | 0.5566 (3) | 0.6801 (3) | 0.1112 (17) | |
| H24A | 0.590665 | 0.541912 | 0.656474 | 0.167* | |
| H24B | 0.652626 | 0.582332 | 0.725925 | 0.167* | |
| H24C | 0.713924 | 0.481527 | 0.680791 | 0.167* | |
| C25 | 0.8677 (4) | 0.7082 (3) | 0.74529 (16) | 0.0678 (10) | |
| H25A | 0.951411 | 0.722287 | 0.759958 | 0.081* | |
| H25B | 0.839097 | 0.636099 | 0.769643 | 0.081* | |
| C26 | 0.7934 (4) | 0.8196 (3) | 0.76588 (18) | 0.0801 (12) | |
| H26A | 0.804123 | 0.833377 | 0.814002 | 0.120* | |
| H26B | 0.709188 | 0.804419 | 0.755044 | 0.120* | |
| H26C | 0.819681 | 0.891737 | 0.741652 | 0.120* | |
| C27 | 0.9144 (3) | 0.7874 (3) | 0.63015 (17) | 0.0609 (10) | |
| H27A | 0.905286 | 0.768693 | 0.581930 | 0.073* | |
| H27B | 0.865104 | 0.859849 | 0.638968 | 0.073* | |
| C28 | 1.0453 (4) | 0.8203 (3) | 0.6455 (2) | 0.0968 (15) | |
| H28A | 1.064308 | 0.897193 | 0.623559 | 0.145* | |
| H28B | 1.096702 | 0.755682 | 0.628966 | 0.145* | |
| H28C | 1.058343 | 0.828854 | 0.693876 | 0.145* | |
| C29 | 0.9416 (3) | 0.5635 (3) | 0.66162 (18) | 0.0599 (9) | |
| H29A | 1.021525 | 0.579523 | 0.681782 | 0.072* | |
| H29B | 0.905901 | 0.496372 | 0.687120 | 0.072* | |
| C30 | 0.9563 (4) | 0.5207 (3) | 0.58923 (19) | 0.0810 (12) | |
| H30A | 1.002975 | 0.445354 | 0.589154 | 0.122* | |
| H30B | 0.997549 | 0.583607 | 0.564185 | 0.122* | |
| H30C | 0.877894 | 0.505702 | 0.568264 | 0.122* | |
| C1 | 0.5837 (3) | 0.1058 (3) | 0.62935 (14) | 0.0395 (7) | |
| C2 | 0.4877 (3) | 0.0172 (3) | 0.62920 (15) | 0.0435 (8) | |
| C3 | 0.3730 (3) | 0.0461 (3) | 0.60651 (15) | 0.0487 (8) | |
| H3A | 0.312492 | −0.014135 | 0.605923 | 0.058* | |
| C4 | 0.3469 (3) | 0.1659 (3) | 0.58428 (15) | 0.0443 (8) | |
| C5 | 0.4352 (3) | 0.2565 (3) | 0.58697 (16) | 0.0508 (9) | |
| H5A | 0.416335 | 0.337348 | 0.573719 | 0.061* | |
| C6 | 0.5507 (3) | 0.2275 (3) | 0.60917 (15) | 0.0482 (8) | |
| H6A | 0.609174 | 0.289699 | 0.611006 | 0.058* | |
| N7 | 0.6961 (2) | 0.0639 (2) | 0.64924 (12) | 0.0463 (7) | |
| S8 | 0.81473 (7) | 0.14575 (7) | 0.64359 (4) | 0.0480 (2) | |
| O9 | 0.9111 (2) | 0.07799 (19) | 0.67799 (12) | 0.0651 (7) | |
| O10 | 0.8031 (2) | 0.27336 (18) | 0.66560 (12) | 0.0650 (7) | |
| C11 | 0.8486 (3) | 0.1503 (3) | 0.55687 (17) | 0.0700 (10) | |
| H11A | 0.923935 | 0.193215 | 0.551381 | 0.105* | |
| H11B | 0.784964 | 0.192962 | 0.531955 | 0.105* | |
| H11C | 0.855381 | 0.067033 | 0.539846 | 0.105* | |
| O12 | 0.5189 (2) | −0.09796 (19) | 0.65561 (12) | 0.0652 (7) | |
| C13 | 0.4449 (3) | −0.2003 (3) | 0.63959 (18) | 0.0470 (8) | |
| C14 | 0.4353 (3) | −0.2436 (3) | 0.57443 (18) | 0.0627 (10) | |
| H14A | 0.471834 | −0.201598 | 0.539124 | 0.075* | |
| C15 | 0.3699 (3) | −0.3516 (3) | 0.56192 (18) | 0.0666 (10) | |
| H15A | 0.362559 | −0.382462 | 0.517737 | 0.080* | |
| C16 | 0.3162 (3) | −0.4132 (3) | 0.6137 (2) | 0.0615 (10) | |
| H16A | 0.272162 | −0.485449 | 0.604827 | 0.074* | |
| C17 | 0.3275 (3) | −0.3681 (3) | 0.6785 (2) | 0.0681 (11) | |
| H17A | 0.291625 | −0.410340 | 0.713952 | 0.082* | |
| C18 | 0.3917 (3) | −0.2604 (3) | 0.69191 (17) | 0.0583 (9) | |
| H18A | 0.398465 | −0.229272 | 0.736042 | 0.070* | |
| N19 | 0.2270 (3) | 0.1945 (3) | 0.55838 (14) | 0.0586 (8) | |
| O20 | 0.1507 (2) | 0.1114 (3) | 0.55356 (16) | 0.0935 (9) | |
| O21 | 0.2031 (2) | 0.3022 (2) | 0.54080 (13) | 0.0770 (8) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N22 | 0.0522 (17) | 0.0362 (15) | 0.0510 (16) | 0.0145 (12) | −0.0039 (14) | 0.0031 (12) |
| C23 | 0.050 (2) | 0.054 (2) | 0.098 (3) | 0.0153 (18) | −0.014 (2) | −0.007 (2) |
| C24 | 0.073 (3) | 0.056 (3) | 0.205 (5) | −0.003 (2) | 0.014 (3) | 0.001 (3) |
| C25 | 0.087 (3) | 0.059 (2) | 0.056 (2) | 0.018 (2) | −0.007 (2) | 0.0037 (18) |
| C26 | 0.119 (4) | 0.061 (2) | 0.061 (2) | 0.024 (2) | 0.015 (2) | −0.0029 (19) |
| C27 | 0.074 (3) | 0.045 (2) | 0.064 (2) | 0.0070 (18) | 0.007 (2) | −0.0001 (17) |
| C28 | 0.072 (3) | 0.068 (3) | 0.151 (4) | −0.002 (2) | 0.021 (3) | −0.006 (3) |
| C29 | 0.058 (2) | 0.0418 (19) | 0.080 (2) | 0.0192 (17) | 0.000 (2) | −0.0001 (17) |
| C30 | 0.097 (3) | 0.055 (2) | 0.091 (3) | 0.024 (2) | 0.009 (3) | −0.011 (2) |
| C1 | 0.040 (2) | 0.0374 (17) | 0.0410 (17) | 0.0008 (15) | −0.0005 (15) | −0.0015 (14) |
| C2 | 0.044 (2) | 0.0340 (18) | 0.0525 (19) | 0.0003 (15) | −0.0052 (16) | 0.0032 (15) |
| C3 | 0.042 (2) | 0.0476 (19) | 0.056 (2) | −0.0055 (16) | −0.0017 (17) | 0.0028 (16) |
| C4 | 0.040 (2) | 0.045 (2) | 0.0471 (18) | 0.0125 (16) | −0.0036 (15) | −0.0040 (15) |
| C5 | 0.057 (2) | 0.0385 (19) | 0.057 (2) | 0.0086 (17) | −0.0053 (18) | −0.0015 (15) |
| C6 | 0.050 (2) | 0.0336 (18) | 0.061 (2) | −0.0008 (15) | −0.0035 (18) | 0.0000 (15) |
| N7 | 0.0420 (16) | 0.0359 (14) | 0.0604 (16) | −0.0043 (12) | −0.0097 (13) | 0.0023 (12) |
| S8 | 0.0430 (5) | 0.0379 (5) | 0.0625 (5) | −0.0019 (4) | −0.0088 (4) | −0.0006 (4) |
| O9 | 0.0511 (15) | 0.0513 (14) | 0.0908 (17) | −0.0002 (11) | −0.0303 (13) | 0.0074 (12) |
| O10 | 0.0650 (16) | 0.0354 (13) | 0.0943 (18) | −0.0065 (11) | −0.0034 (14) | −0.0120 (12) |
| C11 | 0.057 (2) | 0.083 (3) | 0.070 (2) | 0.004 (2) | 0.003 (2) | 0.005 (2) |
| O12 | 0.0570 (15) | 0.0397 (13) | 0.0973 (18) | −0.0101 (11) | −0.0246 (13) | 0.0216 (12) |
| C13 | 0.041 (2) | 0.0308 (17) | 0.069 (2) | 0.0017 (15) | −0.0028 (18) | 0.0121 (17) |
| C14 | 0.069 (3) | 0.060 (2) | 0.059 (2) | −0.005 (2) | 0.005 (2) | 0.0146 (19) |
| C15 | 0.077 (3) | 0.058 (2) | 0.064 (2) | −0.002 (2) | −0.005 (2) | −0.008 (2) |
| C16 | 0.053 (2) | 0.0384 (19) | 0.093 (3) | −0.0068 (16) | 0.000 (2) | 0.004 (2) |
| C17 | 0.074 (3) | 0.052 (2) | 0.079 (3) | −0.014 (2) | 0.020 (2) | 0.006 (2) |
| C18 | 0.069 (2) | 0.048 (2) | 0.058 (2) | −0.0018 (18) | 0.009 (2) | 0.0001 (17) |
| N19 | 0.053 (2) | 0.059 (2) | 0.0638 (18) | 0.0120 (17) | −0.0041 (16) | −0.0079 (16) |
| O20 | 0.0448 (17) | 0.0792 (19) | 0.155 (3) | −0.0002 (15) | −0.0180 (17) | 0.0119 (18) |
| O21 | 0.0763 (19) | 0.0619 (17) | 0.0912 (18) | 0.0275 (14) | −0.0240 (15) | −0.0052 (14) |
Geometric parameters (Å, º)
| N22—C23 | 1.508 (4) | C2—C3 | 1.364 (4) |
| N22—C25 | 1.512 (4) | C2—O12 | 1.383 (3) |
| N22—C29 | 1.514 (3) | C3—C4 | 1.389 (4) |
| N22—C27 | 1.520 (4) | C3—H3A | 0.9300 |
| C23—C24 | 1.536 (5) | C4—C5 | 1.378 (4) |
| C23—H23A | 0.9700 | C4—N19 | 1.436 (4) |
| C23—H23B | 0.9700 | C5—C6 | 1.370 (4) |
| C24—H24A | 0.9600 | C5—H5A | 0.9300 |
| C24—H24B | 0.9600 | C6—H6A | 0.9300 |
| C24—H24C | 0.9600 | N7—S8 | 1.584 (2) |
| C25—C26 | 1.515 (4) | S8—O9 | 1.439 (2) |
| C25—H25A | 0.9700 | S8—O10 | 1.447 (2) |
| C25—H25B | 0.9700 | S8—C11 | 1.755 (3) |
| C26—H26A | 0.9600 | C11—H11A | 0.9600 |
| C26—H26B | 0.9600 | C11—H11B | 0.9600 |
| C26—H26C | 0.9600 | C11—H11C | 0.9600 |
| C27—C28 | 1.508 (5) | O12—C13 | 1.401 (3) |
| C27—H27A | 0.9700 | C13—C14 | 1.363 (4) |
| C27—H27B | 0.9700 | C13—C18 | 1.363 (4) |
| C28—H28A | 0.9600 | C14—C15 | 1.387 (5) |
| C28—H28B | 0.9600 | C14—H14A | 0.9300 |
| C28—H28C | 0.9600 | C15—C16 | 1.364 (5) |
| C29—C30 | 1.508 (4) | C15—H15A | 0.9300 |
| C29—H29A | 0.9700 | C16—C17 | 1.365 (5) |
| C29—H29B | 0.9700 | C16—H16A | 0.9300 |
| C30—H30A | 0.9600 | C17—C18 | 1.379 (4) |
| C30—H30B | 0.9600 | C17—H17A | 0.9300 |
| C30—H30C | 0.9600 | C18—H18A | 0.9300 |
| C1—N7 | 1.365 (4) | N19—O20 | 1.230 (3) |
| C1—C6 | 1.412 (4) | N19—O21 | 1.235 (3) |
| C1—C2 | 1.425 (4) | ||
| C23—N22—C25 | 111.3 (3) | N7—C1—C6 | 127.6 (3) |
| C23—N22—C29 | 111.7 (2) | N7—C1—C2 | 116.6 (3) |
| C25—N22—C29 | 106.5 (2) | C6—C1—C2 | 115.8 (3) |
| C23—N22—C27 | 106.2 (2) | C3—C2—O12 | 123.0 (3) |
| C25—N22—C27 | 110.1 (2) | C3—C2—C1 | 122.0 (3) |
| C29—N22—C27 | 111.2 (2) | O12—C2—C1 | 115.0 (3) |
| N22—C23—C24 | 114.4 (3) | C2—C3—C4 | 119.7 (3) |
| N22—C23—H23A | 108.7 | C2—C3—H3A | 120.2 |
| C24—C23—H23A | 108.7 | C4—C3—H3A | 120.2 |
| N22—C23—H23B | 108.7 | C5—C4—C3 | 120.4 (3) |
| C24—C23—H23B | 108.7 | C5—C4—N19 | 120.2 (3) |
| H23A—C23—H23B | 107.6 | C3—C4—N19 | 119.4 (3) |
| C23—C24—H24A | 109.5 | C6—C5—C4 | 120.0 (3) |
| C23—C24—H24B | 109.5 | C6—C5—H5A | 120.0 |
| H24A—C24—H24B | 109.5 | C4—C5—H5A | 120.0 |
| C23—C24—H24C | 109.5 | C5—C6—C1 | 122.0 (3) |
| H24A—C24—H24C | 109.5 | C5—C6—H6A | 119.0 |
| H24B—C24—H24C | 109.5 | C1—C6—H6A | 119.0 |
| N22—C25—C26 | 115.6 (3) | C1—N7—S8 | 122.7 (2) |
| N22—C25—H25A | 108.4 | O9—S8—O10 | 114.35 (14) |
| C26—C25—H25A | 108.4 | O9—S8—N7 | 106.51 (13) |
| N22—C25—H25B | 108.4 | O10—S8—N7 | 115.20 (13) |
| C26—C25—H25B | 108.4 | O9—S8—C11 | 107.03 (16) |
| H25A—C25—H25B | 107.4 | O10—S8—C11 | 106.68 (16) |
| C25—C26—H26A | 109.5 | N7—S8—C11 | 106.53 (16) |
| C25—C26—H26B | 109.5 | S8—C11—H11A | 109.5 |
| H26A—C26—H26B | 109.5 | S8—C11—H11B | 109.5 |
| C25—C26—H26C | 109.5 | H11A—C11—H11B | 109.5 |
| H26A—C26—H26C | 109.5 | S8—C11—H11C | 109.5 |
| H26B—C26—H26C | 109.5 | H11A—C11—H11C | 109.5 |
| C28—C27—N22 | 115.9 (3) | H11B—C11—H11C | 109.5 |
| C28—C27—H27A | 108.3 | C2—O12—C13 | 119.0 (2) |
| N22—C27—H27A | 108.3 | C14—C13—C18 | 121.5 (3) |
| C28—C27—H27B | 108.3 | C14—C13—O12 | 120.5 (3) |
| N22—C27—H27B | 108.3 | C18—C13—O12 | 117.8 (3) |
| H27A—C27—H27B | 107.4 | C13—C14—C15 | 118.6 (3) |
| C27—C28—H28A | 109.5 | C13—C14—H14A | 120.7 |
| C27—C28—H28B | 109.5 | C15—C14—H14A | 120.7 |
| H28A—C28—H28B | 109.5 | C16—C15—C14 | 120.8 (3) |
| C27—C28—H28C | 109.5 | C16—C15—H15A | 119.6 |
| H28A—C28—H28C | 109.5 | C14—C15—H15A | 119.6 |
| H28B—C28—H28C | 109.5 | C15—C16—C17 | 119.5 (3) |
| C30—C29—N22 | 115.5 (3) | C15—C16—H16A | 120.2 |
| C30—C29—H29A | 108.4 | C17—C16—H16A | 120.2 |
| N22—C29—H29A | 108.4 | C16—C17—C18 | 120.6 (3) |
| C30—C29—H29B | 108.4 | C16—C17—H17A | 119.7 |
| N22—C29—H29B | 108.4 | C18—C17—H17A | 119.7 |
| H29A—C29—H29B | 107.5 | C13—C18—C17 | 119.1 (3) |
| C29—C30—H30A | 109.5 | C13—C18—H18A | 120.5 |
| C29—C30—H30B | 109.5 | C17—C18—H18A | 120.5 |
| H30A—C30—H30B | 109.5 | O20—N19—O21 | 121.5 (3) |
| C29—C30—H30C | 109.5 | O20—N19—C4 | 119.4 (3) |
| H30A—C30—H30C | 109.5 | O21—N19—C4 | 119.1 (3) |
| H30B—C30—H30C | 109.5 | ||
| C25—N22—C23—C24 | −57.0 (4) | N7—C1—C6—C5 | 176.7 (3) |
| C29—N22—C23—C24 | 61.9 (4) | C2—C1—C6—C5 | −3.8 (4) |
| C27—N22—C23—C24 | −176.7 (3) | C6—C1—N7—S8 | −8.2 (4) |
| C23—N22—C25—C26 | −56.6 (4) | C2—C1—N7—S8 | 172.3 (2) |
| C29—N22—C25—C26 | −178.6 (3) | C1—N7—S8—O9 | 172.0 (2) |
| C27—N22—C25—C26 | 60.8 (4) | C1—N7—S8—O10 | 44.0 (3) |
| C23—N22—C27—C28 | −176.8 (3) | C1—N7—S8—C11 | −74.0 (3) |
| C25—N22—C27—C28 | 62.7 (4) | C3—C2—O12—C13 | 20.9 (4) |
| C29—N22—C27—C28 | −55.0 (4) | C1—C2—O12—C13 | −161.0 (3) |
| C23—N22—C29—C30 | 61.3 (4) | C2—O12—C13—C14 | 66.3 (4) |
| C25—N22—C29—C30 | −177.1 (3) | C2—O12—C13—C18 | −118.9 (3) |
| C27—N22—C29—C30 | −57.2 (4) | C18—C13—C14—C15 | −0.2 (5) |
| N7—C1—C2—C3 | −176.0 (3) | O12—C13—C14—C15 | 174.3 (3) |
| C6—C1—C2—C3 | 4.3 (4) | C13—C14—C15—C16 | 0.1 (5) |
| N7—C1—C2—O12 | 5.9 (4) | C14—C15—C16—C17 | −0.3 (6) |
| C6—C1—C2—O12 | −173.7 (3) | C15—C16—C17—C18 | 0.6 (6) |
| O12—C2—C3—C4 | 176.3 (3) | C14—C13—C18—C17 | 0.6 (5) |
| C1—C2—C3—C4 | −1.6 (5) | O12—C13—C18—C17 | −174.1 (3) |
| C2—C3—C4—C5 | −2.0 (4) | C16—C17—C18—C13 | −0.8 (5) |
| C2—C3—C4—N19 | 178.0 (3) | C5—C4—N19—O20 | 176.9 (3) |
| C3—C4—C5—C6 | 2.6 (5) | C3—C4—N19—O20 | −3.1 (4) |
| N19—C4—C5—C6 | −177.4 (3) | C5—C4—N19—O21 | −2.4 (4) |
| C4—C5—C6—C1 | 0.4 (5) | C3—C4—N19—O21 | 177.6 (3) |
Hydrogen-bond geometry (Å, º)
Cg1 and Cg2 are the centroids of the C1–C6 and C13–C18 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C11—H11A···O20i | 0.96 | 2.65 | 3.360 (4) | 131 |
| C11—H11C···O20ii | 0.96 | 2.66 | 3.555 (4) | 156 |
| C17—H17A···N7iii | 0.93 | 2.70 | 3.478 (4) | 142 |
| C24—H24C···O10 | 0.96 | 2.47 | 3.415 (4) | 168 |
| C25—H25A···O9iv | 0.97 | 2.47 | 3.155 (5) | 128 |
| C26—H26C···O9v | 0.96 | 2.58 | 3.541 (4) | 175 |
| C27—H27B···O9v | 0.97 | 2.52 | 3.267 (4) | 134 |
| C14—H14A···Cg1ii | 0.93 | 3.07 | 3.951 (5) | 158 |
| C24—H24A···Cg2v | 0.96 | 2.88 | 3.608 (5) | 134 |
Symmetry codes: (i) x+1, y, z; (ii) −x+1, −y, −z+1; (iii) −x+1, y−1/2, −z+3/2; (iv) −x+2, y+1/2, −z+3/2; (v) x, y+1, z.
Funding Statement
Funding for this research was provided by: Research of Young Scientists grant (BMN) No. 539-T080-B063-23 (University of Gdańsk), DS No. 531-T080-D738-23 (University of Gdańsk), and project ‘Innovation Incubator 4.0’ established by the announcement of the Minister of Science and Higher Education in Poland on 5 June 2020.
References
- Akk, G. & Steinbach, J. H. (2003). J. Physiol. 551, 155–168. [DOI] [PMC free article] [PubMed]
- Banti, C. N., Papatriantafyllopoulou, C., Manoli, M., Tasiopoulos, A. J. & Hadjikakou, S. K. (2016). Inorg. Chem. 55, 8681–8696. [DOI] [PubMed]
- Ben Brahim, K., Ben gzaiel, M., Oueslati, A. & Gargouri, M. (2018). RSC Adv. 8, 40676–40686. [DOI] [PMC free article] [PubMed]
- Bennett, A. & Villa, G. (2000). Expert Opin. Pharmacother. 1, 277–286. [DOI] [PubMed]
- Dupont, L., Pirotte, B., Masereel, B., Delarge, J. & Geczy, J. (1995). Acta Cryst. C51, 507–509.
- Evans, D. J., Hills, A., Hughes, D. L. & Leigh, G. J. (1990). Acta Cryst. C46, 1818–1821.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Fuentes de Arriba, L., Turiel, M. G., Simón, L., Sanz, F., Boyero, J. F., Muñiz, F. M., Morán, J. R. & Alcázar, V. (2011). Org. Biomol. Chem. 9, 8321–8327. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Ikuno, T., Chaikittisilp, W., Liu, Z., Iida, T., Yanaba, Y., Yoshikawa, T., Kohara, S., Wakihara, T. & Okubo, T. (2015). J. Am. Chem. Soc. 137, 14533–14544. [DOI] [PubMed]
- Kleinhaus, A. L. & Prichard, J. (1977). J. Physiol. 270, 181–194. [DOI] [PMC free article] [PubMed]
- Kress, H. G., Baltov, A., Basiński, A., Berghea, F., Castellsague, J., Codreanu, C., Copaciu, E., Giamberardino, M. A., Hakl, M., Hrazdira, L., Kokavec, M., Lejčko, J., Nachtnebl, L., Stančík, R., Švec, A., Tóth, T., Vlaskovska, M. V. & Woroń, J. (2016). Curr. Med. Res. Opin. 32, 23–36. [DOI] [PubMed]
- Lutz, M., Huang, Y., Moret, M.-E. & Klein Gebbink, R. J. M. (2014). Acta Cryst. C70, 470–476. [DOI] [PubMed]
- Oxford Diffraction (2008). CrysAlis CCD and CrysAlis RED. Oxford Diffraction Ltd, Abingdon, England.
- Rybczyńska, M. & Sikorski, A. (2023). Sci. Rep. 13, 17268. [DOI] [PMC free article] [PubMed]
- Sanphui, P., Sarma, B. & Nangia, A. (2011). J. Pharm. Sci. 100, 2287–2299. [DOI] [PubMed]
- Schmidt, J. E., Fu, D., Deem, M. W. & Weckhuysen, B. M. (2016). Angew. Chem. Int. Ed. 55, 16044–16048. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Takekiyo, T. & Yoshimura, Y. (2006). J. Phys. Chem. A, 110, 10829–10833. [DOI] [PubMed]
- Vane, J. R. & Botting, R. M. (1998). Am. J. Med. 104, 2–8.
- Wang, M., Ma, Y., Shi, P., Du, S., Wu, S. & Gong, J. (2021). Cryst. Growth Des. 21, 287–296.
- Warnke, Z., Styczeń, E., Wyrzykowski, D., Sikorski, A., Kłak, J. & Mroziński, J. (2010). Struct. Chem. 21, 285–289.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989024001300/dx2059sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989024001300/dx2059Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989024001300/dx2059Isup3.mol
Supporting information file. DOI: 10.1107/S2056989024001300/dx2059Isup4.cml
CCDC reference: 2332021
Additional supporting information: crystallographic information; 3D view; checkCIF report