Abstract
The yeast mitochondrial HMG-box protein, Abf2p, is essential for maintenance of the mitochondrial genome. To better understand the role of Abf2p in the maintenance of the mitochondrial chromosome, we have isolated a multicopy suppressor (YHM2) of the temperature-sensitive defect associated with an abf2 null mutation. The function of Yhm2p was characterized at the molecular level. Yhm2p has 314 amino acid residues, and the deduced amino acid sequence is similar to that of a family of mitochondrial carrier proteins. Yhm2p is localized in the mitochondrial inner membrane and is also associated with mitochondrial DNA in vivo. Yhm2p exhibits general DNA-binding activity in vitro. Thus, Yhm2p appears to be novel in that it is a membrane-bound DNA-binding protein. A sequence that is similar to the HMG DNA-binding domain is important for the DNA-binding activity of Yhm2p, and a mutation in this region abolishes the ability of YHM2 to suppress the temperature-sensitive defect of respiration of the abf2 null mutant. Disruption of YHM2 causes a significant growth defect in the presence of nonfermentable carbon sources such as glycerol and ethanol, and the cells have defects in respiration as determined by 2,3,5,-triphenyltetrazolium chloride staining. Yhm2p may function as a member of the protein machinery for the mitochondrial inner membrane attachment site of mitochondrial DNA during replication and segregation of mitochondrial genomes.
Maintenance of the mitochondrial genome of eucaryotic organisms is tightly regulated by the products of various nuclear genes to ensure the respiratory competency of the cells and their progeny. A number of nuclear genes have been identified in yeast and other eucaryotic cells that are essential for the mitochondrial genome stability. They can be classified into various functional groups: some encode proteins that are directly involved in the processes of mitochondrial DNA (mtDNA) replication, repair, or recombination (12–14, 41, 52). Other genes encode the components of the mitochondrial translation and transcription apparatus (4, 9, 15), and some genes encode the mitochondrial enzymes for nucleotide biosynthesis (8), a GTP-binding protein (16, 19), a pyrophosphatase (32), and a matrix protease (48, 53). Therefore, various proteins that are essential for mitochondrial function are involved in the maintenance of the mitochondrial chromosome in direct or indirect ways.
The condensation and packaging of mtDNAs are required for optimal functioning of the mitochondrial genome. DNA packaging in mitochondria is accomplished with histones or histone-like proteins, as in eucaryotic nuclei and in bacteria. Caron et al. first purified HM (histone-like protein of mitochondria) as the yeast mitochondrial histone-like protein from isolated mitochondria (3). The HM protein is encoded by the ABF2 gene and has a mass of 20 kDa (6). It binds to mtDNA and is able to introduce supercoil turns into circular relaxed DNA (3, 7, 20, 34); from estimates of its abundance in mitochondria of wild-type cells, there is enough Abf2p to bind to every 30 bp of mtDNA (7). Abf2p is required for maintenance of the mitochondrial genome. Cells with a null allele of ABF2 rapidly lose their rho+ mtDNA when grown in rich glucose medium because they do not require mitochondrial function for growth in a fermentable medium. However, abf2 cells are able to maintain functional mtDNA indefinitely when grown in the presence of nonfermentable carbon sources such as glycerol, which is metabolized by mitochondria (6, 33).
Abf2p contains two high-mobility-group (HMG)-like domains resembling the eucaryotic nuclear nonhistone protein HMG-1 (6). At least one HMG-1 domain of Abf2p is necessary for the maintenance of mtDNA in vivo (20). Other HMG-like proteins, such as human mitochondrial transcription factor (mtTF1) (40) and the yeast nuclear HMG-like nonhistone protein (NHP6A) (20), and the bacterial DNA packaging protein HU (33), which is not a member of the HMG family, can partially complement the mtDNA loss of the abf2 mutant cells when these proteins are targeted to mitochondria. Thus, Abf2p is likely to function in the maintenance of rho+ mtDNA, at least in part, as a consequence of DNA packaging.
However, it has not been understood clearly how Abf2p is involved in the maintenance of the mitochondrial genome. The abf2 mutant cells do not require functional Abf2p for growth in glycerol medium at 30°C, but they cannot grow at all in glycerol at 37°C (33). To better understand the role of Abf2p in mitochondria, we isolated multicopy suppressors from a yeast genomic library that restore the ability of abf2 null mutants to grow at 37°C in the presence of glycerol (21, 34). We first selected and characterized YHM1 from the candidate genes. YHM1 shows the sequence similarity to the mitochondrial carrier protein family (21). Disruption of the YHM1 gene has no obvious effect on the growth of wild-type cells, but the wild-type gene is essential for the growth of the abf2 mutant on glycerol at 30°C (21). However, the in vivo function of YHM1 has not yet been discovered. Another multicopy suppressor, ILV5, was isolated by Zelenaya et al. (59). The ILV5 gene encodes acetohydroxy acid reductoisomerase, which catalyzes a step in branched-chain amino acid biosynthesis in mitochondria. mtDNA is unstable in the ilv5 null mutant cells, leading to the production of [rho−] petite mutants. The most severe instability of [rho+] mtDNA is observed in cells with abf2 ilv5 double mutations (59), as was the phenotype of the abf2 yhm1 double-mutant cells (21). The mechanisms for restoring the defects of the abf2 mutant cells by YHM1 or ILV5 are unclear.
Recently, we have isolated and characterized another multicopy suppressor, called YHM2. Here we report the characterization of YHM2 at the molecular level and discuss a possible role of Yhm2p in mitochondrial DNA metabolism.
MATERIALS AND METHODS
Media and yeast strains.
Rich medium YP (1% yeast extract, 2% peptone) was supplemented with either the fermentable carbon source (2% glucose [YPD]) or the nonfermentable carbon source (3% glycerol [YPG], 3% potassium acetate [YPA], or 3% ethanol [YPE]). Synthetic minimal medium SM (0.67% yeast nitrogen base without amino acids) was supplemented with various carbon sources and nutrients as required. When acetate was used as the carbon source, the pH of the medium was adjusted to 6.5. The differential solid medium YPDGE (1% yeast extract, 2% peptone, 0.1% glucose, 3% glycerol, 2% ethanol, 1.5% Bacto Agar [Difco]) was used for the distinction between respiration-competent cells ([rho+]) and respiration-deficient cells ([rho−] or [rho0]). The yeast strains used in this study are listed in Table 1.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| W303 | MATa/α ade2-1/ade2-1 ura3-1/ura3-1 his3-11,15/his3-11,15 trp1-1/trp1-1 leu2-3,112/leu2-3,112 can1-100/can1-100 | R. Rothstein |
| KLR17 | W303 abf2::URA3/ABF2 | 34 |
| KLR17-3c | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 ABF2 | 34 |
| KLR17-3d | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 abf2::URA3 | 34 |
| KLR70 | KLR17-3d with YEp13 | This study |
| KLR71 | KLR17-3d with pY-D3 | This study |
| KLR72 | KLR17-3d with pY-D3Δ935–1085 | This study |
| KLR73 | KLR17-3d with pY-D3PRM | This study |
| MCY40 | W303 yhm2::TRP1/YHM2 | This study |
| MCY41 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 yhm2::TRP1 | This study |
| MCY42 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 YHM2 | This study |
| MCY41-1 | MCY41 with pY-D3 | This study |
| MCY41-2 | MCY41 with pY-D3PRM | This study |
Cloning of multicopy suppressors of the abf2 mutant.
In a previous study, the abf2 mutant cells were transformed with a yeast genomic library constructed in plasmid YEp13, and the cells were spread on plates containing selective medium (without leucine) and glycerol (21, 34). Five clones (C1, C9, C19, D3, and D5) were able to grow under nonpermissive condition, that is, at 37°C in the presence of glycerol (21, 34). In the present study, the D3 clone, with an open reading frame (ORF) encoding 314 amino acids, was chosen for detailed investigation. The open reading frame encoding 314 amino acids, registered as YMR241w in the SGD database, was named YHM2.
DAPI staining and fluorescence microscopy.
Cells were picked from colonies, fixed with 100% methanol for 5 min, washed with phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 · 7H2O, 1.4 mM KH2PO4 [pH 7.4]), and resuspended in 0.5 μg of DAPI (4′,6-diamidino-2-phenylindole [Sigma]) per ml. After a 1-h incubation with gentle rotation in the dark at room temperature, the cells were gently pelleted, washed three times for 10 min with PBS, and resuspended in 100 μl of PBS. The cells were examined under a fluorescence microscope (Nikon Optiphot-II) with a filter recommended for use with DAPI and photographed at ASA 800 with Kodak T-Max 400 film.
Expression and purification of YHM2 and the mutant proteins in E. coli.
The various recombinant proteins of YHM2 were expressed in Escherichia coli from the wild-type or mutant YHM2 inserted between BamHI and EcoRI of the pRSETA vector (Invitrogen). All recombinant proteins were produced as fusion proteins with a hexahistidine tag at the N terminus. The pRSETA vectors harboring Yhm2-ΔCp (N-terminal half region), Yhm2-ΔNp (C-terminal half region), Yhm2p (full length), or Yhm2-PRMp (mutated at a proline repeat sequence which is in the putative DNA-binding subdomain) were introduced into E. coli BL21(DE3)pLysS. Mutation of the proline repeat region was introduced by a PCR-based technique (18) with an internal mismatch primer set (sense, 5′-GATGGTGGTGGTGGTGGTAATTTGACTGTTGGTAAG-3′; antisense, 5′-ATTACCACCACCACCACCATCTTCCTTCTTAGATTG-3′, where the mismatched positions are underlined). Thus, the sequence QSKKEDPNRPKNLTVGK of amino acids 243 to 259 was changed to QSKKEDGGGGGNLTVGK. The resulting mutant gene was named yhm2-PRM (yhm2 mutated at the proline repeat).
The transformed cells were grown at 37°C in Luria-Bertani medium until the optical density at 600 nm (OD600) reached 0.4, at which time 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added, and growth was continued for an additional 1 h. The cells were pelleted, washed once with ice-cold PBS, suspended in lysis buffer (20 mM sodium phosphate [pH 7.8], 500 mM NaCl, 1 mM phenylmethylsulfonyl fluoride [PMSF]), disrupted by ultrasonication for 3 min, and centrifuged at 15,000 × g for 15 min. The pellet was resuspended in denaturation buffer (6 M guanidine hydrochloride, 20 mM sodium phosphate [pH 7.8], 500 mM NaCl, 1 mM PMSF) and centrifuged at 15,000 × g for 25 min. The supernatant was loaded on a column (1.5 ml) of Ni2+-chelating Sepharose 6B (Pharmacia). The column was washed with 10 bed volumes of denaturation buffer until the OD280 reached baseline and then washed with wash buffer (6 M guanidine hydrochloride, 20 mM sodium phosphate [pH 5.3], 500 mM NaCl). The recombinant proteins with the hexahistidine tag were eluted with elution buffer (6 M guanidine hydrochloride, 20 mM sodium phosphate [pH 4.0], 500 mM NaCl) and dialyzed against dialysis buffer (20 mM Tris-HCl [pH 7.5], 2 mM PMSF) at 4°C overnight. The purity of the recombinant proteins was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and the purified proteins were used for DNA-binding studies.
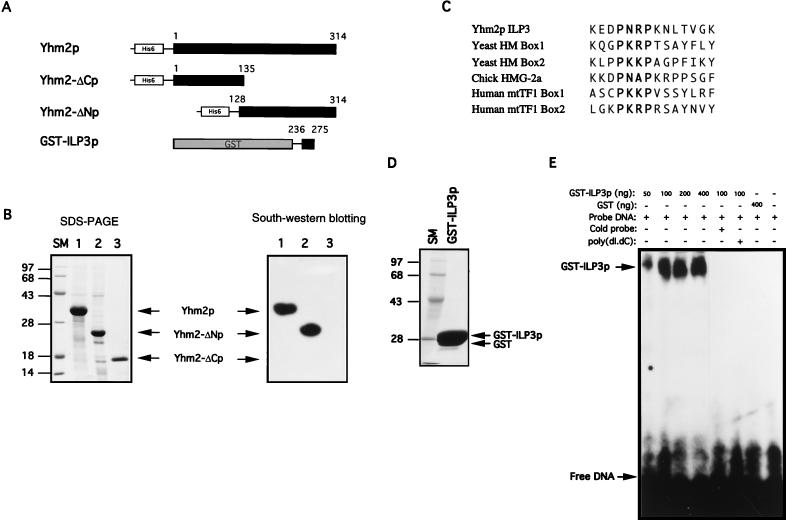
To test whether the most hydrophilic region in the carboxy half of Yhm2p, which is named ILP3 (third internal loop), also binds to DNA, the GST-ILP3p (fusion protein of glutathione S-transferase [GST]) was expressed and purified by the method of Smith and Johnson (47). The pGEX-KG vector harboring ILP3 was introduced into E. coli DH5α. The transformed cells were grown at 37°C in Luria-Bertani medium until the OD600 reached 0.5, at which time 0.5 mM IPTG was added and growth was continued for an additional 1 h. The cells were pelleted, washed once with ice-cold PBS, suspended in lysis buffer (PBS [pH 7.4], 1 mM dithiothreitol [DTT], 2 mM EDTA, 1 mM PMSF), disrupted by ultrasonication for 3 min, and centrifuged at 15,000 × g for 15 min. After addition of Triton X-100 to 0.1%, the soluble fraction of the lysate was loaded on a glutathione-Sepharose 4B column (Pharmacia). The column was washed with 10 bed volumes of lysis buffer–0.1% Triton X-100 until the OD280 reached the baseline. GST-ILP3p was eluted with elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM reduced glutathione) and analyzed by SDS-PAGE. The purity of the isolated fusion proteins was greater than 90% (see Fig. 5D). Since GST itself has no DNA-binding activity, the protein fraction of the GST-ILP3p fusion protein was used for DNA-binding studies without further purification.
FIG. 5.
DNA-binding ability of Yhm2p. (A) Construction of plasmids for expression of full-length and partial fragments of Yhm2p. A hexahistidine peptide sequence was placed at the amino terminus for affinity purification of the protein. For expression of a short segment of ILP3 (from 236 to 275 of Yhm2p [Fig. 3]), the ILP3 sequence was fused with GST. (B) Coomassie blue staining pattern of the proteins purified by affinity chromatography as described in Materials and Methods (left). DNA binding to the proteins immobilized on a nitrocellulose membrane (right). The purified recombinant proteins were fractionated by PAGE and transferred to nitrocellulose membrane. The immobilized protein was refolded in situ and probed with 32P-labeled DNA as described in Materials and Methods. DNA bound to full-length Yhm2p and the carboxy-terminal half of the protein (Yhm2-ΔNp) but not to the amino-terminal half (Yhm2-ΔCp). (C) The amino-acid sequence of ILP3 of Yhm2p is similar to the amino-terminal sequences of various HMG boxes. The proline repeat in bold is important for the DNA-binding activity of the HMG box. (D) Coomassie blue staining of the purified recombinant GST-ILP3 protein. (E) DNA-binding activity of the ILP3 region. DNA binding to the ILP3 region was demonstrated by a DNA mobility shift assay as described in Materials and Methods.
Southwestern blotting.
Purified recombinant proteins were subjected to SDS-PAGE (15% polyacrylamide). The separated proteins were electrotransferred onto nitrocellulose membranes, and the membranes were incubated for 30 min at room temperature in 20 ml of 6 M guanidine hydrochloride–50 mM Tris-HCl (pH 7.0). The immobilized proteins were renatured by incubation of the blot in dilution buffer (50 mM imidazole, 200 mM KCl, 1 mM CaCl2, 0.5% bovine serum albumin, 0.05% Tween 20) containing serially diluted guanidine hydrochloride. Seven to eight incubation steps were carried out until the concentration of guanidine hydrochloride finally reached about 45 mM, starting from 6 M. The incubation time for each step was approximately 10 min. The membranes were subsequently blocked in 1% bovine serum albumin–10 mM HEPES-KOH (pH 7.6) for 1 h at 4°C and then incubated with 32P-end-labelled double-stranded DNA probes (10 fmol/ml, ∼2 × 106 cpm) dissolved in binding buffer (10 mM HEPES-KOH [pH 7.6], 100 mM KCl, 1 mM EDTA, 1 mM DTT) at 4°C for 12 h. The membranes were subsequently washed five times for 10 min each time with binding buffer. Autoradiography was performed at −70°C for 6 to 12 h.
The sequence of the labeled DNA was either 5′-GGCTTTGACGTCAGCCTGGCCT-3′, which contains the cyclic AMP response element (CRE) sequence of the rat tyrosine hydroxylase gene, or 5′-TCCCACTGATGACGTCCATGTGTCATTAGTG-3′, which contains the CRE sequence of the human dopamine β-hydroxylase gene.
DNA mobility shift assay.
The reaction mixture (20 μl), containing variable amounts of GST-ILP3p from 50 to 400 ng and 10 fmol of 32P-end-labeled double-stranded DNA fragment in 4% Ficoll–100 mM KCl–1 mM EDTA–0.5 mM DTT–10 mM HEPES (pH 7.9), was incubated on ice for 1 h and analyzed by electrophoresis on a 6% nondenaturing polyacrylamide gel at 150 V in 0.25× TBE (22.5 mM Tris, 22.5 mM borate, 0.5 mM EDTA [pH 8.3]) at room temperature. The gel was dried and exposed to an X-ray film (Agfa CURIX XP 100 NIF) at −70°C overnight.
Preparation and purification of intact mitochondria, mitochondrial subfractions, and mitochondrial nucleoids.
Mitochondria and mitochondrial nucleoids were prepared according to the method described by Newman et al. (37) with some modifications. Mitochondria were isolated from spheroplasts of 1-liter cultures grown in YPG medium (OD600 = 0.8 to 1.1). All procedures were carried out at 4°C except where noted. Yeast cells were resuspended in 50 ml of 100 mM Tris-HCl–20 mM EDTA (pH 9.3) and shaken with β-mercaptoethanol (5 μl/ml) for 15 min at 30°C. The cells were collected, washed once with SCE buffer (pH 5.8) (600 mM sorbitol, 300 mM mannitol, 20 mM K2HPO4, 20 mM citric acid, 1 mM EDTA), and resuspended in SCE buffer (pH 5.8) (1 ml/g of cells). Yeast lytic enzyme (70,000 U/g; ICN Biochemicals) was added to the cell suspension (0.5 mg/ml), and the cells were treated at 30°C for 30 min with gentle mixing. Spheroplasts were washed twice with 10 ml of SCE buffer (pH 7.0) containing 1 mM spermidine, 7 mM β-mercaptoethanol, and 1 mM PMSF and homogenized by a tight-fitting Dounce homogenizer at 4°C. Cellular debris and unlysed cells were removed by two 5-min centrifugations at 1,600 × g and one 5-min centrifugation at 2,500 × g. Crude mitochondria were collected by centrifugation at 12,000 × g for 20 min. These mitochondria were further purified by flotation.
Isolation of inner and outer mitochondrial membranes was carried out as previously described (5). Purified mitochondria were resuspended in lysis buffer (10 mM Tris-HCl [pH 7.5], 0.1 mM EDTA, 1 μg of aprotinin/ml, 1 μg of leupeptin/ml) at a protein concentration of 5 mg/ml and sonicated on ice three times for 5 s each time (Branson Sonifier 450, maximal output). Total mitochondrial membranes were sedimented at 81,000 × g for 1 h. Inner and outer mitochondrial membrane particles were separated on a sucrose gradient (30 to 50%), and membrane fractions were collected as described previously (5).
To obtain the mitochondrial nucleoids, the purified mitochondria were resuspended in NE2 buffer (250 mM sucrose, 20 mM Tris-HCl [pH 7.6], 2 mM EDTA, 7 mM β-mercaptoethanol) and diluted with an equal volume of 0.5× NE2 buffer to a final concentration of 5 to 7 mg of mitochondrial protein/ml. Spermidine was added to a final concentration of 3 mM, and mitochondria were lysed by adding 20% Nonidet P-40 (NP-40) to a final concentration of 0.5%. After 5 min on ice with gentle mixing, the lysate was fractionated into supernatant and pellet fractions by centrifugation at 12,000 × g for 20 min. The pellet fraction was resuspended as described above and layered on top of step gradients, composed of 3.5 ml of 20%, 2.5 ml of 40%, 1.8 ml of 60%, and 0.9 ml of 70% sucrose in gradient buffer (20 mM Tris-HCl [pH 7.6], 1 mM EDTA, 1 mM spermidine, 7 mM β-mercaptoethanol, 1 mM PMSF), in SW41 tubes and centrifuged at 111,000 × g for 75 min. The gradients were fractionated and analyzed for mtDNA and protein. The crude preparation of mitochondrial nucleoids was further purified by a second sucrose gradient centrifugation using the same procedure. The protein profile and the purity of the final preparation of mitochondrial nucleoids were examined by SDS-PAGE and immunoblot analysis.
Immunoblotting.
The mitochondrial proteins fractionated by SDS-PAGE were transferred to nitrocellulose membrane by electroblotting, and the membrane was probed with affinity-purified rabbit polyclonal antibodies for Yhm2 protein (1:5,000 dilution), monoclonal DA5 antibody for CoxIIIp (Molecular Probes, Inc.; 0.5 μg/ml), or monoclonal 16G9-E6 antibody for porin (Molecular Probes, Inc.; 0.5 μg/ml) and then incubated with anti-rabbit or anti-mouse immunoglobulin G conjugated to alkaline phosphatase (Chemicon; 1:5,000 dilution) by using a published method (44).
Disruption of YHM2.
A 2.6-kb DNA fragment containing the YHM2 gene was ligated into pSP72 digested with EcoRI and XhoI to create pSPYHM2. A 298-bp PvuII-SpeI fragment containing 32% of the ORF of the YHM2 gene was deleted from pSPYHM2 and replaced with a 1.5-kb EcoRI fragment containing the TRP1 gene. The resulting plasmid, pSPyhm2::TRP1, was used as a template for PCR with a primer set to amplify a 2.4-kb DNA fragment containing the disrupted YHM2 gene. The PCR-amplified DNA was then used to transform the diploid yeast strain W303. The yhm2::TRP1 disruption was confirmed by Southern blotting of genomic DNA isolated from Trp+ transformants. Genomic DNA was digested completely with AatII, separated by agarose gel electrophoresis, (0.8% agarose), and probed with a 32P-labeled 0.9-kb DNA fragment containing the YHM2 gene by using published methods (44). Tetrad dissection was carried out to obtain the Trp+ haploid yhm2 mutant (MCY41) and isogenic wild-type (MCY42) strains (Table 1).
RESULTS
YHM2, a multicopy suppressor for the temperature-sensitive defect of the abf2 mutant.
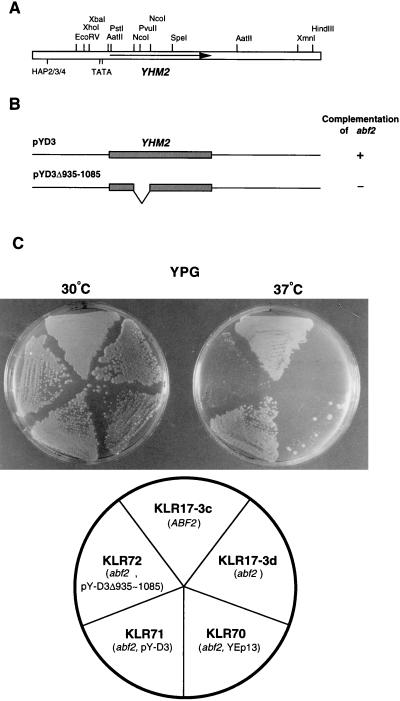
Identification of genomic clone D3, which restores the respiration defect of the abf2 mutant in the presence of glycerol at 37°C, was described in previous reports (21, 34). Plasmid pY-D3 contains a genomic insert of approximately 2.6 kb (Fig. 1A). The whole sequence of the D3 insert was determined, and the possible ORFs and the cis-acting elements of the sequence were searched with a computer program (Fig. 1A). The insert fragment contains six ORFs, the largest of which encodes a protein of 314 amino acid residues. Other ORFs encode proteins with less than 100 amino acids. ORF314 is located in the middle of the D3 fragment and has cis-acting elements. Two putative TATA elements are located 54 and 80 bp upstream from the ATG codon (Fig. 1A). The putative HAP2/HAP3/HAP4 transcription factor-binding sequence, -GATTGGA- (54), is found 559 bp upstream from the ORF.
FIG. 1.
Functional deletion mapping of a 2.6 kb DNA fragment of clone D3. (A) Restriction map of the genomic insert of clone D3. The arrow indicates the YHM2 ORF. The two putative TATA boxes (−54 and −80) and the putative HAP2-HAP3-HAP4 transcription factor-binding site (−559) are indicated. (B) Deletion analysis of the D3 sequence. To identify the gene of interest, the fragment between the two NcoI sites of YHM2 was deleted. The plasmid with the deletion (pY-D3Δ935–1085) was tested for its ability to complement the temperature-sensitive phenotype of abf2 mutant cells. (C) Clone D3 can suppress the temperature-sensitive defect of the abf2 mutant but not the deleted clone D3Δ935–1085. The abf2 mutant cells transformed with YEp13, pY-D3, and pY-D3Δ935–1085 were grown to late log phase in synthetic medium (−Ura, −Leu) containing glucose and were streaked onto YPG plates (3% glycerol as nonfermentable carbon source) and grown at 30 or 37°C.
To determine whether ORF314 encodes the D3 suppressor, we constructed a plasmid containing the D3 fragment with a deletion between the two NcoI sites within the ORF, pY-D3Δ935–1085 (Fig. 1B). This plasmid cannot restore the defect of the abf2 mutant strain (KLR17-3d) (Fig. 1C). Therefore, it appears that ORF314 is indispensable for the restoration activity of the D3 clone. We named this ORF YHM2. The YHM2 gene is located on chromosome XIII according to the sequence determined by the yeast genome-sequencing project (the name of the ORF in the SGD database is YMR241w).
Yhm2p is essential for mitochondrial function in the absence of HM.
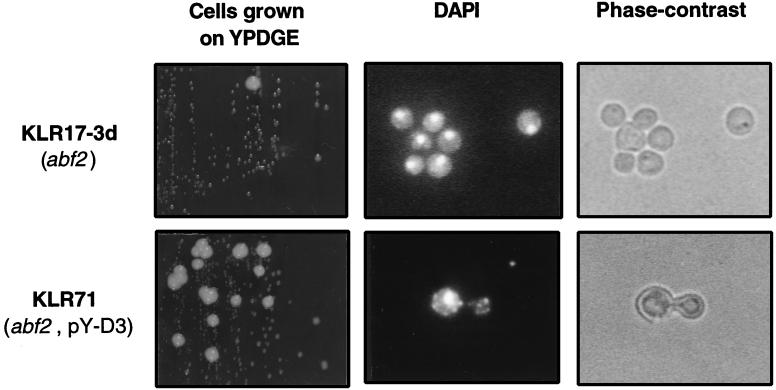
The mitochondrial genome of the abf2 mutant is unstable in media with a high concentration of glucose, such as YPD, and the mutant cells form petite colonies at a high rate (6, 33). Therefore, we investigated how YHM2 suppresses the rapid loss of mtDNA of abf2 mutant cells when they are grown in glucose-rich medium. Strains KLR17-3d (abf2::URA3) and KLR71 (abf2::URA3, pY-D3) were cultivated in YPD liquid medium for 3 days. After being washed twice with sterile distilled water, the cells were streaked on the YPDGE plate and incubated for 4 days at 30°C. The abf2 mutant cells containing the pY-D3 plasmid grew more rapidly and formed larger colonies on the YPDGE plate than did the abf2 mutant cells without the plasmid (Fig. 2). The cells of the large colony formed by each strain were examined by DAPI staining, and small fluorescent spots corresponding to mtDNA nucleoids were observed only in the cells of KLR71 (abf2::URA3, pY-D3) (Fig. 2). The punctate cytoplasmic staining of KLR71 was similar to that of the wild-type [rho+] mitochondria, while strain KLR17-3d (abf2::URA3) showed only diffusely stained [rho+] mtDNA (6, 34, 37). In the cells of small colonies formed by both strains, only the nuclei were stained by DAPI (data not shown). The result suggests that the overproduced Yhm2p has beneficial effects on the maintenance and organization of mtDNA in the mitochondria of strain KLR17-3d (abf2::URA3) under culture conditions that result in rapid loss of the mitochondrial genome.
FIG. 2.
The KLR17-3d (abf2::URA3) strain grown in the presence of glucose loses its mitochondrial genome, whereas the KLR71 (abf2::URA3, pY-D3) strain maintains [rho+] mitochondria. Cells were grown to stationary phase on YPD (2% glucose), streaked on YPDGE (1% yeast extract, 2% peptone, 0.1% glucose, 3% glycerol, 2% ethanol) plates, and then incubated at 30°C. The cells from large colonies of each strain were picked and stained with a DNA-specific dye, DAPI, and visualized by fluorescence microscopy (middle) and phase-contrast microscopy (left). Magnification, ×400. The major fluorescence source in the central region of the cells corresponds to DAPI-stained nuclear DNA. Small and weak fluorescent spots corresponding to mtDNA in the periphery are observed only in cells of KLR71 (abf2::URA3, pY-D3).
Yhm2p is a member of the mitochondrial carrier protein family.
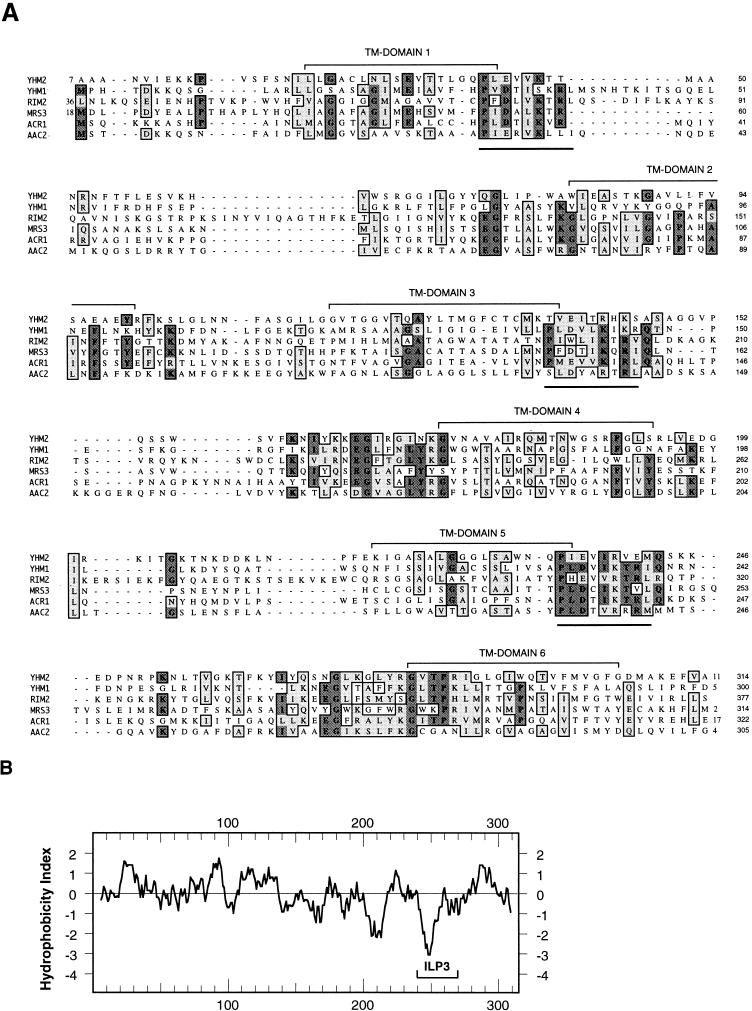
The predicted amino acid sequence encoded by YHM2 has some similarity to the yeast mitochondrial inner membrane carrier protein family (Fig. 3A) (2, 25, 26). Members of the mitochondrial carrier protein family do not have extensive similarity in overall sequence, but they all contain a tripartite structure, composed of three homologous domains of 100 amino acids each, that contains several highly conserved residues that comprise the energy transfer protein signature P-X-(DE)-X-(LIVAT)-(RK)-X-(LRH)-(LIVMFY) (1, 43, 45). This sequence is repeated three times in Yhm2p (underlined in Fig. 3A). The hydropathy profile of Yhm2p suggests that the protein might have some transmembrane domains (Fig. 3B). These data suggest that Yhm2p may be a mitochondrial inner membrane protein.
FIG. 3.
Alignment and hydropathy plot of the deduced amino acid sequence of YHM2. (A) Multiple-sequence alignment for maximal amino acid similarities between Yhm2p and the members of the mitochondrial carrier family obtained with the ClustalW program in MacVector 6.01. The AAC2 gene encodes the mitochondrial ADP/ATP carrier (29), and ACR1 encodes a succinate/fumarate carrier (39). MRS3 encodes the suppressor of the mtRNA splicing defect (55, 56), and RIM2 encodes the suppressor of the pif1 mutant (50). YHM1 was isolated as a suppressor of the abf2 mutant (21, 34). The putative transmembrane (TM) domains are indicated, and the conserved motifs of mitochondrial energy transfer proteins are underlined. (B) Hydrophobicity profile of Yhm2p. The Yhm2p amino acid sequence was analyzed with the DNA Strider 1.2 program by the Kyte and Doolittle method. The most hydrophilic region (ILP3) of YHM2 is indicated.
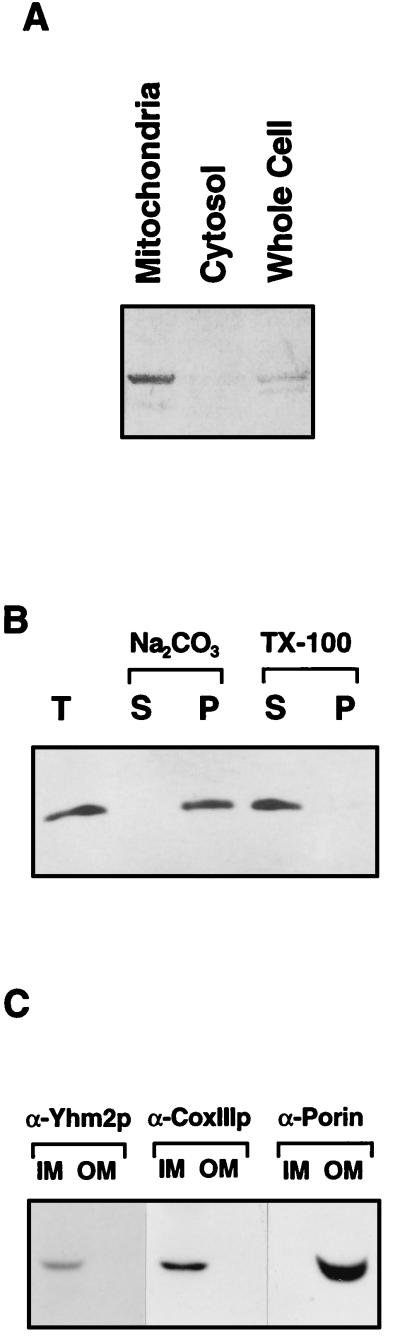
To test this prediction, we isolated mitochondria from wild-type cells and tested the distribution of Yhm2p by immunoblotting with affinity-purified polyclonal anti-Yhm2p antibody. As shown in Fig. 4A, almost all the Yhm2p is localized in mitochondria. To determine whether Yhm2p is the integral membrane protein or loosely associated with the membrane structure, the purified mitochondria were extracted with 100 mM sodium carbonate (pH 12) for 30 min on ice, centrifuged at 100,000 × g for 1 h, and separated into supernatant and pellet fractions. The fractions were analyzed by immunoblot analysis as described above. The result shows that Yhm2p is associated with the membrane fraction under these conditions. Also, 1% Triton X-100 solubilized most of Yhm2p, which confirms that Yhm2p is an integral membrane protein of mitochondria (Fig. 4B). We next sought to determine in which mitochondrial membrane (inner or outer) Yhm2p is located. The purified mitochondria were sonicated briefly, and the mitochondrial membrane was sedimented. The purified mitochondrial membrane was fractionated into inner and outer membranes by sucrose gradient centrifugation, and 5 μg of protein from each fraction was subjected to immunoblot analysis with monospecific anti-Yhm2p antibody and the monoclonal antibodies for the inner or outer membrane-specific marker protein (CoxIIIp or porin, respectively). Yhm2p was detected only in the inner membrane fraction (Fig. 4C). The correct partitioning of CoxIIIp and porin demonstrates the purity of the inner and outer mitochondrial membrane fractions (Fig. 4C).
FIG. 4.
Yhm2p is a mitochondrial inner membrane protein. (A) Western blot analysis of the mitochondrial preparation obtained from wild-type yeast strain W303 shows the presence of Yhm2p in mitochondria. The proteins in whole cells, cytosol, and mitochondria were separated by SDS-PAGE and analyzed by immunoblotting with affinity-purified anti-Yhm2p antibody as described in Materials and Methods. (B) The purified mitochondria was extracted with 100 mM Na2CO3 (pH 12) or 1% Triton X-100, and the proteins in the supernatant and pellet were analyzed in panel A. The result shows that Yhm2p is an integral membrane protein. T, total mitochondria; S, supernatant; P, pellet. (C) Yhm2p is associated with the inner mitochondrial membrane, as determined by immunoblot analysis of inner mitochondrial membrane fractions (IM) and outer mitochondrial membrane fractions (OM) from the wild-type strain with antibodies against Yhm2p (α-Yhm2p), CoxIIIp (α-CoxIII), and porin (α-porin).
Yhm2p has general DNA-binding activity in vitro.
Since Abf2p protein plays an important role in the maintenance of mtDNA by virtue of its general DNA-binding activity (20), and since YHM2 is identified as a suppressor of the abf2 mutation, it is possible that Yhm2p interacts with DNA as well. As shown in the hydropathy plot (Fig. 3B), Yhm2p has two distinctive regions, a hydrophobic amino-terminal half and a very hydrophilic carboxy-terminal half. Each of these two domains of Yhm2p was expressed in E. coli and purified to analyze their DNA-binding properties in vitro. A hexahistidine tag was placed in front of the proteins for affinity purification. Yhm2-ΔCp has the amino acid sequence of positions 1 to 135 of Yhm2p, and Yhm2-ΔNp has the amino acid sequence of positions 128 to 314 (Fig. 5A). The proteins were produced in E. coli, and the purity of the isolated proteins is shown in Fig. 5B (left panel). The full length and the amino-terminal half of Yhm2p are insoluble. Therefore, the proteins transferred to nitrocellulose membrane were refolded in situ by treatment with decreasing concentrations of guanidine hydrochloride, and the DNA-binding activity of the proteins was assayed by blot analysis with 32P-labeled DNA probes in the presence of 100 mM KCl (Fig. 5B [right]). The DNA probes used in this assay were the 22-mer CRE of the rat tyrosine hydroxylase gene and the 31-mer CRE of the human dopamine β-hydroxylase gene, since they were conveniently available in our laboratory. The results showed that the labeled DNA binds to immobilized and renatured Yhm2p (full length) and Yhm2-ΔNp (carboxy-terminal half) but not to Yhm2-ΔCp (amino-terminal half) in the presence of 100 mM KCl (Fig. 5B [right]).
We wondered whether the most hydrophilic sequence in the carboxy-terminal region of Yhm2p can also bind to DNA. Interestingly, this region corresponds to the third internal loop of the mitochondrial carrier protein family which is extruded into the matrix (25, 26) and has the short basic amino acid sequence -KEDPNRPK- (from 246 to 253), which is very similar to the essential sequence of the DNA recognition and binding subdomain of the HMG box (Fig. 5C) (20, 30). The recombinant Abf2p lacking this subdomain neither binds to DNA nor supercoils DNA in vitro and is unable to complement the abf2 mutant in vivo (20). The third internal-loop (ILP3) region was expressed as a GST fusion protein in E. coli because it is rather short (40 amino acids [positions 236 to 275]) to be expressed alone. To study the DNA-binding activity of the GST-ILP3 fusion protein, we used the DNA mobility shift assay since the fusion protein is soluble (Fig. 5D). The purified GST-ILP3 fusion protein bound to the DNA probes in the presence of 100 mM KCl (Fig. 5E). The DNA-binding activity seems to be general, because the DNA-protein interaction was completely inhibited when poly(dI-dC) was added to the reaction mixture as a competitor (Fig. 5E). Thus, the hydrophilic third internal-loop region of the carboxy-terminal half may play an important role in the interaction between Yhm2p and mtDNA.
The putative DNA-binding domain is important for DNA binding and suppression of the abf2 mutant.
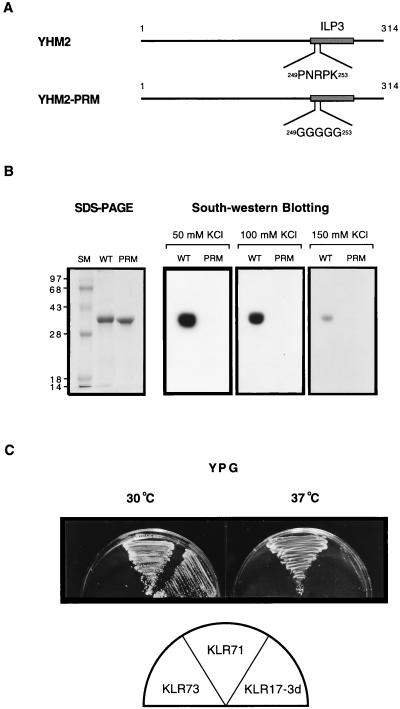
We also investigated whether the DNA-binding activity of Yhm2p is important for the suppression of the abf2 mutant. First, we introduced a mutation into the putative DNA-binding subdomain of the ILP3 region by PCR-based site-directed mutagenesis (Fig. 6A). Mutant Yhm2-PRMp has pentaglycine in the ILP3 region instead of the amino acid sequence -PNRPK- at 249 to 253, which is similar to the proline repeat subdomain of the HMG box (Fig. 5C). Yhm2-PRMp was expressed and purified from E. coli (Fig. 6B), and its DNA-binding activity was determined by the Southwestern method. The strength of DNA binding was analyzed by varying the KCl concentration from 50 to 150 mM in the binding buffer. The result shows that mutant Yhm2-PRMp is not able to bind to DNA under any binding conditions in vitro (Fig. 6B). In contrast, the wild-type Yhm2p bound DNA well at high KCl concentrations (150 mM). We introduced the mutant yhm2-PRM gene (pY-D3PRM) at high copy number into the abf2 mutant strain (KLR17-3d) to investigate whether the mutation interferes with the ability to suppress the abf2 mutant. The transformed strain, KLR73 (abf2::URA3, pY-D3PRM), was unable to utilize nonfermentable carbon sources and could not grow well (Fig. 6C; Table 2). However, this lethal effect of the yhm2-PRM gene is restricted to the abf2 strain, and other strains with this mutant gene showed normal growth (Table 2) and stable expression of Yhm2-PRMp in their mitochondria (data not shown).
FIG. 6.
The putative DNA-binding domain is essential for suppression of the abf2 mutant. (A) Schematic representation of wild-type (YHM2) and mutant (Yhm2-PRM) Yhm2p. The proline repeat sequence in the putative DNA-binding subdomain was changed to pentaglycine by PCR-based site-directed mutagenesis (see Materials and Methods). (B) The putative DNA-binding subdomain is important for Yhm2p binding to DNA. The wild-type (WT) and mutant (PRM) Yhm2p were analyzed for their DNA-binding activity by Southwestern blotting as described in the legend to Fig. 5B. The same amount of each protein (1 μg) was used for both SDS-PAGE (12% polyacrylamide) and Southwestern analysis. To test the difference of DNA-binding affinity between Yhm2p and Yhm2-PRMp, the concentration of KCl in the binding buffer was varied from 50 to 150 mM. The mutant Yhm2-PRMp did not bind DNA under any conditions. (C) The mutation in the putative DNA-binding subdomain abolishes the suppression of the abf2 mutant. KLR17-3d (abf2::URA3) was transformed with the high-copy-number episomal plasmid harboring the yhm2-PRM gene (pY-D3PRM) and tested for growth on YPG at 30 or 37°C. KLR73 (abf2::URA3, pY-D3PRM) grew very poorly at both temperatures. The control strain, KLR71 (abf2::URA3, pY-D3), grew at both temperatures.
TABLE 2.
Generation times of various strains with different carbon source at 30°C
| Strain | Generation time (h)a in:
|
|||||
|---|---|---|---|---|---|---|
| Rich mediumb
|
Synthetic mediumb
|
|||||
| Glucose | Glycerol | Glucose | Glycerol | Acetate | Ethanol | |
| MCY41 (yhm2::TRP1) | 2.2 | 3.4 | 3.4 | 7.5 | 6.4 | 11.8 |
| MCY42 (YHM2) | 2.3 | 3.6 | 3.2 | 4.3 | 5.1 | 5.7 |
| MCY41-1 (yhm2::TRP1/pY-D3) | 2.1 | 3.4 | 3.1 | 3.9 | 5.0 | 5.5 |
| MCY41-2 (yhm2::TRP1/pY-D3PRM) | 2.3 | 3.4 | 3.4 | 7.3 | 6.2 | 12 |
| KLR17-3d (abf2) | 3.3 | 7.1 | ||||
| KLR71 (abf2/pY-D3) | 2.5 | 4.2 | ||||
| KLR73 (abf2/pY-D3PRM) | 3.5 | NDc | ||||
Strains were diluted from overnight YPG cultures and grown in the presence of different carbon sources under aeration at 30°C. Growth was monitored by measuring the OD600 for four to five generations.
Carbon sources are given. See “Media and yeast strains” in Materials and Methods.
ND, not determined. The growth rates of KLR73 on nonfermentable carbon sources could not be determined because the strain grew very poorly.
Yhm2p is a component of the mitochondrial nucleoid structure.
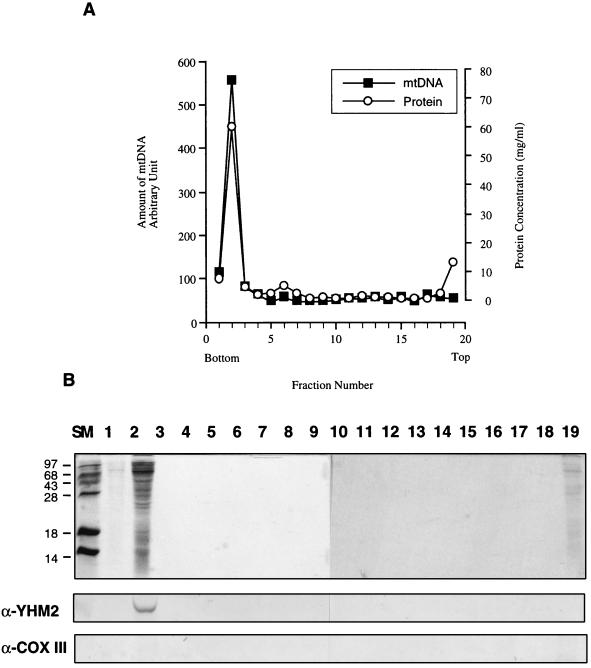
To confirm the DNA-binding activity of Yhm2p in vivo, we purified mitochondrial nucleoids from the pellet of mitochondria extracted with 0.5% NP-40 by using previously described methods (35, 37). Mitochondrial nucleoids were purified by two successive sucrose gradient centrifugations, which minimize contamination by other membrane proteins (37). Figure 7A shows the result of the second sucrose gradient ultracentrifugation. The protein profiles of the fractions were analyzed by SDS-PAGE (15% polyacrylamide). Only the second fraction has the majority of the proteins associated with mtDNA (Fig. 7B [top]). The protein profile of the second fraction was similar to that of the mitochondrial nucleoid preparation described in a previous report (37), except for the absence of a 110-kDa band. The existence of Yhm2p and the purity of mitochondrial nucleoids were determined by immunoblotting with affinity-purified anti-Yhm2p antibody (Fig. 7B [middle]) and monoclonal anti-CoxIIIp antibody (Fig. 7B [bottom]). Monoclonal anti-CoxIIIp antibody was used as a negative control because the protein is not likely to be associated with mtDNA. The results show that Yhm2p is present in the nucleoid from the second fraction whereas CoxIIIp is absent (Fig. 7B).
FIG. 7.
Yhm2p is associated with mitochondrial nucleoids. (A) Sucrose gradient centrifugation of mitochondrial nucleoids. Mitochondria from wild-type yeast cells were lysed in 0.5% NP-40 and centrifuged. The pellet was loaded onto a sucrose gradient and centrifuged. The peak fraction containing mtDNA was reloaded onto a sucrose gradient and centrifuged as described in Materials and Methods. The gradient fractions were analyzed for mtDNA and protein by Southern blotting and the Bradford assay, respectively. (B) The protein profiles of the fractions from second sucrose gradient centrifugation were analyzed by SDS-PAGE (15% polyacrylamide). SM, size markers; the numbers indicate fraction numbers. The presence of Yhm2p and CoxIIIp in the purified mitochondrial nucleoids was determined by Western blot analysis. The anti-CoxIIIp antibody was used as a negative control. As shown in Fig. 4, the anti-CoxIIIp antibody used in this study was active and detected CoxIIIp in the inner mitochondrial membrane fraction.
Yhm2p is essential for mitochondrial function.
To determine whether Yhm2p is essential for viability and mitochondrial function, the YHM2 gene was disrupted in a wild-type strain. We constructed a plasmid in which a large portion (32%) of the YHM2 sequence was deleted and replaced by the selectable marker gene TRP1 as described in Materials and Methods. A PCR-amplified DNA fragment containing the yhm2::TRP1 sequence was used to transform the diploid strain W303, and Trp+ colonies were selected. After dissection of asci, a haploid strain with the yhm2 null allele was obtained. The correct insertion of the TRP1 gene was verified by Southern blotting, and it was confirmed by immunoblotting that the yhm2 mutant strain (MCY41) has no YHM2 gene product (data not shown).
We first examined the difference in the growth rate between MCY41 (yhm2::TRP1) and MCY42 (YHM2). There was no obvious difference in growth when the cells were grown in rich medium such as YPD (2% glucose) and YPG (3% glycerol), but MCY41 (yhm2::TRP1) cells show slower growth than wild-type cells in the minimal media with nonfermentable carbon sources such as glycerol and ethanol (Table 2). Interestingly, in the yhm2 strain which overproduces the mutant Yhm2-PRM protein (MCY41-2), the growth defect was not restored in the presence of a nonfermentable carbon source (Table 2). This result suggests that the DNA-binding activity of Yhm2p is important in vivo.
To compare the stability of the mitochondrial function of various strains, overnight YPG cultures of [rho+] MCY41 (yhm2::TRP1), KLR17-3d (abf2::URA3), KLR71 (abf2::URA3, pY-D3), and wild-type cells were spread on YPD plates and incubated at 30°C for 3 to 5 days. The plates were then overlaid with 2,3,5-triphenyltetrazolium chloride (TTC) agar. In this assay, the respiration-competent cells are stained red with TTC whereas the respiration-incompetent cells are not (38). As expected, colonies of the [rho+] wild-type cells, such as MCY42 (YHM2), stained uniformly red with TTC whereas the colonies of KLR17-3d (abf2::URA3) and MCY41 (yhm2::TRP1) were not stained and the colonies were uniformly white or light pink (data not shown). Thus, the growth defect of MCY41 (yhm2::TRP1) in the media containing nonfermentable carbon sources may result from the defects of its mitochondrial function. Very interestingly, the colonies of KLR71 (abf2::URA3, pY-D3) have distinctive “red islands” in their middle region (data not shown). This abnormal staining pattern may result from the high loss of the episomal plasmid under nonselective conditions. Only the cells with the plasmid stained red with TTC. These results and the DAPI staining results (Fig. 2) suggest that Yhm2p plays an important role in the maintenance of mtDNA.
DISCUSSION
Many attempts have been made to uncover the relationship between Abf2p function and mtDNA metabolism. Disruption of the ABF2 gene causes the yeast cells to produce a petite phenotype at high frequency, and the cells become temperature sensitive for growth on glycerol. The high frequency of the petite phenotype is due to the rapid loss of the mitochondrial chromosome (6, 33). The DNA supercoiling activity of Abf2p and possible interactions with other proteins may contribute to the compaction as well as the maintenance of mtDNA in mitochondria (37). Besides the structural role in the organization of the mtDNA, Abf2p may play some roles in DNA replication, chromosome partitioning, and transcription (7, 34, 40).
Two suppressor genes for the abf2 mutant, YHM1 and ILV5, have been described (21, 59). These gene products, when overexpressed, can efficiently suppress the loss of mtDNA in abf2 null mutants at 37°C. However, the restoration mechanisms of the genes have not yet been resolved (21, 59). The characterization of YHM2 described here, on the other hand, has provided some insight into a possible mechanism for abf2 suppression.
A novel gene, YHM2 was discovered as another high-copy suppressor of the abf2 null mutation. Yhm2p contains 314 amino acid residues and is approximately 34 kDa. Overexpression of this protein restores the maintenance of functional mtDNA in abf2 cells (Fig. 1C and 2). The deduced amino acid sequence of Yhm2p is similar to that of the mitochondrial carrier proteins (Fig. 3A), and we have shown that Yhm2p is an integral inner membrane protein in yeast mitochondria (Fig. 4). YHM2 has unique features different from those of the other abf2 suppressor genes. Yhm2p has general DNA-binding activity like Abf2p in vitro and, moreover, is associated with the mitochondrial nucleoids in vivo (Fig. 5 and 7). The yhm2 mutant has a growth defect on nonfermentable carbon sources such as glycerol, acetate, and ethanol (Table 2), and long-term cultivation in the presence of glucose caused the dysfunction of respiration ability, as determined by the TTC overlay assay.
The copy number of mtDNA is regulated by the carbon source of the medium (49). In the wild-type cells, the amount of mtDNA decreases two- to fivefold when the carbon source is changed from glycerol to glucose at mid-log phase (49). The mechanism related to this phenomenon has not yet been discovered. Recently, we found that yhm2 mutant cells lose significant amounts of their mtDNA even when they are grown in nonfermentable carbon sources and that, when examined by immunoblot analysis, the level of Yhm2p expression is significantly repressed by a high concentration of glucose and low aeration (data not shown). Interestingly, YHM2 has a putative HAP2-HAP3-HAP4 transcription factor-binding sequence upstream from the gene. The regulatory complex HAP2-HAP3-HAP4 is involved in the transcription activation of a number of respiratory genes, such as CYC1, COX4 (11), and HEM1 (24), when the medium is shifted from a fermentable to a nonfermentable carbon source (17). Therefore, YHM2 expression may also be regulated in response to changes in carbon source by this regulatory complex. Thus, it is possible that there are some correlations between mtDNA copy number and YHM2 expression.
How might YHM2 function in mtDNA stability? The details of how YHM2 restores the defect in respiration at 37°C and the mitochondrial genome instability in the abf2 mutant are not clear. However, some possible mechanisms can be deduced from our results. One is that YHM2 stabilizes mtDNA indirectly as a consequence of its activity in the solute transport system of the mitochondrial inner membrane. Yhm2p, when overexpressed, may transport more solutes (of unknown type) into the mitochondrial matrix and increase the stability of mtDNA by an unidentified mechanism. A similar hypothesis was suggested to explain the suppression mechanisms of other mitochondrial carrier proteins, MRS3 and MRS4 (55, 56) and RIM2 (50). MRS3 and MRS4 encode the suppressors of mtRNA splicing defects. RIM2 encodes the suppressor of the pif1 mutant, a yeast mtDNA helicase (28). However, the possibility of carrier functionality acting in suppression has not yet been proven experimentally in any of the above-mentioned cases. Also, as mentioned above, the amino acid sequence of YHM2 shows some similarity to the yeast mitochondrial carrier proteins only in the regions which contain the transmembrane domains. The loop regions between transmembrane domains have highly variable amino acids sequences (Fig. 3). This means that the distinctive amino acid residues may play some unique roles.
Another possibility is that Yhm2p, as a membrane protein, is one of the proteins associated with mtDNA at the DNA attachment sites of the inner membrane. This is supported by the presence of Yhm2p in the nucleoid prepared from detergent-extracted mitochondria and the DNA-binding activity of Yhm2p. Several pieces of evidence supporting a direct interaction between mtDNA and proteins within the mitochondrial inner membrane have been reported. Rat liver mtDNA is associated with the mitochondrial inner membrane on the basis of electron microscopic observation (36). A compact mtDNA-protein structure has been isolated from the rat liver mitochondria and some constituents of these structures are suggested to be derived from the membrane (51). In S. cerevisiae, the mtDNA-protein complex has been characterized and was suggested to be attached to the inner membrane via an RNA molecule and to have a regular structure (42). It has been suggested that in bacteria, the chromosome is attached to the cell membrane and that the attachment may play a role in chromosome segregation, replication, or transcription (10, 31). In another eucaryotic organelle, the chloroplast, its chromosome is also known to bind to membranes (46, 58). Chloroplasts have two distinct membrane structures, the envelope membrane and the thylakoid membrane. The binding of chloroplast DNA to the envelope appears to be involved in replication and segregation, while the binding to the thylakoid membrane appears to be involved in the expression of plastid genes that are necessary for photosynthesis (46, 58). Recent studies indicate that the yeast mitochondrial inner membrane may also have some DNA-binding proteins involved in mtDNA replication and segregation (27, 35, 37). Despite these observations, the proteins involved in the membrane attachment of mtDNA have not been identified. To our knowledge, Yhm2p is the first protein determined as a membrane-bound member of the mitochondrial nucleoid proteins.
Abf2p, as well as the other HMG family proteins, is known to affect DNA conformation by wrapping DNA into negative superhelical turns. This, in turn, influences the interaction of other DNA-binding proteins with the chromosome (37). The mtDNA nucleoids in the abf2 cells grown in glycerol medium are less well organized than those in wild-type cells and are deficient in some proteins normally associated with wild-type nucleoids (37). Therefore, the mitochondrial nucleoids in the abf2 cells may have low binding affinities for the protein groups which may play critical roles for the faithful partition of mitochondrial nucleoids during the mitochondrial division. The abf2 cells overproducing Yhm2p have better defined nucleoid structures than the abf2 cells do (Fig. 2). Although the overall structures of Abf2p and Yhm2p are very different, the highest hydrophilic third internal loop of Yhm2p has a sequence similar to that of the DNA recognition subdomain of the HMG box (Fig. 5C) and has general DNA-binding activity. The mutation of the proline repeat sequence in this subdomain abolishes the ability of YHM2 to suppress the abf2 mutant as well as to bind DNA. Therefore, we suggest that Yhm2p may replace Abf2p at two important points for the maintenance of the stability of mitochondrial genomes. First, Yhm2p may organize the mitochondrial nucleoids into well-defined structures by affecting the conformation of mtDNA so that the mtDNAs can interact with other essential proteins required for mtDNA maintenance. Second, Yhm2p may act as an mtDNA-recruiting protein onto the inner membrane at the matrix side. This is important for mtDNA metabolism because the mitochondrial inner membrane has the protein machineries that implement replication, transcription, and translation (22, 23, 27, 57).
At present, the phenotype of the yhm2 mutant does not provide useful information about the possible role of Yhm2p in mtDNA replication, recombination, and segregation. Detailed biochemical and genetic analyses are required. Also, the double mutation of ABF2 and YHM2 genes may provide useful information about the function of the two proteins.
ACKNOWLEDGMENTS
We are grateful to Seh-Hun Jang for providing yeast plasmids, helpful discussion, and critical reading of the manuscript. We also thank Ronald Butow and Jordan Kolarov for useful comments and advice.
This study was supported in part by Pohang University of Science and Technology and by the Academic Research Fund (GE 96-202) of the Ministry of Education, Republic of Korea.
REFERENCES
- 1.Aquila H, Link T A, Klingenberg M. The uncoupling protein from brown fat mitochondria is related to mitochondrial ADP/ATP carrier. Analysis of sequence homologies and folding of the proteins. EMBO J. 1985;4:2369–2376. doi: 10.1002/j.1460-2075.1985.tb03941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aquila H, Link T A, Klingenberg M. Solute carriers involved in energy transfer of mitochondria form a homologous protein family. FEBS Lett. 1987;212:1–9. doi: 10.1016/0014-5793(87)81546-6. [DOI] [PubMed] [Google Scholar]
- 3.Caron F, Jacq C, Rouviere-Yaniv J. Characterization of a histone-like protein extracted from yeast mitochondria. Proc Natl Acad Sci USA. 1979;76:4265–4269. doi: 10.1073/pnas.76.9.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cote C, Poirier J, Boulet D. Expression of the mammalian mitochondrial genome. Stability of mitochondrial translation products as a function of membrane potential. J Biol Chem. 1989;264:8487–8490. [PubMed] [Google Scholar]
- 5.Daum G, Bohni P C, Schatz G. Import of proteins into mitochondria. Energy-dependent uptake of precursors by isolated mitochondria. J Biol Chem. 1982;257:13028–13035. [PubMed] [Google Scholar]
- 6.Diffley J F X, Stillman B. A close relative of the nuclear chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc Natl Acad Sci USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diffley J F X, Stillman B. DNA binding properties of an HMG1-related protein from yeast mitochondria. J Biol Chem. 1992;267:3368–3374. [PubMed] [Google Scholar]
- 8.Elledge S, Davis R W. Identification and isolation of the gene encoding the small subunit of ribonucleotide reductase from Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2783–2793. doi: 10.1128/mcb.7.8.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fearon K, Mason T L. Structure and function of MRP20 and MRP49, the nuclear genes for 2 proteins of the 54-S-subunit of the yeast mitochondrial ribosome. J Biol Chem. 1992;267:5162–5170. [PubMed] [Google Scholar]
- 10.Firshein W. Role of the DNA/membrane complex in procaryotic DNA replication. Annu Rev Microbiol. 1989;43:89–120. doi: 10.1146/annurev.mi.43.100189.000513. [DOI] [PubMed] [Google Scholar]
- 11.Forsburg S L, Guarente L. Mutational analysis of upstream activation sequence 2 of the CYC1 gene of Saccharomyces cerevisiae: a HAP2-HAP3-responsive site. Mol Cell Biol. 1988;8:647–654. doi: 10.1128/mcb.8.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foury F, Kolodynski J. pif mutation blocks recombination between mitochondrial rho+ and rho− genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1983;80:5345–5349. doi: 10.1073/pnas.80.17.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foury F, Lahaye A. Cloning and sequencing of the PIF gene involved in repair and recombination of yeast mitochondrial DNA. EMBO J. 1987;6:1441–1449. doi: 10.1002/j.1460-2075.1987.tb02385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foury F. Cloning and sequencing of the nuclear gene MIP1 encoding the catalytic subunit of the yeast mitochondrial DNA polymerase. J Biol Chem. 1989;264:20552–20560. [PubMed] [Google Scholar]
- 15.Greenleaf A L, Kelly J L, Lehman I R. Yeast RPO41 gene is required for transcription and maintenance of the mitochondrial genome. Proc Natl Acad Sci USA. 1986;83:3391–3394. doi: 10.1073/pnas.83.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan K L, Farh L, Marshall T K, Deschenes R J. Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1 gene. Curr Genet. 1993;24:141–148. doi: 10.1007/BF00324678. [DOI] [PubMed] [Google Scholar]
- 17.Guarente L, Lalonde B, Gifford P, Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984;36:503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- 18.Higuchi R, Krummel B, Saiki R K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones B A, Fangman W L. Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 1992;6:380–389. doi: 10.1101/gad.6.3.380. [DOI] [PubMed] [Google Scholar]
- 20.Kao L-R, Megraw T L, Chae C-B. Essential role of the HMG domain in the function of yeast mitochondrial histone HM: complementation of HM by the nuclear nonhistone NHP6A. Proc Natl Acad Sci USA. 1993;90:5598–5602. doi: 10.1073/pnas.90.12.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kao L-R, Megraw T L, Chae C-B. SHM1: a multicopy suppressor of temperature-sensitive null mutation in the HMG1-like abf2 gene. Yeast. 1996;12:1239–1250. doi: 10.1002/(sici)1097-0061(19960930)12:12<1239::aid-yea17>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 22.Kazakova T B, Golubkov V I, Ermekova S A. Influence of the functional state of the mitochondrial membranes on processes of transcription of DNA of the mitochondria. Mol Biol. 1972;6:480–487. [PubMed] [Google Scholar]
- 23.Kazakova T B, Mel’nikova M P. The elementary transcription particles of the inner mitochondrial membrane. Mol Biol. 1974;8:107–113. [PubMed] [Google Scholar]
- 24.Keng T, Guarente L. Constitutive expression of the yeast HEM1 gene is actually a composite of activation and repression. Proc Natl Acad Sci USA. 1987;84:9113–9117. doi: 10.1073/pnas.84.24.9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klingen M, Nelson D. Structure-function relationships in the mitochondrial carrier family. In: Papa S, Toger J M, editors. Biochemistry of cell membranes: a compendium of selected topics. Basel, Switzerland: Birkhauser Verlag; 1995. pp. 191–219. [Google Scholar]
- 26.Kuan J, Saier M J. The mitochondrial carrier family of transport proteins: structural, functional, and evolutionary relationships. Crit Rev Biochem Mol Biol. 1993;28:209–233. doi: 10.3109/10409239309086795. [DOI] [PubMed] [Google Scholar]
- 27.Kuroiwa T, Ohta T, Kuroiwa H, Shigeyuki K. Molecular and cellular mechanisms of mitochondrial nuclear division and mitochondriokinesis. Microsc Res Tech. 1994;27:220–232. doi: 10.1002/jemt.1070270304. [DOI] [PubMed] [Google Scholar]
- 28.Lahaye A, Stahl H, Thines-Sempoex D, Foury F. PIF1: a DNA helicase in yeast mitochondria. EMBO J. 1991;10:997–1007. doi: 10.1002/j.1460-2075.1991.tb08034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawson J E, Douglas M G. Separate genes encode functionally equivalent ADP/ATP carrier proteins in Saccharomyces cerevisiae. Isolation and analysis of AAC2. J Biol Chem. 1988;263:14812–14818. [PubMed] [Google Scholar]
- 30.Leblanc B, Read C, Moss T. Recognition of the Xenopus ribosomal core promoter by the transcription factor xUBF involves multiple HMG box domains and leads to an xUBF interdomain interaction. EMBO J. 1993;12:513–525. doi: 10.1002/j.1460-2075.1993.tb05683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leibowitz P J, Schaechter M. The attachment of the bacterial chromosome to the cell membrane. Int Rev Cytol. 1975;41:1–28. doi: 10.1016/s0074-7696(08)60964-x. [DOI] [PubMed] [Google Scholar]
- 32.Lundin M, Baltscheffsky H, Ronne H. Yeast PPA2 gene encodes a mitochondrial inorganic pyrophosphatase that is essential for mitochondrial function. J Biol Chem. 1991;266:12168–12172. [PubMed] [Google Scholar]
- 33.Megraw T L, Chae C-B. Functional complementarity between the HMG1-like yeast mitochondrial histone HM and the bacterial histone-like protein HU. J Biol Chem. 1993;268:12758–12763. [PubMed] [Google Scholar]
- 34.Megraw T L, Kao L-R, Chae C-B. The mitochondrial histone HM: an evolutionary link between bacterial HU and nuclear HMG1 proteins. Biochimie. 1994;76:909–916. doi: 10.1016/0300-9084(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 35.Miyakawa I, Fumoto S, Kuroiwa T, Sando N. Characterization of DNA-binding proteins involved in the assembly of mitochondrial nucleoids in the yeast Saccharomyces cerevisiae. Plant Cell Physiol. 1995;36:1179–1188. [PubMed] [Google Scholar]
- 36.Nass M M K. Mitochondrial DNA. I. Intramitochondrial distribution and structure relations of single- and double-length circular DNA. J Mol Biol. 1969;42:521–528. doi: 10.1016/0022-2836(69)90240-x. [DOI] [PubMed] [Google Scholar]
- 37.Newman S M, Zelenaya T O, Perlman P S, Butow R A. Analysis of mitochondrial DNA nucleoids in wild-type and a mutant strain of Saccharomyces cerevisiae that lacks the mitochondrial HMG box protein Abf2p. Nucleic Acids Res. 1996;24:386–393. doi: 10.1093/nar/24.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogur M, St. John R, Nagai S. Tetrazolium overlay technique for population studies of respiration deficiency in yeast genetics. Science. 1957;125:928–929. doi: 10.1126/science.125.3254.928. [DOI] [PubMed] [Google Scholar]
- 39.Palmieri L, Lasorsa F M, De P A, Palmieri F, Runswick M J, Walker J E. Identification of the yeast ACR1 gene product as a succinate-fumarate transporter essential for growth on ethanol or acetate. FEBS Lett. 1997;417:114–118. doi: 10.1016/s0014-5793(97)01269-6. [DOI] [PubMed] [Google Scholar]
- 40.Parisi M A, Xu B, Clayton D A. A human mitochondrial transcription activator can functionally replace a yeast mitochondrial HMG-box protein in vitro and in vivo. Mol Cell Biol. 1993;13:1951–1961. doi: 10.1128/mcb.13.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reenan R A, Kolodner R D. Isolation and characterization of two Saccharomyces cerevisiae genes encoding homologs of the bacterial HexA and MutS mismatch repair proteins. Genetics. 1992;132:963–973. doi: 10.1093/genetics/132.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rickwood D, Chambers J A A, Baret M. Isolation and preliminary characterization of DNA-protein complexes from the mitochondria of Saccharomyces cerevisiae. FEMS Microbiol Lett. 1981;12:187–190. doi: 10.1016/0014-4827(81)90350-5. [DOI] [PubMed] [Google Scholar]
- 43.Runswick J M, Powell S J, Nyren P, Walker J E. Sequence of the bovine mitochondrial phosphate carrier protein: structural relationship to ADP/ATP translocase and the brown fat mitochondrial uncoupling protein. EMBO J. 1987;6:1367–1373. doi: 10.1002/j.1460-2075.1987.tb02377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Saraste M, Walker J E. Internal sequence repeats and the path of polypeptide in mitochondrial ADP/ATP translocase. FEBS Lett. 1982;144:250–254. doi: 10.1016/0014-5793(82)80648-0. [DOI] [PubMed] [Google Scholar]
- 46.Sato N, Catherine A, Jacques J, Roland D, Kuroiwa T. Detection and characterization of a plastid envelope DNA-binding protein which may anchor plastid nucleoids. EMBO J. 1993;12:555–561. doi: 10.1002/j.1460-2075.1993.tb05687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith D B, Johnson K S. Single-step purification of polypeptide expressed in E. coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki C K, Suda K, Wang N, Schatz G. Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science. 1994;264:273–276. doi: 10.1126/science.8146662. [DOI] [PubMed] [Google Scholar]
- 49.Ulery T L, Jang S H, Jaehning J A. Glucose repression of yeast mitochondrial transcription: kinetics of derepression and role of nuclear genes. Mol Cell Biol. 1994;14:1160–1170. doi: 10.1128/mcb.14.2.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van D E, Jank B, Ragnini A, Schweyen R J, Duyckaerts C, Sluse F, Foury F. Overexpression of a novel member of the mitochondrial carrier family rescues defects in both DNA and RNA metabolism in yeast mitochondria. Mol Gen Genet. 1995;246:426–436. doi: 10.1007/BF00290446. [DOI] [PubMed] [Google Scholar]
- 51.Van T G, McPherson M L. A compact form of rat liver mitochondrial DNA stabilized by bound proteins. J Biol Chem. 1979;254:6044–6053. [PubMed] [Google Scholar]
- 52.Van Dyck E, Foury F, Stillman B, Brill S J. A single-stranded DNA binding protein required for mitochondrial DNA replication in Saccharomyces cerevisiae. EMBO J. 1992;11:3421–3430. doi: 10.1002/j.1460-2075.1992.tb05421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Dyck L, Pearce D A, Sherman F. PIM1 encodes a mitochondrial ATP-dependent protease that is required for mitochondrial function in the yeast Saccharomyces cerevisiae. J Biol Chem. 1994;269:238–242. [PubMed] [Google Scholar]
- 54.Verdier J M. Regulatory DNA-binding protein in yeast: an overview. Yeast. 1990;6:271–297. doi: 10.1002/yea.320060402. [DOI] [PubMed] [Google Scholar]
- 55.Waldherr M, Ragnini A, Jank B, Teply R, Wiesenberger G, Schweyen R J. A multitude of suppressors of group II intron-splicing defects in yeast. Curr Genet. 1993;24:301–306. doi: 10.1007/BF00336780. [DOI] [PubMed] [Google Scholar]
- 56.Wiesenberger G, Link T A, Aschen U, Waldherr M, Schweyen R J. MRS3 and MRS4, two suppressors of mtRNA splicing defects in yeast, are new members of the mitochondrial carrier family. J Mol Biol. 1991;217:23–37. doi: 10.1016/0022-2836(91)90608-9. [DOI] [PubMed] [Google Scholar]
- 57.Wiesenberger G, Fox T D. Pet127p, a membrane-associated protein involved in stability and processing of Saccharomyces cerevisiae mitochondrial RNAs. Mol Cell Biol. 1997;17:2816–2824. doi: 10.1128/mcb.17.5.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu M, Nie Z Q, Yang J. The 18-kD protein that binds to the chloroplast DNA replicative origin is an iron-sulfur protein related to a subunit of NADH dehydrogenase. Plant Cell. 1989;1:551–557. doi: 10.1105/tpc.1.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zelenaya T O, Perlman P S, Butow R A. An enzyme in yeast mitochondria that catalyzes a step in branched-chain amino acid biosynthesis also functions in mitochondrial DNA stability. EMBO J. 1995;14:3268–3276. doi: 10.1002/j.1460-2075.1995.tb07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]