Abstract
Serpentine receptors such as smoothened and frizzled play important roles in cell fate determination during animal development. In Dictyostelium discoideum, four serpentine cyclic AMP (cAMP) receptors (cARs) regulate expression of multiple classes of developmental genes. To understand their function, it is essential to know whether each cAR is coupled to a specific gene regulatory pathway or whether specificity results from the different developmental regulation of individual cARs. To distinguish between these possibilities, we measured gene induction in car1 car3 double mutant cell lines that express equal levels of either cAR1, cAR2, or cAR3 under a constitutive promoter. We found that all cARs efficiently mediate both aggregative gene induction by cAMP pulses and induction of postaggregative and prespore genes by persistent cAMP stimulation. Two exceptions to this functional promiscuity were observed. (i) Only cAR1 can mediate adenosine inhibition of cAMP-induced prespore gene expression, a phenomenon that was found earlier in wild-type cells. cAR1’s mediation of adenosine inhibition suggests that cAR1 normally mediates prespore gene induction. (ii) Only cAR2 allows entry into the prestalk pathway. Prestalk gene expression is induced by differentiation-inducing factor (DIF) but only after cells have been prestimulated with cAMP. We found that DIF-induced prestalk gene expression is 10 times higher in constitutive cAR2 expressors than in constitutive cAR1 or cAR3 expressors (which still have endogenous cAR2), suggesting that cAR2 mediates induction of DIF competence. Since in wild-type slugs cAR2 is expressed only in anterior cells, this could explain the so far puzzling observations that prestalk cells differentiate at the anterior region but that DIF levels are actually higher at the posterior region. After the initial induction of DIF competence, cAMP becomes a repressor of prestalk gene expression. This function can again be mediated by cAR1, cAR2, and cAR3.
Recent years have seen the discovery of critical roles in animal development for serpentine receptors, which are usually coupled to heterotrimeric G proteins. The insect sigaling peptides hedgehog and wingless and their mammalian counterparts sonic hedgehog, desert hedgehog, and indian hedgehog and the wnt factors control a multitude of inductive events during all stages of embryogenesis. The hedgehog signal is detected by two different serpentine receptors, smoothened (1, 40) and patched (21, 38), whereas the wingless or wnt signal is detected by the serpentine receptor D-frizzled-2 (3). In the social amoeba Dictyostelium discoideum, serpentine cyclic AMP (cAMP) receptors (cARs) control induction of cell differentiation during the entire course of development. Starving cells secrete cAMP pulses that induce chemotaxis and expression of genes required for the aggregation process. Cells aggregate to form mounds, which ultimately transform into fruiting structures that consist of a globular spore mass supported by a column of stalk cells. cAMP induces entry into the spore differentiation pathway as well as synthesis of a lipophilic factor, differentiation-inducing factor (DIF), which induces entry into the stalk differentiation pathway (see reference 5). At an early stage of development cAMP synergizes with DIF to induce prestalk genes, but later it becomes an inhibitor of stalk gene expression (2). cARs were shown previously to mediate induction of aggregative genes by cAMP pulses (20) as well as cAMP induction of prespore genes and repression of prestalk genes (31, 37). Remarkably, the target for the latter critical step in cell fate determination is glycogen synthase kinase 3 (GSK-3), a zeste white-3 homolog, which is the target for the effects of wingless and wnt in insects and vertebrates, respectively (7, 34).
Four cARs, showing 54 to 69% amino acid identity, are expressed in a stage- and cell-type-specific manner. cAR1 is predominantly expressed before and during aggregation (18). cAR3 is expressed at late aggregation, and expression is later restricted to the prespore cell population (13, 44). cAR2 and cAR4 are both expressed exclusively in the prestalk cell population after aggregation (19, 30). cAR knockout cell lines were generated to examine the role of the individual cARs in Dictyostelium development. car1 null cells neither aggregate nor express developmental genes but can be triggered to express aggregative and postaggregative genes by stimulation with cAMP (37, 39). car3 null cells aggregate and develop normally (13). car1 car3 double gene disruptants do not aggregate, and developmental gene expression cannot be restored with cAMP, indicating that cAR1 or cAR3 shows functional redundancy and that either one or the other has to be present for gene induction to occur (10, 36). car2 null cells are blocked in the mound stage, while car4 null cells show abnormal slug morphogenesis and culmination. Both lines show reduced expression of prestalk genes and enhanced expression of prespore genes (19, 29).
To understand the function of the four cARs, it is essential to know whether each receptor is coupled to a specific signal transduction pathway that controls a specific cell differentiation event or whether each receptor can activate multiple cell differentiation pathways. In the latter case, it is not the presence of a specific receptor that determines whether a response occurs but the availability of the downstream signaling pathway. To determine whether individual receptors have unique functions in developmental gene expression, we examined gene regulation in cell lines that display about equal levels of cAR1, cAR2, and cAR3 in a car1 car3 mutant background. Our results show that with two exceptions, all three receptors can transduce both the excitation and adaptation components of the different cAMP-regulated gene induction events with almost equal levels of efficiency.
MATERIALS AND METHODS
Materials.
2′,3′-Isopropylidene adenosine (IPA), 5′-N-ethylcarboxyadenosine (NECA), and G418 were obtained from Sigma (St. Louis, Mo.), adenosine 3′,5′-monophosphorothioate Sp-isomer (Sp-cAMPS) was obtained from Biolog Life Science Institute (Bremen, Germany), and DIF was obtained from Affinity Research Products (Exeter, United Kingdom).
Cell lines and culture conditions.
The car1 car3 double mutant cell line RI9 (10) was transformed with the extrachromosomal vector PJK1 (15, 17), with PJK1 harboring a gene fusion of the coding region of either the cAR1 or the cAR2 gene with the actin15 promoter, yielding cell lines act15cAR1 and act15cAR2, or with the integrating vector BS18 harboring a gene fusion of the cAR3 coding region with the actin15 promoter (11), yielding cell line act15cAR3. All cell lines, including wild-type AX3 cells and aca null (27) cells, were grown in standard axenic medium, which was supplemented with 20 μg of G418 per ml for lines transformed with PJK1- or BS18-derived vectors.
Gene induction procedures.
For induction of aggregative and postaggregative gene expression, cells were harvested at the late log phase of development, washed with 10 mM phosphate buffer (pH 6.5), and subsequently shaken at 150 rpm in phosphate buffer supplemented with 0.5 mM MgCl2 and 0.5 mM CaCl2 (DB) at 107 cells/ml and 22°C. Cells were challenged by different regimens of cAMP stimulation for 6 h, washed and resuspended to 5 × 106 cells/ml in DB, and incubated for an additional 8 h as indicated in the figure legends.
For induction of stalk gene expression, cells were incubated in monolayers (2). In short, cells were resuspended in stalk salts (10 mM KCl–2 mM NaCl–1 mM CaCl2 in 10 mM MES [morpholinoethanesulfonic acid] [pH 6.2]) to 5 × 106 cells/ml and incubated at 22°C in 10-ml petri dishes. After 8 h, cAMP was added to a final concentration of 5 mM and incubation was continued for a further 16 h. Cells had then formed tight aggregates, which were dissociated by forcing them through a 21-gauge needle, and cells were incubated in stalk salts at 5 × 106 cells/ml for 8 h, with variables as indicated in the figure legends.
RNA isolation and analysis.
Total cellular RNA was isolated from 2.5 × 107 cells (23), size fractionated on 1.5% agarose gels containing 2.2 M formaldehyde, and transferred to GeneScreen membranes. Northern blot transfers were hybridized to [32P]dATP-labeled DNA probes according to standard procedures and exposed to X-ray films. The optical densities of specific mRNA bands were quantitated with an LKB Ultrascan densitometer.
RESULTS
cAMP-regulated gene expression in constitutive cAR expressors.
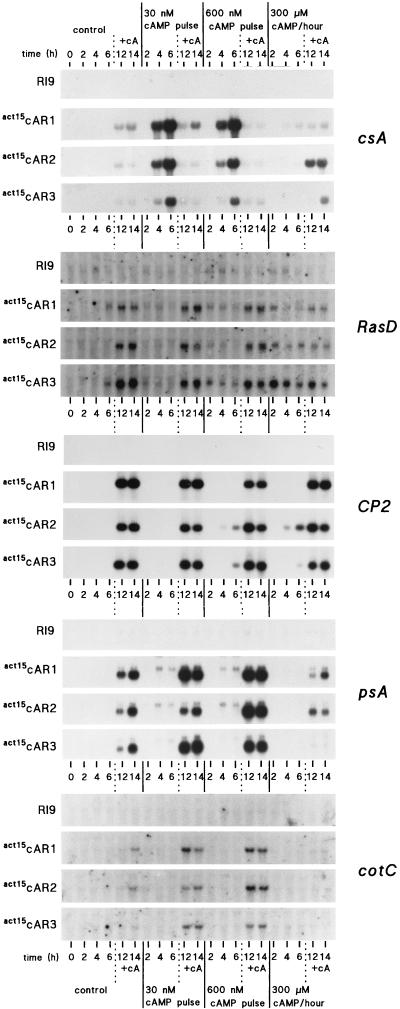
Starving cells secrete cAMP pulses in the nanomolar concentration range, which upregulate expression of aggregative genes such as cAR1 and csA (20, 25). Once cells have aggregated, a micromolar concentration of cAMP is required to first induce non-cell-type-specific genes, such as RasD and CP2 (26, 28), and prespore genes, such as psA and CotC (22, 31). To determine to what extent cAR1, cAR2, or cAR3 can mediate cAMP-induced gene expression, we used three derivatives of the car1 car3 double mutant line RI9 which constitutively express either cAR1, cAR2, or cAR3 under the control of the actin15 promoter. These cell lines are called act15cAR1, act15cAR2, and act15cAR3, respectively, and they express cAR1, cAR2, or cAR3 protein at levels that are similar to the level of expression of cAR1 protein in aggregation-competent wild-type cells (17). To induce aggregative gene expression, cells were stimulated for 6 h with 30 or 600 nM cAMP pulses at 6-min intervals or with a 300 μM cAMP pulse every hour. cAR1, cAR2, or cAR3 shows an affinity of 290 nM, >5 μM, or 490 nM, respectively (12, 16). The 30 nM cAMP pulse was chosen to accomodate the high-affinity cAR1 and cAR3, and the 600 nM pulse was chosen to accomodate the low-affinity cAR2. At higher concentrations cellular phosphodiesterase activity becomes insufficient to degrade cAMP between pulses, which are then detected as a continuous signal. The 300 μM stimulus is perceived as a continuous signal and was used to establish whether the cells adapt. After the initial 6 h of incubation, cells were stimulated for 8 h with 300 μM cAMP to induce postaggregative genes.
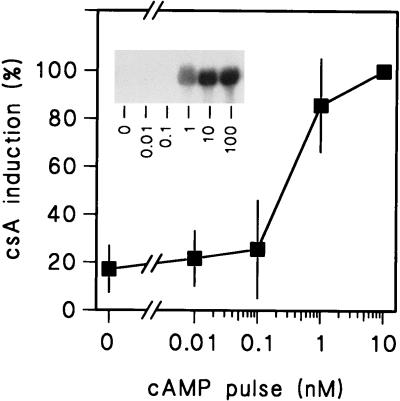
Figure 1 shows that induction of the aggregative gene csA by cAMP pulses occurs with equal efficiencies in act15cAR1, act15cAR2, and act15cAR3 cells but not at all in RI9 cells. Six hours of stimulation with 300 μM cAMP could not induce the csA gene in either mutant, indicating that csA induction in act15cAR1, act15cAR2, and act15cAR3 cells is subject to cellular adaptation. The induction of csA by 30 nM cAMP pulses in the act15cAR2 cells is unexpected, if we consider the low affinities of these receptors. Possibly, the 50% effective concentration (EC50) of cAMP for gene induction is much lower than the dissociation constant (KD) of the receptor that mediates the response. This is the case for the cAMP-induced chemotactic response, which shows an EC50 of 3 nM (41) and is mediated by cAR1, which has a KD of 290 nM. Another possibility is that cAMP pulses cause signal amplification by inducing a cAMP relay response. To measure the actual EC50 for pulse-induced gene expression, we examined the dose-response relationship for this response in aca null cells and in wild-type cells in the presence of the cAMP relay inhibitor caffeine. In the first experiment (data not shown) we chose a range from 1 to 30 nM cAMP and found that csA induction was almost optimal at 1 nM. We then chose a range from 0.01 to 10 nM and found that induction was half-maximal around 0.5 nM cAMP (Fig. 2). This value is about 500-fold lower than the KD of cAR1, which mediates this response. If we take this observation into account, it is not surprising that cAR2 with an affinity of >5 μM can transduce responses to 30 nM cAMP pulses.
FIG. 1.
Induction of aggregative and postaggregative gene expression. RI9, act15cAR1, act15cAR2, and act15cAR3 cells were incubated in DB in the absence of stimuli, with either 30 or 600 nM cAMP pulses delivered at 6-min intervals, or with 300 μM cAMP delivered at 60-min intervals. Cells were then incubated for an additional 8 h with 300 μM cAMP/h. Samples were taken for RNA extraction at 0, 2, 4, 6, 12, and 14 h of incubation. Northern blots were hybridized to 32P-labeled DNA probes for the aggregative gene csA, the postaggregative genes RasD and CP2, and the prespore genes psA and CotC. The experiment was repeated twice with similar results.
FIG. 2.
Dose-response relationship of cAMP pulse-induced gene induction in wild-type cells. AX3 or aca cells were harvested in late log phase and stimulated in the presence (AX3 cells) or absence (aca cells) of 5 mM caffeine and the indicated concentrations of cAMP pulses delivered at 6-min intervals. RNA was isolated after 6 h of incubation and probed with 32P-labeled csA cDNA. The inset shows results of an experiment with aca cells, while the graph indicates means and standard deviations of results from two experiments with AX3 and caffeine and one experiment with aca cells.
The postaggregative genes RasD and CP2 are induced most effectively in the act15cAR1, act15cAR2, and act15cAR3 cell lines in response to 300 μM cAMP after an initial 6-h period of starvation. However, especially for RasD, high concentrations of cAMP can also induce some expression within the first few hours of starvation, which is particularly evident in act15cAR3 cells. In general, the levels of induction of RasD and CP2 are remarkably similar in all act15cAR cell lines.
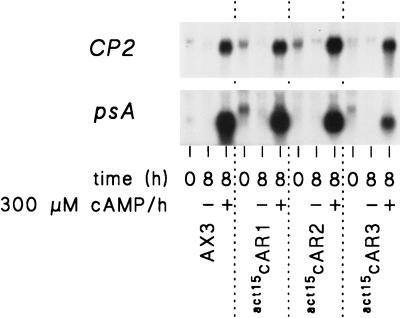
The levels of induction of the prespore genes psA and CotC resemble those of the postaggregative genes in the sense that all three act15cAR cell lines show the same levels of induction by stimulation with 300 μM cAMP after a 6-h period of starvation. However, there is a difference. Prespore gene induction is most efficient in cells that were prestimulated with cAMP pulses. Unlike postaggregative gene induction, prespore gene induction is actually inhibited by prestimulation with 300 μM cAMP in the first 6 h of development. The conditions that induce competence for either postaggregative or prespore gene induction are apparently not the same. The expression levels of prespore and postaggregative genes after 8 h of cAMP stimulation were approximately similar in the wild-type (AX3) and the act15cAR cell lines, and there was no significant gene induction in the absence of cAMP (Fig. 3).
FIG. 3.
Comparison of levels of gene induction in wild-type cells and act15cAR cells. act15cAR1, act15cAR2, act15cAR3, and wild-type AX3 cells were first stimulated for 6 h with 30 nM (act15cAR1, act15cAR3, and AX3) or 600 nM (act15cAR2) cAMP pulses, washed, and subsequently incubated for 0 or 8 h in the presence and absence of 300 μM cAMP per h. mRNA was isolated and hybridized to CP2 and psA cDNAs. The experiment was repeated once with similar results.
Development of act15cAR cell lines.
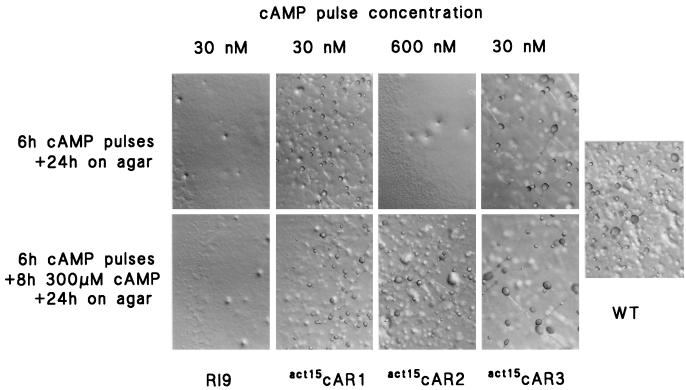
car1 car3 cells cannot aggregate and form fruiting bodies, but development is restored by expression of either cAR1 or cAR3. act15cAR2 cells remain defective in cell aggregation, presumably because these low-affinity receptors cannot initiate spontaneous cAMP oscillations (17). We tested whether cAR2 cells could go through development after being stimulated for 6 h with 600 nM cAMP pulses, and they could not (Fig. 4). When cells were additionally treated for 8 h with 300 μM cAMP, they formed tight aggregates in suspension, which developed into slugs and fruiting bodies of normal sizes and appearance when they were deposited on agar. act15cAR2 cells most likely cannot aggregate and develop, because they are not capable of producing the cAMP concentrations that are required to activate the low-affinity cAR2s. However, they will go normally through the later stages of development after exposure to the appropriate cAMP stimuli to induce expression of developmentally regulated genes.
FIG. 4.
Phenotype of cAR mutants after cAMP treatment. After treatment with either 30 nM cAMP pulses (RI9, act15cAR1, and act15cAR3) or 600 nM cAMP pulses (act15cAR2) for 6 h and after subsequent incubation with 300 μM cAMP for 10 h, aliquots of 10 μl of 108 cells/ml were placed on nonnutrient agar and left to develop at 22°C for 24 h. Wild-type AX3 cells (WT) were placed directly on nonnutrient agar after being harvested from growth medium.
Adenosine regulation of prespore gene expression in act15cAR cell lines.
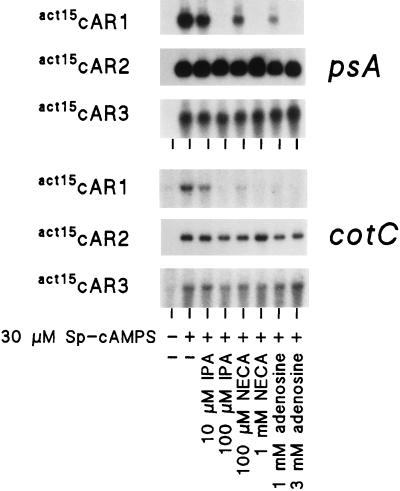
The data presented above yield no clue as to which cAR mediates specific gene regulatory events during normal development. In wild-type cells, cAMP induction of prespore gene expression is inhibited by millimolar concentrations of adenosine and by micromolar concentrations of the adenosine analogs NECA and IPA, which cannot be phosphorylated by an extracellular adenosine kinase (37, 42, 43). Adenosine also inhibits cAMP binding (24, 41), and recent studies showed that only binding of cAMP to cAR1, but not to cAR2 or cAR3, is inhibited by adenosine (17a). To identify the cAR that mediates prespore gene induction and to verify that the inhibitory effects of adenosine are due to inhibition of cAMP binding activity, we tested the effects of adenosine, IPA, and NECA on cAMP induction of prespore gene expression in the act15cAR cell lines. Figure 5 shows that 100 μM IPA, 1 mM NECA, and 3 mM adenosine inhibit psA and CotC induction completely in the act15cAR1 cells but not at all in act15cAR2 and act15cAR3 cells. This result indicates that cAR1 transduces cAMP induction of prespore gene expression during normal development.
FIG. 5.
Effects of adenosine analogues on prespore gene expression. act15cAR1 and act15cAR3 cells were prestimulated for 6 h with 30 nM cAMP pulses, and act15cAR2 cells were prestimulated with 600 nM cAMP pulses. Cells were incubated for an additional 8 h in DB in the absence and presence of 30 μM Sp-cAMPS and the indicated concentrations of adenosine, IPA, or NECA. RNA was isolated and probed with 32P-labeled psA and CotC cDNAs. The experiment was repeated once with similar results.
cAMP regulation of the DIF-inducible gene ecmB.
The DIF-inducible gene ecmB is typically downregulated by nanomolar concentrations of cAMP (2, 37). A micromolar concentration of cAMP was found to have varied effects on ecmB induction. In cells that have just finished aggregation, a micromolar concentration of cAMP stimulates ecmB induction, presumably by inducing competence for ecmB induction by DIF. This effect disappears once cells are competent and a micromolar concentration of cAMP may then just act as a supersaturated inhibitory signal (2, 35, 37). We determined whether the different cARs could mediate the induction of competence for DIF by a micromolar concentration of cAMP and inhibition of the DIF response by nanomolar concentrations of cAMP. RI9, act15cAR1, act15cAR2, and act15cAR3 cells were first incubated for 16 h with 5 mM cAMP to induce competence and then for 8 h with DIF or DIF plus 1 μM Sp-cAMPS (equivalent to 20 to 60 nM cAMP).
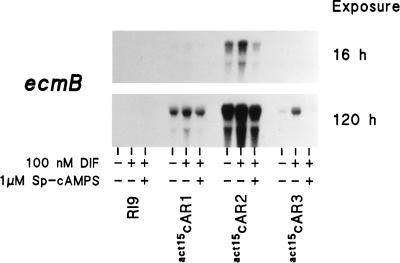
Figure 6 shows that DIF did not induce ecmB in the RI9 cells. In the other cell lines, DIF always induced ecmB to levels above those in unstimulated cells and induction was always inhibited by 1 μM Sp-cAMPS. However, ecmB was induced in act15cAR2 cells to 10-fold higher levels than in either act15cAR1 or act15cAR3 cells. Since the cAR2 gene is still intact in the car1 car3 mutant cell lines and is probably expressed at this developmental stage (except in the RI9 cells, which do not reach this stage), we cannot exclude the possibility that it is actually the single copy of endogenous cAR2 that induces the lower level of competence for DIF induction in the act15cAR1 and act15cAR3 cells.
FIG. 6.
Prestalk gene induction and repression in act15cAR cell lines. RI9, act15cAR1, act15cAR2, and act15cAR3 cells were preincubated for 16 h in monolayers in the presence of 5 mM cAMP, washed, and incubated for 8 h without additives, with 100 nM DIF, and with 100 nM DIF plus 1 μM Sp-cAMPS. RNA was isolated and probed with 32P-labeled ecmB mRNA. Sixteen-hour and 5-day exposures of the same Northern blot are shown. The experiment was repeated once with similar results.
DISCUSSION
cAR1, cAR2, and cAR3 can mediate both adapting and nonadapting gene induction responses.
A remarkable outcome of the experiments presented here is that cAR1, cAR2, and cAR3 show almost complete redundancy of function as far as induction of aggregative, postaggregative, and prespore genes is concerned. On one hand, all three receptors can transduce the effects of cAMP pulses on aggregative gene expression. This response involves activation of an excitatory as well as an inhibitory pathway, since it cannot be induced by continuous stimuli. On the other hand, the three cARs can also transduce the effect of a constant cAMP stimulus, in which adaptation does not play a role. These observations may explain why the phenotypes of null mutants for the individual cARs are not very severe. car1 cells do not aggregate spontaneously, but development can be fully restored by stimulation with cAMP (36, 39). car3 cells show normal development (13). car2 and car4 cells show reduced prestalk differentiation (19, 29), but in none of the null mutants is cAMP or DIF-induced gene expression completely blocked. Our current data indicate that the other cARs may take over the function of the deleted cAR, depending on whether they happen to be expressed at the stage or in the cell type where the deleted cAR has its function.
There is an interesting discrepancy between data presented here and data presented in a previous study with car1 cells. In car1 cells, aggregative genes could not be induced by 30 nM cAMP pulses but could be induced both by 300 nM pulses and a constant 300 μM cAMP stimulus (36). Since cAR3 is the only other cAR expressed at that stage of development, it was concluded that cAR3 mediated the response but that it required higher concentrations of cAMP and was not sensitive to adaptation. We show here that cAR3 can also transduce 30 nM pulses and is susceptible to adaptation. The difference between the two experiments is probably the number of cAR3 receptors. In this study cAR1 and cAR3 expression levels were similar to cAR1 expression levels in aggregating cells (about 100,000 sites/cell). However, their expression levels in car1 cells are so low that cAMP binding activity is undetectable (39). It is plausible that in car1 cells with few cAR3 binding sites, a very high percentage of occupied receptors and therefore high concentrations of cAMP are required to transduce the response. Additionally, the adaptation pathway may require higher numbers of occupied receptors than the excitation pathway and these levels may not have been reached in the car1 cells.
Differences in acquisition of competence for postaggregative and prespore gene induction.
In the act15cAR1, act15cAR2, and act15cAR3 lines, induction of the prespore genes psA and CotC by micromolar concentrations of cAMP requires prestimulation with cAMP pulses. Prestimulation with micromolar concentrations of cAMP inhibits subsequent induction of prespore gene expression. Since in the act15cAR cell lines, the cARs are already present from the onset of starvation, components downstream of cAR must probably be first induced by the pulse regimen. Two signal transduction components, the mitogen-activated protein kinase ERK2 and GSK-3, are specifically required for prespore, but not for postaggregative, gene expression (6, 7). The developmental regulation of GSK-3 has not yet been reported. ERK2 expression is strongly upregulated at the early aggregation stage, but it is not yet known whether the gene is cAMP pulse induced (33). Postaggregative gene expression occurs optimally after a few hours of starvation in the absence of cAMP but does not require cAMP pulses. Expression of these genes depends on expression of the transcription factor G-box binding factor, which is itself induced by a continuous cAMP stimulus (8, 32).
Only cAR1 mediates adenosine inhibition of cAMP-induced prespore gene expression.
Since cAR1, cAR2, and cAR3 are all expressed in postaggregative wild-type cells and all can potentially mediate cAMP induction of prespore gene expression as shown here, it is not clear which cAR actually mediates the response in wild-type cells. During normal development, cAMP-induced prespore gene induction is inhibited by adenosine and more effectively by its analogues IPA and NECA, which cannot be phosphorylated (37, 42, 43). We here show that only cAR1 and not cAR2 or cAR3 can mediate adenosine inhibition of prespore gene expression. This implies that cAR1 mediates prespore gene induction in wild-type cells.
ecmB induction by DIF may require cAR2.
There is another exception to the functional redundancy of cAR1, cAR2, and cAR3. DIF induction of the prestalk gene ecmB is over 10 times more effective in act15cAR2 than in act15cAR1 and act15cAR3 cells and may in the latter two lines be entirely due to endogenous cAR2. cAR2 is probably involved in inducing competence for ecmB induction by DIF, since cAMP itself does not induce ecmB expression. cAR2’s involvement in competence for ecmB induction would also explain why ecmB expression is reduced in car2 mutants (29). The induction of DIF competence by cAR2 may actually solve a conundrum that has puzzled workers in this field for several years. Prestalk gene expression is specifically associated with the anterior region of the slug. However, this region has the highest levels of the DIF-degrading enzyme DIF-dechlorinase and the lowest levels of DIF (4, 9, 14). In wild-type cells, cAR2 is exclusively expressed in the anterior region (29, 30). If only cAR2 can make the cells competent for DIF, then the absolute levels of DIF are much less important in determining where ecmB is turned on than the expression pattern of cAR2. The formation of the prestalk expression pattern is in that case a function of the regulation of cAR2 expression.
ACKNOWLEDGMENT
This work was supported by grant 805-31.051 of the Life Sciences Foundation of The Netherlands Organisation for Scientific Research.
REFERENCES
- 1.Alcedo J, Ayzenzon M, Von Ohlen T, Noll M, Hooper J E. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell. 1996;86:221–232. doi: 10.1016/s0092-8674(00)80094-x. [DOI] [PubMed] [Google Scholar]
- 2.Berks M, Kay R R. Cyclic AMP is an inhibitor of stalk cell differentiation in Dictyostelium discoideum. Dev Biol. 1988;126:108–114. doi: 10.1016/0012-1606(88)90244-8. [DOI] [PubMed] [Google Scholar]
- 3.Bhanot P, Brink M, Harryman Samos C, Hsieh J-C, Wang Y, Macke J P, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- 4.Brookman J J, Jermyn K A, Kay R R. Nature and distribution of the morphogen DIF in the Dictyostelium slug. Development. 1987;100:119–124. doi: 10.1242/dev.100.1.119. [DOI] [PubMed] [Google Scholar]
- 5.Firtel R A. Integration of signaling information in controlling cell-fate decisions in Dictyostelium. Genes Dev. 1995;9:1427–1444. doi: 10.1101/gad.9.12.1427. [DOI] [PubMed] [Google Scholar]
- 6.Gaskins C, Clark A M, Aubry L, Segall J E, Firtel R A. The Dictyostelium MAP kinase ERK2 regulates multiple, independent developmental pathways. Genes Dev. 1996;10:118–128. doi: 10.1101/gad.10.1.118. [DOI] [PubMed] [Google Scholar]
- 7.Harwood A J, Plyte S E, Woodgett J, Strutt H, Kay R R. Glycogen synthase kinase 3 regulates cell fate in Dictyostelium. Cell. 1995;80:139–148. doi: 10.1016/0092-8674(95)90458-1. [DOI] [PubMed] [Google Scholar]
- 8.Hjorth A L, Khanna N C, Firtel R A. A trans-acting factor required for cAMP-induced gene expression in Dictyostelium is regulated developmentally and induced by cAMP. Genes Dev. 1989;3:747–759. doi: 10.1101/gad.3.6.747. [DOI] [PubMed] [Google Scholar]
- 9.Insall R, Nayler O, Kay R R. DIF-1 induces its own breakdown in Dictyostelium. EMBO J. 1992;11:2849–2854. doi: 10.1002/j.1460-2075.1992.tb05352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Insall R H, Soede R D M, Schaap P, Devreotes P N. Two cAMP receptors activate common signaling pathways in Dictyostelium. Mol Biol Cell. 1994;5:703–711. doi: 10.1091/mbc.5.6.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson R L, Vaughan R A, Caterina M J, Van Haastert P J M, Devreotes P N. Overexpression of the cAMP receptor 1 in growing Dictyostelium cells. Biochemistry. 1991;30:6982–6986. doi: 10.1021/bi00242a025. [DOI] [PubMed] [Google Scholar]
- 12.Johnson R L, Van Haastert P J M, Kimmel A R, Saxe III C L, Jastorff B, Devreotes P N. The cyclic nucleotide specificity of three cAMP receptors in Dictyostelium. J Biol Chem. 1992;267:4600–4607. [PubMed] [Google Scholar]
- 13.Johnson R L, Saxe III C L, Gollop R, Kimmel A R, Devreotes P N. Identification and targeted gene disruption of cAR3, a cAMP receptor subtype expressed during multicellular stages of Dictyostelium development. Genes Dev. 1993;7:273–282. doi: 10.1101/gad.7.2.273. [DOI] [PubMed] [Google Scholar]
- 14.Kay R R, Large S, Traynor D, Nayler O. A localized differentiation-inducing-factor sink in the front of the Dictyostelium slug. Proc Natl Acad Sci USA. 1993;90:487–491. doi: 10.1073/pnas.90.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J, Devreotes P N. Random chimeragenesis of G-protein-coupled receptors. Mapping the affinity of the cAMP chemoattractant receptors in Dictyostelium. J Biol Chem. 1994;269:28724–28731. [PubMed] [Google Scholar]
- 16.Kim J, Van Haastert P, Devreotes P N. Social senses: G-protein-coupled receptor signaling pathways in Dictyostelium discoideum. Chem Biol. 1996;3:239–243. doi: 10.1016/s1074-5521(96)90103-9. [DOI] [PubMed] [Google Scholar]
- 17.Kim J Y, Borleis J A, Devreotes P N. Switching of chemoattractant receptors programs development and morphogenesis in Dictyostelium: receptor subtypes activate common responses at different agonist concentrations. Dev Biol. 1998;197:117–128. doi: 10.1006/dbio.1998.8882. [DOI] [PubMed] [Google Scholar]
- 17a.Kim, J. Y., and P. N. Devreotes. Unpublished data.
- 18.Klein P S, Sun T J, Saxe III C L, Kimmel A R, Johnson R L, Devreotes P N. A chemoattractant receptor controls development in Dictyostelium discoideum. Science. 1988;241:1467–1472. doi: 10.1126/science.3047871. [DOI] [PubMed] [Google Scholar]
- 19.Louis J M, Ginsburg G T, Kimmel A R. The cAMP receptor CAR4 regulates axial patterning and cellular differentiation during late development of Dictyostelium. Genes Dev. 1994;8:2086–2096. doi: 10.1101/gad.8.17.2086. [DOI] [PubMed] [Google Scholar]
- 20.Mann S K O, Firtel R A. Two-phase regulatory pathway controls cAMP receptor-mediated expression of early genes in Dictyostelium. Proc Natl Acad Sci USA. 1989;86:1924–1928. doi: 10.1073/pnas.86.6.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marigo V, Davey R A, Zuo Y, Cunningham J M, Tabin C J. Biochemical evidence that patched is the Hedgehog receptor. Nature. 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- 22.Mehdy M C, Firtel R A. A secreted factor and cyclic AMP jointly regulate cell-type-specific gene expression in Dictyostelium discoideum. Mol Cell Biol. 1985;5:705–713. doi: 10.1128/mcb.5.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nellen W, Datta S, Reymond C, Sivertsen A, Mann S, Crowley T, Firtel R A. Molecular biology in Dictyostelium: tools and applications. In: Spudich J A, editor. Methods in cell biology. London, United Kingdom: Academic Press; 1987. pp. 67–100. [DOI] [PubMed] [Google Scholar]
- 24.Newell P C, Ross F M. Inhibition by adenosine of aggregation centre initiation and cyclic AMP binding in Dictyostelium. J Gen Microbiol. 1982;128:2715–2724. [Google Scholar]
- 25.Noegel A, Harloff C, Hirth P, Merkl R, Modersitzki M, Stadler J, Weinhart U, Westphal M, Gerisch G. Probing an adhesion mutant of Dictyostelium discoideum with cDNA clones and monoclonal antibodies indicates a specific defect in the contact site A glycoprotein. EMBO J. 1985;4:3805–3810. doi: 10.1002/j.1460-2075.1985.tb04151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pears C J, Williams J G. Identification of a DNA sequence element required for efficient expression of a developmentally regulated and cAMP-inducible gene of Dictyostelium discoideum. EMBO J. 1987;6:195–200. doi: 10.1002/j.1460-2075.1987.tb04738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitt G S, Milona N, Borleis J, Lin K C, Reed R R, Devreotes P N. Structurally distinct and stage-specific adenylyl cyclase genes play different roles in Dictyostelium development. Cell. 1992;69:305–315. doi: 10.1016/0092-8674(92)90411-5. [DOI] [PubMed] [Google Scholar]
- 28.Reymond C D, Nellen W, Firtel R A. Regulated expression of ras gene constructs in Dictyostelium transformants. Proc Natl Acad Sci USA. 1985;82:7005–7009. doi: 10.1073/pnas.82.20.7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saxe C L, III, Ginsburg G T, Louis J M, Johnson R, Devreotes P N, Kimmel A R. CAR2, a prestalk cAMP receptor required for normal tip formation and late development of Dictyostelium discoideum. Genes Dev. 1993;7:262–272. doi: 10.1101/gad.7.2.262. [DOI] [PubMed] [Google Scholar]
- 30.Saxe C L, III, Yu Y, Jones C, Bauman A, Haynes C. The cAMP receptor subtype cAR2 is restricted to a subset of prestalk cells during Dictyostelium development and displays unexpected DIF 1 responsiveness. Dev Biol. 1996;174:202–213. doi: 10.1006/dbio.1996.0066. [DOI] [PubMed] [Google Scholar]
- 31.Schaap P, Van Driel R. Induction of post-aggregative differentiation in Dictyostelium discoideum by cAMP. Evidence of involvement of the cell surface cAMP receptor. Exp Cell Res. 1985;159:388–398. doi: 10.1016/s0014-4827(85)80012-4. [DOI] [PubMed] [Google Scholar]
- 32.Schnitzler G R, Fischer W H, Firtel R A. Cloning and characterization of the G-box binding factor, an essential component of the developmental switch between early and late development in Dictyostelium. Genes Dev. 1994;8:502–514. doi: 10.1101/gad.8.4.502. [DOI] [PubMed] [Google Scholar]
- 33.Segall J E, Kuspa A, Shaulsky G, Ecke M, Maeda M, Gaskins C, Firtel R A, Loomis W F. A MAP kinase necessary for receptor-mediated activation of adenylyl cyclase in Dictyostelium. J Cell Biol. 1995;128:405–413. doi: 10.1083/jcb.128.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siegfried E, Wilder E L, Perrimon N. Components of wingless signalling in Drosophila. Nature. 1994;367:76–80. doi: 10.1038/367076a0. [DOI] [PubMed] [Google Scholar]
- 35.So J, Weeks G. The effects of presumptive morphogens on prestalk and prespore cell gene expression in monolayers of Dictyostelium discoideum. Differentiation. 1992;51:73–78. [Google Scholar]
- 36.Soede R D M, Insall R H, Devreotes P N, Schaap P. Extracellular cAMP can restore development in Dictyostelium cells lacking one, but not two subtypes of early cAMP receptors (cARs) Development. 1994;120:1997–2002. doi: 10.1242/dev.120.7.1997. [DOI] [PubMed] [Google Scholar]
- 37.Soede R D M, Hopper N A, Williams J G, Schaap P. Extracellular cAMP depletion triggers stalk gene expression in Dictyostelium: disparities in developmental timing and dose dependency indicate that prespore induction and stalk repression by cAMP are mediated by separate signaling pathways. Dev Biol. 1996;177:152–159. doi: 10.1006/dbio.1996.0152. [DOI] [PubMed] [Google Scholar]
- 38.Stone D M, Hynes M, Armanini M, Swanson T A, Gu Q, Johnson R L, Scott M P, Pennica D, Goddard A, Phillips H, Noll M, Hooper J E, de-Sauvage F, Rosenthal A. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 39.Sun T J, Devreotes P N. Gene targeting of the aggregation stage cAMP receptor cAR1 in Dictyostelium. Genes Dev. 1991;5:572–582. doi: 10.1101/gad.5.4.572. [DOI] [PubMed] [Google Scholar]
- 40.Van den Heuvel M, Ingham P W. Smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature. 1996;382:547–551. doi: 10.1038/382547a0. [DOI] [PubMed] [Google Scholar]
- 41.Van Haastert P J M. Binding of cAMP and adenosine derivatives to Dictyostelium discoideum cells. Relationships of binding, chemotactic, and antagonistic activities. J Biol Chem. 1983;258:9643–9648. [PubMed] [Google Scholar]
- 42.Van Lookeren Campagne M M, Schaap P, Van Haastert P J M. Specificity of adenosine inhibition of cAMP-induced responses in Dictyostelium resembles that of the P site of higher organisms. Dev Biol. 1986;117:245–251. [Google Scholar]
- 43.Weijer C J, Durston A J. Influence of cyclic AMP and hydrolysis products on cell type regulation in Dictyostelium discoideum. J Embryol Exp Morphol. 1985;86:19–37. [PubMed] [Google Scholar]
- 44.Yu Y, Saxe C L., III Differential distribution of cAMP receptors cAR2 and cAR3 during Dictyostelium development. Dev Biol. 1996;173:353–356. doi: 10.1006/dbio.1996.0066. [DOI] [PubMed] [Google Scholar]