Abstract
Distinct classes of sporulation-specific genes are sequentially expressed during the process of spore formation in Saccharomyces cerevisiae. The transition from expression of early meiotic genes to expression of middle sporulation-specific genes occurs at about the time that cells exit from pachytene and form the meiosis I spindle. To identify genes encoding potential regulators of middle sporulation-specific gene expression, we screened for mutants that expressed early meiotic genes but failed to express middle sporulation-specific genes. We identified mutant alleles of RPD3, SIN3, and NDT80 in this screen. Rpd3p, a histone deacetylase, and Sin3p are global modulators of gene expression. Ndt80p promotes entry into the meiotic divisions. We found that entry into the meiotic divisions was not required for activation of middle sporulation genes; these genes were efficiently expressed in a clb1 clb3 clb4 strain, which fails to enter the meiotic divisions due to reduced Clb-dependent activation of Cdc28p kinase. In contrast, middle sporulation genes were not expressed in a dmc1 strain, which fails to enter the meiotic divisions because a defect in meiotic recombination leads to a RAD17-dependent checkpoint arrest. Expression of middle sporulation genes, as well as entry into the meiotic divisions, was restored to a dmc1 strain by mutation of RAD17. Our studies also revealed that NDT80 was a temporally distinct, pre-middle sporulation gene and that its expression was reduced, but not abolished, on mutation of DMC1, RPD3, SIN3, or NDT80 itself. In summary, our data indicate that Ndt80p is required for expression of middle sporulation genes and that the activity of Ndt80p is controlled by the meiotic recombination checkpoint. Thus, middle genes are expressed only on completion of meiotic recombination and subsequent generation of an active form of Ndt80p.
Sporulation is a process of cellular differentiation that is initiated in diploid a/α cells of the yeast Saccharomyces cerevisiae in response to nitrogen starvation in the presence of a nonfermentable carbon source. As a cell progresses through the events of meiosis and spore wall formation, a coordinated series of genetic and morphological events generates a tetrad of haploid spores within an ascus (reviewed in references 9 and 30). The sporulation program begins when starved cells exit the mitotic cell cycle and undergo premeiotic DNA replication. This is followed by a lengthy prophase in which homologous chromosomes pair, the synaptonemal complex (SC) is elaborated, and a high level of genetic recombination occurs. At pachytene, the penultimate stage of prophase and the last stage at which cells can return to mitotic growth, the chromosomes are fully synapsed, and the spindle pole bodies lie side-by-side in the nuclear envelope. As cells exit from pachytene, the SC disassembles, chiasmata appear, and the spindle pole bodies separate to form the meiosis I spindle. The two meiotic divisions occur in rapid succession, leading first to segregation of homologous chromosomes and then to segregation of sister chromatids, giving rise to a four-lobed nucleus. The prospore walls, which initiate as a membranous outgrowth from a modified outer plaque of the spindle pole bodies, expand around each nuclear lobe. Ultimately, a haploid nucleus and a portion of maternal cytoplasmic material is engulfed within each prospore. Maturation of the spore wall into a multilayered structure completes the process of spore formation. The successive events of meiosis and spore formation are dependent on the ordered expression of at least four classes of genes, referred to as early, middle, mid-late, and late based on their time of expression during sporulation (reviewed in references 30 and 39). This temporal pattern of gene expression during sporulation in S. cerevisiae provides a simple model for studying the mechanisms by which morphological and genetic changes might control gene expression during development.
Expression of early meiotic genes, such as HOP1 and DMC1, which are involved in pairing and synapsis of homologous chromosomes and meiotic recombination, is regulated by the transcription factors Ime1p and Ume6p (reviewed in reference 30). Repression of early meiotic genes in vegetative cells is mediated by Ume6p, which binds to an URS1 promoter element and recruits a Sin3p-Rpd3p-containing repression complex to the DNA (23). IME1, a key inducer of meiosis, is rapidly upregulated when diploid cells are transferred to sporulation medium. An Ime1p-Rim11p activation complex, which replaces the Sin3p-Rpd3p repression complex (7, 36, 52, 63), then acts in conjunction with other upstream activation sequence (UAS)-bound proteins, such as Abf1p (15), to promote expression of early meiotic genes. Ime2p kinase, an early meiotic gene product, acts in a second pathway to bring about full activation of early meiotic genes (reviewed in reference 39).
As cells enter the meiotic divisions, expression of early meiotic genes ceases and expression of middle sporulation genes begins. Activation of middle sporulation genes, such as SPS1 and SMK1, which contribute to spore wall formation (14, 29), is dependent on Ime2p kinase activity (40, 62). A sporulation-specific activation element, referred to as the middle sporulation element (MSE), has been defined in the promoters of the SPS4 and SPR3 genes, and sequence inspection reveals that similar sites are present in the 5′-flanking sequence of other middle sporulation genes (19, 43).
The transition from expression of early meiotic genes to expression of middle sporulation genes occurs at about the time that cells exit from prophase and form the meiosis I spindle. Commencement of the meiotic divisions, which is dependent on activation of maturation-promoting factor (MPF), is an important regulatory point in the meiotic cell cycle of many organisms (reviewed in reference 44). MPF refers to the complex of cyclin-dependent kinase with its regulatory partner, a B-type cyclin. Several yeast mutants that are defective at intermediate stages of recombination arrest in pachytene, at the end of prophase (2, 4, 37, 38, 48, 68, 70). During meiotic recombination, recombination complexes provide a signal that activates the meiotic recombination checkpoint, preventing exit from pachytene. This pachytene arrest requires the mitotic DNA damage checkpoint genes RAD17, RAD24, and MEC1 (35) and the meiosis-specific recombination genes RED1 and MEK1 (78). Completion of recombination events removes this signal, allowing entry into meiosis I (4, 35, 78, 79). Exit from pachytene also appears to be regulated by other factors (34, 49, 71); for example, mutation of NDT80 leads to pachytene arrest in the absence of any significant defects in meiotic recombination (79).
In this study, we identified SIN3, RPD3, and NDT80 in a genetic screen for regulators of middle sporulation gene expression. The proposal by Xu and colleagues (79) that Ndt80p might control entry into meiosis by modulating the activity of MPF prompted us to test whether MPF activity itself was required for activation of middle sporulation genes. We found that middle sporulation genes were efficiently expressed in cells of a clb1 clb3 clb4 strain, which are deficient in MPF activity and are unable to enter meiosis (13, 18). Thus, entry into meiosis is not a prerequisite for activation of middle sporulation genes. In contrast, we found that mutation of DMC1, which leads to a defect in the strand exchange step of meiotic recombination and triggers pachytene arrest (4, 78), blocked activation of middle sporulation genes. Preventing pachytene arrest by mutation of the meiotic recombination checkpoint gene RAD17 restored middle sporulation gene expression to dmc1 cells. These results suggest that the meiotic recombination checkpoint regulates both entry into meiosis and activation of middle sporulation gene expression. The recent demonstration that Ndt80p is a transcriptional activator that binds to the MSE element (10) accounts for our isolation of NDT80 in a screen for regulators of middle gene expression.
MATERIALS AND METHODS
Yeast strains and genetic procedures.
Table 1 lists the S. cerevisiae strains used in this study. DKB414-1 is a derivative of the MATa spo13 strain DKB414 (provided by D. Bishop) that contained the plasmid pRS315-MATα and was used to screen for mutants. We found that an SK1-derived strain was more proficient than our standard W303-derived laboratory strain at carrying out haploid meiosis and at promoting a high level of expression of a UASSPS4-lacZ reporter gene during sporulation. However, cells from an SK1 strain background lost the ability to grow on acetate more frequently than did cells from a W303 strain background; since growth on nonfermentable carbon is a prerequisite for sporulation, we routinely checked putative mutants for their ability to grow on acetate-containing medium. NKY2296 (79) (provided by N. Kleckner) was transformed with pNKY1212, a plasmid that contains NDT80 (79), and sporulated to obtain the haploid MATα Δndt80 strain NKY2296B. The wild-type diploid strain DKB98 and its isogenic dmc1/dmc1 and dmc1/dmc1 rad17/rad17 derivatives, DKB608 and DKB1170, respectively, were provided by D. Bishop (35). Construction of ime2/ime2 (YKE2) (19) and clb1/clb1 clb3/clb3 clb4/clb4 (CD140; provided by B. Futcher) (13) derivatives of W303 have been described previously.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SK1 derivatives | ||
| DKB414 | MATa ho::LYS2 lys2 ura3 leu2 his4X-ADE2-his4B ade2 spo13::hisG | D. Bishop (4) |
| DKB414-1 | DKB414 + pRS315-MATα | This study |
| DKB414B | MATα derivative of DKB414 | This study |
| DKB414C | DKB414 × DKB414B | This study |
| SRH415B | Δrpd3::URA3 derivative of DKB414B | This study |
| SRH416B | Δsin3::URA3 derivative of DKB414B | This study |
| DKB98 | MATa/MATα lys2/lys2 ho::LYS2/ho::LYS2 ura3/ura3 leu2::hisG/leu2::hisG his4X::LEU2/his4B::LEU2 arg4-NspI/arg4-BglII | D. Bishop (35) |
| DKB608 | MATa/MATα lys2/lys2 ho::LYS2/ho::LYS2 ura3/ura3 leu2::hisG/leu2::hisG arg4-NspI/arg4-NspI Δdmc1::ARG4/Δdmc1::ARG4 | D. Bishop (35) |
| DKB1170 | MATa/MATα lys2/lys2 ho::LYS2/ho::LYS2 ura3/ura3 leu2::hisG/leu2::hisG arg4-BglII/arg4-NspI his4B::LEU2/his4X::LEU2-URA3 Δrad17::hisG/Δrad17::hisG Δdmc1::ARG4/Δdmc1::ARG4 | D. Bishop (35) |
| NKY2296 | MATa/MATα ho::LYS2/ho::LYS2 lys2/lys2 ura3/ura3 leu2/leu2 Δndt80::LEU2/Δndt80::LEU2 | N. Kleckner (79) |
| NKY2296B | Haploid MATα derivative of NKY2296 | This study |
| W303 derivatives | ||
| W3031A | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | S. Lindquist (47) |
| W3031B | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | S. Lindquist (47) |
| LP112 | W3031A × W3031B | S. Lindquist (47) |
| YSH2 | ume6::LEU2/ume6::LEU2 derivative of LP112 | This laboratory (19) |
| YKE2 | ime2::LEU2/ime2::LEU2 derivative of LP112 | This laboratory (19) |
| YKE2α | ime2::LEU2 derivative of W3031B | This laboratory (19) |
| CD140 | clb1::URA3/clb1::URA3 clb3::TRP1/clb3::TRP1 clb4::HIS3/clb4::HIS3 derivative of LP112 | B. Futcher (13) |
Isogenic Δrpd3::URA3 and Δsin3::URA3 derivatives of DKB414B were obtained by integrative transformation (51) with XbaI-linearized pMV130 and SmaI-linearized pMV117, respectively (provided by R. Gaber) (73, 74). The Δspo11::hisG allele was introduced into various strains by integrative transformation with XbaI- and BglII-digested pGB518 (provided by C. Giroux), which contains a Δspo11::hisG-URA3-hisG allele. Appropriate integrative transformation was confirmed by Southern blot analysis of genomic DNA from Ura+ transformants. Strains with the Δspo11::hisG-URA3-hisG allele were grown on SD medium (see below) containing 5-fluoro-orotic acid (5-FOA) to select for cells that had lost the URA3 gene (1). The Δrad17::hisG allele was introduced in the same manner by using BamHI-digested pWL7, which contains the Δrad17::hisG-URA3-hisG allele (provided by T. Weinert).
Yeast transformations were performed by the lithium acetate method (17). Standard genetic methods were used for mating, sporulation, and tetrad analysis (57, 58). Isogenic strains of the opposite mating type were obtained by using pGAL-HO (22) to induce mating-type switching (12). Diploids were obtained by prototrophic selection, taking advantage of either chromosomal markers or markers introduced on plasmids for this purpose.
Media and growth conditions.
SD medium is a minimal medium (2% glucose, 0.7% yeast nitrogen base without amino acids) supplemented with 40 μg of adenine sulfate per ml, 20 μg of l-arginine per ml, 20 μg of l-histidine per ml, 60 μg of l-leucine per ml, 30 μg of l-lysine (mono-HCl) per ml, 20 μg of l-methionine per ml, 50 μg of l-phenylalanine per ml, 200 μg of l-threonine per ml, 40 μg of l-tryptophan per ml, 30 μg of l-tyrosine per ml, and 20 μg of uracil per ml. SD−X medium refers to SD medium that lacks supplement X. Rich medium (YEPD), presporulation medium (YEPA), and sporulation medium (SPO) were as described previously (19) with the exception that for SK1-derived strains YEPA contained 2% potassium acetate, 2% Bacto Peptone, and 1% yeast extract and SPO medium contained 2% potassium acetate supplemented with adenine sulfate, histidine, leucine, lysine, tryptophan, and uracil at the same concentrations as those in SD medium. All yeast cultures were grown at 30°C.
Sporulation of cells of W303-derived strains in liquid culture was described previously (19). For sporulation of SK1-derived strains, cells taken from a YEPA plate were grown to late log phase in YEPD and this culture was used to inoculate YEPA medium (1:100 dilution). When the YEPA culture reached a density of 1.5 × 107 to 2.0 × 107 cells per ml, the cells were harvested by centrifugation, washed once in water, and resuspended in SPO medium at a density of 1.5 × 107 cells per ml. The time of transfer of cells to sporulation medium is referred to as 0 h. The efficiency of ascus formation was assessed by examination of 300 or more cells by light microscopy.
Plasmids.
The multicopy CYC1-lacZ-containing plasmids pLGΔ312(Bgl), pLGΔ312SΔSS(Bgl), and pCYC1-SPS4-lacZ [previously referred to as 29×4/pLGΔ312(Bgl)], have been described previously (19). pSPS4-lacZ, which contains four copies of fragment 29 (nucleotides −145 to −116 of the SPS4 gene) adjacent to a CYC1-lacZ gene that lacks a UAS, was constructed as follows. A SalI-SalI fragment recovered from pCYC1-SPS4-lacZ was cloned into the BglII site of pLGΔ312SΔSS(Bgl) after the restriction endonuclease-generated ends of the vector and the insert had been filled in with the Klenow form of DNA polymerase. Forward orientation of the insert was confirmed by dideoxy sequence analysis. pRS315-MATα was constructed by cloning a 4.2-kb MATα-containing HindIII fragment from pJM9 (provided by A. Mitchell) into the HindIII site of pRS315 (61). pAV79B contains a HOP1-lacZ translational fusion gene (provided by A. Vershon) (72). The YCp50-based plasmids pMV1 and pMV34 contain RPD1 and RPD3, respectively (provided by R. Gaber) (73, 74). p(SPO13)8 is a YCp50-based plasmid which contains the SPO13 gene (provided by C. Giroux) (8). All plasmids were amplified in Escherichia coli DH5α.
Assay for β-galactosidase activity.
β-Galactosidase expression from lacZ reporter genes was monitored by use of a colony overlay assay (3). Ten milliliters of X-Gal-containing agar (0.5% agar, 0.5 M potassium phosphate [pH 7.0], 6% dimethyl formamide, 0.1% sodium dodecyl sulfate, 0.1 to 0.4 mg of 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside [X-Gal] per ml) was poured over colonies on plates, and the plates were incubated at 30°C until blue color development was evident.
Screens for mutants defective in expression of SPS4.
One screen, aimed at the identification of mutants that failed to activate a UASSPS4-lacZ reporter gene during sporulation, was carried out with strain DKB414-1 containing pSPS4-lacZ. Cells that had been mutagenized to ∼50% survival by exposure to ethyl methanesulfonate (EMS) in accordance with the procedure of Lawrence (33) were plated on SD−Leu−Ura medium at 200 to 500 cells per plate and incubated for 4 to 5 days. About 32,000 colonies were patched onto SD−Leu−Ura medium, incubated for 1 to 2 days, and then replica plated to SPO medium. The SPO plates were incubated for 14 to 18 h, and the patches were then overlaid with X-Gal-containing agar (see above). We recovered 258 mutants that failed to form blue patches in this assay and that retained the ability to grow on acetate-containing YEPA medium. To perform secondary tests, these mutant strains were grown on SD medium containing 5-FOA to select for cells that had lost pSPS4-lacZ (5). Each mutant was then cotransformed in duplicate with pRS315-MATα and either pSPS4-lacZ or pAV79B, which contains HOP1-lacZ. The transformants that grew readily on YEPA medium were identified, patched onto SD−Leu−Ura medium, grown for 1 to 2 days, and then patched in duplicate onto SPO medium to retest for expression of UASSPS4-lacZ and to test for expression of HOP1-lacZ. Most mutants were discarded because they expressed UASSPS4-lacZ or because they showed a reduced or undetectable level of HOP1-lacZ expression on transfer to sporulation medium. Three mutant strains, m51, m312, and m320, that had the desired phenotype were identified; they expressed HOP1-lacZ at a high level and UASSPS4-lacZ at a reduced or undetectable level. These strains were found subsequently to contain the mutant alleles das1-1, das3-1, and das1-2, respectively. The gene designation refers to defective in activation of SPS4. We found that DAS1 was identical to NDT80 and that DAS3 was identical to SIN3.
Another screen was aimed at the identification of mutants that expressed a CYC1-UASSPS4-lacZ reporter gene in vegetative cells. Mutagenized cells of strain DKB414-1 that contained pCYC1-SPS4-lacZ were plated onto SD−Leu−Ura medium and incubated for 3 to 4 days to give about 97,000 colonies that were then overlaid with X-Gal-containing agar. We recovered cells from 115 colonies that expressed a high level of β-galactosidase. These strains were grown on SD−Leu−Ura medium, retested for expression of the CYC1-UASSPS4-lacZ reporter gene, and then grown on SD medium containing 5-FOA to select for cells that had lost pCYC1-SPS4-lacZ (5). In secondary tests, 113 of the resulting strains were cotransformed in triplicate with pRS315-MATα and one of the following plasmids: pCYC1-SPS4-lacZ, to retest for expression of the CYC1-UASSPS4-lacZ gene in vegetative cells; pLGΔ312SΔSS(Bgl), to test for expression of a CYC1-lacZ gene that lacks a UAS in vegetative cells; or pSPS4-lacZ, to test for expression of a UASSPS4-lacZ gene in cells transferred to sporulation medium. These tests eliminated 97 strains: 30 were false positives, 2 expressed an elevated level of the CYC1-lacZ gene that lacked a UAS, and 65 expressed a high level of UASSPS4-lacZ and formed spores on transfer to sporulation medium. Of the remaining 16 strains which were defective in expression of UASSPS4-lacZ, 2 (m83 and m96) were transformed with pAV79B and were found to express the HOP1-lacZ reporter gene on transfer to sporulation medium. These strains were found subsequently to contain the mutant alleles das4-1 and das4-2, respectively, with DAS4 being identical to RPD3.
Genetic analysis.
Mutants were placed into complementation groups by using standard techniques (57). Allelism with IME2, SIN3, RPD3, and NDT80 was determined by mating das mutants with YKE2α, SRH416B, SRH415B, and NKY2296B, respectively, and testing the resultant diploids for spore formation.
Cloning DAS4 by complementation.
An a/α das4-1/das4-1 strain that contained pSPS4-lacZ was transformed with a pSB32-based (CEN ARS LEU2) library that consisted of a partial Sau3A digest of genomic DNA (constructed by P. Hieter and provided by B. Andrews) (50). Transformants were selected on SD−Leu−Ura medium and then replica plated to SPO medium. After incubation for 14 to 16 h, colonies that expressed a high level of β-galactosidase were identified by an overlay assay. These strains were retested for expression of UASSPS4-lacZ and examined for spore formation. After complemented strains had been grown on medium containing 5-FOA to select for loss of pSPS4-lacZ, library plasmids were recovered by introducing total yeast genomic DNA into E. coli. Purified library plasmids were retested for their ability to complement the Spo− phenotype of the das4-1/das4-1 strain. Complementing plasmids were subjected to partial dideoxy sequence analysis with primers that flanked the site of insertion of genomic yeast DNA into pSB32. The resultant sequence was used in a BLAST search of the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces) to identify the chromosomal region that was present in the complementing plasmid.
Transposon disruption mutagenesis.
pDAS4-32, which contained yeast genomic DNA that complemented the sporulation defect of an a/α das4-1/das4-1 strain, was mutagenized by transposon-mediated disruption with a HIS3-tagged version of Tn1000 (γδ) as described previously (41, 56). Donor and recipient E. coli strains were provided by M. Donoviel and B. Andrews. The site of transposon insertion in a plasmid that was unable to complement the a/α das4-1/das4-1 mutant was determined by dideoxy sequence analysis with primers that annealed to the ends of the transposon (41, 56). The resultant sequence was used in a BLAST search of the Saccharomyces Genome Database (see above).
FACS analysis.
The relative DNA content of propidium iodide-stained cells was determined by fluorescence-activated cell sorter (FACS) analysis as previously described (14). For each experiment, 10,000 gated events were analyzed with LYSYS II software.
DAPI staining and microscopy.
Cells to be stained with 4′,6-diamidino-2-phenylindole (DAPI) were harvested from about 1 ml of culture, fixed by resuspension in 70% ethanol, and stored at 4°C. Just prior to use, cells were pelleted by centrifugation and resuspended in 50 μl of water. Three microliters of this cell suspension was placed on a glass slide and mixed with 3 μl of a solution that contained 0.6 mg of DAPI per ml of mounting medium (90% glycerol and 10% antiquenching solution [10 mg of p-phenylenediamine per ml of phosphate-buffered saline, pH 9.0]). To monitor the meiotic divisions, the cells were then examined with a Nikon Microphot FXA microscope with Nomarski and fluorescence optics.
RNA isolation and Northern analysis.
RNA was prepared from yeast as described previously (46), except for the experiment shown in Fig. 6. In that case, cells harvested from 50-ml cultures were disrupted by a freeze-thaw procedure (55) and RNA was purified as described previously (55) with the addition of a lithium chloride and an ethanol precipitation of RNA. Northern blot analysis was performed as previously described (19). Gene-specific probes were prepared with the following templates: SMK1, an 800-bp StyI-StyI fragment from pLAKK40 (29); NDT80, a 1.2-kb Eco47III-BamHI fragment from pNKY1212 (79); SPS100, a 750-bp BamHI-NcoI fragment from pE18-B8a (32); and pC4, an uncharacterized control gene (32). Other templates were as described previously (19).
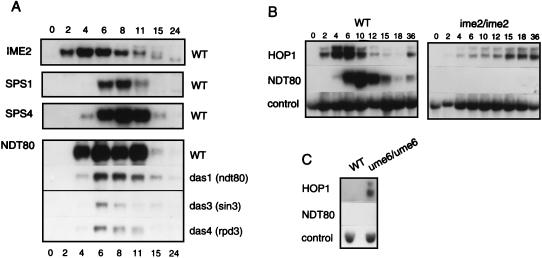
FIG. 6.
Expression of NDT80, a pre-middle sporulation-specific gene, depends on IME2. (A) NDT80 is expressed prior to middle sporulation-specific genes. A set of filters from the experiment shown in Fig. 3, containing RNAs purified from cells of the wild-type strain (DKB414C) (WT) and from mutant das1-1/das1-1 [das1 (ndt80)], das3-1/das3-1 [das3 (sin3)], and das4-1/das4-1 [das4 (rpd3)] strains, was stripped after hybridization with the probe for SMK1-derived transcripts and hybridized with a probe for NDT80-derived transcripts. Northern blot analysis was performed as described in the legend to Fig. 3. The autoradiograms depicting accumulation of IME2, SPS2, and SPS4 transcripts in the wild-type strain are from Fig. 3. The times at which cells were harvested are indicated (in hours) at the top and bottom of the figure. (B) IME2 is required for expression of NDT80 in sporulating cells. RNAs that had been purified previously (19) from cells of the wild-type strain (LP112) (WT) and its isogenic ime2/ime2 derivative (YKE2) (ime2/ime2), as indicated above the panels, were used to prepare filters for Northern blot analysis. The times at which cells were harvested are indicated (in hours) above the panels. The filters were sequentially hybridized with probes for detection of HOP1-, NDT80-, and control PYK1-derived transcripts. Only the portions of the autoradiograms that revealed hybridization with the probes are shown. (C) UME6 is not required for repression of NDT80 in vegetative cells. RNAs that had been purified from cells of the wild-type strain (LP112) (WT) and its isogenic ume6/ume6 derivative (YSH2) (ume6/ume6) growing in YEPD medium, as indicated above the panel, were subjected to Northern blot analysis (19). The autoradiogram of the filter that was hybridized with a probe for HOP1-derived transcripts is from the work of Hepworth et al. (19). A duplicate filter was sequentially hybridized with probes for NDT80- and control PYK1-derived transcripts. Only the portions of the autoradiograms that revealed hybridization with the probes are shown.
RESULTS
Isolation of mutants that are defective in expression of the SPS4 gene.
We used the haploid meiosis strain DKB414-1, which is a spo13 derivative of SK1, to screen for mutations in potential regulators of expression of SPS4, a middle sporulation gene. We previously defined a 15-bp MSE, which we refer to as UASSPS4, in the promoter region of this gene that is sufficient to direct sporulation-specific gene expression (19). The presence of a MATα-containing plasmid in the MATa strain DKB414-1 enables cells to initiate meiosis in response to nutrient starvation. The spo13 mutation causes the cells to bypass what would be a lethal meiosis I division and to enter directly into meiosis II, a mitosis-like division in which sister chromatids are segregated (reviewed in reference 30). Subsequent formation of spore walls generates a dyad of viable haploid spores within each ascus. Strain DKB414-1, therefore, allows identification of recessive mutations affecting sporulation-specific events.
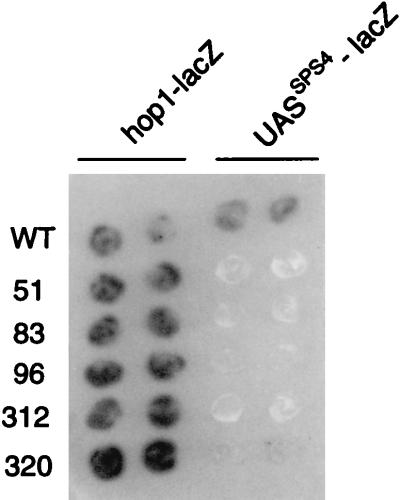
We carried out two screens aimed at the identification of mutants that fail to activate middle sporulation genes. In the first screen, we monitored the expression of the plasmid-borne UASSPS4-lacZ reporter gene, which has four copies of a UASSPS4-containing fragment upstream of a CYC1-lacZ gene that lacks a UAS (see Materials and Methods). This gene was activated midway through sporulation in wild-type cells (data not shown) (see reference 19). Colonies derived from 32,000 survivors of EMS mutagenesis were transferred to sporulation medium and tested for expression of the reporter gene by using an X-Gal overlay assay to monitor β-galactosidase activity (see Materials and Methods). Three mutants, m51, m312, and m320, were chosen for further study because their phenotype was as expected for mutants that entered the sporulation program but could not activate expression of middle sporulation genes: the strains responded appropriately to nutrient starvation and activated early meiotic genes as assessed by expression of the IME1-dependent reporter gene, HOP1-lacZ; they reproducibly failed to activate expression of the UASSPS4-lacZ reporter gene; and they were defective in spore formation (see Materials and Methods) (Fig. 1 and data not shown). Because SPS4 is not required for spore formation (16), we hypothesized that a general defect in expression of middle sporulation genes caused the asporogenous phenotype of the mutant strains.
FIG. 1.
Mutant strains express a HOP1-lacZ reporter gene but not an SPS4-lacZ reporter gene. Duplicate patches of cells from the wild-type strain (DKB414-1) and from mutant m51, m83, m96, m312, and m320 strains were incubated for 18 h on sporulation medium and then overlaid with X-Gal-containing agar to monitor β-galactosidase expression (see Materials and Methods). As indicated at the top of the figure, one set of cells harbored pAV79B, which contains a HOP1-lacZ reporter gene, and another set harbored pSPS4-lacZ, which contains a UASSPS4-lacZ reporter gene.
The second screen took advantage of our previous observation that the CYC1-UASSPS4-lacZ reporter gene, which has four copies of a UASSPS4-containing fragment inserted between the UASCYC1 and the TATACYC1, is not expressed in vegetatively growing cells (19). Because the ability of UASSPS4 to inhibit expression of the reporter gene is position dependent, we hypothesized that a protein that is present in vegetative cells binds to UASSPS4 and prevents UASCYC1-bound activators from functioning via an indirect mechanism such as steric hindrance (19). We therefore screened EMS-mutagenized cells for mutants that allowed expression of the CYC1-UASSPS4-lacZ reporter gene in vegetative cells; we anticipated that such mutants might also be defective in activation of the UASSPS4-lacZ reporter gene during sporulation. Indeed, in a screen of 97,000 colonies, we identified two mutants, m83 and m96, that expressed the CYC1-UASSPS4-lacZ reporter gene in vegetative cells and also fulfilled our criteria for potential regulatory mutants: the two mutants expressed HOP1-lacZ on transfer to sporulation medium but failed to activate the UASSPS4-lacZ reporter gene and were defective in spore formation (see Materials and Methods) (Fig. 1 and data not shown).
Three complementation groups of DAS genes.
The five mutants selected for further study fell into three complementation groups, which we designated DAS1, DAS3, and DAS4 to indicate that the mutants were defective in activation of SPS4 (see Materials and Methods). Two strains, m51 and m320, contained mutant alleles of DAS1, referred to as das1-1 and das1-2, respectively; m312 contained a mutant allele of DAS3 referred to as das3-1; and m83 and m96 contained mutant alleles of DAS4, referred to as das4-1 and das4-2, respectively. We next constructed and sporulated a/α spo13/SPO13 das/DAS diploid strains. The spore progeny from each of 10 tetrads analyzed for each mutant gave 2:2 segregation for a Spo+:Spo− phenotype on mating with the original das strain or its MATa derivative (see Materials and Methods). This suggests that the phenotype of each das strain was caused by a single mutation.
das mutants complete premeiotic DNA synthesis but remain mononucleate.
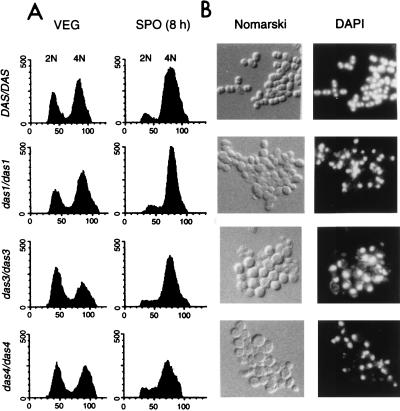
We next monitored the ability of the das mutants to undergo premeiotic DNA synthesis and to complete meiosis. Prior studies have shown that premeiotic DNA synthesis is required for expression of meiotic genes (25, 31). DNA synthesis was assessed by flow cytometric determination of the relative DNA content of cells that had been stained with propidium iodide. For this analysis, we compared the wild-type diploid strain (a/α spo13/spo13 DAS/DAS) with homozygous mutant diploid strains (a/α spo13/spo13 das/das). The FACS scans of cells of the wild-type and mutant strains during vegetative growth gave the expected distribution of cells in the G1 (2N) and G2 (4N) stages of the cell cycle (Fig. 2A, panels VEG). The FACS scans of cells that had been incubated in sporulation medium for 8 h revealed that most cells of the wild-type strain and of the mutant das1-1/das1-1, das3-1/das3-1, and das4-1/das4-1 strains had a 4N DNA content, indicating that they had completed premeiotic DNA synthesis (Fig. 2A, panels SPO).
FIG. 2.
Mutant strains complete premeiotic DNA synthesis but do not segregate their chromosomes. (A) FACS analysis of wild-type and mutant cells during vegetative growth and after transfer to sporulation medium. Aliquots of wild-type DAS/DAS spo13/spo13 cells (DKB414C) and mutant das1-1/das1-1 spo13/spo13, das3-1/das3-1 spo13/spo13, and das4-1/das4-1 spo13/spo13 cells were harvested during logarithmic growth in YEPA medium (VEG) and after incubation for 8 h in sporulation medium (SPO). Cells were fixed and stained with propidium iodide prior to analysis in a Becton-Dickinson FACScan. The peaks marked 2N and 4N represent cells in G1 and G2 portions of the cell cycle, respectively. Relative DNA content (x axis) is plotted versus cell number (y axis). (B) Microscopic appearance of wild-type and mutant cells on transfer to sporulation medium. Photographs are shown of wild-type DAS/DAS spo13/spo13 cells (DKB414C) and mutant das1-1/das1-1 spo13/spo13, das3-1/das3-1 spo13/spo13, and das4-1/das4-1 spo13/spo13 cells that had been incubated in sporulation medium for 24 h, fixed, and then stained with DAPI (see Materials and Methods). Cells were viewed with Nomarski (Nomarski) or fluorescence (DAPI) optics.
We next monitored the ability of the wild-type and mutant strains to complete meiosis by using the fluorescent DNA-specific stain DAPI to visualize the segregation of chromosomes. We examined the microscopic appearance of wild-type (a/α spo13/spo13 DAS/DAS) cells and mutant das/das cells at 24 h after transfer to liquid sporulation medium. In the absence of a functional copy of the SPO13 gene, ∼90% of wild-type cells became binucleate and formed a dyad of spores (Fig. 2B and data not shown). No spores were detected in any of the das mutants, and only 2.6, 4.8, and 12% of das1-1/das1-1, das3-1/das3-1, and das4-1/das4-1 cells, respectively, appeared binucleate (Fig. 2B and data not shown). To ascertain that the failure of the mutant strains to complete meiosis did not depend on mutation of SPO13, we introduced a plasmid-borne version of SPO13 into the mutant strains. Whereas cells of a spo13/spo13 DAS/DAS strain containing this plasmid became tetranucleate on transfer to sporulation medium, albeit not as efficiently as cells of a SPO13/SPO13 strain, most cells of the spo13/spo13 das1-1/das1-1, spo13/spo13 das3-1/das3-1, and spo13/spo13 das4-1/das4-1 strains containing a plasmid-borne version of SPO13 remained mononucleate on sporulation medium (data not shown).
In summary, this analysis indicated that all three mutant strains chosen for study because they were defective in activation of the UASSPS4-lacZ reporter gene completed premeiotic DNA replication but did not complete meiosis.
das mutants are defective in activation of middle sporulation genes.
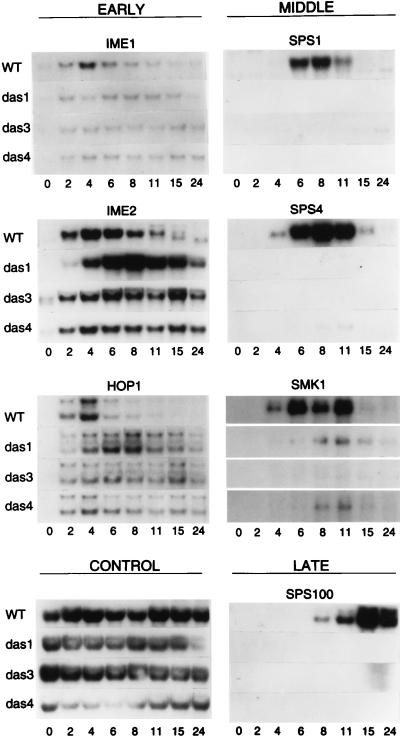
We next tested the hypothesis that the failure of the mutant strains to express the UASSPS4-lacZ reporter gene and to form spores reflected a general defect in middle sporulation gene expression. We monitored transcript accumulation by Northern blot analysis of RNA from wild-type diploid cells (a/α DAS/DAS spo13/spo13) and from mutant das1-1/das1-1, das3-1/das3-1, and das4-1/das4-1 cells during vegetative growth and at various times after transfer to sporulation medium. Blots were probed for the presence of transcripts from the early meiotic genes IME1, IME2, and HOP1, the middle sporulation genes SPS1, SPS4, and SMK1, and the late sporulation gene SPS100.
The early meiotic genes IME1, IME2, and HOP1 were activated in the wild-type and mutant strains shortly after transfer to sporulation medium (Fig. 3). In the wild-type strain, maximal transcript accumulation occurred at about 4 h, and transcript levels then declined from 6 to 11 h of sporulation (Fig. 3). In the das3-1/das3-1 and das4-1/das4-1 strains, the levels of IME1, IME2, and HOP1 transcripts remained relatively constant from 2 to 24 h of sporulation (Fig. 3). In the das1-1/das1-1 strain, IME2 transcripts accumulated to a higher level than in the wild-type strain and maximal accumulation of HOP1 and IME2 transcripts was delayed until about 6 to 8 h of sporulation (Fig. 3). We note that a very low level of IME1 and IME2 transcripts could be detected in most strains during vegetative growth.
FIG. 3.
Expression of sporulation-specific genes in wild-type and das strains. RNA was purified from cells of wild-type strain DKB414C (WT) and mutant das1-1/das1-1 (das1), das3-1/das3-1 (das3), and das4-1/das4-1 (das4) strains, as indicated to the left of the panels. Cells were harvested during logarithmic growth in YEPA medium (0 h) and at various times after transfer to sporulation medium, as indicated (in hours) below the panels. Northern blots of these RNAs were sequentially hybridized with radioactively labeled gene-specific probes (see Materials and Methods), as indicated at the top of the panels. One set of filters was hybridized with probes for transcripts from SPS4, IME2, HOP1, SPS1, IME1, and the control gene. The control probe was prepared with pC4. Two HOP1-hybridizing transcripts have been described previously (20). A duplicate set of filters was used for hybridization with probes for transcripts from SMK1 and SPS100. Only the portions of the autoradiograms that revealed hybridization with the probes are shown.
In the wild-type strain, the middle sporulation genes SPS1, SPS4, and SMK1 were activated at about 4 to 6 h with maximal transcript accumulation occurring at about 8 h of sporulation. The late sporulation gene SPS100 was activated at about 8 h in the wild-type strain with maximal transcript accumulation occurring at about 15 h after transfer of cells to sporulation medium (Fig. 3). Transcripts from these middle and late genes did not accumulate in the das1-1/das1-1, das3-1/das3-1, or das4-1/das4-1 strains, with the exception that a very low level of SMK1-derived transcripts could be detected in RNA from cells of the das1-1/das1-1 and das4-1/das4-1 strains at 8 to 11 h of sporulation (Fig. 3).
In summary, this analysis of transcript accumulation revealed that the das1-1/das1-1, das3-1/das3-1, and das4-1/das4-1 strains had similar transcriptional defects during sporulation. The most striking defect was the failure to activate expression of the middle sporulation genes SPS1, SPS4, and SMK1 and the late sporulation gene SPS100. A minor defect was the continued expression of the early meiotic genes IME1, IME2, and HOP1 at a time at which their expression was turned down in wild-type cells. Although it has been suggested previously that downregulation of IME1 might be a prerequisite for activation of middle sporulation gene expression (40), a recent study indicates that continued expression of IME1 through sporulation does not impede spore formation (34).
DAS1 is NDT80, DAS3 is SIN3, and DAS4 is RPD3.
Having confirmed that the das1-1, das3-1, and das4-1 mutant strains were defective in activation of middle sporulation genes, we proceeded to identify the wild-type DAS genes. We first tested two candidate genes, IME2 and NDT80, for allelism with the das genes. We tested IME2 because it is known to be essential for middle sporulation gene expression (40, 62), including expression of SPS4 (19). We tested NDT80, a key regulatory gene required for the transition from pachytene into the meiosis I division (79), because we had found that the transcriptional defect of the das mutants correlated with a failure to complete meiosis.
We found that das1-1/Δime2, das3-1/Δime2, and das4-1/Δime2 cells that had been transferred to sporulation medium expressed the UASSPS4-lacZ reporter gene and formed spores efficiently, indicating that the das mutations were in genes other than IME2 (data not shown). To test for allelism with NDT80, we introduced a plasmid-borne version of this gene into the das strains. The plasmid-borne NDT80 gene restored both UASSPS4-lacZ expression and spore formation to das1-1/das1-1 cells transferred to sporulation medium, suggesting that das1-1 was an allele of NDT80 (data not shown; see below).
We next isolated complementing plasmids for das4 by screening das4-1/das4-1 cells that had been transformed with a yeast genomic library for restoration of expression of the UASSPS4-lacZ reporter gene during sporulation. We identified two plasmids with overlapping inserts that promoted expression of the UASSPS4-lacZ reporter gene in das4-1/das4-1 and das4-2/das4-2 cells on transfer to sporulation medium and that inhibited expression of the CYC1-UASSPS4-lacZ reporter gene in vegetative cells (data not shown). Transposon mutagenesis of one of the plasmids indicated that mutation of RPD3, one of three genes present in the genomic DNA insert, led to a loss in the ability of the plasmid to complement the das4 mutation (data not shown; see Materials and Methods). As expected, we found that a plasmid-borne version of RPD3 complemented the das4-1 mutation (data not shown).
Mutations in RPD3, which encodes a histone deacetylase (53, 69; reviewed in reference 77), have been shown previously to cause elevated expression of IME2 in vegetative cells and to lead to an asporogenous phenotype (6, 74). Because SIN3/UME4/RPD1 is in the same genetic pathway as RPD3 (64, 73), with sin3 and rpd3 strains having similar phenotypes (73, 74), we tested SIN3 for the ability to complement a mutant allele of DAS3. We found that introduction of a plasmid-borne version of SIN3 into a das3-1/das3-1 strain restored both UASSPS4-lacZ expression and spore formation in cells that had been transferred to sporulation medium (data not shown).
To confirm that NDT80, SIN3, and RPD3 were identical to DAS1, DAS3, and DAS4, respectively, and not low-copy suppressors of the das alleles, we constructed das1-1/Δndt80::LEU2, das3-1/Δsin3::URA3, and das4-1/Δrpd3::URA3 strains (see Materials and Methods). We found that cells of all three diploid strains were unable to form spores on transfer to sporulation medium (data not shown). This complementation analysis supported the assignments of das1-1 as a mutant allele of NDT80, das3-1 as a mutant allele of SIN3/UME4/RPD1, and das4-1 as a mutant allele of RPD3.
In summary, the isolation of mutant alleles of DAS3 and DAS4 in this screen, and their assignment as SIN3 and RPD3, respectively, confirmed previous observations that mutation of these genes leads to misregulation of expression of early meiotic genes and prevents completion of meiosis (6, 65, 73, 74). In addition, the identification of DAS1 as NDT80, a gene that is required for the transition from pachytene into the meiotic divisions, suggests that entry into meiosis and activation of middle sporulation genes might be coregulated.
The B-type cyclins Clb1p, Clb3p, and Clb4p are not required for activation of middle sporulation gene expression.
The middle sporulation genes SPS1 and SMK1 have been characterized in detail and have been shown to play an essential role in spore wall morphogenesis (14, 29). Because spore wall maturation is a postmeiotic event, with the initial prospore membranes emanating from the spindle pole bodies and growing around the four lobes of the postmeiotic nucleus (42; reviewed in reference 30), we considered that the sequence of events during sporulation could be as follows: DNA replication → meiotic recombination → meiotic divisions → middle sporulation genes → spore wall formation. In this cascade, entry into meiosis I would provide a regulatory signal for activation of middle sporulation genes. However, the fact that the mutants that we isolated as defective in expression of middle sporulation genes were also defective in the meiotic divisions suggested that a common regulatory event might promote both entry into meiosis I and the activation of middle genes. To distinguish between these possibilities, we tested whether middle genes were expressed in a strain that arrests at pachytene and does not enter meiosis I due to a reduction in the activity of MPF.
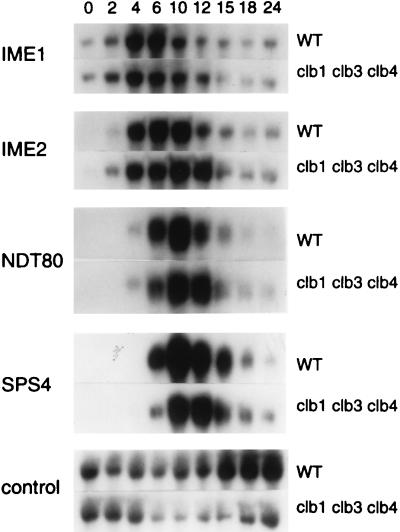
Previous studies have shown that Clb1p, and to a lesser extent Clb3p and Clb4p, is the major meiotic activator of Cdc28p, the kinase component of yeast MPF (13, 18). We therefore monitored gene expression in cells of a clb1/clb1 clb3/clb3 clb4/clb4 strain after transfer to sporulation medium. Because Clb2p is sufficient to regulate the G2/M transition during the mitotic cell cycle, these cells are viable (13). Consistent with previous studies (13, 18), we found that at 36 h of sporulation, a time at which 69% of wild-type cells had formed mature asci, only 2.5% of clb1/clb1 clb3/clb3 clb4/clb4 cells had formed asci (data not shown). As assessed by DAPI staining, 57% of wild-type cells were bi- or tetranucleate at 24 h after transfer to sporulation medium; only 11% of clb1/clb1 clb3/clb3 clb4/clb4 cells were binucleate, and none were tetranucleate at this same time. Despite this defect in meiosis, the expression patterns of IME1, IME2, NDT80, and SPS4 in the clb1/clb1 clb3/clb3 clb4/clb4 strain were indistinguishable from their expression patterns in the wild-type strain (Fig. 4). We therefore concluded that CLB1-, CLB3-, or CLB4-dependent MPF activity and subsequent entry into the meiotic divisions were not required for expression of middle sporulation genes. This suggested that an event occurring prior to entry into meiosis I provided a signal for activation of middle sporulation gene expression.
FIG. 4.
Middle sporulation genes are expressed in a clb1 clb3 clb4 strain. RNA was purified from cells of the wild-type strain (LP112) (WT) and from its isogenic clb1/clb1 clb3/clb3 clb4/clb4 derivative (CD140), as indicated to the right of the figure. Northern blot analysis was performed as described in the legend to Fig. 3. The times at which cells were harvested are indicated (in hours) at the top of the figure; the hybridization probes are indicated to the left of the figure; the control transcript was from the PYK1 gene. Only the portions of the autoradiograms that revealed hybridization with the probes are shown.
Mutation of DMC1 prevents middle sporulation gene expression.
Our experiments indicated that middle sporulation genes were not expressed in cells that arrested in pachytene and failed to enter the meiotic divisions due to mutation of NDT80, a putative regulator of MPF activity (79). In contrast, we found that middle sporulation genes were expressed in clb1 clb3 clb4 cells that failed to enter the meiotic divisions due to reduced MPF activity. Thus, MPF activity and entry into the meiotic divisions are not required for activation of middle sporulation genes. This suggested that pachytene arrest itself might prevent activation of middle sporulation genes. To test this idea, we examined gene expression in a dmc1 strain, which also arrests in pachytene. Whereas pachytene arrest occurs in the das1-1 (ndt80) strain in the absence of any significant defect in meiotic recombination (79), in a dmc1 strain a defect in meiotic recombination results in accumulation of DNA double-strand breaks that triggers the meiotic recombination checkpoint and subsequent pachytene arrest (4).
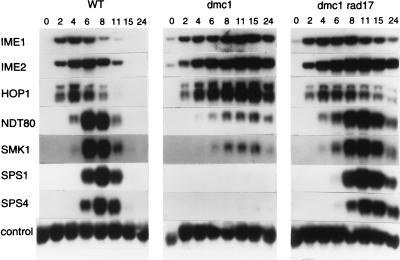
Transcripts from the early genes IME1, IME2, and HOP1 began to accumulate in both the wild-type strain and in the dmc1/dmc1 strain shortly after transfer of cells to sporulation medium (Fig. 5). Transcript accumulation occurred over a longer time period in the dmc1 mutant strain, and peak levels, which occurred at a later time than they did in wild-type cells, were higher. In contrast, accumulation of transcripts from NDT80 and SMK1 was markedly reduced in the dmc1/dmc1 strain, and transcripts from SPS1 and SPS4 were not detected (Fig. 5). We next monitored gene expression in a dmc1/dmc1 rad17/rad17 strain; mutation of the checkpoint gene RAD17 prevents activation of the meiotic recombination checkpoint, thereby allowing dmc1 cells to proceed through the two meiotic divisions with near-normal kinetics (35). In the dmc1 rad17 mutant, expression of all genes tested was similar to that in wild-type cells except that the time period of transcript accumulation was extended and the time of maximal transcript accumulation of middle sporulation genes was delayed by 2 to 3 h (Fig. 5).
FIG. 5.
Sporulation-specific gene expression in wild-type (WT), dmc1/dmc1 (dmc1), and dmc1/dmc1 rad17/rad17 (dmc1 rad17) strains. RNA was purified from cells of the wild-type strain (DKB98) and its isogenic dmc1/dmc1 (DKB608) and dmc1/dmc1 rad17/rad17 (DKB1170) derivatives. Northern blot analysis was performed as described in the legend to Fig. 3. The times at which cells were harvested are indicated (in hours) above the panels. One filter was sequentially hybridized with radioactivity labeled probes for IME1 and IME2 transcripts, and a duplicate filter was sequentially hybridized with probes for SPS1, SPS4, HOP1, NDT80, SMK1, and a control transcript. The control probe was prepared with pC4. Only the portions of the autoradiograms that revealed hybridization with the probes are shown.
In summary, we found that activation of the RAD17-dependent meiotic recombination checkpoint by mutation of dmc1 prevented both entry into meiosis I and activation of middle sporulation genes. We also found that expression of NDT80, which we showed was essential for expression of middle sporulation genes (Fig. 3), was reduced, but nonetheless readily detectable, in a dmc1 strain. This suggested that the Ndt80p produced in the dmc1 strain was unable to activate expression of middle sporulation genes. It is possible that the level of Ndt80p was below a critical threshold in the dmc1-arrested strain or that activation of the meiotic recombination checkpoint in this strain prevented Ndt80p from serving its role in expression of middle sporulation genes (see Discussion and reference 10).
Meiotic arrest of sin3 and rpd3 strains is not suppressed by mutation of SPO11 or RAD17.
We tested whether mutation of SPO11 would overcome the apparent arrest that was observed in das3 (sin3) and das4 (rpd3) strains. Mutation of SPO11 prevents initiation of recombination, thus bypassing defects at intermediate stages of recombination that normally lead to pachytene arrest (2, 4, 27, 37, 38, 48, 68, 70); for example, a spo11/spo11 dmc1/dmc1 strain is able to form spores, albeit inviable ones (4). Whereas cells of a DAS/DAS spo11/spo11 spo13/spo13 strain formed ∼90% dyads on transfer to sporulation medium, cells of isogenic das3-1/das3-1 spo11/spo11 spo13/spo13 and das4-1/das4-1 spo11/spo11 spo13/spo13 strains were unable to form spores (data not shown). This suggests that mutation of SIN3 and mutation of RPD3 prevent the meiotic divisions by a defect other than, or in addition to, a defect in meiotic recombination. Consistent with this, abrogation of the meiotic recombination checkpoint by mutation of the checkpoint gene RAD17 did not allow efficient meiosis to occur in either the das mutants or a Δndt80 strain (data not shown).
Unique regulation of expression of NDT80 and SMK1.
Close examination of the gene expression patterns that we obtained during the course of this study suggested that NDT80 and SMK1 belong to a temporally distinct class of sporulation genes. Activation of these genes occurs after the early meiotic genes are turned on and before middle sporulation genes are first expressed. Additionally, NDT80 and SMK1 are regulated in a unique manner.
Comparison of transcript accumulation from IME2 and NDT80 by Northern blot analysis (Fig. 6A) showed that activation of NDT80 occurred after activation of early meiotic genes, as has been noted previously (79). At 4 h of sporulation, transcripts from the middle sporulation gene SPS1 could not be detected and transcripts from SPS4 were just beginning to appear. At this time, transcript accumulation from NDT80, however, was already close to the maximal level reached at 6 to 11 h of sporulation (Fig. 6A).
We also examined expression of NDT80 in the das1-1/das1-1 (ndt80), das3-1/das3-1 (sin3), and das4-1/das4-1 (rpd3) strains. NDT80 was first expressed in the three mutant strains at about the same time that it was in the wild-type strain. Strikingly, however, NDT80-derived transcripts did not accumulate to any significant level in the three mutant strains (Fig. 6A). The absence of high-level expression of ndt80 in the das1-1/das1-1 (ndt80) strain can be explained by the recent observation that Ndt80p upregulates its own expression (10); we cannot exclude the possibility, however, that the das1-1 allele of NDT80 contained a mutation in its promoter or a mutation that destabilizes its mRNA.
The expression patterns of NDT80 and SMK1 could also be distinguished from those of middle sporulation genes on examination of transcript accumulation in the dmc1/dmc1 and dmc1/dmc1 rad17/rad17 strains (Fig. 5). Whereas NDT80 and SMK1 were expressed at a low level in the dmc1/dmc1 strain, no transcripts could be detected from SPS1 or from SPS4. In the dmc1/dmc1 rad17/rad17 strain, NDT80 and SMK1 transcripts could be detected at 4 h and had accumulated to a significant level by 6 h, a time at which SPS1 and SPS4 transcripts could barely be detected (Fig. 5).
Taking these observations into consideration, we designate NDT80 and SMK1 as pre-middle sporulation genes. Although expression of SMK1 prior to expression of middle genes was not obvious in all of our experiments (e.g., that shown in Fig. 3), a distinction in the time of expression, albeit minor, could be noted in several experiments (Fig. 5) (see reference 75). Because regulation of this temporally distinct class of genes has not been studied, we tested the roles of IME2, a key regulator of middle sporulation gene expression (40), and of UME6, a key regulator of early meiotic gene expression (66), in expression of NDT80. The early meiotic gene HOP1 was expressed in an ime2/ime2 strain after transfer to sporulation medium, although transcript accumulation was delayed and reduced compared to that seen in wild-type cells (Fig. 6B) (19). NDT80 transcripts, however, were not detected in the ime2/ime2 strain (Fig. 6B). Similarly, transcripts from HOP1 were readily detected in RNA from a ume6/ume6 strain growing vegetatively (19), whereas transcripts of NDT80 were not present (Fig. 6C). The findings that Ume6p (Fig. 6C), and Rpd3 and Sin3p (Fig. 6A), were not needed to prevent expression of NDT80 in vegetative cells and that Ime2p was needed for its activation during sporulation suggest that regulation of NDT80 is distinct from the regulation of early meiotic genes.
In summary, we suggest that NDT80 and SMK1 define a class of pre-middle sporulation genes whose transcription is regulated by two mechanisms. An initial low level of IME2-dependent transcription occurs after the activation of the early meiotic genes. Once cells have proceeded through the RAD17-dependent checkpoint, expression of these genes is upregulated.
DISCUSSION
The SPS4 gene of S. cerevisiae belongs to a group of middle sporulation-specific genes that are expressed during the meiotic divisions. A 15-bp regulatory element from the promoter region of SPS4, referred to as UASSPS4, suffices for activation of a heterologous gene midway through sporulation (19). In this study, we identified mutants that had defects in regulation through UASSPS4. The five mutants that we studied in detail defined three complementation groups, referred to as DAS1, DAS3, and DAS4, with similar mutant phenotypes. Cells of the mutant strains completed premeiotic DNA synthesis and expressed early meiotic genes on transfer to sporulation medium but did not complete the meiotic divisions or activate expression of middle sporulation genes. We found that DAS1, DAS3, and DAS4 were identical to the previously characterized genes NDT80, SIN3, and RPD3, respectively.
The roles of these three genes have been well characterized. Mutation of NDT80 has been shown to cause cells to arrest in pachytene in the absence of any defects in meiotic recombination (79). Because the arrest phenotype of an ndt80 mutant is similar to the meiotic phenotype of a cdc28(Ts) mutant (59), Xu and colleagues (79) proposed that NDT80 might encode a regulator of MPF activity. MPF, which consists of Cdc28p kinase and a B-type cyclin as its regulatory partner, controls entry into meiosis I and meiosis II. Indeed, Chu and Herskowitz (10) have recently shown that Ndt80p is a transcriptional activator that induces expression of CLB1, CLB3, and CLB4 during sporulation in addition to activating middle sporulation gene expression.
SIN3 and RPD3 have been identified repeatedly in mutational analyses as modifiers of gene transcription [e.g., as RPD1 (SIN3) and RPD3 (73, 74); as SIN3 (76); as UME4 (SIN3) (65; reviewed in reference 77)]. Recently, it has been shown that Rpd3p, a histone deacetylase, is part of a large multiprotein complex that includes Sin3p (26; reviewed in references 45 and 67). In a mechanism that is conserved among eukaryotes, this complex is recruited to promoters by sequence-specific DNA-binding factors (23). Subsequent histone deacetylation is thought to generate a chromatin structure that represses transcription (24, 54).
Clearly, our screen for regulators of middle sporulation gene expression was not saturating; some genes that might have been identified, such as IME2 and DMC1, were not found. Because expression of the HOP1-lacZ reporter gene is reduced in an ime2 strain (data not shown; see also Fig. 6B), it is possible that any ime2 strain that might have been recovered as a potential das mutant was discarded on the basis of reduced expression of this early meiotic gene. Two of the three genes that we identified, however, were isolated twice. We attribute this bias to the sensitivity of our UASSPS4-lacZ reporter gene. It is likely that strains with mutations that gave a leaky phenotype would have escaped detection. Similarly, mutations in genes that contribute to, but are not essential for, activation of SPS4 would not have been detected.
Pachytene arrest blocks activation of middle sporulation genes.
The observations presented in this study have allowed us to distinguish between two models that we had considered for coordination of the meiotic divisions and middle sporulation gene expression. The first model postulated that entry into the meiotic divisions was required for activation of middle sporulation genes; in this case, expression of middle sporulation genes would depend on activation of MPF. We tested this model by monitoring gene expression in a clb1 clb3 clb4 strain. This strain, which has almost no MPF activity and fails to complete meiosis (13, 18), was proficient in expression of middle sporulation genes (Fig. 4). We therefore conclude that Clb1p-, Clb3p-, and Clb4p-dependent MPF activity and entry into the meiotic divisions is dispensable for activation of middle sporulation genes. Moreover, the observation that CLB1, which encodes the primary meiotic B-type cyclin (13, 18), is expressed as a middle sporulation gene (10) makes it unlikely that MPF would itself serve in activation of middle genes.
The second model postulates that a common signal generated prior to entry into meiosis regulates both commencement of the meiotic divisions and activation of middle sporulation genes. The recovery of NDT80, which is required for the transition from pachytene into meiosis I (79), as a DAS gene was consistent with the notion that entry into meiosis and expression of middle sporulation genes were coordinately regulated. We found that a dmc1 strain, which arrests at pachytene because an accumulation of recombination intermediates activates the meiotic recombination checkpoint (4, 78), failed to express middle sporulation genes. In contrast, a dmc1 rad17 strain, which progresses through the meiotic divisions because of mutation of the checkpoint gene RAD17 (35), did express middle sporulation genes (Fig. 5). For unknown reasons, mature spores do not form in the dmc1 rad17 strain (35; also this study). We propose that the pachytene arrest signal generated by the accumulation of recombination complexes, which prevents entry into the meiotic divisions, also prevents activation of middle sporulation genes.
We note that Clancy and coworkers (11) previously reported that a sporulation-specific glucoamylase activity (SAG) that is encoded by the sporulation gene SGA1 (28, 80) is not expressed in cells that arrest at pachytene. These investigators found that SAG activity was present in a mutant that initiates meiosis I but is blocked in spindle development [spo1(Ts)] and in a mutant that is defective in nuclear migration after meiosis II (spo3(Ts)]. However, SAG activity was not detected in a mutant that arrests at pachytene [pac1(Ts)] nor in a mutant that is defective in premeiotic DNA synthesis [cdc4(Ts)] (11).
Why does mutation of SIN3 or RPD3 confer a meiosis I block?
As mentioned above, SIN3 and RPD3 have been identified repeatedly in mutational analyses as modifiers of gene expression (reviewed in reference 77). Rpd3p, which has histone deacetylase activity, and Sin3p are components of a multiprotein regulatory complex that is recruited to promoters by sequence-specific DNA-binding proteins (26; reviewed in references 45 and 67). Mutation of SIN3 or RPD3 and the resultant histone hyperacetylation could have effects on expression of sporulation genes, on recombination, and on the structure of chromatin and chromosomes. We suggest that a combination of such defects leads to pachytene arrest and that this arrest blocks middle sporulation gene expression. Because neither mutation of SPO11 nor mutation of RAD17 bypassed the arrest caused by mutation of SIN3 or RPD3, we suggest that this arrest is mediated by a mechanism that differs, at least in some aspect, from the meiotic recombination checkpoint (for examples, see references 49 and 71). We note that we found that mutation of SIN3 or RPD3 in some SK1-derived backgrounds led to a delay in meiosis and a reduced efficiency of spore formation rather than a complete block (unpublished observations).
Why does mutation of RPD3 (DAS4) lead to expression of the CYC1-UASSPS4-lacZ reporter gene in vegetative cells?
We identified RPD3 (DAS4) in a screen that took advantage of our previous observation that the CYC1-UASSPS4-lacZ reporter gene, which has four copies of a 29-bp, UASSPS4-containing fragment inserted between the UASCYC1 and the TATACYC1, is not expressed in vegetatively growing cells (19). This 29-bp fragment, which contains UASSPS4 and 14 bp of upstream sequence, promotes 10-fold-higher expression during sporulation than does a 15-bp UASSPS4-containing fragment (19). We hypothesized that a protein that is present in vegetative cells binds to the 29-bp fragment and prevents UASCYC1-bound activators from functioning via an indirect mechanism such as steric hindrance (19). Although the CYC1-UASSPS4-lacZ reporter gene is expressed in rpd3 cells, as well as in sin3 (das3) cells (data not shown), that are growing vegetatively, the SPS4 gene is not expressed in these cells (Fig. 3, SPS4 panel, 0 time point). We speculate, therefore, that the 29-bp fragment contains a sequence that allows Sin3p-Rpd3p-dependent repression to be established in vegetative cells and that this repression, although effective on a heterologous UAS in vegetative cells, is not required for repression of SPS4 itself. Indeed, we have no evidence that the SPS4 gene is subject to negative control in vegetative cells (19). It is possible, however, that the putative DNA-binding protein that contributes to Sin3p-Rpd3p-dependent repression in vegetative cells is also responsible for enhancing Ndt80p-dependent activation of SPS4 during sporulation. We note that this putative DNA-binding protein is not Ume6p, since vegetative repression of the CYC1-UASSPS4-lacZ reporter gene is maintained in a ume6 strain (data not shown). Similarly, as would be expected, vegetative repression of the CYC1-UASSPS4-lacZ reporter gene is maintained in an ndt80 strain (data not shown).
NDT80, a pre-middle sporulation gene.
In the course of our studies, we observed that regulation of expression of NDT80 and SMK1 was distinct from that of middle sporulation genes. In some of our time course experiments, we could readily detect transcripts from NDT80 prior to the appearance of transcripts from SPS4 (for an example, see Fig. 6A). A temporal and regulatory distinction was also apparent on analysis of transcript accumulation in dmc1 and dmc1 rad17 strains (Fig. 5). In the pachytene-arrested dmc1 strain, SPS1 and SPS4 were not expressed but a low level of transcripts could be detected from NDT80 and SMK1 (Fig. 5). In the dmc1 rad17 strain, transcripts from NDT80 and SMK1 could be detected prior to the appearance of transcripts from SPS1 and SPS4 (Fig. 5).
Regulation of NDT80 is clearly distinct from that of early meiotic genes. Whereas repression of early meiotic genes in vegetative cells depends on recruitment of a Sin3p-Rpd3p-containing complex to URS1-bound Ume6p (23), we found that mutation of UME6, SIN3 (DAS3), or RPD3 (DAS4) did not lead to expression of NDT80 in vegetatively growing cells (Fig. 6C and A, NDT80 panel, 0 h time point).
We concluded that NDT80 and SMK1 are members of a temporally distinct class of sporulation-specific genes that is activated after the early meiotic genes but prior to the middle sporulation genes. In this case, the requirement for IME2, an early meiotic gene, for expression of middle sporulation genes could be indirect, reflecting a role of IME2 in expression of NDT80.
Model for coordination of middle sporulation gene expression and the meiotic divisions.
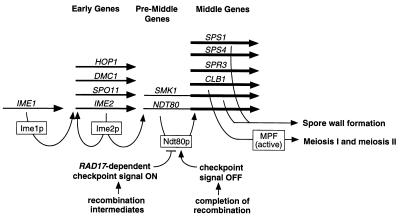
We present the following model for the sequence of events that leads to expression of middle sporulation genes (Fig. 7). We propose that Ime1p-dependent activation of early meiotic genes is followed by Ime2p-dependent activation of pre-middle sporulation genes, such as NDT80 and SMK1. Ime2p, a serine-threonine protein kinase, has been shown to serve initially in IME1-independent activation of early meiotic genes and subsequently in activation of middle sporulation genes (reviewed in reference 39). The manner in which Ime2p carries out these temporally distinct functions is not clear; however, the two roles can be separated genetically (60). We suggest that the primary role of the late function of Ime2p is initial activation of pre-middle sporulation genes, such as NDT80. Because a modest level of NDT80 transcripts accumulated in a dmc1 strain, but middle sporulation genes were not expressed in this strain, we hypothesize that the early NDT80 transcripts are either not translated or, if they are translated, that active Ndt80p is not generated until cells have completed those aspects of meiotic recombination (78) that are monitored by the RAD17-dependent meiotic checkpoint (35). The active form of Ndt80p that we speculate is generated once the cells have progressed through the meiotic recombination checkpoint could result from translation of previously nontranslated transcripts, a posttranslational modification of Ndt80p, interaction with a cofactor, or some other sporulation-specific event. Finally, active Ndt80p would lead to upregulation of expression of pre-middle sporulation genes, including NDT80 itself, and activation of expression of middle sporulation genes, including CLB1 (10). Clb1p would then contribute to activation of MPF and subsequent entry into meiosis I. Hence, the meiotic divisions would be dependent on prior activation of middle sporulation genes, and activation of middle sporulation genes would be dependent on the absence of an arrest signal generated by intermediates of meiotic recombination. In this way, Ndt80p would coordinate entry into meiosis with spore wall formation. This model is consistent with the recent demonstration by Chu and Herskowitz (10) that Ndt80p is an MSE-binding transcriptional activator that activates its own expression. These investigators also noted that the activity of Ndt80p might be regulated by the meiotic recombination checkpoint machinery (10). The proposed autoregulatory loop for NDT80 expression (10) is also consistent with the low level of expression of ndt80 in our das1-1 (ndt80) strain (Fig. 6A). Interestingly, the mei4+ gene of Schizosaccharomyces pombe, which encodes a meiosis-specific transcription factor that is required for meiosis and sporulation-specific gene expression, also appears to be autoregulated (21).
FIG. 7.
Model for the role of Ndt80p in regulation of expression of middle sporulation-specific genes. The long horizontal arrows indicate the relative order of expression of various sporulation-specific genes; curved arrows refer to known or putative regulatory events; boxes indicate gene products. Upon transfer of diploid cells to sporulation medium, Ime1p rapidly activates expression of IME2 and other early meiotic genes. Ime2p has an early role in IME1-independent activation of early meiotic genes and a later role in activation of middle sporulation genes. We propose that the major role of the later function of Ime2p is to promote a low level of expression of pre-middle sporulation genes, such as NDT80. Our results suggest that activation of the RAD17-dependent meiotic recombination checkpoint inhibits the activity of Ndt80p and that an active form of Ndt80p is generated only after cells have completed most prophase events. In particular, meiotic recombination must be completed and any defects in meiotic chromosome metabolism that generate a pachytene arrest signal must be resolved before active Ndt80p is produced. We propose that the active form of Ndt80p is then responsible for upregulation of expression of pre-middle sporulation genes, denoted by a thickening of the horizontal arrows, and for activation of middle sporulation genes, such as SPS1 and SPS4.
Because the signal for pachytene arrest in S. cerevisiae appears to be maintained as long as recombination complexes persist (35, 78), Xu et al. (78, 79) have suggested that the meiotic recombination complex emits an inhibitory signal that prevents entry into meiosis until the recombination process is complete or near completion; it may be that this putative inhibitory signal prevents generation of the proposed active form of Ndt80p (10, 78, 79; also this study). It is interesting to note that meiotic recombination per se is not required to generate an active form of Ndt80p; for example, middle sporulation genes must be expressed in spo11 cells, which enter meiosis I in the absence of recombination.
It will be interesting to investigate whether the meiotic recombination machinery and its checkpoint activities do indeed control the activity of Ndt80p. Future experiments will also determine whether distinct NDT80 promoter elements are required for its initial Ime2p-dependent expression and for its subsequent Ndt80p-dependent upregulation. Finally, investigation of the role of IME2 in the first phase of NDT80 expression may provide insights into how Ime2p serves its key role in regulating spore formation.
ACKNOWLEDGMENTS
We express our appreciation to Shelley Chu and Ira Herskowitz for communicating prior to publication their observation that Ndt80p is an autoregulatory transcriptional activator that binds to MSE elements, and we thank Ira Herskowitz for comments on this manuscript. We thank members of the labs of Brenda Andrews, Doug Bishop, Shelly Esposito, Bruce Futcher, Richard Gaber, Craig Giroux, Nancy Kleckner, Aaron Mitchell, Mike Tyers, Andrew Vershon, and Ted Weinert for providing plasmids and strains; Cheryl Smith of the Flow Cytometry Facility of the Department of Immunology, University of Toronto, for assistance with FACS analysis; and Saul Honigberg, Doug Bishop, and Craig Giroux for helpful discussions. We are very grateful to Chloe Cunningham and Aldo Mistretta for their help in patching cells.
S.R.H. was supported in part by a studentship from the National Sciences and Engineering Research Council (Canada). This work was supported by a Medical Research Council (Canada) grant (MA-6826) to J.S.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 3.Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 4.Bishop D K, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 5.Boeke D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 6.Bowdish K S, Mitchell A P. Bipartite structure of an early meiotic upstream activation sequence from Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2172–2181. doi: 10.1128/mcb.13.4.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowdish K S, Yuan H E, Mitchell A P. Analysis of RIM11, a yeast protein kinase that phosphorylates the meiotic activator IME1. Mol Cell Biol. 1994;14:7909–7919. doi: 10.1128/mcb.14.12.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckingham L E, Wang H-T, Elder R T, McCarroll R M, Slater M R, Esposito R E. Nucleotide sequence and promoter analysis of SPO13, a meiosis-specific gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:9406–9410. doi: 10.1073/pnas.87.23.9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byers B. Cytology of the yeast life cycle. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. pp. 59–96. [Google Scholar]
- 10.Chu S, Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- 11.Clancy M J, Smith L M, Magee P T. Developmental regulation of a sporulation-specific enzyme activity in Saccharomyces cerevisiae. Mol Cell Biol. 1982;2:171–178. doi: 10.1128/mcb.2.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connolly B, White C I, Haber J E. Physical monitoring of mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2342–2349. doi: 10.1128/mcb.8.6.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahmann C, Futcher B. Specialization of B-type cyclins for mitosis or meiosis in S. cerevisiae. Genetics. 1995;140:957–963. doi: 10.1093/genetics/140.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friesen H, Lunz R, Doyle S, Segall J. Mutation of the SPS1-encoded protein kinase of Saccharomyces cerevisiae leads to defects in transcription and morphology during spore formation. Genes Dev. 1994;8:2162–2175. doi: 10.1101/gad.8.18.2162. [DOI] [PubMed] [Google Scholar]
- 15.Gailus-Durner V, Xie J, Chintamaneni C, Vershon A K. Participation of the yeast activator Abf1 in meiosis-specific expression of the HOP1 gene. Mol Cell Biol. 1996;16:2777–2786. doi: 10.1128/mcb.16.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garber A T, Segall J. The SPS4 gene of Saccharomyces cerevisiae encodes a major sporulation-specific mRNA. Mol Cell Biol. 1986;6:4478–4485. doi: 10.1128/mcb.6.12.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grandin N, Reed S I. Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol Cell Biol. 1993;13:2113–2125. doi: 10.1128/mcb.13.4.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hepworth S R, Ebisuzaki L K, Segall J. A 15-base-pair element activates the SPS4 gene midway through sporulation in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:3934–3944. doi: 10.1128/mcb.15.7.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollingsworth N M, Goetsch L, Byers B. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell. 1990;61:73–84. doi: 10.1016/0092-8674(90)90216-2. [DOI] [PubMed] [Google Scholar]
- 21.Horie S, Watanabe Y, Tanaka K, Nishiwaki S, Fujioka H, Abe H, Yamamoto M, Shimoda C. The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol Cell Biol. 1998;18:2118–2129. doi: 10.1128/mcb.18.4.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen R E, Herskowitz I. Directionality and regulation of cassette substitution in yeast. Cold Spring Harbor Symp Quant Biol. 1984;49:97–104. doi: 10.1101/sqb.1984.049.01.013. [DOI] [PubMed] [Google Scholar]
- 23.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 24.Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kao G, Mannix D G, Holaway B L, Finn M C, Bonny A E, Clancy M J. Dependence of inessential late gene expression on early meiotic events in Saccharomyces cerevisiae. Mol Gen Genet. 1989;215:490–500. doi: 10.1007/BF00427048. [DOI] [PubMed] [Google Scholar]
- 26.Kasten M M, Dorlan S, Stillman D J. A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol Cell Biol. 1997;17:4852–4858. doi: 10.1128/mcb.17.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keeney S, Giroux C N, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 28.Kihara K, Nakamura M, Akada R, Yamashita I. Positive and negative elements upstream of the meiosis-specific glucoamylase gene in Saccharomyces cerevisiae. Mol Gen Genet. 1991;226:383–392. doi: 10.1007/BF00260650. [DOI] [PubMed] [Google Scholar]
- 29.Krisak L, Strich R, Winters R S, Hall P, Mallory M J, Dreitzer D, Tuan R S, Winter E. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 1994;8:2151–2161. doi: 10.1101/gad.8.18.2151. [DOI] [PubMed] [Google Scholar]
- 30.Kupiec M, Byers B, Esposito R E, Mitchell A P. Meiosis and sporulation in Saccharomyces cerevisiae. In: Pringle J R, Broach J R, Jones E W, editors. The molecular biology of the yeast Saccharomyces: cell cycle and cell biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 899–1036. [Google Scholar]
- 31.Lamb T, Mitchell A P. Abstracts of the 1996 Yeast Genetics and Molecular Biology Meeting. Bethesda, Md: The Genetics Society of America; 1996. Control of meiotic gene expression by a DNA synthesis checkpoint; p. 156. [Google Scholar]
- 32.Law D T S, Segall J. The SPS100 gene of Saccharomyces cerevisiae is activated late in the sporulation process and contributes to spore wall maturation. Mol Cell Biol. 1988;8:912–922. doi: 10.1128/mcb.8.2.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrence C W. Classical mutagenesis techniques. In: Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. New York, N.Y: Academic Press, Inc.; 1991. pp. 273–281. [Google Scholar]
- 34.Lee R H, Honigberg S M. Nutritional regulation of late meiotic events in Saccharomyces cerevisiae through a pathway distinct from initiation. Mol Cell Biol. 1996;16:3222–3232. doi: 10.1128/mcb.16.6.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lydall D, Nikolsky Y, Bishop D K, Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature (London) 1996;383:840–841. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- 36.Malathi K, Xiao Y, Mitchell A P. Interaction of yeast repressor-activator protein Ume6p with glycogen synthase kinase 3 homolog Rim11p. Mol Cell Biol. 1997;17:7230–7236. doi: 10.1128/mcb.17.12.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKee A H Z, Kleckner N. Mutations in Saccharomyces cerevisiae that block meiotic prophase chromosome metabolism and confer cell cycle arrest at pachytene identify two new meiosis-specific genes SAE1 and SAE2. Genetics. 1997a;146:817–834. doi: 10.1093/genetics/146.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKee A H Z, Kleckner N. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of new gene SAE2. Genetics. 1997b;146:797–816. doi: 10.1093/genetics/146.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell A P. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell A P, Driscoll S E, Smith H E. Positive control of sporulation-specific genes by the IME1 and IME2 products in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2104–2110. doi: 10.1128/mcb.10.5.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgan B A, Conlon F L, Manzanares M, Millar J B A, Kanuga N, Sharpe J, Krumlauf R, Smith J C, Sedgwick S G. Transposon tool for recombinant DNA manipulation: characterization of transcriptional regulators from yeast, Xenopus, and mouse. Proc Natl Acad Sci USA. 1996;93:2801–2806. doi: 10.1073/pnas.93.7.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neiman A M. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J Cell Biol. 1998;140:29–37. doi: 10.1083/jcb.140.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozsarac N, Straffon M J, Dalton H E, Dawes I W. Regulation of gene expression during meiosis in Saccharomyces cerevisiae: SPR3 is controlled by both ABFI and a new sporulation control element. Mol Cell Biol. 1997;17:1152–1159. doi: 10.1128/mcb.17.3.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Page A W, Orr-Weaver T L. Stopping and starting the meiotic cell cycle. Curr Opin Genet Dev. 1997;7:23–31. doi: 10.1016/s0959-437x(97)80105-0. [DOI] [PubMed] [Google Scholar]
- 45.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 46.Percival-Smith A, Segall J. Isolation of DNA sequences preferentially expressed during sporulation in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:142–150. doi: 10.1128/mcb.4.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petko L, Lindquist S. Hsp26 is not required for growth at high temperatures, nor for thermotolerance, spore development, or germination. Cell. 1986;45:885–894. doi: 10.1016/0092-8674(86)90563-5. [DOI] [PubMed] [Google Scholar]
- 48.Prinz S, Amon A, Klein F. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics. 1997;146:781–795. doi: 10.1093/genetics/146.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rose D, Holm C. Meiosis-specific arrest revealed in DNA topoisomerase II mutants. Mol Cell Biol. 1993;13:3445–3455. doi: 10.1128/mcb.13.6.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rose M D, Broach J R. Cloning genes by complementation. In: Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. New York, N.Y: Academic Press, Inc.; 1991. pp. 195–229. [Google Scholar]
- 51.Rothstein R. One-step gene replacement in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 52.Rubin-Berjerano I, Mandel S, Robzyk K, Kassir Y. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol Cell Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rundlett S E, Carmen A A, Suyka N, Turner B M, Grunstein M. Transcription repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 55.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sedgwick S G, Morgan B A. Locating, DNA sequencing, and disrupting yeast genes using tagged Tn1000. Methods Mol Gen. 1994;3:131–140. [Google Scholar]
- 57.Sherman F. Getting started with yeast. In: Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. New York, N.Y: Academic Press, Inc.; 1991. pp. 3–20. [Google Scholar]
- 58.Sherman F, Hicks J. Micromanipulation and dissection of asci. In: Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. New York, N.Y: Academic Press, Inc.; 1991. pp. 21–37. [Google Scholar]
- 59.Shuster E O, Byers B. Pachytene arrest and other meiotic effects of start mutations in Saccharomyces cerevisiae. Genetics. 1989;123:29–43. doi: 10.1093/genetics/123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sia R A L, Mitchell A P. Stimulation of later functions of the yeast meiotic protein kinase Ime2p by the IDS2 gene product. Mol Cell Biol. 1995;15:5279–5287. doi: 10.1128/mcb.15.10.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith H E, Su S S Y, Neigeborn L, Driscoll S E, Mitchell A P. Role of IME1 expression in regulation of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6103–6113. doi: 10.1128/mcb.10.12.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steber C M, Esposito R E. UME6 is a central component of a developmental regulatory switch controlling meiosis-specific gene expression. Proc Natl Acad Sci USA. 1995;92:12490–12494. doi: 10.1073/pnas.92.26.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stillman D J, Dorland S, Yu Y. Epistasis analysis of suppressor mutations that allow HO expression in the absence of the yeast SWI5 transcriptional activator. Genetics. 1994;136:781–788. doi: 10.1093/genetics/136.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strich R, Slater M R, Esposito R E. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc Natl Acad Sci USA. 1989;86:10018–10022. doi: 10.1073/pnas.86.24.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strich R, Surosky R T, Steber C, Dubois E, Messenguy F, Esposito R E. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- 67.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 68.Sym M, Engebrecht J A, Roeder G S. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- 69.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 70.Tishkoff D X, Rockmill B, Roeder G S, Kolodner R D. The sep1 mutant of Saccharomyces cerevisiae arrests in pachytene and is deficient in meiotic recombination. Genetics. 1995;139:495–509. doi: 10.1093/genetics/139.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsui K, Simon L, Norris D. Progression into the first meiotic division is sensitive to histone H2A-H2B dimer concentration in Saccharomyces cerevisiae. Genetics. 1997;145:647–659. doi: 10.1093/genetics/145.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vershon A K, Hollingsworth N M, Johnson A D. Meiotic induction of the yeast HOP1 gene is controlled by positive and negative regulatory sites. Mol Cell Biol. 1992;12:3706–3714. doi: 10.1128/mcb.12.9.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vidal M, Gaber R F. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vidal M, Strich R, Esposito R E, Gaber R F. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol Cell Biol. 1991;11:6306–6316. doi: 10.1128/mcb.11.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wagner M, Pierce M, Winter E. The CDK-activating kinase CAK1 can dosage suppress sporulation defects of smk1 MAP kinase mutants and is required for spore wall morphogenesis in Saccharomyces cerevisiae. EMBO J. 1997;16:1305–1317. doi: 10.1093/emboj/16.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang H, Stillman D J. In vitro regulation of a SIN3-dependent DNA-binding activity by stimulatory and inhibitory factors. Proc Natl Acad Sci USA. 1990;87:9761–9765. doi: 10.1073/pnas.87.24.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolffe A P. Histone deacetylase: a regulator of transcription. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- 78.Xu L, Weiner B M, Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]
- 79.Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamashita I, Fukui S. Transcriptional control of the sporulation-specific glucoamylase gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:3069–3073. doi: 10.1128/mcb.5.11.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]