Abstract
YABBY gene family is a plant-specific transcription factor with DNA binding domain involved in various functions i.e. regulation of style, length of flowers, and polarity development of lateral organs in flowering plants. Computational methods were utilized to identify members of the YABBY gene family, with Carrot (Daucus carota) ‘s genome as a foundational reference. The structure of genes, location of the chromosomes, protein motifs and phylogenetic investigation, syntony and transcriptomic analysis, and miRNA targets were analyzed to unmask the hidden structural and functional characteristics YABBY gene family in Carrots. In the following research, it has been concluded that 11 specific YABBY genes irregularly dispersed on all 9 chromosomes and proteins assembled into five subgroups i.e. AtINO, AtCRC, AtYAB5, AtAFO, and AtYAB2, which were created on the well-known classification of Arabidopsis. The wide ranges of YABBY genes in carrots were dispersed due to segmental duplication, which was detected as prevalent when equated to tandem duplication. Transcriptomic analysis showed that one of the DcYABBY genes was highly expressed during anthocyanin pigmentation in carrot taproots. The cis-regulatory elements (CREs) analysis unveiled elements that particularly respond to light, cell cycle regulation, drought induce ability, ABA hormone, seed, and meristem expression. Furthermore, a relative study among Carrot and Arabidopsis genes of the YABBY family indicated 5 sub-families sharing common characteristics. The comprehensive evaluation of YABBY genes in the genome provides a direction for the cloning and understanding of their functional properties in carrots. Our investigations revealed genome-wide distribution and role of YABBY genes in the carrots with best-fit comparison to Arabidopsis thaliana.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12863-024-01210-4.
Keywords: Bioinformatics, Carrot, Gene family, Genomic analysis, Specific plant transcription factors, YABBY
Introduction
YABBY plant-specific transcription factors (PSTFs) gene family plays an important role in the development of plants i.e. regulation of style length in flowering plants [1] resistance against abiotic stresses [2], polarity development in plant’s lateral organs [3] developmental processes of vegetative and reproductive organs [4], initiating signals responsible for plant hormonal reactions [5] development of vascular organs [6] development of nectary [7] and germination of seed and processes after germination [8, 9]. The DcYABBY genes are members of the YABBY superfamily having functionally important domains i.e., Hmg_box and Hmg_box2. These two domains and the YABBY domain contain highly conserved amino acid residues that function in specific DNA binding [10].
The Carrot (D. carota L.) is a vital biennial vegetable in Apiaceae family. The family Apiaceae also possess several members i.e. Fennel (F. vulgare), celery (A. graveolens), parsley (P. crispum), cilantro (C. sativum) and dill (A. graveolens) [11, 12]. Carrot is a cool-season biennial crop used for domestic, commercial, and medicinal purposes initially and cultivated for over 2000 years. It contains sufficient vitamins and amino acids and helps improve eyesight, lowering cholesterol and improving digestion [13, 14]. Antioxidants like carotenoids & phenolic compounds are found in sufficient amounts in carrot, which are beneficial in several biological processes of the human body [15]. While the amount of carotenoids differs noticeably between different genotypes of carrots, which could be due to the physiological and evolutionary distribution of genomics features [16, 17]. Carrots comprise phenolic components with only one aromatic ring (phenolic acids), 3-O-caffeoylquinic [18]. For new marketable carrot varieties, sweetness was considered a significant factor for acceptance [19]. There is a need to develop highly productive varieties of crops like carrot containing richer nutritional value to enhance the production of healthful foods across the globe [20]. For a balanced, secure, and healthy diet, these foods must be accessible worldwide [21]. Carrot faces several physiological damages due to drought [22, 23]. Therefore, we will also try to find out whether the YABBY transcription factor gene family can solve this problem.
The research aims to discover and describe the genes belonging to the YABBY PSTrFs gene family in the carrot genome using various bioinformatics tools [24]. Concisely, an efficient approach was followed to find the YABBY genes family in carrots. This study unveiled YABBY genes, revealing their chromosomal locations, exon structures, and the presence of cis-regulatory elements, along with conserved domains.. Broad genome-wide assessment of YABBY PSTrF gene family in carrot provides insights to unhide the functional and structural properties which can be used to strengthen the nutritional and food value of other horticulture crops.
Materials and methods
Database search and sequence retrieval
It has been confirmed that the experimental data collection complied with relevant institutional, national, and international guidelines and legislation with appropriate permissions from authorities of the Department of Horticulture, University of the Punjab, Lahore, Lahore 54,300, Pakistan. The amino acid sequences of Plant-Specific Transcription Factors (PSTrFs), specifically YABBY, were obtained from the peptide genome of Arabidopsis thaliana through the Pfam database (Gene ID: PF04690). The YABBY gene’s 164 amino acid sequences were separated from the Arabidopsis thaliana (Accession No. A0A1P8APE2). The following sequences were used in BLAST-P (Basic local protein alignment search tool) for heuristic search against carrot genome using the proteome database at Ensembl plants (https://plants.ensembl.org/index.html) [25–27]. The information on gene IDs, chromosomal position, and sequences of genes and proteins were retrieved. DcYABBY amino acid sequences subjected to motif finder (https://www.genome.jp/tools/motif/) [28, 29] and Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov) National Centre for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov) [30, 31] with customized parameters. The protein sequences that lack in the conserved domain of YABBY proteins were diminished from subjective investigations.
Investigation of physio-chemical characteristics of DcYABBY proteins
The properties of YABBY proteins i.e. length, molecular weight, and theoretical isoelectric point (pI) were predicted using the ProtParam webserver (https://web.expasy.org) [32, 33]. The subcellular localization of the DcYABBY genes was predicted using WoLFPSORT (https://wolfpsort.hgc.jp) [34].
Gene structure analysis
To predict the genomic architecture of carrot YABBY genes, CDS and genomic sequences of DcYABBY genes retrieved from Ensembl plants [26, 27]. These sequences and the Newick format of the carrot phylogenetic tree were subjected to a Gene Structure Display Server (GSDS)(http://gsds.gao-lab.org) [35].
Duplication and syntenic gene analysis
The alignment of protein sequences was conducted using Molecular Evolutionary Genetic Analysis (MEGA) with default parameters. The ratio between the Ka and Ks was predicted using TB tools, and genetic divergence time was calculated using the eq. T = Ks/2r. The “r” signifies a neutral substitution rate (5.2 × 10−9 substitutions per site per year) [36, 37].
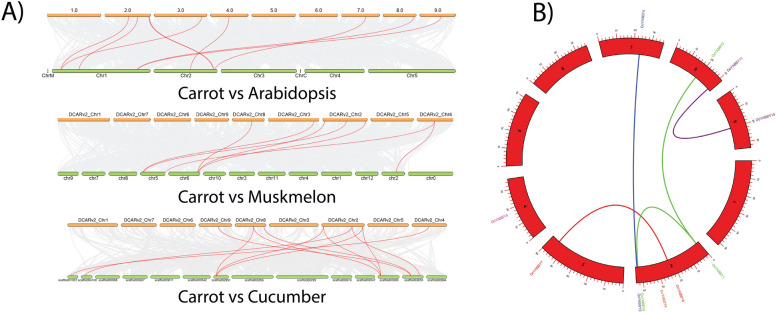
Duplication events of DcYABBY genes were checked with the Multiple Collinearity Scan toolkit (MCScanX) with default settings [38, 39]. Dual synteny analysis of carrot was performed with three crops i.e. Arabidopsis, cucumber, and musk melon. A synteny graph of paralogous of DcYABBY genes was created with circos module using TB tools [40].
Transcriptomic analysis
To check the specific expressions of DcYABBY genes RNA-Seq data was downloaded from NCBI Geo (https://www.ncbi.nlm.nih.gov › geo) [41–43]. A log2 transformation was created to check genes’ expression levels in the Reads per Kilo Base per Million (RPKM) values for different DcYABBY genes. Using TB tool, a heat map was generated to display the expression level of the different genes [40, 44, 45].
Analysis of microRNA target sites
The PmiREN webserver (https://academic.oup.com) was utilized to acquire mature miRNA sequences for carrot species [46, 47]. To identify micro-RNAs targeting DcYABBY genes in carrots, the CDS sequences of DcYABBY genes were inputted into the miRNA and target section of the psRNA Target website (https://bio.tools/psrnatarget). Subsequently, the corresponding complementary miRNAs and their targets were retrieved from this analysis [48, 49].
Results
Identification of the YABBY genes in carrot
In total 22 DcYABBY proteins were identified from proteomic blasts in the carrot genome, and complete domain-possessing sequences were subjected to further investigations. Total 11 sequences of DcYABBY genes were selected for analysis. The range of amino acid length of DcYABBY genes was between 105 and 229 amino acids, while molecular weight was between 12.17 and 25.23 kDa. The DcYABBY8 is the shortest, and DcYABBY1 is the longest protein (Table 1). The pI value of the recognized proteins was extended from 6.82 to 9.16, and it might be due to the increasing number of hydrophobic amino acids. Subcellular localization of these 11 YABBY genes depicted that most of these genes were localized towards the nucleus, including a few to chloroplast and the least in the cytoplasm, as shown in the Fig. 1.
Table 1.
Details of 11 non-redundant YABBY genes identified from the genome of Carrot
| Gene | Accession | Chr | Chromosome Location | Strand | AA | pI | MW |
|---|---|---|---|---|---|---|---|
| DcYABBY1 | DCAR_004921 | 2 | 1,743,987–1,745,265 | + | 229 | 7.71 | 25.08531 |
| DcYABBY2 | DCAR_008543 | 2 | 42,935,336–42,936,404 | – | 227 | 8.13 | 25.1555 |
| DcYABBY3 | DCAR_027801 | 8 | 19,070,569–19,071,054 | + | 228 | 7.71 | 21.29318 |
| DcYABBY4 | DCAR_031517 | 7 | 23,365,804–23,367,132 | + | 192 | 8.62 | 23.66181 |
| DcYABBY5 | DCAR_014892 | 4 | 17,476,581–17,477,177 | + | 209 | 8.99 | 25.3846 |
| DcYABBY6 | DCAR_008464 | 2 | 42,290,491–42,292,134 | + | 231 | 7.71 | 17.73905 |
| DcYABBY7 | DCAR_012254 | 3 | 45,104,893–45,106,507 | – | 155 | 9.33 | 12.17168 |
| DcYABBY8 | DCAR_006190 | 2 | 22,993,021–22,993,857 | – | 105 | 9.22 | 24.39574 |
| DcYABBY9 | DCAR_007074 | 2 | 31,254,793–31,255,348 | + | 219 | 6.82 | 17.87828 |
| DcYABBY10 | DCAR_030050 | 9 | 19,958,066–19,958,530 | + | 166 | 9.16 | 25.23959 |
| DcYABBY11 | DCAR_026683 | 8 | 29,737,553–29,738,146 | – | 229 | 7.71 | 25.23959 |
AA Amino acid: MW Molecular weight: PI Isoelectric point: Chr Chromosome
Fig. 1.
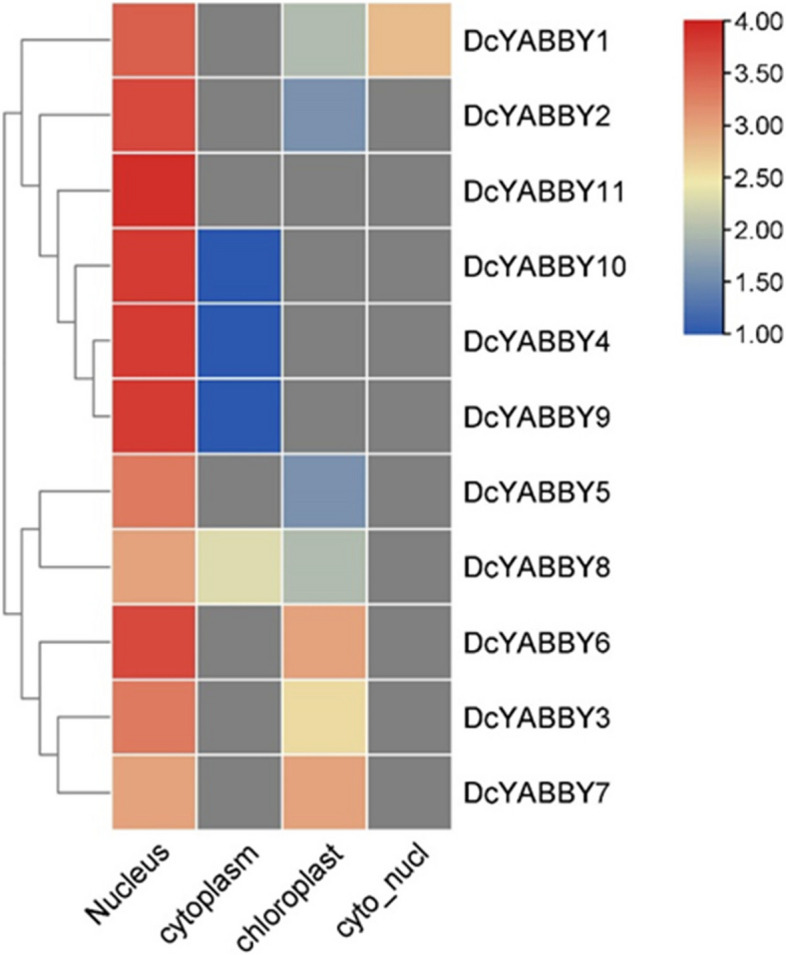
Heat Map representing Sub-cellular localization of all 11 DcYABBY genes to various regions of the plant cell including nucleus, cytoplasm and chloroplast. Grey colour represents absence of respective gene in specific region, white colour is showing minimum functional presence of corresponding gene and Red colour represent maximum value of functionally important gene in that particular region
Gene architecture and conserved motifs analysis
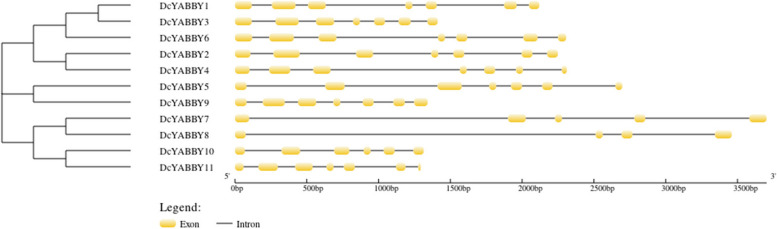
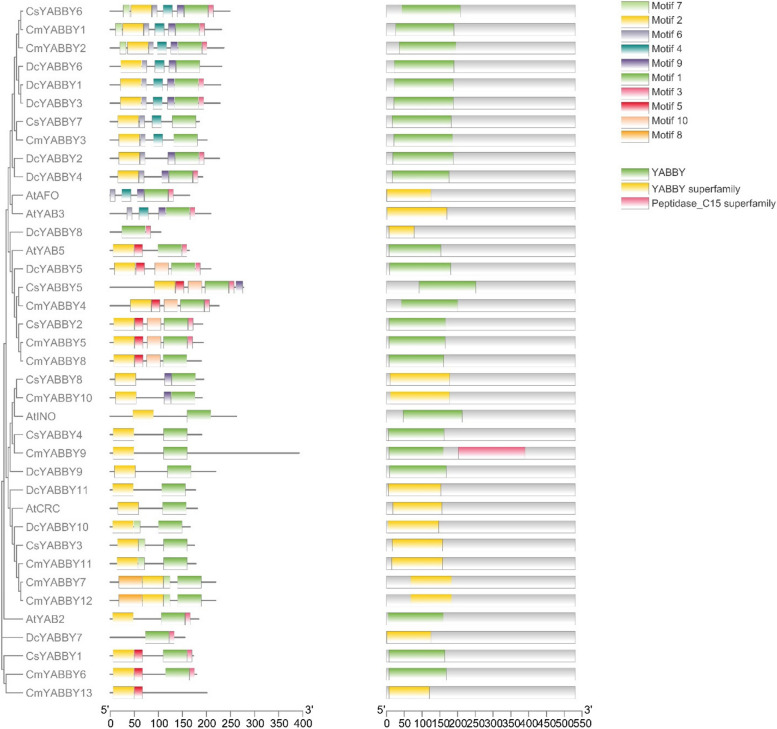
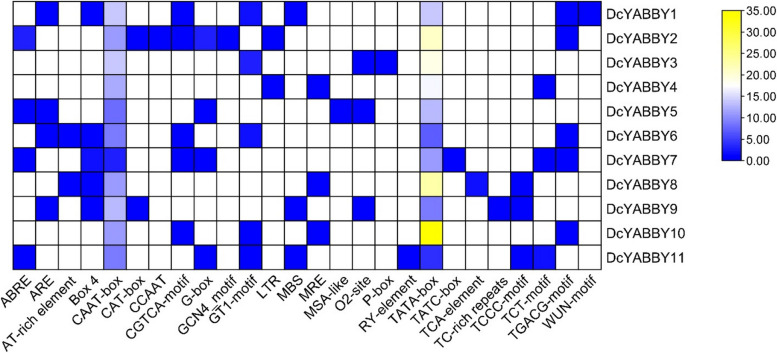
Seven out of eleven genes comprised 7 exons and 6 introns, while two genes contained 6 exons and 5 introns, and one gene comprised 4 exons and 3 introns & the last gene contained 3 exons and 2 introns (Table S5, Fig. 2). The following coincidence and consistency in several introns and exons leads to the clue that these genes share common ancestors and structural and functional features. The genomic architecture showed that DcYABBY8 contained 3 introns (27.27%), DcYABBY7 contains 4 introns (36.36%), and DcYABBY10 have 5 introns (45.45%) while DcYABBY1, DcYABBY2, DcYABBY3, DcYABBY4, DcYABBY5, DcYABBY6, DcYABBY9 and DcYABBY11 contained 6 introns (54.54%) as shown in Fig. 2. There were elucidation and identification of 10 conserved motifs in 11 DcYABBY proteins by the motif identification. The YABBY domain was conserved in all the DcYABBY proteins with several mutations. The motif structure arrangement of the YABBY proteins of Group AtAFO was conserved, and Motif 2, Motif 3, Motif 5, Motif 6, and Motif 14 were structurally conserved. While AtCRC and AtYAB5 have slight variations, AtYAB2 is a much-differentiated family member with eight motifs (Table S3, Fig. 3).
Fig. 2.
The phylogenic representation of intron-exon structure, showing most of large size gene has less number of coding sequences and vice versa. Meanwhile number of introns and exons are conserved throughout the YABBY gene family
Fig. 3.
The distribution of 10 motifs along the 11 YABBY proteins family in carrot. Motifs is conserved throughout the YABBY protein family and are basic structural and functionally important regulator during transient interaction and activation of transcription factors
Phylogenetic analysis
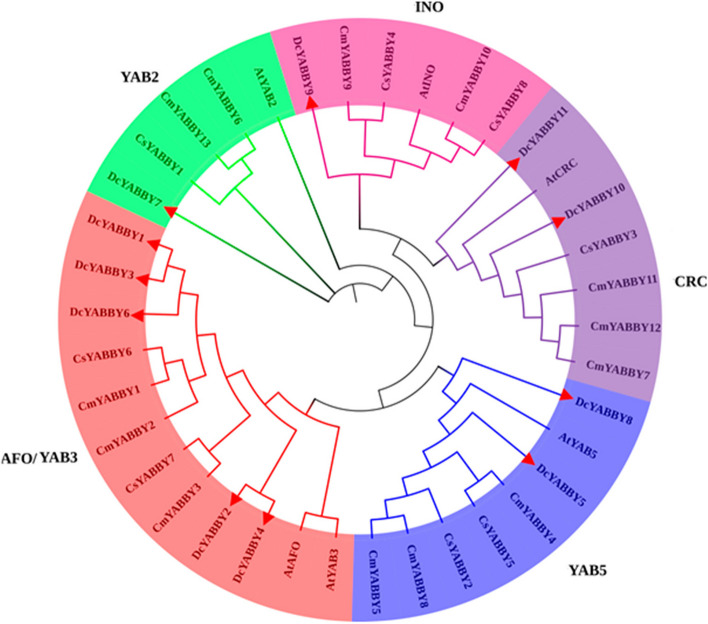
A phylogenetic relationship tree was made among YABBY genes of D. carota, A. thaliana, C. sativus, and C. maxima. D. carota YABBY genes are highlighted with a small red triangle symbol. The figure shows the division of 37 YABBY genes of four different crops. The grouping is based on the typical Arabidopsis phylogenetic grouping system. The results of phylogenetic analysis depicted that 11 DcYABBY proteins were distributed among 5 subgroups named AtINO, AtCRC, AtYAB5, AtAFO/AtYAB3 and AtYAB2 (Fig. 4, Table S4). Group AtINO consists of total 6 YABBY proteins, including 1 from Arabidopsis i.e. AtINO, and the remaining is DcYABBY9, CmYABBY9, CmYABBY10, CsYABBY4, and CsYABBY8. AtCRC group consist of 7 YABBY-like proteins that are AtCRC, DcYABBY10, DcYABBY11, CmYABBY12, CmYABBY11, CmYABBY7 and CsYABBY3. The AtYAB5 group contained 8 YABBY proteins of which 1 is of Arabidopsis AtYAB5, 2 of carrot DcYABBY8, DcYABBY5, 3 of cucumber CmYABBY4, CmYABBY5, CmYABBY8 and 2 of muskmelon CsYABBY5, CsYABBY2. The AtAFO contained 12 YABBY-like proteins in which 2 are of Arabidopsis AtAFO, AtYAB3 while 5 are of carrot, DcYABBY1, DcYABBY2, DcYABBY 3, DcYABBY4, DcYABBY6 and 3 of musk melon CmYABBY1, CmYABBY3, CmYABBY7. The last group AtYAB2, had 5 YABBY-like proteins, 1 of them is of Arabidopsis AtYAB2, 1 from carrot DcYABBY7, 1 of cucumber CsYABBY1, and 2 of musk melon CmYABBY6, CmYABBY13. The members of the same clade represent the same structure and function. Therefore, it has been concluded that sequences of structurally similar proteins have variable and spatio-temporal functional similarity (Fig. 5).
Fig. 4.
Phylogenetic relationship of 37 YABBY genes from four different species i.e. D. carota, A. thaliana, C. sativus, and C. maxima. DcYABBY genes are represented with red triangle
Fig. 5.
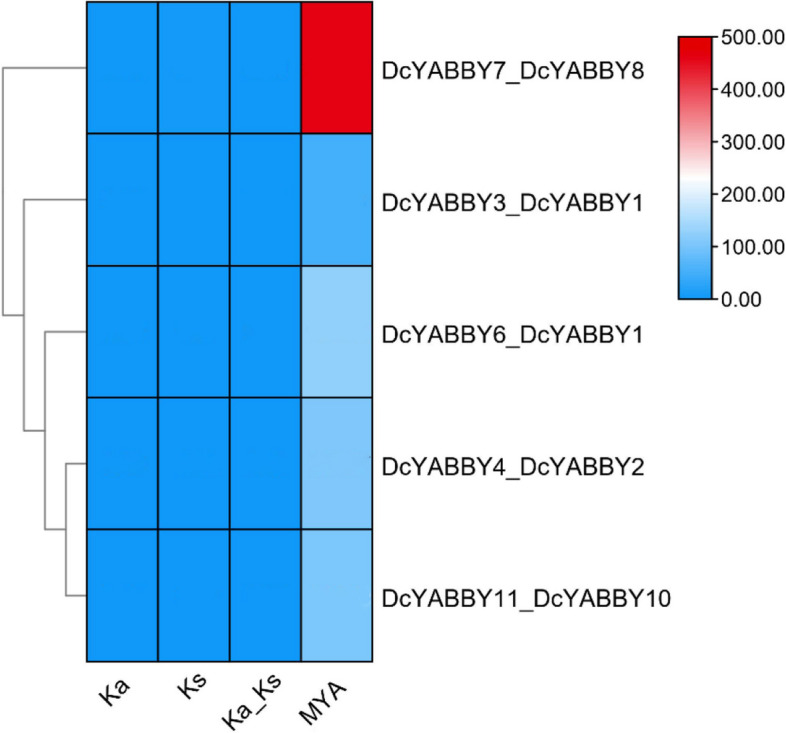
The representation of Ka/Ks the ratio of non-synonymous substitution (Ka) over synonymous substitution (Ks) mutations. The gene duplication over selection and evolutionary pressure to paralogous pairs of potato StYAB genes determined on the basis of Ks and Ka values
Evaluation of gene duplication and gene mapping of carrot YABBY genes
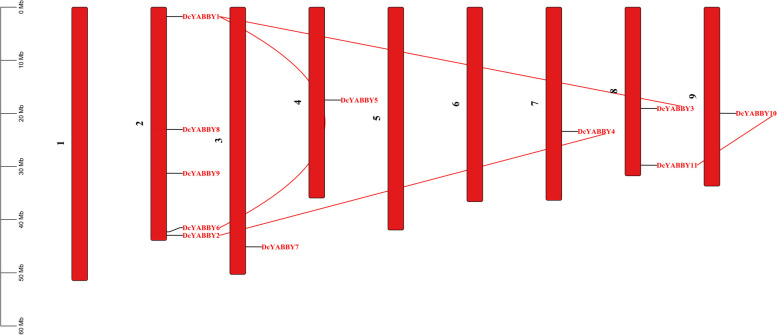
The duplication date of DcYABBY genes was calculated using the TB tool v1.098669 (Fig. 6). The Ka/Ks ratio extended from 0.08888631 in DcYABBY7_DcYABBY8, to 0.1821759 in DcYABBY4_DcYABBY2 pair. The speculative date for segmental duplication date was calculated between 51.0916678 (Mya) for paralogous pair DcYABBY3_DcYABBY1 as highest, to 463.797915 Mya for paralogous pair DcYABBY7_DcYABBY8 as lowest. The Ka/Ks ratios of all the 5 paralogous group pairs were greater than 0.05 and less than 1 ultimatley resulting in a significant divergence during purifying selection period (Fig. 6).
Fig. 6.
Chromosomal mapping showing the paralogues of YABBY genes with putative location. There 11 YABBY genes duplicated during the selection pressure and genomic rearrangement with retaining the ancestral function and gain of stable functional attributes in carrot genome
Analysis of Cis-regulatory elements
Various Cis-regulatory elements with different physiological and biological functions were observed. Many of these include light-responsive elements, specific responsive elements to abscisc acid, salicylic acid and gibberellins, anaerobic induction, meristem expression, seed-specific regulation, zien metabolism, and some defensive regulatory elements (Fig. 7, Table 2). Mainly, ARE element was present in 4 out of 11 DcYABBY genes that have a function in anaerobic induction, 5 DcYABBY genes contained box 4 element, which is a fragment of a conserved DNA module that helps in response to light, that takes part in light responsiveness, ABRE element was contained by 4 DcYABBY genes which have function related to abscisic acid response, 5 DcYABBY genes possessed TGACG element which is a sensitive element to methyl jasmonic acid, 1 DcYABBY gene possessed TCA element linked to respond with salicylic acid, 1 DcYABBY gene displayed wound-responsive WUN motif. While one DcYABBY gene indicated TC-rich repeats that have activity related to stress and defence, 2 DcYABBY genes contained CAT-box, which responds to meristemic expression, and MBS element was possessed by 3 DcYABBY genes which respond to drought induce ability, 2 DcYABBY genes have LTR element which is linked to respond in low-temperature, 1 DcYABBY gene have RY-element which is mainly associated with seed regulation. On the contrary, 1 DcYABBY gene contained GCN4_motif, which takes account of endosperm expression, and 2 DcYABBY genes possess AT-rich elements involved in DNA binding protein ATBP-1.
Fig. 7.
A Dual synteny analysis of Carrot-Arabidopsis, Carrot-Cucumber, and Carrot-Musk melon were performed to unmask the similarity and gene duplication distribution. Genomic regions of afore mentioned plants species have been shown with fine gene duplication and structural sharing among them. B Genome-wide synteny analysis of carrot DcYABBY genes showing paralogous gene pairs in the carrot genome
Table 2.
The spatio-temporal functional distribution of YABBY gene’s Cis-regulatory elements among various tissues and organs during plant biological development process
| Sr # | Cis-Elements | Function | References |
|---|---|---|---|
| 1 | ABRE | cis-acting element involved in the abscisc acid responsiveness | [50] |
| 2 | ARE | cis-acting regulatory element essential for the anaerobic induction | [51] |
| 3 | AT-rich element | binding site of AT-rich DNA binding protein (ATBP-1) | [51] |
| 4 | Box 4 | part of a conserved DNA module involved in light responsiveness | [52] |
| 5 | CAAT-box | common cis-acting element in promoter and enhancer regions | [53] |
| 6 | CAT-box | cis-acting regulatory element related to meristem expression | [54] |
| 7 | CCAAT-box | MYBHv1 binding site | [55] |
| 8 | CGTCA-motif | cis-acting regulatory element involved in the MeJA-responsiveness | [56] |
| 9 | G-box | cis-acting regulatory element involved in light responsiveness | [57] |
| 10 | GT1-motif | light responsive element | [58] |
| 11 | GCN4_motif | cis-regulatory element involved in endosperm expression | [59] |
| 12 | LTR | cis-acting element involved in low-temperature responsiveness | [60] |
| 13 | MBS | MYB binding site involved in drought inducibility | [61] |
| 14 | MRE | MYB binding site involved in light responsiveness | [62] |
| 15 | MSA-like | cis-acting element involved in cell cycle regulation | [63] |
| 16 | O2-site | cis-acting regulatory element involved in zein metabolism regulation | [64] |
| 17 | P-box | gibberellin-responsive element | [65] |
| 18 | RY-element | cis-acting regulatory element involved in seed-specific regulation | [66] |
| 19 | TATA-box | core promoter element around − 30 of transcription start | [67] |
| 20 | TATC-box | cis-acting element involved in gibberellin-responsiveness | [68] |
| 21 | TCA-element | cis-acting element involved in salicylic acid responsiveness | [64] |
| 22 | TC-rich repeats | cis-acting element involved in defense and stress responsiveness | [64] |
| 23 | TCCC-motif | part of a light responsive element | [53] |
| 24 | TCT-motif | part of a light responsive element | [53] |
| 25 | TGACG-motif | cis-acting regulatory element involved in the MeJA-responsiveness | [53] |
The MSA-like element was expressed by 1 DcYABBY gene, which regulates the cell cycle. 1 DcYABBY gene contained CCAT, which is a common binding site for MYBHv1 while 5 DcYABBY genes showed CGTCA-motif that is also involved in methyl jasmonic acid responsiveness, 4 DcYABBY genes have G-box that helps in responding to light, 5 DcYABBY genes contained GT1-motif and 3 DcYABBY genes possessed MRE both of which are light-responsive element, O2-site was possessed by 3 DcYABBY genes which have a very important role in zien metabolism, p-box TATC box and TCA element are only contained by 1 DcYABBY gene and first two are gibberellin responsive elements and the last is salicylic acid responding element. TCCC-motif, TCT, and TGACG-motif contain 3 and 5 DcYABBY genes with varying functions (Figs. 7 and 8).
Fig. 8.
The graphical representation of Cis-regulatory elements of DcYABBY genes with intensity to their function at various levels via each gene’s promoter region. The functional intensity can be defined with red to blue colours from higher to low level during biochemical and physiological plant development respectively
The physiological and biochemical functions with their orthologues in Arabidopsis of DcYABBY genes were studied with the help of gene ontology study (Table 3).
Table 3.
DcYABBY genes have physiological and biochemical functions with their orthologues in Arabidopsis
| Source ID | Gene ID | Gene ontology | A. thaliana Orthologues | Function | Reference |
|---|---|---|---|---|---|
| DCAR_004921 | DcYABBY1 | GO:0007275, GO:0032502 | YABBY3, (AT4G00180) | Transcription cis-regulatory region & DNA-binding transcription factor activity, Ligand binding domain | [69] |
| DCAR_008543 | DcYABBY2 | GO:0007275, GO:0009909, GO:0009933, GO:0009944, GO:0010093, GO:0010154, GO:0010158, GO:0010450, GO:0032502, GO:0045165, GO:0090706, GO:1902183, | YABBY1, (AT2G45190) | Transcription cis-regulatory region & DNA-binding transcription factor activity, Ligand binding domain | [70] |
| DCAR_027801 | DcYABBY3 | GO:0007275, GO:0032502 | YABBY3, (AT4G00180) | Transcription cis-regulatory region binding, DNA-binding transcription factor activity, Ligand binding domain | [71] |
| DCAR_031517 | DcYABBY4 | GO:0007275, GO:0032502 | YABBY1, (AT2G45190) | Transcription cis-regulatory region & DNA-binding transcription factor activity, Ligand binding domain | [72] |
| DCAR_014892 | DcYABBY5 | GO:0007275, GO:0009944, GO:0032502, GO:1902183, GO:2000024 | YABBY5, (AT2G26580) | Gene regulation | [73] |
| DCAR_008464 | DcYABBY6 | GO:0007275, GO:0032502 | YABBY3, (AT4G00180) | Transcription cis-regulatory region & DNA-binding transcription factor activity, Ligand binding domain | [73] |
| DCAR_012254 | DcYABBY7 | GO:0007275, GO:0032502 | YABBY2, (AT1G08465) | Transcription cis-regulatory region binding, DNA-binding transcription factor activity, Ligand binding domain | [74] |
| DCAR_006190 | DcYABBY8 | GO:0007275, GO:0032502 | YABBY2, (AT1G08465) | Transcription cis-regulatory region binding, DNA-binding transcription factor activity, Ligand binding domain | [75] |
| DCAR_007074 | DcYABBY9 | GO:0007275, GO:0032502 | INO, (AT1G23420) | DNA-binding transcription factor activity, Ligand binding domain | [76] |
| DCAR_030050 | DcYABBY10 | GO:0007275, GO:0032502 | YABBY1, (AT2G45190) | Transcription cis-regulatory region binding, DNA-binding transcription factor activity, Ligand binding domain | [77] |
| DCAR_026683 | DcYABBY11 | GO:0007275, GO:0010254, GO:0010582, GO:0032502, GO:0048440, GO:0048479 | CRC, (AT1G69180) | Transcription cis-regulatory region binding, DNA-binding transcription factor activity, Ligand binding domain | [78] |
Transcriptomic analysis of carrot YABBY genes
Regarding gene expression among all the 11 DcYABBY genes, only 1 has been involved in anthocyanin pigmentation in the carrot taproots. DcYABBY9 (DCAR_007074) was expressed in dP2 POP and dP2 NPIP (Fig. 8). The extent of gene expression was slightly varied among these replicates. So it was concluded that DcYABBY9 helps build a dark purple color in the outer phloem of carrot taproot by influencing more anthocyanin pigmentation [41, 42] (Fig. 9).
Fig. 9.
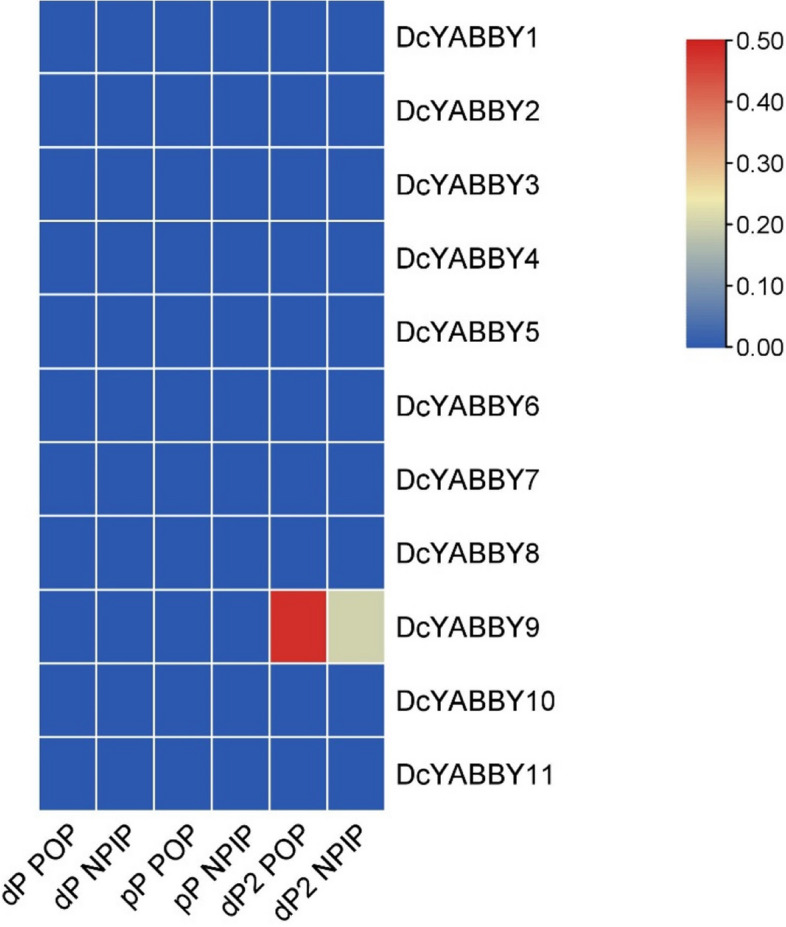
The Heat map of carrot YABBY genes responsible for pigmentation of anthocyanin are represented with higher intensity to low with red to blue colors. dP POP (Dark Purple Outer Phelom), dP NPIP (Dark Purple Inner), pP POP (Pale Purple Outer Phelom) and pP NPIP (Pale Purple Inner Phelom)
Putative miRNA targets in carrot
Consequently, 5 miRNAs target the three genes i.e. DcYABBY2, DcYABBY3 and DcYABBY5 of the total 11 DcYABBY genes. DcYABBY 2 is the gene targeted by 3 mature miRNAs with different PmiREN IDs. On the other hand, DcYABBY 3 and 5 were targeted by 1 of the same mature miRNA (Table 4). None of the mature miRNAs targeted the remaining 8 DcYABBY genes. So, this indicated that DcYABBY 2 was the individual gene targeted by the maximum number of mature miRNAs. While discussing based on groups, AtAFO was targeted by 4 mature miRNAs. In contrast, the minimum number of miRNA targeted groups was AtYAB5, which was targeted by only 1 miRNA (Table 5).
Table 4.
Representation of miRNAs with their targeting genes, length, starts and aligned sequence details
| miRNA ID | Target ID | Length | Start-end | miRNA aligned fragment |
|---|---|---|---|---|
| Dca-MIR408a | DcYABBY5 | 21 | 1–21 | UGCACUGCCUCUUCCCUGGCU |
| Dca-MIR168a | DcYABBY2 | 22 | 1–22 | UCGCUUGGUGCAGGUCGGGACC |
| Dca-MIR168b | DcYABBY2 | 22 | 1–22 | UCGCUUGGUGCAGGUCGGGACC |
| Dca-MIR168c | DcYABBY2 | 22 | 1–22 | UCGCUUGGUGCAGGUCGGGACC |
| Dca-MIR408a | DcYABBY3 | 21 | 1–21 | UGCACUGCCUCUUCCCUGGCU |
Table 5.
Functions of miRNAs and their role in gene regulation during the developmental stages
| miRNA ID | Target gene ID | Function | Reference |
|---|---|---|---|
| Dca-MIR408a | DcYABBY5 | Peptide chain release factor, Plantacyanin, Heat Regulation | [79, 80] |
| Dca-MIR168a | DcYABBY2 | Regulates AGO1 for gene silencing, Response to Bacterial Infection | [79, 81] |
| Dca-MIR168b | DcYABBY2 | Regulates AGO1 for gene silencing, Response to Bacterial Infection | [79, 81] |
| Dca-MIR168c | DcYABBY2 | Regulates AGO1 for gene silencing, Response to Bacterial Infection | [79, 81] |
| Dca-MIR408a | DcYABBY3 | Peptide chain release factor, Plantacyanin, Heat Regulation | [79, 80] |
Discussion
Plant specific Transcription factors (PSTrFs) are important molecules with spatio-temporal function and support during plant development and growth. PSTrFs are key in defining the fate of strong biological development and biochemical actions22. YABBY genes in carrots and other species act as TrFs and provide basic support during the developmental cycle. Phylogenetic and conserved sequences analysis of YABBY TrFs in Arabidopsis thaliana and eggplant of span into five families, including AtINO, AtCRC, AtYAB5, AtAFO/AtYAB3, AtYAB2. The genomic identification of DcYABBY genes has been completed by comparing recently released genomic features from comprehensive plant repository Ensembl plants [26, 27], [82] (Table 1). Phylogenic findings characterize 11 YABBY genes of A. thaliana into five groups AtINO, AtCRC, AtYAB5, AtAFO/AtYAB3, and AtYAB2 (Fig. 5, Table S4). The following distribution leads to new insights into less sequence-level conservation for YABBY carrot genes. The number of YABBY TFs in the carrot is less than other domestic and model plant i.e. rice possesses 30 OsYABBY, Arabidopsis; 36 AtYABBY, tomato; 34 SiYABBY [83] banana; 74 MaYABBY and [84] Chinese cabbage; 76 BrATYABBY [85, 86].
The less correlated number of introns and exons in these families depicts the purifying selection and evolutionary instability with divergent evolution. Higher introns in the plant genome provide information regarding its evolutionary and genomic stability. The genomic architecture and correlation in phylogeny depicted a clear picture of evolutionary correlation among various YABBY gene families [87, 88].
The genomic feature of similar characters possessing genes had the same number of introns and exons at genomic level (Table S2). Same clades of DcYABBY have an almost similar number of exons and introns (Fig. 2) while various clades of different families have different number of introns and exons i.e. Arabidopsis, rice and soybean, suggesting conservation of characteristic sequences among them [89, 90].
The conservation of sequence to function level has been assessed by identification of motif (Fig. 3) sequence among all DcYABBY genes at protein level spanning from 15 to 167 bp (Table S3) amino acids along with frequently existing HMG box domain (Table S5, S9). All members of DcYABBY proteins comprised Motif 1 and motifs named Hmg_box and Hmg_box2 are also residing, and at a functional level, HMG box is responsible for binding the DNA. The sequence-level investigations correspond to similarities at the sequence level, leading to functional and structural correlation. The preservation of evolutionary traits leads to the rearrangement and structuring of domains while maintaining consistent functionality. Confirming these functional similarities, gene ontology (GO) annotation of AtAFO genes in Arabidopsis thaliana has been undertaken. Evolutionary gene expansion might cause arrangements of the YABBY domains to have similar motif patterns in different groups. To recognize the possible function of the Group AtAFO, which contained five DcYABBY genes and several similar motifs, GO annotations of the Group AtAFO genes in Arabidopsis resulting in similarities among DcYABBY genes and AtAFO with transcriptional functions, cis-regulatory region binding, DNA-binding, protein binding and ion channelling (Table 3) [91, 92]. The structural arrangement of the DcYABBY genes was conserved among all the five divided groups of species i.e. Arabidopsis, Cucumber, and Musk melon [93]. Furthermore, an investigation of subcellular localization among DcYABBY proteins using the online web tool WoLF PSORT [34] has been performed and resulted in nuclear localization of DcYABBY proteins to cytoplasm and chloroplast while these all were commonly present in the nucleus (Table S1). Segmental and tandem duplication was observed in the YABBY gene family at various chromosomes, which is a clear picture of genomic rearrangements during the evolutionary process. These rearrangements at the genome level lead to the development of new characters, i.e., conservative sequences and domains for sustaining the functional characteristics of plants [94]. The best-known tandem and segmental duplication in carrot YABBY genes on chromosome 2 (Fig. 7A) and DcYABBY1 with DcYABBY3, DcYABBY2 with DcYABBY4 and DcYABBY10 with DcYABBY11 (Fig. 7B) have been found in this research. Segmental duplications are dominant in chickpea [93] pigeon pea [15, 92], and in the YABBY gene family. These results indicate the main process of gene and conserved region expansion at the genomic level due to duplications of YABBY genes throughout the evolution of eukaryotic plants [95, 96]. The purifying and evolutionary selection at amino acid level and substitution ratio i.e. Ka and Ks (Fig. 5) support these findings that YABBY genes have evolved and retain their function through evolution. Ka/Ks < 1 ratio leads to purifying selection, and positive selection pressure leads to Ka/Ks > 1 values. This selection pressure by the biological clock and environment leads to the rearrangement of specific blocks and domains at the level, resulting in the origination of new characteristics across the species [97]. In current investigations, variation among ratios of Ka /Ks between DcYABBY genes is less and predicted values of Ka/Ks ranges from 0.09 to 0.29 which are less than 1. The aforementioned results showing that sequences of YABBY in all families underwent purifying selection pressure and can only affect few sites during the process of evolution (Fig. 5). The expression profile of DcYABBY genes in several carrot experiments using available RNA sequencing data was analysed, resulting in the conclusion that anthocyanin can accumulate in purple-rooted carrots. Genomic diversity indicates anthocyanin expression either in taproot and tissue specificity confined to phloem’s root or xylem’s tissues. Insilco information and computation i.e. linkage mapping and transcriptomic analysis have been used to assess the hidden facts about anthocyanin pigmentation in inner and outer phloem of carrot taproots in two different genomic backgrounds [41, 42]. Cluster analysis and gene omnibus at NCBI were used to unhide the spatio temporal function of carrot YABBY genes and 1 out of 11 DcYABBY genes involved in anthocyanin pigmentation in the carrot taproot [41, 42]. DcYABBY9 (DCAR_007074) was highly expressed in dP2 POP and dP2 NPIP (Fig. 9) [98]. Except for DcYABBY9, all other genes have no expression or function related to anthocyanin pigmentation. The cis-regulatory analysis also predicts that DcYABBY9 also has a role in light responsiveness, zein metabolism regulation, regulatory function related to meristem expression, involved in drought-induce ability, essential for the anaerobic metabolism during abiotic stress and defence responsiveness (Fig. 8, Table 2). The orthologue of DcYABBY9 in Carrot is AT1G23420 and AtINO, which are in the same group and have a role in DNA and metal ion binding. The orthologues of these three aforementioned Arabidopsis proteins are DcYABBY 10, 11 and 12 which can lead to conclude their similar functions in the Carrot plant as of its orthologues in Arabidopsis.
MicroRNAs are important in plant growth regulation processes extending from developmental to defending against pathogens and sustaining internal immunity [99–102]. MiRNAs are present in most plant species in a conserved manner with specified functions. Most of the DcYABBY genes have transcriptional-associated functions, resulting in the suppression of activity to miRNAs. It is the only reason that three out of 11 DcYABBY genes were targeted by MIR408 and MIR168 family members (Table 5). MIR408 targeted two DcYABBY genes while MIR168 to one gene. These two micro RNAs targeted DcYABBY2, DcYABBY3, and DcYABBY5, respectively. DcYABBY2 was targeted by three miRNAs i.e. MIR168a and MIR168b, which reside on chromosome 1 and MIR168c at chromosome 9 of carrot. Meanwhile DcYABBY3 and DcYABBY5 were both targeted by MIR408a located on chromosome 1. This scenario provides a basis for the conclusion that most of their origin and activity are driven by chromosome 1. MiR408 is abundantly present in different plant species that specifically hits mRNAs related to copper-binding protein. Overexpression of MIR408 was shown to improve phenotypic properties of Arabidopsis by increasing leaf area, plant height, petiole length, flower size, and silique length, which ultimately enhances seed yield and biomass [103]. MiR408 has diverse roles in Arabidopsis, from which we can assume that this micro RNA targeting DcYABBY genes can also play an important role in enriching carrot nutrients. Overexpression of miR408 triggered enhanced drought tolerance in chickpeas by causing plantacyanin transcript suppression, which regulates DREB and other genes related to drought response [104]. In response to miR168, Argonaute (AGO1) is upregulated, activating the RNA silencing complex (RISC) in tomatoes to modulate the small RNA regulatory pathway [105]. The suppression of miR168 by a target mimic (MIM168) not only improves grain yield and shortens rice flowering time but enhances immunity to Magnaporthe oryzae, the causal agent of rice blast disease.
Conclusion
This study comprehensively analyzed DcYABBY PSTrFs genes in the carrot genome. The 11 DcYABBY genes were classified into five groups, and some of the structural and functional properties of each DcYABBY member were characterized. Some of the DcYABBY genes were involved in taproot pigmentation. MiRNA data targeting the DcYABBY gene in anthocyanin pigmentation development in carrot suggest their role in growth and development. The in-depth computational analysis of carrot YABBY proteins revealed in the current study is the first step to undermining the hidden realities of YABBY proteins in carrots and in contrast to other crops. Complex interaction and cooperation at the functional level of YABBY proteins portray their expression level and interaction with different transcription factors. The presence of an almost similar number of YABBY genes i.e. 33 in (tomato), 34 (pepper), and 35 (potato), and a relatively higher number in other plants 78 in soybean and 51 in carrot suggested the variation in YABBY genes at a genomic, structural and functional level.
Supplementary Information
Acknowledgments
The authors extend their appreciation to Researchers Supporting Project Number (RSP2024R165), King Saud University, Riyadh, Saudi Arabia.
Authors’ contributions
MH and MMJ carried out the research and wrote the initial draft of the manuscript. AS, MS, QA, HSUDM, JT, and MAJ carried out the data analysis and provided the resources for research. QA, MAJ, IAS, MZH, MH and DA carried out the final editing of the manuscript. All authors read and approved the final publication of the manuscript.
Funding
No funding was obtained for this study.
Availability of data and materials
The datasets analyzed during the current study are available in the Ensemble plants database (https://plants.ensembl.org/index.html), NCBI-GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181611), and miRBase database (https://mirbase.org/) repository. All data collected or generated has ben provided in manuscript and its supplementary file.
Carrot YABBY genes source accession numbers along with their repository web links.
Carrot YABBY genes source accession numbers along with their repository web links.
| miRNA ID | Target ID | miRNA Repository Web links |
| Dca-MIR408a | DcYABBY5 | https://pmiren.com/singlemirna?Accession=PmiREN034271 |
| Dca-MIR168a | DcYABBY2 | https://pmiren.com/singlemirna?Accession=PmiREN034231 |
| Dca-MIR168b | DcYABBY2 | https://pmiren.com/singlemirna?Accession=PmiREN034232 |
| Dca-MIR168c | DcYABBY2 | https://pmiren.com/singlemirna?Accession=PmiREN034233 |
| Dca-MIR408a | DcYABBY3 | https://pmiren.com/singlemirna?Accession=PmiREN034271 |
Declarations
Ethics approval and consent to participate
Not applicable. All authors have read, understood, and have complied as applicable with the statement on “Ethical responsibilities of authors” as found in the Instructions for Authors and are aware that with minor exceptions, no changes can be made to authorship once the paper is submitted.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mujahid Hussain and Muhammad Mubashar Javed contributed equally to this work.
References
- 1.Ding B, Li J, Gurung V, Lin Q, Sun X, Yuan YW. The leaf polarity factors SGS3 and YABBYs regulate style elongation through auxin signaling in Mimulus lewisii. New Phytol. 2021;232(5):2191–2206. doi: 10.1111/nph.17702. [DOI] [PubMed] [Google Scholar]
- 2.Hussain Q, Asim M, Zhang R, Khan R, Farooq S, Wu J. Transcription factors interact with ABA through gene expression and signaling pathways to mitigate drought and salinity stress. Biomolecules. 2021;11(8):1159. doi: 10.3390/biom11081159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saito K, Takamatsu Y. Periodic breathing in patients with stable obstructive sleep apnea on long-term continuous positive airway pressure treatment: a retrospective study using CPAP remote monitoring data. Sleep and Breathing. 2022;26(3):1181–91. [DOI] [PMC free article] [PubMed]
- 4.Romanova MA, Maksimova AI, Pawlowski K, Voitsekhovskaja OV. YABBY genes in the development and evolution of land plants. Int J Mol Sci. 2021;22(8):4139. doi: 10.3390/ijms22084139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X, Tan C, Cheng X, Zhao X, Li T, Jiang J. miR168 targets Argonaute1A mediated miRNAs regulation pathways in response to potassium deficiency stress in tomato. BMC Plant Biol. 2020;20(1):1–17. doi: 10.1186/s12870-020-02660-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nurani AM, Ozawa Y, Furuya T, Sakamoto Y, Ebine K, Matsunaga S, Ueda T, Fukuda H, Kondo Y. Deep imaging analysis in VISUAL reveals the role of YABBY genes in vascular stem cell fate determination. Plant Cell Physiol. 2020;61(2):255–264. doi: 10.1093/pcp/pcaa002. [DOI] [PubMed] [Google Scholar]
- 7.Phukela B, Geeta R, Das S, Tandon R. Ancestral segmental duplication in Solanaceae is responsible for the origin of CRCa–CRCb paralogues in the family. Mol Gen Genomics. 2020;295(3):563–577. doi: 10.1007/s00438-019-01641-0. [DOI] [PubMed] [Google Scholar]
- 8.Qu C, Zuo Z, Cao L, Huang J, Sun X, Zhang P, Yang C, Li L, Xu Z, Liu G. Comprehensive dissection of transcript and metabolite shifts during seed germination and post-germination stages in poplar. BMC Plant Biol. 2019;19(1):1–15. doi: 10.1186/s12870-019-1862-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sami A, Haider MZ, Shafiq M, Sadiq S, Ahmad F. Genome-Wide Identification and In-silico Expression Analysis of CCO Gene Family in Sunflower (Helianthus annnus). Plant Molecular Boilogy. 2023. [DOI] [PMC free article] [PubMed]
- 10.Li Z, Li G, Cai M, Priyadarshani SV, Aslam M, Zhou Q, Huang X, Wang X, Liu Y, Qin Y. Genome-wide analysis of the YABBY transcription factor family in pineapple and functional identification of AcYABBY4 involvement in salt stress. Int J Mol Sci. 2019;20(23):5863. doi: 10.3390/ijms20235863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaughan J, Geissler C. The new Oxford book of food plants. OUP Oxford; 2009.
- 12.Simpson K, Cerda A, Stange C. Carotenoid Biosynthesis in Daucus carota. In: Carotenoids in Nature: Biosynthesis, Regulation and Function. Edited by Stange C. Cham: Springer International Publishing; 2016. p. 199–217. [DOI] [PubMed]
- 13.Bystrická J, Kavalcová P, Musilová J, Vollmannová A, Tomáš T, LENKOVÁ M: Carrot (Daucus carota L. ssp. sativus (Hoffm.) Arcang.) as source of antioxidants. Acta agriculturae Slovenica 2015, 105(2):303–311.
- 14.Kaack K, Nielsen M, Christensen LP, Thorup-Kristensen K. Nutritionally important chemical constituents and yield of carrot (Daucus carota L.) roots grown organically using ten levels of green manure. Acta Agriculturae Scandinavica, section B-plant. Soil Sci. 2001;51(3):125–136. [Google Scholar]
- 15.Islam MAU, Nupur JA, Shafiq M, Ali Q, Sami A, Shahid MA. In silico and computational analysis of zinc finger motif-associated homeodomain (ZF-HD) family genes in chilli (Capsicum annuum L) BMC Genomics. 2023;24(1):603. doi: 10.1186/s12864-023-09682-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baranski R, Allender C, Klimek-Chodacka M. Towards better tasting and more nutritious carrots: carotenoid and sugar content variation in carrot genetic resources. Food Res Int. 2012;47(2):182–187. doi: 10.1016/j.foodres.2011.05.006. [DOI] [Google Scholar]
- 17.Shafiq M, Manzoor M, Bilal M, Manzoor T, Anees MM, Rizwan M, Haider MZ, Sami A, Haider MS. Genome-wide analysis of plant specific YABBY transcription factor gene family in watermelon (Citrullus lanatus) and Arabidopsis. J Appl Res Plant Sci. 2024;5(01):63–78. [Google Scholar]
- 18.Arscott SA, Tanumihardjo SA. Carrots of many colors provide basic nutrition and bioavailable phytochemicals acting as a functional food. Compr Rev Food Sci Food Saf. 2010;9(2):223–239. doi: 10.1111/j.1541-4337.2009.00103.x. [DOI] [Google Scholar]
- 19.Nookaraju A, Upadhyaya CP, Pandey SK, Young KE, Hong SJ, Park SK, Park SW. Molecular approaches for enhancing sweetness in fruits and vegetables. Sci Hortic. 2010;127(1):1–15. doi: 10.1016/j.scienta.2010.09.014. [DOI] [Google Scholar]
- 20.Mushtaq S, Shafiq M, Tariq MR, Sami A, Nawaz-ul-Rehman MS, Bhatti MHT, Haider MS, Sadiq S, Abbas MT, Hussain M. Interaction between bacterial endophytes and host plants. Front Plant Sci. 2023;13:1092105. doi: 10.3389/fpls.2022.1092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.da Silva Dias JC. Impact of improved vegetable cultivars in overcoming food insecurity. Euphytica. 2010;176(1):125–136. doi: 10.1007/s10681-010-0237-5. [DOI] [Google Scholar]
- 22.Razzaq M, Akram NA, Ashraf M, Naz H. Al-Qurainy F: interactive effect of drought and nitrogen on growth, some key physiological attributes and oxidative defense system in carrot (Daucus carota L.) plants. Sci Hortic. 2017;225:373–379. doi: 10.1016/j.scienta.2017.06.055. [DOI] [Google Scholar]
- 23.Haider MZ, Sami A, Shafiq M, Anwar W, Ali S, Ali Q, et al. Genome-wide identification and in-silico expression analysis of carotenoid cleavage oxygenases gene family in Oryza sativa (rice) in response to abiotic stress. Front Plant Sci. 2023:14. [DOI] [PMC free article] [PubMed]
- 24.Irfan U, Haider M, Shafiq M, Sami A, Ali Q. GENOME EDITING FOR EARLY AND LATE FLOWERING IN PLANTS. Bullet Biolog All Sci Res. 2023;2023(1):45–45. doi: 10.54112/bbasr.v2023i1.45. [DOI] [Google Scholar]
- 25.BlastN G. BLAST: basic local alignment search tool. NUTRITIONAL AND PHYSIOLOGICAL DISORDERS IN HORTICULTURAL CROPS. 2019.
- 26.Bolser D, Staines DM, Pritchard E, Kersey P. Ensembl plants: integrating tools for visualizing, mining, and analyzing plant genomics data. Plant bioinformatics Springer; 2016. pp. 115–140. [DOI] [PubMed] [Google Scholar]
- 27.Bolser DM, Staines DM, Perry E, Kersey PJ. Ensembl plants: integrating tools for visualizing, mining, and analyzing plant genomic data. Plant genomics databases Springer; 2017. pp. 1–31. [DOI] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30(1):42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Almas MH, Shah RA, Tahir SMH, Manzoor M, Shafiq M, Shah MH, Hashmi MM, Ali M, Bhatti MHT, Sami A. The effect of substrate, growth condition and nutrient application methods in morphological and commercial attributes of hybrid rose (Rosa indica L.) cv. Kard J Appl Res Plant Sci. 2023;4(01):356–362. doi: 10.38211/joarps.2023.04.01.44. [DOI] [Google Scholar]
- 30.Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48(D1):D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Almas M, Sami A, Shafiq M. BHATTI M, HAIDER M, Hashmi M, KHALID M: Sale price comparison of saggian flower market: a case study. Bullet Biolog All Sci Res. 2023;2023(1):39–39. doi: 10.54112/bbasr.v2023i1.39. [DOI] [Google Scholar]
- 32.Gasteiger E, Hoogland C, Gattiker A, Duvaud Se, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: The Proteomics Protocols Handbook. Edited by Walker JM. Totowa, NJ: Humana Press; 2005. p. 571–607.
- 33.Meeran M, Sami A, Haider M, Umar M. MULTIVARIATE ANALYSIS FOR MORPHOLOGICAL TRAITS OF AMARANTHUS VIRIDIS. Bullet Biolog All Sci Res. 2023;2023(1):46–46. doi: 10.54112/bbasr.v2023i1.46. [DOI] [Google Scholar]
- 34.Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier C, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35(suppl_2):W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pei Q, Li N, Yang Q, Wu T, Feng S, Feng X, et al. Genome-wide identification and comparative analysis of ARF family genes in three Apiaceae species. Front Genet. 1653;2021 [DOI] [PMC free article] [PubMed]
- 37.Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Li J, Paterson AH. MCScanX-transposed: detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics. 2013;29(11):1458–1460. doi: 10.1093/bioinformatics/btt150. [DOI] [PubMed] [Google Scholar]
- 39.Manzoor M, Hamza A, Javaid A, Anees M, Tariq MR, Firdosi MFH, Intisar A, Sami A, Haider MZ, Haider MS. Bioefficacy of some botanical extracts against Brinjal fruit and shoot borer Leucinodes orbonalis (Guenee); Lepidoptera. Pyrallidae Plant Protection. 2023;7(2):263–272. doi: 10.33804/pp.007.02.4728. [DOI] [Google Scholar]
- 40.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 41.Bannoud F, Bellini C. Adventitious rooting in Populus species: update and perspectives. Front Plant Sci. 2021;12:668837. [DOI] [PMC free article] [PubMed]
- 42.Bannoud F, Carvajal S, Ellison S, Senalik D, Gomez Talquenca S, Iorizzo M, Simon PW, Cavagnaro PF. Genetic and transcription profile analysis of tissue-specific anthocyanin pigmentation in carrot root phloem. Genes. 2021;12(10):1464. doi: 10.3390/genes12101464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clough E, Barrett T. The gene expression omnibus database. Statistical genomics Springer; 2016. pp. 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sami A, Haider M, Iqbal M, Bhatti M, Ahmad S, Khalid M. Deterrence effect of colored diversion sheets on the population density of melon fruit flies bactrocera cucurbitae (coquillett) and yield parameters of bitter gourd (momordica charantia L.) Biolog Agricult Sci Res J. 2023;2023(1):17–17. [Google Scholar]
- 45.Sami A, Haider M, Meeran M, Ali M, Abbas A, Ali Q, Umar M. Exploring morphological traits variation in chenopodium murale: a comprehensive multivariate analysis. Bullet Biolog All Sci Res. 2023;2023(1):43–43. doi: 10.54112/bbasr.v2023i1.43. [DOI] [Google Scholar]
- 46.Guo Z, Kuang Z, Zhao Y, Deng Y, He H, Wan M, Tao Y, Wang D, Wei J, Li L. PmiREN2. 0: from data annotation to functional exploration of plant microRNAs. Nucleic Acids Res. 2022;50(D1):D1475–D1482. doi: 10.1093/nar/gkab811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo Z, Kuang Z, Wang Y, Zhao Y, Tao Y, Cheng C, Yang J, Lu X, Hao C, Wang T. PmiREN: a comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res. 2020;48(D1):D1114–D1121. doi: 10.1093/nar/gkz894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release) Nucleic Acids Res. 2018;46(W1):W49–W54. doi: 10.1093/nar/gky316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai X. Zhao PX: psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39(suppl_2):W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaplan B, Davydov O, Knight H, Galon Y, Knight MR, Fluhr R, Fromm H. Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+−responsive cis elements in Arabidopsis. Plant Cell. 2006;18(10):2733–2748. doi: 10.1105/tpc.106.042713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rieu P, Turchi L, Thévenon E, Zarkadas E, Nanao M, Chahtane H, et al. The F-box protein UFO controls flower development by redirecting the master transcription factor LEAFY to new cis-elements. Nature Plants. 2023:1–15. [DOI] [PubMed]
- 52.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24(4):369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 53.Tatematsu K, Ward S, Leyser O, Kamiya Y, Nambara E. Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol. 2005;138(2):757–766. doi: 10.1104/pp.104.057984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.TANIGUCHI T: Regulation of interferon-β gene: structure and function of cis-elements and trans-acting factors. J Interf Res. 1989, 9(6):633–640. [DOI] [PubMed]
- 55.Kumar GM, Mamidala P, Podile AR. Regulation of Polygalacturonase-inhibitory proteins in plants is highly dependent on stress and light responsive elements. Plant Omics. 2009;2(6):238–249. [Google Scholar]
- 56.Heidari P, Ahmadizadeh M, Najafi-Zarrini H. In silico analysis of Cis-regulatory elements on co-expressed genes. J Biol Environ Sci. 2015;9(25)
- 57.Wingender R, Röhrig H, Höricke C, Schell J. cis-regulatory elements involved in ultraviolet light regulation and plant defense. Plant Cell. 1990;2(10):1019–1026. doi: 10.1105/tpc.2.10.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li D, Zaman W, Lu J, Niu Q, Zhang X, Ayaz A, Saqib S, Yang B, Zhang J, Zhao H. Natural lupeol level variation among castor accessions and the upregulation of lupeol synthesis in response to light. Ind Crop Prod. 2023;192:116090. doi: 10.1016/j.indcrop.2022.116090. [DOI] [Google Scholar]
- 59.Éva C, Moncsek B, Szőke-Pázsi K, Kunos V, Mészáros K, Makai S, Sági L, Juhász A. bZIP transcription factors repress the expression of wheat (Triticum aestivum L.) high molecular weight glutenin subunit genes in vegetative tissues. Acta Physiol Plant. 2023;45(2):1–14. doi: 10.1007/s11738-022-03503-6. [DOI] [Google Scholar]
- 60.Liu Q, Sui X, Wang Y, Zhu M, Zhou Y, Gao F. Genome-wide analyses of Thaumatin-like protein family genes reveal the involvement in the response to low-temperature stress in Ammopiptanthus nanus. Int J Mol Sci. 2023;24(3):2209. doi: 10.3390/ijms24032209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun J, Xu J, Qu W, Han X, Qiu C, Gai Z, Zhai J, Qin R, Liu H, Wu Z. Genome-wide analysis of R2R3-MYB transcription factors reveals their differential responses to drought stress and ABA treatment in desert poplar (Populus euphratica) Gene. 2023;855:147124. doi: 10.1016/j.gene.2022.147124. [DOI] [PubMed] [Google Scholar]
- 62.Weraduwage SM, Sahu A, Kulke M, Vermaas JV, Sharkey TD. Characterization of promoter elements of isoprene-responsive genes and the ability of isoprene to bind START domain transcription factors. Plant Direct. 2023;7(2):e483. doi: 10.1002/pld3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li X, Huang H, Zhang Z-Q. Characterization and expression analysis of bHLH transcription factors reveal their putative regulatory effects on nectar spur development in Aquilegia species. Gene. 2023;852:147057. doi: 10.1016/j.gene.2022.147057. [DOI] [PubMed] [Google Scholar]
- 64.Yu J, Song B, Gu K, Cao B, Zhao K, Wu J, Li J. Genome-wide identification and expression analysis of CAMTA gene family implies PbrCAMTA2 involved in fruit softening in pear. Horticulturae. 2023;9(4):467. doi: 10.3390/horticulturae9040467. [DOI] [Google Scholar]
- 65.Du G, Zhao Y, Xiao C, Ren D, Ding Y, Xu J, Jin H, Jiao H. Mechanism analysis of calcium nitrate application to induce gibberellin biosynthesis and signal transduction promoting stem elongation of Dendrobium officinale. Ind Crop Prod. 2023;195:116495. doi: 10.1016/j.indcrop.2023.116495. [DOI] [Google Scholar]
- 66.Utsugi S, Kawahigashi H, Tagiri A, Kikuchi R, Mishina K, Morishige H, Nakamura S. Conserved cis-acting motifs and localization of transcripts and proteins of MFT2 in barley and rice. bioRxiv. 2023:2023.2001. 2017.524348.
- 67.Ma Z-W, Zhao J-P, Tian J, Zheng C-H. DeeProPre: a promoter predictor based on deep learning. Comput Biol Chem. 2022;101:107770. doi: 10.1016/j.compbiolchem.2022.107770. [DOI] [PubMed] [Google Scholar]
- 68.Bari MA, El-Shehawi AM, Elseehy MM, Naheen NN, Rahman MM, Kabir AH. Molecular characterization and bioinformatics analysis of transporter genes associated with cd-induced phytotoxicity in rice (Oryza sativa L.) Plant Physiol Biochem. 2021;167:438–448. doi: 10.1016/j.plaphy.2021.08.024. [DOI] [PubMed] [Google Scholar]
- 69.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fukuda M, Nishida S, Kakei Y, Shimada Y, Fujiwara T. Genome-wide analysis of long intergenic noncoding RNAs responding to low-nutrient conditions in Arabidopsis thaliana: possible involvement of trans-acting siRNA3 in response to low nitrogen. Plant Cell Physiol. 2019;60(9):1961–1973. doi: 10.1093/pcp/pcz048. [DOI] [PubMed] [Google Scholar]
- 71.Hajheidari M. Huang S-sC: elucidating the biology of transcription factor–DNA interaction for accurate identification of cis-regulatory elements. Curr Opin Plant Biol. 2022;68:102232. doi: 10.1016/j.pbi.2022.102232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Q, Li W, Liu XS, Carroll JS, Jänne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27(3):380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dhatterwal P, Basu S, Mehrotra S, Mehrotra R. Genome wide analysis of W-box element in Arabidopsis thaliana reveals TGAC motif with genes down regulated by heat and salinity. Sci Rep. 2019;9(1):1681. doi: 10.1038/s41598-019-38757-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dey U, Sarkar S, Teronpi V, Yella VR, Kumar A. G-quadruplex motifs are functionally conserved in cis-regulatory regions of pathogenic bacteria: an in-silico evaluation. Biochimie. 2021;184:40–51. doi: 10.1016/j.biochi.2021.01.017. [DOI] [PubMed] [Google Scholar]
- 75.Tripathi RK, Aguirre JA, Singh J. Genome-wide analysis of wall associated kinase (WAK) gene family in barley. Genomics. 2021;113(1):523–530. doi: 10.1016/j.ygeno.2020.09.045. [DOI] [PubMed] [Google Scholar]
- 76.Cheng X, Li M, Abdullah M, Li G, Zhang J, Manzoor MA, Wang H, Jin Q, Jiang T, Cai Y. In silico genome-wide analysis of the pear (Pyrus bretschneideri) KNOX family and the functional characterization of PbKNOX1, an Arabidopsis BREVIPEDICELLUS orthologue gene, involved in cell wall and lignin biosynthesis. Front Genet. 2019;10:632. doi: 10.3389/fgene.2019.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schilling S, Kennedy A, Pan S, Jermiin LS, Melzer R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020;225(1):511–529. doi: 10.1111/nph.16122. [DOI] [PubMed] [Google Scholar]
- 78.Huang W, He Y, Yang L, Lu C, Zhu Y, Sun C, Ma D, Yin J. Genome-wide analysis of growth-regulating factors (GRFs) in Triticum aestivum. PeerJ. 2021;9:e10701. doi: 10.7717/peerj.10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.López-Urrutia E, Bustamante Montes LP. Ladrón de Guevara Cervantes D, Pérez-Plasencia C, Campos-Parra AD: crosstalk between long non-coding RNAs, micro-RNAs and mRNAs: deciphering molecular mechanisms of master regulators in cancer. Front Oncol. 2019;9:669. doi: 10.3389/fonc.2019.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sunkar R, Zhu J-K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16(8):2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu H, Ren G, Chen H, Liu Q, Yang Y, Zhao Q. Predicting lncRNA–miRNA interactions based on logistic matrix factorization with neighborhood regularized. Knowl-Based Syst. 2020;191:105261. doi: 10.1016/j.knosys.2019.105261. [DOI] [Google Scholar]
- 82.AHMAD B, MAHMOOD A, SAMI A, HAIDER m: food choices, clothing patterns and interpersonal relations: effects of social media on YOUTH’S lifestyle. Biolog Agricult Sci Res J. 2023, 2023(1):23–23.
- 83.Cai X, Zhang Y, Zhang C, Zhang T, Hu T, Ye J, Zhang J, Wang T, Li H, Ye Z. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J Integr Plant Biol. 2013;55(6):552–566. doi: 10.1111/jipb.12043. [DOI] [PubMed] [Google Scholar]
- 84.Dong C, Hu H, Xie J. Genome-wide analysis of the DNA-binding with one zinc finger (Dof) transcription factor family in bananas. Genome. 2016;59(12):1085–1100. doi: 10.1139/gen-2016-0081. [DOI] [PubMed] [Google Scholar]
- 85.Ma J, Li M-Y, Wang F, Tang J, Xiong A-S. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics. 2015;16(1):1–15. doi: 10.1186/s12864-015-1242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hussain Z, Muzamil M, Saeed M, Naheed K, Kareem M, Munir A, Haider M. Sami a: trait correlations and implications for yield potential in cotton: a comprehensive study. Biolog Agricult Sci Res J. 2023;2023(1):24–24. [Google Scholar]
- 87.Bondarenko VS, Gelfand MS. Evolution of the exon-intron structure in ciliate genomes. PLoS One. 2016;11(9):e0161476. doi: 10.1371/journal.pone.0161476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haider M, Sami A, Mazhar H, Akram J. NISA B, Umar M, Meeran M: exploring morphological traits variation in Gomphrena globosa: a multivariate analysis. Biolog Agricult Sci Res J. 2023;2023(1):21–21. [Google Scholar]
- 89.Gu X, Zou Y, Su Z, Huang W, Zhou Z, Arendsee Z, Zeng Y. An update of DIVERGE software for functional divergence analysis of protein family. Mol Biol Evol. 2013;30(7):1713–1719. doi: 10.1093/molbev/mst069. [DOI] [PubMed] [Google Scholar]
- 90.Sami A, Haider M, Imran M, Abbas A, Javed M. Synergizing food safety, quality and genetic improvement: the intersection of food microbiology and processing. Bullet Biolog All Sci Res. 2023;2023(1):44–44. doi: 10.54112/bbasr.v2023i1.44. [DOI] [Google Scholar]
- 91.Gupta S, Malviya N, Kushwaha H, Nasim J, Bisht NC, Singh V, Yadav D. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta. 2015;241(3):549–562. doi: 10.1007/s00425-014-2239-3. [DOI] [PubMed] [Google Scholar]
- 92.Malviya N, Gupta S, Singh V, Yadav M, Bisht N, Sarangi B, Yadav D. Genome wide in silico characterization of Dof gene families of pigeonpea (Cajanus cajan (L) Millsp.) Mol Biol Rep. 2015;42(2):535–552. doi: 10.1007/s11033-014-3797-y. [DOI] [PubMed] [Google Scholar]
- 93.Nasim J, Malviya N, Kumar R, Yadav D. Genome-wide bioinformatics analysis of Dof transcription factor gene family of chickpea and its comparative phylogenetic assessment with Arabidopsis and rice. Plant Syst Evol. 2016;302(8):1009–1026. doi: 10.1007/s00606-016-1314-6. [DOI] [Google Scholar]
- 94.Panchy N, Lehti-Shiu M, Shiu S-H. Evolution of gene duplication in plants. Plant Physiol. 2016;171(4):2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Taylor JS, Raes J. Duplication AND divergence: the evolution. Annu Rev Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- 96.Moore RC, Purugganan MD. The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol. 2005;8(2):122–128. doi: 10.1016/j.pbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 97.Morgan CC, Loughran NB, Walsh TA, Harrison AJ, O'Connell MJ. Positive selection neighboring functionally essential sites and disease-implicated regions of mammalian reproductive proteins. BMC Evol Biol. 2010;10(1):1–17. doi: 10.1186/1471-2148-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bülow L, Hehl R. Bioinformatic identification of conserved cis-sequences in coregulated genes. Plant Synthetic Promoters Springer; 2016. pp. 233–245. [DOI] [PubMed] [Google Scholar]
- 99.Carbone F, Bruno L, Perrotta G, Bitonti MB, Muzzalupo I, Chiappetta A. Identification of miRNAs involved in fruit ripening by deep sequencing of Olea europaea L. transcriptome. PLoS One. 2019;14(8):e0221460. doi: 10.1371/journal.pone.0221460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Samad AF, Sajad M, Nazaruddin N, Fauzi IA, Murad AM, Zainal Z, Ismail I. MicroRNA and transcription factor: key players in plant regulatory network. Front Plant Sci. 2017;8:565. doi: 10.3389/fpls.2017.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Spanudakis E, Jackson S. The role of microRNAs in the control of flowering time. J Exp Bot. 2014;65(2):365–380. doi: 10.1093/jxb/ert453. [DOI] [PubMed] [Google Scholar]
- 102.Terzi L, Simpson G. Regulation of flowering time by RNA processing. Nuclear pre-mRNA Processing in Plants. 2008:201–18. [DOI] [PubMed]
- 103.Song Z, Zhang L, Wang Y, Li H, Li S, Zhao H, Zhang H. Constitutive expression of miR408 improves biomass and seed yield in Arabidopsis. Front Plant Sci. 2018;8:2114. doi: 10.3389/fpls.2017.02114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hajyzadeh M, Turktas M, Khawar KM, Unver T. miR408 overexpression causes increased drought tolerance in chickpea. Gene. 2015;555(2):186–193. doi: 10.1016/j.gene.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 105.Sami A, Saeed M, Shafiq M, Abbas SM, Anum A, Haider H, Bhatti MHT, Raza MA, Khan N, Shahid NA. Role of horticulture in disaster risk management. Disaster Risk Reduction in Agriculture Springer; 2023. pp. 393–406. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available in the Ensemble plants database (https://plants.ensembl.org/index.html), NCBI-GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181611), and miRBase database (https://mirbase.org/) repository. All data collected or generated has ben provided in manuscript and its supplementary file.
Carrot YABBY genes source accession numbers along with their repository web links.
Carrot YABBY genes source accession numbers along with their repository web links.
| miRNA ID | Target ID | miRNA Repository Web links |
| Dca-MIR408a | DcYABBY5 | https://pmiren.com/singlemirna?Accession=PmiREN034271 |
| Dca-MIR168a | DcYABBY2 | https://pmiren.com/singlemirna?Accession=PmiREN034231 |
| Dca-MIR168b | DcYABBY2 | https://pmiren.com/singlemirna?Accession=PmiREN034232 |
| Dca-MIR168c | DcYABBY2 | https://pmiren.com/singlemirna?Accession=PmiREN034233 |
| Dca-MIR408a | DcYABBY3 | https://pmiren.com/singlemirna?Accession=PmiREN034271 |