LETTER
The rapid spread of the coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection highlights an urgent need for developing antivirals against coronavirus. An attractive drug target for COVID-19 is the main protease (Mpro), as this conserved enzyme plays a key role in virus replication and host immune evasion (1, 2). Given the urgency of identifying antivirals, repurposing approved drugs as effective Mpro inhibitors represent a promising strategy for COVID-19 treatment (3, 4). Recently, manidipine was identified as a potent Mpro inhibitor with the half-maximal inhibitory concentration (IC50) of 10.4 ± 1.6 µM using fluorescence resonance energy transfer (FRET) assay (5). Nonetheless, a recent study suggested that manidipine is likely to be a promiscuous Mpro inhibitor because it is prone to colloidal aggregation in the enzymatic assay, resulting in the inactivation of Mpro enzyme (6). Considering the potential of manidipine in COVID-19 treatment, a rigorous validation for its Mpro inhibition is necessary.
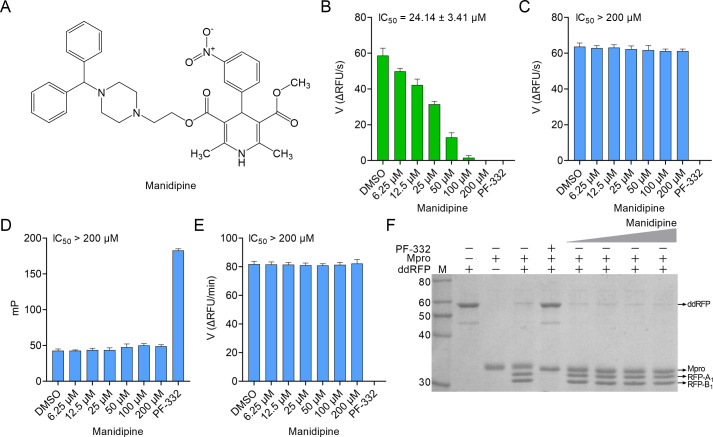
For this purpose, we evaluated the in vitro inhibition of Mpro by manidipine using a robust high-throughput screening (HTS) platform, including FRET, fluorescence polarization (FP), and dimerization-dependent red fluorescent protein (ddRFP) assays (7–9). When we repeated the FRET assay developed by the authors (5), the IC50 value of manidipine was 24.14 ± 3.41 µM (Fig. 1A and B). In clear contrast, manidipine showed no inhibition in the FRET assay when Tween-20 was present in the reaction buffer (IC50 > 200 µM), suggesting that its Mpro inhibition is false positive (Fig. 1C). To further test this possibility, we separately evaluated Mpro inhibition by manidipine using FP and ddRFP assays in the reaction buffer containing 0.05% Tween-20. As expected, manidipine did not inhibit Mpro in both assays (IC50 > 200 µM) (Fig. 1D and E). Consistently, manidipine exhibited no detectable inhibition of Mpro using SDS-PAGE because the ddRFP biosensor could be cleaved into RFP-A1 and RFP-B1 fragments in the presence of manidipine (Fig. 1F). Thus, our results invalidated manidipine as a potential Mpro inhibitor in vitro. Intriguingly, a recent study independently confirmed our results using cell-based assays (10). In the presence of detergent, the aggregation caused by compounds is minimized in the enzymatic assay (11). Therefore, our results demonstrated the necessity for the addition of detergent such as Tween-20 or Triton X-100 into the assay buffer when assessing Mpro inhibitors, which could disrupt colloids and keep Mpro enzyme stable. Moreover, manidipine might have other targets to achieve the anti-SARS-CoV-2 effect, which can be further studied.
Fig 1.
Inhibition of SARS-CoV-2 main protease (Mpro) by manidipine in vitro. (A) The chemical structure of manidipine. (B) Evaluation of the activity of manidipine against Mpro using FRET assay developed by authors (5). (C and D) Evaluation of the activity of manidipine against Mpro using FRET and FP assays in the reaction buffer containing 0.05% Tween-20. In these biochemical assays, nirmatrelvir (PF-332, 1 µM) and DMSO served as the positive and negative controls, respectively. The IC50 value of manidipine was shown. (E) Inhibition of Mpro by manidipine through monitoring the initial velocity of Mpro enzyme reaction initiated by ddRFP biosensor. The IC50 value of manidipine was shown. (F) Gel-based assay of ddRFP biosensor cleavage by manidipine in vitro. In the absence of Mpro inhibitors, the ddRFP biosensor (55 kDa) can be cleaved by Mpro (34 kDa) to generate RFP-A1 (top band, 29 kDa) and RFP-B1 fragments (bottom band, 26 kDa) in the gels. Nirmatrelvir (PF-332) and manidipine were purchased from TargetMol (Shanghai, China). The concentrations of manidipine used in this experiment were 25, 50, 100, or 200 µM. In the ddRFP assay, nirmatrelvir (PF-332, 10 µM) and DMSO served as the positive and negative controls, respectively. All assays were conducted as previously described (7–9, 12). The reaction buffer was 10 mM HEPES, 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), and 0.05% Tween-20 (vol/vol), pH7.0.
In summary, our data indicate that manidipine is not a potential inhibitor against Mpro based on the results from a set of in vitro assays. These results suggest that the colloidal aggregation caused by compounds should be considered when evaluating Mpro inhibitors. It is necessary to verify the inhibitory activity of candidate Mpro inhibitors using diverse biochemical assays due to the limitations of the FRET approach.
ACKNOWLEDGMENTS
This work was supported by University Natural Science Research Project of Anhui Province, China (No. KJ2021A0839); The Young Fellow Program of Wannan Medical College, China (No. wyqnyx202104); and Postgraduate Academic Innovation Program of Anhui Province, China (No. 2022xscx129).
The authors thank Prof. Yanchang Wang (Department of Biomedical Sciences, College of Medicine, Florida State University, Tallahassee, USA) for his insightful comments of the manuscript.
Contributor Information
Chao Shang, Email: shangchao1290@126.com.
Yunyu Chen, Email: chenyunyu1984@163.com.
Miguel Angel Martinez, IrsiCaixa Institut de Recerca de la Sida, Badalona, Barcelona, Spain.
EDITOR'S NOTE
Ed. Note: The authors of the published article did not respond at the time of publication.
REFERENCES
- 1. Anirudhan V, Lee H, Cheng H, Cooper L, Rong L. 2021. Targeting SARS-CoV-2 viral proteases as a therapeutic strategy to treat COVID-19. J Med Virol 93:2722–2734. doi: 10.1002/jmv.26814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu W, Shyr Z, Lo DC, Zheng W. 2021. Viral proteases as targets for coronavirus disease 2019 drug development. J Pharmacol Exp Ther 378:166–172. doi: 10.1124/jpet.121.000688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li G, Hilgenfeld R, Whitley R, De Clercq E. 2023. Therapeutic strategies for COVID-19: progress and lessons learned. Nat Rev Drug Discov 22:449–475. doi: 10.1038/s41573-023-00672-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aherfi S, Pradines B, Devaux C, Honore S, Colson P, Scola BL, Raoult D. 2021. Drug repurposing against SARS-CoV-1, SARS-CoV-2 and MERS-CoV. Future Microbiol 16:1341–1370. doi: 10.2217/fmb-2021-0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kuo C-J, Chao T-L, Kao H-C, Tsai Y-M, Liu Y-K, Wang L-C, Hsieh M-C, Chang S-Y, Liang P-H. 2021. Kinetic characterization and inhibitor screening for the proteases leading to identification of drugs against SARS-CoV-2. Antimicrob Agents Chemother 65:e02577-20. doi: 10.1128/AAC.02577-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. O’Donnell HR, Tummino TA, Bardine C, Craik CS, Shoichet BK. 2021. Colloidal aggregators in biochemical SARS-CoV-2 repurposing screens. J Med Chem 64:17530–17539. doi: 10.1021/acs.jmedchem.1c01547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yan G, Li D, Lin Y, Fu Z, Qi H, Liu X, Zhang J, Si S, Chen Y. 2021. Development of a simple and miniaturized sandwich-like fluorescence polarization assay for rapid screening of SARS-CoV-2 main protease inhibitors. Cell Biosci 11:199. doi: 10.1186/s13578-021-00720-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang J, Yan H, Yan G, Liu X, Wang Y, Chen Y. 2022. Protocol for high-throughput screening of SARS-CoV-2 main protease inhibitors using a robust fluorescence polarization assay. STAR Protoc 3:101794. doi: 10.1016/j.xpro.2022.101794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yan H, Zhang R, Yan G, Liu Z, Liu X, Liu X, Chen Y. 2023. Production of a versatile SARS-CoV-2 main protease biosensor based on a dimerization-dependent red fluorescent protein. J Med Virol 95:e28342. doi: 10.1002/jmv.28342 [DOI] [PubMed] [Google Scholar]
- 10. Ma C, Tan H, Choza J, Wang Y, Wang J. 2022. Validation and invalidation of SARS-CoV-2 main protease inhibitors using the Flip-GFP and protease-Glo luciferase assays. Acta Pharm Sin B 12:1636–1651. doi: 10.1016/j.apsb.2021.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McGovern SL, Helfand BT, Feng B, Shoichet BK. 2003. A specific mechanism of nonspecific inhibition. J Med Chem 46:4265–4272. doi: 10.1021/jm030266r [DOI] [PubMed] [Google Scholar]
- 12. Zhang R, Yan H, Zhou J, Liu X, Chen Y. 2023. Invalidation of geraniin as a potential inhibitor against SARS-CoV-2 main protease. Nat Prod Res:1–4. doi: 10.1080/14786419.2023.2241973 [DOI] [PubMed] [Google Scholar]