Abstract
Objectives
Cardiac arrhythmia is one of the most severe cardiovascular diseases that can be fatal. Therefore, its early detection is critical. However, detecting types of arrhythmia by physicians based on visual identification is time-consuming and subjective. Deep learning can develop effective approaches to classify arrhythmias accurately and quickly. This study proposed a deep learning approach developed based on a Chapman–Shaoxing electrocardiogram (ECG) dataset signal to detect seven types of arrhythmias.
Method
Our DNN model is a hybrid CNN-BILSTM-BiGRU algorithm assisted by a multi-head self-attention mechanism regarding the challenging problem of classifying various arrhythmias of ECG signals. Additionally, the synthetic minority oversampling technique (SMOTE)–Tomek technique was utilized to address the data imbalance problem to detect and classify cardiac arrhythmias.
Result
The proposed model, trained with a single lead, was tested using a dataset containing 10,466 participants. The performance of the algorithm was evaluated using a random split validation approach. The proposed algorithm achieved an accuracy of 98.57% by lead II and 98.34% by lead aVF for the classification of arrhythmias.
Conclusion
We conducted an analysis of single-lead ECG signals to evaluate the effectiveness of our proposed hybrid model in diagnosing and classifying different types of arrhythmias. We trained separate classification models using each individual signal lead. Additionally, we implemented the SMOTE–Tomek technique along with cross-entropy loss as a cost function to address the class imbalance problem. Furthermore, we utilized a multi-headed self-attention mechanism to adjust the network structure and classify the seven arrhythmia classes. Our model achieved high accuracy and demonstrated good generalization ability in detecting ECG arrhythmias. However, further testing of the model with diverse datasets is crucial to validate its performance.
Keywords: Arrhythmia detection, deep learning, electrocardiogram, CNN, BILSTM, BIGRU, SMOTE–Tomek
Introduction
Cardiovascular diseases (CVDs), which impose a significant burden on individuals and societies, are among the leading causes of mortality and morbidity worldwide. 1 Among the various CVDs, arrhythmia is one of the most severe and can be fatal. 2 Arrhythmia refers to irregularities in the rate or rhythm, such as too fast (tachycardia), too slow (bradycardia) or irregular heartbeat patterns. 3 Cardiac arrhythmias vary from benign rhythms to potentially life-threatening rhythm types. 4 Therefore, early detection of arrhythmia is critical for clinical practitioners to save human lives. 5
The electrocardiogram (ECG) is a well-known visual time-series diagnostic method for arrhythmias that records the electrical activity generated by each cardiac cycle in real-time and is now widely used in heart rate detection.6,7 The standard clinical format of the ECG is a 12-lead ECG, including bipolar leads I, II and III; enhanced unipolar leads aVR, aVL and aVF; and unipolar leads V1 through V6. 8 The ECG signals are captured from a different angle activity in both horizontal and vertical planes when the electrodes of an electrocardiograph device are attached to the patient's skin. Ten electrodes are needed to capture 12 leads (signals) and provide an accurate representation of the heart's rhythm.8,9
Correctly detecting the type of arrhythmia is crucial for physicians before administering treatment because it not only helps save a patient's life but also alleviates sequelae, thereby reducing the burden and cost of healthcare. 10 The current standard method for detecting arrhythmia types is visual identification, which can lead to physicians’ subjective biases. Due to the large morphological variances, it is not simple to manually detect ECGs. 11 Additionally, the diagnosis and interpretation of a wide variety of CVDs by ECG mostly rely on physicians’ knowledge and expertise. 12 However, physicians’ diagnosis of cardiac disorders is always associated with levels of imprecision. 4 Therefore, specialists have tried to develop effective approaches to the early diagnosis of cardiac disorders through strong deep learning. Various researchers have attempted to develop effective approaches to accurately and quickly classify arrhythmias using strong deep learning methodologies. 9
Deep learning techniques overcome the constraints imposed by traditional diagnosis methods, enhancing performance and generalization by decreasing pre-processing and feature extraction. 13 Deep learning (DL) is a series of representation layers 14 involving the automated process of detection and extraction of important features and self-learning through training processes, which makes it a very accurate approach.15,16 According to a study, arrhythmia classification by deep learning techniques can achieve higher efficiency and accuracy than expert manual detection.10,17 Among deep learning techniques, especially convolutional neural networks (CNN), recurrent neural networks (RNN), which primarily contain long short-term memory (LSTM), and gated recurrent unit (GRU) networks have been particularly successful in ECG signal processing and were significantly improved. 18
CNNs are among the most widely used DL architectures made to learn the spatial hierarchy of data adaptively by extracting and remembering high-level and low-level patterns to predict the output. 19 One-dimensional convolutional neural networks (1D CNN) were developed to use one-dimensional signals, such as ECG records.20,21 Another deep learning algorithm for ECG analysis is LSTM networks that can learn from sequential data, known as sequence learning. 5 LSTM is an enhancement of the RNN memory performance. Many studies have been conducted on the applicability of LSTM to time-series problems.22,23
Bidirectional LSTM (BiLSTM) is an improvement of LSTM, and the models offer better predictions than LSTMs. 24 Most researchers combined CNN with BiLSTM and achieved satisfactory results in classifying arrhythmias from ECG signals. 10 The GRU network is another type of recurrent neural network, which is similar to an LSTM network but has fewer parameters and converges faster than LSTM. GRU adaptively remembers and forgets its state based on the input signal to the unit. 25 A bidirectional gated recurrent unit (BiGRU) consists of two GRU layers and can process the data in both directions.25,26 In general, different DL techniques based on varied datasets focusing on the automatic detection of ECG arrhythmia classification have been proposed in the past few years. 27
There is usually some degree of imbalance between different classes in any ECG dataset, which is an additional challenge to accurately classify ECG beats. 28 Dataset imbalance affects the model's training. Generally, classes with more samples can dominate the training and cause the classifier to favor classes containing many labeled samples. 29 This leads to low training efficiency because normal ECG beats occupying a large proportion of the dataset are prone to negative effects and model degeneration. 30 To address this problem, the model design must use a loss function in which the cost of misclassifying the minority class is much higher than that of misclassifying the majority class, like the focal loss (FL) function. 31 It is still challenging to deal with an imbalanced dataset using deep learning for classification of multiple types of cardiac arrhythmias, which is highly dependent on data quantity and quality. Furthermore, the high noise and complexity of ECG place a substantial strain on computational resources; therefore, more robust models with fewer parameters are greatly desired. 10
In this study, we designed a deep learning model to classify arrhythmia into seven classes: atrial flutter (AF), atrial fibrillation (AFIB), sinus irregularity (SI), sinus bradycardia (SB), sinus rhythm (SR), sinus tachycardia (ST) and supraventricular tachycardia (SVT) by the 12-lead ECG arrhythmia database collected by Shaoxing People's Hospital (Shaoxing Hospital Zhejiang University School of Medicine) and Chapman University on severely imbalanced data. 32
Our proposed model consists of designing a hybrid model with a fusion of convolutional blocks, a bidirectional RNN (BILSTM-BiGRU) block and an attention mechanism. The second main contribution of this study was providing a solution for the imbalanced data problem by the hybrid synthetic minority oversampling technique (SMOTE)–Tomek dataset balancing algorithm and FL function to resolve the data imbalance problem for detecting seven types of arrhythmias from ECG signals.
Related Literature
Different DL techniques based on varied datasets focusing on the automatic detection of ECG arrhythmia classification have been proposed in the past few years. 27 Nevertheless, the analysis of the findings from each deep learning technique depends on various factors, such as the hardware platform, the model's architecture and compiler optimization, all of which can directly affect the model's training. 33 Deep learning models can extract more distinguishing features automatically and without the need for hand-craft feature extraction to facilitate the detection of the arrhythmia pattern in ECG signals.21,27 In recent years, there has been extensive research using deep learning methods for ECG classification and arrhythmia detection based on ECG datasets, such as the MIT-BIH, Chapman ECG, PTB-XL and several other datasets. 34 In this section, we review the most important deep learning techniques presented for detecting arrhythmia based on ECG signals. Various studies introduced CNN-based deep methods that use 1D filters. Here, several CNN-based methods are explained.
Romdhane et al. designed a CNN classifier using the FL function. The model was trained and evaluated with the MIT-BIH and INCART datasets to identify five arrhythmia categories. The classifier used the dropout and batch normalization mechanism in a coupled-convolution layer structure, obtaining an overall accuracy of 98.41%. 29 Wu et al. proposed a robust and efficient one-dimensional 12-layer convolution neural network (CNN) network structure to classify the five sub-classes of cardiac arrhythmia. The architecture of the CNN network included eight alternating convolutions and average-pooling layers. They were followed by a dropout layer and two fully connected layers in the MIT-BIH arrhythmia database (MITDB), and an overall performance accuracy of 97.2% was achieved. 35 Ahmed et al. developed a novel model consisting of three convolution blocks for ECG arrhythmia classification. Each block contains two 1D-CNN layers, a max-pooling layer, a dropout layer and a batch normalization layer tested on the MITDB. The model achieved overall performance accuracy of 1.00 and 0.99 in the training and testing datasets, respectively. 36 Gao et al. designed a classifier based on effective LSTM recurrence network model with FL to achieve imbalanced ECG signal classification. The model attained an overall accuracy of 99.26% for the categorization of eight beat types in the MITDB, demonstrating outstanding performance. 30 Yildirim et al. proposed a model which included both representation learning and sequence learning tasks. Convolutional layers and sub-sampling layers were used in the representation learning phase. The sequence learning part involved a LSTM unit after the representation of learning layers. They performed two class scenarios, including reduced rhythms (seven rhythm types) and merged rhythms (four rhythm types) according to the records from the Chapman ECG database. The dataset was randomly divided into training, validation and test sets. The model consisting of six convolution layers bearing one-dimensional filters, four max-pooling layers, one LSTM and two fully connected layers categorically used the cross-entropy loss function and Adam optimizer for adjustment. The trained DNN model achieved an accuracy of 92.24% and 96.13% for the reduced and merged rhythm classes, respectively. 5 Rai et al. proposed two distinct deep learning models: CNN and a hybrid model, CNN-LSTM, to detect six types of cardiac arrhythmias. Two standard datasets MIT-BIH arrhythmias database (MITDB) and PTB diagnostic database (PTBDB) were combined, and 23,998 ECG beats were extracted for the model validation. Imbalanced datasets were addressed by using a hybrid data resampling technique called SMOTE and Tomek. The highest overall accuracy was 99.02% on the SMOTE–Tomek sampled dataset by the ensemble technique. 37
Most researchers combined CNN with BILSTM and achieved satisfactory results in classifying arrhythmias from ECG signals. Hassan et al. proposed a novel deep learning model that combined CNN and bi-directional LSTM to classify five classes: non-ectopic (N), supraventricular ectopic (S), ventricular ectopic (V), fusion (F) and unknown (Q) beats. The proposed model was trained, validated and tested using MIT-BIH and St Petersburg data sets separately. This model achieved training, validation and testing accuracies of 100%, 98% and 98%, respectively. 6 Islam et al. proposed the CNN and bidirectional RNN unit (BiGRU–BiLSTM) architecture to generate fusion features. The signals were classified by the fully connected layer and rectified linear unit (ReLU) activation function. The model was measured by the categorical cross entropy (CCE) loss. The experimental findings to detect five classes showed that, for the MIT-BIH-provided ECG data to identify arrhythmias, the proposed BRDC model outperforms the existing models with an accuracy of 99.90%. 38 Islam et al. developed a novel hybrid hierarchical attention-based bidirectional recurrent neural network with dilated CNN (HARDC) to detect five types of arrhythmias from ECG signals using 48 half-hour ECG records of patients investigated at the BIH Arrhythmia. The method achieved an accuracy of 99.60% during training, 99.40% during validation and 99.01% during testing. 39
A study by Rai et al. proposed an automated system for detecting myocardial infarction (MI) using ECG signals. Their approach involved a hybrid model combining CNN and LSTM, along with an ensemble technique. The model was trained and evaluated using the PTBDB and the MITDB. The CNN achieved an impressive classification accuracy of 99.82%, while the hybrid CNN-LSTM and ensemble techniques achieved even higher accuracies of 99.88% and 99.89%, respectively. 40 In another study by Rai et al., a hybrid CNN-LSTM deep learning model utilizing the PTBDB was proposed for the prognosis of MI. The model leveraged ECG beat time intervals and their gradient values as input features. To address the class imbalance, the researchers employed the SMOTE–Tomek link data sampling technique. The CNN-LSTM model achieved a remarkable accuracy of 99.8%. 41 Furthermore, in a study conducted by Rai et al., a hybrid CNN-LSTM deep learning model was proposed for the prediction of cardiac arrhythmias. The model was trained and evaluated using the PTBDB and the MITDB. The model achieved a high accuracy of 99.7%. 42 Sadeghi et al. in 2023 introduced CNNs with SE-Residual blocks and a self-attention mechanism as important components for a more accurate diagnosis of left bundle branch block. This model was evaluated on the PhysioNet Challenge 2020 database, consisting of 10,344 samples of 12-lead ECGs, using a 10-fold cross-validation approach. The model achieved an accuracy of 91.98%, a specificity of 28.99%, a precision of 73.09%, a sensitivity of 82.83%, an F1 score of 77.99% and an area under the curve (AUC) of 0.991. 43
Methods
Study duration and place
The study was conducted at the Iran University of Medical Sciences (IUMS) located in Tehran, the capital of I.R. Iran. The research took place from August 2019 to May 2022.
Nature of the study
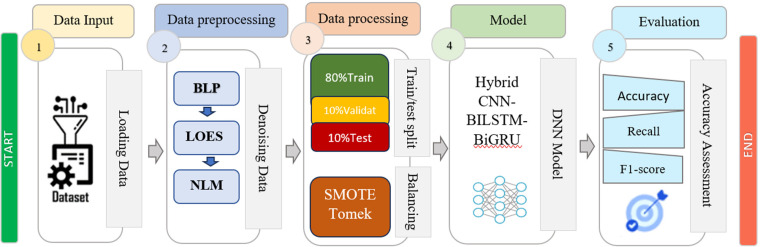
This was a developmental study, carried out quantitatively. The study proposes a deep learning approach developed based on a Chapman–Shaoxing ECG dataset signal to detect seven types of arrhythmias. The basic steps of our proposed methodology are presented using the flowchart in Figure 1.
Figure 1.
The methodology followed in this study.
Data processing
Dataset
This study used the Chapman–Shaoxing ECG dataset. This dataset contains 12-lead ECG signals collected from 10,646 people for 10 s with a frequency of higher than 500 Hz sampling rate under the supervision of Shaoxing People's Hospital (Shaoxing Hospital Zhejiang University School of Medicine) and Chapman University. Each ECG sample in this data set contains 5000 rows and 12 columns, labeled by experts in 11 types of heart rhythms. Each column corresponds to a specific lead, including Lead I, II, III, aVR, aVL, aVF, V1, V2, V3, V4, V5 and V6. With a sampling rate of higher than 500 Hz, each of the 500 rows in a Microsoft Excel column represents the data recorded for one second of the patient's heart function.
The interesting point about this dataset is that the signals are recorded from patients over several days and during different sessions.32,44 Table 1 demonstrates numerical information about the ECG dataset.
Table 1.
Numerical information about the ECG dataset.
| Acronym | Full Name | Frequency N (%) |
|---|---|---|
| SB | Sinus bradycardia | 3889 (36.53) |
| SR | Sinus rhythm | 1826 (17.15 |
| AFIB | Atrial fibrillation | 1780 (16.72) |
| ST | Sinus tachycardia | 1568 (14.73) |
| AF | Atrial flutter | 445 (4.18) |
| SI | Sinus irregularity | 399 (3.75) |
| SVT | Supraventricular tachycardia | 587 (5.51) |
| AT | Atrial tachycardia | 121 (1.14) |
| AVNRT | Atrioventricular node reentrant tachycardia | 16(0.15) |
| AVRT | Atrioventricular reentrant tachycardia | 8 (0.07) |
| SAAWR | Sinus atrium to atrial wandering rhythm | 7 (0.07) |
| Total | 10,646 (100) | |
Bold value represents the best performance.
The Chapman ECG dataset was a valuable resource for our research on arrhythmia detection, primarily due to its unique characteristics. With 12-lead ECG samples and a sampling frequency of 500 Hz, the dataset is well-suited to our research objectives. By utilizing this dataset, we can effectively train algorithms and models for accurate arrhythmia detection using real and reliable data. The selection of the Chapman ECG dataset aligns seamlessly with the timeline of our research proposal, which was approved in 2019, as it was the most suitable option among the available datasets at that time. Moreover, the public availability of the Chapman ECG dataset ensures the reproducibility of our research, facilitating further validation and comparison of the findings. Considering these factors, we are confident that our decision to employ the Chapman ECG dataset as the primary dataset for our research is entirely justified.
Data denoising method
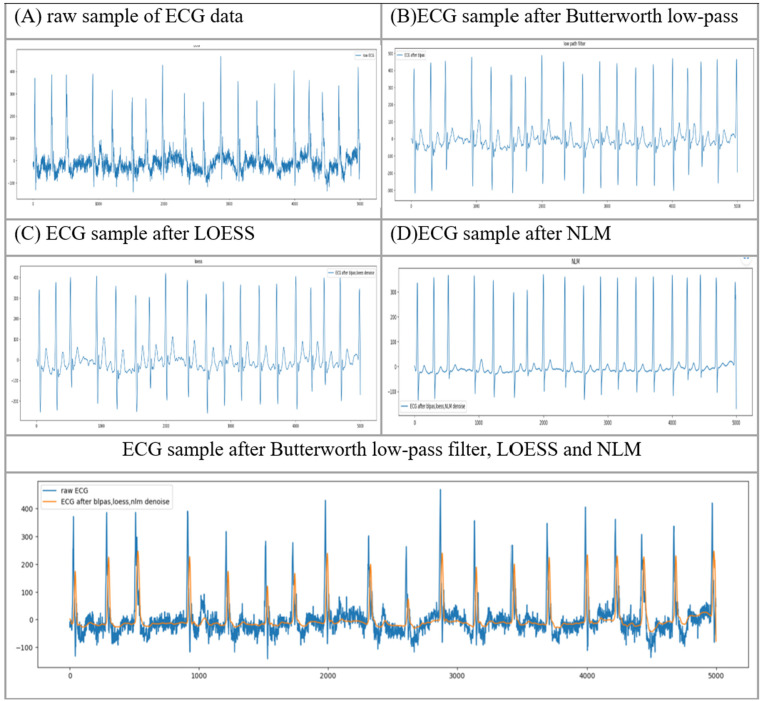
This study implemented the sequential denoising approach to the raw ECG data, according to the approach proposed by Zheng. 32 Initially, a Butterworth low-pass filter was used to remove the signals with a frequency of above 50 Hz. 45 Then, the local polynomial regression smoother (LOESS) was utilized to remove the effects of baseline wandering. 46 Finally, non-local means (NLM) were utilized for residual noise reduction.47,48 These processes were sequentially used to process the raw ECG samples. Figure 2 demonstrates an instance of an ECG signal. Figure 2(a) presents the raw sample of this ECG, Figure 2(b) displays the noise reduction performance after the Butterworth low-pass filter, Figure 2(c) displays the effectiveness of the LOESS smoother and Figure 2(d) indicates this ECG sample after NLM at the end of noise removal preprocessing operations. Generally, the preprocessing operations help normalize signals and reduce complexities. After noise removal preprocessing operations, ECG signals with frequencies above 50 Hz and the baseline wandering effect were removed. Since some ECG samples’ lead values were missing and some contained only zeros, these samples were removed, and a total of 10,618 samples from this dataset were used.
Figure 2.
An instance of ECG signals.
Data balancing
The Chapman–Shaoxing dataset used is an imbalanced ECG dataset. 32 Imbalanced datasets can result in biased and poor performance of the model for minority classes because the model's training is heavily inclined toward the majority class through the backpropagation of the loss function. 10 To prevent the loss function from being disproportionately influenced by the larger sample category in the imbalanced dataset, we implemented the SMOTE–Tomek algorithm. First, SMOTE, which is an oversampling method, increases minority class samples so that the minority class samples are expanded. This synthesizes new plausible examples in the minority class, but the new samples generated by SMOTE also have some limitations.49,50 The newly generated samples have distinct boundary limitations. The Tomek Links algorithm was combined to achieve data balance 50 to avoid this problem caused by oversampling. The Tomek Links algorithm is an under-sampling algorithm for identifying pairs of nearest neighbors in a dataset that have different classes. SMOTE–Tomek is a combined over- and under-sampling using SMOTE and Tomek links. 51 SMOTE–Tomek has been shown to be effective in mitigating the effects of class imbalance. 37 After conducting a review, it was found that none of the previous studies utilized the SMOTE–Tomek technique on the Chapman ECG dataset. However, due to the favorable results reported in using this technique to reduce data imbalance in other studies, we decided to adopt this method. The proven performance of this technique in similar scenarios supports its selection as a suitable approach to address the challenge of class imbalance in the Chapman ECG dataset.
Model architecture
In the proposed approach, we designed a novel hybrid model for automatically detecting seven types of heart arrhythmias, which helps improve overall performance. The overall structure of the proposed deep learning model is implemented by combining two neural networks, CNN and bidirectional RNN (BILSTM-BiGRU). The choice to integrate these two techniques was made with the understanding that their combination provides better outcomes. 52 The model parameters’ settings were adjusted to provide the optimal result. In addition, issues such as which layers should be used and which parts of the model should be placed were time-consuming and difficult processes solved by trying many variations.
Convolutional neural network blocks
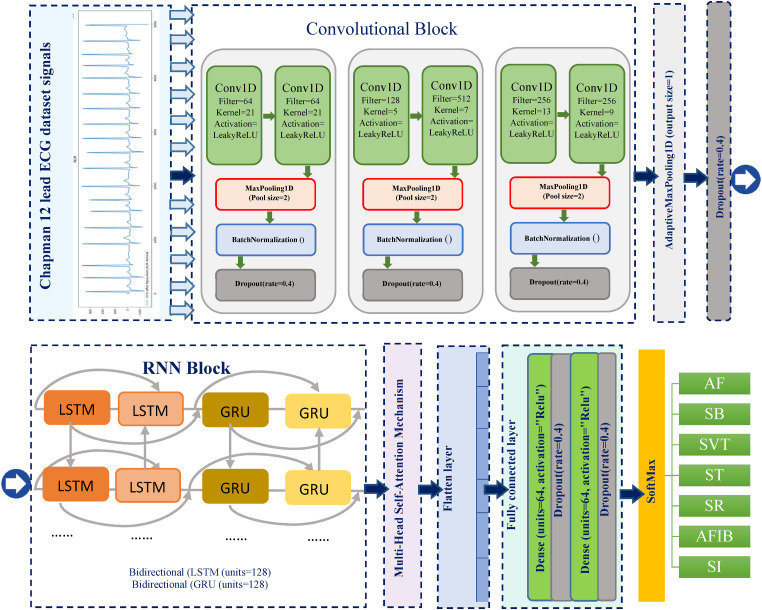
One-dimensional convolutional neural networks (1D CNN) are a variation of two-dimensional CNNs (2D CNNs) developed for using one-dimensional signals such as ECG records.20,21 CNN may be capable of retrieving valuable data whenever the input signal is noisy. The network structure is constructed layer by layer to reflect these performance characteristics. As the amount of network layers increases, features are learned and expressed in a more abstract and concise manner.6,53 The raw data of ECG signals of seven heartbeat types after preprocessing in the training dataset are input into the three convolutional blocks, each of them consisting of two Conv1D layers.
Conv1D can learn to distinguish hierarchical features from raw inputs. 5 To determine the correct parameters, such as the number of filters, kernel size and strides, we used the experience of Yildirim's study. 5 A leaky rectified linear unit (Leaky ReLU) activation function was used in each Conv1D layer. Activation functions are crucial to increasing the expressiveness of neural networks and enhancing the approximation capability between the network's different layers.36,52 To boost the effectiveness of feature extraction from the raw ECG signals, one can add a 1D max-pooling layer of size 2 after each convolutional block to reduce the computational cost. The pooling layer is a significant step and is commonly the next layer of convolution, which is also called the subsampling layer. The max pooling operation divides the input data into rectangular regions and retrieves the maximum value from each subregion, thereby reducing the number of neurons and the dimensionality of the convolution layer output data. This reduction in network complexity helps prevent overfitting and enhance the network's robustness.35,54 A batch normalization layer was used to normalize the data with a dropout layer rate of 40% placed at the end of each block to avoid the over-fitting problem. The dropout layer is used when training the CNNs to temporarily disable some neurons from the network with a certain probability. This reduces the interdependence between neuron nodes, mitigates overfitting and improves the network's generalization capability. 35
Adaptive max pooling
Adaptive pooling reduces the amount of data needed in tuning the downstream portion of CNNs. This technique is utilized to perform multiscale summarization over convolutional feature maps while capturing the required behavior of the feature map itself, decrease the convergence time, and enhance generalization performance. 55
Bidirectional RNN (BILSTM-BiGRU)
After passing through the 1D-CNN layers, the output characteristics are fed into the bidirectional RNN (BILSTM-BiGRU) block consisting of a BiLSTM and a BiGRU to analyze sequential ECG. It is proven that bidirectional RNNs can learn sequential features from ECG data in both forward and backward directions. 56 BiLSTM is an improvement of LSTM that enables additional training by traversing the input data twice, once in the input data (forward) and once in the reverse input data (backward). BiLSTM models offer better predictions than LSTMs, although LSTMs reach equilibrium faster than BiLSTMs. 24 The application of convolutional operations to the input matrix before applying the BiLSTM model is a promising approach. Additional geographical and temporal data properties may be incorporated, thereby increasing the accuracy of predictions. 57 The GRU (gated recurrent unit) network is another type of recurrent neural network that resembles an LSTM network but has fewer parameters and converges faster. GRU remembers and forgets its state adaptively based on the input signal to the unit. In terms of performance, the difference between the two is often small and inconclusive, depending on the task and the dataset. 58 A BGRU consists of two GRU layers and can process data in both directions: one processing the input sequence from left to right and the other from right to left, with two separate hidden layers. The outputs of the two GRU layers are concatenated at each time step, forming a bidirectional representation of the input sequence. BIGRU can capture both the past and future contexts of a sequence and can improve the performance of tasks that require long-term dependencies or complex structures.25,26 Each layer has 128 neuron units and a dropout of 40%, which can alleviate the overfitting concern.
Multi-head self-attention mechanism
RNN compresses all information over time, which can weaken the time difference between input features and affect prediction accuracy. The attention mechanism is used to focus on specific parts of the input sequence based on their importance. 59 The output of the BILSTM-BiGRU layer is transmitted into the self-attention layer. The attention layer is primarily used to focus on a particular network layer. The multi-head self-attention mechanism allows the model to learn relevant information in different subspaces of representation, and it becomes possible to capture a broader range of discriminative features. The self-attention mechanism can process the input data in parallel, reducing the complexity of calculations.60,61
Fully connected layer
Finally, the predicted model consists of a flattening layer and a dropout layer, whose extracted features are used as the input for a dense layer with a Relu activation function and 64 neurons. The dropout layer is set to discard 40% of the input features to prevent overfitting. The output of the dense layer is then fed to a SoftMax layer, which predicts the probability of the ECG heartbeat class.
Model compile
This study utilized FL cross entropy with alpha = 0.25 and gamma = 2 to calculate the loss in the model's training and validation. FL is a modification of cross-entropy (CE) loss where the scaling factor decays to zero as the confidence of the classification increases; as a result, the loss assigned to well-classified examples is down-weighted.30,62 The FL approach reduces the impact of imbalanced heartbeat classes by focusing the loss on minority classes. This technique has been shown to improve the performance of deep learning models on imbalanced datasets. 29
Model checkpoint
The model checkpoint option, provided by Keras, automatically saves the weights of the best suitable model in terms of a specified metric.7,63 We choose to monitor the loss value evaluated on the validation data to keep the model's weights with the monitor. The schematic illustration of the proposed model to classify the ECG signals of this study is illustrated in Figure 3. This diagram depicts in detail the fundamentals of each layer, from the source term to the accurate prediction layer. The arrhythmia goal category is predicted at the end of the process. The structure of the constructed CNN-BILSTM-BiGRU model is depicted in Table 2.
Figure 3.
Schematic illustration of the proposed model to classify ECG signals.
Table 2.
The structure of the constructed CNN-BILSTM-BiGRU model.
| Layer (Type) | Output Shape | Param # |
|---|---|---|
| Conv1D | (None, 453, 64) | 1408 |
| Conv1D | (None, 447, 64) | 28,736 |
| MaxPooling1D | (None, 223, 64) | 0 |
| Batch normalization | (None, 223, 64) | 256 |
| Dropout | (None, 223, 64 | 0 |
| Conv1D | (None, 219, 128) | 41,088 |
| Conv1D | (None, 213, 512) | 459,264 |
| MaxPooling1D | (None, 106, 512) | 0 |
| Batch normalization | (None, 106, 512) | 2048 |
| Dropout | (None, 106, 512) | 0 |
| Conv1D | (None, 94, 256) | 1,704,192 |
| Conv1D | (None, 86, 256) | 590,080 |
| MaxPooling1D | (None, 43, 256) | 0 |
| Batch normalization | (None, 43, 256) | 1024 |
| Adaptive MaxPooling1D | (None, 1, 256) | 0 |
| Dropout | (None, 1, 256) | 0 |
| Bidirectional LSTM | (None, 1, 256) | 394,240 |
| Bidirectional GRU | None, 1, 256) | 296,448 |
| Attention | (None, 1, 256) | 65,537 |
| Flatten | (None, 256) | 0 |
| Dense | (None, 64) | 16,448 |
| Dropout | (None, 64) | 0 |
| Dense | (None, 64) | 4160 |
| Dropout | (None, 64) | 0 |
| Dense | (None, 7) | 455 |
| Total params: 3,605,384. Trainable params: 3,603,720. Non-trainable params: 1664. | ||
Results
A comprehensive experimental analysis is described in this section.
Experimental setup
We have designed a hybrid architecture model to effectively classify imbalanced arrhythmia signals. The experimental analysis was conducted on the Chapman–Shaoxing ECG dataset. We excluded certain classes from the initially published dataset due to an insufficient number of cases. Specifically, the classes atrial tachycardia (121 samples), atrioventricular node reentrant tachycardia (16 samples), atrioventricular reentrant tachycardia (8 samples) and sinus atrium to atrial wandering rhythm (7 samples) were removed from the dataset. Table 3 gives the used dataset, including seven rhythms, in detail.
Table 3.
The used dataset includes seven rhythms
| Acronym Name | Number of Total Samples N (%) | Frequency |
|---|---|---|
| AF | Atrial flutter | 437 |
| SB | Sinus bradycardia | 3884 |
| SVT | Supraventricular tachycardia | 582 |
| ST | Sinus tachycardia | 1563 |
| SR | Sinus rhythm | 1825 |
| AFIB | Atrial fibrillation | 1778 |
| SI | Sinus irregularity | 397 |
| Total | 10,466 | |
Bold value represents the best performance.
To train the proposed model and check the validation, we randomly divided the dataset into three parts: 80% for training, 10% for validation and 10% for testing the model's effectiveness. Training data were balanced by the SMOTE–Tomek method. The model was trained with 200 epochs. The Adam optimizer with a learning rate of 0.001 and the categorical accuracy and F1Score evaluated the training in our proposed system.
ECG signals were classified by the Hybrid CNN-BILSTM-BiGRU Model, and then, the model was trained by using FL. We did not use the early stop criteria to compare the leads’ performances during the same epochs. Subsequently, this model was evaluated only once on the test dataset. The goal was to prevent information leaks into the model due to multiple validation processes, which reduces the reliability of the model even though the model is not directly trained on the validation data. 10 Only a single efficient model was used for all the experiments. The hyperparameters of the model were not altered during training. The total parameters of the proposed model were 3,605,384; there were 3,603,720 trainable parameters and 1664 non-trainable parameters introduced by the batch-normalization layer. Table 4 shows the settings of the model parameters to obtain the best classification accuracy. We verified the effectiveness of the proposed model structure by comparing it with state-of-the-art methods.
Table 4.
Settings of the model parameters.
| Network Layers | Optimizer | Learning Rate | Dropout | Epoch | Batch Size | Cost Function |
|---|---|---|---|---|---|---|
| 25 | Adam | 0.001 | 0.4 | 200 | 128 | Focal loss |
Hardware and software
The constructed deep learning models were trained on a computer with AMD Ryzen 7 5800H with Radeon Graphics running at 3.20 GHz, an NVIDIA GeForce GTX 1650 GPU and 16 GB of memory. The DNN model was constructed using the Keras (v. 2.11.0) deep learning library and the TensorFlow (v. 2.11) framework.
Performance matrices
Our model classified seven classes of arrhythmia. To measure the overall performance of the proposed algorithm, we calculated accuracy, recall, precision and F1-score. These performance metrics are described below:
Accuracy (acc) expresses the percentage of the correctly predicted number in the total dataset and can be used to judge how often the model is correct. True positives (TP) refer to the number of correctly identified positive instances; true negatives (TN) represent the number of correctly identified negative instances and false positives (FP) and false negatives (FN) denote the number of incorrectly identified positive and negative instances, respectively. However, the Chapman–Shaoxing ECG dataset is imbalanced, so accuracy cannot be used as a good performance metric. Therefore, recall or sensitivity is utilized to measure the probability that the model correctly predicts the class as positive. In addition to recall, precision is a useful metric for evaluating the performance of classification models. It measures the frequency at which a model predicts a class as positive relative to the total number of positive instances across all classes. It provides a measure of the model's ability to make accurate positive predictions. The F1-score is a weighted average of recall and precision, offering an overall measure of a model's performance; it considers its ability to correctly identify positive instances and minimize false positives. The formulas for each performance metric are shown in Equations (1)-(4), respectively.
| (1) |
| (2) |
| (3) |
| (4) |
Model performance
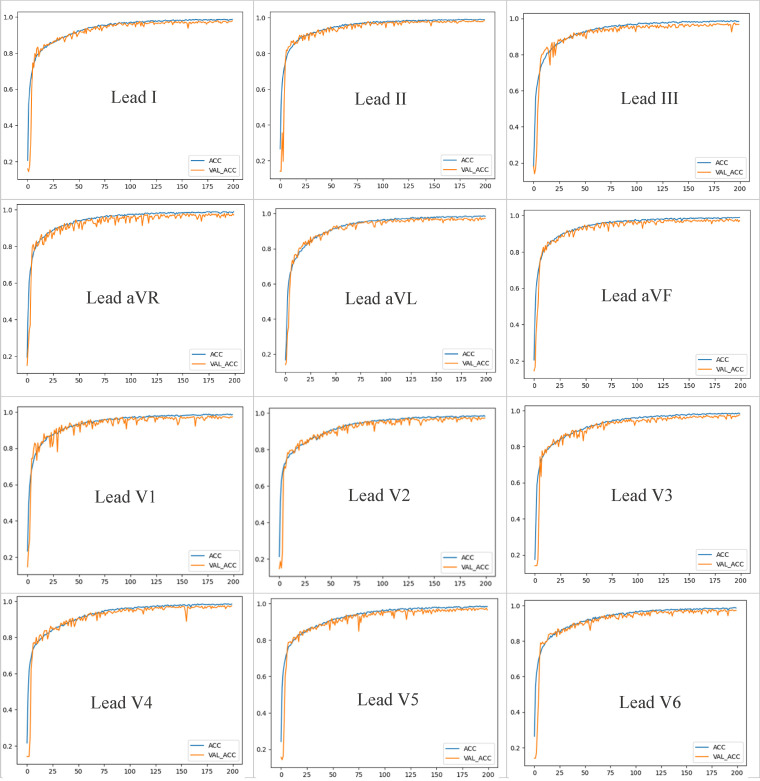
Figure 4 depicts the training and validation graphs obtained from the training process.
Figure 4.
The training and validation graphs obtained from the training process of each lead separately.
The Chapman ECG dataset consists of a 12-lead ECG sample with a sampling frequency rate of 500 Hz for a duration of 10 s. As shown in Figure 4, the models were trained separately for each lead, resulting in a total of 12 models, each dedicated to a specific lead. The dataset distribution ensures an equal number of samples and 5000 data points per sample for each lead. This approach enables individual analysis of the unique features associated with each lead, facilitating the examination and optimization of the model's performance in detecting specific patterns and characteristics within each lead. We present the performance of each lead separately.
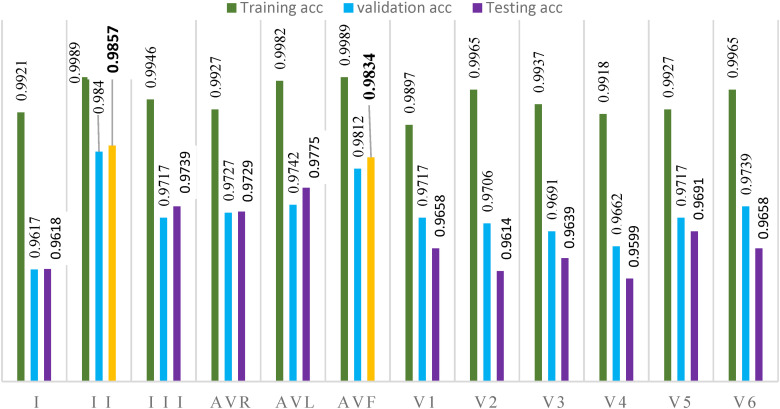
The developed model performed well in examining lead epoch classification. After 200 epochs of training, the categorization accuracy values for the dataset were almost 100%, with a relatively flat curve. It is evident from these graphs that the proposed model exhibited promising results for all lead ECG signals during training. Figure 5 depicts the training, validation and testing performance for arrhythmia classification, respectively. This experimental analysis was accurately calculated. Each evaluation metric is labeled with a different color in these figures, and each bar on each figure represents the accuracy of each ECG lead. The best validation and testing accuracy performance was archived by lead II (0.9845 validation accuracy and 0.9857 testing accuracy). The proposed model achieved a lower performance on lead V4 (0.9662 validation accuracy and 0.9599 testing accuracy).
Figure 5.
Training, validation and testing acc performance for all classes.
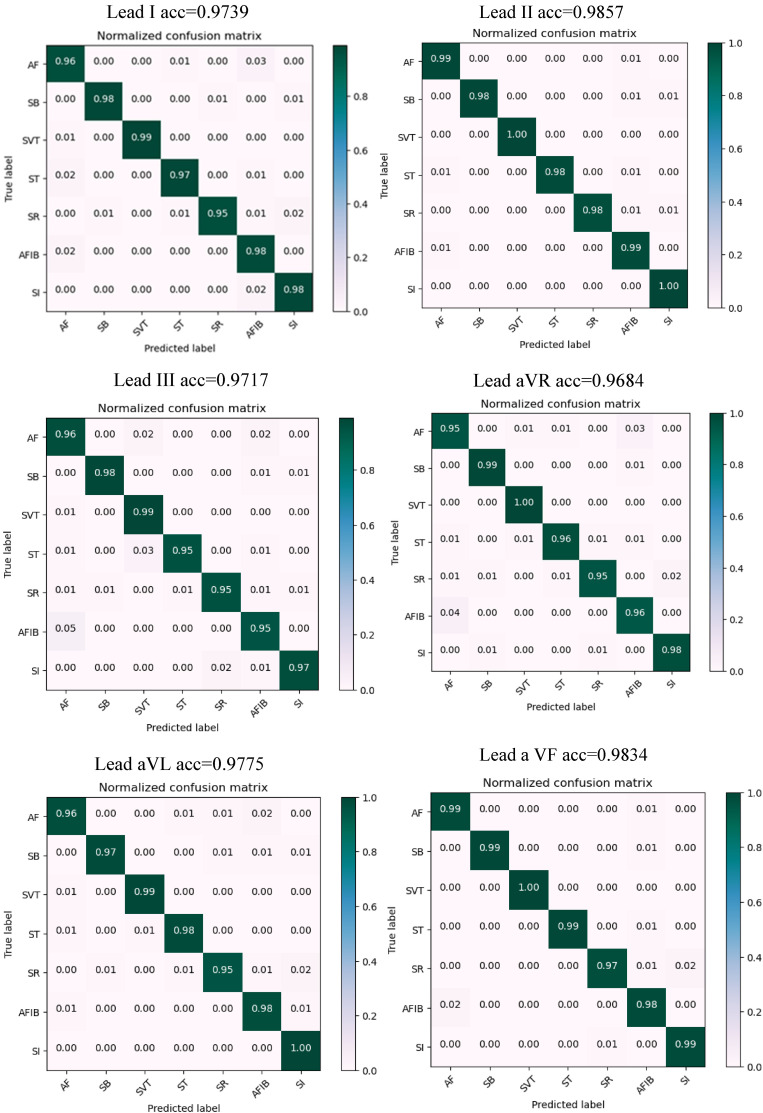
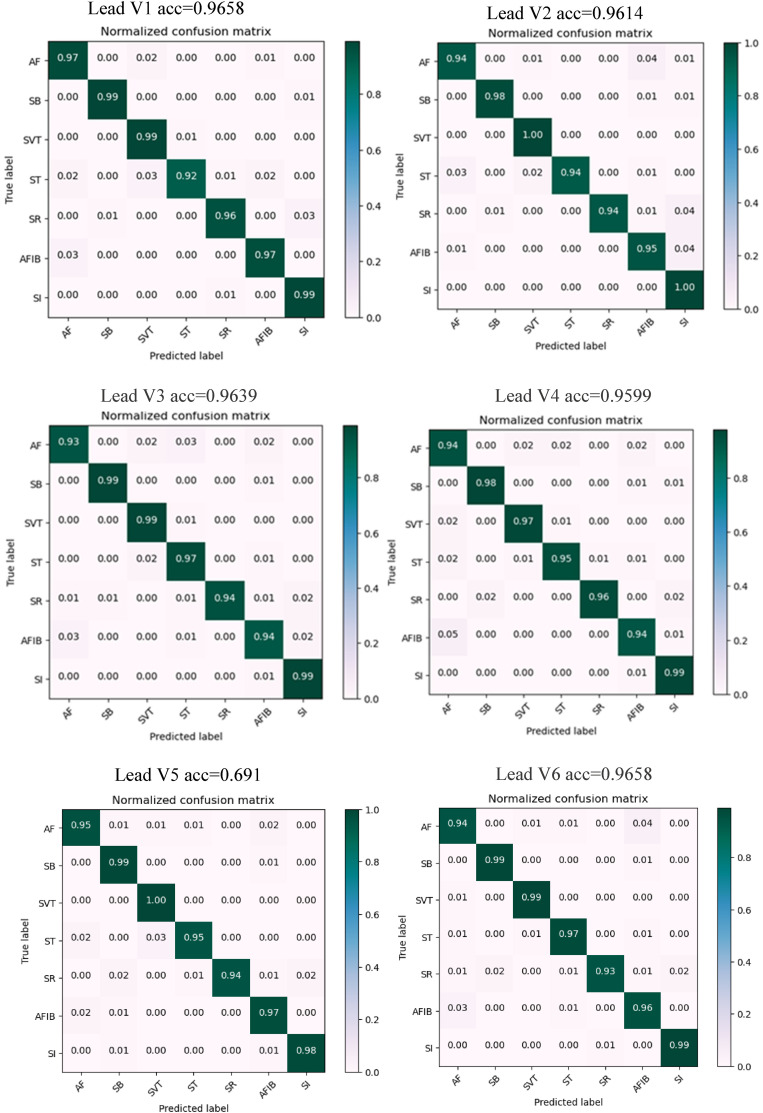
After the training process, the performance of the proposed model was tested on the test sets. The trained model was applied to the test records and yielded promising results on unseen data and confusion matrices obtained during the test process. The confusion matrix plays an essential role in evaluating a model's performance. The testing-based confusion matrix for arrhythmia categorization using the dataset is displayed in Figures 6 and 7, which indicate the number of correctly predicted samples against incorrectly predicted samples. In these figures, the model achieved 0.9857 accuracy on lead II and 0.9599 accuracy on lead V4 signals.
Figure 6.
Testing-based confusion matrix for arrhythmia categorization using leads (I-aVF).
Figure 7.
Testing-based confusion matrix for arrhythmia categorization using leads (V1–V6).
In general, this table shows that the proposed method had an appropriate distribution regarding all classes with no emphasis on a specific class with overfitting and no bad performance concerning a specific arrhythmia class. In this matrix, most wrong states occurred between AF and AFIB arrhythmia classes, as well as SR and SI classes. As far as medical science is concerned, this miscalculation between two rhythm classes is not crucial because the diagnosis of these two arrhythmias can be reached simultaneously.
According to the confusion matrices, we calculated several performance metrics for each lead, including precision, recall and F1-score. The precision performance of each class based on each lead is presented in Table 5.
Table 5.
The precision performance of each class based on each lead.
| AF | SB | SVT | ST | SR | AFIB | SI | |
|---|---|---|---|---|---|---|---|
| Lead I | 0.95.6 | 0.9919 | 0.9921 | 0.9843 | 0.9948 | 0.9395 | 0.9681 |
| Lead II | 0.9792 | 0.9973 | 0.9895 | 0.9974 | 0.9896 | 0.9704 | 0.9781 |
| Lead III | 0.9262 | 0.9761 | 0.9544 | 0.9213 | 0.9716 | 0.9426 | 0.9821 |
| Lead aVR | 0.9401 | 0.9738 | 0.9819 | 0.9740 | 0.9792 | 0.9547 | 0.9750 |
| Lead aVL | 0.9736 | 0.9918 | 0.9868 | 0.9793 | 0.9869 | 0.9653 | 0.9617 |
| Lead aVF | 0.9765 | 1 | 0.9921 | 0.9948 | 0.9795 | 0.9607 | 0.9826 |
| Lead V1 | 0.9462 | 0.9867 | 0.9472 | 0.9888 | 0.9792 | 0.6577 | 0.9586 |
| Lead V2 | 0.9496 | 0.9760 | 0.9692 | 0.9945 | 0.9947 | 0.9358 | 0.9197 |
| Lead V3 | 0.9570 | 0.9893 | 0.9614 | 0.9398 | 0.9973 | 0.9513 | 0.9544 |
| Lead V4 | 0.9084 | 0.9761 | 0.9634 | 0.9735 | 0.9819 | 0.9517 | 0.9658 |
| Lead V5 | 0.9578 | 0.9512 | 0.9644 | 0.9865 | 0.9947 | 0.9556 | 0.9753 |
| Lead V6 | 0.9375 | 0.9787 | 0.9842 | 0.9689 | 0.9841 | 0.9386 | 0.9761 |
Bold values represent the best performance.
The highest precision values were obtained using lead aVF to detect SB (100%). The proposed model achieved the lowest precision performance on lead V4 (0.9084) to detect AF. The recall performance of each class based on each lead is given in Table 6. The highest recall performance values were obtained using lead II and lead aVF to detect SVT (100%). The proposed model achieved the lowest recall performance on lead V3 (0.9252) to detect AFIB. Table 7 shows the F1-score performance of each class based on each lead. The highest F1-score performance values were obtained using lead aVF to detect SVT (99.61%). The proposed model achieved the lowest recall performance on lead V4 (0.9237) to detect AF. It is interesting to know that lead II and lead aVF depicted a better performance in all of these tables. The results are given in Tables 5–7.
Table 6.
The recall performance of each class based on each lead.
| AF | SB | SVT | ST | SR | AFIB | SI | |
|---|---|---|---|---|---|---|---|
| Lead I | 0.9584 | 0.9812 | 0.9947 | 0.9766 | 0.9571 | 0.9676 | 0.9826 |
| Lead II | 0.99 | 0.99 | 1 | 0.98 | 0.97 | 0.9801 | 0.9975 |
| Lead III | 0.9554 | 0.9813 | 0.9947 | 0.9534 | 0.9519 | 0.9403 | 0.9577 |
| Lead aVR | 0.9500 | 0.9947 | 0.9974 | 0.9715 | 0.9544 | 0.9428 | 0.9701 |
| Lead aVL | 0.9685 | 0.9759 | 0.9921 | 0.9844 | 0.9544 | 0.9701 | 0.9975 |
| Lead aVF | 0.9842 | 0.9866 | 1 | 0.9897 | 0.9696 | 0.9751 | 0.9801 |
| Lead V1 | 0.9685 | 0.9920 | 0.9974 | 0.9145 | 0.9496 | 0.9625 | 0.9801 |
| Lead V2 | 0.9396 | 0.9786 | 0.9974 | 0.9326 | 0.9370 | 0.9475 | 0.9975 |
| Lead V3 | 0.9368 | 0.9893 | 0.9868 | 0.9517 | 0.9519 | 0.9252 | 0.9876 |
| Lead V4 | 0.9395 | 0.9840 | 0.9735 | 0.9508 | 0.9620 | 0.9303 | 0.9801 |
| Lead V5 | 0.9553 | 0.9893 | 0.9974 | 0.9482 | 0.9470 | 0.9675 | 0.9801 |
| Lead V6 | 0.9424 | 0.9840 | 0.9894 | 0.9714 | 0.9369 | 0.9526 | 0.9851 |
Bold values represent the best performance.
Table 7.
F1-score performance of each class based on each lead.
| AF | SB | SVT | ST | SR | AFIB | SI | |
|---|---|---|---|---|---|---|---|
| Lead I | 0.9544 | 0.9865 | 0.9934 | 0.9804 | 0.9755 | 0.9533 | 0.9753 |
| Lead II | 0.9843 | 0.9919 | 0.9947 | 0.9882 | 0.9782 | 0.9752 | 0.9877 |
| Lead III | 0.9406 | 0.9787 | 0.9742 | 0.9671 | 0.9616 | 0.9415 | 0.9698 |
| Lead aVR | 0.9450 | 0.9841 | 0.9896 | 0.9728 | 0.9667 | 0.9487 | 0.9726 |
| Lead aVL | 0.9711 | 0.9838 | 0.9894 | 0.9819 | 0.9704 | 0.9677 | 0.9793 |
| Lead aVF | 0.9803 | 0.9933 | 0.9961 | 0.9922 | 0.9746 | 0.9678 | 0.9814 |
| Lead V1 | 0.9572 | 0.9893 | 0.9704 | 0.9502 | 0.9642 | 0.9601 | 0.9692 |
| Lead V2 | 0.9446 | 0.9773 | 0.9831 | 0.9626 | 0.9650 | 0.9416 | 0.9570 |
| Lead V3 | 0.9468 | 0.9893 | 0.9740 | 0.9554 | 0.9741 | 0.9381 | 0.9707 |
| Lead V4 | 0.9237 | 0.9800 | 0.9684 | 0.9620 | 0.9719 | 0.9409 | 0.9729 |
| Lead V5 | 0.9565 | 0.9699 | 0.9806 | 0.9670 | 0.9702 | 0.9615 | 0.9777 |
| Lead V6 | 0.9399 | 0.9813 | 0.9868 | 0.9702 | 0.9599 | 0.9455 | 0.9778 |
Bold values represent the best performance.
Discussion
This paper introduced a new Hybrid CNN-BILSTM-BiGRU algorithm that utilizes FL cross entropy and a multi-head self-attention mechanism to address the challenging problem of classifying various cardiac arrhythmias based on the Chapman–Shaoxing dataset. The SMOTE–Tomek technique was also employed to mitigate the effects of imbalanced data and detect and classify seven types of cardiac arrhythmia based on 12-lead ECG signals. Several studies have proposed models for diagnosing arrhythmias using this public dataset; thus, the advantage is that our results are comparable with those of previous studies on this database.
Numerous hybrid and non-hybrid deep learning models have been proposed in the literature; the distinction of our novel approach lies in the fact that no previous study developed a hybrid CNN-BILSTM-BiGRU algorithm by using FL cross entropy and a multi-head self-attention mechanism based on the Chapman–Shaoxing dataset.
In a comprehensive analysis of studies presented in Table 8, the performance of deep learning models for cardiac arrhythmia detection across various databases was reported. By comparing the performance of our proposed model with the models presented in several other articles, we have demonstrated the effectiveness and competitiveness of our approach in detecting cardiac arrhythmias.
Table 8.
A comparison of the proposed model and some other state-of-the-art methods that used different leads.
| Ref. | Study | Dataset | Number of ECG Records | Rhythm | Method | Performance (Accuracy %) |
|---|---|---|---|---|---|---|
| 64 | Faust et al. | Chapman | 10,093 | 5 | Detrending, ResNet | 98.55 |
| 5 | Yildirim et al. | Chapman | 10,588 10,436 |
7 4 |
CNN-LSTM |
Lead II: 92.24 Lead II: 96.13 |
| 65 | Meqdad et al. | Chapman | 10,646 | 7 | Meta CNN Trees | lead III: 96.92 12-lead fusion:98.29 |
| 66 | Meqdad et al. | Chapman | 10,646 | 7 | CNN Trees | 12-lead fusion:97.60 |
| 67 | Sepahvand et al. | Chapman | 10,436 | 7 | Teacher model Student model |
12-lead: 98.96 98.13 |
| 68 | Andayeshgar et al. | Chapman | 10,494 | 7 | GCN-MI | 12-lead:99.71% |
| 69 | Sadeghi et al. | PhysioNet 2020 | 10,344 | 1 |
SE-Residual blocks and a self-attention mechanism | 91.98 |
| 70 | Gosia Bigam et al. | MIT-BIH | 48 samples half-hour | 5 | CNN-BILSTM | Lead II: 99.51 |
| 38 | Islam et al. | MIT-BIH | 48 samples half-hour | 5 | CNN-BILSTM-BiGRU-CCE | Lead II: 99.90 |
| 71 | Khan | MIT-BIH | 48 samples half-hour | 5 | CNN | Lead II: 92.86 |
| 42 | RAI | MIT-BIH and PTB | 48 samples half-hour | 6 | CNN-LSTM | Lead II: 99.2 |
| The proposed model | Chapman | 10,466 | 7 | CNN-BILSTM-BiGRU-Attention, FL |
Lead II: 98.57
Lead aVF = 98.34 Lead III: 97.17 |
|
Bold values represent the best performance.
For instance, a study 5 based on the Chapman dataset used the CNN model to generate deep spatial features based on raw ECG signals of separate single leads. Then, the output of the CNN model was allocated to the LSTM model to generate the deep temporal features. This model, based on 10,436 patients, achieved a 92.24% accuracy for detecting seven classes, and based on 10,588 patients, it revealed an accuracy of 96.13% for detecting four classes. The algorithm proposed herein yielded a higher accuracy of 6.33% compared to the method proposed in 5 for detecting seven classes.
Another method is based on raw ECG signals of separate single leads. The Detrending-ResNet model was introduced to detect AFIB, atrial flutter (AFL), SVT, ST and SB arrhythmias based on RR interval signals relying on Chapman ECG samples. Its results for the classification of five rhythms manifested an accuracy of 98.55% in the evaluation section. 64 Compared to the method introduced in, 64 our proposed algorithm detected one more class (SI) and yielded a higher accuracy. SR and SI classes are very similar, and, in most models, most wrong states occur between them, which reduces the model's accuracy. Evidence suggests that the proposed model based on raw ECG signals of separately single leads effectively amalgamated the beneficial features of CNN and BILSTM-BIGRU, culminating in superior accuracy, which was further enhanced by an attention mechanism.
Our model processed each lead individually and was trained with single-lead ECG records, but recently, fusion algorithms have been used in the literature to diagnose arrhythmia by fusing 12-lead data. This method has been proposed in.65–68 In a study, 65 after fusing the 12-lead data, CNN models were coded by genetic programming. CNN trees were responsible for learning deep structural features from functional data extracted from 12 leads. According to the evaluation results, the proposed method yielded 97.60% accuracy based on the Chapman dataset. In another study, interpretable meta-structural learning was proposed in combination with CNN models encoded as the evolutionary trees of genetic programming (GP) algorithms. The performance of the proposed classification model constructed for lead III ECG had 96.92% accuracy and, for the fusing of 12-lead ECG, exhibited 98.29% accuracy for the classification of seven types of arrhythmias based on the Chapman dataset recorded from 10,646 patients. We did not combine the performance of all leads, and our proposed model was trained with single-lead ECG records but yielded higher accuracy than the methods proposed in.65,66
The teacher and student models were trained in another recently introduced method. The teacher model was trained with 12-lead ECG records, whereas the student model was trained with single-lead ECG records; the student's accuracy was 98.13%, and the teacher model constructed with the Chapman12-lead ECG dataset yielded a 98.96% accuracy for seven rhythm classes. Regardless of the lead selected from the 12 leads, the student model performed almost similarly to its teacher and exhibited only a small drop of 0.83% in accuracy. Our proposed model yielded a higher accuracy of 0.44% compared to the student method proposed in Sepahvand and Abdali-Mohammadi, 67 regardless of the lead selected from the 12 leads. According to the results displayed in the learning curve graph, the model converged when the number of epochs reached 150, whereas the proposed model in 67 converged when the number of epochs reached 400. In another method, the relationships of 12 ECG leads were measured using mutual information (MI) indices extracted from the ECG leads as an adjacency matrix, illustrated by the developed graph convolutional network (GCN) and included in the ECG-based diagnostic method. The GCNMI structure with 15 layers achieved 99.71% accuracy. 68
In another study, Sadeghi et al. introduced CNNs with SE-Residual blocks and a self-attention mechanism to detect left-bundle branch blocks on the PhysioNet 2020 Challenge database with 10,344 samples from 12-ECGs. This model yielded an accuracy of 98.91%. 69 However, in the present study, we employed a combination model to detect seven types of arrhythmias.
In studies utilizing the MIT-BIH database, only data from lead II are used for the analysis and classification of cardiac rhythms. Gosia Bigam et al. developed a model that includes a CNN in combination with bilateral long-term and short-term memory to detect five types of arrhythmias. This model achieved 99.51% accuracy. 70 In other studies, within the dataset, Islam et al. employed a combination of a dual-structured RNN network with a hierarchical attention mechanism based on dilated CNN to detect five types of arrhythmias. In this study, an adversarial generative network was utilized to address the data imbalance issue, resulting in an accuracy of 99.60%. 38 Another study by Islam et al. introduced the same model architecture without employing the attention mechanism, achieving an accuracy of 99.99%. 39 Besides, Khan et al. utilized CNNs to classify five types of arrhythmias. To address the data imbalance, they employed the SMOTE–Tomek technique and achieved 92.86% accuracy. 71 Rai et al. introduced a combined CNN-LSTM model to detect six types of cardiac arrhythmias based on samples from the MIT-BIH and PTB Diagnostic datasets. The data imbalance issue was addressed using the SMOTE–Tomek technique with an accuracy of 99.2%. 42
Although our proposed model was trained with a single lead and we did not combine the performance of all leads, all lead signals were analyzed; the distinction lies in that, in the present study, the model was trained using all 12 ECG leads separately, whereas these studies solely utilized data from lead II. Although Chapman–Shaoxing is an imbalanced ECG dataset, none of the reviewed studies conducted based on Chapman–Shaoxing reported a suitable method to avoid the effects of class imbalance. For the first time, this study implemented the SMOTE–Tomek technique while also employing cross-entropy loss as a cost function to address the imbalance issue. The results demonstrated that these methods were effective in mitigating the effects of class imbalance in Chapman–Shaoxing. Furthermore, the present study utilized a multi-headed self-attention mechanism for adjusting the network structure and classifying seven arrhythmia classes. Overall, there are several justifications to improve the proposed method compared to previous methods. The accuracy of our model is high (98.57%), and it has a good generalization ability for detecting ECG arrhythmias.
Conclusion
This paper introduced a new Hybrid CNN-BILSTM-BiGRU algorithm that utilizes FL cross entropy and a multi-head self-attention mechanism to address the challenging problem of classifying various cardiac arrhythmias based on ECG data. The SMOTE–Tomek technique was also employed to mitigate the effects of imbalanced data and detect and classify cardiac arrhythmias. The experimental results suggested that the proposed classification model trained with a single lead achieved an accuracy of 98.57% for classifying seven types of arrhythmias in the Chapman ECG dataset using the lead II and samples of 10,466 patients. Furthermore, the model's performance was compared to that of state-of-the-art methods based on deep learning models, and the results demonstrated that it is highly competitive.
We conducted an analysis of all lead signals separately to evaluate the model's effectiveness in diagnosing and classifying different types of arrhythmias. Although we did not combine all the leads, the results indicated that the proposed model's performance can be generalized to 12-lead signals. Our hybrid approach proved to be a significant and accurate method.
The future scope of the research team involves investigating the proposed model's performance on another dataset collected in Isfahan, Iran. This dataset contains 2047 ECG signals collected for 10 or more seconds with a sampling frequency rate higher than 500 Hz, labeled for 11 rhythms, including the same 7 rhythms plus 4 other rhythms, collected from 2020 to 2022. This endeavor aims to provide further insights into the model's performance on diverse datasets and enable a comprehensive comparison with existing approaches. By exploring these additional datasets, we aim to enhance the understanding of our model's effectiveness and potential in various diagnostic scenarios. Furthermore, the research team aims to develop a new deep learning method for detecting arrhythmias by combining the information of 12 leads.
Overall, the hybrid CNN-BILSTM-BiGRU algorithm, assisted by a multi-head self-attention mechanism and the SMOTE–Tomek technique, proved to be a highly effective approach for diagnosing and classifying different types of arrhythmias, representing a significant improvement over current methods. The experimental results showed that the proposed model combined the advantages of CNN and BILSTM-BIGRU and achieved better accuracy with the attention mechanism, which proves the effectiveness of this mechanism.
Acknowledgements
The authors thank the people who assisted them with this study.
Footnotes
Contributorship: Conceptualization by FS; methodology by FS and AR; software by AR and SC; validation by AR and SC; formal analysis by AR and SC; investigation by FS, AR and SC; writing-original draft preparation by SC; writing-review and editing by AR and FS; visualization by AR and SC. The authors have read and agreed to the published version of the manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical approval: The research study obtained ethical approval from the Iran University of Medical Sciences (IUMS) Ethical Review Committee. The approval was granted with reference number IR.Iums.Rec.1400.295.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was funded by the Iran University of Medical Sciences as part of a PhD dissertation, with reference number 1400-1-37-21035.
Guarantor: FS.
Informed consent: The author(s) affirm that the requirement for obtaining written informed consent was waived by the Institutional Review Board. It was further confirmed that all experimental protocols underwent approval by the Iran University of Medical Sciences (IUMS) Ethical Review Committee Board.
ORCID iD: Farahnaz Sadoughi https://orcid.org/0000-0002-7452-0864
References
- 1.Vaduganathan M, Mensah GA, Turco JV, et al. The global burden of cardiovascular diseases and risk: a compass for future health. J Am Coll Cardiol, 2022; 80: 2361–2371. [DOI] [PubMed] [Google Scholar]
- 2.Zhou Y, Suo W, Zhang X, et al. Roles and mechanisms of quercetin on cardiac arrhythmia: a review. Biomed Pharmacother 2022; 153: 113447. [DOI] [PubMed] [Google Scholar]
- 3.Chittoria J, Kamath SS, Mayya V. Detection of Cardiac Arrhythmia Using Machine Learning Approaches. 2022 IEEE Region 10 Symposium (TENSYMP); 2022: IEEE.
- 4.Vollmer M, Sodmann P, Nath Net al. et al. A convolutional neural network for ECG annotation as the basis for the classification of cardiac rhythms. Ulmer Informatik-Berichte 2019; 3: 4. [DOI] [PubMed] [Google Scholar]
- 5.Yildirim O, Talo M, Ciaccio EJ, et al. Accurate deep neural network model to detect cardiac arrhythmia on more than 10,000 individual subject ECG records. Comput Methods Programs Biomed 2020; 197: 105740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassan SU, Mohd Zahid MS, Abdullah TAet al. et al. Classification of cardiac arrhythmia using a convolutional neural network and bi-directional long short-term memory. Digit Health 2022; 8: 20552076221102766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liotto C, Petrillo A, Santini S, et al. A multiclass CNN cascade model for the clinical detection support of cardiac arrhythmia based on subject-exclusive ECG dataset. Biomed Eng Lett 2022; 12: 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irfan S, Anjum N, Althobaiti T, et al. Heartbeat classification and arrhythmia detection using a multi-model deep-learning technique. Sensors 2022; 22: 5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madan P, Singh V, Singh DP, et al. A hybrid deep learning approach for ECG-based arrhythmia classification. Bioengineering 2022; 9: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang M, Liu W, Zhang H. A robust multiple heartbeats classification with weight-based loss based on convolutional neural network and bidirectional long short-term memory. Front Physiol 2022; 13: 2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Acharya UR, Oh SL, Hagiwara Y, et al. A deep convolutional neural network model to classify heartbeats. Comput Biol Med 2017; 89: 389–396. [DOI] [PubMed] [Google Scholar]
- 12.Limaye H, Deshmukh V. ECG Noise sources and various noise removal techniques: a survey. Int J Appl Innov Eng Manage 2016; 5: 86–92. [Google Scholar]
- 13.Alquran H, Alqudah AM, Abu-Qasmieh I, et al. ECG classification using higher order spectral estimation and deep learning techniques. Neural Netw World 2019; 29: 207–219. [Google Scholar]
- 14.Chollet F. Deep learning with Python. 2nd ed. USA: Manning, 2021. [Google Scholar]
- 15.Chou Y-h, Hong S, Zhou Y, et al. Knowledge-shot learning: an interpretable deep model for classifying imbalanced electrocardiography data. Neurocomputing 2020; 417: 64–73. [Google Scholar]
- 16.Loh BC, Then PH. Deep learning for cardiac computer-aided diagnosis: benefits, issues & solutions. Mhealth 2017; 3: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murat F, Yildirim O, Talo M, et al. Application of deep learning techniques for heartbeats detection using ECG signals-analysis and review. Comput Biol Med 2020; 120: 103726. [DOI] [PubMed] [Google Scholar]
- 18.Wang J. An intelligent computer-aided approach for atrial fibrillation and atrial flutter signals classification using modified bidirectional LSTM network. Inf Sci (NY) 2021; 574: 320–332. [Google Scholar]
- 19.Petmezas G, Stefanopoulos L, Kilintzis V, et al. State-of-the-art deep learning methods on electrocardiogram data: systematic review. JMIR Med Inform 2022; 10: e38454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiranyaz S, Avci O, Abdeljaber O, et al. 1D convolutional neural networks and applications: a survey. Mech Syst Signal Process 2021; 151: 107398. [Google Scholar]
- 21.Liu X, Wang H, Li Zet al. et al. Deep learning in ECG diagnosis: a review. Knowl Based Syst 2021; 227: 107187. [Google Scholar]
- 22.Huang R, Wei C, Wang B, et al. Well performance prediction based on long short-term memory (LSTM) neural network. J Pet Sci Eng 2022; 208: 109686. [Google Scholar]
- 23.Yu Y, Si X, Hu Cet al. et al. A review of recurrent neural networks: LSTM cells and network architectures. Neural Comput 2019; 31: 1235–1270. [DOI] [PubMed] [Google Scholar]
- 24.Siami-Namini S, Tavakoli N, Namin AS. The performance of LSTM and BiLSTM in forecasting time series. 2019 IEEE International Conference on Big Data (Big Data); 2019: IEEE.
- 25.Li L, Wan J, Zheng Jet al. et al. Biomedical event extraction based on GRU integrating attention mechanism. BMC Bioinf 2018; 19: 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang R, Chen T, Xiao Fet al. et al. Bi-directional gated recurrent unit recurrent neural networks for failure prognosis of proton exchange membrane fuel cells. Int J Hydrogen Energy 2022; 47: 33027–33038. [Google Scholar]
- 27.Ebrahimi Z, Loni M, Daneshtalab Met al. et al. A review on deep learning methods for ECG arrhythmia classification. Expert Syst Appl: X 2020; 7: 100033. [Google Scholar]
- 28.Luz E, Schwartz WR, Cámara-Chávez Get al. et al. ECG-based heartbeat classification for arrhythmia detection: a survey. Comput Meth Programs Biomed 2016; 127: 144–164. [DOI] [PubMed] [Google Scholar]
- 29.Romdhane TF, Pr MA. Electrocardiogram heartbeat classification based on a deep convolutional neural network and focal loss. Comput Biol Med 2020; 123: 103866. [DOI] [PubMed] [Google Scholar]
- 30.Gao J, Zhang H, Lu Pet al. et al. An effective LSTM recurrent network to detect arrhythmia on imbalanced ECG dataset. J Healthc Eng 2019; 2019: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin R, Qiao K, Wang L, et al. Weighted focal loss: An effective loss function to overcome unbalance problem of chest X-ray14. IOP Conference Series: Materials Science and Engineering; 2018: IOP Publishing.
- 32.Zheng J, Zhang J, Danioko S, et al. A 12-lead electrocardiogram database for arrhythmia research covering more than 10,000 patients. Sci Data 2020; 7: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pham B-T, Le PT, Tai T-C, et al. Electrocardiogram heartbeat classification for arrhythmias and myocardial infarction. Sensors 2023; 23: 2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner P, Strodthoff N, Bousseljot R-D, et al. PTB-XL, a large publicly available electrocardiography dataset. Sci Data 2020; 7: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu M, Lu Y, Yang Wet al. et al. A study on arrhythmia via ECG signal classification using the convolutional neural network. Front Comput Neurosci 2021; 14: 564015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed AA, Ali W, Abdullah TAet al. et al. Classifying cardiac arrhythmia from ECG signal using 1D CNN deep learning model. Mathematics 2023; 11: 562. [Google Scholar]
- 37.Rai HM, Chatterjee K, Dashkevych S. The prediction of cardiac abnormality and enhancement in minority class accuracy from imbalanced ECG signals using modified deep neural network models. Comput Biol Med 2022; 150: 106142. [DOI] [PubMed] [Google Scholar]
- 38.Islam MS, Islam MN, Hashim N, et al. New hybrid deep learning approach using BiGRU-BiLSTM and multilayered dilated CNN to detect arrhythmia. IEEE Access 2022; 10: 58081–58096. [Google Scholar]
- 39.Islam MS, Hasan KF, Sultana S, et al. HARDC : a novel ECG-based heartbeat classification method to detect arrhythmia using hierarchical attention based dual structured RNN with dilated CNN. Neural Netw 2023; 162: 271–287. [DOI] [PubMed] [Google Scholar]
- 40.Rai HM, Chatterjee K. Hybrid CNN-LSTM deep learning model and ensemble technique for automatic detection of myocardial infarction using big ECG data. Appl Intell 2022; 52: 5366–5384. [Google Scholar]
- 41.Rai HM, Chatterjee K, Dubey A, Srivastava P. Myocardial Infarction Detection Using Deep Learning and Ensemble Technique from ECG Signals. Proceedings of Second International Conference on Computing, Communications, and Cyber-Security; 2021 2021//; Singapore: Springer Singapore.
- 42.Rai HM, Chatterjee K, Mukherjee C. Hybrid CNN-LSTM model for automatic prediction of cardiac arrhythmias from ECG big data. 2020 IEEE 7th Uttar Pradesh Section International Conference on Electrical, Electronics and Computer Engineering (UPCON); 2020 27–29 Nov. 2020.
- 43.Alireza S, Alireza R, Farshid H. Diagnosing left bundle branch block in 12-lead electrocardiogram using self-attention convolutional neural networks. medRxiv. 2023:2023.06.25.23291867.
- 44.Zheng J. A 12-lead electrocardiogram database for arrhythmia research covering more than 10,000 patients 2019 [Available from: https://figshare.com/collections/ChapmanECG/4560497/2. [DOI] [PMC free article] [PubMed]
- 45.Pandey H, Tiwari R. An innovative design approach of Butterworth filter for noise reduction in ECG signal processing based applications. Prog Sci Eng Res J PISER 2014; 12: 332–337. [Google Scholar]
- 46.Rahul J, Sora M, Sharma L. Baseline correction of ECG using regression estimation method. 2019 4th International Conference on Internet of Things: Smart Innovation and Usages (IoT-SIU); 2019: IEEE.
- 47.Singh P, Shahnawazuddin S, Pradhan G. An efficient ECG denoising technique based on non-local means estimation and modified empirical mode decomposition. Circuits Syst Signal Process 2018; 37: 4527–4547. [Google Scholar]
- 48.Qian C, Su H, Yu H. Local means denoising of ECG signal. Biomed Signal Process Control 2019; 53: 101571. [Google Scholar]
- 49.Chawla NV, Bowyer KW, Hall LOet al. et al. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 2002; 16: 321–357. [Google Scholar]
- 50.Liu R, Ji C, Niu Jet al. et al. Research on intrusion detection method based on 1D-ICNN-BiGRU. J Phys Conf Ser 2022; 2347: 012001. [Google Scholar]
- 51.Swana EF, Doorsamy W, Bokoro P. Tomek link and SMOTE approaches for machine fault classification with an imbalanced dataset. Sensors (Basel) 2022; 22: 3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng Z, Chen Z, Hu F, et al. An automatic diagnosis of arrhythmias using a combination of CNN and LSTM technology. Electronics (Basel) 2020; 9: 121. [Google Scholar]
- 53.Qian Y, Bi M, Tan Tet al. et al. Very deep convolutional neural networks for noise robust speech recognition. IEEE/ACM Trans Audio, Speech Lang Process 2016; 24: 2263–2276. [Google Scholar]
- 54.Zhang D, Chen Y, Chen Y, et al. An ECG heartbeat classification method based on deep convolutional neural network. J Healthc Eng 2021; 2021: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Abdu-Aguye MG, Gomaa W, Makihara Y, Yagi Y. Adaptive pooling is all you need: an empirical study on hyperparameter-insensitive human action recognition using wearable sensors. 2020 International Joint Conference on Neural Networks (IJCNN); 2020: IEEE.
- 56.Andersen RS, Peimankar A, Puthusserypady S. A deep learning approach for real-time detection of atrial fibrillation. Expert Syst Appl 2019; 115: 465–473. [Google Scholar]
- 57.Zhu C, Liu Q, Meng W, et al. An attention-based cnn-lstm model with limb synergy for joint angles prediction. 2021 IEEE/ASME International Conference on Advanced Intelligent Mechatronics (AIM); 2021: IEEE.
- 58.Cahuantzi R, Chen X, Güttel S. A comparison of LSTM and GRU networks for learning symbolic sequences. arXiv preprint arXiv:210702248. 2021.
- 59.Hu Y, Wong Y, Wei W, et al. A novel attention-based hybrid CNN-RNN architecture for sEMG-based gesture recognition. PLoS One 2018; 13: e0206049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Z, Wang Q. R-transformer network based on position and self-attention mechanism for aspect-level sentiment classification. IEEE Access 2019; 7: 127754–64. [Google Scholar]
- 61.Li X, Ding L, Du Y, et al. Position-enhanced multi-head self-attention based bidirectional gated recurrent unit for aspect-level sentiment classification. Front Psychol 2021; 12: 799926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin T-Y, Goyal P, Girshick R, et al. Focal loss for dense object detection. Proceedings of the IEEE international conference on computer vision; 2017. [DOI] [PubMed]
- 63.Rojas E, Kahira AN, Meneses E, et al. A study of checkpointing in large scale training of deep neural networks. arXiv preprint arXiv:201200825. 2020.
- 64.Faust O, Acharya UR. Automated classification of five arrhythmias and normal sinus rhythm based on RR interval signals. Expert Syst Appl 2021; 181: 115031. [Google Scholar]
- 65.Meqdad MN, Abdali-Mohammadi F, Kadry S. Meta structural learning algorithm with interpretable convolutional neural networks for arrhythmia detection of multisession ECG. IEEE Access 2022; 10: 61410–61425. [Google Scholar]
- 66.Meqdad MN, Abdali-Mohammadi F, Kadry S. A new 12-lead ECG signals fusion method using evolutionary CNN trees for arrhythmia detection. Mathematics 2022; 10: 1911. [Google Scholar]
- 67.Sepahvand M, Abdali-Mohammadi F. A novel method for reducing arrhythmia classification from 12-lead ECG signals to single-lead ECG with minimal loss of accuracy through teacher-student knowledge distillation. Inf Sci (NY) 2022; 593: 64–77. [Google Scholar]
- 68.Andayeshgar B, Abdali-Mohammadi F, Sepahvand M, et al. Developing graph convolutional networks and mutual information for arrhythmic diagnosis based on multichannel ECG signals. Int J Environ Res Public Health 2022; 19: 10707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sadeghi A, Rezaee A, Hajati F. Diagnosing Left Bundle Branch Block in 12-lead Electrocardiogram using Self-Attention Convolutional Neural Networks 2023.
- 70.Ghousia Begum S, Priyadarshi E, Pratap S, et al. Automated detection of abnormalities in ECG signals using deep neural network. Biomed Eng Adv 2023; 5: 100066. [Google Scholar]
- 71.Khan F, Yu X, Yuan Zet al. et al. ECG classification using 1-D convolutional deep residual neural network. PLoS One 2023; 18: e0284791. [DOI] [PMC free article] [PubMed] [Google Scholar]