Abstract
Exposure of yeast cells to increases in extracellular osmolarity activates the HOG1 mitogen-activated protein (MAP) kinase cascade, which is composed of three tiers of protein kinases: (i) the SSK2, SSK22, and STE11 MAP kinase kinase kinases (MAPKKKs), (ii) the PBS2 MAPKK, and (iii) the HOG1 MAP kinase. Activation of the MAP kinase cascade is mediated by two upstream mechanisms. The SLN1-YPD1-SSK1 two-component osmosensor activates the SSK2 and SSK22 MAPKKKs by direct interaction of the SSK1 response regulator with these MAPKKKs. The second mechanism of HOG1 MAP kinase activation is independent of the two-component osmosensor and involves the SHO1 transmembrane protein and the STE11 MAPKKK. Only PBS2 and HOG1 are common to the two mechanisms. We conducted an exhaustive mutant screening to identify additional elements required for activation of STE11 by osmotic stress. We found that strains with mutations in the STE50 gene, in combination with ssk2Δ ssk22Δ mutations, were unable to induce HOG1 phosphorylation after osmotic stress. Both two-hybrid analyses and coprecipitation assays demonstrated that the N-terminal domain of STE50 binds strongly to the N-terminal domain of STE11. The binding of STE50 to STE11 is constitutive and is not affected by osmotic stress. Furthermore, the two proteins relocalize similarly after osmotic shock. It was concluded that STE50 fulfills an essential role in the activation of the high-osmolarity glycerol response pathway by acting as an integral subunit of the STE11 MAPKKK.
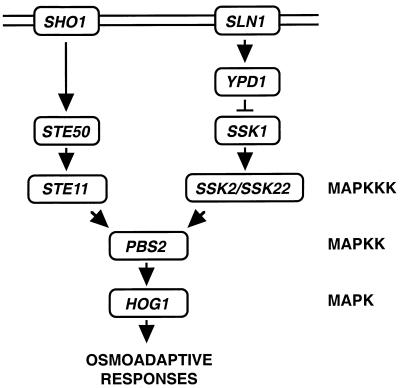
Mitogen-activated protein (MAP) kinase cascades are common signaling modules found in both higher and lower eukaryotic cells. A typical MAP kinase cascade is composed of three tiers of protein kinases, a MAP kinase (MAPK), a MAPK kinase (MAPKK), and a MAPKK kinase (MAPKKK) (27). Yeast cells have several distinct MAP kinase cascades that transduce distinct extracellular stimuli (e.g., mating pheromone, high osmolarity, low osmolarity, and nitrogen starvation) (10, 19). Budding yeast (Saccharomyces cerevisiae) responds to increases in osmolarity in the extracellular environment by activating the HOG1 MAP kinase cascade. This cascade is essential for the survival of yeast in high-osmolarity environments (4, 5). Because a major outcome of the activation of this MAPK pathway is the elevated synthesis of glycerol, this pathway is referred to as the HOG (high-osmolarity glycerol response) pathway (2, 5). Extracellular hyperosmolarity is detected by either of two transmembrane proteins, SLN1 and SHO1 (Fig. 1).
FIG. 1.
Schematic diagram of a current model of the yeast HOG osmoregulatory signal transduction pathway. The arrows do not necessarily indicate direct interactions.
SLN1 is a part of a complex regulatory system with homology to prokaryotic two-component signal transducers. The yeast two-component osmosensor is composed of SLN1, a transmembrane protein that contains an extracellular sensor domain and cytoplasmic histidine kinase and receiver domains, the intermediary protein YPD1, and the response regulator SSK1 (14, 15, 20). The SLN1-YPD1-SSK1 two-component osmosensor works by a multistep phosphorelay mechanism (20). The unphosphorylated form of SSK1 activates SSK2 and SSK22 MAPKKKs by binding to their N-terminal inhibitory domains (17). Once activated, SSK2 and SSK22 phosphorylate the PBS2 MAPKK.
However, the SLN1-YPD1-SSK1 multistep phosphorelay is not the only osmosensing mechanism in yeast. Initially, this was revealed because, unlike highly osmosensitive hog1Δ or pbs2Δ mutants, neither ssk1Δ mutants nor ssk2Δ ssk22Δ double mutants exhibited the expected osmosensitive phenotype. Thus, we conducted a mutant screening based on the assumption that simultaneous inactivation of SSK2, SSK22, and a gene involved in the alternative activation mechanism would create an osmosensitive (Osms) phenotype. Briefly, ssk2Δ ssk22Δ double mutants were mutagenized, and Osms mutants were selected. This screening identified two genes (SHO1 and STE11) whose mutations were synthetically Osms with ssk2 ssk22. SHO1 has four predicted transmembrane segments and a C-terminal cytoplasmic region that contains an SH3 domain. The SH3 domain binds a proline-rich motif in the N-terminal segment of the PBS2 MAPKK. This interaction is a requirement for the activation of the PBS2 MAPKK by the STE11 MAPKKK (13, 16). Thus, PBS2 can be independently activated by two different upstream mechanisms: one involving the SLN1-YPD1-SSK1 two-component osmosensor that activates SSK2 and SSK22 MAPKKKs, and another involving the SHO1 transmembrane protein and the STE11 MAPKKK. Either SSK2, SSK22, or STE11 MAPKKK can activate PBS2 by phosphorylation. Once phosphorylated, PBS2 MAPKK activates the HOG1 MAPK, which induces diverse stress responses.
The mechanism by which SHO1 induces activation of STE11 is not clear. In particular, there remained the possibility that additional elements were required for signal transduction in the SHO1-STE11 branch of the HOG pathway. Therefore, we expanded the synthetic osmosensitive mutant screening to apparent saturation. Thus, we found that in addition to the previously revealed SHO1 and STE11 genes, the STE50 gene is involved in the HOG pathway. Although STE50 was originally identified as a modulator of the pheromone response pathway, ste50 mutations had only moderate effects on mating signal transduction (21, 28). In contrast, we demonstrate in this report that STE50 is absolutely required for the SHO1-STE11-mediated activation of the HOG pathway.
MATERIALS AND METHODS
Yeast strains.
The yeast strains used were FP54 (MATa ura3 leu2 trp1 his3 ste11::HIS3), FP66 (MATa ura3 leu2 trp1 his3 ste50::HIS3), FP67 (MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 ste50::HIS3), FP68 (MATα ura3 leu2 trp1 his3 sho1::TRP1 ste50::HIS3), FP75 (MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 ste11::HIS3), L40 (MATa trp1 leu2 his3 LYS2::lexA-HIS3 URA3::lexA-lacZ), MY007 (MATa ura3 leu2 his3 ssk2::LEU2 ssk22::LEU2 sho1::HIS3), TM101 (MATα ura3 leu2 his3), TM141 (MATa ura3 leu2 trp1 his3), TM252 (MATa ura3 leu2 trp1 ssk2::LEU2 ssk22::LEU2), TM257 (MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2), and TM261 (MATα ura3 leu2 his3 pbs2::LEU2).
Buffers and media.
Tris-maleate buffer contains 50 mM Tris base, 50 mM maleic acid, 7.5 mM ammonium acetate, 0.4 mM MgSO4 and 30 mM CaCl2 (adjusted to pH 6.0 with NaOH). Buffer A consists of 50 mM Tris-HCl (pH 7.5), 15 mM EDTA, 15 mM EDTA, 2 mM dithiothreitol (DTT), 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 5 μg of leupeptin per ml. Buffer B consists of 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 2 mM DTT. Phosphatase inhibitor cocktail consists of 10 mM NaF, 1 mM sodium pyrophosphate, 1 mM sodium vanadate, and 10 mM β-glycerol phosphate. Sodium dodecyl sulfate (SDS) loading buffer consists of 50 mM Tris-HCl (pH 6.8), 100 mM DTT, 2% SDS, 0.1% bromophenol blue, and 10% glycerol. YPD medium contains 10 g of yeast extract, 20 g of tryptone, and 20 g of dextrose per liter. YPGal medium contains 10 g of yeast extract, 20 g of tryptone, and 20 g of galactose per liter.
Isolation of osmosensitive mutants.
Yeast strain TM252 (ssk2Δ ssk22Δ) was mutagenized with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) as follows. TM252 cells were grown in YPD at 30°C to an optical density at 600 nm of 0.3, washed twice in Tris-maleate buffer (pH 6.0), and resuspended in 1/5 of the original volume in the same washing buffer (1). Thirty microliters of 1-mg/ml MNNG solution in 10 mM sodium acetate buffer (pH 5.0) was added to 970 μl of yeast cell suspension in a microcentrifuge tube, and the mixture was incubated at 30°C for 60 min. Cells were sedimented by a brief centrifugation and resuspended in 1 ml of 1% sodium thiosulfate solution (sterilized by filtration). After another centrifugation, cell pellets were resuspended in 5 ml of YPD and incubated at 30°C for 4 h. Glycerol was added to a final concentration of 30%, and aliquots were stored frozen at −80°C. Mutagenized cells were plated on YPD plates (∼500 colonies/plate). After incubation at 30°C for 2 to 3 days, colonies were replica plated onto YPD–1.5 M sorbitol plates. Both the master plates and replica plates were incubated at 30°C for another day or two. Mutants that failed to grow on sorbitol plates were recovered from the master plates. Single colonies were isolated and tested for osmosensitivity on sorbitol plates.
Plasmids.
The yeast expression vector YCpIF16 (PGAL1-hemagglutinin [HA] TRP1+ CEN) allows the expression of HA fusion proteins under control of the GAL1 promoter (9), and p426TEG1 (PTEF1–glutathione S-transferase [GST], URA3+ 2μm) allows the expression of GST fusion proteins under control of the PTEF1 promoter (24). Full-length and several mutant alleles of STE50 and STE11 were cloned into plasmids YCpIF16 and p426TEG1. Plasmid vectors for green fluorescent protein (GFP) tag (pJK50) and Myc3 tag (p1148) were obtained from P. Silver. The tag sequences amplified by PCR were subcloned into pRS416 (URA3+ CEN) and pRS415 (TRP1+ CEN) (23). A STE11ΔSTE50BD mutant containing a deletion of the STE50 binding domain (STE5DBD; amino acids 74 to 152) was made by PCR and verified by DNA sequencing. Yeast expression plasmids for full-length STE50, the N terminus of STE50 (STE50-N; amino acids 1 to 166), and the C terminus of STE50 (STE50-C; amino acids 163 to 346) were constructed by using pYES2 (PGAL1 URA3+ 2μm) (Invitrogen).
Quantitative mating assay.
Cells were grown at 30°C to mid-exponential phase. Mutant strains to be tested (MATa) were mixed with a fourfold excess of an appropriate tester strain (MATα) and immediately filtered through nitrocellulose membranes. The membranes were placed on YPD plates and incubated at 30°C for 4 h to allow mating to proceed. The cells were then washed off the membranes and titrated on selective plates to determine the number of diploid cells formed. Mating efficiency was defined as the number of diploid cells formed divided by the number of experimental (MATa) haploid cells.
Two-hybrid analysis.
The two-hybrid analysis was carried out essentially as described by Durfee et al. (6), using pACTII (11) and pBTM116 (26) as the activation domain plasmid and the LexA DNA binding domain plasmid, respectively. Serial deletion constructs of STE11 and STE50 were prepared by using the Erase-a-Base system (Promega), PCR techniques, and standard DNA procedures. Binding domain plasmids carrying full-length or fragments of STE11 and pACTII plasmids carrying full-length or partial clones of STE50 were cotransformed into the L40 reporter strain. Transformant cells (∼5 × 106) were spotted onto a YPD plate, and after 5 h at 30°C the cells were copied onto a nitrocellulose membrane. β-Galactosidase activity was visualized with the use of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as described elsewhere (26).
In vivo coprecipitation assays.
Cells were grown in the presence of 2% galactose to an optical density at 660 nm of ∼0.5. Cells were treated with a brief osmotic shock (0.4 M NaCl for 5 min) before being harvested by centrifugation. Cells were suspended in buffer A and ground by using glass beads, and supernatant (extract) was prepared by centrifugation. Cell extract (750 μg in buffer A plus 150 mM NaCl) was incubated with 50 μl of glutathione-Sepharose beads overnight at 4°C. Beads were washed extensively with buffer A plus 150 mM NaCl, resuspended in loading buffer, and separated by SDS-polyacrylamide gel electrophoresis. Immunoblotting was done with anti-HA monoclonal antibody 12CA5 (Boehringer Mannheim) and an anti-GST monoclonal antibody (Pharmacia) together with ECL reagent (Amersham). When GFP- and Myc3-tagged STE11 (STE11-GFP and STE50-Myc) were coprecipitated, cells were grown in presence of glucose. Extracts were prepared as before, and STE11-GFP was precipitated by incubation overnight at 4°C with an anti-GFP polyclonal antibody (gift from P. Silver), followed by addition of 50 μl of protein A-Sepharose (Pharmacia) for 1 h at 4°C. Samples were washed with buffer A plus 150 mM NaCl. Following SDS-polyacrylamide gel electrophoresis, immunoblotting was done with anti-Myc monoclonal antibody 9E10 (BabCO) or an anti-GFP polyclonal antibody.
GFP fluorescence microscopy.
GFP was visualized without fixation using a Nikon Optiphot-2 equipped with a MicroMAX charge-coupled device (CCD) camera (Princeton Instruments). Images were taken at a magnification of 100× and converted to Photoshop version 4.0 format (Adobe Systems).
RESULTS
Identification of additional elements required for SSK2/SSK22-independent activation of the HOG1 MAP kinase cascade.
As mentioned in the introduction, ssk1Δ or ssk2Δ ssk22Δ strains are osmoresistant, whereas pbs2Δ strains are highly osmosensitive. This would not be expected if SSK1 (the activator of SSK2/SSK22) and SSK2/SSK2 MAPKKKs are the only means of activating PBS2. Thus, it was predicted that there is an SSK2/SSK22-independent mechanism that can activate PBS2 upon osmotic shock. To identify the elements involved in the alternative osmosensing mechanism, we conducted a mutant screening by which we isolated osmosensitive mutants from a ssk2Δ ssk22Δ double-mutant strain (13, 16). We identified two genes, those encoding the STE11 MAPKKK and the novel transmembrane protein SHO1, mutations of which, combined with the ssk2Δ ssk22Δ double mutation, causes an Osms phenotype. It has been shown that STE11 can activate PBS2 MAPKK both in vivo and in vitro. It was also shown that the C-terminal SH3 domain of SHO1 binds PBS2. However, the mutant screening was probably not saturated, because only one mutant for each of SHO1 and STE11 was isolated.
Thus, to identify additional elements required for the SHO1-STE11 branch of the HOG pathway, an exhaustive screening was performed. The ssk2Δ ssk22Δ strain TM252 was mutagenized with MNNG as described in Materials and Methods; then 200,000 mutagenized cells were grown on YPD plates and replicated onto YPD plates containing 1.5 M sorbitol. By comparing the master and the replica plates, we identified Osms colonies. After single-colony isolation, those candidate mutants were tested again for osmosensitivity (i.e., inability to grow on YPD-sorbitol plates). Thus, 140 Osms mutants were identified. In the next step, we identified the mutants of the already identified genes, namely hog1, pbs2, and sho1, by complementation analysis. As summarized in Table 1, we obtained numerous strains with mutations of these three genes. STE11 mutants could not be directly tested by the complementation test, as they were sterile. However, these mutants could be identified by their sterility in the mating test and by subsequent complementation by an STE11-containing plasmid (Table 1). The seven sterile mutants isolated in this screening were defective in the STE11 gene.
TABLE 1.
Summary of a screening for osmosensitive mutants
| Complementation group | No. of mutants/ 140 cells screened |
|---|---|
| hog1 | 17 |
| pbs2 | 34 |
| sho1 | 7 |
| ste11 | 7 |
| ste50 | 3 |
| gpd1 | 10 |
| X | 17 |
| tat1 | 4 |
| Unclassified mutants | 41 |
Yeast strain TM252 (ssk2Δ ssk22Δ) was mutagenized with MNNG as described in Materials and Methods. Mutagenized cells were grown on YPD plates and replica plated onto YPD–1.5 M sorbitol plates. Mutant cells that failed to grow on sorbitol-containing plates were isolated and classified into complementation groups. Mutants in the gpd1, X, and tat1 complementation groups, as well as the 41 unclassified mutants, were not defective in the hyperosmolarity-induced activation of the HOG1 MAPK and were not characterized further.
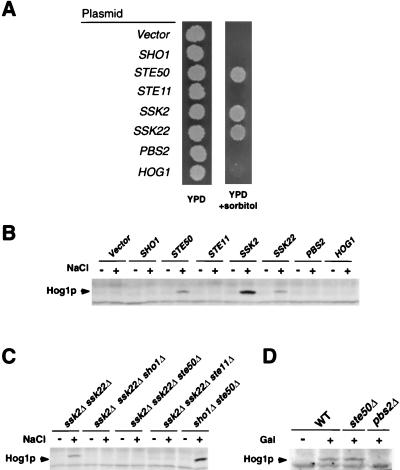
The remaining Osms mutants contained a number of distinct complementation groups. Of those, the largest number of mutants belonged to an unknown gene (X). Two additional complementation groups were identified as gpd1 and tat1 mutants, by complementation cloning. The GPD1 gene encodes glycerol-3-phosphate dehydrogenase, a key enzyme for the synthesis of the compatible osmolyte glycerol (2). The TAT1 gene encodes a valine/tyrosine/tryptophan permease; it is likely that these amino acids also act as compatible osmolytes that maintain the osmotic balance (22). X, GPD1, and TAT1 mutants do not affect the activation of the HOG1 MAPK, because immunoblot analysis of these mutants demonstrated that the HOG1 tyrosine phosphorylation (an indicator of the HOG1 activation) occurred as efficiently as in wild-type cells. Thus, these genes may be important for yeast osmoregulation but not in the activation of the HOG1 MAP kinase cascade itself. To identify mutations that are defective in HOG1 activation among the remaining mutants, we tested individual Osms mutants by immunoblot analysis to see if they are capable of inducing HOG1 tyrosine phosphorylation after osmotic stress. Thus, we found three additional mutants (FOS-41, FOS-174, and FOS-226) that are defective in HOG1 phosphorylation. We isolated the genes that are responsible for their osmosensitivity by complementation using a genomic library. Characterization of the complementing genomic clones by mapping and partial sequencing revealed that they contained either the SSK2, SSK22, or STE50 gene. Figure 2A shows that the osmosensitivity of FOS-41 can be complemented by either SSK2, SSK22, or STE50 carried on a single-copy vector. Essentially identical results were obtained for FOS-174 and FOS-226. The STE50 gene, which encodes a protein of 346 amino acids, was previously implicated in the pheromone response pathway (21, 28).
FIG. 2.
Activation of the HOG1 MAPK by SHO1 requires STE50. (A) Osmosensitivity of FOS-41 (ssk2Δ ssk22Δ ste50-41). FOS-41 was transformed with centromeric plasmids containing the indicated genes. The transformants were spotted on YPD plates with or without 1.5 M sorbitol. (B) Tyrosine phosphorylation of HOG1 induced by high osmolarity. Yeast strain FP67 (ssk2Δ ssk22Δ ste50Δ) was transformed with plasmids carrying the indicated genes. Cells were collected before (−) or 5 min after (+) the addition of NaCl to a final concentration of 0.4 M. Tyrosine-phosphorylated HOG1 (Hog1p) was detected by immunoblot analysis using a monoclonal antibody specific to phosphotyrosine (4G10). (C) High-osmolarity-induced tyrosine phosphorylation of HOG1 in various mutant strains. Cells were treated, and tyrosine-phosphorylated HOG1 (Hog1p) was detected as for panel B. (D) Tyrosine phosphorylation of HOG1 induced by the constitutively active STE11ΔN. Plasmid pGal-STE11ΔN was transformed into wild-type (WT), ste50Δ, and pbs2Δ strains. Cells were grown in synthetic medium containing raffinose, and samples were taken before (−) or 1 h after (+) addition of galactose to induce the expression of STE11ΔN. Tyrosine-phosphorylated HOG1 (Hog1p) was detected by immunoblotting.
Synthetic osmosensitivity of ste50 ssk2 ssk22 triple mutants.
To exclude the possibility that STE50 was merely acting as a multicopy suppressor of FOS-41 and the other two mutants, we generated a disruption mutation of STE50 in an ssk2Δ ssk22Δ background. The ssk2Δ ssk22Δ ste50Δ strain (FP67) was as osmosensitive as FOS-41 (data not shown), indicating that it is very likely that FOS-41 is defective in the STE50 gene. However, because none of the three suspected ste50 mutants were mating defective, we used a ste50Δ strain to reinvestigate the involvement of STE50 in the mating pathway. As shown in Table 2, ste50Δ cells showed only a slight decrease in mating efficiency when mated with an isogenic wild-type strain. Mating deficiency was more severe when both partners were ste50 deficient (but not as defective as the ste11 mutant). Thus, these results are consistent with the apparent mating competence of FOS-41 and the other two mutants. Furthermore, these results confirm that the role of STE50 in the mating pathway is a modulatory one rather than essential, as previously suggested (21, 28).
TABLE 2.
Mating efficiencies
| Relevant genotype
|
Mating efficiency (%)a | |
|---|---|---|
| MATa strain | MATα strain | |
| Wild type | Wild type | 90.3 ± 2.4 |
| ste11Δ | Wild type | <0.0001 |
| ste50Δ | Wild type | 34.7 ± 6.1 |
| ste50Δ | ste50Δ | 0.8 ± 0.2 |
Determined by mixing exponentially growing MATa cells with a fourfold excess of MATα cells (see Materials and Methods) and calculated as [(number of diploid cells formed)/(number of MATa cells)] × 100. Data represents means ± standard deviations of results from three independent experiments.
The ssk2Δ ssk22Δ ste50Δ triple mutant has completely lost the capacity to activate the HOG pathway, as indicated by its inability to tyrosine phosphorylate HOG1 after osmotic stress (Fig. 2B and C). Transformation of the triple-mutant strain with a plasmid carrying either SSK2, SSK22, or STE50 restored HOG1 activation in response to osmotic shock, whereas expression of SHO1, STE11, PBS2, or HOG1 had no apparent effect (Fig. 2B). Any of the three mutations, sho1Δ, ste11Δ, or ste50Δ, when combined with the ssk2Δ ssk22Δ double mutation, disabled the activation of HOG1 upon osmotic shock (Fig. 2C). In a sho1Δ ste50Δ double mutant, in which both SSK2 and SSK22 are functional, HOG1 activation in response to osmotic shock was normal (Fig. 2C). Similarly, sho1Δ ste11Δ double mutants were osmoresistant and capable of activating HOG1 (16). These results suggest that SHO1, STE11, and STE50 are involved in the same upstream branch (the SHO1-STE11 branch) of the HOG pathway.
We then tested whether STE50 is required for STE11 to phosphorylate its substrate PBS2 (for example, STE50 might modulate STE11 affinity to PBS2). If this is the case, it would be expected that STE50 is required for PBS2 phosphorylation even by a constitutively active mutant form of STE11 (STE11ΔN). As previously observed (16), expression of STE11ΔN resulted in PBS2-mediated tyrosine phosphorylation of HOG1 in the absence of osmotic stress (Fig. 2D). Significantly, STE11ΔN could induce HOG1 phosphorylation in a ste50Δ strain, indicating that STE50 is not required for the active STE11 to phosphorylate PBS2. It is therefore more likely that STE50 acts upstream of STE11.
STE50 interacts with STE11 MAPKKK but not with SHO1.
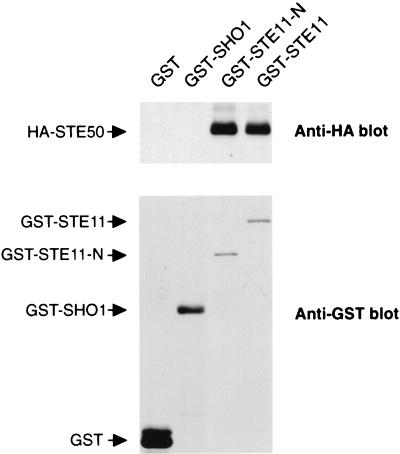
STE50 has some structural similarities to the Schizosaccharomyces pombe Ste4 protein, which has also been implicated in the mating pathway. Ste4 interacts with the N-terminal noncatalytic domain of the Byr2 MAPKKK (a homolog of STE11) (3, 25). It has also been suggested by two-hybrid analysis that STE50 interacts with STE11 (3, 28). To examine whether STE50 was able to interact with STE11 and other components in the SHO1-STE11 branch of the HOG pathway, we performed coprecipitation experiments. Yeast cells were cotransformed with a plasmid that expresses either GST-SHO1 or GST-STE11 and another plasmid that expresses HA-STE50. GST-tagged proteins were precipitated by using glutathione-Sepharose beads, and HA-STE50 in precipitates was probed with an anti-HA monoclonal antibody. As shown in Fig. 3, GST-STE11, but not GST alone or GST-SHO1, coprecipitated HA-STE50. The STE11 N-terminal noncatalytic domain alone (GST-STE11-N; amino acids 1 to 434) also coprecipitated amounts of HA-STE50 equivalent to those of the full-length STE11. The binding of STE11 to STE50 was also observed in sho1Δ and pbs2Δ backgrounds, indicating that neither SHO1 nor PBS2 was required as a bridge between STE11 and STE50 (data not shown). Thus, STE50 interacts with STE11, within the N-terminal regulatory domain of STE11.
FIG. 3.
In vivo binding of STE50 to STE11. Wild-type yeast strain TM141 was cotransformed with a plasmid expressing GST, GST-STE11, GST-STE11-N, or GST-SHO1 and another plasmid expressing HA-STE50. GST proteins were precipitated by using glutathione-Sepharose beads, and the presence of HA-STE50 in precipitates was probed by immunoblotting with anti-HA monoclonal antibody 12CA5 (upper panel). GST fusion proteins in the precipitates were detected by an anti-GST antibody (lower panel).
STE50-N binds to STE50 binding site in the N-terminal domain of STE11.
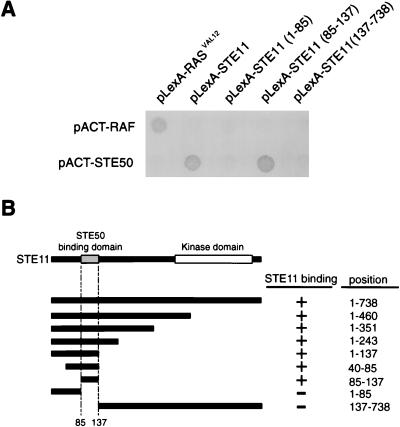
To define the specific regions in STE50 and STE11 that are essential for their interaction, two-hybrid analyses were carried out. To map the STE50 binding site in STE11, various segments of STE11 were fused to the LexA-DNA binding domain, and their interaction with the full-length STE50 fused to the GAL4 activator domain was tested. Figure 4A shows a typical result, and Fig. 4B summarizes the results for various STE11 deletion constructs. These analyses indicated that STE11 residues 85 to 137 are sufficient for binding to STE50, consistent with the coprecipitation experiments. We thus designated this segment STE50BD.
FIG. 4.
Two-hybrid analysis of the STE50-binding domain in STE11. (A) Interactions of various STE11 fragments fused to the LexA-DNA binding domain with the STE50 fragments fused to the GAL4 activator domain. The results, representative of several filter β-galactosidase assays, demonstrate the interactions between STE11 and STE50. Amino acid positions of the STE11 fragments included in the constructs are indicated in parentheses. pACT-STE50 contains the entire STE50 coding sequence. Proteins encoded by the control plasmids pLexA-RASVAL12 and pACT-RAF interact with each other (26). (B) Summary of two-hybrid interaction between STE11 and STE50. Positions of the STE11 segments included in the LexA-DNA binding domain are schematically shown on the left, and their precise amino acid positions are indicated on the right. The presence (+) or absence (−) of interaction is indicated.
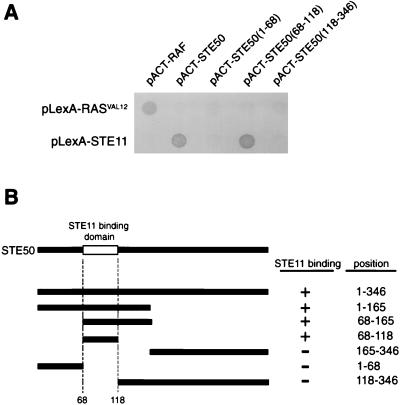
To determine the region in STE50 that is essential for its interaction with the N-terminal domain of STE11, we fused various STE50 segments to the GAL4 activation domain and tested these constructs for interaction with the full-length STE11 fused to the LexA-DNA binding domain. As shown in Fig. 5, an N-terminal segment of STE50 (amino acids 68 to 118) is necessary and sufficient for binding to STE11.
FIG. 5.
Two-hybrid analysis of the STE11BD in STE50. (A) Interactions of various STE50 fragments fused to the GAL4 activator domain with STE11 fused to the LexA-DNA binding domain. The pLexA-STE11 construct contains the entire STE11 coding sequence. Amino acid positions of the STE50 fragments included in the constructs are indicated in parentheses. (B) Summary of the two-hybrid interaction analysis between STE50 and STE11. Positions of the STE50 segments included in the activator domain constructs are schematically shown on the left, and their precise amino acid positions are indicated on the right side. The presence (+) or absence (−) of interaction is indicated.
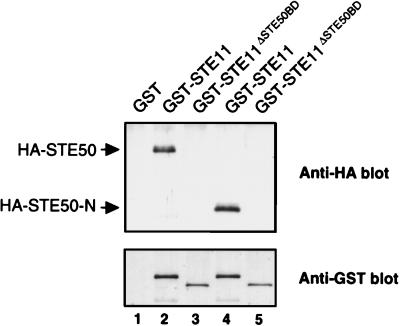
The conclusion from these two-hybrid analyses were confirmed by in vivo coprecipitation experiments. Yeast cells were cotransformed with a plasmid that expresses GST-STE11 or GST-STE11ΔSTE50BD and HA-STE50 or HA-STE50-N. Both HA-STE50 and HA-STE50-N bound to GST-STE11 but not to GST-STE11ΔSTE50BD (Fig. 6). No interaction was observed between STE50-C and GST-STE11 (data not shown). Thus, these in vivo binding assays confirmed the conclusion of the two-hybrid analyses.
FIG. 6.
In vivo binding of STE50-N to the STE50BD in STE11. Wild-type yeast strain TM141 was cotransformed with a plasmid expressing GST, GST-STE11, or GST-STE11ΔSTE50BD (deletion mutant lacking amino acids 74 to 152) and another plasmid expressing HA-STE50 or HA-STE50-N. GST proteins were precipitated by using glutathione-Sepharose beads, and HA-STE50 in precipitates (lanes 1 to 3) or HA-STE50-N (lanes 4 and 5) was probed by immunoblotting with anti-HA monoclonal antibody 12CA5 (upper panel). An antibody against GST was used to detect the GST fusion proteins (lower panel). The GST band in lane 1 is not shown to save space (see Fig. 3).
STE50 binding is essential for STE11 activation by osmotic stress.
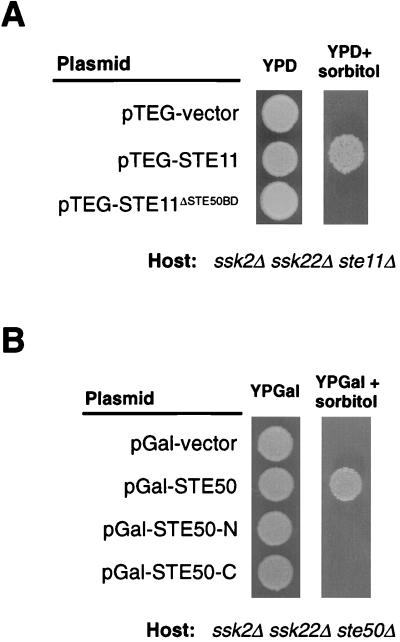
We examine whether STE50 binding to STE11 is necessary for its activation of the HOG pathway under osmotic stresses. For this purpose, we compared wild-type STE11 and the STE11ΔSTE50BD mutant allele for the ability to complement the osmosensitivity of the ssk2Δ ssk22Δ ste11Δ strain (FP75). As shown in Fig. 7A, wild-type STE11 restored growth of FP75 on sorbitol medium whereas STE11ΔSTE50BD did not, suggesting that binding of STE50 to STE11 is essential for activation of the HOC pathway. The STE11DSTE50BD mutant is also mating deficient (data not shown). This appears to be, at least partly, due to a partial overlap between the STE50 and STE5 binding domains in STE11 (18).
FIG. 7.
Expression of STE11 and STE50 mutant alleles in the respective deficient strains. (A) Complementation of the FP75 (ssk2Δ ssk22Δ ste11Δ) osmosensitivity by expression of the wild-type STE11 but not the STE11ΔSTE50BD allele. Host cells were transformed with plasmids that express the indicated STE11 alleles expressed under the constitutive PTEF1 promoter. The transformants were spotted on YPD plates with or without 1.5 M sorbitol. (B) The following segments of STE50 were expressed under the PGAL1 promoter: pGal-STE50 (full-length STE50; amino acids 1 to 346), pGal-STE50-N (amino acids 1 to 166), and pGal-STE50-C (amino acids 165 to 346). Plasmids were transformed into the host, and the transformants were spotted on YPGal plate with or without 1.5 M sorbitol.
We then tested if occupancy of the STE50BD in STE11 was sufficient to allow activation of STE11 under osmotic stress. For this purpose, we examined whether expression of STE50-N, which spans the STE11 binding site, could suppress the osmosensitivity of the ssk2Δ ssk22Δ ste50Δ strain (FP67). As shown in Fig. 7B, expression of neither STE50-N nor STE50-C alone was able to complement the ste50Δ deficiency. Thus, even though the N terminus of STE50 is sufficient for binding to STE11 (Fig. 5 and 6), it is not enough to activate STE11 under osmotic stress.
STE50 is constitutively associated with STE11.
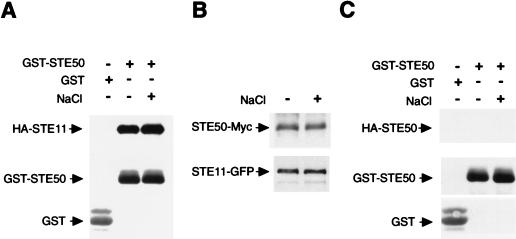
A molecular mechanism to activate MAPKKKs is binding of upstream regulators to N-terminal regulatory regions in MAPKKKs. For example, binding of the SSK1 response regulator to SSK2 MAPKKK induces its kinase activity (17). Thus, a possible role of STE50 is to bind and activate STE11 upon osmotic shock. If this hypothesis is correct, the binding of STE50 to STE11 probably depends on extracellular osmotic conditions. To test this possibility, yeast cells were cotransformed with expression plasmids for GST-STE50 (or GST alone) and HA-STE11. Cells were untreated or treated with a brief osmotic shock before lysis, and the presence of HA-STE11 in the GST-STE50 precipitate was probed. As shown in Fig. 8A, HA-STE11 was precipitated with STE50 equally well when the cells were treated with osmotic shock and when they were not. The same results were obtained when the order of precipitation was reversed, using GST-STE11 and HA-STE50 (data not shown).
FIG. 8.
(A) STE50 binds to STE11 in a constitutive manner. Yeast cells were cotransformed with a plasmid expressing GST-STE50 fusion protein (or control GST) under the constitutive PTEF1 promoter and another plasmid expressing HA-STE11 under the PGAL1 promoter. Cells were grown in the presence of galactose, and samples were taken before (−) or 5 min after (+) the addition of NaCl to a final concentration of 0.4 M. GST proteins were precipitated by using glutathione-Sepharose beads as described in Materials and Methods, and HA-STE11 in precipitates was detected by immunoblotting with an anti-HA antibody. An antibody against GST was used to detect the GST fusion proteins. (B) Yeast cells were cotransformed with single-copy plasmids containing STE50-Myc and STE11-GFP expressed under their own promoters. Cells were grown in the presence of glucose and treated as for panel A, and STE50-Myc and STE11-GFP in precipitates were probed with anti-Myc and anti-GFP antibodies. (C) No homodimerization is observed for STE50. Yeast cells were cotransformed with a plasmid expressing GST-STE50 fusion protein (or control GST) under the constitutive PTEF1 promoter and another plasmid expressing HA-STE50 under the PGAL1 promoter. Cells were grown in the presence of galactose, and samples were treated and analyzed as for panel A.
The above experiments were performed with the strong GAL1 promoter. Because the amounts of STE11 and STE50 proteins in those experiments are much higher than under physiological conditions, we repeated the STE11-STE50 binding test in an assay using a centromeric (low-copy-number) plasmid vector and the native promoters of STE11 and STE50. STE11 was fused to GFP (STE11-GFP), and STE50 was fused to a Myc3 tag (STE50-Myc). Both STE11-GFP and STE50-Myc were functional, because their expression complemented the osmosensitive defects of ste11Δ and ste50Δ mutations, respectively (in the ssk2Δ ssk22Δ background). Yeast cells were cotransformed with STE50-Myc together with STE11-GFP, and cells were subjected to a brief osmotic shock. STE11-GFP was precipitated with an anti-GFP antibody, and the presence of STE50-Myc in precipitates was detected with an anti-Myc monoclonal antibody. Although the levels of protein expression were much lower than in previous experiments, STE11-STE50 interaction was still evident. More important, the level of STE11-STE50 binding was not affected by osmotic shock (Fig. 8B). Thus, our results are consistent with a constitutive association between STE11 and STE50.
Another possible mechanism for MAPKKK activation is signal-induced dimerization of the kinase; for example, induced dimerization of the Raf MAPKKK leads to its activation (7, 12). If STE50 homodimerizes, as previously suggested by a two-hybrid analysis (3), it might mediate STE11 activation, for example, through autophosphorylation. Initially, we tried to demonstrate STE50 dimerization by two-hybrid analysis using pLexA-STE50 and pACT-STE50 constructs. However, because pLexA-STE50 gave a very strong signal by itself, it was not possible to demonstrate specific interaction between the two STE50 constructs (in contrast, the pACT-STE50 constructs used for Fig. 4 and 5 had very low background signals). To test STE50 dimerization more directly, coprecipitation analysis was performed. Yeast cells were cotransfected with expression plasmids for HA-STE50 and GST-STE50 and treated with a brief osmotic shock before cell lysis. The presence of HA-STE50 in the GST-STE50 precipitate was then probed. As shown in Fig. 8C, however, no coprecipitation of HA-STE50 and GST-STE50 was observed, whether cells were treated with a brief osmotic shock or not. Thus, we did not obtain evidence to support dimerization of STE50.
Localization of STE11 and STE50 before and after osmotic stress.
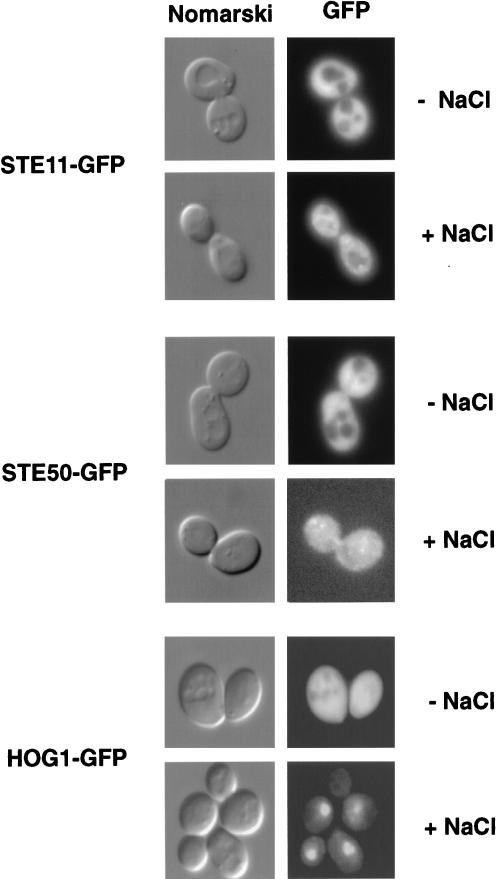
If STE11 and STE50 are constitutively bound, they are expected to be localized similarly within cells. To test the localization of STE11 and STE50, we fused these proteins to GFP and expressed them from their own promoters, so that they were expressed at physiological levels. Both STE11-GFP and STE50-GFP were functional, as indicated by complementation analysis. Under normal conditions (without osmotic stress), both STE11-GFP and STE50-GFP were found diffused throughout the cytoplasm but excluded from nuclei and vacuoles (Fig. 9). When cells were exposed to osmotic stress for 5 min, both proteins rearranged to a punctuate pattern. The expression level and localization of STE11-GFP are indistinguishable between wild-type and ste50 mutant cells (data not shown). As a control to show that these changes are not observed with other proteins, we examined the localization of the HOG1-GFP fusion protein. Before osmotic shock, HOG1-GFP was evenly distributed in both cytoplasm and nucleus (Fig. 9). Upon osmotic shock, HOG1-GFP translocated from the cytoplasm to the nucleus within 5 min. (HOG1 nuclear localization will be described fully elsewhere [8].) Therefore, these data also support the notion that STE50 and STE11 are constitutively bound to each other.
FIG. 9.
Similar intracellular localization patterns of STE50 and STE11. Distributions of STE11-GFP, STE50-GFP, and HOG1-GFP fusion proteins were analyzed by direct fluorescence microscopy. The GFP fusion proteins were expressed under their own promoters in single-copy plasmids. The host cells contain the respective null mutations (ste11Δ, ste50Δ, and hog1Δ) to minimize any dose effect. Pictures were taken before (−) or 5 min after (+) the addition of NaCl to a final concentration of 0.4 M. Nuclear staining by DAPI (4′,6-diamidino-2-phenylindole) showed that both STE11-GFP and STE50-GFP are excluded from nuclei (not shown). They are also excluded from vacuoles, as indicated by Nomarski microscopy.
DISCUSSION
The HOG1 MAP kinase cascade can be activated under osmotic stress by two different and independent upstream mechanisms; a two-component osmosensor (SLN1-YPD1-SSK1) activates the SSK2/SSK22 MAPKKKs, and the transmembrane protein SHO1 mediates activation of the STE11 MAPKKK. The three MAPKKKs (SSK2, SSK22, and STE11) can in turn independently activate the PBS2 MAPKK, the activator of the HOG1 MAPK. While the framework of the HOG1 pathway is now established, there are many mechanistic questions that remained unanswered, including how the transmembrane protein SHO1 activates STE11 MAPKKK. Previously we demonstrated that SHO1 binds directly to the PBS2 MAPKK (13) and that PBS2 also interacts with STE11 (16). However, because SHO1 does not interact directly with STE11 (16), it is difficult to imagine how the activation of STE11 is mediated by SHO1 alone. From these considerations, we suspected that an additional element or elements are required for the osmostress-induced activation of the STE11 MAPKKK. In this report, we described a genetic screening by which we identified STE50 as one such element. This genetic screening was quite exhaustive, since mutants in each complementation group were identified multiple times.
The results of this study demonstrate that STE50 is absolutely required for signal transduction in the SHO1-STE11 branch of the HOG pathway. We also demonstrated that STE50 binds to a specific binding site in the N terminus of STE11. The STE50-STE11 interaction is constitutive and is not affected by environmental osmotic conditions. It seems, therefore, as if STE50 is an integral and essential subunit of the STE11 kinase.
Previously, it was reported that overexpression of STE50 makes cells more sensitive to the presence of mating pheromone, while deletion of STE50 reduced slightly mating efficiency (21, 28). In our strain background also, ste50 mutations had a minimal effect on mating efficiency (Table 2). Therefore, STE50 seems to have only an accessory role, rather than an essential function, in pheromone signal transduction. The difference in levels of significance of STE50 in the two pathways is best illustrated by the different effects of ste11Δ and ste50Δ mutations in these pathways. Disruption of STE11 completely abolishes signaling through both mating and osmosensing pathways, whereas STE50 disruption has a significant effect only in the osmosensing pathway. This difference could be explained by several possible mechanisms. First, STE50 may have two different functions which are specific to each pathway. Alternatively, STE11 may be activated by several different mechanisms in the mating pathway, STE50 being only one of them. This would be consistent with the observation that deletion of STE50 results in only a minor effect on the mating pathway. In contrast, activation of STE11 by osmotic stress may be completely dependent on a mechanism that involves STE50. Finally, it is also possible that the mating and the osmosensing pathway require different levels of the STE11 activity.
The binding of STE50 to STE11 is essential for the activation of STE11 by osmotic stress. It is unlikely, however, that STE50 binding alone activates STE11, because STE50 is constitutively bound to STE11, regardless of the environmental osmotic conditions. It is more likely that the STE50-STE11 interaction is a prerequisite for STE11 to receive an upstream activating signal. STE50-N can bind STE11 but cannot complement the ste50Δ defect, indicating that STE50-C also has an important functional role. One possible role for the C-terminal region is to mediate STE50-STE50 dimerization and thus to indirectly promote STE11 dimerization. However, results of the coprecipitation experiment presented in this report do not support this hypothesis. Another possibility is that STE50-C mediates binding of STE11 to another upstream element that is directly responsible for the activation of STE11 activity. Our failure to identify mutants of such upstream elements suggests either that there is redundancy at that level or that such elements are essential. Attempts to identify STE50 interactors by two-hybrid screening have so far been unsuccessful. It is possible that the interaction of STE50 with the hypothetical upstream element occurs only after osmotic shock.
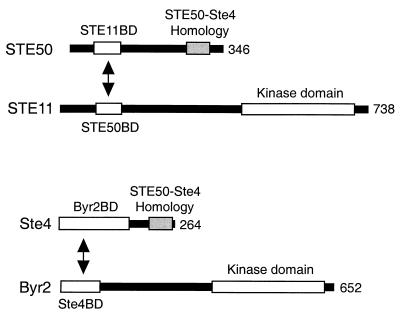
Comparison of the S. pombe Ste4 protein with STE50 gives some support to the latter model. Ste4 seems to serve a role similar to that of STE50 in signal transduction. Ste4 binds to the Byr2 MAPKKK, which is a homolog of STE11 and is involved in the signaling pathway for the mating pheromone or nitrogen starvation. The Ste4 binding domain (Ste4BD) in Byr2 and the Byr2 binding domain (Byr2BD) in Ste4 are both in their N-terminal regions (Fig. 10). Thus, the locations of the interaction domains are very similar to those of STE50-STE11 interaction. A further similarity of the Ste4-Byr2 interaction to the STE50-STE11 interaction is that the Ste4-Byr2 association is constitutive regardless of the presence or absence of mating pheromone or nitrogen starvation (25). Interestingly, however, there are no significant sequence similarities either between STE50BD and Ste4BD or between STE11BD and Byr2 BD: the sequence similarities between Ste4 and STE50 are limited to their C-terminal regions. In other words, their specificities but not the primary sequences, are similar. In contrast, the fact that the C termini of STE50 and Ste4 have some sequence identity (32% identity in a stretch of 59 amino acids) suggests that they may serve as interaction sites for a hypothetical upstream molecule.
FIG. 10.
Schematic diagram of the similarities between STE50 and Ste4. S. cerevisiae STE50 interacts with the STE11 MAPKKK, and S. pombe Ste4 interacts with the Byr2 MAPKKK. The binding domains of these proteins are depicted. Amino acids 267 to 325 of STE50 have 32% identity to residues 204 to 262 of Ste4 (STE50-Ste4 Homology). The positions of the STE11 and Byr2 kinase domains, which are highly homologous to each other, are also shown.
ACKNOWLEDGMENTS
We thank M. Takekawa, Q. Ge, and D. C. Raitt for comments on and critical reading of the manuscript; P. Silver and J. A. Kahana for plasmids and antibody against GFP; and P. Ferrigno for valuable suggestions.
This work was supported by NIH grants GM50909 and GM56699 to H. S. and by a postdoctoral fellowship from la Dirección General de Investigación Científica y Técnica of the Spanish Government to F.P.
REFERENCES
- 1.Adelberg E A, Mandel M, Chen G C C. Optimal conditions for mutagenesis by N-methyl-N′-nitro-N-nitrosoguanidine in Escherichia coli K12. Biochem Biophys Res Commun. 1966;18:788–795. [Google Scholar]
- 2.Albertyn J, Hohmann S, Thevelein J M, Prior B A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr M M, Van Aelst L, Wigler M. Identification of Ste4 as a potential regulator of Byr2 in the sexual response pathway of Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:5597–5603. doi: 10.1128/mcb.16.10.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boguslawski G. PBS2, a yeast gene encoding a putative protein kinase, interacts with the RAS2 pathway and affects osmotic sensitivity of Saccharomyces cerevisiae. J Gen Microbiol. 1992;138:2425–2432. doi: 10.1099/00221287-138-11-2425. [DOI] [PubMed] [Google Scholar]
- 5.Brewster J L, de Valoir T, Dwyer N D, Winter E, Gustin M C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 6.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 7.Farrar M A, Alberola-Ila J, Perlmutter R M. Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature. 1996;383:178–181. doi: 10.1038/383178a0. [DOI] [PubMed] [Google Scholar]
- 8.Ferrigno, P., F. Posas, H. Saito, and P. A. Silver. Submitted for publication.
- 9.Foreman P K, Davis R W. Cloning vectors for the synthesis of epitope-tagged, truncated and chimeric proteins in Saccharomyces cerevisiae. Gene. 1994;144:63–68. doi: 10.1016/0378-1119(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 10.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Elledge S J, Peterson C A, Bales E S, Legerski R J. Specific association between the human DNA repair proteins XPA and ERCC1. Proc Natl Acad Sci USA. 1994;91:5012–5016. doi: 10.1073/pnas.91.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Z, Tzivion G, Belshaw P J, Vavvas D, Marshall M, Avruch J. Oligomerization activates c-Raf-1 through a Ras-dependent mechanism. Nature. 1996;383:181–185. doi: 10.1038/383181a0. [DOI] [PubMed] [Google Scholar]
- 13.Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 14.Maeda T, Wurgler-Murphy S M, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 15.Ota I M, Varshavsky A. A yeast protein similar to bacterial two-component regulators. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- 16.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 17.Posas F, Saito H. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 1998;17:1385–1394. doi: 10.1093/emboj/17.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Posas, F., and H. Saito. Unpublished data.
- 19.Posas F, Takekawa M, Saito H. Signal transduction by MAP kinase cascades in budding yeast. Curr Opin Microbiol. 1998;1:175–182. doi: 10.1016/s1369-5274(98)80008-8. [DOI] [PubMed] [Google Scholar]
- 20.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 21.Rad M R, Xu G, Hollenberg C P. STE50, a novel gene required for activation of conjugation at an early step in mating in Saccharomyces cerevisiae. Mol Gen Genet. 1992;236:145–154. doi: 10.1007/BF00279653. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt A, Hall M N, Koller A. The FK506 resistance-conferring genes in Saccharomyces cerevisiae, TAT1 and TAT2, encode amino acid permeases mediating tyrosine and tryptophan uptake. Mol Cell Biol. 1994;14:6597–6606. doi: 10.1128/mcb.14.10.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sikorski R S, Boeke J D. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 24.Takekawa M, Posas F, Saito H. A human homolog of the yeast Ssk2/Ssk22 MAP kinase kinase kinases, MTK1, mediates stress-induced activation of the p38 and JNK pathways. EMBO J. 1997;16:4973–4982. doi: 10.1093/emboj/16.16.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu H, Barr M, Dong D L, Wigler M. Multiple regulatory domains on the Byr2 protein kinase. Mol Cell Biol. 1997;17:5876–5887. doi: 10.1128/mcb.17.10.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 27.Waskiewicz A J, Cooper J A. Mitogen and stress response pathways: MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr Opin Cell Biol. 1995;7:798–805. doi: 10.1016/0955-0674(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 28.Xu G, Jansen G, Thomas D Y, Hollenberg C P, Rad M R. Ste50p sustains mating pheromone-induced signal transduction in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1996;20:773–783. doi: 10.1111/j.1365-2958.1996.tb02516.x. [DOI] [PubMed] [Google Scholar]