Abstract
Ku antigen is a heterodimer, comprised of 86- and 70-kDa subunits, which binds preferentially to free DNA ends. Ku is associated with a catalytic subunit of 450 kDa in the DNA-dependent protein kinase (DNA-PK), which plays a crucial role in DNA double-strand break (DSB) repair and V(D)J recombination of immunoglobulin and T-cell receptor genes. We now demonstrate that Ku86 (86-kDa subunit)-deficient Chinese hamster cell lines are hypersensitive to ICRF-193, a DNA topoisomerase II inhibitor that does not produce DSB in DNA. Mutant cells were blocked in G2 at drug doses which had no effect on wild-type cells. Moreover, bypass of this G2 block by caffeine revealed defective chromosome condensation in Ku86-deficient cells. The hypersensitivity of Ku86-deficient cells toward ICRF-193 was not due to impaired in vitro decatenation activity or altered levels of DNA topoisomerase IIα or -β. Rather, wild-type sensitivity was restored by transfection of a Ku86 expression plasmid into mutant cells. In contrast to cells deficient in the Ku86 subunit of DNA-PK, cells deficient in the catalytic subunit of the enzyme neither accumulated in G2/M nor displayed defective chromosome condensation at lower doses of ICRF-193 compared to wild-type cells. Our data suggests a novel role for Ku antigen in the G2 and M phases of the cell cycle, a role that is not related to its role in DNA-PK-dependent DNA repair.
Ku is an abundant nuclear protein originally identified as an autoantigen recognized by sera from patients with autoimmune diseases (47). The Ku antigen is a heterodimer comprised of 86- and 70-kDa subunits (Ku86 and Ku70) which interacts preferentially with free DNA ends or particular DNA structures such as hairpins or gaps (46–48). Ku is associated with a catalytic subunit of 450 kDa (DNA-PKCS), forming the DNA-dependent protein kinase (DNA-PK). It is presumed that the Ku antigen tethers the serine-threonine kinase activity of this enzyme to DNA ends, allowing phosphorylation of nearby substrates (22, 28). Accordingly, in vitro phosphorylation of a large number of proteins by DNA-PK is stimulated in the presence of double-strand breaks (DSB) (12, 43; reviewed in reference 3). A series of genetic and biochemical data have converged to establish the crucial role of DNA-PK in DSB repair as well as in V(D)J recombination of immunoglobulin and T-cell receptor genes (reviewed in references 35, 69, and 75). Defective DSB repair and V(D)J recombination in X-ray-sensitive rodent cell lines of the complementation group for XRCC5 were rescued by transfection of the human Ku86 cDNA (9, 57, 59), and mutations that are responsible for the X-ray sensitivity of these cells were mapped to the Ku86 gene, demonstrating unambiguously that Ku86 is the product of XRCC5 (25). An additional role for DNA-PK in DNA replication has been suggested by its ability to phosphorylate replication protein A (10, 52). Furthermore, a role in transcription is indicated by the ability of DNA-PK to inhibit RNA polymerase I activity (39, 41), as well as to modulate the activity of transcription factors through phosphorylation (27).
Recent data provide evidence that the Ku antigen possesses additional functions that are not directly related to its role as part of the DNA-PK enzyme. Indeed, Ku86-null mice display considerable growth defects in addition to an absence of T- and B-lymphocyte maturation (51, 77). Such growth retardation is not observed in SCID mice or foals, which are defective in DNA-PKCS (8, 51, 70). Interestingly, Ku antigen has been shown to possess in vitro intrinsic DNA-dependent ATPase and helicase activity (11, 62).
DNA topoisomerase II catalyzes topological changes in DNA that are essential for normal cell cycle progression. The enzyme can relax supercoiled DNA and can also knot or unknot DNA as well as catenate and decatenate closed circular DNA by passing duplex DNA through an enzyme-mediated DNA gate in an ATP-dependent manner (reviewed in reference 65). Budding yeast topoisomerase II is an essential enzyme for disentangling sister chromatids after DNA replication (31), while the fission yeast enzyme is also involved in chromosome condensation during metaphase (63). Additional evidence for a role of DNA topoisomerase II in chromosome condensation has been provided by in vitro experiments using extracts from Xenopus eggs (2, 30, 50). Although definitive proof that DNA topoisomerase II is required for disentanglement and condensation of sister chromatids in mammalian cells has not been obtained, due to the absence of topoisomerase II mutants, mammalian cells treated with ICRF-193, a novel noncleavable complex-stabilizing type topoisomerase II inhibitor, do not undergo normal chromosome condensation (5, 16, 21, 34).
Two topoisomerase II isoforms, designated topoisomerase IIα and topoisomerase IIβ, exist in mammalian cells. DNA topoisomerase IIα expression is cell cycle dependent and peaks during the G2 and M phases (38, 45, 71). The expression pattern of the β isoform was variably reported to remain constant throughout the cell cycle (38, 71) or to increase during the S, G2, and M phases (45). DNA topoisomerase IIα is a major constituent of the nuclear scaffold and has been implicated directly in the organization of metaphase chromosomes (13, 23, 26, 45). Less is known about the role of the β isoform, which is nucleoplasmic during interphase and diffuses into the cytosol during mitosis (45).
Although DNA topoisomerase IIα clearly plays a catalytic role in decatenation of linked chromatids, its function in chromosome condensation may be stoichiometric (1, 6, 45). DNA topoisomerase II preferentially associates with AT-rich sequences present in the scaffold-associated regions, which are candidate DNA elements for defining the bases of chromatin loops and serving as cis elements of chromosome dynamics (1, 18, 58). Moreover, purified enzyme has been shown to multimerize in vitro, and this aggregation is stimulated by phosphorylation of its C terminus (64).
It is known that X-ray-sensitive cell lines deficient in subunits of the DNA-PK enzyme are hypersensitive to DNA topoisomerase II inhibitors such as etoposide, which stabilize topoisomerase II-DNA cleavage complexes and thus concomitantly induce DSB (44). This hypersensitivity has generally been ascribed to deficiency of these mutant cell lines in DSB repair (29, 36, 66). In contrast to etoposide, ICRF-193 inhibits DNA topoisomerase II activity without inducing any DSB (21, 61). Thus, analysis of cells treated with ICRF-193 allows the effects of DNA topoisomerase II inhibition to be separated from those due to the introduction of DSB. We now demonstrate that topoisomerase II-mediated functions are inhibited at significantly lower doses of ICRF-193 in Ku86-deficient cell lines than in wild-type cells. This difference was not observed when DNA-PKCS-deficient cells were analyzed. Our data indicate a novel role for Ku antigen in the G2 and M phases of the cell cycle, one that appears independent of its participation in DSB repair by DNA-PK.
MATERIALS AND METHODS
Cell lines, plasmids, and reagents.
XR-V15B and XR-V9B mutant cell lines, derived from Chinese hamster V79 cells, have been previously described (25, 76). XR-V16B mutant cells belong to the same complementation group (unpublished results). Stable transfectants (XR-V15B/C2, XR-V15B/C8, XR-V15B/C9, and XR-V15B/pBJ5) were obtained by cotransfection of 1.2 × 106 XR-V15B cells with human Ku86 cDNA inserted into the pBJ5 expression vector (8 μg) (57), or the same vector lacking the insert (8 μg), and pSV40neo (0.8 μg). After 48 h, the cells were diluted and selected for 12 days in medium containing G418 (0.8 mg/ml; Gibco BRL). G418-resistant colonies were isolated, and expression of Ku86 was determined by Western blot assay. The X-ray-sensitive hamster ovary cell line V-3 and its parental cell line AA8 were kindly supplied by G. F. Whitmore (Ontario Cancer Institute, Toronto, Ontario, Canada). All cell lines were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (0.1 mg/ml). Cells were maintained at 37°C under a 5% CO2 humidified atmosphere.
ICRF-193 was kindly provided by A. M. Creighton (St. Bartholomew’s Hospital Medical College, London, England). A stock solution of 1 mg/ml was prepared in dimethyl sulfoxide. Bleomycin (purchased from Roger Ballon) was dissolved at 10 mg/ml in sterile water. Nocodazole was resuspended at 1 mg/ml in dimethyl sulfoxide, caffeine was freshly prepared at 100 mM in sterile phosphate-buffered saline (pH 7.4) (PBS), and hydroxyurea (HU) was dissolved at 200 mM in sterile water (Sigma Chemical Co.).
Drug treatments and cell analysis.
To test the effect of ICRF-193 or bleomycin on exponentially growing cells, 7.5 × 105 (V79B and XR-V15B) or 4.5 × 105 (AA8 and V-3) cells were seeded in 60-mm-diameter dishes and cultured for 16 or 24 h with different drug concentrations. Cells were harvested, and the DNA was stained with propidium iodide solution (0.025 mg of propidium iodide per ml, 0.1% sodium citrate, 0.1 mg of RNase A per ml, 0.1% Triton X-100). To quantify the cells at different stages of the cell cycle, the DNA content of the cells was analyzed by FACscan (Becton Dickinson).
For V79B and XR-V15B cell synchronization experiments, early-S-phase cell populations were obtained by blocking DNA synthesis with HU. Briefly, exponentially growing cells were treated with 1 mM HU for 16 h. Cells were then rinsed for 30 min and recultivated in drug-free medium for 3 h; at this point (50% of cells in late S phase and 50% of cells in G2 phase), different concentrations of ICRF-193 were added to the medium together with nocodazole (40 ng/ml). Cell culture was continued for 3, 4, or 5 h. Treatment with nocodazole blocked the cells which had proceeded through G2 in metaphase. At the end of the treatment, cells were fixed with methanol-acetic acid (3:1) solution and stained with Hoechst 33258. The percentage of cells in mitosis (as indicated by the lack of a nuclear membrane and presence of condensed mitotic chromosomes) was determined by microscopy, counting at least 500 cells for each plate.
Preparation of metaphase chromosomes.
Late S/G2-phase synchronized cells were treated with various concentrations of ICRF-193, 40 ng of nocodazole per ml, and 2 mM caffeine for 5 h. AA8 and V-3 late S/G2 cells were obtained by HU treatment for 16 h and then recultivated in drug-free medium for 5 h. At this point, different concentrations of ICRF-193 were added to the medium together with 80 ng of nocodazole per ml. After 2 h, 2 mM caffeine was added and cell culture was continued for 3 h. Then, mitotic cells were collected by very gentle trypsinization (1 min), resuspended in 10 ml of 75 mM KCl, and left at 37°C for 15 min; 1 ml of fixing solution (methanol-acetic acid, 3:1) was slowly added to the suspension under constant mild agitation, and the cells were centrifuged. After resuspension in 10 ml of fixing solution, cells were centrifuged, kept in 1 to 1.5 ml of fixative, and dispensed onto glass slides. After drying, the samples were stained with Hoechst 33258 solution (1 μg/ml in PBS), and cells were observed by fluorescence microscopy.
PFGE and Southern blot analysis.
In these assays, 1.5 × 106 cells were seeded in 100-mm-diameter dishes and deprived of serum for 24 h (80% cells arrested in G0/G1 phase). The culture medium was replaced by medium containing 10% FCS, and the cells were allowed to progress for 12 h until late S/G2 phase. At this point, different concentrations of ICRF-193 were added to the medium for 4 h. For bleomycin treatment, G0/G1 synchronized cells were cultured in medium supplemented with 10% FCS and different concentrations of bleomycin for 16 h. These drug treatments induced a block in G2 phase similar to that described above. Next, cells were collected and resuspended in PBS at a concentration of 12 × 106 cells/ml. An equal volume of 1% low-melting-point agarose (SeaPlaque GTG; FMC) at 40°C was added, and 100 μl of this mixture was immediately loaded into plug molds, which were kept at 0°C for 15 min. After agarose solidification, gel plugs containing the cells were placed in lysis solution (0.5 M EDTA, 1% sarcosyl, 0.5 mg of proteinase K per ml [pH 9]) at 0°C for 1 h, followed by incubation at 50°C for 48 h. The plugs were then washed four to five times in TE (10 mM Tris-HCl, 1 mM EDTA [pH 8]) at room temperature and electrophoresed in 0.75% agarose (SeaKem Gold; FMC). A third of each plug containing approximately 200,000 lysed cells was loaded in each lane. Pulsed-field gel electrophoresis (PFGE) was carried out with a CHEF-DR II apparatus (Bio-Rad) with a 120° reorientation angle. The gels were run in 0.5× TBE buffer (1× TBE is 89 mM Tris base, 89 mM boric acid, and 2 mM EDTA [pH 8]) at 20°C at 2 V/cm with a switch time changing linearly from 5 min to 2.5 h over 72 h. After the run, the gel was stained with ethidium bromide (1 μg/ml) for 30 min and irradiated at 600 μJ/cm2 to cleave the chromosomal DNA and facilitate transfer onto membranes. DNA was denatured by soaking the gel in a 0.4 N NaOH–1.5 M NaCl solution for 15 min and then transferred onto a nylon membrane for 40 h in the same denaturing solution. The membrane was hybridized with a 32P-labeled V79B genomic DNA probe.
Immunoblotting.
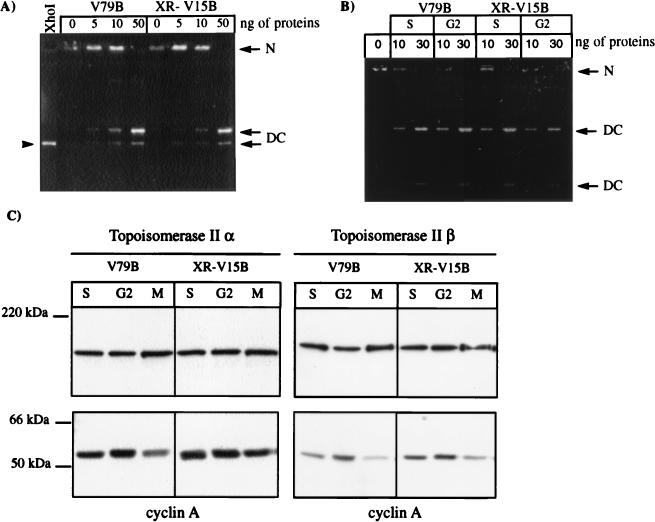
For analysis of DNA topoisomerase IIα levels, whole-cell extracts were obtained from G1/G0-phase cells, S-phase cells (exponentially growing cells treated with 1 mM HU for 16 h), G2-phase cells (early-S-phase cells which were released in fresh medium for 4 h), and M-phase cells (late S/G2-phase cells treated with nocodazole [40 ng/ml] for 5 h). Briefly, cells were harvested by trypsinization and pelleted at 1,200 rpm for 5 min at 4°C, washed with 1 ml of PBS, and lysed by resuspension in cold lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 1% [wt/vol] sodium deoxycholate) containing protease inhibitors (5 μg of aprotinin, 5 μg of leupeptin, and 10 μg of pepstatin A per ml, 1 mM benzamidine, 2 mM phenylmethylsulfonyl fluoride [PMSF]) and 30 mM NaF. Cells were lysed on ice for 30 min and centrifuged at 13,000 rpm for 5 min at 4°C. Protein concentrations in the supernatant were determined by the bicinchoninic acid protein assay (Pierce). For analysis of DNA topoisomerase IIβ levels, cells were trypsinized, sedimented, and lysed in 1% SDS for 5 min at 90°C, and the DNA was finally sheared with a syringe (45). Samples equivalent to 5 × 105 cells were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred electrophoretically to polyvinylidene difluoride membranes (Millipore) at 0.8 mA/cm2 for 1 h in 25 mM Tris-HCl–192 mM glycine–20% methanol. The antibodies used were an anti-DNA topoisomerase IIα polyclonal antibody (Bio Trend), the anti-topoisomerase IIβ monoclonal antibody 3H10, kindly provided by A. Kikuchi (Nagoya University School of Medicine, Nagoya, Japan) (37), a rabbit anti-Ku86 serum (Serotec), an anti-cyclin A monoclonal antibody (Sigma Chemical Co.), and an anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) polyclonal antibody. Antibody binding was detected following incubation with a goat anti-rabbit or anti-mouse horseradish peroxidase-coupled antibody by using enhanced chemiluminescence.
Extraction of nuclear proteins.
Exponentially growing S-, G2- and M-phase synchronized cells were harvested and pelleted at 1,200 rpm for 5 min at 4°C. Cells were resuspended in 3 ml of cold lysis buffer A (10 mM Tris-HCl [pH 7.5], 0.5% Nonidet P-40, 1.5 mM MgCl2, 10 mM KCl, 30 mM NaF, 1 mM PMSF, 10 μg each of aprotinin and leupeptin per ml, 0.5 mM dithiothreitol [DTT], 0.1 mM sodium orthovanadate) and incubated for 10 min at 4°C. The cells were fractionated in a Dounce homogenizer, and the homogenate was centrifuged at 800 × g for 5 min at 4°C. The supernatant was decanted, and the pellet was carefully resuspended in 1 ml of buffer A and centrifuged at 800 × g for 5 min at 4°C. The nuclear pellet was then resuspended in a small volume of cold extraction buffer (50 mM Tris-HCl [pH 7.5], 25% [vol/vol] glycerol, 0.35 M NaCl, 1.5 mM MgCl2, 2 mM EDTA, 30 mM NaF, 10 μg of aprotinin, 10 μg of leupeptin, and 5 μg of pepstatin per ml, 0.5 mM DTT, 1 mM PMSF, 0.1 mM sodium orthovanadate) and left at 4°C for 40 min. The nuclear suspension was centrifuged at 13,000 rpm for 10 min at 4°C. Nuclear proteins in the supernatant were diluted at 0.1 μg/ml with extraction buffer and stored at −70°C.
In vitro topoisomerase II catalytic activity.
Topoisomerase II catalytic activity was quantified by testing the ability of nuclear proteins extracted from V79B and XR-V15B cells to decatenate kinetoplast DNA networks (TopoGen Inc.). DNA cleavage reactions were performed for 15 min at 37°C in a total volume of 20 μl containing 0.23 μg of kinetoplast DNA, 50 mM Tris-HCl (pH 7.5), 120 mM KCl, 10 mM MgCl2, 0.5 mM DTT, 0.5 mM EDTA, 30 μg of bovine serum albumin per ml, 1 mM ATP, and different amounts of nuclear extract (see figure legends). After incubation, 1 μl of 20% SDS and 10 mg of proteinase K per ml were added, and the samples were incubated for 30 min at 37°C, extracted with phenol-chloroform, and migrated on a 1% agarose gel containing ethidium bromide. The gel was photographed on a UV transilluminator. The kinetoplast networks remained at the origin, while the circular or linear monomers generated by topoisomerase II activity migrated into the gel.
RESULTS
Significantly lower doses of the DNA topoisomerase II inhibitor ICRF-193 cause a G2 cell cycle arrest in Ku86-deficient cell lines.
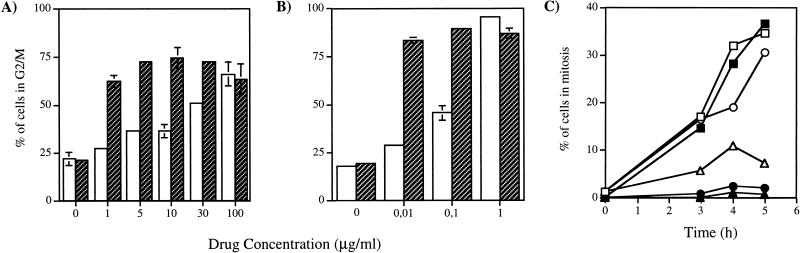
A possible role for the Ku antigen in DNA topoisomerase II-mediated functions during the cell cycle was assessed in assays using Ku-deficient and wild-type cells. The Ku86-deficient XR-V15B Chinese hamster cell line was isolated as an X-ray-hypersensitive derivative after ethylnitrosourea mutagenesis of the V79 cell line (76). The effects of the topoisomerase II inhibitor ICRF-193 and the X-ray-mimetic drug bleomycin were compared during a 16-h treatment of exponentially growing XR-V15B or V79B cells (the latter are wild-type derivatives of V79 cells). Bleomycin treatment leads to a predominant G2 arrest (20). The DNA content of the cells was analyzed by FACscan analysis in order to quantify the percentage of cells at various stages of the cell cycle. Hypersensitivity of XR-V15B cells to agents causing DSB is clearly apparent from the observation that maximal accumulation of the cells in G2/M was obtained at a bleomycin concentration nearly 100-fold lower than the dose necessary to obtain a similar effect in wild-type cells (1 μg/ml for XR-V15B cells, compared to 100 μg/ml for V79B cells) (Fig. 1A). As previously demonstrated for other Ku86-deficient cell lines (29, 36), a similar hypersensitivity of XR-V15B was noted following treatment with the cleavable complex-stabilizing drug etoposide (results not shown). More surprisingly, similar results were obtained with ICRF-193 (Fig. 1B). Indeed, maximal G2/M accumulation was observed at an ICRF-193 concentration of 0.01 μg/ml for XR-V15B cells, compared to 1 μg/ml for V79B cells. This observation was repeated with two other Ku86-deficient cell lines derived from V79 cells, XR-V9B and XR-V16B (results not shown). Thus, Ku86-deficient cells are hypersensitive to agents which induce DSB in DNA, as well as to a topoisomerase II inhibitor which affects enzyme activity without introducing DNA lesions (21, 61).
FIG. 1.
Low doses of ICRF-193 cause a G2 arrest in Ku-deficient cells. Exponentially growing V79B (□) and XR-V15B ( ) cells were incubated with increasing concentrations of bleomycin (A) or ICRF-193 (B) for 16 h. The cells were stained with propidium iodide, and the percentage of cells with a G2/M DNA content was determined by FACscan analysis of 20,000 cells. Each determination is the mean ± standard error of three different experiments. (C) Late S/G2-phase synchronized V79B (open symbols) and XR-V15B (closed symbols) cells were treated with increasing concentrations of ICRF-193 (0 [squares], 0.01 [circles], 1 [triangles] μg/ml) and 40 ng of nocodazole per ml for 3, 4, or 5 h. Cells were fixed and stained with Hoechst 33258 dye. Each time point represents the percentage of mitotic cells in 500 cells determined by fluorescence microscopy. The experiment was repeated three times with similar results.
To determine whether the observed G2/M accumulation was due to a G2 arrest rather than to a slower progression through the G2 and M phases, cells were synchronized in early S phase by HU treatment and allowed to recover for 3 h prior to the addition of ICRF-193 to the medium. Nocodazole was added together with ICRF-193 in order to block in metaphase those cells that proceeded through the G2 checkpoint. The percentage of mitotic cells (indicated by the lack of nuclear membrane and presence of condensed mitotic chromosomes) was then determined after 3, 4, or 5 h of ICRF-193 treatment. As shown in Fig. 1C, less than 3% of XR-V15B cells reached M phase in the presence of ICRF-193, even at concentrations as low as 0.01 μg/ml. Moreover, the proportion of mitotic cells did not increase with longer incubation times (results not shown). At a higher dose (1 μg/ml), less than 10% of V79B cells were able to enter mitosis, but most of these cells had defective chromosome condensation. The G2 arrest that was observed after ICRF-193 treatment is in agreement with findings reported for other cell types (4, 6, 21). Most importantly, these data demonstrate that in the absence of the Ku antigen, cell cycle arrest at G2 occurs following treatment with significantly lower doses of ICRF-193.
DSB are not detected following ICRF-193 treatment.
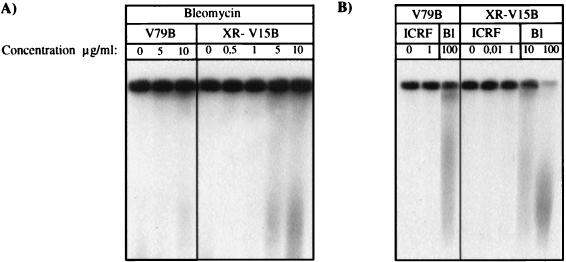
The G2 arrest observed after ICRF-193 treatment may be explained by the operation of a G2 decatenation checkpoint as proposed by Downes and colleagues (21). However, it was also possible that ICRF-193 treatment resulted in low levels of DSB which remained unrepaired in DNA-PK-deficient cells, thereby activating a G2 DNA damage checkpoint. We therefore used PFGE to assess whether there was a low level of chromosome breaks (40). Synchronized V79B or XR-V15B cells were treated with different concentrations of either ICRF-193 or bleomycin. Fragmented chromosomal DNA was allowed to migrate into agarose gels by PFGE and visualized by hybridization with a labeled genomic probe (Fig. 2). In V79B wild-type cells, doses of bleomycin (10 μg/ml) which induced only a minor G2 arrest gave rise to a detectable amount of chromosome damage (Fig. 2A), indicating that the PFGE assay is sensitive enough to detect a level of DNA damage which induced a G2 arrest in only a small population of cells. In contrast, no DNA damage was detected in V79B cells after treatment with ICRF-193 at a dose (1 μg/ml) that induced a maximal G2/M arrest. It is most unlikely that DNA damage is not detected after ICRF-193 treatment due to cross-linking of the DSB by topoisomerase II. Indeed, ICRF-193 has been shown to induce no such complexes between topoisomerase II and DNA in vitro and in vivo (17, 61). These results suggest that the observed G2 arrest was caused principally by inhibition of DNA topoisomerase II decatenation activity without induction of DNA damage. Similarly, no damage was observed after treatment of Ku-deficient cells with different doses of ICRF-193 (0.01 and 1 μg/ml) (Fig. 2B). However, significant chromosome damage was also not detected at a dose of bleomycin (1 μg/ml) which resulted in a G2/M block in XR-V15B cells (Fig. 2A). Thus, from this analysis we cannot completely exclude the possibility that the ICRF-193-induced cell cycle arrest in XR-V15B cells was due to the presence of DSB produced as a secondary consequence of topoisomerase II inhibition.
FIG. 2.
ICRF-193 treatment does not induce DNA damage detectable by PFGE. Late S/G2-phase synchronized V79B and XR-V15B cells were incubated in the presence of increasing concentrations of ICRF-193 for 4 h. For bleomycin treatment, G0/G1 synchronized cells were released into medium supplied with 10% FCS and different drug concentrations for 16 h. Cells were included in 1% low-melting-point agarose at a final density of 6 × 106/ml. DNA from approximately 200,000 lysed cells was migrated by PFGE as described in Materials and Methods. At the end of migration, the DNA was transferred onto a nylon membrane and hybridized with a 32P-labeled probe of V79B genomic DNA. Note that intact chromosomal DNA remained in the wells, while fragmented DNA migrated into the gel. Representative autoradiographs of four experiments are shown. Higher exposures of these membranes did not reveal increased DNA damage in the presence of ICRF-193 compared to controls.
Low doses of ICRF-193 alter chromosome condensation in cells lacking the Ku antigen.
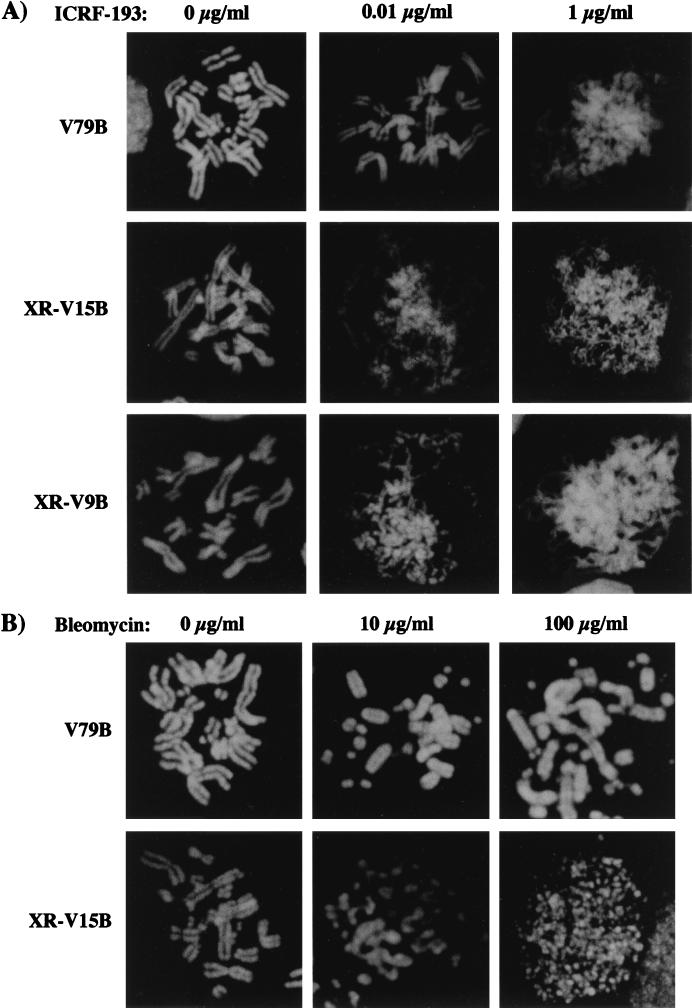
We were next interested in assessing whether another major function of DNA topoisomerase II, the condensation of chromosomes during mitosis, was affected by lower doses of ICRF-193 in Ku-deficient cells. Since chromosome condensation could not be analyzed in the previously described experiments due to G2 arrest, we took advantage of the ability of caffeine to override the G2 checkpoint in the subsequent analysis (4, 21) (Fig. 3). Condensation of metaphase chromosomes in Ku-deficient cells was clearly affected by an ICRF-193 concentration (0.01 μg/ml) which had no effect on chromosome condensation of V79B cells (Fig. 3A). Hypersensitivity of XR-V15B to ICRF-193 was also observed in two other Ku-deficient cell lines, XR-V9B and XR-V16B (Fig. 3A and data not shown). In contrast, fragmented yet normally condensed chromosomes were observed in V79B and XR-V15B cells, when bleomycin-induced G2 arrest was bypassed by caffeine (Fig. 3B; 100 μg/ml for V79B cells; 10 and 100 μg/ml for XR-V15B cells). Thus, Ku-deficient cells showed altered chromosome condensation in the presence of low concentrations of ICRF-193, an effect that is clearly distinct from that observed with doses of bleomycin which induced DSB. Interestingly, at a low dose of ICRF-193 (0.01 μg/ml), chromatid fibers in Ku-deficient cells had a beaded appearance quite different from that observed at higher doses of the drug in both normal and mutant cells, where the chromatid arms were more homogeneously elongated (Fig. 3A) (4, 6, 21). This altered condensation may have been caused by heterogeneous condensation of the chromatids.
FIG. 3.
Ku-deficient cells display altered chromosome condensation in the presence of low doses of ICRF-193. (A) V79B, XR-V15B, and XR-V9B cells were synchronized at late S/G2 phase and then treated with different ICRF-193 concentrations, 40 ng of nocodazole per ml, and 2 mM caffeine for 5 h. Metaphase chromosomes were prepared as described in Materials and Methods and stained with Hoechst 33258 dye for analysis by fluorescence microscopy. Representative images of V79B, XR-V15B, and XR-V9B cells incubated in the absence or presence of 0.01 and 1 μg of ICRF-193 per ml are shown. (B) V79B and XR-V15B cells were synchronized in G0/G1 and released into medium supplied with 10% FCS and different bleomycin concentrations for 17 h; 40 ng of nocodazole per ml and 2 mM caffeine were added 5 h before preparation of metaphase chromosomes. Representative images of V79B and XR-V15B cells incubated in the absence or presence of 10 and 100 μg of bleomycin per ml are shown.
Ku86-deficient cells display normal DNA topoisomerase II in vitro activity and normal α- or β-isoform levels.
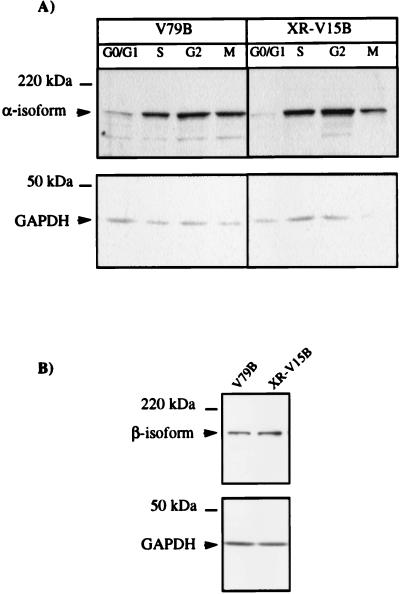
An altered response to DNA topoisomerase II inhibitors has been reported for different cell lines and was usually related to alterations in topoisomerase II expression levels (14, 53, 67) or to the phosphorylation status of the enzyme (19, 60). To rule out the possibility that lower levels of this enzyme could have been responsible for the hypersensitivity of Ku86-deficient cells to ICRF-193, the levels of topoisomerase IIα and -β were determined by Western blotting. As shown in Fig. 4A, and in agreement with previous reports (38, 71), α-isoform levels increased during S phase and were maximal in G2/M phases. More importantly, similar levels of the enzyme were detected in whole-cell extracts of synchronized XR-V15B and V79B cells at different stages of the cell cycle. Similar amounts of topoisomerase IIβ were also found in exponentially growing mutant and wild-type cells (Fig. 4B). No significant differences in expression levels of this isoform were observed during the cell cycle (reference 71 and data not shown).
FIG. 4.
Wild-type and Ku86-deficient cells show similar levels of α and β-isoforms of DNA topoisomerase II. (A) Whole-cell extracts were obtained from G1/G0-, S-, G2-, and M-phase cells (as described in Materials and Methods), and 75-μg aliquots of proteins of the different extracts were analyzed by Western blotting using a polyclonal antibody directed against DNA topoisomerase IIα. (B) A total of 5 × 105 exponentially growing cells were lysed with 1% SDS at 90°C for 5 min and loaded in each lane. DNA topoisomerase IIβ levels were analyzed by using a monoclonal antibody (3H10) directed against this isoform. The lower part of the membranes was probed with an anti-GAPDH antibody to ensure that comparable amounts of protein were loaded in all lanes. Representative results of three assays are shown.
Subsequently, we analyzed DNA topoisomerase II catalytic activity in vitro by kinetoplast DNA decatenation assays. Similar activities were observed when nuclear extracts of exponentially growing wild-type and mutant cells were compared (Fig. 5A). Note that in these assays, the total DNA topoisomerase II activity was measured, and it is thus not possible to differentiate between α- and β-isoform activities. Therefore, we determined the activities present in nuclear extracts of cells synchronized in S and G2 phases of the cell cycle, when the α-isoform level is increasing. In this case also, no significant differences were observed in decatenation activity when comparable amounts of topoisomerases IIα and -β were recovered in nuclear extracts from synchronized V79B and XR-V15B cells (Fig. 5B and C). Similar topoisomerase II activities were also reported when the Ku86-deficient cell lines xrs-1 and xrs-6 were compared with the parental cell line (36, 66). Thus, hypersensitivity to ICRF-193 in Ku86-deficient cell lines cannot be explained by gross abnormalities in the expression of both isoforms or in the catalytic activity of DNA topoisomerase II as assessed in vitro.
FIG. 5.
Ku86-deficient cells display normal DNA topoisomerase II decatenation activity in vitro. (A and B) Nuclear proteins (0, 5, 10, or 50 ng from exponentially growing V79B and XR-V15B cells [A] or 0, 10, or 30 ng from S- and G2-phase synchronized V79B and XR-V15B cells [B]) were used to assess the decatenation activity of DNA topoisomerase II in vitro, using 0.26 μg of kinetoplast DNA networks. After migration in 1% agarose for 1.5 h at 8 V/cm (A) or for 12 h at 1.8 V/cm (B), the gels were observed on a UV transilluminator. The kinetoplast networks (N) remain at the origin of the gel, while the circular or linear monomers generated by topoisomerase II activity migrate into the gel (DC). Kinetoplast DNA digested with XhoI was used as the migration control for linear monomers (arrowhead in panel A). (C) To verify that equivalent amounts of DNA topoisomerase IIα and β were recovered from the nuclear extracts used in the decatenation assays shown in panel B, we determined the α- and β-isoform levels by Western blot assays. Cyclin A levels were analyzed in parallel as an internal control; 10 μg of protein was loaded in each lane.
Transfection of the cDNA for Ku86 restores normal sensitivity of XR-V15B cells to ICRF-193.
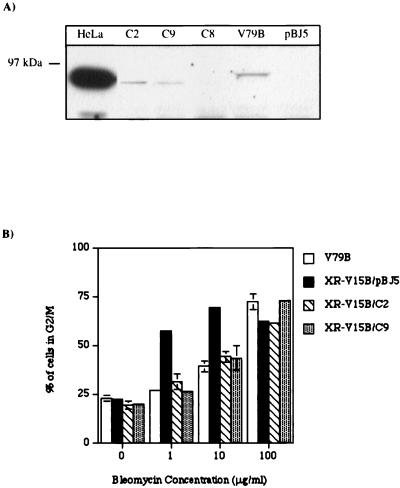
To unequivocally show that the absence of Ku86 was responsible for the hypersensitivity of XR-V15B cells to ICRF-193, we transfected mutant cells with an expression vector containing the human Ku86 cDNA or an empty vector (pBJ5) as a control (57). G418-resistant clones were isolated and analyzed for Ku86 expression by Western blotting. Two clones which expressed a protein that migrated similarly to Ku86 from HeLa cells (XR-V15B/C2 and XR-V15B/C9 [Fig. 6A]) were selected. The human protein reproducibly migrated slightly faster on SDS-PAGE than the protein from Chinese hamster cells. To certify that the human Ku86 protein was functional, the sensitivity of the transfectants to bleomycin was assessed by quantifying the percentage of cells blocked in G2/M (Fig. 6B). Normal sensitivity to bleomycin was fully restored in XR-V15B/C2 and XR-V15B/C9 cells.
FIG. 6.
Transfection of the cDNA for Ku86 restores the normal sensitivity of XR-V15B toward bleomycin. (A) Whole-cell lysates (90 μg of protein) of V79B, XR-V15B/C2 (C2), XR-V15B/C8 (C8), XR-V15B/C9 (C9), and XR-V15B/pBJ5 (pBJ5), and of HeLa cells (15 μg of protein) as a control for the human Ku86 protein, were analyzed by Western blotting using a polyclonal antibody directed against Ku86. (B) Exponentially growing V79B, XR-V15B/pBJ5, XR-V15B/C2, and XR-V15B/C9 cells were incubated with different doses of bleomycin for 16 h. The percentage of cells in G2/M was determined by FACscan analysis. Each determination is the mean ± standard error of two or three different experiments.
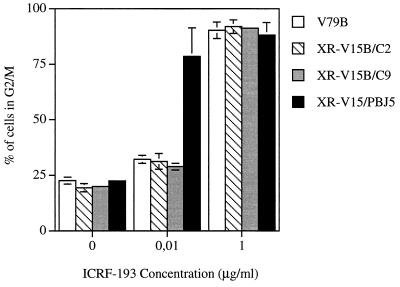
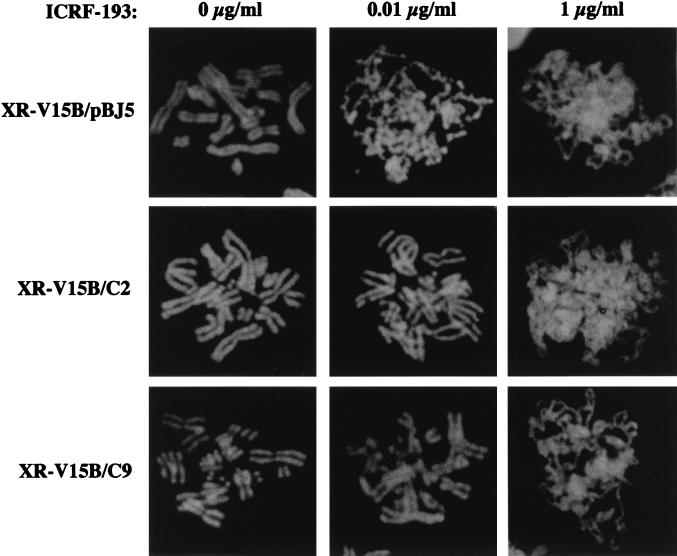
We then monitored the sensitivity of the XR-V15B/C2 and XR-V15B/C9 transfectants to ICRF-193 (Fig. 7). No G2/M arrest was observed at low doses of the drug: wild-type sensitivity was totally restored in the Ku86 transfectants. In contrast, XR-V15B-like hypersensitivity was observed in control pBJ5-transfected cells (Fig. 7). Furthermore, normal chromosome condensation was observed in the transfectants at a dose of the drug (0.01 μg/ml) that clearly affected the condensation of XR-V15B/pBJ5 cells (Fig. 8). In fact, at this dose 80% of V79B, XR-V15B/C2, and XR-V15B/C9 cells showed normal chromosome condensation, compared to only 3% of XR-V15B or XR-V15B/pBJ5 cells. Collectively, these data demonstrate that the absence of Ku86 in XR-V15B cells is responsible for their hypersensitivity to ICRF-193 as manifested by G2/M arrest and defective chromosome condensation after bypass of the G2 arrest.
FIG. 7.
Transfection of the Ku86 cDNA restores a normal sensitivity of XR-V15B cells toward ICRF-193. Exponentially growing V79B, XR-V15B/C2, XR-V15B/C9, and XR-V15B/pBJ5 cells were incubated with different doses of ICRF-193 for 16 h. The percentage of cells with a G2/M DNA content was determined by FACScan analysis. Each determination is the mean ± standard error of three different experiments.
FIG. 8.
Ku86-transfected cells show normal chromosome condensation at lower doses of ICRF-193. Metaphase chromosomes were prepared from synchronized XR-V15B/pBJ5, XR-V15B/C2, and XR-V15B/C9 cells at late S/G2 phase as described in Materials and Methods. Cells were incubated in the absence or presence of 0.01 and 1 μg of ICRF-193 per ml. Representative images of XR-V15B/pBJ5, XR-V15B/C2, and XR-V15B/C9 cells are shown.
DNA-PKCS-deficient cells neither accumulate in G2/M nor display impaired chromosome condensation at lower doses of ICRF-193.
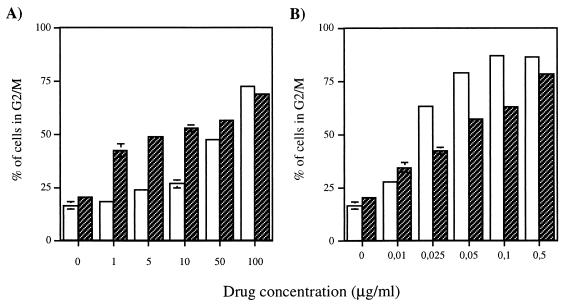
To determine whether the hypersensitivity of Ku-deficient cells to ICRF-193 was due to the absence of DNA-PK enzymatic activity, we assessed ICRF-193 sensitivity in the Chinese hamster ovary cell line V-3, which has a mutation in the DNA-PKCS gene resulting in defective DSB repair and V(D)J recombination (7, 56). First, we used FACscan analysis to determine the DNA content of V-3 and parental wild-type AA8 cells treated with increasing doses of bleomycin during 24 h. V-3 cells accumulated in G2/M following exposure to lower doses of the drug than did wild-type cells (Fig. 9A). There was also a higher fraction of V-3 cells with a DNA content of less than 2N in the presence of bleomycin (data not shown), probably representing apoptotic cells (74). These results confirm the hypersensitivity of V-3 mutant cells to DNA damage-inducing agents. In contrast, no such hypersensitivity was observed after ICRF-193 treatment. There were even slightly more wild-type AA8 than V-3 cells in G2/M at equivalent doses of ICRF-193 (Fig. 9B), a difference partially compensated for by a higher population of mutant cells with a lower than 2N DNA content (data not shown).
FIG. 9.
No difference in the accumulation of cells in G2/M was observed between DNA-PKCS-deficient and wild-type cells in response to ICRF-193. Exponentially growing AA8 (□) and V-3 ( ) cells were incubated with different concentrations of bleomycin (A) or ICRF-193 (B) for 24 h. The cells were harvested and stained with propidium iodide, and the percentage of cells in G2/M was determined by FACscan analysis. Each determination is the mean ± standard error of three different experiments.
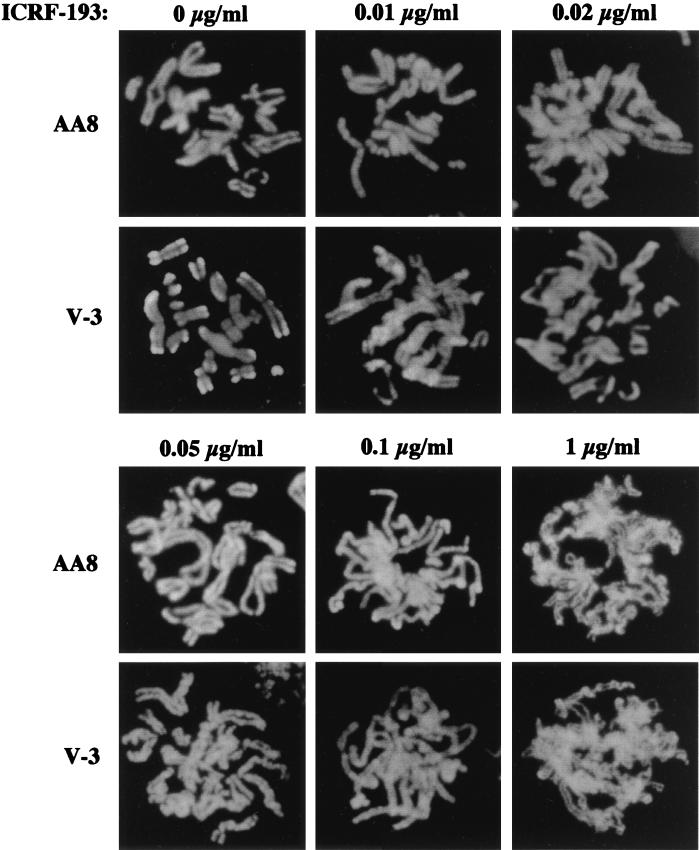
We next analyzed chromosome condensation in the presence of different doses of ICRF-193. Chromosome condensation was progressively affected in both cell lines at increasing concentrations of the drug. Considering that the extent of condensation was not uniform in all cells, even in the absence of ICRF-193, no significant differences between wild-type and V-3 mutant cell lines were observed (Fig. 10).
FIG. 10.
DNA-PKCS-deficient and wild-type cells display similar patterns of chromosome condensation in the presence of ICRF-193. AA8 and V-3 cells were synchronized at late S/G2 phase and then treated with 0, 0.01, 0.02, 0.05, 0.1, and 1 μg of ICRF-193 per ml, 80 ng of nocodazole per ml, and 2 mM caffeine for 5 h. Metaphase chromosomes were prepared as described in Materials and Methods. Representative images of AA8 and V-3 cells are shown.
Together, these results indicate that in contrast to Ku deficiency, an absence of DNA-PK activity does not result in an increased sensitivity to ICRF-193, as assessed by G2/M arrest and chromosome condensation.
DISCUSSION
We show here that Ku-deficient cells are hypersensitive to ICRF-193, a specific inhibitor of DNA topoisomerase II. The response of Ku86-deficient cells to low doses of ICRF-193 differed from that of wild-type cells in at least two respects: (i) they were arrested at the G2/M transition, and (ii) they demonstrated impaired chromosome condensation after forced bypass of the G2 arrest. Both of these defects were corrected by stable transfection of the Ku86 cDNA. These experiments did not differentiate between a specific requirement for the Ku86 or Ku70 subunit or, more globally, for the Ku-antigen heterodimer. Indeed, as the stability of the Ku70 subunit is dependent on the presence of Ku86, restoration of Ku86 results in normal levels of Ku70 (15, 25). A similar hypersensitivity was not observed with cells deficient in DNA-PKCS. Thus, these results indicate a novel role for the Ku antigen during the G2 and M phases of the cell cycle, a role not related to DNA-PK-dependent DNA repair.
The DNA topoisomerase II inhibitor ICRF-193 induces a G2 arrest in the absence of detectable DNA damage.
ICRF-193 variably induces arrest or retardation in the G2 phase depending on the drug concentration and cell type (4, 6, 21, 33, 34). In the case of the V79B wild-type Chinese hamster cells, we observed a G2 arrest at high doses of ICRF-193 in the absence of any detectable DNA damage. In contrast, G2 arrest caused by genotoxic drugs such as bleomycin or etoposide was accompanied by significant DNA damage that could be detected by PFGE (Fig. 2 and unpublished observations). As previously reported by Downes and colleagues (21), operation of either a decatenation checkpoint or a DNA damage checkpoint can cause G2 cell cycle arrest. These authors found that the decatenation, but not the DNA damage checkpoint, could be bypassed by caffeine in HeLa cells (21). However, in BHK or CHO cells, the cell cycle arrest caused by ICRF-193, as well as by the cleavable complex-stabilizing drugs etoposide and VM-26, could be overridden (4, 6). Similarly, in the experiments described here, the G2 arrest in V79B and XR-V15B cells induced by either ICRF-193 or bleomycin could be relieved by caffeine. Thus, we were not able to discriminate between decatenation and DNA damage checkpoints by the use of caffeine.
DNA damage was not detected in G2-arrested Ku86-deficient cells treated with ICRF-193. However, even in the presence of DNA-damaging agents such as bleomycin, mutant cells were blocked in G2 following doses which did not result in detectable DNA damage. Similar findings have been reported for SCID cells, which are deficient in DNA-PKCS (32). Thus, it cannot be excluded from these findings that the ICRF-193-mediated G2 arrest observed in mutant cells was caused by DSB, possibly as a secondary consequence of topoisomerase II inhibition. We therefore performed additional ICRF-193 sensitivity experiments with DNA-PKCS-deficient V-3 cells. As V-3 cells were not found to be hypersensitive to ICRF-193 treatment, it is unlikely that the G2 arrest observed in Ku-deficient cells was due to a deficiency in the repair of DSB. We cannot completely exclude the possibility that ICRF-193 induces DNA lesions which are not repaired by DNA-PK-dependent mechanisms. However, the mechanism by which ICRF-193 interferes with DNA topoisomerase II activity is not easily reconciled with such a hypothesis (54, 61). Thus, it is most likely that the ICRF-193-induced G2 arrest in mutant cells occurred as a consequence of impaired topoisomerase II activity and not because of unrepaired DNA damage.
The reason why Ku-deficient cells arrest in G2 following exposure to low doses of ICRF-193 remains to be determined. Although we found that in vitro decatenation activities were similar in the two cell types, this measure probably reflects only partially the real in vivo activity of topoisomerase II. Further work should be done to compare the latter activity between wild-type and mutant cells.
Bypass of the G2 arrest reveals abnormal chromosome condensation in Ku-deficient cells.
Bypass of the G2 checkpoint by caffeine treatment allowed us to analyze the activity of DNA topoisomerase II in chromosome condensation. We detected a clear inhibition of chromosome condensation in three independently derived Ku-deficient cell lines at the lowest dose of ICRF-193 tested (0.01 μg/ml). At this dose, normal chromosome condensation was observed in wild-type cells as well as in mutant cells transfected with the Ku86 cDNA.
In wild-type cells, ICRF-193 was found to inhibit the compaction of 300-nm-diameter chromatin fibers to 600-nm-diameter chromatids (34). This finding is reconcilable with the observation that topoisomerase II is found to be required in the final phase of compaction during reconstitution of nuclear structures around purified DNA in frog egg extracts (49). Also, partially condensed chromosomes containing two paired arms form at mitosis after ICRF-193 G2 arrest is overridden by 2-aminopurine (6). We observed such figures at high ICRF-193 concentrations in both wild-type and mutant cells. However, at lower concentrations of the drug, chromatid fibers in the mutant cells had a beaded appearance, suggesting that final condensation occurred irregularly along the chromatids. Since DNA topoisomerase IIα has been proposed to play a stoichiometric role during chromosome condensation (1, 6, 45), it is important to note that similar levels of the enzyme are present in Ku-deficient and wild-type cells. These results clearly demonstrate that the sensitivity of Chinese hamster cells to ICRF-193 is modulated by expression of the Ku antigen and not by alterations in DNA topoisomerase II levels.
No difference in chromosome condensation was observed in DNA-PKCS-deficient V-3 and wild-type cells after ICRF-193 treatment. However, we cannot completely exclude the possibility that low residual levels of enzymatic activity, not detectable by biochemical assays, are sufficient to protect V-3 cells against topoisomerase II inhibition by ICRF-193. In that respect, it should be noted that like SCID cells, V-3 cells are capable of normal signal join processing, while XR-V15B cells are defective in the processing of both signal and coding joins (7). Importantly, XR-C1 cells were recently characterized as deficient in DNA-PKCS and were found to be defective in the repair of both types of joins (24). Like V-3 cells, XR-C1 cells show no differences from wild-type cells with respect to chromosome condensation in response to ICRF-193 (our unpublished observations). Thus, available evidence indicates that the hypersensitivity of Ku-deficient cells toward ICRF-193 is specifically due to the absence of the Ku antigen and not to the absence of DNA-PK activity. It will be important to determine which type of molecular modifications are effectively responsible for the differential response of Ku-deficient cells to a DNA topoisomerase II-specific drug.
A novel function for Ku antigen in G2 and M phases of the cell cycle.
Our data suggest a novel role for Ku antigen not related to its function in DNA-PK dependent DNA repair. This role could consist of (i) the direct participation of Ku in the normal functions of DNA topoisomerase II in cell cycle progression or (ii) a checkpoint control function for Ku in the successful separation and condensation of chromatids. What are the indications in favor of the first hypothesis? It is noteworthy that some defects in Ku-deficient cells can be explained by deficiencies in topoisomerase II activity. Ku86-deficient xrs-5 cells have an altered scaffold organization with overcondensed chromosomes, possibly due to larger chromatin loops extending out of the chromosome core (55). Chromatin digestion data also suggest that the matrix attachment regions in xrs-5 cells are different from those in control cells (72), and abnormalities have been detected at the nuclear periphery (73). Additionally, growth defects in Ku86-null mice may be related to abnormalities in topoisomerase II function. This possibility is supported by the observation that fibroblasts derived from the knockout mice have a prolonged G2/M phase (51). Also, the duplication time of Ku-deficient XR-V15B cells is longer than that of the parental cell line (our unpublished observations). It will be important to precisely characterize the differences in topoisomerase II-related phenotypes between wild-type and Ku-deficient cells and to verify that complementation by Ku86 is able to restore wild-type phenotypes to mutant cells.
A possible mechanism by which Ku antigen could interact with DNA topoisomerase II is suggested by the recent observation that yeast topoisomerase II is associated with the Sgs1 helicase and that both topoisomerase and helicase activities are required for faithful chromosome segregation during mitosis (68). It will be interesting to determine whether the helicase activity of the Ku antigen is required for cooperation with DNA topoisomerase II (62).
No major differences between wild-type and mutant cell lines would be expected if Ku participates in G2/M checkpoints. Ku antigen may play a crucial role in the decatenation checkpoint control or more generally in the control of G2-M phase progression. In that respect, it is worth noting that while the G2 arrest observed after ICRF-193 treatment of wild-type cells or mutant cells complemented with Ku86 is reversible, it is irreversible in Ku-deficient cells (our unpublished observations). A similar role for DNA-PK in exiting G2 arrest after induction of DNA damage has recently been proposed (42). An intriguing possibility is that the Ku antigen is implicated in the decatenation checkpoint exit, in association with kinases or other proteins.
Future studies will enable us to determine the exact role of the Ku antigen in the faithful separation and condensation of newly replicated chromosomes in the G2 and M phases of the cell cycle.
ACKNOWLEDGMENTS
We thank A. M. Creighton for the generous gift of ICRF-193, G. F. Whitmore for V-3 cells, A. Kikuchi for monoclonal antibody 3H10 against DNA topoisomerase IIβ, and G. Chu for plasmid pBJ5-Ku86. The expertise of M. Chatenet and F. Clerc for PFGE analysis and V. Marechal for metaphase spreads was invaluable. The comments of N. Taylor and Y. Robbins on the manuscript were particularly helpful. Particular thanks go to E. Moustacchi for hospitality and advice and to U. Hibner and M. Olivier for discussion.
This work was supported by the ACC/SV8 from the Ministère de la Recherche et de l’Enseignement Supérieur and a grant from the Association pour la Recherche contre le Cancer. P.M. was supported by a postdoctoral fellowship from the Spanish Ministery and the European Community.
REFERENCES
- 1.Adachi Y, Kas E, Laemmli U K. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 1989;8:3997–4006. doi: 10.1002/j.1460-2075.1989.tb08582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi Y, Luke M, Laemmli U K. Chromosome assembly in vitro: topoisomerase II is required for condensation. Cell. 1991;64:137–148. doi: 10.1016/0092-8674(91)90215-k. [DOI] [PubMed] [Google Scholar]
- 3.Anderson C W, Lees-Miller S P. The nuclear serine/threonine protein kinase DNA-PK. Crit Rev Eukaryot Gene Expr. 1992;2:283–314. [PubMed] [Google Scholar]
- 4.Anderson H, Roberge M. Topoisomerase II inhibitors affect entry into mitosis and chromosome condensation in BHK cells. Cell Growth Differ. 1996;7:83–90. [PubMed] [Google Scholar]
- 5.Andoh T, Sato M, Narita T, Ishida R. Role of DNA topoisomerase II in chromosome dynamics in mammalian cells. Biotechnol Appl Biochem. 1993;18:165–174. [PubMed] [Google Scholar]
- 6.Andreassen P R, Lacroix F B, Margolis R L. Chromosomes with two intact axial cores are induced by G2 checkpoint override: evidence that DNA decatenation is not required to template the chromosome structure. J Cell Biol. 1997;136:29–43. doi: 10.1083/jcb.136.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blunt T, Finnie N J, Taccioli G E, Smith G C, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine acid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 8.Bosma G C, Custer R P, Bosma M J. A severe combined immunodeficiency mutation in the mouse. Nature. 1983;301:527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 9.Boubnov N V, Hall K T, Wills Z, Lee S E, He D M, Benjamin D M, Pulaski C R, Band H, Reeves W, Hendrickson E A, Weaver D T. Complementation of the ionizing radiation sensitivity, DNA end binding and V(D)J recombination defects of double-strand break repair mutants by the p86 Ku autoantigen. Proc Natl Acad Sci USA. 1995;92:890–894. doi: 10.1073/pnas.92.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brush G S, Anderson C W, Kelly T J. The DNA-activated protein kinase is required for the phosphorylation of replication protein A during simian virus 40 DNA replication. Proc Natl Acad Sci USA. 1994;91:12520–12524. doi: 10.1073/pnas.91.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Q P, Pitt S, Leskyk J, Baril E F. DNA-dependent ATPase from HeLa cells is related to human Ku antigen. Biochemistry. 1994;33:8548–8557. doi: 10.1021/bi00194a021. [DOI] [PubMed] [Google Scholar]
- 12.Carter T, Vancurova I, Sun I, Lou W, DeLeon S. A DNA-activated protein kinase from HeLa cell nuclei. Mol Cell Biol. 1990;10:6460–6471. doi: 10.1128/mcb.10.12.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaly N, Chen X, Dentry J, Brown D L. Organization of DNA topoisomerase II isotypes during the cell cycle of human lymphocytes and HeLa cells. Chromosome Res. 1996;4:457–466. doi: 10.1007/BF02265053. [DOI] [PubMed] [Google Scholar]
- 14.Chen A Y, Liu L F. DNA topoisomerase: essential enzymes and lethal targets. Annu Rev Pharmacol Toxicol. 1994;36:191–218. doi: 10.1146/annurev.pa.34.040194.001203. [DOI] [PubMed] [Google Scholar]
- 15.Chen F, Peterson S R, Story M D, Chen D J. Disruption of DNA-PK in Ku80 mutant xrs-6 and the implications in DNA double-strand break repair. Mutat Res. 1996;362:9–19. doi: 10.1016/0921-8777(95)00026-7. [DOI] [PubMed] [Google Scholar]
- 16.Chen M, Beck W T. Teniposide-resistant CEM cells, which express mutant DNA topoisomerase II alpha, when treated with non-complex-stabilizing inhibitors of the enzyme, display no cross-resistance and reveal aberrant functions of the mutant enzyme. Cancer Res. 1993;53:5946–5953. [PubMed] [Google Scholar]
- 17.Clarke D J, Johnson R T, Downes C S. Topoisomerase II inhibition prevents anaphase chromatid segregation in mammalian cells independently of the generation of DNA strand breaks. J Cell Sci. 1993;105:563–569. doi: 10.1242/jcs.105.2.563. [DOI] [PubMed] [Google Scholar]
- 18.Cockerill P N, Garrard W T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 19.DeVore R F, Corbett A H, Osheroff N. Phosphorylation of topoisomerase II by casein kinase II and protein kinase C: effects on enzyme-mediated DNA cleavage/religation and sensitivity to the antineoplastic drugs etoposide and 4′-(9-acridinylamino)methane-sulfon-m-anisidide. Cancer Res. 1992;52:2156–2161. [PubMed] [Google Scholar]
- 20.Dorr R T. Bleomycin pharmacology: mechanism of action and resistance, and clinical pharmacokinetics. Semin Oncol. 1992;19:3–8. [PubMed] [Google Scholar]
- 21.Downes C S, Clarke D J, Mullinger A M, Gimenez-Abian J F, Creighton A M, Johnson R T. A topoisomerase II-dependent G2 cycle checkpoint in mammalian cells. Nature. 1994;372:467–470. doi: 10.1038/372467a0. . (Erratum, 372:710.) [DOI] [PubMed] [Google Scholar]
- 22.Dvir A, Peterson S R, Knuth M W, Lu H, Dynan W S. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc Natl Acad Sci USA. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Earnshaw W C, Halligan B, Cooke C A, Heck M M, Liu L F. Topoisomerase II is a structural component of mitotic chromosome scaffolds. J Cell Biol. 1985;100:1706–1715. doi: 10.1083/jcb.100.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Errami A, He D M, Friedl A A, Overkamp W J I, Morolli B, Hendrickson E A, Eckardt-Schupp F, Oshimura M, Lohman P H M, Jackson S P, Zdzienicka M Z. XR-C1, a new CHO cell mutant which is defective in DNA-PKcs, is impaired in both V(D)J coding and signal joint formation. Nucleic Acids Res. 1998;26:3146–3153. doi: 10.1093/nar/26.13.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Errami A, Smider V, Rathmell W K, He D M, Hendrickson E A, Zdzienicka M Z, Chu G. Ku86 defines the genetic defect and restores X-ray resistance and V(D)J recombination to complementation group 5 hamster cell mutants. Mol Cell Biol. 1996;16:1519–1526. doi: 10.1128/mcb.16.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gasser S M, Laroche T, Falquet J, Boy de la Tour E, Laemmli U K. Metaphase chromosome structure. Involvement of topoisomerase II. J Mol Biol. 1986;188:613–629. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- 27.Giffin W, Kwast-Welfeld J, Rodda D J, Prefontaine G G, Traykova-Andonova M, Zhang Y, Weigel N L, Lefebvre Y A, Hache R J. Sequence-specific DNA binding and transcription factor phosphorylation by Ku autoantigen/DNA-dependent protein kinase. Phosphorylation of Ser- 527 of the rat glucocorticoid receptor. J Biol Chem. 1997;272:5647–5658. doi: 10.1074/jbc.272.9.5647. [DOI] [PubMed] [Google Scholar]
- 28.Gottlieb T M, Jackson S P. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 29.He D M, Lee S E, Hendrickson E A. Restoration of X-ray and etoposide resistance, Ku-end binding activity and V(D) J recombination to the Chinese hamster sxi-3 mutant by a hamster Ku86 cDNA. Mutat Res. 1996;363:43–56. doi: 10.1016/0921-8777(95)00060-7. [DOI] [PubMed] [Google Scholar]
- 30.Hirano T, Mitchison T J. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J Cell Biol. 1993;120:601–612. doi: 10.1083/jcb.120.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holm C, Goto T, Wang J C, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 32.Huang L C, Clarkin K C, Wahl G M. p53-dependent cell cycle arrests are preserved in DNA-activated protein kinase-deficient mouse fibroblasts. Cancer Res. 1996;56:2940–2944. [PubMed] [Google Scholar]
- 33.Ishida R, Miki T, Narita T, Yui R, Sato M, Utsumi K R, Tanabe K, Andoh T. Inhibition of intracellular topoisomerase II by antitumor bis(2,6-dioxopiperazine) derivatives: mode of cell growth inhibition distinct from that of cleavable complex-forming type inhibitors. Cancer Res. 1991;51:4909–4916. [PubMed] [Google Scholar]
- 34.Ishida R, Sato M, Narita T, Utsumi K R, Nishimoto T, Morita T, Nagata H, Andoh T. Inhibition of DNA topoisomerase II by ICRF-193 induces polyploidization by uncoupling chromosome dynamics from other cell cycle events. J Cell Biol. 1994;126:1341–1351. doi: 10.1083/jcb.126.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson S P, Jeggo P A. DNA double-strand break repair and V(D)J recombination: involvement of DNA-PK. Trends Biochem Sci. 1995;20:412–415. doi: 10.1016/s0968-0004(00)89090-8. [DOI] [PubMed] [Google Scholar]
- 36.Jeggo P A, Caldecott K, Pidsley S, Banks G R. Sensitivity of chinese hamster ovary mutants defective in DNA double strand break repair to topoisomerase II inhibitors. Cancer Res. 1989;49:7057–7063. [PubMed] [Google Scholar]
- 37.Kimura K, Nozaki N, Enomoto T, Tanaka M, Kikuchi A. Analysis of M phase-specific phosphorylation of DNA topoisomerase II. J Biol Chem. 1996;271:21439–21445. doi: 10.1074/jbc.271.35.21439. [DOI] [PubMed] [Google Scholar]
- 38.Kimura K, Saijo M, Ui M, Enomoto T. Growth state- and cell cycle-dependent fluctuation in the expression of two forms of DNA topoisomerase II and possible specific modification of the higher molecular weight form in the M phase. J Biol Chem. 1994;269:1173–1176. [PubMed] [Google Scholar]
- 39.Kuhn A, Gottlieb T M, Jackson S P, Grummt I. DNA-dependent protein kinase: a potent inhibitor of transcription by RNA polymerase I. Genes Dev. 1995;9:193–203. doi: 10.1101/gad.9.2.193. [DOI] [PubMed] [Google Scholar]
- 40.Kysela B P, Michael B D, Arrand J E. Relative contributions of levels of initial DNA damage and repair of double strand breaks to the ionizing radiation-sensitive phenotype of the Chinese hamster cell mutant, XR-V15B. Part I. X-rays. Int J Radiat Biol. 1993;63:609–616. doi: 10.1080/09553009314450791. [DOI] [PubMed] [Google Scholar]
- 41.Labhart P. DNA-dependent protein kinase specifically represses promoter-directed transcription initiation by RNA polymerase I. Proc Natl Acad Sci USA. 1995;92:2934–2938. doi: 10.1073/pnas.92.7.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee S E, Mitchell R A, Cheng A, Hendrickson E A. Evidence for DNA-PK-dependent and -independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol Cell Biol. 1997;17:1425–1433. doi: 10.1128/mcb.17.3.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lees-Miller S P, Chen Y R, Anderson C W. Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Mol Cell Biol. 1990;10:6472–6481. doi: 10.1128/mcb.10.12.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L F. DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- 45.Meyer K N, Kjeldsen E, Straub T, Knudsen B R, Hickson I D, Kikuchi A, Kreipe H, Boege F. Cell cycle-coupled relocation of types I and II topoisomerases and modulation of catalytic enzyme activities. J Cell Biol. 1997;136:775–788. doi: 10.1083/jcb.136.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mimori T, Hardin J A. Mechanism of interaction between Ku protein and DNA. J Biol Chem. 1986;261:10375–10379. [PubMed] [Google Scholar]
- 47.Mimori T, Hardin J A, Steitz J A. Characterization of the DNA-binding protein antigen Ku recognized by autoantibodies from patients with rheumatic disorders. J Biol Chem. 1986;261:2274–2278. [PubMed] [Google Scholar]
- 48.Morozov V E, Falzon M, Anderson C W, Kuff E L. DNA-dependent protein kinase is activated by nicks and larger single-stranded gaps. J Biol Chem. 1994;269:16684–16688. [PubMed] [Google Scholar]
- 49.Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987;48:205–207. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- 50.Newport J, Spann T. Disassembly of the nucleus in mitotic extracts: membrane vesicularization, lamin disassembly, and chromosome condensation are independent processes. Cell. 1987;48:219–230. doi: 10.1016/0092-8674(87)90425-9. [DOI] [PubMed] [Google Scholar]
- 51.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig M C, Li G C. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 52.Pan Z Q, Amin A A, Gibbs E, Niu H, Hurwitz J. Phosphorylation of the p34 subunit of human single-stranded-DNA-binding protein in cyclin A-activated G1 extracts is catalyzed by cdk-cyclin A complex and DNA-dependent protein kinase. Proc Natl Acad Sci USA. 1994;91:8343–8347. doi: 10.1073/pnas.91.18.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pommier Y, Leteurtre F, Fesen M R, Fujimori A, Bertrand R, Solary E, Kolhagen G, Kohn K W. Cellular determinants of sensitivity and resistance to DNA topoisomerase inhibitors. Cancer Investig. 1994;12:530–542. doi: 10.3109/07357909409021413. [DOI] [PubMed] [Google Scholar]
- 54.Roca J, Ishida R, Berger J M, Andoh T, Wang J C. Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc Natl Acad Sci USA. 1994;91:1781–1785. doi: 10.1073/pnas.91.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwartz J L, Cowen J M, Moan E, Sedita B A, Stephens J, Vaughan A T. Metaphase chromosome and nucleoid differences between CHO-K1 and its radiosensitive derivative xrs-5. Mutagenesis. 1993;8:105–108. doi: 10.1093/mutage/8.2.105. [DOI] [PubMed] [Google Scholar]
- 56.Sipley J D, Menninger J C, Hartley K O, Ward D C, Jackson S P, Anderson C W. Gene for the catalytic subunit of the human DNA-activated protein kinase maps to the site of the XRCC7 gene on chromosome 8. Proc Natl Acad Sci USA. 1995;92:7515–7519. doi: 10.1073/pnas.92.16.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smider V, Rathmell W K, Lieber M R, Chu G. Restoration of X-ray resistance and V(D)J recombination in mutant cells by Ku cDNA. Science. 1994;266:288–291. doi: 10.1126/science.7939667. [DOI] [PubMed] [Google Scholar]
- 58.Strick R, Laemmli U K. SARs are cis DNA elements of chromosome dynamics: synthesis of a SAR repressor protein. Cell. 1995;83:1137–1148. doi: 10.1016/0092-8674(95)90140-x. [DOI] [PubMed] [Google Scholar]
- 59.Taccioli G E, Gottlieb T M, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann A R, Alt F W, Jackson S P, Jeggo P A. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 60.Takano H, Kohno K, Ono M, Uchida Y, Kuwano M. Increased phosphorylation of DNA topoisomerase II by antitumor agents bis(2,6-dioxopiperazine) derivatives. Cancer Res. 1991;51:3951–3957. [PubMed] [Google Scholar]
- 61.Tanabe K, Ikegami Y, Ishida R, Andoh T. Inhibition of topoisomerase II by antitumor agents bis(2,6-dioxopiperazine) derivatives. Cancer Res. 1991;51:4903–4908. [PubMed] [Google Scholar]
- 62.Tuteja N, Tuteja R, Ochem A, Taneja P, Huang N W, Simoncsits A, Susic S, Rahman K, Marusic L, Chen J, Zhang J, Wang S, Pongor S, Falaschi A. Human DNA helicase II: a novel DNA unwinding enzyme identified as the Ku autoantigen. EMBO J. 1994;13:4991–5001. doi: 10.1002/j.1460-2075.1994.tb06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- 64.Vassetzky Y S, Dang Q, Benedetti P, Gasser S M. Topoisomerase II forms multimers in vitro: effects of metals, beta-glycerophosphate, and phosphorylation of its C-terminal domain. Mol Cell Biol. 1994;14:6962–6974. doi: 10.1128/mcb.14.10.6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 66.Warters R L, Lyons B W, Li T M, Chen D J. Topoisomerase II activity in DNA double-strand break repair deficient Chinese hamster ovary cell line. Mutat Res. 1991;254:167–174. doi: 10.1016/0921-8777(91)90008-d. [DOI] [PubMed] [Google Scholar]
- 67.Watt P M, Hickson I D. Structure and function of type II DNA topoisomerase. Biochem J. 1994;303:681–695. doi: 10.1042/bj3030681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watt P M, Louis E J, Borts R H, Hickson I D. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 69.Weaver D T. Regulation and repair of double-strand DNA breaks. Crit Rev Eukaryot Gene Expr. 1996;6:345–375. doi: 10.1615/critreveukargeneexpr.v6.i4.20. [DOI] [PubMed] [Google Scholar]
- 70.Wiler R, Leber R, Moore B B, Van Dyk L F, Perryman L E, Meek K. Equine severe combined immunodeficiency: a defect in V(D)J recombination and DNA-dependent protein kinase activity. Proc Natl Acad Sci USA. 1995;92:11485–11489. doi: 10.1073/pnas.92.25.11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woessner R D, Mattern M R, Mirabelli C K, Johnson R K, Drake F H. Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 1991;2:209–214. [PubMed] [Google Scholar]
- 72.Yasui L S, Fink T J, Enrique A M. Nuclear scaffold organization in the X-ray sensitive Chinese hamster mutant cell line, xrs-5. Int J Radiat Biol. 1994;65:185–192. doi: 10.1080/09553009414550221. [DOI] [PubMed] [Google Scholar]
- 73.Yasui L S, Ling-Indeck L, Johnson-Wint B, Fink T J, Molsen D. Changes in the nuclear structure in the radiation-sensitive CHO mutant cell, xrs-5. Radiat Res. 1991;127:269–277. [PubMed] [Google Scholar]
- 74.Yonish-Rouach Y, Borde J, Gotteland M, Mishal Z, Viron A, May E. Induction of apoptosis by transiently transfected metabolically stable wt p53 in transformed cell lines. Cell Death Differ. 1994;1:39–47. [PubMed] [Google Scholar]
- 75.Zdzienicka M Z. Mammalian mutants defective in the response to ionizing radiation-induced DNA damage. Mutat Res. 1995;336:203–213. doi: 10.1016/0921-8777(95)00003-3. [DOI] [PubMed] [Google Scholar]
- 76.Zdzienicka M Z, Tran Q, van der Schans G P, Simons J W. Characterization of an X-ray-hypersensitive mutant of V79 Chinese hamster cells. Mutat Res. 1988;194:239–249. doi: 10.1016/0167-8817(88)90025-9. [DOI] [PubMed] [Google Scholar]
- 77.Zhu C, Bogue M A, Lim D S, Hasty P, Roth D B. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]