Abstract
We describe here a novel targeting gene therapy strategy to direct gene expression responsive to hepatitis C virus (HCV). The goal was approached by engineering a construct containing the antisense sequence of the transgene and internal ribosome entry site of encephalomyocarditis virus flanked by 5′- and 3′-end sequences of HCV cDNA that contain cis-acting replication elements. Thus, expression of the transgene is only promoted when the minus-strand RNA has been synthesized by the functional replication machinery present in infected cells. Reporter assay and strand-specific reverse transcription-PCR showed selective transgene expression in Huh-7 cells harboring an autonomously replicating HCV subgenome but remaining silent in uninfected cells. Furthermore, using the cytosine deaminase suicide gene as a transgene coupled with recombinant adenovirus delivery, we demonstrated that cytosine deaminase was specifically expressed in replicon cells, resulting in marked chemosensitization of replicon cells to the cytotoxic effects of flucytosine. This new targeting strategy could be extended to other single-stranded RNA viruses encoding the unique RNA-dependent RNA polymerase that has no parallel in mammalian cells.
Chronic hepatitis C virus (HCV) infection, which frequently leads to liver cirrhosis and hepatocellular carcinoma (3, 34), remains a major public health problem worldwide. It has been estimated that more than 3% of the world population is infected with HCV. The plus-strand HCV RNA genome is approximately 9,600 nucleotides in length and encodes a polyprotein precursor of about 3,010 amino acids, which is cleaved co- and posttranslationally by cellular and viral proteases to produce structural and nonstructural (NS) proteins (8, 13, 15). One of the NS proteins, NS5B, is an RNA-dependent RNA polymerase (RdRp) that catalyzes the replication of HCV (5, 30).
Current treatment modalities available for HCV infection, including alpha interferon, has limited effectiveness. Only 20 to 30% of alpha interferon-treated patients develop a sustained remission, and increased or prolonged systemic administration is often associated with severe side effects or viral resistance. The combination of ribavirin and interferon is known to be significantly more effective than interferon monotherapy in naïve and relapser patients, but it induces a sustained response only in 41% of patients and in less than 30% of patients infected with genotype 1 (18). The therapeutic potential of small inhibitory molecules that target serine protease, helicase, or RdRp has proved to be promising (4, 6, 10), however, one unavoidable problem of this approach is selection of resistant mutants conferred by single or multiple mutations due to the error-prone nature of RdRp (24, 26, 29). Thus, an alternative approach for treating HCV patients that results in the death of infected cells, thereby limiting or eliminating virus production, while leaving uninfected cells unharmed would have an advantage over the HCV therapies now available.
Here we describe a targeting gene therapy approach that harnesses the viral replication machinery by constructing an HCV-like minigenome that consisted of the antisense sequence of cytosine deaminase (CD) suicide gene and internal ribosome entry site (IRES) element from encephalomyocarditis virus (EMCV) flanked by the 5′- and 3′-end regions of HCV, so that expression of CD is initiated only when the minus-strand RNA has been synthesized by the functional replication components present in infected cells. Using recombinant adenovirus delivery, we demonstrate that CD is specifically expressed in infected cells, resulting in marked chemosensitization of infected cells to the cytotoxic effects of flucytosine (5-FC).
MATERIALS AND METHODS
Cells.
The cell lines Huh-7, HepG2, A549, and 293 were purchased from the American Type Culture Collection (ATCC) and maintained in Dulbecco′s modified Eagle's medium (DMEM, Invitrogen) supplemented with 10% fetal calf serum and 50 u/ml penicillin and streptomycin in a 5% CO2 humidified atmosphere. A Huh-7-derived cell line (Huh-NNRZ) stably replicating the HCV subgenomic replicon was grown in DMEM containing 300 μg/ml G418 (Geneticin, Invitrogen).
Plasmids.
For construction of the reporter vectors pT7cRLs, a fragment containing HCV cDNA (1 to 377) with the T7 promoter directly coupled at the 5′ end was amplified by PCR with primers 5′-tataagcttTAATACGACTCACTATAGCCAGCCCCCGATTGGGGGC-3′ and 5′-tgctctagaTTTGGTTTTTCTTTGAGGTT-3′ (capital letters indicate the sequences originally contained in the target sequence, and lowercase letters indicate the attached sequences which were introduced for the convenience of molecular manipulation, such as restriction sites). The EMCV IRES sequence was amplified from pEMCVRL (42) with primers 5′-cgcggatccAACTAACTAACTAAGCTAGC-3′ and 5′-ctctctagaGTATTATCGTGTTTTTCAAA-3′. The PCR products were digested with HindIII and XbaI or BamHI and XbaI, respectively, and cloned into HindIII/BamHI-digested pUC18 to generate pTCE. The 3′ part of the NS5B coding region connected 3′ untranslated region (UTR) of HCV was amplified by PCR using primers 5′-cgcggatccGGAAACTTGGGGTCCCACCC-3′ and 5′-ataggcgccagcgaggaggctgggaccatgccggccACATGATCTGCAGAGAGGCC-3′, digestedwith BamHI and NarI and cloned, along with the annealed oligonucleotides containing the partial sequence of the HDV ribozyme, sense 5′-CGCCGGCTGGGCAACATTCCGAGGGGACCGTCCCCTCGGTAATGGCGAATGGGACCg-3′ and antisense 5′-aattcGGTCCCATTCGCCATTACCGAGGGGACGGTCCCCTCGGAATGTTGCCCAGCCGG-3′, into BamHI/EcoRI-cut pTCE, generating pTCECD. The Renilla luciferase gene was amplified from pRL-TK (Promega) using primers 5′-ctctctagaATGACTTCGAAAGTTTATGA-3′ and 5′-ctctctagaTTATTGTTCATTTTTGAGAA-3′, digested with XbaI, and inserted into XbaI-digested pTCECD to generate pT7cRLNS5B1 or pT7RLNS5B1. pT7cRLNS5B2 or pT7cRLUTR, which contain the NS5B coding region from nucleotides 9307 to 9371 plus the 3′-UTR or 3′-UTR alone, was constructed similarly.
To construct pmCMVcRLmpA, the HCV minigenome was amplified from pTcRLNS5B1 by primers 5′-atagagctcTCTGGCTAACTGCCAGCCCCCGATTGGGGGC-3′ and 5′-ctcactagtACATGATCTGCAGAGAGGCC-3′, digested with SacI and SpeI and cloned, together with the annealed minimal poly(A) oligonucleotides, sense 5′-CTAGAACTAGTAATAAAGGATCCTTTATTTTCATTGGATCCGTGTGTTGGTTTTTTGTGTGCGGCCGCG-3′ and antisense 5′-AATTCGCGGCCGCACACAAAAAACCAACACACGGATCCAATGAAAATAAAGGATCCTTTATTACTAGTT-3′ into SacI/EcoRI-digested pShuttle (Clontech).
For construction of pmCMVcRLHD, a PCR product amplified from pT7cRLNS5B1 with primers 5′-ATGACTTCGAAAGTTTATGA-3′ and 5′-atacttaagGGTCCCATTCGCCATTACC-3′ was digested with BstBI and AflII and cloned with the SacI-BstBI fragment from pmCMVcRLmpA into SacI/AflII-digested pShuttle. Using pmCMVcRLmpA as a template DNA, the 5′ hammerhead ribozyme sequence was fused to the 5′ UTR by PCR using the primers5′-tatactagtGGGCTGGCCTGATGAGTCCGTGAGGACGAAACATGCATCTCCATGCATGTCGCCAGCCCCCGATTGGGGGC-3′ and 5′-TTTCTCCGCACCCGACATAG-3′, after digestion with SpeI and EcoRI, the fragment was cloned into NheI/EcoRI-digested pShuttle generating pCMVHHcRLmpA. The CD gene was amplified by PCR using primers 5′-gtgtctagaAGGCTAACAATGTCGAATAA−3′ and 5′-atatctagaAGACAGCCGCTGCGAAGGCA−3′, digested with XbaI, and ligated with the XbaI-digested pmCMVcRLmpA to generate pmCMVcCDmpA. The sequences of these constructs were confirmed by nucleotide sequencing.
Adenovirus.
The expression cassette in pmCMVcCDmpA, which is flanked by I-Ceui and PI-SceI sites, was digested with these two restriction enzyme, and ligated to the E1- and E3-deleted Adeno-X viral DNA (I-CeuI and PI-SceI digested) (Adeno-X Expression System, Clontech). The resultant adenoviral DNA (AdmCMVcCDmpA) was digested with PacI and then transfected into low-passage 293 cells. Seven days following transfection, crude virus was prepared from the transfected cells by three cycles of freeze-thawing, and further amplified in 293 cells by several rounds of infection. The purified virus was aliquoted and stored at −80°C before use. The authenticity of recombinant adenoviral DNA was verified before preparing high-titer viral stocks. AdsiNS5B was constructed as described previously (42), the sense-strand sequence of short hairpin RNA is 5′-GAAGGTCACCTTTGACAGA-3′.
In vitro transcription.
Plasmids were linearized at SalI site located immediately downstream of the HDV ribozyme, and these fragments were used as templates for runoff RNA synthesis with T7 RNA polymerase according to the protocol supplied by the manufacturer (Roche). For capped-RNA synthesis, T7 Cap-Scribe (Roche) was used. After transcription, 10 units of RQ DNaseI (Promega) were added to the reaction mixture to digest DNA templates. The mixture was extracted with phenol-chloroform and RNA was precipitated with ethanol-7.5 M ammonium acetate.
Transfection.
Cells were seeded onto 35-mm-diameter tissue culture dishes 24 h before transfection. One microgram of each reporter vector, 1 μg of pAM8-1 (when expression from the T7 promoter was desired), and 0.1 μg of pGL3-Control vector were cotransfected into cells with TransFast Transfection Reagent (Promega). For RNA transfection, 1 μg of each reporter RNA and 0.5 μg of capped firefly luciferase RNA were cotransfected into cells with Lipofectin reagent (Invitrogen). The cells were harvested after 48 h, and cell lysates were assayed for luciferase activity as described below.
Luciferase assay.
Cell lysates were prepared from transfected cells, centrifuged briefly, and 20 μl of the supernatants were used for luciferase assays with Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer′s instructions. Luciferase activities were measured using a TD-20/20 luminometer (Promega).
Strand-specific RT-PCR.
RNAs were isolated from transfected cells with Trizol reagent (Invitrogen) and treated with RNase-free DNase (Promega). The DNA-free RNA was extracted with phenol-chloroform and precipitated with ethanol. The absence of DNA in the RNA templates was confirmed by a control PCR without reverse transcriptase. For reverse transcription (RT), 1 μg of RNA was denatured at 65°C for 2 min, and cDNA synthesis was performed in 20-μl reaction volume with Superscript II reverse transcriptase (Invitrogen) at 42°C for 1 h using a primer complementary to Renilla luciferase gene, 5′-CTTATCTTGATGCTCATAGC-3′. The resulting cDNA was amplified for 35 cycles using primer 5′-ATGACTTCGAAAGTTTATGA-3′ and the primer used in the RT reaction.
Northern blot analysis.
Total RNAs were isolated and purified as described above, separated by denaturing agarose gel electrophoresis, and analyzed by Northern blot using Digoxigenin-labeled sense and antisense Renilla luciferase sequence to detect plus- and minus-strand transcripts.
Real-time RT-PCR.
One microgram of DNase-treated total RNA was reverse-transcribed as described for strand-specific RT-PCR using a primer complementary to NS5B 5′-ACGGAGCGGATGTGGTTGAC-3′. After an incubation at 95°C for 5 min, the resulting cDNA was quantified with SYBR Green according to the protocol supplied by the manufacturer (Takara). PCR was performed with a primer 5′-TGGTCTACGCCACAACATCC-3′ and the primer used in the RT reaction. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA level in each sample was simultaneously quantified to normalize the value of HCV replicon RNA.
CD enzymatic assay.
Huh-NNRZ or Huh-7 cells (1 × 106) were infected with AdmCMVcCDmpA at a multiplicity of infection (MOI) of 0, 10, 20, 50, 100, and 200. After 48 h, cells were harvested, washed once with phosphate-buffered saline, and resuspended in 0.5 ml 10 mM Tris-HCl. The cell suspension were sonicated, briefly centrifuged, and 100 μl supernatant was mixed with 170 μl PBS and 30 μl of 30 mM 5-FC solution. After incubation at 37°C for 24 h, 50 μl of reaction mixture was taken and quenched in 0.95 ml of 0.1 N HCl, the optical densities (ODs) were measured on a UV spectrophotometer at 255 and 290 nM. The amounts of 5-FC and 5-fluorouracil (5-FU) in the reaction mixture were calculated using the equations (0.1191 × OD290 − 0.02485 × OD255)× 20 = mM 5-FC and (0.1849 × OD255 − 0.04907 × OD290) × 20 = mM 5-FU; the conversion of 5-FC to 5-FU was then calculated as [mM 5-FC/(mM 5-FC + mM 5-FU)] × 100% (23).
Cytotoxic assay.
Cells (Huh-NNRZ or Huh-7) were mock infected or infected with AdmCMVcCDmpA (MOI 80). Twenty-four hours later, the medium was changed with fresh DMEM containing 0 or 0.5 mM 5-FC. After an additional 4-day culture, cell viability was measured with cell proliferation reagent WST-1 (Roche) according to the manufacturer's instructions.
RESULTS
Determining cis-acting elements in the NS5B coding region essential for viral minus-strand RNA synthesis.
Synthesis of HCV minus-strand RNA is initiated by recognition of the 3′ end of RNA template by RdRp, which requires a membrane-associated replication complex of viral and cellular proteins and a viral RNA template containing cis-active replication elements. It was known that conserved sequences and structures in the 5′ and 3′ untranslated region (UTR) in HCV RNA function as cis-acting elements essential for viral replication (12, 21, 38). However, the precise mechanism of the initiation of RNA synthesis is not fully understood. Although Oh et al. (31) previously showed that RdRp can utilize the 3′ UTR of HCV RNA as a minimal template in vitro, increasing evidence has supported that the 3′ UTR may not be a good template by itself for efficient RdRp binding and subsequent RNA synthesis, and the 3′ part of NS5B coding region with conserved stem-loop structure may also harbor functional cis-acting element required for viral replication (7, 39).
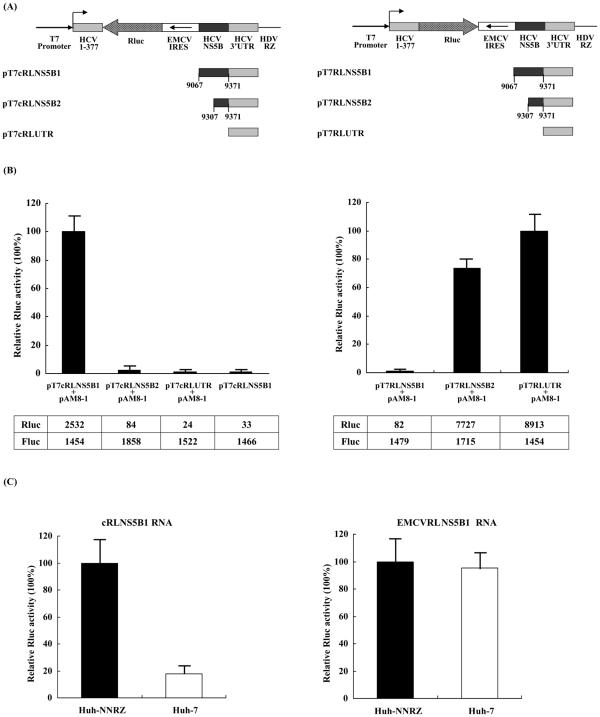
To further define the sequence of the NS5B coding region essential for HCV minus-strand RNA synthesis, we constructed HCV minigenome reporter vectors by inserting the antisense sequence of the Renilla luciferase gene and EMCV IRES between the 5′ end (nucleotides 1 to 377) and differently truncated NS5B coding region-connected 3′ UTR or 3′ UTR alone (Fig. 1A). In view of the consideration that functional HCV minigenome may require authentic 5′ and 3′ ends without overhang, the cassette was juxtaposed precisely at the T7 transcription start site and was followed by self-cleaving HDV ribozyme (36). Huh-NNRZ cells, a Huh-7-derived cell line stably replicating the HCV subgenomic replicon (22, 25, 42), were transfected with each reporter vector, pAM8-1 plasmid expressing T7 RNA polymerase (40) and pGL3-Control vector. The cell lysates were collected and Renilla luciferase activities were measured 48 h after transfection. The firefly luciferase (Fluc) activity from cotransfected pGL3-Control vector was simultaneously measured to normalize the transfection efficiency. As shown in Fig. 1B, the expression of Renilla luciferase, which directly reflects the level of minus-strand RNA synthesized, was only detected in cells transfected with pT7cRLNS5B1, which contains the NS5B coding region from nucleotides 9067 to 9371 upstream of 3′ UTR. Neither of the cells transfected with pT7cRLNS5B2 and pT7cRLUTR, which respectively contain the NS5B coding region from nucleotides 9307 to 9371 plus 3′-UTR and 3′-UTR alone, expressed Renilla luciferase. This indicates that the 3′ part of NS5B coding region from nucleotides 9067 to 9306 contains one or more cis-acting elements, which are absolutely required for HCV minus-strand RNA synthesis. Importantly, the Renilla luciferase activity in Huh-NNRZ cells transfected with pT7cRLNS5B1 alone (not cotransfection with pAM8-1) was almost negligible, providing a evidence that minus-strand RNA detected here does not result from cryptic transcription but from a HCV-dependent trans replication using minigenome transcript as a template (see below). On the basis of this finding, pT7cRLNS5B1 was used for subsequent experiments. For comparison, reporter assay with pT7RLNS5B1, pT7RLNS5B2 and pT7RLUTR, which different from their counterpart in that the Renilla luciferase gene was inserted in the opposite direction (Fig. 1A right), was also conducted. Interestingly, inclusion of the same NS5B coding sequence (9067 ∼ 9306) dramatically inhibited the HCV IRES-directed Renilla luciferase expression in pT7RLNS5B1, suggesting that the 3′-partial NS5B coding region may contain bifunctional element(s) participating in both repression of HCV IRES-dependent translation and initiation of minus-strand RNA synthesis to coordinate these two processes.
FIG. 1.
Determination of cis-acting replication elements in the NS5B coding region. (A) Schematic display of T7-based HCV minigenome reporter constructs. HCV minigenome containing the antisense (left) or sense (right) sequence of the Renilla luciferase gene and the antisense sequence of the EMCV IRES flanked by the 5′ end (1 to 377) and differently truncated NS5B coding region-connected 3′ UTR or 3′ UTR alone was juxtaposed precisely at the T7 transcription start site and followed by the HDV ribozyme gene. (B) The indicated reporter vectors were transfected into Huh-NNRZ cells with or without pAM8-1 expressing T7 RNA polymerase. Relative Renilla luciferase activities in the lysates were determined at 48 h posttransfection. The columns and bars represent the means and standard deviations of three independent triplicate transfections. (C) Huh-NNRZ cells were transfected with in vitro transcribed cRLNS5B1 RNA (left) or EMCVRLNS5B1 RNA (right) together with capped firefly luciferase RNA as an internal control. Relative Renilla luciferase activities in the lysates were determined at 24 h posttransfection. Absolute values of Renilla and firefly luciferase activity are listed below the corresponding bars.
To further rule out the possibility that the minus-strand RNAs which function as mRNAs for Renilla luciferase expression were transcription products by a cryptic promoter element in the 3′ part of NS5B coding region, we monitored the Renilla luciferase activity after transfection with RNAs in vitro transcribed from pT7cRLNS5B1. As a control, transcripts in vitro transcribed from pT7EMCVRLNS5B1, which is identical to pT7cRLNS5B1 except for the sense (not antisense) sequence of EMCV IRES and Renilla luciferase gene inserted between 5′- and 3′-end sequences of HCV, was also included. Capped firefly luciferase RNA was cotransfected as an internal control to normalize the transfection efficiency. As shown in Fig. 1C, Renilla luciferase activity was also detected in Huh-NNRZ cells transfected with cRLNS5B1 RNA, which was significantly higher than that in transfected Huh-7 cells. In contrast, after transfection with EMCVRLNS5B1 RNA, both Huh-NNRZ and Huh-7 cells expressed comparable Renilla luciferase, indicating that the higher level of Renilla luciferase expression in Huh-NNRZ cells was not due to the differences in mRNA stability and/or translation between Huh-NNRZ and Huh-7 cells. In view of this result together with the observation that the Renilla luciferase activity in pT7cRLNS5B1-transfected Huh-NNRZ cells was almost negligible in the absence of T7 RNA polymerase (Fig. 1B), we conclude that the minus-strand RNA was synthesized by HCV-dependent trans replication rather than transcription by a cryptic promoter element in pT7cRLNS5B1.
Specific gene expression in HCV replicon cell line.
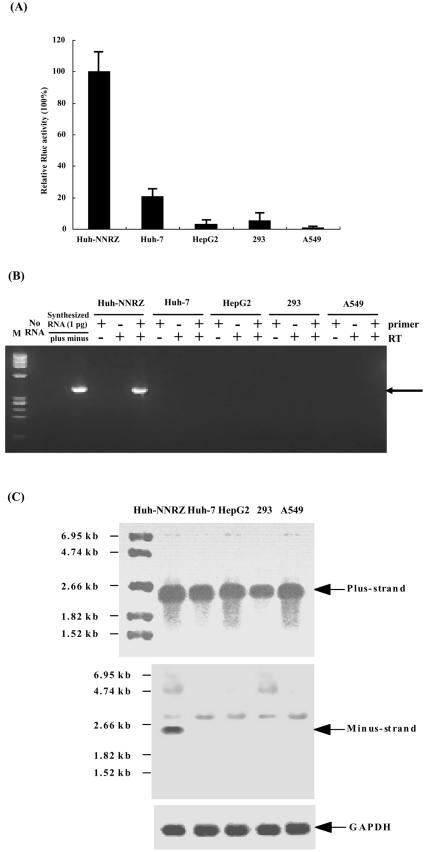
Our targeting strategy in this study is devised by inserting the antisense sequence of transgene and EMCV IRES between 5′- and 3′-end of HCV cDNA, and thus in principle, the foreign gene should be specifically expressed in HCV-infected cells containing functional replication apparatus. To verify this, the reporter vector pT7cRLNS5B1 and pAM8-1 were transfected into Huh-NNRZ, Huh-7, HepG2, 293 and A549 cells, and Renilla luciferase activities in the lysates were determined as described above. Figure 2A shows the only cell line with significant Renilla luciferase activity was Huh-NNRZ. Other cell lines, including both hepatic- and nonhepatic cells, expressed only low or undetectable levels of Renilla luciferase.
FIG. 2.
Specific gene expression in HCV replicon cell line. (A) The indicated cell lines were cotransfected with the pT7cRLNS5B1 and pAM8-1 vectors. Relative Renilla luciferase activities in the lysates were determined as described for Fig. 1. The columns and bars represent the means and standard deviations of two separate triplicate experiments. (B) Total RNAs were prepared from each transfectant. Strand-specific RT-PCR for minus-strand RNA was performed using 1 μg of extracted RNA. The arrow indicates the expected 408-bp PCR products. (C) Northern blot was performed on 5 μg of extracted RNA using digoxigenin-labeled sense and antisense Renilla luciferase RNA probes to detect plus- (upper panel) and minus-strand (middle panel) transcripts. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a loading control. RNA size markers are shown on the left, and the bands corresponding to plus- and minus-strand RNA are indicated on the right.
To further confirm that the transgene expression in pT7cRLNS5B1 is HCV-responsive, we performed strand-specific RT-PCR for minus-strand sequence using the RNA extracted from the transfectants described above. Three different controls (no RNA, no reverse transcriptase, and no primer in the reverse transcription) were included as specificity control and were negative. No minus-strand RNA was detected in the RT-PCR using 1 pg of in vitro transcribed plus-strand control RNA, confirming the strand specificity of the RT-PCR assay (Fig. 2B lane 3). Fully consistent with the results from reporter assay, minus-strand RNA was only detected in Huh-NNRZ cells cotransfected with pT7cRLNS5B1 and pAM8-1, while other transfected cells were negative. Simultaneously, the extracted RNA was subjected to Northern blot analysis using digoxigenin-labeled sense and antisense Renilla luciferase probes to detect plus- and minus-strand transcripts. Similar to the result from strand-specific RT-PCR, minus-strand RNA transcripts of the expected size (2.46 kb in length) were specifically detected in Huh-NNRZ cells transfected with pT7cRLNS5B1 and pAM8-1, although the plus-strand transcripts detected were comparable among different cell lines (Fig. 2C). Taken together, these experiments demonstrate that our approach can selectively direct gene expression in HCV replicon cells containing functional replication components.
Inhibition of transgene expression by short interfering RNA directed against HCV NS5B.
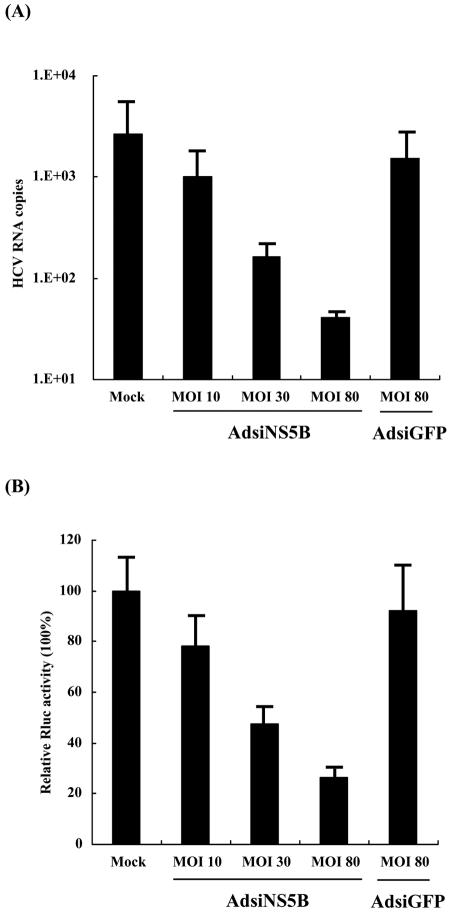
To determine if the transgene expression in pT7cRLNS5B1 was dependent on HCV replicase in the replicon cell, Huh-NNRZ cells were infected with AdsiNS5B at MOIs of 80, 30, and 10 and then cotransfected with pT7cRLNS5B1 and pAM8-1. The cell lysates were assayed for Renilla luciferase expression and total RNAs were subjected to real-time RT-PCR for quantification of HCV replicon RNA levels. As shown in Fig. 3, transduction with AdsiNS5B resulted in a substantial and dose-dependent reduction in the replicon RNA level, whereas the HCV RNA level in cells transduced with irrelevant AdsiGFP, expressing short interfering RNA against green fluorescent protein, even at an MOI of 80, was comparable to that in mock-infected cells. More importantly, Renilla luciferase expression in AdsiNS5B-transduced Huh-NNRZ cells was also dose-dependently inhibited, fully parallel to the HCV RNA levels quantified. Likewise, in an experiment investigating the effect of a nonnucleoside inhibitor of the HCV NS5B polymerase on transgene expression, it was found that Renilla luciferase activity and minus-strand RNA production in replicon cells were dose-dependently reduced after incubation with the NS5B inhibitor (data not shown). These observations provide further support that the transgene expression is strictly dependent on the presence of HCV replicase.
FIG. 3.
Inhibition of transgene expression by silencing of HCV NS5B. Huh-NNRZ cells were infected with AdsiNS5B at an MOI of 80, 30, and 10 and then transfected with pT7cRLNS5B1 and pAM8-1. The transduced cells were harvested at day 2 posttransfection. HCV replicon RNAs were quantified with real-time RT-PCR (A), and relative Renilla luciferase activities were determined as described for Fig. 1 (B). The columns and bars represent the means and standard deviations of two independent experiments.
HCV-dependent gene expression from polymerase II-derived transcripts.
Application of our targeting strategy to gene therapy would require expression from polymerase II RNA polymerase rather than T7-directed transcription. It has been reported for other positive-strand RNA viruses that full-length transcripts produced in the nucleus by polymerase II are delivered in a functional form into the cytoplasm (1, 20). Artificially synthesized RNA transcripts from the cDNA of positive-strand RNA viruses require that both 5′ and 3′ ends accurately reflect those found in the viral genome in order to maximize infectivity (14).
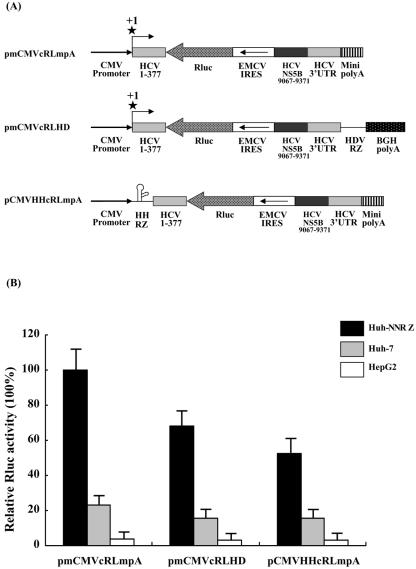
To ensure that the polymerase II-derived transcripts contain a minimal or no overhang at the 5′ end, we adapted two distinct approaches: one by positioning the viral cDNA immediately adjacent to the transcription start site of the cytomegalovirus (CMV) promoter (Fig. 4A, pmCMVcRLmpA and pmCMVcRLHD), and another by inserting a cis-acting hammerhead ribozyme (17) that cleaves to produce authentic 5′ end (pCMVHHcRLmpA). Similarly, to minimize the overhang at the 3′ end of polymerase II-derived transcripts, we used a synthetic, minimal poly(A) signal (37) (pmCMVcRLmpA and pCMVHHcRLmpA) or inserted the self-cleaving HDV ribozyme upstream of the unmodified bovine growth hormone poly(A) signal (pmCMVcRLHD).
FIG. 4.
Polymerase II-derived transcripts are functional templates for minus-strand RNA synthesis. (A) Schematic diagram of polymerase II-directed HCV minigenome reporter constructs. The HCV minigenome containing the antisense sequence of Renilla luciferase and the EMCV IRES flanked by the 5′ end (1 to 377) and 3′ partial NS5B coding region (9067 to 9371)-connected 3′ UTR was placed immediately next to the cytomegalovirus transcription start site or combined with a hammerhead ribozyme and was followed by a minimal poly(A) or HDV ribozyme upstream of the unmodified bovine growth hormone poly(A). (B) Huh-NNRZ, Huh-7, and HepG2 cells were transfected with the indicated reporter vectors. Relative Renilla luciferase activities in the lysates were determined as described for Fig. 1. The columns and bars represent the means and standard deviations of two separate triplicate transfections.
To ascertain whether the polymerase II-derived transcript could act as a functional template for minus-strand RNA synthesis, Huh-NNRZ, Huh-7 and HepG2 cells were transfected with these constructs. Compared with the transfected Huh-7 or HepG2 cells, Renilla luciferase activities in transfected Huh-NNRZ cells were consistently higher, the order being pmCMVcRLmpA > pmCMVcRLHD > pCMVHHcRLmpA (Fig. 4B). This indicates that the polymerase II-derived transcripts, especially that from pmCMVcRLmpA, were competent as templates for minus-strand RNA synthesis, suggesting that 5′ cap structure may not impair the template ability of the polymerase II-derived transcript to replicate. The lower Renilla luciferase activity in cells transfected with pmCMVcRLHD or pCMVHHRZcRLmpA compared with that in the pmCMVcRLmpA transfectant may be attributable to inefficient cleavage by the ribozyme.
Chemosensitizing effect of CD expressed from adenoviraus-delivered HCV-like minigenome.
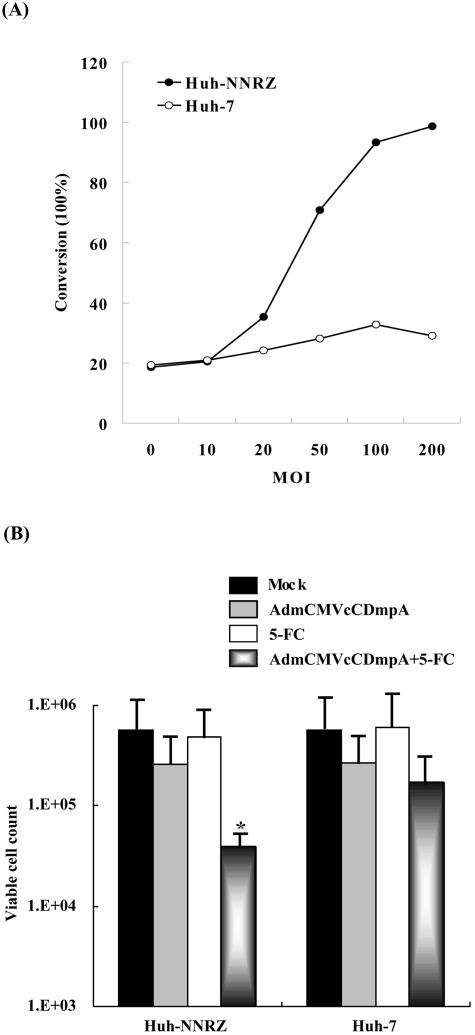
To further investigate the therapeutic feasibility of our targeting approach, we next subcloned the expression cassette from pmCMVcCDmpA, which contains the antisense sequence of the CD suicide gene instead of the Renilla luciferase in pmCMVcRLmpA, into recombinant adenovirus to generate AdmCMVcCDmpA. CD is a prokaryotic enzyme capable of converting the nontoxic prodrug 5-FC to the chemotherapeutic agent 5-fluorouracil (5-FU), which has been widely used in anticancer gene therapy (9, 16). Huh-NNRZ and Huh-7 cells were infected with AdmCMVcCDmpA at an MOI of 0, 10, 20, 50, 100, and 200. After 48 h, cells were harvested and CD enzymatic activity was determined by measuring the conversion of 5-FC to 5-FU. There was a dose-dependent increase in CD activity in Huh-NNRZ cells infected with AdmCMVcCDmpA, while infected Huh-7 cells showed only basal levels of CD activity, demonstrating an HCV-specific CD expression from AdmCMVcCDmpA (Fig. 5A).
FIG. 5.
Adenovirus-delivered HCV-responsive CD expression. (A) Huh-NNRZ and Huh-7 cells were infected with AdmCMVcCDmpA (MOI, 0 to 200). After 48 h, cells were harvested and CD enzymatic activity was determined by measuring the conversion of 5-FC to 5-FU. (B) Huh-NNRZ and Huh-7 cells were mock infected or infected with AdmCMVcCDmpA at an MOI of 80 and maintained in the absence or presence of 5-FC. Viable cells were quantified after 4 days. Representative data are from three separate experiments. *, P < 0.05 compared with the control.
To further verify the data from the enzymatic assay, we next investigated whether Huh-NNRZ cells are sensitized to the cytotoxic effect of 5-FC by transduction of AdmCMVcCDmpA. Huh-NNRZ and Huh-7 cells were infected with AdmCMVcCDmpA at an MOI of 80, followed by the addition of 5-FC (0.5 mM). Controls included AdmCMVcCDmpA alone, 5-FC alone, and no treatment. Four days later, the cytotoxic effects were evaluated with cell proliferation reagent WST-1.
Following a 4-day exposure to 5-FC, the viable cell count of Huh-NNRZ cells transduced with AdmCMVcCDmpA was reduced by 14-fold (compared to the control, P < 0.05), whereas the viability of Huh-NNRZ cells infected with AdmCMVcCDmpA in the absence of 5-FC or treated with 5-FC alone was comparable to that of the control with no treatment (Fig. 5B). In contrast, transduction with AdmCMVcCDmpA did not significantly confer sensitivity to 5-FC on parental Huh-7 cells, indicating that the deaminating reaction converting 5-FC to 5-FU did not occur in uninfected Huh-7 cells due to lack of CD expression. These results further demonstrate that our strategy, by exploiting cis-acting replication elements, renders transgene (CD) expression responsive to the presence of HCV, thereby approaching the goal of selectively killing HCV-infected cells in combination with 5-FC while keeping uninfected cells unharmed.
DISCUSSION
We demonstrate here a proof-of-concept for a new targeting strategy to direct gene expression responsive to HCV by using a construct containing the antisense sequence of transgene and EMCV IRES flanked by the 5′- and 3′-end regions of viral cDNA. Thus, expression of the transgene is initiated only when the minus-strand RNA has been synthesized by the functional replication components present in HCV-infected cells. By using this strategy in combination with recombinant adenovirus delivery, our data showed that expression of the therapeutic gene (CD) depends strictly on the presence of functional HCV replication machinery in replicon cell, resulting in marked chemosensitization of replicon cells to the cytotoxic effects of 5-FC, while having little effect on noninfected cells. In addition to the CD suicide gene, other therapeutic genes such as apoptosis-inducing genes may also be employed in this strategy for eradication of HCV-infected cells. These experiments also show that viral replication components, including RdRp, can act in trans to synthesize minus-strand RNA using the transcript from the engineered construct as a template.
Using HCV minigenome reporter vectors with differently truncated NS5B coding sequences fused upstream of the 3′ UTR, we found that the 3′ part of NS5B coding region from nucleotides 9067 to 9371 is required for HCV minus-strand RNA synthesis, although it is not excluded that this cis-acting sequence could be further reduced by deletion mutagenesis. This result is in agreement with that reported by You et al. (39), who identified a stem-loop (5BSL3.2) at nucleotides 9262 to 9311 and found that both the structure and primary sequence of 5BSL3.2 are essential for HCV RNA replication. Although expression of the transgene from our engineered construct was dependent on the presence of functional HCV replication machinery in replicon cell, an over background level of Renilla luciferase activity was consistently observed in naïve Huh-7 cells, which was higher than in other hepatic and nonhepatic control cells (Fig. 2A). The reason for this “leaky” expression in naïve Huh-7 cells is currently unknown; one possible interpretation is antisense transcription mediated by the T7 polymerase. Further clarification of this HCV-independent expression in naïve Huh-7 cells is under way to determine whether it truly limits the possibility of using cytotoxic gene to selectively kill HCV-infected cells. If this is the case, other “mild” therapeutic genes such as interferons may be alternatively employed in this targeting strategy; directing HCV-responsive interferon expression should be advantageous to avoid toxic side effects.
While a low but detectable Renilla luciferase activity was observed, Renilla luciferase RNA was not detected in Huh-7 cells by Northern blot analysis (Fig. 2C). This discordance may be attributable to a lower sensitivity of the Northern blot compared to the luciferase assay. Similarly, because using RT-PCR for detection of minus-strand RNA is likely to create false positive due to self-priming or random priming of the wrong strand during the RT step, a compromise between sensitivity and strand specificity is required despite efforts to reduce such nonspecific cDNA synthesis. For this reason, to ensure high strand specificity, the sensitivity of the strand-specific RT-PCR procedure used here was limited, which may account for the failure to detect low-level Renilla luciferase coding transcripts in Huh-7 cells. Further experimentation is in progress to elucidate whether the discrepancy in the results of the reporter assay and Northern blot or strand-specific RT-PCR is simply due to the variance in the sensitivity of different assays.
Consistent with those reported previously (27), our study demonstrated that polymerase II-derived transcripts are competent as templates for minus-strand RNA synthesis by the HCV replication complex. Compared with the T7 expression system, polymerase II expressed lower levels of the Renilla luciferase reporter gene, probably due to the lower levels of polymerase II-derived transcript present in transfected cells. Additionally, while rapid amplification of cDNA end analysis of the transcript from pmCMVcRLmpA, which was constructed by trial and error to position the 5′ UTR 4 nucleotides downstream from the putative start site, confirmed that the transcription start site coincided with the first nucleotide of the 5′ UTR, it was shown that this RNA transcript terminated 14 nucleotides downstream of the AAUAA poly(A) signal. Thus, the transcript from pmCMVcRLmpA still contained an overhang of 32 nucleotides at the 3′ end, although it was much shortened by using a minimal poly(A) signal. Further efforts to minimize the overhang at the 3′ end of the polymerase II-derived transcript may be necessary to improve its template activity for minus-strand synthesis.
In an experiment to investigate the duration of reporter gene (Renilla luciferase) expression using pT7cRLNS5B1 (data not shown), we found that Renilla luciferase activity decreased considerably at day 4 and was insignificant at day 6 posttransfection, suggesting that the newly synthesized minus-strand RNA may be incompetent as a template for subsequent plus-strand production. The reason for this is currently unknown, but one possibility is that plus-strand RNA synthesis may require another cis-acting element(s) in addition to those included in our construct. Studies are under way to define this, as well as to identify the minimal trans-acting viral components essential for HCV replication, which will shed light on the development of HCV element-based gene delivery system.
Synthetic minigenomes have been described in a number of minus-stranded RNA viruses of several different families (28, 32) and plus-stranded RNA viruses belonging to the Coronaviridae family (19). Coupled with infectious helper viruses or plasmid-encoded proteins, minigenomes have contributed greatly to the analysis of cis-acting sequences and trans-acting proteins required for viral replication, maturation, and packaging. In addition, coronavirus-derived minigenomes have been used to express transgene in tissue culture and in animals (2). Different from the coronavirus minigenomes with the transgene in the sense orientation, which directs tissue-specific transgene expression by virtue of viral tropism, the HCV minigenome described here contains the transgene and IRES element in the antisense orientation, and the minus-strand RNA synthesized by the replication machinery in HCV-infected cells functions as the mRNA for transgene expression, rendering transgene expression responsive to the presence of HCV. However, when the antisense sequence of the reporter gene was inserted in the minigenome derived from transmissible gastroenteritis virus, the transgene was not expressed (19). Whether the discrepancy between the finding in transmissible gastroenteritis virus and that presented here is simply due to the lack of the IRES element for translation initiation in the former minigenome or due to the intrinsic difference between transmissible gastroenteritis virus and HCV remains to be investigated.
Because the HCV genome RNA and the therapeutic minigenome share the same replication machinery in infected cells, it is possible that replication of the HCV genome may be competitively inhibited when the replication machinery is being actively recruited by the therapeutic HCV minigenome. If this is the case, an additive antiviral effect may be expected. Additionally, given that the HCV-responsive expression construct described here contains the 5′ (nucleotides 1 to 377) region of the HCV genome, which was reported to interact with HCV nucleocapsid (core) protein and play an important role in viral encapsidation (11, 33, 35, 41), it is tempting to speculate that the HCV minigenome may also be packaged inside the virion and budded from infected cells and, if so, it could direct secondary transgene expression by sequential infection, ultimately augmenting its anti-HCV effect.
In summary, we have developed a novel targeting therapeutic approach directed against cells containing the HCV replication apparatus, which may be useful as a prophylactic in the early stage of hepatitis, during limited infection of the liver, or for ex vivo therapy of hepatocytes. It may also reduce virus loads in chronically infected patients and, in combination with interferon and ribavirin therapy, might eradicate HCV from the infected host. In addition to its therapeutic application, the HCV minigenome reported in this study also offers a useful tool for studies aimed at investigation of the molecular mechanisms involved in replication and expression of the HCV genome.
REFERENCES
- 1.Almazan, F., J. M. Gonzale, Z. Penzes, A. Izeta, E. Calvo, J. Plana-Duran, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97:5516-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso, S., I. Sola, J. P. Teifke, I. Reimann, A. Izeta, M. Balasch, J. Plana-Durán, R. J. M. Moormann, and L. Enjuanes. 2002. In vitro and in vivo expression of foreign genes by transmissible gastroenteritis coronavirus-derived minigenomes. J. Gen. Virol. 83:567-579. [DOI] [PubMed] [Google Scholar]
- 3.Alter, M. J., H. S. Margolis, K. Krawczynski, F. N. Judson, A. Mares, W. J. Alexander, P. Y. Hu, J. K. Miller, M. A. Gerber, R. E. Sampliner, E. L. Meeks, and M. J. Beach. 1992. The natural history of community-acquired hepatitis C virus in the United States. N. Engl. J. Med. 327:1899-1905. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager, R. 1997. Candidate targets for hepatitis C vir us-specific antiviral therapy. Intervirology. 40:378-393. [DOI] [PubMed] [Google Scholar]
- 5.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 6.Blight, K. J., A. A. Kolykhalov, K. E. Reed, E. V. Agapov, and C. M. Rice. 1998. Molecular virology of hepatitis C virus: an update with respect to potential antiviral targets. Antivir. Ther. 3:71-81. [PubMed] [Google Scholar]
- 7.Cheng, J.-C., M.-F. Chang, and S. C. Chang. 1999. Specific interaction between the hepatitis C virus NS5B RNA polymerase and the 3′ end of the viral RNA. J. Virol. 73:7044-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choo, Q.-L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, A. Medina-Selby, P. J. Barr, A. J. Weiner, D. W. Bradley, G. Kuo, and M. Houghton. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danielsen, S., M. Kilstrup, K. Barilla, B. Jochimsen, and J. Neuhard. 1992. Characterization of the Escherichia coli codBA operon encoding cytosine permease and cytosine deaminase. Mol. Microbiol. 6:1335-1344. [DOI] [PubMed] [Google Scholar]
- 10.Dhanak, D., K. J. Duffy, V. K. Johnston, J. Lin-Goerke, M. Darcy, A. N. Shaw, B. Gu, C. Silverman, A. T. Gates, M. R. Nonnemacher, D. L. Earnshaw, D. J. Casper, A. Kaura, A. Baker, C. Greenwood, L. L. Gutshall, D. Maley, A. DelVecchio, R. Macarron, G. A. Hofmann, Z. Alnoah, H. Y. Cheng, G. Chan, S. Khandekar, R. W. Keenan, and R. T. Sarisky. 2002. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:38322-38327. [DOI] [PubMed] [Google Scholar]
- 11.Fan, Z., Q. R. Yang, J. S. Twu, and A. H. Sherker. 1999. Specific in vitro association between the hepatitis C viral genome and core protein. J. Med. Virol. 59:131-134. [PubMed] [Google Scholar]
- 12.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ untranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hijikata, M., N. Kato, Y. Ootsuyama, M. Nakagawa, and K. Shimotohno. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. USA 88:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirschowitz, E. A., A. Ohwada, W. R. Pascal, T. J. Russi, and R. G. Crystal. 1995. In vivo adenovirus-mediated gene transfer of the Escherichia coli cytosine deaminase gene to human colon carcinoma-derived tumors induce chemosensitivity to 5-fluorocytosine. Hum. Gene. Ther. 6:1055-1063. [DOI] [PubMed] [Google Scholar]
- 17.Huang, Y., and G. C. Carmichael. 1996. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16:1534-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ideo, G., and A. Bellobuono. 2002. New therapies for the treatment of chronic hepatitis C. Curr. Pharm. Des. 8:959-966. [DOI] [PubMed] [Google Scholar]
- 19.Izeta, A., C. Smerdou, S. Alonso, Z. Penzes, A. Mendez, J. Plana-Duran, and L. Enjuanes. 1999. Replication and packageing of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J. Virol. 73:1535-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khromykh, A. A., A. N. Varnavski, P. L. Sedlak, and E. G. Westaway. 2001. Coupling between replication and packaging of flavivirus RNA: Evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J. Virol. 75:4633-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieft, J. S., K. hou, A. Grech, R. Jubin, and J. A. Doudna. 2002. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 9:370-374. [DOI] [PubMed] [Google Scholar]
- 22.Kishine, H., K. Sugiyama, M. Hijikata, N. Kato, H. Takahashi, T. Noshi, Y. Nio, M. Hosaka, Y. Miyanari, and K. Shimotohno. 2002. Subgenomic replicon derived from a cell line infected with the hepatitis C virus. Biochem. Biophys. Res. Commun. 293:993-999. [DOI] [PubMed] [Google Scholar]
- 23.Kuriyama, S., K. Masui, T. Sakamoto, T. Nakatani, M. Kikukawa, H. Tsujinoue, A. Mitoro, M. Yamazaki, H. Yoshiji, H. Fukui, K. Ikenaka, C. A. Mullen, and T. Tsujii. 1998. Bystander effect caused by cytosine deaminase gene and 5-fluorocytosine in vitro is substantially mediated by generated 5-fluorouracil. Anticancer Res. 18:3399-3406. [PubMed] [Google Scholar]
- 24.Lindenbach, B. D., and C. M. Rice. 2003. Evasive maneuvers by hepatitis C virus. Hepatology 38:769-771. [DOI] [PubMed] [Google Scholar]
- 25.Lohmann, V., F. Korner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 26.Lu, L., T. J. Pilot-Matias, K. D. Stewart, J. T. Randolph, R. Pithawalla, W. He, P. P. Huang, L. L. Klein, H. Mo, and A. Molla. 2004. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 48:2260-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCormick, C. J., L. Challinor, A. Macdonald, D. J. Rowlands, and M. Harris. 2004. Introduction of replication-competent hepatitis C virus transcripts using a tetracycline-regulable baculovirus delivery system. J. Gen. Virol. 85:429-439. [DOI] [PubMed] [Google Scholar]
- 28.Mühlberger, E., M. Weik, V. Volchkov, H. -D. Klenk, and S. Becker. 1999. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J. Virol. 73:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen, T. T., A. T. Gates, L. L. Gutshall, V. K. Johnston, B. Gu, K. J. Duffy, and R. T. Sarisky. 2003. Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob. Agents Chemother. 47:3525-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh, J. W., T. Ito, and M. M. C. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of coping the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh, J. W., G. T. Sheu, and M. M. C. Lai. 2000. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Bio. Chem. 275:17710-17717. [DOI] [PubMed] [Google Scholar]
- 32.Perez, M., A. Sanchez, B. Cubitt, D. Rosario, and J. C. de la Torre. 2003. A reverse genetics system for Borna disease virus. J. Gen. Virol. 84:3099-3104. [DOI] [PubMed] [Google Scholar]
- 33.Ray, R. B., and R. Ray. 2001. Hepatitis C virus core protein: intriguing properties and functional relevance. FEMS Microbiol. Lett. 202:149-156. [DOI] [PubMed] [Google Scholar]
- 34.Saito, I., A. Miyamura, H. Ohbayashi, H. Harada, T. Katayama, S. Kikuchi, Y. Watanabe, S. Koi, M. Onji, Y. Ohta, Q.-L. Choo, M. Houghton, and G. Kuo. 1990. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 87:6540-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimoike, T., S. Mimori, H. Tani, Y. Matsuura, and T. Miyamura. 1999. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 73:9718-9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suh, Y. A., P. K. Kumar, K. Taira, and S. Nishikawa. 1993. Self-cleavage activity of the genomic HDV ribozyme in the presence of various diavalent metal ions. Nucleic Acids Res. 21:3277-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia, H., Q. Mao, H. L. Paulson, and B. L. Davidson. 2002. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 20:1006-1010. [DOI] [PubMed] [Google Scholar]
- 38.Yi, M., and S. M. Lemon. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.You, S., D. D. Stump, A. D. Branch, and C. M. Rice. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 78:1352-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, J., O. Yamada, T. Ito, M. Akiyama, Y. Hashimoto, H. Yoshida, R. Makino, A. Masago, H. Uemura, and H. Araki. 1999. A single nucleotide insertion in the 5′-untranslated region of hepatitis C virus leads to enhanced cap-independent translation. Virology 261:263-270. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, J., O. Yamada, H. Yoshida, T. Iwai, and H. Araki. 2002. Autogenous translational inhibition of core protein: implication for switch from translation to RNA replication in hepatitis C virus. Virology 293:141-150. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, J., O. Yamada, K. Sakamoto, H. Yoshida, T. Iwai, Y. Matsushita, H. Shimamura, H. Araki, and K. Shimotohno. 2004. Down-regulation of viral replication by adenoviral-mediated expression of short interfering RNA against cellular cofactors for hepatitis C virus. Virology 320:135-143. [DOI] [PubMed] [Google Scholar]