Abstract
Endogenous retroviruses (ERVs) are widespread in vertebrate genomes and have been loosely grouped into “classes” on the basis of their phylogenetic relatedness to the established genera of exogenous retroviruses. Four of these genera—the lentiviruses, alpharetroviruses, betaretroviruses, and deltaretroviruses—form a well-supported clade in retroviral phylogenies, and ERVs that group with these genera have been termed class II ERVs. We used PCR amplification and sequencing of retroviral fragments from more than 130 vertebrate taxa to investigate the evolution of the class II retroviruses in detail. We confirm that class II retroviruses are largely confined to mammalian and avian hosts and provide evidence for a major novel group of avian retroviruses, and we identify additional members of both the alpha- and the betaretrovirus genera. Phylogenetic analyses demonstrated that the avian and mammalian viruses form distinct monophyletic groups, implying that interclass transmission has occurred only rarely during the evolution of the class II retroviruses. In contrast to previous reports, the lentiviruses clustered as sister taxa to several endogenous retroviruses derived from rodents and insectivores. This topology was further supported by the shared loss of both the class II PR-Pol frameshift site and the class II retrovirus G-patch domain.
Retroviruses (family Retroviridae) are characterized by a unique replication strategy. The RNA genome of an extracellular retrovirus is first copied into DNA by virus-encoded reverse transcriptase (RT) and is then integrated into the nuclear DNA of the host cell (35). Integration is highly stable and, consequently, infection of germ line cells can lead to vertical transmission of retroviruses from parent to offspring as Mendelian alleles (8). These retroviruses are termed endogenous (to distinguish them from their horizontally transmitted, exogenous counterparts), and they have been identified in almost all vertebrate orders examined (8, 16). Some endogenous retroviruses (ERVs) represent endogenized copies of extant exogenous retroviruses, but the majority are very old and appear to lack closely related exogenous counterparts (8, 16). Analysis of these ERVs in the genomes of humans, mice, and other species indicates a longstanding association between retroviruses and vertebrates, probably dating back several hundred million years, during which retroviruses have repeatedly colonized host genomes (12, 19, 20, 23).
Most ERVs show clear homology to one another and to modern exogenous retroviruses, especially across the RT gene, which is relatively refractory to nonsynonymous substitution. Diverse retrovirus sequences can therefore be aligned in order to investigate phylogenetic relationships, and this has been instrumental in the classification of exogenous retroviruses into seven genera (alpha-, beta-, gamma-, delta-, and epsilonretroviruses; lentivirus; and spumavirus) (12, 26, 34, 37). Although many ERVs have not been assigned to particular genera, there is a growing tendency to group them into classes according to their similarity to exogenous retroviruses (19, 20, 36). Using this system of classification, ERVs clustering with gamma- and epsilonretroviruses are termed class I, those that cluster with lentiviruses, alpha-, beta-, and deltaretroviruses are termed class II, and those that cluster with spumaviruses are termed class III (6, 36). It should be noted that, despite this classification system, most ERVs are only distantly related to known exogenous retroviruses. In particular, the lentiviruses and deltaretroviruses have no closely related endogenous counterparts (16). The distribution and diversity of class I and class III ERVs have both been investigated previously in some detail (6, 16, 23), but this is not the case with the class II ERVs. Only a small number of alpha- and betaretrovirus-related (but no lentivirus or deltaretrovirus-related) elements have been characterized (9, 10, 16). Despite this, sequence analysis has revealed that class II ERVs cluster into a robustly supported clade in retrovirus phylogenies (16). Furthermore, it appears that, in contrast to class I and class III retroviruses, the class II ERVs have a relatively restricted host range, being confined largely to mammals and birds (16). The only exceptions to this are two ERVs, termed python endogenous retroviruses, that were recently identified in boid snakes (18).
Class II ERVs differ from other retroviruses in several features of their genomic organization. All described class II retroviruses produce a Gag-Pol polyprotein via one or two ribosomal frameshifting sites rather than the termination codon suppression mechanism found commonly in other retroviruses (28). One frameshift site is (with the exception of the lentiviruses) located at the protease (PR)-Pol(RT) boundary, whereas the other (encoded by the lentiviruses and the beta- and deltaretroviruses) is situated between Gag and PR (28). Another unusual feature of class II retroviruses is the presence of a short, glycine-rich region related to the G-patch domain found in many RNA binding proteins (2). Within the Mason-Pfizer monkey virus, this region is synthesized with and then cleaved from the PR protein, although its precise function remains to be determined (2, 17).
Here we report the results of widespread sampling within vertebrates for class II ERVs related to the lentiviruses, alpha-, beta-, and deltaretrovirus genera. We show that many features of their genomic organization and host association are remarkably stable, having defined monophyletic origins on phylogenetic trees. We also demonstrate that the exogenous lentiviruses are probably most closely related to several endogenous retroviruses derived from rodents and insectivores.
MATERIALS AND METHODS
Amplification and sequencing.
Genomic DNA was extracted from tissue samples by using a DNeasy tissue extraction kit (Qiagen). PCR amplification of retroviral sequences was performed with primers targeting two highly conserved domains within the retrovirus PR and RT genes. One universal PR primer (5′-GTG/T TTI G/TTI GAC/T ACI GGI G/TC-3′, where I is inosine) was used in conjunction with one of three RT primers designed to amplify retroviruses related to the alpha-, beta-, delta-, and lentivirus genera (5′-GTK TTI KTI GAY ACI GGI KC-3′, 5′-ATI AGI AKR TCR TCC ATR TA-3′, and 5′-AGI AKR TCR TCC ATR TA-3′). Reaction conditions were as described previously (32). Amplification products (800 to 1,000 bp) for each PCR were excised, purified, and cloned before being sequenced in both directions by using an ABI Prism 3700 DNA analyzer. At least five clones were sequenced for each host taxon investigated. The origin of retroviral fragments was confirmed by PCR with separate aliquots of genomic DNA samples and nested primers specific to each retrovirus fragment.
Sequence analysis and alignment.
Novel class II ERV sequences were translated and aligned to previously characterized viruses. Cross-amplified class I and class III viruses were discarded from the data set by constructing neighbor-joining phylogenies and by excluding sequences that clustered with gamma-, epsilon-, or spumaretroviruses. An amino acid alignment was constructed by using (i) the known amino acid sequences of previously described retroviruses and (ii) virtual translations of novel retroviruses that lacked premature frameshifts in the amplified region. This amino acid alignment was used as a template to identify the likeliest locations of indels responsible for frameshifts when the remaining sequences were aligned. Manual adjustments were made on this basis if they were supported by the results of alignment algorithms using the raw nucleotides. Regions lacking clear homology between sequences or where homology could not be unambiguously identified were excluded from the alignment. The final DNA alignment contained 121 taxa spanning 792 bp. The equivalent amino acid alignment spanned 264 residues.
Phylogenetic analysis.
Phylogenetic analyses were performed by using both Bayesian MCMC (Markov Chain Monte Carlo) inference, as implemented in MRBAYES 3.0B4, and maximum-parsimony (MP) and neighbor-joining (NJ) approaches in PAUP 4 (29, 31). For MRBAYES analyses, the nucleic acid alignment and a general time-reversible model with codon position-specific rates was used. Four chains were run well past their asymptotes before 10,000 trees were collected, at one tree per 100 generations. These trees were used to calculate a majority rule phylogram. Nucleic acid-based MP reconstruction was performed by using 30,000 random addition replicates of an unweighted datamatrix, with third-codon positions excluded, holding a single tree in memory during each replicate. The resultant minimum trees were then used as the starting point for a heuristic search, during which a total of seven optimal trees were recovered. MP and NJ analyses were also performed by using an amino acid alignment (to further investigate the topology and sister relationships of the lentiviruses, as discussed below). Amino acid-based MP analysis again used 30,000 random addition replicates, holding one tree in memory during each replicate. Amino acid-based NJ analysis used PAUP defaults, and the tree was bootstrapped by using 1,000 replicates.
Nucleotide sequences and accession numbers.
The novel retrovirus sequences described here have been submitted to the EMBL/GenBank databases and will appear under accession numbers AY820046 to AY820125.
RESULTS
Screening for class II ERV sequences.
PCR screening of 135 vertebrate taxa (49 mammalian taxa; 46 avian taxa; and 40 reptilian, amphibian, or piscine taxa) led to the identification of 84 novel class II ERV fragments (Table 1). The majority (55) of the class II ERVs were derived from avian taxa and were amplified from representatives of every avian order investigated (Table 2). The remaining 29 retrovirus fragments were all mammalian in origin (Table 2). Three additional mammalian sequences (RV-Brown rat I and II and RV-House mouse) were identified via BLAST searches of sequence databanks (1). Class II ERVs were isolated from all mammalian orders investigated, with the exception of Pinnipedia (seals), and Scandentia (tree shrews), for which only one or two genomic DNA samples were screened. We were unable to identify any class II ERVs within reptilian, amphibian, or piscine taxa, although two viruses, derived from boid snakes, have been described (18). A list of species from which we were unable to recover class II ERV sequences is shown in Table S1 in the supplemental material.
TABLE 1.
Screening of vertebrate taxa for class II-related retroviruses
| Host class | No. of:
|
||
|---|---|---|---|
| Taxa screened | Taxa harboring class II viruses | Class II viruses identified | |
| Mammals | 49 | 28 | 32 |
| Birds | 46 | 37 | 55 |
| Reptiles | 14 | 0 | 0 |
| Amphibians | 16 | 0 | 0 |
| Fish | 10 | 0 | 0 |
TABLE 2.
Novel class II ERVs identified in this study
| Host class, order, and family | Host species | Retroviral sequencea |
|---|---|---|
| Aves | ||
| Anseriformes | ||
| Anatidae (swans, geese, and ducks) | White-fronted goose (Anser albifrons) | RV-White-fronted goose (1) |
| North American black duck (Anas rubripes) | RV-Black duck I (0) | |
| RV-Black duck II (0) | ||
| RV-Black duck III (4) | ||
| RV-Black duck IV (0) | ||
| Apterygiformes, Apterygidae (kiwis) | Brown kiwi (Apteryx australis) | RV-Brown kiwi (0) |
| Great spotted kiwi (Apteryx haastii) | RV-Great spotted kiwi (0) | |
| Little spotted kiwi (Apteryx owenii) | RV-Little spotted kiwi I (1) | |
| RV-Little spotted kiwi II (1) | ||
| Casuariformes | ||
| Casuariidae (cassowaries) | Cassowary (Casuarius casuarius) | RV-Cassowary (0) |
| Dromaiidae (emu) | Emu (Dromaius novaehollandiae) | RV-Emu (1) |
| Phoenicopteridae (flamingos) | Chilean flamingo (Phoenicopterus ruber chilensis) | RV-Flamingo (2) |
| Columbiformes, Columbidae (pigeons) | Wood pigeon (Columba palumbus) | RV-Wood pigeon II (0) |
| Falconiformes | ||
| Accipritidae (hawks, eagles, and | Goshawk (Accipiter gentilis) | RV-Goshawk (0) |
| Old World vultures) | Marsh harrier (Circus aeruginosus) | RV-Marsh harrier I (0) |
| Ferruginous hawk (Buteo regalis) | RV-Marsh harrier II (0) | |
| RV-Ferruginous hawk (1) | ||
| Cathartidae (New World vultures) | Turkey vulture (Cathartes aura) | RV-Turkey vulture (0) |
| Falconidae (typical falcons) | Peregrine falcon (Falco peregrinus) | RV-Peregrine falcon (0) |
| Galliformes | ||
| Numididae (guineafowl and | Vulturine guineafowl (Acryllium vulturinum) | RV-Guineafowl I (2) |
| World vultures) | Marsh harrier (Circus aeruginosus) | RV-Guineafowl II (1) |
| Golden pheasant (Chrysolophus pictus) | RV-Golden pheasant (3) | |
| Japanese quail (Coturnix japonica) | RV-Japanese quail (6) | |
| Ring-necked pheasant (Phasianus colchicus) | RV-Ring-necked pheasant II (2) | |
| Blue peacock (Pavo cristatus) | RV-Blue peacock I (4) | |
| RV-Blue peacock II (2) | ||
| Gray partridge (Perdix perdix) | RV-Grey partridge III (6) | |
| Cabot's Tragopan (Tragopan caboti) | RV-Tragopan (0) | |
| Tetraonidae (grouse) | Black grouse (Lyrurus tetrix) | RV-Black grouse (0) |
| Gaviiformes, Gaviidae (loons) | Common loon (Gavia immer) | RV-Common loon I (2) |
| RV-Common loon II (1) | ||
| Gruiformes, Rallidae (rails) | Gray moorhen (Gallinula chloropus) | RV-Gray moorhen I (0) |
| RV-Gray moorhen II (1) | ||
| Passeriformes | ||
| Muscicapidea (thrushes) | Hermit thrush (Catharus guttatus) | RV-Hermit thrush I (2) |
| RV-Hermit thrush II (2) | ||
| RV-Hermit thrush III (0) | ||
| RV-Hermit thrush IV (0) | ||
| Mistle thush (Turdus viscivorus) | RV-Mistle thrush (6) | |
| Paridae (true tits) | Blue tit (Parus caeruleus) | RV-Blue tit I (3) |
| RV-Blue tit II (0) | ||
| RV-Blue tit III (1) | ||
| Corvidae (crows) | Common magpie (Pica pica) | RV-Common magpie II (1) |
| RV-Common magpie III (1) | ||
| RV-Common magpie IV (4) | ||
| Azure-winged magpie (Cyanopica cyana) | RV-Azure-winged magpie (2) | |
| Piciformes | ||
| Picidae (woodpeckers) | Green woodpecker (Picus viridis) | RV-Green woodpecker (2) |
| Rhamphastidae (toucans) | Golden-collared toucanet (Selenidera reinwardtii) | RV-Toucanet I (0) |
| RV-Toucanet II (3) | ||
| Rheiformes, Rheidae (rheas) | Greater rhea (Rhea americana) | RV-Greater rhea (0) |
| Darwin's rhea (Pterocnemia pennata) | RV-Darwin's rhea (0) | |
| Sphenisciformes, Spheniscidae (penguins) | King penguin (Aptenodytes patagonicus) | RV-King penguin (0) |
| Strigiformes, Strigidae (typical owls) | Eastern screech owl (Otus asio) | RV-Eastern screech owl I (4) |
| RV-Eastern screech owl II (5) | ||
| Struthioniformes, Struthionidae (ostriches) | North African ostrich (Struthio camelus) | RV-Ostrich (1) |
| Tinamiformes, Tinamidae (tinamous) | Elegant-crested tinamou (Eudromia elegans) | RV-Elegant-crested tinamou (1) |
| Mammalia | ||
| Artiodactyla | ||
| Bovidae (antelope, cattle, sheep, goats, | American bison (Bison bison) | RV-Bison (1) |
| and relatives) | Musk ox (Ovibos moschatus) | RV-Musk ox (0) |
| Domestic goat (Capra hircus) | RV-Goat (0) | |
| Cervidae (deer) | Caribou (Rangifer tarandus) | RV-Caribou (4) |
| White-tailed deer (Odocoileus virginianus) | RV-White-tailed deer (3) | |
| Giraffidae (giraffe and okapi) | Giraffe (Giraffa camelopardalis) | RV-Giraffe (3) |
| Carnivora | ||
| Felidae (cats) | Cougar (Felis concolor) | RV-Cougar (6) |
| Domestic cat (Felis catus) | RV-Domestic cat (5) | |
| Mustelidae (weasels and relatives) | Small mongoose (Herpestes javanicus) | RV-Small mongoose I (1) |
| RV-Small mongoose II (2) | ||
| RV-Small mongoose III (2) | ||
| Chinese ferret badger (Melogale moschata) | RV-Chinese badger (2) | |
| Cetacea, Delphinidae (dolphins) | Risso's dolphin (Grampus griseus) | RV-Risso's dolphin (1) |
| Insectivora, Erinacidae (hedgehogs) | European hedgehog (Erinaceus europaeus) | RV-European hedgehog (12) |
| Lagomorpha, Leporidae (rabbits and hares) | European rabbit (Oryctolagus cuniculus) | RV-European rabbit (0) |
| Marsupialia, Macropodidae (kangaroos) | Red kangaroo (Macropus rufus) | RV-Red kangaroo (1) |
| Monotremata | ||
| Ornithoryhnchidae (platypus) | Duck-billed platypus (Ornithorhynchus anatinus) | RV-Duck-billed platypus (1) |
| Tachyglossidae (echidna) | Short-beaked echidna (Tachyglossus aculeatus) | RV-Echidna II (2) |
| Primates | ||
| Lorisidae (bush babies) | Slow loris (Nycticebus coucang) | RV-Slow loris (0) |
| Cercopithidae (Old world monkeys) | Black colobus (Colobus angolensis) | RV-Colobus (0) |
| Rodentia | ||
| Muridae (rats and mice) | African grass rat (Arvicanthis ansorgei) | RV-Grass rat I (0) |
| RV-Grass rat II (4) | ||
| House mouse (Mus musculus) | RV-House mouse (ND) | |
| Shrew mouse (Mus pahari) | RV-Shrew mouse (7) | |
| Rice rat (Oryzomys intermedius) | RV-Rice rat (1) | |
| Multimammate rat (Mastomys huberti) | RV-Multimammate rat (2) | |
| Bismark giant rat (Uromys neobritannicus) | RV-Giant rat (1) | |
| Yemeni mouse Myomys yemeni) | RV-Yemeni mouse (5) | |
| Brown rat (Rattus norvegicus) | RV-Brown rat I (ND) | |
| RV-Brown rat II (ND) | ||
| Sciuridae (squirrels and relatives) | Prairie dog (Cynomys ludovicianus) | RV-Prairie dog (0) |
The sum of nonsense mutations (stop codons or frameshifts) within each viral fragment is given in parentheses. ND, not done.
Alignment and phylogeny of the class II retroviruses.
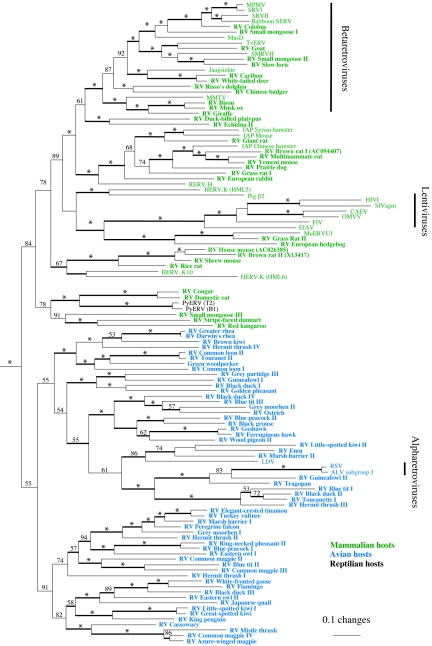
A 792-bp alignment was constructed that included regions of both PR and RT. The region linking these two proteins (between 9 and 150 bp in length, depending on the virus) was highly divergent and therefore excluded. This alignment was subjected to phylogenetic analyses using Bayesian likelihood and MP approaches, as shown in Fig. 1. Although we obtained relatively high posterior probability values using Bayesian likelihood, the backbone of the phylogeny was generally not well supported by bootstrap analysis in MP, with the result that the deeper relationships were only poorly resolved by using this approach. Such lack of support for the more distant relationships often occurs when retroviral phylogenies are based on RT and Pro regions (16, 33) but, despite this, both methods produced similar tree topologies and supported the monophyly of each of the four exogenous retrovirus-containing genera and the intracisternal A-type particle (IAP) elements. Our analyses showed a clear and striking division between viruses harbored by avian hosts and those detected in mammalian taxa. Within Bayesian phylogenies, members of each host vertebrate class were positioned as a single monophyletic clade, whereas MP trees placed the mammalian viruses as monophyletic sister taxa to one of three paraphyletic avian groups (unpublished data). Both methods indicated that the two previously described boid snake viruses should be included within a subclade of the mammalian-derived viruses.
FIG. 1.
ML phylogeny of the class II retroviruses with numbers on each branch indicating percentage posterior probabilities and asterisks showing probabilities of >95. The phylogeny is rooted on several deltaretroviruses (bovine leukemia virus, human T-cell leukemia virus type 1, and human T-cell leukemia virus type 2). Internal branches are thickened if the same topology was recovered in a strict consensus of seven equal-length MP trees. Taxon colors represent the host class of origin with avian hosts in blue, mammalian hosts in green, and reptilian hosts in black (in boldface type if the viruses are described here for the first time).
Avian class II retrovirus sequences have been relatively poorly described to date. The alpharetroviruses are known to be widespread within the Galliformes (including chickens, pheasants, grouse, and ptarmigan), but there are no ERV sequence data from avian species outside of this order (9, 10, 30). Many of the galliform-derived alpharetroviruses have only been partially characterized (via gag gene sequences) (9, 10), and we were therefore unable to include them in our RT-based phylogenies. Our results show that the alpharetroviruses share a clade with a large and diverse array of endogenous retrovirus sequences present within at least 15 avian orders. The viruses we identified as being most closely related to the alpharetrovirus genus, such as RV-Guineafowl II and RV-Tragopan, are also present within galliform hosts, a finding which is consistent with their proposed ancient association with this avian order (9, 10). In silico screening with class II retroviral probes also revealed at least three other ERV lineages within available chicken genome sequences. These lineages clustered with nongalliform avian hosts, including RV-Magpie II and RV-Ostrich (unpublished data).
Betaretroviruses are known to cluster into two subgroups, with one subgroup comprised of viruses present within many primate species, as well as ungulates (such as the Jaagsiekte virus within sheep), rodents (MusD), and marsupials (TvERV within the Brushtail possum) (4, 15, 22, 38). The second subgroup contains the sole representative mouse mammary tumor virus (MMTV) (27). Our results demonstrate that, although betaretroviruses are likely to be restricted to mammals, they are probably widespread throughout this vertebrate class. We found novel examples in several additional mammalian orders, including carnivores and a marine mammal. Furthermore, we identified murine mammary tumor virus-like viruses in several African and North American ungulates.
The sister clade to the betaretroviruses comprises the IAP-related elements, which have been described in a number of rodent species (21), and appear to be abundant within the mouse genome (20). Several novel sequences clustering strongly with the IAP elements were identified during our screening, all derived from rodents or lagomorphs, perhaps suggesting that the IAP elements have a more restricted host range than other class II retroviral groups. Consistent with this, two recent studies have shown that class II viruses related to betaretroviruses and IAP elements are extremely widespread in murid rodents (3, 25). In particular, it appears that there are multiple groups of endogenous class II-related retroviruses, some of which cluster separately with each of SMRV-H (Squirrel monkey retrovirus), Mason-Pfizer monkey virus, Jaagsiekte, and TvERV (3, 25).
Relationship of the lentiviruses to class II ERVs.
An unusual and unexpected feature of the phylogeny shown in Fig. 1 was the placement of the exogenous lentiviruses as sister taxa to several endogenous mammalian viruses from rodents (RV-Grass rat II and MuERVU1) and insectivores (RV-European hedgehog). Nucleic acid-based MP phylogenies also supported this relationship. This is in contrast to previous reports, which have generally placed the lentiviruses toward the base of the class II virus phylogeny, as paraphyletic sister taxa to the exogenous deltaretroviruses (5, 11, 16, 18, 33, 37). Characteristically skewed nucleotide compositions have been described in several retrovirus genera (7). Lentiviruses in particular are notable in being adenine-rich and cytosine-poor across the entire genome. Analysis of the nucleotide composition of RV-European hedgehog, RV-Grass rat II, and MuERVU1 viruses indicated that they did not share this bias (unpublished data). Nevertheless, to exclude the possibility that the observed relationship between these viruses and the lentiviruses was a function of nucleotide composition, we constructed MP and NJ trees by using a protein alignment (which spanned the same residues used in the DNA-based alignment [see Fig. S1 in the supplemental material]). The relationship was retained, with weak bootstrap support, in the case of the NJ analysis (unpublished data). However, the MP analysis placed RV-European hedgehog, RV-Grass rat II, and MuERVU1 as paraphyletic sister taxa to the lentiviruses (unpublished data).
To further investigate the relationship between the lentiviruses and the endogenous rodent- and insectivore-derived viruses, we studied the PR-RT region in more detail. Almost all known class II retroviruses encode PR and Pol (RT, RNase H, and IN) in different reading frames and use ribosomal frameshifting to produce a PR-Pol polyprotein (28). However, this is not the case with the lentiviruses, which encode PR and Pol in the same reading frame (28). We therefore examined our sequences for the presence of ribosomal frameshifting sites. The majority of the mammal-derived viruses and all viruses from avian hosts contained a characteristic thymine-rich region immediately upstream of a −1 frameshift at the boundary of the PR and Pol(RT) proteins (see Fig. 2). However, these features were absent from the RV-European hedgehog, RV-Grass rat II, and MuERVU1 sequences, supporting the hypothesis that these viruses may represent endogenous sister taxa to the lentiviruses.
FIG. 2.
Origin and distribution of the class II PR-RT frameshift mechanism and the G-patch motif. Taxa in boldface do not contain encode the frameshift, whereas the other taxa do (gray type indicates taxa with deletions encompassing the frameshift site). The boxes by each taxon label indicate whether the virus encodes a G-patch domain and whether this domain is degenerate. All avian taxa encode a frameshift site but lack a G-patch domain (not shown).
We then examined the class II sequences for evidence of the G-patch domain which is present (immediately upstream of the PR-RT frameshift site) within certain betaretroviruses and IAP-related elements but is absent from the lentiviruses and avian alpharetroviruses (2, 17). The G-patch domain is a short (typically in the order of 40 amino acids in length) glycine-rich region that is cleaved from PR to generate an RNA-binding protein (p5) that may have a role in splicing or transport of subgenomic retroviral mRNAs (2). We found that the G-patch probably originated relatively early during the evolution of the mammalian class II retroviruses (it was absent from all avian retroviruses) and that it has been lost, or partially deleted, on several occasions since this time (Fig. 2). Furthermore, and consistent with the results from the −1 frameshift site data, both the lentiviruses and the RV-European hedgehog, RV-Grass rat II, and MuERVU1 viruses appear to share a single loss event for this motif.
DISCUSSION
The analyses presented here are consistent with previous reports that suggest a relatively restricted host range for the class II retroviruses compared to both the class I and the class III viruses (16, 23). Despite extensive screening of reptilian, amphibian, and piscine genomes, we were unable to identify any class II retroviruses from these vertebrate classes. However, the presence of such viruses in certain python species (but not other, related boid snakes) demonstrates their presence in at least some reptilian taxa (18). Together, these findings suggest that class II retroviruses probably have only a very limited distribution within lower vertebrates.
The results of our screening also suggest that class II retroviruses are probably present within most mammalian and avian orders (we were able to recover sequences from 24 of the 27 orders investigated). Orders failing to yield class II sequences (Charadriiformes [shorebirds], Ciconiiformes [storks], and Scandentia [tree shrews]) were likely due to the low number of samples screened and the risk of obtaining false-negative results when a PCR-based approach is used. Furthermore, we note that results for individual taxa may not reflect the actual diversity of class II lineages contained in their genomes because only five clones were sequenced from each taxon and factors such as the primer sequence and ERV copy number may influence the results of PCR-based approach to screening. Despite this, it remains possible that class II ERVs may have a patchy distribution across vertebrate orders and families, especially within mammals. We were only able to recover class II sequences from 25 of the 49 mammalian taxa investigated, whereas viral fragments were recovered from 38 of the 46 avian taxa. This finding is consistent with previous liquid hybridization studies of the betaretroviruses and IAP elements (15, 21).
A high proportion of the novel avian ERV fragments encoded open reading frames that were intact, or nearly intact, across the aligned region (excluding the PR/RT frameshift site). Of the 55 sequences identified, 35 encoded one or less in-frame stop-codon or frameshifting mutation (Table 2), suggesting that many of these viruses have been active in the recent past. Thus, exogenous counterparts to these sequences may currently be circulating in avian populations, and we think it likely that exogenous viruses belonging to many of the avian subgroups present within the phylogeny will eventually be isolated. It is possible that retroviruses described previously, but for which pol sequence data are currently unavailable, are in fact members of these groups (9, 10, 13, 14).
In contrast, among the mammalian-derived viruses, novel ERV sequences clustering outside of the betaretrovirus or IAP groups (shown in Fig. 1) were relatively degenerate, displaying multiple in-frame stop codons or frameshifting mutations (unpublished data). Indeed, with the exception of HERV.K (HML2), not a single sequence outside of these two groups was intact across the amplified region. This suggests that many of these mammalian viruses are likely to represent older viral lineages and may therefore be less likely to have extant exogenous counterparts. The relatively intact nature of the betaretroviruses and IAP elements suggests that they may represent some of the more recently active groups of endogenous class II mammalian viruses, as has been suggested previously (3).
One of the most striking results of our phylogenetic analyses was the clustering of viruses derived from mammals and birds into distinct monophyletic groups. These groupings were not strongly supported (and MP analysis placed the mammalian viruses as a paraphyletic sister clade to several groups of avian viruses), but it is unlikely that such a pattern would be obtained by chance. Previous studies have shown intermingling of avian and mammalian class II ERVs in phylogenies (33). However, these studies were based on shorter sequence alignments of a smaller and less diverse range of class II taxa and are therefore likely to be less accurate than the phylogeny presented here. Monophyly of the mammalian and avian derived sequences strongly implies that vertebrate interclass transmission events have been rare in the evolution of the class II retroviruses (although we note that only a very small proportion of endogenous viruses have been examined to date). The most likely exceptions to this are the boid python ERVs, which are sister taxa to two novel viruses derived from Felids. This relationship was supported in both maximum-likelihood (ML) and MP analyses, making it possible that an interclass transmission event from mammals to reptiles has occurred during the evolution of these viruses. The rarity of interclass transmission apparent from our analyses reflects that observed previously with the gammaretroviruses. The gammaretroviruses comprise a genus of exogenous and endogenous viruses that are widespread in tetrapod vertebrates (23, 34) but only very rarely undergo interclass transmission (23, 24). Taken together, these results imply that interclass transmission within the family Retroviridae occurs much less frequently than intraclass transmission.
An intriguing result from our phylogenetic analyses was the positioning of the lentiviruses. Previous reports have suggested they form one of the most basal clades within the class II viruses, as paraphyletic sister taxa to the deltaretroviruses (5, 11, 16, 18, 33, 37). Our analyses (which were based on a larger number of class II sequences than available previously) placed the lentiviruses in a more derived position within the mammalian viruses, as sister taxa to three viral sequences from rodents and insectivores, including the murine element MuERVU1 (previously identified by Bénit et al. [5]). This topology was also observed in MP phylogenies but lacked robust bootstrap support. Further support for this relationship was apparent from analyses investigating the acquisition and loss of viral characters present within the amplified PR-RT region.
The first of these characters, the frameshift event known to occur at the PR-RT boundary in several class II genera, was remarkably stable, being found in the vast majority of class II sequences. Indeed, it appears that ribosomal frameshifting has only been lost on three occasions across the phylogeny. Two of these loss events involve only a single virus (RV Small mongoose I and RV Echidna II), whereas the third is shared by both lentiviruses and RV-Grass rat II, MuERVU1, and RV-European hedgehog. The presence or absence of the G-patch domain (2) also appears to be a relatively stable characteristic. Our phylogenies suggest that the G-patch was acquired early during the evolution of the mammalian class II viruses (it is absent from the avian-derived sequences). Loss (or degeneracy) of the G-patch appears to have occurred more often than loss of ribosomal frameshifting, but a single loss event is, again, shared between the lentiviruses and their three sister taxa.
The lentiviruses are known to encode several accessory genes that are not present within other retroviruses (28). Investigation of MuERVU1, for which the full-length sequence is available, failed to reveal any obvious similarity to these lentivirus-specific genes (unpublished data). However, this might be expected since many of the accessory genes are absent from the more basal lentiviruses (such as EIAV (28) and those that are conserved in all members of the genus (such as tat and rev) are both relatively short and highly divergent. Although our phylogenetic, G-patch and frameshift site analyses all support the conclusion that lentiviruses have distantly related endogenous counterparts, more definitive proof will have to await the detailed comparative analysis of several full-length viral genomes.
Supplementary Material
Acknowledgments
We thank F. Catzeflis, A. Cooper, R. Deaville, K. Fok, L Granjon, G. Jarrell, D. Mindell, J. Patton, J. Sheps, and R. Waugh O'Neill for supplying some of the DNA samples used in this study and R. Belshaw for help with the ML analysis.
R.G. was supported by an NERC studentship.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. L. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind, L., and E. V. Koonin. 1999. G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 24:342-344. [DOI] [PubMed] [Google Scholar]
- 3.Baillie, G. J., L. N. van der Lagemaat, C. Baust, and D. L. Mager. 2004. Multiple groups of endogenous betaretroviruses in mice, rats, and other mammals. J. Virol. 78:5784-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baillie, G. J., and R. J. Wilkins. 2001. Endogenous type D retrovirus in a marsupial, the common brushtail possum (Trichosurus vulpecula). J. Virol. 75:2499-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bénit, L., P. Dessen, and T. Heidmann. 2001. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J. Virol. 75:11709-11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bénit, L., J. B. Lallemand, J. F. Casella, H. Philippe, and T. Heidmann. 1999. ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals. J. Virol. 73:3301-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkhout, B., A. Grigoriev, M. Bakker, and Lukashov. 2002. Codon and amino acid usage in retroviral genomes is consistent with virus-specific nucleotide pressure. AIDS Res. Hum. Retrovir. 18:133-141. [DOI] [PubMed] [Google Scholar]
- 8.Boeke, J. D., and J. P. Stoye. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p. 343-435. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, N.Y. [PubMed]
- 9.Dimcheff, D. E., S. V. Drovetski, M. Krishnan, and D. P. Mindell. 2000. Cospeciation and horizontal transmission of avian sarcoma and leukosis virus gag genes in galliform birds. J. Virol. 74:3984-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dimcheff, D. E., M. Krishnan, and D. P. Mindell. 2001. Evolution and characterization of tetraonine endogenous retrovirus: a new virus related to avian sarcoma and leukosis viruses. J. Virol. 75:2002-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimmic, M. W., J. S. Rest, D. P. Mindell, and R. A. Goldstein. 2002. rtREV: an amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny. J. Mol. Evol. 55:65-73. [DOI] [PubMed] [Google Scholar]
- 12.Doolittle, R. F., D. F. Feng, M. S. Johnson, and M. A. McClure. 1989. Origins and evolutionary relationships of retroviruses. Q. Rev. Biol. 64:1-30. [DOI] [PubMed] [Google Scholar]
- 13.Fujita, D. J., Y. C. Chen, R. R. Friis, and P. K. Vogt. 1974. RNA tumor viruses of pheasants: characterization of avian leukosis subgroups F and G. Virology 60:558-571. [DOI] [PubMed] [Google Scholar]
- 14.Hanafusa, T., H. Hanafusa, C. E. Metroka, W. S. Hayward, C. W. Rettenmier, R. C. Sawyer, R. M. Dougherty, and H. S. Di Stefano. 1976. Pheasant virus: new class of ribodeoxyvirus. Proc. Natl. Acad. Sci. USA 58:1333-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hecht, S. J., K. E. Stedman, J. O. Carlson, and J. C. DeMartini. 1996. Distribution of endogenous type B and type D sheep retrovirus sequences in ungulates and other mammals. Proc. Natl. Acad. Sci. USA 93:3297-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herniou, E., J. Martin, K. Miller, J. Cook, M. Wilkinson, and M. Tristem. 1998. Retroviral diversity and distribution in vertebrates. J. Virol. 72:5955-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hruskova-Heidingsfeldova, O., M. Andreansky, M. Fabry, I. Blaha, P. Strop, and E. Hunter. 1995. Cloning, bacterial expression, and characterization of the Mason-Pfizer monkey virus proteinase. J. Biol. Chem. 270:15053-15058. [DOI] [PubMed] [Google Scholar]
- 18.Huder, J. B., J. Böni, J.-P. Hatt, G. Soldati, H. Lutz, and J. Schüpbach. 2002. Identification and characterization of two closely related unclassifiable endogenous retroviruses in pythons (Python molurus and Python curtus). J. Virol. 76:7607-7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Human Genome Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 20.International Mouse Genome Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 21.Kuff, E. L., and K. K. Leuders. 1988. The intracisternal A-particle family: structure and functional aspects. Adv. Cancer Res. 51:184-276. [DOI] [PubMed] [Google Scholar]
- 22.Mager, D. L., and J. D. Freeman. 2000. Novel mouse type D endogenous proviruses and ETn elements share long terminal repeat and internal sequences. J. Virol. 74:7221-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin, J., E. Herniou, J. Cook, R. W. O'Neill, and M. Tristem. 1999. Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses. J. Virol. 73:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, J., P. Kabat, and M. Tristem. 2002. Cospeciation and horizontal transmission rates in the murine leukemia-related retroviruses, p. 174-194. In R. D. M. Page (ed.), Tangled trees. University of Chicago Press, Chicago.
- 25.McCarthy, E. M., and J. F. McDonald. 2004. Long terminal repeat retrotransposons of Mus musculus. Genome Biol. 5:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClure, M. A., M. S. Johnson, D.-F. Feng, and R. F. Doolittle. 1988. Sequence comparisons of retroviral proteins: relative rates of change and general phylogeny. Proc. Natl. Acad. Sci. USA 85:2469-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore, R., M. Dixon, R. Smith, G. Peters, and C. Dickson. 1987. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol. J. Virol. 61:480-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petropoulos, C. 1997. Retroviral taxonomy, protein structures, sequences, and genetic maps, p. 757-805. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 29.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz, D. E., R. Tizard, and W. Gilbert. 1983. Nucleotide sequence of Rous sarcoma virus. Cell 32:853-869. [DOI] [PubMed] [Google Scholar]
- 31.Swofford, D. L. 1998. PAUP*: phylogenetic analysis using parsimony (* and other methods), 4th ed. Sinauer Associates, Sunderland, Mass.
- 32.Tristem, M. 1996. Amplification of divergent retroelements by PCR. BioTechniques 20:608-612. [DOI] [PubMed] [Google Scholar]
- 33.Tristem, M. 2000. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J. Virol. 74:3715-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: the classification and nomenclature of viruses. Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, Inc., San Diego, Calif.
- 35.Vogt, P. K. 1997. Historical introduction to the general properties of retroviruses, p. 1-25. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, N.Y. [PubMed]
- 36.Wilkinson, D. A., D. L. Mager, and J. C. Leong. 1994. Endogenous human retroviruses, p. 465-535. In J. A. Levy (ed.), The Retroviridae, vol. III. Plenum Press, Inc., New York, N.Y.
- 37.Xiong, Y., and T. H. Eickbush. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9:3353-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.York, D. F., R. Vigne, D. W. Verwoerd, and G. Querat. 1992. Nucleotide sequence of the jaagsiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J. Virol. 66:4930-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.