Abstract
Kinesins are microtubule-based motor proteins that are well known for their key roles in cell biological processes ranging from cell division, to intracellular transport of mRNAs, proteins, vesicles, and organelles, and microtubule disassembly. Interestingly, many of the ~45 distinct kinesin genes in vertebrate genomes have also been associated with specific phenotypes in embryonic development. In this review, we highlight the specific developmental roles of kinesins, link these to cellular roles reported in vitro, and highlight remaining gaps in our understanding of how this large and important family of proteins contributes to the development and morphogenesis of animals.
Introduction:
Microtubules are cytoskeletal elements comprised of tubulin and act as scaffolds for intracellular transport of a wide variety of cargoes, including individual proteins, vesicles, mRNAs, and even organelles. Microtubule-based transport is orchestrated by motor proteins, namely kinesins and dynein, and is integral to differentiation, morphogenesis, and cell survival. Dyneins function exclusively in minus-end directed transport and have been reviewed extensively elsewhere (Hou and Witman, 2015; Reck-Peterson et al., 2018; Viswanadha et al., 2017). By contrast, Kinesins are a large superfamily of proteins that are responsible not only for trafficking cellular cargoes along microtubules but also for controlling microtubule growth and stability. To date, ~45 different vertebrate kinesins have been placed into 15 different subfamilies, generating a wide diversity and specificity of function within kinesins (Supp. Table 1)(Miki et al., 2001).
Most kinesins follow a generalized structure: a highly conserved motor domain that facilitates binding and release of microtubules through ATP hydrolysis, a long neck sequence that allows homo- or heterodimerization, and finally, highly divergent cargo binding domains (Hirokawa and Noda, 2008). The 15 different subfamilies have been further categorized into three broad categories based on the location of the motor domain: N-kinesins, C-Kinesins, and M-Kinesins (Figure 1) (Lawrence et al., 2004; Miki et al., 2001). N-Kinesins contain a N-terminal motor and are plus-end directed in their movement along microtubules (Figure 1A, D). C-Kinesins have a C-terminally located motor domain and are minus-end directed microtubule motors (Figure 1B, D). And finally, M-Kinesins contain a “middle” motor that is more centrally located in the peptide and are described generally as microtubule depolymerizers (Figure 1C, E). Different motor domains are important for the specific microtubule-based functions each kinesin has adopted within the cell (Fig. 2).
Figure 1. Schematic representing motor domain locations in N-, C-, and M-type kinesins.
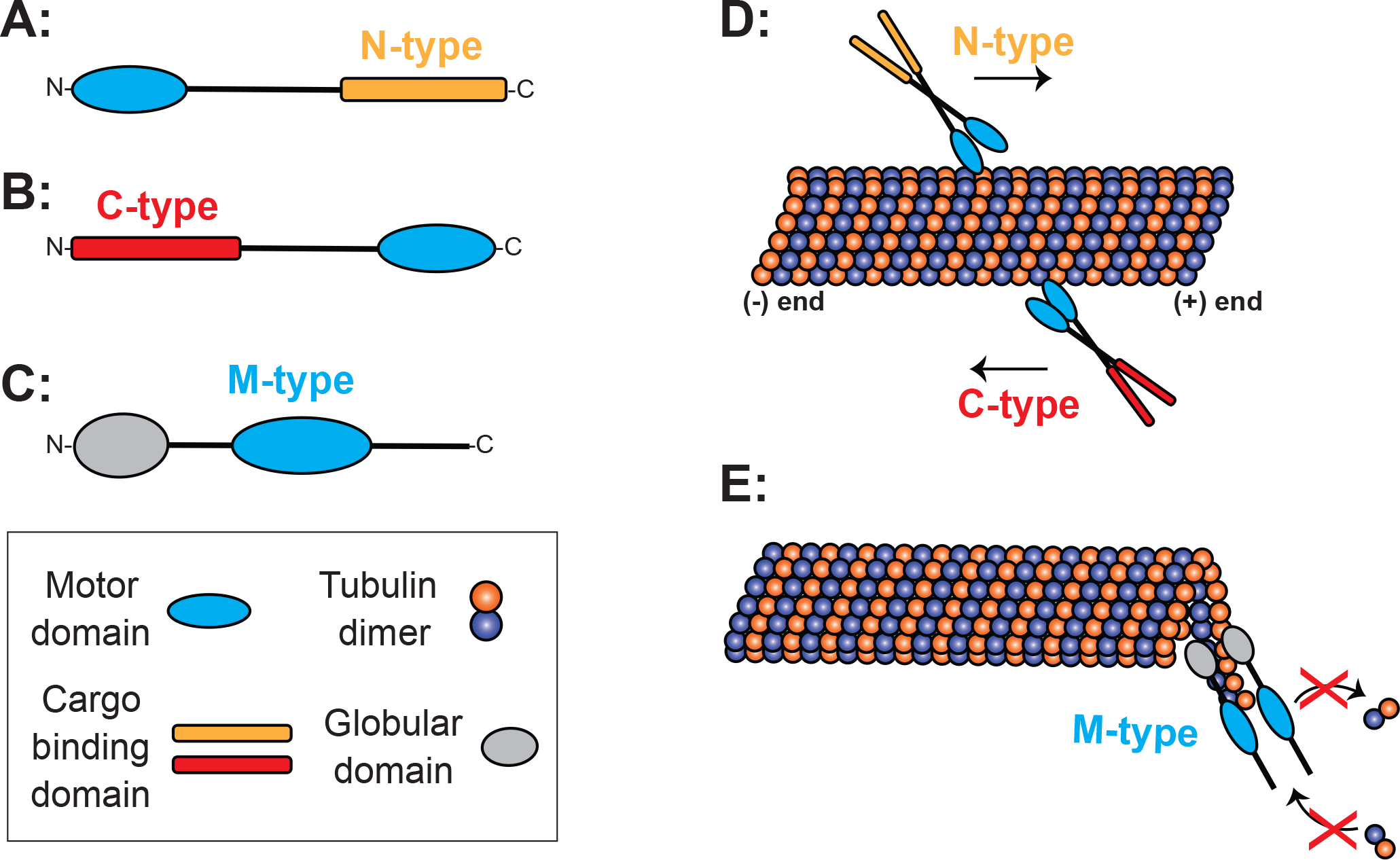
Figure 1A. Schematic of N-type kinesins with N-terminal motor domains. Figure 1B. Schematic of C-type kinesins with C-terminal motor domains. Figure 1C. Schematic of M-type kinesins with motor domains located in the middle of the polypeptide. Figure 1D. Schematic depicting general direction of movement along microtubules of N- and C-type kinesins. N-type are plus end directed, while C-type are minus end directed. Figure1E. Schematic depicting M-type kinesins as microtubule depolymerizing kinesins, regulating plus end growth by depolymerizing microtubules at these locations. Inset acts as legend for figure.
Figure 2. Cellular functions associated with kinesin. Figure 2A.
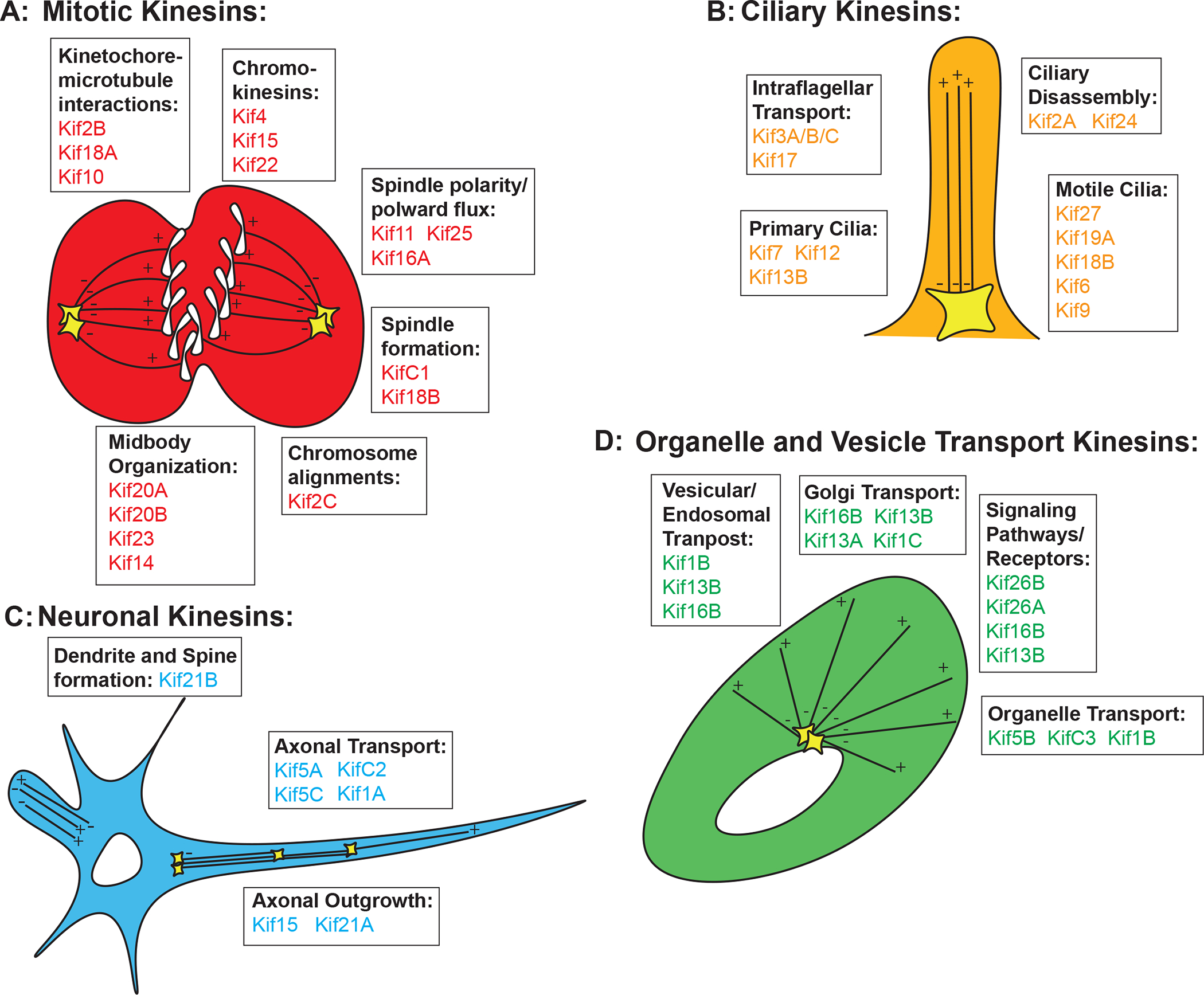
The kinesins found to have a role in mitosis either in vivo and/or in cell culture categorized by the exact functions they have in mitosis. Figure 2B. Kinesins found to have functions in cilia, categorized by the type of cilia and functions they have within cilia. Figure 2C. Kinesins who have a neuronal role categorized by the functions they have in dendrite and axon outgrowth or axonal transport. Figure 2D. Kinesins associated with organelle and vesicle intracellular transport categorized by the type of cargo they transport. Names of kinesins are color coded to match the subcellular functions associated with each kinesin.
How each kinesin interacts with microtubules, and what functions it may display in vitro have been relatively well defined (Supp. Table 1; Cellular roles), and the atomic structures and biophysical properties of kinesins have been reviewed extensively elsewhere (Endow et al., 2010; Verhey and Hammond, 2009; Wang et al., 2015). By contrast, the ways in which kinesins contribute to overall organismal development remain far less well defined, so in this review, we aim to highlight the specific functions of kinesins in embryonic development. Generally, these fall within five broad categories, which we address in turn below: (i) kinesins required for early axis specification by transporting maternal determinants prior to or as the result of fertilization; (ii) mitotic kinesins necessary for successful mitosis and meiosis (Fig. 2A); (iii) ciliary kinesins necessary for the formation, function, and regulation of cilia (Fig. 2B); (iv) kinesins essential for axonal and dendrite formation and function in neuronal tissues (Fig. 2C); and (v) kinesins required for development of specific organs and tissues.
In Supplemental Table 1, we provide a full accounting of all mammalian kinesins, their cell biological roles as established by in vitro studies, and in vivo phenotypes in animal models or humans. In the main text of this review, we will highlight the links between different kinesins’ cellular functions and specific processes in embryogenesis, we note gaps that remain in understanding, and highlight the most exciting questions left to answer in kinesin developmental biology.
Transport of polarizing maternal determinants:
In a variety of different organisms, the embryonic axes are established prior to or as the result of fertilization. Interestingly, the establishment of these embryonic axes are often dependent upon microtubule-based transport. Most notably, axis specification is dependent upon specific members of the N-type kinesins for transport of maternally deposited mRNAs and proteins (Supp. Table 1). N-type kinesins regularly partake in transporting maternal determinants through a variety of different mechanisms that highlight an evolutionarily conserved function. Here, we will review the contribution of kinesins in early axis specification in D. melanogaster, X. laevis, and D. rerio.
Prior to fertilization, the D. melanogaster oocyte establishes an anterior/posterior axis by polarizing maternally deposited mRNAs and proteins. How maternal determinants are polarized in the oocyte has been the subject of much debate for years, however, live imaging revealed oskar mRNA was actively transported along microtubules (Zimyanin et al., 2008). Though microtubules are oriented in every direction in the embryo, there is a slight biased orienting of plus-ends towards the posterior pole (Zimyanin et al., 2008). More detailed analysis later showed that oskar mRNA and Staufen protein are posteriorly localized via multiple modes of activity of the N-terminal plus-end directed Kinesin-1 motor (Brendza et al., 2002, 2000; Clark et al., 1994). In order to properly localize oskar mRNA and Staufen protein, Kinesin-1 transports oskar and Staufen in a two part mechanism: (i) by directly transporting the mRNA and protein as cargo and (ii) by participating in “microtubule sliding” where Kinesin-1 binds microtubules as cargo and walks along neighboring microtubules to produce a “sliding” effect (Lu et al., 2016; Métivier et al., 2019). This microtubule sliding activity of Kinesin-1 in flies is critical for cytoplasmic streaming in Drosophila oocytes that further reinforces oskar and Staufen localization at the posterior pole of the oocyte (Lu et al., 2016).
Interestingly, while Kinesin-1 contributes to overall posterior localization of Staufen and oskar mRNA via these mechanisms, an interesting dynamic occurs between Kinesin-1 and Myosin-V at the posterior pole (Lu et al., 2020). Kinesin-1 actively continues to transport Staufen and oskar mRNA moving them away from the posterior cortex, however, Myosin-V actively works to anchor oskar and Staufen to the posterior cortex (Lu et al., 2020). Eventually Myosin-V wins this battle-of-motor proteins due to the low microtubule density at the posterior pole, anchoring Staufen and oskar mRNA to the posterior cell cortex (Lu et al., 2020). Altogether the activity of Kinesin-1 posteriorly localizes oskar mRNA and Staufen protein in the D. melanogaster oocyte which are each separately critical for axis specification prior to fertilization.
X. laevis, on the other hand, offers an example of post-fertilization axis specification that also requires kinesin activities. In Xenopus, sperm entry triggers the assembly of maternally-deposited cortically-located microtubules to position and anchor their plus ends to the future dorsal cell cortex (Elinson and Rowning, 1988; Olson et al., 2015). These parallel microtubule arrays are essential for the transport of maternally deposited factors to the future dorsal side of the embryo in a process termed “cortical rotation.” Specifically, these microtubules are thought to act as tracks for the dorsal translocation of several regulatory factors, including the Frat1/GBP and Dishevelled proteins, as well as β-catenin, Vg1 and Wnt11 mRNAs (Miller et al., 1999; Schroeder et al., 1999; Tao et al., 2005; Weaver et al., 2003). Strikingly, two sets kinesins transport different mRNA and protein cargoes during these translocation events. Specifically Kinesin light chain (Klc4) was shown to both bind, and be required for transport of Frat1/GBP(Weaver et al., 2003), while the Kinesin-2 complex, Klp3A/3B (the homolog of mammalian Kif3A/3B), specifically transports Vg1 mRNA (Betley et al., 2004). The transport of these maternal mRNAs and proteins by two separate kinesins in X. laevis highlights how kinesins have evolved highly specific interactions with their cargoes. The dorsal translocation activities of these kinesins and the determinants they carry as cargo is central to the establishing the dorsal/ventral axis of the embryo and initiating the program for formation of the Nieuwkoop center and later the Spemann-Mangold organizer (Gerhart et al., 1991; Weaver and Kimelman, 2004; White and Heasman, 2008). In the absence of cortical rotation, the embryo becomes ventralized and fails to form any dorsal structures essential for survival.
Interestingly, Kinesin-1 family members in D. rerio serve similar functions to D. melanogaster and X. laevis in early embryo axis specification. Specifically, Kif5Ba has similar dorsalizing activity to that of Xlc4 and Kif3A/B in X. laevis, while also participating in microtubule organizing activities that have been described with Kinesin-1 in D. melanogaster. Kif5Ba promotes formation of parallel microtubule arrays and delivers wnt8a and Syntabulin to the future dorsal side of the zebrafish embryo (Campbell et al., 2015). This dual function for kinesins in early embryo axis specification is both essential and conserved at some level in vertebrate axis specification. However, how kinesin family members may contribute to early mammalian embryonic axis specification remains an interesting area for future work.
Early embryonic cell cycle progression:
The earliest phase of development in most animals involves a period of remarkably rapid cell division. In order to execute mitosis, cell division is highly dependent upon a complex set of microtubule movements, alignments, and attachments that requires a diverse set of kinesins. Though such cleavage divisions in most embryos lack many of the cell cycle controls in place in adult cells, they nonetheless still require the proper microtubule dynamics, alignment, and attachment to cargoes or adaptors to ensure faithful segregation of chromosomes. A huge number of distinct kinesins have been implicated in diverse aspects of cell division in vitro (Fig. 2), so it is interesting that Kif11, Kif22, and Kif10 (Fig. 2A, 3A) have also been shown to play specific roles in embryonic cleavage and knockout of any one leads to cell cycle arrest and early embryonic death in mice (Supp. Table 1).
Figure 3. Developmental functions of each kinesin with confirmed in vivo studies categorized by organ and tissue.
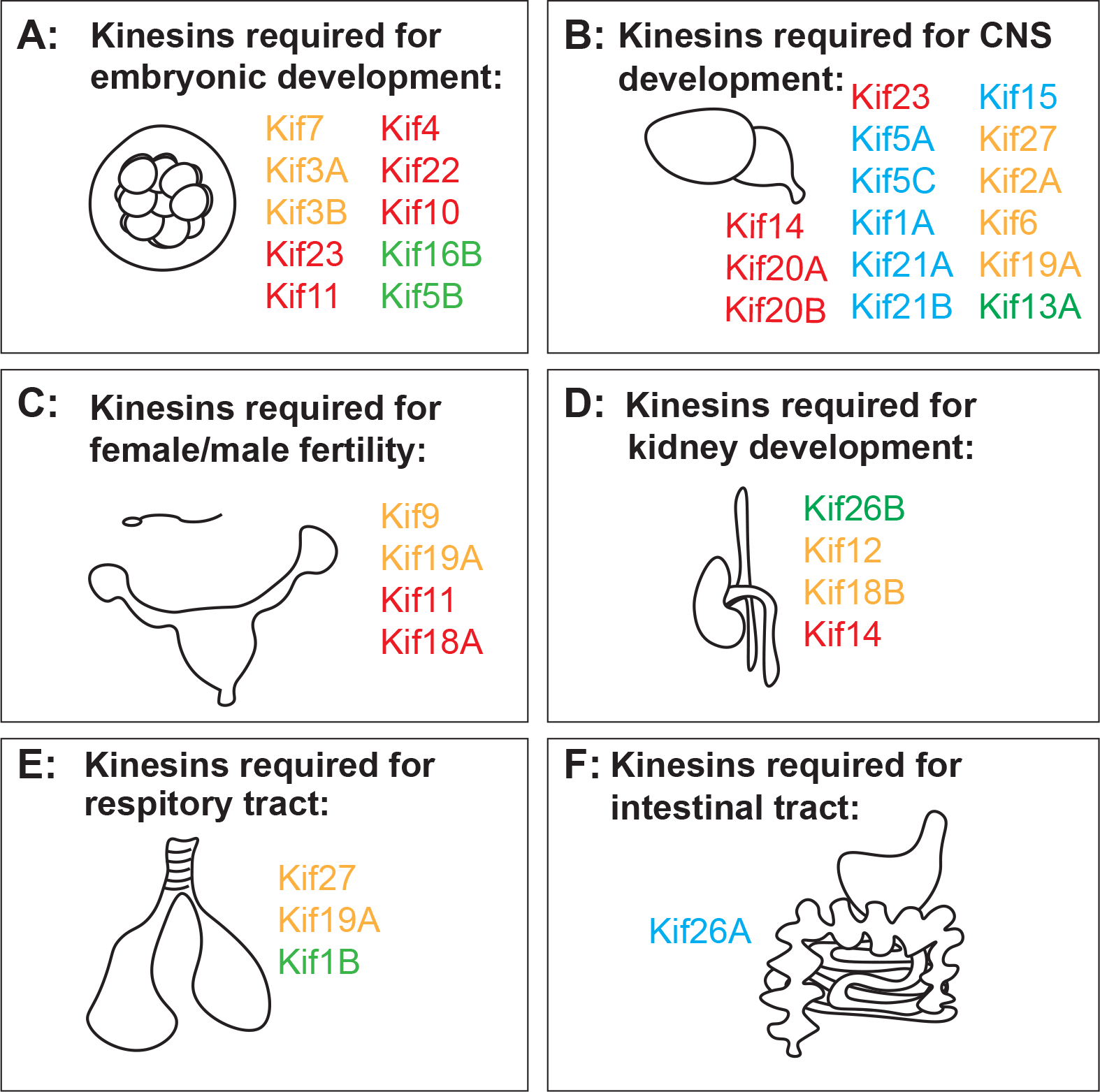
All kinesin names are color coded to match cellular roles defined in Figure1. Red for mitosis, yellow for cilia, blue for neuronal functions, and green for vesicle/endosome transport. Figure 3A. Kinesins required for embryonic development. Figure 3B. Kinesins required for development of brain and central nervous system. Figure 3C. Kinesins required for male and female fertility. Figure 3D. Kinesins required for kidney development. Figure 3E. Kinesins required for respiratory tract development. Figure 3F. Kinesins required for the development of the intestinal tract.
Kif11 has been shown in vitro to be required for spindle bipolarity/poleward flux at metaphase (Fig. 2A) (Heck et al., 1993; Sawin et al., 1992). Kif10 is required at the kinetochore to ensure proper chromosome alignment during mitosis (Fig. 2A) (Brown et al., 1996; Putkey et al., 2002; Wood et al., 1997; Yao et al., 2000; Yen et al., 1992). Finally, Kif22 is classified as a chromokinesin, meaning its cargo binding domain is capable of binding chromosomes. As such Kif22 is required for chromosome movements during prometaphase and metaphase (Fig. 2A) (Antonio et al., 2000; Levesque and Compton, 2001; Ohsugi et al., 2008; Tokai et al., 1996; Tokai-Nishizumi et al., 2005; Zhu et al., 2005). In addition to mammalian kinesins, the microtubule depolymerizing kinesin, KLP10A, related to the Kinesin-13 family in mammals, has dual roles in cell cycle progression in D. melanogaster. In females, KLP10A has been shown to control acentrosomal spindle organization and dynamics in oocytes (Radford et al., 2012), while also controlling centrosomal length in D. melanogaster male germline stem cells (GSCs) (Chen et al., 2016). Given the myriad functions of kinesins in mitosis, these studies highlight the power of intersecting cell biology in vitro with genetics in vivo for understanding kinesin function during development.
Cilia formation and function during development:
Cilia are microtubule-based projections of the cell that function in signaling and fluid flow in many developing organisms. During development, two types of cilia are integral for the organism: primary cilia are essential organelles for processing signaling events that govern embryonic patterning, while motile cilia generate polarized fluid flows essential to both patterning and homeostasis in tubular organs. Cilia rely heavily upon kinesins for their formation, function, and regulation in a variety of different manners. Although ciliary kinesins have been extensively reviewed relatively recently (Lechtreck, 2015; Reilly and Benmerah, 2019; Scholey, 2008), we will briefly review them here with an emphasis on developmental contexts.
Intraflagellar Transport (IFT):
Intraflagellar Transport (IFT) is the process by which ciliary components are actively transported from the basal body, a modified mother centriole, into the cilium and out again. IFT is essential for the proper assembly and maintenance of each and every cilium and it, too, has been reviewed extensively elsewhere (Prevo et al., 2017; Reilly and Benmerah, 2019; Scholey, 2008). The Kinesin-2 family is indispensable for IFT, with Kif3A and Kif3B serving as motors required for anterograde movement of IFT in all cilia types (Fig. 2B, Supp. Table 1). In multiple animal models, loss of Kif3A/B function leads to global ciliogenesis defects in every ciliated cell type (Supp. Table 1). Kif3A and Kif3B mutant mice are embryonic lethal due to exencephaly and laterality defects associated with ciliogenesis defects (Marszalek et al., 1999; Nonaka et al., 1998; Takeda et al., 1999). Interestingly, the two other kinesins of the Kinesin-2 family, Kif17 and Kif3C, have been implicated in IFT in specific tissues in C. elegans and zebrafish (Supp. Table 1), however their roles in IFT in mammals remains unclear (Jiang et al., 2015). Kif17 (osm-3 in C. elegans) has been well-described as an “accessory” IFT motor that aids in assembly of distal segments in specialized cilia types both in C. elegans and in D. rerio (Insinna et al., 2008, 2008; Zhao et al., 2012). Most interestingly, Kif17 is homodimeric and walks at a slower rate than the Kif3A/3B kinesin complex. When bound to an IFT train with Kif3A/3B, a recent study found that the two kinesin complexes find a walking rate between the rate of the faster and slower kinesins (Milic et al., 2017). This unique cooperation between the two sets of motors highlights the complex molecular interactions that can occur between kinesins.
Primary cilia:
Primary cilia act as signaling antenna, transmitting signals like Sonic Hedgehog (Shh) that are vital for proliferation, cell survival, and specification during development (Bangs and Anderson, 2017). Primary cilia are present on mitotically active cells and are continually assembled and disassembled when entering and exiting the cell cycle. Here, we will discuss the kinesins important for Shh signaling and ciliary disassembly.
How the cilium transmits Shh signals detected within the cilium to the rest of the cell still remains an active area of research, but myriad studies have demonstrated that activation requires the localization of Shh signaling components to the ciliary tip. Interestingly, the atypical kinesin Kif7 organizes this compartment of primary cilia by regulating the length of ciliary microtubules at the tip and binding Gli proteins, the transcription factors responsible for downstream Shh signaling (Cheung et al., 2009; He et al., 2014; Liem et al., 2009). Consistent with this cell biological function, Kif7 mutant mice phenocopy mice with mutations in the Shh pathway, dying at the end of gestation with polydactyly and expanded motor neuron fates (Liem et al., 2009).
Both assembly and disassembly of primary cilia are closely linked with the cell cycle, and accordingly, both processes are essential for normal signaling. Cycling cells assemble primary cilia during interphase and disassemble cilia when entering mitosis (Wang and Dynlacht, 2018). For disassembly, two Kinesin-13 family members are essential: Kif2A and Kif24 (Fig. 2B). In general, the Kinesin-13 subfamily acts as M-type depolymerizing kinesins. Kif2A interacts with Polo Like Kinase 1 (PLK1) upon entry into the cell cycle and mediates primary cilia disassembly (Broix et al., 2018; Miyamoto et al., 2015). This kinesin has also been shown to depolymerize axonal microtubules (Homma et al., 2003). Kif2A mutant mice are embryonic lethal with severe brain defects partially due to cell death of neural progenitors due to improper disassembly of cilia upon cell cycle entry (Broix et al., 2018; Homma et al., 2003). Interestingly, the other Kinesin-13 member Kif24, which is mammalian specific, is suggested to play a similar role in ciliary disassembly in cell culture, though no in vivo validation has been reported (Supp. Table 1, Fig. 2B) (Kim et al., 2015; Kobayashi et al., 2011).
Motile cilia and multiciliated cells:
Motile cilia play diverse roles in embryonic development. Single motile cilia are required to move fluid and signals that break the left/right symmetry of the embryo (Little and Norris, 2020). On the other hand, multiciliated cells have dozens of motile cilia located in the brain (i.e. ependymal cells), the trachea, and male/female reproductive tracts where they move fluids to ensure the development and health of those tissues (Brooks and Wallingford, 2014). Additionally, sperm have motile flagella that are essential for sperm movement. A variety of kinesins, including the IFT kinesins, are important for the formation, function, and regulation of motile cilia. In addition, several non-IFT kinesins have been linked to motile cilia specific roles. This set of kinesins are often identified as being direct targets of the master motile cilia transcription factor FoxJ1 (Choksi et al., 2014a, 2014b; Jacquet et al., 2009); these include: Kif6, Kif9, Kif27, Kif19A and Kif18B (Fig. 2B).
Kif6 and Kif9 are the only two members of the Kinesin-9 super family and both have been linked to motile cilia function, albeit through separate and specific functions. This family is thought to consist of N-type motors, although the precise walking and molecular capabilities of these two kinesins remains unexplored. We recently reported that Kif6 is expressed in a highly tissue-specific manner and is specifically required for ciliogenesis of multiciliated ependymal cells in the brain (Fig. 3), while deleterious mutations in Kif6 does not affect other multiciliated tissues in mice or zebrafish (Buchan et al., 2014; Konjikusic et al., 2018). In contrast, Kif9 is not required for ciliogenesis, but rather is necessary for ciliary beating. Klp1 is the Kif9 homologue in the ciliated algae Chlamydomonas, and it localizes to the central apparatus of motile flagella, and its loss leads to structural disruption of the central pair and defective flagellar beating (Bernstein et al., 1994; Yokoyama et al., 2004). Moreover, a recent study reported that Kif9 mutant mice have impaired sperm flagellar motility (Miyata et al., 2020). Intriguingly, this study observed no hydrocephaly in their Kif9 mutant mice, suggesting that Kif9 is dispensable for ependymal cell cilia motility, while analysis of the multiciliated cells of the trachea or oviduct was not reported.
Kif27 is a mammalian-specific paralog of the Kinesin-4 protein Kif7 and has been found to function specifically in motile cilia (Fig. 2B). Mouse Kif27 knockout results in situs inversus, hydrocephaly, and otitis media, all phenotypes linked to motile cilia dysfunction (Vogel et al., 2012). Kif27 thought to be required for assembly of the central pair of microtubules and localizes to basal bodies of trachea multiciliated cells (Wilson et al., 2009; Yue et al., 2018). The precise function of Kif27 remains to be determined, but in vitro, this kinesin is slowly processive and can inhibit microtubule growth (Wilson et al., 2009; Yue et al., 2018)
Finally, the two other kinesins with suggested roles in motile cilia, Kif19A and Kif18B, are part of the Kinesin-8 family, motors that primarily function in microtubule depolymerization. Kif19A localizes to the tip of these multiciliated cells where it depolymerizes microtubules to regulate ciliary length (Niwa et al., 2012; Wang et al., 2016, p. 19). In fact, Kif19A knockout mice present with hydrocephalus and display longer cilia on their ependymal and tracheal/oviductal multiciliated cells, presumably causing disrupted fluid flow in these tissues (Niwa et al., 2012). The other Kinesin-8 family member, Kif18B can promote microtubule catastrophe of the mitotic spindle in cell culture but has also been reported to be a target of FoxJ1 and loss of function in zebrafish showed shorter motile cilia in the pronephros (Choksi et al., 2014a). To date there is no mammalian Kif18B mutant reported to validate a role in mammalian multiciliated cells.
Development of the brain and central nervous system (CNS):
The development of the central nervous system is a complex process requiring proliferation of neural progenitors, extension of long polarized microtubule-based axonal/dendritic processes, transport of support factors along these microtubule-based extensions, and later transport of neurotransmitters/ signals to/from nearby cells. It is no surprise, then, that many of the 45 kinesins have been linked to the CNS in one way or another (Fig. 3C, Supp. Table 1). Although the function of kinesins in the CNS has been reviewed at length (Hirokawa et al., 2010), we will briefly review their roles with a focus on developmental contexts.
Mitosis and Proliferation in the Brain and CNS:
As described above (see early embryonic cleavages), successful completion of mitosis and proliferation/differentiation of cells is critical for the development of any given tissue in the embryo. Cells must not only replicate themselves to form more progenitor cells (a term called “symmetric divisions”), but ultimately divide to replicate two daughter cells with two separate states: progenitor and non-progenitor-like states (“asymmetric divisions”). The development and homeostasis of the brain and CNS is no different. Interestingly, two Kinesin-6 family members play separate and specific roles in mitotic events and proliferation decisions in the CNS. Kif20A is required for the control of symmetric vs. asymmetric cell divisions in the developing CNS, so Kif20A knockout mice are embryonic lethal, displaying smaller brain and body size due to increased apoptosis in the brain caused by improper cell division decisions (Geng et al., 2018). In contrast, Kif20B controls midbody organization during cytokinesis in polarized cortical stem cells. Knockout in mouse leads to perinatal lethality and microcephaly caused by loss of midbody organization (Janisch et al., 2018, 2013). These examples highlight the fact that despite close homology, kinesins have frequently evolved highly divergent roles in mitosis in distinct tissues in the developing brain and CNS.
Axonal outgrowth:
Axons are long, polarized, microtubule-based neuronal projections that send signals from the neuronal cell body to other cells. Axons extend from the cell body, orienting their minus ends towards the cell body, and the plus-ends/growing ends of microtubules towards the tip of an axon (Figure 2C). While the generation and dynamics of the axonal growth cone is heavily dependent upon the actin cytoskeleton and its dynamics (Dent et al., 2011), axons still rely on kinesins for axonal outgrowth in a variety of ways (Hirokawa et al., 2010). Two mammalian kinesins, Kif21A and Kif5A (Fig 2C), that have been linked to the control of axon outgrowth by separate and specific functions. First, Kif21A, a member of the Kinesin-4 family, acts via microtubule growth inhibition at the cell cortex, allowing microtubules to accumulate within the axon rather than at the cortex (van der Vaart et al., 2013). Mutations in Kif21A in human and mouse are associated with Congenital Fibrosis of the Extraocular Muscles Type 1 (CFEOM1) resulting from aberrant axon morphology and reduced responsiveness to inhibitory cues (van der Vaart et al., 2013; Yamada et al., 2003). On the other hand, Kif5A is required for axonal outgrowth through the transport of neurofilaments (Xia et al., 2003, p. 200). Loss of Kif5A in mouse leads to loss of large caliber axons and neurofilament accumulation in neuronal cell bodies (Brenner et al., 2018; Nicolas et al., 2018; Reid et al., 2002). Interesting, in D. melanogaster Kinesin-2 also contributes to axonal outgrowth and microtubule polarity by guiding plus-ends towards axonal outgrowths and actively excluding them from dendrites via the existing microtubule network (Mukherjee et al., 2020). The plus-end directed motion of Kinesin-2 guides growing microtubules as cargo towards existing plus ends, further elongating axons, while preferential excluding them from dendrites where microtubule polarity is minus-ends outward (Mukherjee et al., 2020). Additional kinesins have been linked to axonal outgrowth, but further analysis is warranted (see Supp. Table 1).
Axonal Transport:
Once axonal outgrowth is completed, plus-/minus-end directed transport along fully developed axons is also highly dependent upon kinesins. Several kinesins have evolved specific roles in axonal transport, but unlike Kif5a, are not required for axon outgrowth (Fig 2C). For example, Kif1A and KifC2 are required for bidirectional vesicular transport along axons: Kif1A is a neuronal-specific kinesin required for plus-end directed transport of presynaptic vesicles from the cell body to the axon, and knockout mice die shortly after birth due to motor and sensory neuron deficiencies (Hall and Hedgecock, 1991; Okada et al., n.d.; Otsuka et al., 1991; Stavoe et al., 2016; Yonekawa et al., 1998). Conversely, KifC2, a C-type kinesin in the Kinesin-14B family, is thought to contribute to minus-end directed vesicular transport from the axon back to the cell body, however validation in mice showed no obvious phenotypes as mutants were viable and fertile (Hanlon et al., 1997; Saito et al., 1997). This effect may be due to genetic compensation (see below). Rather than vesicles, Kif13A can directly bind and transport specific proteins within axons (Zhou et al., 2013). Kif13A mutant mice develop normally and are adult-viable but display high-level anxiety phenotypes caused by loss of serotonin receptor transport (Delevoye et al., 2009; Nakagawa et al., 2000; Sagona et al., 2010; Zhou et al., 2013). Finally, the Kinesin-1 family member Kif5C has been exhaustively studied in vitro for its role in intracellular cargo transport, yet Kif5C knockout mice are viable. They do, however, develop with smaller brain sizes due to an overall loss of motor neurons (Kanai et al., 2000). It is not understood how Kif5C contributes to maintenance of motor neurons through axonal transport (Kanai et al., 2000), and a more in-depth analysis is warranted.
Dendrite formation and transport:
Dendrites are the site for receiving neuronal signals. Interestingly, unlike axons, dendrites have a mixed polarity of microtubules, leading to minus ends and plus ends pointing towards and away from the cell bodies. This means that while axons are very polarized, dendrites do not have the stringent traffic patterns observed in axons. Interestingly, though we will not expand on this here, actin also has a role in dendrite formation and maintenance (Konietzny et al., 2017). Dendrites are typically not as long and large as axons, but they are microtubule based and do require kinesins for their formation and function.
Several kinesins have implicated dendritic functions: Kif3B, Kif21B and KifC2 (Fig. 2C). Interestingly, a recent report found human patients with schizophrenia associated with Kif3B mutations (Alsabban et al., 2020). This study further showed Kif3B heterozygous mice displayed schizophrenic phenotypes and abnormal dendritic spine morphology due to loss of specific NMDAR receptor transport (Alsabban et al., 2020). On the other hand, a recent study expressed human missense variants of Kif21B in mouse and found that the mice developed microcephaly and suggested a role for Kif21B in axonal transport (Asselin et al., 2020) (see below in kinesins in human diseases). However, Kif21B has also been shown to be responsible for branching of the dendritic arbor and spine formation (Supp. Table 1; (Joseph R. Marszalek et al., 1999; Muhia et al., 2016). Cell culture experiments support the necessity of KifC2 for dendritic transport, however as discussed above, KifC2 mouse mutants are viable and fertile.
Organogenesis:
The development of each organ in the vertebrate body requires fine regulation of cell division, complex organizations, and specific polarization of multiple cells within the tissue. These processes are heavily reliant on microtubules, so it is not surprising that some kinesins display specific roles in organogenesis (Fig. 3). Here, we will review these roles on an organ by organ basis.
Kidney development:
Several kinesins have associations with development of the kidney in mouse, zebrafish and human (Fig. 3C), and these fall into two separate categories: (i) the necessity for cilia in proper renal function and (ii) the general development of the tissue.
The first category is closely linked to roles in the cilium. Cilia-mediated signaling is essential for mechano-sensation of fluid flow through renal ducts, and ciliary dysfunction leads to Polycystic Kidney Disease (PKD) (Veland et al., 2009). The IFT kinesins have been linked to PKD but we will not discuss their roles here as they are described above (see ciliary kinesins section). Interestingly, Kif12 is a non-IFT kinesin that has a potential link to ciliary function in the mammalian kidney (Fig. 2B, Fig. 3C). Kif12 has been transcriptionally linked to PKD through mouse models (Gong et al., 2009). Additionally, localization of Kif12 has been found at the primary cilia in the mouse kidney though its developmental function has yet to be defined in an animal model (Gong et al., 2009; Mrug et al., 2015).
Two kinesins have discreet non-ciliary functions in kidney development (Fig. 3C). Kif26B is required for migration and polarization of the mesenchyme that surrounds the ureteric bud during development (Guillabert-Gourgues et al., 2016; Uchiyama et al., 2010). Kif26B mutant mice die 24 hours post birth due to kidney agenesis (Uchiyama et al., 2010). On the other hand, human mutations in Kif14 and analysis in developing zebrafish suggest a role for this kinesin in renal hypodysplasia stemming from improper cytokinesis (Carleton et al., 2006; Filges et al., 2014; Gruneberg et al., 2006; Makrythanasis et al., 2018; Moawia et al., 2017; Reilly et al., 2019).
Enteric nervous system (ENS) development:
While many kinesins are required for the development of the CNS in a variety of different manners (see above), one has a particular role in the development of the enteric nervous system (ENS) (Supp. Table 1, Fig. 3D). The ENS is a separate neuronal system which innervates the gut to control the digestive system (Rao and Gershon, 2016). Kif26A is an atypical kinesin in that it lacks the typical ATPase activity of the motor domain. It has been shown to negatively regulate GDNF/Ret signaling important for ENS development but has also been proposed to stably bind microtubules and regulate the length of neurites in the ENS, although this has not been shown in depth (Zhou et al., 2009). Kif26A mutant mice are born at normal mendelian ratios but die at 5 weeks of age with megacolon due to failure of neurite overgrowth in the ENS (Zhou et al., 2009). The mechanism to which Kif26A controls ENS neurite outgrowth may not be fully defined, but it offers another clear example of tissue-specific roles adopted by certain kinesins.
Female/male fertility:
Intriguingly, some kinesins show a specific role in mammalian fertility (Supp. Table 1, Fig. 3F). Among these, Kif18A is the most interesting, as it is required in both female and male fertility. Interestingly, Kif18A male and female mutant mice are born and develop normally, but are infertile (Czechanski et al., 2015) because Kif18A is specifically required for cell cycle progression of germ cells during gonad development (Czechanski et al., 2015; Liu et al., 2010; Mayr et al., 2007; Stumpff et al., 2012, 2008). How this kinesin has evolved such a specific role in gonadal development remains to be explored.
Primitive endoderm/Epiblast:
Kinesins are classically known as microtubule-based transport motors, required for transport of signals, organelles, and cargos across the cell. One kinesin is required for intracellular transport of a specific receptor during early embryonic development, Kif16B (Fig. 2D, 3A). Prior to creation of a mouse knockout, Kif16B was well-described to participate in endosome trafficking and recycling in vitro and in cell culture (Blatner et al., 2007; Hoepfner et al., 2005). However Kif16B knockout in mouse revealed that Kif16B is specifically required for endosomal trafficking of FGFR in the early embryo (Ueno et al., 2011). Upon loss of function, Kif16B mutant embryos fail to form epiblast and primitive endodermal cell lineages due to the loss of FGFR transport, leading to embryonic lethality prior to implantation (Ueno et al., 2011). This phenotype closely mimics phenotypes associated with FGFR mutant mice (Arman et al., 1998). It is the combined in vitro cell culture and in vivo work that defined the developmental role Kif16B plays in the mouse embryo.
Genetic compensation:
Genetic compensation is common in gene families with high sequence homologies (El-Brolosy et al., 2019), as is the case for kinesins. Moreover, recent studies have revealed that genetic compensation plays a significant role in dictating phenotype severity in mutant analysis in vivo (El-Brolosy et al., 2019). We suggest that more in depth investigation of genetic compensation between kinesins could provide important insights into previously undefined developmental roles, so we offer two examples here:
The kinesin-3 family contains two highly homologous kinesins, Kif13A and Kif13B (Hirokawa et al., 2009). Kif13B knockout mice develop completely normally and are viable (Kanai et al., 2014). Knockout of Kif13A alone also produces no severe developmental phenotype, although these mice do display high level anxiety phenotypes (Zhou et al., 2013). Work in cell culture suggested a role of Kif13B in controlling the structure and signaling functions of cilia (Schou et al., 2017), so it was striking that when Kif13A and Kif13B double knockout mice were generated, they displayed perinatal lethality with craniofacial abnormalities but did not display other ciliary related phenotypes such as polydactyly or exencephaly. Because cilia mediated Shh signaling is integral to craniofacial development (Schock and Brugmann, 2017), this genetic experiment suggest the possibility of a highly tissue-specific role for Kif13A/B in craniofacial ciliogenesis.
Within the Kinesin-2 family, studies in zebrafish also reveal a specific genetic compensation between Kif3B and Kif3C (Zhao et al., 2012). Kif3A can dimerize with either Kif3B or Kif3C (Muresan et al., 1998; Nonaka et al., 1998; Yamazaki et al., 1996). However, the role of Kif3C in animal development remains largely unclear. Double knockout of kif3B and kif3C in zebrafish shows a complete loss of photoreceptor and hair cell cilia that is not present in either kif3B or kif3C knockout zebrafish alone (Zhao et al., 2012). In either case, the clear genetic compensation between Kif3 proteins demonstrates that highly homologous kinesins can serve redundant roles. Many other kinesins display high levels of homology within families and could easily function redundantly during development. More in-depth genetic studies of homologous kinesins in vivo should reveal previously undetected roles in animal development.
Kinesins in human genetic disorders:
Given the diversity of kinesin function during developmental and physiology described here, it is no surprise that kinesins are also implicated in human disorders and disease, as was recently reviewed elsewhere (Kalantari and Filges, 2020). In addition to many roles in cancer progression (Rath and Kozielski, 2012), many so-called “Kinesinopathies” are effectively modeled by mouse/zebrafish developmental mutant phenotypes (Supp. Table 1). On the other hand, some kinesins display discrepancies between human disease phenotypes and those reported in animal models (Supp. Table 1), though more detailed analysis is now clarifying these discrepancies and offering insights into previously overlooked kinesin functions.
One recent example involved expression of human missense variants of Kif21B in mouse confirming human microcephaly associations (Asselin et al., 2020). In addition to humanized mutational analysis in vertebrate models, kinesin biologists should not ignore the power of mouse conditional genetics when it comes to in vivo analysis. A recent study used conditional knockout out Kif11 specifically in vascular endothelial cells to overcome early embryonic necessity, which revealed vascular defects in the retina and slower proliferation of the cerebellum, mimicking several symptoms associated with Kif11 mutations in human that were previously overlooked (Wang et al., 2020). While it remains possible that several kinesins have evolved differential functionality in humans in comparison to roles shown in other vertebrates, it is likely that more in depth in vivo studies via humanized mutational analysis or conditional genetic approaches in animal models will continue to offer insights into human disease phenotypes.
Conclusions:
Here, we have outlined the central roles of kinesins during embryonic development. These include regulating cell division and cargo transport in a variety of different tissues and/or cellular compartments, as well as in regulating the length and stability of microtubules in the same or different cell types and tissues. Within subfamilies of kinesins, we observe both redundant roles between family members, and extremely divergent roles. Some kinesin family members play specific roles in specific tissues during development (e.g. Kif6, Kif18A, Kif26B, and Kif26A), while others play multiple roles in several different tissues (Supp. Table 1, Fig. 2). How certain kinesins have evolved such specific roles in tissue development remains to be explored and will elucidate links between kinesin evolution and the evolution of their cargos, transport, and microtubule interactions. Several kinesins clearly are understudied in vivo, and demand our attention (e.g. KifC2, Kif2B, Kif24, Kif25, Kif12, Kif19B, and Kif16A; Supp. Table 1). Additionally, some kinesins have been reported in human disease and disorders yet their etiology remains only poorly defined (e.g. Kif16A, Kif16B, Kif1B, and Kif21B;Supp.Table 1, (Asselin et al., 2020).
Finally, most kinesins have been studied in some capacity with regard to in vivo analysis, but some have no observable phenotype in vertebrate models. Although it remains a possibility that they are dispensable for development, the high homology between certain members of kinesin families leads us to hypothesize that they may function redundantly. As we have outlined, several kinesins have already been subject to such analysis and have redundant roles. We suggest then a more detailed phenotypic analysis for the compensation between kinesins family members may lead to exciting and previously undefined developmental roles.
Supplementary Material
Supplementary Table 1. All 45 mammalian kinesins with reported animal phenotypes, cellular roles, alternate gene names, and citations. Each kinesin is grouped with its family (15 different families). The color coding matches to reported cellular roles, defined in Figure 2. Red is for roles in mitosis, yellow for roles in cilia, blue for roles in neurons, green for intracellular transport of proteins, mRNAs, vesicles and organelles.
Acknowledgements:
We thank E. C. Roberson for critical reading, and members of the Wallingford Laboratory for helpful suggestions. This work and MJK is supported by the Provost Graduate Excellence Fellowship from the Institute of Cell and Molecular Biology, University of Texas at Austin. RSG is supported by the NIAMS (R01AR072009). JBW is supported by the NICHD (R01HD085901) and the NHLBI (R01HL117164).
References:
- Alsabban AH, Morikawa M, Tanaka Y, Takei Y, Hirokawa N, 2020. Kinesin Kif3b mutation reduces NMDAR subunit NR2A trafficking and causes schizophrenia-like phenotypes in mice. The EMBO Journal 39, e101090. 10.15252/embj.2018101090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio C, Ferby I, Wilhelm H, Jones M, Karsenti E, Nebreda AR, Vernos I, 2000. Xkid, a Chromokinesin Required for Chromosome Alignment on the Metaphase Plate. Cell 102, 425–435. 10.1016/S0092-8674(00)00048-9 [DOI] [PubMed] [Google Scholar]
- Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P, 1998. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. PNAS 95, 5082–5087. 10.1073/pnas.95.9.5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin L, Rivera Alvarez J, Heide S, Bonnet CS, Tilly P, Vitet H, Weber C, Bacino CA, Baranaño K, Chassevent A, Dameron A, Faivre L, Hanchard NA, Mahida S, McWalter K, Mignot C, Nava C, Rastetter A, Streff H, Thauvin-Robinet C, Weiss MM, Zapata G, Zwijnenburg PJG, Saudou F, Depienne C, Golzio C, Héron D, Godin JD, 2020. Mutations in the KIF21B kinesin gene cause neurodevelopmental disorders through imbalanced canonical motor activity. Nature Communications 11, 2441. 10.1038/s41467-020-16294-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs F, Anderson KV, 2017. Primary Cilia and Mammalian Hedgehog Signaling. Cold Spring Harb Perspect Biol 9, a028175. 10.1101/cshperspect.a028175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein M, Beech PL, Katz SG, Rosenbaum JL, 1994. A new kinesin-like protein (Klp1) localized to a single microtubule of the Chlamydomonas flagellum. J Cell Biol 125, 1313–1326. 10.1083/jcb.125.6.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betley JN, Heinrich B, Vernos I, Sardet C, Prodon F, Deshler JO, 2004. Kinesin II Mediates Vg1 mRNA Transport in Xenopus Oocytes. Current Biology 14, 219–224. 10.1016/j.cub.2004.01.028 [DOI] [PubMed] [Google Scholar]
- Blatner NR, Wilson MI, Lei C, Hong W, Murray D, Williams RL, Cho W, 2007. The structural basis of novel endosome anchoring activity of KIF16B kinesin. The EMBO Journal 26, 3709–3719. 10.1038/sj.emboj.7601800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza RP, Serbus LR, Duffy JB, Saxton WM, 2000. A Function for Kinesin I in the Posterior Transport of oskar mRNA and Staufen Protein. Science 289, 2120–2122. 10.1126/science.289.5487.2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza RP, Serbus LR, Saxton WM, Duffy JB, 2002. Posterior Localization of Dynein and Dorsal-Ventral Axis Formation Depend on Kinesin in Drosophila Oocytes. Current Biology 12, 1541–1545. 10.1016/S0960-9822(02)01108-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D, Yilmaz R, Müller K, Grehl T, Petri S, Meyer T, Grosskreutz J, Weydt P, Ruf W, Neuwirth C, Weber M, Pinto S, Claeys KG, Schrank B, Jordan B, Knehr A, Günther K, Hübers A, Zeller D, Kubisch C, Jablonka S, Sendtner M, Klopstock T, de Carvalho M, Sperfeld A, Borck G, Volk AE, Dorst J, Weis J, Otto M, Schuster J, Del Tredici K, Braak H, Danzer KM, Freischmidt A, Meitinger T, Strom TM, Ludolph AC, Andersen PM, Weishaupt JH, Weyen U, Hermann A, Hagenacker T, Koch JC, Lingor P, Göricke B, Zierz S, Baum P, Wolf J, Winkler A, Young P, Bogdahn U, Prudlo J, Kassubek J, 2018. Hot-spot KIF5A mutations cause familial ALS. Brain 141, 688–697. 10.1093/brain/awx370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broix L, Asselin L, Silva CG, Ivanova EL, Tilly P, Gilet JG, Lebrun N, Jagline H, Muraca G, Saillour Y, Drouot N, Reilly ML, Francis F, Benmerah A, Bahi-Buisson N, Belvindrah R, Nguyen L, Godin JD, Chelly J, Hinckelmann M-V, 2018. Ciliogenesis and cell cycle alterations contribute to KIF2A-related malformations of cortical development. Hum Mol Genet 27, 224–238. 10.1093/hmg/ddx384 [DOI] [PubMed] [Google Scholar]
- Brooks ER, Wallingford JB, 2014. Multiciliated Cells. Current Biology 24, R973–R982. 10.1016/j.cub.2014.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KD, Wood KW, Cleveland DW, 1996. The kinesin-like protein CENP-E is kinetochore-associated throughout poleward chromosome segregation during anaphase-A. Journal of Cell Science 109, 961–969. [DOI] [PubMed] [Google Scholar]
- Buchan JG, Gray RS, Gansner JM, Alvarado DM, Burgert L, Gitlin JD, Gurnett CA, Goldsmith MI, 2014. Kinesin family member 6 (kif6) is necessary for spine development in zebrafish. Developmental Dynamics 243, 1646–1657. 10.1002/dvdy.24208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PD, Heim AE, Smith MZ, Marlow FL, 2015. Kinesin-1 interacts with Bucky ball to form germ cells and is required to pattern the zebrafish body axis. Development 142, 2996–3008. 10.1242/dev.124586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton M, Mao M, Biery M, Warrener P, Kim S, Buser C, Marshall CG, Fernandes C, Annis J, Linsley PS, 2006. RNA Interference-Mediated Silencing of Mitotic Kinesin KIF14 Disrupts Cell Cycle Progression and Induces Cytokinesis Failure. Molecular and Cellular Biology 26, 3853–3863. 10.1128/MCB.26.10.3853-3863.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Inaba M, Venkei ZG, Yamashita YM, 2016. Klp10A, a stem cell centrosome-enriched kinesin, balances asymmetries in Drosophila male germline stem cell division. eLife 5, e20977. 10.7554/eLife.20977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HO-L, Zhang X, Ribeiro A, Mo R, Makino S, Puviindran V, Law KKL, Briscoe J, Hui C, 2009. The Kinesin Protein Kif7 Is a Critical Regulator of Gli Transcription Factors in Mammalian Hedgehog Signaling. Sci. Signal. 2, ra29–ra29. 10.1126/scisignal.2000405 [DOI] [PubMed] [Google Scholar]
- Choksi SP, Babu D, Lau D, Yu X, Roy S, 2014a. Systematic discovery of novel ciliary genes through functional genomics in the zebrafish. Development 141, 3410. 10.1242/dev.108209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi SP, Lauter G, Swoboda P, Roy S, 2014b. Switching on cilia: transcriptional networks regulating ciliogenesis. Development 141, 1427–1441. 10.1242/dev.074666 [DOI] [PubMed] [Google Scholar]
- Clark I, Giniger E, Ruohola-Baker H, Jan LY, Jan YN, 1994. Transient posterior localization of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Current Biology 4, 289–300. 10.1016/S0960-9822(00)00068-3 [DOI] [PubMed] [Google Scholar]
- Czechanski A, Kim H, Byers C, Greenstein I, Stumpff J, Reinholdt LG, 2015. Kif18a is specifically required for mitotic progression during germ line development. Developmental Biology 402, 253–262. 10.1016/j.ydbio.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye C, Hurbain I, Tenza D, Sibarita J-B, Uzan-Gafsou S, Ohno H, Geerts WJC, Verkleij AJ, Salamero J, Marks MS, Raposo G, 2009. AP-1 and KIF13A coordinate endosomal sorting and positioning during melanosome biogenesis. J Cell Biol 187, 247–264. 10.1083/jcb.200907122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Gupton SL, Gertler FB, 2011. The Growth Cone Cytoskeleton in Axon Outgrowth and Guidance. Cold Spring Harb Perspect Biol 3, a001800. 10.1101/cshperspect.a001800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Günther S, Fukuda N, Kikhi K, Boezio GLM, Takacs CM, Lai S-L, Fukuda R, Gerri C, Giraldez AJ, Stainier DYR, 2019. Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197. 10.1038/s41586-019-1064-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinson RP, Rowning B, 1988. A transient array of parallel microtubules in frog eggs: Potential tracks for a cytoplasmic rotation that specifies the dorso-ventral axis. Developmental Biology 128, 185–197. 10.1016/0012-1606(88)90281-3 [DOI] [PubMed] [Google Scholar]
- Endow SA, Kull FJ, Liu H, 2010. Kinesins at a glance. J Cell Sci 123, 3420–3424. 10.1242/jcs.064113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filges I, Nosova E, Bruder E, Tercanli S, Townsend K, Gibson WT, Röthlisberger B, Heinimann K, Hall JG, Gregory-Evans CY, Wasserman WW, Miny P, Friedman JM, 2014. Exome sequencing identifies mutations in KIF14 as a novel cause of an autosomal recessive lethal fetal ciliopathy phenotype. Clinical Genetics 86, 220–228. 10.1111/cge.12301 [DOI] [PubMed] [Google Scholar]
- Geng A, Qiu R, Murai K, Liu J, Wu X, Zhang H, Farhoodi H, Duong N, Jiang M, Yee J, Tsark W, Lu Q, 2018. KIF20A/MKLP2 regulates the division modes of neural progenitor cells during cortical development. Nature Communications 9, 2707. 10.1038/s41467-018-05152-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart J, Doniach T, Stewart R, 1991. Organizing the Xenopus Organizer, in: Keller R, Clark WH, Griffin F (Eds.), Gastrulation: Movements, Patterns and Molecules, Bodega Marine Laboratory Marine Science Series. Springer US, Boston, MA, pp. 57–77. 10.1007/978-1-4684-6027-8_4 [DOI] [Google Scholar]
- Gong Y, Ma Z, Patel V, Fischer E, Hiesberger T, Pontoglio M, Igarashi P, 2009. HNF-1β Regulates Transcription of the PKD Modifier Gene Kif12. JASN 20, 41–47. 10.1681/ASN.2008020238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U, Neef R, Li X, Chan EHY, Chalamalasetty RB, Nigg EA, Barr FA, 2006. KIF14 and citron kinase act together to promote efficient cytokinesis. J Cell Biol 172, 363–372. 10.1083/jcb.200511061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillabert-Gourgues A, Jaspard-Vinassa B, Bats M-L, Sewduth RN, Franzl N, Peghaire C, Jeanningros S, Moreau C, Roux E, Larrieu-Lahargue F, Dufourcq P, Couffinhal T, Duplàa C, 2016. Kif26b controls endothelial cell polarity through the Dishevelled/Daam1-dependent planar cell polarity–signaling pathway. MBoC 27, 941–953. 10.1091/mbc.E14-08-1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH, Hedgecock EM, 1991. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell 65, 837–847. 10.1016/0092-8674(91)90391-B [DOI] [PubMed] [Google Scholar]
- Hanlon DW, Yang Z, Goldstein LSB, 1997. Characterization of KIFC2, a Neuronal Kinesin Superfamily Member in Mouse. Neuron 18, 439–451. 10.1016/S0896-6273(00)81244-1 [DOI] [PubMed] [Google Scholar]
- He M, Subramanian R, Bangs F, Omelchenko T, Liem KF Jr, Kapoor TM, Anderson KV, 2014. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nature Cell Biology 16, 663–672. 10.1038/ncb2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS, 1993. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol 123, 665–679. 10.1083/jcb.123.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y, 2010. Molecular Motors in Neurons: Transport Mechanisms and Roles in Brain Function, Development, and Disease. Neuron 68, 610–638. 10.1016/j.neuron.2010.09.039 [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, 2008. Intracellular Transport and Kinesin Superfamily Proteins, KIFs: Structure, Function, and Dynamics. Physiological Reviews 88, 1089–1118. 10.1152/physrev.00023.2007 [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S, 2009. Kinesin superfamily motor proteins and intracellular transport. Nature Reviews Molecular Cell Biology 10, 682. [DOI] [PubMed] [Google Scholar]
- Hoepfner S, Severin F, Cabezas A, Habermann B, Runge A, Gillooly D, Stenmark H, Zerial M, 2005. Modulation of Receptor Recycling and Degradation by the Endosomal Kinesin KIF16B. Cell 121, 437–450. 10.1016/j.cell.2005.02.017 [DOI] [PubMed] [Google Scholar]
- Homma N, Takei Y, Tanaka Y, Nakata T, Terada S, Kikkawa M, Noda Y, Hirokawa N, 2003. Kinesin Superfamily Protein 2A (KIF2A) Functions in Suppression of Collateral Branch Extension. Cell 114, 229–239. 10.1016/S0092-8674(03)00522-1 [DOI] [PubMed] [Google Scholar]
- Hou Y, Witman GB, 2015. Dynein and Intraflagellar Transport. Exp Cell Res 334, 26–34. 10.1016/j.yexcr.2015.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insinna C, Pathak N, Perkins B, Drummond I, Besharse JC, 2008. The homodimeric kinesin, Kif17, is essential for vertebrate photoreceptor sensory outer segment development. Dev. Biol. 316, 160–170. 10.1016/j.ydbio.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet BV, Salinas-Mondragon R, Liang H, Therit B, Buie JD, Dykstra M, Campbell K, Ostrowski LE, Brody SL, Ghashghaei HT, 2009. FoxJ1-dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development 136, 4021. 10.1242/dev.041129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janisch KM, McNeely KC, Dardick JM, Lim SH, Dwyer ND, 2018. Kinesin-6 KIF20B is required for efficient cytokinetic furrowing and timely abscission in human cells. MBoC 29, 166–179. 10.1091/mbc.E17-08-0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janisch KM, Vock VM, Fleming MS, Shrestha A, Grimsley-Myers CM, Rasoul BA, Neale SA, Cupp TD, Kinchen JM, Liem KF, Dwyer ND, 2013. The vertebrate-specific Kinesin-6, Kif20b, is required for normal cytokinesis of polarized cortical stem cells and cerebral cortex size. Development 140, 4672–4682. 10.1242/dev.093286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Tam BM, Ying G, Wu S, Hauswirth WW, Frederick JM, Moritz OL, Baehr W, 2015. Kinesin family 17 (osmotic avoidance abnormal-3) is dispensable for photoreceptor morphology and function. The FASEB Journal 29, 4866–4880. 10.1096/fj.15-275677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantari S, Filges I, 2020. ‘Kinesinopathies’: emerging role of the kinesin family member genes in birth defects. Journal of Medical Genetics. 10.1136/jmedgenet-2019-106769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Okada Y, Tanaka Y, Harada A, Terada S, Hirokawa N, 2000. KIF5C, a Novel Neuronal Kinesin Enriched in Motor Neurons. J. Neurosci. 20, 6374–6384. 10.1523/JNEUROSCI.20-17-06374.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Wang D, Hirokawa N, 2014. KIF13B enhances the endocytosis of LRP1 by recruiting LRP1 to caveolae. J Cell Biol 204, 395–408. 10.1083/jcb.201309066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lee K, Choi J-H, Ringstad N, Dynlacht BD, 2015. Nek2 activation of Kif24 ensures cilium disassembly during the cell cycle. Nature Communications 6, 8087. 10.1038/ncomms9087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Tsang WY, Li J, Lane W, Dynlacht BD, 2011. Centriolar Kinesin Kif24 Interacts with CP110 to Remodel Microtubules and Regulate Ciliogenesis. Cell 145, 914–925. 10.1016/j.cell.2011.04.028 [DOI] [PubMed] [Google Scholar]
- Konietzny A, Bär J, Mikhaylova M, 2017. Dendritic Actin Cytoskeleton: Structure, Functions, and Regulations. Front. Cell. Neurosci. 11. 10.3389/fncel.2017.00147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konjikusic MJ, Yeetong P, Boswell CW, Lee C, Roberson EC, Ittiwut R, Suphapeetiporn K, Ciruna B, Gurnett CA, Wallingford JB, Shotelersuk V, Gray RS, 2018. Mutations in Kinesin family member 6 reveal specific role in ependymal cell ciliogenesis and human neurological development. PLOS Genetics 14, e1007817. 10.1371/journal.pgen.1007817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LSB, Goodson HV, Hirokawa N, Howard J, Malmberg RL, McIntosh JR, Miki H, Mitchison TJ, Okada Y, Reddy ASN, Saxton WM, Schliwa M, Scholey JM, Vale RD, Walczak CE, Wordeman L, 2004. A standardized kinesin nomenclature. J Cell Biol 167, 19–22. 10.1083/jcb.200408113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, 2015. IFT–Cargo Interactions and Protein Transport in Cilia. Trends in Biochemical Sciences 40, 765–778. 10.1016/j.tibs.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque AA, Compton DA, 2001. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J Cell Biol 154, 1135–1146. 10.1083/jcb.200106093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem KF, He M, Ocbina PJR, Anderson KV, 2009. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 13377–13382. 10.1073/pnas.0906944106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little RB, Norris DP, 2020. Right, left and cilia: How asymmetry is established. Seminars in Cell & Developmental Biology. 10.1016/j.semcdb.2020.06.003 [DOI] [PubMed] [Google Scholar]
- Liu X, Zhao X, Wang X, Yao Y, Zhang L, Shu R, Ren W, Huang Y, Huang L, Gu M, Kuang Y, Wang L, Lu S, Chi J, Fen J, Wang Y, Fei J, Dai W, Wang Z-G, 2010. Germinal Cell Aplasia in Kif18a Mutant Male Mice Due to Impaired Chromosome Congression and Dysregulated BubR1 and CENP-E. Genes Cancer 1, 26–39. 10.1177/1947601909358184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Lakonishok M, Liu R, Billington N, Rich A, Glotzer M, Sellers JR, Gelfand VI, 2020. Competition between kinesin-1 and myosin-V defines Drosophila posterior determination. eLife 9, e54216. 10.7554/eLife.54216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Winding M, Lakonishok M, Wildonger J, Gelfand VI, 2016. Microtubule–microtubule sliding by kinesin-1 is essential for normal cytoplasmic streaming in Drosophila oocytes. PNAS 113, E4995–E5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrythanasis P, Maroofian R, Stray-Pedersen A, Musaev D, Zaki MS, Mahmoud IG, Selim L, Elbadawy A, Jhangiani SN, Coban Akdemir ZH, Gambin T, Sorte HS, Heiberg A, McEvoy-Venneri J, James KN, Stanley V, Belandres D, Guipponi M, Santoni FA, Ahangari N, Tara F, Doosti M, Iwaszkiewicz J, Zoete V, Backe PH, Hamamy H, Gleeson JG, Lupski JR, Karimiani EG, Antonarakis SE, 2018. Biallelic variants in KIF14 cause intellectual disability with microcephaly. European Journal of Human Genetics 26, 330–339. 10.1038/s41431-017-0088-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS, 1999. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc. Natl. Acad. Sci. U.S.A. 96, 5043–5048. 10.1073/pnas.96.9.5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek Joseph R., Weiner JA, Farlow SJ, Chun J, Goldstein LSB, 1999. Novel Dendritic Kinesin Sorting Identified by Different Process Targeting of Two Related Kinesins: KIF21A and KIF21B. J Cell Biol 145, 469–479. 10.1083/jcb.145.3.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MI, Hümmer S, Bormann J, Grüner T, Adio S, Woehlke G, Mayer TU, 2007. The Human Kinesin Kif18A Is a Motile Microtubule Depolymerase Essential for Chromosome Congression. Current Biology 17, 488–498. 10.1016/j.cub.2007.02.036 [DOI] [PubMed] [Google Scholar]
- Métivier M, Monroy BY, Gallaud E, Caous R, Pascal A, Richard-Parpaillon L, Guichet A, Ori-McKenney KM, Giet R, 2019. Dual control of Kinesin-1 recruitment to microtubules by Ensconsin in Drosophila neuroblasts and oocytes. Development 146. 10.1242/dev.171579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Setou M, Kaneshiro K, Hirokawa N, 2001. All kinesin superfamily protein, KIF, genes in mouse and human. PNAS 98, 7004–7011. 10.1073/pnas.111145398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milic B, Andreasson JOL, Hogan DW, Block SM, 2017. Intraflagellar transport velocity is governed by the number of active KIF17 and KIF3AB motors and their motility properties under load. Proc Natl Acad Sci U S A 114, E6830–E6838. 10.1073/pnas.1708157114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Rowning BA, Larabell CA, Yang-Snyder JA, Bates RL, Moon RT, 1999. Establishment of the Dorsal–Ventral Axis inXenopus Embryos Coincides with the Dorsal Enrichment of Dishevelled That Is Dependent on Cortical Rotation. J Cell Biol 146, 427–438. 10.1083/jcb.146.2.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Hosoba K, Ochiai H, Royba E, Izumi H, Sakuma T, Yamamoto T, Dynlacht BD, Matsuura S, 2015. The Microtubule-Depolymerizing Activity of a Mitotic Kinesin Protein KIF2A Drives Primary Cilia Disassembly Coupled with Cell Proliferation. Cell Reports 10, 664–673. 10.1016/j.celrep.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata H, Shimada K, Morohoshi A, Oura S, Matsumura T, Xu Z, Oyama Y, Ikawa M, 2020. Testis-enriched kinesin KIF9 is important for progressive motility in mouse spermatozoa. The FASEB Journal 34, 5389–5400. 10.1096/fj.201902755R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moawia A, Shaheen R, Rasool S, Waseem SS, Ewida N, Budde B, Kawalia A, Motameny S, Khan K, Fatima A, Jameel M, Ullah F, Akram T, Ali Z, Abdullah U, Irshad S, Höhne W, Noegel AA, Al-Owain M, Hörtnagel K, Stöbe P, Baig SM, Nürnberg P, Alkuraya FS, Hahn A, Hussain MS, 2017. Mutations of KIF14 cause primary microcephaly by impairing cytokinesis. Annals of Neurology 82, 562–577. 10.1002/ana.25044 [DOI] [PubMed] [Google Scholar]
- Mrug M, Zhou J, Yang C, Aronow BJ, Cui X, Schoeb TR, Siegal GP, Yoder BK, Guay-Woodford LM, 2015. Genetic and Informatic Analyses Implicate Kif12 as a Candidate Gene within the Mpkd2 Locus That Modulates Renal Cystic Disease Severity in the Cys1cpk Mouse. PLOS ONE 10, e0135678. 10.1371/journal.pone.0135678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhia M, Thies E, Labonté D, Ghiretti AE, Gromova KV, Xompero F, Lappe-Siefke C, Hermans-Borgmeyer I, Kuhl D, Schweizer M, Ohana O, Schwarz JR, Holzbaur ELF, Kneussel M, 2016. The Kinesin KIF21B Regulates Microtubule Dynamics and Is Essential for Neuronal Morphology, Synapse Function, and Learning and Memory. Cell Reports 15, 968–977. 10.1016/j.celrep.2016.03.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Brooks PS, Bernard F, Guichet A, Conduit PT, 2020. Microtubules originate asymmetrically at the somatic golgi and are guided via Kinesin2 to maintain polarity within neurons. eLife 9, e58943. 10.7554/eLife.58943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan V, Abramson T, Lyass A, Winter D, Porro E, Hong F, Chamberlin NL, Schnapp BJ, 1998. KIF3C and KIF3A Form a Novel Neuronal Heteromeric Kinesin That Associates with Membrane Vesicles. MBoC 9, 637–652. 10.1091/mbc.9.3.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Setou M, Seog D-H, Ogasawara K, Dohmae N, Takio K, Hirokawa N, 2000. A Novel Motor, KIF13A, Transports Mannose-6-Phosphate Receptor to Plasma Membrane through Direct Interaction with AP-1 Complex. Cell 103, 569–581. 10.1016/S0092-8674(00)00161-6 [DOI] [PubMed] [Google Scholar]
- Nicolas A, Kenna KP, Renton AE, Ticozzi N, Faghri F, Chia R, Dominov JA, Kenna BJ, Nalls MA, Keagle P, Rivera AM, van Rheenen W, Murphy NA, van Vugt JJFA, Geiger JT, Van der Spek RA, Pliner HA, Shankaracharya, Smith BN, Marangi G, Topp SD, Abramzon Y, Gkazi AS, Eicher JD, Kenna A, Logullo FO, Simone I, Logroscino G, Salvi F, Bartolomei I, Borghero G, Murru MR, Costantino E, Pani C, Puddu R, Caredda C, Piras V, Tranquilli S, Cuccu S, Corongiu D, Melis M, Milia A, Marrosu F, Marrosu MG, Floris G, Cannas A, Tranquilli S, Capasso M, Caponnetto C, Mancardi G, Origone P, Mandich P, Conforti FL, Cavallaro S, Mora G, Marinou K, Sideri R, Penco S, Mosca L, Lunetta C, Pinter GL, Corbo M, Riva N, Carrera P, Volanti P, Mandrioli J, Fini N, Fasano A, Tremolizzo L, Arosio A, Ferrarese C, Trojsi F, Tedeschi G, Monsurrò MR, Piccirillo G, Femiano C, Ticca A, Ortu E, La Bella V, Spataro R, Colletti T, Sabatelli M, Zollino M, Conte A, Luigetti M, Lattante S, Marangi G, Santarelli M, Petrucci A, Pugliatti M, Pirisi A, Parish LD, Occhineri P, Giannini F, Battistini S, Ricci C, Benigni M, Cau TB, Loi D, Calvo A, Moglia C, Brunetti M, Barberis M, Restagno G, Casale F, Marrali G, Fuda G, Ossola I, Cammarosano S, Canosa A, Ilardi A, Manera U, Grassano M, Tanel R, Pisano F, Mora G, Calvo A, Mazzini L, Riva N, Mandrioli J, Caponnetto C, Battistini S, Volanti P, La Bella V, Conforti FL, Borghero G, Messina S, Simone IL, Trojsi F, Salvi F, Logullo FO, D’Alfonso S, Corrado L, Capasso M, Ferrucci L, Harms MB, Goldstein DB, Shneider NA, Goutman S, Simmons Z, Miller TM, Chandran S, Pal S, Manousakis G, Appel SH, Simpson E, Wang L, Baloh RH, Gibson S, Bedlack R, Lacomis D, Sareen D, Sherman A, Bruijn L, Penny M, Moreno C de AM, Kamalakaran S, Goldstein DB, Allen AS, Appel S, Baloh RH, Bedlack RS, Boone BE, Brown R, Carulli JP, Chesi A, Chung WK, Cirulli ET, Cooper GM, Couthouis J, Day-Williams AG, Dion PA, Gibson S, Gitler AD, Glass JD, Goldstein DB, Han Y, Harms MB, Harris T, Hayes SD, Jones AL, Keebler J, Krueger BJ, Lasseigne BN, Levy SE, Lu Y-F, Maniatis T, McKenna-Yasek D, Miller TM, Myers RM, Petrovski S, Pulst SM, Raphael AR, Ravits JM, Ren Z, Rouleau GA, Sapp PC, Shneider NA, Simpson E, Sims KB, Staropoli JF, Waite LL, Wang Q, Wimbish JR, Xin WW, Gitler AD, Harris T, Myers RM, Phatnani H, Kwan J, Sareen D, Broach JR, Simmons Z, Arcila-Londono X, Lee EB, Van Deerlin VM, Shneider NA, Fraenkel E, Ostrow LW, Baas F, Zaitlen N, Berry JD, Malaspina A, Fratta P, Cox GA, Thompson LM, Finkbeiner Steve, Dardiotis E, Miller TM, Chandran S, Pal S, Hornstein E, MacGowan DJ, Heiman-Patterson T, Hammell MG, Patsopoulos NA, Dubnau J, Nath A, Phatnani H, Musunuri RL, Evani US, Abhyankar A, Zody MC, Kaye J, Finkbeiner Steven, Wyman S, LeNail Alexander, Lima L, Fraenkel E, Rothstein JD, Svendsen CN, Thompson LM, Van Eyk J, Maragakis NJ, Berry JD, Glass JD, Miller TM, Kolb SJ, Baloh RH, Cudkowicz M, Baxi E, Kaye J, Finkbeiner Steven, Wyman SK, LeNail Alex, Lima L, Fraenkel E, Svendsen CN, Thompson LM, Van Eyk JE, Berry JD, Miller TM, Kolb SJ, Cudkowicz M, Baxi E, Benatar M, Taylor JP, Wu G, Rampersaud E, Wuu J, Rademakers R, Züchner S, Schule R, McCauley J, Hussain S, Cooley A, Wallace M, Clayman C, Barohn R, Statland J, Ravits J, Swenson A, Jackson C, Trivedi J, Khan S, Katz J, Jenkins L, Burns T, Gwathmey K, Caress J, McMillan C, Elman L, Pioro E, Heckmann J, So Y, Walk D, Maiser S, Zhang J, Benatar M, Taylor JP, Rampersaud E, Wu G, Wuu J, Silani V, Ticozzi N, Gellera C, Ratti A, Taroni F, Lauria G, Verde F, Fogh I, Tiloca C, Comi GP, Sorarù G, Cereda C, D’Alfonso S, Corrado L, De Marchi F, Corti S, Ceroni M, Mazzini L, Siciliano G, Filosto M, Inghilleri M, Peverelli S, Colombrita C, Poletti B, Maderna L, Del Bo R, Gagliardi S, Querin G, Bertolin C, Pensato V, Castellotti B, Lauria G, Verde F, Fogh I, Tiloca C, Comi GP, Sorarù G, Cereda C, Camu W, Mouzat K, Lumbroso S, Corcia P, Meininger V, Besson G, Lagrange E, Clavelou P, Guy N, Couratier P, Vourch P, Danel V, Bernard E, Lemasson G, Corcia P, Laaksovirta H, Myllykangas L, Jansson L, Valori M, Ealing J, Hamdalla H, Rollinson S, Pickering-Brown S, Orrell RW, Sidle KC, Malaspina A, Hardy J, Singleton AB, Johnson JO, Arepalli S, Sapp PC, McKenna-Yasek D, Polak M, Asress S, Al-Sarraj S, King A, Troakes C, Vance C, de Belleroche J, Baas F, ten Asbroek ALMA, Muñoz-Blanco JL, Hernandez DG, Ding J, Gibbs JR, Scholz SW, Floeter MK, Campbell RH, Landi F, Bowser R, Pulst SM, Ravits JM, MacGowan DJL, Kirby J, Pioro EP, Pamphlett R, Broach J, Gerhard G, Dunckley TL, Brady CB, Kowall NW, Troncoso JC, Le Ber I, Mouzat K, Lumbroso S, Heiman-Patterson TD, Kamel F, Van Den Bosch L, Baloh RH, Strom TM, Meitinger T, Shatunov A, Van Eijk KR, de Carvalho M, Kooyman M, Middelkoop B, Moisse Matthieu, McLaughlin RL, Van Es MA, Weber M, Boylan KB, Van Blitterswijk M, Rademakers R, Morrison KE, Basak AN, Mora JS, Drory VE, Shaw PJ, Turner MR, Talbot K, Hardiman O, Williams KL, Fifita JA, Nicholson GA, Blair IP, Rouleau GA, Esteban-Pérez J, García-Redondo A, Al-Chalabi A, Al Kheifat A, Al-Chalabi A, Andersen P, Basak AN, Blair IP, Chio A, Cooper-Knock Jonathan, Corcia P, Couratier P, de Carvalho M, Dekker A, Drory V, Redondo AG, Gotkine M, Hardiman O, Hide W, Iacoangeli A, Glass J, Kenna K, Kiernan M, Kooyman M, Landers J, McLaughlin R, Middelkoop B, Mill J, Neto MM, Moisse Mattieu, Pardina JM, Morrison K, Newhouse S, Pinto S, Pulit S, Robberecht W, Shatunov A, Shaw P, Shaw C, Silani V, Sproviero W, Tazelaar G, Ticozzi N, van Damme P, van den Berg L, van der Spek R, van Eijk K, van Es M, van Rheenen W, van Vugt J, Veldink J, Weber M, Williams KL, Zatz M, Bauer DC, Twine NA, Rogaeva E, Zinman L, Ostrow LW, Maragakis NJ, Rothstein JD, Simmons Z, Cooper-Knock Johnathan, Brice A, Goutman SA, Feldman EL, Gibson SB, Taroni F, Ratti A, Gellera C, Van Damme P, Robberecht W, Fratta P, Sabatelli M, Lunetta C, Ludolph AC, Andersen PM, Weishaupt JH, Camu W, Trojanowski JQ, Van Deerlin VM, Brown RH, van den Berg LH, Veldink JH, Harms MB, Glass JD, Stone DJ, Tienari P, Silani V, Chiò A, Shaw CE, Traynor BJ, Landers JE, 2018. Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron 97, 1268–1283.e6. 10.1016/j.neuron.2018.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa S, Nakajima K, Miki H, Minato Y, Wang D, Hirokawa N, 2012. KIF19A Is a Microtubule-Depolymerizing Kinesin for Ciliary Length Control. Developmental Cell 23, 1167–1175. 10.1016/j.devcel.2012.10.016 [DOI] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N, 1998. Randomization of Left–Right Asymmetry due to Loss of Nodal Cilia Generating Leftward Flow of Extraembryonic Fluid in Mice Lacking KIF3B Motor Protein. Cell 95, 829–837. 10.1016/S0092-8674(00)81705-5 [DOI] [PubMed] [Google Scholar]
- Ohsugi M, Adachi K, Horai R, Kakuta S, Sudo K, Kotaki H, Tokai-Nishizumi N, Sagara H, Iwakura Y, Yamamoto T, 2008. Kid-Mediated Chromosome Compaction Ensures Proper Nuclear Envelope Formation. Cell 132, 771–782. 10.1016/j.cell.2008.01.029 [DOI] [PubMed] [Google Scholar]
- Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N, n.d. The Neuron-Specific Kinesin Superfamily Protein KIFIA Is a Unique Monomeric Motor for Anterograde Axonal Transport of Synaptic Vesicle Precursors 12. [DOI] [PubMed] [Google Scholar]
- Olson DJ, Oh D, Houston DW, 2015. The dynamics of plus end polarization and microtubule assembly during Xenopus cortical rotation. Developmental Biology 401, 249–263. 10.1016/j.ydbio.2015.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka AJ, Jeyaprakash A, García-Añoveros J, Tang LZ, Fisk G, Hartshorne T, Franco R, Bornt T, 1991. The C. elegans unc-104 4 gene encodes a putative kinesin heavy chain-like protein. Neuron 6, 113–122. 10.1016/0896-6273(91)90126-K [DOI] [PubMed] [Google Scholar]
- Prevo B, Scholey JM, Peterman EJG, 2017. Intraflagellar transport: mechanisms of motor action, cooperation, and cargo delivery. The FEBS Journal 284, 2905–2931. 10.1111/febs.14068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putkey FR, Cramer T, Morphew MK, Silk AD, Johnson RS, McIntosh JR, Cleveland DW, 2002. Unstable Kinetochore-Microtubule Capture and Chromosomal Instability Following Deletion of CENP-E. Developmental Cell 3, 351–365. 10.1016/S1534-5807(02)00255-1 [DOI] [PubMed] [Google Scholar]
- Radford SJ, Harrison AM, McKim KS, 2012. Microtubule-Depolymerizing Kinesin KLP10A Restricts the Length of the Acentrosomal Meiotic Spindle in Drosophila Females. Genetics 192, 431–440. 10.1534/genetics.112.143503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M, Gershon MD, 2016. The bowel and beyond: the enteric nervous system in neurological disorders. Nature Reviews Gastroenterology & Hepatology 13, 517–528. 10.1038/nrgastro.2016.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath O, Kozielski F, 2012. Kinesins and cancer. Nature Reviews Cancer 12, 527–539. 10.1038/nrc3310 [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Redwine WB, Vale RD, Carter AP, 2018. The cytoplasmic dynein transport machinery and its many cargoes. Nature Reviews Molecular Cell Biology 19, 382–398. 10.1038/s41580-018-0004-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E, Kloos M, Ashley-Koch A, Hughes L, Bevan S, Svenson IK, Graham FL, Gaskell PC, Dearlove A, Pericak-Vance MA, Rubinsztein DC, Marchuk DA, 2002. A Kinesin Heavy Chain (KIF5A) Mutation in Hereditary Spastic Paraplegia (SPG10). The American Journal of Human Genetics 71, 1189–1194. 10.1086/344210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly ML, Benmerah A, 2019. Ciliary kinesins beyond IFT: Cilium length, disassembly, cargo transport and signalling. Biology of the Cell 111, 79–94. 10.1111/boc.201800074 [DOI] [PubMed] [Google Scholar]
- Reilly ML, Stokman MF, Magry V, Jeanpierre C, Alves M, Paydar M, Hellinga J, Delous M, Pouly D, Failler M, Martinovic J, Loeuillet L, Leroy B, Tantau J, Roume J, Gregory-Evans CY, Shan X, Filges I, Allingham JS, Kwok BH, Saunier S, Giles RH, Benmerah A, 2019. Loss-of-function mutations in KIF14 cause severe microcephaly and kidney development defects in humans and zebrafish. Hum Mol Genet 28, 778–795. 10.1093/hmg/ddy381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagona AP, Nezis IP, Pedersen NM, Liestøl K, Poulton J, Rusten TE, Skotheim RI, Raiborg C, Stenmark H, 2010. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nature Cell Biology 12, 362–371. 10.1038/ncb2036 [DOI] [PubMed] [Google Scholar]
- Saito N, Okada Y, Noda Y, Kinoshita Y, Kondo S, Hirokawa N, 1997. KIFC2 Is a Novel Neuron-Specific C-Terminal Type Kinesin Superfamily Motor for Dendritic Transport of Multivesicular Body-Like Organelles. Neuron 18, 425–438. 10.1016/S0896-6273(00)81243-X [DOI] [PubMed] [Google Scholar]
- Sawin KE, LeGuellec K, Philippe M, Mitchison TJ, 1992. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 359, 540–543. 10.1038/359540a0 [DOI] [PubMed] [Google Scholar]
- Schock EN, Brugmann SA, 2017. Discovery, Diagnosis, and Etiology of Craniofacial Ciliopathies. Cold Spring Harb Perspect Biol 9, a028258. 10.1101/cshperspect.a028258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM, 2008. Intraflagellar transport motors in cilia: moving along the cell’s antenna. J Cell Biol 180, 23–29. 10.1083/jcb.200709133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schou KB, Mogensen JB, Morthorst SK, Nielsen BS, Aleliunaite A, Serra-Marques A, Fürstenberg N, Saunier S, Bizet AA, Veland IR, Akhmanova A, Christensen ST, Pedersen LB, 2017. KIF13B establishes a CAV1-enriched microdomain at the ciliary transition zone to promote Sonic hedgehog signalling. Nature Communications 8, 14177. 10.1038/ncomms14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder KE, Condic ML, Eisenberg LM, Yost HJ, 1999. Spatially Regulated Translation in Embryos: Asymmetric Expression of Maternal Wnt-11 along the Dorsal–Ventral Axis in Xenopus. Developmental Biology 214, 288–297. 10.1006/dbio.1999.9426 [DOI] [PubMed] [Google Scholar]
- Stavoe AKH, Hill SE, Hall DH, Colón-Ramos DA, 2016. KIF1A/UNC-104 Transports ATG-9 to Regulate Neurodevelopment and Autophagy at Synapses. Developmental Cell 38, 171–185. 10.1016/j.devcel.2016.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L, 2008. The Kinesin-8 Motor Kif18A Suppresses Kinetochore Movements to Control Mitotic Chromosome Alignment. Developmental Cell 14, 252–262. 10.1016/j.devcel.2007.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Wagenbach M, Franck A, Asbury CL, Wordeman L, 2012. Kif18A and Chromokinesins Confine Centromere Movements via Microtubule Growth Suppression and Spatial Control of Kinetochore Tension. Developmental Cell 22, 1017–1029. 10.1016/j.devcel.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Yonekawa Y, Tanaka Y, Okada Y, Nonaka S, Hirokawa N, 1999. Left-Right Asymmetry and Kinesin Superfamily Protein KIF3A: New Insights in Determination of Laterality and Mesoderm Induction by kif3A−/− Mice Analysis. J Cell Biol 145, 825–836. 10.1083/jcb.145.4.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q, Yokota C, Puck H, Kofron M, Birsoy B, Yan D, Asashima M, Wylie CC, Lin X, Heasman J, 2005. Maternal Wnt11 Activates the Canonical Wnt Signaling Pathway Required for Axis Formation in Xenopus Embryos. Cell 120, 857–871. 10.1016/j.cell.2005.01.013 [DOI] [PubMed] [Google Scholar]
- Tokai N, Fujimoto-Nishiyama A, Toyoshima Y, Yonemura S, Tsukita S, Inoue J, Yamamota T, 1996. Kid, a novel kinesin-like DNA binding protein, is localized to chromosomes and the mitotic spindle. The EMBO Journal 15, 457–467. 10.1002/j.1460-2075.1996.tb00378.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokai-Nishizumi N, Ohsugi M, Suzuki E, Yamamoto T, 2005. The Chromokinesin Kid Is Required for Maintenance of Proper Metaphase Spindle Size. MBoC 16, 5455–5463. 10.1091/mbc.e05-03-0244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama Y, Sakaguchi M, Terabayashi T, Inenaga T, Inoue S, Kobayashi C, Oshima N, Kiyonari H, Nakagata N, Sato Y, Sekiguchi K, Miki H, Araki E, Fujimura S, Tanaka SS, Nishinakamura R, 2010. Kif26b, a kinesin family gene, regulates adhesion of the embryonic kidney mesenchyme. PNAS 107, 9240–9245. 10.1073/pnas.0913748107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H, Huang X, Tanaka Y, Hirokawa N, 2011. KIF16B/Rab14 Molecular Motor Complex Is Critical for Early Embryonic Development by Transporting FGF Receptor. Developmental Cell 20, 60–71. 10.1016/j.devcel.2010.11.008 [DOI] [PubMed] [Google Scholar]
- van der Vaart B, van Riel WE, Doodhi H, Kevenaar JT, Katrukha EA, Gumy L, Bouchet BP, Grigoriev I, Spangler SA, Yu KL, Wulf PS, Wu J, Lansbergen G, van Battum EY, Pasterkamp RJ, Mimori-Kiyosue Y, Demmers J, Olieric N, Maly IV, Hoogenraad CC, Akhmanova A, 2013. CFEOM1-Associated Kinesin KIF21A Is a Cortical Microtubule Growth Inhibitor. Developmental Cell 27, 145–160. 10.1016/j.devcel.2013.09.010 [DOI] [PubMed] [Google Scholar]
- Veland IR, Awan A, Pedersen LB, Yoder BK, Christensen ST, 2009. Primary Cilia and Signaling Pathways in Mammalian Development, Health and Disease. NEP 111, p39–p53. 10.1159/000208212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey KJ, Hammond JW, 2009. Traffic control: regulation of kinesin motors. Nature Reviews Molecular Cell Biology 10, 765–777. 10.1038/nrm2782 [DOI] [PubMed] [Google Scholar]
- Viswanadha R, Sale WS, Porter ME, 2017. Ciliary Motility: Regulation of Axonemal Dynein Motors. Cold Spring Harb Perspect Biol 9, a018325. 10.1101/cshperspect.a018325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel P, Read RW, Hansen GM, Payne BJ, Small D, Sands AT, Zambrowicz BP, 2012. Congenital Hydrocephalus in Genetically Engineered Mice. Vet Pathol 49, 166–181. 10.1177/0300985811415708 [DOI] [PubMed] [Google Scholar]
- Wang D, Nitta R, Morikawa M, Yajima H, Inoue S, Shigematsu H, Kikkawa M, Hirokawa N, 2016. Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A. eLife 5, e18101. 10.7554/eLife.18101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Dynlacht BD, 2018. The regulation of cilium assembly and disassembly in development and disease. Development 145. 10.1242/dev.151407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Cao L, Wang C, Gigant B, Knossow M, 2015. Kinesin, 30 years later: Recent insights from structural studies. Protein Science 24, 1047–1056. 10.1002/pro.2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Smallwood PM, Williams J, Nathans J, 2020. A mouse model for kinesin family member 11 (Kif11)-associated familial exudative vitreoretinopathy. Hum Mol Genet 29, 1121–1131. 10.1093/hmg/ddaa018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver C, Farr GH, Pan W, Rowning BA, Wang J, Mao J, Wu D, Li L, Larabell CA, Kimelman D, 2003. GBP binds kinesin light chain and translocates during cortical rotation in Xenopus eggs. Development 130, 5425–5436. 10.1242/dev.00737 [DOI] [PubMed] [Google Scholar]
- Weaver C, Kimelman D, 2004. Move it or lose it: axis specification in Xenopus. Development 131, 3491–3499. 10.1242/dev.01284 [DOI] [PubMed] [Google Scholar]
- White JA, Heasman J, 2008. Maternal control of pattern formation in Xenopus laevis. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 310B, 73–84. 10.1002/jez.b.21153 [DOI] [PubMed] [Google Scholar]
- Wilson CW, Nguyen CT, Chen M-H, Yang J-H, Gacayan R, Huang J, Chen J-N, Chuang P-T, 2009. Fused has evolved divergent roles in vertebrate Hedgehog signalling and motile ciliogenesis. Nature 459, 98–102. 10.1038/nature07883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood KW, Sakowicz R, Goldstein LSB, Cleveland DW, 1997. CENP-E Is a Plus End–Directed Kinetochore Motor Required for Metaphase Chromosome Alignment. Cell 91, 357–366. 10.1016/S0092-8674(00)80419-5 [DOI] [PubMed] [Google Scholar]
- Xia C-H, Roberts EA, Her L-S, Liu X, Williams DS, Cleveland DW, Goldstein LSB, 2003. Abnormal neurofilament transport caused by targeted disruption of neuronal kinesin heavy chain KIF5A. J Cell Biol 161, 55–66. 10.1083/jcb.200301026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Andrews C, Chan W-M, McKeown CA, Magli A, de Berardinis T, Loewenstein A, Lazar M, O’Keefe M, Letson R, London A, Ruttum M, Matsumoto N, Saito N, Morris L, Monte MD, Johnson RH, Uyama E, Houtman WA, de Vries B, Carlow TJ, Hart BL, Krawiecki N, Shoffner J, Vogel MC, Katowitz J, Goldstein SM, Levin AV, Sener EC, Ozturk BT, Akarsu AN, Brodsky MC, Hanisch F, Cruse RP, Zubcov AA, Robb RM, Roggenkäemper P, Gottlob I, Kowal L, Battu R, Traboulsi EI, Franceschini P, Newlin A, Demer JL, Engle EC, 2003. Heterozygous mutations of the kinesin KIF21A in congenital fibrosis of the extraocular muscles type 1 (CFEOM1). Nature Genetics 35, 318–321. 10.1038/ng1261 [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Nakata T, Okada Y, Hirokawa N, 1996. Cloning and characterization of KAP3: a novel kinesin superfamily-associated protein of KIF3A/3B. PNAS 93, 8443–8448. 10.1073/pnas.93.16.8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW, 2000. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nature Cell Biology 2, 484–491. 10.1038/35019518 [DOI] [PubMed] [Google Scholar]
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW, 1992. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature 359, 536–539. 10.1038/359536a0 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, O’Toole E, Ghosh S, Mitchell DR, 2004. Regulation of flagellar dynein activity by a central pair kinesin. PNAS 101, 17398–17403. 10.1073/pnas.0406817101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekawa Y, Harada A, Okada Y, Funakoshi T, Kanai Y, Takei Y, Terada S, Noda T, Hirokawa N, 1998. Defect in Synaptic Vesicle Precursor Transport and Neuronal Cell Death in KIF1A Motor Protein–deficient Mice. J Cell Biol 141, 431–441. 10.1083/jcb.141.2.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Blasius TL, Zhang S, Jariwala S, Walker B, Grant BJ, Cochran JC, Verhey KJ, 2018. Altered chemomechanical coupling causes impaired motility of the kinesin-4 motors KIF27 and KIF7. J Cell Biol 217, 1319–1334. 10.1083/jcb.201708179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Omori Y, Brodowska K, Kovach P, Malicki J, 2012. Kinesin-2 family in vertebrate ciliogenesis. PNAS 109, 2388–2393. 10.1073/pnas.1116035109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Niwa S, Guillaud L, Tong Y, Hirokawa N, 2013. A molecular motor, KIF13A, controls anxiety by transporting the serotonin type 1A receptor. Cell Rep 3, 509–519. 10.1016/j.celrep.2013.01.014 [DOI] [PubMed] [Google Scholar]
- Zhou R, Niwa S, Homma N, Takei Y, Hirokawa N, 2009. KIF26A Is an Unconventional Kinesin and Regulates GDNF-Ret Signaling in Enteric Neuronal Development. Cell 139, 802–813. 10.1016/j.cell.2009.10.023 [DOI] [PubMed] [Google Scholar]
- Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan J-B, Abraham RT, Jiang W, 2005. Functional Analysis of Human Microtubule-based Motor Proteins, the Kinesins and Dyneins, in Mitosis/Cytokinesis Using RNA Interference. MBoC 16, 3187–3199. 10.1091/mbc.e05-02-0167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D, 2008. In Vivo Imaging of oskar mRNA Transport Reveals the Mechanism of Posterior Localization. Cell 134, 843–853. 10.1016/j.cell.2008.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. All 45 mammalian kinesins with reported animal phenotypes, cellular roles, alternate gene names, and citations. Each kinesin is grouped with its family (15 different families). The color coding matches to reported cellular roles, defined in Figure 2. Red is for roles in mitosis, yellow for roles in cilia, blue for roles in neurons, green for intracellular transport of proteins, mRNAs, vesicles and organelles.


