Abstract
Perhaps the best characterized example of an activator-induced chromatin transition is found in the activation of the Saccharomyces cerevisiae acid phosphatase gene PHO5 by the basic helix-loop-helix (bHLH) transcription factor Pho4. Transcription activation of the PHO5 promoter by Pho4 is accompanied by the remodeling of four positioned nucleosomes which is dependent on the Pho4 activation domain but independent of transcription initiation. Whether the requirements for transcription activation through the TATA sequence are different from those necessary for the chromatin transition remains a major outstanding question. In an attempt to understand better the ability of Pho4 to activate transcription and to remodel chromatin, we have initiated a detailed characterization of the Pho4 activation domain. Using both deletion and point mutational analysis, we have defined residues between positions 75 and 99 as being both essential and sufficient to mediate transcription activation. Significantly, there is a marked concordance between the ability of mutations in the Pho4 activation domain to induce chromatin opening and transcription activation. Interestingly, the requirements for transcription activation within the Pho4 activation domain differ significantly if fused to a heterologous bHLH-leucine zipper DNA-binding domain. The implications for transcription activation by Pho4 are discussed.
Activation of transcription by RNA polymerase II (Pol II) can be divided into two steps: chromatin, which can act to repress transcription, must undergo a transition to allow access of the polymerase and transcription activators to the DNA; and a functional preinitiation complex must be assembled to allow RNA Pol II subsequently to catalyze the formation of a nascent RNA molecule. There is evidence for activator function at both stages, with sequence-specific DNA-binding proteins acting both to modulate chromatin structure and to increase recruitment, isomerization, or escape of RNA polymerase from a promoter. For the majority of transcription activators, the DNA-binding and transcription activation functions are found in separate domains within the same protein. Yet in contrast to DNA-binding domains which can be grouped into families based on their structural similarities deduced from physical and biochemical evidence, the classification of activation domains is far more rudimentary, being based largely upon the preponderance of certain amino acids (acidic, glutamine rich, and proline rich) and reflecting the lack of knowledge about the relationship between their structure and function.
The acidic activation domains were the first to be identified and are the most widely studied. Early experiments in which transcriptionally active proteins were created by fusing random Escherichia coli DNA fragments to sequences encoding the Gal4 DNA-binding domain suggested a positive correlation between high net negative charge and activation potential (29). Moreover, in addition to the preponderance of acidic residues, this class of activation domain was predicted to adopt an amphipathic α-helical conformation, a prediction apparently supported by the construction of an artificial activator (Gal-AH) comprising a 15-amino-acid peptide predicted to form such a structure, fused to the Gal4 DNA-binding domain (17). However, in contrast to many other activators, Gal-AH activates poorly unless overexpressed (26) despite apparently having the potential to adopt an α-helical conformation as determined by circular dichroism (CD) analysis (42). Similarly, while the VP16 activation domain has been predicted to adopt an α-helical conformation, evidence from CD analysis as well as nuclear magnetic resonance (NMR) spectroscopy indicates that it is unstructured in an aqueous solution (11, 32). On the other hand, NMR studies using a minimal VP16 activation domain suggested that it may adopt an α-helical conformation on interaction with hTAF31 (41), and, along similar lines, NMR studies have also demonstrated that the activation domain of the cyclic AMP-responsive transcription factor CREB undertakes a random coil-to-helix transition on interaction with the CBP cofactor (34).
The notion that acidic activators adopt an amphipathic α-helical conformation was challenged by CD and mutational analysis of the Gal4 activation domain, which showed that it had the potential to form a β-sheet but not an α-helix (26, 42) and that mutagenesis of the Gal4 activation domain could result in an activating mutant with a net positive charge (26). However, results from Wu et al. (45) obtained by using a combination of mutagenesis and surface plasmon resonance have suggested that while the Gal4 activation domain can interact with TBP and TFIIB, the putative β-sheet in the Gal4 activation domain cannot be required for activation. The concept of a role for acidic residues in the activation process has been further undermined by the observation that mutation of hydrophobic residues in the VP16 activation domain abolishes function even when substitution increases the overall net negative charge (9, 35), while in the Gcn4 transcription factor, bulky hydrophobic residues appear to make a critical contribution to the activation function (12, 20). In summary, despite recent advances, how transcription factors achieve the level of specificity required to target different components of the transcription machinery remains poorly understood.
Considerable progress has been made in understanding activator-target protein interactions required for transcription activation through the core transcription machinery; much less is known of the requirements for modulating chromatin structure. Perhaps the best characterized example of an activator-induced chromatin transition is found in the activation of the Saccharomyces cerevisiae acid phosphatase gene PHO5 by the acidic basic helix-loop-helix (bHLH) transcription factor Pho4, which can bind to two sites within the PHO5 upstream activating sequence (UAS), termed UASp1 and UASp2. The PHO5 gene is highly expressed under low-phosphate conditions and is repressed under high-phosphate conditions. Under high-phosphate conditions, the PHO5 promoter is masked by four precisely positioned nucleosomes with the exception of a nuclease-hypersensitive site located over the upstream Pho4 binding site, UASp1 (1, 13, 44). Under repressing conditions, the Pho4 activator fails to bind either UASp1 or UASp2, despite the fact that UASp1 is not concealed by the positioned nucleosomes. In addition, the activation domain of Pho4 is masked by the Pho80-Pho85 cyclin-cdk complex (18, 22), which also phosphorylates Pho4 (23), resulting in a proportion of Pho4 being located in the cytoplasm (33). On switching to low-phosphate conditions, the Pho80-Pho85 cyclin-cdk complex dissociates from Pho4 (18, 23), unmasking the activation domain and allowing Pho4 to associate with the homeobox factor Pho2. The Pho4-Pho2 complex then binds and activates the PHO5 UAS elements cooperatively (4, 5, 18, 37). As a consequence of Pho4-Pho2 binding to the PHO5 promoter, the four positioned nucleosomes undergo a transition, resulting in the entire promoter becoming nuclease sensitive and Pho4 being bound to both UAS elements. Neither DNA replication nor transcription directed by the PHO5 TATA element are required for the chromatin transition (14, 36). Importantly, however, the Pho4 activation domain is necessary for the chromatin transition to occur (38). Whether the requirements for transcription activation are different from those necessary for the chromatin transition remains a major outstanding question.
In an attempt to understand better the ability of Pho4 to activate transcription and remodel chromatin, we have initiated a detailed characterization of the Pho4 activation domain. Using both deletion and point mutational analysis, we have defined residues between positions 75 and 99 as being both essential and sufficient to mediate transcription activation and demonstrate that the requirements for transcription activation are dependent on the nature of the DNA-binding domain. Point mutations and CD analysis revealed that the N-terminal part of the activation domain may adopt an α-helical structure and that activation has a requirement for aromatic residues. Significantly, there is a marked concordance between the effects of mutations in the Pho4 activation domain on chromatin opening and on transcription activation.
MATERIALS AND METHODS
Genetic and biochemical methods.
The S. cerevisiae strain Y704 (a ade2-1 trp1-1 canR leu2-3 leu2-112 his 3-11,15 ura3 pho4::HIS3) has been described previously (15, 22), as has strain YS33 (α canR leu2-3 leu2-112 his 3-15 ura3Δ5 Δpho4::ura3Δ5 Δpho80::HIS3) (31). Cells were grown and assayed for acid phosphatase activity produced by the PHO5 gene as described previously (38). β-Galactosidase activity was assayed as described previously (18). The values for the β-galactosidase and acid phosphatase assays using the N-terminal and deletion mutants are presented as averages of three independent experiments, each performed in duplicate. The standard deviations calculated for these data were no more than ±10%. All methods for the ClaI accessibility assays have also been described previously (2, 16).
Anti-Pho4 antibody and Western blotting.
Pho4 amino acids 108 to 245 were expressed in E. coli as an in-frame fusion with glutathione S-transferase (GST). After purification on glutathione beads, 100 μg of the GST-Pho4 fusion protein was used to inject rabbits four times at intervals of 1 month. Anti-GST antibodies were removed from the resulting antiserum by incubation with an excess GST protein bound to glutathione-Sepharose beads for 2 h at 4°C. The resulting supernatant was used to probe Western blots for Pho4 expression. Samples for Western blotting were prepared by harvesting yeast cells grown under appropriate conditions by centrifugation, resuspending the pellet in sodium dodecyl sulfate (SDS) lysis buffer, and boiling for 5 min before analysis by SDS-polyacrylamide gel electrophoresis. Following electrophoresis, proteins were transferred to a nitrocellulose membrane, blocked in a buffer containing 5% nonfat milk, and probed with anti-Pho4 antibody for 1 h. After the blots were washed, they were incubated with an anti-rabbit horseradish peroxidase-conjugated secondary antibody and washed extensively, and bands were revealed by using ECL (Amersham) according to the manufacturer’s instructions.
Yeast vectors.
The LexA operator-LacZ reporter has been described previously (22), as have the LexA-fusion protein expression vector and the low-copy-number galactose-inducible Pho4 expression vector, pRS315.KV (18). The Pho4 promoter vector has also been described (38). Pho4 deletion mutants and point mutations introduced into the Pho4-Cpf1 background were made by PCR by using appropriate primers which place BamHI restriction sites at the 5′ and 3′ ends of the coding sequence. The construction of the wild-type (WT) Pho4-Cpf1 fusion was described by Jayaraman et al. (22), and this fusion contains an engineered XhoI restriction site at the junction between the Pho4 and Cpf1 coding sequences. All mutations were verified by sequencing.
CD analysis.
CD spectra were recorded in 10 mM phosphate buffer with a Jasco J-600 spectrophopolarimeter with peptide concentrations of 30 to 50 μM (1-mm path length, fused-silica cuvettes). Spectra are presented as the CD absorption coefficient calculated on a per-residue basis (Δɛmrw).
RESULTS
Defining the Pho4 activation domain.
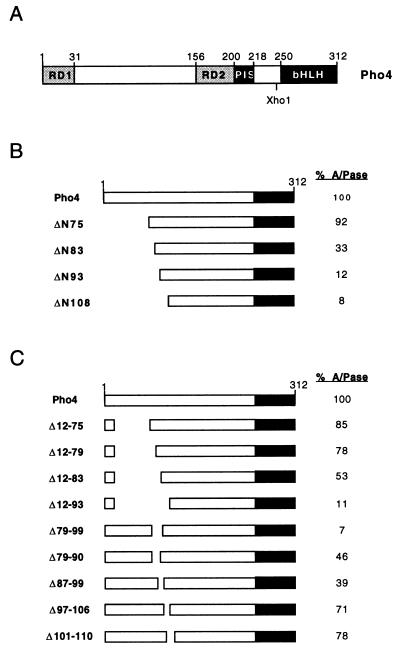
The Pho4 transcription factor comprises a 312-amino-acid protein with a bHLH DNA-binding domain located within the C-terminal 60 amino acids (Fig. 1A). Although previous work has located residues required for transcription activation N-terminal to either position 108 (31) or 118 (22), the results from these studies were inconsistent. Thus, in the exhaustive study of Ogawa and Oshina (31), several regions within the N-terminal 108 amino acids of Pho4 appeared to play a significant role in the ability of Pho4 to activate transcription from the PHO5 promoter. In contrast, Jayaraman et al. (22), using chimeric constructs, failed to identify any requirements for transcription activation N-terminal to amino acid 75. A second region of Pho4 required for transcriptional activation resides between residues 203 and 227 and was termed the oligomerization domain (31). We subsequently identified this region as being essential for interaction and cooperative binding with the Pho2 homeodomain transcription factor (5, 18). As a first step towards understanding the precise requirements for transcription activation by Pho4, we reexamined the boundaries of the activation domain.
FIG. 1.
Defining the Pho4 activation domain. (A) Schematic diagram of the domains of Pho4 as determined by Hirst et al. (18). The repression domains RD1 and RD2 are required for interaction with the Pho80 cyclin, while the Pho2-interacting sequence (PIS) mediates interaction and cooperative DNA binding with Pho2. (B) Activity of a series of Pho4 N-terminal deletion mutants. The indicated mutants were expressed from the GAL10 promoter on a CEN/ARS vector following transformation into strain Y704, which lacks endogenous Pho4. Yeast cells were assayed for acid phosphatase (A/Pase) activity as described previously (38) after induction of Pho4 expression by growth in low-phosphate galactose medium. The level of activation achieved by expression of Pho4 under these conditions is around 60 to 70% of that obtained with endogenous Pho4 in a WT strain under low-phosphate conditions. Results of the acid phosphatase assays using the N-terminal and deletion mutants are presented as an average of three independent experiments, each performed in duplicate. (C) Activity of a series of Pho4 internal deletion mutants. The indicated Pho4 internal deletion mutants were expressed from the PHO4 promoter after transformation of yeast strain YS33 (39), which is deleted for the chromosomal PHO4 and PHO80 genes. Acid phosphatase assays were performed after culture in high-phosphate glucose medium.
To determine the N-terminal boundary of the Pho4 activation domain, a series of N-terminal deletion mutants were constructed and assayed for their ability to activate the endogenous PHO5 acid phosphatase gene under low-phosphate conditions in a strain lacking the endogenous Pho4 protein. The results obtained (Fig. 1B) show that deletion to amino acid 75 (ΔN75) resulted in no significant reduction in acid phosphatase levels. In contrast, removal of an additional eight amino acids (ΔN83) reduced expression around 70% compared to that of the WT protein, while a further deletion to amino acid 93 (ΔN93) activated transcription around 10-fold less efficiently than the full-length protein. A similar level of transcription activation was also observed with the ΔN108 mutant, which lacks all residues N-terminal to position 108. These results would indicate that amino acids located between amino acids 75 and 93 are essential for the function of the Pho4 activation domain.
To confirm that the residues identified by using the N-terminal deletion series were also required in the context of full-length Pho4, yeast cells were transformed with vectors constitutively expressing a series of Pho4 internal deletion mutants and their ability to activate acid phosphatase expression was assessed. The results (Fig. 1C) with mutants Δ12-75, Δ12-79, Δ12-83, and Δ12-93 were in good agreement with those obtained with the N-terminal deletion series. That is, residues 83 to 93 were essential for efficient transcription activation. Consistent with this, removal of residues 79 to 99 failed to activate transcription above the background observed when the ΔN108 mutant was used. Activation of around 40 to 50% of that observed with WT Pho4 was observed with mutants Δ79-90 and Δ87-99, indicating that the regions lying between positions 79 and 87, or 90 and 99, may each contribute to transcription activation. In contrast, no significant effect on the transactivation capacity of Pho4 was detected with mutant Δ97-106 or Δ101-110, indicating that sequences C-terminal to amino acid 97 are not required. In summary, the N-terminal and internal deletion mutants identify a critical region of Pho4 lying between amino acids 79 and 99, which play an essential role in transcription activation by Pho4.
Modulation of chromatin by the Pho4 activation domain.
Under repressing, high-phosphate, conditions, the PHO5 promoter is packaged into an array of four positioned nucleosomes. Upon switching to low-phosphate conditions, the chromatin covering the PHO5 promoter is remodeled, resulting in a 600-bp region of the promoter becoming hypersensitive to nuclease digestion. This chromatin transition is dependent on Pho4 binding to the PHO5 UAS elements and, importantly, is dependent on the Pho4 activation domain (38). However, although the activation domain of Pho4 is required, the alterations in chromatin structure appear to be independent of transcription initiation, since the Pho4-induced chromatin transition occurs even when the PHO5 TATA box has been mutated (14). Having identified residues essential for transcription activation by Pho4, we were now in a position to address a major outstanding question concerning the ability of Pho4 to remodel chromatin, namely, are the requirements within the Pho4 activation domain for transcription initiation different from those required to induce the chromatin transition? In other words, would it be possible to identify activation domain mutations that induced an efficient chromatin transition but which would not support transcription initiation?
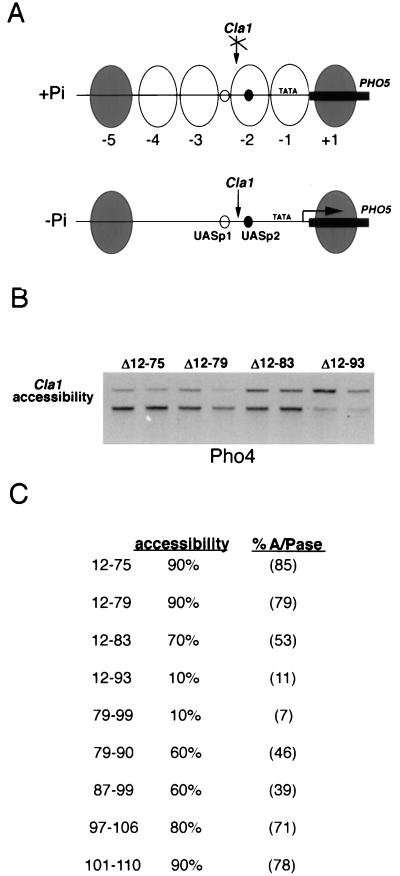
To this end, yeast strain YS33, which lacks both endogenous Pho4 and the repressor Pho80, were transformed with vectors constitutively expressing the series of Pho4 internal deletion mutants and their ability to induce the chromatin transition at the PHO5 promoter was assessed. To assay chromatin disruption at the PHO5 promoter, we isolated yeast nuclei and measured the accessibility of a ClaI site located within positioned nucleosome −2. Although in principle the ability of a specific restriction endonuclease to cut a particular site might reflect only a localized remodeling of chromatin, the ability of ClaI to cut this site within the PHO5 promoter is a reliable and quantitative assay that has been used previously by us (for examples, see references 2, 16, 36, and 44) to provide an accurate reflection of whether the chromatin across the entire PHO5 promoter is in a closed or open conformation. A map of the PHO5 promoter and the relative positions of the four positioned nucleosomes and the ClaI site used for the chromatin opening assays is shown in Fig. 2A, while the results of the ClaI accessibility assays are presented in Fig. 2B and C.
FIG. 2.
Chromatin remodeling by Pho4 deletion mutants. (A) A schematic diagram of the chromatin structure across the PHO5 promoter is shown. Nucleosomes −1, −2, −3, and −4 (open ovals) are remodeled by Pho4 either under conditions of phosphate starvation or in the absence of the Pho80 repressor. The Pho4 binding sites at UASp1 and UASp2 are represented by small open and closed circles, respectively. The locations of the TATA box and the ClaI restriction site used in the chromatin opening assays are also indicated. (B) The indicated series of Pho4 deletion mutants were expressed from the PHO4 promoter in strain YS33, lacking endogenous Pho4 and Pho80, and the chromatin across the PHO5 promoter was analyzed by digestion with ClaI in high-phosphate medium as described previously (16). Each mutant was assayed twice. Essentially, nuclei containing around 10 μg of DNA were digested with an excess of ClaI for 60 min at 37°C. To monitor cleavage by ClaI, DNA was isolated and digested with HaeIII before analysis on a 1% agarose gel and Southern blotting. A 1.38-kb fragment is generated in the absence of cleavage by ClaI, and a 1.07-kb fragment is generated if the ClaI site is accessible. (C) The ClaI accessibility assay results for the entire series of Pho4 deletion mutants are summarized. The results of the acid phosphatase (A/Pase) assays, which were performed in parallel and which are presented in Fig. 1C, are shown in parentheses. Note that previous work has established that endogenous Pho4 opens chromatin at the PHO5 UAS to around 95%, as determined by using the ClaI accessibility assay (2), a level similar to that observed with the plasmid-based Pho4 expression vectors, while the absence of Pho4 results in opening to around 5 to 10%.
Consistent with the results from the N-terminal deletion analysis, efficient chromatin opening (90%) was observed with Pho4 derivatives lacking either residues 12 to 75 or 12 to 79. Mutant Δ12-83, which lacks an additional four amino acids, was also able to open chromatin to 70%. However, Pho4 deleted between amino acids 12 and 93 failed to open chromatin to any significant extent (10%). A similar low level of ClaI accessibility was also observed with mutant Δ79-99, indicating that this region contains residues critical for chromatin modulation. Compared to the Δ79-99 mutant, the presence of residues 91 to 99 (Δ79-90) restores activity to around 50%, as does the presence of residues 79 to 86 (Δ87-99). No significant effect of deletion of residues 97 to 106 or 101 to 110 was observed in the ClaI accessibility assay. Taken together, these results indicate that residues essential for chromatin opening lie between amino acids 79 and 99. A comparison of results from the induction of acid phosphatase expression with those of the ClaI accessibility assay are strikingly concordant; residues essential for induction of the chromatin transition appear to correlate well with those required for transcription activation.
The minimal Pho4 activation domain.
The results described above indicate, first, that residues 79 to 99 are essential for transcription activation and, second, that this region of Pho4 is required both for transcription of the acid phosphatase gene and for modulation of the chromatin located across the PHO5 promoter. However, while the mutants used so far define residues essential for these functions, the question as to whether this region is sufficient remains outstanding.
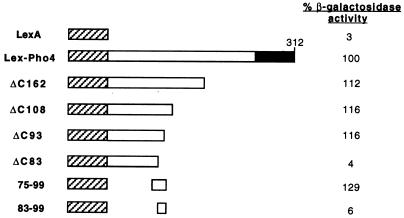
To address this point, full-length Pho4 or C-terminal deletion mutants were expressed as fusions with the bacterial LexA repressor. Fusion to LexA was necessary since C-terminal deletions disrupt the bHLH DNA-binding domain located at the C terminus of Pho4. The results obtained (Fig. 3) by using the Lex-Pho4 fusions and a LacZ reporter driven by the Lex operator show that a deletion removing residues C-terminal to amino acid 162 (ΔC162) activates transcription to WT levels. Similar results were obtained when only the N-terminal 108 amino acids of Pho4 (ΔC108) were fused to LexA. Removal of additional residues in mutant ΔC93 also failed to affect significantly the ability of the chimeric LexA-Pho4 protein to activate transcription. In contrast, further deletion to amino acid 83 (ΔC83) largely abolished the transactivation capacity. Thus, consistent with the N-terminal and internal deletion series, the region lying between amino acids 83 and 93 is essential for transcription activation.
FIG. 3.
Defining the minimal Pho4 activation domain. The indicated series of Pho4 deletion mutants were expressed as LexA fusion proteins from the GAL10 promoter after transformation of strain Y704. β-Galactosidase activity was determined as described previously (15) after overnight growth in galactose. The reporter used has been described elsewhere (22) and contains four Lex operators upstream from the basal CYC1-LacZ reporter.
To determine whether this region was sufficient to activate transcription, the region of Pho4 comprising residues 75 to 99 was fused to LexA and the ability of these residues to activate the Lex operator-LacZ reporter was assayed. As expected from the deletion analysis, these 25 amino acids were sufficient to activate transcription. However, residues 83 to 99 alone failed to activate transcription when fused to LexA. Together with the results from the internal deletion series, the data obtained with LexA fusion proteins suggest that while amino acids 75 to 99 are sufficient to activate transcription, residues N- and C-terminal to position 83 contribute to transcription activation.
Requirements for transcription activation within the Pho4 activation domain are dependent on the DNA-binding domain.
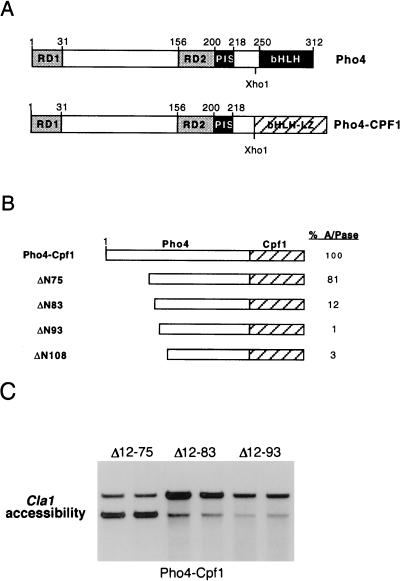
The region of Pho4 between amino acids 75 and 99 contains residues which are both necessary and sufficient for transcription activation. We have previously suggested that the region of Pho4 between residues 74 and 85 may have the potential to form an acidic amphipathic α-helix (22). For other transcription factors, structural studies have demonstrated a key role for α-helices in mediating the interaction between transcription factors and their target proteins (24, 34, 41). However, the Pho4 N-terminal deletion series (Fig. 1) failed to reveal a critical role for residues 75 to 83 in transcription activation by Pho4, perhaps owing to redundancy within the activation domain. To circumvent this problem, we made use of the observation that if the Pho4 DNA-binding domain were replaced with that from the yeast bHLH-leucine zipper (LZ) factor Cpf1 (3, 6, 30), the chimeric protein retained the capacity to bind the endogenous PHO5 UAS elements, but transcription activation by the Pho4-Cpf1 fusion protein, depicted in Fig. 4A, was significantly more sensitive to mutation. The increased sensitivity of a Pho4-Cpf1 chimeric protein to deletions within the activation domain is illustrated in Fig. 4B; for this experiment, a series of N-terminal deletion mutants were constructed and assayed for their ability to activate the endogenous PHO5 acid phosphatase gene under low-phosphate conditions in a strain lacking the endogenous Pho4 protein. The results obtained show that deletion to amino acid 75 (ΔN75) resulted in no more than a 20% reduction in acid phosphatase levels. In contrast, removal of an additional eight amino acids (ΔN83) reduced expression around 10-fold compared to that of the WT Pho4-Cpf1 protein, while a further deletion to amino acid 93 (ΔN93) failed to activate transcription to a measurable extent. A similar inability to activate transcription was also observed with the ΔN108 mutant, which lacks all residues N-terminal to position 108. These results are consistent with those obtained with the WT Pho4 protein, with the exception that the effects of the ΔN83 and ΔN93 mutations are significantly more severe than those in the background of the homologous Pho4 DNA-binding domain (compare Fig. 4B and 1B). Since the ΔN75 and ΔN83 Pho4-Cpf1 proteins were expressed to similar levels, as determined by Western blotting (data not shown), in the context of the Cpf1 DNA-binding domain, effective activation of transcription by Pho4 appeared to require residues which corresponded to the predicted α-helical region, whereas in the context of the natural Pho4 DNA-binding domain, these residues played a relatively minor role.
FIG. 4.
An additional requirement for transcription activation and chromatin opening by Pho4 revealed in the context of the Cpf1 DNA-binding domain. (A) Relative structure of Pho4 and the Pho4-Cpf1 chimeric protein. Note that the two DNA-binding domains are positioned identically relative to the Pho4 N terminus. (B) Effect of deletions in the Pho4 activation domain in the context of a Pho4-Cpf1 fusion protein. The indicated deletion mutants were expressed from the GAL10 promoter and, after transformation into strain Y704 lacking endogenous Pho4, were assayed for acid phosphatase (A/Pase) activity after induction of Pho4 expression in galactose medium. The level of activation achieved by expression of Pho4-Cpf1 under these conditions is around 85% of that obtained with endogenous Pho4 in a WT strain under low-phosphate conditions. (C) Effects of the Pho4-Cpf1 fusion proteins on chromatin opening at the PHO5 promoter as assessed by using the ClaI accessibility assay, which was performed as described in the legend to Fig. 2.
Given the difference in the requirements for activation of transcription depending on whether the Pho4 or Cpf1 DNA-binding domains were used, it was important to examine the ability of the Pho4-Cpf1 deletion mutants to modulate the chromatin structure across the PHO5 promoter. Pho4-Cpf1 deletion mutants Δ12-75, Δ12-83, and Δ12-93 were assayed for their ability to open chromatin across the PHO5 promoter by using the ClaI accessibility assay. The results obtained (Fig. 4C) revealed that the Δ12-75 deletion mutant could open chromatin to 95%, while ClaI accessibility with either the Δ12-83 or Δ12-93 deletion mutant was no more than 10%. Taken together with the results described above, it is evident that the region of Pho4 lying between residues 75 and 83 plays a critical role in transcription activation and chromatin opening in the context of the Pho4-Cpf1 chimera but not when fused to the natural Pho4 DNA-binding domain. Thus, the requirements for transcription activation and chromatin modulation are dependent on the nature of the DNA-binding domain.
Requirements for specific amino acids in activation and chromatin opening by Pho4.
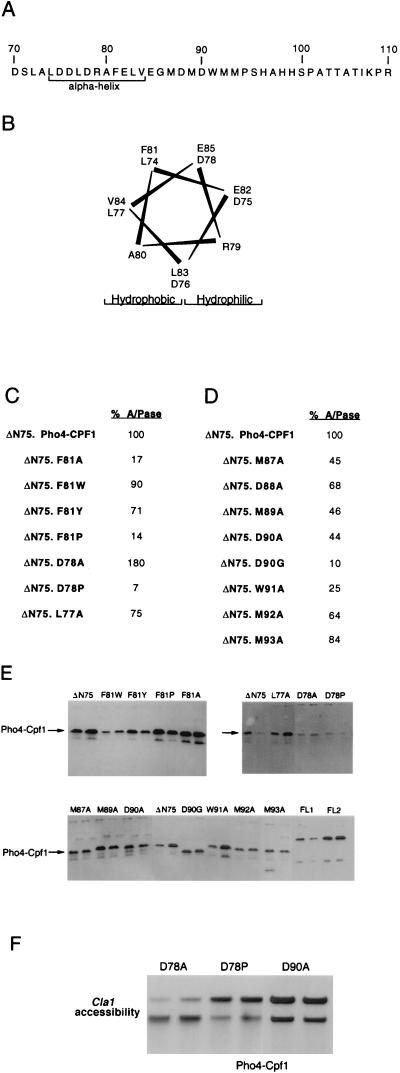
The results obtained by using the Pho4-Cpf1 chimeras revealed that the region of Pho4 between residues 75 and 83 was able to play a critical role in both transcription activation and chromatin opening, while in the context of Pho4 itself, the region between residues 75 and 83 was far less important but the region between residues 83 and 93 was essential. Consistent with the reduced sensitivity of Pho4 to mutation, single amino acid substitutions in the context of Pho4 itself (D78A, D78P, D90A, M92A) failed to affect transcription activation more than twofold (data not shown), most likely owing to redundancy within the activation domain. However, the increased sensitivity of the Pho4-Cpf1 chimera to mutation provided us with an opportunity to explore the specific amino acid requirements within the activation domain. In particular, we wished to determine which residues might contribute to transcription activation and, also, whether evidence from a mutational analysis could lend support to the idea that the region between residues 75 and 83 of Pho4 could adopt an α-helical conformation. The sequence of the Pho4 activation domain and the location of the potential α-helix are shown in Fig. 5A, and a helical wheel analysis (Fig. 5B) illustrates that within the potential amphipathic α-helix, all charged or hydrophilic residues are located on one face while all hydrophobic residues are on the opposite face.
FIG. 5.
Requirement for specific residues in transactivation and chromatin opening. (A) The amino acid sequence across the Pho4 activation domain and location of the predicted α-helix are shown. (B) A helical wheel analysis of residues 74 to 85 showing hydrophobic and hydrophilic faces of the predicted α-helix is presented. (C and D) The indicated mutants, introduced into the context of the ΔN75 Pho4-Cpf1 chimeric protein, were expressed from the GAL10 promoter and, after transformation into strain Y704 lacking endogenous Pho4, were assayed for acid phosphatase (A/Pase) activity after induction of Pho4 expression in low-phosphate galactose medium. (E) Western blots showing relative levels of expression for the indicated Pho4-Cpf1 proteins are shown. The expression of each protein was determined in duplicate from independent yeast cultures by using a specific anti-Pho4 antibody which recognizes the region of Pho4 between residues 108 and 245 that is N-terminal to the DNA-binding domain. Each panel represents the results obtained from a set of samples processed in parallel and blotted and probed at the same time. All panels contain a ΔN75 control, while full-length Pho4-Cpf1 is shown in two duplicate assays (FL1 and FL2). (F) Results of ClaI accessibility assays for the indicated Pho4-Cpf1 mutants performed as described in the legend to Fig. 2 are shown.
A series of point mutations were introduced into the Pho4 activation domain in the context of the ΔN75 Pho4-Cpf1 fusion protein in anticipation that in this background the effects of single point mutations would be evident. In both the VP16 and Gcn4 activation domains, aromatic amino acids play a crucial role (9, 12, 20, 35). In Pho4, a single aromatic residue, F81, lies within the potential α-helix. To determine whether this phenylalanine was important for transcription activation, it was mutated to alanine. Introduction of an alanine would be expected to maintain any potential α-helix while abolishing any interaction dependent on the aromatic nature of the phenylalanine side chain. Compared to the ΔN75 Pho4-Cpf1 derivative, the F81A substitution reduced activity around five- to sixfold (Fig. 5C), despite being expressed to similar levels as determined by Western blotting (Fig. 5E), suggesting that F81 is essential for the full activity of the Pho4 activation domain. In contrast, replacement of F81 with either tryptophan or tyrosine, which like phenylalanine have aromatic side chains, had little effect on transcription activation, acid phosphatase activities being, respectively, 90 and 71% of that obtained with the parental ΔN75 protein. These data indicate that either an aromatic side chain or possibly a bulky hydrophobic residue at position 81 plays a crucial role in the transactivation function.
The region of the Pho4 activation domain containing F81 is predicted to adopt an α-helical conformation. Introduction of a proline residue into this region of Pho4 would most likely disrupt any α-helix and, if an α-helical conformation were important for function, might also be expected to reduce the ability of Pho4-Cpf1 to activate transcription. To test this, we replaced F81 with proline and assayed for the ability of the F81P mutant to activate expression of the PHO5 gene. Consistent with the α-helix being important for function, the F81P mutant activated transcription around sevenfold less well than the parental ΔN75 protein (Fig. 5C) but was expressed to a similar level (Fig. 5E). However, activation by the F81P mutant was not significantly less than that obtained for the F81A mutant, and it could be argued that the sevenfold reduction in activation observed with the F81P mutant simply reflected an absence of an aromatic or large hydrophobic side chain at this position rather than disruption of an α-helix. To distinguish between these possibilities, it was necessary to find residues whose replacement by alanine would not affect the ability to activate transcription.
Our initial attention focused on mutation of the aspartic acid residue at position 78, located on the hydrophilic face within the core of the potential α-helix. Given the key role played by hydrophobic residues in the activation process, it is possible that acidic residues may serve simply to provide a hydrophilic face to the amphipathic helix rather than participating directly in protein-protein interactions. Consistent with this, mutation of D78 to alanine (D78A) failed to decrease the ability of Pho4 to activate transcription and in fact increased the potential for activation around 1.5-fold (Fig. 5C). Since the nature of the amino acid side chain at position 78 did not appear to be a significant factor in determining the capacity for transcription activation, we also introduced a proline residue in the same position. In contrast to the D78A mutation, the D78P mutant activated transcription more than 12-fold less well than the parental ΔN75 protein (Fig. 5C) but was expressed at a level similar to those of the WT and D78A mutant (Fig. 5E). This result is consistent with the region between residues 75 and 85 adopting an α-helical conformation and the α-helix being required for transcription activation.
We also introduced an alanine and proline substitution for leucine at position 77. However, although the L77A mutant activated transcription to almost-WT levels (70%) (Fig. 5C) and was expressed well (Fig. 5E), the substitution with a proline residue at this position resulted in a protein which was poorly expressed as assessed by Western blotting (data not shown) and as such no meaningful information could be gained from the use of the L77P mutant.
The C-terminal half of the Pho4 activation domain between amino acids 87 and 93 is highly methionine rich (Fig. 5A), but despite the presence of two aspartic acid residues at positions 88 and 90, it is not predicted to adopt an α-helical conformation. Nevertheless, this region may be structured in the context of Pho4, and the results from the initial deletion analysis in the context of both Pho4 and LexA implicated this region in transcription activation by Pho4. In an attempt to identify key residues within this region of the activation domain, single alanine substitutions were introduced at each position and the resulting mutants were assayed for their ability to activate PHO5 expression. In particular, given the importance of aromatic residues in transcription activation, we wished to address the contribution of the tryptophan at position 91. The results are shown in Fig. 5D. Mutation of W91 to alanine (W91A) reduced activation of PHO5 fourfold. In contrast, activation was reduced no more than 2.5-fold by introduction of alanines at any of the other six positions, with all proteins being expressed to similar levels (Fig. 5E).
With the exception of W91, the substitution across this region with alanine residues for D88, M89, D90, M92, and M93 failed to dramatically affect the function of the Pho4 transcription activation domain. It was possible, however, that alanines might represent a relatively conservative substitution which, while altering the nature of the amino acid side chains present in this region of Pho4, might nevertheless enable any secondary structure to be conserved. In an attempt to address this possibility, we introduced a glycine residue at position 90. While substitution with alanine at this position resulted in a maximum decrease of Pho4 activity of 2.5-fold, substitution with glycine for aspartic acid at position 90 (D90G mutant) resulted in a 10-fold decrease in PHO5 activity. However, Western blotting (Fig. 5E) revealed that while the D90G mutant was expressed, it was not possible to know whether its slightly faster migration when analyzed by SDS-polyacrylamide gel electrophoresis was caused by an altered conformation or whether it resulted from the removal of a few amino acids by proteolysis. An M92G mutant was poorly expressed (data not shown) and consequently the impact of this mutation on transcription activation could not be assessed.
One of our primary aims in this study was to examine whether mutations within the Pho4 activation domain could be used to separate chromatin opening from transcription activation. The deletion analysis of both Pho4 and the Pho4-Cpf1 chimeric proteins indicated that the two functions were closely linked. To investigate further the requirements for chromatin opening, we also examined three specific point mutations for their ability to modulate chromatin across the PHO5 promoter by using the ClaI accessibility assay. The results are shown in Fig. 5F. Mutant D78A, which was able to activate transcription to WT levels, opened chromatin to around 95%. By contrast, the D78P mutant, which was severely impaired in its ability to activate transcription, failed to open chromatin to more than 15%, and the D90A mutant, which activated PHO5 expression to intermediate levels, between 30 and 45% of that of the WT in different experiments, exhibited an intermediate degree of ClaI accessibility, around 25%. Although we have not tested the entire series of point mutations in the chromatin opening assay, we have yet to find an example by using either the point mutants or the deletion mutants with which efficient chromatin opening was achieved in the absence of transcription activation.
CD analysis of the Pho4 activation domain.
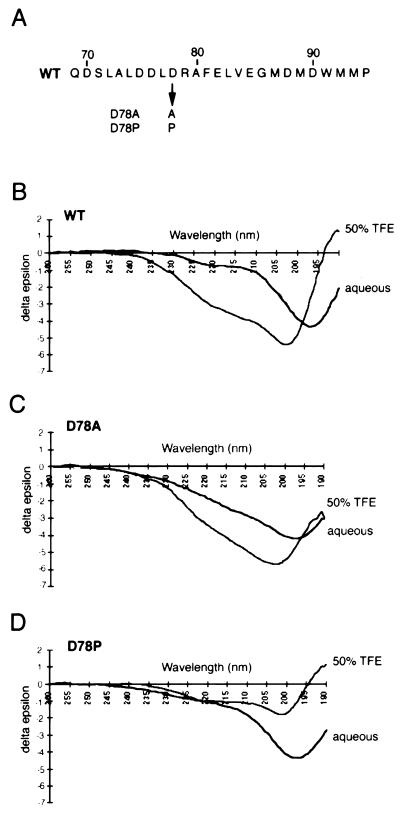
The data described so far would indicate that the Pho4 activation domain comprises residues located between positions 75 and 99, with both the secondary-structure predictions and the mutagenesis indicating that the N-terminal part of this region may adopt an α-helical conformation which contributes to efficient transcription activation. In an attempt to confirm the existence of the potential α-helix, we subjected peptides (Fig. 6A) corresponding to the WT Pho4 activation domain extending from residues 69 to 94 and the D78A and D78P mutants to CD analysis, a technique used to identify secondary structure in proteins and peptides as well as in DNA polymers. The D78A and D78P mutants were chosen since the D78A mutation, which would not be expected to affect any potential α-helical structure, does not reduce the ability of Pho4 to activate transcription while the D78P mutation, which would disrupt any α-helix, severely affects transcription activation. CD analysis was performed with the peptides in either an aqueous buffer or in the presence of 50% trifluoroethylene (TFE), a dehydrating solvent which by providing a less polar environment is useful to reveal hidden structural propensities (for examples, see references 21 and 28). For the WT peptide in aqueous buffer, the marked trough at 197 nm indicates that the peptide is conformationally mobile and, together with the signal at 220 nm, indicates a lack of any significant α-helical content. Similar results have been obtained with a peptide known to adopt an amphipathic α-helical structure (10) and with the activation domains from VP16 (11), CREB (19), and NF-κB (25). By contrast, the same peptide in 50% TFE exhibits a marked decrease in δɛ at 220 nm, consistent with the ca. 30 to 40% of the peptide adopting an α-helical structure. Given that the peptide contains not only the N-terminal α-helical region located between residues 73 and 83 but also residues 84 to 94, which are not expected to participate in any α-helix, we would not anticipate that the WT peptide would in any event contain greater than 50% α-helical structure. A similar result is obtained with a corresponding peptide containing the D78A mutation. In this case, in the presence of 50% TFE, this mutation results again in around 30 to 40% α-helical content, as indicated by the spectrum at 220 nm. By contrast, the α-helical content of the peptide containing the D78P mutation is markedly decreased, to around 10%, with no difference being observed in the value at 220 nm in the presence or absence of TFE. In addition, a peptide extending from amino acids 83 to 106, which lacks the potential α-helical region, failed to reveal any α-helical structure in the presence or absence of TFE (data not shown). The CD analysis is therefore consistent with the N-terminal region of the Pho4 activation domain adopting an α-helical conformation and with the presence of the α-helix in the WT and D78A peptides correlating with the ability of the activation domain to activate transcription.
FIG. 6.
CD analysis of the Pho4 activation domain reveals α-helical structure. (A) Peptides used for CD analysis; (B to D) results of CD analysis of the indicated Pho4 activation domain peptides. Approximate α-helical content (see text) is calculated by using the empirical relationship % helix = (δɛ [at 220 nm] − 0.25)/0.105, as described previously (8).
DISCUSSION
Chromatin plays a major role in the regulation of gene expression. Thus, for a gene to be transcribed, the chromatin assembled across a promoter must adopt a conformation compatible with RNA polymerase and its accessory factors, gaining access to the promoter and initiating transcription. How the modulation of chromatin conformation is coupled to transcription activation remains a largely unresolved question. The ability of the Pho4 transcription factor to induce a chromatin transition and to activate transcription of the PHO5 promoter has provided one of the best systems for studying the concerted regulation of chromatin structure and transcription, with the disruption of the four nucleosomes positioned across the PHO5 promoter being dependent on the presence of the Pho4 activation domain (38). However, the ability of Pho4 to modulate chromatin does not require transcription initiation since deletion of the TATA box, which prevents transcription initiation, still allows the Pho4-dependent chromatin transition to occur (14). This result led to the notion that nucleosome remodeling across the PHO5 promoter may be a prerequisite for transcription activation mediated by the interaction between the Pho4 activation domain and components of the transcriptional machinery.
In this study, we set out to define the requirements within Pho4 for both transcription activation and chromatin modulation with the joint aims of defining the requirements for activation of transcription by Pho4 and of determining whether the requirements for transactivation and chromatin opening within the activation domain were separable. Using a combination of deletion and point mutations, we were able to demonstrate that residues located between amino acids 75 and 99 were both essential and sufficient to catalyze transcription activation, either in the context of Pho4 or when fused to either the LexA or Cpf1 DNA binding domains. Moreover, the analysis of the chromatin structure across the PHO5 promoter using the ClaI accessibility assay revealed a strong correlation between the ability of Pho4 or its derivatives to open chromatin and to activate transcription. In other words, we were unable to identify mutations that were permissive for chromatin opening but deficient in transcriptional activation. This result raises the possibility that chromatin opening and transcription activation are mediated by a single entity that is recruited by the Pho4 activation domain. One candidate for this dual role is the RNA Pol II holoenzyme itself. Thus, when Gal11, a component of the RNA Pol II holoenzyme, is brought to the DNA as a Pho4-Gal11 fusion protein, it is able both to remodel chromatin and to activate transcription from the PHO5 promoter in the absence of a classical activation domain (16). The conclusion from these experiments was that recruitment of the holoenzyme or its associated factors may be sufficient for chromatin remodeling. However, while chromatin opening may be achieved by the artificial targeting of Gal11 to the DNA and recruitment of RNA Pol II and/or associated factors may be sufficient to remodel chromatin, we appreciate fully that this does not necessarily mean that, in its natural environment, the way Pho4 triggers chromatin disruption is by directly recruiting the RNA Pol II holoenzyme to the PHO5 UAS elements.
An alternative possibility is that the Pho4 activation domain may sequentially recruit factors that either remodel chromatin or activate transcription and that these factors have identical, or at least very similar, requirements for interaction with Pho4. The ability of a single activation domain to target multiple distinct factors would not be unreasonable. For example, the activation domain of p53 mediates transcription activation and is targeted directly by the MDM2 repressor, with residues required for interaction with MDM2 also being required for transcription activation (7, 24, 27). At present, our results do not allow us to distinguish between the possibilities outlined above, but the availability of multiple activation domain mutants will be of use in defining physiologically relevant targets for the Pho4 activation domain.
In the course of the work presented here, we found that the effects of activation domain mutations were more severe in the context of the Pho4-Cpf1 chimeric proteins than in the WT Pho4 background. Although other explanations are possible, we view it as likely that in vivo the Cpf1 DNA-binding domain has a lower affinity for the UASp1 element than the Pho4 DNA-binding domain and that a lower affinity for the target sequence may be compensated for in part by the presence of a stronger activation domain. The relationship between the ability to bind DNA and the strength of the activation domain has been pointed out previously (45), and experiments designed to examine DNA binding in vivo have indicated that proteins with identical DNA-binding domains bind DNA with affinities which reflect the strength of the activation domain (40, 43). Thus, activation and DNA-binding domains will cooperate both for transcription activation and DNA recognition, and mutation or alteration of one domain will affect the efficacy of the other. One practical consequence of this is that the requirements for transcription activation which may be identified by mutational analysis are likely to differ depending on the choice of DNA-binding domain. As such, the interpretation of precisely which residues contribute to the activation process may not be straightforward.
Whatever the reason for the increased sensitivity of the Pho4-Cpf1 chimeras to mutagenesis, we were able to exploit this feature to analyze the consequences of single point mutations in the Pho4 activation domain. Together with the data obtained from the deletion mutagenesis, the results appear to be consistent with the activation domain of Pho4 comprising two subdomains, each of which contributes to transcription activation. Although we were unable to obtain any information concerning the potential structure of the C-terminal subdomain, comprising residues 83 to 99, the N-terminal subdomain, located between amino acids 73 and 83, may adopt an α-helical conformation since introduction of alanine for aspartic acid at position 78 failed to affect transcription while substitution with proline for aspartic acid at the same position severely reduced activation.
The conclusion from the mutagenesis was substantiated by the CD analysis of WT and mutant peptides derived from the activation domain. The results showed that the effect of mutations on the ability of the peptides to adopt an α-helical conformation reflected the ability of the same mutations to perturb transcription activation. We note that the peptide corresponding to the activation domain has a propensity to form an α-helix only in 50% TFE, a solvent which provide a less polar environment. Although TFE might be regarded as providing a nonphysiological environment, the information available on activation domains from a number of studies indicates that hydrophobic residues play a key role. Where crystal or NMR structures have been derived from transcription activation domains complexed with repressors, coactivators, or TATA-binding protein-associated factors (TAFs), e.g., p53-MDM2 (24), CREB-CBP (34), or VP16-TAF31 (41), the structures reveal that the key hydrophobic residues are buried in the interface between the interacting proteins and, moreover, that the activation domains in solution are unstructured, adopting an α-helical conformation only on interaction with their target proteins. Thus, the natural environment for a functional transcription activation domain is not an aqueous solution but rather a hydrophobic environment. The purpose of the TFE used in the CD analysis is therefore to provide a more hydrophobic environment which may, to some extent, mimic that of the activation domain when complexed with its target proteins. Indeed, for those activation domains for which both CD and NMR data are available, there is a striking concordance between the results obtained by the two techniques. Thus, for example, the CREB activation domain undergoes a random coil-to-helix transition on interaction with its target, CBP, as determined by NMR (34), with CD analysis revealing the same transition but only in the presence of TFE (19). Similarly, the random coil-helix transition which occurs in the VP16 activation domain on interaction with TAF31 (41) can also be reproduced by using CD analysis (11) but, again, only in the presence of TFE. Although we are fully aware that results from CD analysis should not be taken as conclusive proof of structure, the results obtained are in agreement with the results from the mutagenesis and together are consistent with the N-terminal region of the activation domain having a propensity to adopt an α-helical conformation in vivo, at least when it contacts its appropriate targets.
Although there has long been speculation that activation domains would adopt an α-helical structure, clear evidence that this may be the case has been forthcoming only recently, most likely because the α-helical structure appears to be induced only on binding to an appropriate target, as discussed above. For example, in addition to the CREB-CBP and VP16-TAF31 interactions, the p53 activation domain adopts an α-helical conformation when complexed with the MDM2 repressor (24) and presumably adopts a similar conformation when interacting with target proteins through which p53 mediates transcription activation. It seems likely that many activation domains, including perhaps Pho4, may make use of α-helices to interact with their targets. It is also likely that other structures may be used and that activation domains will contain multiple subregions, each able to contribute to activator-target interactions. In the case of Pho4, the results from the deletion analysis indicated that regions of residues 91 to 98 and 79 to 86, which are present in mutants Δ79-90 and Δ87-99, respectively, each contribute to transcription activation since although these mutants can both activate transcription well, a deletion removing residues 79 to 99 is essentially inactive. The Pho4 activation domain is likely therefore to comprise many residues acting cooperatively to activate transcription, an idea supported by the fact that double point mutations affect transcription activation to a considerably greater degree than single point mutations do (data not shown). However, this should not be taken as evidence that the Pho4 activation domain contacts multiple targets simultaneously. It is equally possible that at any given moment the Pho4 activation domain presents an extended surface for contact with a single target but that the interaction is sufficiently robust to tolerate the loss of a subset of contacts.
Finally, although we have defined specific requirements for transcription activation and chromatin opening on the PHO5 promoter, requirements which presumably reflect the need for Pho4 to interact with or recruit target proteins, it is not evident whether Pho4 or other transcription factors have the same requirements for transcription activation on different promoters. In other words, is it possible that the choice of activator-target protein interactions may be promoter and context dependent? The Pho4 activation domain mutants described here should enable us to address this important question as well as to allow the identification of physiologically relevant targets for the Pho4 activation domain.
ACKNOWLEDGMENTS
We thank Gerard Evan for providing the peptides used for the CD analysis and D. Blaschke for expert assistance.
This work was supported by Marie Curie Cancer Care, the Medical Research Council, the Biotechnology and Biological Sciences Research Council, the Deutsche Forschungsgemeinschaft (SFB 190), and Fonds der Chemischen Industrie.
REFERENCES
- 1.Almer A, Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986;5:2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almer A, Rudolph H, Hinnen A, Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker R E, Masison D C. Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol Cell Biol. 1990;10:2458–2467. doi: 10.1128/mcb.10.6.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbaric S, Münsterkötter M, Svaren J, Hörz W. The homeodomain protein Pho2 and the basic-helix-loop-helix protein Pho4 bind DNA cooperatively at the yeast PHO5 promoter. Nucleic Acids Res. 1996;24:4479–4486. doi: 10.1093/nar/24.22.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbaric S, Münsterkötter M, Goding C, Hörz W. Cooperative Pho2-Pho4 interactions at the PHO5 promoter facilitate Pho4 binding to UASp1 and enhance transactivation by Pho4 at UASp2. Mol Cell Biol. 1998;18:2629–2639. doi: 10.1128/mcb.18.5.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai M, Davis R W. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990;61:437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- 7.Chang J, Kim D H, Lee S W, Choi K Y, Sung Y C. Transactivation ability of p53 transcriptional activation domain is directly related to the binding affinity to TATA-binding protein. J Biol Chem. 1995;270:25014–25019. doi: 10.1074/jbc.270.42.25014. [DOI] [PubMed] [Google Scholar]
- 8.Clark D J, Hill C S, Martin S R, Thomas J O. Alpha-helix in the carboxy-terminal domains of histones H1 and H5. EMBO J. 1988;7:69–75. doi: 10.1002/j.1460-2075.1988.tb02784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cress W D, Triezenberg S J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 10.Degrado W F, Kedzy F J, Kaiser E T. Design, synthesis and characterization of a cytotoxic peptide with melittin-like activity. J Am Chem Soc. 1981;103:679–681. [Google Scholar]
- 11.Donaldson L, Capone J P. Purification and characterization of the carboxy terminal transactivation domain of Vmw65 from herpes simplex virus. J Biol Chem. 1992;267:1411–1414. [PubMed] [Google Scholar]
- 12.Drysdale C M, Duenas E, Jackson B M, Ruesser U, Braus G H, Hinnebusch A G. The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol Cell Biol. 1995;15:1220–1233. doi: 10.1128/mcb.15.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fascher K D, Schmitz J, Hörz W. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 1990;9:2523–2528. doi: 10.1002/j.1460-2075.1990.tb07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fascher K D, Schmitz J, Hörz W. Structural and functional requirements for the chromatin transition at the PHO5 promoter in Saccharomyces cerevisiae upon PHO5 activation. J Mol Biol. 1993;231:658–667. doi: 10.1006/jmbi.1993.1317. [DOI] [PubMed] [Google Scholar]
- 15.Fisher F, Jayaraman P S, Goding C R. C-myc and the yeast transcription factor PHO4 share a common CACGTG-binding motif. Oncogene. 1991;6:1099–1104. [PubMed] [Google Scholar]
- 16.Gaudreau L, Schmid A, Blaschke D, Ptashne M, Hörz W. RNA polymerase holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell. 1997;89:55–62. doi: 10.1016/s0092-8674(00)80182-8. [DOI] [PubMed] [Google Scholar]
- 17.Giniger E, Ptashne M. Transcription in yeast activated by a putative amphipathic alpha helix linked to a DNA binding unit. Nature. 1987;330:670–672. doi: 10.1038/330670a0. [DOI] [PubMed] [Google Scholar]
- 18.Hirst K, Fisher F, McAndrew P C, Goding C R. The transcription factor, the Cdk, its cyclin and their regulator: directing the transcriptional response to a nutritional signal. EMBO J. 1994;13:5410–5420. doi: 10.1002/j.1460-2075.1994.tb06876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua X-Q, Jia W-H, Bullock B P, Habener J F, Weiss M A. Transcriptional activator-cofactor recognition: nascent folding of a kinase-inducible transactivation domain predicts its structure on coactivator binding. Biochemistry. 1998;37:5858–5866. doi: 10.1021/bi9800808. [DOI] [PubMed] [Google Scholar]
- 20.Jackson B M, Drysdale C M, Natarajan K, Hinnebusch A G. Identification of seven hydrophobic clusters in GCN4 making redundant contributions to transcriptional activation. Mol Cell Biol. 1996;16:5557–5571. doi: 10.1128/mcb.16.10.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jasanoff A, Fersht A R. Quantitation of helical propensities from trifluoroethanol titration curves. Biochemistry. 1994;33:2129–2135. doi: 10.1021/bi00174a020. [DOI] [PubMed] [Google Scholar]
- 22.Jayaraman P S, Hirst K, Goding C R. The activation domain of a basic helix-loop-helix protein is masked by repressor interaction with domains distinct from that required for transcription regulation. EMBO J. 1994;13:2192–2199. doi: 10.1002/j.1460-2075.1994.tb06496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaffman A, Herskowitz I, Tjian R, O’Shea E K. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- 24.Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 25.Leinhard Schmitz M, dos Santos Silva M A, Altmann H, Czisch M, Holak T A, Bauerle P A. Structural and functional analysis of the NF-κB p65 C terminus. J Biol Chem. 1994;269:25613–25620. [PubMed] [Google Scholar]
- 26.Leuther K K, Salmeron J M, Johnston S A. Genetic evidence that an activation domain of GAL4 does not require acidity and may form a beta-sheet. Cell. 1993;72:575–585. doi: 10.1016/0092-8674(93)90076-3. [DOI] [PubMed] [Google Scholar]
- 27.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 28.Luo P, Baldwin R L. Mechanism of helix induction by trifluoroethanol: a framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry. 1997;36:8413–8421. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
- 29.Ma J, Ptashne M. A new class of yeast transcriptional activators. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- 30.Mellor J, Jiang W, Funk M, Rathjen J, Barnes C A, Hinz T, Hegemann J H, Philippsen P. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 1990;9:4017–4026. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogawa N, Oshima Y. Functional domains of a positive regulatory protein, PHO4, for transcriptional control of the phosphatase regulon in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2224–2236. doi: 10.1128/mcb.10.5.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Hare P, Williams G. Structural studies of the acidic transactivation domain of the Vmw65 protein of herpes simplex virus using 1H NMR. Biochemistry. 1992;31:4150–4156. doi: 10.1021/bi00131a035. [DOI] [PubMed] [Google Scholar]
- 33.O’Neill E M, Kaffman A, Jolly E R, O’Shea E K. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 34.Radhakrishnan I, Perez-Alvarado G C, Parker D, Dyson H J, Montminy M R, Wright P E. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 35.Regier J L, Shen F, Triezenberg S J. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc Natl Acad Sci USA. 1993;90:883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmid A, Fascher K D, Hörz W. Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication. Cell. 1992;71:853–864. doi: 10.1016/0092-8674(92)90560-y. [DOI] [PubMed] [Google Scholar]
- 37.Shao D, Creasy C L, Bergman L W. Interaction of Saccharomyces cerevisiae Pho2 with Pho4 increases the accessibility of the activation domain. Mol Gen Genet. 1996;251:358–364. doi: 10.1007/BF02172527. [DOI] [PubMed] [Google Scholar]
- 38.Svaren J, Schmitz J, Hörz W. The transactivation domain of Pho4 is required for nucleosome disruption at the PHO5 promoter. EMBO J. 1994;13:4856–4862. doi: 10.1002/j.1460-2075.1994.tb06812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svaren J, Venter U, Hörz W. In vivo analysis of nucleosome structure and transcription factor binding in S. cerevisiae. Methods Mol Genet. 1995;6:153–167. [Google Scholar]
- 40.Tanaka M. Modulation of promoter occupancy by cooperative DNA/binding and activation-domain function is a major determinant of transcriptional regulation by activators in vivo. Proc Natl Acad Sci USA. 1996;93:4311–4315. doi: 10.1073/pnas.93.9.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uesugi M, Nyanguile O, Lu H, Levine A J, Verdine G L. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- 42.Van Hoy M, Leuther K K, Kodadek T, Johnston S A. The acidic activation domains of the GCN4 and GAL4 proteins are not alpha-helical but form beta sheets. Cell. 1993;72:587–594. doi: 10.1016/0092-8674(93)90077-4. [DOI] [PubMed] [Google Scholar]
- 43.Vashee S, Kodadek T. The activation domain of GAL4 protein mediates cooperative promoter binding with general transcription factors in vivo. Proc Natl Acad Sci USA. 1995;92:10683–10687. doi: 10.1073/pnas.92.23.10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venter U, Svaren J, Schmitz J, Schmid A, Hörz W. A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 1994;13:4848–4855. doi: 10.1002/j.1460-2075.1994.tb06811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu R, Reece R J, Ptashne M. Quantitation of putative activator-target affinities predicts transcriptional activating potentials. EMBO J. 1996;15:3951–3963. [PMC free article] [PubMed] [Google Scholar]