Abstract
The formation of cellular condensates, akin to membraneless organelles, is typically mediated by liquid−liquid phase separation (LLPS), during which proteins and RNA molecules interact with each other via multivalent interactions. Gaining a comprehensive understanding of these interactions holds significance in unraveling the mechanisms underlying condensate formation and the pathology of related diseases. In an attempt toward this end, fluorescence microscopy is often used to examine the colocalization of target proteins/RNAs. However, fluorescence colocalization is inadequate to reliably identify protein interaction due to the diffraction limit of traditional fluorescence microscopy. In this study, we achieve this goal through adopting a novel chemical biology approach via the dimerization-dependent fluorescent proteins (ddFPs). We succeeded in utilizing ddFPs to detect protein interaction during LLPS both in vitro and in living cells. The ddFPs allow us to investigate the interaction between two important LLPS-associated proteins, FUS and TDP-43, as cellular condensates formed. Importantly, we revealed that their interaction was associated with RNA binding upon LLPS, indicating that RNA plays a critical role in mediating interactions between RBPs. More broadly, we envision that utilization of ddFPs would reveal previously unknown protein−protein interaction and uncover their functional roles in the formation and disassembly of biomolecular condensates.
INTRODUCTION
Liquid−liquid phase separation (LLPS) plays a vital role in biology, enabling biomolecules to self-assemble into membraneless organelles with diverse specific biological functions. Throughout this process, biomolecules such as proteins and RNAs interact with one another, assembling into condensates. Studying the interactions between biomolecules during LLPS provides valuable insights into the multiplex stages of this fascinating biological process. For example, the intricate details of interactions would help gain a deeper understanding of how biomolecules organize into functional membraneless organelles and participate in various cellular processes, which is essential for investigating the underlying mechanisms of cellular phase separation.
Classical imaging methods used to detect protein−protein interaction mainly consist of the colocalization of fluorophores and the Förster resonance energy transfer (FRET) measurement (Rudolf et al., 2003; Lorkovic et al., 2004). While confocal imaging enables us to visualize the spatial distribution of proteins within cells, but the resolution of an optical microscope is restricted to roughly half the wavelength of light used, owing to light diffraction. As a result, fluorescence microscopy has a resolution limit of about 200 nm. However, protein interaction is typically mediated in close proximity (less than 100 nm), due to nature of intermolecular interaction via electrostatic forces, hydrogen bonding, and hydrophobic effect (Jones and Thornton 1996). Thus, colocalization of proteins using confocal microscopy is not accurate evidence of interaction between proteins. On the other hand, FRET has been used to detect protein−protein interaction by tagging the proteins of interest with donor and acceptor fluorophores. To achieve quantitively measurement of protein interaction, fluorescence lifetime microscopy (FLIM) is often necessary. However, this approach has a high demand for instruments, which may not be readily available in most laboratories (Tramier et al., 2006). Thus, it is desired to develop a new imaging technology that is able to report on and visualize protein interaction during the LLPS approach.
To enable this attempt, we chose FUS and TDP-43 as two representative examples. Both of them have been widely reported in LLPS and bear important pathological correlations, wherein aggregations are the hallmarks of many neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (Fiesel and Kahle 2011; Prasad et al., 2019). FUS and TDP-43, as RNA-binding proteins, participate in the metabolic process of RNA, including splicing, trafficking, miRNA biogenesis, and translation (Ayala et al., 2008; Polymenidou et al., 2011; Tollervey et al., 2011; Carey and Guo 2022). Recent studies show that FUS and TDP-43 colocalize in phase-separated condensates, implying their interaction during the LLPS process (Ling et al., 2010). Furthermore, misfolding of both proteins in aberrantly formed condensates could lead to toxic aggregates in the cytoplasm of neurons, likely due to interacting with distinct sets of proteins or RNAs (Watanabe et al., 2020; Portz et al., 2021). Thus, elucidating the precise nature of interaction between FUS and TDP-43 would help comprehend the underlying mechanism of related processes.
In this work, we applied a set of dimerization-dependent fluorescent proteins (ddFPs) to investigate interaction between proteins associated with LLPS. The adoption of ddFPs offers a robust assay to visualize protein−protein interaction using confocal microscopy. By genetically fusing protein-of-interest with two pairs of ddFPs, we firstly achieved the detection of protein interaction during LLPS both in vitro and in live cells. Subsequently, we applied the ddFP assay to demonstrate the interaction between FUS and TDP-43 during LLPS in response to biological cues. Importantly, we observed that decreased RNA-binding affinity variants of both proteins exhibited reduced their interactions, suggesting that interaction between FUS and TDP-43 was related to RNA binding. Taken together, we confirmed that the ddFPs assay could provide a powerful tool for studying protein−protein interaction during LLPS. Our study provides insights into the interaction between FUS and TDP-43 during LLPS and underscores the crucial role of RNA in facilitating and modulating their interaction. In addition to the cases that we demonstrated in this work, we envision that the ddFP approach could be applied to a wide range of proteins and RNAs that are involved in LLPS, thus allowing the discovery of protein−protein and protein−RNA interactions that mediate the biogenesis of condensates.
RESULTS AND DISCUSSION
The ddFP assay detects protein interaction during in vitro LLPS
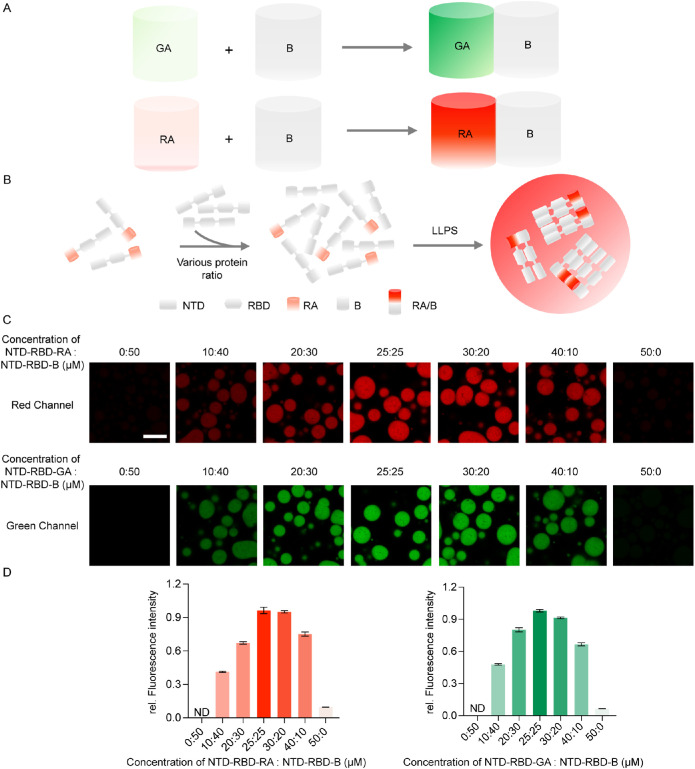
The ddFP was developed as a biosensor to detect protein−protein interaction in living cells. To enable this assay, FPs were mutated into nonfluorescent FP monomers, whose fluorescence would resume when these monomers are placed in proximity to enable the formation of heterodimers (Alford, Abdelfattah et al., 2012; Alford, Ding et al., 2012). As previously reported, one of the monomers (copy A) was mutated to contain a chromophore that is quenched in the monomeric state. While the second monomer (copy B) cannot form a chromophore, it is able to dimerize with the monomer A to activate its fluorescence emission within the AB heterodimers. Such strategies have been applied to generate a green fluorescence version of ddFP named GA/B, as well as a red fluorescent version referred to as RA/B (Figure 1A) (Ding et al., 2015).
FIGURE 1.
The ddFP system can detect protein interaction during in vitro LLPS. (A) Schematic of the ddFP system. (B) The LLPS process of NTD-RBD-RA/B. (C) Fluorescent intensity is ddFP dimer concentration dependent. The total protein concentration is 50 µM, changing the ratio of NTD-RBD-RA/GA and NTD-RBD-B. Fluorescence intensity of droplets decrease with a decrease in concentration of ddFP dimer. (D) Quantification of the fluorescence intensity of the droplets in (C). LLPS of proteins is induced in 20 mM HEPES, 140 mM NaCl, 5% PEG 3350. All measurements were conducted in triplicates and error bars are calculated as standard deviations. Scale bar = 10 µm.
To test whether the ddFP assay was capable of detecting protein interaction during LLPS, we initially chose TDP-43 without its C-terminal domain as a model system. The combination of N-terminal domain (NTD) and RNA-binding domain (RBD) of TDP-43 (NTD-RBD) was shown in previous studies to form liquid droplets via the oligomerization of NTD-RBD (Carter et al., 2021). Thus, we genetically linked GA, B, and RA independently to the C-terminal of NTD-RBD and purified the resulting fusion proteins: NTD-RBD-GA, NTD-RBD-B, and NTD-RBD-RA. When these proteins are subjected to buffer with low salt concentration (50 mM NaCl), we expected that interaction between monomeric NTD-RBD-RA and NTD-RBD-B would increase, resulting in the dimerization of ddFP and formation of droplets that would emit fluorescence (Figure 1B). As expected, NTD-RBD-GA and NTD-RBD-B resulted in droplets with green fluorescence (Figure 1C, upper panel); and the same observation was made for the NTD-RBD-RA and NTD-RBD-B combination (Figure 1C, lower panel). To test whether the fluorescence signal arose from the dimerization of ddFPs, we altered the concentration of dimers by varying the ratio of NTD-RBD-GA/NTD-RBD-RA and NTD-RBD-B. As expected, varying the dimer concentration resulted in droplets displaying different fluorescent intensity, supporting that the fluorescent signal indeed was induced by fluorescent dimers (Figure 1C). Furthermore, we found that the fluorescence intensity reached the peak with a ratio of 1:1 (Figure 1, C and D), suggesting that the fluorescent intensity of droplets was dependent on the concentration of ddFP in dimeric states. To assess the background fluorescence in the absence of LLPS, we mixed NTD-RBD-GA/RA with NTD-RBD-B of 1:1 stoichiometry in high salt condition of 300 mM NaCl, which inhibits the LLPS of NTD-RBD. In the absence of LLPS, we observed minimal fluorescence signal in both the green and red channels (Supplemental Figure S1), indicating that the GA/B or RA/B dimer could only engage to emit fluorescence when the GA or RA are in close proximity with B.
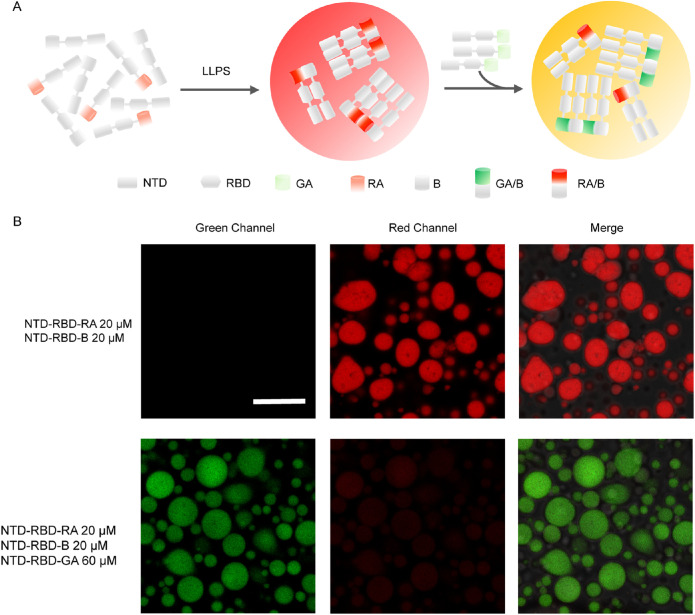
To further confirm the reversibility of GA/B and RA/B interaction, we also performed competitive experiments using two dimers: GA/B and RA/B (Figure 2A). First, droplets consisting of NTD-RBD-RA and NTD-RBD-B were performed to induce the formation of RA/B that emitted red fluorescence. Subsequently, a large amount of NTD-RBD-GA was introduced to the sample. If the RA/B interaction was reversible, NTD-RBD-GA could compete with NTD-RBD-RA, forming GA/B dimers that would emit green fluorescence (Figure 2A). As expected, an obvious transition from red to green fluorescence was observed when the GA/B dimers competed with RA/B dimers (Figure 2B). This result suggested that the dimerization of GA/B and RA/B were reversible and the fluorescence intensity could directly reflect the interaction between GA-tagged proteins or RA-tagged proteins in droplets.
FIGURE 2.
The dimerization of GA/B and RA/B were reversible. (A) Process diagram of competition experiment. After the formation of droplet containing RA/B dimer, abundant NTD-RBD-GA was added to the droplets to compete with NTD-RBD-RA. (B) After adding NTD-RBD-GA, green fluorescence appears and red fluorescence intensity decreases. NTD-RBD-GA competes with NTD-RBD-RA to form fluorescent dimers. Scale bar = 10 µm.
We next asked whether the ddFP assay would work for other proteins, particularly proteins whose phase separation is mediated by the interaction of their IDR regions. To this end, we chose FUS as another model system to prove the ability of ddFP to detect protein interaction during the IDR-induced LLPS. MBP-FUS was purified as a soluble protein, wherein MBP served as a solubilizing tag. When we used the TEV enzyme to cut off MBP tags, FUS would spontaneously form liquid droplets due to the reduction in solubility of proteins (Supplemental Figure S2A). As expected, we discovered green fluorescence in droplets formed by FUS-GA and FUS-B (Figure 4B, upper panel); and the same observation was found for the FUS-RA and FUS-B combinations. (Supplemental Figure S2B, lower panel). Similar to previous experiments, we altered the ratio of FUS-GA/FUS-RA and FUS-B, discovering that the fluorescence intensity was dependent on the concentration of fluorescent dimers (Supplemental Figure S2B). Additionally, we observed that the fluorescence intensity decreased as the ratio of FUS-GA/FUS-RA to FUS-B increased (Supplemental Figure S2C), proving that the dimeric ddFP determines the fluorescent intensity of the droplet. It is worth noting that the fold-of-change for FUS-GA/RA to FUS-B is smaller than that of NTD-RBD, suggesting that the ddFP system would generate different dynamic ranges of fluorescent signals, largely dependent on the protein-of-interest. For instance, the in vitro assay used 10 µM FUS and 50 µM NTD-RBD, thus generating different signals of fluorescence upon dimer formation. In addition, the FUS condensates exhibit a higher mobility than the NTD-RBD condensates, suggesting different interaction strength of dimers. Hence, the stronger NTD-RBD dimer would result in a brighter fluorescence than the FUS dimers.
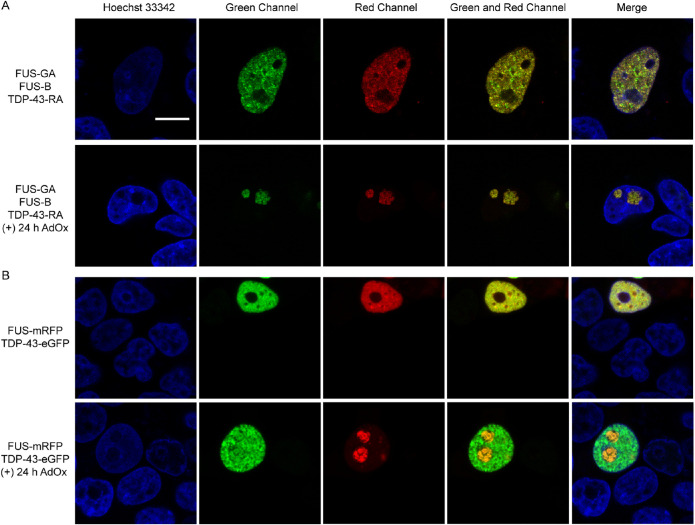
FIGURE 4.
FUS and TDP-43 interact with each other upon LLPS. (A) FUS and TDP-43 distribute in the nucleoplasm (upper panel). Under AdOx treatment, TDP-43 enters the nucleus due to the interaction with FUS (lower panel). (B) FUS-mRFP and TDP-43-eGFP distribute in the nucleoplasm (upper panel). FUS-mRFP and TDP-43-eGFP colocalize in nucleolus after AdOx treatment for 24 h (lower panel). Scale bar = 10 µm.
We also performed competitive experiments to prove the reversibility of the formation of fluorescent dimers (Supplemental Figure S3A). Droplets containing FUS-GA and FUS-B were operated to generate GA/B dimers, emitting green fluorescence. Subsequently, a large amount of FUS-RA was added to compete FUS-B with GA/B dimers, resulting red fluorescence (Supplemental Figure S3A). Obviously, red fluorescence was discovered as a result of the generation of RA/B dimer. We found both green and red fluorescence in droplets, suggesting the concurrence of RA/B and GA/B dimers (Supplemental Figure S3B). This result confirmed that the formation of GA/B and RA/B dimers were reversible and the fluorescence signal could directly reflect the interaction between GA-tagged proteins or RA-tagged proteins in droplets during IDR induced LLPS. All these results collectively suggest that the ddFP system enables investigating protein−protein interaction during in vitro LLPS.
The ddFP system detects protein interaction during LLPS in live cells
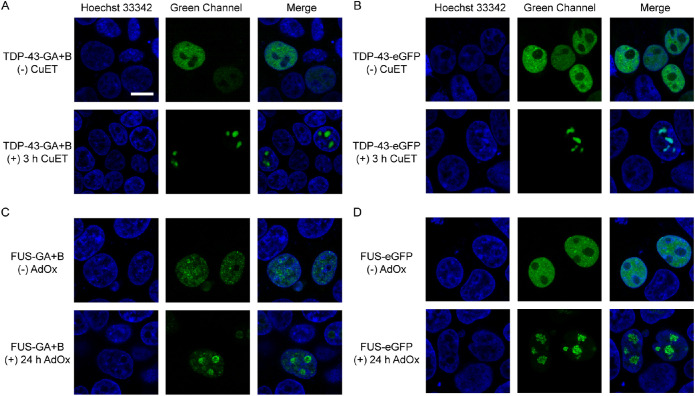
After demonstrating the detection of protein interaction during in vitro LLPS, we attempted to explore whether ddFP can be used in complex cellular environment. To this end, we genetically fused GA or B to the C-terminus of TDP-43, generating TDP-43-GA and TDP-43-B. Previous studies have reported that bis(diethyldithiocarbamate)–copper (CuET) inhibits the p97 AAA- ATPase and causes proteotoxic stress in cells, inducing TDP-43 to form promyelocytic leukemia (PML) nuclear body in the nucleus (Skrott et al., 2017; Jung et al., 2023). We transfected plasmids TDP-43-GA and TDP-43-B into human embryonic kidney (HEK) 293T cells and observed significant green fluorescent droplets after treatment with CuET. This result confirmed that the ddFP system enables detecting protein interaction upon LLPS (Figure 3A). As a control experiment, we fused TDP-43 with the green fluorescent protein eGFP and observed the same phenomenon (Figure 3B).
FIGURE 3.
The ddFP system can detect protein interaction during LLPS in living cells. (A) Cells transfected with TDP-43-GA/B with or without drugs treatment. TDP-43 distribute equally in the nucleus in stress-free condition. When treated with CuET, TDP-43 forms PML. Scale bar = 10 µm. (B) Cells transfected with TDP-43-eGFP exhibited the same phenomenon with that in (A). (C) Cells transfected with FUS-GA/B with or without drugs treatment. FUS is distributed equally in the nucleus in stress-free condition. When treated with AdOx, FUS forms condensate in nucleolus. (D) Cells transfected with FUS-eGFP exhibited the same phenomenon with that in (C). Scale bar = 10 µm.
We further applied the ddFP system to another LLPS-associated protein, FUS. Similarly, we fused GA and B to the C-terminus of FUS to monitor its interaction upon LLPS in cells. As reported before, FUS in nucleoplasm can form nuclear granular structures due to enhanced protein interaction after the treatment of Adenosine dialdehyde (AdOx), a common inhibitor of adenosylmethionine-dependent methyltransferases (Madhavan et al., 1988; Dormann et al., 2012). Compared with the diffuse green fluorescent signals under no pressure condition, we discovered bigger and brighter green fluorescent droplets after treatment with AdOx (Figure 3C). Meanwhile, we also used eGFP to track FUS and acquired similar phenomenon (Figure 3D).
It is worth noting that the green fluorescence signals arose from TDP-43 and FUS exhibited difference before drug treatment. There was significant diffusive fluorescence in cells transfected with TDP-43-GA/TDP-43-B while nearly no green fluorescence signal in samples transfected with FUS-GA/FUS-B (Figure 3, A and C). As the fluorescence signals arise from dimers formed after protein interaction, these differences indicated that no interaction between FUS-GA and FUS-B occurred without AdOx treatment while TDP-43-GA interact with TDP-43-B even without drug treatment. However, both TDP-43-eGFP and FUS-eGFP emitted strong green fluorescence before the formation of droplets (Figure 3, B and D). These results prove that the ddFP system exhibited advantages over traditional FP in detecting protein interaction occurred in the early stage of LLPS, wherein the droplets are still not formed.
FUS and TDP-43 interact during LLPS in live cells
Many evidences have proved that FUS and TDP-43 were both observed in cellular condensates (Kim et al., 2010). We wondered whether FUS and TDP-43 interacted with each other during phase separation in cells. Inspired by previous experiments, we attempted to utilize the ddFP system to explore the interaction between FUS and TDP-43. We introduced two pairs of fluorescent dimers and simultaneously transfected three recombinant plasmids: FUS-GA, FUS-B, and TDP-43-RA into HEK293T cells. In the absence of AdOx, both FUS and TDP-43 exhibited diffusive fluorescence structure in the nucleus (upper panels of Figure 4, A and B). After AdOx treatment to induce phase separation of FUS, we observed red fluorescent signals from the TDP-43-RA/FUS-B dimers colocalized with the green fluorescent droplets of FUS-GA/FUS-B dimers in the nucleolus (Figure 4A, lower panel). This observation indicates that TDP-43-RA and FUS-B reside in proximity to generate red fluorescent RA/B heterodimers, resulting from the LLPS-dependent interaction between FUS and TDP-43. To further confirm this result, we constructed the tandem plasmid that simultaneously expressed FUS-GA, FUS-B, and TDP-43-RA under either CMV or IRES promoters. Similar results were observed that showed that the FUS and TDP-43 exhibited both green and red fluorescence in nucleolus when cells were treated with AdOx (Supplemental Figure S4).
To confirm the interaction between FUS and TDP-43, we conducted the FRET assay, wherein the eYFP and mRFP served as the FRET donor and acceptor, respectively. We cotransfected TDP-43-eYFP and FUS-mRFP and measured the fluorescence lifetime of eYFP using FLIM. When cells were treated with AdOx for 24 h, TDP-43 entered nucleolus with FUS (Supplemental Figure S5A), consistent with observations using the TDP43-RA and FUS-B expression (Figure 4A). In the nucleolus, the fluorescence lifetime of eYFP was determined to be ∼ 2.5 ns (red arrow in Supplemental Figure S5B, quantification in Supplemental Figure S5D), which was significantly shorter than the eYFP lifetime as ∼ 2.8 ns in other locations of nucleus (white arrow in Supplemental Figure S5B, quantification in Supplemental Figure S5D). Consistent to the latter value, the lifetime of eYFP was also measured to be ∼ 2.9 ns in cells that expressed just the TDP-43-eYFP (Supplemental Figure S5C, quantification in Supplemental Figure S5D). These results suggest that FUS and TDP-43 indeed interact upon LLPS in nucleolus, further supporting the capacity of ddFP to detect protein−protein interaction.
To address the concern that the interaction of RA or B monomers may cause the generation of red fluorescence, we selected two classical fluorescent proteins, mRFP and eGFP, that did not interact with each other to track FUS and TDP-43 upon LLPS. Colocalization of green and red fluorescence verified that the interaction between FUS and TDP-43 was the main factor of formation of red fluorescent heterodimers (Figure 4B, lower panel). In addition, we performed immunofluorescence staining assay of FUS and nucleolar marker protein NPM1 to confirm the presence of FUS and TDP-43 in nucleolus upon LLPS. As expected, FUS and TDP-43 were colocalized with NPM1, indicating that both FUS and TDP-43 localize in nucleolus under the treatment of AdOx (Supplemental Figure S6). These results suggest that under drug treatment, FUS forms nuclear granular structures and TDP-43 was recruited into nucleolus due to interaction with FUS.
Because GA and RA could form dimmers with B, it is necessary to test whether the innate affinity between the monomeric units could induce interaction between proteins that otherwise would not form dimers. To evaluate this possibility, we simultaneously expressed FUS-GA, FUS-B, and Halo-RA in cells that were treated with AdOx to induce FUS condensation into nucleolus. As previously demonstrated, FUS condensates could enable formation of the FUS-GA/B dimers and emit green fluorescence (Supplemental Figure S7A). By contrast, Halo-RA exhibited minimal red fluorescence, mostly due to the lack of its interaction with FUS-B. When this experiment was conducted using FUS-eGFP and Halo-mRFP, both green and red fluorescence signals were observed in different cellular locations, indicating minimal interactions (Supplemental Figure S7B). These results suggest that Halo protein could be expressed in cells and the lack of red fluorescence in Supplemental Figure S7A was due to the lack of Halo-RA and FUS-B dimer. Taken together, these results suggest that the strength of GA/RA and B interaction in the ddFP system would not induce artificial protein−protein interactions.
RNA plays a vital role in the interaction of FUS and TDP-43
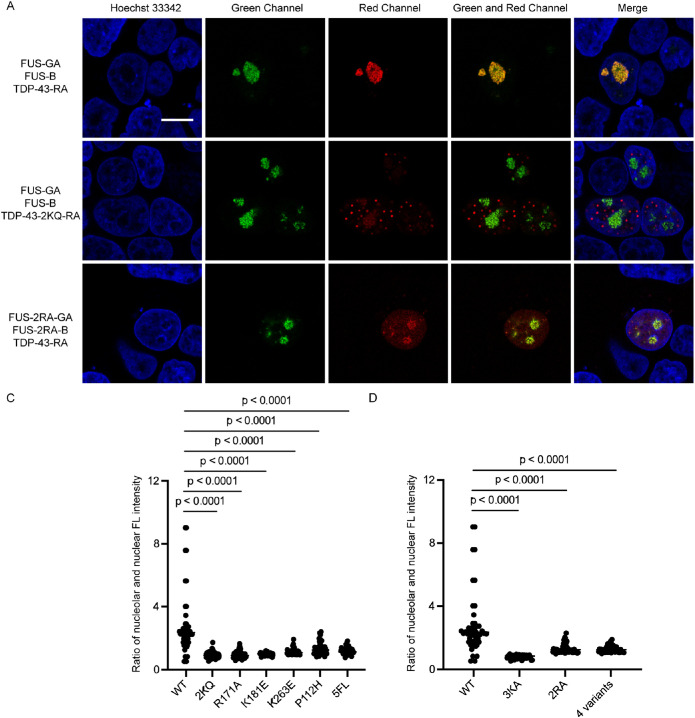
We next asked whether the interaction of FUS and TDP-43 is associated with RNA. To this end, we examined the interaction between FUS and TDP-43 using mutant proteins with RNA-binding deficiency. We firstly chose a TDP-43 variant with low RNA-binding capacity, 2KQ (K145Q/K192Q), which mimics the posttranslational acetylation of two lysine residues by glutamic acid (Cohen et al., 2015). Interestingly, we discovered that the interaction between 2KQ mutant and FUS was significantly reduced (Figure 5A, first and second panels). In addition to 2KQ, we selected another five variants (R171A, K181E, K263E, P112H, and 5FL) that exhibited decreased RNA-binding affinity through different mechanisms (Yu et al., 2021). R171A, K181E, and K263E replace positive charge amino acids by negative residues to reduce electrostatic interactions with nucleic acids (Kovacs et al., 2009; Chen et al., 2019; Sun et al., 2021). P112H mutant of TDP-43 RRM1 disrupts the interactions between Trp113 and nucleic acids (Moreno et al., 2015). By replacing five phenylalanine residues with leucine, the surface of 5FL mutant that binds to RNA is destroyed (Elden et al., 2010). Similar to the result of 2KQ, we noted that the interaction between all these TDP-43 variants and FUS were compromised to different extents (Supplemental Figure S8). To quantify the extent of interaction between FUS and TDP-43, we defined the ratio of nucleolar average red fluorescence intensity to the nuclear fluorescence, confirming that all RNA-binding deficient mutants of TDP-43 show diminished levels of interaction with FUS (Figure 5B). These results suggest that RNA could mediate TDP-43 and FUS interaction.
FIGURE 5.
The interaction of FUS and TDP-43 is associated with RNA binding. (A) Cells transfected with FUS and TDP-43. FUS and TDP-43 have interaction (first panel). Cells transfected with FUS and TDP-43-2KQ. The protein interaction of FUS and TDP-43-2KQ decreases (second panel). Cells transfected with TDP-43 and FUS RNA-binding deficient mutant 2RA. The protein interaction of FUS-2RA and TDP-43 decreases (third panel). Cells were treated with 25 µM AdOx for 24 h to induce LLPS of FUS. (B) Protein interaction was compared by quantifying the ratio of red fluorescence in nucleolar and nuclear based on Figure 5A and Supplemental Figure S8. (C) Quantification of the ratio of red fluorescence in nucleolar and nuclear based on Figure 5B and Supplemental Figure S9. The t test was carried out for 50 cells, p < 0.0001. Scale bar = 10 µm.
To further verify this note, we selected three FUS variants with insufficient RNA-binding affinity (3KA, 2RA, and 4-variants) to detect their interactions with wild-type TDP-43. The 3KA variant harbors three lysine residues mutating to alanine, which disturbs FUS hairpin inserting into the major groove of the RNA (Liu et al., 2013). Replace two arginine residues (R371, R372) by alanine (2RA) influences the FUS loop recognition and reduce RNA binding. Four mutated residues (F288, Y325, K315, K316) on binding surface (4-variants) decrease the RNA-binding capacity (Loughlin et al., 2019). As excepted, these FUS mutants also exhibited a reduced interaction with TDP-43 than the FUS wild-type sequence (third lane of Figure 5A and Supplemental Figure S9). Quantification results also confirms that there is significant difference between these variants and the wild-type FUS (Figure 5C). Meanwhile, GFP/RFP colocalization experiments were carried out to confirm the reduced interactions between the RNA-binding deficient mutants of FUS and TDP-43 (Supplemental Figure S10). Taken together, decreased RNA-binding affinity of FUS or TDP-43 weakens the interaction between these two RNA-binding proteins, further indicating that RNA binding mediates TDP-43 and FUS interaction during LLPS.
CONCLUSION
In summary, we applied the ddFP system to the investigation of protein interaction during phase separation both in vitro and in living cells. The unique working mechanism of the ddFP system allows us to robustly observe protein−protein interaction, overcoming the potential artifact that could be brought by the colocalization of proteins measured by confocal microscopy. We achieved visualizing the interaction between FUS and TDP-43 when they come into proximity during LLPS. Further results revealed that the interaction between FUS and TDP-43 was associated with RNA binding. To confirm the role of RNA in protein−protein interaction, FUS and TDP-43 variants with deficient RNA-binding affinity were used to observe that the interactions between these variants were significantly weakened owing to their insufficient binding to RNA. This discovery offers a new perspective for comprehending the interaction between FUS and TDP-43 during phase separation. We envision that the ddFP system could be applied to investigate more interaction between proteins or RNAs that are associated with the biogenesis of condensates during LLPS.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Plasmid construction
Plasmid MBP-FUS-His was provided by Liu lab from Dalian Institute of Chemical Physics. Plasmid wtTDP-43-tdTOMATAHA (#28205) was purchased from Addgene. Genes of GA-DEVD-GBNES (#61020) and RANES (#61019) were purchased from Addgene and vectors containing FUS and TDP-43 were added using the PIPE method. Plasmids of TDP-43 variants 2KQ, R171A, K181E, K263E, P112H, 5FL were constructed by using quick change methods from wtTDP-43-tdTOMATAHA. FUS plasmids 3KA, 2RA, and 4 variants were mutated from FUS-His. Tandem plasmid that simultaneously expressed FUS-GA, FUS-B, and TDP-43-RA under either CMV or IRES promoters was constructed by Gibson assembly method to add three genes in one vector.
Protein purification
Plasmids NTD-RBD-GA, NTD-RBD-B and NTD-RBD-RA, MBP-FUS-GA, MBP-FUS-B, MBP-FUS-RA were transformed into BL21 (DE3) E. coli cells, respectively. When cells were grown to OD600 of 0.8, 0.5 mM IPTG was added to induce protein expression at 18°C for 24 h. Cultured cells for NTD-RBD- proteins were harvested and resuspended in buffer (20 mM Tris-HCl, 1 M NaCl, 2 mM imidazole, pH = 8.0). Cells expressing recombinant proteins were thawed and lysed by sonication at 4°C. Lysed cells were centrifuged for 30 min at 16,000 rpm twice. The supernatant was collected and loaded onto a 6 ml Ni-NTA column and washed with buffer A (20 mM Tris, 1 M NaCl, 2 mM imidazole, pH = 8.0) for 10-column volume. Proteins were then eluted by gradient addition of buffer B (20 mM Tris-HCl, 500 mM imidazole, pH = 8.0). Proteins were then buffer-exchanged into storage buffer (20 mM HEPES, 300 mM NaCl, 1 mM DTT, pH = 7.5) using a desalting column.
Cultured cells for NTD-RBD- proteins were harvested and resuspended in buffer (20 mM HEPES, 300 mM NaCl, 10% Glycerol, 10 μM ZnCl2, 10 mM imidazole, pH = 8.0). Cells expressing recombinant proteins were thawed and lysed by sonication at 4°C. Lysed cells were centrifuged for 30 min at 16,000 rpm twice. The supernatant was collected and loaded onto a 6 ml Ni-NTA column and washed with buffer A (20 mM HEPES, 300 mM NaCl, 10% glycerol, 10 μM ZnCl2, 10 mM imidazole, pH = 7.5) until UV at 100 mAU and washed with wash buffer (20 mM HEPES, 1 M NaCl, 10% Glycerol, 10 μM ZnCl2, 10 mM imidazole, pH = 7.5) for 10 CV. The supernatant was washed with buffer A until conductivity at plateau. Proteins were then eluted by gradient addition of buffer B (20 mM HEPES, 300 mM NaCl, 10% glycerol, 10 μM ZnCl2, 500 mM imidazole, pH = 7.5). The protein fractions were identified by SDS–PAGE analysis, pooled, and concentrated. The proteins were further loaded on a Mono QTM 10/100 GL MonoBeadsTM column (Cytiva) in buffer QA (20 mM Tris-HCl, 100 mM NaCl, pH = 7.8) and eluted with buffer QB (20 mM Tris, 1M NaCl, pH = 7.8) to remove bound RNA. Proteins were then buffer-exchanged into storage buffer (20 mM HEPES, 300 mM NaCl, 1 mM DTT, pH = 7.5) using a desalting column.
All protein-containing fractions were identified by SDS–PAGE gel analysis, pooled, and concentrated. No significant impurities were identified. MALDI-MS was performed to confirm the mass of proteins (Supplemental Figure S11A) and Circular Dichroism was performed to text second structure of proteins (Supplemental Figure S11B).
Phase separation assay in vitro
Proteins NTD-RBD-GA, NTD-RBD-B, and NTD-RBD-RA were stored at high concentration salt solution (20 mM HEPES, 300 mM NaCl, 1 mM DTT, pH = 7.5). First, NTD-RBD-GA/NTD-RBD-RA and NTD-RBD-B were mixed together in tube 1. Then mixed dilution buffer using low salt solution (20 mM HEPES, pH = 7.5) with 50% PEG 3350 stock solution was added in tube 2. Finally, all the solution in tube 2 was added to tube 1 and mixed well. NTD-RBD- proteins were in phase separation in the final condition: 20 mM HEPES, 140 mM NaCl, 5% PEG 3350; the total protein final concentration was 50 μM. The ratio of NTD-RBD-GA and NTD-RBD-B was changed to get different fluorescent intensity droplets. After all the samples were prepared, 10 μL sample was added to the slide with a spacer, covered with a coverslip, and then turned over and left in an upright position for 10 min, so that the droplets could be adsorbed on the surface of the coverslip.
For FUS-GA/B/RA proteins, TEV protease was added to induce phase separation. The final total protein concentration was 10 μM, and the final LLPS condition was 20 mM HEPES, 200 mM NaCl, 10 µM TEV, 5% PEG 3350. The slides were left in an upright position for 30 min before confocal imaging with Leica Stellaris 5 High-resolution Laser Confocal Fluorescence Microscope with a 63 × oil objective lens.
Cell culture
HEK293T cells were purchased from ATCC® (CRL-3216) and cultured in complete Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies) supplemented with 10% FBS (Life Technologies) and 1 × penicillin streptomycin-glutamine (PSQ, Life Technologies) at 37°C under 5% CO2 atmosphere in a HERAcell Vios 160i LK CO2 incubator (Thermo Fisher Scientific). When the cell density reached about 90%, cell passage was started with 1 × TrypLETM Express (Life Technologies).
Cell imaging assay with confocal microscopy
Cells were seeded on poly-L-lysine−coated coverslips (2.5 × 105 cells) one day prior to the experiment. When ∼ 50% confluency was reached, cells were transiently transfected with 1 µg FUS-GA, 1 µg FUS-B, and 1 µg TDP-43-RA via 6 µl lipofection (X-TremeGene 9, Roche) in 300 µl Opti-MEMTM (Life Technologies), or 1 µg tandem plasmid CMV-FUS-GA-IRES-FUS-B-IRES-TDP-43-RA via 2 µl lipofection in 100 µl Opti-MEMTM. Proteins were transiently expressed for 24 h at 37 °C under 5% CO2 in a HERAcell Vios 160i LK CO2 incubator (Thermo Fisher Scientific). Cells were treated with 25 µM AdOx (Selleck Chem) for 24 h and stained with 1 µM Hoechst 33342 (Enzo) for 10 min before confocal imaging. Cells transfected with 1 µg TDP-43-GA and 1 µg TDP-43-B or 1 µg TDP-43-eGFP only were treated with 5 µM CuET (TCI) for 3 h before imaging. All confocal images of cells were got with Leica Stellaris 5 High-resolution Laser Confocal Fluorescence Microscope.
FRET-FLIM imaging assay
Cells were seeded as described previously by confocal imaging assay and transfected with TDP-43-eYFP only. Cells transfected with TDP-43-eYFP and FUS-mRFP were treated with 25 µM AdOx (Selleck Chem) for 24 h before confocal imaging. All lifetime images were acquired by Leica Stellaris 8 Ultra-high Resolution Confocal Microscope. eYFP was excited by 513 nm laser line. Image analysis was achieved using Leica Application Suite X (LAS X) to get fluorescence lifetime.
Immunofluorescence staining
Cells were transfected by cell imaging assay as described previously. Next day, fixed the cells with 4% formaldehyde (Pierce) for 10 min at room temperature. After washing with PBS three times for 10 min each time, cells were permeabilized with 0.5% Triton X-100 (Sangon Biotech) in PBS for 20 min at room temperature and then washed with PBS three times for 10 min per each time. Cells were then blocked with 3% BSA (Sigma Aldrich, A9085) solution in PBS for 1 h at room temperature, followed by staining with primary antibodies diluted in PBS containing 3% BSA for 1 h. Primary antibody and dilution used was mouse anti-nucleophosmin (Abcam, FC82291, 1:200), After washing with PBS three times for 10 min each time, cells were stained with secondary antibody diluted in PBS containing 3% BSA for 1 h at room temperature in the dark. Secondary antibody was goat anti-mouse IgG secondary antibody, Alexa Fluor 647 (Abcam, ab150115, 1:500). Cells were washed with PBS twice and stained with 1 μM Hoechst 33342 (Enzo) for 10 min. Coverslips were mounted on glass slides with 50% glycerol and sealed with transparent nail polish.
Supplementary Material
Acknowledgments
The authors thank Instrumentation and Service Center for Molecular Sciences at Westlake University for the assistance on fluorescence microscopy. This work is partially supported by the Research Center for Industries of the Future (RCIF) at Westlake University.
Abbreviations used:
- AdOx
Adenosine dialdehyde
- ALS
amyotrophic lateral sclerosis
- BRD
RNA-binding domain
- CUET
bis(diethyldithiocarbamate)–copper
- ddFPs
dimerization-dependent fluorescent proteins
- FRET
Förster resonance energy transfer
- FTD
rontotemporal dementia
- HEK
human embryonic kidney
- LLPS
liquid-liquid phase separation
- NTD
N-terminal domain
- PML
promyelocytic leukemia
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E23-11-0442) on January 17, 2024.
REFERENCES
- Alford SC, Abdelfattah AS, Ding Y, Campbell RE (2012). A fluorogenic red fluorescent protein heterodimer. Chem Biol 19, 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford SC, Ding Y, Simmen T, Campbell RE (2012). Dimerization-dependent green and yellow fluorescent proteins. ACS Synth Biol 1, 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Zago P, D’Ambrogio A, Xu YF, Petrucelli L, Buratti E, Baralle FE (2008). Structural determinants of the cellular localization and shuttling of TDP-43. J Cell Sci 121, 3778–3785. [DOI] [PubMed] [Google Scholar]
- Carey JL, Guo L (2022). Liquid-Liquid Phase Separation of TDP-43 and FUS in Physiology and Pathology of Neurodegenerative Diseases. Front Mol Biosci 9, 826719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter GC, Hsiung CH, Simpson L, Yang H, Zhang X (2021). N-terminal Domain of TDP43 Enhances Liquid-Liquid Phase Separation of Globular Proteins. J Mol Biol 433, 166948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HJ, Topp SD, Hui HS, Zacco E, Katarya M, McLoughlin C, King A, Smith BN, Troakes C, Pastore A, Shaw CE (2019). RRM adjacent TARDBP mutations disrupt RNA binding and enhance TDP-43 proteinopathy. Brain 142, 3753–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TJ, Hwang AW, Restrepo CR, Yuan CX, Trojanowski JQ, Lee VM (2015). An acetylation switch controls TDP-43 function and aggregation propensity. Nat Commun 6, 5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Li J, Enterina JR, Shen Y, Zhang I, Tewson PH, Mo GC, Zhang J, Quinn AM, Hughes TE, et al. (2015). Ratiometric biosensors based on dimerization-dependent fluorescent protein exchange. Nat Methods 12, 195–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Madl T, Valori CF, Bentmann E, Tahirovic S, Abou-Ajram C, Kremmer E, Ansorge O, Mackenzie IR, Neumann M, Haass C (2012). Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J 31, 4258–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. (2010). Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiesel FC, Kahle PJ (2011). TDP-43 and FUS/TLS: cellular functions and implications for neurodegeneration. FEBS J 278, 3550–3568. [DOI] [PubMed] [Google Scholar]
- Jones S, Thornton JM (1996). Principles of protein-protein interactions. Proc Natl Acad Sci USA 93, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Sun J, Hsiung CH, Lance Lian X, Liu Y, Zhang X (2023). Nuclear bodies protect phase separated proteins from degradation in stressed proteome. bioRxiv doi:10.1101/2023.04.19.537522 [Google Scholar]
- Kim SH, Shanware NP, Bowler MJ, Tibbetts RS (2010). Amyotrophic lateral sclerosis-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to co-regulate HDAC6 mRNA. J Biol Chem 285, 34097–34105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs GG, Murrell JR, Horvath S, Haraszti L, Majtenyi K, Molnar MJ, Budka H, Ghetti B, Spina S (2009). TARDBP variation associated with frontotemporal dementia, supranuclear gaze palsy, and chorea. Mov Disord 24, 1843–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Albuquerque CP, Han JS, Lagier-Tourenne C, Tokunaga S, Zhou H, Cleveland DW (2010). ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci USA 107, 13318–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Niu C, Ren J, Zhang J, Xie X, Zhu H, Feng W, Gong W (2013). The RRM domain of human fused in sarcoma protein reveals a non-canonical nucleic acid binding site. Biochim Biophys Acta 1832, 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic ZJ, Hilscher J, Barta A (2004). Use of fluorescent protein tags to study nuclear organization of the spliceosomal machinery in transiently transformed living plant cells. Mol Biol Cell 15, 3233–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin FE, Lukavsky PJ, Kazeeva T, Reber S, Hock EM, Colombo M, Von Schroetter C, Pauli P, Cléry A, Mühlemann O, et al. (2019). The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity. Molecular Cell 73, 490–504.e6. [DOI] [PubMed] [Google Scholar]
- Madhavan GV, McGee DP, Rydzewski RM, Boehme R, Martin JC, Prisbe EJ (1988). Synthesis and antiviral evaluation of 6’-substituted aristeromycins: potential mechanism-based inhibitors of S-adenosylhomocysteine hydrolase. J Med Chem 31, 1798–1804. [DOI] [PubMed] [Google Scholar]
- Moreno F, Rabinovici GD, Karydas A, Miller Z, Hsu SC, Legati A, Fong J, Schonhaut D, Esselmann H, Watson C, et al. (2015). A novel mutation P112H in the TARDBP gene associated with frontotemporal lobar degeneration without motor neuron disease and abundant neuritic amyloid plaques. Acta Neuropathol Commun 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al. (2011). Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portz B, Lee BL, Shorter J (2021). FUS and TDP-43 Phases in Health and Disease. Trends Biochem Sci 46, 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Bharathi V, Sivalingam V, Girdhar A, Patel BK (2019). Molecular Mechanisms of TDP-43 Misfolding and Pathology in Amyotrophic Lateral Sclerosis. Front Mol Neurosci 12, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf R, Mongillo M, Rizzuto R, Pozzan T (2003). Looking forward to seeing calcium. Nat Rev Mol Cell Biol 4, 579–586. [DOI] [PubMed] [Google Scholar]
- Skrott Z, Mistrik M, Andersen KK, Friis S, Majera D, Gursky J, Ozdian T, Bartkova J, Turi Z, Moudry P, et al. (2017). Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature 552, 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Chen W, Chen L, Zheng W (2021). Exploring the molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43 using molecular dynamics simulation and free energy calculation. J Comput Chem 42, 1670–1680. [DOI] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, et al. (2011). Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14, 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramier M, Zahid M, Mevel JC, Masse MJ, Coppey-Moisan M (2006). Sensitivity of CFP/YFP and GFP/mCherry pairs to donor photobleaching on FRET determination by fluorescence lifetime imaging microscopy in living cells. Microsc Res Tech 69, 933–939. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Inami H, Oiwa K, Murata Y, Sakai S, Komine O, Sobue A, Iguchi Y, Katsuno M, Yamanaka K (2020). Aggresome formation and liquid-liquid phase separation independently induce cytoplasmic aggregation of TAR DNA-binding protein 43. Cell Death Dis 11, 909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Lu S, Gasior K, Singh D, Vazquez-Sanchez S, Tapia O, Toprani D, Beccari MS, Yates JR 3rd, Da Cruz S, et al. (2021). HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science 371, eabb4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.