Abstract
Immune cells employ diverse mechanisms for host defense. Macrophages, in response to TLR activation, assemble aggresome-like induced structures (ALIS). Our group has shown TLR4-signaling transcriptionally upregulates p62/sequestome1, which assembles ALIS. We have demonstrated that TLR4-mediated autophagy is, in fact, selective-autophagy of ALIS. We hypothesize that TLR-mediated autophagy and ALIS contribute to host-defense. Here we show that ALIS are assembled in macrophages upon exposure to different bacteria. These structures are associated with pathogen-containing phagosomes. Importantly, we present evidence of increased bacterial burden, where ALIS assembly is prevented with p62-specific siRNA. We have employed 3D-super-resolution structured illumination microscopy (3D-SR-SIM) and mass-spectrometric (MS) analyses to gain insight into the assembly of ALIS. Ultra-structural analyses of known constituents of ALIS (p62, ubiquitin, LC3) reveal that ALIS are organized structures with distinct patterns of alignment. Furthermore, MS-analyses of ALIS identified, among others, several proteins of known antimicrobial properties. We have validated MS data by testing the association of some of these molecules (Bst2, IFITM2, IFITM3) with ALIS and the phagocytosed-bacteria. We surmise that AMPs enrichment in ALIS leads to their delivery to bacteria-containing phagosomes and restricts the bacteria. Our findings in this paper support hitherto unknown functions of ALIS in host-defense.
Host cells employ several strategies to defend against infection. Immune cells upregulate p62/sequestosome, which in turn assembles aggregate-like structures (ALIS). However, their precise nature and contribution to host defense are yet to be established.
The authors find that ALIS contributes by restricting bacterial proliferation. Structured illumination microscopy studies unraveled localization of ALIS constituents, demonstrating their organization. Mass spectrometry revealed numerous immuno-protective molecules including antimicrobial-peptides (AMPs) as ALIS constituents.
These findings suggest that host cells assemble AMP-positive ALIS to facilitate their delivery to pathogen-containing phagosomes to effectively neutralize invading pathogens.
INTRODUCTION
Immune system engages distinct complex signaling pathways to enable higher organisms to survive in the microbial world. This involves recognition of foreign components and mounting of appropriate immune response(s) with minimal damage to the host (Ulevitch, 2004; Takeuchi and Akira, 2010). The recognition of nonself is mediated by antigen presenting cells such as dendritic cells (DCs) and macrophages that serve as sentinels of the mammalian immune system. These sentinel cells express a repertoire of pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), which recognize pathogen-associated molecular patterns (PAMPs) and activate diverse signaling pathways ranging from activation of kinases like IKK, MAP kinases; transcription factors like NF-κB, AP-1; autophagy and assembly of ALIS (Mellman and Steinman, 2001; Vural and Kehrl, 2014). PAMP recognition by TLR4 of DCs and macrophages have been shown to accumulate aggresome-like induced structures (ALIS) in response to lipopolysaccharide (LPS), consisting of LC3 (Lelouard et al., 2002, 2004; Vyas et al., 2008; Fujita et al., 2011). ALIS are distinct from the classical aggresomes, in that they are independent of microtubule organizing centre (MTOC) and lack a vimentin cage (Johnston et al., 1998; Heath et al., 2001). They are also not detergent soluble (insoluble to 2% tritonX-100) (Fujita et al., 2011). D/ALIS are assembled by p62 in infected cells and are also positive for ubiquitin-, p62- and LC3 (Canadien et al., 2005; Herter et al., 2005; Fujita et al., 2011; Thomas et al., 2012).
ALIS-like structures have been reported in different cell lines and upon different types of stress (Lelouard et al., 2004; Bjørkøy et al., 2005). We observed the formation of ALIS in response to LPS and Escherichia coli particles as well as in response to stimulation by many pathogenic and nonpathogenic bacteria, which mediates immune response via different TLRs. However, we also found the ALIS puncta associate with the bacteria inside the cells. This insinuated the involvement and significance of ALIS sequestration in host immune response. The presence of ALIS has the potential to curb bacterial survival inside the host cell, whereas the absence of it, helped the intracellular bacteria to proliferate. ALIS are, essentially, cytosolic structures which, in GFP-LC3 stably expressing background, can be easily visualized as puncta in the cytoplasmic space of the cell (Fujita et al., 2011). Microscopically, we have observed, their size varies from 0.5 to 4 μm in diameter. When p62 and ubiquitin are stained, these colocalize with GFP-LC3-positive puncta (Fujita et al., 2011; Cabe et al., 2018). Given that ALIS are formed in response to stimulation via PRRs, we were curious to find the structural arrangement of the known constituent proteins of ALIS. Although confocal microscopy can provide a resolution of up to 220 nm (laterally; MacDonald et al., 2015), recent advances in imaging techniques have made it easier to observe information at a higher resolution. Thus, we employed superresolution structured illumination microscopy (SIM) which gives a resolution of up to 120 nm in the xy axis (MacDonald et al., 2015; Wegel et al., 2016; Lu et al., 2019). SIM imaging revealed a systematic positioning alignment of the three proteins of ALIS.
To understand and characterize ALIS constituents, we performed proteomic, biochemical and imaging analyses. TLR signaling leads to the production of, along with several cytokines, antimicrobial peptides (AMPs; Flannagan et al., 2009, 2015; Lawrence and Kornbluth, 2012; Stephan et al., 2016; Abuaita et al., 2018; Odendall and Kagan, 2019), thus aiding and activating both innate and adaptive immune systems. We were keen to find whether any of the known AMPs got sequestered along with ALIS upon activation by LPS. We identified several proteins as constituents of ALIS by the mass spectrometry data that are reported in defense response pathways, including AMPs. Here we demonstrate experimentally that three proteins, Bst-2 or Tetherin, IFITM2 and IFITM3 associate with ALIS. We also show that IFITM2 and IFITM3 are upregulated at the protein level upon TLR4 activation. The AMPs as part of the ALIS also associate with the internalized bacteria, thus, suggestively limiting the survival of the pathogen inside the cellular milieu. This study supports a potential role for ALIS in host defense.
RESULTS
Bacterial stimulation induces ALIS assembly and ALIS associates with bacteria-containing phagosomes
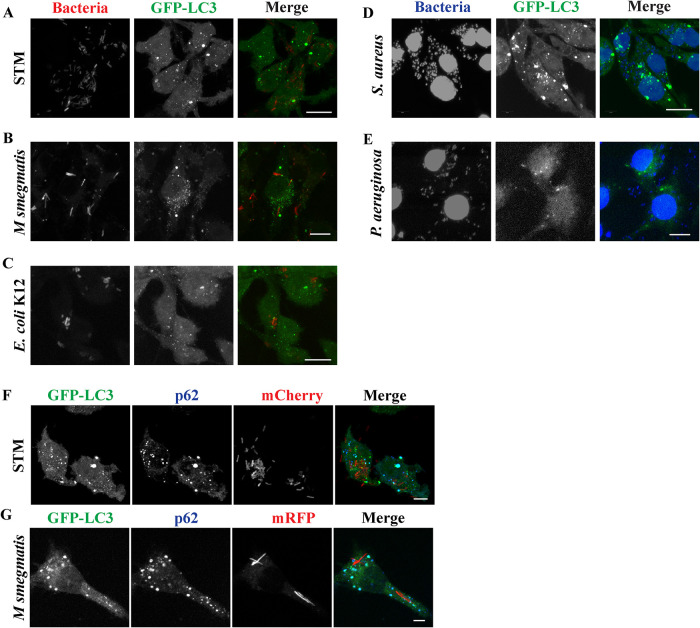
We have used RAW264.7 macrophage cells that stably express GFP-LC3, to analyse ultrastructural features of ALIS. LPS, a component of Gram-negative bacteria cell wall and an endotoxin, has been shown to induce the formation of ALIS in these macrophages (Canadien et al., 2005; Fujita et al., 2011). In response to stimuli by microbial products macrophages form ubiquitin, LC3 and p62-positive punctate structures in the cytoplasm (Canadien et al., 2005; Szeto et al., 2006). Because ALIS formation in macrophages occurs in response to TLR signaling, we were curious to know if macrophages assembled similar structures, as a response to exposure to bacteria, mediated by other PRRs as well. Towards this end, we exposed RAW GFP-LC3 cells to either mCherry expressing Salmonella Typhimurium (STM) or mRFP expressing Mycobacterium smegmatis or mCherry labeled E. coli K12 or pathogenic strain of Staphylococcus aureus (SA5) or Pseudomonas aeruginosa (PA1) (Figure 1A-E). Upon treatment with any of these bacterial strains, cells formed GFP-LC3 puncta similar to those observed in response to LPS as reported previously (Fujita et al., 2011). While STM, E. coli K12, and P. aeruginosa are Gram-negative that are recognized by TLR4; M. smegmatis and S. aureus are Gram-positive bacteria that act through TLR2 (Tapping et al., 2000; Fournier and Philpott, 2005; Fournier, 2013; Pattabiraman et al., 2017) suggesting the engagement of any of these TLRs results in assembly of GFP-LC3 puncta. In macrophages infected by STM-mCherry or M. smegmatis-mRFP these puncta were confirmed to be ALIS by immunostaining the infected cells for p62 (Figure 1, F and G). Both p62 and GFP-LC3 colocalized in ALIS. The pattern of association of the proteins was consistent with that observed in ALIS formed in response to LPS that is positive for p62 and ubiquitin (Supplemental Figure S1A). Another observation was the close association of ALIS with phagosomes formed as a result of infection (Figure 2, A and D) or vacuoles formed upon LPS stimulation (Supplemental Figure S1B). Interestingly, it was noticed that most of the ALIS was associated with the bacteria-containing phagosomes. In many cells these ALIS is present in close vicinity to phagosomal STM or M. smegmatis (Figure 2, A and D). It was also observed that the STM led to ALIS assembly in about 80% of cells upon infection (Figure 2B) and with prolonged incubation of 16 h, the number ALIS associated with the bacteria increased from about 10 to 25% (Figure 2C).
FIGURE 1:
Formation of GFP-LC3 (green) dots in RAW264.7 macrophages. RAW264.7 cells stably expressing GFP-LC3 form GFP-LC3 positive puncta after 6 to 12 h post infection by STM-mCherry (A), Mycobacterium smegmatis-mRFP (B), E. coli K12-mCherry (C), DAPI stained strains of Staphylococcus aureus (D), and Pseudomonas aeruginosa (E). RAW 264.7 cells form ALIS positive for GFP-LC3 (green) and p62 (blue) in response to infection with STM-mCherry (F) or M. smegmatis-mRFP (G). Scale bars, 5μm.
FIGURE 2:
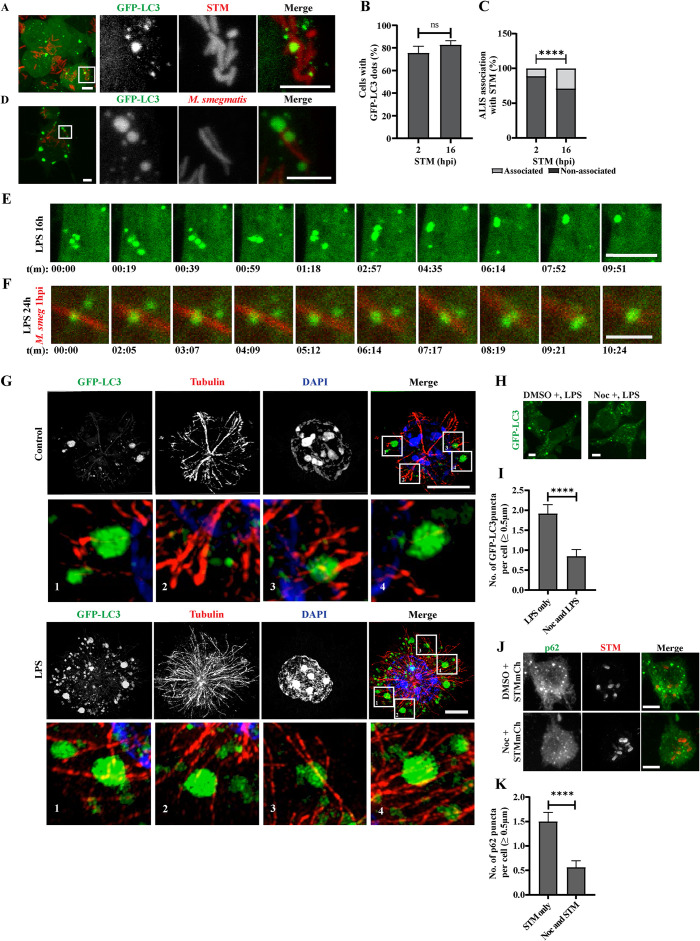
ALIS associates with phagosomes and its assembly depends on microtubules. ALIS associates with bacteria containing phagosomes as detected in macrophage cells after stimulation with STM-mCherry (A) or M. smegmatis-mRFP (D). Inset contrast has been adjusted for better visibility. Graph represents the percentage of cells forming GFP-LC3 dots upon infection with STM (B) and fraction of total associated and non-associated number of ALIS associating with the bacteria upon infection in percentage (C). Statistical significance was calculated on pooled data from experimental replicates using Fisher’s exact test on fraction of total represented as percentage. The experiments were performed in triplicates. A minimum 150 cells were counted for each repeat.
RAW GFP-LC3 macrophages were stimulated with LPS for 16 h (E) or LPS (24 h) followed by infection with M. smegmatis mRFP for 1 h (F). Images from live cells are shown for indicated time points. LPS treated cells with fusion of GFP-LC3 puncta (Supplemental Movie 1) and localization and fusion of GFP-LC3 puncta on bacteria (Supplemental Movies 2 and 3). Occurrence of ALIS fusion is shown in the insets. ALIS association with tubulin was observed in RAW GFP-LC3 cells under control conditions or by stimulating cells with LPS for 12 h and immunostaining for tubulin (G). Inset contrast has been adjusted for better visibility. Same is shown in three-dimensional projection for control cell (Supplemental Movie 4) and LPS treated cell (Supplemental Movie 5). Representative images of RAW GFP-LC3 cells where nocodazole was added before activating the cells by LPS stimulation or STMmCh infection (H and J). Graph represents average number of GFP-LC3 (I) or p62 (K) puncta formed per cells. For microtubule destabilization experiments, statistical significance was calculated on pooled data from experimental replicates using unpaired t test. For calculating the number of puncta per cell, the experiments were performed in duplicates for both LPS and STM. A minimum 50 cells were counted for each repeat for LPS and 40 cells for STMmCh. Scale bars, 5 μm. (P) * < 0.05, (P) ** < 0.005, (P) *** < 0.0005, (P) **** < 0.0001, ns = nonsignificant. Data are represented as mean ± SEM.
Movie S1.
RAW GFP‐LC3 macrophages treated with LPS, sequester ALIS wherein smaller GFP‐LC3 puncta fuse together to form larger structures.
Movie S2.
Smaller GFP‐LC3 structures assemble into larger puncta and associate with the bacteria in macrophage cells pretreated with LPS and infected with M. smegmatis‐mRFP.
Movie S3.
Mature ALIS puncta remain stabilized in macrophages infected with M. smegmatis‐mRFP after LPS pretreatment.
Movie S4.
3D projection of untreated RAW GFP‐LC3 cells immunostained for tubulin (red) and DAPI (blue) to observe ALIS association with microtubules.
Movie S5.
3D projection of ALIS association with microtubules in GFP‐LC3 positive cells treated with LPS for 12 hours followed by immunostaining for tubulin (red) and DAPI (blue).
Because formation of ALIS is robust in LPS treated cells as compared with cells incubated with bacteria, to get comprehensive insight into the ALIS association with the bacteria, we used cells prestimulated with LPS for bacterial infection at low multiplicity of infection (MOI). ALIS formation is a dynamic process (Lelouard et al., 2002) and with time ALIS matures in response to LPS stimulation wherein smaller GFP-LC3 puncta comes together to form larger structures (Figure 2E, Supplemental Movie 1). A similar observation was reported using GFP-Ub and following their association with DALIS by Lelouard et al. (2004). The clustering of small GFP-LC3 puncta around the bacteria was observed and quantified (Supplemental Figure S1C). With time, more such smaller structures assembled around the internalized bacteria. The assembly of ALIS from smaller structures to the larger ones was also observed in macrophage cells infected with M. smegmatis-mRFP, pretreated with LPS (Figure 2F; Supplemental Movie 2; Supplemental Figure S1D; Supplemental Movie 3), wherein larger ALIS remained stabilized. This led us to hypothesize that microtubules play a role for ALIS movement. To establish whether this assembly of ALIS depended on microtubules we explored the association of ALIS and microtubules, by stimulating cells with LPS and immunostaining for tubulin. These cells were analyzed by SR-SIM. ALIS appeared to associate with the tubulin filaments indicating its relevance in forming matured structures (Figure 2G; Supplemental Movies 4 and 5). We also observed that in LPS stimulated macrophages microtubules undergo stabilization (Xu and Harrison, 2015; Supplemental Figure S1E), we could speculate that this might aid in ALIS mobility. To further validate the requirement of microtubule stability for ALIS, cells were stimulated with LPS or STM-mCherry after nocodazole treatment. In these cells the number of GFP-LC3 puncta (≥ 0.5 µm) counted per cell upon nocodazole treatment followed by LPS stimulation decreased significantly, (Figure 2, H and I). In the case of cells treated with nocodazole, after, internalizing STM first, similar observations were made with the number of p62 positive puncta (≥ 0.5 µm) per cell (Figure 2, J and K). This is consistent with previous reports from several other groups that (D)ALIS are motile, and this motility is required for their fusion to form large mature ALIS (Lelouard et al., 2004; Canadien et al., 2005). Nocodazole treatment affects maturation of puncta into structures of size greater than 0.5 µm, without blocking their formation, demonstrating that ALIS are dynamic structures, capable of undergoing microtubule-based movement. Moreover Spartin, a microtubule interacting and trafficking motif-containing protein, associated with ALIS and knockdown of Spartin in RAW 264.7 macrophages affected DALIS formation (Karlsson et al., 2014).
ALIS contributes in compromising bacterial proliferation in macrophages
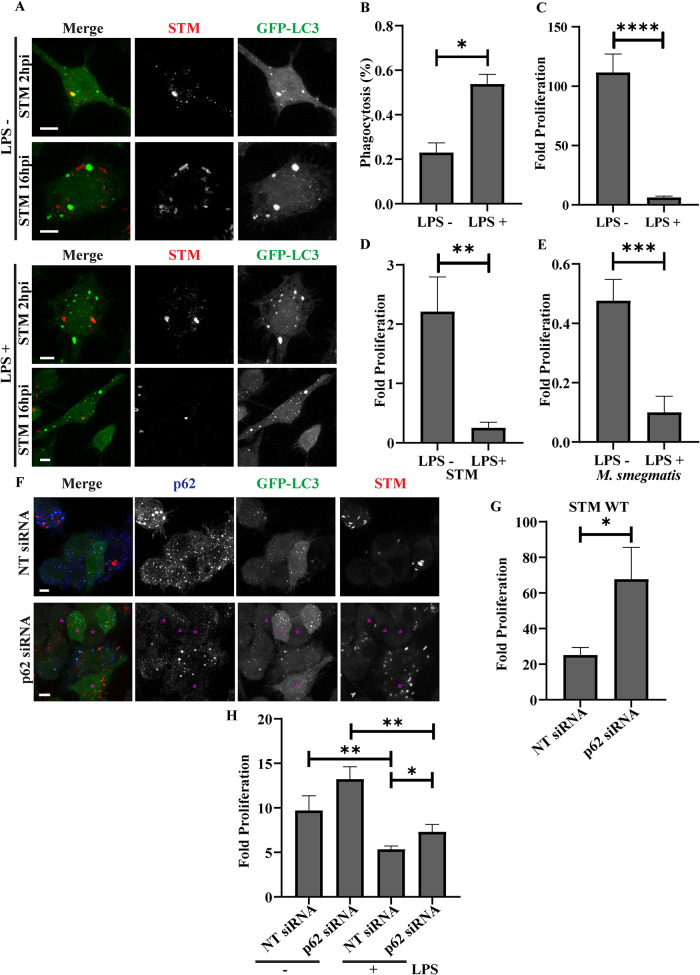
The close association between bacteria and ALIS was intriguing as it suggests a role for ALIS in controlling bacterial growth. To explore this, RAW GFP-LC3 cells were infected with STM-mCherry in LPS prestimulated or unstimulated conditions (Figure 3A). While the internalization of STM was higher in LPS stimulated RAW264.7 macrophages with assembled ALIS, there was a significant decrease in the number of intracellular bacteria as measured by fold proliferation at 16hpi versus 2hpi in these cells as compared with unstimulated cells lacking ALIS assembly (Figure 3, B and C). To validate this interesting observation in primary macrophages, we repeated these experiments in peritoneal macrophages. Intracellular proliferation of STM, M. smegmatis, E. coli K12 or STM WT expressing listeriolysin O (LLO) was observed in peritoneal macrophages (Figure 3, D and E; Supplemental Figure S2, A and B). As expected, in the case of STM WT, M. smegmatis and E. coli K12, a reduction in bacterial proliferation was observed in these cells after LPS stimulation. However, the STM WT::LLO variant managed to escape this effect. LLO is derived from Listeria monocytogenes, which enables these bacteria to evade the vacuolar compartment and remains in the cytoplasm (Hamon et al., 2006; Ruan et al., 2016). Because, STM WT::LLO is also capable of exiting the phagosomal compartment (Chowdhury et al., 2022), its survival in the activated macrophage environment may give clues of ALIS associating with phagosomes in enhancing its cytotoxic effect on the encapsulated bacteria. LPS mediated activation of macrophages can lead to the onset of several proinflammatory pathways capable of killing the phagocytosed bacteria (Viola et al., 2019). Because p62 is essential for the assembly of ALIS and a knockdown of p62 with siRNA results in hampering the ALIS assembly (Fujita et al., 2011; Cabe et al., 2018), we employed p62-siRNA to establish the effect of ALIS in controlling bacterial survival. While lack of ALIS assembly in p62 knockdown cells, did not affect the internalization of bacteria (Figure 3F), the bacterial load was observed to be more in p62-siRNA treated cells compared with nontarget siRNA treated macrophages. These observations were quantified by measuring the mean intensity of fluorescence of STM-mCherry (Supplemental Figure S2C). Intracellular survival assay (ICSA) to determine the fold proliferation of STM in RAW GFP-LC3 macrophages also confirmed the increase in growth of the bacteria in cells without ALIS (Figure 3G). In cell populations with depleted p62, stimulation with LPS along with bacteria still showed increased bacterial proliferation though at much reduced level than in cells without LPS treatment (Figure 3H). These results demonstrate that ALIS contributes to host defense.
FIGURE 3:
ALIS compromises bacterial proliferation in macrophages. Representative images of RAW GFP-LC3 cells infected with STM (immunostained with anti-Salmonella antibody) at 2 hpi and 16 hpi with or without LPS prestimulation (A). Percentage of phagocytosis (B) and fold proliferation (C) of STM in RAW GFP-LC3 cells with or without LPS prestimulation (MOI = 10; n = 3, N = 3). Fold proliferation of STM (D) and M. smegmatis (E) in peritoneal macrophages with or without LPS prestimulation (MOI = 10; n = 3, N = 3).
Representative images showing bacterial load in RAW GFP-LC3 transfected with p62 siRNA as compared with nontarget siRNA (F). Cells with low p62 expression and no GFP-LC3 puncta are marked with a magenta asterisk. Quantification is included in Supplemental Figure S2C. Fold proliferation of STM in RAW GFP-LC3 cells transfected with either nontarget siRNA or p62 siRNA (G). Comparison between fold proliferation of STM in RAW GFP-LC3 transfected with nontarget or p62 specific siRNA and the change in proliferation in cells prestimulated with LPS (H). (MOI = 10; n = 3, N = 1). Statistical significance was calculated on experimental replicates using unpaired t test. (P) * < 0.05, (P) ** < 0.005, (P) *** < 0.0005, (P) **** < 0.0001, ns = non-significant. Data are represented as mean ± SEM. Scale bars, 5 μm.
Structured-illumination microscopy demonstrates ALIS manifesting as a well-organized structure
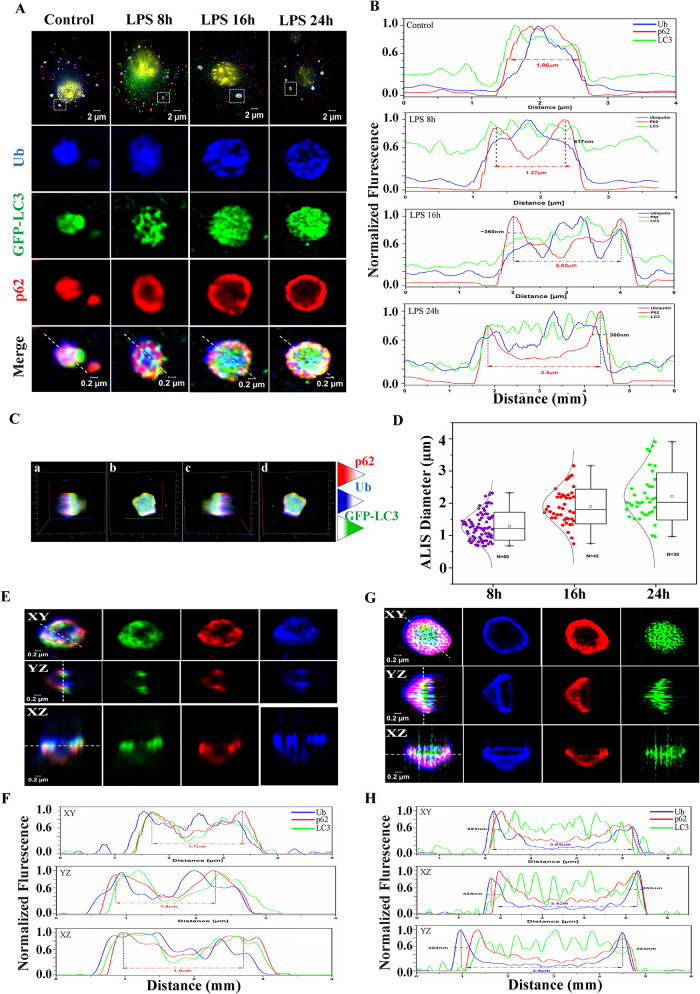
To better understand the formation of ALIS, we looked at its intracellular structural arrangements. To analyze LPS-induced ALIS further, cells stained for p62 or ubiquitin were observed under 3D super-resolution structured illumination microscopy (SR-SIM). In cells immunostained for p62 after 24 h of stimulation, it was observed that p62 organizes in a ring-like pattern, while LC3 is localized mainly in the center of the ALIS structures (Supplemental Figure S3A). Immunostaining for multi-ubiquitin demonstrated colocalization of ubiquitin with LC3, albeit both diffused across ALIS (Supplemental Figure S3B). To understand the arrangement of the three proteins with respect to each other, GFP-LC3 positive cells were stimulated with LPS and immunostained simultaneously for p62 and multi-ubiquitin. A closer inspection by 3D SIM, into the structures indicated an organized arrangement of p62 primarily on the periphery of ALIS, followed by LC3 and ubiquitin filling up the center (Figure 4A). Interestingly, we also observed that ALIS underwent structural maturity with respect to the overall size of the aggresome, with smaller structures (0.5 to 1.5 μm in diameter) representing early stages and larger structures representing more mature ALIS (3.0 μm or more). However, different stages of maturity, represented by variable sizes of ALIS, were observed in 24 h LPS stimulated macrophages. In ALIS of sizes lower than 1.5 μm, LC3 and ubiquitin appeared to interact with p62 at different sites. It was also noticed that at later time points of LPS stimulation (16 h onwards), smaller structures were observed alongside the larger structures (Supplemental Figure S3C). We assessed three individual ALIS of different sizes as compared with diffused puncta observed in unstimulated cells via line profile analysis (Figure 4, A and B). In ALIS corresponding to early stages of maturity, p62, LC3 and ubiquitin appeared throughout ALIS. As represented, most of the ALIS of this profile appeared at approximately 1.0 μm in diameter. As the size of ALIS increased, the orientation of the three proteins with respect to each other also altered. As shown in the data, with increasing size, p62 appeared to acquire more peripheral position, while ubiquitin and GFP-LC3 remained dispersed at the center. However, while the signal for GFP-LC3 was more pronounced at the core, ubiquitin signal seemingly redistributed and dispersed away from the center. We present an ALIS puncta, from 16 h post LPS stimulation, analyzed by reconstituting the SIM images in three-dimension in Figure 4C. ALIS appeared as a cone-like projection. Here, yet again, p62, ubiquitin and GFP-LC3 followed the same pattern of arrangement as observed in earlier images. Rotation by 90 degrees of the three-dimensional projection, p62 appeared across the entire length of the cone of ALIS, as its expression was observed at the base of the cone and continued till the end of the vertex of the cone. LC3 protein could be detected from the center onwards and continued till the vertex of the cone of ALIS. The signal for ubiquitin could be seen from the base of the cone and continued to appear till more than half of the structure.
FIGURE 4:
SIM images representing: Relative positioning of the three proteins, GFP-LC3 (green), p62 (red) and ubiquitin (blue), with respect to each other in ALIS after LPS stimulation in 8, 16, and 24 h as compared with control (untreated) cells (A). Line profile analyses of ALIS at corresponding time points of LPS stimulation (B). Three-dimensional reconstitution from SIM images of a single ALIS observed after coimmunostaining for p62 and ubiquitin in RAW GFP-LC3 cells (a), structure rotated by: 90 degrees (b), 180 degrees (c) and 270 degrees (d) (C). Whisker plot showing the number of ALIS with increased diameter at 8, 16, and 24 h of LPS stimulation, a minimum of 15 cells were used in each time point (D). Two different patterns of organization of the three proteins observed in a mature ALIS (up to 24 h of LPS) along with their respective line profile analyses (E–H).
The mean ALIS diameter across is indicated via the whisker plot (Figure 4D). Two diverse patterns of ALIS at the 24-h time point were observed. Pattern I show (Figure 4, E and F) that two proteins ubiquitin and p62 are partially localized on the periphery core, and the LC3 is localized at the center with a diameter of ∼1.8 μm. Pattern II indicates the proteins are arranged sequentially as an order of three distinct compartments ubiquitin, p62 and LC3 at the center core of the ALIS (Figure 4, G and H) with a diameter of ∼3.2 μm on the XY plane. Significantly, the LC3 is localized as individual spots at the center on XY plane and tube-like rod at the XZ plane. The overall assembly based on p62 and ubiquitin looks like a cone or the bowl-like pattern shows the ubiquitin on the outer layer of the ALIS and p62 is located at the inner layer and the LC3 is spotted as a linear rod-like pattern on central three-dimensional spaces. The SIM imaging and analysis clearly demonstrated that ALIS are assembled in response to infection in immune cells, with individual constituents positioning spatially, in an organized pattern, suggesting a yet to be revealed functional contribution of these structures in host defense.
ALIS enriched lysate fractions were analysed by mass spectrometry and proteomic profiling
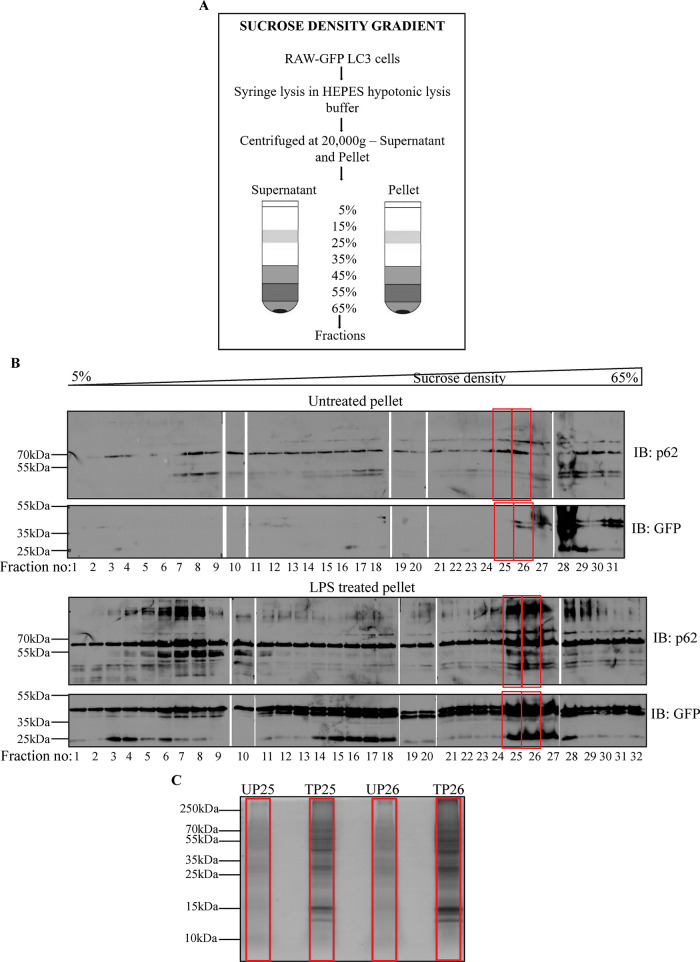
Because ALIS appears to be a well-organized structure, with specific proteins positioning to specific locations, and engages in pathogen clearance, we were interested in identifying components that form ALIS and contribute to its functionality. Towards that end we employed sucrose density gradient (SDG) to separate cellular components from extracts of untreated and LPS treated cells. A discontinuous gradient of sucrose solution was prepared (Strømhaug et al., 1998; Clayton and Shadel, 2014; Páleníková et al., 2021). Samples prepared from supernatant and pellet fractions of RAW264.7 cells stably expressing GFP-LC3, both unstimulated and stimulated with LPS, were layered on top of the gradients separately. Fractions were collected from top to bottom after separation (lowest to highest density; Figure 5A Schematic). The presence of ALIS components in the fractions was assessed by selecting fractions with enriched GFP-LC3 as analyzed by SDS–PAGE and fluorescence spectrophotometer (Supplemental Figure S4, A and B). The fractions were analyzed by immunoblotting with anti-GFP and anti-p62 antibodies (Figure 5B). As expected, GFP gave substantially increased signal across the pellet fractions. Most increase in signal was obtained in fractions 25–28, of stimulated cell lysate pellets. On the other hand, unstimulated pellet fractions did not show any increase in signal in these fractions. Correspondingly, intense bands for p62, along with modified bands were also observed, whereas unstimulated pellet fractions had very low levels of p62 expression across the gradient. The high-density pellet fractions of the sucrose gradient (fractions 25 and 26), positive for enriched amounts of GFP-LC3 and p62 were, thus, selected for proteomic analysis. These samples were again resolved on SDS–PAGE and Coomassie stained (Figure 5C) to observe the enrichment of overall proteins upon LPS stimulation. The individual fraction lanes were prepared and sent for mass spectrometry analysis.
FIGURE 5:
Schematic to represent fractionation of cell lysates by SDG centrifugation (A). Immunoblotting of untreated and LPS treated pellet fractions for GFP and p62 (B). Coomassie stained SDS–PAGE gel indicates an enrichment of proteins in treated fractions as compared with untreated fractions (C).
Mass spectrometry analysis reveals novel components of ALIS
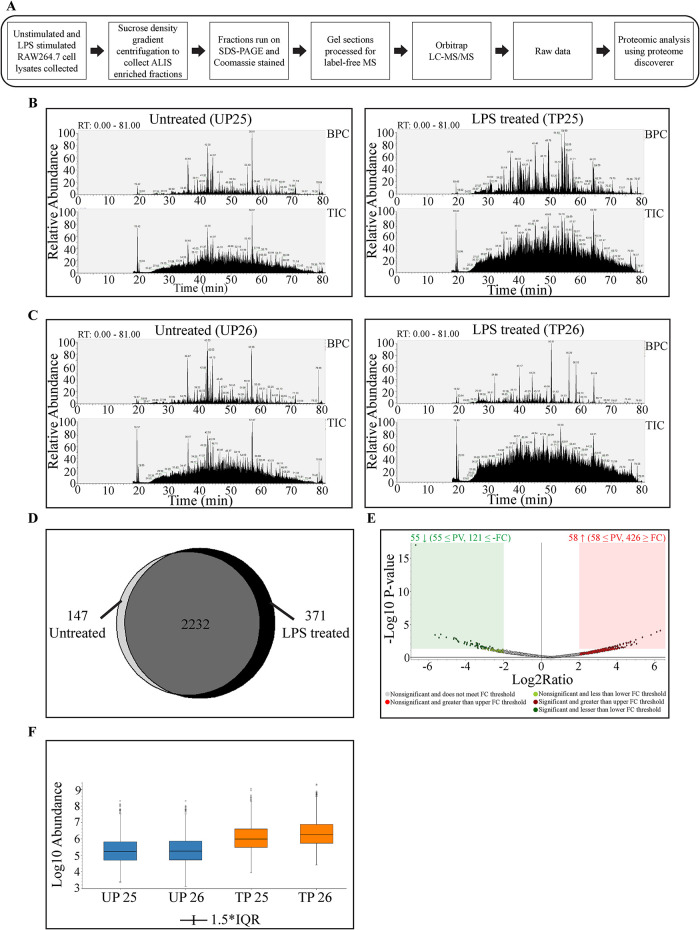
The procedure followed to prepare samples for mass spectrometric analysis is represented schematically in Figure 6A. The mass spectrometry of the selected samples yielded numerous proteins. To begin with, the raw data was analyzed using Xcalibur software. Significant enrichment of peptides, in LPS treated fractions as compared with the untreated fractions, was indicated in total ion chromatogram (TIC) as well as base peak chromatogram (BPC; Figure 6, B and C). Further analysis of the proteomics data was done using Proteome Discoverer 2.2 (PD2.2) which identified peptides grouped into 2752 protein groups. These proteins were then segregated on the basis of origin by generating a Venn diagram wherein the combined list of proteins from untreated pellet fractions 25 and 26 was analyzed against the combined list of proteins from LPS treated pellet fractions 25 and 26 (Figure 6D). We found that 371 were found exclusively in the LPS stimulated fractions, though the majority of the proteins, 2232 of them, were common to both the control and LPS stimulated samples. A volcano plot marking log2 (fold change) versus log10 P value (of 0.05) indicated that 55 proteins were significantly decreased and had a p value less than or equal to 0.05, and 121 had a log2 fold change less than or equal to -2.00. In addition, 58 proteins were significantly increased and had a p value less than or equal to 0.05 with 426 proteins having a log2 fold change greater than or equal to 2.00 (Figure 6E). Sample abundance graph indicating the peak abundance values as box-and-whisker plot for the proteins identified in each of the fractions (UP25, UP26, TP25, and TP26) showed increased values for distribution of abundance values in TP25 and TP26, than UP25 and UP26 (Figure 6F). Thus, a greater number of the ions detected in the TIC and BPC were from this group of proteins, suggesting enrichment upon LPS stimulation.
FIGURE 6:
Bioinformatics analysis of MS data: Schematic to summarize sample preparation for mass spectrometry and data analysis (A). TIC and BPC showing comparison between LPS treated and untreated fractions 25 (B) and 26 (C). Venn diagram showing the distribution of proteins identified in unstimulated cell lysate fractions versus LPS stimulated cell lysate fractions (D). Volcano plot representing the fold change and p value in the proteins identified from LPS and control treatment groups, where x-axis shows fold change and y-axis depicts p value (E). Sample abundance graph indicating the peak intensity values. The second and third quartiles for the data set are represented as rectangles in the graph (IQR = Interquartile Range) (F).
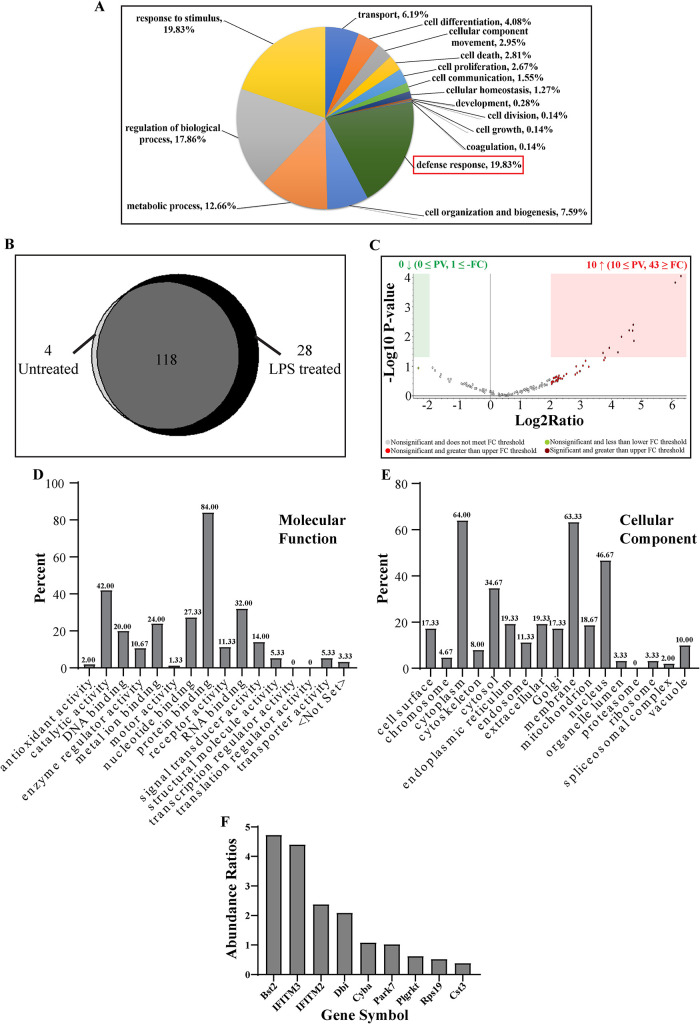
The identified proteins were segregated between LPS treated samples and control samples and analyzed on the basis of either cellular components (Supplemental Figure S4C), molecular functions (Supplemental Figure S4D) or biological process (Supplemental Figure S4E) they are reported to be associated with. The proteins were further classified from LPS stimulated fractions according to their cellular components (Supplemental Figure S4F), molecular function (Supplemental Figure S4G) or biological process (Figure 7A). These included proteins both cytosolic or membrane associated. Presence of membrane associated proteins in these enriched fractions supports recent reports that have shown association of membranous vesicles with (D)ALIS (Kondylis et al., 2013; Montagna et al., 2017). Furthermore, grouping of the molecules according to their molecular function (Supplemental Figure S4, D and G) revealed that many of them are involved in protein binding. This is supported by the idea that ALIS is assembled through interaction between diverse proteins (Zheng et al., 2009; Clausen et al., 2010; Fujita et al., 2011; Liu et al., 2012; Cabe et al., 2018). Lastly, focusing on proteins based on the biological process (Supplemental Figures S4E and 7A), we found that many of the proteins were reported to be involved in defense responses (highlighted in red box). Because these ALIS are formed in response to TLR4 signaling, this supported our hypothesis by indicating the possible recruitment of defense response proteins.
FIGURE 7:
Bioinformatics analysis of defense response proteins: Distribution of proteins from LPS treated fractions based on their biological processes (A). Venn diagram showing the distribution of proteins involved in defense response identified in unstimulated cell lysate fractions versus LPS stimulated cell lysate fractions (B). Volcano plot representing the fold change and p value in the proteins identified from LPS and control treatment groups where x-axis shows fold change and y-axis depicts p value (C). Distribution of proteins based on their molecular functions (D), and cellular components (E). Abundance ratios between proteins from LPS stimulated and unstimulated samples, with molecular masses between 15 to 20 kDa involved in defense response and with membrane binding properties (F).
Using the PD2.2 software, we filtered out the proteins involved in defense response and generated another Venn diagram to compare the number of proteins enriched in LPS stimulated fractions (Figure 7B). Concomitantly, volcano plot revealed the defense response proteins getting enriched substantially after LPS stimulation. Ten were increased and had a p value less than or equal to 0.05 and 43 had a log2 fold change greater than or equal to 2.00. None of the proteins showed a decrease and only one had a log2 fold change less than or equal to -2.00. (Figure 7C). Characterization of all the defense response proteins according to their molecular function and cellular localization indicated that most of the proteins were involved in protein binding function and were vastly membrane associated (Figure 7, D and E). This is consistent with previous reports of ALIS’s association with membranous vesicles and it serving as substrates for autophagy (Kondylis et al., 2013). To screen the proteins further, we went back to our Coomassie stained gel and looked for bands of protein enrichment (Figure 5C). Because many of the antimicrobial proteins are small in molecular mass or are small peptides (Epand and Vogel, 1999; Mahlapuu et al., 2016; Lei et al., 2019; Huan et al., 2020), we started screening for defense response proteins that are membrane associated and small sized (∼15kDa protein band appeared prominently enriched on the gel). The PD2.2 software generated a list of proteins. These proteins were plotted according to the ratios of their abundance values between unstimulated and LPS stimulated. The most highlighted proteins obtained were Bst2 (Tetherin; le Tortorec et al., 2011; Mahauad-Fernandez and Okeoma, 2016; Jin et al., 2017; Sukegawa et al., 2018; Tiwari et al., 2019), IFITM2 and IFITM3 (Bailey et al., 2014; Narayana et al., 2015; Ranjbar et al., 2015; Shi et al., 2017; Sobocinska et al., 2018; Figure 7F).
Antimicrobial peptides association with ALIS validates the MS data
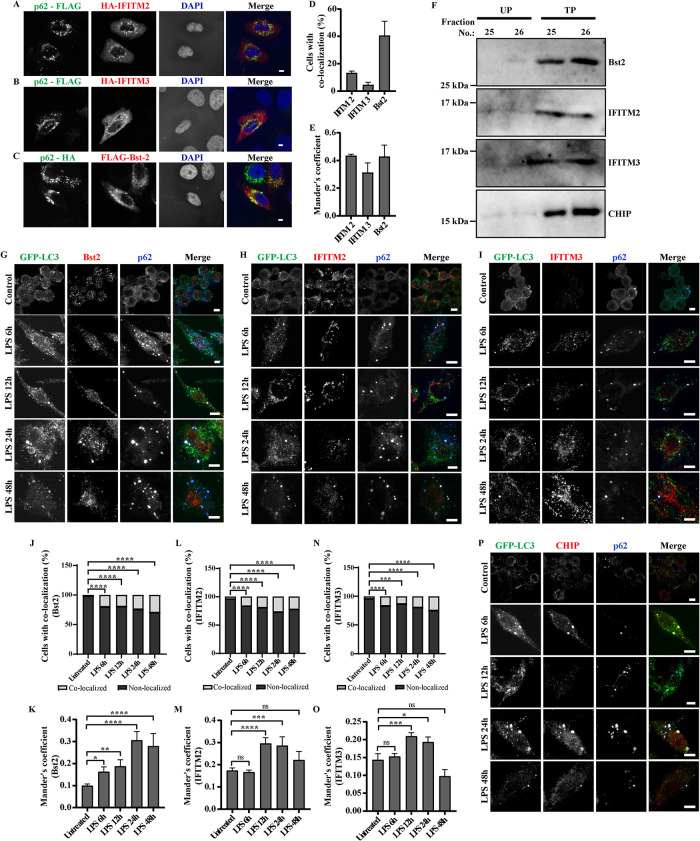
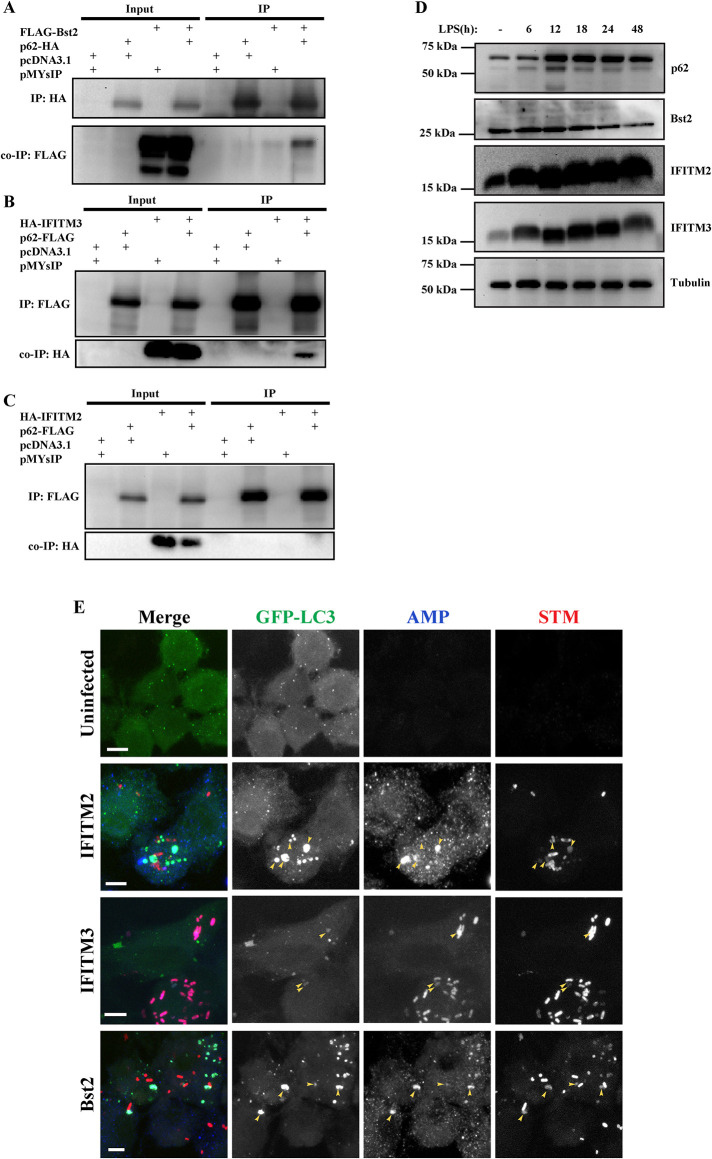
To validate the data from proteomic analysis, association of some of the proteins identified as partners of ALIS from the MS data was investigated. We have demonstrated a substantial increase in p62 protein level upon TLR stimulation and this accumulated endogenous p62 assembles ALIS (Fujita et al., 2011). However, exogenous overexpression of p62 in nonphagocytic cells, like HeLa, also results in formation of ALIS-like structures, though differences in biochemical composition between these structures have not been ruled out (Bjørkøy et al., 2005). Thus, initially we investigated association of AMPs with p62-bodies formed as a result of overexpression of p62 (either p62-FLAG or p62-HA) in HeLa cells. Syk (Tyrosine-protein kinase), IFIT1 (Interferon induced protein with tetratricopeptide repeats1), HBXIP (Hepatitis B virus X-interacting protein), H2B (Histone-2B), IFITM2 (Interferon-induced transmembrane protein2), IFITM3 (Interferon-induced transmembrane protein3) and Bst2 (Bone marrow stromal cell antigen2), with well-established roles in host defense were selected from the mass spectrometry data for this experiment by coexpressing them with either p62-FLAG, p62-HA or untagged p62. Validating the data from the MS analysis, we observed significant colocalization between IFITM2, IFITM3 and Bst2 with p62 puncta, and further confirmed the association using Mander’s coefficient analysis (Figure 8, A–E; Supplemental Figure S5, A–F). Though proteins IFIT1, H2B, HBXIP, and Syk expressed, they appeared to be mostly cytosolic with very little colocalization with p62 bodies, suggesting requirement of additional molecules, or TLR-mediated modifications in a phagocytic environment for stable association of these proteins with ALIS. We also confirmed the enrichment of these endogenous AMPs in ALIS by probing the SDG fractions TP25, 26 and UP25, 26. We also probed for CHIP/STUB1, an already known component of (D)ALIS (Lelouard et al., 2004; Kettern et al., 2011), peptides of which were identified in the mass spectrometry data. In conformity to the high-density pellet fractions of the sucrose gradient are indeed enriched with ALIS, a substantial amount of this protein was noted in fractions 25 and 26 from treated, but not untreated samples. Enrichment of endogenous AMPs, IFITM2, IFITM3, and Bst2 was observed in fractions 25 and 26 from treated pellet samples, IFITM2, IFITM3, and Bst2 in the immunoblots of fractions TP25 and TP26 (Figure 8F).
FIGURE 8:
Validation of association of AMPs identified from the mass spectrometry data with ALIS: HeLa cells cotransfected with pCMV-HA-hIFITM2 and pMY-IP-p62-FLAG (A), pCMV-HA-hIFITM3 and pMY-IP-p62-FLAG (B), and pFLAG-Bst2 and pMY-IP-p62-HA (C). Scale bars, 5 μm. Quantification of colocalization, counted manually and Mander’s correlation, between p62-FLAG with HA-IFITM2 or HA-IFITM3 and p62-HA with FLAG-Bst2 in HeLa cells. A minimum of 50 cells were considered for each experiment. Error bars indicate mean ± SEM from three independent experiments (D and E). Cells were stained for HA and FLAG. Immunoblots of SDG fractions UP25, UP 26, TP 25, and TP26, probed for Bst2, IFITM2, IFITM3, and CHIP (F). Images of RAW264.7 cells stably expressing GFP-LC3 after immunostaining for GFP and endogenous p62, along with endogenous levels of Bst2 (G), IFITM2 (H), IFITM3 (I), and CHIP/STUB1 (P) at 6, 12, 24, and 48 h of LPS stimulation. Scale bars, 5 μm. Quantification of colocalization, counted manually and Mander’s correlation coefficient, between GFP-LC3 dots and Bst2 (J and K), IFITM2 (L and M), IFITM3 (N and O), is shown. The graphs plotted indicate the fraction of total cells with colocalized and nonlocalized calculated for each time point and represented as percentage. Statistical significance was calculated on pooled data from experimental replicates using Fisher’s exact test upon comparing individual time points with untreated data. For IFITM2 and IFITM3, the experiments were performed in triplicates, and in duplicates for Bst2. A minimum 50 cells were counted for each repeat. Mander’s coefficient from individual cells is shown, a minimum of 30 cells were counted. (P) * < 0.05, (P) ** < 0.005, (P) *** < 0.0005, (P) **** < 0.0001, ns = nonsignificant.
To confirm the association of the three AMPs (IFITM2, IFITM3, and Bst2) with ALIS in macrophages, these endogenous proteins were immunostained individually, along with GFP and p62, after different durations of LPS stimulation in RAW GFP-LC3 cells. We observed a clear association of endogenous Bst2, IFITM2, and IITM3 with GFP-LC3 and p62 puncta, in LPS stimulated cells. Mander’s coefficient analysis was employed to assess the correlation of association of these AMPs with GFP/LC3 puncta (Figure 8, G–O). We also analyzed the endogenous expression of CHIP/STUB1 as a positive control and found a similar association (Figure 8P). p62 is essential for the assembly of ALIS upon TLR4 signaling and it directly recruits LC3 and ubiquitin to ALIS through the LIR and UBA motif (Fujita et al., 2011; Cabe et al., 2018). Because IFITM2, IFITM3 and Bst-2 colocalized with p62-bodies, we explored if any of these AMPs coprecipitated with p62 or not. To investigate such an idea, we pulled down either of these AMPs through p62 from extracts of cells expressing p62 along with either IFITM2, IFITM3, or Bst2. Immunoprecipitation (IP) was done using antibody targeting p62 and the precipitates were analyzed for the presence of the respective AMPs. FLAG tagged or HA tagged p62 along with HA-IFITM2, HA-IFITM3 or FLAG-Tetherin were overexpressed, respectively, in HEK293T cells (Figure 9, A–C). Presence of IFITM3 or Tetherin was observed in the immunoprecipitates of p62, however, no IFITM2 was observed with p62 pulldown. These results suggest that IFITM2 association with p62 bodies differs from that of IFITM3 or Tetherin. Further investigations are required to understand these associations and their contributions to host defense.
FIGURE 9:
Interaction between p62 and AMPs: HEK293T cells were transfected with pFLAG-Bst2 and pMY-IP-p62-HA (A), pCMV-HA-hIFITM3 and pMY-IP-p62-FLAG (B) and, pCMV-HA-hIFITM2 and pMY-IP-p62-FLAG (C),. Cell lysates were immunoprecipitated using anti-FLAG or anti-HA antibodies to pull down p62 and the precipitates were probed using anti-FLAG and anti-HA antibodies to detect interaction of AMPs. Changes in the levels of expression of IFITM2, IFITM3, Bst2 and p62 in cell lysate fractions of RAW GFP-LC3 cells with respect to the duration of LPS stimulation (D). Interaction of AMP with STM: RAW GFP-LC3 prestimulated with LPS for 24 h and infected with STM-mCherry and immunostained for endogenous levels of IFITM2, IFITM3 or, Bst2 at 12 hpi (E). Scale bars, 5 μm. Colocalization of GFP-LC3, AMP with phagosomal bacteria is marked by yellow arrows.
LPS is known to upregulate the levels of several defense response genes transcriptionally or translationally (Gunn, 2001; Han et al., 2011; Homann et al., 2011; Blanchet et al., 2013; Diamond and Farzan, 2013; Jones and Okeoma, 2013; Lv et al., 2015; Mahauad-Fernandez and Okeoma, 2016; Pfalzgraff et al., 2016; Ebbensgaard et al., 2018). To evaluate whether LPS treatment could result in increased levels of these AMP proteins in macrophage cells, RAW GFP-LC3 cells were treated with LPS for up to 48 h and cell lysates were collected and separated as 2% Triton X 100 soluble or insoluble fractions (Figure 9D). The three AMPs were predominantly present in the soluble fraction of the lysates. IFITM2 protein levels increased over time upon LPS stimulation up to 48 h. For IFITM3, there was an increase in levels of protein expression up to 24 h which declined by 48 h of LPS stimulation. However, the levels of Bst2 did not alter across time with respect to unstimulated lysates. p62 protein levels increased with increasing duration of LPS stimulation in both soluble and insoluble fractions and declined by 48 h, as reported previously (Fujita et al., 2011). The solubilized fractions from 2% Triton X-100 contained all the three AMPs from LPS stimulated cells. Previous data has shown a detergent-labile association of LC3 with ALIS beyond 0.5% Triton X-100 (Fujita et al., 2011). Thus, we wanted to assess if the AMPs association was affected with varying detergent concentrations. To address this issue, we have done fractionation which shows that insoluble particles, which are presumably ALIS, also enrich these proteins. From the data it is clear that Bst2, IFITM2, and IFITM3 though found in the insoluble fraction, appear to be more labile for detergent treatment than p62, as even 0.5% of Triton X-100 showed only limited cofractionation with ALIS (Supplemental Figure S5G). Similar lability to detergent treatment, of LC3 association with ALIS, was reported earlier. These findings have been summarized in Supplemental Table 1.
Because, ALIS appears to contribute in clearing the intracellular bacteria, it was compelling to investigate whether any of these ALIS-associating AMPs get recruited to the bacteria as well. Thus, RAW GFP-LC3 cells were prestimulated with LPS followed by infection with STM-mCherry. Interestingly, there was a clear association between IFITM2, IFITM3, and Bst2 with the bacterial phagosomes, many of such sites were also positive for GFP-LC3 puncta (Figure 9E). This indicated that the AMPs enriched in ALIS could be delivered to the bacteria-containing phagosomes to eliminate the bacteria.
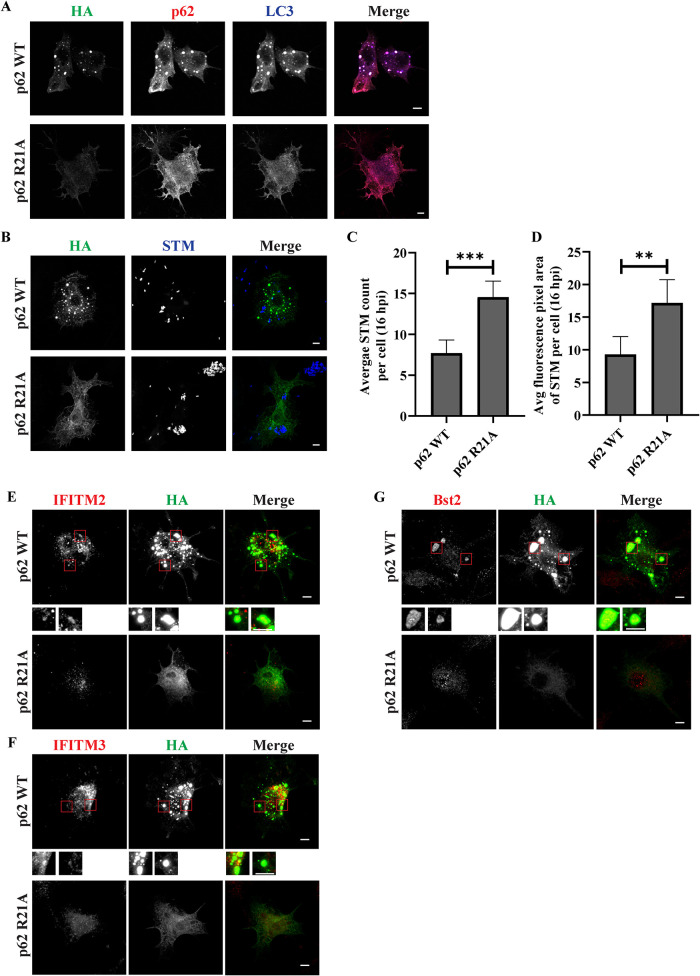
It has been shown that arginine 21 residue is one of the important amino acids for oligomerization of p62, its change to alanine prevents oligomerization of p62 (Misra and Dikic, 2019). To confirm the role of p62 oligomerization on ALIS formation, we transfected RAW264.7 cells with p62 wildtype or R21A mutant with HA-tag and stimulated the cells with LPS. Though wildtype p62 appeared as puncta, p62 R21A as expected appeared cytosolic and did not appear as puncta even after LPS treatment. When these cells were immunostained for endogenous p62 and LC3, no ALIS assembly was noted as compared with cells expressing p62 WT, presumably because of the dominant negative effect of p62 R21A (Bjørkøy et al., 2005; Jakobi et al., 2020; Figure 10A). To estimate intracellular bacterial load in cells expressing R21A variant, macrophage cells transfected with p62 WT or p62 R21A were infected using STM-mCherry following 24 h of LPS treatment. The average number and average fluorescence pixel area of intracellular bacteria were measured. These parameters were significantly higher in cells overexpressing R21A mutant than wild type p62 indicating increased number of bacteria in the presence of an oligomerization-defective p62 and hence in absence of functional ALIS (Figure 10, B–D). Further, we investigated the pattern of endogenous AMPs (IFITM2, IFITM3, and Bst2) in the presence of p62 oligomerization mutant after LPS treatment. Whereas cells expressing wildtype p62 exhibited clear puncta that were positive for the AMPs and also p62, no such association in cells expressing R21A mutant could be observed suggesting failure of the assembly of AMPs into puncta in the absence of functional p62 (Figure 10, E–G).
FIGURE 10:
Oligomerization of p62 is required for AMP colocalization and to restrict bacterial load: RAW264.7 cells were transfected with HA-tagged p62 WT or p62 R21A cDNA constructs. Cells were LPS stimulated for 24 h and immunostained for HA along with p62 and LC3 (A). Transfected and LPS-treated cells were infected with STM-mcherry (STM in pseudocolor blue) and stained for HA 16 h post infection. Cells positively stained for HA were imaged for quantification of bacterial content. Representative images showing bacterial count and fluorescence pixel area in R21A positive cells as compared with p62 WT overexpressing cells (B). Quantification to show average STM count and average fluorescence pixel area of STM signal per cell 16 h post infection (C and D). (MOI = 10). Statistical significance was calculated on data from experimental replicates using unpaired t test. The experiments were performed in triplicates. A minimum 30 cells were counted for each repeat. (P) * < 0.05, (P) ** < 0.005, (P) *** < 0.0005, (P) **** < 0.0001, ns = nonsignificant. Data are represented as mean ± SEM. (N = 3, n ≥ 30). Scale bars, 5 μm. AMP pattern in the presence of p62 R21A: RAW264.7 cells transfected with HA-tagged p62 WT or p62 R21A were treated with LPS and immunostained for endogenous IFITM2, IFITM3, or Bst2. Representative images for localization of IFITM2 (E), IFITM3 (F) and Bst2 (G) in the presence of overexpressed p62 WT or p62 R21A. Scale bars, 5 μm.
DISCUSSION
Macrophages and DCs are amongst the key players of innate immune system recruitment. These are involved in antigen processing and presentation (Ulevitch, 2004; Takeuchi and Akira, 2010). DCs can take up a diverse array of antigens and present them to T cells as peptides bound to both MHC class I and II products. DCs respond to microbial infection by producing cytokines, such as IL-12 and both type I and II interferons, involved in host defense (McNab et al., 2015; Lee and Ashkar, 2018). ALIS formation is observed in DCs in response to inflammatory stimulation (Mellman and Steinman, 2001) with LPS (Lelouard et al., 2002, 2004). Early studies showed that these structures appeared as large ubiquitin positive structures (Lelouard et al., 2002). It was established that ALIS are transient structures, which require continuous protein synthesis. ALIS are considered aggresome-like because despite their appearance they differ from classic aggresomes (Lelouard et al., 2002, 2004; Szeto et al., 2006). Aggresomes are microtubule-dependent inclusion bodies associating with microtubule organizing centre (MTOC). Vimentin, a type-III intermediate filament protein, forms a cage-like structure wrapped around the exterior of the aggresomes (Johnston et al., 1998; Kopito, 2000; Heath et al., 2001). ALIS, on the other hand, do not colocalize with the MTOC and lack a vimentin cage. ALIS are also nonmembranous and detergent-insoluble (Fujita et al., 2011). These were shown to harbor defective ribosomal proteins (DRiPs) as well as neosynthesized proteins which would eventually get targeted by ubiquitination to be processed and presented by MHC I complex (Lelouard et al., 2004; Pierre, 2005). Researchers showed that (D)ALIS serves as sites for storage and processing of antigens (Lelouard et al., 2004; Herter et al., 2005). Lelouard et al, also reported the correlation between DC maturation mediated via PI3K/AKT/mTOR pathway and the appearance of (D)ALIS (Lelouard et al., 2007). Parallel findings confirmed the association of BAG-1 (Bcl-2-associated athanogene 1), a proteasomal chaperone, CHIP (C terminus of Hsc70-interacting protein) an E3 ubiquitin ligase and other proteins of the ubiquitination machinery, with (D)ALIS (Lelouard et al., 2004; Kettern et al., 2011). Further studies identified ALIS formation, in response to TLR4 signaling upon bacterial infection or LPS stimulation in macrophages, indicating their physiological relevance in response to infections (Canadien et al., 2005; Fujita et al., 2011). Fujita et al. also showed that these structures sequestered LC3 and p62 proteins and were eventually targeted for autophagic degradation (Fujita et al., 2011). These studies indicated that ALIS contributes to autophagic and proteasomal machinery (Fujita et al., 2011; Kettern et al., 2011).
ALIS are shown to require p62 induction whereas, because in p62 knockdown cells no ALIS is observed (Fujita et al., 2011; Haldar et al., 2015; Kinsella et al., 2018; Zhang et al., 2019; Paik and Jo, 2020). p62, a multifunctional protein, itself plays a crucial role in many signaling pathways including autophagy, UPS, etc. (Babu et al., 2005; Myeku and Figueiredo-Pereira, 2011; Tian et al., 2014). Involvement of p62 with bacteria has been reported in non-immune or immune cells. As a part of autophagy, p62 recognizes ubiquitinated microbes and delivers them to autophagosomes (Deretic et al., 2013; Gomes and Dikic, 2014). In epithelial cells, p62 protein is shown to get recruited to intracellular STM bacterium, potentially propelling the autophagic machinery to the pathogen (Zheng et al., 2009; Cemma et al., 2011). Upon Salmonella recognition, p62 undergoes phosphorylation and activates Nrf2-Keap1 pathway (Ishimura et al., 2014). Cytosolic or phagosomal Mycobacteria also activate and recruit p62, that restricts bacterial replication and mediates autophagic initiation (Watson et al., 2012; Sakowski et al., 2015; Franco et al., 2017; Zhang et al., 2019, 2023). Recognition and degradation of Shigella and Listeria also depend on p62 and NDP52 dependent autophagic degradation (Mostowy et al., 2011). Coxiella burnetti infections in macrophages also activates p62 mediated Nrf2-Keap1 signaling cascade (Winchell et al., 2018). There are also, contrasting reports of autophagy aiding in microbial survival in the cytosol (Yu et al., 2014; Kimmey et al., 2015). Also, several studies suggest that while p62 is a very crucial protein, loss of it is covered up by several other adaptor proteins such as nuclear dot protein 52 kDa (NDP52), Optineurin and Neighbor of BRCA1 gene 1 (NBR1; Johansen and Lamark, 2011). p62 is involved in several cellular regulation mechanisms such as cell cycle, Nrf2-Keap1 antioxidant pathway, inflammasomal degradation (Shi et al., 2012; Katsuragi et al., 2015) and in activating NF-𝛋B (Sanz, 2000), among many others. All these studies collectively established an important role for p62 in host defense. However, our report, for the first time suggests p62 contributes by enriching AMPs like Bst2, IFITM2 and IFITM3 as part of ALIS in host defense. While this study is in conformity of p62 in immunoprotected role, the mechanism of how this is done adds another layer of complexity to p62 biology.
Thus, in this study the significance of ALIS assembly is explored in host defense with respect to bacteria. Here we demonstrate the association between ALIS and bacterial phagosomes, and the potential protective role of ALIS against intracellular bacteria. For this study, we focused on macrophage response to STM and M. smegmatis. Salmonella Typhimurium is a strain that uses mice as host (Fàbrega and Vila, 2013). STM in mice mimics typhoid symptoms and it causes gastroenteritis in humans (Mathur et al., 2012; Nilsson et al., 2019). M. smegmatis is a nonpathogenic bacterium often used as a model for Mycobacterium tuberculosis under laboratory conditions as they share considerable similarities in their genome and cell wall (He and De Buck, 2010). Both Salmonella and Mycobacterium can employ several mechanisms to evade macrophage onslaught (Deretic et al., 2006; Fàbrega and Vila, 2013). To study the direct effect of mature ALIS in host-defense, macrophages were prestimulated with LPS before bacterial treatment. LPS stimulation not only led to ALIS formation but also substantially decreased the proliferation of all the bacterial strains used. However, this effect was not observed in LLO positive STM, which escapes the phagosome and showed higher survivability even in the presence of ALIS, suggesting ALIS function is restricted to bacteria within the phagosome. This calls for another study using bacteria that actively escape the phagosome like Listeria and Shigella (Ogawa and Sasakawa, 2006; Lam et al., 2013) or that can actively prevent phagosome and lysosome fusion like Legionella (Ivanov and Roy, 2009) and identify the implication of ALIS on these microbes. Another interesting viewpoint could be derived from the work done by Mostowy S. et al. The inability of septin, a cytoskeleton protein, to associate with L. monocytogenes, and thus, evasion of autophagy by the bacteria (Mostowy et al., 2010), could be one of the possibilities why even enhanced ALIS assembly could not restrict STM::LLO proliferation, an observation of hyper-replicating cytosolic STM, coherent with the work done by Yu et al. (Yu et al., 2014). However, a comprehensive study in this aspect is required.
The protective effect of ALIS is further explored in cells where p62 is downregulated with siRNA. These cells without ALIS have also harbored increased numbers of the bacteria that were tested compared with control cells transfected non-target siRNA with functional ALIS.
p62 deficient cells are indeed compromised in handling bacterial infection. And so, the mere downregulation of p62 could contribute to enhanced bacterial proliferation as well. We, here, propose that one of the ways how this is mediated is via ALIS. Unfortunately, at this stage there is no system in place to explore the relative contribution of ALIS independent of p62. p62 mutants, unable to assemble ALIS, could be employed (Cabe et al., 2018) for this purpose. However, it was reported that p62 oligomerization is indeed required for host defense functions (Nakamura et al., 2010; Myeku and Figueiredo-Pereira, 2011; Ciuffa et al., 2015), suggesting that in these cases also, p62 might be functioning by the assembly of ALIS. Thus, we indicate that the close association of ALIS with phagosomal bacteria, the sustenance of ALIS in macrophage cells after infection for up to 40-48 h, sequestration of antimicrobial proteins, the decrease in bacterial proliferation in presence of amplified ALIS formation, and by extension, increase in bacterial survival in the absence of ALIS (under p62 knockdown conditions) cumulatively indicate that ALIS is one of the ways how macrophages, after p62 upregulation, control intracellular pathogen burden. The absence of ALIS in p62 deficient cells, upon infection, leads us to appreciate the role of ALIS assembly mediated by PRR signaling in macrophages and its involvement in controlling bacterial growth.
We experimentally demonstrate that ALIS consists of AMPs (IFITM2, IFITM3 and Bst2) which associate with bacteria containing phagosomes. The association is visibly more pronounced in cells pre-treated with LPS. Because these AMPs have membrane-binding domains (Yoshida et al., 2011; Narayana et al., 2015) this association could be mediated by these membrane-anchoring peptides. In this study, we chose IFITM2, IFITM3 and Bst2 only to validate their association with ALIS and analyze their contribution to host response. Given that the mass spectrometry analysis reveals enrichment of numerous defense response proteins, it would be interesting to validate their association with ALIS and establish their role in bacterial elimination. To further establish that the enriched fractions were indeed positive for ALIS, we also screened for proteins associating with (D)ALIS, as reported in literature. To that end, we could identify BAG6, HSC70 and CHIP/STUB1 (Lelouard et al., 2004; Minami et al., 2010; Kettern et al., 2011) from the proteomics data. We utilized CHIP/STUB1 to validate the structures as ALIS puncta formed in macrophages.
Our data suggests ALIS assembly in infected cells is to protect host cells from pathogens. It is conceivable that the AMPs that constitute ALIS, assemble in a manner that helps them to be delivered to pathogen-containing phagosomes in effective concentrations. Thus, we have employed 3D SR-SIM to get insight into the architecture of ALIS. For this we looked into the association patterns of p62, GFP-LC3 and ubiquitin in ALIS. We speculate that the presence of p62 on the periphery could contribute to the ability of ALIS to interact with and sequester AMPs, and eventually get delivered to and degraded by autophagy. The relative position of LC3 proteins towards the inner core of ALIS also makes these structures distinct from autophagosomes. As expected, we observed that ALIS is an organized structure, signifying that ALIS plays an important role as an active signaling compartment. To explore this further, because purification of ALIS was challenging, we have chosen ALIS enriched fractions using SDG centrifugation. We observed proteins in fractions from LPS treated cells are substantially increased in the same corresponding fraction numbers as in unstimulated cells. To get further insights into the role of ALIS in host-defense, we focused on peptides with antimicrobial properties. These findings were validated by expressing these peptides in RAW264.7 GFP-LC3 positive stable cells and observing their association with ALIS as well as STM. Moreover, some of these peptides are known to carry membrane-associated motifs (Epand and Vogel, 1999; Sobocinska et al., 2018; Liao et al., 2019). Though ALIS itself is a nonmembranous compartment, the presence of these peptides in ALIS makes sense as it has been shown to be engulfed under autophagosomal membranes which are eventually eliminated by autophagy (Fujita et al., 2011). Membrane motifs in these peptides might help in anchoring ALIS to these membranes. Given that, ALIS restricts pathogen load (Canadien et al., 2005), and AMP enriched ALIS anchor to pathogen-containing phagosomes, it is conceivable that ALIS are structures that deliver effective concentrations of AMPs to phagosomes to kill the bacteria. This idea is further supported by the reported association of ALIS with membranous structures in activated macrophages (Kondylis et al., 2013). Furthermore, our studies show that some of the peptides (Bst2 and IFITM3) coprecipitate with p62, but still colocalizes with the ALIS, indicating differences in the nature of association of individual AMPs with the ALIS. It is interesting to explore the extent of contribution of ALIS in pathogen elimination. Given that engagement of TLRs results in activation of multiple intra- and intercellular immune response pathways (El-Zayat et al., 2019), ALIS renders as one of the mechanisms that macrophages employ to tackle bacterial load, which itself is assembled in response to TLR activation (Lelouard et al., 2002; Canadien et al., 2005; Zheng et al., 2009; Cooney et al., 2010; Mesquita et al., 2012; Kondylis et al., 2013). We found that cells lacking ALIS have higher bacterial proliferation. However, because of lack of alternative experimental strategies that would effectively regulate ALIS assembly, without altering other cellular functions, we had to rely on p62 siRNA, a main constituent of ALIS, for this study. Ultrastructural analyses to visualize the positioning of these AMPs within ALIS would be very interesting using Cryo TEM or CLEM (Correlative light Electron microscopy) procedure. Though this study focuses on AMPs, it is most certain that proteins other than AMPs could also play an important role in the assembly of ALIS and its elimination via autophagy. These proteins await further exploration of the mass spectrometry data. In conclusion, our findings demonstrate that ALIS is not an aggregate of proteins but an organized structure with potential contribution to programmed host defense mechanisms.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Ethics statement
The animal experiments were approved by the Institutional Animal Ethics Committee (IAEC) at Indian Institute of Science, Bangalore, India (Registration No: 48/1999/CPCSEA) and the guidelines followed were provided by the Committee for the Purpose of Control and Supervision of Experiments on Animals CPCSEA. The CPCSEA was established under Chapter 4, Section 15(1) of the Prevention of Cruelty to Animals Act 1960. Ethical clearance number for this study is CAF/Ethics/853/2021.
Cell culture, stable cell preparation and preparation of primary macrophages
RAW264.7, RAW GFP-LC3, HeLa-Kyoto and HEK293T cells (ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM with 4.5g/l glucose, L-glutamine, 3.7 g/l sodium bicarbonate and sodium pyruvate) (AL007A, HiMedia) supplemented with heat-inactivated 10% FBS (P30-3302, PAN biotech; 10270-106, Life Technologies) and 1% penicillin-streptomycin (A001A, HiMedia). Cells were incubated in a humidified atmosphere at 37°C in 5% CO2. RAW264.7 cells stably expressing GFP-LC3 were prepared by using pMYs-IRES-GFP, a retroviral expression vector as reported previously (Fujita et al., 2011). Salmonella enterica serovar Typhimurium (STM WT) wild type strain ATCC 14028s or ATCC 14028s constitutively expressing m-cherry protein (pFPV-mCherry) were used in all experiments. The Mycobacterium smegmatis MC2 155 strain and pBEN vector were a gift from Prof. Ajay Kumar (RGCB, Thiruvananthapuram). M. smegmatis were transformed by electroporation with a pBEN-mRFP plasmid for constitutive cytosolic expression of mRFP. Staphylococcus aureus (SA5) and Pseudomonas aeruginosa (PA1) strains were used for infecting macrophage cells. All the bacterial strains were grown on Luria–Bertani (LB) agar plates containing required antibiotics and colonies were inoculated in LB media. STM and STM WT::LLO harvested from peritoneal macrophages for ICSA were plated on Salmonella-Shigella (SS) agar (M108, HiMedia) plates.
Peritoneal macrophages (PMs) were isolated from 4–6 wk old C57BL/6 mice. All mice used (C57BL/6) were bred and housed at the Central Animal Facility, Indian Institute of Science, Bangalore, India. Mice were injected with 2 ml of 4% Brewer thioglycollate solution per mouse into the peritoneal cavity and primed for 4 d. On the 5th day, macrophages were harvested from the mouse peritoneal cavity using 1X phosphate-buffered saline (PBS). Cells were further cultured in RPMI medium 1640 (12167Q, Lonza) containing 10% FBS and 1% penicillin-streptomycin. The Institutional Animal Ethics Committee approved all the animal experiments, and the National Animal Care Guidelines were strictly followed. Cells were cultured in a humidified atmosphere at 37°C in 5% CO2.
For infection of PMs or RAW264.7 cells, 3 × 105 and 2 × 105 cells were seeded, respectively, in a 24-well plate. Bacterial strains grown overnight in LB broth (OD600 0.3) were taken. The MOI 10 was used for most of the infection studies. MOI of 20-25 was used for imaging of bacteria inside the macrophages.
For microtubule disruption experiments, nocodazole (25µm) was added to cells 1 h prior to stimulation with LPS and images, under live setup, were taken after 6 h of LPS treatment. For STM infection, macrophages were incubated with bacteria for 25 min to allow internalization. Fresh media containing nocodazole was added to cells to disrupt microtubules after infection was established. Cells were fixed and immunostained for GFP (Living Colors), p62 (ProGen) and Salmonella (Thermo). Images were collected after 6 h of LPS stimulation or STM infection and number of ALIS puncta were analyzed as compared with cells with LPS or STM and DMSO as control. Percent phagocytosis and ICSA: RAW264.7 cells were infected with STM (WT) at MOI of 10. The infected cells were centrifuged at 800 rpm for 5 min, followed by incubating the infected cells at 37°C in the presence of 5% CO2 for 25 min. Next, the cells were washed with PBS to remove unattached extracellular bacteria and subjected to 100 μg/ml gentamicin (1405-41-0-50827, SRL) treatment for 1 h. Cells were, then, maintained in the presence of 25 μg/ml concentration of gentamicin throughout the experiment. The cells were lysed with 0.1% triton-X-100 at 2 h and 16 h postinfection. The lysates were plated on SS-agar or LB-agar media, and the corresponding colony forming units (CFU) value was determined at 2 and 16 h. The percent phagocytosis and intracellular proliferation of bacteria (fold proliferation) was determined using the formulae:
Percent phagocytosis = [CFU at 2 h]/[CFU of preinoculum]*100
Fold proliferation = [CFU at 16 h]/[CFU at 2 h]
Plasmids, Antibodies, and Reagents
The following plasmids used were obtained from Addgene, H2B-pDendra2(N) (Addgene plasmid#: 75283), pMSCV-mCherry-Syk (Addgene plasmid#: 50045), pcDNA3.1 3xFlag IFIT1 (Addgene plasmid#: 53554), pCMV-HA-hIFITM2 (Addgene plasmid#: 58398), pFLAG-Tetherin (Addgene plasmid#: 41070), pRK5-FLAG-HBXIP (Addgene plasmid#: 42326). The pCMV-HA-mIFITM3 construct was generated by cloning the coding sequence of IFITM3 from cDNA prepared from RAW264.7 GFP-LC3 cells, obtained by PCR amplification using primers:
FP-5′-GGATCCGAATTCAGATGAACCACACTTCTCAAGCC-3′ and
RP-5′-GGATCCGAATTCAGATGAACCACACTTCTCAAGCC-3′
and cloned into EcoRI/SalI sites of pCMV-HA-hIFITM2 vector.
The antibodies used in this study are: p62 (PM045, MBL and GP62-C, ProGen), LC3 (PD014, MBL), Multi Ubiquitin (D058-3, MBL), tubulin (T6199, Sigma), GFP (632375, Mouse Living Colours Clonetek), HA (H6908, Sigma and sc-7392, Santa Cruz Biotechnology), FLAG (F3165 and A8592 Sigma Aldrich), IFITM2 (21 7 96 -1-AP, Proteintech), IFITM3 (11714-1-AP, Proteintech), Bst-2 (NBP2-27154SS, Novus Biologicals), CHIP/STUB1 (C3B6, Cell Signaling Technology), Tubulin (E7, DSHB), Antimouse HRP (61-6520, Invitrogen), antirabbit HRP (65-6120, Invitrogen), antiguinea pig HRP (SAB3700337-2MG, Sigma), donkey anti-Rabbit IgG (H+L) Alexa Fluor 488 (A21206, Invitrogen), goat antirabbit IgG (H+L) Alexa Fluor 568 (A11036, Invitrogen), goat anti-Rabbit IgG (H+L) Alexa Fluor 660 (A21074, Invitrogen), goat anti-Mouse IgG (H+L) Alexa Fluor 488 (A11001,Invitrogen), goat anti-Mouse IgG (H+L) Alexa Fluor 568 (A11031, Invitrogen), goat anti-Mouse IgG (H+L) Alexa Fluor 660 (A21055, Invitrogen), goat anti-Guinea pig IgG (H+L) Alexa Fluor 568 (A11075, Invitrogen), goat anti-Guinea pig IgG (H+L) Alexa Fluor 647 (A21450, Invitrogen). Reagents used were: Lipopolysaccharide (0111:B4, L4391, Sigma), Lipofectamine 3000 reagent (L3000-015, Invitrogen), Polyethylenimine, Linear, MW 25000, Transfection Grade (23966, Polysciences), Nocodazole (SML1665, Sigma-Aldrich).
Immunofluorescence and Microscopy
To visualize internalized STM, bacteria were stained using Salmonella Polyclonal (PA1-20811, Thermo) antibody (1:1000) and Alexa Fluor 568 goat antirabbit antibody (1:1000), wherever mentioned. Staphylococcus and Pseudomonas were stained using DAPI at 1 µg/ml. Cells were stimulated with different bacterial strains for the indicated durations of time. To observe colocalization of p62, ubiquitin with GFP-LC3 in ALIS, RAW GFP-LC3 cells (2 × 105) were grown on coverslips in 24-well plates for 18 h. LPS (1 μg/ml) was added to stimulate the cells for the indicated durations of time. Cells were fixed for 20 min with 4% paraformaldehyde (PFA) in PBS. Permeabilization was done using 0.1% Triton X-100 in PBS for 10 min followed by blocking in 3% bovine serum albumin (BSA) diluted in PBS for 1 h. Cells were coincubated with anti-Ub (MBL) and anti-p62 (ProGen) antibodies at 1:1000 dilution each in 3% BSA solution for 1 h. Following this Alexa Fluor 568 goat anti-Mouse (1:1000) and Alexa Fluor 647 goat anti-Guinea pig (1:300) secondary antibodies were added to the coverslips for 1 h. The coverslips were mounted on slides using ProLong Gold antifade reagent (P36930, Invitrogen).
HeLa Kyoto cells (1 × 105) were grown on coverslips for 18–24 h and cotransfected with either pMYs-IP-p62-FLAG with pCMV-HA-IFITM2, pMYs-IP-p62-FLAG with pCMV-HA-IFITM3 or pMYs-IP-p62-HA with pFLAG-Tetherin using Lipofectamine 3000 reagent. Cells were coimmunostained using anti-HA (Sigma) and anti-FLAG (Sigma) antibodies (1:1000) for 1 h followed by secondary antibody incubation of Alexa Fluor 568 goat anti-Mouse, Alexa Fluor 568 goat anti-Rabbit, Alexa Fluor 488 goat anti-Mouse or Alexa Fluor 488 donkey anti-Rabbit. All secondary antibodies were used at 1:1000 dilution for 1 h. Cells were washed with PBS and the nucleus were stained using DAPI at 10 μg/ml for 30 s. Coverslips were washed with PBS and mounted on glass slides using ProLong Gold antifade reagent.
For immunostaining endogenous levels of the AMPs and CHIP/STUB1, unstimulated and LPS stimulated RAW GFP-LC3 cells were probed with anti-IFITM2 (Proteintech), anti-IFITM3 (Proteintech), anti-Bst-2 (Novus Biologicals) or anti-STUB1 antibodies, along with anti-GFP (Living Colors) and anti-p62 (ProGen) antibodies (1:1000, 1:500 for CHIP/STUB1), in 3% BSA blocking solution containing 0.01% saponin in PBS for 20 min. These cells were then incubated with Alexa Fluor 568 goat anti-Rabbit (1:1000) for AMPs and CHIP/STUB,1 and Alexa Fluor 488 goat anti-mouse for GFP and Alexa Fluor 647 goat antiguinea pig for p62. Cells were washed with PBS and mounted on glass slides using ProLong Gold antifade reagent.
RAW GFP-LC3 cells grown on coverslips, were infected with STM-mCherry either under LPS prestimulated or unstimulated conditions. PFA fixed cells were incubated with indicated antibodies diluted to 1:1000 in 3% BSA blocking buffer containing 0.01% saponin. Corresponding secondary antibodies were added in the same buffer at 1:1000 dilution. Cells were thoroughly washed with PBS before mounting on glass slides. RAW GFP-LC3 cells were reverse transfected with mentioned siRNA using Lipofectamine 3000.
For analyzing the effect of p62 mutant on bacterial survival and AMPs, RAW264.7 cells were seeded on coverslips. Cells were reverse transfected using Lipofectamine 3000. After 24 h of transfection, LPS treatment was given to cells for 24 h. Cells were infected with STM-mCherry and fixed after the mentioned hours post infection for immunostaining using 4% PFA. Imaging was done using Olympus FV3000 laser scanning microscope at 63X. Images used for representation were intensity adjusted using software FV31S-SW, Adobe photoshop keeping the same settings for all samples.
Imaging was done with a Zeiss LSM 880 confocal system at 63X or 100x/1.4 Oil DIC Plan-APOCHROMAT objective. Images used for representation were intensity adjusted using ZEN black software (ZEISS) digital imaging suite, Adobe photoshop keeping the same settings for all samples.
For structured illumination microscopy, RAW GFP-LC3 cells were immunostained upon LPS stimulation with anti-p62 (MBL), -ubiquitin (MBL), and -α-tubulin (Sigma) antibodies (1:1000). Alexa Fluor 568 goat anti-Mouse (1:1000) and Alexa Fluor 660 anti-Rabbit (1:300) were used as secondary antibodies. SR-SIM images were acquired using ELYRA PS.1 (Carl Zeiss, Germany). SIM images were captured using high-NA oil immersion objective (Plan-Apochromat 63x/1.4). This three-dimensional SR-SIM system can achieve a resolution of ∼120 nm along lateral (X–Y axis) and ∼280 nm along the axial (Z-axis) direction. Each image was acquired with exposure time of 80 and 100 ms and captured with five different angular orientations of illumination for each Z- plane, with Z spacing of 110 nm between planes using sCMOS camera (pco.edge 5.5, 2560 × 2160 pixels at 6.5 μm pixel size, FOV ∼80 × 80 μm2). Laser lines at 405, 488, 561 and 647 nm (class 3B lasers) were used for excitation. SIM images were processed with the SIM module of the Zen BLACK software (Carl Zeiss, Germany) and analyzed further for three-dimensional reconstruction with a 180-frame rotation series along the Y-axis. The total thickness of the sample was about 4 to 5 μm leading to the acquisition of 25–30 slices. The orthogonal view and three-dimensional image reconstruction was generated using ZEN Blue software. For quantitative analysis of ALIS, acquired images were analyzed with Zen software (Carl Zeiss, Germany). It provides line intensity histograms and a three-dimensional-rendering image. The absolute intensity values from SIM images were normalized using Origin pro-software to a range between 0 and 1 and plotted against distance (μm). To quantitatively address the diameter of ALIS structure by means of maximum absolute fluorescence intensity corresponds to various protein distributions. The line profiles were drawn perpendicular to ALIS acquired by SIM. Based on visual inspection of the thickness or the distribution of the p62, ubiquitin and LC3 expression we measured the diameter of ALIS based on these intensity values.
All incubations were performed at room temperature.
Co-Immunoprecipitation (Co-IP)
HEK293T cells were grown to 80–85% confluency in 6-well plates and transfected with either pMYs-IP-p62-FLAG with pCMV-HA-IFITM2, pMYs-IP-p62-FLAG with pCMV-HA-IFITM3 or pMYs-IP-p62-HA with pFLAG-Tetherin using PEI reagent (DNA [1]:PEI reagent [3]). Cells were collected by trypsinization after 24 h of transfection and lysed by incubating for 20 min on ice in IP lysis buffer (Tris-Cl pH 7.5 20 mM, EDTA 1 mM, NaCl 50 mM, TritonX-100 0.5%, phenylmethylsulfonyl fluoride [PMSF, P7626, Sigma] 2 mM, protease inhibitor cocktail (PIC, P8340, Sigma) 1X and phosphatase inhibitors 2 and 3 [P5726 and P044, Sigma] 1X each). Cell suspensions were subjected to centrifugation at 15,000 rpm for 15 min at 4°C. Cell lysate supernatants were then incubated with either anti-FLAG or anti-HA antibodies to bind with p62-FLAG or p62-HA respectively for 4 h. The protein-antibody complex was then incubated with Protein A/G PLUS-Agarose (sc-2003, Santa Cruz) for 2 h. All incubations were done at 4°C on the tube rotator. These beads were washed with IP buffer and boiled in Laemmli buffer at 95°C for 15 min.
ALIS enrichment using SDG
Unstimulated and LPS treated RAW GFP-LC3 cells were collected and incubated in 1 ml HEPES hypotonic buffer containing HEPES 10 mM, MgCl2 1.5 mM, KCl 10 mM, PMSF 0.2 mM, dithiothreitol 0.5 mM, PIC 1X (P8340, Sigma Aldrich) for 20 min in ice. Cells were syringe lysed using a 26G needle. The lysed cells were centrifuged at 20,000g for 15 min at 4°C temperature. The supernatant was collected and the pellet was resuspended in the same lysis buffer. 5, 15, 25, 35, 45, 55, and 65 % (wt/vol) sucrose solutions were prepared in the HEPES hypotonic buffer. Discontinuous SDG was prepared manually from 5 to 65% with the densest layer at the bottom. The supernatants and resuspended pellets were fractionated by layering on top of the SDG and centrifuging at 30,000 rpm for 5 h in SW40Ti Rotor. 1 ml fractions were collected from top to bottom of the gradient. Laemmli buffer was added to the fractions and the samples were boiled at 95°C for 15 min. Proteins specific to ALIS were detected by resolving the fractions on SDS–PAGE and probing for p62 (MBL) and GFP (Living Colors).
Immunoblotting
For confirming the presence of AMPs in the SDG fractions, samples were prepared using Laemmli buffer from fractions 25 and 26 of treated and untreated pellet samples. The samples were boiled at 95°C for 15 min. Proteins identified from mass spectrometry were detected by resolving the fractions on SDS–PAGE and probing for IFITM2 (Proteintech), IFITM3 (Proteintech), Bst2 (Novus Biologicals), and STUB1 (CST).
For detecting endogenous levels of AMPs, following LPS treatment, RAW GFP-LC3 cells were lysed in lysis buffer containing Tris (pH 8.0) 50 mM, NaCl 150 mM, EDTA 1 mM, glycerol 10%, Triton X-100 2%, PMSF 2 mM, PIC 1X, phosphatase inhibitors (2 and 3) 1X. Cell lysates were centrifuged at 20,000g for 15 min at 4°C and the soluble fraction was collected. Insoluble fraction was dissolved in the buffer containing 1% SDS. Protein levels in the samples were estimated using BCA reagents (23225, Pierce BCA Protein Assay Kit). Laemmli buffer was mixed with the fractions and the samples were boiled at 95°C for 15 min.
To identify detergent labile AMPs, four types of lysis buffers were prepared with 0.2, 0.5, 1, and 2% Triton X-100. Soluble cell extracts, from RAW GFP LC3 macrophages stimulated with LPS for 18 h, were collected after incubation with these buffers and centrifugation at 20,000g for 15 min at 4°C. These samples were mixed with Laemmli buffer and the detergent insoluble pellets from each lysate were resuspended in Laemmli buffer and boiled.
Samples were first resolved on SDS–PAGE and then transferred to PVDF membranes (IPVH00010, Merck) at 90V. The proteins were detected by incubating the membranes with the indicated antibodies. The signals were visualized using chemiluminescent HRP substrate (WBKLS0500, Merck) on ChemiDoc XRS+ imaging system using QuantityOne software. All primary antibodies were used at 1:1000 dilutions and secondary antibodies were used at 1:5000 dilutions. Contrast for some blots was adjusted uniformly for better visibility. Original blots/gels are presented in Supplemental Information.
Identification of AMPs in ALIS enriched fractions
Fraction numbers 25 and 26 from untreated pellet and LPS treated pellet lysates were electrophoresed on 12% SDS–PAGE gel. The protein bands were stained using Coomassie brilliant blue R-250 (MB153, HiMedia). Each of the fraction lanes from the gel were cut as a single sample and sent to Taplin Mass Spectrometry Facility at Harvard Medical School to identify proteins. The gel sections were subjected to trypsin in-gel digestion followed by microcapillary LC/MS/MS analysis (Shevchenko et al., 1964). The facility then performed a protein database search, data analysis and reported the data. To identify the proteins detected from the samples, raw files were analyzed through Xcalibur and Proteome Discoverer 2.2 software (Washburn, 2015).
Statistical analysis
The statistical tests and significance are mentioned in the respective figure legends. GraphPad PRISM 8 was used to perform statistical analyses. No statistical methods were used to determine the sample size. The sample sizes have been mentioned in the figure legends. The “n” denotes the technical replicates, whereas “N” has been used to demonstrate biological replicates. Sample sizes were chosen on the basis of preliminary experiments so as to provide sufficient power for statistical comparison.
Data Availability
The mass spectrometry data from this manuscript have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (www.ebi.ac.uk/pride) with the dataset identifier PXD031163 and 10.6019/PXD031163.
Supplementary Material
Acknowledgments
We thank IISER TVM for the fellowship of AB. This project is partly funded by Department of Biotechnology grant (BT/PR21325/BRB/10/1554/2016), Department of Science and Technology-Science and Engineering Research Board (DST-SERB) grant (EMR/2016/008048) and IISER TVM intramural funding awarded to S.M.S.; DAE SRC fellowship (DAE00195), DST (FIST), and UGC to D.C., D.C. acknowledges the ASTRA Chair professorship grant from IISc and TATA innovation fellowship grant; financial support from Department of Science and Technology [VI-D&P/411/2012-2013/TDT (G)] and Department of Biotechnology [BT/PR28626BRB/10/1672/2018], Govt of India to S.M.. We kindly acknowledge, Dr Pradyot Prakash, Prof & Head, Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, Varanasi, the Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University for the Staphylococcus and Pseudomonas strains. We thank Prof. Ajay Kumar (RGCB, Thiruvananthapuram) for Mycobacterium smegmatis and pBEN vector. We also thank Leah Susan, Parul Jain, Anusha D. Bhat and Rydham Goyal for technical help. We extend our gratitude to Ross Tomaino at Taplin Mass Spectrometry Facility for generating the spectra and the expert help and advice.
Abbreviations used:
- ALIS
aggresome-like induced structures
- TLR
Toll-like receptor
- STM
Salmonella Typhimurium
- SIM
Structured illumination microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E23-09-0347) on January 3, 2024.
REFERENCES
- Abuaita BH, Schultz TL, O’Riordan MX (2018). Mitochondria-derived vesicles deliver antimicrobial reactive oxygen species to control phagosome-localized Staphylococcus aureus. Cell Host Microbe 24, 625–636.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu JR, Geetha T, Wooten MW (2005). Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem 94, 192–203. [DOI] [PubMed] [Google Scholar]
- Bailey CC, Zhong G, Huang IC, Farzan M (2014). IFITM-family proteins: The cell’s first line of antiviral defense. Annu Rev Virol 1, 261–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, Stenmark H, Johansen T (2005). p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171, 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet FP, Stalder R, Czubala M, Lehmann M, Rio L, Mangeat B, Piguet V (2013). TLR-4 engagement of dendritic cells confers a BST-2/tetherin-mediated restriction of HIV-1 infection to CD4+ T cells across the virological synapse. Retrovirology 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabe M, Rademacher DJ, Karlsson AB, Cherukuri S, Bakowska JC (2018). PB1 and UBA domains of p62 are essential for aggresome-like induced structure formation. Biochem Biophys Res Commun 503, 2306–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canadien V, Tan T, Zilber R, Szeto J, Perrin AJ, Brumell JH (2005). Cutting Edge: Microbial products elicit formation of dendritic cell aggresome-like induced structures in macrophages. J Immunol 174, 2471–2475. [DOI] [PubMed] [Google Scholar]
- Cemma M, Kim PK, Brumell JH (2011). The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria-associated microdomains to target Salmonella to the autophagy pathway. Autophagy 7, 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury AR, Sah S, Varshney U, Chakravortty D (2022). Salmonella Typhimurium outer membrane protein A (OmpA) renders protection from nitrosative stress of macrophages by maintaining the stability of bacterial outer membrane. PLoS Pathog 18, e1010708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffa R, Lamark T, Tarafder AK, Guesdon A, Rybina S, Hagen WJH, Johansen T, Sachse C (2015). The selective autophagy receptor p62 forms a flexible filamentous helical scaffold. Cell Rep 11, 748–758. [DOI] [PubMed] [Google Scholar]
- Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Øvervatn A, Stenmark H, Bjørkøy G, Simonsen A, Johansen T (2010). p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 6, 330–344. [DOI] [PubMed] [Google Scholar]
- Clayton DA, Shadel GS (2014). Purification of mitochondria by sucrose step density gradient centrifugation. Cold Spring Harb Protoc 2014, 1115–1117. [DOI] [PubMed] [Google Scholar]
- Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJP, Campbell BJ, Jewell D, Simmons A (2010). NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med 16, 90–97. [DOI] [PubMed] [Google Scholar]
- Deretic V, Singh S, Master S, Harris J, Roberts E, Kyei G, Davis A, de Haro S, Naylor J, Lee HH, Vergne I (2006). Mycobacterium tuberculosis inhibition of phagolysosome biogenesis and autophagy as a host defence mechanism. Cell Microbiol 8, 719–727. [DOI] [PubMed] [Google Scholar]
- Deretic V, Saitoh T, Akira S (2013). Autophagy in infection, inflammation and immunity. Nat Rev Immunol 13, 722–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Farzan M (2013). The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol 13, 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbensgaard A, Mordhorst H, Aarestrup FM, Hansen EB (2018). The role of outer membrane proteins and lipopolysaccharides for the sensitivity of escherichia coli to antimicrobial peptides. Front Microbiol 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Zayat SR, Sibaii H, Mannaa FA (2019). Toll-like receptors activation, signaling, and targeting: An overview. Bull Natl Res Cent 43, 187. [Google Scholar]
- Epand RM, Vogel HJ (1999). Diversity of antimicrobial peptides and their mechanisms of action. Biochim Biophys Acta - Biomembr 1462, 11–28. [DOI] [PubMed] [Google Scholar]
- Fàbrega A, Vila J (2013). Salmonella enterica serovar Typhimurium skills to succeed in the host: Virulence and regulation. Clin Microbiol Rev 26, 308–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannagan RS, Cosío G, Grinstein S (2009). Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol 7, 355–366. [DOI] [PubMed] [Google Scholar]
- Flannagan RS, Heit B, Heinrichs DE (2015). Antimicrobial mechanisms of macrophages and the immune evasion strategies of Staphylococcus aureus. Pathogens 4, 826–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier B (2013). The function of TLR2 during staphylococcal diseases. Front Cell Infect Microbiol 4, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier B, Philpott DJ (2005). Recognition of Staphylococcus aureus by the innate immune system. Clin Microbiol Rev 18, 521–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco LH, Nair VR, Scharn CR, Xavier RJ, Torrealba JR, Shiloh MU, Levine B (2017). The ubiquitin ligase smurf1 functions in selective autophagy of mycobacterium tuberculosis and anti-tuberculous host defense. Cell Host Microbe 21, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita KI, Maeda D, Xiao Q, Srinivasula SM (2011). Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc Natl Acad Sci USA 108, 1427–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LC, Dikic I (2014). Autophagy in antimicrobial immunity. Mol Cell 54, 224–233. [DOI] [PubMed] [Google Scholar]
- Gunn JS (2001). Bacterial modification of LPS and resistance to antimicrobial peptides. J Endotoxin Res 7, 57–62. [PubMed] [Google Scholar]
- Haldar AK, Foltz C, Finethy R, Piro AS, Feeley EM, Pilla-Moffett DM, Komatsu M, Frickel EM, Coers J (2015). Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. Proc Natl Acad Sci USA 112, E5628–E5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon M, Bierne H, Cossart P (2006). Listeria monocytogenes: A multifaceted model. Nat Rev Microbiol 4, 423–434. [DOI] [PubMed] [Google Scholar]
- Han JH, Lee S, Park YS, Park JS, Kim KY, Lim JS, Oh KS, Yang Y (2011). IFITM6 expression is increased in macrophages of tumor-bearing mice. Oncol Rep 25, 531–536. [DOI] [PubMed] [Google Scholar]
- He Z, De Buck J (2010). Cell wall proteome analysis of Mycobacterium smegmatis strain MC2 155. BMC Microbiol 10, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath CM, Windsor M, Wileman T (2001). Aggresomes resemble sites specialized for virus assembly. J Cell Biol 152, 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herter S, Osterloh P, Hilf N, Rechtsteiner G, Höhfeld J, Rammensee H-G, Schild H (2005). Dendritic cell aggresome-like-induced structure formation and delayed antigen presentation coincide in influenza virus-infected dendritic cells. J Immunol 175, 891–898. [DOI] [PubMed] [Google Scholar]
- Homann S, Smith D, Little S, Richman D, Guatelli J (2011). Upregulation of BST-2/Tetherin by HIV infection in vivo. J Virol 85, 10659–10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan Y, Kong Q, Mou H, Yi H (2020). Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front Microbiol 11, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura R, Tanaka K, Komatsu M (2014). Dissection of the role of p62/Sqstm1 in activation of Nrf2 during xenophagy. FEBS Lett 588, 822–828. [DOI] [PubMed] [Google Scholar]
- Ivanov SS, Roy CR (2009). Modulation of ubiquitin dynamics and suppression of DALIS formation by the legionella pneumophila Dot/Icm system. Cell Microbiol 11, 261–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobi AJ, Huber ST, Mortensen SA, Schultz SW, Palara A, Kuhm T, Shrestha BK, Lamark T, Hagen WJH, Wilmanns M, et al. (2020). Structural basis of p62/SQSTM1 helical filaments and their role in cellular cargo uptake. Nat Commun 11, 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Tian S, Luo M, Xie W, Liu T, Duan T, Wu Y, Cui J (2017). Tetherin suppresses Type I Interferon signaling by targeting MAVS for NDP52-mediated selective autophagic degradation in human cells. Mol Cell 68, 308–322.e4. [DOI] [PubMed] [Google Scholar]
- Johansen T, Lamark T (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR (1998). Aggresomes: A cellular response to misfolded proteins. J Cell Biol 143, 1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PH, Okeoma CM (2013). Phosphatidylinositol 3-kinase is involved in Toll-like receptor 4-mediated BST-2/tetherin regulation. Cell Signal 25, 2752–2761. [DOI] [PubMed] [Google Scholar]
- Karlsson AB, Washington J, Dimitrova V, Hooper C, Shekhtman A, Bakowska JC (2014). The role of spartin and its novel ubiquitin binding region in DALIS occurrence. Mol Biol Cell 25, 1355–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuragi Y, Ichimura Y, Komatsu M (2015). P62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J 282, 4672–4678. [DOI] [PubMed] [Google Scholar]
- Kettern N, Rogon C, Limmer A, Schild H, Höhfeld J (2011). The Hsc/Hsp70 Co-Chaperone network controls antigen aggregation and presentation during maturation of professional antigen presenting cells. PLoS One 6, e16398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmey JM, Huynh JP, Weiss LA, Park S, Kambal A, Debnath J, Virgin HW, Stallings CL (2015). Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature 528, 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella RL, Nehls EM, Stallings CL (2018). Roles for autophagy proteins in immunity and host defense. Vet Pathol 55, 366–373. [DOI] [PubMed] [Google Scholar]
- Kondylis V, Van Nispen Tot Pannerden HE, Van Dijk S, Ten Broeke T, Wubbolts R, Geerts WJ, Seinen C, Mutis T, Heijnen HFG (2013). Endosome-mediated autophagy: An unconventional MIIC-driven autophagic pathway operational in dendritic cells. Autophagy 9, 861–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito RR (2000). Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol 10, 524–530. [DOI] [PubMed] [Google Scholar]
- Lam GY, Cemma M, Muise AM, Higgins DE, Brumell JH (2013). Host and bacterial factors that regulate LC3 recruitment to Listeria monocytogenes during the early stages of macrophage infection. Autophagy 9, 985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence DW, Kornbluth J (2012). E3 ubiquitin ligase NKLAM is a macrophage phagosome protein and plays a role in bacterial killing. Cell Immunol 279, 46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AJ, Ashkar AA (2018). The dual nature of type I and type II interferons. Front Immunol 9, 2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Sun LC, Huang S, Zhu C, Li P, He J, Mackey V, Coy DH, He QY (2019). The antimicrobial peptides and their potential clinical applications. Am J Transl Res 11, 3919–3931. [PMC free article] [PubMed] [Google Scholar]
- Lelouard H, Gatti E, Cappello F, Gresser O, Camosseto V, Pierre P (2002). Transient aggregation of ubiquitinated proteins during dendritic cell maturation. Nature 417, 177–182. [DOI] [PubMed] [Google Scholar]
- Lelouard H, Ferrand V, Marguet D, Bania J, Camosseto V, David A, Gatti E, Pierre P (2004). Dendritic cell aggresome-like induced structures are dedicated areas for ubiquitination and storage of newly synthesized defective proteins. J Cell Biol 164, 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelouard H, Schmidt EK, Camosseto V, Clavarino G, Ceppi M, Hsu HT, Pierre P (2007). Regulation of translation is required for dendritic cell function and survival during activation. J Cell Biol 179, 1427–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Goraya MU, Yuan X, Zhang B, Chiu SH, Chen JL (2019). Functional involvement of interferon-inducible transmembrane proteins in antiviral immunity. Front Microbiol 10, 1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XD, Ko S, Xu Y, Fattah EA, Xiang Q, Jagannath C, Ishii T, Komatsu M, Eissa NT (2012). Transient aggregation of ubiquitinated proteins is a cytosolic unfolded protein response to inflammation and endoplasmic reticulum stress. J Biol Chem 287, 19687–19698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Banetta L, Young LJ, Smith EJ, Bates GP, Zaccone A, Kaminski Schierle GS, Tunnacliffe A, Kaminski CF (2019). Live-cell super-resolution microscopy reveals a primary role for diffusion in polyglutamine-driven aggresome assembly. J Biol Chem 294, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv M, Zhang B, Shi Y, Han Z, Zhang Y, Zhou Y, Zhang W, Niu J, Yu XF (2015). Identification of BST-2/tetherin-induced hepatitis B virus restriction and hepatocyte-specific BST-2 inactivation. Sci Rep 5, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald L, Baldini G, Storrie B (2015). Does Super-Resolution fluorescence microscopy obsolete previous microscopic approaches to protein Co-Localization? Methods Mol Biol 1270, 255–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahauad-Fernandez WD, Okeoma CM (2016). The role of BST-2/Tetherin in host protection and disease manifestation. Immunity, Inflamm Dis 4, 4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M, Håkansson J, Ringstad L, Björn C (2016). Antimicrobial peptides: An emerging category of therapeutic agents. Front Cell Infect Microbiol 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur R, Oh H, Zhang D, Park SG, Seo J, Koblansky A, Hayden MS, Ghosh S (2012). A mouse model of salmonella typhi infection. Cell 151, 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A (2015). Type I interferons in infectious disease. Nat Rev Immunol 15, 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Steinman RM (2001). Dendritic cells: Specialized and regulated antigen processing machines. Cell 106, 255–258. [DOI] [PubMed] [Google Scholar]
- Mesquita FS, Thomas M, Sachse M, Santos AJM, Figueira R, Holden DW (2012). The Salmonella deubiquitinase Ssel inhibits selective autophagy of cytosolic aggregates. PLoS Pathog 8, e1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami R, Hayakawa A, Kagawa H, Yanagi Y, Yokosawa H, Kawahara H (2010). BAG-6 is essential for selective elimination of defective proteasomal substrates. J Cell Biol 190, 637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra M, Dikic I (2019). RNA binding to p62 impacts selective autophagy. Cell Res 29, 512–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagna D, Sommi P, Necchi V, Vitali A, Montini E, Turin I, Ferraro D, Ricci V, Solcia E (2017). Different polyubiquitinated bodies in human dendritic cells: IL-4 causes PaCS during differentiation while LPS or IFNα induces DALIS during maturation. Sci Rep 7, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S, Bonazzi M, Hamon MA, Tham TN, Mallet A, Lelek M, Gouin E, Demangel C, Brosch R, Zimmer C, et al. (2010). Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe 8, 433–444. [DOI] [PubMed] [Google Scholar]
- Mostowy S, Sancho-Shimizu V, Hamon MA, Simeone R, Brosch R, Johansen T, Cossart P (2011). p62 and NDP52 proteins target Intracytosolic Shigella and listeria to different autophagy pathways. J Biol Chem 286, 26987–26995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myeku N, Figueiredo-Pereira ME (2011). Dynamics of the degradation of ubiquitinated proteins by proteasomes and autophagy: Association with sequestosome 1/p62. J Biol Chem 286, 22426–22440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kimple AJ, Siderovski DP, Johnson GL (2010). PB1 domain interaction of p62/sequestosome 1 and MEKK3 regulates NF-κB activation. J Biol Chem 285, 2077–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayana SK, Helbig KJ, McCartney EM, Eyre NS, Bull RA, Eltahla A, Lloyd AR, Beard MR (2015). The interferon-induced transmembrane proteins, IFITM1, IFITM2, and IFITM3 inhibit hepatitis C virus entry. J Biol Chem 290, 25946–25959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson OR, Kari L, Steele-Mortimer O (2019). Foodborne infection of mice with Salmonella Typhimurium. PLoS One 14, e0215190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odendall C, Kagan JC (2019). Host-encoded sensors of bacteria: Our windows into the microbial world. Microbiol Spectr 7, 10.1128/microbiolspec.bai-0011-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Sasakawa C (2006). Intracellular survival of Shigella. Cell Microbiol 8, 177–184. [DOI] [PubMed] [Google Scholar]
- Paik S, Jo EK (2020). An interplay between autophagy and immunometabolism for host defense against mycobacterial infection. Front Immunol 11, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Páleníková P, Harbour ME, Ding S, Fearnley IM, Van Haute L, Rorbach J, Scavetta R, Minczuk M, Rebelo-Guiomar P (2021). Quantitative density gradient analysis by mass spectrometry (qDGMS) and complexome profiling analysis (ComPrAn) R package for the study of macromolecular complexes. Biochim Biophys Acta - Bioenerg 1862, 148399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman G, Panchal R, Medvedev AE (2017). The R753Q polymorphism in Toll-like receptor 2 (TLR2) attenuates innate immune responses to mycobacteria and impairs MyD88 adapter recruitment to TLR2. J Biol Chem 292, 10685–10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalzgraff A, Heinbockel L, Su Q, Gutsmann T, Brandenburg K, Weindl G (2016). Synthetic antimicrobial and LPS-neutralising peptides suppress inflammatory and immune responses in skin cells and promote keratinocyte migration. Sci Rep 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre P (2005). Dendritic cells, DRiPs, and DALIS in the control of antigen processing. Immunol Rev 207, 184–190. [DOI] [PubMed] [Google Scholar]
- Ranjbar S, Haridas V, Jasenosky LD, Falvo JV, Goldfeld AE (2015). A role for IFITM proteins in restriction of mycobacterium tuberculosis infection. Cell Rep 13, 874–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y, Rezelj S, Bedina Zavec A, Anderluh G, Scheuring S (2016). Listeriolysin O membrane damaging activity involves arc formation and lineaction – Implication for Listeria monocytogenes escape from phagocytic vacuole. PLoS Pathog 12, e1005597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakowski ET, Koster S, Portal Celhay C, Park HS, Shrestha E, Hetzenecker SE, Maurer K, Cadwell K, Philips JA (2015). Ubiquilin 1 Promotes IFN-γ-Induced Xenophagy of Mycobacterium tuberculosis. PLoS Pathog 11, e1005076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L (2000). The atypical PKC-interacting protein p62 channels NF-kappa B activation by the IL-1-TRAF6 pathway. EMBO J 19, 1576–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1964). Techniques in protein chemistry. J Am Chem Soc 86, 1653–1653. [Google Scholar]
- Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH (2012). Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol 13, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G, Schwartz O, Compton AA (2017). More than meets the I: The diverse antiviral and cellular functions of interferon-induced transmembrane proteins. Retrovirology 14, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobocinska J, Roszczenko-Jasinska P, Ciesielska A, Kwiatkowska K (2018). Protein palmitoylation and its role in bacterial and viral infections. Front Immunol 8, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan A, Batinica M, Steiger J, Hartmann P, Zaucke F, Bloch W, Fabri M (2016). LL37:DNA complexes provide antimicrobial activity against intracellular bacteria in human macrophages. Immunology 148, 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strømhaug PE, Berg TO, Fengsrud M, Seglen PO (1998). Purification and characterization of autophagosomes from rat hepatocytes. Biochem J 335, 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukegawa S, Miyagi E, Bouamr F, Farkašová H, Strebel K (2018). Mannose Receptor 1 restricts HIV particle release from infected macrophages. Cell Rep 22, 786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto J, Kaniuk NA, Canadien V, Nisman R, Mizushima N, Yoshimori T, Bazett-Jones DP, Brumell JH (2006). ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy 2, 189–199. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S (2010). Pattern recognition receptors and inflammation. Cell 140, 805–820. [DOI] [PubMed] [Google Scholar]
- Tapping RI, Akashi S, Miyake K, Godowski PJ, Tobias PS (2000). Toll-like Receptor 4, but not toll-like Receptor 2, is a signaling receptor for Escherichia and Salmonella Lipopolysaccharides. J Immunol 165, 5780–5787. [DOI] [PubMed] [Google Scholar]
- Thomas M, Mesquita FS, Holden DW (2012). The DUB-ious lack of ALIS in Salmonella infection: A Salmonella deubiquitinase regulates the autophagy of protein aggregates. Autophagy 8, 1824–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z, Wang C, Hu C, Tian Y, Liu J, Wang X (2014). Autophagic-lysosomal inhibition compromises ubiquitin-proteasome system performance in a p62 dependent manner in cardiomyocytes. PLoS One 9, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari R, De La Torre JC, Mcgavern DB, Nayak D (2019). Beyond tethering the viral particles: Immunomodulatory functions of tetherin (BST-2). DNA Cell Biol 38, 1170–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Tortorec A, Willey S, Neil SJD (2011). Antiviral inhibition of enveloped virus release by Tetherin/BST-2: Action and counteraction. Viruses 3, 520–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulevitch RJ (2004). Therapeutics targeting the innate immune system. Nat Rev Immunol 4, 512–520. [DOI] [PubMed] [Google Scholar]
- Viola A, Munari F, Sánchez-Rodríguez R, Scolaro T, Castegna A (2019). The metabolic signature of macrophage responses. Front Immunol 10, 1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vural A, Kehrl JH (2014). Autophagy in macrophages: Impacting inflammation and bacterial infection. Scientifica (Cairo) 2014, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas JM, Van Der Veen AG, Ploegh HL (2008). The known unknowns of antigen processing and presentation. Nat Rev Immunol 8, 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP (2015). The H-Index of “An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database.” [DOI] [PubMed]
- Watson RO, Manzanillo PS, Cox JS (2012). Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150, 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegel E, Göhler A, Lagerholm BC, Wainman A, Uphoff S, Kaufmann R, Dobbie IM (2016). Imaging cellular structures in super-resolution with SIM, STED and Localisation Microscopy: A practical comparison. Sci Rep 6, 27290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchell CG, Dragan AL, Brann KR, Onyilagha FI, Kurten RC, Voth DE (2018). Coxiella burnetii subverts p62/sequestosome 1 and activates Nrf2 signaling in human macrophages. Infect Immun 86, e00608–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Harrison RE (2015). Down-regulation of stathmin is required for the phenotypic changes and classical activation of macrophages. J Biol Chem 290, 19245–19260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Kao S, Strebel K (2011). Identification of residues in the BST-2 TM domain important for antagonism by HIV-1 Vpu using a gain-of-function approach. Front Microbiol 2, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HB, Croxen MA, Marchiando AM, Ferreira RBR, Cadwell K, Foster LJ, Brett Finlay B (2014). Autophagy facilitates Salmonella replication in HeLa cells. MBio 5, e00865–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH (2009). The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol 183, 5909–5916. [DOI] [PubMed] [Google Scholar]
- Zhang R, Varela M, Vallentgoed W, Forn-Cuni G, van der Vaart M, Meijer AH (2019). The selective autophagy receptors Optineurin and p62 are both required for zebrafish host resistance to mycobacterial infection. PLoS Pathog 15, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Han L, Ma Q, Wang X, Yu J, Xu Y, Zhang X, Wu X, Deng G (2023). RIP3 impedes Mycobacterium tuberculosis survival and promotes p62-mediated autophagy. Int Immunopharmacol 115, 109696. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data from this manuscript have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (www.ebi.ac.uk/pride) with the dataset identifier PXD031163 and 10.6019/PXD031163.