Abstract
Sequence motifs (L domains) have been described in viral structural proteins. Mutations in these lead to a defect at a late stage in virus assembly and budding. For several viruses, recruitment of an endosomal sorting complexes required for transport 1 subunit (Tsg101), a component of the class E vacuolar protein sorting (EVPS) machinery, is a prerequisite for virion budding. To effect this, Tsg101 interacts with the PT/SAP L domain. We have identified candidate L-domain motifs, PSAP, PPPI, and YEIL, in the prototypic foamy virus (PFV) Gag protein, based on their homology to known viral L domains. Mutation of the PSAP and PPPI motifs individually reduced PFV egress, and their combined mutation had an additive effect. When PSAP was mutated, residual infectious PFV release was unaffected by dominant negative Vps4 (an ATPase involved in the final stages of budding), and sensitivity to dominant negative Tsg101 was dramatically reduced, suggesting that the PSAP motif functions as a conventional class E VPS-dependent L domain. Consistent with this notion, yeast two-hybrid analysis showed a PSAP motif-dependent interaction between PFV Gag and Tsg101. Surprisingly, PFV release which is dependent on the PPPI motif was Vps4-independent and was partially inhibited by dominant negative Tsg101, suggesting that PPPI functions by an unconventional mechanism to facilitate PFV egress. Mutation of the YEIL sequence completely abolished particle formation and also reduced the rate of Gag processing by the viral protease, suggesting that the integrity of YEIL is required at an assembly step prior to budding and YEIL is not acting as an L domain.
Viruses rely on using normal cellular processes for their replication, assembly, and egress from the infected cell. An essential stage in this process for enveloped viruses is budding through a cellular membrane to acquire their envelope, and, in the case of budding through the plasma membrane, to exit the cell. The major structural proteins of some enveloped viruses (e.g., retrovirus Gag proteins, filovirus matrix proteins, and rhabdovirus M proteins) are both necessary and sufficient for virus-like particle assembly and egress. These proteins contain late-budding domains (L domains), which play an essential role in virus particle release. Mutations in these sequences lead to a ‘late’ budding defect in which virus particles assemble but are unable to complete the final stages in budding from cellular membranes and, hence, remain tethered to the cell (11).
To date, three classes of L domain have been identified in a number of retrovirus Gag proteins, including those of human immunodeficiency virus type 1 (14, 19), murine leukemia virus (52), and equine infectious anemia virus (39), as well as in the structural proteins of Ebola virus (16, 30), rabies virus (17), and arenaviruses (35). These are characterized by the sequence motifs PT/SAP, PPXY, and YPDL/LXXLF, which can apparently occur individually or in close proximity within viral structural proteins (5, 15, 19, 29, 37, 39, 41, 46). A number of recent studies have shown that at least two of these motifs bind to components of the class E vacuolar protein sorting (VPS) pathway which functions during vesicular budding into multivesicular bodies (MVB) (5, 7, 12, 30-32, 38, 41, 42). This process likely involves the sequential recruitment of a number of multiprotein complexes, termed endosomal sorting complexes required for transport (ESCRT), to endosomal membranes. Initially, an ubiquitin-based sorting signal on cargo proteins is recognized by the hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) (2, 22, 28, 38). This recruits ESCRT-I, which further recruits the ESCRT-II/III complexes either directly or via an interaction with AIP-1/ALIX (a mammalian orthologue of the yeast VPS factor Bro1) (31, 41, 44). A final step prior to membrane fusion requires the disassembly of ESCRT complexes catalyzed by the homomultimeric ATPase Vps4 (1).
Several studies have established that the PT/SAP motif functions during viral budding by recruiting the class E VPS pathway via a direct interaction with a component of the ESCRT-I complex, Tsg101 (tumor-susceptibility gene product 101) (7, 12, 30, 42). Conversely, the binding partner for the YPDL domain was initially suggested to be the AP-2 adaptor complex involved in clathrin-mediated endocytosis from the plasma membrane (40). However, more recent studies have suggested that the functional interaction is more likely to be with a class E VPS factor, namely, AIP-1/ALIX (32, 41). Additionally, expression of dominant negative forms of Vps4 inhibits the budding of all retroviruses tested thus far (12, 32, 41). It thus seems likely that the mechanism of particle budding in viruses containing L domains is making use of the cellular pathways normally used to sort cargo into MVBs. This model is particularly attractive since MVB formation and virus budding are the only known examples of sorting/budding of cellular membranes away from the cytoplasm. However, while the PPXY motif is thought to interact with the WW domains of the Nedd4-like E3 ubiquitin ligases (16, 23, 48), precisely how this leads to a dependence on class E VPS factors for budding in most, but apparently not all, cases (20) is unknown.
The prototypic foamy virus is unusual among retroviruses in that it has a number of unique features to its replication cycle (25). The provirus genome organization is similar to that of other complex retroviruses, but there are important differences in the way the Pol protein is expressed (3, 9, 27, 51), the nature of the nucleic acid in the virion (25, 51), the regulation of gene expression (26), and the structure and function of the Env protein (24). Some of these features would appear to resemble the hepadnavirus replication strategy more closely than that of classical retroviruses (51). The mechanism of foamy virus particle assembly and budding further shows important deviations from that of other retroviruses. These include differences in the minimal structural protein requirement for particle assembly and budding, particularly a requirement for Env in addition to Gag, and the site of budding which occurs primarily in an intracellular compartment, thought to be related to the endoplasmic reticulum (ER) (10, 36). The prototypic foamy virus (PFV) Env protein contains an ER retrieval signal within its cytoplasmic C terminus (13), and the N-terminal 15 amino acids are essential for virus particle formation and budding (24).
The majority of virus particles remain cell associated in vitro. Standard procedures, such as cell lysis by sonication, do nothing to facilitate virion egress. This is relevant to the fact that foamy viruses offer an as yet untapped alternative vector system for gene therapy, given their lack of disease association, broad host range, larger than average retroviral packaging capacity, and ability to integrate into cells. Currently, their major disadvantage is the modest titer that foamy virus-derived vectors have compared to other viral vectors. An understanding of why this virus tends to remain cell associated and the mechanisms that might be employed to enhance their release would undoubtedly strengthen the case for their exploitation as vectors.
In this study, we identified three sequences with homology to known L domains, PSAP, PPPI and YEIL, in the PFV Gag protein and show that at least one of these acts as an interface with the class E VPS pathway to facilitate PFV egress. Specifically, the PSAP motif functions as a conventional Tsg101 and Vps4-dependent L domain and the PPPI motif appears to facilitate PFV egress by an unconventional mechanism that may share some features with that of classical L domains. However, the YEIL sequence does not function as an L domain, but its integrity is nevertheless important for PFV assembly and/or Gag processing.
MATERIALS AND METHODS
Cell lines, plasmids, and mutagenesis.
Human fibrosarcoma (HT1080), dog fibroblast (D17), human embryonic kidney (293T) cells, and baby hamster kidney cells stably transfected with the lacZ gene under the transcriptional control of the PFV long terminal repeat (BHLL cells) (4) were cultivated in Dulbecco's modified Eagle minimal essential medium supplemented with 10% fetal calf serum, penicillin, streptomycinm and 2 mM l-glutamine.
The PFV vector was produced using the vector plasmid pMH71 (18) and pczHFVenv encoding the HFV envelope protein (36). The wild-type PFV was produced using the construct pczHSRV2 (33). Amino acid mutations were introduced into the Gag proteins of MH71 and pczHSRV2 by site-directed mutagenesis using a QuikChange XL kit (Stratagene). Following mutagenesis, the integrity of all constructs was confirmed by sequencing.
Plasmids expressing dominant negative forms of Vps4, Vps4B/SKD1, Tsg101, and AIP-1/ALIX in mammalian cells, namely, pCR3.1/GFP-VPS4 223 M, pDSRED-VPS4B E228Q (a gift from Wes Sundquist, University of Utah), pCR3.1/YFP-Tsg101 (1-157), and pCR3.1/YFP-ALIX(d1-176), have been described previously (12, 31, 32). Plasmids based on pGBKT7 and expressing a GAL-4 DNA binding domain fused to Tsg101 or AIP-1/ALIX have been previously described (32), as have pVP16-based plasmids expressing VP16 activation domains fused to Tsg101, AIP-1/ALIX, and EIAV-p9. Additional plasmids expressing GAL4 or VP16 fused to wild-type and mutant forms of PFV Gag were generated by PCR amplification of the latter from the pMH71-based constructs and insertion of the amplicons into pGBKT7 and pVP16, respectively.
PFV vector and virus production.
The PFV vector and virus, and their mutated forms, were produced by transient transfection of 293T cells using polyethylenimine (34). Briefly, 293T cells were seeded into 12-well plates at 3 × 105 cells per well and incubated overnight at 37°C. For comparison of virion particle production by wild-type and L-domain mutants, the cells were transfected with 1.5 μg MH71 and 0.5 μg pczHFVenv or 3 μg pczHSRV2. To analyze the effect of dominant negative Tsg101, Vps4, Vps4B/SKD1, or AIP-1/ALIX proteins on PFV egress, 293T cells were cotransfected with 20 ng pCR3.1/YFP-Tsg101 (1-157), 20 ng wild-type or mutant pCR3.1/GFP-VPS4, 20 ng pDSRED-C1/DSREDVps4B-E228Q, or 100 ng pCR3.1/YFP-ALIX(d1-176). The supernatants were harvested 24, 48, or 72 h after transfection and clarified through a 0.45-μm-pore-size filter. For Western blot analysis, vector or viral particles were further purified by centrifugation (twice for 10 min at 2,000 × g followed by centrifugation through a 20% sucrose cushion at 100,000 × g for 25 min). All centrifugation steps were performed at 4°C and transfected cells or viral pellets resuspended in Laemmli polyacrylamide gel electrophoresis-sodium dodecyl sulfate (PAGE-SDS) lysis buffer (30).
Vector and virus infectivity assays.
D17 cells were used as target cells to measure eGFP-expressing PFV vector titers, and BHLL cells were used to titer replication-competent virus. The cells were seeded into 96-well plates at 3 × 103 cells per well and incubated at 37°C overnight. All retroviral supernatant stocks were serially diluted 10-fold (to 10−5) in growth medium, and then 75 μl of each dilution was added to triplicate wells. Transduced cells were incubated for 3 days at 37°C and fixed in 4% formaldehyde and the eGFP-expressing D17 cells detected using fluorescence microscopy. The PFV-infected BHLL cells expressing β-galactosidase were detected by the insoluble X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside assay, as described previously (4).
Western blot analysis.
Virion and cell lysates were separated on 10% acrylamide gels and proteins transferred to nitrocellulose membranes (Amersham). The blots were incubated with human anti-PFV serum (diluted 1:500) overnight at 4°C, probed with peroxidase-conjugated anti-human secondary antibody (Sigma), and developed with metal-enhanced diaminobenzidine (Pierce).
Electron microscopy (EM) analysis.
293T cells in 35-mm-diameter dishes were transfected with plasmids to produce wild-type vector and incubated at 37°C for 48 h. The cells were fixed for 30 min in 200 mM cacodylate-0.5% glutaraldehyde and then washed in 200 mM cacodylate and postfixed in 1% osmium tetroxide and 1.5% potassium ferrocyanide for 1 h. After extensive washing in water, the cells were incubated in 0.5% magnesium uranyl acetate overnight at 4°C, dehydrated in ethanol and propylene oxide, and embedded in epon resin. Ultrathin sections were cut and collected on transmission electron microscopy grids, and lead citrate was added as a contrast agent. Sections were analyzed using an FEI Technai G2 transmission electron microscope, and digital images were captured using Soft Imaging Software and processed in Adobe Photoshop 7.
Yeast two-hybrid assays.
Yeast Y190 cells were transformed with pGBKT7 and pVP16 derivatives and transformants selected on medium lacking tryptophan and leucine. Protein-protein interactions were measured by induction of β-galactosidase reporter activity, using assays described previously (30).
RESULTS
Prototypic foamy virus Gag protein contains functional L domains.
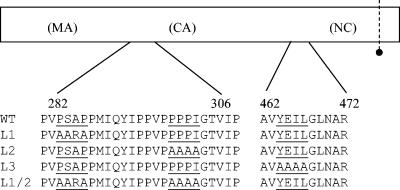
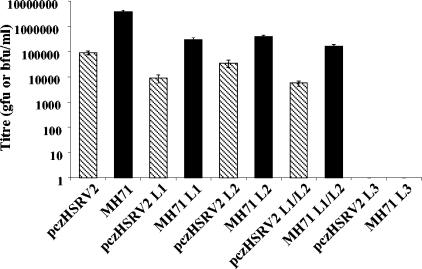
Three classes of L-domain sequence, based on the peptide sequences PT/SAP, PPPY, and YPDL/LXXLF, have been described in retroviruses, two of which are known to bind class E VPS factors. An analysis of the PFV Gag protein sequence identified candidate L-domain motifs based on sequence similarity (Fig. 1). To test whether these motifs were required for virion budding, each candidate L domain was mutated separately or in combination (Fig. 1). Mutations were introduced into both the intact proviral plasmid pczHSRV2 and the PFV vector construct pMH71. These mutant plasmids were then transfected into 293T cells, either alone for pczHSRV2-based constructs or with pczHFVenv, which expresses PFV Env, for the production of pMH71-based vectors. The viral or vector titer in the corresponding culture supernatants was determined (Fig. 2). Mutation of the L1 domain, from PSAP to AARA, resulted in titer reductions of 10.1-fold for the intact virus and 13.4-fold for the PFV vector. Mutation of the L2 domain, from PPPI to AAAA, resulted in a reduction in titer by 2.5-fold for the virus and 10-fold for the vector. Mutating both L1 and L2 motifs reduced the titer by 15.9-fold for virus and 24.1-fold for vector and mutation of the L3 domain from YEIL to AAAA reduced the titer to below the detection threshold (<10 infectious particles/ml) in both cases.
FIG. 1.
Schematic representation of the PFV Gag protein. Possible positions of the matrix (MA), capsid (CA), and nucleocapsid (NC) regions are indicated in brackets. The positions of putative L domains PSAP, PPPI, and YEIL are underlined along with the mutations made. The dotted line indicates the major proteolytic cleavage site for the viral protease.
FIG. 2.
The PSAP and PPPI motifs are required for optimal release of PFV vector and wild-type virus. The solid columns show titers of vector released from 293T cells transfected with either the wild-type (MH71), PSAP-mutated (MH71 L1), PPPI-mutated (MH71 L2), PSAP/PPPI dual-mutated (MH71 L1/L2), or YEIL-mutated (MH71 L3) vector constructs, all cotransfected with the pczHFVenv construct. Vector was titrated on D17 cells. The hashed columns show titers of virus released from 293T cells transfected with either the wild-type (pczHSRV2), PSAP-mutated (pczHSRV2 L1), PPPI-mutated (pczHSRV2 L2), PSAP/PPPI dual-mutated (pczHSRV2 L1/L2), or YEIL-mutated (pczHSRV2 L3) virus constructs. Virus is titrated on BHLL cells. Results shown are the means and standard errors of three independent experiments.
Mutation of PFV potential L domains results in defects of virus particle budding but not in Gag synthesis or particle infectivity.
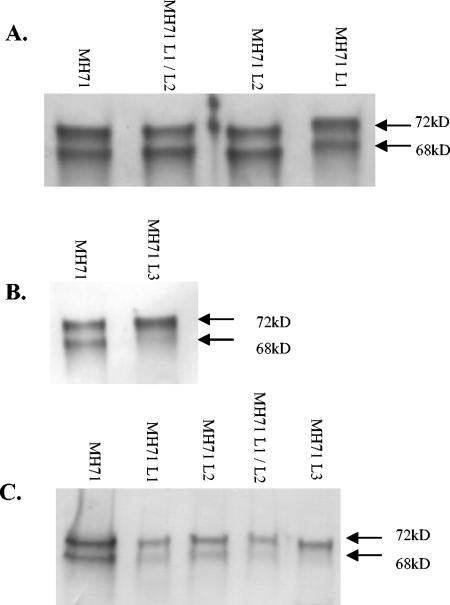
Reductions in titer could reflect a reduced level of Gag expression, a failure of virus particles to bud from the cell, or budding of normal quantities of less infectious particles. In order to differentiate between these possibilities, lysates and culture supernatants from cells expressing wild-type and mutant PFV vectors were analyzed by SDS-PAGE and Western blotting for the presence of PFV proteins. This analysis showed that in all cases, the mutant forms of Gag were expressed as full-length proteins at levels comparable to those of wild-type Gag (Fig. 3A). Normally, a significant proportion of the full-length 72-kDa PFV Gag undergoes a posttranslational maturation, with a 4-kDa fragment being removed by the viral-encoded protease to yield a 68-kDa form. The degree and rate of this cleavage appears normal for Gag with mutated L1, L2, and L1 and L2 domains (Fig. 3A). However, Gag processing was significantly reduced when the L3 domain was mutated (Fig. 3B).
FIG. 3.
Western blots showing the PFV Gag proteins for vector and L-domain mutants. (A) Gag proteins from lysates of 293T cells transfected with the pczHFVenv construct and either the wild-type (MH71), PSAP/PPPI dual-mutated (MH71 L1/L2), PPPI-mutated (MH71 L2), or PSAP-mutated (MH71 L1) vector constructs and harvested at day 3 posttransfection. Normal Gag processing is seen for the L1, L2, and L1/L2 double mutants. (B) Gag in lysates from cells 3 days posttransfection with the pczHFVenv construct and either the wild-type (MH71) or YEIL mutation (MH71 L3) where little of the 68-kDa cleavage product is observed. In this case, the 293T cells were transfected with half the normal amount of MH71 or MH71 L3 construct. For panels A and B, identical results were obtained when lysates were prepared on days 1 and 2 posttransfection. (C) Gag proteins of sucrose-cushion purified vector particles from the wild-type and L-domain mutant vector preparations. A reduced amount of Gag is seen in the mutant vector preparations consistent with the reduced titer. In the case of the L3 mutant, only unprocessed Gag is detected in the culture supernatant.
Having shown that the reduction in titer was not simply a reflection of reduced expression of mutant Gag, we analyzed the level of Gag released to the culture supernatant that could be pelleted through a sucrose cushion. In each case, mutation of the candidate L domain resulted in a reduction in the fraction of Gag that was present in the virion pellets (Fig. 3C). Although the Western blot analysis was not quantitative, these data suggest that the reduced PFV titers obtained upon L1 and L2 mutation predominantly reflect a reduced level of particle egress, rather than release of noninfectious particles. In the case of the L3 mutant, where no infectious particles could be detected, some Gag was detectable in the pellet. This was all in the unprocessed form, in agreement with a previous study which showed that PFV particles containing unprocessed Gag are noninfectious (8). No particles were detected in supernatants from cells transfected with wild-type Gag in the absence of expression of Env (data not shown).
PFV budding is inhibited by a blockage of the VPS pathway.
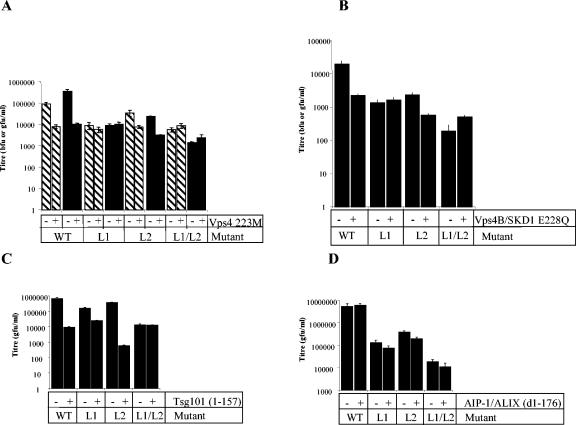
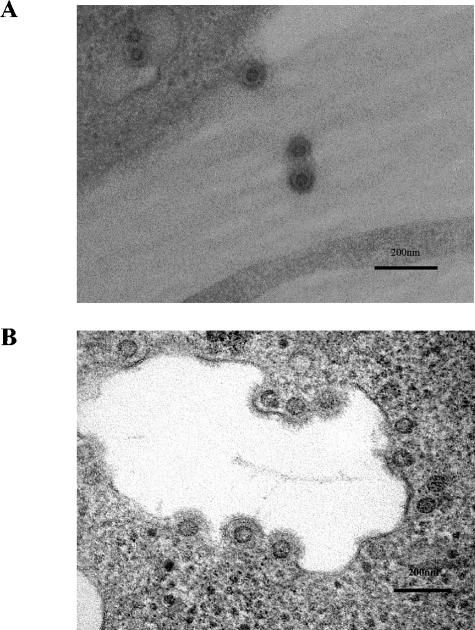
One of the final steps in budding via the class E VPS pathway requires the assembled ESCRT protein complexes to be actively removed by the action of the ATPase Vps4. Thus, expression of a dominant negative form of this enzyme results in inhibition of class E VPS-dependent viral budding. For a number of retroviruses, this leads to the accumulation of virions that appear, in electron micrographs, to be blocked at a late stage in budding. We, therefore, coexpressed dominant negative forms of Vps4 and the closely related Vps4B/SKD1 (50) as GFP or DS-RED fusion proteins in 293T cells transfected with PFV virus and vector constructs. Unfused GFP or DS-RED was used as a control for nonspecific effects. Dominant negative Vps4 reduced the level of PFV virus and vector production by 11.5-fold and 34.8-fold, respectively (Fig. 4A), and dominant negative Vps4B/SKD1 reduced vector production by 8.7-fold (Fig. 4B). EM analysis of 293T cells cotransfected with PFV vector and dominant negative Vps4 revealed a phenotype similar to that seen for other retroviruses known to require an intact VPS pathway during budding, namely, an accumulation of assembled virus particles arrested at a late stage of budding (Fig. 5B). In addition, we frequently observed fully assembled, but not yet enveloped, capsids in close proximity to cell membranes, as well as apparently partially enveloped forms. Taken together, these results show that inhibition of the VPS pathway inhibits PFV budding and perhaps envelopment.
FIG. 4.
Effect of the dominant negative Vps4, Tsg101 or AIP-1/ALIX proteins on release of PFV virus or vector. Hatched bars indicate titers of virus produced from 293T cells transfected with pczHSRV2 (wild type or mutant), solid bars show titers of vector from cells transfected with MH71 (wild type or mutant). All vector constructs were cotransfected with pczHFVenv. In all cases, negative controls were also cotransfected with pCR3.1GFP or pDSRED-C1. (A) Effect of expression of dominant negative Vps4 (VPS4-223 M-GFP). (B) Expression of dominant negative Vps4B/SKD1 E228Q. (C) Expression of dominant negative truncated Tsg101 (pCR3.1-YFP-Tsg101 [1-157]). (D) Expression of dominant negative truncated AIP-1/ALIX [pCR3.1-YFP-ALIX(d1-176)].
FIG. 5.
EM analysis of 293T cells transfected with PFV vector constructs. (A) Wild-type PFV vector constructs. (B) Wild-type PFV vector constructs cotransfected with dominant negative Vps4.
We then asked if the budding of particles with mutated L1 and L2 domains was also sensitive to expression of dominant negative Vps4. Only PFV with an intact L1 domain was sensitive to dominant negative Vps4, with infectious virus or vector production being inhibited by 4.6-fold and by 7.5-fold, respectively (Fig. 4A). An identical result was obtained with dominant negative Vps4B/SKD1 (Fig. 4B).
L domains of the PT/SAP (L1) type interact directly with the Tsg101 component of the ESCRT-I complex (30). We therefore examined the effect of a dominant negative, PT/SAP-binding form of Tsg101 consisting of the N-terminal 157 amino acids on budding of PFV vector (Fig. 4C). Consistent with the aforementioned inhibition by dominant negative Vps4, wild-type PFV vector, as well as mutants in which Gag had an intact L1 domain, were inhibited by the overexpression of dominant negative Tsg101. Constructs in which both L1 and L2 domains were mutated were completely insensitive to dominant negative Tsg101. Surprisingly, when Gag had a mutated L1 (PSAP) domain but an intact L2 (PPPI), residual infectious virus release was inhibited to a significant degree (approximately 10-fold) by dominant negative Tsg101. However, the degree of inhibition was 10- to 100-fold less than that for Gag with an intact L1 domain. Analysis of interactions between PFV Gag and Tsg101 using yeast 2-hybrid assays showed a clear interaction between wild-type Gag and Tsg101. This interaction was dependent on the L1 (PSAP) motif, and no interaction between L2 (PPPI) and Tsg101 was observed (Table 1).
TABLE 1.
Direct interaction of PFV Gag with Tsg101 shown by yeast two-hybrid assaya
| GAL4 (DNA binding) fusion | VP16 (activation domain) fusion | β-Galactosidase activity (OD540)b |
|---|---|---|
| None | None | <0.01 |
| PFV Gag (WT) | None | <0.01 |
| PFV Gag (L1) | None | <0.01 |
| PFV Gag (L2) | None | <0.01 |
| PFV Gag (L1 + L2) | None | <0.01 |
| PFV Gag (WT) | Tsg101 | 1.056 |
| PFV Gag (L1) | Tsg101 | <0.01 |
| PFV Gag (L2) | Tsg101 | 0.938 |
| PFV Gag (L1 + L2) | Tsg101 | <0.01 |
| Tsg101 | None | <0.01 |
| Tsg101 | PFV Gag (WT) | 0.927 |
| Tsg101 | PFV Gag (L1) | <0.01 |
| Tsg101 | PFV Gag (L2) | 1.167 |
| Tsg101 | PFV Gag (L1 + L2) | <0.01 |
| PFV Gag | AIP1/ALIX | <0.01 |
| AIP1/ALIX | PFV Gag | <0.01 |
| AIP1/ALIX | EIAV p9 | 3.21 |
Yeast (Y190) cells were transformed with the indicated GAL4 DNA binding domain and VP16 activation domain fusion protein expression plasmids. Transformants were selected on media lacking tryptophan and leucine, and β-galactosidase levels in cell lysates were determined. WT, wild type.
OD540, optical density at 540 nm.
Using yeast two hybrid assays, we tested whether PFV Gag containing an intact or mutated PPPI motif could bind to the WW domains from an variety of HECT ubiqutin ligases, including Nedd4, Nedd4-L, Bul-1, WWP1, and Itch. These results were universally negative (data not shown).
The L3 (YPXL/LXXLF) L domain type apparently recruits components of the class E VPS pathway at a stage downstream of Tsg101, via an interaction with the protein AIP-1/ALIX (23, 32, 41), which bridges the ESCRT-I and ESCRT-III complexes. We, therefore, determined whether the expression of a dominant negative form of AIP-1/ALIX (lacking amino acids 1 through 176) inhibited PFV vector budding. While this protein has previously been shown to inhibit potently YPDL-dependent EIAV budding (32), PFV release was not significantly affected, irrespective of whether the other candidate L domains were intact or mutated (Fig. 4D). Additionally, no interaction between PFV Gag and AIP-1/ALIX was observed using the yeast two-hybrid assay (Table 1).
DISCUSSION
Three classes of L-domain sequences based on the peptide motifs PT/SAP, PPPY, and YPDL/LXXLF within the structural proteins of retroviruses and several other enveloped viruses interact with cellular machinery involved in vesicle budding (reviewed in reference 11). Our examination of the PFV Gag sequence revealed three tetrapeptide motifs with significant homology to each of the three known classes of viral L domains, specifically, PSAP, PPPI, and YEIL. While the PSAP and PPPI motifs are present in only PFV and the other primate foamy viruses, the YXXL domain is conserved in all known foamy virus Gag proteins. In the context of both intact PFV and in a PFV-based vector system, mutation of each individual candidate L domain had no effect on levels of Gag protein synthesis, but resulted in a significant reduction in the yield of virus particles. Mutation of either the PSAP or PPPI domain reduced viral titer by 2.5- to 10-fold, an effect similar in magnitude to that observed upon mutation of the L domains of other retroviruses (5, 15, 16, 45, 47). L domains frequently occur in combination and bind separate cofactors. We, therefore, constructed PFV containing a Gag protein with mutations in both PSAP and PPPI domains. This had a greater effect on viral particle release than did each single mutation, confirming that these domains act in a nonredundant fashion to promote the formation of PFV virions.
The known L domains of enveloped viruses hijack the cellular machinery normally used in budding of vesicles into MVBs. This involves the sequential recruitment of ESCRT complexes, with the AAA-ATPase Vps4 subsequently being required for their dissociation from the membrane and recycling. PT/SAP based L domains bind directly to the Tsg101 component of the ESCRT-I complex (12, 42), YPXL motifs bind AIP-1/ALIX (32, 41), and PPPY motifs the WW domains of Nedd4-like ubiquitin ligases (48, 49), respectively. We found that expression of dominant negative versions of the Tsg101 component of ESCRT -I and Vps4 significantly reduced the budding of PFV. This suggests that, like other retroviruses, PFV budding is dependent, at least in part, on the class E VPS machinery. However, expression of a dominant negative form of AIP-1/ALIX had no effect on PFV budding, suggesting that it is not required in the budding of PFV. Consistent with these observations, PFV Gag bound to Tsg101 but not to AIP-1/ALIX. Of note, ESCRT complex assembly does not require AIP-1/ALIX in the yeast Saccharomyces cerevisiae (6) or human cells (44).
An analysis of the action of dominant negative Tsg101 and Vps4 proteins on mutant PFV budding revealed discrepancies between the current models of L-domain exploitation of ESCRT machinery and that apparently used by PFV. Current models predict that if both PSAP and PPPI domains act by recruiting components of the class E VPS pathway, then a dominant negative form of Vps4 should be inhibitory for both. However, for both replication-competent PFV and the PFV-based vector, dominant negative Vps4 and Vps4B/SKD1 reduced virion yield only in the presence of an intact PSAP motif. Dominant negative Tsg101 inhibited both PSAP and PPPI domain function. This might suggest that the PPPI motif is not a true L domain and facilitates PFV egress by some mechanism completely independent of the class E VPS pathway. However, this interpretation is difficult to reconcile with the observation that the presence of the PPPI motif both stimulates PFV egress and confers at least some degree of sensitivity to dominant negative Tsg101 in the absence of PSAP. Conversely, absence of the PPPI motif apparently renders PSAP-dependent PFV egress hypersensitive to dominant negative Tsg101. This finding contrasts with previous studies which showed that PPXY-dependent MuLV budding was unaffected by dominant negative Tsg101 fragments (7, 31) and insensitive to Tsg101 depletion using siRNA (12). These observations hint at a functional relationship between the PPPI motif and Tsg101, but the PFV PPPI clearly does not bind directly to Tsg101 in the yeast two-hybrid assays. It is possible that either the presence of an isoleucine in the PFV PPPI motif (as opposed to a tyrosine in conventional PPXY L domains), and/or that the cellular location of PFV budding (ER rather than plasma membrane or endosomal membranes) changes the nature of the factors recruited by the PFV PPPI domain and confers its partial dependence on Tsg101. It will be interesting to determine why PPPI-dependent PFV budding appears Tsg101 dependent but Vps4 independent and whether PFV budding shares features with that of Rhabdoviruses which has been reported to be Vps4 independent (20).
PFV Gag also contains the sequence YEIL, which is homologous to the YPDL-based L domain found in EIAV. Mutation of this sequence completely abolished the ability of the PFV Gag protein to support infectious particle formation. While the YPDL motif from EIAV binds to AIP-1/ALIX (12, 32, 41), the PFV YEIL motif lacks this activity, since we find no direct interaction of PFV Gag and AIP-1/ALIX in yeast two-hybrid assays. Predictably therefore, a dominant negative form of AIP-1/ALIX failed to inhibit PFV budding. While this class of L domain was originally described as ‘YXXL’ rather than ‘YPXL’ (39) it has been shown that the proline plays an important role in EIAV budding (39) and binding of AIP-1/ALIX (43). Thus we conclude that PFV budding probably proceeds independently of AIP-1/ALIX.
At present, it is therefore unclear what role the YEIL motif plays in PFV assembly. It may simply be required for the structural integrity of Gag monomers or multimers at an earlier stage in the assembly process. Indeed, the virally encoded protease cleaves 4 kDa from the C terminus of PFV Gag (8, 10). This occurs prior to budding and is thought to be required for the viral particles to be infectious (8). Mutation of the PSAP and PPPI domains had no effect on this processing. However, mutation of YEIL significantly reduced the kinetics of processing. The YEIL domain lies in a region of sequence previously shown to contain a splice acceptor sight for the production of the Pol mRNA transcript (21, 51). These studies found that minor nucleotide changes in this region, which conserve the Gag coding sequence, abolish the expression of Pol and prevent the processing of Gag. In our hands, mutation of the YEIL domain to AAAA does not completely abolish processing of Gag but slows its kinetics, possibly reflecting a reduced expression of Pol.
In conclusion, we have identified three motifs within PFV Gag with significant homology to known L domains. One of these, PSAP, functions as a conventional L domain by binding to Tsg101. A second, PPPI, which does not bind directly Tsg101, also plays a role in virus assembly or budding and confers sensitivity to dominant negative Tsg101. The third candidate L-domain motif, YEIL, is required for virus assembly but appears to act by a mechanism distinct from that of known L domains. Differences in the mechanism by which these domains function in PFV compared to other retroviruses may reflect differences in budding mechanisms across specific cellular membranes. Elucidation of the reasons for these differences provides scope for future study.
Acknowledgments
We thank Wes Sundquist for providing dominant negative Vps4B/SKD1 constructs and comments on the manuscript and also Mike Hollinshead for assistance with electron microscopy.
This work was supported by grants from the Wellcome Trust and the Jefferiss Research Trust to M.O.M. and the NIH (R01A152774 and R01A150111) to P.D.B.
REFERENCES
- 1.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bache, K. G., A. Brech, A. Mehlum, and H. Stenmark. 2003. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J. Cell Biol. 162:435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin, D. N., and M. L. Linial. 1998. The roles of Pol and Env in the assembly pathway of human foamy virus. J. Virol. 72:3658-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieniasz, P. D., A. Rethwilm, R. Pitman, M. D. Daniel, I. Chrystie, and M. O. McClure. 1995. A comparative study of higher primate foamy viruses, including a new virus from a gorilla. Virology 207:217-228. [DOI] [PubMed] [Google Scholar]
- 5.Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, M. D. Los Santos, A. Rein, and S. P. Goff. 2003. PPPYEPTAP motif is the late domain of human T-cell leukemia virus type 1 GaG and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 77:11882-11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowers, K., J. Lottridge, S. B. Helliwell, L. M. Goldthwaite, J. P. Luzio, and T. H. Stevens. 2004. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic 5:194-210. [DOI] [PubMed] [Google Scholar]
- 7.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eastman, S. W., and M. L. Linial. 2001. Identification of a conserved residue of foamy virus Gag required for intracellular capsid assembly. J. Virol. 75:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enssle, J., I. Jordan, B. Mauer, and A. Rethwilm. 1996. Foamy virus reverse transcriptase is expressed independently from the Gag protein. Proc. Natl. Acad. Sci. USA 93:4137-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer, N., M. Heinkelein, D. Lindemann, J. Enssle, C. Baum, E. Werder, H. Zentgraf, J. G. Muller, and A. Rethwilm. 1998. Foamy virus particle formation. J. Virol. 72:1610-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 13.Goepfert, P. A., K. Shaw, G. Wang, A. Bansal, B. H. Edwards, and M. J. Mulligan. 1999. An endoplasmic reticulum retrieval signal partitions human foamy virus maturation to intracytoplasmic membranes. J. Virol. 73:7210-7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinkelein, M., M. Rammling, T. Juretzek, D. Lindemann, and A. Rethwilm. 2003. Retrotransposition and cell-to-cell transfer of foamy viruses. J. Virol. 77:11855-11858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irie, T., J. M. Licata, J. P. McGettigan, M. J. Schnell, and R. N. Harty. 2004. Budding of PPxY-containing rhabdoviruses is not dependent on host proteins TGS101 and VPS4A. J. Virol. 78:2657-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jordan, I., J. Enssle, E. Guttler, B. Mauer, and A. Rethwilm. 1996. Expression of human foamy virus reverse transcriptase involves a spliced pol mRNA. Virology 224:314-319. [DOI] [PubMed] [Google Scholar]
- 22.Katzmann, D. J., C. J. Stefan, M. Babst, and S. D. Emr. 2003. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindemann, D., T. Pietschmann, M. Picard-Maureau, A. Berg, M. Heinkelein, J. Thurow, P. Knaus, H. Zentgraf, and A. Rethwilm. 2001. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linial, M. L. 1999. Foamy viruses are unconventional retroviruses. J. Virol. 73:1747-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lochelt, M. 2003. Foamy virus transactivation and gene expression, p. 27-62. In A. Rethwilm (ed.), Foamy viruses. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 27.Lochelt, M., and R. M. Flugel. 1996. The human foamy virus pol gene is expressed as a Pro-Pol polyprotein and not as a Gag-Pol fusion protein. J. Virol. 70:1033-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, Q., L. W. Q. Hope, M. Brasch, C. Reinhard, and S. N. Cohen. 2003. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc. Natl. Acad. Sci. USA 100:7626-7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin-Serrano, J., D. Perez-Caballero, and P. D. Bieniasz. 2004. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 78:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 31.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin-Serrano, J., A. Yaravoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moebes, A., J. Enssle, P. D. Bieniasz, M. Heinkelein, D. Lindemann, M. Bock, M. O. McClure, and A. Rethwilm. 1997. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 71:7305-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patton, G. S., O. Erlwein, and M. O. McClure. 2004. Cell cycle dependence of foamy virus vectors. J. Gen. Virol. 85:2925-2930. [DOI] [PubMed] [Google Scholar]
- 35.Perez, M., R. C. Craven, and J. C. de la Torre. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. USA 100:12978-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pietschmann, T., M. Heinkelein, M. Heldmann, H. Zentgraf, A. Rethwilm, and D. Lindemann. 1999. Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 73:2613-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pornillos, O., J. E. Garrus, and W. I. Sundquist. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12:569-579. [DOI] [PubMed] [Google Scholar]
- 38.Pornillos, O., D. S. Higginson, K. M. Stray, R. D. Fisher, J. E. Garrus, M. Payne, G. P. He, H. E. Wang, S. G. Morham, and W. I. Sundquist. 2003. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J. Cell Biol. 162:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72:10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 42.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA. 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vincent, O., L. Rainbow, J. Tilburn, H. N. Arst, Jr., and M. A. Peñalva. 2003. YPXL/I is a protein interaction motif recognized by Aspergillus PalA and its human homologue, AIP1/Alix. Mol. Cell. Biol. 23:1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 45.Wang, H. T., N. J. Machesky, and L. M. Mansky. 2004. Both the PPPY and PTAP motifs are involved in human T-cell leukemia virus type 1 particle release. J. Virol. 78:1503-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yasuda, J., M. Nakao, Y. Kawaoka, and H. Shida. 2003. Nedd4 regulates egress of Ebola virus-like particles from host cells. J. Virol. 77:9987-9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshimori, T., F. Yamagata, A. Yamamoto, N. Mizushima, Y. Kabeya, A. Nara, I. Miwako, M. Ohashi, M. Ohsumi, and Y. Ohsumi. 2000. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol. Biol. Cell 11:747-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, S. F., D. N. Baldwin, S. R. Gwynn, S. Yendapalli, and M. L. Linial. 1996. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271:1579-1582. [DOI] [PubMed] [Google Scholar]
- 52.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]