Abstract
Synaptic plasticity is a process that shapes neuronal connections during neurodevelopment and learning and memory. Autophagy is a mechanism that allows the cell to degrade its unnecessary or dysfunctional components. Autophagosomes appear at dendritic spines in response to plasticity-inducing stimuli. Autophagy defects contribute to altered dendritic spine development, autistic-like behavior in mice, and neurological disease. While several studies have explored the involvement of autophagy in synaptic plasticity, the initial steps of the emergence of autophagosomes at the postsynapse remain unknown. Here, we demonstrate a postsynaptic association of autophagy-related protein 9A (Atg9A), known to be involved in the early stages of autophagosome formation, with Rab11, a small GTPase that regulates endosomal trafficking. Rab11 activity was necessary to maintain Atg9A-positive structures at dendritic spines. Inhibition of mTOR increased Rab11 and Atg9A interaction and increased the emergence of LC3 positive vesicles, an autophagosome membrane-associated protein marker, in dendritic spines when coupled to NMDA receptor stimulation. Dendritic spines with newly formed LC3+ vesicles were more resistant to NMDA-induced morphologic change. Rab11 DN overexpression suppressed appearance of LC3+ vesicles. Collectively, these results suggest that initiation of autophagy in dendritic spines depends on neuronal activity and Rab11a-dependent Atg9A interaction that is regulated by mTOR activity.
Autophagy in neurons is involved in neuronal connectivity and NMDA receptor dependent changes in synaptic plasticity. Early steps of autophagy in the postsynaptic compartment and their regulation are not known.
With live- and fixed-cell imaging in hippocampal neurons and coimmunoprecipitation from synaptoneurosomes, the authors show a new role of Rab11a GTPase and its interaction with Atg9A, dependent on mTOR and NMDA stimulation, for the emergence of LC3+ structures at the postsynapse.
This study provides new insight in the spatiotemporal dynamics, mechanism and regulation of autophagy initiation in dendritic spines and its effect on neuroplasticity.
INTRODUCTION
Macroautophagy or autophagy is the biological process of sequestering and degrading proteins or even whole organelles in bulk (Yin et al., 2016). Mammalian/mechanistic target of rapamycin (mTOR) kinase regulates autophagy in mammalian cells (Noda and Ohsumi, 1998; Ganley et al., 2009; Jung et al., 2009). Autophagosomes are double-membrane structures formed during autophagy that fuse with lysosomes to digest autolysosome contents (Yin et al., 2016).
Autophagy plays an essential role in neuronal development and synaptic function, which can be altered in neurological disease (Stavoe and Holzbaur, 2019). Long-term mTOR inhibition increases the number of autophagosomes in the neuronal soma of the mouse cortex (Mizushima et al., 2004; Boland et al., 2008). Autophagy occurs continuously in axons (Maday et al., 2012, Maday and Holzbaur, 2016) and contributes to synaptic plasticity – defined as a process that changes the strength of synaptic connections (Bosch and Hayashi, 2012; Lai and Ip, 2013; Borczyk et al., 2019). Silencing or enhancing global synaptic activity in cultured neurons alters the movements of autophagosomes in dendritic shafts (Kulkarni et al., 2021). One form of synaptic plasticity is long-term depression (LTD), defined as the prolonged weakening of the strength of synaptic connections (Malenka and Bear, 2004). Currently, several studies report the connection of LTD to neuronal autophagy. Autophagosomes in neuronal soma, dendrites, and dendritic spines are at low levels under basal conditions and increase in number upon the induction of NMDA-chemical LTD (cLTD; Shehata et al., 2012; Kallergi et al., 2022). Postsynaptic scaffold proteins and glutamate receptors are degraded via autophagy, which modulates synaptic plasticity (Nikoletopulou et al., 2017; Compans et al., 2021; Kallergi et al., 2022). However, it is not known whether the autophagosomes can originate at the postsynapse.
Studies in nonneuronal cells provide insight into the mechanism of autophagy initiation. Other proteins, such as Unc-51 like autophagy activating kinase 1 (Ulk1), autophagy-related protein (Atg) 5, Atg13, and other Atg family proteins, participate in autophagy initiation (Yang and Klionsky, 2010). In neurons, Ulk1 puncta increased in dendrites upon NMDA-cLTD treatment (Kallergi et al., 2022). In axons, autophagosomes emerge from endoplasmic reticulum domains positive for double FYVE-containing protein 1 and mature during retrograde axonal transport (Maday et al., 2012; Maday and Holzbaur, 2016).
Recent studies report that Rab11, a guanosine triphosphatase (GTPase), is associated with recycling endosomes in nonneuronal cells, which act as a membrane source for autophagosomes (Longatti et al., 2012; Welz et al., 2014). Rab11-positive recycling endosomes colocalized with Ulk1 and Atg9A, dispatching the membrane, causing the emergence of autophagosomes (Welz et al., 2014). Atg9A is essential for mammalian autophagy by interacting with the autophagy initiation site (Orsi et al., 2012) and trafficking through recycling endosomes (Imai et al., 2016). Loss of axonal Atg9A caused defective autophagosome maturation in axons in AP-4 deficiency syndrome (Ivankovic et al., 2020). Puri et al. (2018) proposed that Rab11-positive recycling endosomes in HeLa cells form a platform for assembling autophagy machinery and autophagosome formation.
Much remains unknown about the cell biological steps preceding the phagophore formation via the Ulk1 complex and the emergence of autophagosomes within dendritic spines, which are morphologically and functionally distinct postsynaptic compartments crucial for neuroplasticity, memory, and learning. Here, we demonstrate a novel role for Rab11 in the initiation of autophagy at the postsynapse. Rab11-positive vesicles are present in dendritic spines, and we observed that their mobility is reduced upon treatment with the mTOR inhibitor INK128. Atg9A was shown to be highly prevalent at the dendritic spines, which was not previously known, and we observed that Rab11 is necessary for the postsynaptic accumulation of Atg9A. Rab11 coimmunoprecipitated with Atg9A and microtubule-associated protein 1A/1B-light chain 3 (LC3), an autophagosome membrane-associated protein marker. Live cell imaging of the same neurons over time revealed the dynamics of postsynaptic LC3 positive vesicle formation upon mTOR inhibition and NMDA stimulation, which corresponded in a spatiotemporal manner to Rab11 and Atg9A accumulation and association. Finally, Rab11 activity was necessary for the emergence of LC3 positive vesicles in the dendritic spines upon mTOR inhibition and NMDA stimulation. Taken together our study provides new insight in the spatiotemporal dynamics, mechanism, and physiologic regulation of autophagosome initiation in dendritic spines that occurs via a Rab11-dependent interaction with Atg9A.
RESULTS
Rab11a mobility decreases at dendritic spines upon mTOR inhibition
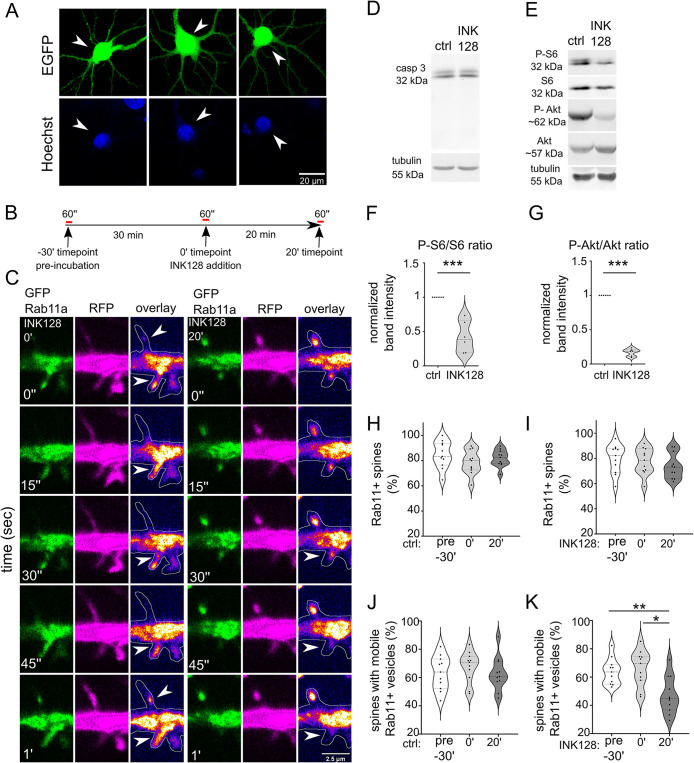
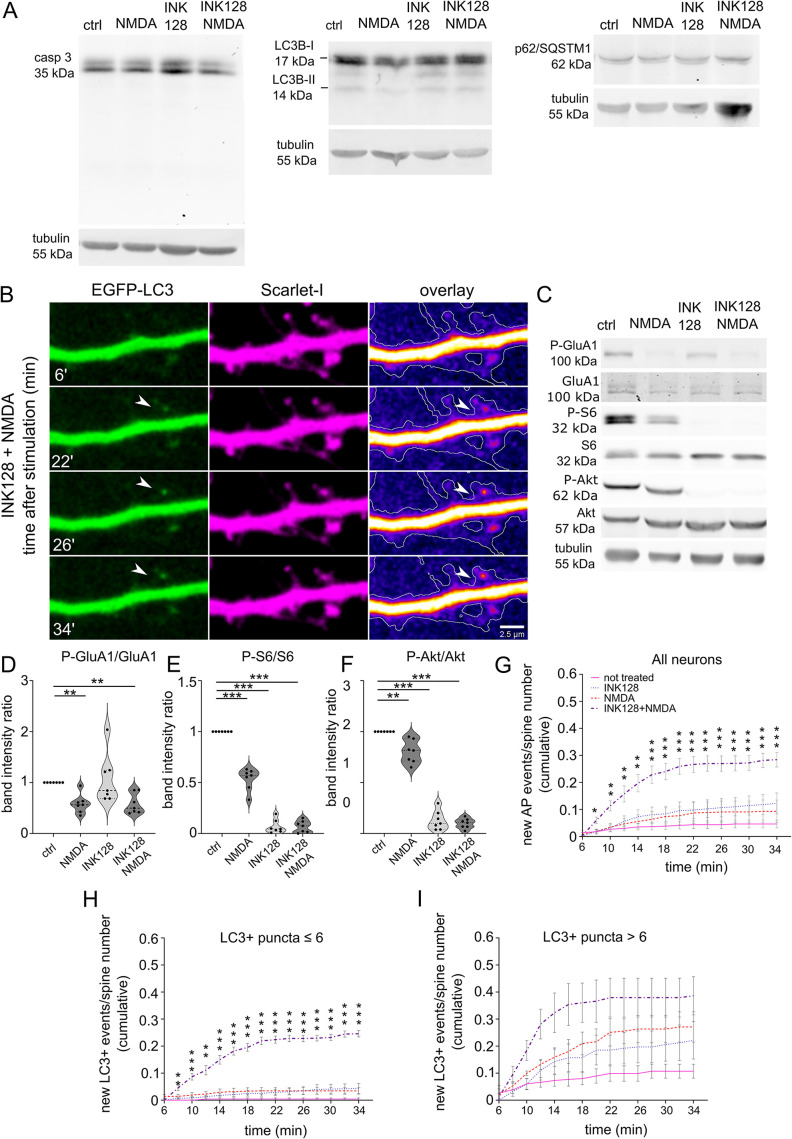
An earlier study showed that Rab11-positive recycling endosomes contribute to autophagy (Longatti et al., 2012). A lack of nutrients inhibits mTOR complex 1 (mTORC1), one of two protein complexes formed by mTOR, and the suppression of mTORC1 activity induces autophagy, which was demonstrated in both yeast and nonneuronal cells (Noda and Ohsumi, 1998; Levine and Klionsky, 2004). Considering these findings in nonneuronal cells, we performed a Rab11 mobility assay to investigate the influence of mTOR kinase on Rab11a at dendritic spines. To suppress mTOR, we applied INK128, an ATP-competitive inhibitor of mTORC1 and mTORC2. mTORC1 is an inhibitor of autophagy, and mTORC2 likely influences autophagy through Akt activation (Mammucari et al., 2007), protein kinase Cα/β (Renna et al., 2013), or changes in mitochondrial homeostasis (Aspernig et al., 2019). In our experiments, we used embryonic rat hippocampal neurons that had mature dendritic spines (day in vitro [DIV] 21-25) with a gentle lipofectamine-based transfection protocol suited to older neurons which allows them to be transfected without showing signs of cytotoxicity (Figure 1A). For the Rab11 mobility assay, we transfected neurons with plasmids encoding green fluorescent protein (GFP)-Rab11a and red fluorescent protein (RFP) to visualize transfected neurons’ morphology (including dendrites and dendritic spines). We performed time-lapse imaging of transfected cells using a confocal spinning-disk microscope the next day. We imaged each neuron three times: at baseline (preincubation; unpublished data), after 30 min of incubation on the microscope stage to monitor for potential signs of neuronal toxicity and any changes in imaging conditions (e.g., temperature; 0 min. timepoint), then after 20 min of incubation either without (control) or with INK128 (300 nM; Figure 1, B and C). INK128 (300 nM) did not cause any detectable cytotoxicity (WB using an antibody against caspase 3) but effectively decreased phosphorylation of S6 (serines 235/236) and Akt (serine 473), commonly used as indicators of mTORC1 and mTORC2 activity, respectively (Figure 1, D–G). We counted the number of spines in which Rab11a-positive vesicles were present (Figure 1, H and I). Of these spines, an average of ∼66% contained mobile Rab11a that entered or exited the dendritic spine at some point over 1 min or moved from the dendritic spine head to the neck (Figure 1, J and K; Supplemental Movies 1 and 2). The remaining spines contained only Rab11a vesicles that did not move, and resided primarily at the spine head (Figure 1, J and K). The 30-min preincubation step did not affect Rab11a dynamics. Twenty minutes of incubation with INK128 decreased the number of dendritic spines that contained mobile Rab11a by ∼30% (Figure 1, J and K) compared with the respective control. Altogether, these data indicated that mTOR inhibition, which is known to induce autophagy, resulted in the partial immobilization of Rab11a in dendritic spines.
FIGURE 1:
mTOR inhibition decreases Rab11 mobility in dendritic spines. (A) Representative images of neurons transfected, using Lipofectamine 2000, at ∼ 3 wk in culture, with plasmid encoding EGFP (arrowheads) stained with Hoechst33258. The image shows healthy, nonshrunk nuclei of transfected cells. (B) Diagram of treatment and imaging for experiments in Figure 1. Neurons were recorded for 1 min (preincubation recording; pre-30′), then incubated for 30 min to control potential phototoxicity or overheating in the microscope stage. Then, neurons were rerecorded (0′) and incubated for 20 min without (ctrl 20′) or with 300 nM mTOR inhibitor INK128 (INK128 20′) for the final recording. (C) Representative time-lapse images of dendrites of rat hippocampal neurons that were cotransfected on DIV22 with plasmids encoding GFP-Rab11a and RFP and treated the next day with INK128 (300 nM). The figure shows Rab11a vesicles (green), RFP (magenta), an overlay with GFP-Rab11a (fire pseudocolor), and the outline based on the RFP channel (hand-drawn ROI). Arrows indicate dendritic spines with motile vesicles. Imaging occurred ∼17–24 h after transfection. Neurons were imaged three times, always for 1 min, at the baseline (unpublished data, in D and E; INK 128 pre-30′; see panels I and K), 30 min later just before treatment (INK128 0′) and after 20′ incubation with INK128 (INK128 20′). Control neurons (unpublished data in C) were imaged as described above without any treatment (ctrl pre-30′, ctrl 0′, and ctrl 20′, see H and J). Scale bar = 2.5 µm. (D) Western blot analysis of INK128 treatment effects on caspase 3 (casp 3) cleavage in cultured neurons. Primary hippocampal rat neurons (DIV21) were treated with INK128 (300 nM) for 20 min. Extracts were blotted against casp 3 with tubulin as a loading control. (E–G) Western blot analysis of mTORC1 and mTORC2 inhibition by INK128. The same extracts as in (D) were blotted against phosphorylated (Ser235/S236, P-S6) and total S6 and phosphorylated (Ser473, P-Akt) and total Akt to evaluate mTORC1 and mTORC2 suppression, respectively, with tubulin as a loading control. Data are presented as mean gray values for each phosphorylated protein normalized to total protein. n = 6 both for ctrl and INK128. ***p ≤ 0.001 (one-sample t test) (H) Percentage of Rab11a-positive spines in control neurons. (I) Percentage of Rab11a-positive spines in INK128-treated neurons. (J) Percentage of spines with mobile Rab11a in control neurons. (K) Percentage of spines with mobile Rab11a in INK128-treated neurons. The data are presented as a mean percentage of Rab11a positive spines normalized to all spines (H and I) or the mean percentage of Rab11a positive spines with mobile vesicles normalized to all Rab11a positive spines for all analyzed cells (J and K). n = 11 cells for treated and untreated neurons from four independent experiments. *p ≤ 0.05, **p ≤ 0.01 (repeated-measures ANOVA followed by Tukey’s post hoc test, all columns were compared with all columns).
Movie S1.
Fragment of a dendrite of a 21 DIV hippocampal neuron transfected with plasmids encoding GFP‐Rab11a (green) and RFP (magenta) over 1 min. Arrows point to examples of spines where mobile vesicles are present.
Movie S2.
GFP‐Rab11a channel of the same neuron as in Movie 1. Arrows point to examples of spines where mobile vesicles are present.
Co-occurrence of Rab11 and Atg9A increases upon mTOR inhibition
The mobile pool of Rab11a in dendritic spines was previously assigned to recycling endosomes and the delivery of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)-type ionotropic glutamate receptors to postsynaptic sites (Esteves da Silva et al., 2015). The presence of stationary Rab11a, which is present in membranous structures, combined with the earlier findings on the potential contribution of Rab11 to autophagy, prompted us to investigate whether Rab11-positive vesicles participate in this process at dendritic spines upon mTOR inhibition. We evaluated changes in the co-occurrence of Rab11 with autophagy- and recycling endosome-associated markers in response to INK128 application. A frequently used autophagy marker is LC3, which is recruited to the forming APs and is present until mature APs reach lysosomes. LC3-positive APs are sparse at dendritic spines, which is speculated to result from their dynamic behavior and short lifespan (Shehata et al., 2012). Little is known about preautophagosomal structures and the steps upstream of phagophore formation in the dendrites and dendritic spines. Therefore, we used another marker, Atg9A, to visualize early endomembrane structures where autophagy can originate. Atg9A is a transmembrane protein that initiates autophagy and can migrate through recycling endosomes to autophagosome nucleation sites (Imai et al., 2016). It transiently colocalizes with autophagosomes in HEK293 cells (Orsi et al., 2012).
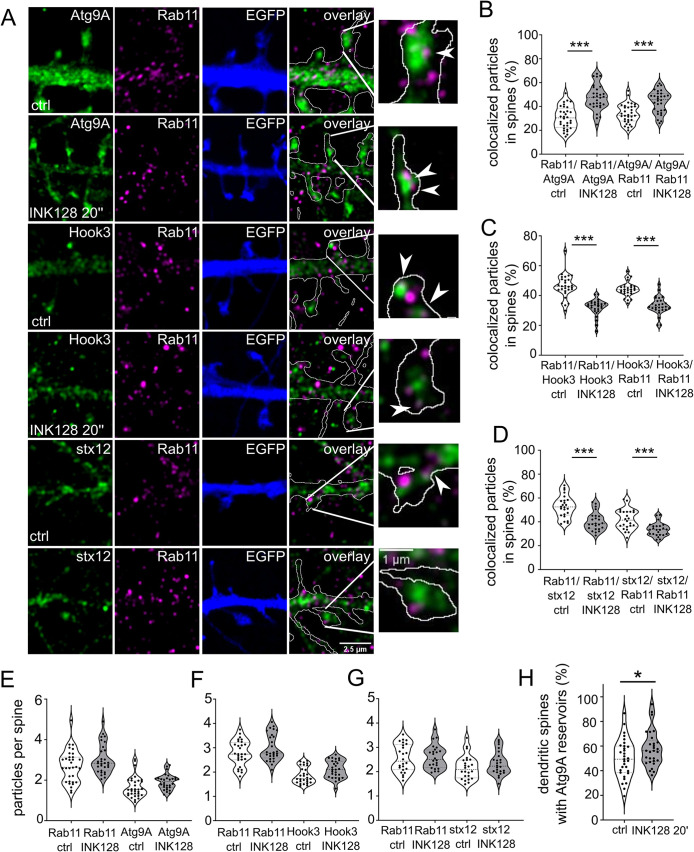
DIV21-25 rat hippocampal neurons were transfected with plasmid encoding EGFP to trace dendrites and spines of individual cells more accurately, treated with INK128 as described above, and fixed for immunofluorescence and super-resolution imaging. We performed double staining for Rab11 using three different markers. Besides Atg9A, as the marker of preautophagic vesicles, we stained neurons with Hook3 and syntaxin-12, which are associated with endocytic recycling. Hook3 is a cargo adaptor protein known to form a motile complex with dynein and dynactin in microtubular transport (Schroeder and Vale, 2016). Endosomal transport in dendritic spines mostly depends on actin filaments and actin-myosin interactions. However, microtubules were recently shown to dynamically enter and recede from dendritic spines in an activity-dependent manner, thus accelerating Rab11-positive vesicle transport (Gu et al., 2008; Hu et al., 2008; Jaworski et al., 2009; Esteves da Silva et al., 2015). Another marker associated with endocytic recycling is syntaxin-12 (also known as syntaxin 13), which binds to Rab11 through Glutamate receptor-interacting protein 1 (GRIP1)-associated protein 1 (GRASP-1) (Hoogenraad et al., 2010). Following antibody staining, we performed Airyscan super-resolution imaging to visualize the investigated proteins in dendritic spines. We used the EGFP channel to separate the transfected neuron from the processes originating in other nontransfected neurons (Figure 2A). Based on it, we isolated dendritic spines of all shapes with their spine neck and head, then performed an object-based colocalization analysis.
FIGURE 2:
mTOR inhibition targets Rab11 to Atg9A-positive reservoirs. (A) Representative Airyscan images of dendrites of neurons that were transfected with a plasmid encoding EGFP (blue) on DIV22 to allow accurate dendritic spine tracing and immunostained the next day for native Rab11 (magenta) and Atg9A, Hook3, or syntaxin 12 (stx12; green). The overlay channel shows Rab11 (magenta), Atg9A, Hook3, or stx12 (green), and the EGFP channel as an outline. The plasmid was expressed overnight. The neurons were incubated with INK128 (300 nM) for 20 min, fixed, and immunostained. Scale bar = 2.5 µm. The rightmost photomicrographs represent individual dendritic spines indicated in the overlay panels. Arrows indicate exemplary colocalized particles. Scale bar = 1 µm. (B) Colocalization of Rab11 with Atg9A in dendritic spines of neurons that were treated as in (A). (C) Colocalization of Rab11 with Hook3 in dendritic spines of neurons that were treated as in (A). (D) Colocalization of Rab11 with stx12 in dendritic spines of neurons that were treated as in (A). (E) Number of Rab11 and Atg9A particles per spine in neurons treated as in (A). (F) Number of Rab11 and Hook3 particles per spine in neurons treated as in (A). (G) Number of Rab11 and stx12 particles per spine in neurons treated as in (A). Data are presented as percentages of colocalized particles normalized to all particles in each channel (B, C, and D) or the number of total particles normalized to the number of dendritic spines (D, E, and F) for all neurons. The number of cells for Atg9A immunofluorescence analysis: n = 32 (control) and 30 (INK128) neurons from four independent experiments. The number of cells for Hook3 immunofluorescence analysis: n = 28 for both variants from three independent experiments. The number of cells for stx12 immunofluorescence analysis: n = 24 for both variants from three independent experiments. ***p ≤ 0.001 (one-way ANOVA followed by Sidak’s post hoc test). (H) Semiquantitative estimate of the prevalence of Atg9A large vesicles (reservoirs) in the dendritic spines. Same neurons as in B were evaluated for the presence of Atg9A-positive structures > 0.150 µm2 in the spines. The result was normalized to the number of dendritic spines and shown as a percentage. Data are presented as mean. n = 32 and 30 cells for control and INK128 variants from four independent experiments. *p < 0.05 (unpaired t test).
Atg9A was common in dendritic spines, with either punctate staining or prominent structures primarily in heads of mushroom-shaped spines and occasionally at the base of the spine and in dendrites (Figure 2A). At the baseline, over 40% of dendritic spines had large Atg9A structures (i.e., > 150 μm2), called Atg9 reservoirs. INK128 treatment slightly increased the fraction of such spines (Figure 2H). The colocalization of Rab11 with Atg9A and vice versa increased from ∼30 to ∼45% after 20 min of INK128 treatment, while the average particle number per total number of spines did not significantly change (Figure 2, B and E). The reverse was true for Hook3, in which colocalization decreased from ∼45 to ∼32% (Figure 2, C and F). Rab11 colocalization with syntaxin-12 decreased by 12% upon INK128 treatment; this difference was smaller with syntaxin-12 versus Rab11 colocalization (Figure 2, D and G). These results suggest that most syntaxin-12 co-occurs with Rab11 (a characteristic of recycling endosomes), while Rab11 changes its co-occurrence with syntaxin-12 upon INK128 treatment. Thus, the association between Rab11 and recycling endosomes was diminished. Altogether, the more significant co-occurrence of Rab11 and Atg9A and decrease in colocalization with the mobility marker Hook3 and recycling endosome marker syntaxin-12 suggest that Rab11 partly shifts to compartments that contribute to early autophagy upon mTOR inhibition. Overall, these results suggest that Rab11 plays a role in initiating autophagy in dendritic spines.
Rab11a activity is necessary for Atg9A maintenance at dendritic spines
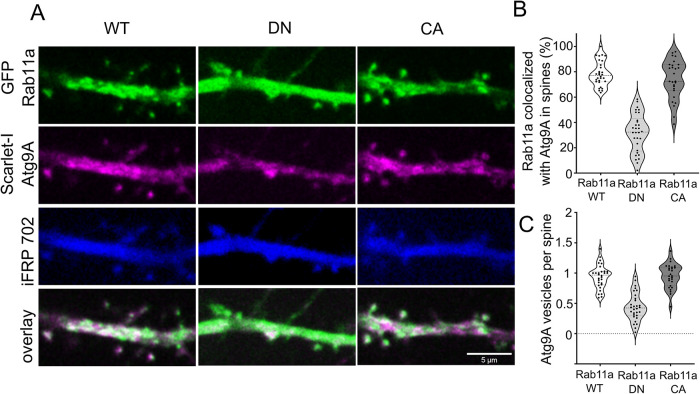
Rab11 and Atg9A co-occurred at dendritic spines, and their colocalization changed upon mTOR inhibition. We also observed that large Atg9A-positive structures are widespread at the dendritic spines. We next sought to confirm that the interaction between Rab11 and Atg9A has functional consequences in dendritic spines. The expression of a dominant-negative (DN) variant of Rab11a (S25N) blocks the activity of endogenous Rab11a, halts endocytic recycling, and changes the morphology of dendritic spines (Esteves da Silva et al., 2015). In mouse embryonic fibroblasts, Atg9A moved through recycling endosomes that delivered it to preautophagosomal sites (Imai et al., 2016), suggesting that Rab11 might be needed for Atg9A to reach its destination. Therefore, we investigated whether Rab11a activity is necessary to maintain Atg9A at dendritic spines. We transfected DIV21-25 rat embryonic hippocampal neurons with plasmids encoding iRFP702 to fill the dendrites using Scarlet-I-Atg9A, and one of the three variants of GFP-Rab11a (wildtype [WT], DN Rab11a variant S25N (Hunyady et al., 2002), or constitutively active [CA] Rab11a variant S20V; Pasqualato et al., 2004). The next day, we analyzed live neurons’ Scarlet-I-Atg9A distribution and colocalization with GFP-Rab11a variants (by object-based colocalization analysis). Scarlet-I-Atg9A was present in dendritic spines of GFP-Rab11a WT and GFP-Rab11a CA transfected neurons, where it colocalized with GFP-Rab11 were no significant differences between analyzed variants (Figure 3, A–C). Supplemental Movies 3, 4, and 5 that correspond to Figure 3. show co-occurrence of Rab11 with Atg9A in the Rab11 WT overexpressing neuron. In Rab11a DN overexpressing neurons, however, Scarlet-I-Atg9A presence in dendritic spines decreased from ∼0.9 to 0.45 particles per dendritic spine number (i.e., all dendritic spines in focus in the field of view) compared with Rab11a WT. Also, Scarlet-I-Atg9A/GFP-Rab11a colocalization in dendritic spines greatly diminished in Rab11a DN transfected cells (from ∼80 to ∼30%; Figure 3, A–C). Of note, GFP-Rab11a DN was present and diffusely distributed in all dendritic spines of transfected neurons (Figure 3A); therefore, the results of colocalization, in this case, depend only on the dendritic distribution of Atg9A and not its co-occurrence with the recycling endosomes. Altogether, these data showed that Rab11a activity was needed for Atg9A maintenance at dendritic spines.
FIGURE 3:
Rab11a GTPase activity is necessary for the presence of Atg9A at dendritic spines. (A) Representative images of DIV22 neurons that were cotransfected with plasmids encoding GFP-Rab11a, DN Rab11a, or constitutively active (CA) Rab11a (green) together with Scarlet-I-Atg9A (magenta) and iRFP702 (blue). Imaging occurred 17–24 h after transfection. Scale bar = 5 µm. (B) Colocalization rate (percentage of colocalized vesicles) of transfected Rab11 variant and Atg9a at dendritic spines. (C) The number of Atg9a particles per spine of neurons transfected with the indicated Rab11 variants. Data are presented as the mean percentage of colocalized Rab11a particles normalized to all particles (B) or the number of Atg9A particles normalized to the number of spines for all cells (C). Number of analyzed cells: n = 30 for Rab11a WT, n = 30 for DN Rab11, and n = 27 for CA Rab11a from three independent experiments. ***p ≤ 0.001 (one-way ANOVA followed by Sidak’s post hoc test).
Movie S3.
Fragment of a dendrite of a neuron transfected with plasmids encoding GFP‐Rab11a (green) and Scarlet‐I‐Atg9A (magenta) over 100 sec. Arrows point to Atg9A co‐occurring in the same spines as Rab11.
Movie S4.
Atg9A channel of the same movie as in Movie 3. Arrows point to Atg9A co‐occurring in the same spines as Rab11.
Movie S5.
Rab11 channel of the same movie as in Movie 3. Arrows point to Rab11 co‐occurring in the same spines as Atg9A.
Rab11 coimmunoprecipitates with Atg9A in mouse synaptoneurosomes
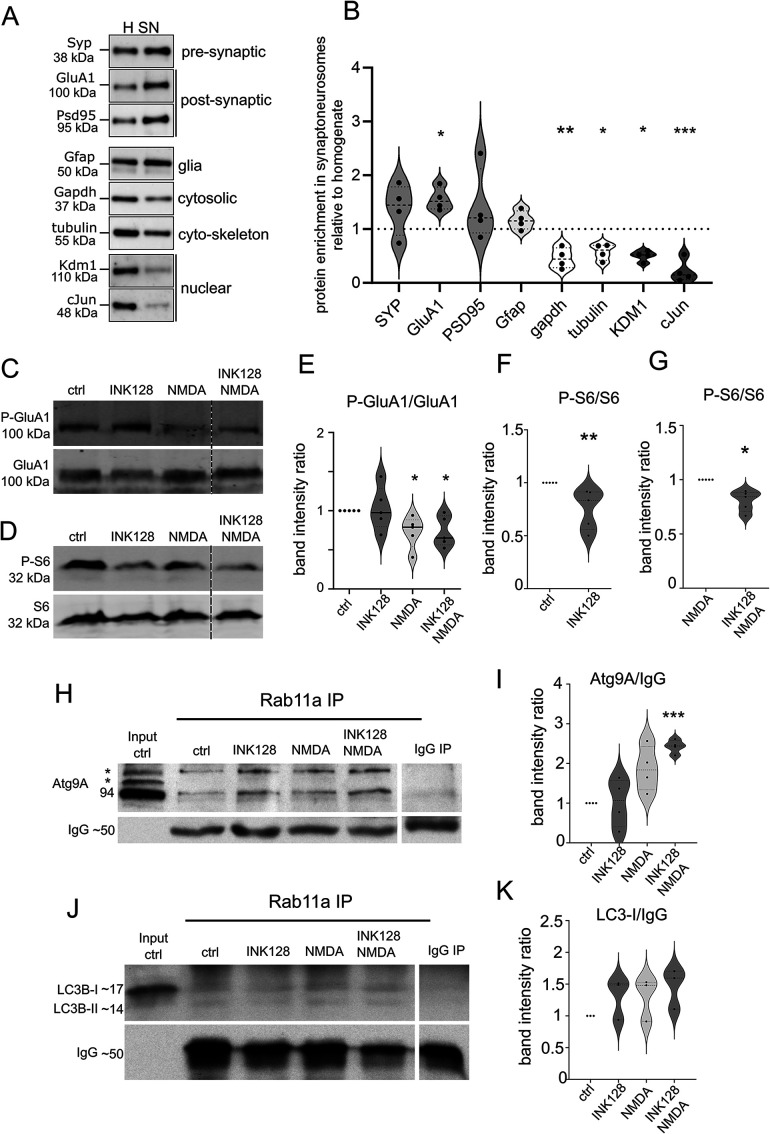
Because we demonstrated that Rab11 and Atg9A partially colocalize, we sought to investigate whether these two proteins occur in the same protein complex. To this end, we additionally performed Rab11 coimmunoprecipitation with an anti-Rab11 antibody. Because Atg9A is not indicative of ongoing autophagy and only provides the capacity to begin the autophagy, we were also interested in an established autophagy marker, namely LC3. Therefore, we sought to investigate whether LC3B, an isoform of LC3 that is widely expressed in the brain, is coprecipitated by Rab11, which would confirm the study by Puri et al. (2018) in HeLa cells. As starting material, we used synaptoneurosomes, a biochemical model of the synapse (Hollingsworth et al., 1985; Villasana et al., 2006) that previously worked in our hands (Dziembowska et al., 2012; Janusz et al., 2013; Jasińska et al., 2016; Kuzniewska et al., 2017, 2020). Synaptoneurosomes are a brain tissue preparation enriched in pre- and postsynaptic vesicles, able to react to stimulation and partially recapitulate downstream processes that occur under the stimulation of the whole neuron ([Hollingsworth et al., 1985; Villasana et al., 2006; Dziembowska et al., 2012; Janusz et al., 2013; Jasińska et al., 2016; Kuzniewska et al., 2017, 2020). The enrichment of synaptic proteins in the synaptoneurosomes is presented in Figure 4, A and B. Other researchers reported the participation of autophagy in cLTD. Considering the results of our earlier experiments with INK128, we also investigated whether the degree of Atg9A coimmunoprecipitation with Rab11 in synaptoneurosomes changes upon NMDA stimulation that in live neurons or slices evoked cLTD (Lee et al., 1998; Li et al., 2004) or mTOR suppression.
FIGURE 4:
Rab11a interacts with Atg9A in mouse synaptoneurosomes. (A and B) Western blot analysis of the fractions obtained during synaptoneurosomes (SN) preparation. Enrichment of both pre- and postsynaptic markers and depletion of cytosolic and nuclear markers in the SN fraction relative to the homogenate was observed. Glial marker (Gfap) was present, but not enriched (n = 4 biological replicates, paired t test, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001). (C and E) Western blot analysis of NMDA stimulation in synaptoneurosomal preparations. Synaptoneurosomes were prepared as in Figure 4A, and levels of protein extracts were blotted against phosphorylated (Ser845; P-GluA1) and total GluA1 to evaluate GluA1 dephosphorylation. Dashed lines indicate membrane image cuts. Data are presented as mean gray values for each phosphorylated protein normalized to total protein. n = 5 for all samples. *p < 0.05, **p < 0.01 (one-sample t test). (D, F, and G) Western blot analysis of mTORC1 inhibition by INK128. Synaptoneurosome extracts were blotted against phosphorylated P-S6 (Ser235/236; C, D) and S6 to evaluate mTORC1 complex suppression. Dashed lines show membrane image cuts. Data are presented as mean gray values for each phosphorylated protein normalized to total protein. n = 5 for all samples. *p < 0.05, **p < 0.01 (one-sample t test). (H) Western blot analysis of Atg9A coimmunoprecipitation with Rab11a from mouse synaptoneurosomes. Samples were first subjected to the treatments and then coimmunoprecipitated with anti-Rab11a antibody (see Materials and Methods). The blot shows the control immunoprecipitation sample following the input fraction (precleared control synaptoneurosome extract). The samples were then treated with INK128 (600 nM) alone, NMDA (50 µM) alone, or NMDA+INK128. The bands marked as * corresponds to posttranslational modifications of Atg9A. (I) Atg9A signal intensity ratios of the respective samples. The lower band (Atg9A without posttranslational modifications) was used for quantification. Mean gray values were normalized to IgG as a loading control and then to the control sample. n = 4. ***p ≤ 0.001 (one-sample t test). (J) Western blot analysis of LC3B coimmunoprecipitation with Rab11a from mouse synaptoneurosomes prepared and treated as in (H). (K) LC3B western blot and signal intensity ratios, calculated as above. The band corresponding to LC3-I was used for quantitation to assess LC3B association with Rab11/Atg9A complex regardless of autophagic flux. n = 3.
Synaptoneurosomes were treated with INK128 (600 nM), NMDA (50 μM), or a combination of both drugs. Treatment with NMDA was used to stimulate NMDA-type glutamate receptors. Instead of washing out the NMDA after 5 min of stimulation, synaptoneurosomes were diluted five times to decrease the concentration of NMDA below levels shown to evoke cLTD (Lee et al., 1998) and then incubated for 25 min. Interestingly, this kind of treatment decreased the level of serine 845 phosphorylated GluA1 (P-GluA1), a subunit of AMPA receptors (Figure 4, C and E), which is considered the biochemical hallmark of cLTD (Kameyama et al., 1998; Lee et al., 1998). While this in simplified model we cannot perform true cLTD, this would suggest that the biochemical pathways active downstream of NMDA receptors are partially stimulated. Figure 4, H–K shows that both Atg9A and LC3B coimmunoprecipitated with Rab11. INK128 alone had no apparent influence on the Rab11/Atg9A interaction in synaptoneurosomes likely due to its weak effect on P-S6 (Ser235/236) in our synaptoneurosome preparations (Figure 4, D, F, and G). NMDA treatment tended to increase Rab11-Atg9A immunoprecipitation, but a significant difference was achieved only when combined with INK128 (Figure 4I), suggesting that Rab11 associates more efficiently with Atg9A at the synapse upon NMDA stimulation. However, no such increase was observed with the LC3-I form of LC3B (Figure 4, J and K). However, coimmunoprecipitation of both LC3-I and its digested LC3-II form with Rab11 hinted at the potential of an LC3+ vesicle forming at the dendritic spine. These findings prompted us to evaluate the emergence and dynamics of LC3-positive vesicles in the dendritic spines of live hippocampal neurons upon conditions shown by others to induce cLTD.
Autophagy is enabled in dendritic spines of rat hippocampal neurons by simultaneous mTOR inhibition and NMDA treatment
We have yet to determine whether the same treatments that increase the Atg9A and Rab11 interaction induce autophagosome formation in dendritic spines. Therefore, we returned to live primary neuronal hippocampal cultures. We counted LC3 puncta as a proxy of autophagosome formation at the dendritic spines as they appeared in real-time after stimulation, without applying any treatments that block autophagosome progression to downstream structures. We transfected cells with a pMX-IP-EGFP-LC3 plasmid, taking advantage of the retroviral long terminal repeat as a weak promoter: therefore, having less potential to overexpress LC3 to the degree at which it forms aggregates. We used the EGFP-tagged version of LC3 despite the recent report of high background in dendrites (Kulkarni et al., 2021) because we have not observed this unspecific staining in spines. We performed confocal spinning-disk imaging and treatments, that is, application of INK128 (mTOR inhibitor) or/and NMDA, proven previously to induce cLTD-like changes in neurons cultured in vitro, for example, AMPA receptors internalization (Lee et al., 2002; Hsin et al., 2010; Li et al., 2010), the next day after transfection. Our treatments triggered neither cell death nor increased autophagy at the whole-cell level (Figure 5A), thus suggesting that the newly formed LC3+ vesicles observed in the dendrites and dendritic spines approximated a physiological reaction. For quantitative measurement of the autophagic response at the postsynapse, new vesicles were counted as one event, regardless of whether they remained at the spine, withdrew to the dendrite, or shuttled for several minutes between the spine and dendrite. New LC3+ vesicles appeared at dendritic spines as soon as 10–15 min upon NMDA application and washout in the presence of INK128 (Figure 5, B and G; Supplemental Movies 6 and 7). New LC3+ vesicles were less common in the other stimulation variants, that is, NMDA or INK128 alone (Figure 5G). We recapitulated the same stimulation conditions and performed western blotting for proteins that strongly increase or are digested upon excitotoxic cell death (casp-3, LC3B, p62/SQSTM1). We did not measure physiological autophagic flux because no inhibitor or autophagosome acidification was used. No indication of toxicity was found. We have shown representative blots out of three biological replicates. A biochemical approximation of cLTD induction by NMDA treatment was demonstrated by decreased levels of phosphorylated GluA1 (P-S845; Kameyama et al., 1998; Lee et al., 1998), while INK128 resulted in a substantial decrease in mTOR activity (F). Our observations revealed that the neurons reacted differently to treatments depending on whether they already had multiple LC3+ puncta at the dendritic spines before the stimulation. To tease out this difference, we subdivided neurons into two groups: neurons with low count of LC3+ vesicles at the start of the experiment (i.e., ≤ six dendritic spines that contained LC3+ puncta, Figure 5H) and neurons with high count of LC3+ vesicles at the start of the experiment (i.e., > six dendritic spines that contained LC3+ puncta; Figure 5I). Altogether, our results revealed that NMDA stimulation induced the emergence of LC3+ autophagosome-like puncta at dendritic spines shortly after application and that a large subpopulation of them reacted when enabling conditions (i.e., low mTOR activity) were met.
FIGURE 5:
Simultaneous mTOR inhibition and NMDA stimulation induces the emergence of LC3+ vesicles at dendritic spines in rat hippocampal cultured neurons. (A) Western blot analysis of levels of caspase 3 (casp 3), LC3B, and p62 in primary hippocampal rat neurons (DIV21) treated with INK128 (300 nM) or DMSO for 15 min and then NMDA (50 µM) for 3 min followed by the medium changed, and incubation with DMSO or INK128 (300 nM) for next 35 min. Tubulin is shown as a loading control. (B) Representative images of a dendrite of a rat hippocampal neuron cotransfected with plasmids encoding EGFP-LC3 and Scarlet-I on DIV22 were treated the next day with INK128 (300 nM) and NMDA (50 µM). The arrow points to a new LC3+ vesicle emerging upon treatment. Scale bar = 2.5 µm. (C–F) Western blot analysis of NMDA application and mTORC1 and mTORC2 inhibition by INK128. Extracts treated as in (A) were blotted against phosphorylated (Ser845; P-GluA1) and total GluA1 to evaluate GluA1 dephosphorylation considered a marker for cLTD, P-S6 (Ser235/236), and S6 to evaluate mTORC1 complex suppression and P-Akt (Ser473) and Akt to evaluate mTORC2 suppression. Tubulin level serves as a loading control. Data are presented as mean gray values for each phosphorylated protein normalized to total protein. n = 7 for all samples. **p ≤ 0.01, ***p ≤ 0,001 (one-sample t test). (G) Neurons that were transfected as in (B) were either untreated or pretreated with INK128 (300 nM) for 15 min and then treated with NMDA for 3 min. The medium was then changed, and the neurons were incubated with or without INK128 (300 nM). Cumulative analysis of EGFP-LC3 vesicles showed that they appeared at dendritic spines at 2-min intervals, starting 6 min after NMDA application (shown as the total number of events normalized to the number of visible spines). Data are presented as a mean of cumulative events normalized to the number of spines for all time points and cells. Error bars show the standard error of the mean. Comparisons refer to the control cells in each time point, respectively. n = 14 control cells. n = 12 INK128-treated cells. n = 10 NMDA-treated cells. n = 11 INK128+NMDA-treated cells. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (two-way ANOVA followed by Dunnett’s post hoc test). (H and I) Cumulative analysis of EGFP-LC3 vesicles of the same neurons as in (G), split into two groups with initial LC3+ puncta number ≤ 6 (H) or > 6 (I). Data are presented as a mean of cumulative events normalized to the number of spines for all time points and cells. Comparisons refer to the control cells in each time point, respectively. Error bars show the standard error of the mean. In (H): n = 11 for control cells, n = 7 for INK128-treated cells, n = 7 for NMDA-treated cells, n = 8 for INK128+NMDA-treated cells. In (I): n = 3 for control cells, n = 5 for INK128-treated cells, n = 3 for NMDA-treated cells, n = 3 for INK128+NMDA treated cells. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (two-way ANOVA followed by Dunnett’s post hoc test).
Movie S6.
Fragment of a dendrite of a hippocampal neuron transfected with plasmid encoding EGFP‐LC3 (green) and Scarlet‐I (magenta) upon INK128+NMDA treatment over 44 min. Arrow points to the emerging EGFP‐LC3‐positive vesicle. EGFP‐LC3 channel is shown in fire LUT.
Movie S7.
Fragment of the same dendrite as in Movie 6, both channels.
EGFP-LC3B-negative spines are more dynamic in response to NMDA treatment
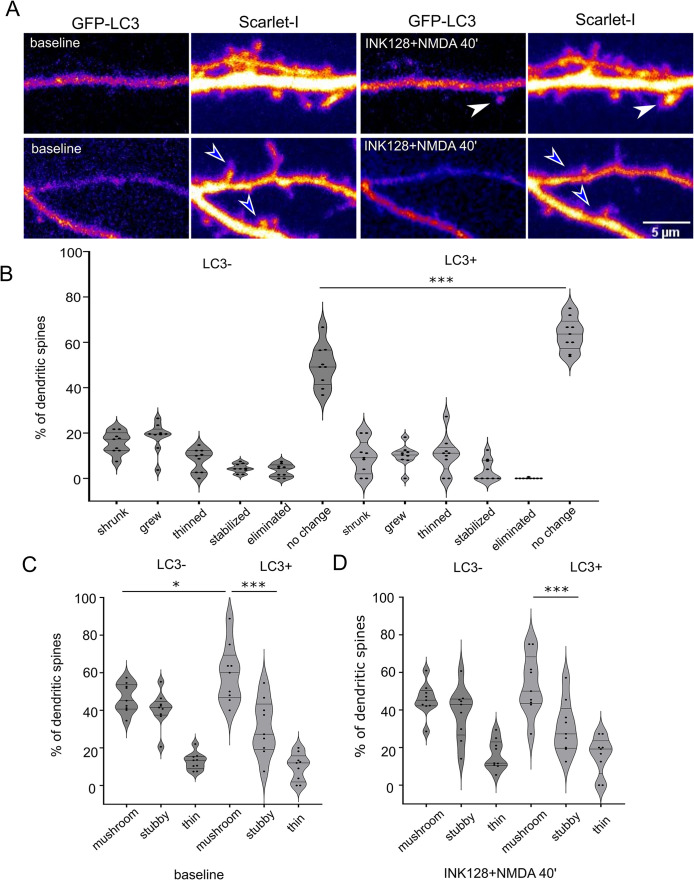
Next, we learned the fate of dendritic spines that formed LC3+ puncta in response to INK128 and NMDA. To this end, we experimented in an analogous way to that above, except that the imaging was prolonged (i.e., to at least 40 min after NMDA washout, which sums to ∼ 46 min after NMDA application) to observe the change in the shape of the dendritic spines (Figure 6A). Analysis of dendritic spine length showed that dendritic spines in which EGFP-LC3 was absent in response to INK128 and NMDA administration changed their shape significantly more often than EGFP-LC3-positive ones (Figure 6B; Supplemental Movies 8, 9, and 10). We have sorted the dendritic spines into three categories: mushroom, stubby, and thin (Figure 6, C and D). Mushroom spines, considered the most stable, mature spines and least prone to plasticity changes (Swanger et al., 2011) were prevalent in the LC3+ group and this did not change after the stimulation. Therefore, we consider that although counterintuitive, the appearance of LC3+ puncta does not immediately lead to drastic spine pruning; on the contrary, it correlates with their increased structural stability.
FIGURE 6:
Dendritic spines that contain EGFP-LC3-positive vesicles after NMDA application are less susceptible to shape change. (A) Representative images of dendrites of a rat hippocampal neuron cotransfected on DIV22 with plasmids encoding GFP-LC3 and Scarlet-I that were treated with INK128 and NMDA the next day. Scale bar = 5 µm. Before treatment, neurons were recorded for 10 min (baseline). Then neurons were pretreated with INK128 (300 nM) for 15 min and next stimulated with NMDA (50 µM) for 3 min (INK128+NMDA 40′). Afterward, media was replaced with conditioned media containing INK128. Neurons were recorded every 2 min for at least 46 min, starting 6 min after NMDA application. Dendritic spines were evaluated for the presence of LC3-positive vesicles throughout the movie. White arrows show examples of dendritic spines where LC3+ puncta appeared. Blue arrowheads show examples of dendritic spines without LC3+ puncta that underwent pruning. (B) Percentage of spines that underwent following changes: grew (over 25% increase in length compared with the baseline and/or change from stubby to mushroom), shrunk (over 25% decrease in length and/or change from mushroom to stubby), thinned (changed type from stubby or mushroom to thin spines), stabilized (thin changed to stubby or mushroom), eliminated (loss of spine confirmed visually over the whole timecourse) or did not change, evaluated for the emergence of LC3 puncta through the time-lapse. Data are presented as percentages of LC3- or LC3+ spines for all cells. n = 9 cells from four independent experiments. One way ANOVA with Sidak’s post hoc test. Columns were compared pairwise in all categories between LC3- and LC3+ spines. There were slightly more spines that grew in LC3- variant than in the LC3+ variant (p = 0.2), which was not shown on the graph for clarity. (***p ≤ 0.001). (C and D) Percentage of spines classified as mushroom, stubby, or thin (see Materials and Methods), measured in the frame in focus minimum 40 min after the NMDA application, for LC3- and LC3+ puncta, respectively. Data are presented as percentages of LC3- or LC3+ spines for all cells. n = 9 cells from four independent experiments One way ANOVA with Sidak post hoc test. All columns were compared with all columns and selected comparisons were presented on the graph. (**p ≤ 0.01, ***p ≤ 0.001).
Movie S8.
Fragment of a dendrite of a hippocampal neuron transfected with plasmid encoding EGFP‐LC3 (green) and Scarlet‐I (magenta) upon INK128+NMDA treatment over 44 min. White arrow points to the emerging EGFP‐LC3‐positive vesicle. Yellow arrow points to LC3+ vesicle exiting the dendritic spine. EGFP‐LC3 channel is shown.
Movie S9.
Fragment of the same dendrite as in Movie 6, Scarlet‐I channel. Except for moving in the medium, neither dendritic spine indicated with the arrow changes shape.
Movie S10.
Fragment of the same dendrite as in Movie 6, both channels.
Rab11a activity is necessary for LC3 positive vesicle emergence at dendritic spines upon simultaneous mTOR inhibition and NMDA treatment
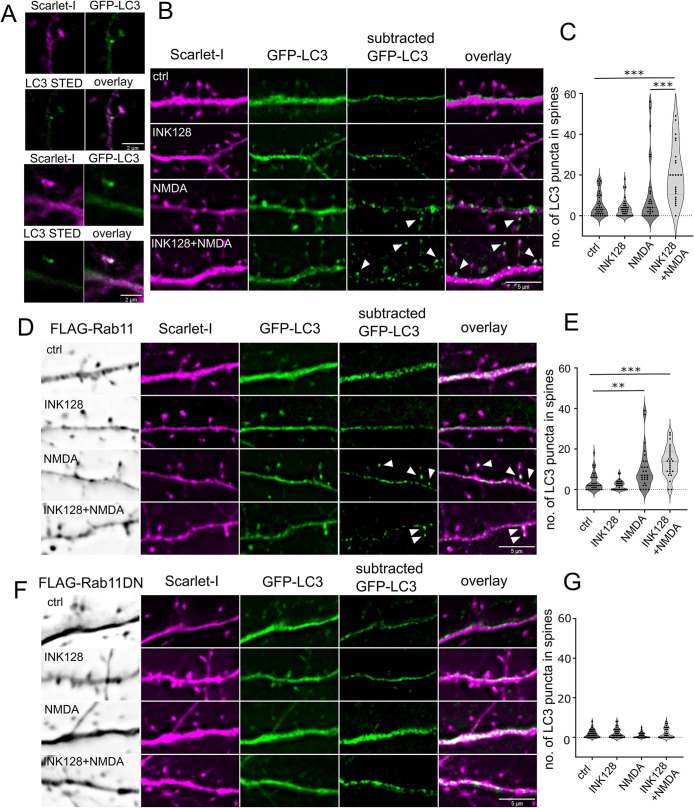
In final series of our experiments, we investigated causative link between Rab11 and local autophagy at the dendritic spines. To this end, we have replicated transfections with EGFP-LC3 and Scarlet-I expressing plasmids with concurrent overexpression of 3xFLAG-Rab11 wild type, dominant negative 3xFLAG-Rab11DN (S25N) or pcDNA to keep the same transfection proportions. This was followed by the same series of treatments as before with the addition of Baf1 A (100 nm), an inhibitor of autophagosome acidification, poststimulation. Cells were fixed after 40 min and anti-FLAG immunostained in far red channel. We learned that the experiment recapitulates our results described earlier in Figure 5. LC3+ puncta number in the dendritic spines was strongly increased in the INK128+ NMDA treatment variant. Interestingly, in the NMDA stimulation variant a small subpopulation of cells reacted as well, but the effect was much stronger upon mTOR inhibition (Figure 7, B and C). LC3+ puncta exhibited in the spines upon stimulation had a round, autophagosome like shape when detected with STED confocal microscope (Figure 7A). When Rab11 wild type was overexpressed, NMDA was sufficient to cause LC3+ puncta appearance at the dendritic spines, although the effect was still trending higher in the case of INK128-NMDA treatment (Figure 7, D and E). When dominant negative Rab11 was overexpressed, it suppressed appearance of LC3+ puncta in all treatment variants (Figure 7, F and G). Thus, we could conclude that Rab11 activity is needed for LC3+ positive vesicle emergence upon stimulation.
FIGURE 7:
Rab11 activity is necessary for LC3+ dendritic spine puncta increase upon NMDA application and mTOR inhibition. (A) Example confocal and STED images (20 nm resolution) of LC3+ puncta in the dendritic spine neck and head, respectively. The same cells were evaluated as in (B) The examples come from the INK128 + NMDA treatment variant. Scale bar = 2 µm. (B and C) 22 DIV neurons were transfected with plasmids expressing EGFP-LC3, Scarlet-I and pcDNA as a supplemental plasmid. Neurons were either untreated or pretreated with INK128 (300 nM) for 15 min and then treated with 20 µM NMDA for 5 min. The medium was then changed, and the neurons were incubated with or without INK128 (300 nM) with the addition of Baf1A (100 nm) as an inhibitor of autophagosome acidification. Following the experiment neurons were fixed and immunostained for FLAG-tag together with Rab11 WT and DN overexpressing neurons. Results present number of spherical/ovoid LC3+ puncta over ∼0.47 µm in diameter in the dendritic spine head or neck per cell in view. Scale bar = 5 µm. N = 4 independent experiments, n for each variant: ctrl = 36, INK128 = 35, NMDA = 29, INK128+NMDA = 26. Outliers (5) were removed with ROUT test with Q = 0.1%. Outlier removal with ROUT test did not change which comparisons were significant, nor p intervals. One way ANOVA with Tukey posttest, all columns were compared with all columns. Comparison of INK128 with INK128 NMDA variant was also significant (p ≤ 0.001) but not shown on the graph for clarity (* p ≤ 0.05, *** p ≤ 0.001). (D and E). Neurons from the same experiments with FLAG-Rab11 overexpression instead of pcDNA. Scale bar = 5 µm. N = 4 independent experiments, n for each variant: ctrl = 34, INK128 = 34, NMDA = 28, INK128+NMDA = 28. Outliers (6) were removed with ROUT test with Q = 0.1%. Outlier removal with ROUT test did not change, which comparisons were significant, nor p intervals. One way ANOVA with Tukey post hoc test, all columns were compared with all columns. Comparisons of INK128 with NMDA and INK128 NMDA variant were also significant (p ≤ 0.001) but not shown on the graph for clarity (**p ≤ 0.01, ***p ≤ 0.001). (F and G). Neurons from the same experiments with FLAG-Rab11 DN overexpression instead of pcDNA. Scale bar = 5 µm. N = 4 independent experiments, n for each variant: ctrl = 36, INK128 = 36, NMDA = 31, INK128+NMDA = 34. Outliers (8) were removed with ROUT test with Q = 0.1%. One way ANOVA with Tukey post hoc test, all columns were compared with all columns. Outlier removal with ROUT test did not change, which comparisons were significant, nor p intervals.
DISCUSSION
Here we investigated the initial steps that lead to the autophagy at the dendritic spines, a morphologically and functionally distinct compartment of the neuron, where, following neuronal stimulation, localized processes responsible for neuroplasticity occur. We believe this is the first evidence to show that Rab11-Atg9A interaction predates local autophagy understood as emergence of LC3 puncta in the dendritic spines and facilitates their appearance upon using a neuronal stimulation paradigm that in other works was shown to evoke cLTD. Taken together our study provides new insight in the spatiotemporal dynamics, mechanism and physiologic regulation of autophagy initiation in dendritic spines.
The present study showed that Rab11 GTPase co-occurs with Atg9A at dendritic spines, and this co-occurrence increases upon mTOR suppression. We identified an Atg9A reservoir at dendritic spines, and we propose that it serves as a site of nucleation for autophagosomes. We argue that Rab11 regulates the maintenance of this reservoir and, through interaction with Atg9A and potentially other autophagy-associated proteins, is necessary for autophagosome emergence at the dendritic spines induced by the neuronal activity. We also propose that autophagosome formation at the postsynapse is a common process that occurs after cLTD and can be enabled by low mTOR kinase activity.
The entrance of Rab11 into dendritic spines in the context of neuroplasticity, namely chemical LTP, was described previously. Recycling endosomes participated in delivering AMPA receptors to the plasma membrane in LTP (Park et al., 2004) and directed dendritic spine growth (Park et al., 2006). Later, Rab11-positive endosomes were shown to recycle AMPA receptors to the postsynapse (Esteves da Silva et al., 2015). We found that the suppression of mTOR activity diminished the movement of Rab11-positive vesicles, but this did not influence their presence at the postsynapse in the short term (i.e., up to 40 min). We also showed that Rab11 colocalized with Atg9A (Figure 2), similar to nonneuronal cells, and confirmed their biochemical association by coimmunoprecipitation (Figure 4). This interplay between Rab11 and Atg9A depends on both mTOR activity and NMDA receptor stimulation. Rab11 and Atg9A colocalization increased within 20 min upon mTOR inhibition in neuronal cultures, while colocalization with other proteins related to endocytic transport and recycling decreased (Figure 2). In synaptoneurosomes, a preparation considered a biochemical model of the synapse, the increase in the Rab11/Atg9A association depended on NMDA application (Figure 4). These findings suggested that Rab11 partially switched from recycling endosomes to immobilized compartments, which are potentially autophagosome nucleation sites, and that stationary Rab11-positive vesicles could participate in autophagy initiation.
The hypothesis that Rab11 acts upstream of autophagy was tested in several studies of nonneuronal cells. Rab11 was required for autophagy in HEK293A cells and contributed to the early steps of autophagosome formation via Atg9A and serine/threonine-protein kinase Ulk1 (Longatti et al., 2012). Recycling endosomes have a role in Atg9A and Atg16L compartment fusion, leading to autophagy (Puri et al., 2013). Also, Atg9A-containing membranes are generated at Rab11 reservoirs in HeLa cells and mouse fibroblasts (Takahashi et al., 2016). Finally, the Rab11a compartment may serve as a platform to assemble other proteins for autophagy machinery (Puri et al., 2018). These studies showed that Rab11 is crucial for autophagy, notably autophagy initiation in mammalian cells. Until completion of this work, the reverse has not been suggested, thus we explored this relationship unidirectionally.
Rab11 might play a dual role in initiating autophagy in dendritic spines as a vehicle for transporting autophagy proteins and as an autophagy platform. We used primary neurons to show that the presence of Atg9A reservoirs at dendritic spines in the long term (i.e., after overnight transfection) depends on Rab11 GTPase activity, suggesting that the postsynaptic Atg9A pool must be regularly replenished by recycling endosomes. Our results are consistent with findings from others in cell lines (Longatti et al., 2012; Puri et al., 2013; Imai et al., 2016).
One issue is whether the Rab11/Atg9A assembly increase at dendritic spines is equivalent to autophagosome formation. We applied the same treatments, mTOR suppression with INK128, and NMDA stimulation individually and together in a live imaging experiment with EGFP-LC3 transfected neurons to resolve this question. We showed that Rab11/Atg9A colocalization increases upon mTOR suppression in cultured neurons. However, INK128 treatment alone did not cause the formation of nascent LC3+ puncta in the dendritic spines of neurons with a low baseline number of autophagosomes at the spines. Thus, we can infer that increased colocalization of Rab11/Atg9A is not equivalent to autophagy. NMDA stimulation alone also did not cause the emergence of LC3+ puncta in the dendritic spines of neurons with a low baseline number of LC3+ vesicles. However, simultaneous mTOR suppression and NMDA stimulation induced the emergence of LC3+ vesicles at the dendritic spines in the case of neurons with low baseline LC3+ puncta count at the spines. We can speculate that when baseline autophagy in cells was already at a higher level, new autophagosomes arrived at spines regardless of which treatment was applied. This observation may explain the differences between our work and research performed by Shehata et al. (2012) or Kallergi et al. (2022), in which cLTD was sufficient for increased autophagosome presence in the dendrites and dendritic spines. In fact, our experiment with fixed neurons showed a similar trend with NMDA stimulation alone and the effect was much stronger with INK 128 pretreatment. Because neurons with high basal LC3+ vesicle count reacted to any treatment in live experiments, it is easy to see how they could obscure the population with low basal LC3+ vesicle count in other studies.
In the present study, we were also able to observe the dynamics and location of the nascent LC3+ puncta. Most new vesicles at the dendritic spines emerged within 10–18 min after NMDA treatment. Using 2-min intervals to prevent bleaching prevented us from discerning which vesicles originated in situ and which ones arrived there. We observed potential incidences of LC3+ vesicles both emerging in situ and “visiting” the spine. However, the synaptoneurosome coimmunoprecipitation experiment supported the hypothesis that autophagosomes can arise at the postsynapse because LC3B is present within the Rab11 complex in the preparation enriched in dendritic spines. This does not exclude the formation of the AP in the dendritic shaft or in the cell body as indicated by other researchers (Shehata et al., 2012; Kallergi et al., 2022). Although we have not focused on Atg9A reservoirs in the dendrites, we speculate that neurons are likely to possess protein processing compartments located in their other parts, including shaft and branching points.
Our study showed that local formation of LC3+ puncta depends on Rab11 activity. Overexpression of Rab11DN variant strongly suppressed emergence of LC3+ puncta at the dendritic spines in all treatment variants. On another note, neurons overexpressing wild type form of Rab11 reacted to NMDA treatment alone, even without INK128 treatment that in our other experiments helped to keep Rab11 vesicles in the spine. This, again, underscores the participation of Rab11 recycling endosomes in local autophagy.
Overall, our results suggest that dendritic spines have the potential to perform cLTD-dependent autophagy on demand. We propose that this process is dependent on Rab11 and mediated by the Atg9A reservoir. Unknown is whether this compartment corresponds to the spine apparatus or other endomembranes. The lower basal levels of mTOR activity may be a prerequisite but are not the cause of local autophagy at postsynaptic sites. We propose a model in which the assembly of mTOR-dependent preautophagy machinery predates and is necessary for the selective autophagosome formation upon neuronal stimulation. Our results suggest that under physiological conditions autophagosomes do not lead to immediate spine pruning, as hypothesized by others (Kallergi et al., 2022). In our study, autophagosomes emerged often in the pool of mushroom spines, and these are not prone to rapid changes in shape (Swanger et al., 2011). Autophagosome function at the postsynapse remains speculative.
Dendritic autophagy removes AMPA receptors, an essential step for LTD (Shehata et al., 2012; Kallergi et al., 2022). Moreover, recent studies reported that cLTD-induced autophagosomes contain many more proteins than AMPAR subunits, including elements vital for neuronal plasticity and spine structure, for example, PSD-95 (Compans et al., 2021; Kallergi et al., 2022). These findings suggest that cLTD-triggered spine content remodeling may be essential to the morphological change of initially stable spines. Kulkarni et al. (2021) showed that dendritic autophagy is also triggered rapidly by neuronal stimulation known to increase synaptic strength and spine growth. Thus, questions arise whether autophagy occurs in proximity to “plastic” less stable spines under such conditions (Kallergi et al., 2022). We might also speculate whether autophagosomes render the spines less reactive and less prone to stimulation-induced changes, while other spines rapidly change shape upon cLTD. These questions, however, remain beyond the scope of this study. Recent advances in microscopy bring new opportunities to investigate complex endomembrane machinery found in the dendrites and at the postsynapse, its interplay, and its influence on neuroplasticity, and there’s a potential for elucidating these gaps in knowledge in the near future.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Primary hippocampal cell cultures, treatments, and transfections
The animals used to obtain neurons for further experiments were sacrificed according to protocols that followed the European Community Council Directive 2010/63/EU. Alternatively, dissected hippocampi were ordered from TransnetYX (former BrainBits) and processed according to the same protocol (Jaworski et al., 2005). Rat embryonic primary hippocampal cell cultures were prepared and transfected using Lipofectamine 2000 (Thermo Fisher Scientific, Invitrogen, catalogue no. 11668019) as described previously (Urbanska et al., 2012; Janusz et al., 2013). Transfection protocol was formulated to accommodate mature neurons. Briefly, freshly made complete neuronal media (Janusz et al., 2013) were added to each well in 1:1 ratio, then half of the cell-conditioned and fresh media mix was saved. Neurons were then incubated with DNA/Lipofectamine 2000 mix prepared as per manufacturer’s instructions in the proportion of 0.9 µg DNA and 1.8 µl Lipofectamine 2000 per well in a 12-well plate, added dropwise. Following 1.5–2 h incubation, neurons were rinsed once with Neurobasal A, then media was replaced with the saved mix of cell-conditioned and fresh complete media. All media were warmed and equilibrated in the cell culture incubator before addition to the cells. DIV21-25 neurons with mature, stabilized dendritic spines were used for all the experiments. INK128 (300 nM; Cayman Chemical, catalogue no. 11811-5) was used for mTOR inhibition. Neurons were stimulated with NMDA (50 µM; Tocris Biosciences, catalogue no. 0114/50) for 3 min followed by immediate washout (Shehata et al., 2012). Alternatively, neurons were stimulated with 20 µM NMDA for 5 min followed by immediate washout and subsequent incubation with bafilomycin A (100 nM, Cell Signaling Technology, catalogue no.54645) (Lee et al., 1998; Shehata et al., 2012; Kallergi et al., 2022) The neurons were then imaged for up to 60 min in live experiments or fixed after 25 min following INK128 application or 40 min following NMDA application.
Antibodies
For Western blot (WB), immunofluorescence (IF), and immunoprecipitation (IP), the following primary antibodies were used: rabbit antiphospho-Akt (Ser473; Cell Signaling Technology, catalogue no. 4060, 1:1000 for WB), mouse anti-Akt (Cell Signaling, catalogue no. 2920, 1:1000 for WB), rabbit anti-Atg9A antibody (Thermo Fisher Scientific, catalogue no. PA5-21043, 1:200 for IF, 1:100 for WB), rabbit anti-caspase 3 antibody (Cell Signaling, catalogue no. 9662; 1:1000 for WB), mouse monoclonal anti-FLAG(R) M2 antibody (Sigma, F1804, 1:200 for IF) rabbit anti-phospho-GluA1 (Ser845; Millipore, catalogue no. 04-1073, 1:500 for WB), mouse anti-GluA1 (Santa Cruz Biotechnology, catalogue no. sc-55509, 1:100 for WB), rabbit anti-Hook3 antibody (Proteintech, catalogue no. 15457-1-AP, 1:100 for IF), rabbit anti-LC3B antibody (Thermo Fisher Scientific, catalogue no. PA1-46286, 1:200 for WB), rabbit anti-LC3B antibody (Sigma Aldrich, catalogue no. L7543, 1:2000 for WB), rabbit anti-SQSTM1/p62 polyclonal antibody (Cell Signaling, catalogue no. 9662; 1:1000 for WB), rabbit anti-syntaxin 12 antibody (Proteintech, catalogue no. 14259-1-AP, 1:100 for IF), mouse anti-Rab11 antibody (BD Biosciences, catalogue no. 610656, Clone 47/Rab11, 1:50 for IF), and rabbit anti-Rab11a antibody (Thermo Fisher Scientific, catalogue no. 71-5300, 5 µg for IP), rabbit anti-phospho ribosomal protein S6 (Ser235/236; Cell Signaling Technology, catalogue no. 4858, 1:1000 for WB), mouse antiribosomal protein S6 (Cell Signaling Technology, catalogue no. 2317, 1:1000 for WB), mouse anti-α-tubulin (Sigma-Aldrich, catalogue no. T5168, 1:5000 for WB). For synaptoneurosome analysis with WB, following primary antibodies were used: synaptophysin, Merck Millipore, catalogue no. MAB329; GluA1, Thermo Scientific, catalogue no. PA1-37776; PSD95, Merck Millipore, catalogue no., MAB1598; Gfap Proteintech, catalogue no. 16825-1-AP; Gapdh Merck Millipore catalogue no. MAB374; β3-Tubulin catalogue no. 5568, Cell Signaling; KDM1/LSD1, Abcam catalogue no. ab129195; c-Jun, Cell Signaling Technology, catalogue no. 9165. Although all were raised against synthetic Rab11a peptide, the mouse monoclonal (used for published results for Figure 2) and rabbit polyclonal from Thermo potentially could also react with Rab11b. In the present study, we did not attempt to resolve this distinction. The following secondary antibodies were used for immunofluorescence and Western blot: antirabbit Alexa Fluor 405 (Thermo Fisher Scientific, catalogue no. A-31556), antirabbit Alexa Fluor 488 (Thermo Fisher Scientific, catalogue no. A-11034), antirabbit Alexa Fluor 647 (Thermo Fisher Scientific, catalogue no. A-31573), and antimouse Alexa Fluor 555 (Thermo Fisher Scientific, catalogue no. A-31570). Peroxidase AffiniPure donkey antirabbit IgG (H+L) (Jackson ImmunoResearch; catalogue no. 711-035-152), peroxidase AffiniPure donkey antimouse IgG (H+L; Jackson ImmunoResearch; catalogue no. 715-035-150), IRDye 680RD donkey anti-Mouse IgG (LiCOR Biosystems catalogue no. 926-68072), IRDye 680RD donkey anti-Rabbit IgG (LiCOR Biosystems catalogue no. 926-68073), IRDye 800CW donkey anti-Mouse IgG (LiCOR Biosystems catalogue no. 926-32212), IRDye 800CW donkey anti-Rabbit IgG (LiCOR Biosystems catalogue no. 926-32213) and VeriBlot for immunoprecipitation detection reagent (HRP; Abcam; catalogue no. ab131366). Secondary antibodies for Western blot were diluted at either 1:5000 (HRP-conjugated) or 1:10000 (IRDye-conjugated).
Plasmids
The following plasmids were used for the transfection of cultured cells: pMXs-IP-EGFP-LC3 (Hara et al., 2008; gift from Noboru Mizushima; Addgene plasmid no. 38195; http://n2t.net/addgene:38195; RRID: Addgene_38195), pMXs-puro-RFP-ATG9A (Koyama-Honda et al., 2013), (gift from Noboru Mizushima; Addgene plasmid no. 60609; http://n2t.net/addgene:60609; RRID: Addgene_60609), piRFP702-N1 (Shcherbakova and Verkhusha, 2013; gift from Vladislav Verkhusha; Addgene plasmid no. 45456; http://n2t.net/addgene:45456; RRID: Addgene_45456), pmScarlet-i_C1 (Bindels et al., 2017; gift from Dorus Gadella; Addgene plasmid no. 85044; http://n2t.net/addgene:85044; RRID: Addgene_85044), mRFP-Rab11a, and GW1-GFP-Rab11a (Esteves da Silva et al., 2015; gift from Casper Hoogenraad). GW1-GFP-Rab11S25N and Rab11S20V were obtained by QuickChange mutagenesis using the following primers: Rab11S25N (5′-GAGATTCTGGTGTTGGAAAGAATAATCTCCTGTCTCGATTTAC-3′ and 5′-GTAAATCGAGACAGGAGATTATTCTTTCCAACACCAGAATCTC-3′), Rab11S20V (5′-ACTACCTCTTTAAAGTTGTCCTTATTGGAGATGTTGGTGTTGGAAAGAGTAAT-3′ and 5′-ATTACTCTTTCCAACACCAACATCTCCAATAAGGACAACTTTAAAGAGGTAGT-3′). Plasmid pmScarlet-i-Atg9A was prepared by the sequence- and ligation-independent cloning (SLIC; Li and Elledge, 2012) of Atg9A cDNA from pMXs-puro-RFP-ATG9A to pmScarlet-i_C1. The plasmid containing the insert was digested with BsrGI, and the backbone was digested with XhoI. The SLIC primers were the following: 5′-GTACAAGTCCGGACTCAGATCTCGAATGGCGCAGTTTGACACTG-3′ and 5′-TGCAGAATTCGAAGCTTGAGCTCGAACGCGTGAATTCGTTCTAGAAAC-3′.
3xFLAG plasmids were prepared courtesy to Jie Jiang’s lab as follows: 3xFLAG-Rab11 (human) and 3xFLAG-Rab11S25N g-block sequences were ordered from IDT company, then cloned into pJ289_GW1-EGFP plasmid by the excision of EGFP and replacement with g-block sequence.
Imaging of live neurons
For the Rab11 assays, neurons were cultured on MatTek multiwell glass-bottom plates (P12G-1.0-14-F), treated with 1 M HCl for 15 min, rinsed three times with phosphate-buffered saline (PBS), rinsed three times with distilled H2O, and coated with poly-D-lysine and laminin (Jaworski et al., 2005). For the autophagy assays, neurons were cultured on 18-mm diameter glass slides in 12-well plates prepared and coated according to routine laboratory protocols (Jaworski et al., 2005).
Imaging occurred using the Andor Revolutions XD spinning disk microscope with 63x lens and 1.6 Optovar (final resolution of 68 nm) or without Optovar in LC3+ experiments (final resolution of 105 nm) at 1004 × 1002-pixels, equipped with a double incubation chamber with an external incubation cage and a heating insert with CO2 delivery. The CO2 valve was opened at least an hour before imaging, and the setup was warmed to 37°C overnight. In the case of MatTek multiwell glass-bottom plates, cells were imaged in the same culture media they were cultured in (Neurobasal/B27 Supplement/Glutamax, all from Thermo Fisher Scientific). In the case of neurons cultured on the coverslips, Chamlide magnetic chamber (Quorum Technologies) was used with 500 µl of the culture media from the same well. All subsequent media changes and washes were conducted using warm culture media from the same plate. The temperature was calibrated in the incubation cage and the sample before the experiments. Both temperature and CO2 flow were monitored in the incubation cage throughout the experiment. Cells were equilibrated for 30 min on the microscope stage and monitored for any changes before imaging.
For the Rab11 mobility assay, neurons were transfected with plasmids encoding RFP and GFP-Rab11a. The next day, time-lapse live imaging occurred using an Andor confocal spinning disk microscope at 300 ms per frame (63x objective, 1.4 NA, Optovar magnification changer 1.6, calibrated pixel size = 68 nm). Imaging began no later than 18 h after transfection. Neurons were recorded for 1 min at the start of the experiment (preincubation), after 30 min, and then for the third time after 20 min after INK128 (300 nM) or mock treatment. All dendritic spines in the field of view were marked with separate regions of interest (ROIs) in ImageJ software. Each spine was analyzed for the presence of Rab11a vesicles. Briefly, vesicles that entered or left the spine or at least reached the base of the spine neck were classified as mobile. Immobile vesicles remained in the spine head and did not travel. Dendritic spines were classified into spines that contained mobile or immobile vesicles. Spines that harbored both types were classified as spines that contained mobile vesicles. The same dendritic spines were analyzed at -30 min (preincubation), 0 min, and 20 min time points.
For the Rab11a and Atg9A assays, neurons were transfected with plasmids encoding either GFP-Rab11a or its mutants (DN GFP-Rab11 and CA GFP-Rab11), supplemented with ScarletI-Atg9A and iRFP702 to visualize dendrites and dendritic spines. The next day, three-channel snapshots were taken at the dendritic spine level. Dendritic spines that were in focus were marked as ROIs and counted. All ROIs were converted into a mask. Masked channels were then subjected to object-based colocalization in ImageJ using the JACoP plugin (Bolte and Cordelières, 2006).
For the LC3+ vesicle emergence assay, neurons were transfected with plasmids encoding EGFP-LC3 and Scarlet-I to visualize the cell body, dendrites, and spines. The next day, cells were preincubated with INK128 (300 nM) for 15 min or kept without treatment for 15 min. NMDA (50 µM) was added for 3 min, then the medium was aspirated and replaced with either conditioned medium or conditioned medium supplemented with INK128 (300 nM). Imaging began 3 min after medium replacement (i.e., 6 min after stimulation). For all four variants (control, INK128 only, NMDA only, and INK128+NMDA), full-stack images (Z-step = 0.6 µm) were taken at 2-min intervals before stimulation 40 min after stimulation or, in the case of the control, 40 min after first imaging. Z-stacks were processed as maximum Z projections using Andor software. For the analysis, neurons with high EGFP-LC3 expression, neurons with aberrant dendritic spine morphology, or neurons that degraded over time were rejected. Neurons were analyzed using ImageJ to determine the number of dendritic spines and new puncta that appeared after stimulation or in the equivalent period.
Neurons were subjected to the INK128 preincubation and subsequent NMDA stimulation in the presence of INK128, as described above, to analyze dendritic spine morphology. Images were collected every 2 min for 10 min before and after the stimulation. Imaging concluded after at least 46 min following the stimulation. The last frame taken before the stimulation (baseline) and the last frame at 46 min after the stimulation were used to analyze dendritic spine length. Spine length was measured with line ROI for each visible spine in the Scarlet-I channel. Dendritic spines before and after stimulation were compared. Those with more than 25% change in length (either by elongation or shrinking) were arbitrarily classified as those that changed shape.
Afterward, the images were analyzed in the EGFP-LC3 channel. During the movie, poststimulation, dendritic spines that did not exhibit any LC3+ puncta were classified as spines with no autophagosomes. During the movie poststimulation, dendritic spines that exhibited LC3+ puncta were classified as spines with autophagosomes. Dendritic spines possessing LC3+ puncta at baseline were not counted.
Afterward, we classified the shape of each spine pre and post stimulation. We used the manual method due to the technical caveats of our equipment. We used Image J to measure length, minimum neck width (N) and maximum head width (H) for each spine. We then sorted the dendritic spines into three categories: mushroom, stubby, and thin. We used the classification used by other authors (Swanger et al., 2011; Kim et al., 2022) as follows:
Mushroom = head to neck ratio ≥ 1.5
Stubby = head to neck ratio ≤ 1.5, length below 2 µm
Thin = head to neck ratio ≤ 1.5, length above 2 µm
As the threshold for dendritic spine length to classify them into thin spines we chose 2 µm due to exaggeration of dendritic spine length in live microscopy settings. The maximum length of a spine was 10 µm for the same reason.
Then, we sorted the spines into following categories poststimulation:
Grew: increase in length over 25% and/or change the category from stubby to mushroom via increase in head width
Shrunk: decrease in length over 25% and/or change from mushroom to stubby via decrease in head width
Thinned: change of mushroom or stubby spines into thin
Stabilized: change of thin spines into mushroom or stubby
Eliminated: was confirmed visually over the whole time course.
Due to the dendritic spines moving in the medium, the best focus frame after frame 17 (34 min post stimulation) or later (until frame 25) was selected, to ensure the change was true and not an effect of spine moving out of focus. Transient “spinules” (very thin, extremely dynamic spine projections) that emerged from the spines for one frame and did not become permanent changes were weakly visible and not accounted for.
We presented the result as a percentage of spine shape change in two categories, LC3+ negative or positive, in each cell.
Immunofluorescence and superresolution Airyscan imaging
Primary rat hippocampal neurons were cultured on glass coverslips in 24-well plates, treated as described above, and fixed for 15 min with 4% paraformaldehyde, followed by 2 min of –20°C methanol postfixation to retrieve membrane epitopes. The glass coverslips were then rinsed in PBS and blocked for 30 min in immunofluorescence (IF) solution 1 (0.1% saponin, 0.2% gelatin, and 5 mg/ml bovine serum albumin (BSA in PBS), followed by a single wash and subsequent incubation in IF solution 2 (0.2% gelatin and 0.01% saponin in PBS) with the primary antibody in a cold room overnight. The coverslips were then rinsed three times in IF solution 2 and incubated in IF solution 2 with a secondary fluorescent antibody. The coverslips were then rinsed three times in PBS and mounted in Prolong Gold antifade reagent (Thermo Fisher Scientific, catalogue no. P10144). The coverslips were imaged on a Zeiss LSM 800 microscope with the Airyscan module (63x objective, 1.4 NA, 1.3⋅ digital magnification) and then processed using the Airyscan module at default settings. The final image resolution was 28 nm/pixel. For colocalization, the GFP channel was masked to analyze only areas that contained dendritic spines of all shapes, including neck and head spines. Spines within ∼ 10 µm to the cell soma were omitted. Object-based colocalization was then performed based on centers of mass (Rab11 channel) – particles coincidence using the JACoP plugin (Bolte and Cordelières, 2006) and two focal planes that contained dendritic spines that were in focus out of five focal planes that encompassed the whole neuron.
Immunofluorescence, widefield, and STED confocal imaging
Primary rat hippocampal neurons were cultured on 18-mm glass coverslips in 12-well plates, transfected with plasmids encoding EGFP-LC3 and Scarlet-I to visualize the cell body, dendrites, and spines, and either pcDNA, or plasmids endocing 3xFLAG-Rab11 or 3xFLAG-Rab11DN mutant, then treated as in live experiments, with the exception of NMDA stimulation at 20 µM for 5 min and poststimulation incubation with Baf1A (100 nM, Cell Signaling Technology, catalogue no.54645), then fixed 40 min post stimulation for 15 min with 4% paraformaldehyde. The glass coverslips were then rinsed in PBS and blocked for 45 min in blocking buffer (5% Normal Donkey Serum, 0.1% BSA, 0.1% Triton-X in PBS), then stained with the primary antibody diluted with blocking buffer overnight in the fridge. The coverslips were then rinsed three times in PBS and incubated with the secondary antibody for 90 min before three final washes with PBS and embedding in ProLong Glass Antifade Mountant (Thermo Fisher Scientific, catalogue no. P36980).
The coverslips were imaged on Nikon Eclipse TiE Inverted Microscope with AURA III Light Engine (Lumencor) and the usage of following fluorescent cubes: "C-FL GFP, Hard Coat, High Signal-to-Noise, Zero Shift, Excitation: 470/40 nm(450–490 nm), Emission: 525/50 nm (500–550 nm), Dichroic Mirror: 495 nm, C-FL DS Red, Hard Coat, High Signal-to-Noise, Zero Shift, Excitation: 545/30 nm (530–560 nm), Emission: 620/60 nm (590–630 nm), Dichroic Mirror: 570 nm, C-FL CY5, Hard Coat, High Signal-to-Noise, Zero Shift, Excitation: 620/60 nm (590–650 nm), Emission: 700/75 nm (663–738 nm), Dichroic Mirror: 660 nm.” CFI60 Apochromat TIRF 60X Oil Immersion Objective Lens N.A. 1.49 was used. Stacks of seven focal planes with 0.2 µm interval were collected. Final pixel resolution was 108 nm.
The aforementioned experiment with 3xFLAGRab11 and 3xFLAG-Rab11DN overexpression was blinded at image acquisition and unblinded at analysis. Collected images were then deconvolved with AutoQuant software and processed in Image J as follows: First, to account for fluorescence intensity discrepancies, deconvolved 32-bit image stacks were converted to 16 bit and subjected to histogram matching (Image → Adjust > Bleach correction → Histogram matching). Then, Scarlet-I channel was subtracted from EGFP-LC3 channel (Process → Image Calculator → Subtract). Finally, the subtracted channel was Z-projected as sum of slices (Image → Stackes → Z project) and analyzed for distinct spherical/ovoid puncta with diameter ∼0.47 µm or more that overlapped with the dendritic spines. Alternatively, Z-projection was performed first after the removal of out-of-focus planes, and then the same procedure followed. Because fluorescence intensity was changed by this method, the analysis was purely object based. Manual counting was applied.
For confirmation that the acquired LC3+ puncta were consistent with LC3+ vesicles, coverslips from the GFP-LC3+ transfected neurons from INK128+NMDA stimulation variant were then imaged with Abberior STED Facility Line confocal microscope at Emory University Integrated Cellular Imaging Core. Laser lines used were 485 and 561 for excitation, then subsequently 595 for pulsed STED depletion. The objective used was UPLXAPO60XO, 60x, NA 1.42, oil and the final image resolution was 20 nm. Images were collected with 15x line averaging.
Synaptoneurosome isolation, treatment, and protein coimmunoprecipitation
Cortical and hippocampal synaptoneurosomes from the brain of adult mice were prepared according to our previously published protocol (Janusz et al., 2013; Jasińska et al., 2016; Kuzniewska et al., 2017, 2020). Briefly, Krebs buffer referred to as synaptoneurosome buffer (2.5 mM CaCl2, 1.18 mM KH2PO4, 118.5 mM NaCl, 24.9 mM NaHCO3, 1.18 mM MgSO4, 3.8 mM MgCl2, and 212.7 mM glucose) was supplemented with air for 30 min at 4°C. Next, pH was calibrated at 7.4 with dry ice. Buffer was supplemented with Protease Inhibitor Cocktail (Sigma-Aldrich) and 100 U/ml mammalian placental RNase inhibitor (Fermentas). Hippocampi and cortices were isolated from the brains of mice killed by cervical dislocation. One hemisphere (∼50 mg) was homogenized in 1.5 ml Krebs buffer using Dounce homogenizer with 10–12 strokes. The whole procedure was performed on ice, in cold room. Homogenates were passed through a series of presoaked with synaptoneurosome buffer nylon mesh filters consecutively 100, 60, 30, and 10 μm (Merck Millipore) in cold room to 50-ml polypropylene tube, centrifuged at 1000 × g for 15 min at 4°C, and washed. Final pellets were diluted in 500 µl synaptoneurosome buffer per brain, pooled, and divided into 250 µl aliquots. Fresh synaptoneurosomes were then incubated at 37°C for 15 min with INK128 (600 nM) or without and then stimulated for 5 min with NMDA (50 µM). After 5 min, all the variants were diluted five times with a preheated synaptoneurosome buffer to reduce the concentration of NMDA under the threshold known to induce cLTD (Lee et al., 1998) with or without INK128 (600 nM) and incubated for 25 min. After this time, samples were either lysed for WB analysis of GluA1 and S6 phosphorylation level or were centrifuged at 1000 × g for 15 min, resuspended in immunoprecipitation buffer (10 mM HEPES, 1.5 mM MgCl2, 2 mM EDTA, 0.6% CHAPS, 120 mM NaCl, and protease inhibitors [benzamidine, aprotinin, Pefabloc; Sigma-Aldrich]), and allowed to incubate on ice for 30–60 min. Extracts were precleared with Dynabeads G (Invitrogen) and precipitated with 5 µg anti-Rab11 antibody or total rabbit IgG. Finally, the beads were washed three to five times with immunoprecipitation buffer, and protein was eluted with Laemmli buffer at 98°C for 10 min. Protein samples were separated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE), followed by Western blot (WB) analysis.
Western blot
SDS–PAGE occurred in gradient gels (12/15%, 1 mm thick) to analyze IP results. The gels were run at 120 V for ∼ 1.5 h until the dye reached the bottom of the gel. Then, the gels were wet transferred at 400 mA for 90 min to methanol-activated 0.2 µm pore size PVDF membrane (Bio-Rad). Blots were blocked in 5% nonfat milk in Tris-buffered saline (0.5 mM Tris-HCl [pH 7.5] and 1.5 mM NaCl) with 0.1% Tween-20 (TBST) for 1 h and then incubated overnight in a cold room in primary antibody diluted in 5% milk in TBST. Following incubation, they were washed three times for 10 min each in TBST and then incubated for 1.5–2 h in HRP-conjugated secondary antibody diluted in 2.5% milk in TBST. Finally, blots were developed for ∼2–3 min in a freshly made chemiluminescence developing buffer (100 mM Tris [pH 8.5], 1.25 mM 3-aminoftalhydraside, 200 μM coumaric acid, and 0.01% hydrogen peroxide). Signals were detected on radiographic film after digital processing, and band intensities were measured as mean gray values using Image J software. Mean gray values of Atg9A and LC3 were normalized to mean gray values of IgG as an external loading control and then to the control (untreated) immunoprecipitation sample. Mean gray values of every other sample are presented as ratios. Quantitative analysis of S6, Akt, and GluA1 phosphorylation levels was performed using the Infrared Odyssey Imaging System (LI-COR Biosciences) described previously (Urbanska et al., 2018, 2019).
Alternatively, for synaptoneurosome preparation analysis, equal amounts of proteins from homogenate and synaptoneurosomal fraction were resolved on SDS–PAGE (10%, TGX Stain-Free FastCast Acrylamide Solutions, BioRad). Next, proteins were transferred to PVDF membranes (pore size 0.45 µm, Immobilon-P, Merck Millipore) using Trans-Blot Turbo Blotting System (BioRad; 170-4155). Membranes were blocked for 1 h at room temperature in 5% nonfat dry milk in PBS-T (PBS with 0.01% Tween-20), followed by overnight incubation at 4°C with primary antibodies in 5% milk in PBS-T. Blots were washed three times for 5 min with PBS-T and incubated for 1 h at room temperature with HRP-conjugated secondary antibody (1:10,000 in 5% milk). Next, the membranes were washed three times for 5 min with PBS-T and HRP signal was detected using Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare) on Amersham Imager 600 using automatic detection settings.
Statistical analysis
The number of cells and culture batches analyzed, and a description of statistical tests used are provided in the figure legends. All the analyses employed GraphPad Prism 8–10 software.
Acknowledgments
We thank Bartosz Tarkowski for help with the SLIC primer design, Alina Zielinska and Kinga Kuchcinska for technical support, and Michael Arends for proofreading the manuscript. We also thank Magdalena Dziembowska for her comments on the manuscript. The Polish National Science Centre Opus supported this work with grants nos. 2012/07/E/NZ3/00503 and 2017/27/B/NZ3/01358 to J.J.. A.B. was partly supported by a Polish National Science Centre Opus grant no. 2016/21/B/NZ3/03639 to J.J.. A.T. was partly financed by a Polish National Science Centre Opus grant no. 2017/25/N/NZ3/01280. J.J., M.U., and J.Z. are partly or wholly financed by the TEAM grant from the Foundation for Polish Science (POIR.04.04.00-00-5CBE/17-00). We thank members of the Bassell lab for technical support and helpful discussions. We thank Jie Jiang and the members of his lab for cloning for us the 3xFLAG-Rab11 plasmids constructs. Research by A.J.K. done at Emory University was supported by National Institutes of Health 1R01MH109026 (G.J.B.).
Abbreviations used:
- ANOVA
analysis of variance
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- AP
autophagosomes
- Atg
autophagy-related protein
- CA
constitutively active
- cLTD
chemical long-term depression
- DIV
day in vitro
- DN
dominant-negative
- EGFP
enhanced green fluorescent protein
- GTPase
guanosine triphosphatase
- HEK293A
human embryonic kidney 293A
- HRP
horseradish peroxidase
- LC3
microtubule-associated protein 1A/1B-light chain 3
- mTOR
mammalian/mechanistic target of rapamycin
- mTORC1
mTOR complex 1
- NMDA
N-methyl- d-aspartate
- PBS
phosphate-buffered saline
- Rab11
Ras-related protein Rab-11A
- RFP
red fluorescent protein
- ROI
region of interest
- SDS–PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- WB
Western blot.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www .molbiolcell.org/cgi/doi/10.1091/mbc.E23-02-0060) on January 31, 2024.
REFERENCES
- Aspernig H, Heimbucher T, Qi W, Gangurde D, Curic S, Yan Y, Donner von Gromoff E, Baumeister R, Thien A (2019). Mitochondrial Perturbations Couple mTORC2 to Autophagy in C. elegans. Cell Reports 29, 1399–1409.e5. [DOI] [PubMed] [Google Scholar]
- Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, Gadella TWJ (2017). mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods 14, 53–56. [DOI] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA (2008). Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci 28, 6926–6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelières FP (2006). A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224, 213–232. [DOI] [PubMed] [Google Scholar]
- Borczyk M, Śliwińska MA, Caly A, Bernas T, Radwanska K (2019). Neuronal plasticity affects correlation between the size of dendritic spine and its postsynaptic density. Sci Rep 9, 1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Hayashi Y (2012). Structural plasticity of dendritic spines. Curr Opin Neurobiol 22, 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compans B, Camus C, Kallergi E, Sposini S, Martineau M, Butler C, Kechkar A, Klaassen RV, Retailleau N, Sejnowski TJ, et al. (2021). NMDAR-dependent long-term depression is associated with increased short-term plasticity through autophagy-mediated loss of PSD-95. Nat Commun 12, 2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowska M, Milek J, Janusz A, Rejmak E, Romanowska E, Gorkiewicz T, Tiron A, Bramham CR, Kaczmarek L (2012). Activity-dependent local translation of matrix metalloproteinase-9. J. Neurosci , 32, 14538–14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves da Silva M, Adrian M, Schätzle P, Lipka J, Watanabe T, Cho S, Futai K, Wierenga CJ, Kapitein LC, Hoogenraad CC (2015). Positioning of AMPA Receptor-Containing Endosomes Regulates Synapse Architecture. Cell Rep 13, 933–943. [DOI] [PubMed] [Google Scholar]
- Ganley IG, Lam DH, Wang J, Ding X, Chen S, Jiang X (2009). ULK1·ATG13·FIP200 Complex Mediates mTOR Signaling and Is Essential for Autophagy. J Biol Chem 284, 12297–12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Firestein BL, Zheng JQ (2008). Microtubules in Dendritic Spine Development. J Neurosci 28, 12120–12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S-I, Natsume T, Guan J-L, Mizushima N (2008). FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181, 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth EB, McNeal ET, Burton JL, Williams RJ, Daly JW, Creveling CR (1985). Biochemical characterization of a filtered synaptoneurosome preparation from guinea pig cerebral cortex: cyclic adenosine 3’:5’-monophosphate-generating systems, receptors, and enzymes. J Neurosci 5, 2240–53. J.Neurosci. 05-08-02240.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad CC, Popa I, Futai K, Sanchez-Martinez E, Wulf PS, van Vlijmen T, Dortland BR, Oorschot V, Govers R, Monti M, et al. (2010). Neuron Specific Rab4 Effector GRASP-1 Coordinates Membrane Specialization and Maturation of Recycling Endosomes. PLOS Biology 8, e1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin H, Kim MJ, Wang C-F, Sheng M (2010). Proline-rich tyrosine kinase 2 regulates hippocampal long-term depression. J Neurosci 30, 11983–11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Viesselmann C, Nam S, Merriam E, Dent EW (2008). Activity-dependent dynamic microtubule invasion of dendritic spines. J Neurosci 28, 13094–13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunyady L, Baukal AJ, Gáborik Z, Olivares-Reyes JA, Bor M, Szaszák M, Lodge R, Catt KJ, Balla T (2002). Differential PI 3-kinase dependence of early and late phases of recycling of the internalized AT1 angiotensin receptor. J Cell Biol 157, 1211–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Hao F, Fujita N, Tsuji Y, Oe Y, Araki Y, Hamasaki M, Noda T, Yoshimori T (2016). Atg9A trafficking through the recycling endosomes is required for autophagosome formation. J Cell Sci 129, 3781–3791. [DOI] [PubMed] [Google Scholar]
- Ivankovic D, Drew J, Lesept F, White IJ, López Doménech G, Tooze SA, Kittler JT (2020). Axonal autophagosome maturation defect through failure of ATG9A sorting underpins pathology in AP-4 deficiency syndrome. Autophagy, 16, 391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janusz A, Milek J, Perycz M, Pacini L, Bagni C, Kaczmarek L, Dziembowska M (2013). The Fragile X mental retardation protein regulates matrix metalloproteinase 9 mRNA at synapses. J Neurosci 33, 18234–18241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasińska M, Miłek J, Cymerman IA, Łęski S, Kaczmarek L, Dziembowska M (2016). miR-132 Regulates Dendritic Spine Structure by Direct Targeting of Matrix Metalloproteinase 9 mRNA. Mol Neurobiol, 53, 4701–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M (2005). Control of dendritic arborization by the phosphoinositide-3’-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci 25, 11300–11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Kapitein LC, Gouveia SM, Dortland BR, Wulf PS, Grigoriev I, Camera P, Spangler SA, Di Stefano P, Demmers J, et al. (2009). Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron 61, 85–100. [DOI] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro S-H, Kim Y-M, Otto NM, Cao J, Kundu M, Kim D-H (2009). ULK-Atg13-FIP200 Complexes Mediate mTOR Signaling to the Autophagy Machinery. Mol Biol Cell 20, 1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallergi E, Daskalaki A-D, Kolaxi A, Camus C, Ioannou E, Mercaldo V, Haberkant P, Stein F, Sidiropoulou K, Dalezios Y, et al. (2022). Dendritic autophagy degrades postsynaptic proteins and is required for long-term synaptic depression in mice. Nat Commun 13, 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameyama K, Lee HK, Bear MF, Huganir RL (1998). Involvement of a postsynaptic protein kinase A substrate in the expression of homosynaptic long-term depression. Neuron 21, 1163–1175. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Woo Y, Kim HJ, Goo BS, Nhung TTM, Lee SA, Suh BK, Mun DJ, Kim JH, Park SK (2022). Retinoic acid-induced protein 14 controls dendritic spine dynamics associated with depressive-like behaviors. eLife, 11, e77755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama-Honda I, Itakura E, Fujiwara TK, Mizushima N (2013). Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy 9, 1491–9. [DOI] [PubMed] [Google Scholar]
- Kulkarni VV, Anand A, Herr JB, Miranda C, Vogel MC, Maday S (2021). Synaptic activity controls autophagic vacuole motility and function in dendrites. J Cell Biol 220, e202002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzniewska B, Chojnacka M, Milek J, Dziembowska M (2017). Preparation of polysomal fractions from mouse brain synaptoneurosomes and analysis of polysomal-bound mRNAs. J Neurosci Methods, 293, 226–233. [DOI] [PubMed] [Google Scholar]
- Kuzniewska B, Cysewski D, Wasilewski M, Sakowska P, Milek J, Kulinski TM, Winiarski M, Kozielewicz P, Knapska E, Dadlez M, et al. (2020). Mitochondrial protein biogenesis in the synapse is supported by local translation. EMBO reports, 21, e48882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai K-O, Ip NY (2013). Structural plasticity of dendritic spines: The underlying mechanisms and its dysregulation in brain disorders. Biochim Biophys Acta 1832, 2257–2263. [DOI] [PubMed] [Google Scholar]
- Lee HK, Kameyama K, Huganir RL, Bear MF (1998). NMDA induces long-term synaptic depression and dephosphorylation of the GluR1 subunit of AMPA receptors in hippocampus. Neuron 21, 1151–1162. [DOI] [PubMed] [Google Scholar]
- Lee SH, Liu L, Wang YT, Sheng M (2002). Clathrin adaptor AP2 and NSF interact with overlapping sites of GluR2 and play distinct roles in AMPA receptor trafficking and hippocampal LTD. Neuron 36, 661–674. [DOI] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ (2004). Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6, 463–477. [DOI] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ (2012). SLIC: a method for sequence- and ligation-independent cloning. Methods Mol Biol 852, 51–59. [DOI] [PubMed] [Google Scholar]
- Li R, Dozmorov M, Hellberg F, Tian Y, Jilderos B, Wigström H (2004). Characterization of NMDA induced depression in rat hippocampus: involvement of AMPA and NMDA receptors. Neuroscience Letters 357, 87–90. [DOI] [PubMed] [Google Scholar]
- Li Z, Jo J, Jia J-M, Lo S-C, Whitcomb DJ, Jiao S, Cho K, Sheng M (2010). Caspase-3 activation via mitochondria is required for long-term depression and AMPA receptor internalization. Cell 141, 859–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA (2012). TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol 197, 659–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Holzbaur ELF (2016). Compartment-specific regulation of autophagy in primary neurons. J Neurosci 36, 5933–5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Wallace KE, Holzbaur ELF (2012). Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol 196, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC, Bear MF (2004). LTP and LTD: An Embarrassment of Riches. Neuron 44, 5–21. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, et al. (2007). FoxO3 Controls Autophagy in Skeletal Muscle In Vivo. Cell Metabolism 6, 458–471. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y (2004). In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15, 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoletopulou V, Sidiropoulou K, Kallergi E, Dalezios Y, Tavernarakis N (2017). Modulation of Autophagy by BDNF Underlies Synaptic Plasticity. Cell Metab 26, 230–242.e5. [DOI] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y (1998). Tor, a Phosphatidylinositol Kinase Homologue, Controls Autophagy in Yeast. J Biol Chem 273, 3963–3966. [DOI] [PubMed] [Google Scholar]
- Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, Tooze SA (2012). Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell 23, 1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Penick EC, Edwards JG, Kauer JA, Ehlers MD (2004). Recycling endosomes supply AMPA receptors for LTP. Science 305, 1972–1975. [DOI] [PubMed] [Google Scholar]
- Park M, Salgado JM, Ostroff L, Helton TD, Robinson CG, Harris KM, Ehlers MD (2006). Plasticity-induced growth of dendritic spines by exocytic trafficking from recycling endosomes. Neuron, 52, 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualato S, Senic-Matuglia F, Renault L, Goud B, Salamero J, Cherfils J (2004). The Structural GDP/GTP Cycle of Rab11 Reveals a Novel Interface Involved in the Dynamics of Recycling Endosomes. J Biol Chem 279, 11480–11488. [DOI] [PubMed] [Google Scholar]
- Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC (2013). Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 154, 1285–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri C, Vicinanza M, Ashkenazi A, Gratian MJ, Zhang Q, Bento CF, Renna M, Menzies FM, Rubinsztein DC (2018). The RAB11A-Positive Compartment Is a Primary Platform for Autophagosome Assembly Mediated by WIPI2 Recognition of PI3P-RAB11A. Dev Cell 45, 114–131.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renna M, Bento CF, Fleming A, Menzies FM, Siddiqi FH, Ravikumar B, Puri C, Garcia-Arencibia M, Sadiq O, Corrochano S, et al. (2013). IGF-1 receptor antagonism inhibits autophagy. Hum Mol Genet 22, 4528–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder CM, Vale RD (2016). Assembly and activation of dynein-dynactin by the cargo adaptor protein Hook3. J Cell Biol 214, 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova DM, Verkhusha VV (2013). Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat Methods 10, 751–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M, Matsumura H, Okubo-Suzuki R, Ohkawa N, Inokuchi K (2012). Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. J Neurosci 32, 10413–10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavoe AKH, Holzbaur ELF (2019). Autophagy in Neurons. Annu Rev Cell Dev Biol 35, 477–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanger SA, Yao X, Gross C, Bassell GJ (2011). Automated 4D analysis of dendritic spine morphology: applications to stimulus-induced spine remodeling and pharmacological rescue in a disease model. Mol Brain, 4, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Tsotakos N, Liu Y, Young MM, Serfass J, Tang Z, Abraham T, Wang H-G (2016). The Bif-1-Dynamin 2 membrane fission machinery regulates Atg9-containing vesicle generation at the Rab11-positive reservoirs. Oncotarget 7, 20855–20868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanska M, Gozdz A, Macias M, Cymerman IA, Liszewska E, Kondratiuk I, Devijver H, Lechat B, Van Leuven F, Jaworski J (2018). GSK3β Controls mTOR and Prosurvival Signaling in Neurons. Mol Neurobiol 55, 6050–6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanska M, Gozdz A, Swiech LJ, Jaworski J (2012). Mammalian target of rapamycin complex 1 (mTORC1) and 2 (mTORC2) control the dendritic arbor morphology of hippocampal neurons. J Biol Chem 287, 30240–30256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanska M, Kazmierska-Grebowska P, Kowalczyk T, Caban B, Nader K, Pijet B, Kalita K, Gozdz A, Devijver H, Lechat B, et al. (2019). GSK3β activity alleviates epileptogenesis and limits GluA1 phosphorylation. EBioMedicine 39, 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villasana LE, Klann E, Tejada-Simon MV (2006). Rapid isolation of synaptoneurosomes and postsynaptic densities from adult mouse hippocampus. J Neurosci Methods 158, 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz T, Wellbourne-Wood J, Kerkhoff E (2014). Orchestration of cell surface proteins by Rab11. Trends Cell Biol 24, 407–415. [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ (2010). Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 22, 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Pascual C, Klionsky D (2016). Autophagy: machinery and regulation. Microb Cell 3, 588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]