Abstract
Fatty acids stored in triacylglycerol-rich lipid droplets are assembled with a surface monolayer composed primarily of phosphatidylcholine (PC). Fatty acids stimulate PC synthesis by translocating CTP:phosphocholine cytidylyltransferase (CCT) α to the inner nuclear membrane, nuclear lipid droplets (nLD) and lipid associated promyelocytic leukemia (PML) structures (LAPS). Huh7 cells were used to identify how CCTα translocation onto these nuclear structures are regulated by fatty acids and phosphorylation of its serine-rich P-domain. Oleate treatment of Huh7 cells increased nLDs and LAPS that became progressively enriched in CCTα. In cells expressing the phosphatidic acid phosphatase Lipin1α or 1β, the expanded pool of nLDs and LAPS had a proportional increase in associated CCTα. In contrast, palmitate induced few nLDs and LAPS and inhibited the oleate-dependent translocation of CCTα without affecting total nLDs. Phospho-memetic or phospho-null mutations in the P-domain revealed that a 70% phosphorylation threshold, rather than site-specific phosphorylation, regulated CCTα association with nLDs and LAPS. In vitro candidate kinase and inhibitor studies in Huh7 cells identified cyclin-dependent kinase (CDK) 1 and 2 as putative P-domain kinases. In conclusion, CCTα translocation onto nLDs and LAPS is dependent on available surface area and fatty acid composition, as well as threshold phosphorylation of the P-domain potentially involving CDKs.
PC synthesis is stimulated by CCTα translocation to the nuclear envelope and nuclear LDs. How enzyme association with these two compartments is spatially and temporally regulated is poorly understood.
Oleate stimulated the rapid dephosphorylation and association of CCTα with the nuclear envelope, followed by gradual recruitment to nuclear LDs in proportion to droplet expansion driven by Lipin1.
CCTα association with nuclear LDs was inhibited by palmitate and, 70% phosphorylation of its serine-rich domain. These data show that transition of CCTα from the nuclear envelope to nuclear LDs is coupled to Lipin1 activity and dephosphorylation to a critical threshold.
INTRODUCTION
Excess fatty acids and cholesterol are esterified to triacylglycerol (TAG) and steryl esters and stored in cytoplasmic lipid droplets (cLD) that are emulsified and stabilized by a surface monolayer of phospholipids and associated proteins. The principal functions of cLDs are to store and release fatty acids and sterols in response to the changing nutrient and growth demands of a cell, and to protect against lipid metabolic stress (Walther and Farese, 2012). cLD biogenesis is initiated by the polymerization of seipin and lipid droplet assembly factor 1 (LDAF1) on endoplasmic reticulum (ER) and the subsequent assembly of a metabolic complex of lipid biosynthetic enzymes and regulatory factors that promote cLD maturation and budding (Yan et al., 2018; Chung et al., 2019; Kim et al., 2022). ER-resident and cytosolic proteins that associate with the surface of LDs control the function of LDs with respect to nutrient provision, steroidogenesis, and innate immunity (Kory et al., 2016; Shen et al., 2016; Henne et al., 2018; Bosch et al., 2021; Song et al., 2022).
Recent studies have established the presence of nuclear lipid droplets (nLDs) that are structurally related but functionally distinct from their cytoplasmic counterparts (Fujimoto, 2022; McPhee et al., 2022). High resolution imaging identified nLDs in human and rat liver nuclei (Layerenza et al., 2013; Uzbekov and Roingeard, 2013), and later in the budding yeast Saccharomyces cerevisiae (Romanauska and Kohler, 2018), Drosophila melanogaster (Jacquemyn et al., 2021), and Caenorhabditis elegans (Mosquera et al., 2021). The biogenesis of nLDs occurs by at least two mechanisms. In hepatoma cells, nLDs originate from very low-density lipoprotein (VLDL) precursors in the ER lumen that migrate into and rupture the type I nucleoplasmic reticulum (NR) at sites that are enriched in isoform II of the promyelocytic leukemia (PML) protein but depleted of lamina (Soltysik et al., 2019). The second mechanism, described in yeast and U2OS cells, involves de novo biogenesis of nLDs at the inner nuclear membrane (INM) where many of the enzymes required for TAG synthesis are localized, including glycerol-3-phosphate acyltransferase (GPAT) 3 and 4, 1-acyl-glycerol-3-phosphate acyltransferase (AGPAT) 1, Lipin-1, and diacylglycerol acyltransferase (DGAT) 1 and 2 (Lee et al., 2020; Soltysik et al., 2021). Interestingly, nLD formation on the INM of U2OS cells does not require seipin (Soltysik et al., 2021). Consistent with a de novo origin at the INM, nLDs also contain the TAG and phospholipid precursors diacylglycerol (DAG) and phosphatidic acid (PA; Lee et al., 2020; Soltysik et al., 2021). nLDs shrink in size when cells are deprived of exogenous fatty acids (Soltysik et al., 2021) but the relevant lipases have not been identified.
While nLDs are widespread in nature, they are restricted to specific cell types, are fewer in number than cLDs and are often observed under specific genetic and nutritional conditions. Thus, nLDs are unlikely to be involved in energy homeostasis but could participate in nuclear responses to lipid metabolic stress. Clues to nLD function comes from their association with the nuclear proteins PML and CTP:phosphocholine cytidylyltransferase (CCT) α. PML forms the structural scaffold of PML nuclear bodies (PML NBs), which recruit and regulate the activity of >150 client proteins that mediate responses to DNA damage, oxidative stress, apoptosis, and transcription (Dellaire et al., 2006; Corpet et al., 2020). In Huh7 cells, the PMLII isoform is required for assembly of 40–50% of total nLDs (Ohsaki et al., 2016). Knockout of the PML gene in U2OS cells resulted in a similar reduction in total nLDs indicating the presence of a unique subset of lipid-associated PML structures (LAPS; Lee et al., 2020). Moreover, LAPS in U2OS cells are depleted of canonical proteins found in PML NBs, such as death domain associated protein (DAXX), small ubiquitin-like modifier (SUMO) and SP100. Thus, the accretion of nLDs shifts PML to LAPS at the expense of PML NBs, potentially inhibiting PML NB functions but generating a novel structure that ostensibly is involved in the nuclear response to lipid overload and toxicity.
Approximately 30% of nLDs and LAPS in U2OS cells contain CCTα, which catalyzes the rate-limiting and regulated step in PC synthesis (Lee et al., 2020). In Huh7 cells, CCTα is activated on nLDs and LAPS resulting in increased PC synthesis to reduce ER stress, and enhanced storage of TAG and secretion in VLDL (Ohsaki et al., 2016; Soltysik et al., 2019). The loss of LAPS in PML-knockout U2OS cells caused reduced CCTα association with the residual nLD pool and a 40% reduction in PC synthesis (Lee et al., 2020). These data indicate that nLDs and LAPS ameliorate the effects of fatty acid overload and ER stress by acting as platforms for the recruitment of CCTα to activate PC synthesis by the CDP-choline pathway. The association of CCTα with nLDs and LAPS is dependent on its membrane-binding M-domain, which is inhibited by phosphorylation of the adjacent serine-rich P-domain (Lee et al., 2020). However, it is unknown if this regulatory mechanism involves global or site-specific changes in P-domain phosphorylation or applies to other metabolic conditions under which nLDs and LAPS are formed. The results presented here show that the monounsaturated fatty acid oleate is the preferred substrate for nLD and LAPS formation and CCTα activation, which is enhanced by Lipin1 expression but opposed by global increases in P-domain phosphorylation by cyclin dependent kinases (CDK).
RESULTS
Lipin1 expression increases CCTα-positive nLDs
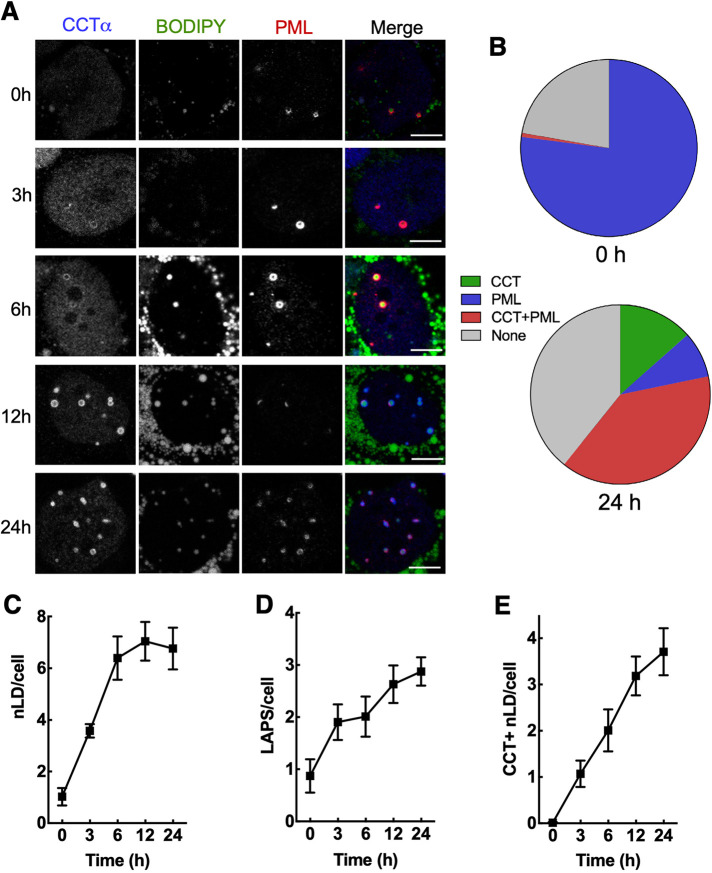
PC synthesis is increased in oleate-treated U2OS and Huh7 cells as a result of recruitment and activation of CCTα on nLDs and LAPS (Ohsaki et al., 2016; Lee et al., 2020). How CCTα recruitment is regulated and the relative contribution of nLDs and LAPS is unknown. To address this, we used confocal immunofluorescence to investigate the temporal sequence of CCTα recruitment to BODIPY-positive nLDs, and LAPS in Huh7 cells treated with oleate for up to 24 h (Figure 1). In this study, nLDs refer to the total pool of BODIPY-positive nLDs, and LAPS defines the portion that is PML-positive. Untreated Huh7 cells contained a small number of nLDs (∼1 per cell), the majority of which (∼80%) were LAPS devoid of CCTα (Figure 1, A and B). Treatment with oleate for 24 h increased total nLDs and LAPS, with the latter containing the majority of CCTα (Figure 1, A and B). Quantitation of immunofluorescence images over the 24 h oleate-treatment period (Figure 1A) revealed that total nLDs per cell increased up to 6 h before plateauing (Figure 1C). However, the number of LAPS (Figure 1D) and CCTα-positive nLDs (Figure 1E) increased continually for 24 h. At 24 h, 60% of nLDs were CCTα-positive, with the majority being CCTα-positive LAPS (Figure 1B). The proportion of nLDs that were devoid of both PML and CCTα increased to ∼30% by 24 h (Figure 1B). These data show that nLDs and LAPS are omnipresent in Huh7 but only after oleate treatment does CCTα translocate to preexisting or newly made nLDs and LAPS.
FIGURE 1:
Temporal association of CCTα with nLDs and LAPS in oleate-treated Huh7 cells. (A) Huh7 cells were treated with oleate (0.4 mM) for the indicated times and immunostained with antibodies against CCTα and PML. LDs were visualized with BODIPY 493/503 (bar, 5 μm). (B) mean percent distribution of CCTα and PML on nLDs at 0 and 24 h. (C–E) quantification of total BODIPY-positive nLDs (panel C), LAPS (panel D) and CCTα-positive nLDs (panel E). Results are the mean and SEM of 9–15 confocal images (10–20 cells per field) from three independent experiments.
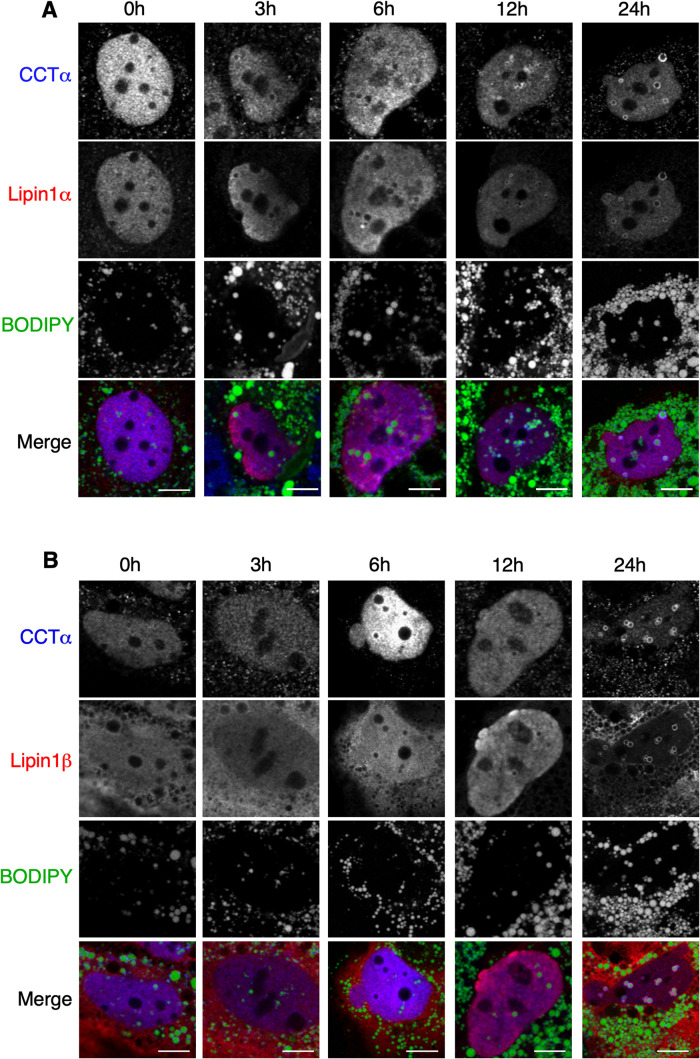
The PA phosphatase Lipin1α and 1β isoforms associate with nLDs and LAPS in response to oleate treatment (Lee et al., 2020) and when nuclear import is stimulated by mTORC1 inhibition in U2OS cells (Soltysik et al., 2021). Lipin1 expression in U2OS cells increases nLD formation by providing the DAG precursor for TAG synthesis (Soltysik et al., 2021). To test whether Lipin affects CCTα association with nLDs and LAPS, Huh7 cells transiently expressing V5-tagged Lipin1α or 1β were treated with oleate for 24 h. Representative confocal images of Huh7 cell nuclei show that Lipin isoforms did not appear on nLDs until 6–12 h after oleate addition (Figure 2, A and B). Endogenous CCTα also appeared on nLDs at 6–12 h and extensively colocalized with Lipin1α and 1β by 24 h. Lipin1α and 1β did not associate with cLDs despite partial cytosolic localization of both isoforms. Quantitation of Lipin1 localization during oleate treatment revealed that the 1α isoform was predominately nuclear and its distribution between the nucleus and cytoplasm did not change significantly with oleate treatment (Figure 3A). Lipin1β was predominantly cytoplasmic but there was an appreciable shift to the nucleus after 24-h oleate treatment. Consistent with its prominent nuclear localization (Peterfy et al., 2005; Figure 3A), Lipin1α appeared on nLDs more rapidly but by 24 h both isoforms populated a similar number of nLDs (∼6 per cell, Figure 3B). Compared to nonexpressing cells in the same field, Huh7 cells expressing Lipin1α had a significant twofold increase in nLDs after 6 h of oleate exposure (Figure 3C). A significant increase in nLDs in cells expressing Lipin1β cells was not observed until 24 h. In parallel with the increase in nLDs, CCTα-positive nLDs were also significantly increased twofold in cells expressing Lipin1α or 1β at 12 and 24 h (Figure 3D), evidence that recruitment of CCTα to nLDs reflects the increase in nLD biogenesis afforded by Lipin1 expression.
FIGURE 2:
Confocal immunofluorescence imaging of Lipin-1α and -1β. Huh7 cells transiently expressing Lipin1α (panel A) or 1β (panel B) were treated with oleate (0.4 mM) for the indicated times and immunostained with antibodies against V5 and CCTα. LDs were visualized with BODIPY 493/503 (bar, 5 μm).
FIGURE 3:
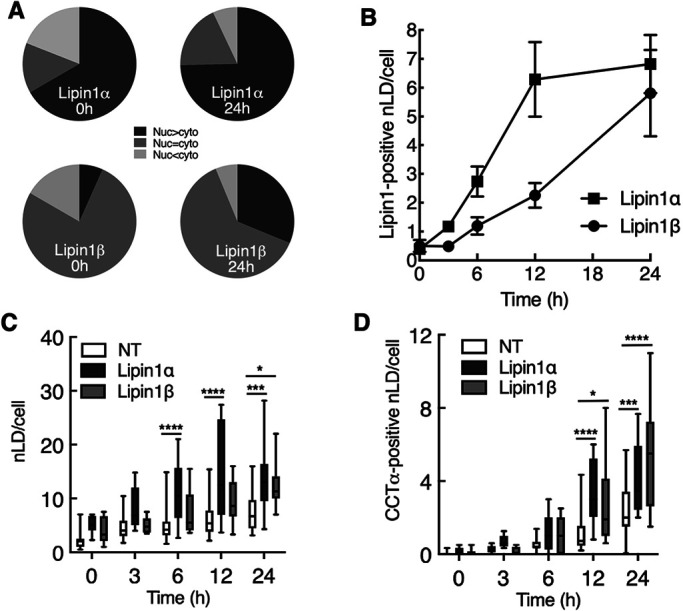
Lipin1 overexpression in Huh7 cells increases nLD and LAPS with associated CCTα. Confocal immunofluorescence images shown in Figure 2 were used for quantification. (A) percentage distribution of Lipin1α and 1β following oleate treatment for 0 and 24 h. Lipin1 was classified as more nuclear than cytoplasmic (Nuc > cyto), equally partitioned (Nuc = cyto) or more cytoplasmic (Nuc < cyto). (B) time-course for association of Lipin1α and 1β with nLDs. (C and D) total nLDs (panel C) and CCTα-positive nLDs were quantified in cells expressing Lipin1α and 1β, and in nontransfected (NT) cells in the same fields. Results in panels B–D were derived from 3–5 confocal images per time point (10–20 transfected cells per image) from 2–3 independent experiments. Panels C and D are presented as box and whisker plots showing the mean and 5–95th percentile. Statistical differences between NT, Lipin-1α, and -1β cells was determined using two-way ANOVA and Tukey's multiple comparison; *p < 0.01 ***p < 0.001 ****p < 0.0001.
Palmitate inhibits CCTα translocation to nLDs and LAPS
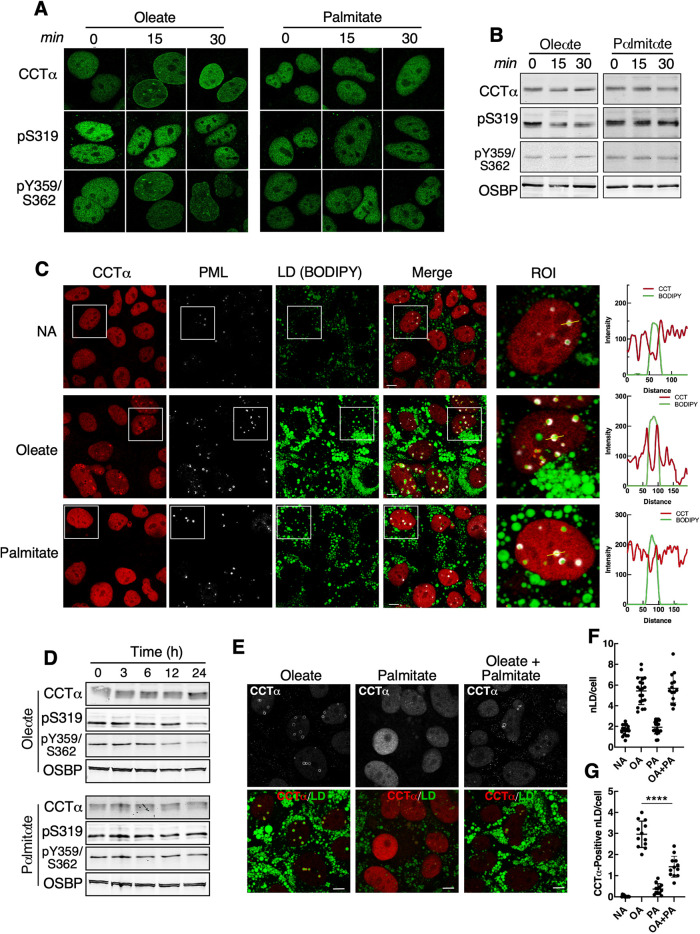
The biogenesis of nLDs has been studied almost exclusively in the context of the monounsaturated fatty acid oleate, which is efficiently stored in LDs. Palmitate is poorly incorporated into LDs (Listenberger et al., 2003; Leamy et al., 2016), and induces lipotoxicity and the ER unfolded protein response by increasing the saturation of membrane lipids (Borradaile et al., 2006; Kitai et al., 2013; Hirsova et al., 2016). To determine whether palmitate activates CCTα, its translocation to the nuclear envelope (NE), nLDs and LAPS was monitored in Huh7 cells exposed to equal concentrations of oleate or palmitate complexed with bovine serum albumin (BSA). Treatment with oleate for 15 or 30 min caused CCTα translocation to the NE as well as nucleoplasmic puncta (Figure 4A). CCTα-pY359/S362 was also detected on the NE after oleate treatment but CCTα-pS319 remained in the nucleoplasm. This is consistent with partial dephosphorylation of S319 but not Y359/S362 following 15- and 30-min exposure to oleate (Figure 4B). In contrast, palmitate did not induce CCTα translocation to the NE in Huh7 cells, or effect the localization or phosphorylation of CCTα-pS319 or -pY359/S362 (Figure 4, A and B). Next, Huh7 cells were treated with oleate or palmitate for 24 h, and the presence of CCTα on LAPS and nLDs was visualized by immunofluorescence confocal microscopy (Figure 4C). Oleate-treated Huh7 cells had numerous LAPS and nLDs coated in CCTα (Figure 4C, and associated line-plots). Comparatively, palmitate-treatment did not increase nLDs and LAPS, which were devoid of CCTα (see quantitation in Figure 4, F and G). Similarly, palmitate did not promote the translocation of Lipin1α or 1β to LAPS or nLDs (Supplemental Figure 1), which was strongly induced by the same concentration of oleate (Figures 2 and 3). CCTα translocation to nLDs and LAPS in oleate-treated cells was accompanied by the partial dephosphorylation at S319 and S359/Y362 after 12 h (Figure 4D). Dephosphorylation of S319 and S359/Y362 was not observed in palmitate-treated cells (Figure 4D), consistent with the lack of translocation to LAPS and nLDs shown in Figure 4C. Compared to cells treated with oleate (0.4 mM), the addition of palmitate (0.1 mM) did not affect total nLDs (Figure 4, E and F) but suppressed the translocation of CCTα to nLDs by >50% (Figure 4, E and G). We conclude that palmitate poorly induces nLD and LAPS formation and antagonizes oleate-induced recruitment of CCTα to nLDs or LAPS.
FIGURE 4:
LAPS and nLDs formed by palmitate treatment do not recruit CCTα. (A) Huh7 cells treated with oleate (0.4 mM) or palmitate (0.4 mM) for 0, 15, and 30 min were immunostained with antibodies against CCTα, CCTα-pS319, and CCTα-pY359/pS362. (B) immunoblot of lysates from cells treated as described in Panel A. (C) Huh7 cells treated with no addition (NA), oleate (0.4 mM), or palmitate (0.4 mM) for 24 h were immunostained for CCTα and PML, and LDs were visualized with BODIPY493/503 (bar, 5 μm). RGB line plots for CCTα and BODIPY are for selected LAPs (yellow line) in the region of interest (ROI) panel. (D) immunoblot analysis of CCTα phosphorylation in Huh7 cells treated with oleate or palmitate for up to 24 h. (E) immunostaining of Huh7 cells that were treated with oleate (0.4 mM), palmitate (0.4 mM) or oleate plus palmitate (0.3 and 0.1 mM, respectively) for 24 h (bar, 5 μm). (F) quantification of nLDs in cells treated with no addition (NA), oleate (0.4 mM, OA), palmitate (0.4 mM, PA), oleate and palmitate (0.4 and 0.1 mM) for 24 h. (G) quantification of CCTα-positive nLDs as described in panel F. Results in panels F and G are from three experiments that quantified 12–16 images (12–16 cells/field). Significance was determined using a Students t test; ****p < 0.0001.
Threshold phosphorylation of the CCTα P-domain controls translocation to LAPS and nLDs
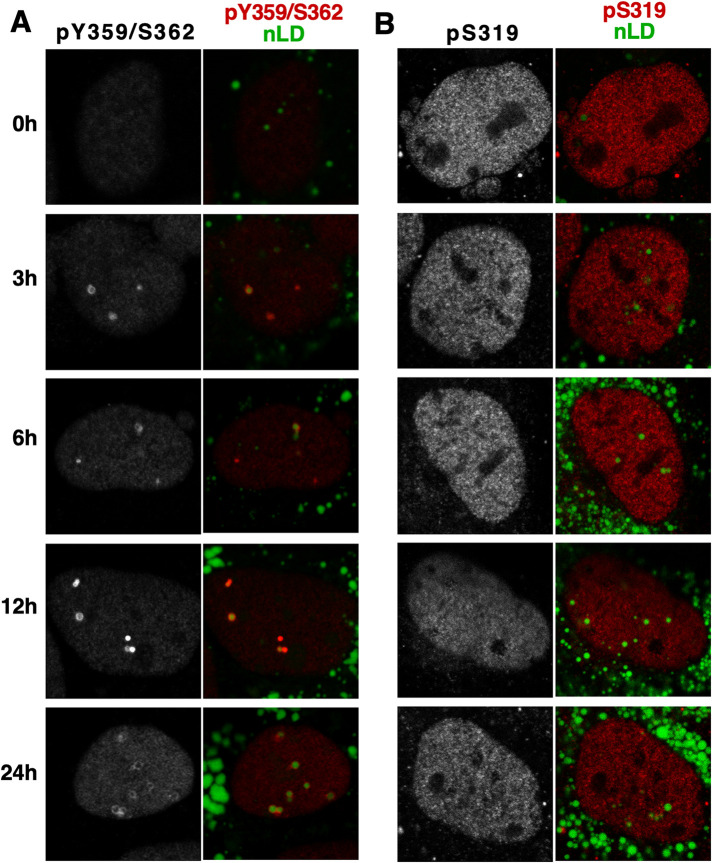
Activation of CCTα on nLDs and LAPS by insertion of the amphipathic M-domain into the surface monolayer is inhibited by phosphomimetic mutation of 16 serine residues in the P-domain or enhanced by deletion of the P-domain (Lee et al., 2020). In U2OS and Caco2 cells, CCTα S319 was dephosphorylated upon nLD association, while the phosphorylation of the C-terminal Y359/S362 site was unaffected (Yue et al., 2020). Huh7 cells treated with oleate for 24 h showed a progressive increase in CCTα-pY359/pS362-positive nLDs (Figure 5A), but CCTα-pS319 was nucleoplasmic and decreased slightly at 24 h (Figure 5B). Immunoblotting of Huh7 lysates from an oleate time course showed that CCTα-pS319 and -pY359/pS362 were reduced by 24 h (Figure 4D).
FIGURE 5:
CCTα phosphorylated on S319 and Y359/S362 differentially associates with nLDs. (A and B) Huh7 cells were treated with oleate for the indicated times and immunostained with antibodies against the CCTα-pY359/pS362 (panel A) and CCTα-pS319 (panel B). LDs were visualized with BODIPY493/503.
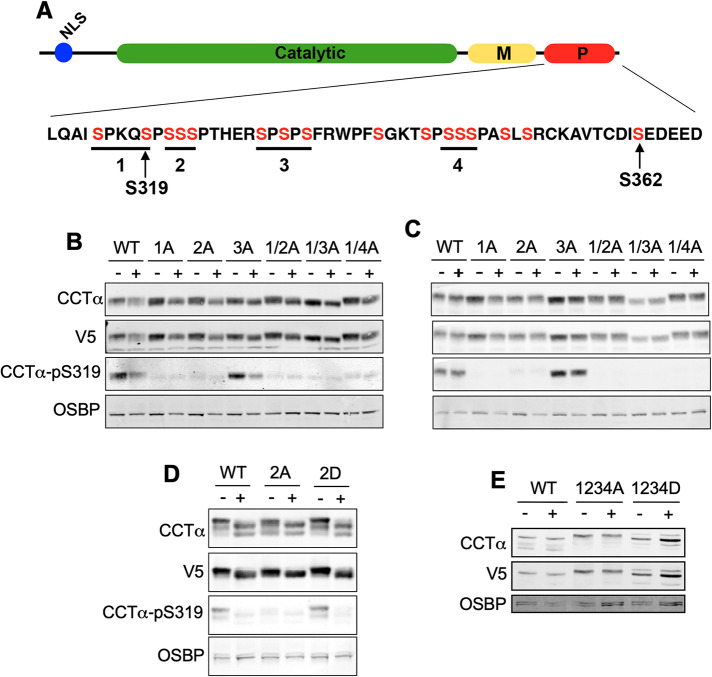
Results with Huh7 (Figure 5) and other cells (Yue et al., 2020) showed that P-domain sites are differentially dephosphorylated during or after association with the NE and nLDs. However, systematic mutagenesis of the P-domain has not been undertaken to determine whether bulk or site-specific phosphorylation regulates CCTα localization to nLDs and LAPS. To address this question, four separate tracts of two to three serine residues in V5-tagged rat CCTα were mutated individually or in combination to alanine or aspartate residues, resulting in a series of mutants in which from two to 11 serine residues were converted to a dephospho- or phosphomimetic state (Figure 6A). U2OS cells were used to analyze the CCTα phosphomutants because they express relatively less endogenous CCTα (Supplemental Figure 2A) and form nLDs and LAPS. Temperature-sensitive CHO-MT58 cells (Esko et al., 1981) were used to monitor the expression of CCTα phosphosite mutants because endogenous CCTα was undetectable by immunoblotting at the nonpermissive temperature (Supplemental Figure 2B). Immunoblots of lysates from U2OS (Figure 6, B, D, and E) and CHO-MT58 cells (Figure 6C) using a V5 antibody indicated similar expression levels for the transfected CCTα phosphosite mutants. Treatment of U2OS cells expressing wild-type, single or double serine-to-alanine mutants with oleate for 24 h caused a slight shift to lower mass species indicative of dephosphorylation (Figure 6B). Immunoblotting for CCTα-pS319 showed that this site was dephosphorylated in oleate-treated U2OS cells expressing wild-type CCTα and, as expected, was absent in mutants in which this was mutated to alanine (Figure 6B). pS319 was not dephosphorylated in oleate-treated CHO-MT58 cells that do not form nLDs (Figure 6C).
FIGURE 6:
Analysis of CCTα P-domain phosphomutants reveals hierarchical phosphorylation of S319. (A) Domain structure of CCTα showing the serine residues (in red) in tracts 1–4 of the P-domain that were mutated singly or in combination to alanine (1A–4A) or aspartate (1D–4D). (B) U2OS cells transiently expressing wild-type (WT) or CCTα with the indicated serine-to-alanine mutations were treated with (+) or without (-) oleate for 24 h. Total cell lysates were immunoblotted with antibodies against V5, CCTα, CCTα-pS319, and OSBP (load control). (C) CHO-MT58 cells were transiently transfected, treated with oleate and cell lysates were immunoblotted as described in Panel B. (D) Lysates from CHO-MT58 cells expressing WT, CCTα-2A or CCTα-2D and treated with or without oleate for 24 h were immunoblotted as described in Panel B. (E) lysates from CHO-MT58 cells expressing WT, CCTα-1234A, or -1234D were immunoblotted for CCTα and V5. Results in panels B–E are representative experiments that were repeated at least two other times.
Unexpectedly, the CCTα-pS319 signal was absent in U2OS and CHO-MT58 cells expressing CCTα-2A (Figure 6, B and C), a site adjacent to S319 that does not overlap with the pS319 antibody epitope. Phosphorylation of S319 was not affected by the CCTα-3A mutation (Figure 7, B and C). To determine whether S319 phosphorylation in CCTα-2A was inhibited due to a lack of negative charge at that site, the S319 phosphorylation status of CCTα-2D was determined in control and oleate-treated U2OS cells (Figure 6D). Unlike CCTα-2A, S319 was phosphorylated in CCTα-2D in untreated cells and dephosphorylated in cells treated with oleate. This result indicates that S319 phosphorylation requires prior phosphorylation at S321-323, further evidence in support of hierarchical phosphorylation of the P-domain (Cornell et al., 1995).
FIGURE 7:
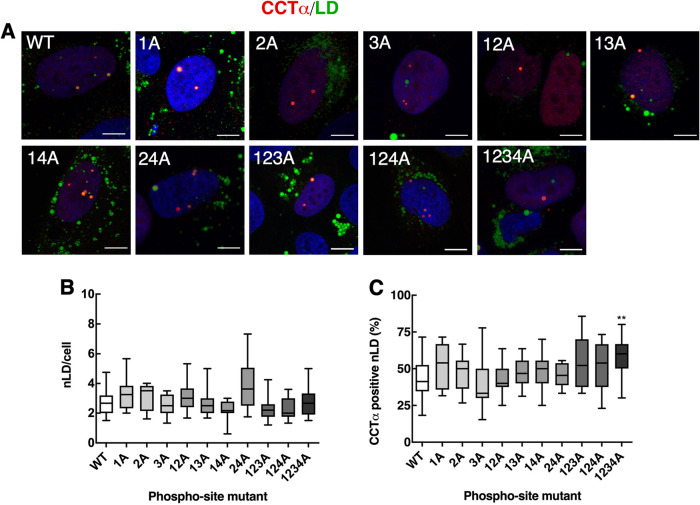
CCTα association with nLDs is enhanced by P-domain serine-to-alanine mutations. (A) U2OS cells transiently expressing wild-type (WT) and CCTα serine-to-alanine mutants were treated with oleate for 24 h. Cells were immunostained with antibodies against V5 (CCT) (red), and LDs were visualized with BODIPY493/503 (green) and the nuclei with propidium iodide (blue) (bar, 5 μm). (B) quantitation of total nLDs in cells expressing WT and CCTα mutants. (C) percentage of CCTα-V5-positive nLDs. Quantitation in panels B and C used 15 confocal images (∼ 15–20 cells per field) from three independent experiments. Results are presented as box and whisker plots showing the mean and 5–95th percentile. Significance was determined using one-way ANOVA and Tukey's multiple comparison versus wild-type. **p < 0.01.
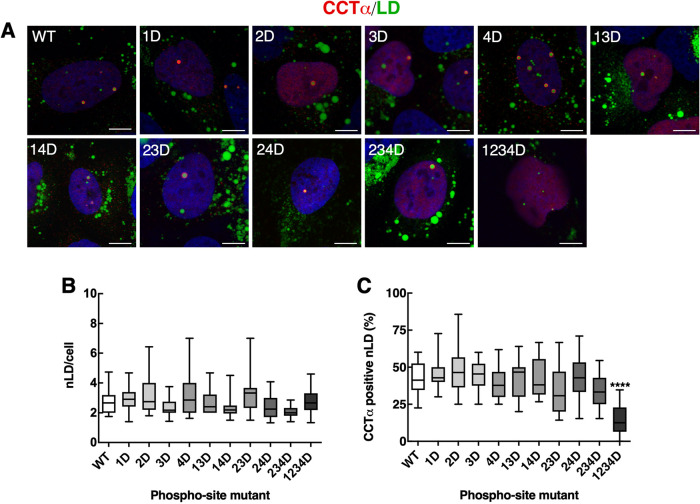
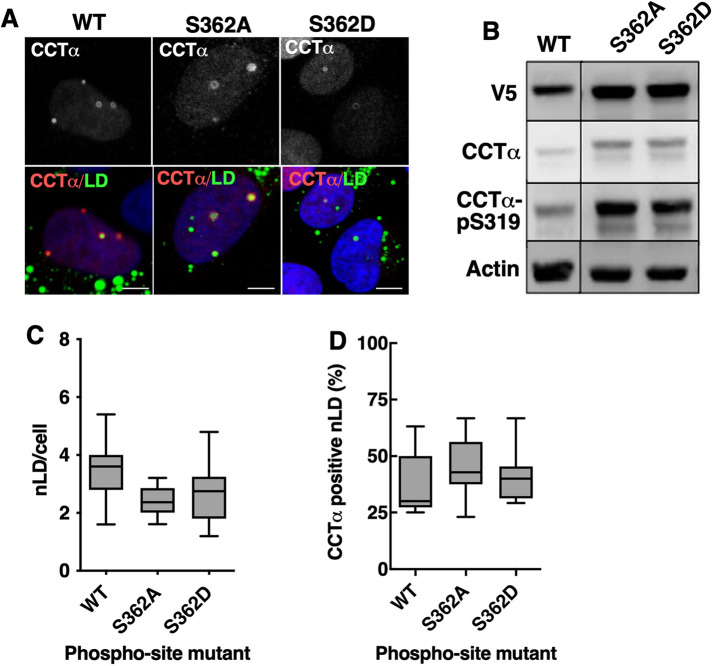
Next, we used immunofluorescence confocal microscopy to determine whether combinations of CCTα serine-to-alanine or -aspartate mutations affected localization to nLDs in U2OS cells treated with oleate for 24 h. Because the level of overexpression of CCTα-V5 mutants was six- to eightfold over endogenous CCTα (Supplemental Figure 2A), results would not be impacted by dimerization with the endogenous enzyme. Mutation of all 11 targeted CCTα serine-to-alanine sites had no clear effect on the total number of nLDs compared with wild-type (Figure 7, A and B). CCTα association with nLDs was also unaffected by single, double, or triple serine-to-alanine mutations (Figure 7C). However, when all four tracts (11 serine residues) were mutated in CCTα-1234A was there a significant 25% increase in association with nLDs (Figure 7C), indicating that reducing the bulk negative charge of the P-domain enhances CCTα association. The opposite trend was observed when the same experiments were repeated using CCTα serine-to-aspartate mutants; the total number of nLDs per cell was unaffected by expression of any mutant (Figure 8, A and B), and only CCTα-1234D had a significantly reduced association with nLDs (Figure 8, A and C). We also determined whether S362A and S362D mutations at the C-terminal end of the P-domain (Figure 6A) affected the association of CCTα with nLDs in U2OS cells. Both mutants were phosphorylated on S319 (Figure 9B). Quantification of confocal images from oleate-treated U2OS cells indicated that neither S362 mutation affected total nLDs (Figure 9, A and C) or association of CCTα with nLDs (Figure 9, A and B). Collectively these data show that CCTα association with nLDs is enhanced or inhibited by a threshold phosphorylation mechanism that is triggered when 11/16 serine residues have a neutral or negative charge, respectively.
FIGURE 8:
CCTα association with nLDs is inhibited by P-domain negative charge density. (A) U2OS cells transiently expressing wild-type (WT) or CCTα aspartate mutants were treated with oleate for 24 h and immunostained with antibodies against V5 (CCT) (red), LDs were visualized with BODIPY 493/503 (green) and the nuclei with propidium iodide (blue) (bar, 5 μm). (B) quantification of the total nLDs in cells expressing WT and CCTα mutants. (C) percentage of CCTα-V5-positive nLDs. Quantification in panels B and C used 15 confocal images (∼ 15–20 cells) from three independent experiments. Results are presented as box and whisker plots showing the mean and 5–95th percentile. Significance was determined using one-way ANOVA and Tukey's multiple comparison versus wild-type. ****p < 0.0001.
FIGURE 9:
CCTα-S362 phosphomutations do not affect association with nLDs. (A) U2OS cells transiently expressing wild-type (WT) or V5-tagged CCTα-S362A or -S362D mutants were treated with oleate for 24 h. Cells were immunostained with antibodies against V5 (CCT), LDs were visualized with BODIPY 493/503 and the nuclei with propidium iodide (blue) (bar, 5 μm). (B) lysates from U2OS cells transiently expressing CCTα -S362A or -S362D and treated with oleate for 24 h were immunoblotted with antibodies against V5, CCTα, and CCTα-pS319. (C) total nLDs in cells expressing the CCTα mutants were quantified. (D) percentage of CCTα-positive nLDs. Results in Panels C and D were quantified from three independent experiments and expressed as box and whisker plots showing mean and 5–95th percentile.
Phosphorylation of the P-domain by CDK
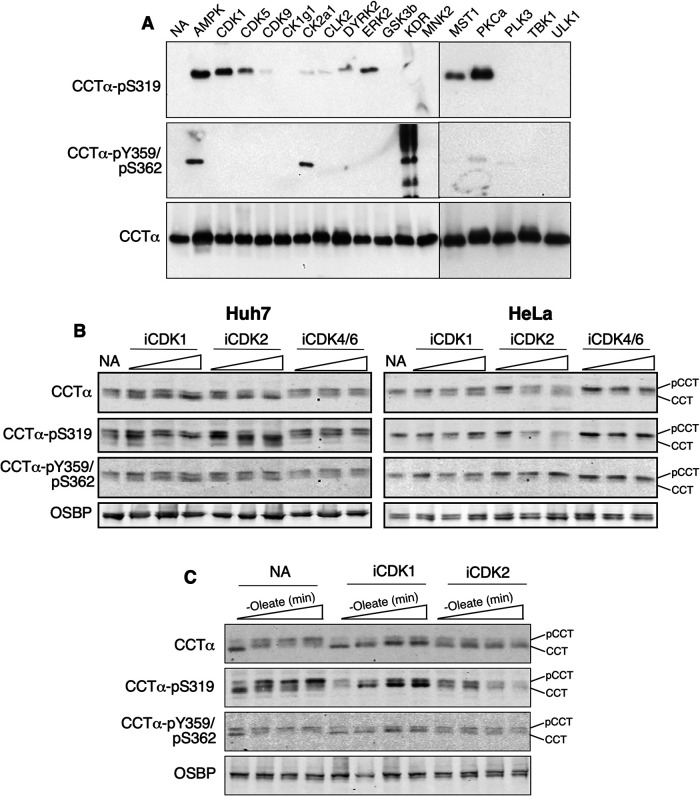
The 11 serine residues that were mutated in CCTα (Figure 6A) are predicted to be substrates for proline-directed kinases. To identify the kinase(s) responsible for phosphorylation of these sites we incubated dephosphorylated recombinant rat CCTα with purified candidate kinases and ATP. Phosphorylation of CCTα was detected by immunoblotting with antibodies against pS319, a predicted proline-directed site, and pY359/pS362, predicted protein-tyrosine kinase and casein kinase 2 (CK2) sites (Y359 is missing in rat CCTα). In addition to detection of CCTα, these antibodies also detected phosphorylation of several of the kinases and their activating proteins (Supplemental Figure 3, B–D). S319 was phosphorylated by AMP-activated protein kinase (AMPK), cyclin-dependent kinases (CDK) 1 and 5, ERK2, MST1, and PKCα, while AMPK and CK2a1 phosphorylated the S362 site (Figure 10A). GSK3β did not phosphorylate S319, likely because it was unable to phosphorylate S327 that would then convert S319 into a consensus GSK3 site.
FIGURE 10.
Phosphorylation of P-domain residues by CDKs. (A) after incubating dephosphorylated recombinant rat CCTα with the indicated kinases and ATP, phosphorylation of S319 and Y359/S362 was determined by immunoblotting. (B) Huh7 and HeLa cells received no addition (NA) or were incubated with inhibitors of CDK1, CDK2, or CDK4/6 (1, 5, and 10 μM) for 6 h and total cell lysates were immunoblotted using the indicated antibodies. (C) Huh7 cells were cultured in serum-free medium for 1 h, followed by serum-free media containing oleate (0.4 mM) for 30 min. Cells then received medium without oleate but supplemented with no addition, CDK1 or CDK2 inhibitors (10 μM). After 0, 30, 60, and 120 min, cell lysates were prepared and immunoblotted with the indicated antibodies. Results are the representative of three experiments. pCCT, phosphorylated CCT; CCT, dephosphorylated CCT.
Eukaryotic CDKs comprise a large family of proline-directed kinases that phosphorylate intrinsically disordered serine-rich domains in proteins involved in cell cycle regulation and transcription (Brown et al., 1999; Echalier et al., 2010; Malumbres, 2014). The CCTα P-domain is an intrinsically disordered structure with numerous serine-proline motifs (Figure 6A; Dennis et al., 2011) and is temporally phosphorylated during cell division (Jackowski, 1994), underscoring its potential as a CDK substrate. In addition, CDK1 and 5 did not phosphorylate S362, which was phosphorylated by CK2a1 (Figure 10A; Cornell et al., 1995). To confirm that CCTα was a CDK substrate, Huh7 and HeLa cells were treated with increasing concentrations of CDK1 (RO-3306), CDK2 (KO3861), or CDK4/6 (PD0332991) inhibitors for 6 h and the phosphorylation of S319 and S362 was determined by immunoblotting (Figure 10B). Under these basal conditions it was apparent that the CDK1 and 2 inhibitors caused a shift in CCTα to the low mass dephosphorylated species that was accompanied by a reduction in the pS319 signal. This trend was most prominent with the CDK2 inhibitor while the CDK4/6 inhibitor had no effect on CCTα phosphorylation. Phosphorylation of Y359/S362 was unaffected by the inhibitors.
To better assess the role of CDKs in phosphorylation, Huh7 cells were treated with oleate to promote CCTα translocation and dephosphorylation at the INM (Figure 4, A and B). This was followed by removal of oleate in the presence of CDK inhibitors to assess their effect on CCTα rephosphorylation (Figure 10C). In Huh7 cells that received no inhibitors (NA), oleate removal caused a progressive increase in phosphorylation as indicted by a shift to higher mass CCTα species that were enriched in pS319. This shift to higher mass phosphorylated species was partially prevented by the CDK1 inhibitor. Similar to results in Figure 10B, the CDK2 inhibitor was more effective in blocking rephosphorylation of S319 following oleate removal and prevented the appearance of the higher mass phosphorylated CCTα. CDK inhibitors had no effect on the Y359/S362 sites after oleate removal. Although the mammalian CDK family and associated cyclin activators is vast, these initial experiments support a role for CDK1 and 2 in phosphorylation of CCTα.
DISCUSSION
The synthesis of PC by the CDP-choline pathways is regulated by the reversible association of CCTα with the INM in response to lipid activators such as oleate (Lagace and Ridgway, 2005) or PC deficiency (Haider et al., 2018; Dorighello et al., 2023). This mode of regulation was recently extended to include the association of CCTα with the surface of nLDs and LAPS by virtue of its membrane-binding M-domain (Lee et al., 2020). nLDs and LAPS are formed in response to fatty acid storage in TAG, indicating that they provide a platform from which to drive PC synthesis as a response to fatty acid overload. To understand the role of CCTα and PC synthesis in this stress response, we identified factors that control the association of CCTα with nLDs and LAPS in Huh7 cells.
CCTα translocated to the INM of oleate-treated cells Huh7 cells as early as 15 min and began to appear on nLDs and LAPS after 3 h, followed by steady enrichment and preferred association with LAPS over 24 h. This temporal relationship indicates that the initial activation of CCTα is in response to enrichment of oleate or an oleate-derived lipid activator in the INM followed by a shift to nLDs and LAPS as these structures accumulated in the nucleoplasm. This shift in CCTα localization reflects an increased requirement for PC synthesis to accommodate the storage and/or secretion of excess fatty acids as TAG. Accordingly, we found that CCTα does not associate with preexisting LAPS in untreated Huh7 cells. CCTα on the INM could associate with nLDs or LAPS as they form in situ, but a more likely scenario is dynamic dissociation and reassociation cycle that is typical of proteins with amphipathic helices (Olarte et al., 2021).
Lipin1 associates with cLDs and LAPS and plays an indispensable role in their biogenesis by providing the substrate for TAG synthesis (Lee et al., 2020; Soltysik et al., 2021). Increased nuclear localization of Lipin1 by inhibition of mTOR in U2OS cells resulted in increased nLD formation that was dependent on the PA phosphatase activity of the enzyme (Soltysik et al., 2021). Here we showed that the increase in nLDs due to overexpression Lipin1α and 1β was accompanied by a proportional increase in CCTα association. The effect was greater for Lipin1α, consistent with its increased nuclear and nLD localization compared with Lipin1β. Lipin1α or 1β and CCTα occupied the same nLDs (Figure 2), and CCTα association with nLDs and LAPS was not correlated with the presence of DAG measured with a fluorescent reporter (Lee et al., 2020). These results indicate that CCTα association with nLDs and LAPS is coupled to Lipin1 activity and primarily driven by availability of a surface monolayer for M-domain insertion. Because LD surface area is related to the rate of TAG synthesis and storage, this would provide a feedforward mechanism to coordinate the rates of PC and TAG synthesis to maintain nLD stability. A similar mechanism was proposed to explain the localization of CCT1 to the surface of cLD in insect cells during storage of excess oleate in TAG (Krahmer et al., 2011).
Compared to oleate, the saturated fatty acid palmitate induced fewer nLDs and LAPS, and failed to recruit CCTα onto the INM, nLDs, or LAPS. Palmitate also inhibited oleate-dependent translocation of CCTα to nLDs when combined at a 1:4 molar ratio but without effecting nLD formation. This indicates that palmitate inhibits CCTα translocation by modifying the fatty acid content of nLDs rather than reducing nLD surface area. CCTα is activated by membrane negative curvature stress induced by lipids such as PE that are enriched in unsaturated fatty acids and create gaps in the membrane or monolayer surface into which the domain M amphiphilic inserts (Attard et al., 2000; Davies et al., 2001). The enrichment of phospholipids in palmitate would promote tighter packing of phospholipids and inhibit domain M insertion. Because the ER stress response to palmitate is caused in part by increased saturation of membrane phospholipids (Leamy et al., 2014), lack of activation of CCTα and PC synthesis in palmitate-treated cells could be a mechanism to prevent the further incorporation of saturated phospholipids into membranes (Piccolis et al., 2019).
Insertion of the α-helical M-domain of CCTα into membranes results in the removal of the N-terminal auto-inhibitory region from the active site and derepression of catalytic activity (Ramezanpour et al., 2018). M-domain association with membranes is negatively regulated by phosphorylation of the adjacent C-terminal P-domain, which in humans and rodents contains 16 phosphorylation sites. Similarly, CCTα association with nLDs is enhanced by deletion of the P-domain, and dephosphorylation at S319 but not Y359/S262 is associated with increased nLD association (Lee et al., 2020; Yue et al., 2020). A temporal analysis of CCTα phosphorylation in Huh7 cells revealed that S319 was dephosphorylated after treatment with oleate 15 and 30 min, was rephosphorylated by 3 h and did not decrease again until after 12 h. Throughout the entire time course CCTα-pS319 was not detected on the INM, nLDs, or LAPS. A potential explanation is that the S319 is rephosphorylated by 3 h as fatty acids are packaged into TAG that is incorporated into cLDs, nLDs, and LAPS. The gradual association of CCTα with nLDs and LAPS between 3–12 h appears to involve a pool of enzyme that is already dephosphorylated at S319, and only after 12 h does the CCTα-pS319 fraction start to dephosphorylate again. Unlike U2OS and Caco2 cells (Yue et al., 2020), Y359/S362 was partially dephosphorylated after 24 h oleate exposure in Huh7 cells, perhaps reflecting cell-specific responses to long-term fatty acid exposure.
Due to the complexity of the P-domain, it is unknown which phosphorylation site(s) are regulatory and the kinase(s) and phosphatases(s) involved. To address this problem, we conducted additive mutagenesis of small clusters of serine-proline phosphorylation sites, as well as a putative casein kinase II site at the C-terminus (S362). In the case of serine-to-aspartate mutants, it was not until all four sites encompassing 11 serine residues were mutated that there was a significant inhibition of CCTα association with nLDs. Together with results for serine-to-alanine mutants, this evidence favors a model in which the net-charge of the P-domain defines its regulatory status. Thus, a threshold of P-domain phosphorylation is required to generate sufficient negative charge to attract the M-domain or repulse membrane lipids, leading to CCTα disengagement from a membrane or monolayer. Similarly, a critical level of dephosphorylation of nucleoplasmic CCTα would relieve inhibition and promote membrane or monolayer association. The mutagenesis studies also supplied evidence that phosphorylation of the P-domain could be sequential. Phosphorylation of S319 in site 1A did not occur unless the adjacent site 2A SSSP motif was negatively charged due to aspartate mutations. Although additional phosphospecific antibodies are needed to confirm these results at other sites, it indicates that the sequence of P-domain phosphorylation could occur in a C-to-N-terminal direction. Because the P-domain is a disordered structure, it is feasible that phosphorylation at C-terminal sites triggers its reorganization and exposure of other sites to phosphorylation.
Of the kinases that phosphorylated S319 in vitro, CDK1, CDK5, and ERK were considered viable candidates based on their specificity for proline-directed phosphorylation sites, which make up the majority of serine phosphorylation sites in the P-domain (Figure 6A). Although AMPK phosphorylated both S319 and S362, it has not been implicated in PC metabolism (Jacobs et al., 2007), and RNA interference and pharmacological inhibition of AMPK in cultured cells did not affect S319 phosphorylation (result on not shown). In contrast, specific inhibitors of CDK1 and CDK2 promoted a shift to dephospho-species of CCTα on SDS–PAGE and reduced pS319 under basal conditions and after removal of oleate from cultured Huh7 cells. Inhibition of CDK2 was more effective in this regard, causing an almost complete inhibition of S319 phosphorylation following removal of oleate from Huh7 cells. Our data showing that CDK1 and/or CDK2 are CCTα kinases is supported by the results of a recent CRISPR screen that identified checkpoint kinase 1 (Chk1) as a positive effector of PC biosynthesis (Tsuchiya et al., 2023). Chk1 regulates genotoxic stress and cell cycle progression by suppressing CDC25-dependent activation of CDK1 and CDK2 at inter-S and G2/M mitotic checkpoints (Neizer-Ashun and Bhattacharya, 2021). Inhibition of Chk1 resulted in increased CCT phosphorylation and reduced PC synthesis, which was partially prevented by coapplication of a CDK1 inhibitor (Tsuchiya et al., 2023). The activation of CCTα during G1 is primarily driven by the enrichment of lipid activators derived from lipid degradation in nuclear membranes (Jackowski, 1996; Ng et al., 2004). CCTα phosphorylation then increases steadily from S phase through to G2/M, mirroring the decline in CCTα activity and PC synthesis (Jackowski, 1994). Based on the cell cycle activity of CCTα, CDK1 or CDK2 would be active during S and G2/M phase to suppress CCTα activity and PC synthesis once the demand for new membrane synthesis has been fulfilled. Our results showing that the addition and removal of oleate from unsynchronized cells initiates rapid changes in CCTα phosphorylation indicates that the activity of CDK and unknown phosphatase(s) is also regulated by acute changes in the abundance of lipid activators that are distinct from modes of cell cycle regulation of CDK activity described above.
Our analysis reveals that CCTα activation in response to oleate occurs in stages, with translocation to the INM followed by its appearance on nLDs and LAPS in conjunction with Lipin1. The shift of CCTα from nuclear membranes to a lipid droplet monolayer reflects a change in demand for PC to accommodate the storage of excess fatty acids in TAG. This sequence of events was inhibited by palmitate, which is poorly incorporated into LDs, and by bulk phosphorylation of the P-domain, possibly involving CDK1 and/or CDK2.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell culture and transfections
U2OS cells were cultured in DMEM containing 10% fetal bovine serum (FBS). Huh7 cells were cultured in low-glucose (1000 mg/l) DMEM containing 10% FBS. CHO-MT58 cells that express a temperature-sensitive allele of CCTα were cultured at 33°C in DMEM with 5% FBS and 34 μg/ml proline (Esko et al., 1981). Oleate/ BSA complexes (6:1 mol/mol) were prepared as previously described (Goldstein et al., 1983). Palmitate/BSA complexes (6:1 mol/mol) were prepared using the same protocol except the palmitate solution was dissolved at 70°C. Cells were treated with 0.4 mM oleate or palmitate unless otherwise stated. Stock solutions of CDK1 (RO-3306, Sigma Aldrich), CDK2 (KO3861, Sigma Aldrich), and CDK4/6 (PD0332991, Sigma Aldrich) inhibitors were prepared in DMSO and used at concentrations previously described (Vassilev et al., 2006; Alexander et al., 2015).
U2OS, Huh7, and CHO-MT58 cells were transfected at 60–70% confluency with a mixture of Lipofectamine 2000 and plasmids (pGFP-C1(2)δ-2xNLS [Lee et al., 2020], pLIPIN-1α-V5 or pLIPIN-1β-V5 [Bou Khalil et al., 2009] or pCMV-CCTα phospho-mutants) according to the manufacturer's instructions (Thermo Fisher Scientific). Experiments were initiated 16–24 h posttransfection.
Mutagenesis of P-domain phosphorylation sites
Four separate tracts of serine residues and S362 encoded in the P-domain of pCMV-CCTα-V5 (see Figure 6A) were mutated to alanine or aspartate residues using the primers listed in Supplemental Table 1. These four P-domain mutants were then used to generate double, triple and quadruple mutants using the same mutagenic primer pairs (specific primer sets were required for tract 2 mutagenesis of pCMV-CCTα-1A-V5 and pCMV-CCTα-1D-V5 due to sequence overlap). Following the annealing of mutagenic primers to plasmid templates and 20–30 rounds of extension using Phusion DNA polymerase (Cell Signaling Technology), products were incubated with DpnI to cleave the methylated DNA template and transformed into E.coli. All mutations were verified by sequencing.
SDS–PAGE and immunoblotting
After removal of media, cells were washed twice with cold phosphate-buffered saline (PBS), scraped in PBS, collected by centrifugation, and lysed with a detergent buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.25% (wt/vol) deoxycholate, 1% (vol/vol) NP40, 1 mM EDTA, protease inhibitor cocktail, 1 mM sodium metavanadate and 2.5 mM sodium fluoride) for 15 min on ice. Lysates were centrifuged at 13,000×g for 10 min at 4°C, and the protein concentration of the supernatants was measured using a Pierce BCA protein assay kit. SDS–PAGE buffer (62.5 mM Tris-HCl pH 6.8, 10% glycerol, 2% SDS, 0.05% bromophenol blue, and 5% β-mercaptoethanol) was added to lysates, which were then heated at 95°C for 3 min, separated on SDS–PAGE and transferred to nitrocellulose membranes. Proteins were detected on nitrocellulose membranes by immunoblotting at room temperature for 60 min or overnight at 4°C using Tris-buffer saline-Tween (TBS-T; 20 mM Tris, pH of 7.4, 500 mM NaCl, and 0.05% Tween 20): LICOR Odyssey blocking buffer (4:1, vol/vol). Antibodies against the following proteins and phosphorylation sites were used: β-actin (Sigma Aldrich, AC-15), CCTα (Gehrig et al., 2009), CCTα-pS319 (Kinexus, PN546; Yue et al., 2020), CCTα-pY359/pS362 (Kinexus, PN548; Yue et al., 2020), Lipin1 (Abcam, ab181389), OSBP (Zhao and Ridgway, 2017) and V5 (BioRad, MCA1360GA or Invitrogen, PA1-993). After removal of primary antibodies, membranes were washed twice and incubated for 1 h at room temperature with IRdye800CW- or IRdye680LT-secondary antibodies (LICOR Biosciences). After washing in TBS-T, membranes were scanned using a LICOR Odyssey and fluorescence quantified using associated software (v3.0).
Immunofluorescence imaging
Cells cultured on glass coverslips (0.13–0.16 mm) were fixed with 4% (wt/vol) paraformaldehyde in PBS for 15 min at room temperature, washed twice with 5 mM NH4Cl and permeabilized with 0.5% Triton X-100 in PBS at 4°C for 15 min. Coverslips were blocked at room temperature for 1 h using in PBS with 1% BSA and incubated with anti-PML (E1, Santa Cruz Biotechnology) or the antibodies described above overnight at 4°C. After removal of primary antibodies, coverslips were incubated with goat antimouse and goat antirabbit secondary antibodies conjugated to Alexafluor-488, -594, or -647 (Thermo Fisher Scientific) at room temperature for 1 h. LDs were visualized using BODIPY493/503 (Thermo Fisher Scientific) or LipidTox Red (Invitrogen) diluted in PBS. Nuclei were visualized using DAPI diluted in PBS. Coverslips were mounted on glass slides using Mowiol and imaged using a Zeiss LSM710 or Leica SP8 laser scanning confocal microscopes with HC Plan-Apochromat 63x (1.4 NA) oil immersion objectives and solid state 405, 488, 552, and 638-nm lasers. The association of proteins with nLDs and LAPS was quantified in confocal sections (0.8–1 μm) that contained 10–20 nuclei/image. Using Leica application suite X annotations software, BODIPY493/503-positive nLD in the 488 channel were manually counted within the nuclear DAPI mask. Association of PML, CCTα, and Lipin1 with nLDs was determined after overlaying their respective 552 or 638 channels. Some images were quantified by automated counting using ImageJ and binary masks for the nucleus (DAPI) with no significant difference.
Identification of CCTα kinases
To identify candidate CCTα kinases, those with consensus phosphorylation sites in the CCTα P-domain sites were selected based on the Kinexus Kinase Phosphosite Predictor algorithm (www.phosphonet.ca). The candidate kinases were then tested for their ability to phosphorylate S319 and Y359/S362 in recombinant rat CCTα expressed in Sf21 cells (Taneva et al., 2003). First, S319 was dephosphorylated in recombinant CCTα by λphosphatase (New England BioLabs, P07535; Supplemental Figure 3A). CCTα (100 μg) was incubated with of λphosphatase (60U) in 50 mM HEPES, 100 mM NaCl, 2 mM dithiothreitol, 1 mM MnCl2, 0.01% Brij 35, pH 7.5 for 1 h at 30°C. Next, dephosphorylated CCTα (600 ng) was incubated with the purified kinases (10–50 ng) listed in Supplemental Table 2 (SignalChem Life Sciences) using specified buffers and 10 mM MgCl2 in a final volume of 60 μl. Assays were initiated by addition of ATP (0.5 mM), incubated at 25°C for 30 min and terminated by addition of 20 μl of 4xSDS-PAGE lysis buffer. Samples were denatured at 100°C for 2 min, separated by SDS–PAGE and transferred to nitrocellulose membranes. pS319 and pY359/S362 were detected by immunoblotting as described in the previous sections.
Supplementary Material
Acknowledgments
Thanks to Robert Douglas for assistance in tissue culture. This work was supported by a Project Grant from the Canadian Institutes of Health Research (PJT62390) to G.D. and N.D.R., an endowment from the Lockwood Trust to N.D.R. J.F. was the recipient of a Research Nova Scotia graduate studentship award. M.M. is the recipient of a IWK graduate studentship award.
Abbreviations used:
- CCT
CTP:phosphocholine cytidylyltransferase
- CDK
cyclin-dependent kinase
- Chk1
checkpoint kinase 1
- DAG
diacylglycerol
- cLD
cytoplasmic lipid droplet
- ER
endoplasmic reticulum
- INM
inner nuclear membrane
- LAPS
lipid-associated PML structure
- nLD
nuclear lipid droplet
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PML NB
promyelocytic leukemia nuclear body
- TAG
triacylglycerol
- VLDL
very low-density lipoprotein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E23-09-0354) on January 3, 2024.
REFERENCES
- Alexander LT, Mobitz H, Drueckes P, Savitsky P, Fedorov O, Elkins JM, Deane CM, Cowan-Jacob SW, Knapp S (2015). Type II inhibitors targeting CDK2. ACS Chem Biol 10, 2116–2125. [DOI] [PubMed] [Google Scholar]
- Attard GS, Templer RH, Smith WS, Hunt AN, Jackowski S (2000). Modulation of CTP:phosphocholine cytidylyltransferase by membrane curvature elastic stress. Proc Natl Acad Sci USA 97, 9032–9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borradaile NM, Han X, Harp JD, Gale SE, Ory DS, Schaffer JE (2006). Disruption of endoplasmic reticulum structure and integrity in lipotoxic cell death. J Lipid Res 47, 2726–2737. [DOI] [PubMed] [Google Scholar]
- Bosch M, Sweet MJ, Parton RG, Pol A (2021). Lipid droplets and the host-pathogen dynamic: FATal attraction? J Cell Biol 220, e202104005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou Khalil M, Sundaram M, Zhang HY, Links PH, Raven JF, Manmontri B, Sariahmetoglu M, Tran K, Reue K, Brindley DN, Yao Z (2009). The level and compartmentalization of phosphatidate phosphatase-1 (lipin-1) control the assembly and secretion of hepatic VLDL. J Lipid Res 50, 47–58. [DOI] [PubMed] [Google Scholar]
- Brown NR, Noble ME, Endicott JA, Johnson LN (1999). The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol 1, 438–443. [DOI] [PubMed] [Google Scholar]
- Chung J, Wu X, Lambert TJ, Lai ZW, Walther TC, Farese RV Jr. (2019). LDAF1 and Seipin form a lipid droplet assembly complex. Dev Cell 51, 551–563 e557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell RB, Kalmar GB, Kay RJ, Johnson MA, Sanghera JS, Pelech SL (1995). Functions of the C-terminal domain of CTP: phosphocholine cytidylyltransferase. Effects of C-terminal deletions on enzyme activity, intracellular localization and phosphorylation potential. Biochem J 310, 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet A, Kleijwegt C, Roubille S, Juillard F, Jacquet K, Texier P, Lomonte P (2020). PML nuclear bodies and chromatin dynamics: catch me if you can!. Nucleic Acids Res 48, 11890–11912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SM, Epand RM, Kraayenhof R, Cornell RB (2001). Regulation of CTP: phosphocholine cytidylyltransferase activity by the physical properties of lipid membranes: an important role for stored curvature strain energy. Biochemistry 40, 10522–10531. [DOI] [PubMed] [Google Scholar]
- Dellaire G, Ching RW, Ahmed K, Jalali F, Tse KC, Bristow RG, Bazett-Jones DP (2006). Promyelocytic leukemia nuclear bodies behave as DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS1 and the kinases ATM, Chk2, and ATR. J Cell Biol 175, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis MK, Taneva SG, Cornell RB (2011). The intrinsically disordered nuclear localization signal and phosphorylation segments distinguish the membrane affinity of two cytidylyltransferase isoforms. J Biol Chem 286, 12349–12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorighello G, McPhee M, Halliday K, Dellaire G, Ridgway N (2023). Differential contributions of phosphotransferases CEPT1 and CHPT1 to phosphatidylcholine homeostasis and lipid droplet biogenesis. J Biol Chem 299, 104578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echalier A, Endicott JA, Noble ME (2010). Recent developments in cyclin-dependent kinase biochemical and structural studies. Biochim Biophys Acta 1804, 511–519. [DOI] [PubMed] [Google Scholar]
- Esko JD, Wermuth MM, Raetz CR (1981). Thermolabile CDP-choline synthetase in an animal cell mutant defective in lecithin formation. J Biol Chem 256, 7388–7393. [PubMed] [Google Scholar]
- Fujimoto T (2022). Nuclear lipid droplets - how are they different from their cytoplasmic siblings? J Cell Sci 135, jcs259253. [DOI] [PubMed] [Google Scholar]
- Gehrig K, Morton CC, Ridgway ND (2009). Nuclear export of the rate-limiting enzyme in phosphatidylcholine synthesis is mediated by its membrane binding domain. J Lipid Res 50, 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Basu SK, Brown MS (1983). Receptor mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol 98, 241–260. [DOI] [PubMed] [Google Scholar]
- Haider A, Wei YC, Lim K, Barbosa AD, Liu CH, Weber U, Mlodzik M, Oras K, Collier S, Hussain MM, et al. (2018). PCYT1A regulates phosphatidylcholine homeostasis from the inner nuclear membrane in response to membrane stored curvature elastic stress. Dev Cell 45, 481–495 e488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Reese ML, Goodman JM (2018). The assembly of lipid droplets and their roles in challenged cells. EMBO J 37, e98947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsova P, Ibrabim SH, Gores GJ, Malhi H (2016). Lipotoxic lethal and sublethal stress signaling in hepatocytes: relevance to NASH pathogenesis. J Lipid Res 57, 1758–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowski S (1994). Coordination of membrane phospholipid synthesis with the cell cycle. J Biol Chem 269, 3858–3867. [PubMed] [Google Scholar]
- Jackowski S (1996). Cell cycle regulation of membrane phospholipid metabolism. J Biol Chem 271, 20219–20222. [DOI] [PubMed] [Google Scholar]
- Jacobs RL, Lingrell S, Dyck JRB, Vance DE (2007). Inhibition of hepatic phosphatidylcholine synthesis by 5-aminoimidazole-4-carboxamide-1-beta-4-ribofuranoside is independent of AMP-activated protein kinase activation. J Biol Chem 282, 4516–4523. [DOI] [PubMed] [Google Scholar]
- Jacquemyn J, Foroozandeh J, Vints K, Swerts J, Verstreken P, Gounko NV, Gallego SF, Goodchild R (2021). Torsin and NEP1R1-CTDNEP1 phosphatase affect interphase nuclear pore complex insertion by lipid-dependent and lipid-independent mechanisms. EMBO J 40, e106914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Chung J, Arlt H, Pak AJ, Farese RVJ, Walther TC, Voth GA (2022). Seipin transmembrane segments critically function in triglyceride nucleation and lipid droplet budding from the membrane. Elife 11, e75808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitai Y, Ariyama H, Kono N, Oikawa D, Iwawaki T, Arai H (2013). Membrane lipid saturation activates IRE1alpha without inducing clustering. Genes Cells 18, 798–809. [DOI] [PubMed] [Google Scholar]
- Kory N, Farese RV, Jr., Walther TC (2016). Targeting fat: Mechanisms of protein localization to lipid droplets. Trends Cell Biol 26, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, Guo Y, Wilfling F, Hilger M, Lingrell S, Heger K, Newman HW, Schmidt-Supprian M, Vance DE, Mann M, et al. (2011). Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab 14, 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace TA, Ridgway ND (2005). The rate-limiting enzyme in phosphatidylcholine synthesis regulates proliferation of the nucleoplasmic reticulum. Mol Biol Cell 16, 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layerenza JP, Gonzalez P, Garcia de Bravo MM, Polo MP, Sisti MS, Ves-Losada A (2013). Nuclear lipid droplets: a novel nuclear domain. Biochim Biophys Acta 1831, 327–340. [DOI] [PubMed] [Google Scholar]
- Leamy AK, Egnatchik RA, Shiota M, Ivanova PT, Myers DS, Brown HA, Young JD (2014). Enhanced synthesis of saturated phospholipids is associated with ER stress and lipotoxicity in palmitate treated hepatic cells. J Lipid Res 55, 1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy AK, Hasenour CM, Egnatchik RA, Trenary IA, Yao CH, Patti GJ, Shiota M, Young JD (2016). Knockdown of triglyceride synthesis does not enhance palmitate lipotoxicity or prevent oleate-mediated rescue in rat hepatocytes. Biochim Biophys Acta 1861, 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Salsman J, Foster J, Dellaire G, Ridgway ND (2020). Lipid-associated PML structures assemble nuclear lipid droplets containing CCTalpha and Lipin1. Life Sci Alliance 3, e202000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Jr., Ory DS, Schaffer JE (2003). Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci USA 100, 3077–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M (2014). Cyclin-dependent kinases. Genome Biol 15, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee MJ, Salsman J, Foster J, Thompson J, Mathavarajah S, Dellaire G, Ridgway ND (2022). Running ‘LAPS’ around nLD: Nuclear lipid droplet form and function. Front Cell Dev Biol 10, 837406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera JV, Bacher MC, Priess JR (2021). Nuclear lipid droplets and nuclear damage in Caenorhabditis elegans. PLoS Genet 17, e1009602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neizer-Ashun F, Bhattacharya R (2021). Reality CHEK: Understanding the biology and clinical potential of CHK1. Cancer Lett 497, 202–211. [DOI] [PubMed] [Google Scholar]
- Ng MN, Kitos TE, Cornell RB (2004). Contribution of lipid second messengers to the regulation of phosphatidylcholine synthesis during cell cycle re-entry. Biochim Biophys Acta 1686, 85–99. [DOI] [PubMed] [Google Scholar]
- Ohsaki Y, Kawai T, Yoshikawa Y, Cheng J, Jokitalo E, Fujimoto T (2016). PML isoform II plays a critical role in nuclear lipid droplet formation. J Cell Biol 212, 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olarte MJ, Swanson JMJ, Walther TC, Farese RV, Jr. (2021). The CYTOLD and ERTOLD pathways for lipid droplet-protein targeting. Trends Biochem Sci doi: 10.1016/j.tibs.2021.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterfy M, Phan J, Reue K (2005). Alternatively spliced lipin isoforms exhibit distinct expression pattern, subcellular localization, and role in adipogenesis. J Biol Chem 280, 32883–32889. [DOI] [PubMed] [Google Scholar]
- Piccolis M, Bond LM, Kampmann M, Pulimeno P, Chitraju C, Jayson CBK, Vaites LP, Boland S, Lai ZW, Gabriel KR, et al. (2019). Probing the global cellular responses to lipotoxicity caused by saturated fatty acids. Mol Cell 74, 32–44 e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezanpour M, Lee J, Taneva SG, Tieleman DP, Cornell RB (2018). An auto-inhibitory helix in CTP:phosphocholine cytidylyltransferase hi-jacks the catalytic residue and constrains a pliable, domain-bridging helix pair. J Biol Chem 293, 7070–7083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanauska A, Kohler A (2018). The inner nuclear membrane is a metabolically active territory that generates nuclear lipid droplets. Cell 174, 700–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WJ, Azhar S, Kraemer FB (2016). Lipid droplets and steroidogenic cells. Exp Cell Res 340, 209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltysik K, Ohsaki Y, Tatematsu T, Cheng J, Fujimoto T (2019). Nuclear lipid droplets derive from a lipoprotein precursor and regulate phosphatidylcholine synthesis. Nat Commun 10, 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltysik K, Ohsaki Y, Tatematsu T, Cheng J, Maeda A, Morita SY, Fujimoto T (2021). Nuclear lipid droplets form in the inner nuclear membrane in a seipin-independent manner. J Cell Biol 220, e202005026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Mizrak A, Lee CW, Cicconet M, Lai ZW, Tang WC, Lu CH, Mohr SE, Farese RV, Jr., Walther TC (2022). Identification of two pathways mediating protein targeting from ER to lipid droplets. Nat Cell Biol 24, 1364–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneva S, Johnson JE, Cornell RB (2003). Lipid-induced conformational switch in the membrane binding domain of CTP:phosphocholine cytidylyltransferase: a circular dichroism study. Biochemistry 42, 11768–11776. [DOI] [PubMed] [Google Scholar]
- Tsuchiya M, Tachibana N, Nagao K, Tamura T, Hamachi I (2023). Organelle-selective click labeling coupled with flow cytometry allows pooled CRISPR screening of genes involved in phosphatidylcholine metabolism. Cell Metab 35, 1072–1083. [DOI] [PubMed] [Google Scholar]
- Uzbekov R, Roingeard P (2013). Nuclear lipid droplets identified by electron microscopy of serial sections. BMC Res Notes 6, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L (2006). Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci USA 103, 10660–10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RV, Jr. (2012). Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 81, 687–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Qian H, Lukmantara I, Gao M, Du X, Yan N, Yang H (2018). Human SEIPIN binds anionic phospholipids. Dev Cell 47, 248–256 e244. [DOI] [PubMed] [Google Scholar]
- Yue L, McPhee MJ, Gonzalez K, Charman M, Lee J, Thompson J, Winkler DFH, Cornell RB, Pelech S, Ridgway ND (2020). Differential dephosphorylation of CTP:phosphocholine cytidylyltransferase upon translocation to nuclear membranes and lipid droplets. Mol Biol Cell 31, 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Ridgway ND (2017). Oxysterol-binding protein-related protein 1L regulates cholesterol egress from the endo-lysosomal System. Cell Rep 19, 1807–1818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.