Abstract
The ubiquitination of transmembrane receptors regulates endocytosis, intracellular traffic, and signal transduction. Bone marrow−derived macrophages from myeloid Cbl–/– and Cbl-b–/– double knockout (DKO) mice display sustained proliferation mirroring the myeloproliferative disease that these mice succumb to. Here, we found that the ubiquitin ligases Cbl and Cbl-b have overlapping functions for controlling the endocytosis and intracellular traffic of the CSF-1R. DKO macrophages displayed complete loss of ubiquitination of the CSF-1R whereas partial ubiquitination was observed for either single Cbl–/– or Cbl-b–/– macrophages. Unlike wild type, DKO macrophages were immortal and displayed slower CSF-1R internalization, elevated AKT signaling, and a failure to transport the CSF-1R into the lumen of nascent macropinosomes, leaving its cytoplasmic region available for signaling. CSF-1R degradation depended upon lysosomal vATPase activity in both WT and DKO macrophages, with this degradation confined to macropinosomes in WT but occurring in distributed/tubular lysosomes in DKO cells. RNA-sequencing comparison of Cbl–/–, Cbl-b–/– and DKO macrophages indicated that while the overall macrophage transcriptional program remained intact, DKO macrophages had alterations in gene expression associated with growth factor signaling, cell cycle, inflammation and senescence. Cbl-b–/– had minimal effect on the transcriptional program whereas Cbl–/– led to more alternations but only DKO macrophages demonstrated substantial changes in the transcriptome, suggesting overlapping but unique functions for the two Cbl-family members. Thus, Cbl/Cbl-b-mediated ubiquitination of CSF-1R regulates its endocytic fate, constrains inflammatory gene expression, and regulates signaling for macrophage proliferation.
Macrophages form macropinosomes in response to signaling from the CSF-1R. The CSF-1R is sequestered in the lumen of macropinosomes, however, the mechanism regulating this step and its importance in CSF-1R signaling is unknown.
The authors found that overlapping ubiquitination activity of Cbl and Cbl-b are critical for sequestration of the CSF-1R within macropinosomes, shaping CSF-1R signaling and controlling macrophage proliferation and preventing senescence.
These findings define a new function for macropinosomes and Cbl and Cbl-b in regulating growth factor signaling with implications for cancer and immunology.
INTRODUCTION
Cbl and Cbl-b are E3 ubiquitin ligases that regulate the endocytic traffic and signaling of receptor tyrosine kinases (RTKs) and immunoreceptors including EGFR, PDGFR, c-Kit, CSF-1R, TCR and FcγR (Matsuo et al., 1996; Erdreich‑Epstein et al., 1999; Schmidt and Dikic, 2005; Marois et al., 2011; Mohapatra et al., 2013; Voisinne et al., 2016). Upon receptor activation, Cbl and Cbl-b are recruited to phosphotyrosine residues on the cytoplasmic tail of RTKs by the adaptor Grb2 or through direct interaction mediated by their tyrosine kinase-binding domains (Miyake et al., 1997; Ettenberg et al., 1999; Pennock and Wang, 2008). Loss-of-function mutations in the Cbl gene have been identified in more than 10% of patients with juvenile myelomonocytic leukemia, a myeloproliferative disease (Loh et al., 2009; Makishima et al., 2009; Sanada et al., 2009) as well as in a smaller proportion of adult myeloid neoplasms (Nadeau et al., 2012). Mice lacking either Cbl or Cbl-b in the hematopoietic compartment survive with normal to mild developmental defects, whereas mice carrying the double knockout for Cbl and Cbl-b (DKO) develop severe myeloproliferative disease with a substantially shortened life-span averaging 65 days (Naramura et al., 2010; An et al., 2015, 2016; Goetz et al., 2016). This strong association with myeloproliferative disease suggests that Cbl and Cbl-b function redundantly to control the growth of myeloid cells (Naramura et al., 2010; An et al., 2015, 2016; Goetz et al., 2016).
Ubiquitination of RTKs mediated by Cbl and Cbl-b regulates receptor endocytosis and intracellular traffic and thus controls the subcellular location and degradation. Polyubiquitinated RTKs such as EGFR and VEGFR are recognized by the endosomal sorting complex required for transport complex (ESCRT) for sorting in multivesicular bodies and targeted for lysosomal degradation (Katzmann et al., 2002; Lu et al., 2003; Haglund and Dikic, 2012). In addition to ubiquitin ligase activity, Cbl can act as an adaptor or scaffold protein by binding SH2- and SH3-containing signaling proteins including Crk, Fyn, Lck, PI3K, and Shc (Miyake et al., 1997; Andoniou et al., 2000; Rao et al., 2002a, 2002b; Duan et al., 2004; Ghosh et al., 2004).
The CSF-1R is an RTK that supports proliferation and differentiation of macrophages (Chitu and Stanley, 2017). CSF-1R signaling is of interest for modulation of macrophage-dependent immune functions (Edwards et al., 2019), cancer therapy (Cannarile et al., 2017), and as a model for high-frequency oncogenic paralogs such as Kit and Flt3 (Ridge et al., 1990; Sapi et al., 2004). Although the signaling of the CSF-1R has been extensively studied, little is known about the contributions of endocytosis and the effects of subcellular compartmentalization on CSF-1R signaling (Lou et al., 2014). In macrophages, phosphorylated CSF-1R follows an unconventional endocytic pathway that involves small-vesicle endocytosis followed by subsequent traffic to macropinosomes formed in response to CSF-1R signaling. It has been recognized for some time that the CSF-1R traffic in macrophages is unique and understanding whether CSF-1R traffic regulates its signaling will be important for delineating how CSF-1R signaling is regulated by endocytosis (Lee et al., 1999; Huynh et al., 2012; Lou et al., 2014).
By analyzing CSF-1R signaling and traffic in macrophages from WT, Cbl –/–, Cbl-b –/–, and Cbl/Cbl-b DKO mice, we have found that Cbl and Cbl-b both contribute to the regulation of CSF-1R internalization, are required for delivery of the CSF-1R to the macropinosome and influence the nature and duration of downstream signaling. Importantly, these changes in DKO bone marrow−derived macrophages (BMDM) drive shifts in the macrophage transcriptional program that are sufficient to maintain elevated cell growth and escape from senescence. Mechanistically, these changes are driven by Cbl and Cbl-b-mediated receptor ubiquitination that drives intralumenal budding into macropinosomes and thus modifies the character of CSF-1R signaling to regulate growth control.
RESULTS
Growth phenotype in DKO macrophages
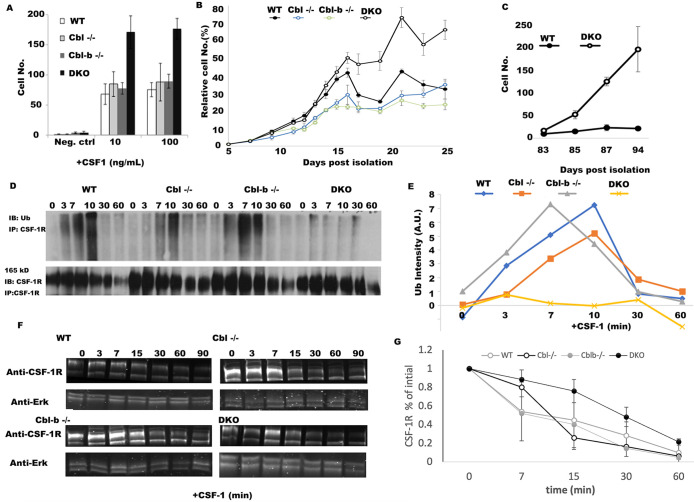
Given the severe myeloproliferative disease observed in myeloid DKO mice (Naramura et al., 2010; An et al., 2015, 2016; Goetz et al., 2016), we sought to determine the extent to which Cbl and Cbl-b constrain the proliferation of macrophages via CSF-1R-mediated signaling. BMDM from WT and Cbl–/–, Cbl-b–/– and DKO mice all demonstrated CSF-1-dependent growth, with DKO cells displaying a potent hyperproliferative phenotype at day 6 (Figure 1A). Analysis of their growth kinetics revealed that initial growth rates of macrophages from WT, Cbl –/–, Cbl-b–/–, and DKO mice macrophages during 5-15 d in culture were similar; however, beyond day 15 in culture, the DKO macrophages continued to proliferate, whereas the macrophages of other genotypes began to senesce (Figure 1B). Previous work using Cbl–/– macrophages demonstrated a mild hyperproliferative phenotype that was not detected here (Lee et al., 1999). Given their strong hyperproliferative phenotype, we sought to determine whether the DKO BMDM escape senesce. For this purpose, we continued to culture the WT and DKO cells for 94 d postisolation. The growth rates between days 83 and 94 illustrate that while WT macrophages can be maintained, they were senescent, whereas the DKO cells continue to proliferate (Figure 1C). Thus, Cbl and Cbl-b redundantly regulate CSF-1-dependent growth and senescence in macrophages, providing an explanation for the myeloproliferative phenotype observed in Cbl/Cbl-b-DKO mouse.
FIGURE 1:
Cbl and Cbl-b DKO macrophages evade exit from the cell cycle and have minimal CSF-1R ubiquitination. (A) BMDM growth measured at day 6 postisolation (n = 5 fields of view and n = 3 replicates, error bar = std. dev.). (B) Growth curves, measured by alamar blue for WT, Cbl–/–, Cbl-b–/– and DKO BMDMs measured over 25 d from 1,000 cells starting at day 5 postisolation. CSF-1−containing media was added every other day followed by replating to a density of 1,000 cells/well (indicated by arrows, n = 6 replicates, error bars = std. dev.). (C) The growth rates between day 83 to day 94 of DKO and WT cells (n = 3 replicates, error bars are std. dev.). (D) CSF-1R immunoprecipitation and ubiquitin blot across BMDM of different genotypes following stimulation with CSF-1 (minutes after stimulation). (E) Quantification of D (data is representative of 3 independent experiments). F. CSF-1R western blot and quantification in BMDM stimulated with CSF-1 (after 0 min). Erk1/2 was used as a loading control. G. Quantification of CSF-1R degradation across three experiments (mean +/- std. dev.).
Cbl and Cbl-b redundantly promote the CSF-1-induced ubiquitination of CSF-1R and its signaling complex, but are not required for CSF-1R degradation
We speculated that accelerated CSF-1-dependent growth observed in DKO macrophages was a result of the loss of ubiquitination of the CSF-1R. Immunoprecipitation of the CSF-1R followed by ubiquitin blotting showed nearly complete loss of CSF-1 induced ubiquitination of the CSF-1R in DKO (Figure 1, D and E). In contrast, WT and Cbl-b–/– displayed strong ubiquitination at 3, 7, 10 min post CSF-1 stimulation (Figure 1, D and E). Interestingly, Cbl–/– macrophages had a delay and partial reduction in ubiquitination relative to WT, consistent with earlier work (Lee et al., 1999). CSF-1R ubiquitination in Cbl-b–/– BMDM was almost indistinguishable from WT (Figure 1E). These patterns suggest a more prominent contribution of Cbl to ubiquitination of the CSF-1R at early time points and Cbl-b mediating CSF-1R ubiquitination with a slower kinetic. Taken together, our results suggest that Cbl and Cbl-b both promote the ubiquitination of the CSF-1R, with Cbl being particularly important for the early phases.
Western blotting for total CSF-1R showed that a significant proportion of the CSF-1R was degraded upon CSF-1 stimulation of macrophages of all four genotypes, including the DKO (Figure 1D and F). Together, these results suggest that while Cbl and Cbl-b are essential to promote the CSF-1R ubiquitination, the CSF-1-induced receptor degradation can proceed despite marked loss of ubiquitination.
Cbl and Cbl-b regulate CSF-1R endocytosis and transport to the lumen of nascent macropinosomes
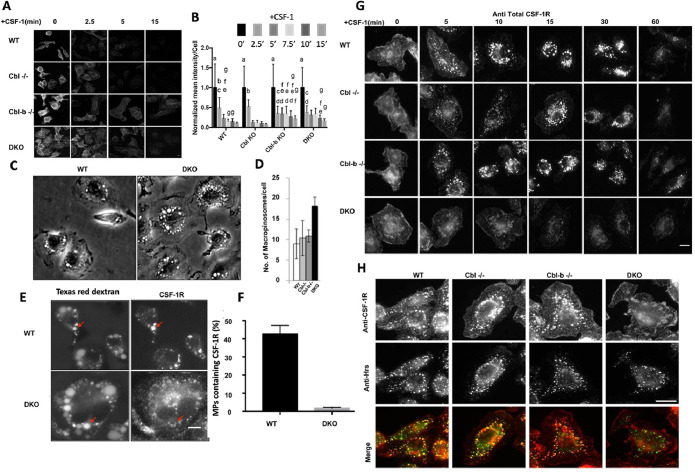
Endocytosis and subsequent subcellular traffic of the ligand-activated CSF-1R are known to contribute to its signaling and signal attenuation (Huynh et al., 2012). Given prior work demonstrating a mild CSF-1R endocytic defect in Cbl–/– macrophages (Lee et al., 1999), we first sought to examine the endocytosis of CSF-1R in macrophages of various genotypes. Immunofluorescence analysis of cell surface CSF-1R following CSF-1 stimulation indicated that the receptor was rapidly internalized across the genotypes at 2.5 min post CSF-1 exposure with slightly less complete clearance in Cbl-b–/– and DKO macrophages at later time points (Figure 2, A and B). These results indicate that Cbl and Cbl-b have minor roles in the endocytosis of activated CSF-1R, with Cbl-b appearing to have a role at later times.
FIGURE 2:
Cbl and Cbl-b facilitate CSF-1R internalization and translocation to the macropinosome lumen. (A) Immunofluorescence of cell surface CSF-1R in WT and DKO macrophages at indicated CSF-1 stimulation period. (B) Quantification of surface CSF-1R at different time points, scale bars are SD with n = 100, same letters are not significantly different (p< 0.05) by 2-way ANOVA followed by Tukey HSD comparison of means. (C) Phase contrast images illustrating the number of macropinosomes in DKO BMDM. (D) Quantified number of macropinosomes per cell (error bars = std. dev., n = 100 macrophages across two experiments). (E) Delivery of the CSF-1R to the macropinosome visualized labeling by sequential imaging of Texas-red dextran (macropinosome) and CSF-1R immunofluorescence following permeabilization (scale bar = 5 μm). (F) Quantification of the percentage of macropinosomes that contain luminal CSF-1R (error bar = std. dev. with n = 50 from two independent experiments). (G) CSF-1Rimmunofluorescence delivery to macropinosomes and degradation (scale bar = 10 μm). (H). Colocalization of CSF-1R and Hrs 10 min after CSF-1 exposure (scale bar = 10 μm).
Previously, we determined that the activated CSF-1R traffics into the lumen of newly formed macropinosomes following the endocytic uptake via small vesicular carriers that include clathrin-coated pits (Lou et al., 2014). DKO macrophages exhibited nearly twice as many macropinosomes per cell as in WT macrophages while the numbers in Cbl–/– and Cbl-b–/– macrophages were comparable to those in WT (Figure 2C, D), suggesting a role for Cbl and Cbl-b in regulating CSF-1R signaling for micropinocytosis. Given that only a modest CSF-1R endocytosis defect was observed in DKO macrophages, and that the CSF-1R can signal from endosomes (Huynh et al., 2012), we speculated that Cbl and Cbl-b may be responsible for directing the endosomal traffic of the CSF-1R to macropinosomes. We quantified the delivery of the CSF-1R to newly-formed macropinosomes by sequential imaging of the Texas-red dextran to identify macropinosomes followed by fixation and permeabilization of cells and staining of the same cells for CSF-1R (Lou et al., 2014). Strong colocalization of the CSF-1R in Texas-red dextran macropinosomes was observed in WT; however, DKO macrophages displayed much lower quantities of CSF-1R localizing to macropinosomes (Figure 2E, F, Figure S2). Confocal imaging of total immunostained CSF-1R in macrophages across different genotypes showed that trafficking of the CSF-1R was altered in DKO macrophages (Figure 2G). Specifically, WT, Cbl–/– or Cbl-b–/– macrophages rapidly transported the CSF-1R into small (<1 μm) endosomal compartments at 5 min that were then delivered to large newly formed macropinosomes and accumulated in their lumen (bright circular structures, Figure 2G). However, DKO macrophages trafficked the CSF-1R through dispersed vesicles that were less efficient at delivering the CSF-1R to macropinosomes and did not result in CSF-1R accumulation in their lumen (Figure 2G). Thus, Cbl and Cbl-b work together to direct the transport of the CSF-1R into the macropinosome lumen. Consistent with the western blot data showing unaltered ligand-induced CSF-1R degradation in DKO macrophages (Figure 1, F and G), the CSF-1R immunofluorescence signals showed a similar decay at late time points across all genotypes (Figure 2G).
Cbl and Cbl-b regulate Hrs and CSF-1R association in macrophages
We surmised that the delivery of the CSF-1R into the macropinosome lumen may involve the Endosomal Sorting Complex Required for Transport (ESCRT) recognition of the ubiquitin-interacting protein Hrs (Hurley, 2008). Indeed, immunofluorescence analysis demonstrated that while Hrs strongly colocalized with the CSF-1R in WT, Cbl–/– or Cbl-b–/– BMDM, colocalization was markedly reduced in DKO BMDM (Figure 2H), indicating that CSF-1R ubiquitination by Cbl and Cbl-b was required for recognition by the ESCRT and subsequent luminal budding into macropinosomes.
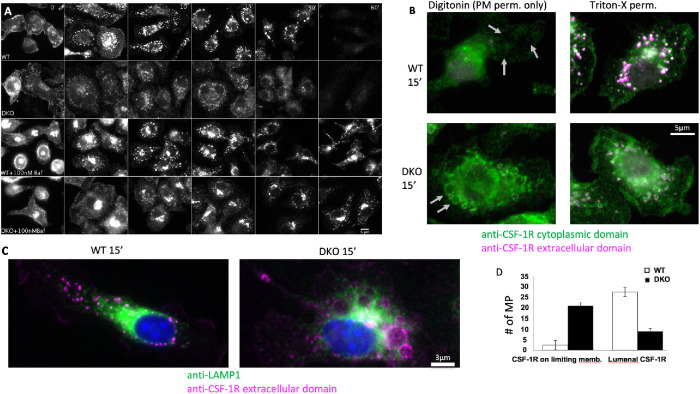
Cbl and Cbl-b are essential for sequestration and subsequent degradation of the CSF-1R within macropinosomes in a lysosome-dependent manner
These results support the idea that Cbl- and Cbl-b-mediated ubiquitination enables rapid sequestration of the CSF-1R into the lumen of macropinosomes via ESCRT-mediated luminal budding and subsequent fusion of the macropinosome with the lysosome. Inhibition of the vacuolar ATPase with bafilomycin A1 for 30 min protected the CSF-1R from degradation in both WT and DKO BMDM (Figure 3A). Lysosomal neutralization was verified by loss of lysosomal pHrodoRed-Dextran signal (Supplemental Figure S4). Proteasome inhibition by Bortezomib had no effect on the rate of CSF-1R degradation (Supplemental Figure S3). We also noticed that bafilomycin A1 treatment caused some of the CSF-1R to redistribute into a perinuclear region (Figure 3A, 0 min), which was similar to an effect caused by treating BMDM with the drug EIPA (Lou et al., 2014). We speculated that this relocalization was associated with alkalinization of lysosomes and/or the extracellular environment. Indeed, culturing macrophages in media of pH 5−9 for 15 min recapitulated this alteration in CSF-1R endocytosis and traffic (Supplemental Figure S5). Regardless of this effect, the CSF-1R was observed to traffic normally to the macropinosome lumen in WT cells but showed defective transport (including the limiting membranes of macropinosomes) in DKO cells, while bafilomycin A1 blocked CSF-1R degradation in both cases, indicating that lysosomes mediate the degradation of the CSF-1R (Figure 4A).
FIGURE 3:
Cbl and Cbl-b are essential for sequestration of the CSF-1R and subsequent degradation within macropinosomes. (A) Immunofluorescence of the CSF-1R with and without inhibition of the vacuolar ATPase with 30 min pretreatment with bafilomycin A1 followed by CSF-1 exposure time course (images are identically window leveled and were collected under identical conditions). (B) Immunofluorescence staining of the intracellular and extracellular epitopes of the CSF-1R with selective permeabilization of the plasma membrane with digitonin or complete membrane permeabilization with Triton-x 100 at 15 min post CSF-1 stimulation. Gray arrows denote locations of macropinosomes. Green staining in during digitonin indicates cytosolic accessibility of the CSF-1R. (C) Distribution of LAMP1-positive lysosomes in relation to the CSF-1R contained with the lumen (WT) or limiting membrane of the macropinosome (DKO) at 15 min post CSF-1 stimulation. (D) Quantification of the CSF-1R localization from 30 macropinosomes for each WT and DKO based on morphology in the Triton-X 100 staining condition.
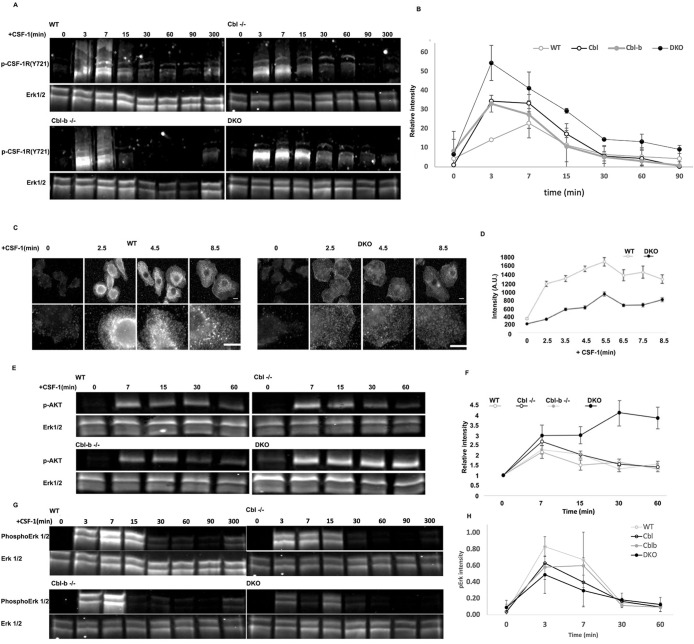
FIGURE 4:
Cbl and Cbl-b limit CSF-1R and AKT phosphorylation while supporting Erk1/2 and broad tyrosine phosphorylation. (A) Immunoblot of CSF-1R phosphorylation (pY721) following CSF-1 stimulation for 0−300 min. Note the higher molecular weight products consistent with ubiquitination observed in all blots except DKO. (B) Quantification of the phospho-CSF-1R signal intensity from three immunoblots (all molecular weights of CSF-1R), normalized to total Erk1/2 (mean+/– std. dev.) (C and D) Immunofluorescence and quantification of pan phosphotyrosine in WT and DKO BMDM following CSF-1 stimulation, illustrated broadly impaired tyrosine phosphorylation in the DKO. Error bar is standard error of mean, n = 50 cells per genotype. (E) AKT(S473) phosphorylation following CSF-1 stimulation. (F) Quantification of pAKT(S473), mean+/-std. dev. from two independent experiments. (G) Western blot of Erk1/2 phosphorylation following CSF-1 stimulation. (H) Quantification of pErk (mean +/- std. dev., n = 3 experiments).
To determine whether Cbl and Cbl-b were required for sequestration of the cytoplasmic tail, we took advantage of our previously developed method to costain with antibodies specific for the extracellular and cytoplasmic domains of the CSF-1R following either selective permeabilization of the plasma membrane using digitonin or complete permeabilization using triton-X100 (Lou et al., 2014). At 15-min post CSF-1 stimulation, under digitonin permeabilization, WT cells sequestered the CSF-1R cytoplasmic tail away from the cytosol and were negative for both antibodies, whereas the CSF-1R cytoplasmic tail was accessible in DKO in macropinosomes and lysosomes (Figure 3, B and D). Permeabilization with triton-X 100 revealed that both intracellular and extracellular domains of the CSF-1R were localized within the macropinosome lumen in WT cells. DKO cells, however, displayed the same overall pattern as that observed with digitonin permeabilization, which was largely limited to the macropinosome limiting membrane and some tubular structures (Figure 3B). Indeed, at 15 min, tubular lysosomes were observed to interact and envelop macropinosomes that either had the CSF-1R within their lumen (WT) or on their limiting membrane (DKO) (Figure 3C). The CSF-1R could also be observed in tubular structures that colocalized with lysosomes suggesting that lysosome−macropinosome kiss and run events (Racoosin and Swanson, 1993) were responsible for the delivery of the lysosomal hydrolyses responsible for CSF-1R degradation. These results indicate that Cbl and Cbl-b are not required for CSF-1R degradation by lysosomal hydrolases; however, they are essential for the intralumenal sequestration of the CSF-1R from the cytosol implying their importance for regulating signal transduction.
Cbl and Cbl-b regulate CSF-1R phosphorylation and limit downstream AKT and ERK signaling
Given the continued proliferation of DKO macrophages beyond when WT cells exhibited senescence, together with the loss of CSF-1R ubiquitination and its traffic to the macropinosome, we sought to determine the impact of DKO on CSF-1R signaling. Western blotting for CSF-1R pY721 showed that the CSF-1R in WT, Cbl–/– and Cbl-b–/– macrophages ran as a high molecular weight smear, that was not present in the DKO cells (Figure 4A). This molecular weight shift is consistent with Cbl and Cbl-b mediated ubiquitination (Figure 1D). Quantification of CSF-1R phosphorylation which included the shifted (smeared) molecular weight species, indicated that the peak pY721 levels were elevated and remained higher for longer in DKO cells, with Cbl–/– macrophages exhibiting a similar but less robust elevation and Cbl-b–/– macrophages only showing a moderate elevation of peak phosphorylation compared with WT cells (Figure 4B). These results indicated an inverse relationship between the levels of ubiquitination and phosphorylation of the CSF-1R and support the conclusion that Cbl and Cbl-b have additive, but potentially distinct roles with Cbl more prominently involved in attenuating CSF-1R phosphorylation.
Given the CSF-1R hyperphosphorylation in DKO macrophages, we considered the possibility that downstream targets would also be hyperphosphorylated. Surprisingly, immunofluorescence staining with an anti-phosphotyrosine antibody revealed lower total tyrosine phosphorylation in CSF-1 stimulated DKO compared with WT macrophages (Figure 4, C and D). Furthermore, tyrosine phosphorylation in CSF-1-stimulated WT macrophages was localized to punctate intracellular structures reminiscent of early endosomes and macropinosomes, consistent with previous observations that the CSF-1R continues to signal from endosomes (Huynh et al., 2012). Thus, Cbl and Cbl-b attenuation of CSF-1R phosphorylation does not translate directly into increased downstream signaling, but rather Cbl and Cbl-b both constrain CSF-1R signaling and scaffolding of the downstream signaling directly or through ubiquitin-driven processes.
The altered pattern of tyrosine phosphorylation upon Cbl/Cbl-b DKO suggested that Cbl and Cbl-b may shape the character of CSF-1R signaling. Measurements of p-AKT signaling, which is mainly initiated at the cell surface (Katz et al., 2007), demonstrated that CSF-1 stimulation led to increased p-AKT(S473) signals that peaked at 7 min in WT, Cbl–/– and Cbl-b–/– cells and then came down quickly reaching baseline levels by 15min (Figure 4, E and F). In contrast, DKO cells exhibited a markedly elevated and sustained induction of p-AKT(S473). This pattern is consistent with an extended residence of activated CSF-1R at the plasma membrane, resulting in hyperphosphorylation of AKT. In contrast, induction of p-Erk as a measure of activation of the Erk kinase, which signals downstream of the CSF-1R, reduced and less sustained in DKO macrophages (Figure 4G). These results are consistent with previous observations that CSF-1R activation of the MAPK/ERK signaling is amplified on endosomes (Huynh et al., 2012) and suggest that regulation of subcellular CSF-1R traffic by Cbl and Cbl-b or their potential adapter roles contribute to Erk pathway activation. Overall, our findings suggest that Cbl and Cbl-b function together to control the subcellular distribution of CSF-1R signaling and thus determine the nature of CSF-1R signaling via Erk and Akt.
Cbl and Cbl-b regulate gene expression downstream of the CSF-1R
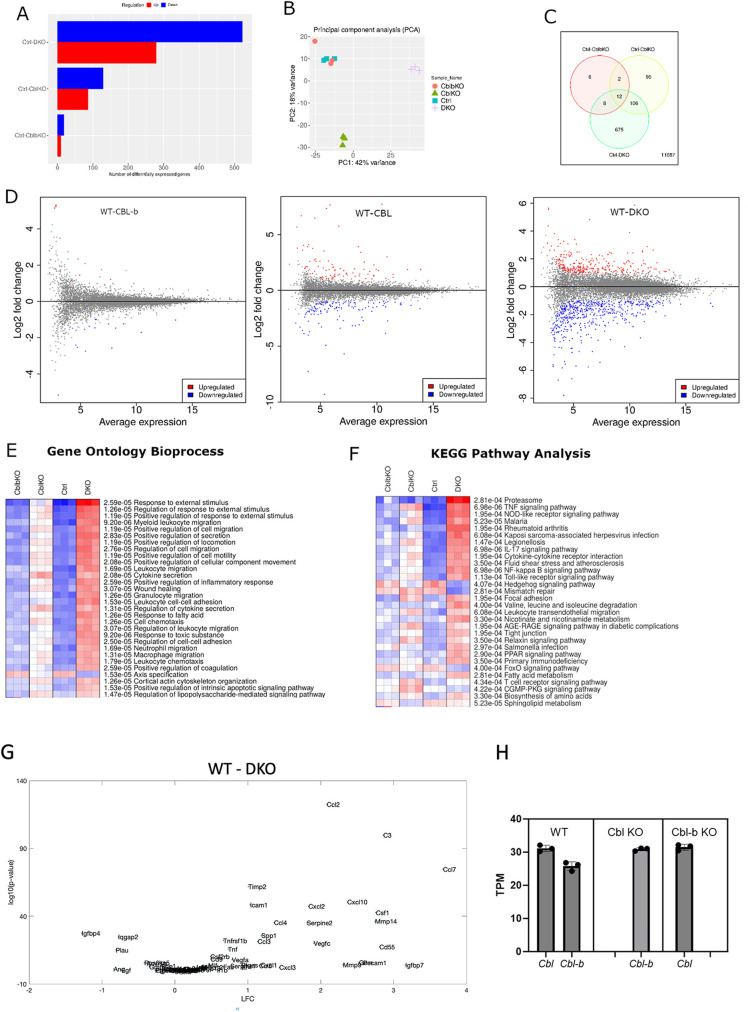
We conducted RNA-sequencing experiments on day 8 macrophages to delineate the consequences of Cbl and Cbl-b regulation of CSF-1R traffic and signaling on gene expression that constrains cell proliferation and growth. We anticipated that transcripts associated with increased responsiveness to CSF-1 and signaling via the AKT pathway might be upregulated. Differential expression analysis indicated that 520 genes were upregulated and 280 were downregulated in DKO macrophages when compared with WT controls and a smaller number (200) of total genes were differentially expressed in Cbl–/– and only 28 in Cbl-b–/– relative to WT (Figure 5). Thus, consistent with the changes in ubiquitination, traffic, and phosphorylation, Cbl had a stronger influence than Cbl-b in regulating gene expression, but Cbl-b largely compensated for Cbl–/–, as could be seen in that the DKO cells displayed many more alterations in gene expression levels than either single knockout. Principle component analysis indicated that WT and Cbl-b–/– macrophages were similar in gene expression character and that Cbl–/– and DKO displayed different principal components with PC1 and PC2 accounting for 60% of the total variance (Figure 5B). A Venn diagram captures the number of genes differentially expressed by comparison with the WT with a false discovery rate (FDR) of 0.05 (a statistic derived from the DESeq2, predicting ∼5% of the differential genes would be false positive (Love et al., 2014), Figure 5C). Of these, the WT/DKO pair showed 675 unique differentially expressed genes, vastly exceeding that of WT paired with Cbl–/– or Cbl-b–/–. The large number of genes whose expression is altered in the DKO macrophages is consistent with a redundant contribution of Cbl and Cbl-b to shaping the macrophage transcriptome. MA plots illustrate the effect of this differential gene expression and highlight broader alterations of the macrophage transcriptional program in the DKO than in the single KOs (Figure 5D). We considered the possibility that relative expression levels of Cbl and Cbl-b may determine the changes in the transcriptomic profile in KO cells. However, computing the transcripts per million reads (TPM) indicated that in our system they are expressed at similar levels and only an small (∼15%) increase in expression was observed for Cbl-b in Cbl–/– macrophages, with no change in Cbl transcripts in Cbl-b–/– cells, indicating little or no expression compensation (Figure 5H). Thus, we conclude that Cbl and Cbl-b cooperate to constrain signaling from the CSF-1R and likely other receptors to shape certain components of the macrophage transcriptional program.
FIGURE 5:
Differential gene expression analysis of Cbl, Cbl-b, and DKO BMDMs. (A) Differential genes expression (DESeq2) across genotypes as compared WT(ctrl) (FDR = 0.05). (B) Principal component analysis indicated that gene expression in Cbl-b closely matched WT, whereas Cbl and DKO displayed more unique variations in gene expression. (C) Venn diagram illustrating the number of differentially expressed genes shared and unique to each genotype comparison. (D) MA plots comparing each mutant with WT. Statistically significant expression differences are color coded. (E and F) Gene ontology bioprocess and KEGG pathway analysis of genes with FDR of < 0.05. G. DQseq2 analysis comparing WT and DKO genes contained within the mouse Saul_Sen_Mayo GSEA molecular signatures database.
To gain more in-depth insights into the gene expression changes resulting from combined Cbl and Cbl-b KO, we conducted pathway analyses using the iDEP as a broad multiplatform analytical tool (Ge et al., 2018). Gene ontology analysis across BioProcess revealed that the DKO macrophages were dramatically altered relative to WT, including pathways corresponding to responses to external stimulus, leukocyte, and leukocyte cell migration (Figure 5E). A KEGG pathway analysis identified alterations in Proteasome degradation, presumably reflecting altered traffic of cargo proteins whose ubiquitination is regulated by Cbl/Cbl-b, as well as a broad number of inflammation-related cytokine and innate immunity pathways (Figure 5F). In view of the pathway analysis results, we carried out Gene set enrichment analysis (GSEA) for genes associated with senescence (Saul et al., 2022). Indeed, many of the macrophage genes associated with senescence were observed as being upregulated in the DKO cells (Figure 5G). While counterintuitive given the hyperproliferative phenotype, this gene set likely reflects upregulation of macrophage genes in tissues bearing an aged cell burden, as opposed to genes that directly contribute to senescence, suggesting that the sustained growth of DKO cultures triggers an environment that shares similarity with the in vivo situation, which may require macrophage proliferation to compensate.
DISCUSSION
The Cbl family of E3 ubiquitin ligases are recognized as important negative regulators of receptor tyrosine kinase signaling pathways, primarily through their activities to promote ubiquitin modification and subsequent endocytic degradation of target receptors (Thien and Langdon, 2001; Schmidt and Dikic, 2005; Mohapatra et al., 2013). In this work, we shed new light on the mechanisms by which the individual and additive functions of Cbl and Cbl-b regulate growth factor signaling in a prototypical myeloid cell, the macrophage. This work provides insight into why Cbl or Cbl-b single knockout in mice are nonlethal, while Cbl and Cbl-b double knock out mice develop lethal myeloproliferative disease (Naramura et al., 2010; An et al., 2015, 2016; Goetz et al., 2016). Specifically, we found that Cbl and Cbl-b have overlapping function in controlling macrophage growth and ubiquitination of CSF-1R, leading to CSF-1R traffic to macropinosomes where it associates with Hrs and is packaged into the macropinosome lumen to suppress signaling (Lou et al., 2014). The control of CSF-1R ubiquitination and traffic by Cbl and Cbl-b regulates CSF-1R signaling through AKT and Erk ultimately regulating the macrophage transcriptional program and controlling proliferation and exit from the cell cycle.
Previously we characterized the transport of the CSF-1R into the lumen of macropinosomes where it is degraded (Lou et al., 2014). Single knockouts in either Cbl or Cbl-b had a modest effect on this pathway. However, DKO macrophages lost the ability to transport the CSF-1R into the macropinosome presumably via action of the ESCRT which in turn led to amplified AKT signaling. Ubiquitination of the CSF-1R by Cbl and Cbl-b was not required for its degradation, yet the lysosome function was. We conclude that there are redundant mechanisms that traffic ligand-stimulated CSF-1R to the lysosome although with prolonged exposure of the cytoplasmic tail to the cytosol. Thus, the import of activated CSF-1R into macropinosomes is a critical step in attenuating its signaling (Lou et al., 2014). The CSF-1R can signal from endosomes (Huynh et al., 2012), indicating that Cbl and Cbl-b direction of the CSF-1R to the macropinosome acts as an important step in endosomal CSF-1R signaling and signal termination. The loss of this activity in DKO macrophages is consistent with potentiated AKT signaling, presumably at the plasma membrane, as larger amounts of CSF-1R (pY721) were observed with slower internalization from the surface (Lee et al. 1999), consistent with the reported function of CSF-1R Y721 phosphorylation in the activation of AKT signaling pathway at the plasma membrane (Katz et al., 2007). Thus, our studies indicate that in addition to directing CSF-1R traffic to macropinosomes that Cbl and Cbl-b are important constraining AKT signaling.
Consistent with prior work previously implicating Cbl in regulating CSF-1R signaling (Lee et al., 1999), we observed that Cbl KO macrophages had a stronger phenotype for ubiquitination, signaling and gene expression changes over Cbl-b–/–. Our findings reproduce some of these phenotypes, however these are mild when compared with the phenotype of DKO macrophages, leading to the conclusion that either Cbl or Cbl-b protein is sufficient to regulate CSF-1-depndent macrophage growth and that their overlapping functions protect against sustained proliferation of myeloid cells, which is also consistent with the myeloproliferative phenotype observed in the DKO mice. Moreover, Cbl and Cbl-b were recently found to also constrain the growth of type 1 conventional dendritic cells (Xu et al., 2022). The 8 tyrosine residues on the intracellular domain of CSF-1R can contribute to activation of different signaling pathways (Pixley and Stanley, 2004) that may be influenced by the timing of ubiquitination. Specifically, Y559 recruitment of Cbl via SFK-mediated ubiquitination of CSF-1R is thought to be important for phosphorylation of other tyrosine residues on CSF-1R, while Y697 recruitment of Cbl via Grb2 is important for ERK signaling pathway activation and tyrosine phosphorylation (Pixley and Stanley, 2004; Stanley and Chitu, 2014). Our work suggests that Cbl and Cbl-b likely have overlapping, but different capacities for these two roles, with Cbl showing a slightly dominant effect on CSF-1R signaling, despite similar expression levels of these two genes (Figure 5H).
In summary, we find that Cbl and Cbl-b have overlapping functions in ligand-induced CSF-1R ubiquitination, endocytosis, transport into the lumen of macropinosomes. Future studies under more limiting growth factor conditions may help reveal more significant differences since Cbl/Cbl-b combined deletion is known to render cells more sensitive to lower levels of growth factors (Naramura et al. 2010). Together, these shape the signaling and transcriptional regulation of the CSF-1R and constrain macrophage proliferation. Thus, Cbl and Cbl-b play a critical role in controlling CSF-1R signaling and macrophage-related growth and likely contribute to the regulation of other myeloid growth factor receptors, potentially including the GM-CSF receptor (Xu et al., 2022).
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Reagents
Dulbecco’s Modified Eagle Medium (#SH30022, GE Healthcare Life Sciences), FBS (#SH30088, GE Healthcare Life Sciences) and L-cell supernatant were used for cell culture. Recombinant human CSF-1 (#:574806, BioLegend) was used for CSF-1R stimulation. Alamar blue reagent (#DAL1025, Thermo Fisher Scientific) was used for growth assay of macrophages. CSF-1R antibody (SC692, Santa Cruz Biotechnology, AFS98, eBioscience), pCSF-1R (Y721-mouse/Y723-human) antibody (49C10, Cell Signaling Technology), p-ERK antibody (9101, Cell Signaling Technology), pAKT antibody (4691, Cell Signaling Technology), p-Tyrosine antibody (P-Tyr-100, Cell Signaling Technology), goat anti-mouse Dylight 600, goat anti-rabbit Dylight 800, goat anti-Rat Dylight 594, goat anti-Rabbit Dylight 488 conjugated secondary antibodies (Thermo Fisher Scientific) were used for western blot or immunofluorescence. Texas-Red Dextran (70 kD) (#D1864, Thermo Fisher Scientific) was used to label macropinosomes. GeneJET RNA Purification Kit (K0732, Thermo Fisher Scientific) and Turbofree DNA free Kit (AM1907, Thermo Fisher Scientific) were used to purify total RNA for RNA sequencing. ToxinSensorTM Gel Clot Endotoxin Assay Kit (L00351, GenScript) was used to assess any LPS contamination of the media.
Generation of WT, Cbl –/–, Cbl-b –/–, and DKO macrophages
Bone marrow cells were isolated from mouse femurs of WT, Cbl–/–, Cbl-b–/–, and Cbl/Cbl-b-DKO mice and cultured in bone marrow medium, which consists of 20% FBS and 30% L-cell supernatant, a source of CSF-1 (Stanley et al., 1976; Waheed and Shadduck, 1979), and 50% Dulbecco’s Modified Growth Medium containing 4.5 g/L glucose, 110 mg/l sodium pyruvate, 584 mg/l L-glutamine, 1 IU/ml penicillin, and 100 μg/ml streptomycin (Naramura et al., 2010). Genotypes were validated by PCR on BMDM DNA using primers for Cbl: forward-TTG CAG GTC AGA TCA ATA GTG G, reverse-TGG GAG CCT AAT GTG TCT TGC and Cbl-b: forward-TCC TGT TGG GGA AAT ACT TTC ATA, reverse-TGC ATC CTG AAT AGC ATC AA (Supplemental Figure S1).
Growth assay of macrophages
Day 6 postisolation macrophages were differentiated in bone marrow medium, and their growth was monitored by counting five sites per sample in triplicates under microscope with DMEM+10% FBS containing 100 ng/mL CSF-1. The Alamar blue assay was used to measure the growth of wildtype and mutant macrophages between days 5−25 postisolation. Briefly, six replicate wells of day 5 macrophages were plated on nontissue culture−treated 96-well plates in DMEM containing 10% FBS with no CSF-1, 10 ng/ml CSF-1, or 100 ng/ml CSF-1. A total of 1,000 cells/well were originally plated, and cells were replated after reaching 90% confluence. The Alamar blue dye was applied to cells every other day from day 5 to 25. Alamar blue dye reaction with cells was done in a 37o C CO2 incubator for 40 min. The reaction product was measured using 550 nm excitation and 585 nm emission to calculate the cell number per well, and data were normalized to cell number on day 5. To assay growth from day 81 to day 94 of culture, macrophages were plated on 6-well plates with triplicate wells, and the cell number was recorded in 5 fields of view and plotted against days of cell culture.
Immunofluorescence
For immunofluorescence analysis of intracellular targets, cells were fixed with 4% (PFA) for 10 min, and permeabilized with 0.1% Triton X-100 at room temperature for 15 min for p-ERK staining or fixed and permeabilized with cold methanol at −20 °C for CSF-1R or phospho-tyrosine staining. For analysis of surface CSF-1R, cells were fixed with 2% PFA for 5 min and used without permeabilization. The samples were blocked with 2.5% BSA in PBS. The samples were incubated with primary antibodies for 1 h at room temperature or overnight at 4°C, washed and secondary antibody added for 1 h incubation at room temperature.
Western blot analysis
Macrophages were lysed in M-Per solution (Cat#78501 Thermo Fisher Scientific) at different times following CSF-1 stimulation, lysates were centrifuged for 10 min at 12,000 rpm at 4°C and cleared supernatants analyzed for total protein using the BCA reagent (#P123221, Thermo Fisher Scientific). 20-µg aliquots of total lysate protein were loaded on 8% SDS–PAGE gels and resolved at 150V for 1 h. Protein Transfer to nitrocellulose membrane was achieved at 300 amps for 30 min. The membrane was blocked in 5% BSA in PBST (PBS with 0.1% Tween 20) or TBST (20 mM Tris pH 7.5, 150 mM NaCl, 0.1% Tween 20) solution at room temperature for 30 min. The membranes were incubated with the primary antibody overnight at 4°C, followed by secondary antibody at room temperature for 1 h.
Microscopy and data analysis
Immunofluorescence images were acquired on a Leica CTR4000 inverted microscope equipped with an QICAM 16-bit color camera via Micromanager software and 60 X oil lens. Exposure time was optimized according to sample brightness and held constant for all the samples within a staining group. Imaging parameters were kept constant for each experimental condition. Cell Nucleus was masked with HCS (#H10325, Thermo Fisher Scientific), and Pholloidin-Dylight647 (#A22287, Thermo Fisher Scientific) labeling of actin cytoskeleton was used to mask cells. CellProfiler (Broad Institute) was used to measure the fluorescence intensity from individual cells over the region defined by the phalloidin stain and HSC.
Macropinosomes were labeled by incubating cells with Texas red-dextran (70 kD) or FITC-dextran (150 kD) and CSF-1. Sequential immunostaining of the CSF-1R was performed on a custom-built Till iMic microscope (FEI) using 60 X water objective, or on a High content microscope (ImageExpress Pro XLS, Molecular Devices) using 60 X air objective.
RNA Sequencing by Illumina
Bone marrow derived macrophages from WT, Cbl–/–, Cbl-b–/–, Cbl/Cbl-b-DKO mice were cultured in DMEMmedium supplemented with CSF-1 (100 ng/ml) until day 8 in 6-well nontissue-treated plates. Total RNA was purified from each sample and indexed libraries prepared using the Illumina Truseq kit. The samples were sequenced on a NextSeq500 using a high output Illumina chip with 150-nt single-end read lengths. Reads were filtered based on quality control set up and mapped to Mouse genome using the RNA-sequencing pipeline in CLCBio genomic workbench 9.0.1 (QIAGEN) and counts were normalized to calculate the expression level as RPKM (Reads Per Kilobase of transcript per Million reads mapped). Raw total exon counts were also used to do principal component analysis, MA plot of differentially expressed genes, gene ontology analysis, KEGG pathway analysis, and cancer gene analysis in macrophages across different genotypes in iDEP with cutoff values of FDR < 0.05 (Ge et al., 2018). All data are available via the NIH Gene Expression Omnibus at GSE130527.
Supplementary Material
Acknowledgments
This material is based on the work supported by the National Science Foundation, EPSCoR Cooperative Agreement #IIA-1355423, the South Dakota Research and Innovation Center, BioSNTR, and by the State of South Dakota BOR CRGP to ADH as well as by a Nebraska Research Initiative seed grant to HB. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. We also thank Shuangling Zhang for technical assistance. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number U01AI148153 and National Institute of General Medical Sciences of the National Institutes of Health under Award Number P20GM135008. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations used:
- BCR
B cell receptor
- BMDM
bone marrow derived macrophage
- BSA
bovine serum albumin
- CSF-1
colony stimulating factor 1
- CSF-1R
colony stimulating factor 1 receptor
- DKO
double knockout
- EGFR
epidermal growth factor receptor
- EIPA
5-(N-ethyl-N-isopropyl)-amiloride
- ESCRT
endosomal sorting complex required for transport
- FcγR
Fc-γ receptor
- FDR
false discovery rate
- GM-CSF
granulocyte macrophage colony stimulating factor
- GSEA
gene set enrichment analysis
- KEGG
kyoto encyclopedia of genes and genomes
- PBS
phosphate buffered saline
- PC
principle component
- PCR
polymerase chain reaction
- PFA
paraformaldehyde
- PI3K
phosphoinositide 3-kinases
- RTK
receptor tyrosine kinase
- TCR
T cell receptor
- TPM
transcripts per million
- VEGFR
vascular endothelial growth factor receptor
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E23-09-0345) on January 3, 2024.
REFERENCES
- Andoniou CE, Lill NL, Thien CB, Lupher ML, Ota S, Bowtell DD, Scaife RM, Langdon WY, Band H (2000). The Cbl proto-oncogene product negatively regulates the Src-family tyrosine kinase Fyn by enhancing its degradation. Mol Cell Biol 20, 851–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An W, Nadeau SA, Mohapatra BC, Feng D, Zutshi N, Storck MD, Arya P, Talmadge JE, Meza JL, Band V, Band H (2015). Loss of Cbl and Cbl-b ubiquitin ligases abrogates hematopoietic stem cell quiescence and sensitizes leukemic disease to chemotherapy. Oncotarget 6, 10498–10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An W, Mohapatra BC, Zutshi N, Bielecki TA, Goez BT, Luan H, Iseka F, Mushtaq I, Storck MD, Band V, Band H (2016). VAV1-Cre mediated hematopoietic deletion of CBL and CBL-B leads to JMML-like aggressive early-neonatal myeloproliferative disease. Oncotarget 7, 59006–59016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannarile MA, Weisser M, Jacob W, Jegg A-M, Ries CH, Rüttinger D (2017). Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J Immunother Cancer 5, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitu V, Stanley ER (2017). Regulation of Embryonic and Postnatal Development by the CSF-1 Receptor. Curr Top Dev Biol 123, 229–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L, Reddi AL, Ghosh A, Dimri M, Band H (2004). The Cbl family and other ubiquitin ligases: destructive forces in control of antigen receptor signaling. Immunity 21, 7–17. [DOI] [PubMed] [Google Scholar]
- Edwards DK, Watanabe-Smith K, Rofelty A, Damnernsawad A, Laderas T, Lamble A, Lind EF, Kaempf A, Mori M, Rosenberg M, et al. (2019). CSF1R inhibitors exhibit antitumor activity in acute myeloid leukemia by blocking paracrine signals from support cells. Blood 133, 588–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdreich-Epstein A, Liu M, Kant AM, Izadi KD, Nolta JA, Durden DL (1999). Cbl functions downstream of Src kinases in Fc gamma RI signaling in primary human macrophages. J Leukoc Biol 65, 523–534. [DOI] [PubMed] [Google Scholar]
- Ettenberg SA, Keane MM, Nau MM, Frankel M, Wang LM, Pierce JH, Lipkowitz S (1999). cbl-b inhibits epidermal growth factor receptor signaling. Oncogene 18, 1855–1866. [DOI] [PubMed] [Google Scholar]
- Ge SX, Son EW, Yao R (2018). iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics 19, 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AK, Reddi AL, Rao NL, Duan L, Band V, Band H (2004). Biochemical basis for the requirement of kinase activity for Cbl-dependent ubiquitinylation and degradation of a target tyrosine kinase. J Biol Chem 279, 36132–36141. [DOI] [PubMed] [Google Scholar]
- Goetz B, An W, Mohapatra B, Zutshi N, Iseka F, Storck MD, Meza J, Sheinin Y, Band V, Band H (2016). A novel CBL-Bflox/flox mouse model allows tissue-selective fully conditional CBL/CBL-B double-knockout: CD4-Cre mediated CBL/CBL-B deletion occurs in both T-cells and hematopoietic stem cells. Oncotarget 7, 51107–51123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Dikic I (2012). The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci 125, 265–275. [DOI] [PubMed] [Google Scholar]
- Hurley JH (2008). ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol 20, 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J, Kwa MQ, Cook AD, Hamilton JA, Scholz GM (2012). CSF-1 receptor signalling from endosomes mediates the sustained activation of Erk1/2 and Akt in macrophages. Cell Signal 24, 1753–1761. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD (2002). Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol 3, 893–905. [DOI] [PubMed] [Google Scholar]
- Katz M, Amit I, Yarden Y (2007). Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim Biophys Acta 1773, 1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Wang Y, Dominguez MG, Yeung YG, Murphy MA, Bowtell DD, Stanley ER (1999). The Cbl protooncoprotein stimulates CSF-1 receptor multiubiquitination and endocytosis, and attenuates macrophage proliferation. EMBO J 18, 3616–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh ML, Sakai DS, Flotho C, Kang M, Fliegauf M, Archambeault S, Mullighan CG, Chen L, Bergstraesser E, Bueso-Ramos CE, et al. (2009). Mutations in CBL occur frequently in juvenile myelomonocytic leukemia. Blood 114, 1859–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou J, Low-Nam ST, Kerkvliet JG, Hoppe AD (2014). Delivery of CSF-1R to the lumen of macropinosomes promotes its destruction in macrophages. J Cell Sci 127, 5228–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Hope LW, Brasch M, Reinhard C, Cohen SN (2003). TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc Natl Acad Sci USA 100, 7626–7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima H, Cazzolli H, Szpurka H, Dunbar A, Tiu R, Huh J, Muramatsu H, O’Keefe C, Hsi E, Paquette RL, et al. (2009). Mutations of e3 ubiquitin ligase cbl family members constitute a novel common pathogenic lesion in myeloid malignancies. J Clin Oncol 27, 6109–6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marois L, Vaillancourt M, Paré G, Gagné V, Fernandes MJG, Rollet-Labelle E, Naccache PH (2011). CIN85 modulates the down-regulation of Fc gammaRIIa expression and function by c-Cbl in a PKC-dependent manner in human neutrophils. J Biol Chem 286, 15073–15084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Hazeki K, Hazeki O, Katada T, Ui M (1996). Specific association of phosphatidylinositol 3-kinase with the protooncogene product Cbl in Fc gamma receptor signaling. FEBS Lett 382, 11–14. [DOI] [PubMed] [Google Scholar]
- Miyake S, Lupher ML, Andoniou CE, Lill NL, Ota S, Douillard P, Rao N, Band H (1997). The Cbl protooncogene product: from an enigmatic oncogene to center stage of signal transduction. Crit Rev Oncog 8, 189–218. [DOI] [PubMed] [Google Scholar]
- Mohapatra B, Ahmad G, Nadeau S, Zutshi N, An W, Scheffe S, Dong L, Feng D, Goetz B, Arya P, et al. (2013). Protein tyrosine kinase regulation by ubiquitination: critical roles of Cbl-family ubiquitin ligases. Biochim Biophys Acta 1833, 122–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau S, An W, Palermo N, Feng D, Ahmad G, Dong L, Borgstahl GE, Natarajan A, Naramura M, Band V, Band H (2012). Oncogenic Signaling by Leukemia-Associated Mutant Cbl Proteins. Biochem Anal Biochem Suppl 6, 7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramura M, Nandwani N, Gu H, Band V, Band H (2010). Rapidly fatal myeloproliferative disorders in mice with deletion of Casitas B-cell lymphoma (Cbl) and Cbl-b in hematopoietic stem cells. Proc Natl Acad Sci USA 107, 16274–16279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennock S, Wang Z (2008). A tale of two Cbls: interplay of c-Cbl and Cbl-b in epidermal growth factor receptor downregulation. Mol Cell Biol 28, 3020–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pixley FJ, Stanley ER (2004). CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol 14, 628–638. [DOI] [PubMed] [Google Scholar]
- Racoosin EL, Swanson JA (1993). Macropinosome maturation and fusion with tubular lysosomes in macrophages. J Cell Biol 121, 1011–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao N, Ghosh AK, Douillard P, Andoniou CE, Zhou P, Band H (2002a). An essential role of ubiquitination in Cbl-mediated negative regulation of the Src-family kinase Fyn. Signal Transduct 2, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao N, Miyake S, Reddi AL, Douillard P, Ghosh AK, Dodge IL, Zhou P, Fernandes ND, Band H (2002b). Negative regulation of Lck by Cbl ubiquitin ligase. Proc Natl Acad Sci USA 99, 3794–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge SA, Worwood M, Oscier D, Jacobs A, Padua RA (1990). FMS mutations in myelodysplastic, leukemic, and normal subjects. Proc Natl Acad Sci USA 87, 1377–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S, Tamura A, Honda H, Sakata-Yanagimoto M, Kumano K, et al. (2009). Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature 460, 904–908. [DOI] [PubMed] [Google Scholar]
- Sapi E, Kosinsky RL, Atkinson EJ, Doolittle ML, Zhang X, LeBrasseur NK, Pignolo RJ, Robbins PD, Niedernhofer LJ, Ikeno Y, et al. (2004). The role of CSF-1 in normal physiology of mammary gland and breast cancer: an update. Exp Biol Med (Maywood) 229, 1–11. [DOI] [PubMed] [Google Scholar]
- Saul D, Kosinsky RL, Atkinson EJ, Doolittle ML, Zhang X, LeBrasseur NK, Pignolo RJ, Robbins PD, Niedernhofer LJ, Ikeno Y, et al. (2022). A new gene set identifies senescent cells and predicts senescence-associated pathways across tissues. Nat Commun 13, 4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MHH, Dikic I (2005). The Cbl interactome and its functions. Nat Rev Mol Cell Biol 6, 907–918. [DOI] [PubMed] [Google Scholar]
- Stanley ER, Chitu V (2014). CSF-1 receptor signaling in myeloid cells. Cold Spring Harb Perspect Biol 6, a021857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley ER, Cifone M, Heard PM, Defendi V (1976). Factors regulating macrophage production and growth: identity of colony-stimulating factor and macrophage growth factor. J Exp Med 143, 631–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thien CB, Langdon WY (2001). Cbl: many adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell Biol 2, 294–307. [DOI] [PubMed] [Google Scholar]
- Voisinne G, García-Blesa A, Chaoui K, Fiore F, Bergot E, Girard L, Malissen M, Burlet-Schiltz O, Gonzalez de Peredo A, Malissen B, Roncagalli R. (2016). Co-recruitment analysis of the CBL and CBLB signalosomes in primary T cells identifies CD5 as a key regulator of TCR-induced ubiquitylation. Mol Syst Biol 12, 876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waheed A, Shadduck RK (1979). Purification and properties of L cell-derived colony-stimulating factor. J Lab Clin Med 94, 180–193. [PubMed] [Google Scholar]
- Xu F, Liu C, Dong Y, Wu W, Xu J, Yan Y, Shao Y, Hao C, Yang Y, Zhang J (2022). Ablation of Cbl-b and c-Cbl in dendritic cells causes spontaneous liver cirrhosis via altering multiple properties of CD103+ cDC1s. Cell Death Discov 8, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.