Abstract
Alkylpurine-DNA-N-glycosylase (APNG) null mice have been generated by homologous recombination in embryonic stem cells. The null status of the animals was confirmed at the mRNA level by reverse transcription-PCR and by the inability of cell extracts of tissues from the knockout (ko) animals to release 3-methyladenine (3-meA) or 7-methylguanine (7-meG) from 3H-methylated calf thymus DNA in vitro. Following treatment with DNA-methylating agents, increased persistence of 7-meG was found in liver sections of APNG ko mice in comparison with wild-type (wt) mice, demonstrating an in vivo phenotype for the APNG null animals. Unlike other null mutants of the base excision repair pathway, the APNG ko mice exhibit a very mild phenotype, show no outward abnormalities, are fertile, and have an apparently normal life span. Neither a difference in the number of leukocytes in peripheral blood nor a difference in the number of bone marrow polychromatic erythrocytes was found when ko and wt mice were exposed to methylating or chloroethylating agents. These agents also showed similar growth-inhibitory effects in primary embryonic fibroblasts isolated from ko and wt mice. However, treatment with methyl methanesulfonate resulted in three- to fourfold more hprt mutations in splenic T lymphocytes from APNG ko mice than in those from wt mice. These mutations were predominantly single-base-pair changes; in the ko mice, they consisted primarily of AT→TA and GC→TA transversions, which most likely are caused by 3-meA and 3- or 7-meG, respectively. These results clearly show an important role for APNG in attenuating the mutagenic effects of N-alkylpurines in vivo.
Alkylpurine-DNA-N-glycosylase (APNG) is one of a growing list of enzymes responsible for the recognition and excision of altered bases in the first step of the base excision repair pathway (49, 56). In the simplest form of base excision repair, the resulting abasic site is then repaired by the sequential action of an apurinic-apyrimidinic (AP) endonuclease that generates a single-strand break, the removal of the 5′-terminal deoxyribose phosphate residue, insertion of a single nucleotide by DNA polymerase β, and finally ligation of the repaired patch by DNA ligase I or XRCC1-DNA ligase III (55, 56).
Mammalian APNGs have been shown to be active against a wide range of modified bases in vitro, many structurally unrelated to 3-methyladenine (3-meA), the substrate after which the enzyme was first named (32). In particular, APNG appears to be the only glycosylase in mammalian cells that can release hypoxanthine from DNA, a promutagenic base resulting from the spontaneous deamination of adenine (22, 47). Likewise, the highly mutagenic adduct 1,N6-ethenoadenine (41), which is produced by metabolic products of the environmental hepatocarcinogens vinyl chloride and ethyl carbamate, is released by APNG (22, 48); indeed, according to one report, the recombinant human enzyme reacted 10- to 20-fold more efficiently with this adduct than with 3-meA in an in vitro assay (14). These results, together with the recent findings that 1,N6-ethenoadenine can also be formed endogenously by the interaction of lipid peroxidation products with DNA (12, 16) and that APNG is again the sole glycosylase in mammalian cells responsible for its removal (22), raise the question of the true physiological substrates of APNG.
The complexity of such a broad spectrum of activity and the profound differences in rate of repair of particular DNA lesions by numerous bacterial, yeast, and mammalian homologues of APNG, has complicated the task of defining specific biological roles for APNG in mammalian cells. Thus, there are conflicting reports as to the efficacy of mammalian APNG in protecting cells against the cytotoxic effects of bifunctional alkylating agents. Although increased levels of DNA repair activities, including APNG, were observed in nitrogen mustard-resistant B lymphocytes (20) and N-(2-chloroethyl)-N-nitrosourea (CNU)-resistant gliomas (34), overexpression of human APNG in Chinese hamster ovary cells afforded no protection against the cytotoxic effects of CNU or nitrogen mustards (5). Similarly, while the overexpression of the Escherichia coli tag gene, which codes for a 3-meA-DNA glycosylase with a narrow substrate specificity (1), reduced the toxic and mutagenic effects of methylating agents in Chinese hamster V79 cells (27, 28), no difference in the cytotoxicities of these or other alkylating agents was found in tag-transfected NIH 3T3 murine fibroblasts (23). Furthermore, while the expression of the Saccharomyces cerevisiae MAG gene in glycosylase-deficient S. cerevisiae cells or E. coli cells protected against methylating-agent and CNU cytotoxicity (9, 35), overexpression of human APNG in Chinese hamster ovary cells resulted in greater sensitivity to cell killing by methyl methanesulfonate (MMS) and an increased frequency of chromosomal aberrations (13). One explanation proposed for the latter result is imbalanced repair; that is, abasic sites resulting from the action of APNG, or the single-strand breaks resulting from the subsequent action of AP-endonuclease on these sites, may be ultimate genotoxic lesions responsible for the biological effects observed.
In order to determine the normal physiological role of APNG, its substrate specificity in vivo, and therefore the ability of APNG to modulate the biological effects of various alkylating agents and other genotoxins, we used the technique of gene targeting in murine embryonic stem (ES) cells to generate mice with a null mutation in the gene encoding APNG. Recently we published some aspects of the biochemical phenotype of our knockout (ko) mice (22), and those described for another APNG ko strain recently reported by Engelward et al. (18) appear to be similar. Here, we affirm that the APNG ko mice have a mild phenotype and an apparently normal life span, and that, far from being susceptible to the cytotoxic properties of 3-meA, which is known to block DNA replication by chain termination in E. coli (3, 31), they are virtually indistinguishable from wild-type (wt) controls in a number of different biological assays. Importantly, however, we have found an increase in the frequency of mutations at the hypoxanthine guanine phosphoribosyltransferase gene (hprt) in splenic T lymphocytes from APNG ko mice following treatment with MMS. On sequencing the mutants, we found that most of the single-base-pair changes were AT→TA transversions, implicating 3-meA as the likely promutagenic lesion.
MATERIALS AND METHODS
Gene targeting in ES cells.
Genomic clones containing the APNG gene were isolated from a 129sv DNA lambda library (λFIXII; Stratagene). The gene-targeting construct was based on plasmid pPNT-1 (a kind gift of V. Tybulewicz, Medical Research Council—National Institute for Medical Research, London, England), which contains both the neo and herpes simplex virus (HSV) tk genes under the control of the murine pgk-1 promoter. Initially a 3.4-kb NotI-XhoI fragment containing a sequence upstream of exon 1 was ligated 5′ of the neo cassette. The vector was completed by the ligation of a 0.9-kb BamHI-EcoRI fragment, containing an intron sequence between exons 2 and 3, into pPNT-1 between neo and HSV tk. The vector was linearized with NotI, and 107 ES cells (MESC 20, mouse strain 129/OLA) (6) were electroporated with 40 μg of vector at 240 V and 500 μF in ice-cold phosphate-buffered saline (PBS). ES cells colonies resistant to 250 μg of G418 (Gibco BRL) ml−1 and 2 μM ganciclovir (a kind gift of Syntex Pharmaceuticals Ltd., Maidenhead, England) were expanded, and DNA was isolated as described by Tybulewicz et al. (52) for analysis by Southern blotting. Targeted cells were microinjected into blastocysts from C57BL/6J mice and then implanted into the uteri of pseudopregnant (C57BL/6J × DBA2)F1 females. The resulting chimeras were mated with C57BL/6J females to produce mice heterozygous for the null mutation. These F1 mice were then crossed to obtain F2 offspring, which were wt, heterozygous for the targeted allele, and homozygous for the targeted allele, and these were used for the initial characterization of the phenotype (see Fig. 1 to 3). Heterozygotes were backcrossed to C57BL/6J mice for a further three generations, and the F5 heterozygotes were used to generate mice for the remainder of the experiments.
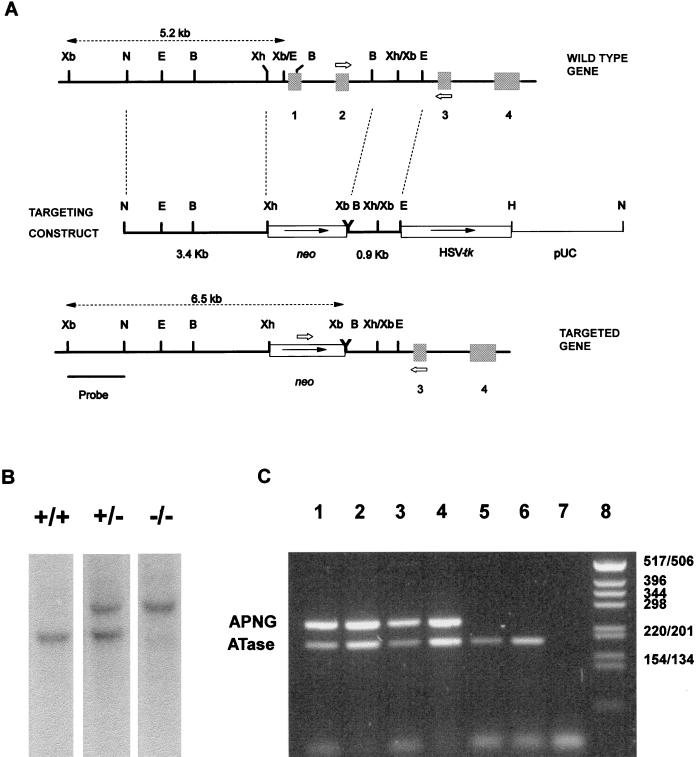
FIG. 1.
(A) Strategy for the disruption of the murine APNG gene. The genomic structures of the wt and mutated genes, together with the gene-targeting construct, linearized at a unique NotI site, are shown. For the targeting construct, a 3.4-kb NotI-XhoI genomic fragment was ligated upstream of a pgk-neo cassette and a 0.9-kb BamHI-EcoRI fragment was placed between the neo and pgk-HSV tk expression cassette. Upon homologous recombination a 2.5-kb fragment of the gene containing exons 1 and 2 is replaced by pgk-neo, and HSV tk is deleted. Identification of correctly targeted ES cell clones was achieved in the first instance by PCR using oligonucleotides complementary to sequences in exons 2 and 3 and to neo (open arrows), and then by Southern blotting following XbaI digestion of genomic DNA. By using a 1.4-kb XbaI-NotI fragment external to the targeted region, a 5.2-kb band indicates the presence of the wt gene, and a 6.5-kb band indicates the presence of the mutant gene. Restriction enzyme sites: B, BamHI; E, EcoRI; H, HindIII; N, NotI; Xb, XbaI; Xh, XhoI. (B) Southern blot analysis of XbaI-digested tail DNA from wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mutant mice, showing fragments of the expected sizes after hybridization to the probe. (C) Multiplex RT-PCR for APNG and ATase on RNA prepared from testes of wt mice (+/+; lanes 1 and 2) and mice heterozygous (+/−; lanes 3 and 4) or homozygous (−/−; lanes 5 and 6) for the mutant APNG gene. Lanes 1, 3, and 5, 1 μl of cDNA; lanes 2, 4, and 6, 2 μl of cDNA; lane 7, no cDNA; lane 8, DNA ladder.
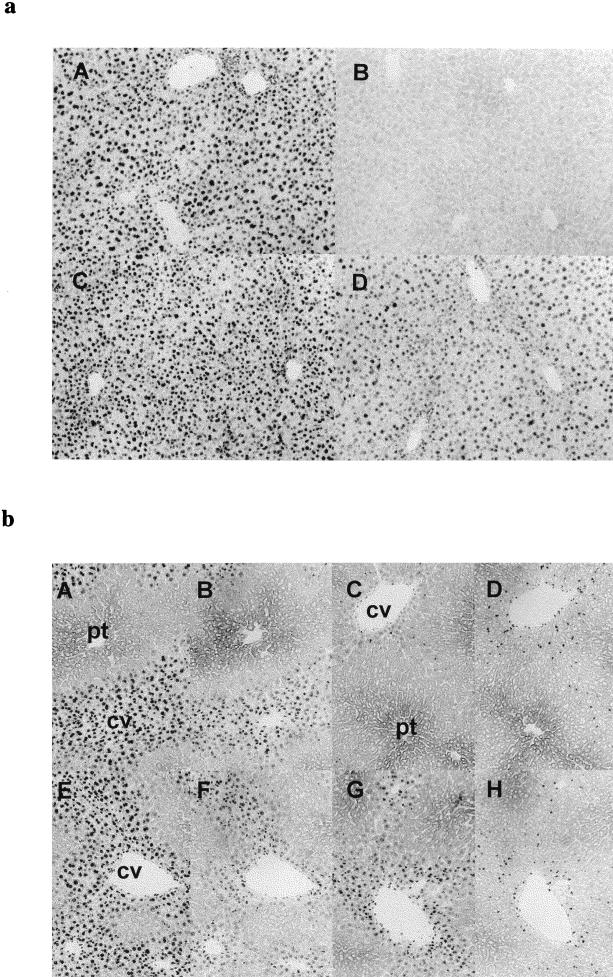
FIG. 3.
Persistence of 7-meG and O6-meG in liver sections from APNG wt and ko mice following treatment with 60 mg of MNU kg−1 (a) or 6 mg of NDMA kg−1 (b). (a) Panels A and B, 7-meG staining in wt liver 24 h and 7 days after MNU treatment, respectively. Panels C and D, 7-meG staining in ko liver 24 h and 7 days after MNU treatment, respectively. (b) Panels A through D, wt liver; panels E through H, ko liver. Panels A and E show 7-meG staining, and panels B and F show O6-meG staining, 24 h after NDMA treatment. Panels C and G show 7-meG staining, and panels D and H show O6-meG staining, 7 days after NDMA treatment. cv, central vein; pt, portal tract.
Multiplex reverse transcription-PCR (RT-PCR).
Total cellular RNA was prepared from the testes of F2 wt, heterozygous, and homozygous mutant mice by using RNAzol (Cinna/Biotecx Laboratories Inc.). For reverse transcription, 1 μg of random primers was incubated with 1.25 μg of the RNA at 65°C for 5 min and then placed in ice. Polymerization was carried out at 42°C for 1 h in a total volume of 25 μl of 50 mM Tris-HCl (pH 8.3)–75 mM KCl–3 mM MgCl2–10 mM dithiothreitol–8 mM deoxynucleoside triphosphates (dNTPs) containing 20 U of rRNAsin (Promega) and 200 U of Moloney murine leukemia virus reverse transcriptase (Promega). One and two microliters of each reaction mixture were then amplified by using primers specific to exon 2 (5′-GCG GAG TAT CTA CTT CTC CAG CCC-3′) and exon 3 (5′-CAT GAA CAT GCC ACG GTT CCG G-3′) of APNG in a total volume of 50 μl containing 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% Triton X-100, 1.5 mM MgCl2, 0.1 mg of bovine serum albumin (BSA) ml−1, 1 mM dNTPs, 0.1 pM each primer, and 0.625 U of Taq DNA polymerase (Promega). Primers to exons 4 and 5 of O6-alkylguanine-DNA alkyltransferase (ATase) were also included as a positive control. Cycle conditions were 94°C for 3 min, 60°C for 3 min, and 72°C for 3 min for 1 cycle, followed by 30 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 2 min. Ten microliters of each reaction product was then subjected to electrophoresis through 2% NuSieve agarose (Flowgen) in Tris-borate-EDTA buffer.
Assay of APNG activity.
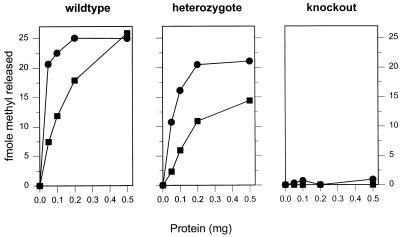
Sonicated tissue extracts were prepared as described previously (15), and protein concentration was determined by a Coomassie blue G-250 binding assay (4). For the determination of APNG activity, increasing amounts of extract or BSA were incubated with 3H-methylated calf thymus DNA substrate (18 Ci mmol−1) for 1 h at 37°C in a total volume of 100 μl containing 50 mM Tris-HCl (pH 8.3), 3 mM dithiothreitol, and 2 mM EDTA. DNA and protein were precipitated by the addition of 30 μl of precipitant (2 M NaCl, 1 mg of BSA ml−1, 0.5 mg of calf thymus DNA ml−1) followed by 250 μl of absolute ethanol and incubation in solid CO2 for at least 20 min. The precipitated material was removed by centrifugation (20 min at 15,000 × g), the supernatant was aspirated, dried in vacuo, and redissolved in 200 μl of buffer A (0.1 M triethylamine acetate–1% acetonitrile [pH 7.0]), and 3-meA and 7-methylguanine (7-meG) markers (approximately 1 nmol of each) were added. Chromatography was carried out with an analytical Hypersil 5-μm octyldecyl silane high-performance liquid chromatography column eluted isocratically with buffer A at a flow rate of 1.0 ml min−1. Under these conditions, 3-meA and 7-meG (Sigma) eluted at 9.6 and 12.3 min, respectively. Fractions were collected at 1-min intervals, and the amount of radioactivity in each was determined by scintillation counting. Radioactive peaks corresponding to the elution positions of the marker compounds were integrated, and the negative control (BSA) values (for 3-meA, 292 dpm; for 7-meG, 937 dpm) were deducted to account for thermal depurination. The values obtained (see Fig. 2) were plotted against the amount of protein used for extracts from wt, heterozygote, and null mice.
FIG. 2.
Release of 3-meA (•) and 7-meG (■) from a 3H-methylated calf thymus DNA substrate by testis extracts prepared from wt, APNG null heterozygote, and APNG ko mice.
Immunohistochemistry.
Groups of four APNG wt and ko mice for each time point were injected intraperitoneally (i.p.) with 60 mg of N-methyl-N-nitrosourea (MNU) kg of body weight−1 freshly dissolved in citrate buffer (pH 6.5) or with 6 mg of N-nitrosodimethylamine (NDMA) kg−1 in 0.9% NaCl. Animals were sacrificed by cervical dislocation up to 7 days later, and equivalent liver slices were fixed in 70% ethanol for at least 24 h.
An antiserum was raised against 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine (imidazole ring-opened 7-methylguanosine), conjugated to keyhole limpet hemocyanin (KLH) (Pierce), in prebled Half-Lop rabbits. Primary intramuscular injections of the conjugate (two injections, consisting of 0.5 mg of KLH conjugate in 50% TiterMax adjuvant; Stratech Scientific Ltd.) were followed by two booster injections, each of 0.5 mg of KLH conjugate in 20% TiterMax, at intervals of 4 weeks. Sera from test bleeds were tested by enzyme-linked immunosorbent assay (ELISA) for antigenicity against imidazole ring-opened 7-methylguanosine conjugated to BSA. Total immunoglobulin G (IgG) from the resulting antiserum was purified by protein G chromatography. Details of the anti-O6-methylguanine (O6-meG) antiserum have been published previously (46).
For the immunohistochemical detection of 7-meG, ethanol-fixed liver slices were embedded in paraffin, and 3-μm-thick sections were cut and mounted onto 3-aminopropyltriethoxysilane-coated slides. After dewaxing through xylene and absolute ethanol, the slides were washed in 40% ethanol and then incubated at 55°C for 20 min in preheated 40% ethanol containing 50 mM NaOH in order to ring-open 7-meG in the DNA. Slides were neutralized in 40% ethanol containing 5% acetic acid for 30 s, washed for 5 min in 40% ethanol, and rinsed thoroughly in Tris-buffered saline (pH 7.5) (TBS). Endogenous avidin and biotin binding sites were blocked with the ABC Blocking kit (Vector Laboratories Ltd.), and the slides were incubated with normal swine serum (1:20 dilution) in TBS to minimize nonspecific IgG binding. A 1:400 dilution of the anti-imidazole ring-opened 7-methylguanosine IgG in 10% normal mouse serum was then applied, and the slides were incubated at 4°C overnight. After equilibration to room temperature, the slides were washed in TBS and incubated with a biotinylated swine anti-rabbit antibody (1:4,000 dilution; Dako) for 30 min, and this was removed by a wash in TBS. Streptavidin complexed with biotinylated horseradish peroxidase was then applied (prepared as directed by the manufacturer, Dako), and the slides were incubated for a further 30 min. Finally, visualization of the antibody-antigen complexes was achieved by 3,3-diaminobenzidine (Sigma) staining for 5 min. Detection of O6-meG was performed as described above, except that the dilution of the primary antibody was 1:4,000.
Cell growth inhibition studies.
Embryos from wt and ko matings were isolated at 14 days postcoitum, and fibroblasts were brought into culture. For cell survival assays, primary embryonic fibroblasts (PEF) were plated in a 96-well tissue culture dish at a density of 2,000 cells per well and were incubated overnight at 37°C. Test compounds were prepared in dimethyl sulfoxide (DMSO) and diluted in cell culture medium just before application to the cells, in triplicate. Cells were grown for 6 days, after which time 50 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (3 mg ml−1 in PBS) was added and cells were incubated for a further 3 h at 37°C. The medium was aspirated, the purple formazan crystals formed in cells with intact mitochondria were dissolved in 200 μl of DMSO, and after 10 min, the optical densities at 540 and 690 nm of each well were read in a Titertek Multiskan ELISA plate reader. Cell growth was calculated as a percentage of the absorbance determined for the control (untreated) cells.
Number of leukocytes in peripheral blood.
Two groups of 17 mice (wt and ko) were prebled from the tail vein, and the number of leukocytes in 20 μl of blood was determined by using a Sysmex Microcellcounter (TOA Medical Electronics Co. Ltd., Kobe, Japan). The mice were then injected i.p. with 30 mg of 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU) kg−1 in 0.9% NaCl containing 3% ethanol, and the number of leukocytes was determined up to 25 days after treatment.
Micronucleus assay.
Groups of at least four female APNG wt and ko mice were injected i.p. with the agent of choice or with the appropriate control for that agent. Twenty-four hours later, the animals were sacrificed by cervical dislocation, one femur was removed, and the bone marrow cells were flushed out with fetal bovine serum (FBS). Cells were pelleted by centrifugation and resuspended in a minimum volume of serum, and duplicate slides were prepared. Following air drying and fixation in methanol, slides were stained with acridine orange as described by Tinwell and Ashby (51). Micronucleated polychromatic erythrocytes (PCE) were scored from at least 1,000 PCE per slide, and the ratio of PCE to normoerythrocytes was monitored to ensure that scoring was carried out at subtoxic doses.
Mutational analysis of splenic T lymphocytes.
The culture medium used for priming and cloning of T lymphocytes consisted of RPMI 1640 with 25 mM HEPES, 2 mg of NaHCO3 ml−1, and 2 mM l-glutamine, supplemented with 30% of the serum-free lymphocyte medium AIM-V (Gibco BRL), 10% lymphokine-activated killer cell supernatant (50), 15% heat-inactivated FBS (Fetal Clone I; HyClone), 50 μM 2-mercaptoethanol, 1 mM sodium pyruvate, 4 mM l-Glutamax (Gibco BRL), and antibiotics (100 U of penicillin ml−1 and 100 μg of streptomycin sulfate ml−1; Gibco BRL).
Isolation and priming of T lymphocytes.
T lymphocytes were isolated from the spleen by passing the spleen through a sterile 100-μm nylon mesh (Falcon) and were frozen in RPMI medium supplemented with 10% DMSO and 40% FBS by using a programmable cell freezer (Planar Products Ltd.). Frozen cells were thawed at 37°C and immediately stored on ice. Subsequently, 10 ml of RPMI supplemented with 40% FBS was slowly added to the cells. Cells were primed in 15 ml of culture medium supplemented with 4 μg of concanavalin A (ConA; Pharmacia) ml−1 for 44 h at 37°C under a 5% CO2 atmosphere.
Selection of 6-TG-resistant mutants.
Stimulated T lymphocytes were counted and reseeded in culture medium containing 1 μg of ConA ml−1 at a density of 6 cells per well in three round-bottom 96-well microtiter plates. These plates also contained 5 × 103 Sp2/0 feeder cells (mouse lympoblastoid cells irradiated with 30 Gy of X rays) per well. hprt-deficient mutants were selected in culture medium containing 6-thioguanine (6-TG; 2.5 μg ml−1) and ConA (1 μg ml−1) at a cell density of 2 × 105 cells, together with 5 × 103 feeder cells per well (seven plates; 672 wells). After incubation for 6 to 8 days at 37°C under a 5% CO2 atmosphere, plates were scored for colony growth by using a stage-inverted microscope. Cloning efficiencies and mutant frequencies were calculated as described elsewhere (50).
Molecular characterization of 6-TG-resistant clones.
6-TG-resistant clones were selected and diluted 1:3 in culture medium containing 6-TG (2.5 μg ml−1). After 3 days of culture, the cells were used for isolation of total cytoplasmic RNA as previously described (53). One-third of total cytoplasmic RNA was subjected to hprt cDNA synthesis essentially according to the method of Jansen et al. (24), except that 20 pmol of an oligonucleotide primer specific for mouse hprt mRNA was used (5′-CGACGTTGTAAAACGACGGCCAGTGCAGATTCAACTTGCGCTC-3′). Three microliters of hprt cDNA was amplified in vitro by PCR in a reaction mixture consisting of 10 μl of 10× Taq buffer (100 mM Tris-HCl [pH 8.3], 15 mM MgCl2, 500 mM KCl, 2 mg of BSA ml−1, 10 μl of dNTPs (2.5 mM dATP, 2.5 mM dCTP, 2.5 mM dGTP, 2.5 mM dTTP), 1 μl of the mouse hprt-specific primer (20 pmol μl−1), 1 μl of a 5′-biotinylated, hprt-specific oligonucleotide (5′-GGCTTCCTCCTCAGACCGCT-3′), 1 U of Taq polymerase (Amplitaq; Cetus), and 74.8 μl of sterile water. The PCR was performed in a Perkin-Elmer thermocycler for 35 cycles of 1 min at 93°C, 1 min at 55°C, and 3 min at 72°C. Subsequently, 5′-biotinylated PCR products were isolated from NuSieve agarose after electrophoresis, bound to Dynabeads coated with streptavidin, and subjected to DNA sequence analysis according to the work of Rossi et al. (42) by using sequencing primers for mouse hprt cDNA.
RESULTS
APNG ko mice are viable.
APNG is active against a range of alkylpurines and deaminated adenine as well as, potentially, oxidatively damaged purines in DNA. In order to examine the role of these lesions and of APNG in cellular responses to genotoxic agents, exons 1 and 2 of the murine APNG gene were deleted by homologous recombination in murine ES cells (Fig. 1A). The targeting vector was designed to disrupt the expression of APNG while leaving intact DNase-hypersensitive sites both 5′ to and internal to the APNG gene, which may be involved in the regulation of other genes in the locus where the APNG gene is situated (26). Three germ line chimeras were obtained, and crossing of the resultant APNG heterozygotes yielded homozygous null animals at the expected Mendelian ratio. Genotyping was carried out by PCR or Southern blotting (Fig. 1B). Further confirmation of the null status of the animals was obtained by multiplex RT-PCR (Fig. 1C). When APNG-specific primers were used, a PCR product of 240 bp was detected only when RNA from wt mice and APNG heterozygotes was used. No PCR product was obtained when RNA from APNG homozygous nulls was used. Multiplex RT-PCR with ATase-specific primers yielded a PCR product of 187 bp with RNA from all three genotypes.
APNG ko mice lack 3-meA and 7-meG glycosylase activity.
APNG activity in wt, heterozygous, and null mice was determined by an in vitro assay for the release of 7-meG and 3-meA from 3H-methylated substrate DNA. Crude cell extracts of testes from wt and heterozygous animals released both adducts; the activity in the heterozygote extracts was somewhat lower than that in the wt extracts. Essentially no release was seen with extracts from homozygous null testes (Fig. 2).
Increased persistence of 7-meG, but not of O6-meG, can be observed in the livers of APNG ko mice.
Since the conditions of the in vitro assay might not allow the detection of other repair functions acting on the same lesions that could compensate for the absence of APNG in vivo, we examined the persistence of 7-meG up to 7 days after administration of MNU or NDMA, using immunohistochemical techniques to probe ethanol-fixed sections of liver. Thus, while similar staining patterns for 7-meG were observed in liver sections from wt and APNG ko mice 24 h after treatment with MNU (Fig. 3a, panels A and C), differential staining was observed after 7 days: while the lesion had been essentially eliminated from wt hepatocyte nuclei, hepatocytes in the APNG ko liver still stained strongly for 7-meG (Fig. 3a, panels B and D). The high level of DNA damage in hepatocytes around the central vein, in contrast to those cells proximal to the portal tract, following NDMA treatment (19) enabled us to probe these sections additionally, with an antibody against O6-meG, a quantitatively minor lesion induced in nucleic acids by this agent (39). Thus, while similar intensities of 7-meG staining were obtained as for the MNU-treated animals after 24 h and 7 days (Fig. 3b; compare panels A and E with panels C and G), no differential pattern of staining was observed for O6-meG (Fig. 3b; compare panels B and F with panels D and H). These results indicate that the levels of O6-meG are similar in the liver cells of wt and APNG ko mice and hence that the differences observed for 7-meG are not a consequence of differences in drug dosage. The results also indicate that the activity of ATase, the principal repair mechanism for O6-meG lesions, is not changed by the deletion of APNG, and they support the in vitro observation that APNG is the principal mechanism of repair for 7-meG lesions.
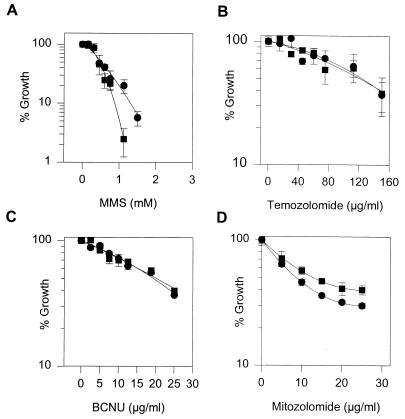
APNG null PEF do not show increased sensitivity to the cytotoxic effects of temozolomide or CNUs.
In order to examine the consequences of APNG deletion on the cytotoxic effects of alkylating agents, we established PEF lines from wt and ko mice and exposed them to increasing doses of MMS and the antitumor methylating agent temozolomide, as well as the chloroethylating agents mitozolomide and BCNU. Deletion of APNG from these cells has very little impact on the growth-inhibitory effects of these compounds (Fig. 4). Only with MMS were the ko PEF routinely more sensitive than the wt cells, and then only at the higher doses. This enhanced cell killing is most likely due to the nonrepair of 3-meA in the absence of significant levels of cytotoxic O6-meG formed by MMS, in contrast to temozolomide, where no differential cytotoxicity was observed (Fig. 4A and B). For the CNUs and temozolomide, the O6-alkylguanine adduct is thought to be the principal cytotoxic lesion (2, 10), and given the limitations of the assay, the results suggest that in PEF, the action of APNG does not substantially attenuate the toxic effects of these compounds (Fig. 4C and D).
FIG. 4.
Cell survival of PEF derived from wt (•) and APNG ko (■) mice after treatment with MMS, temozolomide, BCNU, or mitozolomide.
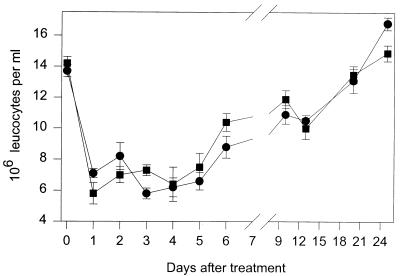
APNG deletion has no deleterious effect on the number of leukocytes following BCNU treatment.
To study the cytotoxicity of CNUs in APNG ko mice in vivo, we measured the number of leukocytes in the peripheral blood of wt and APNG ko mice following treatment with a single i.p. injection of BCNU (30 mg kg−1). This compound has been studied extensively with respect to its hematopoietic toxicity and was thus considered ideal for this study (10). However, as Fig. 5 shows, there was no significant difference in the cytotoxicity of BCNU between the two strains of mice. In both, the number of leukocytes reached a nadir at 3 days after treatment, and both recovered to pretreatment levels at the same rate. A preliminary investigation into the effect of BCNU on hematopoietic progenitors (granulocyte-macrophage colony-forming cells, and CFU spleen assays) has revealed that the deletion of APNG has no deleterious effect on the toxicity of BCNU on these cells (data not shown).
FIG. 5.
Numbers of leukocytes in peripheral blood from wt (•) and APNG ko (■) mice following treatment with 30 mg of BCNU/kg. Data are presented as means ± standard errors for groups of 17 mice of each strain.
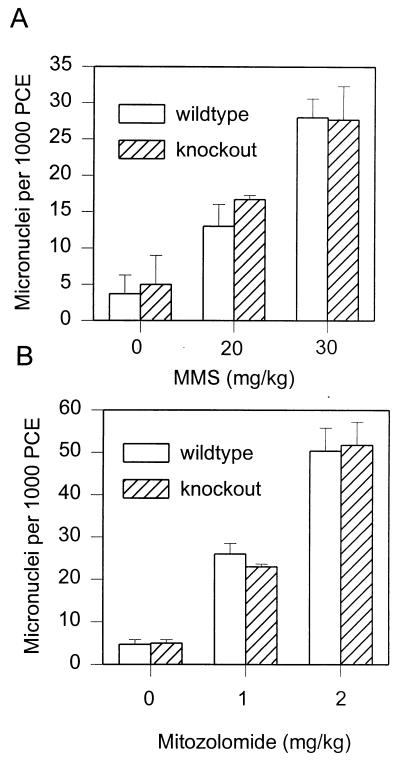
Deletion of APNG does not increase the clastogenicity of MMS or mitozolomide in bone marrow PCE.
The dose response for micronucleus induction in bone marrow PCE was unchanged in wt and ko mice 24 h after administration of MMS or mitozolomide (Fig. 6). Thus, the increased persistence of N-alkylpurines in the ko mice does not alter the clastogenicity of alkylating agents in this tissue compared with that in wt mice.
FIG. 6.
Induction of micronuclei in PCE of wt and APNG ko mice following treatment with increasing amounts of MMS or mitozolomide. Data are presented as means ± standard errors for groups of at least four animals per point.
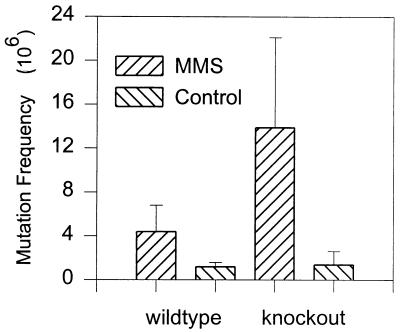
Increased frequency of MMS-induced hprt gene mutations in APNG ko mice.
To study the effect of APNG deficiency on methylating-agent-induced mutagenesis, wt and APNG ko mice were given a single dose of MMS (150 mg kg−1) and hprt mutant frequencies were determined in splenic T lymphocytes. While the spontaneous, background levels of mutation at the hprt locus were essentially the same in cells isolated from the ko mice and those from the wt mice, MMS induced greater than threefold more hprt mutants in APNG ko mice than in wt mice (Fig. 7). This increase was highly significant (P < 0.009 by Student’s t test). These results show that at the hprt locus, MMS induced mutagenic DNA lesions that are substrates for APNG-mediated repair and also that the APNG null background had no effect on the spontaneous-mutation frequency.
FIG. 7.
Induction of hprt mutations in splenic T lymphocytes from wt and APNG ko mice after treatment with 150 mg of MMS/kg. Control groups contained four animals, and treated groups contained a minimum of eight animals per group (P < 0.009 for treated wt mice versus treated ko mice).
To determine the molecular nature of MMS-induced mutations, 59 hprt mutant T lymphocytes from five APNG ko mice and 28 hprt mutants from six wt mice were analyzed by RT-PCR and DNA sequencing (Tables 1 and 2). In the ko mice, 39 of 59 hprt mutants (66%) gave a PCR product representing hprt cDNA. Seventy-seven percent of these mutants showed a single-base change in the coding region, mostly AT→TA transversions (47%). GC→TA transversions were also found frequently (27%), whereas transition-type mutations were present in only 20% of the mutants containing a single-base-pair change. Most of the remaining hprt mutants gave PCR products which are probably the result of mis-splicing of the hprt message. Most of the hprt mutants in the ko mice represent MMS-induced events, since the mean frequency of hprt mutants among MMS-exposed animals is 10-fold higher than the spontaneous background level.
TABLE 1.
Mutational spectrum of hprt gene mutations occurring in splenic T lymphocytes of APNG ko mice exposed in vivo to MMS (150 mg/kg)
| Positiona (mutation) | Target sequence,b 5′→3′ | Amino acid change | No. of mutants in animal:
|
||||
|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | |||
| Transversions | |||||||
| 1 (AT→TA) | GTC (A) TGC | Met→Leu | 0 | 1 | 0 | 0 | 0 |
| 62 (AT→TA) | ATT (T) GTT | Leu→stop | 3 | 0 | 0 | 0 | 0 |
| 133 (AT→TA) | GAC (A) GGA | Arg→Trp | 0 | 0 | 0 | 1 | 0 |
| 466 (AT→TA) | CCC (A) AAA | Lys→stop | 0 | 1 | 0 | 0 | 0 |
| 473 (AT→TA) | TGG (T) TAA | Val→Asp | 0 | 0 | 2 | 0 | 0 |
| 499 (AT→TA) | AAA (A) GGA | Arg→Trp | 0 | 1 | 0 | 0 | 0 |
| 556 (AT→TA) | GAC (A) AGT | Lys→stop | 0 | 1 | 0 | 0 | 0 |
| 595 (AT→TA) | TAC (T) TCA | Phe→Ile | 0 | 1 | 0 | 0 | 0 |
| 623 (AT→TA) | TCA (T) TAG | Ile→Asn | 0 | 0 | 0 | 1 | 0 |
| 643 (AT→TA) | GCC (A) AAT | Lys→stop | 0 | 0 | 0 | 0 | 1 |
| 655 (AT→TA) | GCC (T) AAG | Stop→Lys | 0 | 0 | 1 | 0 | 0 |
| 116 (AT→CG) | CTC (A) TGG | His→Pro | 0 | 2 | 0 | 0 | 0 |
| 40 (GC→TA) | GAT (G) AAC | Glu→stop | 0 | 0 | 2 | 0 | 0 |
| 97 (GC→TA) | TTG (G) AAA | Glu→stop | 0 | 0 | 0 | 1 | 0 |
| 118 (GC→TA) | CAT (G) GAC | Gly→stop | 0 | 1 | 0 | 0 | 0 |
| 419 (GC→TA) | CTG (G) TAA | Gly→Val | 0 | 0 | 1 | 0 | 0 |
| 580 (GC→TA) | CTT (G) ACT | Asp→Tyr | 2 | 0 | 0 | 0 | 0 |
| 626 (GC→TA) | TTA (G) TGA | Ser→Ile | 0 | 1 | 0 | 0 | 0 |
| Transitions | |||||||
| 491 (AT→GC) | TGC (T) GGT | Leu→Pro | 0 | 0 | 1 | 0 | 0 |
| 533 (AT→GC) | ACT (T) TGT | Phe→Ser | 0 | 0 | 0 | 1 | 0 |
| 655 (AT→GC) | GCC (T) AAG | Stop→Gln | 0 | 0 | 1 | 0 | 0 |
| 145 (GC→AT) | AGA (C) TTG | Leu→Phe | 0 | 0 | 1 | 0 | 0 |
| 551 (GC→AT) | TTC (C) AGA | Pro→Leu | 2 | 0 | 0 | 0 | 0 |
| Mis-splicing | Exon 3 | 0 | 1 | 0 | 0 | 1 | |
| Exon 7 | 0 | 1 | 0 | 0 | 0 | ||
| Exon 8 | 0 | 0 | 0 | 1 | 0 | ||
| Exon 6c | 0 | 0 | 0 | 0 | 1 | ||
| Double molecules | 0 | 3 | 0 | 0 | 0 | ||
| 636 (AT→GC + frameshift [−1]) | TGG (AA) AAG | 1 | 0 | 0 | 0 | 0 | |
Position 1 is the first base of the start codon in the hprt coding sequence.
Nontranscribed strand. Bases in parentheses are sites where MMS-induced mutations were found.
Cryptic splice.
TABLE 2.
Mutational spectrum of hprt gene mutations occurring in splenic T lymphocytes of wt mice exposed in vivo to MMS (150 mg/kg)
| Positiona (mutation) | Target sequence,b 5′→3′ | Amino acid change | No. of mutants in animal:
|
|||||
|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | |||
| Transversions | ||||||||
| 1 (AT→TA) | GTC (A) TGC | Met→Leu | 0 | 0 | 1 | 0 | 0 | 0 |
| 203 (AT→TA) | TGC (T) CAA | Leu→His | 0 | 0 | 0 | 4 | 0 | 0 |
| 40 (GC→TA) | GAT (G) AAC | Glu→stop | 0 | 0 | 0 | 0 | 0 | 1 |
| Transitions | ||||||||
| 1 (AT→GC) | GTC (A) TGC | Met→Val | 0 | 0 | 2 | 0 | 1 | 0 |
| 47 (GC→AT) | CAG (G) TTA | Gly→Asp | 1 | 0 | 0 | 0 | 0 | 0 |
| 551 (GC→AT) | TTC (C) AGA | Pro→Leu | 0 | 0 | 0 | 0 | 1 | 0 |
| Cryptic splices | Exon 9 | 0 | 0 | 0 | 0 | 0 | 5 | |
| Exon 6 | 0 | 2 | 0 | 0 | 0 | 0 | ||
| Additions | ||||||||
| 131 (triplet) | +TGG | 0 | 0 | 1 | 0 | 0 | 0 | |
| 397/398 (+G) | ATT (G) TTG | +1 Frameshift | 0 | 0 | 1 | 0 | 0 | 0 |
Position 1 is the first base of the start codon in the hprt coding sequence.
Nontranscribed strand. Bases in parentheses are sites where MMS-induced mutations were found.
In wt mice, 20 of 28 hprt mutants (71%) yielded PCR products following RT-PCR of hprt cDNA. Fifty-five percent of these mutants contained a single-base-pair change in the coding region, but unlike the results obtained for the ko mice, the proportion of transversion-type mutations was similar (55%) to that of transition-type mutations (45%) (Table 2), although these figures are based on a limited number of samples. Since the frequency of hprt mutants among MMS-exposed wt mice is only threefold higher than the spontaneous background level, at least one-third of the mutants are due to spontaneously occurring events. Insufficient numbers of mutants from untreated animals were obtained to allow for analysis of the spontaneous background in a similar way.
DISCUSSION
Genotoxic agents and endogenous cellular processes can result in the production of a great many different types of lesions in DNA, and usually a single agent can generate a wide spectrum of damage. A variety of DNA repair processes have evolved in living organisms, presumably to protect cells against the potential adverse biological effects of such damage, and the specificity of these processes is strongly suggestive of the likely genotoxic effects of the substrate lesions. However, the identification of the lesion or lesions that are important in the carcinogenic effects of environmental or endogenous agents or in the cytotoxic effects of antitumor agents is complicated by the number of different lesions present. For alkylating agents, O6-alkylguanine (O6-alkG) is known to be the principal cytotoxic, clastogenic, and promutagenic lesion formed, and a specific DNA repair protein, ATase, has evolved to minimize the consequences of cellular exposure to this lesion (49, 56). Less is known about the genotoxic potency of the N-alkylpurines, the main products of many methylating and chloroethylating agents reacting with DNA (39), which are repaired by base excision repair. The generation of APNG ko mice, as described here and elsewhere (18), should, for the first time, allow an analysis of the deleterious properties of N-alkylpurines under in vivo conditions.
3-meA is a cytotoxic lesion in bacteria and in mammalian cells, protruding into the minor groove and blocking DNA replication by causing premature chain termination (3, 31). Evidence for the toxicity of 3-meA in mammalian cells has come from transfection studies and from the deletion of APNG in murine cells by gene targeting (18, 22, 27). The fact that the APNG ko mice are viable and, to date, show no significant detrimental effects compared to their wt counterparts seems to suggest that 3-meA either (i) is formed spontaneously only at a low frequency, (ii) is cleared rapidly from the cell by spontaneous depurination (33), or (iii) is efficiently removed by another repair mechanism in the absence of APNG. Although at this time we cannot rule out the last possibility, we were unable to detect release of 3-meA by extracts from APNG ko testes in our in vitro assay (Fig. 2).
The lack of any substantial increase in sensitivity to the cytotoxic effects of methylating agents and CNUs in PEF is apparently at odds with the results of Engelward et al. (17), who found an increase in the sensitivity of APNG null ES cells to a variety of alkylating agents. However, ES cells, in contrast to PEF, are known to contain high levels of p53 and to be extremely sensitive to DNA-damaging agents (44). This is borne out in the higher alkylating-agent concentrations and UV doses used by these authors in PEF to elicit a response similar to that in ES cells (17, 18). Furthermore, as O6-alkG is likely to be the principal determinant of an agent’s cytotoxicity, it is interesting that only with agents that did not give rise to significant quantities of O6-alkG [i.e., MMS and MeOSO2(CH2)2-lexitropsin] was a difference between the wt and ko PEF observed (Fig. 4A) (18). Therefore, it is likely that in vivo, 3-meA does not normally present a cytotoxic danger to the cell due to its relatively short half-life (33).
No evidence for a protective effect of APNG against the cytotoxic effects of CNUs was observed in our cell culture studies or following administration of BCNU in vivo (Fig. 4 and 5). Again, it is likely that DNA interstrand cross-links which form from the initial O6-alkG monoadducts are the main toxic lesions (2). However, the studies presented here would suggest that murine APNG, unlike its S. cerevisiae counterpart (when overexpressed in prokaryotes and lower eukaryotes [35]), does not play a role in reducing the toxic effects of these agents, and further investigation into the ability of cell extracts prepared from wt and APNG ko mice to release 7-chloroethylguanine and 7-hydroxyethylguanine is required. Again, this result is in contrast to that reported for ES cells (17). Similarly, we could find no difference in cell growth after treatment with mitomycin C, a compound not known to produce DNA lesions that are APNG substrates, in contrast to results reported for ES cells (data not shown and reference 17).
To examine whether or not increased chromosome damage in the APNG ko mice would result from treatment with MMS or a CNU analogue, mitozolomide, we measured the induction of micronuclei in bone marrow PCE. The results indicated no increased induction of micronuclei in the PCE of the ko mice compared with the wt mice. The results suggest that, in this tissue, the N-alkylpurines are not important clastogenic lesions and that the observed clastogenic effects are mediated through O6-alkG lesions. The role of the latter has been shown in studies with ATase, which have shown a protective effect of the overexpression of this repair protein on clastogenic events in Chinese hamster cells (54) and in transduced hematopoietic progenitors (11).
A small but highly significant difference between ko and wt mice was found in hprt gene mutations in splenic T lymphocytes exposed to MMS. The finding that MMS was at least threefold more mutagenic in APNG ko mice than in wt mice suggests that MMS induces mutagenic DNA lesions that are normally substrates for APNG. 3-meA is a good candidate for such a lesion, since (i) this DNA adduct is one of the major reaction products formed in nucleic acids following exposure to MMS (39), (ii) 3-meA is a substrate for APNG-mediated repair in eukaryotes (49), and (iii) the spectrum of MMS-induced hprt gene mutations in APNG ko splenocytes results predominantly in AT→TA transversions. The last point is in contrast to the results of Klungland et al. (29), who found that in both an E. coli tag-expressing Chinese hamster cell line and the vector-only-transfected control cells, GC→AT transitions in the hprt gene predominated after MMS or ethyl methanesulfonate treatment. Similar results have been found in Chinese hamster ovary cells (40). Mutations at AT base pairs were found at very low frequencies, and it was concluded that 3-meA was not directly mutagenic. However, the cell lines used in these studies lack ATase activity. This implies that the highly mutagenic lesion O6-meG, which gives rise to GC→AT transitions (45), is not repaired and therefore dominated the induction of mutations. Furthermore, expression of the tag gene in these cells did reduce the mutagenicity of MMS by a factor of 2, implicating 3-meA as the likely mutagenic lesion (28). Furthermore, a role for 3-ethyladenine in the induction of mutations by ethylating agents has also been proposed by Op het Veld et al. (40). It is likely that 3-meA induces AT→TA transversions by mispairing with adenine during replication. Alternatively, MMS-induced AT→TA transversions may be due to preferential insertion of adenines during DNA replication across abasic sites formed by unstable 3-meA: the half life of 3-meA in DNA under in vitro conditions of pH 7.0 and 37°C is 24 h (33). Preferential incorporation of adenine at abasic sites during DNA replication was one explanation for the high proportion of AT→TA transversions in the spectrum of MMS-induced vermilion mutations in naturally repair-deficient, postmeiotic male germ cells of Drosophila melanogaster (38). It should be noted, however, that in mammalian cells, mostly insertions of dGMP (7), dTMP (25), or nonspecifically, each of the four deoxyribonucleoside monophosphates (8) are found opposite abasic sites or chemical analogues in single-stranded or double-stranded DNA.
N-methylated guanines, which make up more than 85% of the DNA adducts formed by MMS in vivo (39), may also be candidates for mutagenic lesions that are substrates for APNG-mediated repair, albeit to a lesser extent than 3-meA. Although it represents only about 0.7% of the methylated bases formed by MMS in vivo (39), 3-methylguanine (3-meG) must be a good candidate for the increased frequency of GC→TA transversions observed in the ko cells. As 7-meG makes up the majority of the N-methylated guanines formed by MMS and has a reported half-life of 72 h in vivo (33) (Fig. 3), it seems unlikely that the deletion of APNG would have much effect on any deleterious effect of this lesion, unless it was targeted to active genes. Indeed, the deletion of APNG may have a protective effect by reducing the number of abasic sites produced upon release of 7-meG. Furthermore, the facts that no miscoding properties have been reported for 7-meG and that the E. coli tag gene product specifically releases 3-meA and 3-meG (1) suggest that the latter is a biologically relevant lesion.
However, 3-meA and N-methylated guanines are only weakly mutagenic compared with O6-meG and O4-methylthymine, which are formed at very low frequencies following the exposure of DNA to MMS and give rise to GC→AT and AT→GC transitions, respectively (45). Since both O6-meG and O4-methylthymine are found at very low frequencies in the spectra of MMS-induced mutations in splenocytes of both wt and ko mice, it can be concluded that these DNA lesions are efficiently repaired in these cells. Therefore, MMS is only weakly mutagenic in wt mice, since both O- and N-methylated bases are efficiently repaired before mutation fixation can occur.
The present study has shown that APNG protects mammalian cells in vivo against the mutagenic effects of MMS, which are possibly due to 3-meA and 3-meG or 7-meG, and in this respect, it would be interesting to know if APNG ko mice are also more susceptible to MMS-induced carcinogenesis than wt mice. However, following alkylating-agent exposure, an effect of APNG deficiency on other biological end points was not apparent. Therefore, these results suggest that those lesions that are substrates for APNG are not likely to contribute substantially to the cytotoxicity of the alkylating antitumor agents but might be a factor in their long-term carcinogenic effects.
If the potentially harmful effects of the N-3-methylated purines are normally attenuated by the lability of their N-glycosidic bonds, why have mammalian cells evolved APNG? One explanation may be that their main function is to reduce the cellular burden of the highly promutagenic lesion 1,N6-ethenoadenine (41). Supporting this is the recent finding that a separate glycosylase exists in mammalian cells to remove the equally promutagenic 3,N4-ethenocytosine (36), although it is not known whether this glycosylase can, additionally, remove other damaged bases (21). These adducts were first described in the RNA of rats exposed to vinyl chloride (30); however, they have recently been found in liver DNA from untreated rodents and humans (37), possibly arising from endogenously formed lipid peroxidation (12, 16). An additional activity of APNG in vitro is the removal from DNA of promutagenic deoxyinosine residues (22, 47), which arise by the deamination of adenine, either spontaneously or through the interaction of reactive nitrogen species with adenine (43). However, to date we have not observed any significant increase in the spontaneous mutation rate at the hprt locus of the APNG ko mice (Fig. 7) or any marked increase in spontaneous cancer incidence over a 24-month period with our ko mice (unpublished observations). Therefore, we are currently targeting our research to these areas in order to address this apparent enigma.
ACKNOWLEDGMENTS
We thank D. G. Brown for the gift of the MESC 20 ES cells, N. Chinnasamy for help with the bone marrow micronucleus assay, and L. H. F. Mullenders for suggestions and helpful comments on the manuscript.
This work was supported by the Cancer Research Campaign (CRC). M.C.H. was the recipient of an award from the EU Concerted Action on DNA Repair and Cancer.
REFERENCES
- 1.Bjelland S, Bjørås M, Seeberg E. Excision of 3-methylguanine from alkylated DNA by 3-methyladenine DNA glycosylase I of Escherichia coli. Nucleic Acids Res. 1993;21:2045–2049. doi: 10.1093/nar/21.9.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodell W J, Aida T, Berger M S, Rosenblum M L. Repair of O6-(2-chloroethyl) guanine mediates the biological effects of chloroethylnitrosoureas. Environ Health Perspect. 1985;62:119–126. doi: 10.1289/ehp.8562119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boiteux S, Huisman O, Laval J. 3-Methyladenine residues in DNA induce the SOS function sfiA in Escherichia coli. EMBO J. 1984;3:2569–2573. doi: 10.1002/j.1460-2075.1984.tb02175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Bramson J, O’Connor T, Panasci L. Effect of alkyl-N-purine DNA glycosylase overexpression on cellular resistance to bifunctional alkylating agents. Biochem Pharmacol. 1995;50:39–44. doi: 10.1016/0006-2952(95)00114-f. [DOI] [PubMed] [Google Scholar]
- 6.Brown D G, Willington M A, Findlay I, Muggleton-Harris A L. Criteria that optimize the potential of murine embryonic stem cells for in vitro and in vivo developmental studies. In Vitro Cell Dev Biol. 1992;28A:773–778. doi: 10.1007/BF02631066. [DOI] [PubMed] [Google Scholar]
- 7.Cabral Neto J B, Gentil A, Caseira Cabral R E, Sarasin A. Mutation spectrum of heat-induced abasic sites on a single-stranded shuttle vector replicated in mammalian cells. J Biol Chem. 1992;267:19718–19723. [PubMed] [Google Scholar]
- 8.Cabral Neto J B, Caseira Cabral R E, Le Page A M F, Sarasin A, Gentil A. Coding properties of a unique apurinic/apyrimidinic site replicated in mammalian cells. J Biol Chem. 1994;240:416–420. doi: 10.1006/jmbi.1994.1457. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Derfler B, Maskati A, Samson L. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 1990;9:4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinnasamy N, Rafferty J A, Hickson I, Ashby J, Tinwell H, Margison G P, Dexter T M, Fairbairn L. O6-Benzylguanine potentiates the in vivo toxicity and clastogenicity of temozolomide and BCNU in mouse bone marrow. Blood. 1997;89:1566–1573. [PubMed] [Google Scholar]
- 11.Chinnasamy N, Rafferty J A, Hickson I, Lashford L S, Thatcher N, Margison G P, Dexter T M, Fairbairn L J. Chemoprotective gene therapy. II. Multilineage in vivo protection of haemopoiesis against the effects of an anti-tumour agent by expression of a mutant human O6-alkylguanine-DNA-alkyltransferase. Gene Ther. 1998;5:842–847. doi: 10.1038/sj.gt.3300699. [DOI] [PubMed] [Google Scholar]
- 12.Chung F-L, Chen H-J C, Nath R G. Lipid peroxidation as a potential endogenous source for the formation of exocyclic DNA adducts. Carcinogenesis. 1996;17:2105–2111. doi: 10.1093/carcin/17.10.2105. [DOI] [PubMed] [Google Scholar]
- 13.Coquerelle T, Dosch J, Kaina B. Overexpression of N-methylpurine-DNA glycosylase in Chinese hamster ovary cells renders them more sensitive to the production of chromosomal aberrations by methylating agents—a case of imbalanced DNA repair. Mutat Res. 1995;336:9–17. doi: 10.1016/0921-8777(94)00035-5. [DOI] [PubMed] [Google Scholar]
- 14.Dosanjh M K, Roy R, Mitra S, Singer B. 1,N6-Ethenoadenine is preferred over 3-methyladenine as substrate by a cloned human N-methylpurine-DNA glycosylase (3-methyladenine-DNA glycosylase) Biochemistry. 1994;33:1624–1628. doi: 10.1021/bi00173a002. [DOI] [PubMed] [Google Scholar]
- 15.Elder R H, Margison G P, Rafferty J A. Differential inactivation of mammalian and Escherichia coli O6-alkylguanine-DNA alkyltransferases by O6-benzylguanine. Biochem J. 1994;298:231–235. doi: 10.1042/bj2980231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Ghissassi F, Barbin A, Nair J, Bartsch H. Formation of 1,N6-ethenoadenine and 3,N4-ethenocytosine by lipid peroxidation products and nucleic acid bases. Chem Res Toxicol. 1995;8:278–283. doi: 10.1021/tx00044a013. [DOI] [PubMed] [Google Scholar]
- 17.Engelward B P, Dreslin A, Christensen J, Huszar D, Kurahara C, Samson L. Repair-deficient 3-methyladenine DNA glycosylase homozygous mutant mouse cells have increased sensitivity to alkylation-induced chromosome damage and cell killing. EMBO J. 1996;15:945–952. [PMC free article] [PubMed] [Google Scholar]
- 18.Engelward B P, Weeda G, Wyatt M D, Broekhof J L, De Wit J, Donker I, Allan J M, Gold B, Hoeijmakers J H J, Samson L D. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc Natl Acad Sci USA. 1997;94:13087–13092. doi: 10.1073/pnas.94.24.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan C-Y, Butler W H, O’Connor P J. Cell and tissue specific localisation of O6-methylguanine in the DNA of rats given N-nitrosodimethylamine: effects of protein deficient diets. Carcinogenesis. 1989;10:1967–1970. doi: 10.1093/carcin/10.10.1967. [DOI] [PubMed] [Google Scholar]
- 20.Geleziunas R, McQuillan A, Malapetsa A, Hutchinson M, Kopriva D, Wainberg M A, Hiscott J, Bramson J, Panasci L. Increased DNA synthesis and repair-enzyme expression in lymphocytes from patients with chronic lymphocytic leukemia resistant to nitrogen mustards. J Natl Cancer Inst. 1991;83:557–564. doi: 10.1093/jnci/83.8.557. [DOI] [PubMed] [Google Scholar]
- 21.Hang B, Chenna A, Rao S, Singer B. 1,N6-Ethenoadenine and 3,N4-ethenocytosine are excised by separate human DNA glycosylases. Carcinogenesis. 1996;17:155–157. doi: 10.1093/carcin/17.1.155. [DOI] [PubMed] [Google Scholar]
- 22.Hang B, Singer B, Margison G P, Elder R H. Targeted deletion of alkylpurine-DNA-N-glycosylase in mice eliminates repair of 1,N6-ethenoadenine and hypoxanthine but not of 3,N4-ethenocytosine or 8-oxoguanine. Proc Natl Acad Sci USA. 1997;94:12869–12874. doi: 10.1073/pnas.94.24.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imperitori L, Damia G, Taverna P, Garattini E, Citti L, Boldrini L, D’Incalci M. 3T3 NIH murine fibroblasts and B78 murine melanoma cells expressing the Escherichia coli N3-methyladenine-DNA glycosylase I do not become resistant to alkylating agents. Carcinogenesis. 1994;15:533–537. doi: 10.1093/carcin/15.3.533. [DOI] [PubMed] [Google Scholar]
- 24.Jansen J G, Vrieling H, van Zeeland A A, Mohn G R. The gene encoding hypoxanthine-guanine phosphoribosyltransferase as target for mutational analysis: PCR cloning and sequencing of the cDNA from the rat. Mutat Res. 1992;266:105–116. doi: 10.1016/0027-5107(92)90178-5. [DOI] [PubMed] [Google Scholar]
- 25.Kamiya H, Suzuki M, Komatsu Y, Miura H, Kikuchi K, Sekaguchi T, Murata N, Masutani C, Hanaoka F, Ohtsuka E. An abasic site analogue activates a c-Ha-ras gene by a point mutation at modified and adjacent positions. Nucleic Acids Res. 1992;20:4409–4415. doi: 10.1093/nar/20.17.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kielman M F, Smits R, Bernini L F. Structure of the mouse 3-methyladenine DNA glycosylase gene and exact localization upstream of the α-globin gene cluster on chromosome 11. Mamm Genome. 1995;6:499–504. doi: 10.1007/BF00356165. [DOI] [PubMed] [Google Scholar]
- 27.Klungland A, Fairbairn L, Watson A J, Margison G P, Seeberg E. Expression of the E. coli 3-methyladenine DNA glycosylase I gene in mammalian cells reduces the toxic and mutagenic effects of methylating agents. EMBO J. 1992;11:4439–4444. doi: 10.1002/j.1460-2075.1992.tb05544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klungland A, Bjoras M, Hoff E, Seeberg E. Increased removal of 3-alkyladenine reduces the frequencies of hprt mutations induced by methyl- and ethylmethane sulfonate in Chinese hamster fibroblast cells. Nucleic Acids Res. 1994;22:1670–1674. doi: 10.1093/nar/22.9.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klungland A, Lakke K, Hoff E, Seeberg E. Spectrum of mutations induced by methyl and ethyl methanesulfonate at the hprt locus of normal and tag expressing Chinese hamster fibroblasts. Carcinogenesis. 1995;16:1281–1285. doi: 10.1093/carcin/16.6.1281. [DOI] [PubMed] [Google Scholar]
- 30.Laib R J, Gwinner L M, Bolt H M. DNA alkylation by vinyl chloride metabolites: etheno derivatives or 7-alkylation of guanine? Chem Biol Interact. 1981;37:219–231. doi: 10.1016/0009-2797(81)90179-4. [DOI] [PubMed] [Google Scholar]
- 31.Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1984;150:77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- 32.Lindahl T. New class of enzymes acting on damaged DNA. Nature. 1976;259:64–66. doi: 10.1038/259064a0. [DOI] [PubMed] [Google Scholar]
- 33.Margison G P, O’Connor P J. Biological implications of the instability of the N-glycosidic bond of 3-methyldeoxyadenosine in DNA. Biochim Biophys Acta. 1973;331:349–356. doi: 10.1016/0005-2787(73)90021-x. [DOI] [PubMed] [Google Scholar]
- 34.Matijasevic Z, Bodell W J, Ludlum D B. 3-Methyladenine DNA glycosylase activity in a glial cell line sensitive to the haloethylnitrosoureas in comparison with a resistant cell line. Cancer Res. 1991;51:1568–1570. [PubMed] [Google Scholar]
- 35.Matijasevic Z, Boosalis M, Mackay W, Samson L, Ludlum D B. Protection against chloroethylnitrosourea cytotoxicity by eukaryotic 3-methyladenine DNA glycosylase. Proc Natl Acad Sci USA. 1993;90:11855–11859. doi: 10.1073/pnas.90.24.11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moriya M, Zhang W, Johnson F, Grollman A P. Mutagenic potency of exocyclic DNA adducts: marked differences between Escherichia coli and simian kidney cells. Proc Natl Acad Sci USA. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nair J, Barbin A, Guichard Y, Bartsch H. 1,N6-Ethenodeoxyadenosine and 3,N4-ethenodeoxycytidine in liver DNA from humans and untreated rodents detected by immunoaffinity/32P-postlabelling. Carcinogenesis. 1995;16:613–617. doi: 10.1093/carcin/16.3.613. [DOI] [PubMed] [Google Scholar]
- 38.Nivard M J M, Pastink A, Vogel E W. Molecular analysis of mutations induced in the vermilion gene of Drosophila melanogaster by methyl methanesulfonate. Genetics. 1992;131:673–682. doi: 10.1093/genetics/131.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Connor P J, Capp M J, Craig A W, Lawley P D, Shah S A. Differences in the patterns of methylation in rat liver ribosomal ribonucleic acid after reaction in vivo with methyl methanesulphonate and N,N-dimethylnitrosamine. Biochem J. 1972;129:519–524. doi: 10.1042/bj1290519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Op het Veld C W, Zdzienicka M Z, Vrieling H, Lohman P H M, van Zeeland A A. Molecular analysis of ethyl methanesulfonate-induced mutations at the hprt gene in the ethyl methanesulfonate-sensitive Chinese hamster cell line EM-C11 and its parental line CH09. Cancer Res. 1994;54:3001–3006. [PubMed] [Google Scholar]
- 41.Pandya G, Moriya M. 1,N6-Ethenodeoxyadenosine, a DNA adduct highly mutagenic in mammalian cells. Biochemistry. 1996;35:11487–11492. doi: 10.1021/bi960170h. [DOI] [PubMed] [Google Scholar]
- 42.Rossi A M, Tates A D, van Zeeland A A, Vrieling H. Molecular analysis of mutations affecting hprt mRNA splicing in human T-lymphocytes in vivo. Environ Mol Mutagen. 1992;19:7–13. doi: 10.1002/em.2850190103. [DOI] [PubMed] [Google Scholar]
- 43.Routledge M N, Wink D A, Keefer L K, Dipple A. DNA sequence changes induced by two nitric oxide donor drugs in the SupF assay. Chem Res Toxicol. 1994;7:628–632. doi: 10.1021/tx00041a007. [DOI] [PubMed] [Google Scholar]
- 44.Sabapathy K, Klemm M, Jaenisch R, Wagner E F. Regulation of ES cell differentiation by functional and conformational modulation of p53. EMBO J. 1997;16:6217–6229. doi: 10.1093/emboj/16.20.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saffhill R, Margison G P, O’Connor P J. Mechanisms of carcinogenesis induced by alkylating agents. Biochim Biophys Acta. 1985;823:111–145. doi: 10.1016/0304-419x(85)90009-5. [DOI] [PubMed] [Google Scholar]
- 46.Saffhill R, Fida S, Bromley M, O’Connor P J. Promutagenic alkyl lesions are induced in the tissue DNA of animals treated with ionising radiation. Hum Toxicol. 1988;7:311–317. doi: 10.1177/096032718800700403. [DOI] [PubMed] [Google Scholar]
- 47.Saparbaev M, Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc Natl Acad Sci USA. 1994;91:5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saparbaev M, Kleibl K, Laval J. Escherichia coli, Saccharomyces cerevisiae, rat and human 3-methyladenine DNA glycosylases repair 1,N6-ethenoadenine when present in DNA. Nucleic Acids Res. 1995;23:3750–3755. doi: 10.1093/nar/23.18.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singer B, Hang B. What structural features determine repair enzyme specificity and mechanism in chemically modified DNA? Chem Res Toxicol. 1997;10:713–732. doi: 10.1021/tx970011e. [DOI] [PubMed] [Google Scholar]
- 50.Tates A D, van Dam F J, de Zwart F A, van Teylingen C M M, Natarajan A T. Development of a cloning assay with high cloning efficiency to detect induction of 6-thioguanine-resistant lymphocytes in spleen of adult mice following in vivo inhalation exposure to 1,3-butadiene. Mutat Res. 1994;309:299–306. doi: 10.1016/0027-5107(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 51.Tinwell H, Ashby J. Comparison of acridine orange and Giemsa stains in several mouse bone marrow micronucleus assays, including a triple dose study. Mutagenesis. 1989;4:476–481. doi: 10.1093/mutage/4.6.476. [DOI] [PubMed] [Google Scholar]
- 52.Tybulewicz V L J, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl protooncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 53.Vrieling H, Simons J W I M, van Zeeland A A. Nucleotide sequence determination of point mutations at the mouse HPRT locus using in vitro amplification of HPRT mRNA sequences. Mutat Res. 1988;198:107–113. doi: 10.1016/0027-5107(88)90046-2. [DOI] [PubMed] [Google Scholar]
- 54.White G R M, Ockey C H, Brennand J, Margison G P. Chinese hamster cells harbouring the Escherichia coli O6-alkylguanine alkyltransferase gene are less susceptible to sister chromatid exchange induction and chromosome damage by methylating agents. Carcinogenesis. 1986;7:2077–2080. doi: 10.1093/carcin/7.12.2077. [DOI] [PubMed] [Google Scholar]
- 55.Wilson D M, III, Thompson L H. Life without DNA repair. Proc Natl Acad Sci USA. 1997;94:12754–12757. doi: 10.1073/pnas.94.24.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood R D. DNA repair in eukaryotes. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]