Abstract
We identified new residues within a 101-amino-acid stretch of the murine leukemia virus capsid that differentially modulate resistance and susceptibility to the mouse Fv1 and human Ref1 genes. Among these residues, aspartate 92 and histidine 117 are both required for Fv1b resistance, whereas the latter is sufficient to confer Ref1 resistance.
The Friend virus susceptibility gene 1 (Fv1) restricts replication of N-tropic and B-tropic murine leukemia viruses (MLV) in laboratory mouse strains harboring the b or n allele, respectively (15, 29). Several other alleles are present in wild mice (18, 26). Fv1 products are related to retroviral capsids (CA) with closest homologies to human (HERV-L) and mouse (MuERV-L) endogenous retroviral sequences (2, 5). The Fv1 restriction does not block reverse transcription but prevents proviral DNA integration through unknown mechanisms. Restriction at early stages of retroviral replication has been described in other mammalian species, including monkeys and humans (3, 6, 8, 11, 14, 30, 32). The most studied restriction genes in primates are the human Ref1 (32) and simian Lv1 (4, 8) genes. The primate TRIM5α protein, a member of the tripartite motif protein superfamily, unrelated to Fv1, has recently been shown to be an early-stage restrictive factor of primate lentiviruses (31) associated with Ref1 and Lv1 activities (13, 17, 25, 34). Ref1 and Lv1, in contrast to Fv1, target a wide range of mammalian lentiviruses and have been shown to restrict N-tropic but not B-tropic MLV (11, 32).
MLV CA proteins are the main target of the Fv1, Ref1, and Lv1 restrictions (9, 12, 22-24). Residue 110 of MLV CA has been identified as the discriminating target determinant between N-tropic, Ref1-susceptible MLV and B-tropic, Ref1-resistant MLV in murine (18) as well as other mammalian (17, 25, 32, 34) cells. However, under certain conditions, changes at CA residues 105 (10) and 114 (16) have also been reported to modulate Fv1 restriction.
NB-tropic MLV strains, such as the prototypic Friend and Moloney MLV, are resistant to all tested Fv1 alleles. Moloney MLV has also been documented to be resistant to Ref1 and Lv1 (25, 31, 34), but the status of Friend MLV with respect to these primate restriction systems has not been described. Intriguingly, Friend MLV is NB-tropic despite the presence of the N-tropic hallmark, an arginine at position 110 (Arg110), and conserved residues at positions 105 and 114 (Fig. 1A). Therefore, evasion of the mouse restriction systems, as observed with NB-tropic Friend MLV, appears to involve a residue(s) other than residue 110. To determine the sensitivity of Friend MLV to Ref1 and more precisely map CA determinants affecting resistance to the mouse Fv1b and Fv1n alleles, we derived Friend MLV CA-based constructs and mutants. Here, we demonstrate that Friend MLV is resistant to the human Ref1 restriction and identify CA residues that condition N, B, NB, and Ref1 tropisms. We thereby describe residue combinations that differentially modulate susceptibility and resistance to Fv1 and Ref1 restrictions.
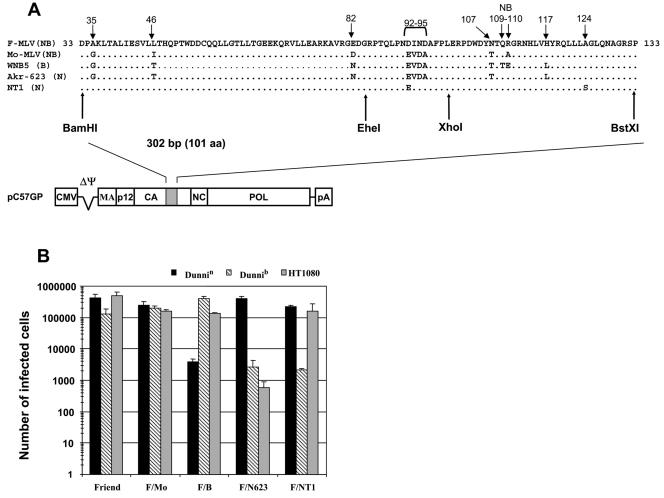
FIG. 1.
Sequences, expression vectors, and tropism associated with prototypic MLV capsids. (A) Sequences of the capsid fragment used in the swaps are shown. Amino acid sequence alignment is shown for NB-tropic Friend MLV strain 57 and Moloney MLV (F-MLV and Mo-MLV, GenBank accession no. X02794 and J02255, respectively), B-tropic WNB5 MLV (GenBank accession no. K01190), N-tropic Akr-623 MLV (GenBank accession no. J01998), and the NT1 N-tropic Tennant strain of Friend MLV (this study; GenBank accession no. AY883167). NB, B, or N tropism is indicated in parentheses next to the MLV strain. The amino acid sequence encoded by the BamHI-BstXI 302-bp cassette (positions 1362 to 1664 in the Friend MLV clone 57 sequence), corresponding to amino acids 33 to 133 of the swapped capsid fragment, is shown. Positions corresponding to the EheI and XhoI internal restriction sites are also indicated. Distinctive residues including the NB determinant at positions 109 and 110 and the DIND motif (see text) at positions 92 to 95 are indicated. Dots represent residues identical to the Friend MLV 57 sequence. Each cassette was introduced in the CA gene of the pC57GP gag-pol expression vector from the NB-tropic strain 57 of Friend MLV. CMV, cytomegalovirus promoter; MA, matrix; CA, capsid; NC, nucleocapsid; pA, polyadenylation signal. (B) Evaluation of Fv1 and Ref1 MLV restrictions with parental CA cassettes. Virions were produced by transfecting human 293T cells with the vector shown in panel A and complementing vectors as described in the text. Viral supernatants were standardized on permissive Mus dunni Fv1−/− cells (dunni). Sensitivity to Fv1 and Ref1 restrictions was assayed on dunnin and dunnib cells stably expressing the Fv1n gene or the Fv1b gene, respectively, and on Ref1-positive human HT1080 cells. Cells were infected with serial dilutions of viral supernatant in the presence of Polybrene (8 μg/ml), and 2 days after infection, target cells were fixed and stained for alkaline phosphatase activity (20), and FFU per milliliter were counted. Mean viral titers ± standard errors of the means are shown and were calculated from at least three independent experiments using FFU values obtained in the linear portion of the titration curve.
A 101-amino-acid fragment recapitulates MLV capsid susceptibilities to Fv1 and Ref1 targets.
Early studies of Fv1 MLV target determinants mapped differences between N- and B-tropic MLV to a 302-bp fragment encoding a CA sequence comprised between residues 33 and 133. Residues 109 and 110 were reported to be the MLV NB determinant (9) (Fig. 1A). We constructed a Friend MLV-based Gag-Pol expression vector (pC57GP) that allowed allelic exchanges of this 302-bp MLV CA cassette between BamHI and BstXI restriction sites. These sites were either naturally present or introduced by PCR-directed mutagenesis, in prototypic MLV strains of different NB tropisms. Additional EheI and XhoI sites were also used to swap smaller fragments (Fig. 1A). Prototypic CA sequences were derived from the N-tropic Akr-623 and B-tropic WNB5 MLV clones (7, 19) (kind gifts of A. Rein) and from the NB-tropic Friend MLV 57 and Moloney MLV 8.2 clones (see reference 27 for details). We also PCR amplified and sequenced a BamHI-BstXI fragment from the historical N-tropic Tennant isolate of the Friend complex (NT1; a kind gift of S. Gisselbrecht). NT1 differs from NB-tropic Friend MLV 57 at positions 92 and 124, with glutamate and serine residues substituted for aspartate and alanine, respectively (Fig. 1A).
Single-round infectious retroviral particles with MLV cores were harvested from 293T cell supernatants 48 h after cotransfection of one of the gag-pol vectors with the pCSI-G vesicular stomatitis virus G protein expression vector (1) and the pLAPSN retroviral vector carrying the neo and human placental alkaline phosphatase reporter genes (20). Viral tropism was assayed on Fv1-nonrestrictive Mus dunni (dunni) cells or on dunni cells stably transfected with Fv1n (dunnin) or Fv1b (dunnib) expression vectors (Fv1 plasmids were a kind gift of J. Stoye). Ref1 restriction was tested on human HT1080 cells (Fig. 1B) and TE671 cells as indicated.
All reported virion preparations had similar infectious levels as measured on nonrestrictive dunni cells, with titers ranging from 105 to 106 focus-forming units (FFU)/ml. All parental MLV prototypic tropisms were reproduced when using our chimeric CA-based vector system. Thus, Fv1b and Fv1n restrictions resulted in a 20- to 400-fold drop in titers. Strong Ref1-mediated restriction, with an average 200-fold drop in titers, was also observed with N-MLV virions (Fig. 1B and 2A). The NB-tropic F-MLV construct was resistant to both Fv1b and Fv1n, as well as to Ref1. Surprisingly though, the Fv1b-susceptible NT1-derived construct was resistant to Ref1-mediated restriction (Fig. 1B). This Ref1 resistance of an N-tropic MLV was observed on HT1080 and TE671, human cell lines that exert strong Ref1 restriction on other retroviruses (32), as well as on dunni cells stably transduced with human TRIM5α expressed from the pLXSN retroviral expression vector (dunni/TRIM5α) (data not shown). This is the first identified CA sequence that distinguishes between Fv1b and Ref1 susceptibilities.
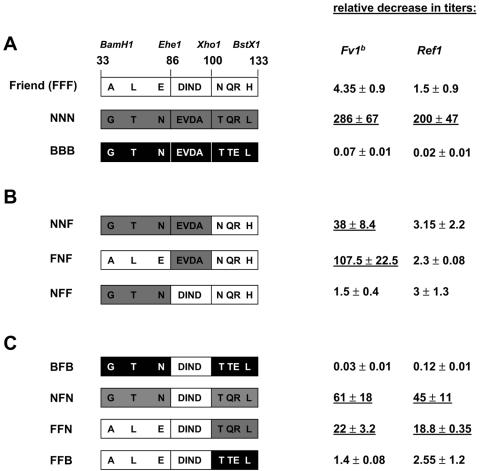
FIG. 2.
Fv1 and Ref1 restrictions of MLV particles harboring chimeric CA. Allelic fragment exchanges between Friend NB-tropic MLV (F, open boxes), N-tropic Akr-623 MLV (N, grey boxes), and B-tropic WNB5 MLV (B, black boxes) were generated using BamHI, EheI, XhoI, and BstXI restriction sites. Fragment combinations are designated by a three-letter code corresponding to the parental MLV fragments delineated by residues 33 to 86, 87 to 100, and 101 to 133, respectively. Infections were performed as described in Fig. 1. Relative decreases in titers with Fv1b are shown as mean ratios ± standard errors of the means of viral titers obtained on dunnin cells over dunnib cells whereas relative decreases in titers with Ref1 are shown as mean ratios ± standard errors of the means of titers obtained on dunnin cells over Ref1-positive HT1080 human cells. Data were calculated from at least three independent experiments. Underlined values indicate chimeras that are susceptible to Fv1b or Ref1 restrictions.
MLV resistance to Fv1 is due to multiple determinants that modulate position 110.
Arg110 is the discriminating determinant between N-tropic, Ref1-susceptible MLV and B-tropic, Ref1-resistant MLV (17, 18, 25, 32, 34). However, the fact that NB-tropic and N-tropic Friend MLV strains encoding Arg110 are resistant to Fv1b, Ref1, or both indicated that other residues in the 101-amino-acid-long capsid region govern Fv1b and Ref1 susceptibilities. We therefore further evaluated the bases for the Friend MLV Fv1 and Ref1 resistance with smaller domain swaps.
A domain swap introducing N-MLV residues 33 to 100, excluding the canonical 110 residue, into Friend MLV was sufficient to render the latter susceptible to Fv1b restriction (chimera NNF, Fig. 2B). Friend MLV CA residues 92 to 95 consist of a DIND motif corresponding to an EVDA motif in prototypic N-tropic MLV (Fig. 1A). The swap of this motif was sufficient to convert Friend MLV to N-tropism as shown by a full Fv1b susceptibility of the corresponding construct (FNF, Fig. 2B). Thus, the DIND motif plays a key role in suppressing the accessibility of the N-tropic target in Friend MLV. Notably though, swapping of this motif did not render Friend MLV susceptible to Ref1, and this resistance was maintained even following a larger swap from residues 33 to 100 (NNF, Fig. 2B).
We further assessed the suppressive effect of the Friend MLV DIND motif in a parental B-MLV (BFB) or N-MLV (NFN) context. The sole substitution of this DIND motif was not sufficient to modulate Fv1 or Ref1 resistance in either context (Fig. 2C). When this swap was combined with that of the upstream fragment, B-tropic virions became resistant to the Fv1n restriction. Therefore, glycine, threonine, and/or asparagine at positions 35, 46, and 82, respectively, conditions CA targeting by Fv1. Nevertheless, this contribution was not as potent in the N-tropic context, since FFN particles remained susceptible to Fv1b and Ref1 restrictions, albeit with a slightly decreased susceptibility (Fig. 2C). The prominent exposure of Asp82, according to the recently published structure of the N-AKV CA amino-terminal domain (21) (Fig. 4B), is in favor of a role for this residue in Fv1 tropism, as also suggested by others (28).
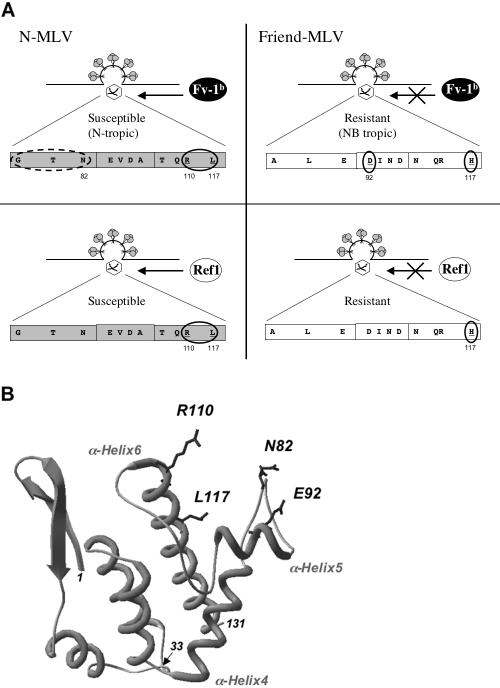
FIG. 4.
Schematic representation of the Fv1 and Ref1 restriction targets. (A) Target and masking determinants for Fv1b (top) and Ref1 (bottom) restrictions in the MLV CA. Key residues involved in the susceptibility of N-MLV to Fv1b and Ref1 restriction genes are circled (left panels, R110 and L117). The dashed circle indicates a fragment found to modulate the restriction level (see text). Residues sufficient to make the Friend MLV CA target inaccessible to Fv1b and/or Ref1 restrictions are circled (right panels, combined D92 and H117 for Fv1b; H117 for Ref1). (B) Three-dimensional structure of the amino-terminal domain of N-Akv CA (adapted from reference 21). Amino-terminal (position 1) and carboxy-terminal (position 131) extremities of the crystallized fragment are indicated as well as position 33 corresponding to the BamHI cloning site shown in Fig. 1 and α-helices 4, 5, and 6. Representation of asparagine 82 (N82), aspartate 92 (E92), arginine 110 (R110), and leucine 117 (L117) side chains highlights their potential accessibility to restriction factors.
Mutation of histidine to leucine at position 117 is sufficient to render Friend MLV susceptible to Fv1b and Ref1 restrictions.
In contrast to Friend MLV virions, FFN virions (Fig. 2C) were susceptible to both Fv1b and Ref1 restrictions. As the difference between these viruses was limited to amino acid changes at positions 107 and 117, we introduced individual substitutions at these positions in the Friend MLV background. Infections on restrictive cell lines showed that the presence of leucine at position 117, F(H117L), was sufficient to induce a susceptibility to Fv1 and Ref1 restrictions (Fig. 3A). Charge rather than aromatic properties was crucial for resistance, as mutation of His117 to a lysine in Friend MLV maintained this property whereas mutation to either phenylalanine or tyrosine reversed resistance (not shown).
FIG. 3.
Residues responsible for resistance of Friend MLV 57 (Friend) to Fv1b and Ref1. (A) Point mutations were introduced in the NB-tropic Friend MLV CA at positions 107, F(N107T), and 117, F(H117L), to match the N-tropic Akr-623 sequence and (B) at positions 92, F(D92E), and 124, F(A124S), to match the N-tropic F/NT1 sequence. F/NT1 corresponds to the Friend MLV57 CA into which the NT1 residues 33 to 133 have been swapped. All infections were performed as indicated in Fig. 1, and relative mean decreases in titers were calculated from at least three independent experiments as in Fig. 2.
Loss of Fv1b but not Ref1 restriction in the presence of aspartate 92.
As indicated above, suppression of the canonical residue 110 as a target for Fv1b is due to the combined presence of His117 and a DIND motif. Interestingly, the N-tropic NT1 strain harbors an EIND motif. The aspartate-to-glutamate change was sufficient to confer Fv1b susceptibility with no effect on Ref1 resistance, as observed with the F(D92E) mutant (Fig. 3B). When introduced alone, the second amino acid difference between the Friend MLV and NT1 strain CA at position 124 did not influence either Fv1 or Ref1 susceptibility [F(A124S), Fig. 3B]. Therefore, Glu92 was key to the recognition of the Fv1b target in the presence of His117, while it did not modulate Ref1 restriction. Similar data were obtained using dunni/TRIM5α cells.
In conclusion, while Arg110 is required for both Fv1b and Ref1 restrictions, Leu117 is also a major target determinant for both genes (Fig. 4A, left panels). Furthermore, Asp92 is required for efficient resistance to Fv1b when combined with His117. This is in agreement with a recent report using a different model of Fv1 restriction (28). However, we found that Ref1 resistance was not affected by Asp92 (Fig. 4A, right panels). This is compatible with the recently reported crystal structure of the CA amino-terminal domain of an N-tropic MLV in which residue 92, on the one hand, and residues 110 and 117, on the other, are located in two separate neighboring helices, α-helices 5 and 6, respectively (21) (Fig. 4B). Our results suggest that CA MLV recognition by Fv1 involves both α-helices, which are exposed to the outside of the amino-terminal CA hexamer (21), whereas Ref1/TRIM5α recognition appears to involve mainly α-helix 6. While α-helices 4 and 6 appear structurally conserved between human immunodeficiency virus type 1 and N-Akv CA, the interconnecting sequence, which includes α-helix 5 of N-Akv CA, displays a variable succession of loops and helices (21). Interestingly, the latter sequence comprises residue 92 of the MLV CA, which we identified as a major Fv1-modulating determinant, and the human immunodeficiency virus type 1 CA CypA-binding loop involved in Lv1 restriction (12, 23, 33), thus providing differential target patterns recognized by these restrictive cellular mechanisms.
Nucleotide sequence accession number.
The GenBank accession number of the BamHI-BstXI fragment from the historical N-tropic Tennant isolate of the Friend complex is AY883167.
Acknowledgments
We thank N. Taylor for helpful discussions and critical reading of the manuscript and L. Boone, F.-L. Cosset, S. Gisselbrecht, A. Rein, J. Stoye, and G. Towers for providing us with materials. We also thank Muriel Audit, Dorothée Molle, and Valérie Thibert for their participation in the preparation of some of the reagents used in this study and all members of our laboratory for their continuous input.
A.L. is funded by the Ministère de l'Education Nationale de la Recherche et de la Technologie (MENRT). M.S. and J.-L.B. are supported by the Institut National de la Santé et de la Recherche Médicale (INSERM). This work was partly supported by grants from the Agence Nationale de la Recherche sur le Sida (ANRS), to M.S. and J.-L.B., and the Association Française contre les Myopathies (AFM), to M.S.
REFERENCES
- 1.Battini, J. L., J. E. Rasko, and A. D. Miller. 1999. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. USA 96:1385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benit, L., N. De Parseval, J. F. Casella, I. Callebaut, A. Cordonnier, and T. Heidmann. 1997. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J. Virol. 71:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besnier, C., L. Ylinen, B. Strange, A. Lister, Y. Takeuchi, S. P. Goff, and G. J. Towers. 2003. Characterization of murine leukemia virus restriction in mammals. J. Virol. 77:13403-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 6.Bieniasz, P. D. 2003. Restriction factors: a defense against retroviral infection. Trends Microbiol. 11:286-291. [DOI] [PubMed] [Google Scholar]
- 7.Boone, L. R., F. E. Myer, D. M. Yang, C. Y. Ou, C. K. Koh, L. E. Roberson, R. W. Tennant, and W. K. Yang. 1983. Reversal of Fv-1 host range by in vitro restriction endonuclease fragment exchange between molecular clones of N-tropic and B-tropic murine leukemia virus genomes. J. Virol. 48:110-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DesGroseillers, L., and P. Jolicoeur. 1983. Physical mapping of the Fv-1 tropism host range determinant of BALB/c murine leukemia viruses. J. Virol. 48:685-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doi, K., A. Kawana, A. Iwamoto, H. Yoshikura, and T. Odawara. 1997. One base change is sufficient for host range conversion of murine leukemia virus from B to NB tropism. Arch. Virol. 142:1889-1894. [DOI] [PubMed] [Google Scholar]
- 11.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatziioannou, T., S. Cowan, U. K. Von Schwedler, W. I. Sundquist, and P. D. Bieniasz. 2004. Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid. J. Virol. 78:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann, W., D. Schubert, J. LaBonte, L. Munson, S. Gibson, J. Scammell, P. Ferrigno, and J. Sodroski. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73:10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jolicoeur, P. 1979. The Fv-1 gene of the mouse and its control of murine leukemia virus replication. Curr. Top. Microbiol. Immunol. 86:67-122. [DOI] [PubMed] [Google Scholar]
- 16.Jung, Y. T., and C. A. Kozak. 2000. A single amino acid change in the murine leukemia virus capsid gene responsible for the Fv1nr phenotype. J. Virol. 74:5385-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak, C. A., and A. Chakraborti. 1996. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology 225:300-305. [DOI] [PubMed] [Google Scholar]
- 19.Levin, J. G., S. C. Hu, A. Rein, L. I. Messer, and B. I. Gerwin. 1984. Murine leukemia virus mutant with a frameshift in the reverse transcriptase coding region: implications for pol gene structure. J. Virol. 51:470-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, D. G., R. H. Edwards, and A. D. Miller. 1994. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc. Natl. Acad. Sci. USA 91:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mortuza, G. B., L. F. Haire, A. Stevens, S. J. Smerdon, J. P. Stoye, and I. A. Taylor. 2004. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature 431:481-485. [DOI] [PubMed] [Google Scholar]
- 22.Ou, C. Y., L. R. Boone, C. K. Koh, R. W. Tennant, and W. K. Yang. 1983. Nucleotide sequences of gag-pol regions that determine the Fv-1 host range property of BALB/c N-tropic and B-tropic murine leukemia viruses. J. Virol. 48:779-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owens, C. M., B. Song, M. J. Perron, P. C. Yang, M. Stremlau, and J. Sodroski. 2004. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J. Virol. 78:5423-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owens, C. M., P. C. Yang, H. Gottlinger, and J. Sodroski. 2003. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J. Virol. 77:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5α mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi, C. F., F. Bonhomme, A. Buckler-White, C. Buckler, A. Orth, M. R. Lander, S. K. Chattopadhyay, and H. C. Morse III. 1998. Molecular phylogeny of Fv1. Mamm. Genome 9:1049-1055. [DOI] [PubMed] [Google Scholar]
- 27.Richardson, J., A. Corbin, F. Pozo, S. Orsoni, and M. Sitbon. 1993. Sequences responsible for the distinctive hemolytic potentials of Friend and Moloney murine leukemia viruses are dispersed but confined to the Ψ-gag-PR region. J. Virol. 67:5478-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevens, A., M. Bock, S. Ellis, P. LeTissier, K. N. Bishop, M. W. Yap, W. Taylor, and J. P. Stoye. 2004. Retroviral capsid determinants of Fv1 NB and NR tropism. J. Virol. 78:9592-9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoye, J. P. 1998. Fv1, the mouse retrovirus resistance gene. Rev. Sci. Technol. 17:269-277. [DOI] [PubMed] [Google Scholar]
- 30.Stoye, J. P. 2002. An intracellular block to primate lentivirus replication. Proc. Natl. Acad. Sci. USA 99:11549-11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 32.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 34.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]