Abstract
Nidoviruses produce an extensive 3′-coterminal nested set of subgenomic (sg) mRNAs, which are used to express structural proteins and sometimes accessory proteins. In arteriviruses and coronaviruses, these mRNAs contain a common 5′ leader sequence, derived from the genomic 5′ end. The joining of the leader sequence to different segments derived from the 3′-proximal part of the genome (mRNA bodies) presumably involves a unique mechanism of discontinuous minus-strand RNA synthesis in which base pairing between sense and antisense transcription-regulating sequences (TRSs) plays an essential role. The leader TRS is present in the loop of a hairpin structure that functions in sg mRNA synthesis. In this study, the minimal sequences in the 5′-proximal region of the Equine arteritis virus genome that are required for sg RNA synthesis were delimited through mutagenesis. A full-length cDNA clone was engineered in which this domain was duplicated, allowing us to make mutations and monitor their effects on sg RNA synthesis without seriously affecting genome replication and translation. The leader TRS present in the duplicated sequence was used and yielded novel sg mRNAs with significantly extended leaders. Our combined findings suggest that the leader TRS hairpin (LTH) and its immediate flanking sequences are essential for efficient sg RNA synthesis and form an independent functional entity that could be moved 300 nucleotides downstream of its original position in the genome. We hypothesize that a conformational switch in the LTH region regulates the role of the 5′-proximal region of the arterivirus genome in subgenomic RNA synthesis.
Many eukaryotic plus-strand RNA (+RNA) viruses generate subgenomic (sg) mRNAs as a strategy to regulate the expression of one or several viral proteins (21). In addition to internal initiation of translation, sg mRNA synthesis provides a mechanism to express genes positioned downstream of the 5′-proximal gene in polycistronic genomes of +RNA viruses of eukaryotes. Different mechanisms are employed by +RNA viruses to achieve the transcription of sg mRNAs (47). The generation of an extensive 3′-coterminal nested set of sg mRNAs to express structural and/or accessory proteins is a common property of viruses belonging to the order Nidovirales (Coronaviridae, Arteriviridae, and Roniviridae) (6, 34). In the case of arteriviruses and coronaviruses, all transcripts contain a common 5′ sequence element, the so-called “leader,” which is colinear with the 5′-proximal part of the genome and is fused to different regions derived from the 3′-proximal third of the genome, which are termed “mRNA bodies” (Fig. 1A). For two other nidovirus subgroups, roniviruses and toroviruses (with the exception of sg mRNA2 of the latter), sg mRNAs without a common 5′ leader sequence have been described (3, 32, 45).
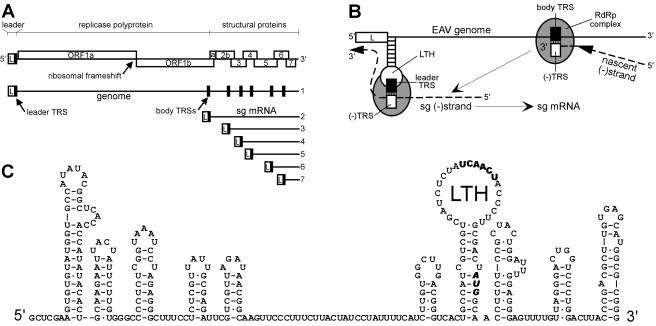
FIG. 1.
(A) Schematic overview of the genome organization and expression of EAV. The replicase gene (ORFs 1a and 1b), the structural protein genes (2a, 2b, 3, 4, 5, 6, and 7), and the leader (L) are indicated. The black boxes in the genomic RNA indicate the positions of the leader TRS and (major) body TRSs. A coterminal nested set of sg mRNAs 2 to 7 is depicted below the genome. (B) Discontinuous minus-strand extension model for nidovirus sg RNA synthesis (31). After attenuation of the RdRp complex at a body TRS in the plus-strand genomic template, the nascent strand is translocated to the leader TRS region. Following (−) body TRS to (+) leader TRS base pairing, minus-strand RNA synthesis resumes to add the complement of the leader sequence. Subsequently, the sg minus strand serves as a template for sg mRNA production. (C) RNA secondary-structure model of the 5′-proximal 313 nt of the EAV RNA genome (38). The leader TRS and the replicase translation initiation codon are depicted in boldface.
For sg RNAs of coronaviruses and arteriviruses, and also the largest sg RNA of toroviruses (45), the fusion of the sg RNA body to the common leader sequence has been postulated to involve the discontinuous extension of minus-strand RNA synthesis, which presumably yields sg minus-strand templates that are used to produce the sg plus strands (31). After attenuation of RNA synthesis, the nascent minus strand is thought to be translocated to the 5′-proximal region of the genomic template, where RNA synthesis is resumed to add the complement of the genomic leader sequence, thus completing the minus-strand template for sg mRNA synthesis (Fig. 1B). Key elements in the joining of leader and body are short conserved transcription-regulating sequences (TRSs) that are present both at the 3′ end of the leader sequence (leader TRS) and at the 5′ end of each of the body regions in the 3′-proximal region of the genome (body TRSs) (Fig. 1A). Base pairing between the leader TRS (in the plus strand) and the complement of the body TRSs (in the nascent minus strand) has been shown to be essential for sg mRNA synthesis in arteriviruses and coronaviruses (14, 26, 27, 43, 49).
Many details of the mechanism that regulates the fusion of sequences that are noncontiguous in the genome during sg RNA synthesis of arteriviruses and coronaviruses remain to be elucidated. It was proposed that the process may mechanistically resemble similarity-assisted copy choice RNA recombination (1, 2, 26). As in the case of RNA recombination, the higher-order structure of the template (and possibly also of the nascent strand) may play an important role, in particular in the regions where leader TRS and body TRSs reside. For Equine arteritis virus (EAV), the arterivirus prototype, we previously predicted the presence of a hairpin structure in the 5′-proximal region of the genome that contains the leader TRS in its loop (43). Subsequently, the presence and importance of this leader TRS hairpin (LTH) was supported by phylogenetic analysis, RNA structure-probing experiments, and a limited mutagenesis study (Fig. 1C) (38). Although the last analysis was hampered by side effects of the mutations at the level of genome replication, the proposed role of the LTH in sg mRNA synthesis was generally supported by the fact that altering its conformation decreased the synthesis of sg mRNAs, while the introduction of compensatory mutations restored sg mRNA production to a certain extent. The identification of potential LTHs in predicted structures of the 5′-proximal genome regions of other arteriviruses and coronaviruses supported the importance of the structure for the base-pairing interaction between the sense leader TRS and antisense body TRS (2, 38). Furthermore, the proposed common ancestry of the viral enzymes involved suggested that common principles may underlie the mechanisms for sg RNA synthesis in both virus groups (4).
Here, we have focused on the role of the 5′-proximal region of the EAV genome in sg RNA synthesis. This region, including the leader TRS and LTH, was successfully duplicated in the context of an EAV full-length cDNA clone, allowing the introduction of mutations in signals for sg RNA synthesis without notably interfering with genome replication and translation. Rigorous changes in the 5′-proximal domain of the genome did not result in apparent defects at the level of genome replication, implying that genome translation was not significantly affected. Still, an analysis of the translation of RNAs carrying wild-type (wt) or mutant EAV 5′ genomic sequences suggested that the virus probably conforms to the conventional “ribosome-scanning” model of translational initiation. A deletion mutagenesis study of the 5′-proximal domain revealed that the LTH and its immediate flanking sequences are required for efficient sg RNA synthesis. This region can fold into an extended conformation of the previously described LTH, which may function as an independent entity that regulates the production of all sg mRNAs and is hypothesized to be involved in a conformational RNA switch in the 5′-proximal region of the genome.
MATERIALS AND METHODS
RNA transfection, infection, and protein analysis.
Baby hamster kidney cells (BHK-21; ATCC CCL10) were maintained (37) and used for propagation of EAV and RNA transfection experiments as described previously (41). Virus titration using plaque assays was done as described previously (22). Immunofluorescence assays with EAV-specific antisera for nsp3 (98.E3) and nucleocapsid protein N (monoclonal antibody 3E2) were done on EAV RNA-transfected and EAV-infected cells (39). Prior to Western blot analysis of nsp1 expression using the anti-nsp1 monoclonal antibody 12A4 (46), cells were lysed at 15 h posttransfection (p.t.) according to the method described previously (5). Proteins were separated by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a Hybond-P polyvinylidene difluoride transfer membrane (Amersham Biosciences), essentially as described before (33). The nsp1 band was visualized using the ECL Plus Western Blotting Detection System (Amersham Biosciences) according to the manufacturer's instructions.
Mutagenesis of an EAV full-length cDNA clone.
Mutations were introduced into shuttle plasmids by standard site-directed PCR mutagenesis. After sequence analysis, restriction fragments containing the desired mutations were transferred to the full-length clone pEAV211, a derivative of pEAV030 (41) containing a few translationally silent engineered restriction sites. Virus derived from clone pEAV211 has a wt phenotype, as determined by comparison with pEAV030-derived virus in growth curve experiments and plaque assays (data not shown).
Construct pD5Dwt (Fig. 2A) contained a tandem repeat of the 5′-proximal 301 nucleotides (nt) of the EAV genome, which was generated by inserting a PCR cassette at nt 306 using an engineered EcoRI site and an MluI site (nt 589) of a pEAV211-derived clone, in which the original open reading frame 1a (ORF1a) translation initiation codon had been changed to UAG (35). Because this mutation introduced a 2-base-pair mismatch in the LTH stem, the base-pairing potential was restored by making two compensating mutations in the opposite side of the stem (38). Replicase translation in D5Dwt was initiated at the start codon present in the duplicated sequence. In addition, the original leader TRS in the genomic 5′-proximal sequence was inactivated (5′-UCAACU-3′ → 5′-AGUUGU-3′; mutated nucleotides are underlined) (43). In some pD5Dwt variants, the replicase codons present within the duplicate (encoding the N-terminal 29 amino acids) were changed as much as possible without changing the encoded protein sequence (constructs containing this set of mutations have a lower-case “c” in their names, e.g., pD5Dc): 5′-AUG GCU ACG UUU AGU GCA ACA GGU UUC GGU GGC UCA UUC GUA CGC GAU UGG AGU UUA GAU CUU CCG GAU GCA UGC GAA CAC GGG GCC-3′).
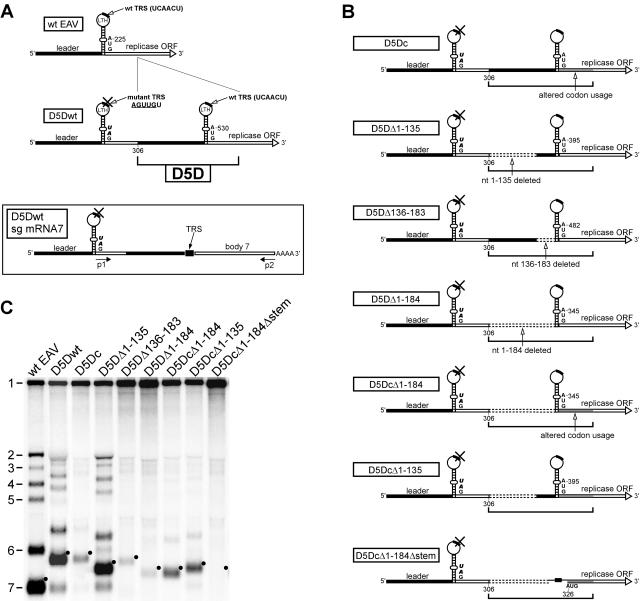
FIG. 2.
Duplication of 5′-proximal sequences in the EAV genome as a strategy to delimit sequences required for sg RNA synthesis. (A) Schematic representation of the 5′-proximal regions of the genomes of wt virus and construct D5Dwt. The sg mRNA 7 transcript produced by D5Dwt is shown below, with the locations of RT-PCR primers used to amplify the leader-to-body junction region of this transcript. The RNA region corresponding to the leader sequence is depicted as black bars; replicase-coding sequences are depicted as white bars. The replicase translation initiation codon, nucleotide numbers (counting from the 5′ end of the genome), LTH, and D5D are indicated. (B) Schematic overview of the 5′-proximal region of the genome of D5Dwt derivatives carrying internal deletions in the D5D. When the codon usage of the replicase-coding region was altered, this is depicted as a gray bar. (C) Hybridization analysis of virus-specific RNA isolated at 15 h p.t. from BHK-21 cells transfected with wt RNA, D5Dwt RNA, or RNA transcribed from the pD5Dwt derivatives depicted in panel B. The positions of the mRNAs of wt virus are indicated: 1, genomic RNA; 2 to 7, sg mRNAs. The sg mRNA7 bands are indicated with black dots.
Several deletions were made in the duplicate of the EAV 5′ sequence in constructs pD5Dwt and pD5Dc. The region corresponding to nt 1 to 135 of the genome was deleted, yielding pD5DΔ1-135 and pD5DcΔ1-135, respectively. The region directly upstream of the LTH (corresponding to nt 136 to 183 of the genome) was removed from pD5Dwt to create pD5DΔ136-183. In addition, nt 1 to 184 were deleted from the duplicated sequence, generating pD5DΔ1-184 and pD5DcΔ1-184, respectively. From the latter construct, the LTH stem was removed, generating pD5DcΔ1-184Δstem. Three constructs were generated in an adapted version of pD5Dc, pD5Dcm, in which the replicase ORF initiated 27 nt downstream of the normal AUG codon present in the duplicated sequence. The latter start codon was changed to UAG, and the base pairing in the LTH stem was restored as described above. The region encompassing nt 1 to 163 or 1 to 184 was deleted from the duplicate, yielding pD5DcmΔ1-163 or pD5DcmΔ1-184, respectively. Furthermore, a derivative of pD5DcmΔ1-163 was generated in which the original wt leader TRS was not inactivated by mutagenesis and which therefore contained two wt leader TRSs (pD5DcmΔ1-163\2xwt).
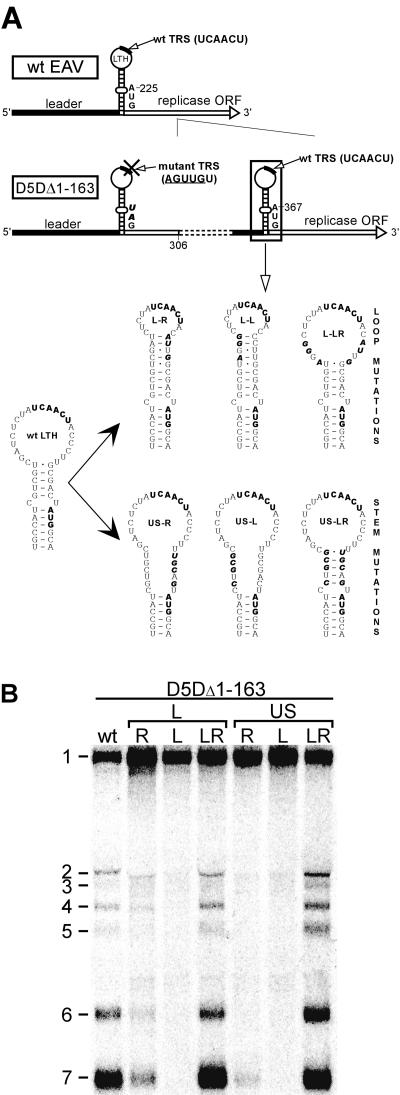
To investigate the function of the LTH by mutagenesis, construct pD5DΔ1-163 was used with an intact (pD5DΔ1-163\wt) or mutant LTH in the duplicate. To disrupt the upper part of the LTH stem, the 5′ side of the stem was mutated (5′-GUCGU-3′ → 5′-CUGCG-3′), creating pD5DΔ1-163\US-L, or the 3′ side of the stem was mutated (5′-GCGAC-3′ → 5′-UGCAG-3′) to create pD5DΔ1-163\US-R. Combining these mutations resulted in pD5DΔ1-163\US-LR. The LTH loop was partly closed by introducing mutations in the 5′ side of the loop (5′-CGAU-3′ → 5′-AGGG-3′), creating pD5DΔ1-163\L-L, or the 3′ side of the loop (5′-CCUU-3′ → 5′-AUUG-3′), creating pD5DΔ1-163\L-R. Combining these mutations resulted in pD5DΔ1-163\L-LR.
To investigate the presence and role of the predicted extended version of the LTH stem, construct pD5DcmΔ1-163 was used as a basis. The entire 5′ side of the stem region was mutated (5′-AUCUUGUGGCUUGACGGGU-3′ → 5′-UAGGUCAUCGUUCUCUCCA-3′), creating pD5DcmΔ1-163\CL, or the entire 3′ side of the stem was mutated (5′-ACCUUCUCCGCUACUGGAU-3′ → 5′-UGGGUCAGCCGGUGUUCUA-3′), creating pD5DcmΔ1-163\CR. Combining these mutations resulted in pD5DcmΔ1-163\CLR. Also, a smaller base-paired region directly below the LTH was probed by mutating the 5′ side of the stem (5′-AUCUUGUGGCUUGACGGGU-3′ → 5′-AUCUUGUGGCUUGACUCCA-3′), creating pD5DcmΔ1-163\AL, or by mutating the 3′ side of the stem (5′-ACCUUCUCCGCUACUGGAU-3′ → 5′-UGGGUCUCCGCUACUGGAU-3′) to create pD5DcmΔ1-163\AR. Combining these mutations resulted in pD5DcmΔ1-163\ALR. Finally, a second smaller base-paired region was changed by mutating the 5′ side of the stem (5′-AUCUUGUGGCUUGACGGGU-3′ → 5′-AUCUUCAUCGUUGACGGGU-3′), creating pD5DcmΔ1-163\BL, or the 3′ side of the stem (5′-ACCUUCUCCGCUACUGGAU-3′ → 5′-ACCUUCUCCCGGUGUGGAU-3′), creating pD5DcmΔ1-163\BR. Combining these mutations resulted in pD5DcmΔ1-163\BLR.
RNA isolation and analysis.
Intracellular RNA from BHK-21 cells was isolated at 15 h p.t., separated in denaturing agarose-formaldehyde gels, and detected by hybridization to a 32P-labeled probe (5′-TTGGTTCCTGGGTGGCTAATAACTACTT-3′), which is complementary to the 3′ end of the EAV genome and recognizes both the genome and all sg mRNAs (43). Dried gels were exposed to phosphorimager screens, which were scanned with a Personal Molecular Imager FX (Bio-Rad) after exposure. Band intensities were quantitated with Quantity One v4.2.2 (Bio-Rad). The same RNA was also used as a template for primer extension analysis, essentially as described previously (38), and for reverse transcription (RT) reactions and subsequent PCRs. PCR products were sequenced using standard protocols.
RNA secondary-structure prediction.
RNA secondary structures were predicted using the genetic algorithm of STAR v4.4 (10) and the energy minimization program of Mfold Web server (48).
Translation assays.
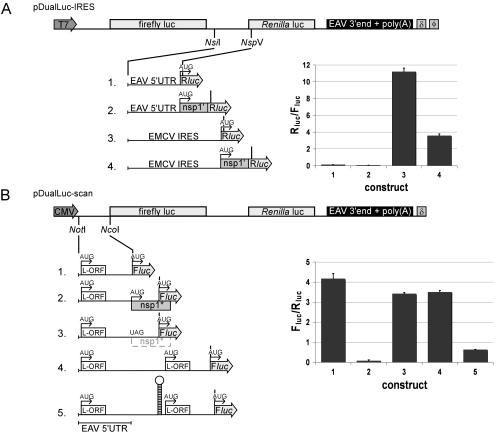
To test for internal ribosomal entry site (IRES) activity of specific EAV sequences, cDNA cassettes were placed in between the Photinus pyralis (firefly) and Renilla reniformis (Renilla) luciferase genes of a pDualLuc reporter plasmid (29). Bicistronic mRNA transcripts were initiated from a T7 RNA polymerase promoter and contained the firefly (Fluc) and Renilla (Rluc) luciferase genes, the 3′-terminal region of the EAV genome starting from nucleotide 11,737 and including a poly(A) tail, and the hepatitis virus δ ribozyme (see Fig. 7A). In construct pDualLuc-IRES1, the EAV 5′ untranslated region (UTR) (nt 1 to 224) was tested, with the replicase translation initiation codon being the starting point for Rluc expression. The 3′ end of the EAV 5′ UTR was extended with sequences encoding the N-terminal part of nsp1 (nt 225 to 287) in construct pDualLuc-IRES2. For both plasmids, a positive control was generated in which the EAV 5′ UTR was replaced with the encephalomyocarditis virus (EMCV) IRES element (12, 13), yielding pDualLuc-IRES3 and pDualLuc-IRES4, respectively. Rabbit kidney (RK-13) cells (5) were infected with vaccinia virus recombinant vTF7-3 (9), which expresses the T7 RNA polymerase, at a multiplicity of infection of 10 and transfected with 1 μg of plasmid using Lipofectamine PLUS (Invitrogen) at 2 h postinfection (p.i.). The cells were lysed at 8 h p.i. and assayed for Fluc and Rluc activities. Enzymatic activities were measured using a luminometer (Turner Designs TD-20/20) and performed with the Dual-luciferase Reporter assay according to the manufacturer's protocol (Promega).
FIG. 7.
Analysis of translation initiation at the 5′ end of the EAV genome by using bicistronic reporter constructs and dual luciferase translation assays. (A) To assess the possibility of the presence of an IRES element, EAV 5′-proximal sequences were cloned in pDualLuc-IRES between the Fluc and Rluc reporter genes. The EMCV IRES element was used as a positive control. nsp1′ indicates the N-terminal in-frame extension of the Renilla luciferase with the N-terminal 21 amino acids of nsp1. The T7 promoter (arrow) and terminator (φ), the 3′-terminal region of the EAV genome starting from nucleotide 11,737 and including a poly(A) tail, and the hepatitis virus δ ribozyme (δ) are indicated. The numbers before each construct on the left correspond to the bar in the graph showing the relative expression of Rluc versus Fluc. The error bars indicate standard deviations. (B) Analysis of ribosome scanning after cap-dependent translation initiation. Sequence elements were cloned into pDualLuc-scan upstream of the Fluc reporter gene. The intraleader (L-ORF; see the text) and the position of the EAV 5′ UTR are indicated. nsp1* corresponds to an upstream ORF (out of frame with Fluc) that encodes the N-terminal 53 amino acids of nsp1. The cytomegalovirus promoter (arrow) is indicated. The numbers before each construct on the left correspond to the bar in the graph showing the relative expression of Fluc versus Rluc.
To assess whether cap-dependent translation initiation of the EAV genome is followed by classical linear ribosome scanning, different cassettes were cloned upstream of the Fluc gene in pDualLuc-IRES3. Because the capping efficiency of the vTF7-3-driven expression system is relatively poor, the T7 promoter was replaced with a cytomegalovirus immediate-early promoter (see Fig. 7B). The EAV 5′ UTR (nt 1 to 224) was placed upstream of the Fluc gene, yielding pDualLuc-scan1. In construct pDualLuc-scan2, the EAV 5′ UTR was extended until nt 303 (including the 5′ end of the replicase gene), containing an extra AUG codon at its 3′ end to drive Fluc expression. Subsequently, the replicase AUG was mutated into UAG (pDualLuc-scan3). Finally, the 5′ UTR of D1-301 (see above) was inserted (pDualLuc-scan4) and a stable hairpin was introduced into this region at nt 306 to block scanning ribosomes, generating pDualLuc-scan5 (40). RK-13 cells were transfected with 1 μg of plasmid and were lysed at 20 h p.t. and assayed for Fluc and Rluc activities.
RESULTS
Leader-to-body fusion can occur at a different site in the EAV genome.
In a recent mutagenesis study, the loop and stem regions of the EAV LTH were targeted to disrupt their primary and secondary structures. It was found that mutants in which the LTH conformation was predicted to be maintained showed the highest level of sg RNA synthesis. From this analysis, the EAV LTH was concluded to be generally important for transcription (38), but it should be noted that the analysis of many of the mutants in this study was hampered by side effects at the level of genome replication, for which signals apparently (and not unexpectedly) overlap with those for sg RNA synthesis.
If the LTH were an independent entity in sg RNA synthesis, functioning as a sort of acceptor “hot spot” for a copy choice recombination-like process, it might be possible to move it to a different position in the genome while maintaining its function in leader-to-body fusion. Due to the compact organization of the EAV genome, locations where such an insertion would not interrupt one of the viral genes were not readily available. Thus, we chose to duplicate the 5′-proximal 301 nucleotides of the genome, which were previously found to suffice for efficient genome replication and sg RNA synthesis (35). The duplicated sequence (“duplicated 5′-proximal domain,” or D5D) consisted of the 5′-untranslated region and the first 29 codons of the replicase gene (nsp1-coding region) and was inserted at nucleotide position 306 of the EAV genome. In the construct with this 5′-proximal tandem repeat (D5Dwt) (Fig. 2A), the native leader TRS and replicase translation initiation codon were inactivated (UCAACU to AGUUGU and AUG to UAG, respectively; see Materials and Methods). Thus, translation initiation should now occur from the D5D AUG codon (at nt 530 instead of 224 of the genome). For leader-to-body fusion, the D5D LTH was available, and its use should result in addition of a 516- instead of 211-nt common leader sequence to all sg RNAs.
Construct D5Dwt was tested in a transfection experiment, and immunofluorescence assays showed that transfection efficiencies were comparable to those of the wt control and that this construct was able to replicate its genome RNA. In addition, nucleocapsid protein was readily detectable, which is indicative of the synthesis of sg mRNA7 (data not shown). A hybridization analysis of total intracellular RNA isolated at 15 h p.t. indeed confirmed the synthesis of a complete set of sg RNAs that were each approximately 300 nt longer than those of the wt control (Fig. 2C; compare the two leftmost lanes). To exclude the possibility that input RNA contributed to the genomic RNA signal in hybridizations, the replication-deficient SGA mutant (42), carrying a fatal amino acid substitution in the RdRp domain, was included in the analysis. Even after prolonged exposure, the transfected RNA of this mutant could not be detected (data not shown).
Because the efficiencies of D5Dwt and wt virus genome replication were very similar (see below), the ratio of subgenomic RNA versus genome RNA could be used to measure the transcriptional capability of the D5Dwt mutant. Quantitation of viral RNAs following hybridization analysis revealed that transcription levels for this mutant reached about 70% of those of the wt virus. The use of the LTH duplicate for leader-to-body fusion was verified by a specific RT-PCR analysis, which confirmed the presence of an extended sg mRNA7 that was not present in cells transfected with the wt control. Direct sequence analysis of the PCR product confirmed that in this novel sg mRNA the leader-to-body fusion indeed mapped to the leader TRS in the LTH duplicate (Fig. 2A and data not shown).
Not unexpectedly, the tandem repeat of two almost identical ∼300-nt sequences in D5Dwt was prone to homologous recombination, rapidly leading to wt recombinant virus and explaining the background of wt-size sg mRNAs in the hybridization analysis (Fig. 2C). Nevertheless, our data confirmed that the 5′-proximal 301 nt of the EAV genome contains essential signals for efficient sg mRNA synthesis and that these signals can be moved 300 nt more distal from the genomic 5′ end without a profound loss of function.
The flanking sequences of the LTH are required for sg RNA synthesis.
The amount of D5Dwt genomic RNA detected in hybridization analysis (Fig. 2C) indicated that its replication, and therefore probably also its translation (see below), were not notably altered compared to the wt genome, a remarkable finding given the >2-fold extension of the 5′ UTR of the viral genome. Consequently, D5Dwt offered an excellent opportunity to analyze the minimal sequence requirements for EAV sg RNA synthesis. Construct pD5Dwt was used as a starting point to make deletions in the D5D sequence, although straightforward deletions downstream of the LTH could not be made, since the replicase-coding region overlaps with the 3′ side of the LTH stem. To circumvent this problem, the codons in the D5D that specified the N-terminal 29 amino acids of nsp1, the N-terminal subunit of the replicase, were altered in some of the mutants. This allowed us to investigate the presence of RNA signals in the sequence downstream of the LTH without changing nsp1. The corresponding mutant, D5Dc (Fig. 2B), which combined a maximum number of translationally silent mutations in the 5′ end of the replicase gene with an intact LTH structure (see Materials and Methods), produced only small amounts of sg mRNA (Fig. 2C), suggesting that RNA sequences downstream of the LTH are relevant for sg RNA synthesis.
To assess the function of the sequences upstream of the LTH, a set of three deletion mutants was engineered. The borders of the deletions were based on the previously established RNA secondary structure (38), which defined a 5′-proximal domain consisting of five hairpins followed by a single-stranded polypyrimidine-rich stretch, which in turn is followed by the hairpin that flanks the LTH at its 5′ side (Fig. 1C). Remarkably, upon deletion of the first domain (corresponding to nt 1 to 135 of the genome) from construct D5Dwt, sg RNA levels were somewhat increased compared to those of D5Dwt (mutant D5DΔ1-135) (Fig. 2B). As expected, the sg mRNAs of D5DΔ1-135 were slightly smaller due to its smaller leader sequence compared to D5Dwt (Fig. 2C). This result indicated that nucleotides 1 to 135 of the D5D sequence were dispensable for transcription. On the other hand, the deletion of the domain comprising the single-stranded stretch and the hairpin upstream of the LTH blocked efficient sg RNA synthesis (mutant D5DΔ136-183) (Fig. 2). A combination of both deletions (mutant D5DΔ1-184), which removed the entire D5D sequence upstream of the LTH, had a similar effect on transcription (Fig. 2C).
The above-mentioned results suggested that flanking sequences on either side of the LTH duplicate are important for efficient sg RNA synthesis. Consequently, we expected that removal of sequences on both sides of the LTH would completely abolish transcription. Surprisingly, however, mutant D5DcΔ1-184 (Fig. 2B) produced moderate levels of sg mRNAs, exceeding the amounts produced by mutants D5DΔ1-184 and D5Dc, which lacked sequences upstream or downstream of the LTH duplicate, respectively (Fig. 2C). These paradoxical results may be explained by the fact that the RNA structures required for transcription are less disturbed in this mutant (see Discussion). Alternatively, the presence of RNA structures induced by the unnaturally rearranged sequences may influence LTH structure or function. The important role of sequences downstream of the LTH was confirmed by altering the replicase codons in this region in mutant D5DΔ1-135, which showed the highest level of transcription in this deletion study so far (Fig. 2B, mutant D5DcΔ1-135). As in the case of the D5Dwt-D5Dc pair, mutations in this region were found to reduce sg RNA transcription (Fig. 2C).
To this point, our analysis indicated that the leader TRS in the context of the LTH alone could be utilized for leader-to-body fusion to a limited extent only and that up- and downstream sequences clearly enhanced the efficiency of this process. To directly assess the role of the LTH structure itself, the stem region was removed from mutant D5DcΔ1-184 to obtain mutant D5DcΔ1-184 Δstem (Fig. 2B). Although the entire 21-nt LTH loop sequence, including the leader TRS, was retained in this construct, hybridization analysis revealed that this deletion abolished sg RNA synthesis completely (Fig. 2C). In addition, by sensitive RT-PCR analysis, only trace amounts of sg mRNA could be detected (data not shown). This finding clearly supported the importance of the LTH itself and its postulated role in sg RNA synthesis.
The minimal region required for efficient sg RNA synthesis may fold into an extended version of the LTH.
During our recent RNA secondary-structure analysis of the 5′-proximal 313 nucleotides of the EAV genome, the equivalent of the D5D sequence, we came across two possible conformations of the LTH, differing in the lengths of their stems (Fig. 1C and 3A) (38). The formation of the extended conformation of the LTH (eLTH) involves the base pairing of sequences that flank the LTH, and since these were found to be necessary for efficient sg RNA synthesis (see above), the eLTH conformation may in fact represent the structure that guides discontinuous RNA synthesis.
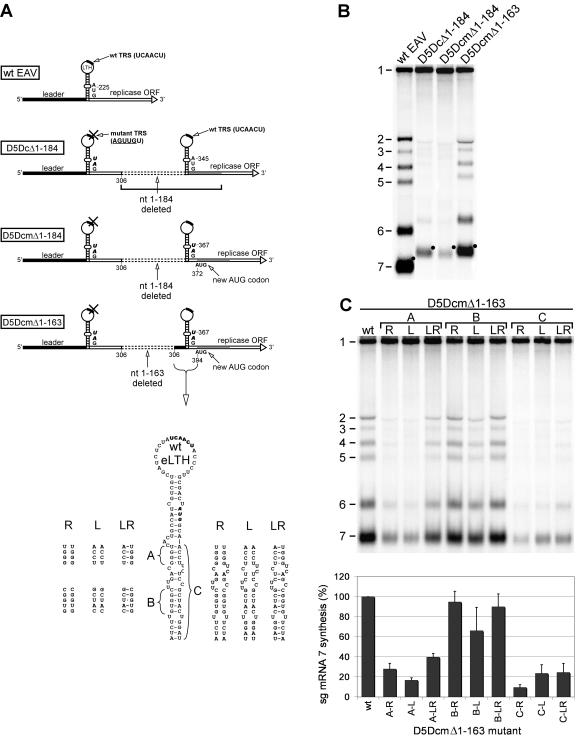
FIG. 3.
Analysis of the relevance of an LTH structure with extended stem for EAV sg RNA synthesis. (A) Schematic representation of the constructs that revealed the importance of LTH-flanking sequences for optimal transcriptional activity (see the legend to Fig. 2). Construct D5DcmΔ1-163 was used to test the importance of the eLTH by site-directed mutagenesis of the indicated regions A, B, and C (see the text). (B) Hybridization analysis of virus-specific RNAs isolated at 15 h p.t. from BHK-21 cells transfected with wt RNA or constructs D5DcΔ1-184, D5DcmΔ1-184, and D5DcmΔ1-163. Note the striking upregulation of sg RNA synthesis in the last construct. (C) Hybridization analysis of virus-specific RNA isolated at 15 h p.t. from BHK-21 cells transfected with mutants in which regions A, B, and C of the eLTH stem were targeted (panel A). For each region, the right side (R), left side (L), or both sides (LR) of the predicted stem were mutated, with potential restoration of base pairing occurring in each of the LR mutants. The bar graph below represents the quantitation of the intensities of the sg mRNA 7 bands, which were normalized to the intensity of the genome band.
Experimental support for this hypothesis was obtained when the upstream flanking sequences of the LTH region in mutant D5DcΔ1-184 were extended in the 5′ direction. This was done in a derivative of D5DcΔ1-184 (D5DcmΔ1-184) in which the 3′ side of the LTH had been rendered noncoding by moving the replicase translation initiation codon 27 nt downstream (Fig. 3A). This change somewhat further impaired sg RNA synthesis of D5DcΔ1-184 (Fig. 3B) but allowed the free mutagenesis of LTH-flanking sequences (see below) without affecting the replicase ORF. Remarkably, the inclusion of additional flanking sequences at the 5′ side of the LTH (corresponding to nt 164 to 184 of the genome) (Fig. 3A) in construct D5DcmΔ1-163 dramatically increased the synthesis of sg mRNAs (compare the two rightmost lanes of Fig. 3B) to levels comparable to those of D5Dwt (Fig. 2C).
The data presented in the previous paragraph suggested that the formation of the eLTH structure might be relevant or, alternatively, that the sequence of nt 164 to 184 of the genome was important for sg RNA synthesis. These possibilities were further investigated by mutagenesis of sequences predicted to be involved in formation of the lower part of the eLTH stem (Fig. 3A). In view of its excellent transcriptional activity, construct D5DcmΔ1-163 was used as a starting point for this analysis. Two regions in the predicted stem extension (A and B in Fig. 3A) were changed, and in addition, the entire predicted stem-extending region was targeted (C in Fig. 3A). All mutants were analyzed in three independent experiments, which revealed that genome replication was not affected by the introduction of mutations in this region of the RNA. Thus, we could use the ratio of sg mRNA7 (the most abundant sg transcript) to genome RNA to quantify transcription levels (Fig. 3C).
The predicted 4-bp stem A, just below the original LTH, was disrupted by changing either its 3′ or its 5′ side to the sequence present at the opposite side, resulting in mutants D5DcmΔ1-163\AR and D5DcmΔ1-163\AL, respectively (Fig. 3A). Both mutations decreased transcription levels four to five times compared to the parental constructs (Fig. 3C). By combining these mutations in the double-mutant D5DcmΔ1-163\ALR, the base-pairing potential was restored, and this resulted in a 1.5- to 2-fold increase in sg RNA synthesis compared to that of the single mutants, although the original transcription levels were not reached (Fig. 3C). The large stem B region was subjected to a similar analysis (Fig. 3A and C), but the transcription levels of the single mutants D5DcmΔ1-163\BR and D5DcmΔ1-163\BL were only slightly affected compared to the level achieved by the parental construct. The transcription levels of the double mutant D5DcmΔ1-163\BLR were in the range of those of the single mutants (Fig. 3C). Finally, the entire predicted eLTH stem (stem C) (Fig. 3A) was altered while keeping the bulges intact. Opening the stem by changing its complete 3′ side (mutant D5DcmΔ1-163\CR) reduced transcription to approximately 10% of the level of the control construct, whereas changing the opposite side of stem C (D5DcmΔ1-163\CL) reduced it to approximately 20% (Fig. 3C). The combination of these mutations in mutant D5DcmΔ1-163\CLR resulted in a twofold increase of sg RNA synthesis compared to D5DcmΔ1-163\CR, but no significant upregulation was observed compared to D5DcmΔ1-163\CL.
Taken together, the data presented above demonstrated that the LTH-surrounding sequences clearly influence sg RNA synthesis and that in particular stem A (Fig. 3A) may contribute to the formation of an extended stem at the base of the originally proposed LTH structure.
Subgenomic mRNA synthesis requires an intact LTH.
In a previous mutagenesis study (38), aimed at providing experimental support for the importance of the LTH structure, many of the mutations introduced into the wt genome severely affected genome replication, thus preventing a straightforward interpretation of the results with regard to sg RNA synthesis. The D5D construct and its derivatives, as described in the previous paragraphs, provided a solid alternative to study transcription-related RNA signals. Thus, previously engineered LTH stem and loop mutations were transferred to construct D5DΔ1-163 (Fig. 4A), in which nt 164 to 301 of the genome were duplicated.
FIG. 4.
Site-directed mutagenesis underlines the importance of the EAV LTH structure for sg RNA synthesis. (A) Schematic representation of the mutations introduced into the LTH structure of the parental construct pD5DΔ1-163 (see the legend to Fig. 2). (B) Hybridization analysis of virus-specific RNA isolated at 15 h p.t. from BHK-21 cells transfected with D5DΔ1-163 RNA or RNAs from the mutants depicted in panel A.
As expected, all D5DΔ1-163-derived LTH mutants displayed efficient genome replication. Subgenomic RNA synthesis, on the other hand, was seriously affected by mutations that partly closed the LTH loop (Fig. 4, D5DΔ1-163\L-R and D5DΔ1-163\L-L) and was almost blocked when the upper part of the LTH stem was destabilized (Fig. 4, D5DΔ1-163\US-R and D5DΔ1-163\US-L). The combination of these sets of mutations in one construct, restoring either the loop size (mutant D5DΔ1-163\L-LR) or the base-pairing possibilities in the upper part of the stem (mutant D5DΔ1-163\US-LR), rescued sg RNA synthesis to levels close to that of the parental construct (Fig. 4B). In conclusion, the results confirmed and extended our previous mutagenesis study performed in the context of a wt full-length clone, which suggested that an intact LTH has an important function in sg RNA synthesis.
Replication competence and stability of mutants with duplicated 5′-proximal sequences.
Potentially, all mutants described in the previous paragraphs were genetically unstable due to the presence of direct repeats in the 5′-proximal regions of their genomes. This was immediately apparent when long sequence stretches had been duplicated, like the entire 5′-proximal 301 nucleotides of the genome in the case of D5Dwt (Fig. 2C). Via a single crossover, the genome of this kind of mutant could be restored to wt sequence. The fact that wt-size sg mRNAs could be observed in a first-cycle analysis strongly suggested the independent occurrence of many such recombination events.
The contribution of recombination-derived wt virus to the genomic RNA detected in hybridizations was assessed in three experiments by using primer extension analysis on intracellular RNA isolated from cells transfected with one of a selected set of constructs: the wild-type control, D5Dwt, D5Dc, and D5DcmΔ1-163. In RNA samples from cells transfected with D5Dwt or D5Dc, both containing the largest duplication used in this study, approximately 10% of the genome RNA originated from recombinant wt virus (data not shown). In contrast, wt-size genome RNA was not detected for mutant D5DcmΔ1-163, suggesting that recombination events were rare (data not shown). Subsequently, the same RNA was subjected to hybridization analysis and the amount of genomic RNA was quantified and corrected for transfection efficiencies. The three mutants were found to produce equal or even larger amounts of genomic RNA than the wt virus, indicating that genome replication was not affected (data not shown).
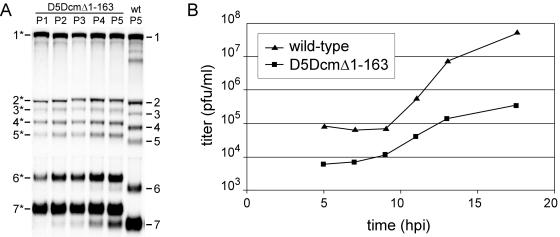
The properties of the most stable mutant, D5DcmΔ1-163 (Fig. 3), were evaluated in five subsequent passages, and its growth characteristics were compared to those of the wt virus. This mutant harbored the smallest duplication that still allowed the same level of sg mRNA synthesis as in the original D5Dwt construct (Fig. 2). BHK-21 cells were transfected with in vitro-transcribed D5DcmΔ1-163 RNA, and the cell culture medium was harvested at 24 h p.t. and used for serial undiluted passaging on fresh BHK-21 cells for four subsequent rounds. Hybridization analysis of intracellular RNA isolated from these cell cultures showed that mutant D5DcmΔ1-163 was relatively stable and that after five passages the majority of the sg mRNAs were still derived from mutant genomes (Fig. 5A). In addition, the replication kinetics of mutant D5DcmΔ1-163 was compared to that of the wt virus, and it showed no apparent delay in virus production, although the overall virus yield after the first infection cycle (∼15 h p.i. at 37°C) was about 10- to 100-fold reduced (Fig. 5B).
FIG. 5.
Characterization of the stability and growth of D5DcmΔ1-163 virus, a viable virus mutant with an extended common leader sequence on all its mRNAs. (A) Gel hybridization analysis of D5DcmΔ1-163-specific RNA isolated at 15 h p.t. (P1) or postinfection (P2 to P5) from BHK-21 cells. After transfection of BHK-21 cells, medium was harvested at 24 h p.t. and subsequently passaged four times in undiluted form. RNA from a wt control is shown in the rightmost lane. The positions of the wt and D5DcmΔ1-163 (indicated with asterisks) mRNAs are indicated. (B) Growth curve of plaque-purified wt and D5DcmΔ1-163 virus. BHK-21 cells were infected at a multiplicity of infection of 5, medium was harvested at the indicated time points, and virus titers were subsequently determined by plaque assays.
Efficient synthesis of a double-nested set of mRNAs.
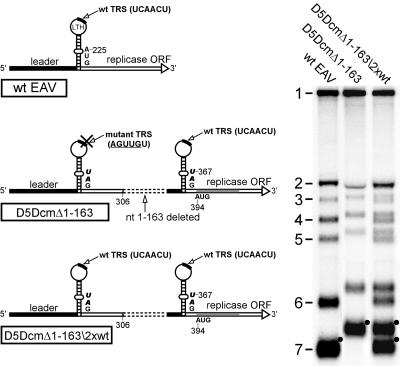
In all constructs described thus far, the authentic leader TRS was inactivated to avoid production of wt sg mRNAs. In a construct carrying a double wt leader TRS, each presented in the context of an LTH structure, we anticipated competition for leader-to-body fusion events. Such a construct would thus provide an indication of how the functionality of the LTH duplicate is related to that of the wt structure.
Construct D5DcmΔ1-163 was used to address this question, because the sizes of the (predicted) sets of sg mRNAs were suitable to achieve good separation upon agarose gel electrophoresis. Construct D5DcmΔ1-163\2xwt was created by restoring the authentic leader TRS to its wt 5′-UCAACU-3′ sequence (Fig. 6A). The presence of two wt leader TRSs (the authentic one and the copy in the D5D sequence) indeed resulted in the synthesis of a perfect double set of six sg mRNAs each (Fig. 6B). Surprisingly, compared to construct D5DcmΔ1-163, the transcription level of the set of “larger” mRNAs was hardly affected by the activation of the production of the set of “smaller” mRNAs. The production of the latter set, in turn, was quite efficient compared to the synthesis of the same mRNAs by the wt virus (Fig. 6B). Despite a small reduction in genome replication (to about 80% of wt levels), the total amount of sg mRNA produced by D5DcmΔ1-163\2xwt was similar to that of the wt virus, and both leader TRSs yielded approximately equal amounts of sg mRNAs. This underlined once again that the LTH in the D5D sequence element indeed is an independent and efficient alternative signal for leader-to-body fusion during EAV sg RNA synthesis.
FIG. 6.
A D5D derivative with two wt LTH domains, including two wt leader TRSs, produces a perfect double-nested set of sg mRNAs. A schematic representation of the wt genome and constructs D5DcmΔ1-163 and D5DcmΔ1-163\2xwt is shown on the left (see the legend to Fig. 2). On the right, a hybridization analysis of intracellular RNA at 15 h p.t. is shown.
Initiation of EAV genome translation.
At 223 nt, the 5′ UTR of the wt EAV genome is relatively long. Also, it contains a short “intraleader” ORF (L-ORF) of 37 codons, starting with an AUG codon at nt 14 to 16. In view of its proximity to the 5′ end of the genome, it is unclear whether this ORF is actually translated. Its product—if any—was previously found to be dispensable for replication in cell culture (23). A remarkable finding during the analysis of the mutants described in the previous paragraphs was the fact that the 5′ UTR of the EAV genome could apparently be extended to 529 nt without a notable effect on replicase expression, a conclusion that was based on Western blot analysis of nsp1 expression in transfected cells and immunofluorescence assays (data not shown). In addition, some of these mutant 5′ UTRs now contained two copies of the L-ORF upstream of the replicase translation initiation codon, with the second one no longer being positioned close to the 5′ end of the genome.
The presence of a cap structure has been reported for another arterivirus (30), and cap analogue is known to be an essential component of in vitro transcription reactions to make infectious full-length RNA (20, 41). Although this suggests that translation is initiated via a cap-dependent mechanism, the observations summarized above prompted us to investigate the mechanism of EAV genome translation in more detail. The possibility of translation initiation via an IRES element, a well-known mechanism proposed to be used by several virus groups (36) and a growing list of cellular mRNAs (11), was evaluated in comparison to classical cap-dependent initiation followed by linear ribosome scanning, with initiation occurring at the most 5′-proximal AUG codon (15, 17).
The IRES activity of the EAV 5′ UTR (nt 1 to 224) was tested using a dual-luciferase reporter construct in which this element was inserted between the firefly and Renilla luciferase genes (pDualLuc-IRES1) (Fig. 7A). To exclude the possibility that coding sequences downstream of the EAV 5′ UTR are an essential part of a possible IRES element, the 5′ end of ORF1a (nt 225 to 287) was added to the 5′ UTR and fused in frame to the Renilla gene, thus creating an N-terminal extension of 21 amino acids (pDualLuc-IRES2). The ratio of Rluc over Fluc expression was taken as a measure for IRES activity. Since in both cases no detectable IRES activity was found compared to the well-established activity of the EMCV IRES element (pDualLuc-IRES3 and pDualLuc-IRES4), the results obtained with the above-mentioned constructs strongly argued against the presence of an IRES element in the 5′-proximal region of the EAV genome (Fig. 7A).
Using the EMCV IRES-mediated Rluc expression of construct pDualLuc-IRES3 as an internal standard, we subsequently analyzed whether the 5′ UTR of the EAV genome is scanned by ribosomes, presumably after cap-dependent translation initiation. When the EAV 5′ UTR preceded the Fluc gene (pDualLuc-scan1), strong Fluc expression (Fluc/Rluc ratio) was observed (Fig. 7B). Subsequently, a small (53-codon) ORF was introduced upstream of the Fluc gene by extending the EAV 5′ UTR with the 5′ end of ORF1a (nt 227 to 303). Scanning ribosomes would be expected to translate this upstream ORF, and as a result, fewer ribosomes would be available for the translation of the downstream Fluc gene (16). Indeed, Fluc expression by this mutant was decreased dramatically (pDualLuc-scan2) (Fig. 7B). In line with the scanning hypothesis, Fluc expression was restored to normal levels when the upstream AUG was converted to UAG (pDualLuc-scan3) (Fig. 7B). The influence of the short L-ORF duplicate in the 529-nt-long 5′ UTR of mutant D5D (Fig. 2A) was evaluated by inserting this 5′ UTR upstream of the Fluc gene (pDualLuc-scan4). Remarkably, Fluc expression was hardly affected by the increased size of this 5′ UTR, nor was it affected by the presence of two copies of the L-ORF (Fig. 7B). This result indicated that, at least in vitro, the large 5′ UTR of D5D-derived constructs did not negatively influence translation of the replicase. Finally, we evaluated the effect of the insertion of a stable hairpin structure upstream of the Fluc ORF, a change that should reduce translation efficiency because such structures have to be unfolded before scanning ribosomes can proceed (15, 28). At nt 300 of the 529-nt D5D 5′ UTR, we inserted a stable hairpin (ΔG = −60 kcal/mol) that was previously shown to inhibit chloramphenicol acetyltransferase expression by 92 to 98%, depending on the position of this hairpin in the insulin-like growth factor II leader 1 (40). In our hands, the presence of this hairpin in pDualLuc-scan5 reduced Fluc expression by 80%, suggesting that indeed this hairpin interfered with ribosome scanning (Fig. 7B). In conclusion, the experiments performed with the pDualLuc vectors strongly suggested that EAV genome translation is not IRES mediated and that translational initiation conforms to the conventional “ribosome-scanning” model (15, 17).
DISCUSSION
In addition to providing experimental support for an important role of the previously defined leader TRS-presenting hairpin in sg RNA synthesis, the constructs used in this study have revealed a remarkable functional flexibility of the 5′-proximal region of the EAV genome. Genome replication was not significantly affected in D5Dwt and its derivatives with duplicated 5′-proximal sequences. Even more striking, and logically the basis of efficient genome replication, was the fact that increasing the size of the genomic 5′ UTR by more than twofold did not apparently cripple genome translation, despite the abundant presence of RNA structures (Fig. 1C) and, in some cases, an additional L-ORF. However, this short L-ORF did not affect expression of the Fluc reporter (Fig. 7B). The simplest explanation is that conventional scanning ribosomes do not recognize the L-ORF AUG codon, because it is located in a weak sequence context for translation initiation. The collective data from our translation experiments argued against IRES-mediated translation initiation (Fig. 7A). Besides, the region flanking the novel replicase translation initiation codon in the D5D was altered extensively during our mutagenesis studies and would likely have affected RNA structures with IRES activity. The translational flexibility of the system permitted the expansion of the 5′ UTR of the genome and all sg mRNAs in D5Dwt and its derivatives. Some of these (e.g., D5DcmΔ1-163) (Fig. 3 and 5) were even stable for several passages and replicated to titers 10- to 100-fold lower than those reached by the wt virus. The successful uncoupling of functions normally relying on overlapping RNA signals and the type of mutants developed in this study will be useful for further dissection of this multifunctional domain of the EAV genome.
In previous studies, we analyzed the functions of the EAV leader TRS, as well as body TRSs, in sg RNA synthesis. Whereas multiple functions were attributed to the body TRSs, it was proposed that the primary role of the leader TRS is to serve as a “base-pairing target” in the recombination-like fusion process that results in the joining of leader and body sequences (Fig. 1B) (26). Upon mutagenesis of the leader TRS, the presence of sufficient base-pairing potential appeared to be the only prerequisite for sg RNA synthesis, and a direct correlation between the stability of the leader TRS-body TRS duplex and transcription levels was observed, both for arteriviruses and coronaviruses (27, 49). Although TRS-like sequences are present at other positions in the EAV genome, we have never obtained evidence for productive base pairing of a body TRS to an alternative acceptor TRS. Clearly, its 5′-proximal position makes the leader TRS unique, possibly also because 5′-3′ interactions in the genome may bring body and leader TRSs into each other's vicinity (18). However, our previous studies of the RNA secondary structure of this region of the genome suggested additional specific features that might influence leader TRS function, and preliminary evidence supporting the importance of the structural context provided by the LTH was obtained (38). The present study confirms and extends these observations and demonstrates that the LTH is an independent structural entity that can be moved to direct leader-to-body fusion at another position in the 5′-proximal region of the EAV genome. Mutagenesis of the LTH in the context of the D5D constructs strongly supported the importance of the LTH as a transcription-guiding element (Fig. 4). We also delimited the LTH-flanking sequences required for efficient EAV transcription by deletion mutagenesis of the D5D sequence (Fig. 2). The activity of the LTH structure was clearly influenced by the presence of both upstream and downstream flanking sequences (Fig. 2). These sequences were predicted to fold into a base-paired region that may extend the LTH stem (Fig. 3). Mutations in this region clearly affected transcription, although the attempts to obtain direct support for base-pairing interactions of these sequences were not very convincing (Fig. 3). In this regard, the best results were obtained with domain A (Fig. 3), the top part of the LTH stem extension, for which a certain level of restoration of sg RNA synthesis was observed in the LR double mutant. An additional role for the RNA primary structure or the formation of misfolded alternatives of the eLTH may explain the fact that the results with domain B and C mutants were less convincing (Fig. 3). Alternatively, the stability of the LTH stem closest to the leader TRS may be very critical for its function (Fig. 4), and the contribution of lower stem regions may be small (Fig. 3). Subtler single-nucleotide changes may be used in future studies to probe the functions of the LTH stem regions in more detail.
Other support for the importance of an eLTH fold comes from the theoretical analysis of RNA structures in some of our mutants and in other arteriviruses. For example, compared to mutant D5DΔ1-184 (Fig. 2), sg mRNA synthesis by construct D5DcΔ1-184 (Fig. 3) was remarkably increased. In the latter construct, coding sequences downstream of the LTH were mutated. The entirely artificial combination of this set of translationally silent mutations downstream of the LTH and the Δ1-184 deletion upstream was found to give rise to an alternative potential LTH extension, mimicking the wt structure and with similar stability (Fig. 8A). During our analysis of other arterivirus genomes, the LTH was predicted to contain either a long or a short stem region (Fig. 8B) (38). In the structure prediction for the short version of the LTH, alternative hairpins were predicted at the expense of the formation of the lower part of the eLTH stem. The folding of the eLTH, in turn, is supported by a covariation observed at the bottom of the Porcine reproductive and respiratory syndrome virus (PRRSV) structure (Fig. 8B). The two alternative predicted LTH conformations did tolerate the sequence variations found in different strains for EAV, Lactate dehydrogenase-elevating virus, and PRRSV (only one Simian hemorrhagic fever virus sequence was available). Additionally, the phylogenetic and probing data obtained during our analysis of the 5′-proximal region of the EAV genome did not exclude alternative conformations, although the data favored the short LTH to a certain extent (38).
FIG. 8.
(A) RNA secondary-structure prediction of the LTH and flanking sequences of EAV strain Bucyrus and construct D5DcΔ1-184. The arrow signifies a possible switch between the two possible conformations of the EAV LTH region. Structure stability is indicated with a free-energy value. (B) RNA-secondary structure predictions of the LTH regions of different arteriviruses, with possibilities for the formation of an extended LTH stem in each of them. Depicted are LDV strain Plagemann, PRRSV strains VR-2332 and Lelystad, and simian hemorrhagic fever virus strain LVR 42-0/M6941. A box indicates base pair covariations between two PRRSV strains. The position where each predicted stem extension of the LTH starts is indicated with an arrowhead. The replicase translation initiation codon is shown in boldface italic font, and the leader TRS is indicated in boldface font.
The discussion above supports our hypothesis that the eLTH structure may be important for sg RNA synthesis and also suggests that the type and stability of the stem may be a crucial factor. Nevertheless, biochemical evidence for the presence of these structures remains to be obtained. In addition, protein factors may recognize the different LTH conformations and thereby, e.g., influence sg RNA synthesis. Whether the alternative structures predicted for arteriviruses represent a conformational switch in this multifunctional region of the genome remains to be investigated. Examples of such conformational switches that are based on alternative RNA secondary structures, either self-induced (24) or modulated by other molecules (19), are widespread in RNA virology. Conformational transitions in viral RNAs were suggested to regulate RNA function at different stages of the viral replication cycle (7, 8, 25, 44). In the case of arteriviruses, a conformational switch in the genomic leader region could regulate, e.g., functions involved in translation, replication, and/or sg mRNA synthesis.
Acknowledgments
We thank Udeni Balasuriya and Jim Maclachlan (University of California, Davis) for providing monoclonal antibodies. Chantal Reusken kindly provided some of the pDualLuc reporter plasmids. We appreciated helpful discussions with Willy Spaan and Gijs Versteeg.
E. van den Born was supported by Council for Chemical Sciences grant CW 99-010 from The Netherlands Organization for Scientific Research.
REFERENCES
- 1.Brian, D. A., and W. J. M. Spaan. 1997. Recombination and coronavirus defective interfering RNAs. Semin. Virol. 8:101-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, R. Y., R. Krishnan, and D. A. Brian. 1996. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J. Virol. 70:2720-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowley, J. A., C. M. Dimmock, and P. J. Walker. 2002. Gill-associated nidovirus of Penaeus monodon prawns transcribes 3′-coterminal subgenomic mRNAs that do not possess 5′-leader sequences. J. Gen. Virol. 83:927-935. [DOI] [PubMed] [Google Scholar]
- 4.den Boon, J. A., E. J. Snijder, E. D. Chirnside, A. A. de Vries, M. C. Horzinek, and W. J. M. Spaan. 1991. Equine arteritis virus is not a togavirus but belongs to the coronavirus-like superfamily. J. Virol. 65:2910-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Vries, A. A., E. D. Chirnside, M. C. Horzinek, and P. J. Rottier. 1992. Structural proteins of equine arteritis virus. J. Virol. 66:6294-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Vries, A. A., M. C. Horzinek, P. J. Rottier, and R. J. de Groot. 1997. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin. Virol. 8:33-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diez, J., M. Ishikawa, M. Kaido, and P. Ahlquist. 2000. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA 97:3913-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dirac, A. M., H. Huthoff, J. Kjems, and B. Berkhout. 2002. Regulated HIV-2 RNA dimerization by means of alternative RNA conformations. Nucleic Acids Res. 30:2647-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gultyaev, A. P., F. H. van Batenburg, and C. W. Pleij. 1995. The computer simulation of RNA folding pathways using a genetic algorithm. J. Mol. Biol. 250:37-51. [DOI] [PubMed] [Google Scholar]
- 11.Hellen, C. U. T., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 12.Jackson, R. J., and A. Kaminski. 1995. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA 1:985-1000. [PMC free article] [PubMed] [Google Scholar]
- 13.Jang, S. K., M. V. Davies, R. J. Kaufman, and E. Wimmer. 1989. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J. Virol. 63:1651-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joo, M., and S. Makino. 1992. Mutagenic analysis of the coronavirus intergenic consensus sequence. J. Virol. 66:6330-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozak, M. 1989. The scanning model for translation—an update. J. Cell Biol. 108:229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak, M. 1991. Structural features in eukaryotic messenger-RNAs that modulate the initiation of translation. J. Biol. Chem. 266:19867-19870. [PubMed] [Google Scholar]
- 17.Kozak, M. 2002. Pushing the limits of the scanning mechanism for initiation of translation. Gene 299:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai, M. M. 1998. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology 244:1-12. [DOI] [PubMed] [Google Scholar]
- 19.Mandal, M., and R. R. Breaker. 2004. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 5:451-463. [DOI] [PubMed] [Google Scholar]
- 20.Meulenberg, J. J. M., J. N. A. Bos-De Ruijter, R. van de Graaf, G. Wensvoort, and R. J. M. Moormann. 1998. Infectious transcripts from cloned genome-length cDNA of porcine reproductive and respiratory syndrome virus. J. Virol. 72:380-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 22.Molenkamp, R., S. Greve, W. J. M. Spaan, and E. J. Snijder. 2000. Efficient homologous RNA recombination and requirement for an open reading frame during replication of equine arteritis virus defective interfering RNAs. J. Virol. 74:9062-9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molenkamp, R., H. van Tol, B. C. Rozier, Y. van der Meer, W. J. M. Spaan, and E. J. Snijder. 2000. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J. Gen. Virol. 81:2491-2496. [DOI] [PubMed] [Google Scholar]
- 24.Nagel, J. H. A., and C. W. Pleij. 2002. Self-induced structural switches in RNA. Biochimie 84:913-923. [DOI] [PubMed] [Google Scholar]
- 25.Olsthoorn, R. C., S. Mertens, F. T. Brederode, and J. F. Bol. 1999. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 18:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasternak, A. O., E. van den Born, W. J. M. Spaan, and E. J. Snijder. 2001. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 20:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasternak, A. O., E. van den Born, W. J. M. Spaan, and E. J. Snijder. 2003. The stability of the duplex between sense and antisense transcription-regulating sequences is a crucial factor in arterivirus subgenomic mRNA synthesis. J. Virol. 77:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelletier, J., and N. Sonenberg. 1985. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic messenger-RNA reduces translational efficiency. Cell 40:515-526. [DOI] [PubMed] [Google Scholar]
- 29.Reusken, C. B. E. M., T. J. Dalebout, P. Eerligh, P. J. Bredenbeek, and W. J. M. Spaan. 2003. Analysis of hepatitis C virus/classical swine fever virus chimeric 5′ NTRs: sequences within the hepatitis C virus IRES are required for viral RNA replication. J. Gen. Virol. 84:1761-1769. [DOI] [PubMed] [Google Scholar]
- 30.Sagripanti, J. L., R. O. Zandomeni, and R. Weinmann. 1986. The cap structure of simian hemorrhagic fever virion RNA. Virology 151:146-150. [DOI] [PubMed] [Google Scholar]
- 31.Sawicki, S. G., and D. L. Sawicki. 1995. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 380:499-506. [DOI] [PubMed] [Google Scholar]
- 32.Snijder, E. J., M. C. Horzinek, and W. J. M. Spaan. 1990. A 3′-coterminal nested set of independently transcribed messenger-RNAs is generated during Berne virus replication. J. Virol. 64:331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snijder, E. J., A. L. M. Wassenaar, and W. J. M. Spaan. 1994. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J. Virol. 68:5755-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spaan, W. J. M., H. Delius, M. Skinner, J. Armstrong, P. Rottier, S. Smeekens, B. A. van der Zeijst, and S. G. Siddell. 1983. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 2:1839-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tijms, M. A., L. C. van Dinten, A. E. Gorbalenya, and E. J. Snijder. 2001. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus. Proc. Natl. Acad. Sci. USA 98:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vagner, S., B. Galy, and S. Pyronnet. 2001. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2:893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Berlo, M. F., M. C. Horzinek, and B. A. van der Zeijst. 1982. Equine arteritis virus-infected cells contain six polyadenylated virus-specific RNAs. Virology 118:345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van den Born, E., A. P. Gultyaev, and E. J. Snijder. 2004. Secondary structure and function of the 5′-proximal region of the equine arteritis virus RNA genome. RNA 10:424-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Meer, Y., H. van Tol, J. K. Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72:6689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Velden, A. W., K. van Nierop, H. O. Voorma, and A. A. M. Thomas. 2002. Ribosomal scanning on the highly structured insulin-like growth factor II-leader 1. Int. J. Biochem. Cell Biol. 34:286-297. [DOI] [PubMed] [Google Scholar]
- 41.van Dinten, L. C., J. A. den Boon, A. L. M. Wassenaar, W. J. M. Spaan, and E. J. Snijder. 1997. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc. Natl. Acad. Sci. USA 94:991-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Dinten, L. C., S. Rensen, A. E. Gorbalenya, and E. J. Snijder. 1999. Proteolytic processing of the open reading frame 1b-encoded part of arterivirus replicase is mediated by nsp4 serine protease and is essential for virus replication. J. Virol. 73:2027-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. M. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:12056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Meerten, D., G. Girard, and J. van Duin. 2001. Translational control by delayed RNA folding: identification of the kinetic trap. RNA 7:483-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Vliet, A. L., S. L. Smits, P. J. Rottier, and R. J. de Groot. 2002. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 21:6571-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner, H. M., U. B. R. Balasuriya, and M. N. James. 2003. The serologic response of horses to equine arteritis virus as determined by competitive enzyme-linked immunosorbent assays (c-ELISAs) to structural and non-structural viral proteins. Comp. Immunol. Microbiol. Infect. Dis. 26:251-260. [DOI] [PubMed] [Google Scholar]
- 47.White, K. A. 2002. The premature termination model: a possible third mechanism for subgenomic mRNA transcription in (+)-strand RNA viruses. Virology 304:147-154. [DOI] [PubMed] [Google Scholar]
- 48.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuniga, S., I. Sola, S. Alonso, and L. Enjuanes. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78:980-994. [DOI] [PMC free article] [PubMed] [Google Scholar]