Abstract
The Nipah virus V and W proteins, which are encoded by the P gene via RNA editing, have a common N-terminal domain but unique C-terminal domains. They localize to the cytoplasm and nucleus, respectively, and have both been shown to function as inhibitors of JAK/STAT signaling. Here we report that V and W proteins also block virus activation of the beta interferon (IFN-β) promoter and the IFN regulatory factor 3 (IRF3)-responsive IFN-stimulated gene 54 promoter. Surprisingly, only W protein shows strong inhibition of promoter activation in response to stimulation of Toll-like receptor 3 (TLR3) by extracellular double-stranded RNA. This activity is dependent on the nuclear localization of W protein. Within the unique C-terminal domain of W protein, we have identified a nuclear localization signal (NLS) that requires basic residues at positions 439, 440, and 442. This NLS is responsible for mediating the preferential interaction of W protein with karyopherin-α 3 and karyopherin-α 4. Nuclear localization of W protein therefore enables it to target both virus and TLR3 pathways, whereas the cytoplasmic V protein is restricted to inhibiting the virus pathway. We propose that this discrepancy is in part due to the V protein being less able to block signaling in response to the kinase, TBK-1, whereas both V and W can prevent promoter activation in response to IKKɛ. We demonstrate that, when the TLR3 pathway is stimulated, the levels of phosphorylated IRF3 are reduced in the presence of W protein but not V protein, confirming the differential effects of these proteins and illustrating that W protein-mediated inhibition is due to a loss of active IRF3.
A major role of the host's innate immune response upon viral infection is to sense the presence of the invading virus and to respond by establishing an antiviral state within the infected cell. This response is mediated primarily by alpha/beta interferon (IFN-α/β) and can be divided into three basic stages (reviewed in references 22 and 56). First, virus infection stimulates the production of IFN-β, which is released from the infected cell. In the second stage, the released IFN binds to the IFN-α/β receptor and initiates a signaling cascade (termed the JAK/STAT pathway) that results in the transcriptional upregulation of IFN-stimulated genes (ISGs). The products of these genes are antiviral proteins (e.g., PKR and Mx) that function in a variety of ways to halt virus replication in the infected cells and prevent infection of neighboring uninfected cells.
Induction of IFN-β synthesis is a hallmark of virus infection, but it can also be induced by treating cells with double-stranded RNA (dsRNA) alone. dsRNA is produced as a replication product during most virus infections, and therefore it is thought to be the virus-associated molecule that triggers IFN synthesis. This implicates Toll-like receptor 3 (TLR3) as a sensor of virus infection, since it has been shown to specifically recognize dsRNA, an event that leads to IFN-β induction (2). TLR3 signaling is mediated by association of its Toll/interleukin-1 receptor (TIR) domain with a TIR domain-containing adaptor protein called TRIF (46, 73, 74). Signaling downstream of TRIF activates nuclear factor κB (NF-κB) and IFN regulatory factor 3 (IRF3) (20, 54), two transcription factors that are also activated in response to viral infection and are essential for activation of the IFN-β promoter. Whereas NF-κB activation also occurs in response to other TLRs, IRF3 activation is specific to TLRs that utilize TRIF as an adaptor, i.e., TLR3 and TLR4 (1). IRF3 is activated by phosphorylation of its C-terminal domain by the recently described kinases, IKKɛ and TBK-1 (20, 41, 58). This places the kinases upstream of IRF3 and downstream of TRIF and TLR3 in the signaling cascade. However, despite the similarity between the signaling molecules involved in TLR3- and virus-mediated activation of the IFN-β promoter, several lines of evidence from gene knockout studies support the notion that the virus pathway is independent of TLR3 and TRIF (18, 33). Further evidence comes from the recent identification of a cytoplasmic RNA helicase, RIG-I, that is required for virus induction of IFN-β and functions in the absence of TRIF and TLR3 (76). Another RNA helicase, mda-5, has also been shown to be involved in the induction of IFN-β (2a).
To counteract the antiviral effects of IFN, many viruses have devised ways of downregulating IFN synthesis. The discovery of the signaling components involved in this pathway will aid in the identification of the precise molecules that are targeted by the viral IFN antagonist proteins. Among RNA viruses, the influenza virus NS1 protein (63), the Ebola virus VP35 protein (5), and the NS3/4A protein of hepatitis C virus (21) have all been shown to block activation of IRF3 and therefore likely target an upstream component in the pathway. The NSs proteins of bunyamwera virus and rift valley fever virus also inhibit IFN production but act downstream of IRF3 by targeting the RNA polymerase transcription machinery (7, 65, 72). Although the majority of paramyxoviruses target the IFN signaling (or JAK/STAT) pathway, a few members have also been found to limit the production of IFN: the rubulaviruses, simian virus 5 (SV5), and human parainfluenza virus 2, encode this function in their V proteins (30, 51), whereas both V and C proteins of the respirovirus, Sendai virus (SeV), appear to be involved (37). Measles virus (a morbillivirus) has also been reported to prevent IFN synthesis, but it is unclear which gene product encodes this function (44). Finally, the pneumoviruses, human and bovine respiratory syncytial virus, target the IFN production pathway through either one or both of their nonstructural proteins, NS1 and NS2 (9, 55, 61, 66).
Nipah virus and Hendra virus are the only two members of the Henipavirus genus within the Paramyxovirinae subfamily, and they are characterized by their ability to infect multiple mammalian species (27, 34, 67). In humans, Nipah virus is a highly virulent pathogen causing severe encephalitis that resulted in high fatality rates in both the Malaysian outbreak in 1999 (14) and the 2004 outbreak in Bangladesh (12, 19). The P gene of Nipah virus is predicted to encode four proteins, namely, P, V, W, and C (27, 67). The V and W transcripts are produced as a result of RNA editing, where one or two extra G residues, respectively, are inserted at the editing site during viral transcription. This causes a frameshift so that the resulting V and W proteins share the same N-terminal domain with P but have unique C-terminal domains. The C protein is encoded by an alternate open reading frame (ORF) in the 5′ end of the P gene. All four of the P gene products have been demonstrated to have IFN antagonist activity (47, 53, 59). The target of the C protein is not known, but the P, V, and W proteins act on the IFN signaling or JAK/STAT pathway by interacting with STAT1 through a domain in their common N terminus (52, 53, 59). This prevents the phosphorylation and activation of STAT1 and hence the transcriptional upregulation of ISGs. Although they act by the same mechanism, V and W proteins do so from different cellular compartments, with V protein in the cytoplasm and W protein in the nucleus (59). V protein has been described to shuttle between the cytoplasm and nucleus and a nuclear export signal has been identified that is required for its accumulation in the cytoplasm (52, 53). We describe here the presence of a nuclear localization signal (NLS) in the C terminus of the W protein. It would therefore appear that the virus is purposefully targeting V and W proteins to different cellular compartments. In the course of investigating the potential role of V and W proteins in preventing the production of IFN, as has been described for the V proteins of SV5 and SeV (30, 37, 51), we discovered that nuclear localization imparts a unique function on W protein, enabling it to inhibit both TLR3- and virus-induced signaling pathways, whereas the cytoplasmic V protein predominantly targets the virus pathway.
MATERIALS AND METHODS
Cells and virus.
HeLa and 293T cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. Virus-infected cells were maintained in Dulbecco modified Eagle medium supplemented with 0.3% bovine serum albumin. SeV strain Cantell was grown in 10-day-old embryonated chicken eggs at 37°C for 48 h.
Expression plasmids and antibodies.
Unless otherwise stated, all constructs are cloned into the mammalian expression vector, pCAGGS (45). The Nipah virus V and W protein expression plasmids have been described previously (47, 59). The WBR1, WBR2, WBR3, WBR4, WBR12, and WBR34 constructs, described in Results, were made by site-directed mutagenesis by using the QuikChange site-directed mutagenesis kit from Stratagene. The V-SV40 NLS and WBR3-SV40 NLS constructs contain the NLS sequence (PKKKRKV) of the SV40 T antigen at their C terminus. All V and W constructs have an N-terminal hemagglutinin (HA) tag. The karyopherin-α 1 and karyopherin-α 2 constructs were described previously (68). The full-length clone of karyopherin-α 3 was purchased from the American Type Culture Collection, whereas karyopherin-α 4 was amplified by reverse transcription-PCR (RT-PCR) from RNA extracted from A549 cells. All karyopherin-α constructs have an N-terminal FLAG tag. The cDNAs for TLR3 and TRIF were amplified by RT-PCR from RNA extracted from human dendritic cells. The expression plasmids for TBK-1 and IKKɛ (in pcDNA3.1−) were kindly provided by John Hiscott (Lady Davis Institute, McGill University, Montreal, Quebec, Canada). The IRF3 monoclonal antibody (17C2) was generated by immunizing mice with a bacterially expressed glutathione S-transferase (GST) fusion protein of the C-terminal 100 amino acids (328 to 427) of human IRF3 and was used at 1:2,000 dilution. The antibody recognizing IRF3 phosphorylated at serine 396 (Upstate, Lake Placid, NY) was used at a 1:1,000 dilution. The P56 antibody was kindly provided by Ganes Sen (Cleveland Clinic Foundation, Cleveland, Ohio) and was used at a 1:2,000 dilution.
Reporter gene assays.
Transfections of 293T cells were performed by using a calcium phosphate method (Mammalian Transfection Kit; Stratagene), according to the manufacturer's instructions. Approximately 5 × 105 cells were transfected with 0.3 μg of the IFN-β promoter or ISG54 promoter chloramphenicol acetyltransferase (CAT) reporter constructs, 0.3 μg of a constitutive Renilla luciferase reporter construct (pRL-tk)[Promega], and 2.5 μg of the relevant expression plasmid. Where indicated, expression plasmids for TLR3 (250 ng), TRIF (10 ng), TBK-1 (100 ng), or IKKɛ (100 ng) were included in the transfection mix. When we measured promoter activation by TRIF, TBK-1, or IKKɛ, cells were harvested 24 h posttransfection and analyzed for CAT and luciferase activities. For measuring TLR3 signaling, poly(I-C) was added to the medium 24 h posttransfection at a final concentration of 25 μg/ml. Fresh medium was added 7 h posttreatment and, after overnight incubation, the cells were harvested and analyzed for CAT and luciferase activities. When we analyzed the response to intracellular dsRNA, the cells were transfected with 20 μg of poly(I-C) by using Lipofectamine 2000 (Invitrogen) 24 h after the initial transfection. Cells were harvested 24 h later and analyzed for CAT and luciferase activities. When we measured promoter activation in response to virus, cells were infected with 25 μl of SeV (10 hemagglutination units/ml) at 24 h posttransfection. Cells were harvested and analyzed for CAT and luciferase activities 24 h postinfection. The CAT activity was quantified by using a phosphorimager and normalized against the luciferase activity.
Immunofluorescence.
HeLa cells were transfected with 2 μg of the indicated HA-tagged Nipah virus expression plasmid by using Lipofectamine 2000 (Invitrogen). Transfected cells were seeded onto glass coverslips and, 24 h posttransfection, were fixed and permeabilized in ice-cold methanol. Fixed cells were probed with an anti-HA rabbit polyclonal antibody (Sigma) and were analyzed by fluorescence microscopy after incubation with a fluorescein isothiocyanate-conjugated secondary antibody (Jackson Immunoresearch).
Immunoprecipitation.
293T cells were transfected with 5 μg of the indicated FLAG-tagged karyopherin-α plasmid or empty vector and 5 μg of the indicated HA-tagged W plasmid by using Lipofectamine 2000 (Invitrogen). At 24 h posttransfection, the cells were washed in cold phosphate-buffered saline (PBS) and lysed in 500 μl of extract buffer (50 mM Tris [pH 7.5], 280 mM NaCl, 0.5% IGEPAL, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, 1 mM dithiothreitol, 0.1 mM sodium vanadate, and protease inhibitors [Complete; Roche]) on ice for 30 min. Extracts were centrifuged at 13,000 rpm for 15 min in a Heraeus Biofuge Pico microcentrifuge, and the supernatant was collected. Agarose beads conjugated with anti-FLAG antibody (Sigma) were washed in PBS and added to the cell extracts, which were then incubated overnight at 4°C with rotation. The beads were washed three times with extract buffer and boiled in 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Eluted proteins and those in the lysate were separated by SDS-PAGE on a 10% gel and transferred to nitrocellulose membrane. The precipitated proteins were probed with an anti-HA antibody (Sigma), and the lysate proteins were probed with an anti-FLAG antibody (Sigma). After incubation with peroxidase-labeled secondary antibodies, the blots were analyzed by chemiluminescence.
GST-pulldown assay.
To create a GST fusion, the C terminus of W protein (Wc) was cloned into the pGEX6P1 vector (Amersham Biosciences, Inc.) between the EcoRI and XhoI sites. The GST-Wc fusion protein was purified from Escherichia coli by using glutathione-Sepharose beads (Amersham Biosciences, Inc.) according to the manufacturer's instructions. Extracts from 293T cells to be used in the pulldown were prepared as follows: 7 × 106 cells were washed in cold PBS and then lysed in 1 ml of lysis buffer (20 mM HEPES [pH 7.4], 150 mM NaCl, 0.5% NP-40, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 10% glycerol, protease inhibitors [Complete; Roche]) for 30 min at 4°C. Lysates were sonicated three times for 5 s on ice and then centrifuged at 13,000 rpm for 15 min in a Heraeus Biofuge Pico microcentrifuge. Supernatants were collected, and the protein concentration was determined by Bradford assay. The lysates were precleared with glutathione-beads (80 μl of a 50% slurry per ml of lysate) for 1 h at 4°C. For the pulldown, 2 to 2.5 mg of precleared cell extract was added to 10 μg of purified GST-Wc or GST alone. After overnight incubation at 4°C, 40 μl of glutathione beads (Amersham) in a 50% slurry were added, and incubation was continued for a further 2 h. The beads were pelleted by centrifugation at 3,000 rpm, the supernatant removed, and the beads washed 10 times in 1 ml of lysis buffer for 5 min. Protein was eluted in 20 μl of glutathione elution buffer from Sigma (prepared according to manufacturer's instructions), boiled in 30 μl of 2× SDS-PAGE loading buffer, and then separated by SDS-PAGE on a 16-by-16-cm 10% gel. Protein bands were visualized by Coomassie blue staining, and bands that were unique to the GST-Wc sample were excised and analyzed by mass spectrometry (The Rockefeller University Proteomics Resource Center, New York, NY).
Quantitative RT-PCR analysis.
Approximately 5 × 105 293T cells were transfected with 2.5 μg of the indicated expression plasmid either with or without 10 ng of TRIF expression plasmid. At 12 h posttransfection the cells were harvested, and the total RNA was isolated by using the Absolutely RNA RT-PCR miniprep kit (Stratagene). Quantitative RT-PCR was performed by the Quantitative PCR Shared Research Facility at Mount Sinai using a previously described SYBR green in an ABI 7900 HT protocol (78). Individual transcripts in each sample were assayed three times, and the mean threshold cycle (CT) was used to calculate the relative fold changes for each gene. Three housekeeping genes (Rps11, actin, and tubulin) were used for global normalization in each experiment, as described previously (25). Primer sequences are as follows: IFNβ (sense-GTCAGAGTGGAAATCCTAAG and antisense-ACAGCATCTGCTGGTTGAAG), ISG56 (sense-TCGGAGAAAGGCATTAGATC and antisense-GACCTTGTCTCACAGAGTTC), and ISG54 (sense-CGTGGGAACCTGGTGACTAA and antisense-TCGTTCCAAGCATACCGTGA).
RESULTS
Identification of the NLS in the W protein.
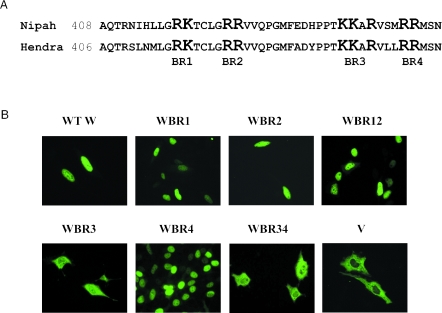
Previously we reported that the Nipah virus V and W proteins have distinctly different localization patterns, with V in the cytoplasm and W in the nucleus (59) (see Fig. 1B). Since these proteins differ only in their C termini, we predicted that an NLS most probably lies within the unique C terminus of W protein. Analysis of this sequence revealed four basic regions that are conserved between Nipah and Hendra viruses (Fig. 1A). Stretches of basic residues are characteristic of NLSs, so to assess whether any one of these regions (or a combination thereof) determines nuclear localization of W, they were mutated to alanine, and the effect on localization examined. Mutation of basic region (BR) 1, 2, or 4 had no effect on the localization of W protein, whereas a mutated BR3 resulted in a significant loss of nuclear accumulation, with a concomitant increase in the cytoplasmic staining (Fig. 1B). Therefore, BR3 (or the “KKAR” motif) serves as an NLS for W protein.
FIG. 1.
Identification of the NLS in the Nipah virus W protein. (A) Alignment of the amino acid sequence of the W protein C-terminal domain for Nipah and Hendra viruses. The four regions rich in basic residues (BR1, BR2, BR3, and BR4) are highlighted. For identification of the NLS, the highlighted residues in each region were mutated to alanine. (B) Localization of the WT V and W proteins and the mutant W proteins as detected by indirect immunofluorescence with an antibody against the HA epitope. The nomenclature of the mutant proteins indicates the basic region in which residues were mutated.
The W NLS mediates preferential binding to karyopherin-α 3 and karyopherin-α 4.
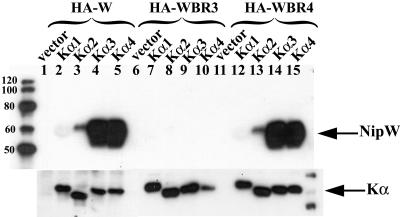
To identify cellular proteins that interact with the C-terminal domain of W, this region was fused to GST, and the purified GST fusion protein was incubated with whole-cell lysate from 293T cells. The interacting cellular proteins were isolated as described in the Materials and Methods and were identified by mass spectrometry. Two of these interactors were identified as karyopherin-α 3 (importin-α 4) and karyopherin-α 4 (importin-α 3), which are closely related members of a family of proteins that mediate nuclear import by binding directly to the NLS of the target protein. The interaction with full-length W protein was verified by immunoprecipitation of W protein with an antibody against FLAG-tagged versions of karyopherin-α 3 and karyopherin-α 4 (Fig. 2). No interaction was detected when the NLS mutant form of W protein (WBR3) was expressed, whereas WBR4 (which still localizes to the nucleus) showed a similar level of binding as wild-type (WT) W protein. These data further confirm that BR3 is an NLS since it is required for the interaction of W protein with karyopherin-α and thus nuclear import. Interestingly, W protein showed a much reduced affinity for binding to karyopherin-α 1 and karyopherin-α 2 (Fig. 2), suggesting that W protein has a preference for using karyopherins 3 and 4 to mediate transport into the nucleus.
FIG. 2.
The preferential interaction of the W protein with karyopherin-α 3 and karyopherin-α 4 is dependent on the C-terminal NLS. 293T cells were transfected with the indicated FLAG-tagged karyopherin-α (Kα) construct together with HA-tagged constructs of either WT W, WBR3 or WBR4. Protein complexes were immunoprecipitated with an antibody against the FLAG epitope. Coimmunoprecipitation of protein W was analyzed by SDS-PAGE and immunoblotting with an anti-HA antibody (upper panel), while expression of karyopherin-α in the lysates was confirmed by immunoblotting with an anti-FLAG antibody (lower panel).
Both V and W proteins inhibit virus activation of IRF3-responsive promoters.
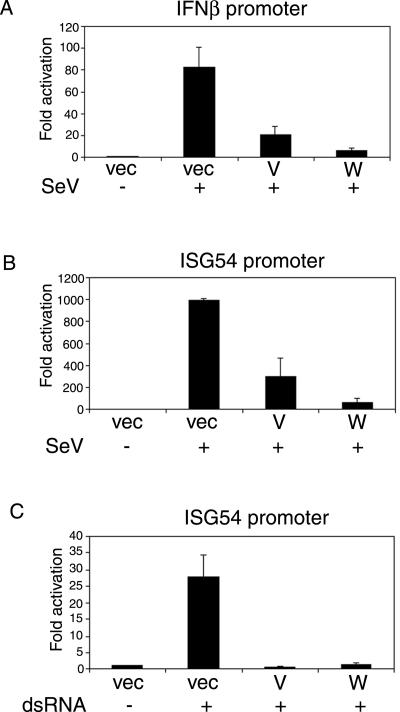
The V and W proteins have been described as IFN antagonist proteins because of their ability to block signaling from the IFN receptor (the JAK/STAT pathway) (47, 53). This pathway is targeted by the majority of paramyxovirus IFN antagonists described to date. However, in addition to targeting IFN signaling, the V proteins of SV5 and hPIV2 have also been reported to inhibit the induction of IFN-β (30, 51). To investigate whether the V and W proteins of Nipah virus have an effect on the IFN production pathway, we used a reporter assay to measure activation of the IFN-β promoter in response to SeV infection. In the presence of either V or W protein, activation of the promoter was reduced 4- to 13-fold (Fig. 3A), indicating that both proteins can potentially limit transcription of the IFN-β gene in response to virus infection. IRF3 is one of the key transcription factors involved in activation of the IFN-β promoter (35, 71, 77). To study the IRF3-dependent pathway in isolation, we used the ISG54 promoter, which is directly responsive to activated IRF3 (26) and is strongly activated in response to virus infection. As was seen with the IFN-β promoter, both V and W proteins efficiently inhibited activation of the ISG54 promoter in response to SeV infection (Fig. 3B), suggesting that they block the IRF3-dependent pathway. dsRNA, produced during viral replication, is believed to serve as the signal that initiates the IFN response within an infected cell. The presence of intracellular synthetic dsRNA can therefore mimic a virus infection. Similar to the effect seen with SeV infection, V and W proteins blocked activation of the ISG54 promoter in response to transfected dsRNA (Fig. 3C). Therefore, V and W proteins are likely targeting the signaling pathway that is activated upon recognition of intracellular dsRNA.
FIG. 3.
Nipah virus V and W proteins inhibit IRF3-responsive promoter activation by virus and intracellular dsRNA. (A and B) 293T cells were transfected with either the IFNβ-CAT (A) or ISG54-CAT (B) reporter plasmids and the indicated expression construct. Transfected cells were either mock infected or infected with 25 μl of SeV (10 hemagglutination units/ml). (C) 293T cells were first transfected with the ISG54-CAT reporter plasmid and the indicated expression plasmids and then, 1 day later, were either mock transfected or transfected with 20 μg of poly(I-C). CAT and luciferase activities were assayed 1 day postinfection or poly(I-C) transfection, and all data are expressed as the fold activation (compared to untreated control) of relative CAT activity as determined by normalization to a constitutively expressed Renilla luciferase control. Error bars indicate the means ± the standard deviations (SD) of two (A and B) or three (C) experiments.
W protein blocks dsRNA/TLR3 signaling to IRF3-responsive promoters.
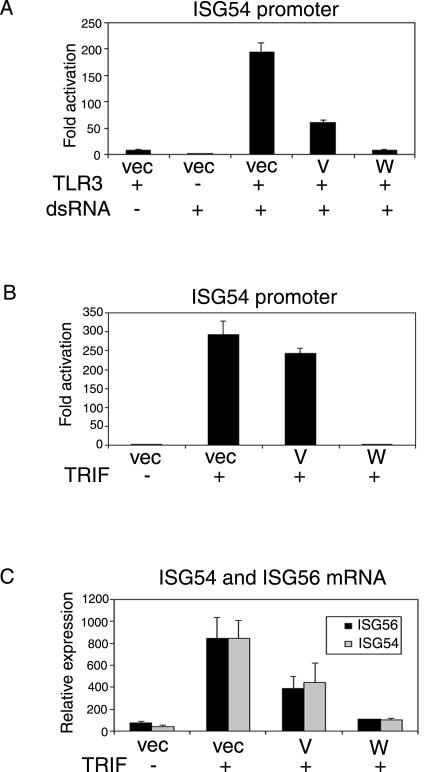
In recent years much has been learned about TLR signaling pathways and how they contribute to the innate immune response. TLR3 specifically recognizes and responds to dsRNA (2); however, 293T cells do not express TLR3, so the promoter activation seen in Fig. 3 is most likely TLR3 independent. To reconstitute TLR3 signaling in 293T cells and to assess whether V and W are able to inhibit this pathway, cells were transfected with a TLR3 expression plasmid and synthetic dsRNA [poly(I-C)] was added to the culture medium 24 h later. As can be seen in Fig. 4A, both TLR3 and dsRNA are required for activation of the IRF3-responsive promoter (ISG54). In the presence of W protein, this activation was strongly inhibited (20-fold), but the V protein was much less effective (3-fold) in blocking TLR3 signaling (Fig. 4A). This difference between the V and W proteins is greater than that seen with inhibition of signaling from intracellular dsRNA (Fig. 3). TLR3 uses an adapter molecule, called TRIF, which mediates the downstream signal (46, 73, 74), and overexpression of TRIF therefore activates the same signaling cascade. When TRIF was used as an activator, the differential effects of V and W on activation of the promoter were even more pronounced (Fig. 4B). This pattern was also seen when the IRF-responsive mouse IFN-α4 promoter was used (data not shown). Quantitative analysis of endogenous ISG54 and ISG56 mRNA confirmed that W protein has a stronger inhibitory effect on TRIF-activated expression of these genes than does V protein (Fig. 4C). Since both V and W proteins can inhibit the JAK/STAT pathway equally well, this inhibitory activity of W protein on the TLR3 pathway must be independent of IFN signaling. These data provide the first evidence that V and W proteins have functional differences.
FIG. 4.
The Nipah virus W protein, but not the V protein, inhibits the TLR3 signaling pathway. (A) 293T cells were transfected with the ISG54-CAT reporter construct, empty vector, or the indicated Nipah expression vectors and 250 ng of a TLR3 expression plasmid. One day posttransfection, the cells were either mock treated or treated by the addition of poly(I-C) (25 μg/ml) to the medium for 7 h. The cells were incubated overnight then harvested and assayed for CAT, and luciferase activities. (B) 293T cells were transfected with the ISG54-CAT reporter construct, empty vector or the indicated Nipah expression vectors and 10 ng of a TRIF expression plasmid. The cells were harvested 24 h posttransfection and assayed for CAT and luciferase activities. The data are expressed as the fold activation of relative CAT activity as determined by normalization to a constitutively expressed Renilla luciferase control. (C) 293T cells were transfected with empty vector or the indicated Nipah expression vectors and 10 ng of a TRIF expression plasmid. The cells were harvested 12 h posttransfection, and the total RNA was analyzed by quantitative RT-PCR for levels of ISG56 (▪) and ISG54 (░⃞) mRNA. Error bars indicate the means ± the SD of three experiments.
Nuclear localization of W protein is required to inhibit TLR3 signaling.
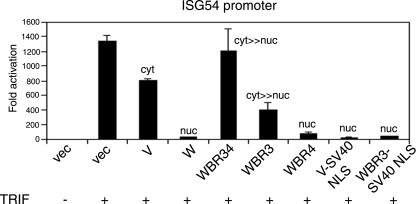
The finding that the W protein, but not V protein, inhibits TLR3 signaling was intriguing since these two proteins are ca. 90% identical at the amino acid level, differing only in their short C-terminal domains. Another difference between the V and W proteins is their localization. To explore the possibility that nuclear localization of W protein is important for blocking TLR3 signaling, NLS mutant forms of W protein (WBR3 and WBR34) were tested for their ability to block TRIF activation of the ISG54 promoter. In contrast to WT W protein, these cytoplasmic forms of W protein were impaired in their ability to inhibit signaling and behaved more like the V protein (Fig. 5). The WBR4 protein, which still localizes to the nucleus, blocked activation of the promoter, as well as the WT W protein. To address the question of whether nuclear localization per se is required for inhibition or whether it is the actual residues in the W NLS that are important, a mutant form of V was constructed where an artificial NLS (derived from the SV40 T antigen) was added onto the C terminus of the protein. A similar mutant form of WBR3 was made. These two proteins both localize to the nucleus (data not shown), indicating that the SV40 NLS is functional. Both of these proteins are now also able to inhibit TRIF activation of the ISG54 promoter (Fig. 5). Therefore, the specific inhibitory effect of W is dependent on its nuclear localization and not on any other sequence within its unique C-terminal domain.
FIG. 5.
Inhibition of TLR3 signaling by the W protein is dependent on its nuclear localization. Activation of the ISG54 promoter by overexpression of TRIF was performed as described in Fig. 4B. The V-SV40 NLS and WBR3-SV40 NLS constructs contain the NLS from the SV40 T antigen at the extreme C terminus of the V and WBR3 ORFs, respectively. The subcellular localization of the proteins encoded by the V and W expression constructs is indicated above the corresponding bar for fold activation. Error bars indicate the means ± the SD of two experiments.
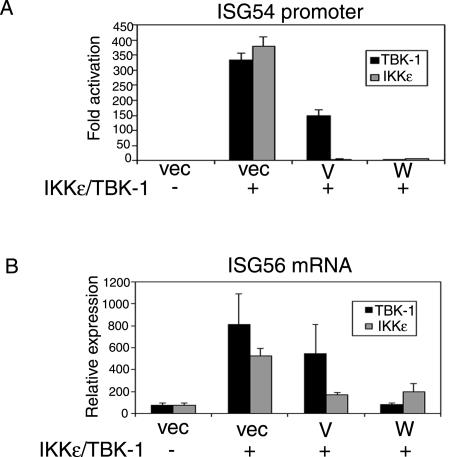
Both V and W proteins can inhibit promoter activation by IKKɛ, but only W inhibits activation by TBK-1.
TBK-1 and IKKɛ have been identified as the kinases that phosphorylate IRF3 after virus infection, as well as after stimulation of TLR3 (20, 58). Overexpression of these kinases activates the downstream signaling pathway, leading to IRF3 activation. We investigated the effect of V and W proteins on this pathway to ask whether both V and W proteins could block activation or if the inhibitory activity would be restricted to W protein. Both kinases activated the promoter strongly but, surprisingly, rather than seeing the same effect of V and W proteins on the two kinases, there was a marked difference seen with TBK-1 versus IKKɛ. Although both V and W proteins inhibited activation in response to IKKɛ, only W protein showed significant inhibition in response to TBK-1 (Fig. 6A). A similar trend was observed when the levels of ISG56 mRNA were measured (Fig. 6B). Thus, V and W proteins are able to block the signaling pathway that initiates upon activation of the kinases and leads to gene activation. The fact that V is able to inhibit IKKɛ-induced gene activation to a much greater extent than that of TBK-1 implies that these kinases are probably involved in two separate pathways.
FIG. 6.
The V protein inhibits promoter activation in response to IKKɛ but not TBK-1. (A) 293T cells were transfected with the ISG54-CAT reporter construct, empty vector, or the Nipah expression plasmids and 100 ng of either TBK-1 (▪) or IKKɛ (░⃞) expression plasmids, as indicated. One day posttransfection, cells were harvested and assayed for CAT and luciferase activities. The data are presented as the fold activation of relative CAT activity, as determined by normalization to a constitutively expressed Renilla luciferase control. (B) 293T cells were transfected with empty vector or the Nipah expression plasmids and 100 ng of either TBK-1 (▪) or IKKɛ (░⃞) expression plasmids, as indicated. The cells were harvested 12 h posttransfection, and total RNA was analyzed by determining the quantitative RT-PCR for levels of ISG56 mRNA. Error bars indicate means ± the SD of three experiments.
Expression of the W protein induces loss of phosphorylated IRF3.
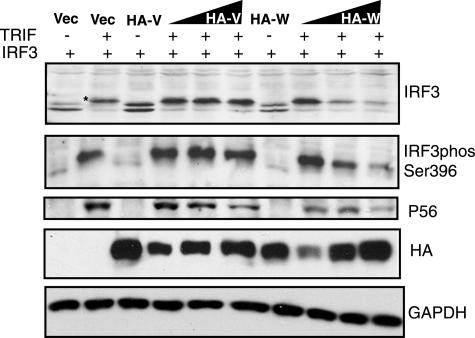
One of the key events in activation of IRF3 is phosphorylation of residues in its C-terminal domain. We examined the phosphorylation status of IRF3 upon activation with TRIF in the presence of increasing amounts of V or W protein (Fig. 7). In the absence of TRIF, IRF3 resolved as a doublet that was also seen when V or W proteins were expressed. In the presence of TRIF, all IRF3 shifted to a higher-molecular-weight form (indicated by the star), which represents hyperphosphorylated IRF3. Increasing expression of the V protein had no effect on IRF3 activation by TRIF; however, increasing amounts of W resulted in a dose-dependent loss of the phosphorylated form of IRF3. This effect was mirrored when activated IRF3 was detected by an antibody that specifically recognizes the phosphorylated serine residue 396 of IRF3, and the loss of phosphorylated IRF3 was concurrent with reduced levels of P56 expression. This suggests that it is the loss of activated IRF3 that is causing the inhibition of gene expression in the presence of W protein.
FIG. 7.
The W protein induces loss of phosphorylated IRF3. 293T cells were transfected with 1 μg of IRF3, 250 ng of TRIF, and either empty vector or the HA-tagged V and W expression plasmids as indicated. In the absence of TRIF, 4 μg of HA-V or HA-W plasmid was added. In the presence of TRIF, increasing amounts of HA-V or HA-W plasmids (1, 2, and 4 μg) were added as indicated by the wedges. Cells were harvested 24 h posttransfection, and lysates were analyzed by Western blotting for IRF3, phosIRF3 Ser396, P56, HA-V, HA-W, and GAPDH. The hyperphosphorylated form of IRF3 induced by TRIF is indicated by an asterisk in the IRF3 panel.
DISCUSSION
The W ORF of Nipah and Hendra viruses is distinct in that the +2 frame extends for 43 amino acids after the editing site, giving the W protein a substantial unique C-terminal domain. In contrast, the W ORF of other paramyxoviruses such as SeV and Newcastle disease virus ends shortly after the editing site (15, 62). Our finding that the Nipah virus W protein localizes to the nucleus (59) due to the presence of this C-terminal domain immediately suggested that this strategy of relocalizing a protein that is otherwise highly similar to its cytoplasmic counterpart (V) may allow it to perform unique functions. Furthermore, it is considered unusual for a virus that replicates exclusively in the cytoplasm to encode a nuclear protein, and it is thought that the role of these proteins is probably to interfere with host cell nuclear functions (32).
In the present study we have shown that the “KKAR” sequence within the unique C terminus of the Nipah W protein is essential for nuclear accumulation of W and thus acts as an NLS. This sequence is conserved in Hendra virus W and therefore most likely serves as an NLS for this protein as well. It is noticeable that there is still a small degree of nuclear staining visible in cells expressing the W NLS mutants (WBR3 and WBR34) and that their localization is not identical to that of the V protein. This implies that there may be a second, weaker NLS elsewhere in the protein. In light of the fact that a nuclear export signal has been described for the V protein (52), this is perhaps not unexpected as this protein must of course be imported before it can be exported. Although the export signal is located in the common N-terminal domain, it is apparently not functional in the W protein, perhaps due to conformational differences between V and W proteins. Therefore, it seems that export predominates for V protein, whereas localization of W protein is determined by the NLS at its C terminus, potentially aided by another NLS in the shared N-terminal domain. The importance of the C-terminal NLS was verified by the finding that a W protein lacking this NLS is no longer able to interact with karyopherin-α. Karyopherin-α (also known as importin-α) is part of the nuclear import machinery and is responsible for recognizing the cargo protein and binding directly to its NLS (reviewed in reference 13). Six mammalian homologues of karyopherin-α have been identified, and they can be grouped into three subfamilies (75). Karyopherin-α 3 and karyopherin-α 4 fall into one of these subfamilies, and the W protein shows a marked preference for binding to these two members over karyopherin-α 1 and karyopherin-α 2, which belong to the remaining two subfamilies. It remains to be seen whether this preference has any functional implications. However, in view of the data indicating that W blocks activation of IRF3-responsive promoters, it is interesting that the IRF3 NLS displays the same preference for binding karyopherin-α 3 and karyopherin-α 4 as does the W protein (39).
We initially discovered that the Nipah virus V, W, and P proteins have anti-IFN activity when we showed that expression of the individual proteins could prevent activation of an antiviral state within the cell and thereby rescue replication of a green fluorescent protein-expressing Newcastle disease virus (47, 59). Since all three proteins can interact with STAT1 and prevent its activation in response to IFN (53, 59), it was thought that inhibition of IFN signaling is the reason for these proteins' ability to block the induction of an antiviral state. However, this finding does not exclude the possibility that additional IFN pathways are being targeted.
Several viruses are known to target multiple IFN pathways in an effort to maximize their ability to evade the innate immune response, and these effects are sometimes mediated by the same viral protein (reviewed in references 4, 22, and 36). Although inhibition of IFN signaling will block amplification of the IFN response in the infected cell, it will not prevent the induction of an antiviral state within neighboring, noninfected cells and therefore will not facilitate virus spread. One would imagine that limiting the production of IFN from the infected cell would be more advantageous in this regard, as this would not alert the surrounding cells to the invading virus. In fact, some paramyxoviruses, which typically target the IFN signaling pathway, have also been shown to inhibit IFN production. For example, SV5, which inhibits IFN signaling by targeting STAT1 for degradation, has been demonstrated to be a poor inducer of IFN-β (30, 51, 69, 70), and the V protein of SV5 is responsible for mediating inhibition of both IFN signaling (16) and IFN production (30, 51, 70). Similarly, we report here that both Nipah virus V and W proteins can inhibit virus-induced activation of the IFN-β promoter. If we assume that the signaling pathways activated by Nipah virus are analogous to those of SeV, we would predict that expression of the V and W proteins in a Nipah virus-infected cell will limit virus-activated transcription of the IFN-β gene. The same effect is seen on the ISG54 promoter, which is responsive to IFN signaling but, after virus infection, can also be activated directly by IRF3 in the absence of IFN production (26, 43). The fact that the same effect is observed with both ISG54 and IFN-β promoters, the latter of which is unresponsive to IFN signaling, indicates that this response is probably independent of IFN signaling. Therefore, V and W proteins can block activation of an IRF3-responsive promoter, whether activated by virus or by transfected dsRNA.
dsRNA, which is produced during the replication cycle of most viruses, is believed to serve as a foreign molecule that triggers the innate immune response of the host and, since dsRNA is the ligand for TLR3, it was proposed that TLR3 is a prime sensor of viral infections (2). Indeed, both virus infection and dsRNA stimulation of TLR3 lead to activation of IRF3, NF-κB, and transcriptional upregulation of the IFN-β gene but, whereas the two pathways probably share several signaling components, data from TLR3−/− mice indicate that TLR3 is not required for activation of the IFN response by all viruses (18, 33). Our data show that the Nipah virus V protein is able to block the signaling pathway in response to virus and intracellular dsRNA but has a much weaker effect on the extracellular dsRNA/TLR3 pathway (this weak effect is most likely due to its transient nuclear localization). The implication of this finding is that these pathways are distinct and that whatever V protein is targeting in the virus pathway is either not part of the TLR3 pathway or is not critical. W protein, on the other hand, blocks both pathways equally well which suggests that it is targeting a shared component of the two pathways. This is most likely a nuclear component, since W protein-mediated inhibition of the TLR3 pathway is dependent on its nuclear localization, which is determined by the NLS in its C terminus. However, the domain that interacts with the putative nuclear component lies within the common N-terminal region that W protein shares with V protein, as demonstrated by the fact that a V protein that is artificially localized to the nucleus gains the ability to block the TLR3 pathway.
The IKK-related kinases, TBK-1 and IKKɛ, have been identified as the kinases that are responsible for phosphorylating IRF3 in response to both virus infection and dsRNA stimulation of TLR3 (20, 58), and therefore the virus and TLR3 pathways seem to converge at the level of the kinases. However, we found that the presence of V protein inhibits IRF3-dependent gene activation in response to IKKɛ but not TBK-1, which corresponds with the effect of V protein on virus and TLR3 signaling, respectively. This leads us to the hypothesis that, in 293T cells at least, TLR3 signaling is TBK-1 dependent but virus activation of gene expression is predominantly IKKɛ mediated (Fig. 8). TBK-1 is probably also involved in the virus pathway to some extent, which may explain why W protein has a slightly stronger inhibitory effect on this pathway than V protein, since it is able to block the signaling pathways in response to both TBK-1 and IKKɛ. There has been some debate over the need for both kinases and whether or not they are redundant. This is primarily because IKKɛ is only highly expressed in immune cells (low levels were detected in 293T cells [data not shown]) and is IFN inducible (60), whereas TBK-1 is constitutively expressed in most cell types (50). Recent studies with TBK-1−/− mice have begun to address this issue. In TBK-1-deficient embryonic fibroblasts IRF3 activation is impaired in response to lipopolysaccharide, poly(I-C), TRIF, or virus infection (31, 49, 64). However, in TBK-1−/− embryonic fibroblasts infected with vesicular stomatitis virus the levels of activated IRF3 were found to increase over time, and this was associated with increased levels of IKKɛ, presumably induced by virus infection (64). Perry et al. (49) also reported that high levels of IKKɛ are present in bone marrow macrophages (BMMs) and that, when TBK-1−/− BMMs are infected with SeV, IRF3 is activated normally, as is IFN-β gene expression. In contrast, there is no IRF3 activation in TBK-1−/− BMMs stimulated with poly(I-C) or LPS (49). This is consistent with our model, where TBK-1 is essential for activation of IRF3-responsive genes in response to TLR3 stimulation, whereas TBK-1 and IKKɛ play a more redundant role in the virus pathway depending on their relative expression levels in different cell types.
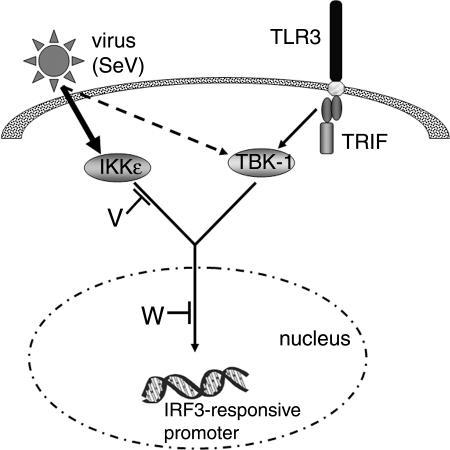
FIG. 8.
Illustration of the virus and TLR3 activated signaling pathways, and the points at which the Nipah virus V and W proteins are predicted to exert their inhibitory activity. Stimulation of TLR3 by dsRNA activates a signaling cascade that is mediated by the adapter, TRIF, and results in activation of the kinase TBK-1. TBK-1 phosphorylates the transcription factor, IRF3, leading to activation of IRF3-responsive promoters. The Nipah virus W protein targets a nuclear component of this signaling pathway that results in the loss of phosphorylated IRF3. Virus infection also results in the activation of IRF3-responsive promoters, and the nuclear portion of this pathway is probably shared with that of the TLR3 pathway, so the W protein is also able to inhibit virus-activated signaling. The upstream signaling events in the virus pathway are distinct from those in the TLR3 pathway in that signaling is mediated predominantly by IKKɛ (thick arrow), although a minor contribution of TBK-1 cannot be excluded (broken arrow). The Nipah virus V protein is only able to prevent gene activation in response to IKKɛ and therefore is restricted to blocking the virus pathway, presumably downstream of IKKɛ.
The fact that V and W proteins interfere with signaling that initiates upon activation of the kinases suggests that the cellular targets could be either the kinases themselves or something downstream of them. IRF3 is a prime candidate for the latter since it is directly activated by the kinases and is required for gene activation. Using TRIF as an activator of the TLR3 signaling pathway, we found that the presence of W results in a dose-dependent loss of phosphorylated IRF3 but does not appear to affect basal levels of IRF3. Phosphorylation of serine 396 in the C-terminal domain of IRF3 has been demonstrated to occur in vivo, and this residue represents the minimal phosphoacceptor site required for activation of IRF3 (57). Expression of W protein results in a substantial reduction in the amount of IRF3 that is phosphorylated at this site, indicating a loss of the active form of IRF3. This correlates well with a reduction in the levels of P56 protein, confirming the inhibition of ISG56 expression (Fig. 4C). This phenomenon is specific to W protein, which is consistent with the finding that only W protein inhibits TLR3 signaling and also demonstrates that this inhibition is due to a loss of activated IRF3. We suggest that W protein destabilizes the phosphorylated form of IRF3, either directly or indirectly and that this event occurs in the nucleus.
The question of whether inhibition of the TLR3 pathway is of any significance for Nipah virus will need to be addressed by comparison of Nipah virus infections in TLR3−/− versus WT systems. It is possible that inhibition of TLR3 reflects the fact that W protein targets a downstream signaling molecule that is shared by both virus- and TLR3-activated pathways. However, despite the finding that lack of TLR3 expression has no detrimental effect on the host's ability to mount an immune response against some virus infections (18), certain viruses have been shown to activate TLR pathways through their envelope glycoproteins (8). Within the paramyxovirus family, these include the F protein of respiratory syncytial virus, which stimulates TLR4 (29, 40), and the hemagglutinin protein of measles virus, which is associated with TLR2 activation (6). These viruses would therefore have reason to interfere with the TLR signaling pathway, particularly that of TLR4, which also leads to induction of IFN-β. In fact, since TRIF is also involved in TLR4 signaling (73), the Nipah virus W protein is quite possibly able to block both TLR4- and TLR3-activated gene expression. Vaccinia virus has been shown to encode three proteins that inhibit TLR signaling, indicating the importance of the TLR responses in the host defense against vaccinia virus (10, 17, 28). Interestingly, one of these proteins, N1L, is known to be a strong determinant of virulence (3, 38) and has recently been shown to associate with TBK-1 (17).
In their role as antagonists of the JAK/STAT1 pathway and of the IFN production pathway, the Nipah virus V and W proteins can be termed multifunctional IFN antagonists and are anticipated to act a virulence factors, as do several other viral IFN antagonists (11, 23, 24, 42, 48). The ingenious inclusion of an NLS in the C terminus of the W protein also gives the virus the potential to inhibit TLR-induced activation of the IFN response if TLRs are stimulated during Nipah virus infection. This differential localization of the V and W proteins therefore imparts functional differences on proteins that are otherwise 90% identical and gives a virus with limited coding capacity the ability to inhibit multiple arms of the host's innate immune response.
Acknowledgments
This study was supported by NIH grants to P.P. and C.F.B. C.F.B. is an Ellison Medical Foundation New Scholar in Global Infectious Diseases. P.P. is an Ellison Medical Foundation Senior Scholar.
We thank John Hiscott (McGill University, Montreal, Quebec, Canada) for providing the TBK-1 and IKKɛ expression plasmids, Ganes Sen (Cleveland Clinic Foundation, Cleveland, Ohio) for the P56 antibody, and Neva Morales for technical support.
REFERENCES
- 1.Akira, S., and K. Takeda. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4:499-511. [DOI] [PubMed] [Google Scholar]
- 2.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 2a.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-β promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed]
- 3.Bartlett, N., J. A. Symons, D. C. Tscharke, and G. L. Smith. 2002. The vaccinia virus N1L protein is an intracellular homodimer that promotes virulence. J. Gen. Virol. 83:1965-1976. [DOI] [PubMed] [Google Scholar]
- 4.Basler, C. F., and A. Garcia-Sastre. 2002. Viruses and the type I interferon antiviral system: induction and evasion. Int. Rev. Immunol. 21:305-337. [DOI] [PubMed] [Google Scholar]
- 5.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bieback, K., E. Lien, I. M. Klagge, E. Avota, J. Schneider-Schaulies, W. P. Duprex, H. Wagner, C. J. Kirschning, V. Ter Meulen, and S. Schneider-Schaulies. 2002. Hemagglutinin protein of wild-type measles virus activates Toll-like receptor 2 signaling. J. Virol. 76:8729-8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billecocq, A., M. Spiegel, P. Vialat, A. Kohl, F. Weber, M. Bouloy, and O. Haller. 2004. NSs protein of rift valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78:9798-9806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boehme, K. W., and T. Compton. 2004. Innate sensing of viruses by Toll-like receptors. J. Virol. 78:7867-7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bossert, B., S. Marozin, and K. K. Conzelmann. 2003. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J. Virol. 77:8661-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowie, A., E. Kiss-Toth, J. A. Symons, G. L. Smith, S. K. Dower, and L. A. O'Neill. 2000. A46R and A52R from vaccinia virus are antagonists of host IL-1 and Toll-like receptor signaling. Proc. Natl. Acad. Sci. USA 97:10162-10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bridgen, A., F. Weber, J. K. Fazakerley, and R. M. Elliott. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. USA 98:664-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler, D. 2004. Fatal fruit bat virus sparks epidemics in southern Asia. Nature 429:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chook, Y. M., and G. Blobel. 2001. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11:703-715. [DOI] [PubMed] [Google Scholar]
- 14.Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazek, P. E. Rollin, S. R. Zaki, W. Shieh, C. S. Goldsmith, D. J. Gubler, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, P. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. Mahy. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432-1435. [DOI] [PubMed] [Google Scholar]
- 15.Curran, J., R. Boeck, and D. Kolakofsky. 1991. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 10:3079-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DiPerna, G., J. Stack, A. G. Bowie, A. Boyd, G. Kotwal, Z. Zhang, S. Arvikar, E. Latz, K. A. Fitzgerald, and W. L. Marshall. 2004. Poxvirus protein N1L targets the IκB kinase complex, inhibits signaling to NF-κB by the tumor necrosis factor superfamily of receptors, and inhibits NF-κB and IRF3 signaling by Toll-like receptors. J. Biol. Chem. 279:36570-36578. [DOI] [PubMed] [Google Scholar]
- 18.Edelmann, K. H., S. Richardson-Burns, L. Alexopoulou, K. L. Tyler, R. A. Flavell, and M. B. Oldstone. 2004. Does Toll-like receptor 3 play a biological role in virus infections? Virology 322:231-238. [DOI] [PubMed] [Google Scholar]
- 19.Enserink, M. 2004. Emerging infectious diseases: Nipah virus (or a cousin) strikes again. Science 303:1121. [DOI] [PubMed] [Google Scholar]
- 20.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 21.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Sastre, A. 2002. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes Infect. 4:647-655. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 24.Garcin, D., M. Itoh, and D. Kolakofsky. 1997. A point mutation in the Sendai virus accessory C proteins attenuates virulence for mice, but not virus growth in cell culture. Virology 238:424-431. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez-Maeso, J., T. Yuen, B. J. Ebersole, E. Wurmbach, A. Lira, M. Zhou, N. Weisstaub, R. Hen, J. A. Gingrich, and S. C. Sealfon. 2003. Transcriptome fingerprints distinguish hallucinogenic and nonhallucinogenic 5-hydroxytryptamine 2A receptor agonist effects in mouse somatosensory cortex. J. Neurosci. 23:8836-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harcourt, B. H., A. Tamin, T. G. Ksiazek, P. E. Rollin, L. J. Anderson, W. J. Bellini, and P. A. Rota. 2000. Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 271:334-349. [DOI] [PubMed] [Google Scholar]
- 28.Harte, M. T., I. R. Haga, G. Maloney, P. Gray, P. C. Reading, N. W. Bartlett, G. L. Smith, A. Bowie, and L. A. O'Neill. 2003. The poxvirus protein A52R targets Toll-like receptor signaling complexes to suppress host defense. J. Exp. Med. 197:343-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haynes, L. M., D. D. Moore, E. A. Kurt-Jones, R. W. Finberg, L. J. Anderson, and R. A. Tripp. 2001. Involvement of Toll-like receptor 4 in innate immunity to respiratory syncytial virus. J. Virol. 75:10730-10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 31.Hemmi, H., O. Takeuchi, S. Sato, M. Yamamoto, T. Kaisho, H. Sanjo, T. Kawai, K. Hoshino, K. Takeda, and S. Akira. 2004. The roles of two IκB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J. Exp. Med. 199:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiscox, J. A. 2003. The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res. 95:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Honda, K., S. Sakaguchi, C. Nakajima, A. Watanabe, H. Yanai, M. Matsumoto, T. Ohteki, T. Kaisho, A. Takaoka, S. Akira, T. Seya, and T. Taniguchi. 2003. Selective contribution of IFN-alpha/beta signaling to the maturation of dendritic cells induced by double-stranded RNA or viral infection. Proc. Natl. Acad. Sci. USA 100:10872-10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hooper, P., S. Zaki, P. Daniels, and D. Middleton. 2001. Comparative pathology of the diseases caused by Hendra and Nipah viruses. Microbes Infect. 3:315-322. [DOI] [PubMed] [Google Scholar]
- 35.Juang, Y. T., W. Lowther, M. Kellum, W. C. Au, R. Lin, J. Hiscott, and P. M. Pitha. 1998. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc. Natl. Acad. Sci. USA 95:9837-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 37.Komatsu, T., K. Takeuchi, J. Yokoo, and B. Gotoh. 2004. C and V proteins of Sendai virus target signaling pathways leading to IRF-3 activation for the negative regulation of interferon-beta production. Virology 325:137-148. [DOI] [PubMed] [Google Scholar]
- 38.Kotwal, G. J., A. W. Hugin, and B. Moss. 1989. Mapping and insertional mutagenesis of a vaccinia virus gene encoding a 13,800-Da secreted protein. Virology 171:579-587. [DOI] [PubMed] [Google Scholar]
- 39.Kumar, K. P., K. M. McBride, B. K. Weaver, C. Dingwall, and N. C. Reich. 2000. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol. 20:4159-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurt-Jones, E. A., L. Popova, L. Kwinn, L. M. Haynes, L. P. Jones, R. A. Tripp, E. E. Walsh, M. W. Freeman, D. T. Golenbock, L. J. Anderson, and R. W. Finberg. 2000. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 1:398-401. [DOI] [PubMed] [Google Scholar]
- 41.McWhirter, S. M., K. A. Fitzgerald, J. Rosains, D. C. Rowe, D. T. Golenbock, and T. Maniatis. 2004. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. USA 101:233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mebatsion, T., S. Verstegen, L. T. De Vaan, A. Romer-Oberdorfer, and C. C. Schrier. 2001. A recombinant newcastle disease virus with low-level V protein expression is immunogenic and lacks pathogenicity for chicken embryos. J. Virol. 75:420-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakaya, T., M. Sato, N. Hata, M. Asagiri, H. Suemori, S. Noguchi, N. Tanaka, and T. Taniguchi. 2001. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem. Biophys. Res. Commun. 283:1150-1156. [DOI] [PubMed] [Google Scholar]
- 44.Naniche, D., A. Yeh, D. Eto, M. Manchester, R. M. Friedman, and M. B. Oldstone. 2000. Evasion of host defenses by measles virus: wild-type measles virus infection interferes with induction of alpha/beta interferon production. J. Virol. 74:7478-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 46.Oshiumi, H., M. Matsumoto, K. Funami, T. Akazawa, and T. Seya. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4:161-167. [DOI] [PubMed] [Google Scholar]
- 47.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patterson, J. B., D. Thomas, H. Lewicki, M. A. Billeter, and M. B. Oldstone. 2000. V and C proteins of measles virus function as virulence factors in vivo. Virology 267:80-89. [DOI] [PubMed] [Google Scholar]
- 49.Perry, A. K., E. K. Chow, J. B. Goodnough, W. C. Yeh, and G. Cheng. 2004. Differential requirement for TANK-binding kinase-1 in type I interferon responses to Toll-like receptor activation and viral infection. J. Exp. Med. 199:1651-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pomerantz, J. L., and D. Baltimore. 1999. NF-κB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 18:6694-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez, J. J., C. D. Cruz, and C. M. Horvath. 2004. Identification of the nuclear export signal and STAT-binding domains of the Nipah virus V protein reveals mechanisms underlying interferon evasion. J. Virol. 78:5358-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodriguez, J. J., J. P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76:11476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sato, S., M. Sugiyama, M. Yamamoto, Y. Watanabe, T. Kawai, K. Takeda, and S. Akira. 2003. Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-κB and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 171:4304-4310. [DOI] [PubMed] [Google Scholar]
- 55.Schlender, J., B. Bossert, U. Buchholz, and K. K. Conzelmann. 2000. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74:8234-8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 57.Servant, M. J., N. Grandvaux, B. R. tenOever, D. Duguay, R. Lin, and J. Hiscott. 2003. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 278:9441-9447. [DOI] [PubMed] [Google Scholar]
- 58.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 59.Shaw, M. L., A. Garcia-Sastre, P. Palese, and C. F. Basler. 2004. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 78:5633-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shimada, T., T. Kawai, K. Takeda, M. Matsumoto, J. Inoue, Y. Tatsumi, A. Kanamaru, and S. Akira. 1999. IKK-i, a novel lipopolysaccharide-inducible kinase that is related to IκB kinases. Int. Immunol. 11:1357-1362. [DOI] [PubMed] [Google Scholar]
- 61.Spann, K. M., K. C. Tran, B. Chi, R. L. Rabin, and P. L. Collins. 2004. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 78:4363-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steward, M., I. B. Vipond, N. S. Millar, and P. T. Emmerson. 1993. RNA editing in Newcastle disease virus. J. Gen. Virol. 74(Pt. 12):2539-2547. [DOI] [PubMed] [Google Scholar]
- 63.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.tenOever, B. R., S. Sharma, W. Zou, Q. Sun, N. Grandvaux, I. Julkunen, H. Hemmi, M. Yamamoto, S. Akira, W. C. Yeh, R. Lin, and J. Hiscott. 2004. Activation of TBK1 and IKKvarepsilon kinases by vesicular stomatitis virus infection and the role of viral ribonucleoprotein in the development of interferon antiviral immunity. J. Virol. 78:10636-10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas, D., G. Blakqori, V. Wagner, M. Banholzer, N. Kessler, R. M. Elliott, O. Haller, and F. Weber. 2004. Inhibition of RNA polymerase II phosphorylation by a viral interferon antagonist. J. Biol. Chem. 279:31471-31477. [DOI] [PubMed] [Google Scholar]
- 66.Valarcher, J. F., J. Furze, S. Wyld, R. Cook, K. K. Conzelmann, and G. Taylor. 2003. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J. Virol. 77:8426-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, L., B. H. Harcourt, M. Yu, A. Tamin, P. A. Rota, W. J. Bellini, and B. T. Eaton. 2001. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 3:279-287. [DOI] [PubMed] [Google Scholar]
- 68.Wang, P., P. Palese, and R. E. O'Neill. 1997. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. J. Virol. 71:1850-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wansley, E. K., J. M. Grayson, and G. D. Parks. 2003. Apoptosis induction and interferon signaling but not IFN-beta promoter induction by an SV5 P/V mutant are rescued by coinfection with wild-type SV5. Virology 316:41-54. [DOI] [PubMed] [Google Scholar]
- 70.Wansley, E. K., and G. D. Parks. 2002. Naturally occurring substitutions in the P/V gene convert the noncytopathic paramyxovirus simian virus 5 into a virus that induces alpha/beta interferon synthesis and cell death. J. Virol. 76:10109-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 72.Weber, F., A. Bridgen, J. K. Fazakerley, H. Streitenfeld, N. Kessler, R. E. Randall, and R. M. Elliott. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 76:7949-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamamoto, M., S. Sato, H. Hemmi, K. Hoshino, T. Kaisho, H. Sanjo, O. Takeuchi, M. Sugiyama, M. Okabe, K. Takeda, and S. Akira. 2003. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science 301:640-643. [DOI] [PubMed] [Google Scholar]
- 74.Yamamoto, M., S. Sato, K. Mori, K. Hoshino, O. Takeuchi, K. Takeda, and S. Akira. 2002. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J. Immunol. 169:6668-6672. [DOI] [PubMed] [Google Scholar]
- 75.Yoneda, Y. 2000. Nucleocytoplasmic protein traffic and its significance to cell function. Genes Cells 5:777-787. [DOI] [PubMed] [Google Scholar]
- 76.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 77.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yuen, T., E. Wurmbach, R. L. Pfeffer, B. J. Ebersole, and S. C. Sealfon. 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]